Adney et al. describe the complete and comprehensive codon substitution mutagenesis of human retrotransposon LINE-1 using a synthetic DNA approach. This experiment is the first of its kind for any transposon...
Keywords: LINE-1, L1, retrotransposon, scanning mutagenesis
Abstract
Long Interspersed Nuclear Element-1 (LINE-1, L1) is the only autonomous active transposable element in the human genome. The L1-encoded proteins ORF1p and ORF2p enable the element to jump from one locus to another via a “copy-and-paste” mechanism. ORF1p is an RNA-binding protein, and ORF2p has endonuclease and reverse transcriptase activities. The huge number of truncated L1 remnants in the human genome suggests that the host has likely evolved mechanisms to prevent full L1 replication, and thereby decrease the proliferation of active elements and reduce the mutagenic potential of L1. In turn, L1 appears to have a minimized length to increase the probability of successful full-length replication. This streamlining would be expected to lead to high information density. Here, we describe the construction and initial characterization of a library of 538 consecutive trialanine substitutions that scan along ORF1p and ORF2p to identify functionally important regions. In accordance with the streamlining hypothesis, retrotransposition was overall very sensitive to mutations in ORF1p and ORF2p; only 16% of trialanine mutants retained near-wild-type (WT) activity. All ORF1p mutants formed near-WT levels of mRNA transcripts and 75% formed near-WT levels of protein. Two ORF1p mutants presented a unique nucleolar-relocalization phenotype. Regions of ORF2p that are sensitive to mutagenesis but lack phylogenetic conservation were also identified. We provide comprehensive information on the regions most critical to retrotransposition. This resource will guide future studies of intermolecular interactions that form with RNA, proteins, and target DNA throughout the L1 life cycle.
APPROXIMATELY 45% of the human genome consists of retroelements, three of which are highly active non-LTR retrotransposon families in modern humans: L1, Alu, and SVA (Sine, short, interspersed nuclear element; VNTR, variable number of terminal repeats; Alu, name of a mobile element). These mobile genetic elements use a copy-and-paste mechanism called retrotransposition to propagate themselves within the host genome. The long interspersed element-1s (LINE-1s or L1s) are the only autonomously active human mobile element (Ostertag and Kazazian 2001; Brouha et al. 2003). Alu and SVA elements depend on L1-encoded proteins to execute retrotransposition and are thus considered nonautonomous.
There are roughly 500,000 copies of L1, making up ∼17% of the human genome (Lander et al. 2001). The vast majority of these are severely 5′ truncated and have diverged from the L1 consensus sequence, suggesting that they are very old and incapable of retrotransposition (Szak et al. 2002; Beck et al. 2010). About 15% of genomic L1Ta copies (Szak et al. 2002) and 6% of newly recovered experimentally induced elements are full-length (Gilbert et al., 2002, 2005; Symer et al. 2002), but the latter value is probably an undercount due to less-efficient recovery of full-length elements. Nevertheless, ∼90 L1 elements per diploid human genome remain retrotransposition competent and ongoing L1 activity continues to shape the evolution of mammalian genomes (Kazazian 2004; Huang et al. 2012; Faulkner and Garcia-Perez 2017).
The enormous number of 5′ truncated LINEs is a genomic feature of diverse species, but despite this they are not well understood mechanistically. The pervasiveness of 5′ truncation may reflect the action of anti-retrotransposon factors that play an active role in minimizing retrotransposon length. If these assumptions are correct, minimization of L1 length might help reduce the opportunity for truncations. As a consequence, L1 would become streamlined and highly enriched for sequences that are key for retrotransposition.
L1 activity plays important roles in both normal development and pathology. There is evidence that L1 activity is highest in the germline and somatic insertion events are also reported in a variety of tissues, notably the brain, as well as during early development (Ostertag et al. 2002; Muotri et al. 2005; An et al. 2006; Kano et al. 2009; O’Donnell et al. 2013; Carreira et al. 2014). Insertions into coding regions can cause human disease (Hancks and Kazazian 2016) and increased L1 expression (and in some cases retrotransposition) is also observed in various cancers (Lee et al. 2012; Rodić et al. 2014; Doucet-O’Hare et al. 2015; Ardeljan et al. 2017; Burns 2017; Nguyen et al. 2018). L1 activity has been reported to correlate with aging, stress, DNA damage, and telomere shortening, all of which are processes that are likely normally regulated to keep the mutagenic capacity of L1 jumping in check (Gorbunova et al. 2014; Van Meter et al. 2014; De Cecco et al. 2019). Therefore, better understanding of the mechanisms of L1 retrotransposition should provide new insights and opportunities in the fields of genome evolution, development, cancer biology, aging, and neurodegeneration.
The full-length human L1 element specifies production of a 6-kb long transcript that encodes two proteins, ORF1p and ORF2p (Ostertag and Kazazian 2001), which are both essential for retrotransposition. ORF1p is a 40-kDa protein with both nucleic acid-binding and chaperone activities (Kolosha and Martin 1997; Martin and Bushman 2001). ORF2p is a 150-kDa protein that has endonuclease (EN) (Feng et al. 1996), reverse transcriptase (RT) (Mathias et al. 1991), and nucleic acid-binding (Piskareva et al. 2013) activities. Upon translation of L1, ORF1p and ORF2p are thought to bind the same RNA molecule from which they were transcribed through a poorly understood process called cis-preference, also thought to require the 3′ poly(A) tail of L1 RNA (Boeke 1997; Wei et al. 2001; Kulpa and Moran 2006; Doucet et al. 2015). ORF1p is translated quite efficiently, but ORF2p translation occurs at much lower levels, through an unconventional process that is also poorly understood (Alisch et al. 2006). The L1 RNA, ORF1p, and ORF2p complex is referred to as the L1 ribonucleoprotein (RNP) complex and is likely to be the direct intermediate in retrotransposition (Martin 1991; Hohjoh and Singer 1996; Kulpa and Moran 2005; Doucet et al. 2010; Taylor et al. 2013, 2018). L1 insertion at the target genomic locus occurs via target-primed reverse transcription (TPRT) (Luan et al. 1993; Feng et al. 1996; Cost et al. 2002). While some key amino acid sequences have been elucidated (Mathias et al. 1991; Feng et al. 1996; Weichenrieder et al. 2004; Khazina et al. 2011; Christian et al. 2016; Ade et al. 2018; Khazina and Weichenrieder 2018), there is still much more that remains to be understood about the various L1 protein motifs and how they contribute to the L1 life cycle.
ORF1p consists of an unstructured N-terminal region followed by three structured domains (Figure 1A), including a coiled coil (a domain consisting of an extended series of heptad repeats; the human ORF1p contains 14 of these), an RNA-recognition motif (RRM) domain, and a C-terminal domain (CTD). The structure of human ORF1 has been well characterized by X-ray crystallography (Khazina et al. 2011; Khazina and Weichenrieder 2018), culminating in a near-full-length structural model used extensively in this report (Khazina and Weichenrieder 2018). The coiled-coil domain causes ORF1p to trimerize (Martin and Bushman 2001; Khazina et al. 2011), and the RRM and CTD domains are jointly responsible for single-stranded RNA binding (Januszyk et al. 2007; Khazina and Weichenrieder 2009; Khazina et al. 2011). Recent work has shown that the extended coiled-coil domain structure is metastable, particularly its N-terminal half, which contains a single “stammer” insertion (residues M91, E92, and L93) in one of the heptad repeats. This stammer is thought to lead to metastability of ORF1p because the distal part of the homotrimeric coiled coil can sample a partially unstructured state that may allow ORF1p trimers to interact with one another and form higher-order structures (Khazina and Weichenrieder 2018).
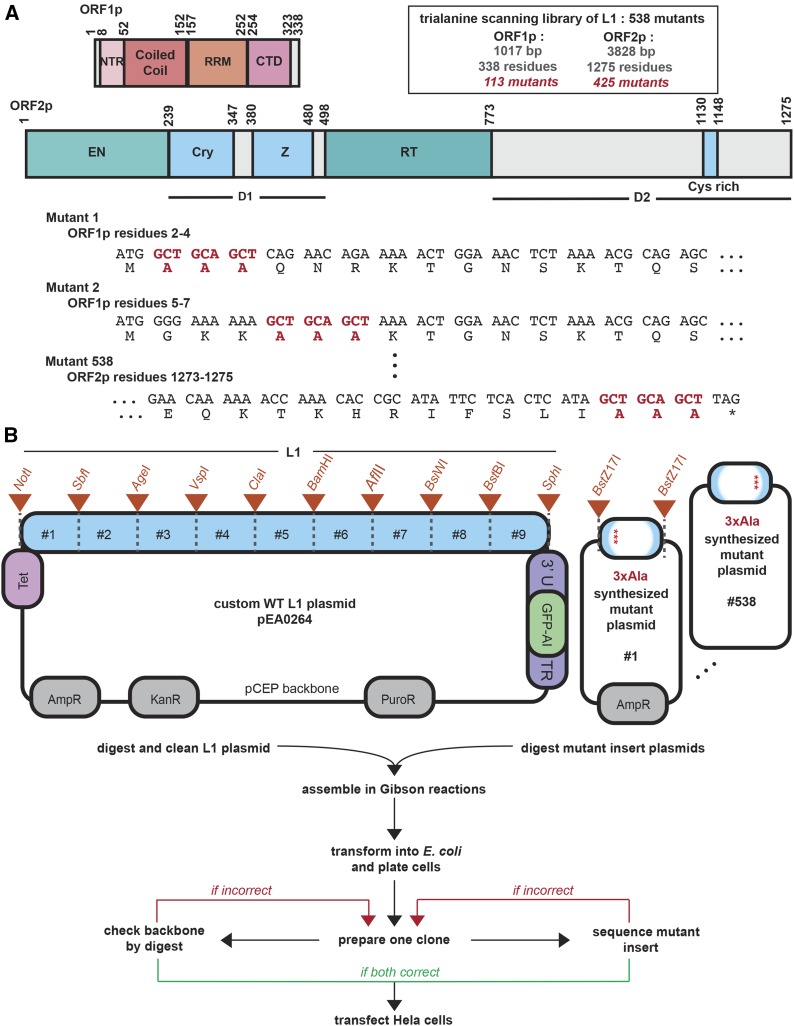
Figure 1.
