Discovered over a half-century ago, lipoteichoic acid (LTA) is an abundant polymer found on the surface of Gram-positive bacteria. Although LTA is essential for the survival of many Gram-positive species, knowledge of how LTA contributes to bacterial physiology has remained elusive. Recently, LTA-deficient strains have been generated in some Gram-positive species, including the human oral commensal Streptococcus gordonii. The significance of our research is that we utilized an LTA-deficient strain of S. gordonii to address why LTA is physiologically important to Gram-positive bacteria. We demonstrate that in S. gordonii, LTA plays an important role in the presentation of many cell surface-associated proteins, contributing to cell envelope homeostasis, cell-to-cell interactions in biofilms, and adhesion to eukaryotic cells. These data may broadly reflect a physiological role of LTA in Gram-positive bacteria.
KEYWORDS: Gram-positive bacteria, LTA, Streptococcus gordonii, cell wall, lipoteichoic acid, surface proteins
ABSTRACT
Lipoteichoic acid (LTA) is an abundant polymer of the Gram-positive bacterial cell envelope and is essential for many species. Whereas the exact function of LTA has not been elucidated, loss of LTA in some species affects hydrophobicity, biofilm formation, and cell division. Using a viable LTA-deficient strain of the human oral commensal Streptococcus gordonii, we demonstrated that LTA plays an important role in surface protein presentation. Cell wall fractions derived from the wild-type and LTA-deficient strains of S. gordonii were analyzed using label-free mass spectroscopy. Comparisons showed that the abundances of many proteins differed, including (i) SspA, SspB, and S. gordonii 0707 (SGO_0707) (biofilm formation); (ii) FtsE (cell division); (iii) Pbp1a and Pbp2a (cell wall biosynthesis and remodeling); and (iv) DegP (envelope stress response). These changes in cell surface protein presentation appear to explain our observations of altered cell envelope homeostasis, biofilm formation, and adhesion to eukaryotic cells, without affecting binding and coaggregation with other bacterial species, and provide insight into the phenotypes revealed by the loss of LTA in other species of Gram-positive bacteria. We also characterized the chemical structure of the LTA expressed by S. gordonii. Similarly to Streptococcus suis, S. gordonii produced a complex type I LTA, decorated with multiple d-alanylations and glycosylations. Hence, the S. gordonii LTA appears to orchestrate expression and presentation of cell surface-associated proteins and functions.
IMPORTANCE Discovered over a half-century ago, lipoteichoic acid (LTA) is an abundant polymer found on the surface of Gram-positive bacteria. Although LTA is essential for the survival of many Gram-positive species, knowledge of how LTA contributes to bacterial physiology has remained elusive. Recently, LTA-deficient strains have been generated in some Gram-positive species, including the human oral commensal Streptococcus gordonii. The significance of our research is that we utilized an LTA-deficient strain of S. gordonii to address why LTA is physiologically important to Gram-positive bacteria. We demonstrate that in S. gordonii, LTA plays an important role in the presentation of many cell surface-associated proteins, contributing to cell envelope homeostasis, cell-to-cell interactions in biofilms, and adhesion to eukaryotic cells. These data may broadly reflect a physiological role of LTA in Gram-positive bacteria.
INTRODUCTION
Bacteria interact directly with their environment through surface structures that decorate the cell envelope. The cell envelope of most Gram-positive bacteria consists of the exterior, thick peptidoglycan layer of the cell wall and the interior cytoplasmic membrane. Essential to cell physiology, outward-reaching structures anchored to the cell envelope mediate surface attachment and interspecies interactions, protect against environmental stresses, and help maintain cellular homeostasis (1).
Among the surface polymers of the cell envelope are exterior-reaching glycolipid polymers, including teichoic acids (TAs), which contain phosphodiester-linked polyol repeat units (2). Discovered almost 60 years ago (3), TAs can be found attached to the cell wall (wall teichoic acid [WTA]) or to the cell membrane (lipoteichoic acid [LTA]). Based on their chemical composition, five types of LTAs (types I to V) have been characterized (reviewed in reference 4).
LTA polymers are abundant and comprise a significant percentage of the dry weight of the bacterial cell wall (5). Cells lacking LTA are often nonviable or display severe growth defects (6). LTA contributes to surface hydrophobicity (7, 8), Mg2+ ion scavenging (9), and cell division (10). LTA may also function as an adhesion-promoting molecule (adhesin), mediating interactions with other bacterial and/or eukaryotic cells (11, 12). Thus, LTA is presumed to play an important role in the overall biology of Gram-positive bacteria. The precise physiological function(s) of LTA in the bacterial cell, however, remains unknown.
Recently, a viable LTA-defective strain of the human oral commensal Streptococcus gordonii was generated and studied for its role in the production of nitric oxide by murine macrophages (13) and of interleukin-8 by human periodontal ligament cells (14). A commensal bacterium and pioneer colonizer in oral biofilms, including dental plaque (15), S. gordonii belongs to the viridans group of oral streptococci (16). In dental plaque, S. gordonii is considered beneficial, antagonizing its cariogenic relative, Streptococcus mutans (17). In the bloodstream, however, endogenous strains of S. gordonii have been associated with systemic infections, including infective endocarditis (18, 19).
To investigate how LTA impacts the physiology of S. gordonii cells, we constructed a viable LTA-deficient strain. Here, we report that LTA plays a significant role in surface protein biogenesis, affecting the presentation of several cell wall-associated proteins. These proteins are involved in surface attachment, cell division, and peptidoglycan synthesis, ultimately affecting biofilm formation and the ability of S. gordonii to bind oral keratinocytes but not coaggregation with other bacterial species.
RESULTS
Mutant confirmation.
An LTA-deficient strain of S. gordonii DL1 was constructed by allelic replacement of an open reading frame (S. gordonii 1377 [SGO_1377]) which encodes the LTA synthase (LtaS) homolog in S. gordonii. SGO_1377 replacement (ΔltaS) was confirmed by PCR (Fig. 1A) and whole-genome sequence analysis (see Fig. S1 in the supplemental material). A monoclonal antibody against LTA was used to confirm the loss of LTA in the SGO_1377 deletion background (Fig. 1B). LTA synthesis was restored when SGO_1377 was reinserted into the chromosome at the attB site (ΔltaSc) (Fig. 1B). SGO_1377 is 39% identical and 57% similar to LtaS of Staphylococcus aureus Newman. Since it is required for LTA synthesis by S. gordonii, SGO_1377 is referred to here as LtaS.
FIG 1.
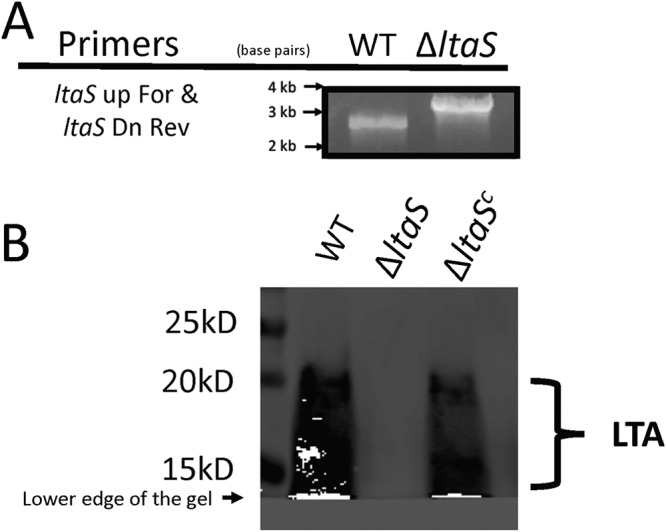
ltaS deletion leads to loss of LTA. (A) ltaS deletion was confirmed by PCR amplification of the wild-type (WT) and the LTA-deficient (ΔltaS) genomic DNA with the primer pair ltaS up For and ltaS Dn Rev. (B) Mouse anti-LTA antibody was used to detect LTA presence on cell wall fractions from the WT strain, the ΔltaS strain, and the complemented LTA deletion strain (ΔltaSc) using Western blotting.
Whole-genome sequencing confirms ltaS deletion. The image shows the region of the S. gordonii genome at the ltaS locus in the WT or ΔltaS strains. The ΔltaS strain showed an absence of sequence alignment at the correct locus when aligned with the reference genome and WT strain. Download FIG S1, TIF file, 0.2 MB (230.4KB, tif) .
Copyright © 2019 Lima et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S. gordonii LTA structure.
