Abstract
Macrophages (MΘs) are key immune infiltrates in solid tumors and serve as major drivers behind tumor growth, immune suppression, and inhibition of adaptive immune responses in the tumor microenvironment (TME). Bromodomain and extraterminal (BET) protein, BRD4, which binds to acetylated lysine on histone tails has recently been reported to promote gene transcription of pro-inflammatory cytokines, but has rarely been explored for its role in IL4-driven MΘ transcriptional programming and MΘ-mediated immunosuppression in the TME. Herein, we report that BET bromodomain inhibitor JQ1, blocks association of BRD4 with promoters of arginase and other IL4-driven MΘ genes, which promote immunosuppression in TME. Pharmacological inhibition of BRD4 using JQ1 and/or PI3K using dual PI3K/BRD4 inhibitor SF2523 (previously reported by our group as a potent inhibitor to block tumor growth and metastasis in various cancer models) suppresses tumor growth in syngeneic and spontaneous murine cancer models; reduces infiltration of myeloid-derived suppressor cells (MDSCs); blocks polarization of immunosuppressive MΘs; restores CD8+ T-cell activity and stimulates anti-tumor immune responses. Finally, our results suggest that BRD4 regulates the immunosuppressive myeloid tumor microenvironment and BET inhibitors and dual PI3K/BRD4 inhibitors are therapeutic strategies for cancers driven by the MΘ-dependent immunosuppressive TME.
Keywords: macrophages, adaptive immunity, JQ1, BRD4, BMDMs
Introduction:
Small molecule epigenetic modulators have recently gained significant attention because of their potential as therapeutic agents (1-3). The bromodomain and extraterminal domain (BET) family of epigenetic “reader” proteins (BRDT, BRD2, BRD3 and BRD4) play important roles in histone acetylation-dependent transcriptional regulation (4). Various BET inhibitors successfully block BET protein recruitment and BET-mediated transcriptional activity (5,6). JQ1 and analogs were the first drug developed that specifically blocked interactions between multiple BET proteins (BRD2/3/4) and acetylated histones (7), and has been used to treat various myelomas and NUT midline carcinomas (8). Recent studies highlight that these BET inhibitors promote anti-tumor effects not only by inducing apoptosis and cell cycle arrest, but also by modulating innate and adaptive immune responses (9). The importance of BET proteins in innate macrophage (MΘ) inflammatory responses is well documented and recent studies suggest that BET protein inhibitors, I-BET and JQ1 can suppress pro-inflammatory cytokine production in MΘs (10,11). In context of adaptive immune responses, BRD4 inhibitors were recently shown to suppress expression of immune checkpoint ligand, PDL1, and this study has opened new avenues to explore BET inhibitors with immune modulating agents (12). In this regard, BET bromodomain inhibitors in combination with anti-PD1 leads to synergistic responses in a murine model of non-small lung cancer with KRAS activation (13) and in mice bearing Myc-driven lymphomas (9). BET inhibitors can also restore an immune-active environment in malignant pleural mesothelioma by reducing the expansion of myeloid derived suppressor cells (MDSCs) and by increasing intratumoral CD8+ T-cells (14). Taken together, these studies illustrate the strong role of BET bromodomain proteins in regulating innate and adaptive immune responses. However, the roles of these proteins have not been explored within the context of MΘ-mediated immunosuppression. MΘs play a crucial role in tumor progression, immunosuppression, and inhibition of anti-tumor adaptive immunity (15). Considering the role of BET proteins as important players in transcriptional elongation, we hypothesized that BRD4 plays a significant role in IL4-induced MΘ transcriptional programming and MΘ-mediated immunosuppression which leads to the inhibition of anti-tumor adaptive responses.
Another target for negative regulation of anti-tumor adaptive responses recently reported by our and other labs is the PI3K signaling pathway in particular, p110γ and p110δ in MΘs and regulatory T-cells, respectively (16-19). Our lab has reported that p110γ in MΘs promotes expression of pro-tumorigenic MΘs in the tumor microenvironment (TME) and our pan-PI3K inhibitor, SF1126, suppressed tumor growth and inhibited the HIF1α-VEGF signaling axis and other proangiogenic factors secreted by MΘs (16). This finding prompted us to test our novel dual PI3K/BRD4 inhibitor, SF2523, in the control of macrophage-mediated immunosuppression and activation of adaptive immune responses in the TME. SF2523 is a dual PI3K/BRD4 inhibitor generated from the TP series (20), which orthogonally hit PI3K and BRD4 to block expression, activation, and stabilization of Myc, leading to reduced tumor growth and metastasis in various preclinical models (21-23). The rationale for using this dual PI3K/BRD4 inhibitor in the present study is based on the hypothesis that targeting two crucial signaling pathways that promote MΘ-mediated immunosuppression and inhibit adaptive immune responses will provide greater benefit to patients. Our data demonstrate that BET bromodomain inhibitor, JQ1, and novel dual PI3K/BRD4 inhibitor, SF2523, demonstrate great potency in suppressing IL4 induced MΘ immunomodulatory responses, myeloid cell-mediated tumor growth, and metastasis in murine syngeneic and spontaneous tumor models. Taken together, these studies establish a role for BET proteins in MΘ-mediated immunosuppression and justify testing of BET protein–targeting drugs or dual PI3K/BRD4 drugs in cancers driven by immunosuppressive MΘs.
Materials and Methods
Isolation of bone marrow-derived macrophages, inhibitor experiments
Bone marrow-derived macrophages (BMDMs) were isolated as described previously (16,24). For IL4 and LPS stimulations, on day 7, BMDMs were treated with 20 ng/ml IL4 or 100 ng/ml LPS or 20ng/ml IFNγ for 10 hrs. For the inhibitor experiment, 7 day old MΘs were treated with 500 nM or 1 ¼M JQ1 or SF2523 for an hour followed by stimulation IL4 or LPS. JQ1 was provided by Dr. James Bradner.
Quantification of gene expression and RNA sequencing
Total RNA was isolated from BMDMs and sorted TAMs using the Qiagen RNAeasy kit (Qiagen, Hilden, Germany). cDNA (2 ¼L) was amplified by RT-PCR reactions as described before (24). Relative expression levels were normalized to Gapdh expression according to the formula, < 2^(Ct gene of interest-Ct Gapdh) > (25). For RNA sequencing, RNA libraries were prepared and sequenced on Illumina HiSeq2000 using standard Illumina protocols described in Supplementary methods. RNA sequencing data can be accessed using GEO accession number GSE123154.
