Abstract
Cannabis sativa L. has been utilized for a long time as a traditional herbal medicine in Korea. Dry fruits, achenes, each containing a single seed of Cannabis, are currently prescribed as Ma In (Cannabis Semen), a laxative. As each achene is enclosed by a bract, in which tetrahydrocannabinol (THC), the main psychological active compound in Cannabis is synthesized; achene is easily contaminated by THC from bract remnants. Therefore, it is safer to harvest achenes from Cannabis with a low THC content. Seeds of hemp, a low THC Cannabis, were recently classified as possible sources of new pharmacologically active compounds. Thus, a proper method to select appropriate Cannabis plants with low THC among cultivars in South Korea for medicinal purpose is necessary. As a result of cross-selection, Cannabis L. cultivar “Cheungsam” (CH) with the lowest THC content among cultivars cultivated in South Korea has been developed. In this study, we developed two DNA markers to reliably discriminate CH from other local cultivars with higher THC contents. We developed primer sets CHF3/CHR2 to amplify the 642 bp DNA marker of CH based on differences in the nucleotide sequences of the THCA synthase gene, which encodes a key enzyme in THC synthesis. We then developed a CHF1/CHR3 primer set to amplify the 401 bp DNA marker of CH based on the differences in both the content of very long chain fatty acids (VLCFs) and the sequence of the putative 3-ketoacyl-CoA synthase (KCS) gene encoding enzymes synthesizing VLCFs among local cultivars.
1. Introduction
Cannabis sativa L. has been cultivated for a long time for the supply of fibers, foods, and medicines [1]. Owing to selective breeding for centuries, over 700 different varieties and cultivars of C. sativa have been described and many more are thought to exist [2]. Varieties and cultivars of Cannabis are generally classified into two groups based on their purpose or tetrahydrocannabinol (THC) content (the primary psychological active component in Cannabis) and not on taxonomy [3]. The industrial nondrug Cannabis, known as hemp, has been cultivated for its fibers and seeds for thousands of years [4, 5], whereas the drug type, known as marijuana or hashish, is cultivated to obtain the intoxicant compound [6]. The THC content in the drug type is 1–3.7% (dry weight) [6], whereas in hemp, it is less than 0.2% [7] or 0.35% [8].
Achenes of C. sativa L. are currently prescribed as Ma In (Cannabis Semen; Huomaren (Cannabis Fructus) in Chinese), an herbal medicine used in Korea [9–11]. Ma In is used as a laxative; it stimulates the intestinal mucosa to excrete waste products through the large intestine and is especially formulated for old and pregnant individuals [12]. Each achene is enclosed by a persistent floral bract, which has a large number of glandular trichomes synthesizing various cannabinoids, including THC. The THC levels in Cannabis are highest in the bracts [13]. Therefore, achenes are easily contaminated by THC from bract remnants, confirmed using monoclonal antibodies against THC [14]. Achenes are suitable sources of Ma In that could be harvested from low THC Cannabis varieties or cultivars. Recently, various pharmacologically active compounds have been uncovered in seeds of low THC Cannabis plants, which have been classified as hemp; low THC Cannabis achenes are evaluated as sources of new compounds, which have health benefits [15]. Therefore, the selection of ideal Cannabis plants cultivated in South Korea is necessary to supply the proper sources of Ma In and develop new biologically active compounds.
Owing to cross fertilization, a new Cannabis cultivar, ‘Cheungsam' (CH), which has a THC content of 0.34%, has been developed in Korea [16]. Cheungsam was selected because it contained low THC concentrations and exhibited the best agronomic characteristics under South Korean environmental conditions among F1s of Korean local cultivars with high levels of THC (around 1.7%) and introduced low THC accessions (0.08–0.28%) from the Center for Plant Breeding and Reproduction Research (CPRO) in the Netherlands [16, 17]. Cheungsam is currently the lowest THC Cannabis among cultivars cultivated in South Korea.
Kojoma et al. [18] suggested that the substitution of amino acids caused by variations in nucleotides could result in an intense reduction in the activities of tetrahydrocannabinolic acid (THCA) synthases, a key enzyme used to synthesize THC and the level of THC in Cannabis. Doh [19] found variations in the composition and fatty acid content of achenes, especially in very long chain fatty acids (VLCFAs) synthesized by VLCFA elongase (3-ketoacyl-CoA synthase; KCS) among Korean cultivars. In this study, we developed two DNA markers to discriminate, indubitably, CH from other local South Korean cultivars with high THC content based on differences in the nucleotide sequences of the THCA synthase and putative KCS genes encoding key enzymes in the synthesis of VLCFAs for the supply of Cannabis achenes or seeds appropriate for medicinal use.
2. Materials and Methods
2.1. Plant Materials
Achenes of CH and local Cannabis cultivars from South Korea were collected from provinces, such as Dangjin (central western province), Boseung (south western province), Jecheon (central province), Jeogseon, and Andong (central eastern provinces) (Table 1). As the same cultivar was found in both Jungseon and Andong provinces, they will hereafter be referred to as “JU.” The samples listed in Table 1 are 1 g of each cultivar randomly selected from harvested seeds. Achenes used in this study were deposited as specimens at the department of herbology in the College of Korea Medicine, Wonkwang University.
Table 1.