L1 architecture and the design of the trialanine scan. (A) The human L1 proteins are depicted in detail. The residue positions of characterized domains are shown for ORF1p and ORF2p. The library consists of 538 mutants. The design of the trialanine mutants for the first two and the last mutant of the library are shown at the DNA and protein sequence levels. Start and stop codons were not mutated. The trialanine mutants are consecutive and nonoverlapping. (B) The parental L1 plasmid, pEA0264, is diagrammed in the upper left, featuring the engineered restriction sites. Orange triangles annotate the boundaries (designed unique restriction sites) of nine chunks. In the upper right, each of the 538 synthesized mutant plasmids were identical, excepting the 3xAla 600-bp fragment provided between the BstZ17I restriction sites. The pipeline for building the library is outlined below the plasmid schematics. An efficient two-piece Gibson assembly approach followed by a two-part quality control procedure was used to build each mutant L1 construct in the library. GFP-AI, GFP-AI fluorescent retrotransposition reporter construct; AmpR, ampicillin resistance; Cry, Cryptic motif; CTD, C-terminal domain; E. coli, Escherichia coli; EN, endonuclease; KanR, kanamycin resistance; L1, long interspersed element-1; NTR, N-terminal region; PuroR, puromycin resistance; RRM, RNA-recognition motif; RT, reverse transcriptase; Tet, Tet promoter; WT, wild-type; Z, Z-domain region.
ORF2p also has regions of well-characterized structure and function. Functionally, the most thoroughly understood regions are the enzymatic EN and RT domains (Mathias et al. 1991; Feng et al. 1996). Other less-functionally defined motifs include the recently described Cryptic (Cry) sequence (Christian et al. 2016), the Z-domain region (Clements and Singer 1998), and the C-terminal segment (CTS), which harbors a cysteine-rich motif (Fanning and Singer 1987) that is important for retrotransposition. There is a crystal structure of the EN domain (Weichenrieder et al. 2004), but the remainder of ORF2p remains structurally uncharacterized. In this work, we refer to two large, poorly characterized regions of ORF2p as Desert 1 (D1: the region between the EN and Z domains, which contains the Cry sequence) and Desert 2 (D2: the region that lies after the RT domain, and contains the CTS and cysteine-rich motif) (Figure 1A).
The L1 RNP also interacts with various host factors. RNP composition is complex and dynamic in that its intracellular location and composition changes throughout the L1 life cycle (Taylor et al. 2013, 2018; Mita et al. 2018). Extensive research has gone into identifying and characterizing retrotransposition host factors, as well as factors that influence retrotransposition (Niewiadomska et al. 2007; Beauregard et al. 2008; Suzuki et al. 2009; Arjan-Odedra et al. 2012; Dai et al. 2012; Goodier et al. 2012, 2013; Peddigari et al. 2013; Taylor et al. 2013, 2018; Pizarro and Cristofari 2016; Attig et al. 2018; Liu et al. 2018; Attig and Ule 2019). Different host factors could inhibit or facilitate L1 activity, and it is likely that ORF1p and ORF2p have coevolved with these factors. This host-specific coevolution could lead to essential amino acid sequences that are not well conserved.
L1 employs these endogenous activities and interactions with host factors to progress through a multistage life cycle. L1 RNA must be transcribed, exported, and protected from degradation. ORF1p and ORF2p must be translated, folded, and coassembled with L1 RNA. This RNP must incorporate or exclude host factors. Finally, the RNP must be imported to the nucleus and ORF2p must mediate TPRT at a target locus. Mutating L1 affects DNA, RNA, and protein primary sequences, and thus may affect any of the steps listed above. While excellent work has begun to dissect the molecular details of this life cycle, the functional significance of most ORF1p and ORF2p residues remain unknown. Therefore, we set out to build and characterize a scanning trialanine mutant library to determine how disruption of L1’s sequence may impact its cellular activities. We built 538 mutants of a human L1 and characterized this ordered library by measuring retrotransposition efficiency, ORF1 RNA and protein abundance, and ORF1p cellular localization. We also compared conservation and retrotransposition efficiency throughout ORF2p, which helped identify which areas in the poorly characterized ORF2p deserts were most interesting for further study. This first comprehensive scanning mutagenic library of any transposable element provides a map that indicates which residues are critical or dispensable for the L1 life cycle.
Materials and Methods
Design and construction of the trialanine scanning mutagenic library
A major goal was to create a pipeline in which an ordered (as opposed to pooled) library could be efficiently assembled. The original vector backbone, extensively reengineered in our laboratory, was based on the pCEP4 origin of plasmid replication/Epstein-Barr nuclear antigen (EBNA) - based vector that replicates autonomously in primate cells (pCEP4 Catalog no. V044-50; Thermo Fisher Scientific), which we refer to as pCEP-puro [puromycin (puro); the original hygromycin resistance (HygroR) cassette was replaced with a puro resistance (PuroR) cassette]. This was the basic backbone of the parental L1-containing plasmid, pEA0264, into which each trialanine mutant was cloned (Figure 1B). We added a kanamycin resistance (KanR) cassette to the vector backbone to facilitate subcloning of synthetic fragments delivered in an ampicillin resistance (AmpR) vector. pEA0264 contained a human L1-rp cassette, expressed from the tetracycline (tet) - inducible, minimal - cytomegalovirus (CMV) promoter, which is called the Tet promoter. The construct did not include the native L1 5′-UTR sequence. The full native L1-rp 3′-UTR sequence was present, and also contained the GFP-AI fluorescent retrotransposition reporter construct (Ostertag et al. 2000). Because the L1-rp native 3′-UTR has a weak poly(A) addition signal, we also included a downstream SV40 poly(A) addition signal from pCEP4.
As described in the text, unique restriction sites were designed such that they fell only within L1 and not in the vector backbone, and were spaced roughly equally, about every 600 bp. This entailed both removing and adding (“silently,” when in a coding region) restriction enzyme cut sites from throughout the plasmid backbone and the L1-rp cassette using the GeneDesign online tool (Richardson et al. 2006). The library was optimized to facilitate downstream combinatorial cloning and manipulation of the individual mutants. The logic behind the design, and the construction of pEA0264 and the full mutant library derived from it, has been extensively described in detail (Adney 2018).
The 538 trialanine mutants were generated using Gibson assembly (Gibson et al. 2009), as shown in Figure 1B. Each mutant was contained within one of nine “chunks” of synthetic DNA, which effectively replaced the wild-type (WT) chunk. An efficient, high-throughput protocol was developed to assemble the library, perform quality control, and prepare tissue culture-grade DNA for subsequent experiments [Supplemental Material, Figures S1 and S2, and Adney (2018)].
96-well retrotransposition assay
Retrotransposition was measured as outlined in Figure S3 using HeLa-M2 cells (Hampf and Gossen 2007). The protocol for the following has been described in detail (Adney 2018), but in brief: on day 1, 25,000 HeLa cells were seeded per well in 50 µl DMEM in a 96-well plate and transfected with 60 ng DNA ∼1 hr later. On day 2, puro was added to each well to select for cells containing plasmid; on day 3, the cells were split to a black-walled 96-well tissue culture plate and doxycycline (dox) was added to induce expression of the L1 cassette; and on day 6, the cells were fixed and stained for analysis. The plates were imaged at NYU Langone’s High Throughput Biology Laboratory for data analysis, discussed below. Figure S3 also shows controls done to prove the robustness and reproducibility of this technique.
