Abstract
Lineage specification in early development is the basis for the exquisitely precise body plan of multicellular organisms. It is therefore critical to understand cell fate decisions in early development. Moreover, for regenerative medicine, the accurate specification of cell types to replace damaged/diseased tissue is strongly dependent on identifying determinants of cell identity. Long noncoding RNAs (lncRNAs) have been shown to regulate cellular plasticity, including pluripotency establishment and maintenance, differentiation and development, yet broad phenotypic analysis and the mechanistic basis of their function remains lacking. As components of molecular condensates, lncRNAs interact with almost all classes of cellular biomolecules, including proteins, DNA, mRNAs, and microRNAs. With functions ranging from controlling alternative splicing of mRNAs, to providing scaffolding upon which chromatin modifiers are assembled, it is clear that at least a subset of lncRNAs are far from the transcriptional noise they were once deemed. This review highlights the diversity of lncRNA interactions in the context of cell fate specification, and provides examples of each type of interaction in relevant developmental contexts. Also highlighted are experimental and computational approaches to study lncRNAs.
Keywords: long noncoding RNAs, miRNAs, competing endogenous RNAs, k-mers, cell fate specification
LINEAGE specification decisions in early development provide a blueprint of the body plan in multicellular organisms. Model systems such as embryonic stem (ES) cells are often employed in the study of early cell fate decisions. Understanding cell fate is also critical for regenerative medicine, as cell-based approaches pose significant therapeutic promise. Toward this end, induced pluripotent stem (iPS) cells, which display characteristics of ES cells, and can be patient-derived, have the potential to be differentiated into a myriad of different cell types.
Understanding determinants of cell fate is critical both for understanding early development, and to guide lineage commitment of pluripotent stem cells to enable the replacement of diseased cell types in patients. While central transcriptional regulators of pluripotency including OCT4, SOX2, and NANOG, which maintain the pluripotent state, and specification factors such as SOX1, MEOX1, and SOX17 (Kan et al. 2004; Shimoda et al. 2007; Wang et al. 2013) are relatively well understood, many key cell-fate determinants remain functionally undefined. Importantly, recent developments in transcriptomics have demonstrated that, although the majority of the mammalian genome is transcribed, protein coding sequences amount to <2% of transcribed genomic sequence (Dinger et al. 2008; Alexander et al. 2010; Harrow et al. 2012), with the number of noncoding RNA (ncRNA) genes equaling, or possibly even outnumbering, protein-coding genes based on estimates from GENCODE and FANTOM (Hon et al. 2017; Frankish et al. 2019). Already, certain noncoding transcripts, including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs; designated as transcripts >200 nt) have been implicated in cell fate decisions for unspecialized cells, including pluripotent stem cells; however, the vast majority of ncRNAs remain understudied.
Using pluripotent cells and their derivatives for illustrations, this review centers on lncRNAs, with a focus on the multimodal interactions through which they regulate cell fate specification. These interactions identify lncRNAs as important factors in early developmental processes and suggest that they should be considered in the design of regenerative medicine strategies.
LncRNAs: Interactions as Functional Determinants
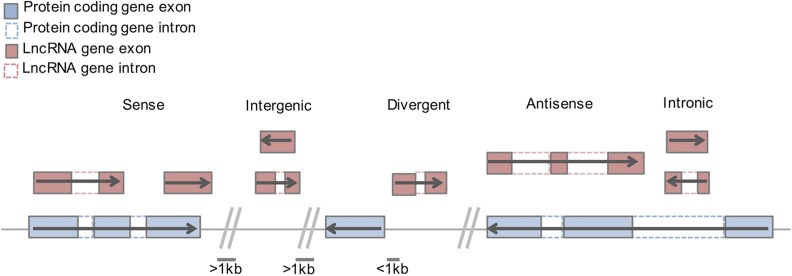
The presence and number of lncRNAs appear to correlate with organismal complexity, and their expression patterns show subcellular, cellular, and tissue specificity, which suggests context-dependent roles, particularly in the determination of cell fate (Mattick 2001). Different classes of lncRNAs are defined based on transcription direction and location (for example: sense, bidirectional, antisense, intronic, intergenic; Figure 1) relative to other genes (Mattick and Rinn 2015). While this location classification might be suggestive of mechanism, lncRNA genomic location does not always strictly dictate function (Mattick and Rinn 2015; Quinn and Chang 2016). On the other hand, intermolecular interactions with other RNAs, proteins and chromatin have revealed emerging functional themes, and demonstrated the far-reaching regulatory potential of lncRNAs. Here, we examine the implication of these interactions in cell fate determination and early developmental processes.
Figure 1.
General principles illustrating lncRNA subtypes and genomic origin. LncRNAs may originate from various regions in the genome, including proximal, distal, and overlapping, with respect to protein coding genes. Sense and antisense lncRNAs may, or may not, fully overlap with protein coding genes. Divergent and intergenic lncRNAs are arbitrarily distinguished based on distance from the nearest protein coding gene.
Much of the function of lncRNAs depend on their ability to base pair to other RNAs or DNA through conventional or Hoogsteen base pairing, to form complex intramolecular and intermolecular secondary and higher order structures (Mercer and Mattick 2013). The structures formed by lncRNAs regulate and direct interaction with RNA-binding proteins (RBPs) to regulate, negatively or positively, their cellular targets. These proteins are central to lncRNA mechanisms of action and regulation of their function (Rinn and Ule 2014).
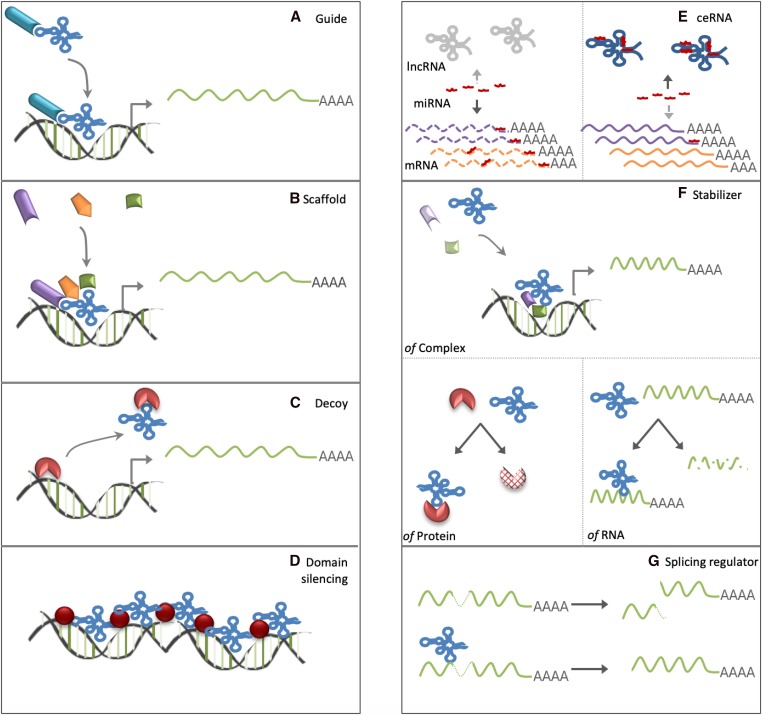
Localization of a lncRNA transcript can be suggestive of its functional role and contribution to gene regulation. While cytoplasmic lncRNAs tend to function post-transcriptionally, many nuclear lncRNAs regulate gene expression at the transcriptional level (Rinn and Chang 2012; Mercer and Mattick 2013). Within the nucleus, expression of a gene requires chromatin decompaction, particularly in heterochromatic regions. The compaction state is determined by chemical modifications of nucleosomal histone proteins, controlled by histone-modifying enzymes. LncRNAs have been shown to interact with chromatin modifiers (readers, writers, erasers) and remodelers to facilitate changes in the chromatin’s biochemical and accessibility landscape at specific gene loci through both cis- and trans-acting mechanisms (Figure 2) (Rinn and Chang 2012).
Figure 2.
Schematic illustration of the different modes of action for lncRNAs. Localization of lncRNAs to the nucleus or cytoplasm can dictate different mechanisms of action. Based on their ability to bind to DNA and interact with proteins, lncRNAs can guide transcription regulators and epigenetic modulators (A); act as scaffolds to assemble chromatin regulatory factors (B); titrate away regulators of transcription by acting as decoys (C); regulate domain- or chromosome-wide chromatin state to regulate transcriptional output (D); act as ceRNAs to capture regulatory factors such as miRNAs away from target genes (E); contribute to the stabilization of protein complexes, proteins, and mRNAs (F); and influence alternative splicing (G).
Additionally, lncRNAs have also been shown to have a number of post-transcriptional and cytoplasmic functions in many developmental processes. These include functioning in mRNA stability and translation regulation through protein, miRNA, and mRNA interactions (Figure 2) (Batista and Chang 2013; Yoon et al. 2013; Quinn and Chang 2016). These interactions provide a basis for lncRNA functional classification, but can also be targeted to direct cell fate.
Interactions with proteins: chromatin regulation
The plasticity of pluripotent stem cells is related to the high ratio of euchromatin to heterochromatin (Gaspar-Maia et al. 2011), making more chromatin accessible to transcription factors, RNA polymerase, and other proteins necessary for transcription. Pluripotent stem cells also have a high proportion of poised chromatin (Fisher and Fisher 2011), which facilitates the rapid gene derepression required for lineage commitment. Extensive binding of lncRNAs to epigenetic regulators that control chromatin accessibility defines one category of lncRNA function (Mercer and Mattick 2013). Studies using ES cells and other cell types have shown that ∼30% of intergenic lncRNAs were bound by at least one epigenetic regulator (Khalil et al. 2009), indicating widespread impact of lncRNAs on cell identity at the transcriptional level.
LncRNAs can act in cis by binding to neighboring genes and facilitating recruitment of chromatin modifier/remodelers to the target locus (Bassett et al. 2014). The act of lncRNA transcription can also have a cis-regulatory function in gene expression, and influence genome organization (Bassett et al. 2014; Engreitz et al. 2016; Melé and Rinn 2016). LncRNAs can function in trans as well, either by serving as a recruitment or scaffolding factor on which chromatin modifying proteins assemble, or by modulating the stability of the chromatin regulatory protein complex (Rinn and Chang 2012; Bassett et al. 2014).
Cis-regulatory lncRNAs control expression of neighboring genes. In the context of cell fate specification, these lncRNA genes are often located adjacent to key developmental regulators that determine cell fate and organismal development (Bassett et al. 2014; Engreitz et al. 2016; Melé and Rinn 2016). This mechanism is commonly used for antisense and divergent lncRNAs that are typically <5 kb from, and transcribed in the opposite direction relative to, a transcribed gene. For example, Evx1as and its neighboring protein-coding gene, Evx1, demonstrate highly correlated expression in murine ES cells (Luo et al. 2016). Depletion of Evx1as indicated unidirectional regulation of the protein-coding neighbor by the lncRNA where Evx1as bound to its own promoter and facilitated binding of Mediator to activate transcription at the locus (Luo et al. 2016). This example highlights how lncRNAs can act in cis, tethered to their promoter, to modify gene expression near their transcription site.
Another illustration of cis influence of a lncRNA is exemplified by Chaserr’s regulation of Chd2—a chromatin remodeler with roles in cell differentiation in mice (Rom et al. 2019). Chaserr’s transcript is produced upstream of the transcription start site of Chd2, where it collaborates with CHD2 protein to repress Chd2’s expression in a negative feedback loop to maintain cellular levels of CHD2 (Rom et al. 2019).
yylncT—a member of the divergent subclass of lncRNAs known as yin yang (yy) lncRNAs—supports expression of its gene neighbor, Brachyury (T) by localizing to its locus during mesoderm commitment in human ES cells (Frank et al. 2019). yylncRNAs are primarily encoded from genomic loci of key cell-fate regulators, thus mirroring their developmental expression patterns, and, as a class, they illustrate a broad mechanism through which lncRNAs safeguard cell-fate decisions (Frank et al. 2019).