The list of C. sativa achenes used in this study.
| Sample | Locality | Date of collection | Voucher no. |
|---|---|---|---|
| CH1 | Dangjin | 2009.7 | WCSCH01 |
| CH2 | 2009.7 | WCSCH02 | |
| CH3 | 2009.7 | WCSCH03 | |
| CH4 | 2009.7 | WCSCH04 | |
| CH5 | 2009.7 | WCSCH05 | |
| CH6 | 2009.7 | WCSCH06 | |
| CH7 | 2008.7 | WCSCH07 | |
| CH8 | 2008.7 | WCSCH08 | |
| CH9 | 2008.7 | WCSCH09 | |
| CH10 | 2008.7 | WCSCH10 | |
|
| |||
| BO1 | Boseung | 2009.7 | WCSBO01 |
| BO2 | 2009.7 | WCSBO02 | |
| BO3 | 2009.7 | WCSBO03 | |
| BO4 | 2009.7 | WCSBO04 | |
| BO5 | 2009.7 | WCSBO05 | |
| BO6 | 2009.7 | WCSBO06 | |
| BO7 | 2008.7 | WCSBO07 | |
| BO8 | 2008.7 | WCSBO08 | |
| BO9 | 2008.7 | WCSBO09 | |
| BO10 | 2008.7 | WCSBO10 | |
|
| |||
| JU1 | Jeongseon/Andong | 2008.7 | WCSJU01 |
| JU2 | 2008.7 | WCSJU02 | |
| JU3 | 2008.7 | WCSJU03 | |
| JU4 | 2008.7 | WCSJU04 | |
| JU5 | 2008.7 | WCSJU05 | |
| JU6 | 2008.7 | WCSJU06 | |
| JU7 | 2008.7 | WCSJU07 | |
| JU8 | 2008.7 | WCSJU08 | |
| JU9 | 2008.7 | WCSJU09 | |
| JU10 | 2008.7 | WCSJU10 | |
|
| |||
| JE1 | Jecheon | 2008.7 | WCSJE01 |
| JE2 | 2008.7 | WCSJE02 | |
| JE3 | 2008.7 | WCSJE03 | |
| JE4 | 2008.7 | WCSJE04 | |
| JE5 | 2008.7 | WCSJE05 | |
| JE6 | 2008.7 | WCSJE06 | |
| JE7 | 2008.7 | WCSJE07 | |
| JE8 | 2008.7 | WCSJE08 | |
| JE9 | 2008.7 | WCSJE09 | |
| JE10 | 2008.7 | WCSJE10 | |
2.2. Preparation of Genomic DNA
The genomic DNA of each sample was extracted using the NucleoSpin® Plant II (Macherey-Nagel, Duerern, Germany) according to the manufacturer's instructions.
2.3. Polymerase Chain Reaction (PCR) Amplification
2.3.1. Amplification of THCA Gene
The amplifications of THCA synthase genes by PCR were conducted using a T-personal cycler (Biometra, Goettingen, Germany) according to the protocol by Sirikantaramas et al. [20]. Briefly, 1.2 pmol of forward primer (5-TGAAGAAAAAAAATGAATTGCTCAGCATTTTCC-3′) and reverse primer (5′-TCTATTTAAAGATAATTAATGATGATGCGGTGG-3′), 1 U Taq polymerase (ABgene, Epson, UK), and 20 ng of genomic DNA extracted from each sample were used. The 35-cycle PCR process consisted of pre-denaturation for 5 min at 95°C, denaturation for 30 s at 95°C, annealing for 30 s at 58°C, extension for 1 min at 72°C, and a final reaction step for 7 min at 72°C. The amplified PCR product was separated on 1.5% agarose gel electrophoresis after staining with Safe-white™ (ABM, Canada) and analyzed using MyImage (Seoulin Biotechnology, Seoul, Korea).
2.3.2. Amplification of Putative KCS Gene
For the PCR amplification of putative KCS genes in C. sativa, forward primer HKCS F (5′-ATGACGTCCATTAACGTAAAGCTCC-3′) and reverse primer HKCS R (5′-TTAGGACCGACCG TTTTGGGC-3′) were designed based on the nucleotide sequences of KCS genes deposited in the NCBI GenBank. Approximately 2.4 pmol of both primers, 1 U Taq polymerase (ABgene, Epson, UK), and 20 ng of genomic DNA were used for PCR amplification. The 35-cycle PCR process consisted of pre-denaturation for 5 min at 95°C, denaturation for 30 s at 95°C, annealing for 30 s at 57°C, extension for 40 s at 72°C, and a final extension for 5 min at 72°C. The amplified PCR product was separated on 1.5% agarose gel electrophoresis after staining with Safe-white™ (ABM, Canada) and analyzed using MyImage (Seoulin Biotechnology, Seoul, Korea).
2.3.3. Amplification of DNA Markers
For the amplification of a 642 bp DNA marker for the discrimination of CH from other Korean local cultivars based on the nucleotide sequences of THCA genes, forward primer CHF3 (5′-TAGTACTCATGACTCACTTCAG-3′) and reverse primer CHR2 (5′-GTGTAATTATTAGGACTCG-CAG-3′) were used. The CHF3/ISR (5′-TCCACCATGAAAAATTGAAGA-3′) primer set was used to amplify 100 bp size internal standards (IS) to evaluate the PCR process. For the amplification of the 401 bp sized DNA marker for cultivar CH based on the nucleotide sequences of putative KCS genes, forward primer CHF1 (5′-TAGCTCATTCAGTCCGACGC-3′) and reverse primer CHR3 (5′-CGATTGTGCCTCTATCGG-3′) were used. The CHF1/ISR2 (5′-GAATGAGATTAAGCCGGC-3′) primer set was used as IS to amplify the 121 bp product. Briefly, 2.4 pmol of the CHF3/CHR2/IS R and CHF1/CHR3/ISR2 primer sets were used with 1 U Taq polymerase (ABgene, Epson, UK) and 20 ng of genomic DNA. The 23-cycle PCR amplification process of both DNA markers included pre-denaturation for 5 min at 95°C, denaturation for 30 s at 95°C, annealing for 30 s at 54°C, and an extension process for 1 min at 72°C.