Quantification of retrotransposition
96-well black imaging plates (3603; Corning product) were imaged on an Arrayscan VTI using the following parameters: 5 × magnification, 2 × 2 binning, and four fields per well. Image analysis was performed using the Target Activation Bioapplication (Thermo Fisher Scientific Cellomics Scan version 6.6.0, build 8153). DAPI-positive nuclei were identified using a dynamic isodata thresholding algorithm after minimal background subtraction. DAPI-positive objects were used to identify cell nuclei and to delineate nuclear borders. A “circle” (x = 2 µm) greater than the nuclear border was drawn for each cell and the GFP expression within this area was quantified. Cells expressing cytoplasmic GFP represented retrotransposition-positive cells (since the limits of fluorescence were set so that no cells were considered positive for preparations of control cells lacking GFP). The reported parameters are explained as follows: Total = total number of DAPI nuclei counted and GFP+ = above GFP threshold;
Statistical analysis of retrotransposition frequency
Once all retrotransposition efficiency data were acquired, we set thresholds for which trialanine mutants had a “strong effect” (depleting activity) and which had “WT activity”. First, to set the lower threshold, we looked at mutants containing ORF2p residues known to be critical for retrotransposition and thought to be catalytic (N14, E43, D145, D205, H230, and D702), which all showed a strong effect with retrotransposition frequencies < 20% of WT, providing a good calibration of the lowest-activity category. By setting a conservative threshold at 25%, we allowed for some biological variation in any given mutant’s interexperimental variation in retrotransposition level.
Second, we did a statistical analysis to set the range of WT, which meant taking all the data into consideration and establishing what we did not consider to significantly deviate from WT activity (100%). We first made sure that we did not see any major batch effects between experiments; none were noted. When the data were divided into four groups based on their activities with an equal number of mutants in each group, as expected, the error decreased as the median increased. We estimated the error distribution for different number of replicates in the four regions by randomly resampling the data points with replacement. Using the error distribution for the group with the highest activity that contained the WT data points, we estimated a confidence interval (CI) for what represented WT activity. For mutants with four replicate measurements, the 99% CI was estimated at 78–126% of the reference WT plasmid’s activity, and we used 80% as a conservative lower limit for WT activity. No mutant’s activity averaged > 125%, indicating that we did not isolate any strong “gain-of-function” mutant in this library.
Immunoblot assays and statistical analysis of ORF1p mutant protein abundance
Each ORF1p mutant was tested for protein production in two separate biological replicates. The HeLa-M2 cells were treated and harvested in a six-well plate format, and protein was extracted and measured by quantification on a Western blot, as previously described (Adney 2018). ORF1 protein is expressed in HeLa-M2 cells, thus 13% of the ORF1p signal in cells expressing pEA0264 WT ORF1p in these experiments does not come from the plasmid. Thus, we first normalized by adjusting for this endogenous expression of ORF1 protein so that we only compared protein expression off the L1-containing plasmid (thus, although there may be ORF1p signal in the blot, the true amount of expression of a mutant can be 0% in the data depicting ORF1p expression of each of the mutants compared to the WT plasmid pEA0264 at 100%). Based on a statistical analysis of each ORF1p mutant’s protein abundance, computed in the same manner as described above for the retrotransposition activity thresholds, the 99% CI estimated 50% of WT protein abundance as the lower limit. Hence, the protein levels for each mutant are referred to as either “high,” which refers to WT ORF1p abundance, or “low,” which refers to a protein abundance that was < 50% that of WT (significantly depleted).
Measurement of total RNA abundance
The RNA level of an ORF1p mutant was calculated by comparing the total RNA to the total plasmid DNA for a given ORF1p trialanine mutant, and then normalizing that to the WT value. For these measurements, we took a pooled approach in which we transfected anywhere from 1 to 14 mutants into one well of cells. Cell lysate was prepared from transfected HeLa cells, and total plasmid for DNA sequencing (DNA-seq) or total RNA for RNA sequencing (RNA-seq) was isolated, and the respective libraries were prepared and sequenced as described (Adney 2018) using 36-bp paired-end reads on an Illumina NextSeq 500. For analysis of pooled samples, we designed a custom series of L1 reference sequences corresponding to each L1 trialanine mutant. The references were designed for each mutant: (1) with the mutant sequence (9 bp) located at the center of a 75-bp sequence and (2) the exact same sequence that was fully WT. The 36-bp reads only required 1 bp of overlap with the mutant sequence to map well. We then compared read counts, as previously described (Adney 2018).
Quantification of ORF1p cellular localization
Transfected HeLa-M2 cells were prepared in a 96-well plate, fixed, and stained (with the anti-ORF1p antibody, the nucleolus using an anti-fibrillarin antibody, and Hoechst 33342) for imaging analysis as described (Adney 2018). Images were obtained using an Andor Yokogawa CSU-X confocal spinning disk on a Nikon (Garden City, NY) TI Eclipse microscope and fluorescence was recorded with an sCMOS Prime 95B camera (Photometrics) with a 100 × objective (pixel size: 0.11 m). Five random fields of view were imaged per construct per experiment. One DAPI image and a six-step 6-μm Z-stack in the ORF1p channel were acquired for each field of view. Images were acquired using Nikon Elements software and analyzed using ImageJ/Fiji. Each channel was z-projected using “Sum Slices.” The data were blinded and manually scored for nucleolar localization by a naïve investigator who recorded the number of nuclei in the image, the number of nuclei that had the nucleolar phenotype, and the approximate nucleolar-to-cytoplasmic ORF1p intensity ratio of the positive cells. Nucleolar phenotype was qualitatively evaluated by normalizing a given cell’s nucleolar ORF1p intensity to its cytoplasmic ORF1p intensity and comparing it to the same ratio in cells transfected with the WT construct. Nucleoli were identified by DAPI and were confirmed by fibrillarin immunofluorescence (IF) in a subset of experiments. The frequency of the nucleolar phenotype was evaluated over ≥ 20 cells per construct. A given mutant was considered positive for the nucleolar phenotype if its phenotype rate was > 1 SD above the mean phenotype rate across all constructs tested.
Generation of alignments to evaluate conservation in ORF2p
ORF2 sequences were translated and aligned from a compilation of L1 nucleotide sequences (Boissinot and Sookdeo 2016), which are listed in Table S7. Fifty-five of the sequences, including L1-rp ORF2, were run through multiple sequence alignment analysis, followed by measurements of percent identity using Geneious (v 11.1.2; build 2018-03-01 15:52; Java version 1.8.0-162-b12 64 bit: restricted R11 license). An alignment of a representative subset of these sequences is presented in Figure S5. The program produced the percent identity score at each residue. Since we were working with three-residue windows, we used the percent identity value corresponding to the residue with the highest identity score for each trialanine mutant. We binned the identity score quantities into four bins, spanning 0–29%, 30–69%, 70–99%, and 100%. We then compared these categories to the three bins of retrotransposition efficiency explained in the text (no retrotransposition, reduced retrotransposition, and WT levels of retrotransposition). Then, the status of each mutant by each of these two measures was analyzed.
Data availability
All strains are available upon request. Supplemental material available at figshare: https://doi.org/10.25386/genetics.9978590.
Results and Discussion
Retrotransposition efficiency is extremely sensitive to ORF mutations
To determine amino acid sequences in ORF1p and ORF2p that are critical for L1 function, we undertook a scanning mutagenesis study, producing a library of 538 trialanine mutants scanning human L1. These L1 proteins consisted of 338 and 1275 residues, respectively (Figure 1A). To obtain a complete mutagenic scan of the ORFs, we designed an ordered library of 113 mutants for ORF1p and 425 mutants for ORF2p, totaling 538 mutants, each of which had three consecutive residues mutated to alanine (each referred to as a trialanine mutant). The mutants tiled through the proteins, did not overlap, and did not include start or stop codons (Figure 1A and Table S1). The identities of the final constructs that made up the library are detailed in the first column of Table S2.
We used a human L1 sequence (L1-rp, accession number AF148856), derived from a retinitis pigmentosa patient cell line, that is known to be retrotransposition competent (Kimberland et al. 1999). We used the nonendogenous, dox-inducible, Tet-minimal CMV reporter to drive L1 expression in place of the 5′-UTR promoter sequence (O’Donnell et al. 2013; Taylor et al. 2013). We tested the ability of each mutant to retrotranspose using a retrotransposition assay (Figure S3A), which is the most stringent test for function; any aspect of the L1 life cycle that was impacted by our mutations should be evident. Retrotransposition efficiency values are listed in Table S2, and Figure 2 summarizes the retrotransposition efficiency of each mutant relative to WT and maps this value along the length of the ORFs, highlighting key motifs and previously studied essential residues.