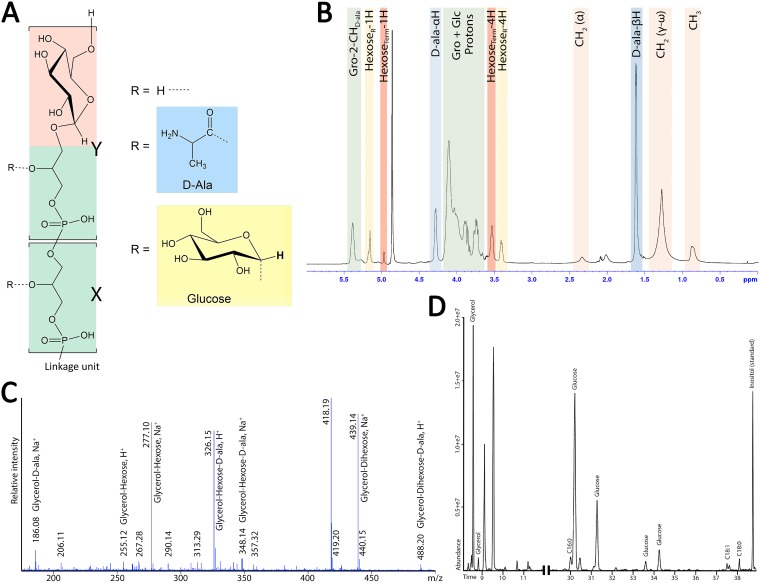
Five types of LTAs (types I to V) have been described based on the chemical architecture of their repeating units (e.g., polyglycerolphosphate [type I], complex glycosylglycerol-phosphate [types II and III], glycosyl-ribitolphosphate [type IV], and glycosyl-phosphate [type V]) (4). The S. gordonii LTA is consistent with the Streptococcus suis type I LTA decorated with d-alanines and glycosyls as analyzed by one-dimensional (1D) 1H nuclear magnetic resonance (NMR) (Fig. 2A). The signals at 5.16 ppm and 3.41 ppm are consistent with hexose anomeric H-1 and H-4 chemical shifts. The second set of signals at 4.97 ppm and 3.54 ppm suggests alternative glycosylation corresponding to either a different sugar moiety or a different position of the LTA backbone (Fig. 2B). To better resolve the structure, purified LTA monomers were subjected to electrospray ionization-mass spectrometry (ESI-MS). Consistent with the presence of hexose substituents, ion signals corresponding to glycerol-hexose were observed at m/z 255.12 (H+) and 277.10 (Na+). Abundant ion signals were also observed, corresponding to multiple glycerol modifications (e.g., m/z 326.15 [glycerol-hexose-d-ala, H+], 348.14 [glycerol-hexose-d-ala, Na+], and 439.14 [glycerol-dihexose, Na+]) (Fig. 2C). Moreover, data from glycosyl compositional analysis indicate that glucose is the sole, highly abundant LTA-associated monosaccharide residue, strongly suggesting that the glycerol repeat units contain at least two unique glycosidic linkages involving glucose (Fig. 2D). As is common with LTA lipid anchors, fatty acid heterogeneity was observed in the forms of C16:0, C18:1, and C18:0.
FIG 2.
LTA structure. (A) Proposed structure of S. gordonii type I LTA with glycerol phosphate (X) and terminal glucose-glycerol phosphate (Y) repeat units. (B) NMR spectrum of purified LTA extracted from S. gordonii containing two distinct anomeric proton signals. (C) Purified S. gordonii LTA was also subjected to electrospray ionization-mass spectrometry after monomerization by hydrofluoric acid. Multiply modified glycerol signals are observed at m/z 326.15, 348.14, 439.14, and 488.20. (D) Carbohydrate composition analysis of monomerized LTA by GC/MS (carbohydrate composition) identified glucose as the sole monosaccharide residue in the S. gordonii LTA, comprising 43.2% (mol%) versus 56.8% of glycerol.
The 1H NMR spectrum, including the two distinct α-glucose anomeric proton signals at 4.97 ppm and 5.16 ppm, is consistent with the type I LTA structure recently characterized in S. suis serotype 2 strain 89-1591 (ST25) (20). The ST25 LTA is a complex type I LTA. Terminal glycerol-glucose repeating units cap a simple type I glycerol phosphate repeating unit. Both repeat units can still be modified by d-alanylation or glycosylation on the glycerol C2-OH position, hence, the presence of the signals of the multiply modified glycerols.
Growth and morphological effects. (i) Growth.
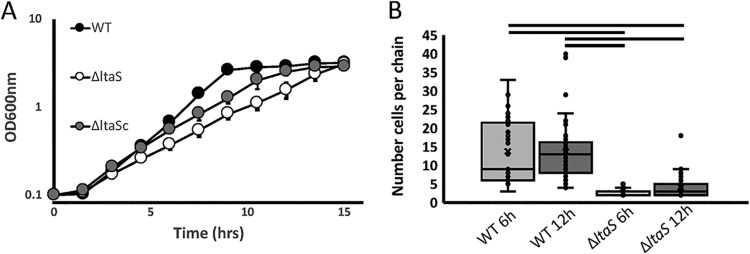
The ΔltaS strain grew more slowly than the wild type (WT) on solid media. An additional day of incubation was needed before colonies were visualized on the antibiotic-selection plates. The ΔltaS cells showed a longer doubling time (∼145 min) than the WT cells (∼85 min), whereas the two strains grew to similar densities at the stationary phase (15 h) in FMC liquid medium (Fig. 3A), suggesting that although loss of LTA affects cell division, the mutant and WT can grow to similar cell densities. Complementation of the ΔltaS strain (strain ΔltaSc) partially rescued the growth defect (Fig. 3A).
FIG 3.
ltaS deletion affects growth and chain length. (A) Growth curve of S. gordonii strains (WT, ΔltaS, and ΔltaSc) as determined by optical density (λ = 600 nm). Data represent means of results from three independent biological replicates (± standard deviations [SD]). (B) Number of cells per chain of S. gordonii in samples from WT and ΔltaS strains collected after 6 and 12 h of growth visualized by light microscopy. Each data point represents one chain. Bars on top of the graph show statistical significance (P < 0.05) as determined by analysis of variance (ANOVA).
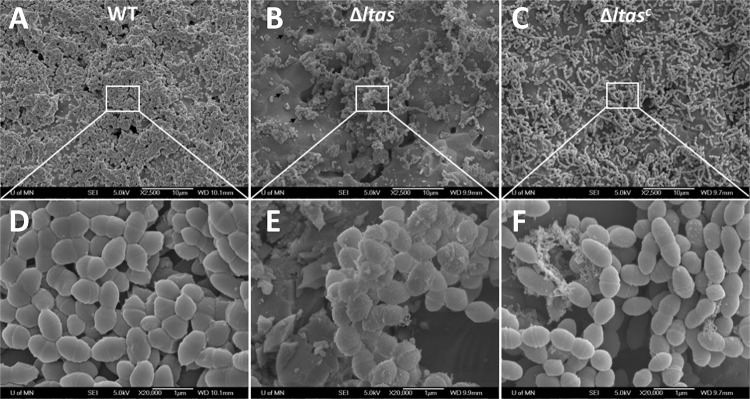
(ii) Coccal chain size and cellular morphology. At 6 h (mid-exponential phase) and 12 h (stationary phase) of growth, the ΔltaS mutant grew in shorter chains than the WT (Fig. 3B). Since LTA is an abundant component of the Gram-positive cell wall, changes in morphology due to loss of LTA were visualized in biofilms using scanning electron microscopy (SEM). After growth on saliva-coated hydroxyapatite disks for 12 h, the WT strains formed a thick biofilm that covered most of the disk surface (Fig. 4A), while the ΔltaS strain failed to cover the disk surface (Fig. 4B). Complementation of the ΔltaS strain partially restored the biofilm phenotype (Fig. 4C).
FIG 4.
SEM of S. gordonii biofilms. S. gordonii strains (WT, ΔltaS, and ΔltaSc) were allowed to form biofilm on saliva-coated hydroxyapatite disks for 12 h. Biofilms were visualized at ×2,500 (A [WT strain], B [ΔltaS strain], and C [ΔltaSc strain]) and ×20,000 (D [WT strain], E [ΔltaS strain], and F [ΔltaSc strain]).
At higher magnification (×20,000), the surface of the WT cells appeared smooth and uniform (Fig. 4D; see also Fig. S2A), whereas the cell surface of the ΔltaS strain appeared irregular, and amorphous extracellular material was apparent (Fig. 4E; see also Fig. S2B). The cell surface of the complemented strain was more similar to the cell surface of the WT strain than to that of the ΔltaS strain (Fig. 4F). The ΔltaS cells appeared rounder than the WT cells (Fig. S2A and B), perhaps due to a weaker peptidoglycan layer. Consistent with a decrease in cell wall strength, the ΔltaS mutant was significantly more sensitive to exposure to hypotonic shock than the WT (Fig. S3).
S. gordonii strains (WT and ΔltaS) were allowed to form biofilm on saliva-coated hydroxyapatite disks for 6 h. Biofilms were visualized at ×5,000. Download FIG S2, TIF file, 0.8 MB (939.3KB, tif) .
Copyright © 2019 Lima et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Equal numbers of WT and ΔltaS S. gordonii cells collected during mid-exponential phase were exposed to sterile PBS or water for 30 min. Next, the cells were plated on TH agar (A) and the colonies were enumerated to determine the percentage of surviving cells compared to the initial inoculum (B). Bars represent means of results from six biological replicates ± SD. Download FIG S3, JPG file, 0.03 MB (31.7KB, jpg) .
Copyright © 2019 Lima et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Functional effects. (i) Surface attachment and biofilm.