Chromatin immunoprecipitation
Twenty four hours after LPS/IL4 stimulation and/or JQ1 treatment, BMDMs were fixed in 1% formaldehyde at 37°C for 10 min, then subjected to ChIP as previously published and described in Supplementary methods (11). ChIP and input DNA were analyzed by Real Time PCR using primers described in Supplementary Table S1. The fold difference was calculated as 2^[Ct(input) 2 Ct(ChIP)], and fold enrichment over an unrelated IgG Ab was assessed.
In vivo tumor growth and metastasis experiments
All procedures involving animals were approved by the University of California San Diego Animal Care Committee, which serves to ensure that all federal guidelines concerning animal experimentation are met. Lewis lung carcinoma (LLC) cells, CT26 colon adenocarcinomas and B16 melanomas were obtained from the American Type Culture Collection (ATCC), no further cell line authentication was performed by authors. All cells were cultured in DMEM media containing 10% FBS and tested for mycoplasma before implanting in animals. LLC cells or B16 (1 × 105) were injected subcutaneously into syngeneic 4–6 week old C57Bl/6 mice or 1 × 105 CT26 tumors were injected subcutaneously in Balb/c or nude mice and were treated with 40 mg/kg of JQ1 or 40mg/kg SF2523 when tumors reached a tumor volume of 100 mm3. For CD8 depletion, mice were treated with 200¼g of anti-CD8 (clone YTS 169.4) or an isotype control (LTF-2) from Bio-X-cell administered ip on day −3, 0, 3, 6 and 10 day of tumor inoculation.
B16 F10 luciferase melanoma cells (5 × 105) were injected intravenously and mice were treated with 40 mg/kg SF2523 as previously described (19). For spontaneous metastasis, 1 × 106 Panc02 were implanted orthotopically into the pancreas of syngeneic mice and were treated with 40 mg/kg SF2523 as described before (24). In some experiments, 9 week old PyMT+ female mice (26) (with spontaneous breast tumors) were treated with 40 mg/kg SF2523 (thrice weekly) for 4 weeks (n=10). Total tumor burden was obtained from detecting the total mammary gland mass in PYMT+ mouse.
Isolation of single cells from tumors and flow cytometry
Tumors were isolated, minced and then enzymatically dissociated in collagenase digestion cocktail at 37°C for 30–45 min and cells were prepared for magnetic bead purification of CD11b, Gr1 or CD90.2 cells for flow cytometry as reported before (24) and described in supplementary methods. Arginase activity was measured in MΘs sorted from tumors as previously described (24). In vitro cytotoxicity assay was performed using Cytotox non-radioactive cytotoxicity assay kit as described in Supplementary methods.
Results
BRD4 promotes immunosuppressive MΘ polarization
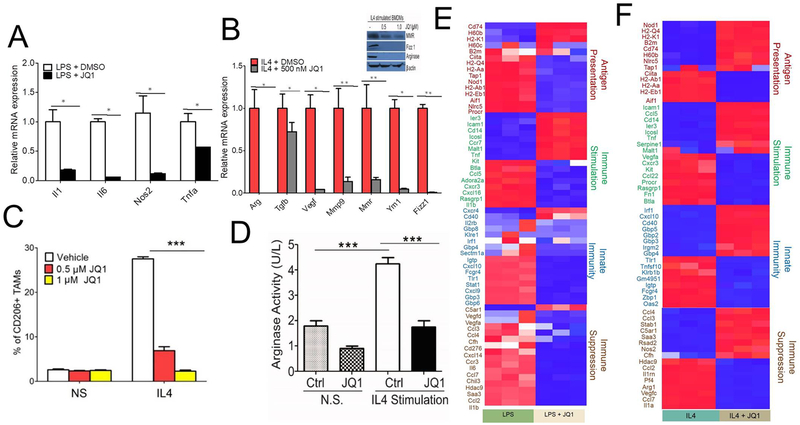
BET bromodomain proteins have recently been reported to play an important role in mouse MΘ inflammatory responses (11), but their role in modulating the expression of IL4-induced immunosuppressive genes has never been investigated. LPS induces MΘ expression of TH1 cytokines, IL1, IL6, and TNF alpha, whereas IL4 signaling stimulates TH2 response characterized by enhanced expression of arginase, scavenging molecules, and mannose and galactose receptors (27). To determine whether BRD4 controls transcriptional changes in MΘs, we exposed BMDMs to LPS and IL4 and concomitantly treated them with JQ1. Consistent with previously published reports (11), we found that JQ1 suppressed LPS-induced expression of pro-inflammatory genes viz. Il1, Il6, Nos2, and Tnfa in MΘs compared to control (Fig. 1A). Similarly, JQ1 suppressed IL4-induced expression of Arg, Tgfb, Vegf, Mmp9, Mmr, Ym1 and Fizz1 in JQ1-treated BMDMs compared to control (Fig. 1B). Moreover, western blot analysis of IL4 stimulated JQ1-treated BMDMs showed dose-dependent suppression of protein expression of MMR, FIZZ1, and Arginase (Fig. 1B). These results were also verified by inhibition of CD206 expression in JQ1 treated BMDMs as revealed by FACS analysis (Fig. 1C and Supplementary Fig. S1). Likewise, IL4-stimulated BMDMs showed a two-fold increase in arginase activity as compared to non-stimulated ones, and this increase is blocked by treating MΘs with 1¼M JQ1 (Fig. 1D). To further investigate the effect of BRD4 on immunosuppressive MΘ polarization, RNA-seq was performed on LPS-stimulated, or IL4-stimulated BMDMs treated with JQ1. Stimulation of BMDMs with LPS or IL4 upregulated numerous genes involved in antigen presentation, immune stimulation, innate immunity, and immune suppression (Supplementary Fig. S2). Pre-treatment of BMDMs with JQ1 shortly before LPS or IL4 stimulation resulted in the downregulation of 6484 of the 7822 significant (p ≤0.05) LPS-inducible genes and 7470 of 8979 significant (p ≤0.05) IL4-inducible genes (Supplementary Table S2). Most genes related to innate immunity, antigen presentation, and immune suppression upregulated by LPS were downregulated by JQ1 (Fig. 1E). However, in BMDMs treated with JQ1 and IL4, the genes related to antigen presentation and innate immunity were upregulated and immunosuppressive genes were downregulated (Fig. 1F). These results confirm that the BET protein inhibitor, JQ1, had relatively broad anti-inflammatory and immunomodulatory effects on MΘs.