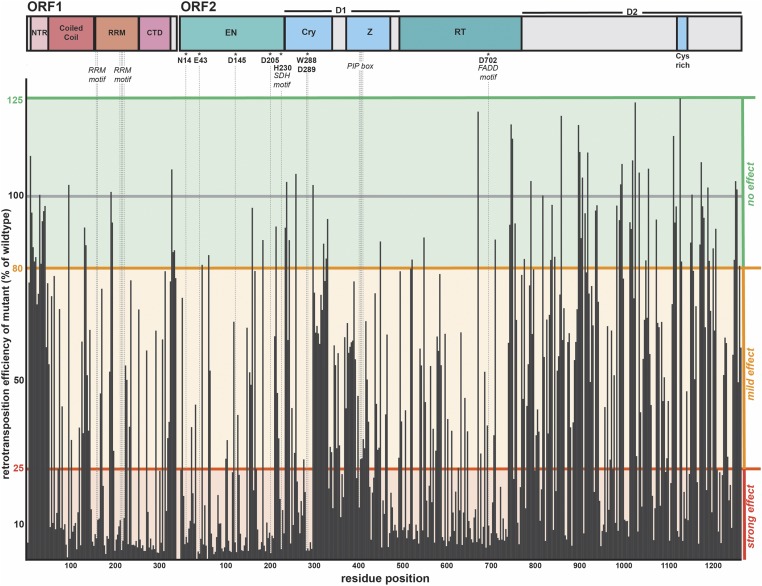
Figure 2.
The retrotransposition efficiency of each trialanine mutant. Along the top are the schematics of ORF1p and ORF2p, highlighting domain boundaries, as well as well-characterized motifs and essential residues. The residue position is indicated along the x-axis. The y-axis denotes the percentage of WT activity of each mutant. Each mutant’s retrotransposition was normalized to WT measurements made in the same experiment on the same plate. WT retrotransposition frequency was set to 100% (gray bar). Statistically (Materials and Methods), values ranging between 80 and 125% were within the WT range of activity, in which the trialanine mutation had no effect (green background). A mutant was classified as mild effect for values ranging between > 25 and < 80% (orange background), and strong effect for values ≤ 25% (red background). Cry, Cryptic motif; CTD, C-terminal domain; EN, endonuclease; NTR, N-terminal region; PIP, PCNA-interacting protein; RRM, RNA-recognition motif; RT, reverse transcriptase; WT, wild-type; Z, Z-domain region.
Retrotransposition efficiency was extremely sensitive to ORF1p and ORF2p mutations, consistent with expectations for a “streamlined” and highly conserved element. About 50% of the trialanine mutants had a strong effect, 34% had a mild effect, and only 16% retained WT activity. A significant fraction of the total mutants (25% of ORF1p and 12% of ORF2p mutants) had activity ≤ 5% of WT. None of the mutants caused a significant increase in activity (> 125% of WT). ORF1p and ORF2p had similar frequencies of deleterious mutations, with obvious clusters of strong effect in the more conserved domains of the proteins (Table 1).
Table 1. Impact on retrotransposition efficiency organized by protein domain.
| Percentage of 3xAla mutations in each retrotransposition efficiency category | |||
|---|---|---|---|
| Domain | Strong | Mild | None |
| OFR1p | |||
| Full-length protein | 53 | 31 | 16 |
| NTR | 6 | 35 | 59 |
| Coiled coil | 56 | 35 | 9 |
| RRM | 70 | 24 | 6 |
| CTD | 55 | 32 | 13 |
| ORF2p | |||
| Full-length protein | 48 | 35 | 17 |
| EN | 74 | 19 | 8 |
| D1 | 38 | 47 | 15 |
| Z | 36 | 61 | 3 |
| RT | 62 | 28 | 10 |
| D2 | 32 | 41 | 27 |
The percentages of 3xAla mutants showing a strong, mild, or no effect on retrotransposition efficiency are represented for both ORF1p and ORF2p. The values for the full-length protein and then for each domain are shown. CTD, C-terminal domain; EN, endonuclease; NTR, N-terminal region; RRM, RNA-recognition motif; RT, reverse transcriptase; Z, Z-domain region.
Overlaying retrotransposition levels of the mutants on solved WT crystal structures gives a visual representation of each mutant’s impact, for example the EN domain of ORF2p (Figure S4). The mapping of mutant phenotypes onto the full-length ORF1p structure model will be presented visually in the next section, together with protein abundance data.
Mobile elements that remain active in the human genome inspire comparison to host–parasite arms races (Daugherty and Malik 2012). While L1 is not simply a parasite and does play important roles, L1 elements also pose a strong risk to the host due to their strong mutagenic capacity, and so the element can be considered to be analogous to a parasite with respect to the evolution of its DNA sequence. The host is likely under strong selection to reduce retrotransposition while L1 must evolve a robust life cycle to avoid extinction. This type of antagonistic selection tends to minimize genome sizes in obligate-parasitic organisms (Wolf and Koonin 2013). In addition, the biochemistry of the L1 life cycle may drive genome minimization. The huge number of truncated L1 remnants in the human genome suggests that the RT step is frequently not processive enough to drive successful retrotransposition in the host environment. This may be an intrinsic limitation of the RT enzyme, but it is also likely that the host has evolved mechanisms that actively promote 5′ truncation. Thus, shortening the L1 genome would increase its probability of propagation. The net result would be an increase in information density in the protein-coding regions in the element. The high density of critical regions for retrotransposition that we found provides strong evidence for this streamlining hypothesis.
Most ORF1 mutants are expressed robustly
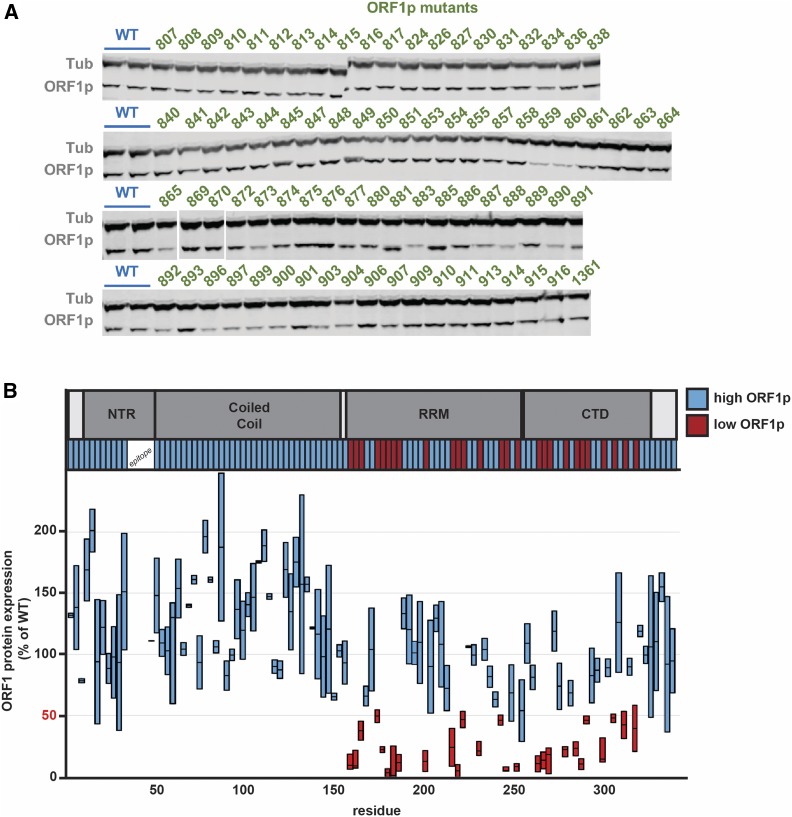
We quantified the relative protein levels of the ORF1p mutants individually by immunoblotting (Figure 3A and Table S3) using a monoclonal antibody targeting endogenous human ORF1 (Rodić et al. 2014). Due to substantial variations (twofold) in ORF1p levels in replicate immunoblot experiments we treated the average protein abundance for each mutant as binary, with a conservative cutoff: high (> 50% that of WT) or low (< 50%). Only 24% of the mutants (27/113) resulted in ORF1p reduction to < 50% that of WT. All of these mutants with low ORF1p also showed loss of retrotransposition activity. Retrotransposition and protein abundance data are summarized in Table S4. Trialanine mutants that disrupted the epitope that our antibody recognizes could not be assessed by western blot; however, since all these mutants showed WT or close-to-WT levels of retrotransposition, we can confidently surmise that they were well expressed. Figure 4 summarizes the effects of protein levels and retrotransposition activity mapped onto the ORF1p crystal structure. Of the mutants that showed low protein levels, all mapped to the RRM and CTD domains (16 and 11 mutants, respectively; Figure 3B). We speculate that these mutants interfere with the folding of these highly structured domains.
Figure 3.
Protein abundance of mutants of ORF1p. (A) Representative immunoblots for WT pEA0264 and the ORF1p mutants. Samples were prepared from six-well plates of HeLa cells, the clarified lysates of which were probed with anti-ORF1 and anti-Tub antibodies. HeLa cells lacking a plasmid reproducibly expressed ORF1p at a level of 13% of pEA0264. (B) The ORF1p schematic is shown at the top. Results from immunoblot analyses for each ORF1p mutant are represented on the plot. Two measurements are shown for each mutant, quantified from independent experiments. These values were background subtracted to remove signal corresponding to endogenous ORF1p expression. Protein levels are plotted on the y-axis and residue positions are indicated on the x-axis. We observed some variability and thus plotted the range for each mutant as a bar, with a horizontal bar marking the mean. We refer to protein abundance in binary terms, as either high (+) or low (−), using 50% (marked in red) as the threshold. The mutants are color coded in the bar below the ORF1p schematic to highlight which regions had high (blue) or low (red) protein levels. CTD, C-terminal domain; NTR, N-terminal region; RRM, RNA-recognition motif; Tub, tubulin; WT, wild-type.