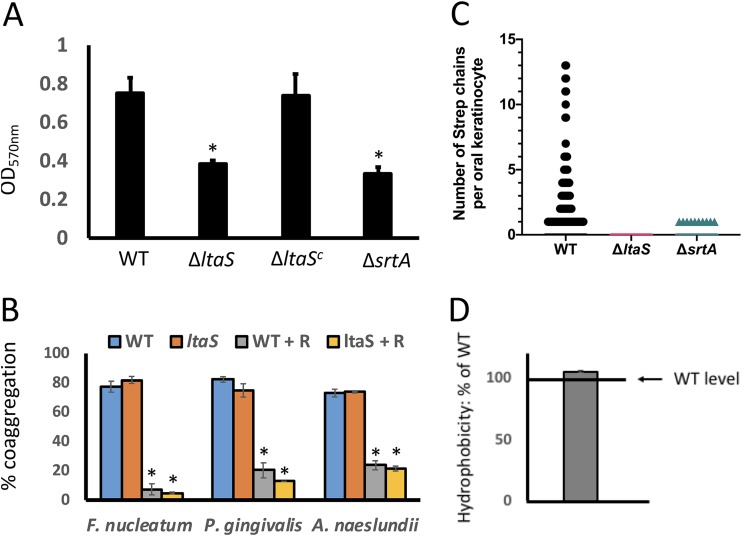
Surface attachment and subsequent biofilm formation are vital for bacteria to persist in the oral environment (21–23) and require presentation of surface-associated proteins. Since the surface of the ΔltaS mutant appeared irregular in comparison to that of the WT (Fig. 4), we investigated whether the ability of S. gordonii to attach to and/or form biofilms on saliva-coated surfaces was affected. Whereas the WT strain and ΔltaS mutant showed similar initial attachment results (4.4 × 107 ± 6.1 × 106 CFU and 6.9 × 107 ± 1.8 × 107 CFU, respectively), the biofilms formed by the ΔltaS strain were ∼67% less dense than those formed by the WT strain after overnight incubation (Fig. 4A). The reduction in biofilm biomass by the ΔltaS mutant was similar to that previously seen with a ΔsrtA strain, which fails to express many of the cell wall-anchored adhesive proteins (adhesins) (24) (Fig. 5A).
FIG 5.
LTA is involved in biofilm formation and interaction with oral keratinocytes. (A) Quantification of biofilm biomass of S. gordonii WT, ΔltaS, ΔltaSc, and ΔsrtA strains on saliva-coated polystyrene wells. Bars represent means of results from six biological replicates ± SD. *, P < 0.05. (B) Quantitative coaggregation of S. gordonii WT and ΔltaS strains with F. nucleatum, P. gingivalis, and A. naeslundii with or without 50 mM l-arginine (R). Bars represent means of results from six biological replicates ± SD. *, P < 0.05. (C) Number of S. gordonii cell chains from the WT, ΔltaS, and ΔsrtA strains found associated with immortalized human oral keratinocytes (OKF6/TERT-2). A total of 2,921 cells derived from three separate experiments were visualized. (D) Relative hydrophobicity levels of S. gordonii ΔltaS strain, compared to the WT, as determined by association with hexadecane. The bar represents the mean of results from six biological replicates ± SD.
(ii) Coaggregation.
S. gordonii may also contribute to the maturation of oral biofilms through direct cell-to-cell contacts with other members of the community, including Fusobacterium nucleatum, Porphyromonas gingivalis, and Actinomyces naeslundii (for a review, see reference 25). To determine whether LTA contributes to direct binding to other members of the oral community, we performed in vitro coaggregation assays. The loss of LTA was not reflected in any change in coaggregation phenotype or inhibition by l-arginine (Fig. 5B).
(iii) Adherence to immortalized oral keratinocytes.
S. gordonii and other closely related oral streptococci attach to host cells (for a review, see reference 26). As modeled using group A streptococcus (GAS), LTA has been proposed to mediate initial interactions with human epithelial cells (27). Thus, we tested S. gordonii for LTA-dependent attachment to immortalized oral keratinocytes. The WT strain bound oral keratinocytes, whereas the ΔltaS and ΔsrtA strains did not (Fig. 5C).
(iv) Surface hydrophobicity.
Although LTA has been reported to contribute to surface hydrophobicity in GAS (7), we observed no differences in surface hydrophobicity between the ΔltaS and WT strains of S. gordonii (Fig. 5D).
LTA deficiency affects expression and presentation of cell wall-associated proteins.
LTA is an abundant component of the cell envelope of Gram-positive bacteria (4), and the ltaS deletion appeared to affect cell surface morphology (Fig. 4D to F) and select functions (Fig. 5). Therefore, we investigated the effect of ltaS deletion on the protein profiles of the membrane and cell wall. Cell membrane and cell wall fractions were collected and resolved on SDS-PAGE gels. The protein presentations in the WT and ΔltaS cell membrane fractions were similar, whereas the protein profiles of the cell wall fractions were markedly different (Fig. 6).
FIG 6.
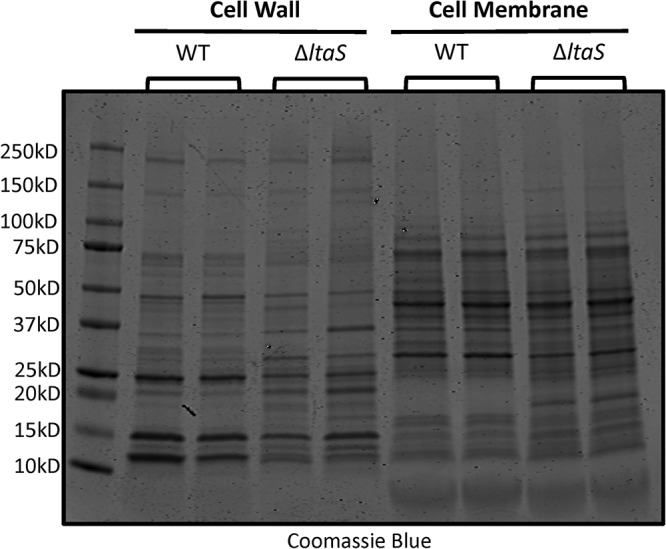
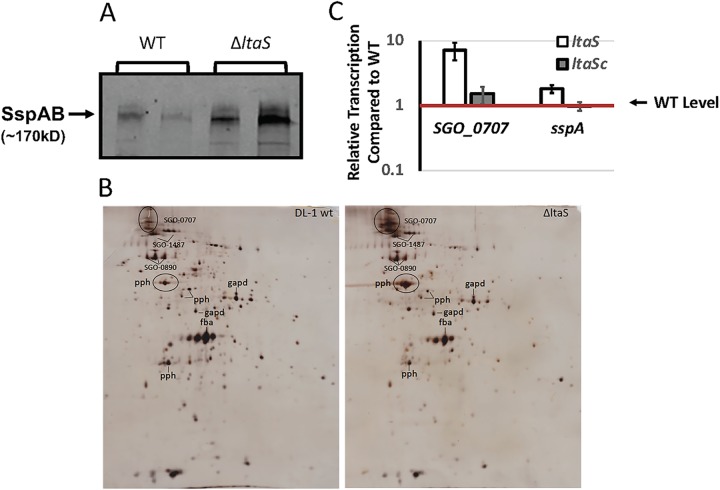
ltaS deletion affects cell wall-associated protein expression. Proteins (60 μg) from cell wall and cell membrane fractions from two biological replicates of WT and ΔltaS strains were resolved on a 4%-to-20%-gradient SDS-PAGE gel and stained with GelCode Blue Safe protein stain.
Using label-free mass spectrometry (MS) analysis, we identified 80 proteins that were differentially expressed in the cell walls of the WT and the ΔltaS strains. The analysis showed that 14 were more abundant in the ΔltaS strain whereas 66 were more abundant in the WT (Table 1). Of particular interest, 5 LPXTG family proteins were more abundant in the cell wall fraction of the ΔltaS strain (SGO_0430, SGO_0707, SGO_0890, SspA, and SspB). Using anti-P1 serum, which binds both SspA and SspB, we confirmed that the ΔltaS strain showed greater SspAB abundance in the cell wall than the WT (Fig. 7A). Similarly, the ΔltaS strain showed greater SGO_0707 abundance than the WT in the cell wall fraction as visualized using 2D gel electrophoresis (Fig. 7B). Therefore, in the WT strain, LTA appears to dampen SspAB and SGO_0707 presentation in the cell wall. To determine whether suppression occurs during or after transcription, we performed qRT-PCR analysis. The expression levels of SGO_0707 and sspA in the ΔltaS strain were 7-fold and 2-fold greater than those seen with the WT, respectively (Fig. 7C). Hence, expression of SGO_0707 and sspA appears to be transcriptionally suppressed in the WT. Complementation of the ΔltaS strain (ltaSc) restored transcription of SGO_0707 and sspA to WT levels.
TABLE 1.
