Abstract
RNA‐binding proteins (RBPs) participate in all steps of gene expression, underscoring their potential as regulators of RNA homeostasis. We structurally and functionally characterize Mip6, a four‐RNA recognition motif (RRM)‐containing RBP, as a functional and physical interactor of the export factor Mex67. Mip6‐RRM4 directly interacts with the ubiquitin‐associated (UBA) domain of Mex67 through a loop containing tryptophan 442. Mip6 shuttles between the nucleus and the cytoplasm in a Mex67‐dependent manner and concentrates in cytoplasmic foci under stress. Photoactivatable ribonucleoside‐enhanced crosslinking and immunoprecipitation experiments show preferential binding of Mip6 to mRNAs regulated by the stress‐response Msn2/4 transcription factors. Consistent with this binding, MIP6 deletion affects their export and expression levels. Additionally, Mip6 interacts physically and/or functionally with proteins with a role in mRNA metabolism and transcription such as Rrp6, Xrn1, Sgf73, and Rpb1. These results reveal a novel role for Mip6 in the homeostasis of Msn2/4‐dependent transcripts through its direct interaction with the Mex67 UBA domain.
Keywords: Mex67, Mip6, mRNA export, Msn2/4, RNA‐binding protein
Subject Categories: Membrane & Intracellular Transport, RNA Biology
The RNA binding protein Mip6 interacts with the ubiquitin‐associated domain of Mex67 and attenuates the nuclear export of mRNAs regulated by the stress response Msn2/4 transcription factors.

Introduction
mRNA export from the nucleus is a crucial step in gene expression in order to ensure efficient protein production. In eukaryotic cells, mRNA export is connected to upstream and downstream events that modulate mRNA fate through the action of a repertoire of factors and multiprotein complexes 1, 2, 3, 4. Among these factors, the yeast export receptor heterodimer Mex67‐Mtr2 (TAP‐p15 in humans), which is loaded onto transcripts early during transcription 5, 6, is crucial for mRNA export, conferring competency on messenger ribonucleoparticles (mRNPs) for docking at the nuclear pore complex (NPC). In addition to Mex67‐Mtr2, mRNPs incorporate adaptor proteins that modulate mRNA metabolism and that can act positively or negatively in mRNA export 7, 8, 9. The best known of these proteins are the RNA‐binding proteins Nab2, Hrb1, Gbp2, and Npl3 that interact with Mex67‐Mtr2 10, 11, 12. Furthermore, some RNA‐binding proteins that are also known as “guard proteins” can bind to pre‐mRNA, acting as a tag of the unprocessed state 13 and preventing undesirable Mex67 contacts, thereby functioning as a mechanism for blocking uncontrolled RNA export 10, 14, 15. This quality control mechanism might also be used to regulate export routes under specific circumstances such as stress during cellular growth.
Cells have evolved a sophisticated stress response that includes a specific gene expression program 16, 17, 18, 19. Under stress, splicing of most housekeeping genes is disrupted and their export to the cytoplasm is blocked 16, 18. Simultaneously, the export and translation of stress‐responsive genes is enhanced, and their increased expression becomes essential for cell survival 20, 21. Recent work has investigated the role of “guard proteins” in controlling Mex67 binding to housekeeping and stress‐responsive transcripts upon stress 15. These data showed that Mex67 is directly recruited to stress‐responsive transcripts through interaction with the transcription factor Hsf1 at early steps during transcription. This recruitment is independent of adaptor proteins and facilitates the export of these transcripts, bypassing the quality control mechanisms. However, such independence from adaptor (or guard) proteins that is observed for Hsf1 targets might not be universal for all types of stress‐responsive mRNAs and furthermore cannot be assumed for the variety of stresses in which Hsf1 is not directly involved. An example of the latter case is Msn2/4 transcription factor‐dependent transcription 15 that is involved in the environmental stress response (ESR) that senses different kinds of stresses including heat shock, glucose starvation, and oxidative and osmotic stress 22, 23, 24, 25. Binding of Msn2/4 to specific DNA sequences leads to a transcriptional change that mediates cell survival 26, 27. Whether Msn2/4 transcripts are independent of adaptor proteins or whether they require specific factors for their metabolism is currently unknown.
Mip6 (Mex67 Interacting Protein 6) was discovered in 1997 during a yeast two‐hybrid (Y2H) screen using Mex67 as bait 28. This putative RNA‐binding protein bears four RRMs and was suggested by Hurt and colleagues to interact with the C‐terminal domain of Mex67. However, it is only recently that further functional characterization of Mip6 was performed, when the role of this factor in different fields started to attract attention. In a recent screen in yeast for dosage‐sensitive proteins that have a propensity for liquid–liquid phase separation (LLPS), it was found that, at least for Mip6, liquid–liquid demixing is associated with toxicity when the protein is highly overexpressed 29. Under these conditions, Mip6 changes its cellular localization and accumulates into liquid‐like cytoplasmic foci. Apart from increasing interest in understanding the physiological consequences of liquid–liquid demixing in the cell, this study did not address the functional role of Mip6 in yeast. A more recent study proposed a specific role for Mip6 during sporulation 30. Mip6, together with its paralogue Pes4, was suggested to act as regulators of late translation, protection, and mRNA localization within the NDT80 regulon that are translated at different times during sporulation.
In this work, we showed that Mip6 interacts functionally and/or physically with factors involved in several steps of mRNA metabolism such as Mex67, Rrp6, Xrn1, Sgf73, and Rpb1 and we further characterized its interaction with the general exporter Mex67 both at functional and at structural levels. The functional and physical interaction between Mip6 and these factors has been characterized by using a multidisciplinary approach, combining biophysical, biochemical, molecular, and microscopical techniques. The data show that Mip6 and Mex67 interact directly and that this interaction is important for Mip6 in vivo function, which is related to the Msn2/4 regulatory network attenuating the expression of their target genes under non‐stress. These data indicate that Mip6 is a new regulator of stress responses.
Results
Mip6 interacts with key components of the mRNA export machinery
In searching for novel interacting partners of Sus1, we purified Sus1 in the absence of Ubp8, when it is preferentially associated only with the deubiquitination module (DUB) and with TREX2 31. In addition to expected factors, we found that the putative RNA‐binding protein Mip6 co‐purified with Sus1‐TAP (Fig 1A). Mip6 was first reported as the Mex67 Interacting Protein 6 (MIP6) in a two‐hybrid screen using Mex67 as a bait 28. We further confirmed that Mip6 co‐purified with Mex67 from yeast cells (Fig 1B) and that this interaction is not affected by RNase A treatment (Fig 1C), indicating that it is not RNA‐dependent. In addition to its observed physical interaction with Mex67, the lack of MIP6 led to a negative synthetic growth phenotype when combined with mex67‐5, a thermosensitive allele of Mex67 28, 32, at 30°C and at 33°C (Fig 1D). Thus, Mip6 interacts genetically and physically with the general eukaryotic export factor Mex67 and physically with Sus1.
Figure 1. Mip6 interacts with components of the mRNA export machinery.

- Sus1‐TAP precipitation from WT and ubp8∆ strains. Sus1‐TAP and co‐precipitating proteins were analyzed on an SDS 4–12% gradient polyacrylamide gel and stained with Coomassie Blue. Bands corresponding to Sus1‐TAP and Mip6, identified by mass spectrometry, are indicated.
- Immunoprecipitation with the TAP antibody using no‐tag or Mip6‐TAP‐expressing cells. Mip6‐TAP and Mex67 were detected in INPUT and IP samples by Western blotting.
- As in (B) in the mip6Δ strain transformed with Mip6‐GFP‐expressing plasmid with and without RNase A treatment at the indicated concentrations.
- Growth of WT, and of mip6Δ, mex67‐5, and mip6Δmex67‐5 mutants. Precultures were diluted in liquid YPD medium. The 10‐fold dilutions of cells were then spotted onto YPD plates and incubated for 2 days at the indicated temperatures.
The Mex67 UBA domain specifically recognizes Mip6 RRM4
To better characterize the Mip6‐Mex67 physical interaction, we used biophysical and structural methods. Pull‐down assays showed co‐purification of recombinant Mip6 RRM1‐4 and Mex67(528–599) (Appendix Fig S1 and Fig EV1A). This Mex67(528–599) construct includes the C‐terminal part of Mex67 containing the UBA domain that was found to interact with Mip6 by Y2H 28 (Appendix Fig S1). In contrast, the Mip6 construct lacking the fourth RRM (Mip6 RRM1‐3) lost the ability to interact with Mex67(528–599) (Fig EV1A), indicating that the UBA site of Mex67 binds to Mip6 RRM4. This binding was confirmed by size‐exclusion chromatography (Fig EV1B).
Figure EV1. Mex67 interacts with Mip6 RRM4.
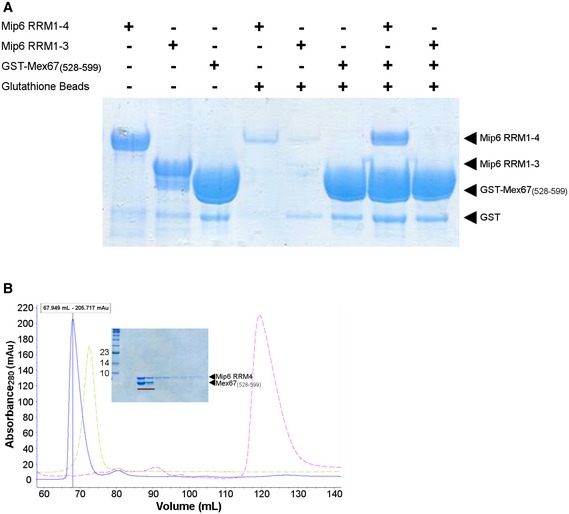
- Mip6 RRM4 mediates the interaction of Mip6 with Mex67(528–599). Purified recombinant GST‐tagged Mex67(528–599) was bound to pre‐equilibrated Glutathione‐Agarose beads as indicated. After washing, the beads were incubated with purified untagged recombinant Mip6 RRM1‐4 or RRM1‐3 as indicated, were washed again, and were then loaded onto a 10% SDS–PAGE gel that was subsequently stained with Coomassie Blue.
- Stable complex of Mex67(528–599) and Mip6 RRM4(401–480) during gel filtration. Superposition of the chromatogram representing the size‐exclusion elution profile of the complex formed between Mex67(528–599) and Mip6 RRM4(401–480), obtained from a 16/60 Superdex 75 gel filtration column at a volume of 67.9 ml, onto the elution profile previously obtained for each of the proteins separately. The elution peak of the complex is shown in blue, and the corresponding fractions visualized by Coomassie staining of a 10% SDS–PAGE gel are marked with a red bar (inset). The light green peak corresponds to the separate elution peak of Mex67(528–599), while the pink peak corresponds to that of the truncated Mip6 RRM4(401–480) construct.
Crystallization screening to study the interaction between Mip6 RRM4 and Mex67(528–599) was set up using both proteins in a 1:1 molar ratio. However, only crystals of the Mex67 molecule were obtained. The determined X‐ray structure of the obtained Mex67(528–599) crystal (Table EV1, PDB: 6EXZ) contains one molecule of Mex67 in an asymmetric unit that comprises the UBA domain plus the previously unreported presence of 16 N‐terminal residues. Mex67(528–599) folding is conserved compared to the previous four‐helix bundle solution structure of the Mex67 UBA 33, 34. However, our Mex67(528–599) construct (hereafter named Mex67 N+UBA) revealed the additional presence of two novel short α‐helices (helix N1 and N2) (Fig 2A upper panel). This new structure represents a more precise picture of the UBA fold and provides a more complete structural description of the Mex67 C‐terminal region than was previously available.
Figure 2. The Mex67 C‐terminal domain structure and its interaction with Mip6.