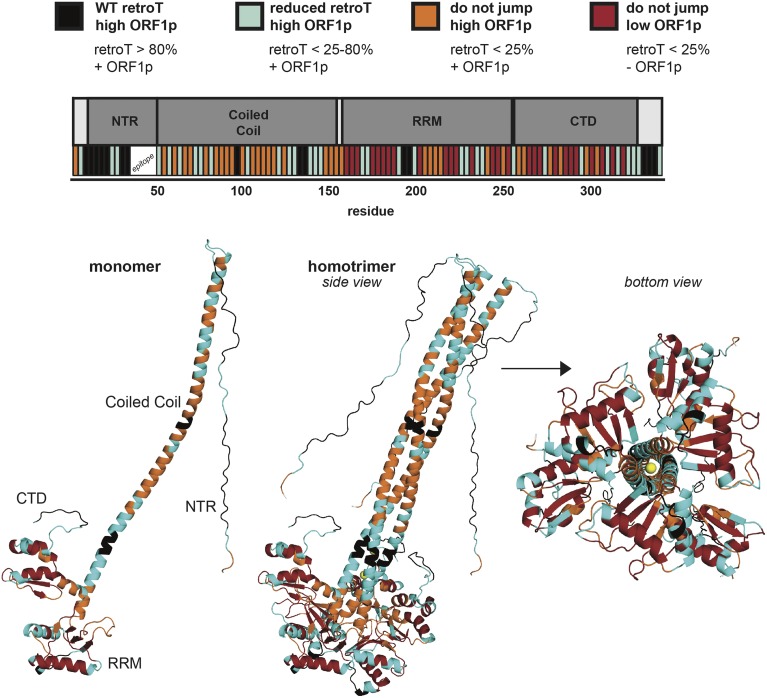
Figure 4.
Retrotransposition and protein abundance of mutants mapped onto a three-dimensional model of trimeric ORF1p. The model is based on available crystal structures (Khazina and Weichenrieder 2018). The mutants are divided into four categories and color coded, shown at the top. This provides a visual representation of retrotransposition efficiency and protein abundance, both along the linear schematic of ORF1 with the corresponding color-coded bars, as well as projected onto the WT ORF1p monomer and trimer structures. The color code is as follows: high ORF1p and WT retrotransposition (black), high ORF1p and reduced retrotransposition (cyan), high ORF1p and no retrotransposition (orange), low ORF1p and no retrotransposition (red), chloride ions noted in the structure of Khazina et al. (2011) (yellow), and the initial methionine (not mutated, white). CTD, C-terminal domain; NTR, N-terminal region; RRM, RNA-recognition motif; WT, wild-type.
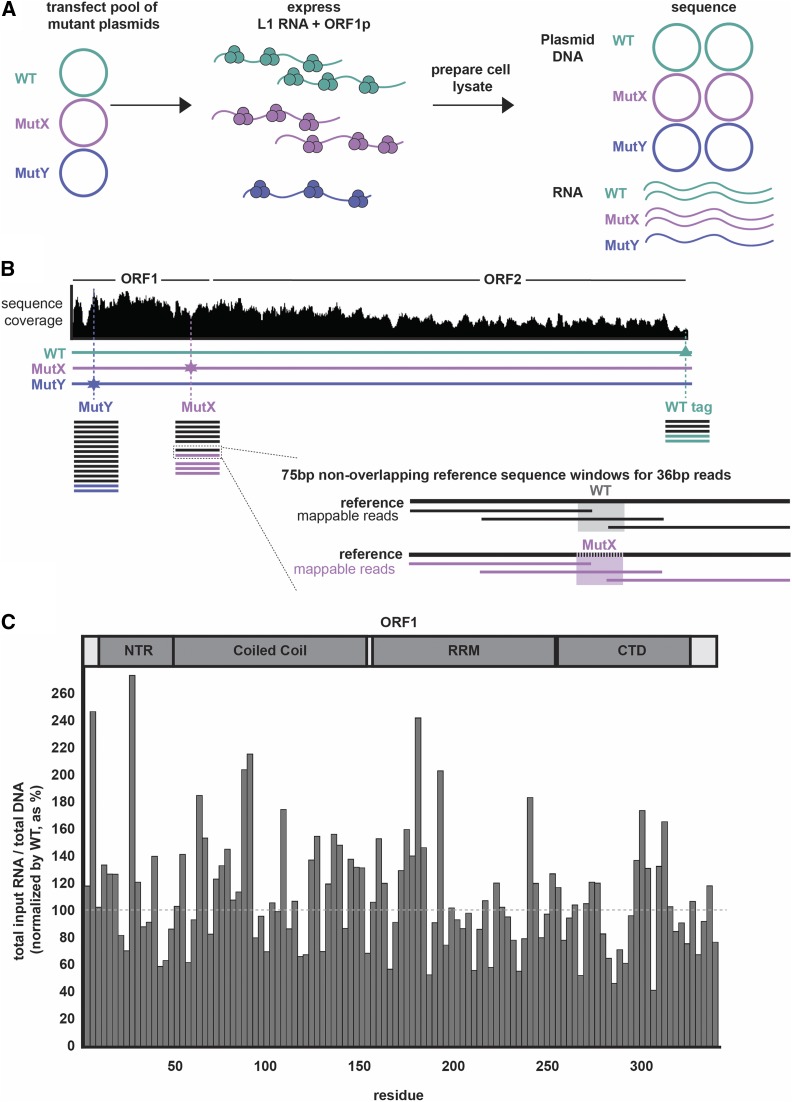
Next, we wished to evaluate the effect of each mutant on L1 RNA stability. To do so, we designed a pooled RNA-seq and DNA-seq experiment to evaluate the impact of ORF1p mutations on RNA abundance. The experimental design is shown in Figure 5, A and B: hypothetical Mut X has abundant RNA while Mut Y has low-abundance RNA. DNA-seq reads were used to normalize the transfection efficiency of each plasmid. Sequencing reads containing the unique 9 bp of each 3xAla insertion were used to determine RNA and DNA levels. The WT plasmid and RNA were used as internal controls for WT L1 behavior. In this way, we determined the relative RNA abundance of every mutant.
Figure 5.
The protocol for a pooled approach to sequence plasmid DNA and total RNA of L1 mutants. (A) The workflow for transfecting a pool of two mutants and the WT plasmids (thus coexpressing three constructs), preparing cell lysate, and sequencing the isolated pools of L1 plasmid DNA and total RNA is shown. In this depiction, at the end of the experiment, all three plasmids have an equal abundance of plasmid DNA copies. MutX and WT show equal L1 RNA abundance, while that of MutY is reduced by one-half relative to WT. (B) Sequence coverage across L1 coding region is uneven. This diagram depicts how reads were mapped and read count was normalized to both the sequencing depth at a given window as well as to the internal WT plasmid control. (analysis was done the same for both DNA and RNA; the schematic represents read ratios of the RNA). (C) The RNA abundance (normalized to plasmid DNA abundance) of each ORF1p mutant is shown as a percent of WT. The fraction of total mutant L1 RNA in the lysate is shown, normalized to the WT level. The gray dashed line indicates WT levels at 100%. CTD, C-terminal domain; L1, long interspersed element-1; Mut, mutant; NTR, N-terminal region; RRM, RNA-recognition motif; WT, wild-type.
We pooled several mutants at once with the WT construct for a total of eight pools (named pools 1–8; Table S5) and expressed each of them in human cells. The data are reported as the RNA abundance of each mutant in Figure 5C. Notably all mutants had near-WT levels of RNA abundance, and no mutant had < 60% that of WT, indicating that RNA abundance (reflecting transcription efficiency and stability) is unlikely to explain ablation of retrotransposition in many of the ORF1p mutants. However, a formal demonstration of this will require replicates, an expensive experiment for what is likely to be a negative result.