2.3.4. Determination of Nucleotide Sequence-Amplified Products by PCR
PCR products separated on the agarose gel were cloned using the pGEM®-T Vector System I (Promega, Madison, WI, USA). The nucleotide sequences of cloned PCR products were determined by the Sanger sequencing method [21].
2.3.5. Alignment of Nucleotide Sequences and Construction of the Dendrogram
Nucleotide sequences were aligned using the Clustal W multiple sequence alignment program in Bioedit v.7.0.9 (http://mbio.ncsu.edu/BioEdit/page2.html). A THCA synthase dendrogram was constructed using DNADist in Bioedit. As an outer-group of a dendrogram, nucleotide sequences of genes encoding CBDA synthase of C. sativa (accession number AB292682.1) and berberine bridge enzymes of Eschscholtzia californica (AF005655.1) were used.
3. Results
3.1. Discrimination of CH from Korean Local Cultivars Based on Nucleotide Sequences of THC Synthase Genes
To distinguish CH, which is proper Cannabis, from Cannabis plants cultivated in South Korea for medicinal purposes, we developed DNA markers of CH. First, we tested whether CH could be efficiently separated from other Korean local cultivars with high THC content based on differences in nucleotide sequences of the THCA synthase gene in 40 samples (Table 1). Using primers designed by Sirikantaramas et al. [20], we amplified and determined the nucleotide sequences of 1,665 bp THCA synthase genes in CH and other cultivars (data not shown). Determined sequences were deposited in the NCBI GenBank (MN422084-MN422092). As shown in Figure 1, there were differences in the nucleotide sequences of THCA synthase genes among examined cultivars and within each cultivar. We detected 20 nucleotide variations between CH and BO, and eight nucleotide variations between JE and JU. There was no nucleotide variation within cultivar JE or JU. In contrast, two nucleotide variations were detected in two samples (B5 and B8) of BO and three nucleotide variations were detected in three samples (CH4, CH6, and CH8) of CH (Figure 1). To confirm whether CH could be differentiated from other cultivars based on differences in the sequences of THCA synthase genes despite the presence of nucleotide variations within CH or BO, we constructed a dendrogram based on the nucleotide sequences of THCA synthase genes (Figure 2). The nucleotide sequences of THCA synthase genes determined in this study and deposited in NCBI GenBank, which were isolated in Cannabis varieties or cultivars and classified into drug or nondrug types, were used to construct the dendrogram. The group composed of CH samples significantly differed from other groups consisting of local cultivars on the dendrogram (Figure 2). Therefore, we expected to be able to develop a DNA marker to distinguish CH from local cultivars based on differences in the nucleotide sequences of THCA synthase genes. We designed primer set CHF3/CHR2 based on variations in the 893rd and 1493rd nucleotides to amplify 642 bp DNA markers, which appeared solely in the CH samples (Figure 1). We also designed the primer set CHF3/ISR to amplify the 100 bp size internal standard (IS) and confirm the PCR amplification (Figure 2). As shown in Figure 3, 642 bp PCR products appeared uniquely, as expected, in randomly chosen CH samples listed in Table 1.
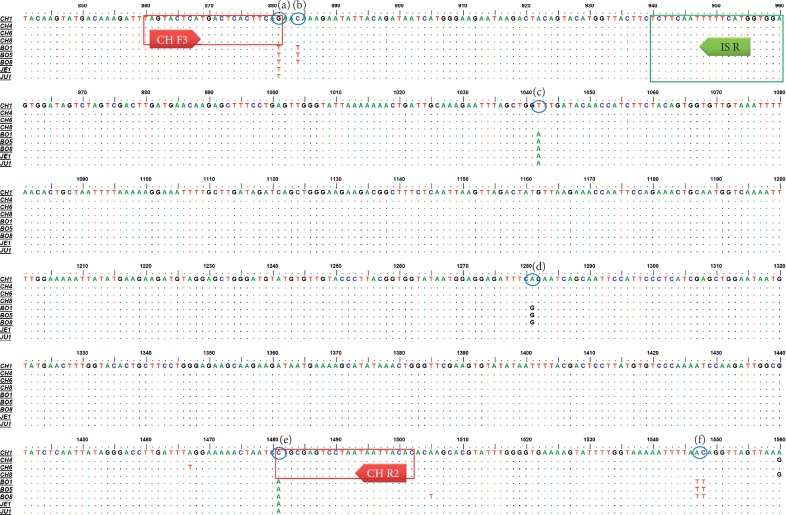
Figure 1.
Multiple alignment of the partial nucleotide sequences of tetrahydrocannabinolic acid (THCA) synthase genes from samples listed in Table 1. The dots indicate the consensus nucleotide; bold arrows indicate the primers used to amplify DNA markers of CH and the internal standard; the boxes represent nucleotide sequences and the positions of primers CHF3, CHR2, and ISR. (a)–(c) represent residues that might encode the regions located in the substrate-binding pocket beside to the FAD cofactor, and (d)–(f) represent residues that might encode the regions located on the surface of THCA synthase. The regions encoded by residues were predicted on the basis of the uncovered crystal structure of THCA synthase (PDB ID 3VTE) from Cannabis sativa L.