List of differentially expressed proteins identified by mass spectrometry
| Name | Locus | Annotation |
|---|---|---|
| Proteins more abundant in ΔltaS strain | ||
| SGO_1247 | 5’-Nucleotidase family protein | |
| AckA | SGO_1916 | Acetate kinase |
| SGO_1069 | Aminopeptidase | |
| SGO_0843 | Carboxypeptidase | |
| SGO_0911 | Hypothetical protein | |
| SGO_0430 | LPXTG cell wall surface protein | |
| SGO_0707 | LPXTG cell wall surface protein | |
| SGO_0890 | LPXTG cell wall surface protein, collagen-binding domain | |
| SGO_1176 | Peptide methionine sulfoxide reductase | |
| PgK | SGO_0209 | Phosphoglycerate kinase |
| DegP | SGO_2150 | Serine protease |
| SspA | SGO_0210 | Streptococcal surface protein A |
| SspB | SGO_0211 | Streptococcal surface protein B |
| SGO_1177 | Thioredoxin family protein | |
| Proteins more abundant in wild type | ||
| SGO_0953 | 2-Iminobutanoate/2-iminopropanoate deaminase | |
| ThiJ | SGO_1434 | 4-Methyl-5(beta-hydroxyethyl)-thiazole monophosphate synthesis protein |
| SGO_1860 | 5'-Nucleotidase, lipoprotein e(P4) family | |
| SGO_1342 | ABC transporter, ATP-binding protein | |
| ButA | SGO_1096 | Acetoin dehydrogenase |
| SGO_1862 | Alkaline shock protein | |
| SGO_0578 | Amino acid ABC transporter, permease protein | |
| SGO_0982 | Amino acid ABC transporter, amino acid-binding protein | |
| SGO_0104 | Arabinogalactan oligomer/maltooligosaccharide transport system substrate-binding protein | |
| ArcA | SGO_1593 | Arginine deiminase |
| SGO_1082 | Basic membrane protein A | |
| SGO_1630 | Branched-chain amino acid transport system substrate-binding protein | |
| SGO_1626 | Branched-chain amino acid transport system substrate-binding protein | |
| FtsE | SGO_1440 | Cell division transport system ATP-binding protein |
| SGO_0823 | Cof family protein | |
| SGO_0785 | Cof family protein | |
| SGO_0059 | Conserved hypothetical protein | |
| SGO_0957 | Conserved hypothetical protein | |
| AtpF | SGO_1546 | F-type H+-transporting ATPase subunit B |
| PrsA | SGO_1572 | Foldase protein |
| GlcK | SGO_1144 | Glucokinase |
| GdhA | SGO_0276 | Glutamate dehydrogenase |
| SGO_1036 | Glutamine transport system ATP-binding protein | |
| SGO_1037 | Glutamine transport system substrate-binding protein | |
| Gap | SGO_0207 | Glyceraldehyde 3-phosphate dehydrogenase |
| SGO_0390 | Glycerol-3-phosphate dehydrogenase | |
| SGO_0164 | Glycerol-3-phosphate dehydrogenase | |
| SGO_0832 | Hypothetical protein | |
| SGO_1065 | Hypothetical protein | |
| SGO_1677 | Hypothetical protein | |
| SGO_0378 | Hypothetical protein | |
| SGO_1232 | l-Lactate dehydrogenase | |
| SGO_0652 | Lon-like protease | |
| SGO_0372 | Malolactic enzyme | |
| RgfB | SGO_0506 | Maltose 6'-phosphate phosphatase |
| SGO_1283 | Oxidoreductase | |
| Pbp1a | SGO_0586 | Penicillin-binding protein 1A |
| Pbp2a | SGO_2010 | Penicillin-binding protein 2A |
| PgdA | SGO_0948 | Peptidoglycan-N-acetylglucosamine deacetylase |
| ManB | SGO_1215 | Phosphoglucomutase |
| SGO_1149 | Pneumococcal vaccine antigen A-like protein | |
| SGO_0457 | Polar amino acid transport system substrate-binding protein | |
| Wzd | SGO_2016 | Polysaccharide export protein |
| Bta | SGO_1216 | Possible bacteriocin transport accessory protein |
| SGO_0599 | PPM family protein phosphatase | |
| PepQ | SGO_0771 | Proline dipeptidase |
| SGO_1580 | PTS, cellobiose-specific IIB componenta | |
| PtcC | SGO_1576 | PTS, cellobiose-specific IIC component |
| SGO_1763 | Putative aldouronate transport system substrate-binding protein | |
| SGO_1799 | Putative endopeptidase | |
| SGO_0004 | Putative lipoprotein | |
| SGO_0233 | Putative lipoprotein | |
| SGO_0060 | Putative membrane protein | |
| SGO_0140 | Putative multi-antimicrobial-extrusion family transporter | |
| ArcT | SGO_1589 | Putative transaminase/peptidase |
| LytR | SGO_0535 | Putative transcriptional regulator |
| Pyk | SGO_1339 | Pyruvate kinase |
| SGO_0667 | Rhodanese family protein | |
| SGO_1338 | Signal peptidase I | |
| SrtB | SGO_2104 | Sortase B |
| SGO_1110 | Surface antigen SCP-like domain | |
| SGO_0482 | ThiJ/PfpI family protein | |
| InfA | SGO_1963 | Translation initiation factor IF-1 |
| TpiA | SGO_0762 | Triosephosphate isomerase |
| SGO_1864 | X-Pro aminopeptidase | |
| SGO_0521 | YidC/OxaI family membrane protein insertase | |
PTS, phosphotransferase system.
FIG 7.
ltaS deletion affects SspAB and SGO_0707 expression. (A) Protein (60 μg) isolated from the cell wall fraction of two biological replicates of the WT and ΔltaS strains was resolved on a 4%-to-20%-gradient SDS-PAGE gel and transferred to a nitrocellulose membrane. Levels of SspAB were detected by Western immunoblot analysis performed with the anti-P1 antibody. (B) Cell wall fractions (20 μg) of the WT and ΔltaS strains were separated by isoelectric focusing (pH 4 to 7) in the first dimension and 7% SDS-PAGE in the second dimension. The identity of the major spots is shown. (C) Relative expression levels of SGO_0707 and sspA were determined during the exponential-growth phase in chemically defined medium (FMC) by qPCR analysis of total RNA extracted from the WT, ΔltaS, and ΔltaSc strains at 37°C. Bars represent means of results from six biological replicates ± SD.
DISCUSSION
LTA has long been viewed as an important component of the Gram-positive cell envelope, since inhibition of LTA synthesis leads to the death or impaired growth of many species (28–32). Why LTA is physiologically important to the cell, however, remains obscure. To start addressing this issue, we took advantage of a viable LTA-deficient strain of the human oral commensal S. gordonii. Our data implicate LTA as an important contributor to surface protein biogenesis, affecting the abundance of as many as 80 proteins under the conditions tested (Table 1). This widespread effect in surface protein presentation could easily explain the pleiotropic phenotypes associated with LTA loss.
LTA synthesis and structure.
Differing from the complex LTA structure of Streptococcus pneumoniae, which displays a type IV LTA, the structure of the unusual type I LTA produced by S. gordonii is decorated with d-alanine and glucose, based on comparative 1D 1H NMR, ESI-MS, and carbohydrate compositional analyses (Fig. 2). Whereas LtaS is essential for many species, Bacillus subtilis and Bacillus anthracis encode four LtaS homologs which seem to perform overlapping and or redundant activities (reviewed in reference 6). S. gordonii differs from those species in that there is only one ltaS ortholog (SGO_1377) identified in its genome, and its deletion leads to complete loss of LTA as determined by Western immunoblotting (Fig. 1). Less obvious to us, however, are the components involved in the synthesis and translocation of the glycolipid anchor from the inner to the outer leaflet of the S. gordonii membrane. In some bacteria, or in mutant strains with deletions in the glycolipid synthesizing enzymes, LTA can be polymerized directly onto diacylglycerol (DAG) groups (33–35). In other bacteria, such as S. aureus, B. subtilis, S. agalactiae, and Enterococcus faecalis, LTA is polymerized on a Glc2-DAG glycolipid anchor (reviewed in reference 36). We searched the S. gordonii genome for homologs of the glucosyltransferases involved in Glc2-DAG synthesis in S. aureus, B. subtilis, S. agalactiae, and E. faecalis. We identified two proximal glucosyltransferase genes (SGO_0774 and SGO_0775) with sequence similarities to GBS0683 and ladA of S. agalactiae. We are currently assessing the roles of SGO_0774 and SGO_0775 in Glc2-DAG and LTA synthesis and function.
Whereas Glc2-DAG synthesis occurs inside the cell, the addition of glycerol phosphate subunits onto the glycolipid anchor (and therefore LTA synthesis) occurs outside (37). In S. aureus, the membrane permease LtaA is required for the translocation of glycolipids across the membrane (30). In S. gordonii and other species, no enzyme with comparable function has been identified.
A role in peptidoglycan maintenance and envelope stress response (ESR).
In many Gram-positive bacteria, LTA depletion is associated with cell growth defects (reviewed in reference 6). In S. aureus, for example, depletion of LtaS results in aberrant positioning of division septa, pointing to a link between LTA and cell division (32). Septum formation and cytokinesis are crucial steps in bacterial growth and are often regulated by FtsE and FtsX (38–41). In S. gordonii, loss of LTA decreases FtsE abundance, potentially affecting septum formation and contributing to the growth defect in our LTA-deficient strain (Fig. 3A). Interestingly, S. aureus, B. subtilis and Listeria monocytogenes cells are elongated when LTA synthesis is disrupted (reviewed in reference 6), whereas S. gordonii appears to show the opposite phenotype (Fig. 3B).
Peptidoglycan biosynthesis and remodeling are also important steps in cell division and in the overall maintenance of the cell wall (reviewed in reference 42). Cell wall maintenance requires the activity of multiple enzymes, including members of the widely conserved penicillin-binding protein (PBP) family. In S. aureus, LtaS interacts with Pbp1 and Pbp2 among other peptidoglycan biosynthesis and remodeling proteins (43). Similar interactions may also occur in S. gordonii, since Pbp1a and Pbp2a are less abundant in cell wall fractions derived from our LTA-deficient strain (Table 1). In S. aureus, depletion of Pbp1 induces incomplete septation and alterations in cell morphology but does not appear to alter peptidoglycan cross-linking (44, 45), whereas depletion of Pbp2 results in the presence of peptidoglycan that is significantly less extensively cross-linked (46, 47) and therefore weaker. Thus, the cell-rounding phenotype displayed by the ΔltaS strain (see Fig. S2A and B in the supplemental material) and its increased sensitivity to osmotic pressure (Fig. S3A and B) might result from the decrease in the level of Pbp1a and/or Pbp2a in the cell wall.