- Ribbon depiction of the crystal structure of Mex67 N+UBA (PDB: 6EXZ). The UBA domain (helices H1–H4) is folded as in previously reported structures (PDB ID: 1OAI). The new N‐terminus (shaded light blue) is formed by two short alpha helices (N1 and N2 that fold back into helices H1 and H2). The red bars in the histogram below summarize the analysis of Mip6 RRM4‐Mex67 N+UBA binding by NMR titration experiments monitored on the Mex67 N+UBA 1H‐15N HSQC spectrum. Peak intensity ratios between free Mex67 N+UBA and a mixture containing Mip6 RRM4 and Mex67 N+UBA (at 15% saturation on Mip6) were plotted against the Mex67 sequence (red bars). The green bars in the histogram are the results of equivalent analysis of the Mip6 RRM4 W442A/Mex67 N+UBA mixture (also at 15% saturation on the Mip6 mutant), and show that W442A abolished the interaction. The signal intensity decreases in the red histogram are mapped on the structure of Mex67 N+UBA. The level of red color saturation directly corresponds to the decrease in signal intensity and reveals the putative Mex67 N+UBA binding site of Mip6 RRM4.
- Biophysical characterization of the interaction between Mex67(528–599) and Mip6. The upper panel shows the ITC binding isotherms of Mex67(528–599) (N+UBA) titrated into Mip6 RRM1‐4(111–480) (green) or Mip6 RRM4(401–480) (red). The lower panel shows the ITC binding isotherms of Mex67 N+UBA titrated into Mip6 RRM4 (red), using the same data as the upper panel, or the Mip6W442A mutant (blue).
- Immunoprecipitation of the empty plasmid pADH1pr‐GFP, Mip6‐GFP, or Mip6‐ΔRRM4‐GFP (left) and of pADH1pr‐GFP, Mip6‐GFP, or Mip6W442A‐GFP mutant (right) expressed from the indicated plasmids in an mip6Δ strain. Mip6‐GFP and an empty plasmid were used as positive and negative controls, respectively, in both panels. Mip6‐GFP, including its mutant constructions, and Mex67 were detected by Western blotting using the indicated antibodies. The bar graphs at right indicate the quantification of Mex67 IP signal intensity normalized to Mip6. Mean ± SEM from 2 to 5 biological replicates was represented; significant differences from unpaired Student's t‐test with Welch's correlations and one‐tailed were represented (*P‐value < 0.05; ***P‐value < 0.001).
- Growth of WT, Mip6‐GFP, and Mip6W442A‐GFP strains. Precultures were diluted in liquid YPD medium, and 10‐fold dilutions of cells were spotted onto YPD plates. Plates were incubated for 2 days at the indicated temperatures.
To quantify the strength of the interaction between the Mex67 N+UBA and Mip6, we performed isothermal titration calorimetry (ITC). ITC showed a similar value between 1 and 2 μM for the apparent dissociation constant (KD) for Mex67 N+UBA and an Mip6 RRM1–4 or an Mip6 RRM4 construct (Fig 2B upper panel). A value of n = 0.96, in agreement with a 1:1 stoichiometry, was obtained for the interaction between Mex67 N+UBA and Mip6 RRM4.
Next, we analyzed the Mex67‐Mip6 interaction by nuclear magnetic resonance (NMR) to gather structural information about the Mip6 RRM4–Mex67 N+UBA interaction interface. We assigned the NMR spectra of each domain (Appendix Fig S2A and B) and performed titration experiments. However, binding occurred in a chemical exchange regime that caused severe line broadening effects above 15% saturation (Appendix Fig S3); thus, no direct chemical shift mapping could be obtained. However, the same chemical exchange process caused peak intensity losses in the 1H‐15N HSQC spectra (Fig 2A red histogram) at low complex saturation (> 15%) that could be mapped on the crystal structure (Fig 2A upper panel, red shading). This mapping suggested that the Mip6 binding interface is at the confluence of helices N1, N2, and H2 of the Mex67 N+UBA. To evaluate the possible involvement of the Mex67 N‐terminal extension (helices N1 and N2) in Mip6 recognition, we quantified the energetic contribution by biolayer interferometry (BLI). Experiments using GST‐Mex67‐UBA and GST‐Mex67‐N+UBA constructs provided similar KD values (Appendix Fig S4), and no detectable binding was observed with a construct including only the N‐terminal extension (GST‐Mex67481–544), suggesting that the Mex67 N‐terminal extension did not contribute to Mex67 UBA binding and that the changes detected by NMR might be due to subtle conformational rearrangements. In summary, these results show a direct interaction between Mip6 RRM4 and Mex67 UBA domains, likely involving the alpha helix H2.
A loop in Mip6 RRM4 including W442 is responsible for Mex67 binding specificity
To identify the Mip6 RRM4 structural elements directly involved in Mex67 recognition, we analyzed the effect of titration of Mex67 onto the Mip6 RRM4 1H‐15N HSQC spectra. Most of the RRM structural core signals (β–sheet and α‐helices) remained unchanged by Mex67 titration, in contrast to variations induced by Mex67 titration that were found in a set of unassigned signals that likely correspond to loop residues (Fig EV2A). Furthermore, of the two tryptophans in the Mip6 RRM4 domain, only W442 was strongly affected by Mex67 binding and therefore was directly involved in the binding (Fig EV2A). This W442 residue is located in an unusually long loop (loop 1) and can be mutated (mip6 RRM4 W 442 A) without compromising the structural integrity of RRM4, as the mutant NMR spectra were almost identical to those of the wild type (hereafter WT) (Fig EV2B). In contrast to the WT, the NMR spectrum of mip6 RRM4 W 442 A was insensitive to Mex67 titrations (Fig 2A, green histogram), which confirms that Mip6 W442 is essential for Mip6‐Mex67 interaction (Fig EV2C). Additional ITC experiments further demonstrated that this single point mutation in Mip6 (W442A) is critical for Mex67 recognition in vitro (Fig 2B lower panel).
Figure EV2. Mip6 RRM4 NMR analysis.

- Left panel: Superposition of 1H‐15N HSQC spectra of the titration of unlabeled Mex67 N+UBA onto the Mip6 RRM4: black (free protein), dark red (50% saturated with unlabeled Mex67 N+UBA), and red (100% saturated). Signals corresponding to the side chain crosspeaks of tryptophans have been marked to show their differential behavior upon Mex67 interaction. Many signals disappear upon titration, most of which were not assigned and likely belong to loops. Right panel: the chemical shift mapping and their representation over the structure of the Mip6 RRM4 model: Shades of blue show non‐interacting residues, shades of red show interacting residues, and unassigned residues are shown in gray.
- Superposition of the 1H‐15N HSQC spectra of Mip6 RRM4 (purple) and the Mip6 RRM4 W442A mutant (green). Note the disappearance of the W442 side chain crosspeak in the mutant. The majority of the peaks remain at the same position in the mutant, showing that the structures are very similar. Crosspeaks in the mutant are sharper than those in the WT, showing that it is less prone to aggregation.
- NMR‐monitored experiments of the titration of unlabeled Mip6 RRM4 W442A into 15N‐labeled Mex67 N+UBA. There are almost no changes upon the addition of equimolecular amounts of the Mip6 mutant in contrast to the WT case (Appendix Fig S3).
To analyze the in vivo relevance of Mip6 RRM4 and its W442 residue in Mip6‐Mex67 interaction, Mip6 pull‐down experiments were performed using WT cells transformed with plasmids expressing Mip6‐GFP, Mip6‐ΔRRM4‐GFP, or Mip6W442A‐GFP. As shown in Fig 2C (left panel), a strong reduction in the level of Mex67 co‐purifying with Mip6 was observed for the Mip6‐ΔRRM4 mutant compared to WT. Similarly, point mutation of W442A also reduced association of Mip6 with Mex67 to background levels (Fig 2C, right panel). The combined structural and biophysical data show that Mex67 directly interacts with the Mip6 RRM4 through W442 in loop 1, by using the interface of helix 2 (in Mex67). This binding mode can be effectively disrupted in vitro and in vivo by a single point mutation of tryptophan 442 in the Mip6 protein to alanine.
Although deletion of MIP6 does not affect normal yeast growth, we created a genomic version of Mip6W442A‐GFP using a Mip6‐GFP strain to compare the cell growth phenotypes of WT (Mip6‐GFP) and mutant Mip6W442A‐GFP strains. A dot spot assay showed a growth defect at all tested temperatures for the Mip6W442A‐GFP mutant but not for Mip6‐GFP (Fig 2D). We concluded that Mip6 W442A mutation, which altered Mip6 interaction with Mex67, affects cellular growth, likely causing a toxic effect.
Binding of Mip6 RRM4 to Mex67 N+UBA prevents its association with RNA
The above experiments showed that Mip6 RRM4 interacts with the Mex67 UBA. We next tested whether this Mip6 domain could also bind RNA. Polyuridylic acid–agarose bead (poly(U)) pull‐downs with different Mip6 constructs showed that Mip6 constructs expressing RRM1/2 or RRM3/4 bound similar to poly(U) (Fig 3A and Appendix Fig S1). The poly(U) binding of Mip6 RRM4 alone was similar to that of Mip6 RRM3/4 (Fig 3A), whereas the binding of Mip6 RRM3 was lower than that of the other constructs (Fig 3A). We tested binding to other RNA homopolymers (i.e., poly(A)) but unfortunately could not extract clear conclusions about protein selectivity.
Figure 3. Binding of Mip6 RRM4 to Mex67 N+UBA prevents its association with RNA.

- Purified recombinant Mip6 constructs were bound to pre‐equilibrated poly(U) agarose beads. After washing, beads were boiled with SDS sample loading buffer and loaded onto a 10% SDS–PAGE gel that was subsequently stained with Coomassie Blue Input (I), wash (W), and eluate (E), which are shown for each pull‐down. BSA was used as a negative control. The asterisk indicates GST from the purification.
- As in (A) using preincubated complex of Mex67(528–599) with Mip6 RRM4 (left) or Mip6 RRM1/4 construct (right). Mex67(528–599) shows no poly(U) binding when preincubated with Mip6.
Because Mip6 RRM4 acts both as a bona fide RNA and as a protein (Mex67 N+UBA) recognition domain, we further investigated crosstalk between these two events. Pull‐down experiments showed that, although Mip6 RRM4 and RMM1/4 bind effectively to poly(U) agarose beads (Fig 3A), notably, their complexes of Mip6 RRM4 or Mip6 RRM1/4 with Mex67 N+UBA do not (Fig 3B). Thus, Mip6 RRM4 can bind both a protein Mex67 and poly(U), but apparently not simultaneously in vitro.
Mip6 shuttles between the nucleus and the cytoplasm partially via Mex67 interaction
To gain insights into the cellular role of Mip6, we investigated whether Mip6, similar to Mex67, shuttles between the nucleus and the cytoplasm. Unfortunately for the purpose of such analysis, an endogenous Mip6‐GFP protein was undetectable by confocal microscopy, probably due to its low abundancy 30. We therefore expressed Mip6 under the control of the ADH1 promoter (ADH1pr) either as a plasmid (p‐ADH1pr‐Mip6‐GFP) or at its genomic locus (g‐ADH1pr‐Mip6‐GFP), both of which resulted in increased Mip6 expression levels (Fig 4A). Highly overexpressed Mip6 from the GAL1 promoter was correlated with toxicity 29. However, overexpression of Mip6 under the ADH1 promoter from the p‐ADH1pr‐Mip6‐GFP plasmid resulted in a non‐toxic phenotype, allowing the use of this plasmid for functional studies (Fig 4B). Under this overexpression condition, Mip6‐GFP is homogeneously distributed between the nucleus and the cytoplasm (Fig 4C).
Figure 4. Mip6 shuttles between the nucleus and the cytoplasm in a Mex67‐dependent manner.

-
AWestern blotting of whole‐cell extracts obtained from strains expressing Mip6‐GFP under the control of the ADH1 promoter from a plasmid (1) or at the MIP6 genomic locus (2). A WT strain was used as negative control, and Pgk1 levels were used as loading control.