Fig. 1. BET inhibitor JQ1 suppresses IL4 induced macrophage transcriptional programming:
A & B, RTPCR analysis of mRNA for immunostimulatory (A) or immunosuppressive genes (B) in BMDMs treated with 500 nM JQ1 followed by stimulation with 100ng/ml LPS + 20ng/ml IFNγ (A) or 20ng/ml IL4 (B) for 24 hrs, * P<0.05, ** P<0.01, student’s t–test. Inset in Fig. B shows western blot analysis of MMR, Fizz, and Arginase protein expression in IL4 stimulated BMDMs. C, FACS analysis of CD11b+F4/80+CD206+ MΘs in JQ1-treated BMDMs, two-way ANOVA with Boniferroni posttests, *** P<0.001. D, Arginase activity of JQ1-treated BMDMs. one-sided ANOVA with Tukey’s posthoc test where ** P<0.01. E-F, Heat map of log2 fold differences from sample wise mean expression for genes that were significantly differentially expressed (at an FDR of 0.05) in LPS-stimulated versus LPS-stimulated JQ1-treated BMDMs (E) or IL4-stimulated vs. IL4-stimulated JQ1-treated BMDMs (F) (n=3).
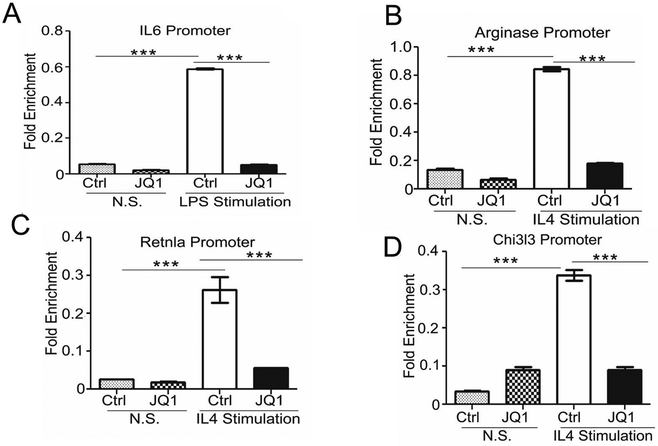
JQ1 treatment disrupts association of BRD4 with the regulatory chromatin of IL6, Arg, YM1, and Retnla promoters
Findings thus far corroborate that active cytokine promoters are packaged by acetylated histone proteins, and JQ1 functions as an anti-inflammatory compound by altering BET association with acetylated histone–packaged cytokine promoters (11), so we investigated if BRD4 physically associates with the gene promoters of anti-inflammatory MΘs. For these studies, we used anti-BRD4 antibodies to precipitate sheared chromosomal DNA from BMDMs stimulated with or without LPS/IL4 in the presence of JQ1. Similar to the previous report (11), our results validate that BRD4 modestly bound the IL6 promoter in LPS-stimulated cells and JQ1 blocked association of BRD4 with IL6 promoter (Fig. 2A). Interestingly, BRD4 modestly bound the Arg, Chi3l3/YM1, and Retnla/Fizz1 promoters in nonstimulated cells (Fig. 2 B-D); however, when cells were activated with IL4, we observed significant amounts of Arg, Chi3l3/YM1, and Retnla/Fizz1 promoter DNA precipitated with anti-BRD4 antibodies. Importantly, JQ1 treatment prevented association of both BRD4 with the Arg, Chi3l3/YM1, and Retnla/Fizz1 promoters (Fig. 2 B-D). These data suggest that the IL4-inducible association of BET proteins with the Arg, Chi3l3/YM1, and Retnla/Fizz1 promoters is significantly decreased in the presence of JQ1, which explains how JQ1 directs the association of BRD4 with IL4 driven genes and has a role in immunomodulation. Taken together, we identify BRD4 as a novel target that controls the expression of anti-inflammatory and immunosuppressive cytokines which promote immunosuppression.
Fig. 2. BRD4 binding to the IL6, Retnla, Chi3l3 and Arg transcriptional start site is abrogated by JQ1.
A-D, BRD4 ChIP analysis was performed at IL6 (A), Retnla (B), Chi3l3 (C) and Arg (D) transcriptional start site in BMDMs treated with JQ1 (1 μM), data was analyzed by one-sided ANOVA with Tukey’s posthoc test, *** P<0.001.