The cell wall and the cell membrane (together referred to as the cell envelope) defend the cell against environmental insults. Thus, it is not surprising that most bacterial species employ multiple systems to monitor cellular integrity (reviewed in reference 48). As a major component of the Gram-positive cell wall and therefore the cell envelope, loss of LTA could impact cell envelope integrity and trigger an envelope stress response (ESR). In fact, LTA has been previously proposed to help stabilize the Gram-positive cell membrane (49).
In Gram-negative bacteria, the DegP serine protease is an important component of the ESR, degrading misfolded or aggregated cell envelope proteins (reviewed in reference 50). We observed that DegP abundance was greater in cell wall fractions from the ltaS mutant than in those from the WT (Table 1), perhaps signaling that the number of misfolded proteins had increased. The mechanism behind the increase in DegP abundance in the ltaS mutant remains unexplored. In Escherichia coli, degP transcription is regulated by the stress response two-component system (TCS) CpxAR (51). Despite significant progress in the past decade, ESR studies in Gram-positive bacteria still lag those in Gram-negative species. At this point, it is unclear if and how Gram-positive bacteria can sense the accumulation of misfolded proteins in their cell envelope. Accumulation of misfolded proteins could affect membrane composition triggering the activation of a TCS. In B. subtilis, for example, at least three of the four TCSs involved in ESR have sensor kinases with very short (<10 amino acids) extracellular loops connecting their two transmembrane domains (52). This domain architecture has been identified in at least one TCS in S. gordonii, SGO_1180 (53). Consistent with the hypothesis that SGO_1180 might be involved in mitigating an ESR induced by the loss of LTA, a ΔltaS ΔSGO_1180 double mutant seems to be lethal (B. P. Lima, unpublished data), suggesting that the ability of S. gordonii to cope with the loss of LTA requires SGO_1180.
LTA and biofilm formation.
LTA contributes to biofilm formation in many species (8, 54–56). How loss of LTA affects biofilm formation, however, seems to differ from species to species. In S. aureus, for example, LTA is required for attachment to polystyrene surfaces, affecting the overall biofilm formation (8). In Enterococcus faecalis, however, loss of LTA does not affect surface attachment but severely affects biomass accumulation after the initial attachment (54). Similarly, our data show that loss of LTA in S. gordonii leads to a significant defect in biofilm formation (Fig. 4 and 5A) without any detectable impact in the initial attachment to saliva-coated polystyrene plates, suggesting a defect in a biofilm maturation step.
Concluding remarks.
Although an LTA-deficient strain of S. gordonii has been constructed previously (13, 14, 57), the immune response to LTA was the primary focus of those studies. Here, we show that S. gordonii produces a complex type I LTA that shows abundant d-alanylation and glycosylation. This LTA plays an important role in cell surface protein biogenesis, key to cell envelope homeostasis.
MATERIALS AND METHODS
Bacterial strains and media.
All bacterial strains and plasmids used in this study are listed in Table 2. S. gordonii DL1 was grown in brain heart infusion (BHI) broth or on agar plates (Difco, Sparks, MD) or in chemically defined media (FMC) (58) at 37°C in 5% CO2 as indicated. Actinomyces naeslundii ATCC 12104 was grown in BHI broth or agar plates (Difco, Sparks, MD) at 37°C in 5% CO2. Fusobacterium nucleatum strain ATCC 23726 was grown anaerobically at 37°C in Columbia broth or on Columbia agar plates (BD Difco, Detroit, MI) supplemented with 5% defibrinated sheep blood (HemoStat Laboratories, Dixon, CA). Porphyromonas gingivalis A7436 was grown anaerobically on Columbia broth supplemented with 5% defibrinated sheep blood, 1 μg/ml hemin, and 1 μg/ml menadione. Escherichia coli was grown aerobically at 37°C in Luria-Bertani (LB) broth or on agar plates (BD Difco, Detroit, MI). When necessary, 50 μg/ml spectinomycin was added for selection of E. coli.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description (purpose)a | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli NEB 5-alpha | Competent E. coli—DH5α derivative | NEB |
| Streptococcus gordonii DL1 | S. gordonii wild type | 75 |
| Streptococcus gordonii ΔltaS::JHMD1 | DL1 with in-frame deletion of SGO_1377 | This study |
| Streptococcus gordonii ΔltaSc | Complemented ΔltaS::JHMD1; SGO_1377 reinserted into the attB locus | This study |
| Streptococcus gordonii ΔsrtA | DL1 with a markerless deletion of srtA | 53 |
| Fusobacterium nucleatum ATCC 23726 | Wild type | ATCC |
| Actinomyces naeslundii ATCC 12104 | Wild type | ATCC |
| Porphyromonas gingivalis 4612 | Wild type | 76 |
| Plasmids | ||
| pJHMD1 |
S. gordonii ldh promoter—ermAM-pheS*(A316G) in pUC57; Ampr (markerless deletion system) |
53 |
| pFW5 | S. gordonii shuttle vector, Specr | 77 |
| pBPL11 | pFW5 derivative containing ltaS ORF and its promoter (complementation) | This study |
Ampr, ampicillin resistance; ORF, open reading frame; Specr, spectinomycin resistance.
Construction of LTA-deficient mutant.
The ΔltaS strain was constructed by allelic replacement of the nucleotide sequence that includes ltaS (SGO_1377) using the JHMD1 cassette from pJHMD1 as described previously (53). An ∼500-bp region immediately upstream and downstream of ltaS was amplified with primers ltaS-UP (Forw 5′-AAGAAAAGAGAGCATAGTCCC-3′ and Rev 5′-CTATGCTATGAGTGTTATCGTTTCTCGTTTTTTCACAAAAGTACTTCCTTG-3′) and ltaS-Down (Forw 5′-GTTATCTATTATTTAACGGGAGGAAATAAAGCAATACTTTGTCACACC-3′ and Rev 5′-TATACGAATTTATCCAAAAAAC-3′). Chromosomal DNA from S. gordonii was used as a template. The JHMD1 replacement sequence was amplified with primers JHMD1-Forw (5′-CGAGAAACGATAACACTCATAGCATAG-3′) and JHMD1-Rev (5′-TTATTTCCTCCCGTTAAATAATAGATAAC-3′) (53) and fused to the upstream and downstream flanking regions by splice overlap extension PCR (SOE-PCR) as described previously (59). The PCR product was then transformed into WT S. gordonii and plated on Todd-Hewitt (TH) agar plates containing 5 μg/ml erythromycin and incubated under anaerobic conditions (10% H2, 10% CO2, 80% N2) at 37°C as described previously (53).
LTA purification and characterization.
S. gordonii was grown in FMC medium at 37°C in 5% CO2. LTA was purified from the WT biomass (8 liters of stationary-phase culture) and analyzed as described previously (60, 61). As judged by NMR, the purified LTA preparations did not contain high levels of lipoprotein or phospholipid, but the presence of trace levels of these contaminants cannot be ruled out. For de-d-alanylation, purified LTA (0.5 mg) was incubated in 5 mM ammonium bicarbonate (pH 8.5) for 24 h at room temperature, followed by freeze-drying to remove ammonium bicarbonate. LTA was then resuspended and dialyzed against MilliQ distilled water (dH2O) using cellulose ester dialysis membranes (Spectrum Labs) (molecular weight cutoff [MWCO], 0.5 to 1.0 kDa) to remove free d-alanines. LTA (1 mg) was monomerized by treatment at room temperature for 20 h with 100 μl of 47% hydrofluoric acid (HF), which was then evaporated under a filtered air stream and resuspended in 200 μl of ammonium bicarbonate. LTA monomers were further neutralized with dilute ammonium hydroxide and lyophilized.
LTA monomers were suspended in electrospray ionization mass spectrometry (ESI-MS) mobile phase (50% acetonitrile, 50% aqueous 10 mM ammonium acetate) and analyzed using a Waters 2695 high-performance liquid chromatography (LC) system and flow injection at a rate of 0.15 ml/min. Nitrogen drying gas was used at a setting of 300°C at 7 liters/min with capillary voltage of 2.8 kV.
Samples were subjected to acidic methanolysis and analyzed for glycosyl composition by combined gas chromatography/mass spectrometry (GC/MS) analyses of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides as described previously (62).
Whole-genome sequencing.
S. gordonii WT and ΔltaS strains were grown in BHI broth at 37°C and 5% CO2 for 18 h. Genomic DNA was isolated using a Wizard genomic DNA isolation kit (Promega). To sequence paired-end reads, the University of Minnesota Genomics Center created 5 dually indexed Nextera XT libraries from 500 ng genomic DNA. The libraries were combined into a single pool and sequenced using a MiSeq PE Nano sequencer (v2; 250 bp), generating ≥1 million pass filter reads for the run. All expected barcodes were detected and well represented. The mean quality scores were ≥Q30 for all libraries. Reads were mapped against the S. gordonii genome (NC_009785_1) using Burrows-Wheeler Aligner version: 0.7.17-r1188. Using the CLC Genomics suite, binary alignment map (BAM) files were used to extract consensus sequences with a minimum coverage of 5 reads without the inclusion of the reference strain. Single nucleotide polymorphisms (SNPs) distinguishing the WT and ΔltaS strains were manually identified using Integrative Genome Viewer.
ΔltaS mutant complementation.