A handful of cis-acting lncRNAs are also known to repress gene expression over long genomic distances. In the most extreme example, the lncRNA Xist silences gene expression over the entire 165 million base pair X chromosome early during the development of female mammals, as part of the dosage compensation process called X-chromosome Inactivation (XCI) (recently reviewed by Sahakyan et al. 2018).
Xist induces stable gene silencing through two parallel pathways. In the first, Xist silences actively transcribed genes through an incompletely defined mechanism that involves the protein SPEN and the RNA element Repeat A, which is a tandem repeat located at the 5′ end of Xist. In parallel, and subsequent to Repeat-A-mediated silencing, Xist induces the spread of Polycomb Repressive Complexes (PRCs) over transcriptionally inactive chromatin (Nesterova et al. 2019; Żylicz et al. 2019). In a mechanistic sense, this spread of PRCs over the X is likely a major means by which Xist orchestrates stable silencing that is inherited through subsequent cell divisions (Wang et al. 2001; Kalantry et al. 2006; Sahakyan et al. 2018). Other cis-repressive lncRNAs that depend on PRCs for their silencing functions, such as Kcnq1ot1, Airn, Morrbid, and Haunt, may utilize similar mechanisms to bring PRCs to chromatin (Regha et al. 2007; Terranova et al. 2008; Yin et al. 2015; Kotzin et al. 2016; Schertzer et al. 2019). Indeed, both Kcnq1ot1 and Airn were recently shown to require the Xist cofactor HNRNPK to induce the spread of PRCs in mouse trophoblast stem cells (Schertzer et al. 2019).
In addition, the PRCs, particularly PRC2, have been shown to interact with many RNAs, and the functional consequence of this interaction has not always been clear. For example, the PRC2 component SUZ12 has been shown to interact with a lncRNA to repress a differentiation-inducing transcriptional program in human ES cells. Here, the lncRNA tsRMST uses multiple mechanisms, including coregulation with SUZ12 and NANOG, to block expression of lineage specification genes and impede WNT5A-induced epithelial-mesenchymal transition (Yu and Kuo 2016). SUZ12 and the central pluripotency regulator SOX2 also interacts with lncRNA_ES1 and lncRNA_ES2 to contribute to pluripotency maintenance (Ng et al. 2012) through unclear mechanisms.
Other lncRNAs, at least partially through their interaction with PRC2, have been implicated in processes supporting lineage commitment. For example, the lncRNA Braveheart interacts with PRC2, and, perhaps in part due to a consequence of this interaction, Braveheart directs murine pluripotent cells to a cardiac fate by moderating a mesoderm and cardiac-specific transcription factor network (Klattenhoff et al. 2013). Nevertheless, through a specific structured element, Braveheart interacts with the CNBP/ZNF9 nucleic acid binding protein, and at least a portion of Braveheart function can be ascribed to the CNBP/ZNF9 interaction (Xue et al. 2016). Moreover, and surprisingly, even though depletion of Braveheart results in myogenic defects through its control of central cardiomyogenic regulators including Mesp1, Hand1, Nkx2.5, and Tbx20, and general loss of sarcomere gene expression, Braveheart null mice were grossly phenotypically normal (Han et al. 2018). Conversely, genetic ablation of the lncRNA Fendrr, which has also been shown to interact with PRC2, results in mouse embryonic lethality at around E13.75 due to myocardial defects (Grote et al. 2013). Here, it has been proposed that Fendrr’s interaction with both PRC2 and Trithorax Group/MLL complexes modulates chromatin signatures in control of lateral mesoderm differentiation (Grote et al. 2013). PRC2 function has also been reported to be regulated in trans, in mouse ES cells and human iPS cells, by the relatively abundant lncRNAs Rian, Mirg, and Meg3/Gtl2, which are produced from an imprinted cluster (Kaneko et al. 2014). Examples of these lncRNAs with clear roles for repressing transcription in cis also exist (Sanli et al. 2018). Additionally, many lncRNAs produced from the developmentally important Hox gene clusters also bind PRC2, the most notable of which may be the lncRNA HOTAIR. The extent to which lncRNA/PRC2 interactions in the Hox clusters contribute to gene regulation in mammalian development, whether the regulation occurs in cis or in trans, and what the mechanisms are, however, remain unclear (Li et al. 2013, 2016; Tsai et al. 2010; Amândio et al. 2016; Selleri et al. 2016; Portoso et al. 2017).
Importantly, both crosslinking immunoprecipitation (CLIP) studies of PRC2 as well as those that have studied the RNA-binding properties of PRC2 in vitro have found that PRC2 binds RNA with little sequence specificity and nanomolar affinity (Kaneko et al. 2013, 2014; Davidovich et al. 2015; Wang et al. 2017). Collectively, these studies suggest that one function of lncRNAs, and perhaps chromatin-bound RNAs in general, is to tether PRC2 to transcriptionally active regions of chromatin. This tethering may keep PRC2 in a poised state, in close proximity to future target genes, where it can initiate stable gene silencing upon receipt of the appropriate cues (Kaneko et al. 2013, 2014; Davidovich et al. 2015; Wang et al. 2017). High-affinity and nonspecific interactions with RNA may also govern PRC1 function in an analogous fashion (Bernstein et al. 2006; Bonasio et al. 2014).
LncRNAs have been shown to interact with a wide variety of chromatin modulatory factors. In addition to PRCs, these include histone methylases (Hendrickson et al. 2016). The H3K4 methylase MLL family is necessary for activating the expression of certain genes (Yang et al. 2014). WDR5 is a protein-subunit of MLL, recruiting the complex to target sites for activation (Yang et al. 2014). WDR5 engages with several lncRNAs that have been implicated in the self-renewal of ES cells (Yang et al. 2014). When the RNA-binding site of WDR5 was mutated in mouse ES cells, rendering it unable to bind lncRNAs, WDR5 was significantly less stable (Yang et al. 2014). This loss of stability resulted in a severe decrease in H3K4me3 marks on the promoters of pluripotency-related genes, and a loss of the ES cell state in 50% of colonies (Yang et al. 2014).
Interactions between lncRNAs and chromatin readers are exemplified by the interaction between DIGIT and BRD3 in the regulation of endoderm differentiation (Daneshvar et al. 2016, 2019). Here, DIGIT supports BRD3 recruitment to H3K18ac at regions in the genome enriched during endoderm differentiation in human ES cells (Daneshvar et al. 2019).
In addition to chromatin remodelers and modifiers, analysis of lncRNA interactomes has also identified transcription factors as key players in lncRNA function. Panct1—a nuclear-functioning sense lncRNA transcribed from an intron of the gene coding for its protein interacting partner, TOBF1—exemplifies this type of trans interaction in mouse ES cells (Chakraborty et al. 2017). Panct1 was shown to facilitate the binding of TOBF1 to pluripotency marker promoters by way of sequence-directed binding to Oct-Sox motifs, thus recruiting transcription factors such as Oct4 to promote target expression (Chakraborty et al. 2017). This interaction illustrates the role of a lncRNA in efficient binding of pluripotency-associated transcription factors to their target sites without direct interaction (Chakraborty et al. 2017).
The aforementioned examples demonstrate the breadth of lncRNA–protein interactions that influence transcription in developmental processes, and on which cell specification is at least partly dependent. Many of the described lncRNA interactions involve key transcription factors and epigenetic regulators that affect developmental progression. Modulation of specific lncRNAs could therefore be a viable avenue for specific regulation of target expression in developmental contexts.
Interactions with proteins: stability and sequestration
LncRNAs enhance or repress protein function through a variety of mechanisms including sequestration, binding support, and degradation, as in the aforementioned case of WDR5 and its reliance on lncRNAs for stability (Yang et al. 2014). Conversely, in other contexts, lncRNAs have been shown to be dependent on their protein partners for stability to carry out their functions, and regulate their half-life. This is the case for lncR492—a noncoding transcript that inhibits neural differentiation in mouse ES cells (Winzi et al. 2018). Knockdown of lncR492 resulted in increased expression of neural markers such as Pax6 and Nestin during differentiation. Proteomic analysis indicated that lncR492 directly interacts with HuR—a mRNA binding protein with functions in the (de)stabilization of mRNA transcripts. Overexpression and knockdown of HuR moderated the expression of lncR492, increasing and reducing the prevalence of the lncRNA, respectively—a pattern that suggests HuR supports the stability of the lncR492 transcript. Finally, both lncR492 and HuR positively influence WNT signaling, which has a known inhibitory effect on neural differentiation (Haegele et al. 2003), outlining the axis by which lncR492 and HUR function (Winzi et al. 2018).
LncRNAs can impede protein function through binding and sequestration. In the context of pluripotency, the chromatin mark H3K56 acetylation activates core pluripotency-related genes and is required for the maintenance of the undifferentiated ES cell state. SIRT6 is a chromatin-binding protein that removes this chromatin modification and functionally represses pluripotency-related genes to promote exit from the stem cell state (Etchegaray et al. 2015). LncPRESS1 functions as a molecular decoy for SIRT6, sequestering the protein, which binds to the 3′-end of the lncRNA in human ES cells. This interaction in the nucleus prevents SIRT6 from binding promoters of pluripotency-related genes and repressing transcription via deacetylation (Jain et al. 2016). Another regulatory interaction in this network is illustrated by P53, which antagonizes lncPRESS1, freeing SIRT6 to further repress pluripotency markers in human ES cells (Jain et al. 2016).
Emerging data indicate that regulatory lncRNA–protein interactions occur in specialized microenvironments that display characteristics of phase-separated particles (Hnisz et al. 2017; Daneshvar et al. 2019). These liquid-like condensates are able to exchange molecules dynamically with their surroundings (Bergeron-Sandoval et al. 2016; Boeynaems et al. 2018; Lu et al. 2018). The preceding interactions demonstrate the role lncRNA-protein interactions play in integral processes throughout differentiation and development. Such interactions can be investigated as points of manipulation for control of differentiation processes, especially when they localize to distinct microdroplets, and when the mechanisms for the interactions have been clearly defined.
Interactions with RNA
Similar to the prevalence of lncRNA–protein interactions, lncRNA–RNA interactions are prolific and have widespread effects on cell identity. Interestingly, lncRNAs interplay with multiple other RNA types, from mRNA to other ncRNA, including miRNAs and circular RNAs. Through different mechanisms, these lncRNA-RNA interactions can affect lncRNA function through repressing or supporting downstream targets.
Canonical functions of mRNAs are determined by their availability and potential to be translated. In addition to long-characterized protein factors, the half-life of a mRNA is determined by various co- and post-transcriptional regulatory factors, including lncRNAs. LncRNA interaction with mRNAs or mRNA-regulatory factors can stabilize or facilitate the degradation of the mRNA molecules, increasing or decreasing translational output (Faghihi et al. 2008, 2010; Gong and Maquat 2011). For example, Sirt1-AS interacts with Sirt1 mRNA to promote its stability, thereby inhibiting myogenic differentiation in favor of myoblast proliferation in mice (Wang et al. 2016).