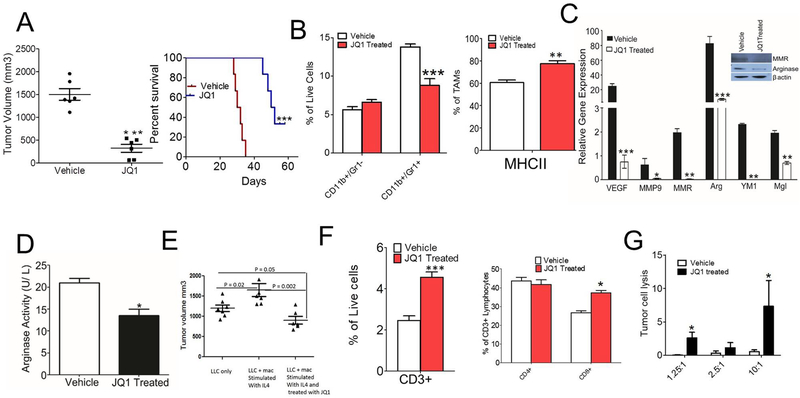
BET protein inhibition with JQ1 blocks MΘ polarization and immune suppression
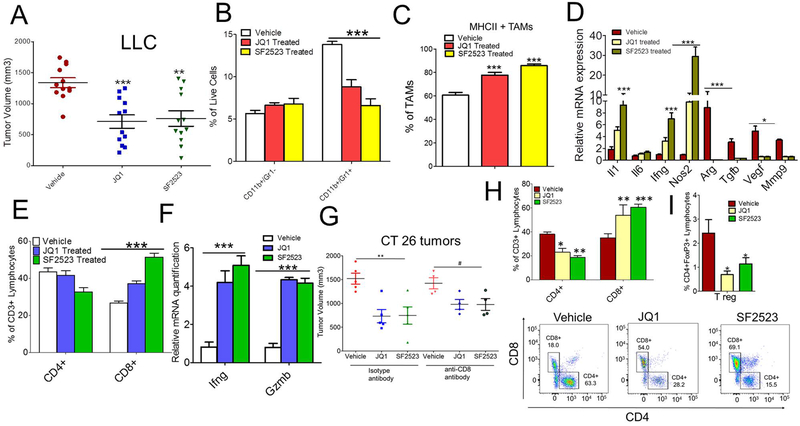
Based on data in Fig. 1, which show that JQ1 reduces expression of immunosuppressive MΘs in vitro, we investigated whether JQ1 blocks immunosuppression in TME. For this study, we injected C57BL/6 mice with LLC tumor cells, and the mice were treated with 50 mg/kg JQ1, when tumor volume reached 100 mm3. The tumor growth was suppressed in the JQ1 treated group as revealed by increased survival of mice treated with 50mg/kg JQ1 (Fig. 3A). JQ1 has no effect on proliferation of LLC tumor cells in vitro (Supplementary Fig. S3). Recent reports have shown that tumor-associated myeloid cells (TAMs), specifically CD11b+Gr1− monocytes and CD11b+Gr1+ granulocytes or MDSCs, are highly infiltrated in the TME where they inhibit innate and adaptive immune responses and promote tumor immune escape (28). Thus, we determined if BRD4 contributes to myeloid cell trafficking in tumors. We observed a significant reduction in the population of granulocytes or MDSCs infiltrating in the LLC (Fig. 3B, p < 0.01) tumors treated with JQ1 with no significant decrease in the CD11b+Gr1− monocyte population infiltrating the tumor (Fig. 3B). However, we noted a significant increase in infiltration of MHCII+ TAMs in the JQ1-treated group (Fig. 3B). We next investigated whether BRD4 inhibition alters expression of genes associated with immune suppression and tumor progression. The expression of immunosuppressive genes were significantly lower in TAMs isolated from JQ1 treated LLC tumors (Fig. 3C). These results were further verified by western blot analyses that displayed reduced protein expression of MMR and arginase in MΘs isolated from JQ1-treated tumors (Fig. 3C). Moreover, arginase activity was also found to be lower in the MΘs isolated from JQ1-treated tumors (Fig. 3D). In order to validate the hypothesis that the reduction in LLC tumor size observed in JQ1-treated mice was specifically due to its effect on IL4-driven immunosuppressive MΘs, adoptive transfer of IL4-stimulated MΘs treated with or without JQ1 was performed. The tumor growth increased in the mice injected with admix of LLC cells and IL4-stimulated MΘs compared to that of LLC alone as well as the admix of LLC cells and IL4-stimulated MΘs treated with JQ1 (Fig. 3E). To understand the mechanisms underlying the immunosuppression mediated by pharmacological inhibition of BRD4, we evaluated whether JQ1 affects activation of tumor infiltrating lymphocytes (TILs), which play an important role in anti-tumor immune response. Flow cytometric analysis of LLC tumors demonstrate that number of CD3+ and CD8+ T-cells were significantly enhanced in the LLC tumors treated with JQ1 (Fig. 3F, Supplementary Fig. S3). To investigate if inhibition of JQ1 activates T-cell-mediated anti-tumor immune response in vivo, TILs were isolated from tumors implanted in JQ1 treated mice and tumor cell cytotoxicity assay was performed. We found that CD90+ T-cells isolated from JQ1 treated LLC tumors significantly lyse LLC tumor cells at 1.25:1 and 10:1 ratio of T-cells: LLC cells (Fig. 3G). Taken together, our results demonstrate that JQ1 suppresses tumor growth by promoting recruitment of activated T-cells and reducing expression of immunosuppressive cytokines by MΘs.
Fig. 3: BET protein inhibition with JQ1 reduces macrophage-mediated immunosuppression and promotes anti-tumor adaptive immune response.
A, Left panel shows tumor volume of LLC cells injected into C57BL/6 mice and treated with 50 mg/kg JQ1. Graphs present mean ± SD of 5 mice in each group, unpaired t-test, *** p< 0.001. Right panel shows Kaplan Meier survival curve for JQ1 treated mice. B, Fig. shows CD11b+Gr1− and CD11b+Gr1+ cells infiltrated in LLC tumors treated with or without JQ1 (n=3), 2 way ANOVA with Bonferroni posttests *** p< 0.001 (left panel), two-tailed Student’s t-test (right panel) C, Quantitative PCR analysis of mRNA for immunosuppressive genes in the CD11b+ cells isolated from treated and untreated tumors described in A.,* p< 0.05, ** p< 0.01, *** p<0.001, two-tailed Student’s t-test. Fig. in inset shows western blot analysis of MMR, Fizz1 and Arginase in CD11b cells isolated from treated and untreated tumors described in A. D, Arginase activity assay done on CD11b+ cells isolated from JQ1 treated tumors described in A. E, Representative tumor volumes of mice injected with LLC either alone or in admix with IL4-stimulated MΘs treated with or without JQ1, one way ANOVA with Tukey’s posthoc test,* p< 0.05, ** p< 0.01, *** p<0.001. F, Flow cytometric quantification of CD3+ (left panel), CD4+ and CD8+ T cells (right panel) in the LLC tumors treated with JQ1 (n=5, p<.0001, t test). G, LLC tumor cell cytotoxicity induced by T cells isolated from LLC tumors from wild-type and JQ1 treated tumors. Ratio represents no. of T cells: LLC cells used in the assay, data analyzed by two-tailed Student’s t-test.