-
BDot spot growth assay of WT‐ and mip6∆‐transformed strains. Culture dilutions were done in SC‐URA plates and were incubated for 3 days at 30°C.
-
CA confocal image of GFP‐Mip6‐expressing yeast cells from a plasmid (1) growing at 30°C. Scale bar: 5 μm.
-
D, EConfocal images of Mip6‐GFP localization in the following strains co‐expressing p‐ADH1pr‐Mip6‐GFP: (D) the shuffle mex67Δ strain transformed with Mex67‐HA or Mex67∆UBA‐HA and grown at 30°C; (E) the double mutant mip6∆mex67‐5 strain grown at 30°C or at 30°C followed by incubation at 39°C for 120 min;
-
FConfocal images of Mip6‐GFP localization in mip6∆ cells expressing p‐ADH1pr‐Mip6W442A‐GFP or p‐ADH1pr‐Mip6‐ΔRRM4‐GFP and grown at 30°C.
To analyze how interaction with Mex67 impacts Mip6 shuttling, we analyzed Mip6 localization in yeast cells expressing one of two different Mex67 mutants. First, a strain expressing Mex67ΔUBA 35, which is unable to interact in vitro with Mip6 (see Appendix Fig S4), was transformed with Mip6‐GFP. As shown in Fig 4D, both the GFP‐fluorescence intensity reflecting Mip6 protein expression levels and the percentage of cells showing Mip6 nuclear localization increased in cells with Mex67 lacking the UBA domain. Second, a strain expressing the mex67‐5 conditional allele, which is inactivated by heat, was transformed with Mip6‐GFP. At the permissive temperature (30°C), Mip6‐GFP was distributed between the nucleus and the cytoplasm in mex67‐5, with minor nuclear accumulation (Fig 4E). Shifting of the cells to 39°C for 2 h, which completely inactivates the mex67‐5 mutant, resulted in the nuclear accumulation of Mip6‐GFP (Fig 4E). These results suggest that Mex67 is required for the transport of Mip6 from the nucleus to the cytoplasm.
A caveat of the previous experiments is that deletion of the Mex67 UBA domain or its complete inactivation in mex67‐5 cells after 2‐h treatment at 39°C affects many other cellular processes such as mRNA export or interaction of Mex67 with other essential proteins. To circumvent these indirect effects, we checked the localization of the Mip6 point mutant W442A and that of the Mip6‐ΔRRM4 mutant. As shown above, both mutations reduced the interaction of Mip6 with Mex67 in vivo (see Fig 2C). As expected, both mutations resulted in a partial nuclear retention of Mip6 (Fig 4F). Notably, none of these mutations affected Mip6‐GFP protein expression levels (Appendix Fig S5). In summary, these results show that Mip6 shuttles between the nucleus and the cytoplasm and that the interaction of Mip6 with Mex67 is important for this process.
Mip6 accumulates in stress granules under stress conditions
Mip6 was shown to partially localize to P‐body‐like assemblies when overexpressed at levels that inhibited growth, as for instance, under the GAL1 promoter 29, and to localize along the prospore membrane in diploid cells 30. We noticed that increasing the temperature to 39°C for 20 min led to partial accumulation of Mip6 into cytoplasmic granules in WT cells (Fig 5A, left panel). This effect was not a consequence of toxicity, since no growth defect associated with Mip6 expression was evident under stress conditions (Fig 5A, right panel). Notably, Mip6 interaction with Mex67 was not affected by heat shock (Fig EV3). To assess whether the granules into which Mip6 accumulates were stress‐induced granules (SG) 36, we followed Mip6 granule formation upon different types of stresses. Figure 5B shows that Mip6 accumulates in cytoplasmic granules upon heat shock, sodium azide treatment, and glucose starvation. These results demonstrated that Mip6 localization is affected by several types of stress.
Figure 5. Mip6 accumulates in cytoplasmic stress‐induced granules.

- Confocal images of Mip6‐GFP localization in yeast cells incubated at 30°C or after incubation at 39°C for 20 min. DAPI was used to stain nuclei. Scale bar: 5 μm (left panel). Growth assay of WT and mip6∆ cells expressing p‐ADH1pr‐GFP or p‐ADH1pr‐Mip6‐GFP plasmids (right panel). SC‐URA plates were incubated for 3 days at 37°C.
- Mip6‐GFP localization in mip6∆ cells grown under the different stress conditions indicated in the figure. Scale bar: 5 μm.
- As in (B) but analyzed at different time points of growth at 39°C. Scale bar: 5 μm.
- Confocal analysis of the colocalization of Mip6‐GFP with Pab1‐RFP (upper) or Pbp1‐mCherry (lower) after incubation at 42°C for 30 min or at 39°C for 20 min, respectively. Scale bar: 5 μm. Merge shows the composite images of the two colors. Arrowheads indicate colocalization events.
- Effect of cycloheximide (CHX) on Mip6 granule formation when added during heat shock at 39°C for 20 min (CHX + 39°C 20′) or when incubated for 10 min after 20 min of heat shock (39°C 20′ + CHX 10′). Scale bar: 5 μm.
Figure EV3. Mip6 interaction with Mex67 is not affected by heat shock.

Immunoprecipitation of plasmid‐expressed Mip6‐GFP in a mip6Δ strain incubated at 30°C or after incubation at 39°C for 20 min. An empty plasmid was used as negative control. Mip6‐GFP and Mex67 were detected by Western blotting using the indicated antibodies.
Further study of the kinetics of temperature‐induced Mip6 granule formation showed that their formation was reversible when the stress condition was maintained over a longer period (Fig 5C). This finding was consistent with the notion that the granules containing Mip6 are SG, since these granules disassemble when cells are adapted to stress 37, 38. In addition, Mip6 partially colocalized with the well‐known SG components Pbp1 at 39°C and Pab1 at 42°C (Fig 5D). Lastly, Mip6‐containing granules were sensitive to the addition of cycloheximide, which is a property of stress granules and P‐bodies 39 (Fig 5E). In conclusion, when non‐toxic levels of Mip6 are expressed under the control of the ADH1 promoter, Mip6 accumulates in cytoplasmic stress‐induced granules upon various stresses.
Mip6 binds RNA in vivo, preferentially binding to Msn2/4‐dependent transcripts
Since Mip6 binds RNA in vitro (see Fig 3), we explored whether Mip6 could also bind RNAs in vivo using the photoactivatable ribonucleoside‐enhanced crosslinking and immunoprecipitation (PAR‐CLIP) technique 40. Analysis of PAR‐CLIP reads is shown in Dataset EV1. Aggregating reads across genes (metagene analysis) revealed a significant enrichment of Mip6 toward the 3′ end region of coding sequences (CDS) (Fig 6A). This profile is similar to that of other mRNP‐associated factors that bind co‐transcriptionally 41. Analysis of the top 50 Mip6 targets revealed that Mip6 binds to some heat‐shock mRNAs, including HSP12, HSP31, and HSP10, that are expressed at a low level. This finding might indicate a role for Mip6 in the metabolism of stress‐responsive transcripts, as has been recently suggested for Mex67 15. To evaluate this possibility, we repeated the PAR‐CLIP experiments after 20 min of heat‐shock treatment at 39°C, conditions under which Mip6 localization is also affected. Although the binding pattern of Mip6 at 39°C was similar to that observed at 30°C (Fig 6A, Dataset EV1), we sought to determine whether Mip6 associates preferentially with specific transcript classes under these two conditions. For this purpose, transcripts were ranked by the magnitude of their binding to Mip6 at 30°C and at 39°C. In order to avoid any binding bias caused by the differential expression levels of these genes at 30°C vs. 39°C, the binding values were normalized by their expression levels under each condition 42 (see Materials and Methods). After this normalization, differential binding of Mip6 at 30°C vs. 39°C was examined using NOISeq. NOISeq is a non‐parametric approach for the identification of differentially expressed genes from next‐generation sequencing count data 43. Differential binding was found for 488 transcripts: A total of 110 mRNAs showed higher binding at 30°C, whereas preferential binding at 39°C was observed for 378 mRNAs (Fig 6B, Dataset EV2). Gene set enrichment analysis (GSEA) was performed in which specific groups of genes related to heat stress that are under the control of the transcription factors Hsf1 44 (HSE) and Msn2/4 25 (STRE) were also included in the gene sets. These analyses revealed that, while Mip6 binding at 39°C includes binding to transcripts related to “ribosome” and to “cytoplasmic translation”, Mip6 binding at 30°C showed significant enrichment of only two sets of genes, “ketone metabolism” and “Msn2/4‐dependent genes” (Fig 6C, Dataset EV3). The latter finding suggests that Mip6 association with Msn2/4 targets is enriched under non‐stress conditions. Moreover, we found that the percentage enrichment of Msn2/4‐dependent genes in Mip6 targets at 30°C was above 50% (Fig 6D, Appendix Table S1). These findings suggest a possible specific role for Mip6 in the metabolism of Msn2/4 transcripts. To study whether Msn2/4 affected Mip6 localization, we analyzed Mip6‐GFP localization in msn2Δ, msn4Δ, and msn2Δmsn4Δ mutants. As shown in Fig 6E, the presence of Msn2/4 is required for the correct nuclear/cytoplasmic transport of Mip6 at 30°C and after incubation at 39°C, although Mip6 still accumulates into SG at 39°C.
Figure 6. Mip6 RNA binding in vivo shows preferential binding to Msn2/4‐dependent transcripts.
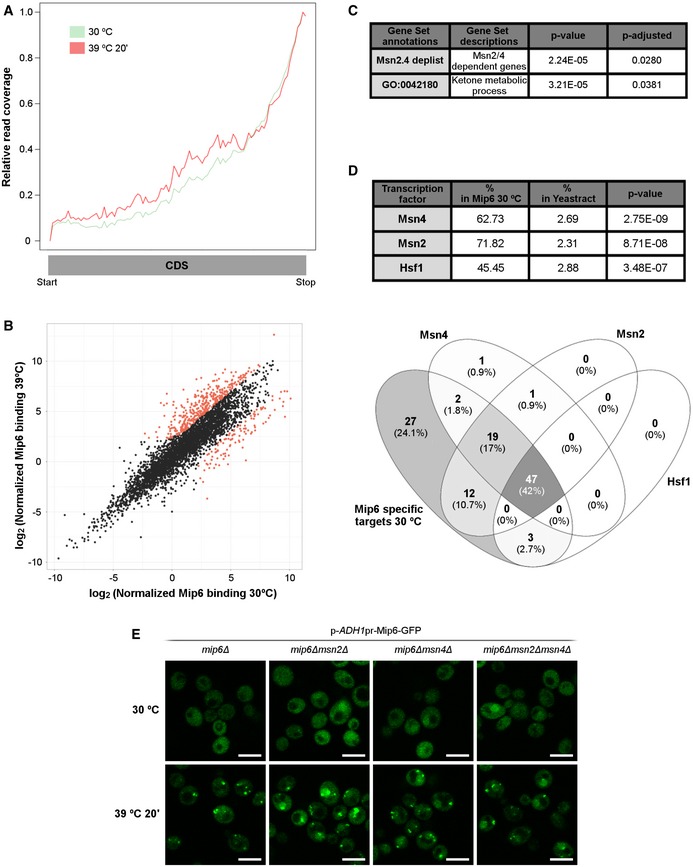
- Read coverage profiles throughout the CoDing Sequence (CDS) of Mip6 RNA binding at 30°C (green) and after heat‐shock induction for 20 min (red).
- Scatter plot describing the relationship between Mip6 RNA binding at 30°C and that after incubation at 39°C for 20 min. Red points indicate genes (488 from 4,121) showing statistically significant differences (diffNoiseq_rna_prob > 0.9; Dataset EV2).
- Significative GSEA results obtained from 110 genes that showed preferential binding at 30°C (P‐adjusted < 0.05).