LncRNAs have also been shown to alter the translation output of mRNAs by affecting alternative splicing. A prime example of this mechanism of action is the interaction of Zeb2-NAT—an antisense lncRNA—with its sense transcript, protein-coding Zeb2. Zeb2-NAT was demonstrated to bind Zeb2’s 5′ UTR, which contains an intron where the internal ribosome entry site for the ZEB2 protein resides (Beltran et al. 2008). Without protection of the first intron by lncRNA binding, ZEB2 protein levels are significantly diminished. In mice, Zeb2-NAT and ZEB2 prevent fibroblasts from being effectively reprogrammed to the pluripotent state, possibly by supporting senescence due to E-cadherin downregulation (Beltran et al. 2008; Bernardes de Jesus et al. 2018). Conversely, under conditions of decreased Zeb2-NAT expression, the mouse fibroblasts readily transitioned to ES cell-like cells in media conditions that support the pluripotent state (Bernardes de Jesus et al. 2018). Further, mouse ES cells in Zeb2-NAT knockdown conditions were able to maintain the pluripotent state in differentiation-inducing contexts (Bernardes de Jesus et al. 2018).
Perhaps even more impactful, based on their numerous targets, is the influence of lncRNAs on miRNAs. These small ncRNAs (∼22 nt long) are key post-transcriptional regulatory factors that influence target transcript translational repression and/or degradation (Heinrich and Dimmeler 2012). miRNA function has been implicated in the establishment and maintenance of ES cell pluripotency and differentiation (Heinrich and Dimmeler 2012). Generally, miRNAs and associated proteins assemble to form the RNA-induced silencing complex (RISC), in which the miRNA serves as a guide to target specific mRNAs, which are degraded in proportion to the degree of complementarity with the miRNA (Gregory et al. 2005). LncRNAs can affect the efficiency of these processes as well, since they can dictate the abundance of individual miRNAs by supporting miRNA stability or by causing their degradation, for example through template-mediated degradation (Fuchs Wightman et al. 2018). Additionally, lncRNAs can alter miRNA function by behaving as a sponge or competing endogenous RNA (ceRNA) that sequesters the miRNA, thus preventing degradation of the miRNA target genes to support or promote exit from the stem cell state (Liu et al. 2014, see Table 1).
Table 1. Paradigmatic modes of lncRNA interaction and functional output using ES cells, differentiation and developmental processes as models for cell fate specification.
| lncRNA | Mode of action | Interactor(s) | Result | Reference |
|---|---|---|---|---|
| Airn | Transcriptional | Igf2r | Silences Igf2r cluster to guide development | Santoro et al. (2013) |
| AK028326 | Transcriptional | Oct4 | Positively regulates Oct-4 to promote self-renewal | Sheik Mohamed et al. (2010) |
| Apela RNA | Transcriptional | p53, hnRNPL | Interacts with hnRNPL to repress p53-induced apoptosis | M. Li et al. (2015) |
| Braveheart | Transcriptional | PRC2 (Suz12), MesP1 | Regulates cardiac lineage commitment in ES cells | Klattenhoff et al. (2013) |
| Chaserr | Transcriptional | CHD2 | Influences cell differentiation through regulation of Chd2 | Rom et al. (2019) |
| Deanr1 | Transcriptional | SMAD2/3, FoxA2 | Recruits SMAD2/3 to the FOXA2 promoter to promote endoderm differentiation | Jiang et al. (2015) |
| DIGIT | Transcriptional | GSC, BRD3 | Supports endoderm differentiation | Daneshvar et al. (2016) |
| Evx1as | Transcriptional | Evx1 | Promotes Evx1 expression to regulate mesendodermal differentiation | Luo et al. (2016) |
| Fendrr | Transcriptional | PRC2, Trithorax group | Targets promoters for proper heart and body wall formation | Grote et al. (2013) |
| FIRRE | Transcriptional | CTCF | Preservation of silencing of inactive X chromosome | Yang et al. (2015) |
| Haunt | Transcriptional | HOXA | Inhibits HOXA expression and ES cell differentiation, whereas the Haunt locus is an enhancer for HOXA | Yin et al. (2015) |
| HERVH | Transcriptional | Oct4 | Recruits Oct4 to maintain pluripotency | Lu et al. (2014) |
| HOTTIP | Transcriptional | WDR5 | Binds WDR5 to activate developmental regulators | Wang et al. (2011) |
| LncPress1 | Transcriptional | SIRT6 | Binds SIRT6 to promote expression of pluripotency-related genes | Jain et al. (2016) |
| lncR492 | Transcriptional | HuR | Associates with HuR to promote pluripotency | Winzi et al. (2018) |
| lncRNA_ES1 | Transcriptional | SUZ12, SOX2 | Interacts with SUZ12 and SOX2 to prevent differentiation | Ng et al. (2012) |
| lncRNA_ES2 | Transcriptional | SUZ12, SOX2 | Interacts with SUZ12 and SOX2 to prevent differentiation | Ng et al. (2012) |
| Meg3/Gtl2 | Transcriptional | PRC2 (JARID2) | Regulates recruitment of PCR2 to chromatin in iPS cells | Kaneko et al. (2014) |
| Panct1 | Transcriptional | TOBF1 | Regulates recruitment of TOBF1 to Oct-Sox Motifs to support the pluripotent state | Chakraborty et al. (2017) |
| pRNA | Transcriptional | TIP5, TTF1 | Interaction with TIP5, TTF1 contributes to heterochromatin formation required for differentiation | Savić et al. (2014) |
| RMST | Transcriptional | SOX2 | Associates with SOX2 to regulate neural differentiation | Ng et al. (2013) |
| tsRMST | Transcriptional | PRC2, NANOG, WNT | Associates with PRC2, NANOG, WNT to repress differentiation | Yu and Kuo (2016) |
| TUNA (megamind) | Transcriptional | NCL, PTBP1, hnRNP-K | Associates with specified RBPs to activate pluripotency genes and neural differentiation genes | Lin et al. (2014) |
| yyT | Transcriptional | Brachyury | Regulates Brachyury (T) in mesoderm specification | Frank et al. (2019) |
| AK048794 | ceRNA | miR-592 | Sponges miR-592 to support pluripotency | Zhou et al. (2016) |
| Cyrano | ceRNA | mir-7 | Inhibit mir-7 to support self-renewal | Smith et al. (2017) |
| H19 | ceRNA | let-7 microRNAs | Modulates let-7 to impede muscle differentiation | Kallen et al. (2013) |
| HPAT5 | ceRNA | let-7 microRNAs | Modulates let-7 to promote pluripotency | Durruthy-Durruthy et al. (2016) |
| linc-ROR | ceRNA | miR-145 | Inhibits miR-145 suppression of self-renewal genes | Wang et al. (2013) |
| lncRNA-1064 | ceRNA | miR-200c | Inhibits miR-200c to regulate neural differentiation | Weng et al. (2018) |
| MD1 | ceRNA | miR-133, miR-135 | Supports differentiation by inhibiting miR-133, miR-135 | Cesana et al. (2011) |
| T-UCstem1 | ceRNA | miR-9, PRC2 | Maintains self-renewal by modulating miR-9 and PRC2 | Fiorenzano et al. (2018) |
| HOTAIR | Scaffold | PRC2, LSD1 | Originally proposed to coordinate PRC2 and LSD1 complexes for proper embryonic development, although contested in the more recent literature | Tsai et al. (2010), Li et al. (2013), Amândio et al. (2016), Li et al. (2016), Selleri et al. (2016), Portoso et al. (2017) |
| XIST | Scaffold | X chromosome, PRC2 | Inactivates X chromosome for dosage control | (Brown et al. 1991) |
| Cyrano | Multimodal/Other | Stat3, signaling network | Supports ES cell maintenance | Smith et al. (2018) |
| Tug1 | Multimodal/Other | Lin28A, Fragile X mental retardation protein | Regulates various differentiation processes including osteogenesis, neuronal differentiation and spermatogenesis | Young et al. (2005), Guo et al. (2018), 1; He et al. (2018), 1; Lewandowski et al. (2019) |
| Zeb2-NAT | Other | Zeb2 | Facilitates Zeb2 processing to regulate EMT and pluripotency | Beltran et al. (2008); Bernardes de Jesus et al. (2018) |
Linc-RoR (Regulator of Reprogramming) exemplifies the ceRNA mechanism in the context of reprogramming and the maintenance of pluripotency. Deviation from precise linc-RoR levels results in the differentiation of human ES cells to mesoderm and/or endoderm if its levels are depleted, or the inability of cells to properly differentiate if linc-RoR levels are elevated. Linc-ROR was shown to be a ceRNA for miR-145-5p, suppressing the miRNA’s negative regulation of stem cell regulatory factors such as OCT4 and SOX2 (Loewer et al. 2010; Wang et al. 2013).
While linc-ROR guides reprogramming, lncRNA-1064 was shown to support neural differentiation (Weng et al. 2018). Knockdown of lncRNA-1064 led to a decrease in neural lineage markers and reduced neural differentiation of mouse ES cells in vitro and in vivo in a murine teratoma model (Weng et al. 2018). Mechanistic analysis revealed lncRNA-1064 contains multiple miRNA target sites, with the most favorable being for mir-200c (Weng et al. 2018). lncRNA-1064-mediated sequestration of miR-200c transcripts enables ZEB1/2 to reach their gene targets to signal for neural differentiation (Weng et al. 2018). This interaction is similar to that of lncRNA AK048794, which functions as a ceRNA with miR-592 (Zhou et al. 2016). In mouse ES cells, miR-592 was found to bind the 3′ UTR of FAM91A1, reducing its protein levels and those of pluripotency regulators Oct4, Sox2, and Nanog, although the downstream mechanism is less clear. Altogether, these studies describe instances of lncRNAs functioning as ceRNAs, sequestering miRNAs, and preventing degradation of miRNA targets to either support or undermine pluripotency. Often, lncRNAs acting as ceRNAs can be a part of a multi-component network, such as the case of AK048794 (Zhou et al. 2016).
Because of their prevalence and well-delineated mechanism, lncRNA control of miRNA levels and availability could be harnessed to modulate miRNA levels in therapeutic contexts, particularly for cases where a wholesale loss of the target miRNA would be phenotypically disadvantageous (Kleaveland et al. 2018). Effective ceRNA activity would require a minimum threshold level for the lncRNA. Therefore, one important consideration pertaining to ceRNA/sponge function may be the relative abundance of the lncRNA, miRNA, and mRNA target. This is because the number of miRNA and target molecules would typically outnumber that of a lncRNA (Palazzo and Lee 2015). Another key consideration is whether, and how, RNA structure changes in response to the binding of proteins, other RNAs, or even small molecule metabolites to regulate the activity of ceRNA, as this could influence access to specific sites/sequences on the RNA. This is an aspect of lncRNA regulation that has escaped sustained attention so far, but is likely to be an important aspect of ceRNA, and, more generally, lncRNA regulation.