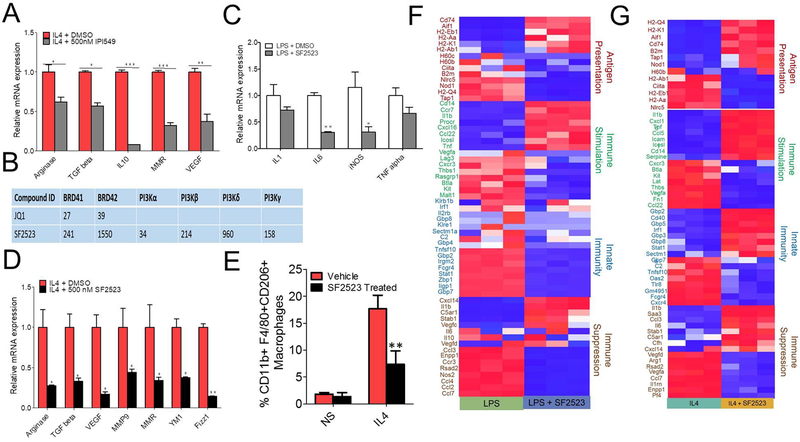
SF2523, a dual PI3K/BRD4 inhibitor, suppresses IL4-induced MΘ polarization
Recently, our and other laboratories reported that PI3Kγ is one target that is a major driver of tumor growth and immune suppression, and that inhibitors of this kinase promote adaptive immune responses in syngeneic mouse models (16,17,29). The PI3Kγ inhibitor, IPI549, decreases IL4-induced MΘ polarization (Fig. 4A), and pan-PI3K/BRD4 inhibitor, SF1126, reduces tumor growth and immunosuppressive MΘ polarization in the LLC tumor model (Supplementary Fig. S4 A and B) (16). Considering the role of both PI3K and BRD4 in MΘ-mediated immunosuppression and inhibition of adaptive immune responses, we examined if the dual PI3K/BRD4 inhibitor, SF2523, which showed potent in vivo activity in reducing tumor growth in various in vivo models (21), would have any effect on tumor growth, immunosuppression, and activation of adaptive immune responses. The chemical structure of SF2523 was provided before [21]. SF2523 is a dual PI3K/BRD4 inhibitor which hits both targets, BRD4 and PI3K, with nM potency compared to JQ1 which only hits BRD4 (Fig. 4B). We first investigated if SF2523 can decrease LPS- or IL4-induced MΘ polarization. SF2523 significantly decreased the mRNA expression of IL6 and iNos in LPS induced BMDMs and gene expression of Arg, Tgfb, Vegf, Mmr, Ym1, and Fizz1 in IL4-induced BMDMs (Fig. 4C & D). SF2523 also reduced CD206 expression in IL4-stimulated BMDMs as revealed by FACS analysis (Fig. 4E). RNA-seq experiments suggest that SF2523 treatment decreased expression of genes related to innate immunity, antigen presentation, and immune suppression although the cells were exposed to M1-polarizing LPS (Fig. 4F). However, most of the genes related to antigen presentation and innate immunity were upregulated and immunosuppressive genes were downregulated with SF2523 treatment in IL4-induced BMDMs (Fig. 4G). Overall, SF2523 exposure down regulated 4183 of 7822 significant (p ≤0.05) LPS-inducible genes and 3755 of 8979 significant (p ≤0.05) IL4-inducible genes (Supplementary Table S2).
Fig. 4: SF2523 a dual PI3K/BRD4 inhibitor suppresses LPS induced inflammatory gene expression as well as IL4 induced M2 macrophage response.
A, BMDMs were treated with 500 nM IPI-549 followed by stimulation with 20ng/ml IL4, RNA isolation and RTPCR analysis of genes promoting immunosuppression in TME, data analyzed by unpaired Student’s t-test ,* p< 0.05, ** p< 0.01, *** p<0.001 B, Enzymatic inhibition profiles of JQ1 and SF2523 against different targets. C-G, BMDMs were treated with 500 nM SF2523 followed by stimulation with 100ng/ml LPS + 20ng/ml IFNγ (C,F) or 20ng/ml IL4 (D,E,G) for 24 hrs and were used in RNA isolation for real time PCR analysis (C,D) or RNA -seq (F,G) or FACS analysis of CD11b+F4/80+CD206+ MΘs (E), one way ANOVA with Tukey’s post-hoc test, *** P<0.001.
SF2523 reduces tumor growth and immunosuppression
Our published report (21) provided proof of concept that SF2523 reduced tumor growth and metastasis in orthotopic Panc02 mouse model efficiently with less toxicity than in mice treated with two individual inhibitors combined JQ1 and BKM120 to inhibit BRD4 and PI3K signaling, respectively (21). The high efficacy and low toxicity of SF2523 prompted us to test if SF2523 could decrease MΘ-induced immunosuppression responses in vivo. For this, we injected LLC syngeneic tumor cells in C57Bl/6 mice and investigated its effect on tumor growth and immunosuppression in vivo. The administration of a 40 mg/kg dose of SF2523, three times a week, significantly reduced tumor growth in WT animals implanted with LLC (Fig. 5A), with no effect of SF2523 on the proliferation of LLC cells in vitro (Supplementary Fig. S5A). The reduction in tumor growth is comparable to the tumor reduction observed in mice treated with 40 mg/kg JQ1 (Fig. 5A). Next, we determined if SF2523 treatment can block recruitment of CD11b+Gr1+ MDSCs in the tumor. Interestingly, both JQ1 and SF2523 decreased MDSC recruitment in TME with no significant difference in recruitment of CD11b+Gr1− monocytes (Fig. 5B). However, the MHCII+ TAMs are highly recruited in the JQ1- and SF2523-treated group (Fig. 5C). Moreover, the expression levels of Arg, Mmp9, Tgfb, and Vegf are significantly higher in CD11b+ cells isolated from LLC tumors of untreated mice while usually under expressed in cells isolated from JQ1- or SF2523-treated mice (Fig. 5D). Interestingly, the expression of immunostimulatory genes viz., Il1, Il6, Ifng, and Nos2 are higher in the JQ1 and SF2523 treated group as compared to vehicle (Fig. 5D). We next determined if SF2523 treatment increased infiltration of CD8+ T-cells in TME. Both JQ1-and SF2523-treated tumors increased the infiltration and activation of CD8+ T-cells in the tumor as shown by increased mRNA expression of Ifng and Gzmb (Fig. 5E & F). Moreover, JQ1 and SF2523 both suppressed tumor growth in CT26 colon adenocarcinoma and B16 melanoma model (Fig. 5G and Supplementary Fig. S5B). In order to distinguish whether JQ1 and SF2523 reduces tumor growth due to its effect on immune cell compartment and not on tumor compartment, we tested the effect of JQ1 and SF2523 on CT26 tumor growth in immunodeficient NSG mice and immunocompromised Balb/c mice. JQ1 and SF2523 administration did not reduce CT26 tumor growth in NSG mice (Supplementary Fig. S5C), but blocked CT26 tumor growth in Balb/c mice (Fig. 5G), validating our results that JQ1 and SF2523 reduces tumor growth due to its effect on MΘs. These results were further verified in a parallel experiment where CD8 depletion didn’t effect CT26 tumor growth in JQ1- or SF2523-treated Balb/c mice (Fig. 5G & Supplementary Fig. S5D). There is significant increase in infiltration of CD8+ T cells in the JQ1- and SF2523-treated CT26 tumors and simultaneous decrease in CD4+ T cells and CD4+FoxP3+ regulatory T (Treg) cells in JQ1 and SF2523 treated tumors (Fig. 5H & I). All together, these results implicate both JQ1 and SF2523 as effective inhibitors of reducing tumor growth and activators of adaptive immune responses.