Figure 2.
Dendrogram based on the nucleotide sequences of tetrahydrocannabinolic acid (THCA) synthase genes of samples listed in Table 1 and deposited in NCBI GenBank. The deposited nucleotide sequences of THC synthase in drug-type Cannabis (AB212829.1, AB212832.1, AB212834.1, AB212835.1, AB2128837.1, and AB212838.1) and hemp (AB212830.1, AB212831.1, AB212833.1, AB212836.1, AB212839.4, AB212840.1, and AB212841.1) were used to construct the dendrogram. As outer-groups of a dendrogram, nucleotide sequences of genes encoding CBDA synthase of C. sativa (AB292682.1) and berberine bridge enzyme of E. californica (AF005655.1) were used.
Figure 3.
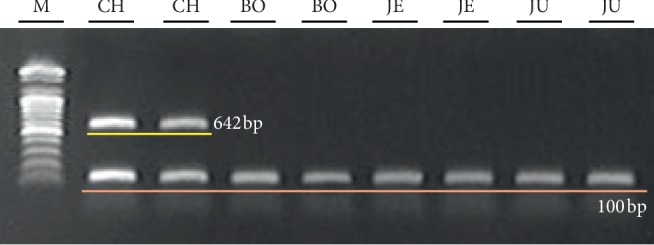
Polymerase chain reaction products of the primer sets, CHF3/CHR2 and CHF3/ISR, presented in Figure 1 from randomly chosen samples in Table 1 for the discrimination of “Cheungsam” cultivar. M: 100 bp ladder.
3.2. Amplification of Putative KCS Genes in CH and Korean Local Cultivars
We uncovered variations in the composition and concentration of fatty acids in seeds contained in achenes among CH and other local Korean cultivars, especially in VLCFs [19]. Therefore, we inferred that the variations in the nucleotide sequences of KCS genes encoding key enzymes of VLCFA biosynthesis could be another criterion used to select appropriate Cannabis from among South Korean cultivars for the medicinal applications. To the best of our knowledge, KCS genes participating in the synthesis of VLCFAs in seeds of Cannabis sativa L. have not been isolated as of yet. We aligned and compared the nucleotide sequences of seven KCS genes isolated from various plants, such as Arabidopsis and Brassica, deposited in the NCBI GenBank to determine the conserved regions of KCS genes, in which the consensus in the nucleotides among plants is shown (Figure 4). We also designed the primer set HKCS R/HKCS to amplify 1,563 bp regions in seven KCS genes (Figure 4). We amplified putative KCS genes in CH and local cultivars using this primer set, determined the nucleotide sequences of these PCR products, and deposited the sequences in the NCBI GenBank (MN422080-MN422083). There were no variations in the sequences of amplified PCR products within CH and other cultivars. Among cultivars, four nucleotide variations between CH and BO, and two nucleotide variations between JE and JU were detected. The partial nucleotide sequence of amplified PCR products of CH and other local cultivars is represented in Figure 5.
Figure 4.
Multiple alignment of the nucleotide sequences of 3-ketoacyl-CoA synthase (KCS) genes deposited in NCBI GenBank. The nucleotide sequences of KCS genes in Arabidopsis thaliana (EU29142.1), Brassica oleracea (AF490460.1), Brassica napus (AF490459.1 and AF490462.1), Brassica rapa (AF490461.1), Cardamine graeca (EU871788.1), and Lunaria annua (EU871787.1) were aligned. The dots indicate the consensus nucleotide; bold arrows indicate the primers to amplify putative KCS synthase genes. The boxes represent nucleotide sequences as well as the positions of primers.
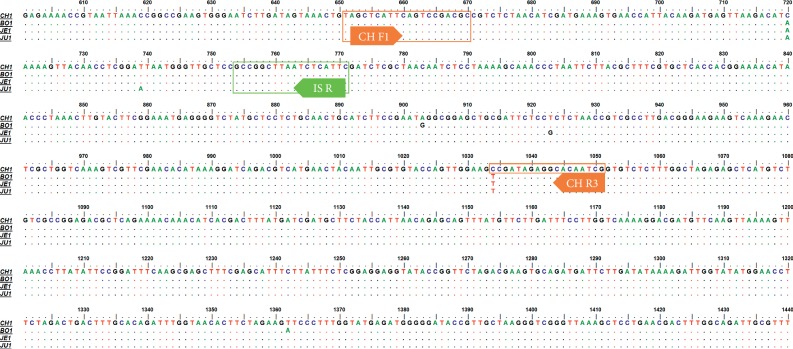
Figure 5.
Multiple alignment of the nucleotide sequences of partial putative 3-ketoacyl-CoA synthase (KCS) synthase genes amplified by the HKCS F/HKCS R primer set presented in Figure 4 from samples in Table 1. The dots indicate the consensus nucleotide; bold arrows indicate the primers to amplify DNA markers of CH and the internal standard; the boxes represent nucleotide sequences as well as the positions of primers CHF1, CHR3, and ISR.
3.3. Discrimination of CH from Korean Local Cultivars Based on the Nucleotide Sequences of Putative KCS Genes
To distinguish CH definitively from other South Korean local cultivars, we developed another DNA marker of CH based on the difference in the sequences of amplified putative KCS genes among cultivars. We designed the primer set CHF1/CHR3 to amplify the 401 bp DNA marker, which appeared only in samples of CH based on the substitution of C at the 1034th nucleotide of the amplified PCR product in CH to T in local cultivars (Figure 5). We also designed the primer set CHF1/ISR, which amplified the 121 bp internal standard for confirmation of the PCR process (Figure 5). As shown in Figure 6, 401 bp PCR products were amplified only in CH samples, as expected.