Multimodal interactions
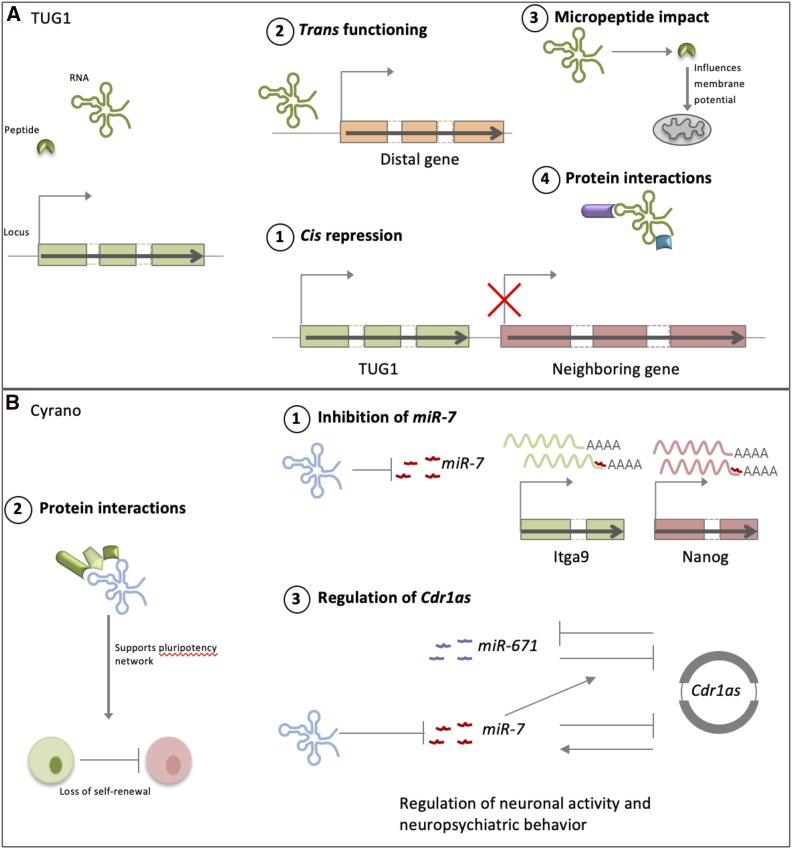
Since lncRNA functions can vary widely, it could be expected that their functions are multifaceted (Figure 3) and not necessarily dependent on a single mechanism even in a single cell type. Previously characterized as a lncRNA, Tug1, is produced from a highly conserved locus (Young et al. 2005), and is involved in many developmental processes including photoreceptor specification, axonal differentiation, and osteogenesis regulation, where some functions have been found to be mediated by protein interactions with key cell-fate regulators such as LIN28 (Young et al. 2005; Guo et al. 2018; He et al. 2018). While knockout mice are viable, they display sterility with complete penetrance due to defects in spermatogenesis (Lewandowski et al. 2019). Intriguingly, in addition to having two distinct noncoding functions, one of Tug1’s functions is dependent on an encoded micropeptide. Specifically, the Tug1 locus represses downstream genes in a cis-manner, while Tug1 RNA itself regulates genes that then are dysregulated upon knockout. Additionally, the 5′ conserved region of the Tug1 gene encodes a peptide, TUG1-BOAT that influences mitochondrial membrane potential (Lewandowski et al. 2019).
Figure 3.
By engaging in diverse interaction patterns, a single lncRNA can impact multiple cellular processes. LncRNAs harness different mechanisms and access multiple networks of interacting partners in a context-specific manner. (A)TUG1 can (1) regulate neighboring genes in cis, (2) function in trans to regulate target genes, (3) be translated into a micropeptide that regulates mitochondrial membrane potential, and (4) interact with proteins such as Lin28A to regulate various cell fate decisions. (B) Cyrano’s multifaceted functions are illustrated by (1) its ability to inhibit miR-7-mediated repression of Itga9 and Nanog, (2) interact with proteins to support maintenance of the self-renewing pluripotent state, and (3) function in a multi-RNA regulatory network to impact Cdr1as expression, and, ultimately, neuronal activity and neuropsychiatric behavior in mice.
The lncRNA Cyrano (OIP5-AS1, 1700020I14Rik, linc-oip5, Oip5os1) is another illustration of multimodal function. While lncRNAs generally display limited sequence conservation even between closely related species, a 300 nt region shows very high conservation in tetrapods, with ∼100 nucleotides conserved between zebrafish and humans (Ulitsky et al. 2011). Uniquely, there is a sequence stretch that has nearly perfectly complementarity to miR-7 in all tetrapods examined, which regulates miR-7 degradation (Kleaveland et al. 2018). The conserved sequence folds into a conserved secondary structure within which embeds the mir-7 binding sequence and partially masks it by base pairing (G. Varani unpublished results). How the RNA structure affects protein recruitment and degradation remains unclear, however, and it might be that the structure itself is incidental to miR-7 degradation. Still, it provides tantalizing suggestions that secondary and higher order structure might be essential components of lncRNA regulatory activities.
Cyrano shows rare maternal and zygotic expression during early development (Karlic et al. 2017), and in various mammalian cells it is a proliferation regulator (Smith et al. 2017; Deng et al. 2018; X. Liu et al. 2018; Naemura et al. 2018). Using proteomic analyses, Cyrano was found to interact with a developmental/signaling protein network, through which it partially supports mouse ES cell characteristics (Smith et al. 2018). Further, Cyrano inhibition of mir-7, which targets the pluripotency regulator Nanog, as well as Itga9, contributes to regulation of stemness and cellular adhesion (Smith et al. 2017).
Cyrano has also been shown to exist in a multi-component RNA network with the circular RNA Cdr1as and several miRNAs, where it maintains appropriate Cdr1as levels in the brain to control neuronal activity and neuropsychiatric behavior in mice (Piwecka et al. 2017; Kleaveland et al. 2018). However, despite displaying strong expression and significant molecular and cellular phenotypes, Cyrano is an example of a lncRNA that does not show an overt developmental phenotype with differing knockout strategies (Han et al. 2018; Kleaveland et al. 2018), which raises the question of whether there could be compensatory mechanisms for its function.
LncRNA Properties that Underlie Their Intermolecular Interactions
Sequence
A challenge in the lncRNA field is identifying the function of a lncRNA based on analysis of its sequence content alone, which is most easily accessible. This challenge stems from the fact that most proteins interact with RNA through sequence motifs that are degenerate and have hidden structural preferences (Dominguez et al. 2018). Compounding the difficulty is that the order of protein-binding modules within a lncRNA is likely to be less important than the mere presence of the binding modules themselves. Thus, two lncRNAs may encode identical functions through different sequence solutions.
An additional obstacle lies in the fact that existing sequence alignment algorithms, which, in large part, have been designed to detect linear sequence relationships between evolutionarily related nucleic acid or protein species, often fail to detect significant homology between lncRNAs (Altschul et al. 1990; Rice et al. 2000; Edgar and Batzoglou 2006; Wheeler and Eddy 2013). In order to address this problem, Kirk and colleagues recently developed a method called SEEKR (Sequence evaluation through k-mer representation) to quantify nonlinear sequence similarity between lncRNAs (Kirk et al. 2018). Rather than evaluating similarity between lncRNAs based on the extent of linear sequence homology, SEEKR functions by counting the abundance of all possible combinations of sequence substrings at a given length, k, within a lncRNA, and then scaling these abundances by the extent to which they differ from the mean abundance of each k-mer in the group of lncRNAs being analyzed. The extent of nonlinear similarity between lncRNAs as defined by SEEKR was found to correlate significantly with lncRNA subcellular localization and with protein binding, although the ability to predict either of these two properties from k-mer content alone was minimal. Using a transgenic assay to monitor the ability of a lncRNA to induce Xist-like repression, it was found that k-mer content, but not linear sequence homology, strongly correlated with the ability of lncRNAs to induce this type of repression. In a subsequent study, Sprague and colleagues found substantial levels of nonlinear sequence similarity between functional domains in Xist, and domains in the lncRNA Rsx, a marsupial lncRNA that has been proposed to be a functional analog of Xist that arose through convergent evolution (Grant et al. 2012; Sprague et al. 2019). Collectively, these data support the notions that different lncRNAs can encode similar function through different spatial arrangements of related, but not necessarily identical, sequence motifs, and that k-mer based classification provides an approach to detect such similarities. The weak-to-modest predictive power of k-mer content in most scenarios hints at ubiquitous and difficult-to-model roles for RNA structure in lncRNA function (Kirk et al. 2018). Nevertheless, k-mer based classification schemes, which have been broadly used in other biological contexts (Blaisdell 1989; Burge et al. 1992; Kari et al. 2015; Lees et al. 2016; Pandey et al. 2018), represent promising avenues that may ultimately aid in the functional classification of lncRNAs from sequence content alone, much in the way that functional domains can now be routinely identified in proteins (UniProt Consortium 2015).
Structure
LncRNAs function through sequence-specific interactions with proteins that recognize stretches of sequence, as well as with other RNAs or DNAs by base pairing through Watson–Crick or Hoogsteen structures or by forming triple helices. However, intermolecular recognition is often dependent on, or regulated by, specific secondary and tertiary structural features of the lncRNA. Intriguingly, when chemical modification techniques have been used to probe lncRNA structure, it has generally revealed high levels of base pairing, more than in mRNAs and comparable to the ribosome or self-splicing introns—contexts in which secondary structure is complex and essential (Somarowthu et al. 2015; Hawkes et al. 2016). Conversely, in cells, RBPs such as hnRNPs (Dreyfuss et al. 1993) might keep lncRNAs less tightly folded. Furthermore, the observation of secondary structure does not necessarily imply function, especially in the absence of clear evolutionary conservation through covariation, as observed for the ribosome. Similarities shared with the ribosome and RNA enzymes could indicate a lncRNA architecture composed of structured domains, possibly flexibly connected, that establish interactions with other RNAs, chromatin, or specific protein complexes to bring them within functional proximity. While this modular structure hypothesis is appealing because it would provide for intricate functional specificities even in the absence of sequence conservation, it remains to be investigated at the molecular level.
It also remains to be investigated the extent to which secondary structures are functional, and whether they coalesce to form tertiary and higher order structures and interactions. Thus far, relatively few lncRNAs have been characterized at the secondary structure level, including H19 (Hurst and Smith 1999; Juan 2000), Xist (Wutz et al. 2002; Fang et al. 2015; Lu et al. 2016; Smola et al. 2016; F. Liu et al. 2017), Braveheart (Xue et al. 2016), HOTAIR (Somarowthu et al. 2015), COOLAIR (Hawkes et al. 2016), or lincRNA-p21 (Chillón and Pyle 2016) and several others.
A few studies indicate nevertheless that lncRNA structure is important for regulation, and the principles learned through these examples might be more broadly applicable to other lncRNAs. One study showed that regulation of transcription of the E-cadherin gene is regulated by a sense lncRNA in epithelial cells that is independently transcribed upstream of the promoter (Pisignano et al. 2017). The structure of this RNA is controlled by a SNP that modifies local RNA secondary structure and affects loading of epigenetic enzymes that then regulate the downstream promoter. How common regulation through conformational switching is remains unclear, but RNA is structurally malleable and physically well-suited for this mechanism of gene regulation. Riboswitches, for example, are RNA structures that toggle between distinct conformational states upon binding of small molecules and are widespread in bacterial gene regulation (Mandal and Breaker 2004). Riboswitch-like mechanisms of regulation might be present in lncRNAs as well, but their prevalence and function remain to be investigated. A study indicates regulation by conformational switching is provided by the roX lncRNA, which targets the MSL complex to the Drosophila melanogaster X chromosome as part of the dosage compensation process that occurs in male flies. MSL is recruited to roX lncRNA by a conserved stem-loop structure; once bound by the MSL-component MLE, this stem-loop unfolds to form an alternate RNA structure that appears to trap MSL on roX (Ilik et al. 2013, 2017; Quinn et al. 2016). In the case of Xist, structured regions, but also regions notable for their absence of structure, likely serve to recruit different subsets of proteins along the length of the lncRNA (Wutz et al. 2002; Fang et al. 2015; Smola et al. 2016; F. Liu et al. 2017). Here, toggling between different conformational states might define the subset of proteins that associate with the lncRNA under distinct cellular conditions.