Fig. 5. SF2523 reduces tumor growth, immunosuppression and promotes anti-tumor adaptive immune responses.
A, Tumor volume of LLC inoculated subcutaneously in WT mice (n=12/group) treated with 40mg/kg JQ1 or SF2523. Data analyzed by one way ANOVA using Tukey’s post-hoc test ** P<0.01, *** P<0.001. B, CD11b+Gr1− and CD11b+Gr1+ cells infiltrated in LLC tumors treated with JQ1 or SF2523 (n=3), two way ANOVA with Bonferroni posttests *** p<0.001 C, MHCII+ TAMs infiltrated in the JQ1 or SF2523 treated LLC tumors, data in B and C, one-way ANOVA using Tukey’s post-hoc test,* p< 0.05, ** p< 0.01, *** p<0.001. D, Relative expression levels of immune regulatory genes in CD11b+ cells isolated from JQ1 or SF2523 treated LLC tumors mentioned in A subpanel. E, Flow cytometric quantification of CD3+, CD4+ and CD8+ T cells in the LLC tumors treated with JQ1 or SF2523. F, Relative mRNA expression of Ifng and Gzmb from JQ1- and SF2523-treated tumors. G, Tumor volume of CT26 tumors depleted with anti-CD8 antibody or isotype control antibody and treated with 40mg/kg JQ1 or 40 mg/kg SF2523. H & I, Flow cytometric quantification of CD4+, CD8+ (H, upper and lower panel) and CD4+FOXP3+ (I) T cells in the CT26 tumors treated with JQ1 or SF2523
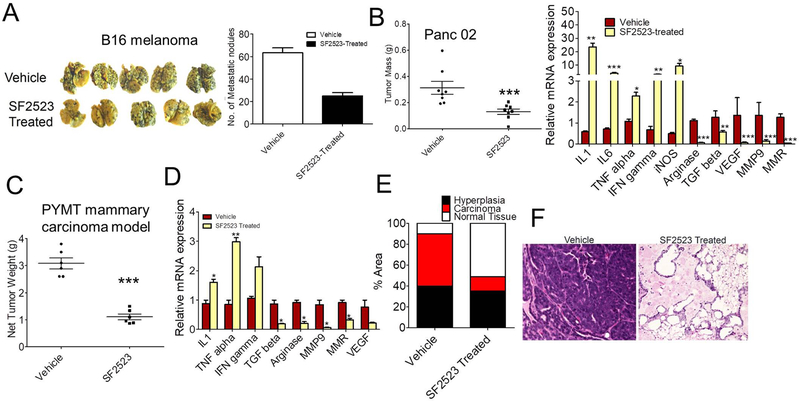
SF2523 reduces tumor metastasis in orthotopic and spontaneous mouse models
Given we demonstrated that SF2523 reduced tumor immunosuppression and activated adaptive immune responses (Fig. 5), we next examined if SF2523 can block metastasis in both experimental as well as spontaneous metastasis mouse models. B16F10 is considered a reliable method to study experimental metastasis in the C57BL/6 background (30). We observed a marked increase in luciferase signal coming from metastasized lungs of WT mice compared to low signal intensity in SF2523-treated mice as well as 75% reduction in metastatic nodules seen in SF2523-treated mice injected with B16F10 luciferase melanoma (p < 0.01; Fig. 6A and Supplementary Fig. S6). In order to study the effect of SF2523 on spontaneous metastasis, we used orthotopic Panc02 model and PYMT breast cancer model. SF2523 decreased tumor growth and immunosuppressive macrophage polarization in the orthotopic Panc02 mouse model (Fig. 6B). Net tumor weight and immunosuppressive macrophage polarization was strongly reduced in mammary tumors from the SF2523-treated mice (Fig. 6C-D). Tumor progression was also inhibited in SF2523-treated PYMT tumor model animals as tumors in these animals exhibited substantially less carcinoma and more normal tissue than control-treated animals (Fig. 6E-F).
Fig. 6. SF2523 reduces experimental as well as spontaneous breast tumor metastasis.
A, Experimental metastasis of B16 melanoma cells in WT mice treated with or without SF2523 (n=5). Left panel shows the photographs of the lungs removed on day 15. Right panel shows mean number of tumor nodules visible on the surface of the lungs in vehicle and SF2523-treated mice. B, Left panel shows tumor mass of Panc02 tumors orthotopically implanted in WT mice and treated with 40 mg/kg SF2523. Right panel shows gene expression of immune genes from CD11b+ cells isolated from SF2523 treated mice. C-D, Tumor burden (C) and mRNA expression of immunosuppressive genes (D) in spontaneous breast tumors from 13 week old control and SF2523-treated mice FVB PyMT+ mice (n=10). *p<0.01 vs. control. E, Fig. shows percent area of normal, hyperplastic, and carcinoma tissue from whole mounts of 4th mammary glands from C (n=5). F, H&E-stained mammary glands from Fig. E, scale bars, 40 μm.