Figure 6.
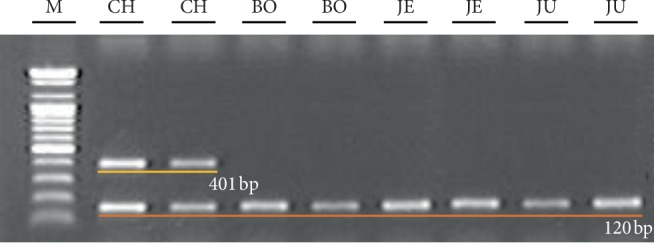
Polymerase chain reaction products of the primer sets, CH F1/CH R3 and CH F1/IS R2, presented in Figure 5 from randomly chosen samples in Table 1 for the discrimination of “Cheungsam” cultivar. M: 100 bp ladder.
4. Discussion
4.1. CH Is Distinguishable from Korean Local Cultivars by the Differences in the Sequence of the THCA Synthase Gene
The chemical features of Cannabis, specifically the THC content, are used to classify varieties and cultivars into drug and nondrug types (hemp) [7, 18]. However, the content of THC varies depending on the tissue or organ type [22]. It is also dependent on the developmental stage of the plant and environmental conditions, such as light and nutrients, under which it develops [23–25]. Moreover, long-term storage of Cannabis results in partial degradation of THC by light and oxygen [26]. In the former USSR and current Europe, 0.2% THC was the maximum permitted content for Cannabis to be classified as hemp [7, 27]. Apropos of this, currently, 0.3% and 0.35% THC are the maximum permitted content in Canada and New Zealand, respectively [8, 27]. In contrast, the relative content of THC to cannabidiol (CBD) (THC/CBD ratio), both of which are converted from the same precursor, cannabigerol, is believed to remain constant throughout all the developmental stages, relatively unaffected by environmental factors [25, 28]. When the ratio of THC/CBD is more than 1, Cannabis is classified as a drug type [7]. Our study was restricted to achenes, where THC and/or CBD was not detected unless and until contamination derived from bracts occurred [29]. Therefore, we were not able to compare the THC and THC/CBD ratio in CH and the other examined local cultivars. According to a report by Moon et al. [16] who developed CH, the THC and CBD levels in CH were 0.34% and 1.34%, respectively. Therefore, CH is on the border of being a drug and nondrug based on its absolute THC content. However, by the criterion of the THC: CBD ratio, CH is classified as hemp. Hillig and Mahlberg [30] and Hilling [31] classified landraces cultivated in eastern Asia, including Korea, into one putative taxon, C. indica biotype based on the two-species concept of Cannabis taxonomy or one of putative taxa comprising the C. indica gene pool based on the polytypic concept of Cannabis composed 3 species (C. sativa, C. indica, and C. ruderalis). Hillig and Mahlberg [30] showed that average THC levels were significantly higher in C. indica than in C. sativa. According to these results, almost all cultivars in South Korea could be drug-type Cannabis except CH. This possibility was further supported by the dendrogram presented in Figure 2. Cultivars examined in this study, excepting CH, were classified into subgroups of a drug-type group.
Seeds of hemp have been sources of essential amino acids and fatty acids for a long time [32]. However, the diverse pharmacological activities of hemp seeds in relation to human health, such as immunomodulatory effects, cardiovascular benefits, and anti-neuroinflammatory activities, were discovered recently [6, 32, 33]. Hemp is now considered a source of new bioactive compounds [15]. Therefore, CH might be an excellent source of not only Ma In but also for the development of new pharmacological compounds among cultivars examined from South Korea.
Although various DNA profiling methods, such as ITS and trnL–F intergenic spacer, have been applied to distinguish between CH and other cultivars, we were unable to find an effective method (data not shown).
Kojoma et al. suggested that types of Cannabis plants could be determined by the DNA polymorphism in THCA synthase gene [18]. A single nucleotide polymorphism (SNP) assay for the discrimination of drug from nondrug type Cannabis [34] and a DNA marker to predict THC content were developed [22] based on that suggestion.
Based on the uncovered crystal structure of THCA synthase of Cannabis sativa L. (PDB ID 3VTE), we predicted that residues which showed variations in the nucleotide sequences among cultivars especially marked as (a)–(c) in Figure 1 might encode region(s) located in the substrate binding pocket next to FAD cofactor. Other residues such as (e)–(f) might encode region(s) on the surface of THCA synthase. Therefore, variations in the nucleotide in these residues could affect the activity of THCA synthase in cultivar “CH” and finally decreased the content of THC. In this study, we applied variation in the nucleotide sequences of both residues (a) and (f) to design primers (Figure 1) to amplify 642 bp DNA markers to select “CH” (Figure 3)
4.2. Discrimination of CH from Korean Local Cultivars Based on the Differences in the Sequences of Putative KCS Gene
Doh [19] examined fatty acids in seeds of South Korea Cannabis cultivars. The main fatty acids were VLCFAs, such as C18 : 1, C18 : 2, and C18 : 3, as well as LCFAs (long chain fatty acids) such as C16 : 0 as reported previously [35–37]. Other VLCFAs, such as C20 : 0 and C22 : 2, were detected as minor fatty acids. Doh (2012) also found variations in the content and amount of fatty acids especially with respect to minor VLCFAs between CH and other local cultivars (data not shown).