Resource Toolkit for LncRNA Interaction Profiling
RNAs are largely dependent on proteins for their production, processing, transport, and localization. Based on reagent availability and ease-of-study, approaches to investigate RNA–protein interactions have historically been protein-centric. As the diversity and functionality of lncRNAs emerged and expanded, these tools, including RNA-immunoprecipitation (RIP), CLIP and its variations, including HITS-CLIP, PAR-CLIP, iCLIP, and Fast-iCLIP, have begun to reveal the magnitude of protein–lncRNA interactions (Ule et al. 2005; Hafner et al. 2010; König et al. 2010; Flynn et al. 2015; J.-H. Li et al. 2015; Zarnegar et al. 2016). Similarly, efforts are ongoing to define the complete repertoire of RNA-binding proteins using proteomics-based methods such as OOPS, R-DeeP, XRNAX, and DIF-FRAC, which often incorporate RNA dependency in their analysis (Mallam et al. 2018; Caudron-Herger et al. 2019; Queiroz et al. 2019; Trendel et al. 2019), and reveal surprisingly widespread RNA-dependent protein functionality, even for well characterized proteins such as CTCF (Caudron-Herger et al. 2019).
The expanding RNA functionalities highlight the need for RNA-centered methods to empirically determine binding partners of lncRNAs (Table 2). New computational resources that enable queries on previously identified interactors, and those that allow for prediction of new interacting candidates have therefore seen remarkable growth in just the last few years.
Table 2. Methods to Investigate lncRNA function in cell fate determination.
| Approach | Type | Readout | Output |
|---|---|---|---|
| Differential expression analysis | Experimental | Indirect | Gene expression differences between cell types with differentiation, or in a condition of interest |
| Expression correlation (+/− target or effector) | Experimental | Indirect | Network generation to identify similarly expressed gene clusters, including candidate target molecules or possible upstream regulators |
| Affinity purification/proximity ligation and deep sequencing (CHART/ChIRP/RAP/MARGI/GRID-Seq/CHAR-Seq/Radicl-Seg/PIRCh-Seq) | Experimental | Direct | Chromatin targets |
| Affinity purification and mass spectrometry/Western blot (CHART/ChIRP/RAP) | Experimental | Direct | Protein interactions |
| Single molecule fluorescence in situ hybridization (smFISH) +/− immunofluorescence | Experimental | Direct | Subcellular localization and colocalization with targets or effectors |
| Conservation analysis | Experimental | Direct | Applicability of function across species |
| Structural analysis | Experimental | Direct | Secondary structure and functional domain identification; 3D structure |
| Bioinformatics tools | Computational | Indirect | Predict interactions, assess k-mer content, make structural inferences |
Experimental methods
Capture hybridization analysis of RNA targets:
Capture hybridization analysis of RNA targets (CHART) methodology allows for the identification of chromatin and protein interactors of lncRNAs. First, regions that are accessible for probe-based isolation are mapped using RNase H-dependent digestion. DNA oligonucleotides can then be used to isolate the RNA in an affinity purification step, followed by high-throughput sequencing to determine DNA segments contacted by the lncRNA, or mass spectrometry, to determine the protein interactome of the candidate lncRNA (Simon et al. 2011).
Chromatin isolation by RNA purification:
Similar to CHART, chromatin isolation by RNA purification (ChIRP) or dChIRP (Chu et al. 2011; Quinn et al. 2014) uses a probe-based affinity approach built on biotinylated oligonucleotides that tile the lncRNA. Isolated interactors that bind the lncRNA (ChIRP), or bind to a specific domain (dChIRP), can be analyzed by high-throughput sequencing, or by methods that detect proteins, such as mass spectrometry or Western blotting.
RNA antisense purification:
The RNA antisense purification (RAP) method (McHugh et al. 2015), instead of using short probes (∼20 nt) as in CHART or ChIRP, utilizes longer probes of ∼60 nt to increase the stability of the interaction in affinity pulldowns.
RNA pulldown:
The above-mentioned methods to investigate lncRNA-partner molecules in intact cells were preceded by probe-based isolation of RNAs in cell extracts in pulldown experiments similar to protein coimmunoprecipitation.
Mapping RNA-genome interactions:
Mapping RNA-genome interactions (MARGI) uses proximity ligation to connect chromatin-associated RNAs to their genomic targets, thus revealing native RNA-chromatin interactions. Variations in the approach were designed to differentiate between direct interactions (diMARGI), mediated by protein or RNA-tethered interactions, of RNA–DNA chimeras, or passive interactions (pxMARGI) (Sridhar et al. 2017).
Global RNA interaction with DNA by deep sequencing:
Global RNA interaction with DNA by deep sequencing (GRID-Seq) harnesses in situ ligation to identify genome-wide contacts between RNA and chromatin. The developers of this method included mouse ES cells and found distinct cis- and trans- chromatin interacting RNAs tied to cell-specific gene expression patterns. GRID-Seq particularly enriches for chromatin interactions with nascent RNAs (Li et al. 2017).
Chromatin associated RNA sequencing:
Chromatin associated RNA sequencing (CHAR-Seq) is an in situ proximity ligation approach coupled with enzymatic chromatin digestion to detect RNA–DNA contacts genome-wide. After sequencing, CHAR-Seq maps the genomic interacting sites of multiple classes of chromatin-associated RNAs including nascent transcripts, ncRNAs involved in regulation of dosage compensation, and trans-interacting RNAs involved in RNA processing (Bell et al. 2018).
RNA and DNA interacting complexes ligated and sequenced:
RNA and DNA interacting complexes ligated and sequenced (RADICL-Seq) identifies genome-wide RNA-chromatin interactions in cross-linked nuclei. Thus far, it has been applied to two cell types—mouse ES cells and mouse oligodendrocyte progenitors—that can differentiate toward multiple cell fates. This approach revealed cell-type specific RNA–chromatin interactions, and was able to identify unique genome occupancy patterns for different classes of transcripts (Bonetti et al. 2019).
Profiling interacting RNAs on chromatin:
Profiling interacting RNAs on chromatin (PIRCh-Seq) is an antibody-dependent approach that profiles RNA–chromatin interactions, with less enrichment of nascent RNAs cotranscriptionally tethered by RNA polymerases to chromatin. PIRCh-Seq has been used to identify the chromatin-associated transcriptome in both human and mouse ES cells and fibroblasts, as well as mouse neuronal progenitors, where the authors found cell- and allele-specific RNA–chromatin interactions (Fang et al. 2019).
Microscopy:
As previously discussed, lncRNA localization can provide functional clues. The advent of single molecule imaging approaches facilitates the localization of RNA relative to other interacting molecules. For example, labeled FISH probes used in single molecule fluorescence in situ microscopy (smFISH) (Cabili et al. 2015; Dunagin et al. 2015), followed by immunofluorescence microscopy with three-dimensional and quantitative fluorescence image analysis allows for the visualization of colocalized lncRNA and protein interactors within subcellular domains (Lino Cardenas et al. 2018). Variations, including merFISH (Chen et al. 2015) and seqFISH (Shah et al. 2016), depend on barcoding in sequential rounds of hybridization to enable the detection of many transcripts simultaneously. The resulting data complexity creates the need for computational tools such as trendsceek (Edsgärd et al. 2018) to analyze these data. We can expect lncRNA monitoring in spatial transcriptomics to increase as different functionalities continue to emerge.
Bioinformatics
NPInter:
Hao et al. (2016) is a repository of functional interactions between noncoding RNAs and interacting partners including small and large RNAs, DNA and proteins. At the core of NPInter is a manual curation process based on published literature, with a primary focus on experimentally verified physical interactions, supplemented with in silico predictions supported by high-throughput sequence data. The latest version contains >900,000 interactions between noncoding RNAs and other biomolecules from 22 organisms. For RNAs, accession IDs from NONCODE, Ensembl, and RefSeq are supported, and integration with the UCSC Genome Browser facilitates visualization of binding sites for human, mouse, and yeast genomes.
POSTAR:
Hu et al. (2017), Zhu et al. (2018) is a database that enables exploration of post-transcriptional regulatory interactions, based primarily on ∼1200 CLIP-Seq data sets. The aim of POSTAR is to contribute a better understanding of how RBPs impact post-transcriptional regulatory processes in six species. Its integration with the UCSC Genome Browser facilitates rapid visualization of RBP binding sites within transcripts.
RAID:
The RAID database (Zhang et al. 2014; Yi et al. 2017), formerly CLIPdb, incorporates experimentally derived and computationally predicted RNA interactions from the published literature, as well as other databases. RAID includes data for 60 species and >1.2 million individual RNA–protein and 4 million RNA–RNA interactions, respectively. A score that is based on the evidence supporting the interaction indicates the confidence in each interaction.
RNA–protein interaction prediction:
Using protein and RNA sequence data, the family of RPISeq machine learning qualifiers (Muppirala et al. 2011) provides RNA–protein interaction probabilities. Different versions of the tool provide the probability of interaction between a specific RNA and protein, a specific RNA and up to 100 proteins, or a specific protein and up to 100 RNAs. Additionally, the sequence of a specific protein can be used to query the RPIntDB, which contains >30,000 individual RNA–protein interactions.
RNA–protein interaction predictor:
RPI-Pred (Suresh et al. 2015) is a Support Vector Machine based prediction tool that uses RNA and protein sequence information and protein structural fragment data. Users can also test multiple candidates including assessing the potential of a single RNA to interact with multiple proteins or multiple RNAs interacting with a single protein.
StarBase:
This RNA-centric resource (Yang et al. 2011; Li et al. 2014) details interactions between various classes of long and short RNAs, as well as between RNAs and proteins as extracted from CLIP-Seq (PAR-CLIP, HITS-CLIP, iCLIP, CLASH), degradome-seq, and RNA–RNA interactome data. Users are also able to impute downstream effects of these interactions based on accompanying gene expression data.
Combined approaches
Structure determination and analysis:
The secondary structure of RNA can be predicted from thermodynamic principles, but inaccuracy in the parameters means that experimental input is required to generate a reliable model. This is most often provided in the form of constraints on secondary structure generated from either evolutionary considerations (conservation of base pairs, ideally by covariation) and/or direct experimental mapping of secondary structure using either enzymatic, or, most often, chemical techniques such as SHAPE and dimethyl sulfate (DMS) mapping (Kirk et al. 2018), psoralen crosslinking (Lu et al. 2016), and high throughput ligation followed by deep-sequencing (Ramani et al. 2015). Detection can be achieved efficiently through deep-sequencing, although capillary electrophoresis, and even polyacrylamide gels, can be used at much lower cost when studying single lncRNAs. Because folding in vitro and in cells might differ because of kinetic constraints on cotranscriptional RNA folding and the presence of RBPs (Leamy et al. 2016), techniques are being developed to probe RNA secondary structures in cellular contexts as well. Here, the primary limitation is sensitivity and the requirement to have sufficient RNA for detection, which could require overexpression since most lncRNAs are present at relatively low copy number. Higher resolution methods such as SAXS (Small Angle X-ray Scattering) (Rambo and Tainer 2013) or X-ray crystallography and NMR currently have very low throughput and can be used only to investigate a few paradigmatic RNAs or systems of particularly high biological interest.
Conclusion and Perspectives
This review summarizes multiple lines of evidence showing that lncRNAs regulate cellular plasticity and cell fate determination, often through combination of multiple mechanisms. By adding a further layer of complexity to gene regulation, they broadly contribute to gene expression regulation to either (i) maintain a blanket undifferentiated state, (ii) promote exit toward a specified cell type, (iii) reprogram cells to a pluripotent ground state, or (iv) contribute to cell specification control in organismal development. LncRNAs perform these complex functions in integrated networks with a diverse set of cellular players with which they interact physically and/or functionally.