- Upper panel: Table obtained using YEASTRACT 87 showing the percentages of Mip6 targets significantly enriched at 30°C, ranked by dependence on the transcription factors Msn2, Msn4, and Hsf1. Lower panel: a Venn diagram representing the intersection of Mip6‐specific targets at 30°C controlled by Msn2, Msn4, and Hsf1 transcription factors.
- Confocal images of Mip6‐GFP localization in mip6∆, mip6∆msn2∆, mip6∆msn4∆, and mip6∆msn2∆msn4∆ cells incubated at 30°C or after incubation at 39°C for 20 min. Scale bar: 5 μm.
Mip6 is required for the metabolism of Msn2/4‐dependent transcripts
The above PAR‐CLIP results showed that Mip6 preferentially associates with Msn2/4‐dependent transcripts under non‐stress conditions. Since these genes are expressed at a low level under standard conditions, it is possible that Mip6 modulates their expression levels. To assess the role of Mip6 in the expression of specific Msn2/4‐dependent genes, we measured the steady‐state levels of HSP12, CTT1, and SSA4 at 25°C in WT and mip6Δ cells. These three genes were overexpressed in the absence of Mip6 in cells growing at 25°C (Fig 7A, upper panel). Upon stress, these genes remained overexpressed in mip6Δ compared to WT cells (Fig 7A, lower panel). The induction patterns of these three genes upon heat stress (Fig EV4A) were very similar to the published profiles 45, showing the changes in their transcription rate and mRNA over time at 37°C (Fig EV4B). To examine whether this effect of Mip6 is more general, we compared the expression levels of other Msn2/4‐dependent transcripts that are enriched in Mip6 binding (Appendix Table S1) in mip6Δ vs. WT cells under standard temperature and/or stress conditions. For this comparison, gene expression levels were represented as the number of reads from RNA‐seq experiments (Nuño‐Cabanes et al; the complete study will be published elsewhere). Notably, the levels of most of these Msn2/4‐dependent transcripts were increased in mip6Δ vs. WT cells under both conditions assayed (Fig 7B). These results support a role for Mip6 in the maintenance of appropriate levels of Msn2/4 transcripts.
Figure 7. Mip6 is required for the metabolism of Msn2/4‐dependent transcripts.

-
AHSP12, CTT1, and SSA4 expression, measured using qRT–PCR in WT and mip6∆ strains.
-
BExpression of Msn2/4‐dependent targets in WT and mip6∆ strains, using data obtained in RNA‐seq experiments.
-
C, DAs in (A), in cells expressing Mip6‐GFP or Mip6W442A‐GFP (C) or in WT, mip6∆, rrp6∆, or mip6∆rrp6∆ cells (D).
-
EGrowth of WT, and of mip6Δ, rrp6Δ, and mip6Δrrp6Δ mutants. Precultures were diluted in liquid YPD medium. The 10‐fold dilutions of cells were then spotted onto YPD plates and incubated for 3 days at 30°C or 7 days at 37°C.
-
FImmunoprecipitation with the TAP antibody using Sgf73‐MYC, Mip6‐TAP, or Mip6‐TAP/Sgf73‐MYC expressing cells. Mip6‐TAP and Sgf73‐MYC were detected in INPUT and IP by Western blotting with the mentioned antibodies. Asterisk indicates an unspecific band from the TAP antibody.
-
GFISH analysis using a Cy3‐labeled specific oligo targeting SSA4 expression in mip6∆ and mex67‐5 cells after incubation at 42°C for 30 min. DAPI was used to stain nuclei. Scale bar: 5 μm. Quantification is shown in Appendix Fig S7.
Figure EV4. Expression pattern of HSP12, CTT1, and SSA4 is modulated by heat stress.

- HSP12, CTT1, and SSA4 expression profiles measured using qRT–PCR in WT and mip6∆ strains incubated at 39°C. Mean values normalized to WT from two biological replicates were shown.
- Transcription rate (TR) and mRNA amount (RA) of HSP12, CTT1, and SSA4 transcripts in WT cells incubated at 37°C 45.
Since Mip6 binds Mex67, we hypothesized that the effect of Mip6 on the expression of Msn2/4 targets might depend on its ability to interact with Mex67. To test this hypothesis, the expression of HSP12, CTT1, and SSA4 in the mip6‐W 442 A mutant was compared with that in WT cells under standard and stress conditions. The results shown in Fig 7C demonstrated that tryptophan 442 of Mip6 is required for the correct expression of HSP12 and CTT1, suggesting that Mip6‐Mex67 interaction is necessary to maintain WT levels of Msn2/4 targets under both standard and stress conditions.
The higher expression level of Msn2/4 targets in mip6Δ vs. WT cells could be a consequence of reduced mRNA degradation mediated by the nuclear exosome 46, 47. To study whether Mip6 displays a functional relationship with the nuclear exosome, MIP6 was deleted in the rrp6Δ mutant and we compared the levels of HSP12, CTT1, and SSA4 in the mip6Δrrp6Δ double mutant to those in the single mutants and in WT cells. As shown in Fig 7D, a significant enrichment of the target mRNAs was observed at 25°C in the double mutant compared to the single mutants and WT cells, suggesting cooperation between Mip6 and Rrp6. This phenotype was also evident at 39°C except for SSA4 (Fig 7D). Deletion of MIP6 from rrp6Δ partially rescued the growth defect of the rrp6Δ mutant at the restrictive temperature (Fig 7E). In contrast, no cooperative effect on RNA levels or genetic interaction was detected when mip6Δ was combined with deletion of the cytoplasmic exosome decay factor SKI7 48 (Appendix Fig S6A and B).
During the revision of this work, it was shown that the DUB subunit Sgf73 cooperates with Rrp6 as a RNA surveillance factor during mRNA export 49. Similar to rrp6Δ, deletion of SGF73 partially suppresses the growth defect of the single mutant rrp6Δ at 37°C. Since we had discovered Mip6 as a protein interacting with Sus1 (another DUB subunit), we asked whether Mip6 interacts with Sgf73. As shown in Fig 7F, physical interaction between Mip6 and Sgf73 was confirmed by Co‐IP in vivo. Remarkably, tagging both proteins in the same strain leads to a significant destabilization (Fig 7F, low panel). Additionally, we investigated whether Mip6 is required for mRNA export. No poly(A) + RNA export defect was observed in cells lacking Mip6 (Appendix Fig S7A); however, deletion of MIP6 lowered the export efficiency of SSA4 at 42°C when compared to a WT strain, being partially required for at least SSA4 mRNA export (Fig 7G, and Appendix Fig S7B and C).
Mip6 functionally interacts with Rpb1 and Xrn1
To assess whether Mip6 reduces the levels of Msn2/4 targets by stimulating their degradation or by repressing their transcription, we analyzed Msn2/4 targets upon repression of transcription by using rpb1‐1, which is a thermosensitive mutant of the largest RNA polymerase II (RNAPII) subunit. We compared HSP12, CTT1, and SSA4 expressions in rpb1‐1 with those in the double mutant mip6Δrpb1‐1 that we generated. There was an increased response of this set of genes within 30 min after heat inactivation of RNAPII in the rpb1‐1 allele mutant (Fig 8A, blue lines). These results are in agreement with previous reports 50 and with the observation that RNAPII is partially associated with stress‐responsive genes for up to 30 min after its thermal inactivation 51. No significant differences in the expression levels of the three transcripts were observed between mip6Δrpb1‐1 and rpb1‐1 cells after the 20‐min time point despite the higher initial levels reported in mip6Δrpb1‐1 cells (Fig 8A). Interestingly, the steady‐state levels of the three mRNAs falls at the same rate in both strains after reaching its maximum level, suggesting that Mip6 does not stimulate their degradation. The initial elevated levels of HSP12, CTT1, and SSA4 in mip6Δrpb1‐1 vs. rpb1‐1 cannot therefore be due to an mRNA stabilizing effect but rather suggest a role for Mip6 during transcription.
Figure 8. Mip6 is functionally linked to Rpb1 and Xrn1.

- HSP12, CTT1, and SSA4 expression measured using qRT–PCR in rpb1‐1 and mip6∆rpb1‐1 strains after incubation at 37°C for 20, 60, and 120 min. Mean values normalized to rpb1‐1 ± SEM from three biological replicates were indicated. Significant differences from one‐tailed unpaired Student's t‐test with Welch's corrections were represented (*P‐value < 0.05; **P‐value < 0.01).
- Growth of WT, mip6Δ, rpb1‐1, and mip6Δrpb1‐1 strains (upper panel) and of WT, mip6Δ, xrn1Δ, and mip6Δxrn1Δ strains (lower panel). Precultures were diluted in liquid YPD medium. The 10‐fold dilutions of cells were then spotted onto YPD plates and incubated for 4 days (30°C) and 6 days (35°C) for rpb1‐1 mutants and for 2 days (30°C) or 5 days (37°C) for xrn1Δ mutants.
- Confocal images of Mip6‐GFP localization in mip6Δ (left), mip6Δrpb1‐1 (middle), and mip6Δxrn1Δ (right) cells incubated at 30°C (upper panel) or after incubation at 39°C for 20 min (lower panel). Scale bar: 5 μm.
- Immunoprecipitation of the empty plasmid pADH1pr‐GFP or Mip6‐GFP expressed from the indicated plasmids in a mip6Δ strain. Mip6‐GFP and Rpb1 were detected in INPUT and IP by Western blotting using the indicated antibodies. Signal intensity was quantified and set to 1 for INPUT and IP of the empty plasmid pADH1pr‐GFP. The relative levels of Rpb1 to the negative control are indicated below.
To obtain data to support the functional interaction between Mip6 and factors involved in both mRNA transcription (Rpb1) and degradation (Xrn1), we compared the growth of mip6Δrpb1‐1 and mip6Δxrn1Δ mutants with that of single mutants and WT cells. A slight growth recovery in the double mutants mip6Δrpb1‐1 and mip6Δxrn1Δ over the singles rpb1‐1 and xrn1Δ was observed at 35 and 37°C, respectively (Fig 8B). We observed that both factors were required, directly or indirectly, for the correct shuttling of Mip6 between the nucleus and the cytoplasm (Fig 8C), as inactivation of Rpb1 at 37°C led to partial nuclear retention of Mip6 and deletion of XRN1 affected Mip6 localization even at 30°C. This observation is consistent with Xrn1's role in P‐bodies and SG formation 52. Last, we performed mass spectrometric analysis (LC‐MS/MS) of proteins that co‐purified with a TAP‐tagged version of Mip6 (complete results of this purification will be published elsewhere) and confirmed that Mip6‐TAP specifically co‐purified with Mex67 (Appendix Fig S8). In this analysis, we also identified Rbp1 and Rpb2 (Appendix Fig S8), the two largest subunits of RNAPII, in the pool of high molecular mass Mip6 co‐purifying proteins. We validated Mip6‐Rpb1 interaction, by Western blot detection of Rpb1 in the purified Mip6‐GFP extract (Fig 8D). This result suggests that Mip6 might contact the transcriptional machinery.
Discussion
The metabolism of mRNAs is a fundamental process for the survival of eukaryotic cells. The coordinated action of many factors acting at different steps of RNA metabolism serves as a mechanism to regulate RNA homeostasis in a specific cellular context, and RBPs are crucial elements in this process. Furthermore, RBPs play critical roles in mRNA metabolism in pathological states in humans. This study functionally and biophysically characterized the RBP Mip6, providing new insights into its binding mode with the conserved mRNA exporter Mex67. Furthermore, we demonstrated that Mip6 participates in the homeostasis of Msn2/4 dependent transcripts, which are relevant to stress responses.