The ΔltaS strain was complemented by insertion of the endogenous ltaS gene with its native promoter into the attB site of the S. gordonii genome. The ltaS gene and its promoter were amplified from the WT S. gordonii strain using primer pair latS-For (5′-ATTCTAAATTATATCAAATATTGAGAAATATTT C3’) and ltaS-Rev (5′-TCATTGCTTACTAGAAGAAG-3’). The entire attB sequence was amplified using primer pair attB-Forw (5′-AAGGCATTTGTCTTTAATTCTACTG-3′) and attB-Rev (5′-AACCTGATTTATCAGGAAGC3-3’). The attB fragment was fused to ltaS and inserted by Gibson assembly (New England Biolabs, Ipswich, MA) into pDL278, which had been digested previously with enzymes BamHI and SalI (New England Biolabs, Ipswich, MA). The resulting plasmid was named pBPL11.
Saliva collection and preparation for adhesion and biofilm assays.
Stimulated whole saliva was collected and pooled from at least three healthy, medication-free adult volunteers using protocols that were reviewed and approved by the Institutional Review Boards from the University of Minnesota and Malmo University. Saliva was processed as described previously (53) and sterilized by exposure to UV irradiation for 30 min (Spectroline UV Crosslinker FB-UVXL-1000; Spectronics, Westbury, NY). After a 48-h incubation at 37°C in 5% CO2, sterilization was confirmed by plating saliva on BHI agar and enumerating CFU.
Adhesion to saliva-coated surfaces.
The bottoms of 12-well plates (Corning Costar catalog no. 3513) were coated with 500 μl of sterilized saliva. To assess surface attachment, 1 ml of WT S. gordonii or ΔltaS cells (adjusted to an optical density at 600 nm [OD600] of 1 in FMC broth) was added to each saliva-coated well. and the plates were incubated at 37°C for 45 min in 5% CO2 (∼1/2 S. gordonii doubling time under the conditions tested), avoiding rounds of cell division. After incubation, the FMC medium was aspirated and each well was washed twice with 1 ml of sterile phosphate-buffered saline (PBS), which was removed by aspiration. Adherent cells were scraped from the bottom of the wells using cell scrapers (Sarstedt, Inc., Newton, NC, USA) (25-mm overall length), resuspended in 1 ml of sterile PBS, serially diluted in 10-fold dilutions, and enumerated as CFU on BHI agar plates.
Biofilm assay.
To assess biofilm formation, the S. gordonii WT and ΔltaS strains were inoculated (200 μl of a 1:100 dilution of an overnight culture) into saliva-coated 96-well round-bottom plates and grown in FMC broth at 37°C for 16 to 18 h in 5% CO2. Biofilm formation was assessed by crystal violet retention as described previously (53).
Coaggregation.
Interspecies coaggregation assays were performed in coaggregation buffer (CAB) containing 150 mM NaCl, 1 mM Tris, 0.1 mM CaCl2, and 0.1 mM MgCl2 as described previously (63, 64). Coaggregation was inhibited by adding 50 mM l-arginine to the suspensions of S. gordonii strains and vortex mixing after adding the partner strains.
Adherence to immortalized oral keratinocytes.
Immortalized human oral keratinocytes (OKF6/telomerase reverse transcriptase 2 [TERT-2]) were grown as described previously (65) with modifications. Briefly, keratinocytes (1 × 105 cells) were seeded on a sterile Fisherbrand microscope cover glass in wells of Costar 3524 24-well plates in 500 μl Gibco K-SFM and grown for 24 h at 37°C in 5% CO2.
The WT, ΔsrtA, and ΔltaS strains were grown overnight at 37°C in 5% CO2 and adjusted to a multiplicity of infection (MOI) of 100:1, with 10 μl of bacteria added to each well containing keratinocytes. Cultures were incubated for 30 min at 4°C to minimize internalization of bacteria, the medium was aspirated, wells were washed with 500 μl PBS and aspirated, and a LIVE/DEAD BacLight bacterial viability kit (Life Technologies) was added to each well according to the manufacturer’s protocol. After a 15-min incubation at room temperature in the dark, the PBS containing the stain was aspirated from the wells. To visualize and count attached bacteria, mounting oil (Life Technologies) (2 μl) was placed onto a glass slide (Matsunami Glass Industries), and the glass slide was covered with a cover glass from the experimental wells (described above) and secured in place using clear nail polish.
Hexadecane binding assay.
Hydrophobicity was assessed by hexadecane binding as described previously (66), with minor modifications. S. gordonii was grown in FMC medium overnight at 37°C in 5% CO2. An overnight culture was adjusted to an OD600 of 1.0, 1 ml was placed into a glass tube, and 100 μl of hexadecane (Sigma-Aldrich, St. Louis, MO) was added. The mixture was subjected to vigorous vortex mixing for 2 min and then allowed to stand for 10 min at room temperature to facilitate phase separation. The OD600 of the lower aqueous phase was recorded. Percent hydrophobicity was calculated using the following formula: percent hydrophobicity = [1 − (OD600 after vortex mixing/OD600 before vortex mixing)] × 100.
Cell fractionation.
S. gordonii was fractionated as described previously (67) with minor modifications. Cells were grown in FMC medium and pelleted, and protoplasts were harvested as follows. Protoplasts and supernatants were separated by centrifugation at 12,000 × g for 20 min at 4°C. Supernatants were collected and dialyzed against 2 liters of 0.2% (wt/vol) Na2EDTA for 4 h followed by 2 liters of distilled water overnight and concentrated by centrifugation using an Amicon Ultra-15 Centrifugal Filter Untracel (Tullagreen, Carrigtwohill, Co. Cork, Ireland) (10K), and the product was designated the cell wall fraction. Protoplasts were resuspended in 10 ml of bacterial lysis buffer (BugBuster [EMD Millipore Corp., Billerica, MA]) and lysed by sonication (75 W for 8 min), and insoluble materials were removed by centrifugation (12,000 × g, 20 min, 4°C). Supernatants were collected, and membrane fractions were separated from cytoplasmic fractions using ultracentrifugation at 120,000 × g for 90 min at 4°C. Pellets and supernatants were collected separately. The supernatant was designated the cytoplasmic fraction. The pellet was resuspended in lysis buffer and designated the membrane fraction. Protein concentrations for each fraction were determined using Pierce bicinchoninic acid (BCA) protein assay (Thermo Scientific, Rockford, IL). To compare protein profiles, 60 μg of total protein from each fraction was resolved by 4% to 20% gradient SDS-PAGE and stained with GelCode Blue Safe protein stain (Thermo Scientific, Rockford, IL).
Label-free quantification. (i) In-gel trypsin digestion.
A total of 60 μg of protein from each biological replicate was resolved by SDS-PAGE and digested as described previously (68).
(ii) Liquid chromatography and mass spectrometry. Triplicate WT and ΔltaS samples were analyzed in random order. Approximately 1 μg of peptide mixture was injected for each sample. After trypsin digestion, the peptide mixtures were resolved using capillary liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a Velos Orbitrap system (Thermo Scientific, Waltham, MA) (69) modified as follows. The capillary column dimensions were 100-μm internal diameter by 14-cm length; the flow rate for direct column load was 1.1 μl/min; the minimum signal intensity for precursor ion trigger was 15,000 counts (lock mass was not used); the dynamic exclusion (DE) duration was 30 s.
(iii) Database searching and quantification. We analyzed the tandem MS data in PEAKS Studio 8.5 (Bioinformatics Solutions, Inc., Waterloo, Ontario, CA) with the Quant module for protein detection and label-free quantification. The database search parameters were set as follows: no corrections for charge or precursor; no merge options and no spectrum filter applied; de novo precursor mass error tolerance of 50.0 ppm and fragment mass error tolerance of 0.1 Da; de novo enzyme trypsin and variable modifications for methionine oxidation and carbamidomethyl cysteine; maximum of 3 variable posttranslational modifications (PTMs) per peptide; PEAKS DB (database search) precursor mass error tolerance of 50.0 ppm and de novo fragment mass error tolerance of 0.1 Da; monoisotopic search type with trypsin enzyme, 1 missed cleavage site and amino acid modifications identical to de novo settings; S. gordonii (taxon ID 1302) protein database from NCBI reference sequence (downloaded 16 October 2017) merged with the common laboratory contaminant proteins from http://www.thegpm.org/crap/; PEAKS PTM parameters for deamidation (NQ), dioxidation (M), pyro-glutamic acid from Q, and N-terminal acetylation and oxidation (HW) with a maximum of 3 variable modifications per peptide; false-discovery-rate estimation enabled. For quantification, we set the following parameters: PEAKSQ significance method; protein signification value of ≥20; minimum fold change = 2; total ion chromatogram for the normalization method. For comparisons, the six samples (triplicates for each condition) were assembled into 2 groups. Details of the retention alignment algorithm (70) and PEAKS Q significance scores were as described previously (71).
Two-dimensional polyacrylamide gel electrophoresis.
Two-dimensional (2D) polyacrylamide gel electrophoresis resolved a volume corresponding to 20 μg of cell wall protein as described previously (72). The protein concentration was determined using a 2D Quant kit (GE Healthcare Life Sciences). All gels were run in triplicate. Only protein spots detected in all replicates were identified.
Identification of proteins on 2D gels using LC-MS/MS.
Proteins of interest were excised manually from Coomassie brilliant blue-stained gels and trypsin digested, and peptides were separated using LC and characterized using MS/MS as described previously (72). Mass lists were used as the input for Mascot MS/MS ion searches of the NCBInr database using the Matrix Science Web server.