Progress toward the phenotypic assessment of lncRNA depletion occurs through loss-of-function and gain-of-function approaches (Liu and Lim 2018), facilitated by the advent of CRISPR/Cas9 technologies. Indeed, high-throughput screens using CRISPR interference identified >300 lncRNAs that impacted iPS cell growth, with a smaller subset influencing pluripotency maintenance as determined by OCT4 expression (S. J. Liu et al. 2017). Functional ablation approaches include poly-A signal insertion proximal to the transcription start site, although a drawback of this insertion is residual background expression, as well as deletion of the lncRNA locus, which results in total loss of lncRNA function, but which may also affect unannotated regulatory elements. Even the genetic manipulation of smaller sequences such as promoters or single exons for well-annotated intergenic lncRNAs should be carried out with caution to avoid modifying regulatory genomic sequences. It should also be noted that DNA-targeting approaches have resulted in differing phenotypes, as exemplified by Fendrr. Studies using a reporter gene replacement strategy for Fendrr found abnormalities in lung development and lethality at a later time point (Sauvageau et al. 2013; Lai et al. 2015), compared with the heart and body wall abnormalities resulting in prenatal lethality in earlier investigations (Grote et al. 2013). Regardless of the specific technical approach for DNA sequence manipulation, it will be important to study the impact of lncRNA expression ablation on the function and regulation of the interacting proteins, RNAs, and chromatin in development. Specific molecular targeting of the lncRNA itself using CRISPR/Cas13 (Abudayyeh et al. 2017; Cox et al. 2017) could facilitate such investigations.
Intriguingly, at least several lncRNAs displaying differing levels of conservation, such as Malat1, Neat1, Cyrano, Braveheart, Evx1as, and Visc-2, and found to have profound molecular or cellular functions, had no overt developmental phenotype in knockout animals (Han et al. 2018). This suggests either a primary role for lncRNAs in fine-tuning developmental functions, distinct roles in specific cellular processes requiring situational study, or yet unearthed compensatory functions, potentially by related/familial lncRNAs. Another explanation for the absence of animal phenotypes could be off-target effects of knockdown approaches using RNA interference or antisense oligonucleotide-dependent depletion (Matsui and Corey 2017). Increased use of gene editing approaches such as CRISPR/Cas9 will help to clarify lncRNA functionality in cell and animal models.
Improvements are needed to allow study of lncRNA interactions at the single cell and single molecule level. While technically feasible to a limited extent using imaging technologies, these methods remain specialized and low throughput. These studies would allow the determination of cell fate as single cell expression and epigenetic studies have indicated substantial heterogeneity even within clonal cellular subpopulations. Perhaps dynamic lncRNA interactions contribute to this heterogeneity to dictate differing cell fates as well.
Related to this heterogeneity, it is still unclear whether a classification system that would allow prediction of lncRNA interactions will be found, but, if such a system existed, it is unlikely to be based on broad segments of sequence conservation because these are generally absent in lncRNAs. However, it might be possible to base classification on the identification of shorter sequence stretches (k-mers) or structural features of the lncRNA that facilitate interactions with other biomolecules. Identification of the relevant structural elements would provide insight into lncRNA interactions with noncanonical RNA binding proteins as well, including those without conserved and/or overt RNA binding domains.
There are untapped opportunities for progress in the mechanistic analysis of lncRNA function for better understanding of specific developmental processes and some downstream applications, including personalized therapeutics. For example, cancers typically progress through the acquisition of stemness features (Malta et al. 2018) and undifferentiated tumors have poor prognosis because they share immortality and repopulation capacity characteristics with stem cells. LncRNAs have emerged as central oncogenic and tumor suppressive factors involved in misregulated cancer pathways (Berger et al. 2018; Chiu et al. 2018), and many lncRNAs, such as Cyrano, have been have been shown to support cellular proliferation (Smith et al. 2017; Deng et al. 2018; X. Liu et al. 2018; Naemura et al. 2018). Studies of lncRNAs in stem cell contexts will not only enable better understanding of mammalian development and differentiation, but may also eventually facilitate better treatment of cancer and degenerative diseases.
Acknowledgments
Grant support: National Institutes of Health (NIH) R01 GM101974 to T.M., NIH R03 HD093977 to K.N.S, NIH R01GM121806 to J.M.C., as well as R35 GM126942 and RO1 GM 103834 to G.V.
Footnotes
Communicating editor: J. Rine
Literature Cited
- Abudayyeh O. O., Gootenberg J. S., Essletzbichler P., Han S., Joung J. \et al. , 2017. RNA targeting with CRISPR–Cas13. Nature 550: 280–284. 10.1038/nature24049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander R. P., Fang G., Rozowsky J., Snyder M., and Gerstein M. B., 2010. Annotating non-coding regions of the genome. Nat. Rev. Genet. 11: 559–571. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amândio A. R., Necsulea A., Joye E., Mascrez B., and Duboule D., 2016. Hotair is dispensible for mouse development. PLoS Genet. 12:e1006232 10.1371/journal.pgen.1006232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Akhtar A., Barlow D. P., Bird A. P., Brockdorff N., et al. , 2014. Considerations when investigating LncRNA function in vivo. eLife 3: e03058 10.7554/eLife.03058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P. J., and Chang H. Y., 2013. Long noncoding RNAs: cellular address codes in development and disease. Cell 152: 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, J. C., D. Jukam, N. A. Teran, V. I. Risca, O. K. Smith, et al., 2018 Chromatin-associated RNA sequencing (ChAR-Seq) maps genome-wide RNA-to-DNA contacts. eLife 7: pii: e27024. 10.7554/eLife.27024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M., Puig I., Pena C., Garcia J. M., Alvarez A. B. et al. , 2008. A natural antisense transcript regulates Zeb2/Sip1 gene expression during snail1-induced epithelial-mesenchymal transition. Genes Dev. 22: 756–769. 10.1101/gad.455708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A. C., Korkut A., Kanchi R. S., Hegde A. M., Lenoir W. et al. , 2018. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell 33: 690–705.e9. 10.1016/j.ccell.2018.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron-Sandoval L.-P., Safaee N., and Michnick S. W., 2016. Mechanisms and consequences of macromolecular phase separation. Cell 165: 1067–1079. 10.1016/j.cell.2016.05.026 [DOI] [PubMed] [Google Scholar]
- Bernardes de Jesus B., Marinho S. P., Barros S., Sousa-Franco A., Alves-Vale C. et al. , 2018. Silencing of the LncRNA Zeb2-NAT facilitates reprogramming of aged fibroblasts and safeguards stem cell pluripotency. Nat. Commun. 9: 94 https://www.nature.com/articles/s41467-017-01921-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Duncan E. M., Masui O., Gil J., Heard E. et al. , 2006. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 26: 2560–2569. 10.1128/MCB.26.7.2560-2569.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisdell B. E., 1989. Effectiveness of measures requiring and not requiring prior sequence alignment for estimating the dissimilarity of natural sequences. J. Mol. Evol. 29: 526–537. 10.1007/BF02602924 [DOI] [PubMed] [Google Scholar]
- Boeynaems S., Alberti S., Fawzi N. L., Mittag T., Polymenidou M. et al. , 2018. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28: 420–435. 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R., Lecona E., Narendra V., Voigt P., Parisi F. et al. , 2014. Interactions with RNA direct the polycomb group protein SCML2 to chromatin where it represses target genes. eLife 3: e02637 10.7554/eLife.02637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti A., Agostini F., Suzuki A. M., Hashimoto K., Pascarella G. et al. , 2019. RADICL-Seq identifies general and cell type-specific principles of genome-wide RNA-chromatin interactions. bioRxiv. 10.1101/681924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Ballabio A., Rupert J. L., Lafreniere R. G., Grompe M., et al. , 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44. 10.1038/349038a0 [DOI] [PubMed] [Google Scholar]
- Burge C., Campbell A. M., and Karlin S., 1992. Over- and under-representation of short oligonucleotides in DNA sequences. Proc. Natl. Acad. Sci. USA 89: 1358–1362. 10.1073/pnas.89.4.1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N., Dunagin M. C., McClanahan P. D., Biaesch A., Padovan-Merhar O. et al. , 2015. Localization and abundance analysis of human LncRNAs at single-cell and single-molecule resolution. Genome Biol. 16: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron-Herger M., Rusin S. F., Adamo M. E., Seiler J., Schmid V. K. et al. , 2019. R-deeP: proteome-wide and quantitative identification of RNA-dependent proteins by density gradient ultracentrifugation. Mol. Cell 75: 184–199.e10. 10.1016/j.molcel.2019.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O. et al. , 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369. 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty D., Paszkowski-Rogacz M., Berger N., Ding L., Mircetic J. et al. , 2017. LncRNA Panct1 maintains mouse embryonic stem cell identity by regulating TOBF1 recruitment to Oct-Sox sequences in early G1. Cell Rep. 21: 3012–3021. 10.1016/j.celrep.2017.11.045 [DOI] [PubMed] [Google Scholar]
- Chen K. H., Boettiger A. N., Moffitt J. R., Wang S., and Zhuang X., 2015. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348: aaa6090 10.1126/science.aaa6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillón I., and Pyle A. M., 2016. Inverted repeat Alu elements in the human LincRNA-P21 adopt a conserved secondary structure that regulates RNA function. Nucleic Acids Res. 44: 9462–9471. 10.1093/nar/gkw599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H. S., Somvanshi S., Patel E., Chen T. W., Singh V. P. et al. ; Cancer Genome Atlas Research Network , 2018. Pan-cancer analysis of LncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep. 23: 297–312.e12. 10.1016/j.celrep.2018.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C., Qu K., Zhong F. L., Artandi S. E., and Chang H. Y., 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44: 667–678. 10.1016/j.molcel.2011.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. B. T., Gootenberg J. S., Abudayyeh O. O., Franklin B., Kellner M. J. et al. , 2017. RNA editing with CRISPR-Cas13. Science 358: 1019–1027. 10.1126/science.aaq0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar K., Pondick J. V., Kim B.-M., Zhou C., York S. R. et al. , 2016. DIGIT is a conserved long noncoding RNA that regulates GSC expression to control definitive endoderm differentiation of embryonic stem cells. Cell Rep. 17: 353–365. 10.1016/j.celrep.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar K., Behfar Ardehali M., Klein I. A., Kratkiewicz A. J., Zhou C. et al. , 2019. LncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. bioRxiv. 10.1101/547513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P. P., Hendrix D. A., Apostolou E., Buchner A. H., Canver M. C. et al. , 2015. PRC2 is required to maintain expression of the maternal Gtl2-Rian-Mirg locus by preventing de novo DNA methylation in mouse embryonic stem cells. Cell Rep. 12: 1456–1470. 10.1016/j.celrep.2015.07.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C., Wang X., Cifuentes-Rojas C., Goodrich K. J., Gooding A. R. et al. , 2015. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell 57: 552–558. 10.1016/j.molcel.2014.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Deng H., Liu C., Liang Y., and Wang S., 2018. Long non-coding RNA OIP5–AS1 functions as an oncogene in lung adenocarcinoma through targeting MiR-448/Bcl-2. Biomed. Pharmacother. 98: 102–110. 10.1016/j.biopha.2017.12.031 [DOI] [PubMed] [Google Scholar]
- Dinger M. E., Amaral P. P., Mercer T. R., Pang K. C., Bruce S. J. et al. , 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18: 1433–1445. 10.1101/gr.078378.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D., Freese P., Alexis M. S., Su A., Hochman M. et al. , 2018. Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell 70: 854–867.e9. 10.1016/j.molcel.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M. J., Pinol-Roma S., and Burd C. G., 1993. HnRNP proteins and the biogenesis of MRNA. Annu. Rev. Biochem. 62: 289–321. 10.1146/annurev.bi.62.070193.001445 [DOI] [PubMed] [Google Scholar]