Mip6 RRM4 interacts directly with the Mex67 UBA domain: structural and functional insights
Mip6 architecture is typical of that of an RNA‐binding protein, combining well‐folded RNA‐binding motifs (four RRMs in two tandems) with intrinsically disordered low complexity regions. RRMs are among the most abundant RNA‐binding domains but can also recognize proteins 53, 54, 55. Given that Mip6 displays such dual binding, we focused on characterization of the molecular details of its interaction with Mex67 and its RNA‐binding properties. The main results allowed us to: (i) describe the poly(U) binding properties of the RRM domains of Mip6; (ii) obtain a new structure of the Mex67 UBA with an additional N‐terminal 16 residues that form two additional short α‐helices that do not seem to make significant contributions to the Mip6 binding; and (iii) characterize the key elements involved in Mip6/Mex67 interaction (a loop in Mip6 RRM4 containing tryptophan 442, and helix H2 of the Mex67 UBA).
Although RRMs can bind both RNA and protein, they sometimes work as protein recognition modules with no RNA‐binding activity 54, 56. Most RRMs that bind proteins, such as the non‐canonical U2AF homology motif family (UHM) of RRMs, lack the conserved aromatic residues within RNP1 and RNP2 sequences that are important for RNA recognition 57. Others, e.g., Gbp2 and the Hrb1 C‐terminal RRMs, have their RNA‐binding surfaces blocked by non‐canonical structural elements 54, 56. In the paradigmatic case of PABP‐eIF4G, the second RRM of PABP can recognize both poly(A) and eIF4G peptide 54. Most of the protein‐binding RRMs use the helical interface for binding. In the case of Mip6 RRM4, binding of Mex67 involves a long loop connecting strand β1 and helix α2, in which W442 plays a key role. To our knowledge, this represents a novel RRM‐mediated protein recognition mode. Similar to the structurally related PABP, Mip6 RRM4 can also bind both a protein Mex67 (eIF4G in the case of PABP) and RNA, but apparently not simultaneously.
Although the NMR data suggest that canonical RNA interfaces (RNP1, RNP2) are not altered upon Mex67 binding (Fig EV2), a preconfigured Mip6 RRM4/Mex67 UBA complex is no longer able to interact with poly(U) (Fig 3B). Unexpectedly, a Mip6 construct with all RRMs behaves similarly in vitro to RRM4 alone, and no Mip6/Mex67 complex is observed in the retained poly(U) agarose fraction. Given that Mip6 RRM1‐4 and Mex67 N+UBA can indeed interact in vitro in the absence of RNA (Fig EV1), the results obtained using poly(U) agarose suggest that poly(U) might disrupt the Mex67/Mip6 protein–protein interaction and Mex67 N+UBA is removed during washes of the poly(U) agarose. RRM4 of the Mip6 RRM1‐4 construct likely binds to poly(U), thereby preventing Mip6 interaction with Mex67. Our data indicate that poly(U) has some effect on the ability of Mex67 N+UBA to interact with Mip6 in vitro, suggesting a possible regulatory or switch mechanism in vivo for RNA or Mex67 recognition. This might suggest a possible regulatory or switch mechanism in vivo for Mip6 concerning RNA or Mex67 binding. Further research is needed to fully understand the complex mechanisms behind Mip6 regulation.
Concerning the Mex67‐Mip6 binding interface, we have shown that Mip6 binding to Mex67 alters the Mex67 NMR spectra, suggesting that Mip6 binding affects various Mex67 helices (Fig 2A). These could be caused by direct contact and/or by conformation changes. Given the BLI data (Appendix Fig S4), it seems that the changes seen in NMR for helices N1 and N2 are caused by subtle conformation changes that have no significant impact on binding energetics. The combination of the two techniques allows a more precise definition of the interface at the Mex67 side that would involve helix H2. In these lines, Mip6 mapping on the Mex67 structure (Fig 2A) seems to be structurally compatible with the binding of other UBA Mex67 protein targets such as FG‐rich nucleoporins (NUPs) and/or ubiquitinated Hpr1 34, 58, 59. Hpr1 is part of the THO and TREX complexes and participates in different steps of gene expression 60, 61. Hpr1 is ubiquitinated during transcription, and its degradation is regulated by its association with Mex67 34. To determine when Mip6 interacts with Mex67 during this process will be a challenge for future experiments.
Mip6 participates in the homeostasis of Msn2/4‐dependent transcripts
Our in vivo results show that Mip6 binds preferentially to transcripts that depend on Msn2/4 factors under non‐stress conditions and that these transcripts are present at higher levels in mip6Δ compared to WT cells (Fig 7A). The magnitude of this increase in mip6Δ vs. WT cells is similar under stress, suggesting that the main impact of Mip6 on these RNAs occurs in basal conditions. We are starting to elucidate the possible mechanism behind these observed results. One possibility is that Mip6 could potentially be a novel factor participating in transcript buffering, which is a balancing act between the synthesis and degradation of mRNAs during stress responses.
A recent study proposed a new mode of action for Mex67 alone in immediate mRNA export, which bypasses nuclear quality control upon stress 15. We found here that Mip6 displays a functional relationship with the nuclear exosome subunit Rrp6 and that the double mutant mip6∆rrp6∆ grows better at restrictive temperature. It is well established that, under specific circumstances (as for instance stress), in the absence of mRNA biogenesis quality control, escape of premature transcripts to the cytoplasm is less detrimental for cell growth than no export at all 13, 62. These results are in agreement with the data reported by the Tollervey group, showing that global mRNA expression levels were affected upon inhibition of nuclear exosome components but not in strains lacking the cytoplasmic exosome co‐factor Ski7 63. Our results suggest that Mip6 cooperates with Rrp6 in regulating the nuclear fate of mRNA. During revision of this work, a novel role was proposed for the DUB subunit Sgf73 in orchestrating mRNA export under stress in combination with the central surveillance factor Rrp6 49. The authors proposed a model in which Sgf73, in conjunction with Rrp6, oversees the nucleocytoplasmic translocation of export‐competent mRNPs. Upon heat stress, Sgf73 initiates the non‐canonical mRNA export pathway, which is subject to minimal quality control, and facilitates the export of selective transcripts. In this respect, it is interesting that in this study we found that Mip6 co‐purified with Sus1 in ubp8Δ cells, in which Sus1 is only associated with the DUB and TREX‐2, but not with SAGA (Spt‐Ada‐Gcn5 acetyltransferase) 31, 64. Further work will be required to address whether Mip6 has a fundamental role in this non‐canonical export pathway, and to determine its functional connections with other DUB or SAGA subunits.
Increases in transcription are balanced by adjustment in global rates of RNA degradation, but the underlying molecular mechanisms are not well understood 65, 66. One problem with analyses of mRNA regulation during stress is that the synthesis and degradation of stress‐responsive RNAs cannot be studied through conventional approaches of transcriptional block using rpb1‐1 or thiolutin 50 since both methods alter mRNA stability and, rather than blocking, they induce the transcription of heat‐shock genes 67, 68, 69. However, since RNAPII is mostly inactivated after 30–45 min of incubation at 37°C in rpb1‐1 51 , our results show that after transcriptional shutoff the Msn2/4‐dependent transcripts decay at the same rate in both mutant (Fig 8A) regardless of their basal levels. This suggests that increased basal mRNA steady‐state levels in mip6∆ mutants could be explained by a higher synthesis rate. In this respect, genes regulated by the stress activators Msn2 and Msn4 were found to be strongly biased toward being SAGA dominated 70, pointing to a prominent role for SAGA (and/or DUB subunits) in their synthesis. Further investigation is needed to fully delineate the complex network involving these factors and to determine whether Mip6 lowers the transcription of target genes by repressing SAGA activity under non‐stress. In contrast, upon stress, the cell requires high production of these transcripts, and therefore, the export of Mip6 through Mex67 binding could serve as a fast way to exclude Mip6 from the nucleus. It is interesting that mip6∆ and mip6‐W 442 A cells show a similar increase in the levels of Msn2/4 mRNAs (Fig 7) and both Mip6 mutants increased Mip6 nuclear accumulation (Fig 4), since Mip6 can likely no longer access Mex67 for export. Thus, Mip6 nuclear retention might have a double impact on these transcripts, both stimulating higher transcriptional levels and lowering degradation rates due to their improper export to the cytoplasm. In this respect, recent work has shown that changes in cytoplasmic mRNA decay can directly impact RBP localization, providing a mechanism to connect distal stages of gene expression 71. More work is required to understand how Mip6 could stimulate both activities, since the observed partial suppression of rpb1‐1 thermosensitive growth at 35°C (Fig 8B) fits well with a role for Mip6 as a transcriptional repressor rather than an active role in mRNA degradation.
Our current work provides a starting point for future investigations into how Mip6 attenuates the expression of stress genes under non‐stress conditions. A Protein Data Bank (PDB) search for sequence similarity to Mip6 RRMs in databank solved structures gave the highest score (36% identity) for the similarity between Mip6 RRM1/2 and RRM1/2 of human cytoplasmic poly(A)‐binding proteins (PABPCs) (Appendix Fig S9). PABPCs have recently been shown to be involved in mediating mRNA decay‐driven repression of RNAPII promoter recruitment 71. It would be very interesting to address whether Mip6 is functionally related to human PABPCs.
Materials and Methods
Yeast strains, DNA recombinant work, and microbiological techniques
The yeast strains and the primers used in this study are listed in Appendix Tables S2 and S3, respectively. Standard methods were followed for plasmid construction, yeast growth, transformation, and gene deletions/replacements 72. To create the Mip6W442A‐GFP mutant strain, a region from the MIP6 locus (442–659) was deleted from the WT. This mutant was then transformed with a PCR product that included the point mutation within the primers and the sequence of GFP at the C‐terminus. All strains were confirmed by PCR analysis, sequencing, and/or Western blotting. Plasmids containing Mip6 and Mex67 constructs were cloned by standard molecular biological methods using the primers listed in Appendix Table S3. Mip6ΔRRM4 and Mip6W442A point mutation plasmids were generated by site‐directed mutagenesis using the QuikChange strategy (Agilent).
Yeast cells were grown in liquid culture at the appropriate temperatures in Yeast Peptone Dextrose (YPD) or Synthetic Complete (SC) selected media. Cultures were diluted to 0.2–0.4 OD600, and serial dilutions (1:10) were spotted onto YPD or SC‐URA plates. Cells were incubated at different temperatures and photographed after 2–7 days of incubation.
TAP purifications, immunoprecipitations, and Western blotting experiments
The tagged Sus1‐tandem affinity purification (TAP) and Mip6‐TAP proteins in WT and mutant strains were purified as described previously in Refs 72 and 73, respectively. The different purifications were analyzed using Coomassie‐stained gels and by immunoblotting using specific antibodies (Appendix Table S4). Samples were analyzed and detected using reagents from ECL, Amersham, or blots were developed using fluorescence detection by the Odyssey Infrared Imaging System from LI‐COR Biosciences.
For protein purifications and immunoprecipitation (IP), yeast cells expressing Mip6‐TAP were grown in 50 ml rich medium to a 0.5 OD600. Cells were harvested, washed with water, and resuspended in 250 μl of lysis buffer (50 mM HEPES–KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP‐40, 1 mM PMSF, and protease inhibitors). An equal volume of glass beads was added. Breakage was achieved by four pulses of vortexing at 4°C each lasting for 1 min. The clarified extracts were immunoprecipitated for 120 min at 4°C using IgG Sepharose six Fast Flow resin (GE Healthcare) (Fig 1A) or Dynabeads Pan Mouse IgG (Invitrogen) (Fig 1B), were washed three times with 1 ml of lysis buffer for 10 min, and were resuspended in 50 μl of SDS–PAGE sample buffer. Western blot analysis was performed using the indicated antibodies. For Mip6‐GFP‐Mex67 and Mip6‐GFP‐Rpb1 co‐immunoprecipitations, GFP‐TRAP®_A beads were used as recommended by the manufacturer (ChromoTek). RNase A treatments were performed at indicated concentrations during the incubation of the beads with the protein extracts. Immunoprecipitation of Mip6‐TAP‐Sgf73‐MYC was done as follows: Logarithmically growing yeast cells in selective medium at 30°C were resuspended in lysis buffer containing 10 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 2 mM PMSF, and protease inhibitors (EDTA‐free Protease Inhibitor Cocktail Tablets, Sigma‐Aldrich). Cells were broken by mixing with glass beads. The cell extract was obtained by two consecutive centrifugations, the first 20 min at 14,000 × g and the second 10 min at 14,000 × g. Glycerol was added to a final concentration of 10%. An amount of 5 mg of total proteins were incubated with IgG Sepharose Beads (GE Healthcare) on a rotor at 4°C for 2 h. Beads were pelleted and washed three times with IPP500 buffer (10 mM Tris–HCl pH 7.5, 500 mM NaCl, 5 mM MgCl2) followed by 2 rounds of three washes each. Additional washes were done with the same buffer excepting the NaCl concentration that was either decreased to 150 mM or removed. Proteins bound to the beads were eluted with SDS–PAGE sample buffer by boiling for 5 min.