Western immunoblot analysis.
S. gordonii cultures were harvested and fractionated as described above, and 10 μg of protein from each fraction was analyzed using Western immunoblotting. To detect S. gordonii LTA, mouse monoclonal antibody against Gram-positive bacterial LTA (G43J; Thermo Fisher Scientific, Rockford, IL) was used at a 1:50 (vol/vol) dilution. To detect S. gordonii SspA and SspB surface proteins, rabbit anti-S. mutans P1 serum diluted 1:1,000 (vol/vol) was incubated overnight at 4°C as described previously (24). Blots were incubated for 1 h at room temperature with IRDye 680RD goat anti-mouse and IRDye 800CW goat anti-rabbit secondary antibody (Li-Cor, Lincoln, NE), respectively, and immunoreactive proteins were visualized. Total protein transferred onto the nitrocellulose membrane was quantified using REVERT total protein stain (Li-Cor, Lincoln, NE) according to the manufacturer’s instructions.
Total RNA purification and RT-qPCR.
Overnight cultures were diluted 1:100 (vol/vol) in 5 ml of sterile FMC media in 15-ml polypropylene conical vials (Sarstedt) and incubated overnight at 37°C in 5% CO2. Cultures were pelleted at 4,300 × g at 4°C, and supernatants were removed. Total RNA purification, cDNA synthesis, and multiplex real-time quantitative PCR (RT-qPCR) were performed as described previously (53).
Scanning electron microscopy.
Biofilms were allowed to form on saliva-coated hydroxyapatite disks (Clarkson Chromatography Products Inc., South Williamsport, PA) (9.65 mm by 1.52 mm) for 12 h, dehydrated, and processed as described previously (73). The samples were viewed with a field-emission-gun scanning electron microscope (FE-SEM) (6500; JEOL, Tokyo, Japan) (74).
ACKNOWLEDGMENTS
We thank members of the Herzberg laboratory for discussions and/or critical reading of the manuscript. We thank James Rheinwald for providing the immortalized human oral keratinocytes (OKF6/TERT-2) reported here. We also thank Neil Hunter, the University of Sydney, NSW, for providing the anti-P1 serum used in this study. We thank LeeAnn Higgins and Todd Markowski at the University of Minnesota Center for Mass Spectrometry and Proteomics for their help with the mass spectrometry analysis of ΔltaS and WT cell wall fractions. Finally, we would like to thank Tania Laremore (Proteomics and Mass Spectrometry Core Facility, The Pennsylvania State University) for help with ESI mass spectrometry experiments.
The study was supported by a grant from the National Institutes of Health (NIDCR R01 DE025618) (M.C.H.) with a research supplement (B.P.L.), by NIDCR grant K08 DE027705 (B.P.L.), and by the Pennsylvania State University’s Eberly College of Science (T.C.M.). This work was also supported by the Swedish Research Council and by the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, U.S. Department of Energy (grant DE-SC0015662) to Parastoo Azadi at the Complex Carbohydrate Research Center. The funding agencies had no role in the study design, data collection, interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward JB. 1981. Teichoic and teichuronic acids: biosynthesis, assembly, and location. Microbiol Rev 45:211–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong JJ, Baddiley J, Buchanan JG, Davision AL, Kelemen MV, Neuhaus FC. 1959. Composition of teichoic acids from a number of bacterial walls. Nature 184:247–248. doi: 10.1038/184247a0. [DOI] [PubMed] [Google Scholar]
- 4.Percy MG, Grundling A. 2014. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol 68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 5.Fischer W. 1988. Physiology of lipoteichoic acids in bacteria. Adv Microb Physiol 29:233–302. doi: 10.1016/s0065-2911(08)60349-5. [DOI] [PubMed] [Google Scholar]
- 6.Schneewind O, Missiakas D. 2014. Lipoteichoic acids, phosphate-containing polymers in the envelope of gram-positive bacteria. J Bacteriol 196:1133–1142. doi: 10.1128/JB.01155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miorner H, Johansson G, Kronvall G. 1983. Lipoteichoic acid is the major cell wall component responsible for surface hydrophobicity of group A streptococci. Infect Immun 39:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fedtke I, Mader D, Kohler T, Moll H, Nicholson G, Biswas R, Henseler K, Gotz F, Zahringer U, Peschel A. 2007. A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol Microbiol 65:1078–1091. doi: 10.1111/j.1365-2958.2007.05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert PA, Hancock IC, Baddiley J. 1977. Occurrence and function of membrane teichoic acids. Biochim Biophys Acta 472:1–12. doi: 10.1016/0304-4157(77)90012-0. [DOI] [PubMed] [Google Scholar]
- 10.Cleveland RF, Holtje JV, Wicken AJ, Tomasz A, Daneo-Moore L, Shockman GD. 1975. Inhibition of bacterial wall lysins by lipoteichoic acids and related compounds. Biochem Biophys Res Commun 67:1128–1135. doi: 10.1016/0006-291x(75)90791-3. [DOI] [PubMed] [Google Scholar]
- 11.Simpson WA, Courtney HS, Ofek I. 1987. Interactions of fibronectin with streptococci: the role of fibronectin as a receptor for Streptococcus pyogenes. Rev Infect Dis 9(Suppl 4):S351–S359. doi: 10.1093/clinids/9.supplement_4.s351. [DOI] [PubMed] [Google Scholar]
- 12.Granato D, Perotti F, Masserey I, Rouvet M, Golliard M, Servin A, Brassart D. 1999. Cell surface-associated lipoteichoic acid acts as an adhesion factor for attachment of Lactobacillus johnsonii La1 to human enterocyte-like Caco-2 cells. Appl Environ Microbiol 65:1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HY, Baik JE, Ahn KB, Seo HS, Yun CH, Han SH. 2017. Streptococcus gordonii induces nitric oxide production through its lipoproteins stimulating Toll-like receptor 2 in murine macrophages. Mol Immunol 82:75–83. doi: 10.1016/j.molimm.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Kim AR, Ahn KB, Kim HY, Seo HS, Kum KY, Yun CH, Han SH. 2017. Streptococcus gordonii lipoproteins induce IL-8 in human periodontal ligament cells. Mol Immunol 91:218–224. doi: 10.1016/j.molimm.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Rosan B, Lamont RJ. 2000. Dental plaque formation. Microbes Infect 2:1599–1607. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- 16.Garnier F, Gerbaud G, Courvalin P, Galimand M. 1997. Identification of clinically relevant viridans group streptococci to the species level by PCR. J Clin Microbiol 35:2337–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol 190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douglas CW, Heath J, Hampton KK, Preston FE. 1993. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol 39:179–182. doi: 10.1099/00222615-39-3-179. [DOI] [PubMed] [Google Scholar]
- 19.Veloso TR, Amiguet M, Rousson V, Giddey M, Vouillamoz J, Moreillon P, Entenza JM. 2011. Induction of experimental endocarditis by continuous low-grade bacteremia mimicking spontaneous bacteremia in humans. Infect Immun 79:2006–2011. doi: 10.1128/IAI.01208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gisch N, Auger JP, Thomsen S, Roy D, Xu J, Schwudke D, Gottschalk M. 2018. Structural analysis and immunostimulatory potency of lipoteichoic acids isolated from three Streptococcus suis serotype 2 strains. J Biol Chem 293:12011–12025. doi: 10.1074/jbc.RA118.002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boles BR, Horswill AR. 2011. Staphylococcal biofilm disassembly. Trends Microbiol 19:449–455. doi: 10.1016/j.tim.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol Mol Biol Rev 73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nobbs AH, Vajna RM, Johnson JR, Zhang Y, Erlandsen SL, Oli MW, Kreth J, Brady LJ, Herzberg MC. 2007. Consequences of a sortase A mutation in Streptococcus gordonii. Microbiology 153:4088–4097. doi: 10.1099/mic.0.2007/007252-0. [DOI] [PubMed] [Google Scholar]
- 25.Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. 2010. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 26.Rohde M, Chhatwal GS. 2013. Adherence and invasion of streptococci to eukaryotic cells and their role in disease pathogenesis. Curr Top Microbiol Immunol 368:83–110. doi: 10.1007/82_2012_281. [DOI] [PubMed] [Google Scholar]
- 27.Hasty DL, Ofek I, Courtney HS, Doyle RJ. 1992. Multiple adhesins of streptococci. Infect Immun 60:2147–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev 67:686–723. doi: 10.1128/mmbr.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mauel C, Young M, Margot P, Karamata D. 1989. The essential nature of teichoic acids in Bacillus subtilis as revealed by insertional mutagenesis. Mol Gen Genet 215:388–394. doi: 10.1007/bf00427034. [DOI] [PubMed] [Google Scholar]
- 30.Grundling A, Schneewind O. 2007. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J Bacteriol 189:2521–2530. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter SG, Elli D, Kim HK, Hendrickx AP, Sorg JA, Schneewind O, Missiakas D. 2013. Small molecule inhibitor of lipoteichoic acid synthesis is an antibiotic for Gram-positive bacteria. Proc Natl Acad Sci U S A 110:3531–3536. doi: 10.1073/pnas.1217337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oku Y, Kurokawa K, Matsuo M, Yamada S, Lee BL, Sekimizu K. 2009. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J Bacteriol 191:141–151. doi: 10.1128/JB.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwasaki H, Shimada A, Ito E. 1986. Comparative studies of lipoteichoic acids from several Bacillus strains. J Bacteriol 167:508–516. doi: 10.1128/jb.167.2.508-516.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer W. 1990. Bacterial phosphoglycolipids and lipoteichoic acids, vol 6 Springer, Boston, MA. [Google Scholar]
- 35.Ganfield MC, Pieringer RA. 1980. The biosynthesis of nascent membrane lipoteichoic acid of Streptococcus faecium (S. faecalis ATCC 9790) from phosphatidylkojibiosyl diacylglycerol and phosphatidylglycerol. J Biol Chem 255:5164–5169. [PubMed] [Google Scholar]