- Dunagin M., Cabili M. N., Rinn J., and Raj A., 2015. Visualization of LncRNA by single-molecule fluorescence in situ hybridization. Methods Mol. Biol. 1262:3–19. 10.1007/978-1-4939-2253-6_1 [DOI] [PubMed] [Google Scholar]
- Durruthy-Durruthy J., Sebastiano V., Wossidlo M., Cepeda D., and Cui J., 2016. The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat. Genet. 48: 44–52. 10.1038/ng.3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., and Batzoglou S., 2006. Multiple sequence alignment. Curr. Opin. Struct. Biol. 16: 368–373. 10.1016/j.sbi.2006.04.004 [DOI] [PubMed] [Google Scholar]
- Edsgärd D., Johnsson P., and Sandberg R., 2018. Identification of spatial expression trends in single-cell gene expression data. Nat. Methods 15: 339–342. 10.1038/nmeth.4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J. M., Haines J. E., Perez E. M., Munson G., Chen J. et al. , 2016. Local regulation of gene expression by LncRNA promoters, transcription and splicing. Nature 539: 452–455. 10.1038/nature20149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray J.-P., Chavez L., Huang Y., Ross K. N., Choi J. et al. , 2015. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat. Cell Biol. 17: 545–557. 10.1038/ncb3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi M. A., Modarresi F., Khalil A. M., Wood D. E., Sahagan B. G. et al. , 2008. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14: 723–730. 10.1038/nm1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi M. A., Zhang M., Huang J., Modarresi F., Van der Brug M. P., 2010. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11: R56 10.1186/gb-2010-11-5-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Ma Q., Chu C., Huang B., Li L. et al. , 2019. Functional classification of noncoding RNAs associated with distinct histone modifications by PIRCh-seq. bioRxiv. 10.1101/667881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R., Moss W. N., Rutenberg-Schoenberg M., and Simon M. D., 2015. Probing Xist RNA structure in cells using targeted structure-seq. PLoS Genet. 11: e1005668 . 10.1371/journal.pgen.1005668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorenzano A., Pascale E., Gagliardi M., Terreri S., Papa M. et al. , 2018. An ultraconserved element containing LncRNA preserves transcriptional dynamics and maintains ESC self-renewal. Stem Cell Rep. 10: 1102–1114. 10.1016/j.stemcr.2018.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C. L., and Fisher A. G.. 2011. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr. Opin. Genet. Dev. 21: 140–46. 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Flynn R. A., Martin L., Spitale R. C., Do B. T., Sagan S. M. et al. , 2015. Dissecting noncoding and pathogen RNA-protein interactomes. RNA 21: 135–143. 10.1261/rna.047803.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Ahuja G., Bartsch D., Russ N., Yao W. et al. , 2019. YylncT defines a class of divergently transcribed LncRNAs and safeguards the T-mediated mesodermal commitment of human PSCs. Cell Stem Cell 24: 318–327.e8. 10.1016/j.stem.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Frankish A., Diekhans M., Ferreira A.-M., Johnson R., Jungreis I. et al. , 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47: D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Wightman F., Giono L. E., Fededa J. P., and de la Mata M., 2018. Target RNAs strike back on microRNAs. Front. Genet. 9: 435 10.3389/fgene.2018.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Meshorer E., and Ramalho-Santos M., 2011. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12: 36–47 (erratum: Nat. Rev. Mol. Cell Biol. 12: 273). 10.1038/nrm3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., and Maquat L. E., 2011. LncRNAs transactivate STAU1-mediated MRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant J., Mahadevaiah S. K., Khil P., Sangrithi M. N., Royo H. et al. , 2012. Rsx is a metatherian RNA with Xist-like properties in X–chromosome inactivation. Nature 487: 254–258. 10.1038/nature11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R. I., Chendrimada T. P., Cooch N., and Shiekhattar R., 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640. 10.1016/j.cell.2005.10.022 [DOI] [PubMed] [Google Scholar]
- Grote P., Wittler L., Hendrix D., Koch F., Währisch S. et al. , 2013. The tissue-specific LncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24: 206–214. 10.1016/j.devcel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Chen X., Xing R., Wang M., Zhu X. et al. , 2018. Interplay between FMRP and LncRNA TUG1 regulates axonal development through mediating SnoN-Ccd1 pathway. Hum. Mol. Genet. 27: 475–485. 10.1093/hmg/ddx417 [DOI] [PubMed] [Google Scholar]
- Haegele L., Ingold B., Naumann H., Tabatabai G., Ledermann B. et al. , 2003. Wnt signalling inhibits neural differentiation of embryonic stem cells by controlling bone morphogenetic protein expression. Mol. Cell. Neurosci. 24: 696–708. 10.1016/S1044-7431(03)00232-X [DOI] [PubMed] [Google Scholar]
- Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J. et al. , 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Luo S., Peng G., Lu J. Y., Cui G., et al. , 2018. Mouse knockout models reveal largely dispensable but context-dependent functions of LncRNAs during development. J. Mol. Cell Biol. 10: 175–178. . 10.1093/jmcb/mjy003 [DOI] [PubMed] [Google Scholar]
- Hao Y., Wu W., Li H., Yuan J., Luo J. et al. , 2016. NPInter v3.0: an upgraded database of noncoding RNA-associated interactions. Database (Oxford) 2016: pii: baw057. 10.1093/database/baw057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J. M., Tapanari E., Diekhans M. et al. , 2012. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res 22: 1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes E. J., Hennelly S. P., Novikova I. V., Irwin J. A., Dean C. et al. , 2016. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep 16: 3087–3096. 10.1016/j.celrep.2016.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Yang S., Gu X., Li M., Wang C. et al. , 2018. Long noncoding RNA TUG1 facilitates osteogenic differentiation of periodontal ligament stem cells via interacting with Lin28A. Cell Death Dis. 9: 455 10.1038/s41419-018-0484-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich E.-M., and Dimmeler S., 2012. MicroRNAs and stem cells: control of pluripotency, reprogramming, and lineage commitment. Circ. Res. 110: 1014–1022. 10.1161/CIRCRESAHA.111.243394 [DOI] [PubMed] [Google Scholar]
- Hendrickson D.G., Kelley D. R., Tenen D., Bernstein B., and Rinn J. L., 2016. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 17: 28 10.1186/s13059-016-0878-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., and Sharp P. A., 2017. A phase separation model for transcriptional control. Cell 169: 13–23. 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C.-C., Ramilowski J. A., Harshbarger J., Bertin N., Rackham O. J. L. et al. , 2017. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543: 199–204. 10.1038/nature21374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Yang Y. T., Huang Y., Zhu Y., and Lu Z. J., 2017. POSTAR: a platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res. 45: D104–D114. 10.1093/nar/gkw888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L. D., and Smith N. G. C., 1999. Molecular evolutionary evidence that H19 MRNA is functional. Trends Genet. 15: 134–135. 10.1016/S0168-9525(99)01696-0 [DOI] [PubMed] [Google Scholar]
- Ilik I. A., Quinn J. J., Georgiev P., Tavares-Cadete F., Maticzka D. et al. , 2013. Tandem stem-loops in RoX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol. Cell 51: 156–173. 10.1016/j.molcel.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilik I. A., Maticzka D., Georgiev P., Gutierrez N. M., Backofen R. et al. , 2017. A mutually exclusive stem–loop arrangement in RoX2 RNA is essential for X–chromosome regulation in Drosophila. Genes Dev. 31: 1973–1987. 10.1101/gad.304600.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A. K., Xi Y., McCarthy R., Allton K., Akdemir K. C. et al. , 2016. LncPRESS1 is a P53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol. Cell 64: 967–981. 10.1016/j.molcel.2016.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Liu Y., Liu R., Zhang K., and Zhang Y., 2015. The LncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 11: 137–148. 10.1016/j.celrep.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan V., 2000. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 28: 1221–1227. 10.1093/nar/28.5.1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S., Mills K. C., Yee D., Otte A. P., Panning B. et al. , 2006. The Polycomb group protein Eed protects the inactive X–chromosome from differentiation-induced reactivation. Nat. Cell Biol. 8: 195–202. 10.1038/ncb1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen A. N., Zhou X. B., Xu J., Qiao C., Ma J., et al. , 2013. The imprinted H19 LncRNA antagonizes let-7 microRNAs. Mol. Cell 52: 101–112. 10.1016/j.molcel.2013.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L., Israsena N., Zhang Z., Hu M., Zhao L.-R. et al. , 2004. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev. Biol. 269: 580–594. 10.1016/j.ydbio.2004.02.005 [DOI] [PubMed] [Google Scholar]
- Kaneko, S., J. Son, S. S. Shen, D. Reinberg, and R. Bonasio, 2013 PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 20: 1258–1264. 10.1038/nsmb.2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Bonasio R., Saldaña-Meyer R., Yoshida T., Son J. et al. , 2014. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 53: 290–300. 10.1016/j.molcel.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kari L., Hill K. A., Sayem A. S., Karamichalis R., Bryans N. et al. , 2015. Mapping the space of genomic signatures. PLoS One 10: e0119815 . 10.1371/journal.pone.0119815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlic R., Ganesh S., Franke V., Svobodova E., Urbanova J. et al. , 2017. Long non-coding RNA exchange during the oocyte-to-embryo transition in mice. DNA Res. 24: 129–141 (erratum: DNA Res. 24: 219–220). 10.1093/dnares/dsw058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. M., Guttman M., Huarte M., Garber M., Raj A. et al. , 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 106: 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk J. M., Kim S. O., Inoue K., Smola M. J., Lee D. M. et al. , 2018. Functional classification of long non-coding RNAs by k-Mer content. Nat. Genet. 50: 1474–1482. 10.1038/s41588-018-0207-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C. A., Scheuermann J. C., Surface L. E., Bradley R. K., Fields P. A. et al. , 2013. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152: 570–583. 10.1016/j.cell.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleaveland B., Shi C. Y., Stefano J., and Bartel D. P., 2018. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174: 350–362.e17. 10.1016/j.cell.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J., Zarnack K., Rot G., Curk T., Kayikci M. et al. , 2010. ICLIP reveals the function of HnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17: 909–915. 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzin J. J., Spencer S. P., McCright S. J., Uthaya Kumar D. B., Collet M. A. et al. , 2016. The long non-coding RNA morrbid regulates bim and short-lived myeloid cell lifespan. Nature 537: 239–243. 10.1038/nature19346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K. M., Gong G., Atanasio A., Rojas J., Quispe J. et al. , 2015. Diverse phenotypes and specific transcription patterns in twenty mouse lines with ablated LincRNAs. PLoS One 10: e0125522 . 10.1371/journal.pone.0125522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy K. A., Assmann S. M., Mathews D. H., and Bevilacqua P. C., 2016. Bridging the gap between in vitro and in vivo RNA folding. Q. Rev. Biophys. 49: e10 10.1017/S003358351600007X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees J. A., Vehkala M., Välimäki N., Harris S. R., Chewapreecha C. et al. , 2016. Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes. Nat. Commun. 7: 12797 10.1038/ncomms12797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski J. P., Dumbović G., Watson A. R., Hwang T., Jacobs-Palmer E. et al. , 2019. The locus is essential for male fertility. bioRxiv. 10.1101/562066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. H., Liu S., Zhou H., Qu L. H., and Yang J. H., 2014. StarBase v2.0: decoding MiRNA-CeRNA, MiRNA-NcRNA and protein-RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Res. 42: D92–D97. 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. H., Liu S., Zheng L. L., Wu J., Sun W. J. et al. , 2015. Discovery of protein-LncRNA interactions by integrating large-scale CLIP-seq and RNA-seq datasets. Front Bioeng Biotechnol. 2: 88 10.3389/fbioe.2014.