The biosynthesis of VLCFs is catalyzed by acyl-CoA elongase, a membrane-bound enzyme complex composed of 3-ketoacyl-CoA synthase (KCS), 3-ketoacyl-CoA reductase (KCR), 3-hydroacyl-CoA dehydratase, and trans-2,3-enoyl-CoA reductase (ECR) [38]. The initial and rate-limiting step in fatty acid elongation is catalyzed by KCS. FAE1(KCS19), a KCS gene, which is believed to participate in the biosynthesis of C22 : 1 fatty acid in seeds, was first isolated in A. thaliana [39, 40]. Subsequently, 21 KCS genes, including KCS2, involved in the biosynthesis of specific VLCFAs, a precursor of aliphatic suberin in roots and seeds, were isolated in A. thaliana [41]. Therefore, we speculated that the amount and/or composition of VLCFA in seeds of Cannabis might, at least in part, be determined by the activity of specific KCS genes. The activity of KCS may be influenced by DNA polymorphism in KCS genes, and this DNA polymorphism could be used to distinguish CH from other local cultivars for the supply of sources of Ma In and developing new compounds.
As mentioned, it is well known that KCS are encoded by the multigene family. The number of members is increased up to 30 in divergent angiosperm species [42]. Therefore, for the amplification of putative KCS gene(s) participating in the synthesis of VLCFA in seeds of Cannabis in CH and other Korean cultivars by PCR, we compared the nucleotide sequences of seven KCS genes such as FAE1 in Arabidopsis to analyze the presence of variables and/or consensus regions in the nucleotide sequences among KCS (Figure 4). The nucleotide sequences of KCS genes in Cardamine graeca and Lunaria annua, in which nervonic acid (C24 : 1) is synthesized in seeds [43, 44] and in genus Brassica containing erucic acid (C22 : 1) as major fatty acids in seed oils [45], were also compared. As shown in Figure 5, regions such as those encoding N terminal (from 1st to 39th nucleotide) and the middle of the sequence (from 556th to 596th nucleotide, from 622nd to 642nd nucleotide, and from 1, 252nd to 1, 274th nucleotide) of the KCS genes were well conserved. The 1, 509th to 1, 527th nucleotide of KCS genes were highly conserved in the region encoding the C terminal. Based on the consensus nucleotide sequences encoding N and C terminals in examined KCS genes, we designed the primer sets, HKCS F/HKCS R, to amplify 1,563 bp size putative KCS genes in examined Cannabis plants (Figure 4). When we compared the nucleotide sequences of those PCR products with sequences deposited in NCBI, FAE1, and KCS in Arabidopsis (U29142.1, AY096568.1, AY074285.1, AF053345.1, and AK226331.1) and KCS in Brassica (AY642539.1, XM 013749750.1, NM 001316053.1, and AY642539.1), 82–87% and 80–85% sequence identities, respectively, were observed. Further studies are necessary to confirm that the amplified products in this study are the expected KCS genes that participate in the synthesis of VLCFAs in Cannabis seeds. As presented in Figure 5, there was variation in the nucleotide sequences among amplified putative KCS genes in CH and other local cultivars used to design the CHF1/CHR3 primer set to amplify 401 bp DNA markers to discriminate CH from other local cultivars (Figure 6). When these two developed DNA markers in this study are combined, we can select proper CH more definitively to supply resources for medicinal purposes among Cannabis cultivated in South Korea.
5. Conclusion
Cannabis sativa L. ‘Cheungsam' (CH), in which the THC content is the lowest among examined cultivars in South Korea, is the most appropriate Cannabis plant for the supply of achenes as herbal medicine, Ma In, and sources for developing new pharmacological compounds in seeds. In this study, we developed two specific DNA markers of CH to distinguish it from other local cultivars with higher THC contents. A 642 bp DNA marker of CH based on the differences in the nucleotide sequences of genes encoding THCA synthase, a key enzyme for THC synthesis, was amplified by the primer set CHF3/CHR2 designed in this study. The other 401 bp DNA maker of CH based on differences in the nucleotide sequences of putative 3-ketoacyl-CoA synthase (KCS), which plays a role in the synthesis of VLCFA, was amplified by the primer set CHF1/CHR3.
Data Availability
The nucleotide sequence data, besides those deposited in the NCBI GenBank, that support the findings of this study are available from the first or corresponding author upon request.
Disclosure
This study was based on the results of previous studies supported by Agricultural R&D Promotion Center in Republic of Korea (No. 308006-03). Eui Jeong Doh is presently at Research Center of Traditional Korean Medicine, Wonkwang University, Iksan 54538, Republic of Korea.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this study.