Protein expression and purification
Recombinant protein constructs were expressed in bacteria (Appendix Table S5). In general, cells were grown at 37°C up to an OD600 of 0.6 and induced under similar conditions. Cells were harvested by centrifugation, frozen in liquid nitrogen, and stored at −80°C until further use. After lysis (by sonication) and clarification by ultracentrifugation, GST‐ and His‐fusion proteins were bound either to a 5 ml GSTrap HP or to a 5 ml HiTrap column (GE Healthcare), respectively, using different buffers (Appendix Table S6), were washed (for the 6xHis‐TEV protease (txAHTEV‐tag)), and were eluted in batch (GST pGEX‐6‐P2‐ and txAHTEV‐tags) or gradient (pET28‐NKI/LIC 6His/3C‐tag) modes. When required, the fusion tags were removed with 3C protease overnight at 4°C (GST pGEX‐6‐P2‐ and pET28‐NKI/LIC 6His/3C‐tag) or with TEV protease overnight at 4°C (txAHTEV‐tag) and were further purified on 5 ml GSTrap or HisTrap columns to remove the free tags, unreacted fusion proteins, and proteases. Protein‐containing fractions were further purified using size‐exclusion chromatography HiLoad Superdex 75 columns equilibrated in final buffers according to their later use (see next section). Fractions containing pure protein were pooled, concentrated, flash‐frozen in liquid nitrogen, and stored at −80°C until use.
Crystallization and data collection
Mex67(528–599) structure
The crystallization screening plates were set up by mixing Mex67(528–599) with Mip6 RRM3/4(313–480) in a 1:1 ratio at 21°C using the sitting drop vapor diffusion method. Crystals obtained containing Mex67(528–599) only were directly cryo‐cooled in liquid nitrogen, and X‐ray diffraction data were collected using the Diamond light source (Harwell, UK) I04 beamline. The best crystals obtained in 35% Tacsimate pH 7.0 diffracted up to 1.3 Å. Data were automatically processed in Diamond by the xia2 pipeline and were then scaled using Aimless from CCP4 74. The UBA domain of the Mex67 human homolog (PDB ID: 1OAI) was used as an initial model for molecular replacement utilizing the Phaser‐MR module from Phenix. Initial phases were used to build the rest of the structure using AutoBuild from Phenix and additional manual building. Cycles of Phenix refine and COOT were used for further structural improvements and refinements. The Mex67(528–599) structure was deposited in the Protein Data Bank under PDB ID: 6EXZ. The detailed final refinement statistics are shown in Table EV1.
Nuclear magnetic resonance
Nuclear magnetic resonance (NMR) experimental data were acquired at 25°C in a Bruker AV‐800 US2 spectrometer equipped with a TCI (1H, 13C, 15N) cryoprobe. Mip6 and Mex67 samples were prepared at 0.1–1 mM concentration in 25 mM potassium phosphate (pH 6.5), 150 mM NaCl, and 1 mM DTT buffer containing 10% deuterium oxide for the locking signal. The 1H‐15N HSQC spectra of Mip6 RRM4, Mip6 RRM4 W442A, and the Mex67 C‐terminus were assigned with the triple resonance approach using the series of 3D HNCA, 3D HNCO, 3D HN(CO)CA, and 3D CBCA(CO)NH acquired in double‐labeled samples (15N/13C) and were analyzed with CCPN software 74. NMR titrations were performed as described in Results section and were analyzed by chemical shift perturbations or by integration of crosspeaks volumes.
Isothermal titration calorimetry
For each isothermal titration calorimetry (ITC) experiment, purified proteins were simultaneously dialyzed in buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM β‐mercaptoethanol) using Slide‐A‐Lyzer Dialysis Cassettes (Thermo Scientific). Nano ITC using TA instruments was then employed to perform the measurements. All experiments were performed at 15°C and consisted of 20–30 serial injections with degassed samples. The concentration of proteins used either in the syringe or in the cell, plus the number and volume of injections (2–2.4 μl), depended on the requirement of each experiment. Data fitting and representation were performed using NanoAnalyze v.3.7.0 software (TA Instruments). Data are averaged from at least two independent experiments.
Biolayer interferometry
The biolayer interferometry system (BLItz, Pall ForteBio) was used to evaluate molecular interactions and to calculate binding affinities. To calculate the affinity of protein binding to RNA, a 5′ biotinylated poly(U) RNA of 15 residues (Sigma‐Aldrich) at a concentration of 50–100 μg/ml was immobilized on Streptavidin biosensors (ForteBio). The streptavidin biosensors were hydrated for 10 min before use in a buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM β‐mercaptoethanol, and 0.5 mg/ml BSA. At least three increasing concentrations (0.25–20 μM) of each prey protein in a 4 μl volume were utilized to estimate the binding affinity. The prey proteins were diluted in the same hydration buffer that was used throughout the experiment containing 0.5 mg/ml BSA in order to eliminate non‐specific binding of the prey protein to the Streptavidin biosensor.
To assess the binding of GST‐Mex67(528–599), GST‐Mex67 UBA(545–599), or GST‐Mex67∆UBA(481–544) constructs to Mip6 RRM3/4(313–480), the bait protein at a concentration of 50 μg/ml was immobilized on anti‐GST biosensors (ForteBio). Different concentrations of the prey protein were then used to evaluate the association and dissociation steps. BSA (0.5 mg/ml) was used in the buffer throughout the experiment to reduce non‐specific binding. Curve fitting, and association (Ka) and dissociation (Kd) constant calculations were done using BLItz Pro 1.2 software. The duration of the loading, association, and dissociation steps were adjusted for each experiment depending on the saturation of the signal obtained.
Polyuridylic (poly(U)) agarose bead binding experiment
Lyophilized polyuridylic acid‐agarose powder (10 mg; Sigma‐Aldrich; P8563) was weighed, washed with diethyl pyrocarbonate (DEPC)‐treated RNase‐free water, and left to swell for a few minutes. The beads were then equilibrated twice with binding buffer containing 50 mM HEPES pH 7.5, 100 mM NaCl, 0.1 mg/ml BSA, and 2.5 mM MgCl2. Purified protein (or protein complex) (200 μg) diluted in the reaction buffer was loaded onto the beads and mixed at 4°C for 30–60 min with constant rotation. The beads were spun down and washed 3–4 times with 0.5–1 ml binding buffer without BSA. Subsequently, 30 μl of 6× SDS–PAGE loading buffer was added and the sample was boiled for 5 min at 95°C. The samples were then analyzed by 10% SDS–PAGE followed by Coomassie Blue staining.
Glutathione‐Sepharose bead pull‐down
Purified GST‐fused protein (100–200 μg) was incubated with 70–100 μl of Glutathione‐Sepharose 4B beads (GE Healthcare), which was previously washed and equilibrated with buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 1 mM β‐mercaptoethanol, 5% glycerol), for around 30 min at 4°C with constant rotation. The GST beads were then washed with 1 ml of the equilibration buffer. The prey protein was then added to the beads in equimolar concentrations, and the beads were rotated for an additional 30 min. Supernatants were then discarded, and the unbound protein fraction was removed by washing the beads 3–4 times with 1 ml of fresh equilibration buffer each time. Finally, the bead‐bound protein fraction was eluted by adding 30 μl of 6× SDS–PAGE sample buffer and was boiled for 10 min at 95°C. Samples were then loaded onto an SDS–PAGE gel, which was then stained with Coomassie Blue.
Microscopy
Yeast cultures were grown to 0.2–0.3 OD600 in the appropriate SC media. Cultures were incubated with 0.5 μg/ml DAPI solution for 2 h to stain nuclei. Cells were harvested and resuspended in fresh medium with DAPI under specific stress conditions for heat‐shock and NaN3 stresses. For the glucose depletion assay, yeast cells were collected by centrifugation, washed three times in fresh SC medium ±2% glucose, resuspended in fresh liquid medium with DAPI, and incubated at 30°C for 30 min. After stress conditions were applied, cells were harvested, resuspended in PBS 1×, and spotted on slides for rapid examination at room temperature. All images were acquired using a Leica SP8 confocal system with an inverted DMI 6000 microscope running LAS X software (Leica microsystems). For colocalization pictures in yeast, images were obtained with DAPI, mCherry, and/or RFP immediately after GFP imaging. ImageJ (http://rsbweb.nih.gov/ij/) was used to obtain equal contrast and adjust all images. Cycloheximide treatments (100 μg/ml) were performed with minor modifications as previously described 39.
Fluorescence in situ hybridization
In situ hybridization using a probe targeting SSA4 mRNA or poly(A) + RNA was done as previously described with minor modifications 72, 75. Briefly, yeast cells were grown in 50 ml of YPD medium at 30°C at 0.1–0.2 OD600 and then temperature‐shifted to 39°C for 20 min for poly(A) + RNA FISH or 42°C for 30 min for SSA4 mRNA FISH both by diluting with an equal volume of 59°C media. Cells were immediately fixed in 4% formaldehyde for 15 min at 39°C (poly(A)+) or 42°C (SSA4 mRNA) followed by 30 min at room temperature. The fixative was removed by two rounds of centrifugation, and the cells were washed with 0.1 M potassium phosphate pH 6.4. Cells were resuspended in ice‐cold washing buffer (1.2 M sorbitol and 0.1 M potassium phosphate pH 6.4), and subsequently, the cell wall was digested with 0.5 mg/ml of zymolyase 100T. Following digestion, cell samples were applied onto poly‐Lysine‐coated slide wells. Non‐adhering cells were removed by aspiration, the cells were washed twice in wash buffer, once in 0.1 M KH2PO4/K2HPO4 pH 6.5 and 0.1% NP‐40 and once in 0.1 M KH2PO4/K2HPO4 pH 6.5, and were finally incubated with cold 70% ethanol for 30 min at −20°C. Prior to probe addition, the ethanol was drained from the cells and the cells were washed twice at 20°C for 5 min per wash in 2 × SSC and once for 10 min in 40% formamide/2 × SSC, 0.1% Triton X‐100, before overnight incubation at 37°C with 10 μl of the probe mix. For each well, 300 ng of probe was mixed with 5 μl of solution I (80% formamide, 10 mM NaHPO4 pH 7.0, 2 μg/μl salmon sperm DNA, 2 μg/μl yeast tRNA), denatured for 5 min at 95°C, and finally mixed with 5 μl of solution II (4 × SSC, 20 mM vanadyl ribonucleoside, 4 μg/μl BSA, 0.1 U/μl RNasin). Probe removal was followed by the following washing steps: (i) twice in 40% formamide/2× SSC for 10 min per wash at 37°C; (ii) once in 2× SSC/0.1% Triton X‐100 for 10 min at 20°C; (iii) twice in 1 × SSC for 10 min per wash at 20°C; and (iv) twice in 1× PBS for 5 min per wash at 20°C. Slides were mounted using ProLong™ Gold Antifade Mountant with DAPI (Invitrogen). The Cy3‐LNA SSA4 probe was detected using the DMI 6000 inverted fluorescence microscope. Pictures were taken using a Leica TCS‐SP4‐AOBS confocal microscope.