Discussion
It is well established that MΘs represent the highly infiltrated leukocytes in the tumor compartment, and play a major role in inhibition of adaptive immune responses (15). Therapeutic strategies that are focused at blocking myeloid cell-mediated immunosuppression have shown great efficacy in preclinical mouse models and have been suggested as an effective strategy to suppress tumor metastasis. In the present study, we reveal that BET bromodomain inhibitors, which have an established role in blocking inflammation (10,11,31), can also suppress anti-inflammatory macrophage polarization in the TME (Fig. 1-3). Furthermore, based on this data and the literature supporting the role of PI3K signaling in immunosuppressive MΘ polarization and tumor inflammation (16,17), we evaluated the first-in-class dual PI3K/BRD4 inhibitor, SF2523, in IL4-induced MΘ polarization, and tumor metastasis. SF2523 is a rationally designed dual inhibitor can hit both PI3K and BRD4 signaling efficiently, with less toxicity as compared to combining individual inhibitors of PI3K and BRD4 signaling (21-23) and has the potential to reduce the tumor burden and increase immune effector response, which open opportunities to explore SF2523 in the treatment of MΘ-driven cancers.
We used multiple approaches inclusive of RTPCR analysis, RNA-Seq, and chromatin immunoprecipitation on primary BMDMs, to validate that JQ1 and SF2523 suppresses LPS- and IL4-induced MΘ transcriptional programming (Fig. 1 and 2). RNA-seq results suggest that SF2523 could be more effective at preserving pro-inflammatory MΘ polarization than JQ1 because it only down regulates 53.48% (4183/7822) of LPS-associated genes versus JQ1’s 82.89% (6484/7822). In particular, SF2523 down regulates a lower proportion of genes related to undesirable IL4-induced MΘ polarization (3755/8979, 41.82%) than JQ1 (7470/8979, 83.19%), indicating that BET bromodomain inhibitors, like JQ1 and PI3K/BRD4 dual inhibitors, use different mechanisms of action to manipulate MΘs into a more anti-tumor state.
Our ChIP-seq data clearly suggest that BRD4 physically associates with promoters of anti-inflammatory genes and JQ1 can disrupt this interaction (Fig. 2). Previous reports clearly suggest that histone methylation and acetylation are important for anti-inflammatory MΘ polarization (32,33). A recent study by Ishii et al. suggested that M2- MΘ marker genes are regulated epigenetically by the modification of histones, H3 lysine-4 (H3K4) and H3 lysine-27 (H3K27), via methylation; and the latter methylation marks are removed by the H3K27 demethylase Jumonji domain containing 3 (Jmjd3) (32). This study provides evidence of JMJD3 binding at M2 genes such as Arg1, Retnla, and Chi3l3. In addition to methylation, alternative activation is also mediated by a transcriptional program that is influenced by histone acetylation. Recently, a study reported the effect of a BET inhibitor, I-BET151, on human monocyte and MΘ responses (34). However, this study didn’t focus on the effect of BET bromodomain inhibition on IL4-stimulated M2 MΘs nor highlight any BRD4 binding with M2 MΘ promoters. Our data demonstrate direct BRD4 binding with the promoters of Arg, Chi3l3, and Retnla in IL4-stimulated macrophages, and that this association is blocked in the presence of JQ1.
Our results suggest that both JQ1 and SF2523 inhibit tumor growth and MΘ-mediated immunosuppression, and activate adaptive immune responses (Fig. 3-6). Consistent to our results, recent studies have shown that BET inhibitors reduce the expansion of MDSCs, increase activation of intratumoral CD8+ T-cells and promote anti-tumor immunity (12,14).
Although, the results in Fig. 5A, shows that blocking both PI3K and BRD4 signaling has no additive effect on tumor volume, but our data in Fig. 5 B-D shows that in comparison to JQ1, SF2523 is more effective in reducing infiltration of MDSCs and expression of immunosuppressive genes in macrophages infiltrated in tumor. Moreover in both LLC and CT26 tumors, it increases infiltration of CD8+ T cells and expression of immunostimulatory genes in macrophages compared to JQ1 (Fig. 5). Based on our previous (21) and current report, we can suggest that SF2523 might be more efficacious in Myc driven cancers, to suppress Myc activation and to provide sustained anti-tumor immunity. These data open new avenues to test BET inhibitors and dual PI3K/BRD4 inhibitors to treat cancers driven by the MΘ-dependent immunosuppressive TME.
Supplementary Material
Acknowledgements
JQ1 was provided by Dr. James Bradner. This work was supported by grants from NCI R01 CA94233–09 and FDA RO1 FD-04385 to D.L. Durden.
Footnotes
Disclosure of potential conflict of interest
J.R.G., G.A.M., and D.L.D. are consultants of SignalRx Pharmaceuticals and have financial conflicts of interest regarding the SF2523 compound used in this manuscript.