References
- 1.Brand E. J., Zhao Z. Cannabis in Chinese medicine: are some traditional indications referenced in ancient literature related to cannabinoids? Frontiers in Pharmacology. 2017;8 doi: 10.3389/fphar.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hazekamp A., Fischedick J. T. Cannabis-from cultivar to chemovar. Drug Testing and Analysis. 2012;4(7-8):660–667. doi: 10.1002/dta.407. [DOI] [PubMed] [Google Scholar]
- 3.Siniscalco Gigliano G. Preliminary data on the usefulness of internal transcribed spacer 1 (ITS1) sequence in Cannabis sativa L. identification. Journal of Forensic Sciences. 1999;44(3):457–475. doi: 10.1520/jfs14497j. [DOI] [PubMed] [Google Scholar]
- 4.Grotenhermen F., Karus M. Industrial hemp is not marijuana: comments on the durg potential fiber Cannabis. Journal of Industrial Hemp. 1998;5:96–101. [Google Scholar]
- 5.Kojoma M., Iida O., Makino Y., Sekita S., Satake M. DNA fingerprinting of Cannabis sativa using inter-simple sequence repeat (ISSR) amplification. Planta Medica. 2002;68(1):60–63. doi: 10.1055/s-2002-19875. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Leyva D., Pierce G. N. The cardiac and haemostatic effects of dietary hempseeds. Nutrition & Metabolism. 2010;7 doi: 10.1186/1743-7075-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Meijer E. P. M., van der Kamp H. J., van Eeuwijk F. A. Characterisation of Cannabis accessions with regard to cannabinoid content in relation to other plant characters. Euphytica. 1992;62(3):187–200. doi: 10.1007/bf00041753. [DOI] [Google Scholar]
- 8.McPartland J. M., Cutler S., McIntosh D. J. Hemp production in Aotearoa. Journal of Industrial Hemp. 2004;9(1):105–115. doi: 10.1300/j237v09n01_12. [DOI] [Google Scholar]
- 9.Ministry of Food and Drug Safety. The Korean Herbal Pharmacopoeia. Monographs, Part I, 4th edition, partial amendment 2017. [Google Scholar]
- 10.The Ministry of Health. Labour and Welfare, The Japanese Pharmacopoeia. 17th. 2016. p. p. 1919. Notification No. 64. [Google Scholar]
- 11.Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China. 2015. 2015. p. p. 80. Monographs, Part I. [Google Scholar]
- 12.Korea Institute of Oriental Medicine. Korea Oriental Medicine Classical Data Base. 2019. https://mediclassics.kr/books/8/volume/4#content_1421. [Google Scholar]
- 13.Small E., Marcus D. Tetrahydrocannabinol levels in hemp (Cannabis sativa) germplasm resources. Economic Botany. 2003;57(4):545–558. doi: 10.1663/0013-0001(2003)057[0545:tlihcs]2.0.co;2. [DOI] [Google Scholar]
- 14.Kim E.-S., Mahlberg P. G. Immunochemical localization of tetrahydrocannabinol (THC) in cryofixed glandular trichomes of Cannabis (Cannabaceae) American Journal of Botany. 1997;84(3):336–342. doi: 10.2307/2446007. [DOI] [PubMed] [Google Scholar]
- 15.Yan X., Tang J., dos Santos Passos C., et al. Characterization of lignanamides from hemp (Cannabis sativa L.) seed and their antioxidant and acetylcholinesterase inhibitory activities. Journal of Agricultural and Food Chemistry. 2015;63(49):10611–10619. doi: 10.1021/acs.jafc.5b05282. [DOI] [PubMed] [Google Scholar]
- 16.Moon Y. H., Lee B. H., Jeong B. C., Kim Y. U., Kim GY G. Y. Breeding history of non-drug type hemp variety “Cheungsam” and it’s characteristics. Korean Journal International Agriculture. 2002;14(2):119–126. [Google Scholar]
- 17.Moon Y. H., Song Y. S., Jeong B. C., Bang J. K. Cluster analysis and growth characteristics of hemp (Cannabis sativa L.) Germplasm. Korean Journal Corp Science. 2006;51(5):483–490. [Google Scholar]
- 18.Kojoma M., Seki H., Yoshida S., Muranaka T. DNA polymorphisms in the tetrahydrocannabinolic acid (THCA) synthase gene in “drug-type” and “fiber-type” Cannabis sativa L. Forensic Science International. 2006;159(2-3):132–140. doi: 10.1016/j.forsciint.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Doh E. J. Seoul, Korea: Konkuk University; 2012. Identification of medicinal plants by molecular biological techniques; pp. 80–83. Doctoral Dissertation. [Google Scholar]
- 20.Sirikantaramas S., Taura F., Tanaka Y., Ishikawa Y., Morimoto S., Shoyama Y. Tetrahydrocannabinolic acid synthase, the enzyme controlling marijuana psychoactivity, is secreted into the storage cavity of the glandular trichomes. Plant and Cell Physiology. 2005;46(9):1578–1582. doi: 10.1093/pcp/pci166. [DOI] [PubMed] [Google Scholar]
- 21.Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staginnus C., Zörntlein S., de Meijer E. A PCR marker linked to a THCA synthase polymorphism is a reliable tool to discriminate potentially THC-rich plants of Cannabis sativa L. Journal of Forensic Sciences. 2014;59(4):919–926. doi: 10.1111/1556-4029.12448. [DOI] [PubMed] [Google Scholar]
- 23.Vanhove W., van Damme P., Meert N. Factors determining yield and quality of illicit indoor cannabis (Cannabis spp.) production. Forensic Science International. 2011;212(1-3):158–163. doi: 10.1016/j.forsciint.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Toonen M., Ribot S., Thissen J. Yield of illicit indoor cannabis cultivation in The Netherlands. Journal of Forensic Sciences. 2006;51(5):1050–1054. doi: 10.1111/j.1556-4029.2006.00228.x. [DOI] [PubMed] [Google Scholar]