PAR‐CLIP
Photoactivatable ribonucleoside‐enhanced crosslinking and immunoprecipitation (PAR‐CLIP) experiments were performed as previously described 40. Briefly, cells were grown overnight at 30°C in YPD. Cells at an OD600 of 0.1 were seeded into sterile crosslinking media (SC‐URA supplemented with 2% glucose, 40 mg/l adenine, 60 μM uracil, and 1 μM biotin) and incubated until an OD600 of 0.6–0.8 was reached. 4‐Thiouracil (4tU; 4 mM) was then added and incubated at 30°C for 15 min. The cells were then further Incubated for 30 min at 30°C (non‐stressed condition) or for 10 min at 30°C followed by 20 min at 39°C (heat‐stressed condition). The subsequent procedures for irradiation with UV, protein and RNA purification, reverse transcription, PCR, and gel extraction of cDNA libraries were as described previously 40. The primers used in this assay are listed in Appendix Table S3.
Bioinformatic analysis
cDNA libraries were paired‐end‐sequenced. Demultiplexed sequencing data were preprocessed as described previously 76. Raw reads were trimmed of the 3′ adaptor using the Sarah Wheelan wrapper 40. Sequences were condensed to eliminate PCR artifacts and aligned to the SacCer3 genome (R64‐1‐1) by using Bowtie 1.1.1 77. Subsequent filtering of mapped reads was performed to discard sequences without PAR‐CLIP‐specific T to C mismatches. Consequently, only reads resulting from mRNA‐RBP crosslinking events remained for further analysis. In‐house scripts and a Pysam python library 78 were used to find PAR‐CLIP‐specific mismatches. Subsequently, read coverages over the gene body were calculated by using the geneBody_coverage.py function from the RSeqQC toolkit 79. Gene models described in gtf file version 2.2 were used for gene coverage and binding‐level computation. Binding levels per gene were computed by using the HTSeq count tool 80. RPKM 81 and trimmed mean of M‐values 82 normalization approaches were applied for correction of binding values both within and between samples. To further correct binding estimates by gene expression levels, RNA‐seq data from previous studies were used 42. RNA‐seq reads were aligned to the reference genome with TopHat2 83, and gene quantification and normalization were computed following the same pipeline applied for PAR‐CLIP data. Corrected Mip6 binding levels were obtained by dividing nominal binding values by gene expression values. Differential binding for both absolute and relative binding estimations was analyzed using the non‐parametric noiseq function from the NOISeq package 43. A probability of 0.9 was set as the threshold to call genes differentially bound to Mip6 between conditions. Overrepresentation analysis (ORA) and gene set enrichment analysis (GSEA) were performed by using the GOseq 84 and Mdgsa 85 package, respectively. Gene Ontology functional annotation was retrieved from the Ensembl Biomart web site 86. tRNA and rRNA genes were discarded in all bioinformatic analyses.
RNA extraction, reverse transcription, and quantitative PCR (qRT–PCR) analyses
Yeast cultures were grown at 25°C (qRT–PCR) or 30°C (RNA‐seq) in YPD to a 0.5–0.8 OD600. Heat‐shock treatment at 39°C for 20 min was performed except for rpb1‐1 mutants that were incubated at 37°C for 120 min. Total RNA was obtained by hot phenol/chloroform extraction. Total RNA (10 μg) was treated with DNase‐I and purified by phenol/chloroform extraction. Reverse transcription (RT) was performed with 1 μg of DNase‐I‐treated RNA following standard procedures with random hexamers and SuperScript III reverse transcriptase. Specific pairs of primers were used to amplify the qRT–PCR products of transcripts SCR1, HSP12, CTT1, and SSA4 using 3 μl of cDNA as a template (previously diluted 1:10). For qPCR, SYBR® Premix Ex Taq™ (Tli RNaseH Plus) was used in 10 μl of the final volume. Each sample was analyzed in triplicate, and qPCR was performed in a LightCycler® Roche 96. Real‐time PCR amplification efficiency was verified from the given slopes in the LightCycler® Roche 96. The ∆∆C t method was used to analyze qPCR results. The HSP12, CTT1, and SSA4 mRNA normalized levels were calculated using SCR1 mRNA levels as a reference and are expressed in relation to the transcript level from WT 25°C, Mip6‐GFP 25°C, or ski7Δ 25°C samples in each case. Four biological replicates were used for RNA‐seq experiments.
Statistics
Purified Mex67 from GFP‐TRAP experiments (Fig 2C) was calculated from at least two different experiments (np‐ADH1pr‐Mip6‐GFP = 5, np‐ADH1pr‐Mip6W442A‐GFP = 5; np‐ADH1pr‐Mip6‐ΔRRM4‐GFP = 2) and represented as Arbitrary Units (A. U.) ± SEM. Unpaired Student's t‐tests with Welch's corrections and one‐tailed (*P‐value < 0.05; **P‐value < 0.01; ***P‐value < 0.001) were used to test significant differences. Mip6 nuclear retention (Fig 4) was calculated from at least three different experiments (4D: nMex67‐HA = 844, nMex67ΔUBA‐HA = 612; 4E: nmip6Δ 30°C = 1,666, nmip6Δ 39°C = 1,567, nmip6Δmex67‐5 30°C = 989, nmip6Δmex67‐5 39°C = 871; 4F: np‐ADH1pr‐Mip6‐GFP = 1,606, np‐ADH1pr‐Mip6W442A‐GFP = 2,017, np‐ADH1pr‐Mip6‐ΔRRM4‐GFP = 1,400 cells) and represented the percentage of cells with nuclear retention ± SEM. One‐tailed binomial statistical tests were performed to confirm differences (***P‐value < 0.001). qRT–PCR data normalized to WT 25°C (Fig 7A and D), Mip6‐GFP 25°C (Fig 7C), or ski7Δ 25°C (Appendix Fig S6) levels are shown as means ± SEM. One‐tailed unpaired Student's t‐tests with Welch's corrections (*P‐value < 0.05; **P‐value < 0.01; ***P‐value < 0.001) were used to test significant differences from at least three biological replicates (nWT; mip6Δ = 6, nMip6‐GFP; Mip6W442A‐GFP; rrp6Δ; mip6Δrrp6Δ; ski7Δ; mip6Δski7Δ = 3) by using GraphPad Prism 5 software (t‐values and degrees of freedom in Appendix Table S7). For differential expression analyses in RNA‐seq experiments, LIMMA R package was used. The complete description will be published elsewhere. FISH data (Fig 7G and Appendix Fig S7B) (nWT 30°C = 100, nWT 42°C = 81, nmip6Δ 30°C = 95, nmip6Δ 42°C = 129, nmex67‐5 30°C = 84, nmex67‐5 42°C = 94) were quantified with ImageJ, and significant differences were determined using the Mann–Whitney U‐test (Appendix Fig S7C; *P‐value < 0.05; **P‐value < 0.01; ***P‐value < 0.001). Preparation of figures and tables was accomplished using Microsoft Excel, Microsoft Word, GraphPad Prism 5, and GIMP 2 software.
Author contributions
SR‐N supervised and led the study; MM‐E and M‐EG designed new experiments, discussed the results, and wrote the first draft of the manuscript together with SR‐N. They also performed the in vivo experiments (localization, in situ analyses, creation of yeast plasmids and genetic work, IPs, TAP experiments, RT–PCR analyses), with the contribution of CN‐C, AT‐C, and PP‐G. JC, JM, and MM‐E performed the PAR‐CLIP experiments, and AC, LF, and MM‐E analyzed the data; BC‐A, JMP‐C, NM, and JB performed recombinant protein production for NMR and X‐ray analyses. NM performed the BLI assays, RNA‐binding experiments, and crystallographic studies with the help of JB; BC and JMP‐C performed the NMR experiments; and MM‐E, M‐EG, JB, JMP‐C, and SR‐N wrote the final version of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Dataset EV1
Dataset EV2
Dataset EV3
Review Process File
Acknowledgements
We are grateful to Dr. C. Dargemont (University Paris Diderot) for antibodies and strains; Dr. E. Hurt (University of Heidelberg) for Mex67 plasmids; Dr. R. Parker (University of Colorado) for plasmids; A. Gagete and A. Ruiz for their contribution to initial aspects of this project; J. Serrano and J. Martínez from the Rodríguez‐Navarro Lab for the help provided to MM‐E. Special thanks to Dr Martínez‐Beneyto for her contribution in creating the synopsis image. P. Ordoño, I. Ruíz, and S. Masiá from the Bravo Lab for technical assistance. MM‐E, CN‐C, and AT‐C were supported by the GVA (Val I+D: ACIF/2015/025) PROMETEO (PROM/2016/093), and GVA (Val I+D: ACIF/2019/212). NM was partially supported by the European Union Erasmus Mundus Programme. NMR experiments were performed in the “Manuel Rico” NMR laboratory (LMR) (CSIC). Crystal tests were performed at XALOC beamline at ALBA Synchrotron with the collaboration of ALBA staff. The authors would like to thank Diamond Light Source for beamtime (proposal mx‐10121), and the staff of beamline I04 for assistance with crystal testing and data collection. This study was supported by funds to SR‐N from the Spanish MINECO, MICIIN (BFU2014‐57636, BFU2015‐71978, PGC2018‐099872‐B‐I00), and the Generalitat Valenciana (PROM/2012/061, ACOMP2014/061 and PROMETEO 2016/093), and to JB from the Spanish MINECO (SAF2015‐67077‐R, SAF2017‐89901‐R) and J.MP‐C from the Spanish MINECO (CTQ2018‐84371) and Comunidad de Madrid (B2017/BMD‐3770).
EMBO Reports (2019) 20: e47964
Data availability
PAR‐CLIP datasets are available at GEO as GSE126236 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126236.