References:
- 1.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014;124(1):30–9 doi 10.1172/JCI6973869738 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crea F, Fornaro L, Bocci G, Sun L, Farrar WL, Falcone A, et al. EZH2 inhibition: targeting the crossroad of tumor invasion and angiogenesis. Cancer Metastasis Rev 2012;31(3–4):753–61 doi 10.1007/s10555-012-9387-3. [DOI] [PubMed] [Google Scholar]
- 3.He Y, Korboukh I, Jin J, Huang J. Targeting protein lysine methylation and demethylation in cancers. Acta Biochim Biophys Sin (Shanghai) 2012;44(1):70–9 doi 10.1093/abbs/gmr109gmr109 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hewings DS, Rooney TP, Jennings LE, Hay DA, Schofield CJ, Brennan PE, et al. Progress in the development and application of small molecule inhibitors of bromodomain-acetyl-lysine interactions. J Med Chem 2012;55(22):9393–413 doi 10.1021/jm300915b. [DOI] [PubMed] [Google Scholar]
- 5.Gosmini R, Nguyen VL, Toum J, Simon C, Brusq JM, Krysa G, et al. The discovery of I-BET726 (GSK1324726A), a potent tetrahydroquinoline ApoA1 up-regulator and selective BET bromodomain inhibitor. J Med Chem 2014;57(19):8111–31 doi 10.1021/jm5010539. [DOI] [PubMed] [Google Scholar]
- 6.Picaud S, Wells C, Felletar I, Brotherton D, Martin S, Savitsky P, et al. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci U S A 2013;110(49):19754–9 doi 10.1073/pnas.13106581101310658110 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature 2010;468(7327):1067–73 doi 10.1038/nature09504nature09504 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, Vakoc CR. Targeting Cancer Cells with BET Bromodomain Inhibitors. Cold Spring Harb Perspect Med 2017;7(7) doi a026674 [pii] 10.1101/cshperspect.a026674cshperspect.a026674 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogg SJ, Vervoort SJ, Deswal S, Ott CJ, Li J, Cluse LA, et al. BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep 2017;18(9):2162–74 doi S2211–1247(17)30176–6 [pii] 10.1016/j.celrep.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature 2010;468(7327):1119–23 doi 10.1038/nature09589nature09589 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol 2013;190(7):3670–8 doi 10.4049/jimmunol.1202838jimmunol.1202838 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, et al. BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep 2016;16(11):2829–37 doi S2211–1247(16)31096–8 [pii] 10.1016/j.celrep.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adeegbe DO, Liu S, Hattersley MM, Bowden M, Zhou CW, Li S, et al. BET Bromodomain Inhibition Cooperates with PD-1 Blockade to Facilitate Antitumor Response in Kras-Mutant Non-Small Cell Lung Cancer. Cancer Immunol Res 2018;6(10):1234–45 doi 10.1158/2326-6066.CIR-18-00772326-6066.CIR-18-0077 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riganti C, Lingua MF, Salaroglio IC, Falcomata C, Righi L, Morena D, et al. Bromodomain inhibition exerts its therapeutic potential in malignant pleural mesothelioma by promoting immunogenic cell death and changing the tumor immune-environment. Oncoimmunology 2018;7(3):e1398874 doi 10.1080/2162402X.2017.13988741398874 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity 2014;41(1):49–61 doi 10.1016/j.immuni.2014.06.010S1074-7613(14)00230-1 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi S, Singh AR, Zulcic M, Durden DL. A macrophage-dominant PI3K isoform controls hypoxia-induced HIF1alpha and HIF2alpha stability and tumor growth, angiogenesis, and metastasis. Mol Cancer Res 2014;12(10):1520–31 doi 10.1158/1541-7786.MCR-13-06821541-7786.MCR-13-0682 [pii]. [DOI] [PubMed] [Google Scholar]
- 17.Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S, et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature 2016;539(7629):437–42 doi 10.1038/nature19834nature19834 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid MC, Avraamides CJ, Dippold HC, Franco I, Foubert P, Ellies LG, et al. Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3kgamma, a single convergent point promoting tumor inflammation and progression. Cancer Cell 2011;19(6):715–27 doi 10.1016/j.ccr.2011.04.016S1535-6108(11)00160-7 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi S, Singh AR, Zulcic M, Durden DL. A Macrophage-Dominant PI3K Isoform Controls Hypoxia-Induced HIF1alpha and HIF2alpha Stability and Tumor Growth, Angiogenesis, and Metastasis. Mol Cancer Res 2014. doi 1541–7786.MCR-13–0682 [pii] 10.1158/1541-7786.MCR-13-0682. [DOI] [PubMed] [Google Scholar]
- 20.Morales GA, Garlich JR, Su J, Peng X, Newblom J, Weber K, et al. Synthesis and cancer stem cell-based activity of substituted 5-morpholino-7H-thieno[3,2-b]pyran-7-ones designed as next generation PI3K inhibitors. J Med Chem 2013;56(5):1922–39 doi 10.1021/jm301522m. [DOI] [PubMed] [Google Scholar]
- 21.Andrews FH, Singh AR, Joshi S, Smith CA, Morales GA, Garlich JR, et al. Dual-activity PI3K-BRD4 inhibitor for the orthogonal inhibition of MYC to block tumor growth and metastasis. Proc Natl Acad Sci U S A 2017;114(7):E1072–E80 doi 10.1073/pnas.16130911141613091114 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen G, Jiang M, Pu J. Dual inhibition of BRD4 and PI3K by SF2523 suppresses human prostate cancer cell growth in vitro and in vivo. Biochem Biophys Res Commun 2018;495(1):567–73 doi S0006–291X(17)32240–4 [pii] 10.1016/j.bbrc.2017.11.062. [DOI] [PubMed] [Google Scholar]
- 23.Zhu H, Mao JH, Wang Y, Gu DH, Pan XD, Shan Y, et al. Dual inhibition of BRD4 and PI3K-AKT by SF2523 suppresses human renal cell carcinoma cell growth. Oncotarget 2017;8(58):98471–81 doi 10.18632/oncotarget.2143221432 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi S, Singh AR, Zulcic M, Bao L, Messer K, Ideker T, et al. Rac2 controls tumor growth, metastasis and M1-M2 macrophage differentiation in vivo. PLoS One 2014;9(4):e95893 doi 10.1371/journal.pone.0095893PONE-D-13-51894 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 26.Davie SA, Maglione JE, Manner CK, Young D, Cardiff RD, MacLeod CL, et al. Effects of FVB/NJ and C57Bl/6J strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic Res 2007;16(2):193–201 doi 10.1007/s11248-006-9056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 2014;6:13 doi 10.12703/P6-1313 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol 2016;37(3):208–20 doi 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Henau O, Rausch M, Winkler D, Campesato LF, Liu C, Cymerman DH, et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature 2016;539(7629):443–7 doi 10.1038/nature20554nature20554 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khanna C, Hunter K. Modeling metastasis in vivo. Carcinogenesis 2005;26(3):513–23 doi 10.1093/carcin/bgh261bgh261 [pii]. [DOI] [PubMed] [Google Scholar]
- 31.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol 2009;9(10):692–703 doi 10.1038/nri2634nri2634 [pii]. [DOI] [PubMed] [Google Scholar]
- 32.Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 2009;114(15):3244–54 doi 10.1182/blood-2009-04-217620blood-2009-04-217620 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullican SE, Gaddis CA, Alenghat T, Nair MG, Giacomin PR, Everett LJ, et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev 2011;25(23):2480–8 doi 10.1101/gad.175950.11125/23/2480 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Qiao LBI. Effect and mechanism of BET bromodomain inhibition in macrophage transcriptional programming. Inflammation & Cell Signaling 2015;2: (e600). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.