- 25.Pacifico D., Miselli F., Carboni A., Moschella A., Mandolino G. Time course of cannabinoid accumulation and chemotype development during the growth of Cannabis sativa L. Euphytica. 2008;160(2):231–240. doi: 10.1007/s10681-007-9543-y. [DOI] [Google Scholar]
- 26.Harvey D. J. Stability of cannabinoids in dried samples of cannabis dating from around 1896–1905. Journal of Ethnopharmacology. 1990;28(1):117–128. doi: 10.1016/0378-8741(90)90068-5. [DOI] [PubMed] [Google Scholar]
- 27.United Nations Office on Drug and Crime Vienna. Recommended Methods for the Identification and Analysis of Cannabis and Cannabis Products: Manual for Use by National Drug Analysis Laboratories. New York, NY, USA: United Nations; 2009. p. p. 20. [Google Scholar]
- 28.De Backer B., Maebe K., Verstraete A. G., Charlier C. Evolution of the content of THC and other major cannabinoids in drug-type cannabis cuttings and seedlings during growth of plants. Journal of Forensic Sciences. 2012;57(4):918–922. doi: 10.1111/j.1556-4029.2012.02068.x. [DOI] [PubMed] [Google Scholar]
- 29.Pate D. W. Chemical ecology of cannabis. Journal of International Hemp. 1994;2(29):32–37. [Google Scholar]
- 30.Hillig K. W., Mahlberg P. G. A chemotaxonomic analysis of cannabinoid variation in Cannabis (Cannabaceae) American Journal of Botany. 2004;91(6):966–975. doi: 10.3732/ajb.91.6.966. [DOI] [PubMed] [Google Scholar]
- 31.Hillig K. W. Genetic evidence for speciation in Cannabis (cannabaceae) Genetic Resources and Crop Evolution. 2005;52(2):161–180. doi: 10.1007/s10722-003-4452-y. [DOI] [Google Scholar]
- 32.Montserrat-de la Paz S., Marín-Aguilar F., García-Giménez M. D., Fernández-Arche M. A. Hemp (Cannabis sativa L.) seed oil: analytical and phytochemical characterization of the unsaponifiable fraction. Journal of Agricultural and Food Chemistry. 2014;62(5):1105–1110. doi: 10.1021/jf404278q. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y., Wang S., Lou H., Fan P. Chemical constituents of hemp (Cannabis sativa L.) seed with potential anti-neuroinflammatory activity. Phytochemistry Letters. 2018;23:57–61. doi: 10.1016/j.phytol.2017.11.013. [DOI] [Google Scholar]
- 34.Rotherham D., Harbison S. A. Differentiation of drug and non-drug Cannabis using a single nucleotide polymorphism (SNP) assay. Forensic Science International. 2011;207(1-3):193–197. doi: 10.1016/j.forsciint.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Calloway J. C., Laakkonen T. T. Cultivation of cannabis oil seed varieties in Finland. Journal of International Hemp. 1996;3(1):32–34. [Google Scholar]
- 36.Deferne J. L., Pate D. W. Hemp seed oil: a source of valuable essential fatty acids. Journal of International Hemp. 1996;3(1):4–7. [Google Scholar]
- 37.Leizer C., Ribnicky D., Poulev A., Dushenkov S., Raskin I. The composition of hemp seed oil and its potential as an important source of nutrition. Journal of Nutraceuticals, Functional & Medical Foods. 2000;2(4):35–53. doi: 10.1300/j133v02n04_04. [DOI] [Google Scholar]
- 38.Joubès J., Raffaele S., Bourdenx B., et al. The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Molecular Biology. 2008;67(5):547–566. doi: 10.1007/s11103-008-9339-z. [DOI] [PubMed] [Google Scholar]
- 39.Millar A. A., Kunst L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. The Plant Journal. 1997;12(1):121–131. doi: 10.1046/j.1365-313x.1997.12010121.x. [DOI] [PubMed] [Google Scholar]
- 40.James D. W., Lim E., Keller J., Plooy I., Ralston E., Dooner H. K. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. The Plant Cell. 1995;7(3):309–319. doi: 10.2307/3869853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franke R., Höfer R., Briesen I., et al. The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. The Plant Journal. 2009;57(1):80–95. doi: 10.1111/j.1365-313x.2008.03674.x. [DOI] [PubMed] [Google Scholar]
- 42.Gue H. S., Zhang Y. M., Sun X. Q., Li M. M., Hang Y. Y., Xue J. Y. Evolution of the KCS gene family in plants: the history of gene duplication, sub/neofunctionalization and redundancy. Molecular Genetics and Genomics. 2016;291:739–752. doi: 10.1007/s00438-015-1142-3. [DOI] [PubMed] [Google Scholar]
- 43.Jart A. The fatty acid composition of various cruciferous seeds. Journal of the American Oil Chemists’ Society. 1978;55(12):873–875. doi: 10.1007/bf02671410. [DOI] [Google Scholar]
- 44.Taylor D. C., Francis T., Guo Y., et al. Molecular cloning and characterization of a KCS gene from Cardamine graeca and its heterologous expression in Brassica oilseeds to engineer high nervonic acid oils for potential medical and industrial use. Plant Biotechnology Journal. 2009;7(9):925–938. doi: 10.1111/j.1467-7652.2009.00454.x. [DOI] [PubMed] [Google Scholar]
- 45.Sharafi Y., Majidi M. M., Goli S. A. H., Rashidi F. Oil content and fatty acids composition in Brassica Species. International Journal of Food Properties. 2015;18(10):2145–2154. doi: 10.1080/10942912.2014.968284. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequence data, besides those deposited in the NCBI GenBank, that support the findings of this study are available from the first or corresponding author upon request.