Two coiled-coil mutants have significant relocalization to the nucleolus
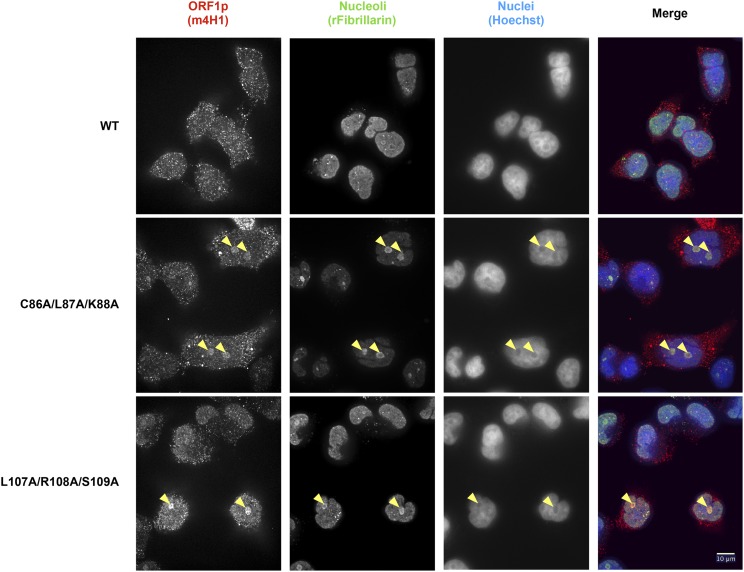
We evaluated whether any ORF1p mutants that block retrotransposition might do so by interfering with proper subcellular localization. Nucleocytoplasmic trafficking is key to the L1 life cycle and our previous studies revealed relocalization of ORF1p from the cytoplasm to the nucleus during the M/G1 phase of the cell cycle (Mita et al. 2018). We therefore used IF to probe the localization of the 40 ORF1p trialanine mutants that produce normal levels of protein but have decreased retrotransposition activity (Table S6). The vast majority of the ORF1p mutants localized primarily to the cytoplasm, just like WT ORF1p. However, we observed that two ORF1p mutants, CLK86-88AAA and LRS107-109AAA, displayed strong nucleolar localization in a subset of cells (Figure 6 and Table S6). This striking relocalization phenotype was seen in 8% and 24% of total cells for the CLK86-88AAA and LRS107-109AAA mutants, respectively, as compared to < 1% of cells expressing WT ORF1p. We did not observe a correlation between nucleolar localization and total ORF1p fluorescence in a given cell.
Figure 6.
Immunofluorescence analysis reveals intriguing nucleolar localization of a small subset of ORF1p mutants. Representative images of immunostained HeLa-M2 cells expressing WT L1 (top) or L1 ORF1p mutants [CLK86-88AAA (middle) and LRS107-109AAA (bottom)]. Yellow arrowheads indicate nucleoli with diffuse ORF1p localization in the mutant construct transfections. Cells were stained with mouse anti-ORF1p (left) and rabbit anti-fibrillarin (middle left) antibodies, and Hoechst 33342 (middle right). Antibody target names are reported above the corresponding pictures and colored according to the colors used in the merged pictures (right). Bar, 10 μm. L1, long interspersed element-1; WT, wild-type.
Both mutants of interest reside in the coiled-coil domain of ORF1p. A C86S substitution was previously shown to strongly reduce retrotransposition, which was surprising given the poor conservation of C86 across primate L1 sequences and its position on the surface of the coiled coil (Khazina and Weichenrieder 2018). Our data with CLK86-88AAA recapitulate the sharp decrease in retrotransposition and suggest a defect in intracellular localization as a potential mechanism. Additionally, a three-residue stammer insertion (residues 91–93) in a portion of the heptad-repeat structure of the ORF1p coiled coil was proposed to contribute to the structural malleability of the coiled coil N-terminal to the stammer (Khazina and Weichenrieder 2018). They proposed a model in which the stammer introduces flexibility into the coiled coil that then allows for ORF1p trimers to adopt an open conformation and form intertrimer interactions between ORF1p N-termini. These intertrimer interactions were suggested to drive higher-order ORF1p structures, such as linear arrays and a larger meshwork of trimers. The stammer lies between our two trialanine mutants of interest. It is conceivable that the CLK86-88AAA and LRS107-109AAA mutants change the flexibility of the ORF1p coiled coil in similar ways, thus interfering with the L1 life cycle and increasing the propensity of the protein to localize to the nucleolus. While the reasons for nucleolar localization will require further investigation, we speculate that the localization of a subset of ORF1p mutants to the nucleolus could be the result of altered binding affinities for nucleic acid or protein partners.
Previous work on LINE-1 proteins and the nucleolus demonstrated localization of WT ORF1p to the nucleolus in close to 50% of 143B TK cells (Goodier et al. 2004). However, this localization was tag-dependent, and was seen either in ORF1p-only expression constructs or in bicistronic constructs with two internal ribosome entry sites (IRESs), which complicates interpretation. Further exploration of ORF1p-only constructs identified an E165G ORF1p mutant that has enhanced nucleolar localization, and also indicated that nucleolar localization is likely RRM-dependent since actinomycin D treatment abolished nucleolar localization of WT and E165G ORF1p without changing cytoplasmic foci formation (Goodier et al. 2007). Taken together, we expect that ORF1p localization to the nucleolus might be a physiological step in the L1 life cycle, but more likely that accumulation of ORF1p in the nucleolus may instead be a cause or consequence of L1 transposition defects. Notably, other work showed that the L1 RNA itself interacts with nucleolin, a nucleolar protein, and promotes transcriptional program changes that are necessary for embryonic development in mice (Percharde et al. 2018). However, while L1 RNA was predominantly nuclear in these mouse embryonic stem cells, ORF1p was mostly cytoplasmic. It is possible that in our cell system, endogenous nucleolin captures more ORF1p-bound L1 RNA and that ORF1p mutations alter the ability of nucleolin to bind to ORF1p-decorated L1 RNA. Interestingly, nucleolin was previously identified as a factor that specifically promotes ORF2p translation, and nucleolin knockdown was found to decrease L1 retrotransposition rates (Peddigari et al. 2013). Thus, localization of ORF1p mutants to the nucleolus may be indicative of an imbalance of L1 RNA interactions with ORF1p and nucleolin, which could in turn lead to a decrease in L1 retrotransposition rates.
A cluster of transposition-defective mutations in a nonconserved domain of ORF2
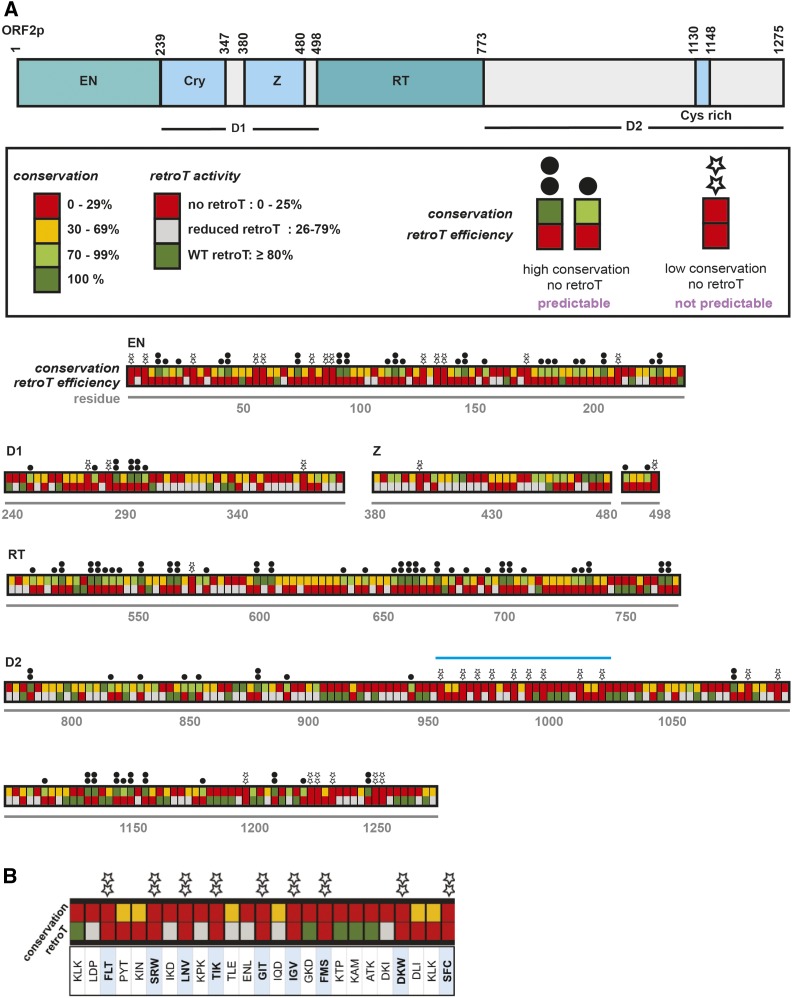
Similar to ORF1p, where some of the least-conserved portions of the protein are functionally essential (Khazina and Weichenrieder 2018), there could also be such regions in ORF2p, which might not be detectable purely through sequence analysis, but only using functional analysis. Therefore, we not only mapped retrotransposition efficiency onto the crystallized endonuclease domain of L1 ORF2p (Figure S4), but also correlated retrotransposition efficiency with sequence conservation all along the ORF2p sequence. To this aim, we aligned the human ORF2 protein sequence to 14 diverse mammalian sequences as well as others from more distant vertebrates (Figure S5 and Table S7 and Table S8). As expected, we found highly conserved sequences to be important for retrotransposition. Importantly, however, we also identified clusters of functionally crucial residues in the less-conserved regions.