References
- 1. Garcia‐Oliver E, Garcia‐Molinero V, Rodriguez‐Navarro S (2012) mRNA export and gene expression: the SAGA‐TREX‐2 connection. Biochem Biophys Acta 1819: 555–565 [DOI] [PubMed] [Google Scholar]
- 2. Reed R (2003) Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol 15: 326–331 [DOI] [PubMed] [Google Scholar]
- 3. Rodriguez‐Navarro S, Hurt E (2011) Linking gene regulation to mRNA production and export. Curr Opin Cell Biol 23: 302–309 [DOI] [PubMed] [Google Scholar]
- 4. Rondon AG, Jimeno S, Aguilera A (2010) The interface between transcription and mRNP export: from THO to THSC/TREX‐2. Biochem Biophys Acta 1799: 533–538 [DOI] [PubMed] [Google Scholar]
- 5. Dieppois G, Iglesias N, Stutz F (2006) Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol 26: 7858–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kelly SM, Corbett AH (2009) Messenger RNA export from the nucleus: a series of molecular wardrobe changes. Traffic 10: 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dunn EF, Hammell CM, Hodge CA, Cole CN (2005) Yeast poly(A)‐binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev 19: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E (2000) REF, an evolutionary conserved family of hnRNP‐like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zenklusen D, Vinciguerra P, Strahm Y, Stutz F (2001) The yeast hnRNP‐Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol 21: 4219–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hackmann A, Wu H, Schneider UM, Meyer K, Jung K, Krebber H (2014) Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat Commun 5: 3123 [DOI] [PubMed] [Google Scholar]
- 11. Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, Dargemont C, Stutz F (2010) Ubiquitin‐mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev 24: 1927–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lei EP, Krebber H, Silver PA (2001) Messenger RNAs are recruited for nuclear export during transcription. Genes Dev 15: 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zander G, Krebber H (2017) Quick or quality? How mRNA escapes nuclear quality control during stress. RNA Biol 14: 1642–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ling SH, Song H (2010) Mechanistic insights into mRNA export through structures of Dbp5. RNA Biol 7: 23–27 [DOI] [PubMed] [Google Scholar]
- 15. Zander G, Hackmann A, Bender L, Becker D, Lingner T, Salinas G, Krebber H (2016) mRNA quality control is bypassed for immediate export of stress‐responsive transcripts. Nature 540: 593–596 [DOI] [PubMed] [Google Scholar]
- 16. Bond U (2006) Stressed out! Effects of environmental stress on mRNA metabolism. FEMS Yeast Res 6: 160–170 [DOI] [PubMed] [Google Scholar]
- 17. Morano KA, Grant CM, Moye‐Rowley WS (2012) The response to heat shock and oxidative stress in Saccharomyces cerevisiae . Genetics 190: 1157–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rollenhagen C, Hodge CA, Cole CN (2007) Following temperature stress, export of heat shock mRNA occurs efficiently in cells with mutations in genes normally important for mRNA export. Eukaryot Cell 6: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saavedra C, Tung KS, Amberg DC, Hopper AK, Cole CN (1996) Regulation of mRNA export in response to stress in Saccharomyces cerevisiae . Genes Dev 10: 1608–1620 [DOI] [PubMed] [Google Scholar]
- 20. Galy V, Gadal O, Fromont‐Racine M, Romano A, Jacquier A, Nehrbass U (2004) Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- 21. Zid BM, O'Shea EK (2014) Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514: 117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Amoros M, Estruch F (2001) Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene‐ and stress type‐dependent manner. Mol Microbiol 39: 1523–1532 [DOI] [PubMed] [Google Scholar]
- 23. Gorner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schuller C (2002) Acute glucose starvation activates the nuclear localization signal of a stress‐specific yeast transcription factor. EMBO J 21: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hasan R, Leroy C, Isnard AD, Labarre J, Boy‐Marcotte E, Toledano MB (2002) The control of the yeast H2O2 response by the Msn2/4 transcription factors. Mol Microbiol 45: 233–241 [DOI] [PubMed] [Google Scholar]
- 25. Martinez‐Pastor MT, Marchler G, Schuller C, Marchler‐Bauer A, Ruis H, Estruch F (1996) The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15: 2227–2235 [PMC free article] [PubMed] [Google Scholar]
- 26. Lee P, Kim MS, Paik SM, Choi SH, Cho BR, Hahn JS (2013) Rim15‐dependent activation of Hsf1 and Msn2/4 transcription factors by direct phosphorylation in Saccharomyces cerevisiae . FEBS Lett 587: 3648–3655 [DOI] [PubMed] [Google Scholar]
- 27. Sadeh A, Baran D, Volokh M, Aharoni A (2012) Conserved motifs in the Msn2‐activating domain are important for Msn2‐mediated yeast stress response. J Cell Sci 125: 3333–3342 [DOI] [PubMed] [Google Scholar]
- 28. Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A) + RNA and nuclear pores. EMBO J 16: 3256–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bolognesi B, Lorenzo Gotor N, Dhar R, Cirillo D, Baldrighi M, Tartaglia GG, Lehner B (2016) A concentration‐dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep 16: 222–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin L, Zhang K, Sternglanz R, Neiman AM (2017) Predicted RNA binding proteins Pes4 and Mip6 regulate mRNA levels, translation, and localization during sporulation in budding yeast. Mol Cell Biol 37: e00408–e00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kohler A, Pascual‐Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez‐Navarro S (2006) The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase‐mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell 17: 4228–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scarcelli JJ, Hodge CA, Cole CN (2007) The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J Cell Biol 178: 799–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grant RP, Neuhaus D, Stewart M (2003) Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1A resolution. J Mol Biol 326: 849–858 [DOI] [PubMed] [Google Scholar]
- 34. Hobeika M, Brockmann C, Iglesias N, Gwizdek C, Neuhaus D, Stutz F, Stewart M, Divita G, Dargemont C (2007) Coordination of Hpr1 and ubiquitin binding by the UBA domain of the mRNA export factor Mex67. Mol Biol Cell 18: 2561–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gwizdek C, Iglesias N, Rodriguez MS, Ossareh‐Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C (2006) Ubiquitin‐associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci USA 103: 16376–16381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khong A, Parker R (2018) mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J Cell Biol 217: 4124–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wallace EW, Kear‐Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM et al (2015) Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162: 1286–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamamoto Y, Izawa S (2013) Adaptive response in stress granule formation and bulk translational repression upon a combined stress of mild heat shock and mild ethanol stress in yeast. Genes Cells 18: 974–984 [DOI] [PubMed] [Google Scholar]
- 39. Teixeira D, Sheth U, Valencia‐Sanchez MA, Brengues M, Parker R (2005) Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schaughency P, Merran J, Corden JL (2014) Genome‐wide mapping of yeast RNA polymerase II termination. PLoS Genet 10: e1004632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Santos‐Pereira JM, Garcia‐Rubio ML, Gonzalez‐Aguilera C, Luna R, Aguilera A (2014) A genome‐wide function of THSC/TREX‐2 at active genes prevents transcription‐replication collisions. Nucleic Acids Res 42: 12000–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McKinlay A, Araya CL, Fields S (2011) Genome‐wide analysis of nascent transcription in Saccharomyces cerevisiae . G3 (Bethesda) 1: 549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tarazona S, Furio‐Tari P, Turra D, Pietro AD, Nueda MJ, Ferrer A, Conesa A (2015) Data quality aware analysis of differential expression in RNA‐seq with NOISeq R/Bioc package. Nucleic Acids Res 43: e140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Amin J, Ananthan J, Voellmy R (1988) Key features of heat shock regulatory elements. Mol Cell Biol 8: 3761–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Castells‐Roca L, Garcia‐Martinez J, Moreno J, Herrero E, Belli G, Perez‐Ortin JE (2011) Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PLoS One 6: e17272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fox MJ, Gao H, Smith‐Kinnaman WR, Liu Y, Mosley AL (2015) The exosome component Rrp6 is required for RNA polymerase II termination at specific targets of the Nrd1‐Nab3 pathway. PLoS Genet 11: e1004999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH (2001) Quality control of mRNA 3′‐end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- 48. Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T (2001) Ski7p G protein interacts with the exosome and the Ski complex for 3′‐to‐5′ mRNA decay in yeast. EMBO J 20: 4684–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim M, Choi Y, Kim H, Lee D (2019) SAGA DUBm‐mediated surveillance regulates prompt export of stress‐inducible transcripts for proteostasis. Nat Commun 10: 2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Talarek N, Bontron S, De Virgilio C (2013) Quantification of mRNA stability of stress‐responsive yeast genes following conditional excision of open reading frames. RNA Biol 10: 1299–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim TS, Liu CL, Yassour M, Holik J, Friedman N, Buratowski S, Rando OJ (2010) RNA polymerase mapping during stress responses reveals widespread nonproductive transcription in yeast. Genome Biol 11: R75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sheth U, Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Muto Y, Yokoyama S (2012) Structural insight into RNA recognition motifs: versatile molecular Lego building blocks for biological systems. Wiley Interdiscip Rev RNA 3: 229–246 [DOI] [PubMed] [Google Scholar]
- 54. Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, Gehring K (2012) Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol Cell 48: 375–386 [DOI] [PubMed] [Google Scholar]
- 55. Santiveri CM, Mirassou Y, Rico‐Lastres P, Martinez‐Lumbreras S, Perez‐Canadillas JM (2011) Pub1p C‐terminal RRM domain interacts with Tif4631p through a conserved region neighbouring the Pab1p binding site. PLoS One 6: e24481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martinez‐Lumbreras S, Taverniti V, Zorrilla S, Seraphin B, Perez‐Canadillas JM (2016) Gbp2 interacts with THO/TREX through a novel type of RRM domain. Nucleic Acids Res 44: 437–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kielkopf CL, Lucke S, Green MR (2004) U2AF homology motifs: protein recognition in the RRM world. Genes Dev 18: 1513–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hobeika M, Brockmann C, Gruessing F, Neuhaus D, Divita G, Stewart M, Dargemont C (2009) Structural requirements for the ubiquitin‐associated domain of the mRNA export factor Mex67 to bind its specific targets, the transcription elongation THO complex component Hpr1 and nucleoporin FXFG repeats. J Biol Chem 284: 17575–17583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Strasser K, Bassler J, Hurt E (2000) Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol 150: 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chavez S, Garcia‐Rubio M, Prado F, Aguilera A (2001) Hpr1 is preferentially required for transcription of either long or G+C‐rich DNA sequences in Saccharomyces cerevisiae . Mol Cell Biol 21: 7054–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez‐Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R et al (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- 62. Bresson S, Tollervey D (2018) Surveillance‐ready transcription: nuclear RNA decay as a default fate. Open Biol 8: 170270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Houalla R, Devaux F, Fatica A, Kufel J, Barrass D, Torchet C, Tollervey D (2006) Microarray detection of novel nuclear RNA substrates for the exosome. Yeast 23: 439–454 [DOI] [PubMed] [Google Scholar]
- 64. Pascual‐Garcia P, Govind CK, Queralt E, Cuenca‐Bono B, Llopis A, Chavez S, Hinnebusch AG, Rodriguez‐Navarro S (2008) Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev 22: 2811–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Munchel SE, Shultzaberger RK, Takizawa N, Weis K (2011) Dynamic profiling of mRNA turnover reveals gene‐specific and system‐wide regulation of mRNA decay. Mol Biol Cell 22: 2787–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Perez‐Ortin JE, Alepuz P, Chavez S, Choder M (2013) Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J Mol Biol 425: 3750–3775 [DOI] [PubMed] [Google Scholar]
- 67. Herruer MH, Mager WH, Raue HA, Vreken P, Wilms E, Planta RJ (1988) Mild temperature shock affects transcription of yeast ribosomal protein genes as well as the stability of their mRNAs. Nucleic Acids Res 16: 7917–7929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Adams CC, Gross DS (1991) The yeast heat shock response is induced by conversion of cells to spheroplasts and by potent transcriptional inhibitors. J Bacteriol 173: 7429–7435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- 70. Huisinga KL, Pugh BF (2004) A genome‐wide housekeeping role for TFIID and a highly regulated stress‐related role for SAGA in Saccharomyces cerevisiae . Mol Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- 71. Gilbertson S, Federspiel JD, Hartenian E, Cristea IM, Glaunsinger B (2018) Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. Elife 7: e37663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Oliete‐Calvo P, Serrano‐Quilez J, Nuno‐Cabanes C, Perez‐Martinez ME, Soares LM, Dichtl B, Buratowski S, Perez‐Ortin JE, Rodriguez‐Navarro S (2018) A role for Mog1 in H2Bub1 and H3K4me3 regulation affecting RNAPII transcription and mRNA export. EMBO Rep 19: e45992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- 74. Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59: 687–696 [DOI] [PubMed] [Google Scholar]
- 75. Thomsen R, Nielsen PS, Jensen TH (2005) Dramatically improved RNA in situ hybridization signals using LNA‐modified probes. RNA 11: 1745–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jamonnak N, Creamer TJ, Darby MM, Schaughency P, Wheelan SJ, Corden JL (2011) Yeast Nrd1, Nab3, and Sen1 transcriptome‐wide binding maps suggest multiple roles in post‐transcriptional RNA processing. RNA 17: 2011–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang L, Wang S, Li W (2012) RSeQC: quality control of RNA‐seq experiments. Bioinformatics 28: 2184–2185 [DOI] [PubMed] [Google Scholar]
- 80. Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- 82. Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA‐seq data. Genome Biol 11: R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA‐seq: accounting for selection bias. Genome Biol 11: R14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Montaner D, Dopazo J (2010) Multidimensional gene set analysis of genomic data. PLoS One 5: e10348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida‐King J, Staines D, Derwent P, Kerhornou A et al (2011) Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database 2011: bar030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Teixeira MC, Monteiro PT, Palma M, Costa C, Godinho CP, Pais P, Cavalheiro M, Antunes M, Lemos A, Pedreira T et al (2018) YEASTRACT: an upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae . Nucleic Acids Res 46: D348–D353 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Dataset EV1
Dataset EV2
Dataset EV3
Review Process File
Data Availability Statement
PAR‐CLIP datasets are available at GEO as GSE126236 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126236.