- 36.Reichmann NT, Grundling A. 2011. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett 319:97–105. doi: 10.1111/j.1574-6968.2011.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer W. 1994. Lipoteichoic acid and lipids in the membrane of Staphylococcus aureus. Med Microbiol Immunol 183:61–76. doi: 10.1007/bf00277157. [DOI] [PubMed] [Google Scholar]
- 38.Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. 2011. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci U S A 108:E1052–E1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sham LT, Jensen KR, Bruce KE, Winkler ME. 2013. Involvement of FtsE ATPase and FtsX extracellular loops 1 and 2 in FtsEX-PcsB complex function in cell division of Streptococcus pneumoniae D39. mBio 4:e00431-13. doi: 10.1128/mBio.00431-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sham LT, Barendt SM, Kopecky KE, Winkler ME. 2011. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc Natl Acad Sci U S A 108:E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meier EL, Daitch AK, Yao Q, Bhargava A, Jensen GJ, Goley ED. 2017. FtsEX-mediated regulation of the final stages of cell division reveals morphogenetic plasticity in Caulobacter crescentus. PLoS Genet 13:e1006999. doi: 10.1371/journal.pgen.1006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reichmann NT, Piçarra Cassona C, Monteiro JM, Bottomley AL, Corrigan RM, Foster SJ, Pinho MG, Gründling A. 2014. Differential localization of LTA synthesis proteins and their interaction with the cell division machinery in Staphylococcus aureus. Mol Microbiol 92:273–286. doi: 10.1111/mmi.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereira SF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. 2009. Evidence for a dual role of PBP1 in the cell division and cell separation of Staphylococcus aureus. Mol Microbiol 72:895–904. doi: 10.1111/j.1365-2958.2009.06687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pereira SF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. 2007. Role of PBP1 in cell division of Staphylococcus aureus. J Bacteriol 189:3525–3531. doi: 10.1128/JB.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinho MG, Filipe SR, de Lencastre H, Tomasz A. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J Bacteriol 183:6525–6531. doi: 10.1128/JB.183.22.6525-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Łeski TA, Tomasz A. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol 187:1815–1824. doi: 10.1128/JB.187.5.1815-1824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jordan S, Hutchings MI, Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev 32:107–146. doi: 10.1111/j.1574-6976.2007.00091.x. [DOI] [PubMed] [Google Scholar]
- 49.Gutberlet T, Frank J, Bradaczek H, Fischer W. 1997. Effect of lipoteichoic acid on thermotropic membrane properties. J Bacteriol 179:2879–2883. doi: 10.1128/jb.179.9.2879-2883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raivio TL. 2005. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol 56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 51.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev 9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 52.Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- 53.Hall JW, Lima BP, Herbomel GG, Gopinath T, McDonald L, Shyne MT, Lee JK, Kreth J, Ross KF, Veglia G, Herzberg MC. 7 May 2019. An intramembrane sensory circuit monitors sortase A-mediated processing of streptococcal adhesins. Sci Signal doi: 10.1126/scisignal.aas9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theilacker C, Sava I, Sanchez-Carballo P, Bao Y, Kropec A, Grohmann E, Holst O, Huebner J. 2011. Deletion of the glycosyltransferase bgsB of Enterococcus faecalis leads to a complete loss of glycolipids from the cell membrane and to impaired biofilm formation. BMC Microbiol 11:67. doi: 10.1186/1471-2180-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. 2004. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol 186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lazarevic V, Soldo B, Medico N, Pooley H, Bron S, Karamata D. 2005. Bacillus subtilis alpha-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl Environ Microbiol 71:39–45. doi: 10.1128/AEM.71.1.39-45.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim HY, Kim AR, Seo HS, Baik JE, Ahn KB, Yun CH, Han SH. 2018. Lipoproteins in Streptococcus gordonii are critical in the infection and inflammatory responses. Mol Immunol 101:574–584. doi: 10.1016/j.molimm.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 58.Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun 11:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie Z, Okinaga T, Qi F, Zhang Z, Merritt J. 2011. Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl Environ Microbiol 77:8025–8033. doi: 10.1128/AEM.06362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kho K, Meredith TC. 24 May 2018. Salt-induced stress stimulates a lipoteichoic acid-specific three-component glycosylation system in Staphylococcus aureus. J Bacteriol doi: 10.1128/JB.00017-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kho K, Meredith TC. 5 November 2018. Extraction and analysis of bacterial teichoic acids. Bio Protoc doi: 10.21769/BioProtoc.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santander J, Martin T, Loh A, Pohlenz C, Gatlin DM III, Curtiss R III. 2013. Mechanisms of intrinsic resistance to antimicrobial peptides of Edwardsiella ictaluri and its influence on fish gut inflammation and virulence. Microbiology 159:1471–1486. doi: 10.1099/mic.0.066639-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaplan CW, Lux R, Haake SK, Shi W. 2009. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol 71:35–47. doi: 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lima BP, Shi W, Lux R. 2017. Identification and characterization of a novel Fusobacterium nucleatum adhesin involved in physical interaction and biofilm formation with Streptococcus gordonii. Microbiologyopen 6:e00444. doi: 10.1002/mbo3.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vacharaksa A, Asrani AC, Gebhard KH, Fasching CE, Giacaman RA, Janoff EN, Ross KF, Herzberg MC. 2008. Oral keratinocytes support non-replicative infection and transfer of harbored HIV-1 to permissive cells. Retrovirology 5:66. doi: 10.1186/1742-4690-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ofek I, Whitnack E, Beachey EH. 1983. Hydrophobic interactions of group A streptococci with hexadecane droplets. J Bacteriol 154:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herzberg MC, Erickson PR, Kane PK, Clawson DJ, Clawson CC, Hoff FA. 1990. Platelet-interactive products of Streptococcus sanguis protoplasts. Infect Immun 58:4117–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thu YM, Van Riper SK, Higgins L, Zhang T, Becker JR, Markowski TW, Nguyen HD, Griffin TJ, Bielinsky AK. 2016. Slx5/Slx8 promotes replication stress tolerance by facilitating mitotic progression. Cell Rep 15:1254–1265. doi: 10.1016/j.celrep.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin-Moshier Y, Sebastian PJ, Higgins L, Sampson ND, Hewitt JE, Marchant JS. 2013. Re-evaluation of the role of calcium homeostasis endoplasmic reticulum protein (CHERP) in cellular calcium signaling. J Biol Chem 288:355–367. doi: 10.1074/jbc.M112.405761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin H, He L, Ma B. 2013. A combinatorial approach to the peptide feature matching problem for label-free quantification. Bioinformatics 29:1768–1775. doi: 10.1093/bioinformatics/btt274. [DOI] [PubMed] [Google Scholar]
- 71.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 72.Davies JR, Svensater G, Herzberg MC. 2009. Identification of novel LPXTG-linked surface proteins from Streptococcus gordonii. Microbiology 155:1977–1988. doi: 10.1099/mic.0.027854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rudney JD, Chen R, Lenton P, Li J, Li Y, Jones RS, Reilly C, Fok AS, Aparicio C. 2012. A reproducible oral microcosm biofilm model for testing dental materials. J Appl Microbiol 113:1540–1553. doi: 10.1111/j.1365-2672.2012.05439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erlandsen SL, Kristich CJ, Dunny GM, Wells CL. 2004. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J Histochem Cytochem 52:1427–1435. doi: 10.1369/jhc.4A6428.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Lei Y, Nobbs A, Khammanivong A, Herzberg MC. 2005. Inactivation of Streptococcus gordonii SspAB alters expression of multiple adhesin genes. Infect Immun 73:3351–3357. doi: 10.1128/IAI.73.6.3351-3357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun 63:3878–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Podbielski A, Spellerberg B, Woischnik M, Pohl B, Lutticken R. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137–147. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole-genome sequencing confirms ltaS deletion. The image shows the region of the S. gordonii genome at the ltaS locus in the WT or ΔltaS strains. The ΔltaS strain showed an absence of sequence alignment at the correct locus when aligned with the reference genome and WT strain. Download FIG S1, TIF file, 0.2 MB (230.4KB, tif) .
Copyright © 2019 Lima et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S. gordonii strains (WT and ΔltaS) were allowed to form biofilm on saliva-coated hydroxyapatite disks for 6 h. Biofilms were visualized at ×5,000. Download FIG S2, TIF file, 0.8 MB (939.3KB, tif) .
Copyright © 2019 Lima et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Equal numbers of WT and ΔltaS S. gordonii cells collected during mid-exponential phase were exposed to sterile PBS or water for 30 min. Next, the cells were plated on TH agar (A) and the colonies were enumerated to determine the percentage of surviving cells compared to the initial inoculum (B). Bars represent means of results from six biological replicates ± SD. Download FIG S3, JPG file, 0.03 MB (31.7KB, jpg) .
Copyright © 2019 Lima et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.