Until now, conservation of functional residues summarized as short amino acid-sequence motifs has been integral to identifying regions of ORF2p indispensable for L1 activity. However, there are “desert” regions (D1 and D2) in ORF2p that have no structural motifs and no clear conservation. Our unbiased scanning approach helps us reach beyond the most-studied regions of ORF2p and creates a framework for prioritizing functional regions for further study. Figure 7 summarizes both the conservation and retrotransposition frequencies of each ORF2p mutant. A few previously noted amino acid sequence motifs were confirmed to be essential by this analysis, such as the Cry motif in the D1 region and the Cys-rich motifs in the D2 region. However, there were also regions that lacked amino acid-sequence conservation but showed profound retrotransposition defects. We denoted these positions with stars in Figure 7. This analysis revealed a “star cluster” contained in the window of residues F952–C1020. This is a previously mechanistically uninvestigated region with a high density of amino acid sequences of this type. A larger region containing the star cluster has previously been shown to have species specificity: the substitution of the human L1 sequence with the corresponding mouse L1 sequence fully depleted activity (Wagstaff et al. 2011). This region is of special interest for further characterization.
Figure 7.
Trends in amino acid conservation and sensitivity to mutation across ORF2p. (A) The schematic for the ORF2p domains is along the top. This is a graphical representation and interpretation of the conservation data displayed in Table S8 (mammals and all vertebrates; the “mammals alone” column is excluded). For each trialanine mutant, we took the value corresponding to the residue with the highest conservation to represent the mutant. As shown in the box, conservation and retroT are color-coded. The boxes are stacked to compare conservation and activity. Two dots and one black dot above a mutant mean that, as expected, there was strong conservation (100% and 70–99%, respectively) and no retroT. Stars above the mutant mean that there is low conservation (0–29%) and no retroT, highlighting areas that may be important in ORF2p not predicted by conservation alone. The star cluster region is indicated with a light blue bar, which is shown (B) zoomed in and in detail with the three WT amino acids (in single letter format) corresponding to each mutant. Cry, Cryptic motif; EN, endonuclease; retroT, retrotransposition; RT, reverse transcriptase; WT, wild-type; Z, Z-domain region.
We report here the most comprehensive ordered and arrayed amino acid substitution library for any retrotransposon, DNA transposon, or retrovirus. We anticipate that this resource will be of substantial interest to students of these elements and may serve as a model for future libraries of this type.
Acknowledgments
We thank Elena Khazina and Oliver Weichenrieder for the structural coordinates of their composite L1ORF1p model, and for sharing information before publication; Zoltán Ivics for support of M.T.O. during his visit to our laboratory; and Kathleen Burns (reader), David Graham, Jeremy Nathans, and Roger Reeves for input throughout the project and for serving on the PhD thesis committee of E.M.A. This work was supported in part by National Institutes of Health grants P50 GM-107632 to J.D.B., and P01 AG051449 to John Sedivy and J.D.B.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.9978590.
Communicating editor: P. Geyer
Literature Cited
- Ade C. M., Derbes R. S., Wagstaff B. J., Linker S. B., White T. B. et al. , 2018. Evaluating different DNA binding domains to modulate L1 ORF2p-driven site-specific retrotransposition events in human cells. Gene 642: 188–198. 10.1016/j.gene.2017.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adney, E. M., 2018 Comprehensive scanning mutagenesis of a human retrotransposon identifies motifs essential for function. Ph.D. Thesis, Johns Hopkins University School of Medicine, Baltimore. [Google Scholar]
- Alisch R. S., Garcia-Perez J. L., Muotri A. R., Gage F. H., and Moran J. V., 2006. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 20: 210–224. 10.1101/gad.1380406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W., Han J. S., Wheelan S. J., Davis E. S., Coombes C. E. et al. , 2006. Active retrotransposition by a synthetic L1 element in mice. Proc. Natl. Acad. Sci. USA 103: 18662–18667. 10.1073/pnas.0605300103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardeljan D., Taylor M. S., Ting D. T., and Burns K. H., 2017. Th human long interspersed element-1 retrotransposon: an emerging biomarker of neoplasia. Clin. Chem. 63: 816–822. 10.1373/clinchem.2016.257444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjan-Odedra S., Swanson C. M., Sherer N. M., Wolinsky S. M., and Malim M. H., 2012. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology 9: 53 10.1186/1742-4690-9-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attig J., and Ule J., 2019. Genomic accumulation of retrotransposons was facilitated by repressive RNA-binding proteins: a hypothesis. Bioessays 41: e1800132 10.1002/bies.201800132 [DOI] [PubMed] [Google Scholar]
- Attig J., Agostini F., Gooding C., Chakrabarti A. M., Singh A. et al. , 2018. Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell 174: 1067–1081.e17. 10.1016/j.cell.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauregard A., Curcio M. J., and Belfort M., 2008. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 42: 587–617. 10.1146/annurev.genet.42.110807.091549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C. R., Collier P., Macfarlane C., Malig M., Kidd J. M. et al. , 2010. LINE-1 retrotransposition activity in human genomes. Cell 141: 1159–1170. 10.1016/j.cell.2010.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., 1997. LINEs and Alus--the polyA connection. Nat. Genet. 16: 6–7. 10.1038/ng0597-6 [DOI] [PubMed] [Google Scholar]
- Boissinot S., and Sookdeo A., 2016. The evolution of LINE-1 in vertebrates. Genome Biol. Evol. 8: 3485–3507. 10.1093/gbe/evw247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B., Schustak J., Badge R. M., Lutz-Prigge S., Farley A. H. et al. , 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 100: 5280–5285. 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns K. H., 2017. Transposable elements in cancer. Nat. Rev. Cancer 17: 415–424. 10.1038/nrc.2017.35 [DOI] [PubMed] [Google Scholar]
- Carreira P. E., Richardson S. R., and Faulkner G. J., 2014. L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J. 281: 63–73. 10.1111/febs.12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian C. M., Deharo D., Kines K. J., Sokolowski M., and Belancio V. P., 2016. Identification of L1 ORF2p sequence important to retrotransposition using Bipartile Alu retrotransposition (BAR). Nucleic Acids Res. 44: 4818–4834. 10.1093/nar/gkw277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements A. P., and Singer M. F., 1998. The human LINE-1 reverse transcriptase: effect of deletions outside the common reverse transcriptase domain. Nucleic Acids Res. 26: 3528–3535. 10.1093/nar/26.15.3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost G. J., Feng Q., Jacquier A., and Boeke J. D., 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21: 5899–5910. 10.1093/emboj/cdf592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Taylor M. S., O’Donnell K. A., and Boeke J. D., 2012. Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation. Mol. Cell. Biol. 32: 4323–4336. 10.1128/MCB.06785-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty M. D., and Malik H. S., 2012. Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 46: 677–700. 10.1146/annurev-genet-110711-155522 [DOI] [PubMed] [Google Scholar]
- De Cecco, M., T. Ito, A. P. Petrashen, A. E. Elias, N. J. Skvir et al., 2019 L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566: 73–78 (erratum: Nature 572: E5). 10.1038/s41586-018-0784-9 [DOI] [PMC free article] [PubMed]
- Doucet A. J., Hulme A. E., Sahinovic E., Kulpa D. A., Moldovan J. B. et al. , 2010. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 6: e1001150. 10.1371/journal.pgen.1001150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet A. J., Wilusz J. E., Miyoshi T., Liu Y., and Moran J. V., 2015. A 3′ poly(A) tract is required for LINE-1 retrotransposition. Mol. Cell 60: 728–741. 10.1016/j.molcel.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-O’Hare T. T., Rodić N., Sharma R., Darbari I., Abril G. et al. , 2015. LINE-1 expression and retrotransposition in Barrett’s esophagus and esophageal carcinoma. Proc. Natl. Acad. Sci. USA 112: E4894–E4900. 10.1073/pnas.1502474112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T., and Singer M., 1987. The line-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res. 15: 2251–2260. 10.1093/nar/15.5.2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G. J., and Garcia-Perez J. L., 2017. L1 mosaicism in mammals: extent, effects, and evolution. Trends Genet. 33: 802–816. 10.1016/j.tig.2017.07.004 [DOI] [PubMed] [Google Scholar]
- Feng Q., Moran J. V., Kazazian H. H. Jr., and Boeke J. D., 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87: 905–916. 10.1016/S0092-8674(00)81997-2 [DOI] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. III et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gilbert N., Lutz-Prigge S., Moran J. V., 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell. 110: 315–25. 10.1016/s0092-8674(02)00828-0 [DOI] [PubMed] [Google Scholar]
- Gilbert N., Lutz S., Morrish T. A., and Moran J. V., 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25: 7780–95. 10.1128/MCB.25.17.7780-7795.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J. L., Ostertag E. M., Engleka K. A., Seleme M. C., and Kazazian H. H., 2004. A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 13: 1041–1048. 10.1093/hmg/ddh118 [DOI] [PubMed] [Google Scholar]
- Goodier J. L., Cheung L. E., and Kazazian H. H. Jr, 2012. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 8: e1002941 10.1371/journal.pgen.1002941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J. L., Zhang L., Vetter M. R., and Kazazian H. H., 2007. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. and Cell. Biol. 27: 6469–6483; 10.1128/MCB.00332-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J. L., Cheung L. E., and Kazazian H. H., 2013. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 41: 7401–7419. 10.1093/nar/gkt512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V., Boeke J. D., Helfand S. L., and Sedivy J. M., 2014. Sleeping dogs of the genome. Science 346: 1187–1188. 10.1126/science.aaa3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampf M., and Gossen M., 2007. Promoter crosstalk effects on gene expression. J. Mol. Biol. 365: 911–920. 10.1016/j.jmb.2006.10.009 [DOI] [PubMed] [Google Scholar]
- Hancks D. C., and Kazazian H. H. Jr, 2016. Roles for retrotransposon insertions in human disease. Mob. DNA 7: 9 10.1186/s13100-016-0065-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H., and Singer M. F., 1996. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15: 630–639. 10.1002/j.1460-2075.1996.tb00395.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. R., Burns K. H., and Boeke J. D., 2012. Active transposition in genomes. Annu. Rev. Genet. 46: 651–675. 10.1146/annurev-genet-110711-155616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszyk K., Li P. W., Villareal V., Branciforte D., Wu H. et al. , 2007. Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1. J. Biol. Chem. 282: 24893–24904. 10.1074/jbc.M702023200 [DOI] [PubMed] [Google Scholar]
- Kano H., Godoy I., Courtney C., Vetter M. R., Gerton G. L. et al. , 2009. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23: 1303–1312. 10.1101/gad.1803909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H. H., Jr, 2004. Mobile elements: drivers of genome evolution. Science 303: 1626–1632. 10.1126/science.1089670 [DOI] [PubMed] [Google Scholar]
- Khazina E., and Weichenrieder O., 2009. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc. Natl. Acad. Sci. USA 106: 731–736. 10.1073/pnas.0809964106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazina E., and Weichenrieder O., 2018. Human LINE-1 retrotransposition requires a metastable coiled coil and a positively charged N-terminus in L1ORF1p. Elife 7: e34960. 10.7554/eLife.34960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazina E., Truffault V., Büttner R., Schmidt S., Coles M. et al. , 2011. Trimeric structure and flexibility of the L1 ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 18: 1006–1014. 10.1038/nsmb.2097 [DOI] [PubMed] [Google Scholar]
- Kimberland M. L., Divoky V., Prchal J., Schwahn U., Berger W. et al. , 1999. Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 8: 1557–1560. 10.1093/hmg/8.8.1557 [DOI] [PubMed] [Google Scholar]
- Kolosha V. O., and Martin S. L., 1997. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl. Acad. Sci. USA 94: 10155–10160. 10.1073/pnas.94.19.10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa D. A., and Moran J. V., 2005. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retro-transposition. Hum. Mol. Genet. 14: 3237–3248. 10.1093/hmg/ddi354 [DOI] [PubMed] [Google Scholar]
- Kulpa D. A., and Moran J. V., 2006. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 13: 655–660. 10.1038/nsmb1107 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C. et al. , 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 (erratum: Nature 411: 720) (erratum: Nature 412: 565). doi: 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lee E., Iskow R., Yang L., Gokcumen O., Haseley P. et al. , 2012. Landscape of somatic retrotransposition in human cancers. Science 337: 967–971. 10.1126/science.1222077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Lee C. H., Swigut T., Grow E., Gu B. et al. , 2018. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553: 228–232. 10.1038/nature25179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D. D., Korman M. H., Jakubczak J. L., and Eickbush T. H., 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605. 10.1016/0092-8674(93)90078-5 [DOI] [PubMed] [Google Scholar]
- Martin S. L., 1991. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 11: 4804–4807. 10.1128/MCB.11.9.4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L., and Bushman F. D., 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21: 467–475. 10.1128/MCB.21.2.467-475.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias S. L., Scott A. F., Kazazian H. H., Boeke J. D., and Gabriel A., 1991. Reverse transcriptase encoded by a human transposable element. Science 254: 1808–1810. 10.1126/science.1722352 [DOI] [PubMed] [Google Scholar]
- Mita P., Wudzinska A., Sun X., Andrade J., Nayak S. et al. , 2018. LINE-1 protein localization and functional dynamics during the cell cycle. Elife 7: e30058. 10.7554/eLife.30058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri A. R., Chu V. T., Marchetto M. C., Deng W., Moran J. V. et al. , 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435: 903–910. 10.1038/nature03663 [DOI] [PubMed] [Google Scholar]
- Nguyen T. H. M., Carreira P. E., Sanchez-Luque F. J., Schauer S. N., Fagg A. C. et al. , 2018. L1 retrotransposon heterogeneity in ovarian tumor cell evolution. Cell Rep. 23: 3730–3740. 10.1016/j.celrep.2018.05.090 [DOI] [PubMed] [Google Scholar]
- Niewiadomska A. M., Tian C., Tan L., Wang T., Sarkis P. T. et al. , 2007. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 81: 9577–9583. 10.1128/JVI.02800-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K. A., An W., Schrum C. T., Wheelan S. J., and Boeke J. D., 2013. Controlled insertional mutagenesis using a LINE-1 (ORFeus)gene-trap mouse model. Proc. Natl. Acad. Sci. USA 110: E2706–E2713. 10.1073/pnas.1302504110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag E. M., and Kazazian H. H., 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35: 501–538. 10.1146/annurev.genet.35.102401.091032 [DOI] [PubMed] [Google Scholar]
- Ostertag E. M., Prak E. T., DeBerardinis R. J., Moran J. V., and Kazazian H. H. Jr, 2000. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28: 1418–1423. 10.1093/nar/28.6.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag E. M., DeBerardinis R. J., Goodier J. L., Zhang Y., Yang N. et al. , 2002. A mouse model of human L1 retrotransposition. Nat. Genet. 32: 655–660. 10.1038/ng1022 [DOI] [PubMed] [Google Scholar]
- Peddigari S., Li P. W., Rabe J. L., and Martin S. L., 2013. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res. 41: 575–585. 10.1093/nar/gks1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M., Lin C. J., Yin Y., Guan J., Peixoto G. A. et al. , 2018. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174: 391–405.e19. 10.1016/j.cell.2018.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskareva O., Ernst C., Higgins N., and Schmatchenko V., 2013. The carboxy-terminal segment of the human LINE-1 ORF2 protein is involved in RNA binding. FEBS Open Bio 3: 433–437. 10.1016/j.fob.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro J. G., and Cristofari G., 2016. Post-transcriptional control of LINE-1 retrotransposition by cellular host factors in somatic cells. Front. Cell Dev. Biol. 4: 14 10.3389/fcell.2016.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S. M., Wheelan S. J., Yarrington R. M., and Boeke J. D., 2006. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 16: 550–556. 10.1101/gr.4431306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N., Sharma R., Sharma R., Zampella J., Dai L. et al. , 2014. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 184: 1280–1286. 10.1016/j.ajpath.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J., Yamaguchi K., Kajikawa M., Ichiyanagi K., Adachi N. et al. , 2009. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 5: e1000461 10.1371/journal.pgen.1000461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer D. E., Connelly C., Szak S. T., Caputo E. M., Cost GJ G. J., et al. , 2002. Human L1 retrotransposition is associated with genetic instability in vivo. Cell. 110: 327–38. 10.1016/s0092-8674(02)00839-5 [DOI] [PubMed] [Google Scholar]
- Szak, S. T., O. K. Pickeral, W. Makalowski, M. S. Boguski, D. Landsman et al., 2002 Molecular archeology of L1 insertions in the human genome. Genome Biol. 3: research0052. 10.1186/gb-2002-3-10-research0052 [DOI] [PMC free article] [PubMed]
- Taylor M. S., LaCava J., Mita P., Molloy K. R., Huang C. R. et al. , 2013. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 155: 1034–1048. 10.1016/j.cell.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. S., Altukhov I., Molloy K. R., Mita P., Jiang H. et al. , 2018. Dissection of affinity captured LINE-1 macromolecular complexes. Elife 7: e30094. 10.7554/eLife.30094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M., Kashyap M., Rezazadeh S., Geneva A. J., Morello T. D. et al. , 2014. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 5: 5011 10.1038/ncomms6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff B. J., Barnerssoi M., and Roy-Engel A. M., 2011. Evolutionary conservation of the functional modularity of primate and murine LINE-1 elements. PLoS One 6: e19672 10.1371/journal.pone.0019672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Gilbert N., Ooi S. L., Lawler J. F., Ostertag E. M. et al. , 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21: 1429–1439. 10.1128/MCB.21.4.1429-1439.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O., Repanas K., and Perrakis A., 2004. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure 12: 975–986. 10.1016/j.str.2004.04.011 [DOI] [PubMed] [Google Scholar]
- Wolf Y. I., and Koonin E. V., 2013. Genome reduction as the dominant mode of evolution. Bioessays 35: 829–837. 10.1002/bies.201300037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains are available upon request. Supplemental material available at figshare: https://doi.org/10.25386/genetics.9978590.