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liu B., Wapinski O. L., Tsai M. -C., Qu K. et al. , 2013. Targeted disruption of hotair leads to homeotic transformation and gene derepression. Cell Rep. 5: 3–12. 10.1016/j.celrep.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Helms J. A., and Chang H. Y., 2016. Comment on ‘hotair is dispensable for mouse development.’ PLoS Genet. 12: e1006406 10.1371/journal.pgen.1006406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Gou H., Tripathi B. K., Huang J., Jiang S. et al. , 2015b An apela RNA-containing negative feedback loop regulates P53-mediated apoptosis in embryonic stem cells. Cell Stem Cell 16: 669–683. 10.1016/j.stem.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhou B., Chen L., Gou L. T., Li H. et al. , 2017. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 35: 940–950. 10.1038/nbt.3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N., Chang K. Y., Li Z., Gates K., Rana Z. A. et al. , 2014. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 53: 1005–1019. 10.1016/j.molcel.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino Cardenas C. L., Kessinger C. W., Cheng Y., MacDonald C., MacGillivray T. et al. , 2018. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat. Commun. 9: 1009 https://www.nature.com/articles/s41467-018-03394-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Somarowthu S., and Pyle A. M., 2017. Visualizing the secondary and tertiary architectural domains of LncRNA RepA. Nat. Chem. Biol. 13: 282–289. 10.1038/nchembio.2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. J., and Lim D. A., 2018. Modulating the expression of long non‐coding RNAs for functional studies. EMBO Rep. 19:pii: e46955. 10.15252/embr.201846955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. J., M. A. Horlbeck, S. W. Cho, H. S. Birk, M. Malatesta et al., 2017b CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355: pii: aah7111. 10.1126/science.aah7111. 10.1126/science.aah7111 [DOI] [PMC free article] [PubMed]
- Liu X. H., Sun M., Nie F. Q., Ge Y. B., Zhang E. B. et al. , 2014. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging MiR-331-3p in gastric cancer. Mol. Cancer 13: 92 10.1186/1476-4598-13-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zheng J., Xue Y., Yu H., Gong W. et al. , 2018. PIWIL3/OIP5–AS1/MiR-367–3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics 8: 1084–1105. 10.7150/thno.21740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S., Cabili M. N., Guttman M., Loh Y. H., Thomas K. et al. , 2010. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 42: 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Yu D., Hansen A. S., Ganguly S., Liu R. et al. , 2018. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558: 318–323. 10.1038/s41586-018-0174-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Sachs F., Ramsay L., Jacques P. É., Göke J. et al. , 2014. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol. 21: 423–425. 10.1038/nsmb.2799 [DOI] [PubMed] [Google Scholar]
- Lu Z., Zhang Q. C., Lee B., Flynn R. A., Smith M. A. et al. , 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Lu J. Y., Liu L., Yin Y., Chen C. et al. , 2016. Divergent LncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18: 637–652. 10.1016/j.stem.2016.01.024 [DOI] [PubMed] [Google Scholar]
- Mallam A. L., Sae-Lee W., Schaub J. M., Tu F., Battenhouse A. et al. , 2018. Systematic discovery of endogenous human ribonucleoprotein complexes. bioRxiv. 10.1101/480061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malta T. M., Sokolov A., Gentles A. J., Burzykowski T., Poisson L. et al. , 2018. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173: 338–354.e15. 10.1016/j.cell.2018.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., and Breaker R. R., 2004. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 5: 451–463. 10.1038/nrm1403 [DOI] [PubMed] [Google Scholar]
- Matsui M., and Corey D. R., 2017. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 16: 167–179. 10.1038/nrd.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S., 2001. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2: 986–991. 10.1093/embo-reports/kve230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S., and Rinn J. L., 2015. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22: 5–7. 10.1038/nsmb.2942 [DOI] [PubMed] [Google Scholar]
- McHugh C. A., Chen C. K., Chow A., Surka C. F., Tran C. et al. , 2015. The Xist LncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–236. 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M., and Rinn J. L., 2016. ‘Cat’s Cradling’ the 3D genome by the act of LncRNA transcription. Mol. Cell 62: 657–664. 10.1016/j.molcel.2016.05.011 [DOI] [PubMed] [Google Scholar]
- Mercer T. R., and Mattick J. S., 2013. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20: 300–307. 10.1038/nsmb.2480 [DOI] [PubMed] [Google Scholar]
- Muppirala U. K., Honavar V. G., and Dobbs D., 2011. Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics 12: 489 10.1186/1471-2105-12-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naemura M., Kuroki M., Tsunoda T., Arikawa N., Sawata Y. et al. , 2018. The long noncoding RNA OIP5-AS1 is involved in the regulation of cell proliferation. Anticancer Res. 38: 77–81. 10.21873/anticanres.12194 [DOI] [PubMed] [Google Scholar]
- Nesterova T. B., Wei G., Coker H., Pintacuda G., Bowness J. S. et al. , 2019. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nature Comm. 10: 3129 10.1038/s41467-019-11171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. Y., Johnson R., and Stanton L. W., 2012. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31: 522–533. 10.1038/emboj.2011.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.-Y., Bogu G. K., Soh B. S., and Stanton L. W., 2013. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51: 349–359. 10.1016/j.molcel.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Palazzo A. F., and Lee E. S., 2015. Non-coding RNA: what is functional and what is junk? Front. Genet. 6: 2 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Bender M. A., Johnson R., Patro R., and Berger B., 2018. Squeakr: an exact and approximate k-mer counting system. Bioinformatics 34: 568–575. . 10.1093/bioinformatics/btx636 [DOI] [PubMed] [Google Scholar]
- Pisignano G., Napoli S., Magistri M., Mapelli S. N., Pastori C. et al. , 2017. A promoter-proximal transcript targeted by genetic polymorphism controls E-cadherin silencing in human cancers. Nat. Commun. 8: 15622 10.1038/ncomms15622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka, M., P. Glažar, L. R. Hernandez-Miranda, S. Memczak, S. A. Wolf et al., 2017 Loss of a mammalian circular RNA locus causes MiRNA deregulation and affects brain function. Science 357: pii: eaam8526. 10.1126/science.aam8526. 10.1126/science.aam8526 [DOI] [PubMed]
- Portoso M., Ragazzini R., Brenčič Ž., Moiani A., Michaud A. et al. , 2017. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 36: 981–994. 10.15252/embj.201695335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz R. M. L., Smith T., Villanueva E., Marti-Solano M., Monti M. et al. , 2019. Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat. Biotechnol. 37: 169–178 (erratum: Nat. Biotechnol. 37: 692). 10.1038/s41587-018-0001-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. J., and Chang H. Y., 2016. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17: 47–62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- Quinn J. J., Ilik I. A., Qu K.,Georgiev P., Chu C. et al. , 2014. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat. Biotechnol. 32: 933–940. 10.1038/nbt.2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. J., Zhang Q. C., Georgiev P., Ilik I. A., Akhtar A. et al. , 2016. Rapid evolutionary turnover underlies conserved LncRNA-genome interactions. Genes Dev. 30: 191–207. 10.1101/gad.272187.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V., Qiu R., and Shendure J., 2015. High-throughput determination of RNA structure by proximity ligation. Nat. Biotechnol. 33: 980–984. 10.1038/nbt.3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo R. P., and Tainer J. A., 2013. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu. Rev. Biophys. 42: 415–441. 10.1146/annurev-biophys-083012-130301 [DOI] [PubMed] [Google Scholar]
- Regha K., Sloane M. A., Huang R., Pauler F. M., Warczok K. E. et al. , 2007. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell 27: 353–366. 10.1016/j.molcel.2007.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I., and Bleasby A., 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16: 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Rinn J. L., and Chang H. Y., 2012. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81: 145–166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L., and Ule J., 2014. Oming in on RNA-protein interactions. Genome Biol. 15: 401 10.1186/gb4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom A., Melamed L., Goldrich M. J., Kadir R., Golan M. et al. , 2019. Regulation of CHD2 expression by the long noncoding RNA is essential for viability. bioRxiv. 10.1101/536771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A., Yang Y., and Plath K., 2018. The role of Xist in X–chromosome dosage compensation. Trends Cell Biol. 28: 999–1013. 10.1016/j.tcb.2018.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanli I., Lalevée S., Cammisa M., Perrin A., Rage F. et al. , 2018. Meg3 non-coding RNA expression controls imprinting by preventing transcriptional upregulation in Cis. Cell Rep. 23: 337–348. 10.1016/j.celrep.2018.03.044 [DOI] [PubMed] [Google Scholar]
- Santoro F., Mayer D., Klement R. M., Warczok K. E., Stukalov A. et al. , 2013. Imprinted Igf2r silencing depends on continuous airn LncRNA expression and is not restricted to a developmental window. Development 140: 1184–1195. 10.1242/dev.088849 [DOI] [PubMed] [Google Scholar]
- Sauvageau M., Goff L. A., Lodato S.,Bonev B., Groff A. F. et al. , 2013. Multiple knockout mouse models reveal LincRNAs are required for life and brain development. eLife 2: e01749 10.7554/eLife.01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savić N., Bär D., Leone S., Frommel S. C., Weber F. A. et al. , 2014. LncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15: 720–734. 10.1016/j.stem.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Schertzer M. D., Braceros K. C. A., Starmer J., Cherney R. E., Lee D. M. et al. , 2019. LncRNA-induced spread of polycomb controlled by genome architecture, RNA abundance, and CpG island DNA. Mol. Cell 75: 523–537.e10. 10.1016/j.molcel.2019.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleri L., Bartolomei M. S., Bickmore W. A., He L., Stubbs L. et al. , 2016. A hox-embedded long noncoding RNA: is it all hot air? PLoS Genet. 12: e1006485 10.1371/journal.pgen.1006485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Lubeck E., Zhou W., and Cai L., 2016. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse Hippocampus. Neuron 92: 342–357. 10.1016/j.neuron.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik Mohamed J., Gaughwin P. M., Lim B., Robson P., and Lipovich L.. 2010. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16: 324–337. 10.1261/rna.1441510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda M., Kanai-Azuma M., Hara K., Miyazaki S., Kanai Y. et al. , 2007. Sox17 plays a substantial role in late-stage differentiation of the extraembryonic endoderm in vitro. J. Cell Sci. 120: 3859–3869. 10.1242/jcs.007856 [DOI] [PubMed] [Google Scholar]
- Simon M. D., Wang C. I., Kharchenko P. V., West J. A., Chapman B. A. et al. , 2011. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA 108: 20497–20502. 10.1073/pnas.1113536108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. N., Starmer J., Miller S. C., Sethupathy P., and Magnuson T., 2017. Long noncoding RNA moderates microRNA activity to maintain self-renewal in embryonic stem cells. Stem Cell Rep. 9: 108–121. 10.1016/j.stemcr.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. N., Starmer J., and Magnuson T., 2018. Interactome determination of a long noncoding RNA implicated in embryonic stem cell self-renewal. Sci. Rep. 8: 17568 10.1038/s41598-018-34864-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola M. J., Christy T. W., Inoue K., Nicholson C. O., Friedersdorf M. et al. , 2016. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist LncRNA in living cells. Proc. Natl. Acad. Sci. USA 113: 10322–10327. 10.1073/pnas.1600008113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarowthu S., Legiewicz M., Chillón I., Marcia M., Liu F. et al. , 2015. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 58: 353–361. 10.1016/j.molcel.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague D., Waters S. A., Kirk J. M., Wang J. R., Samollow P. B. et al. , 2019. Nonlinear sequence similarity between the Xist and Rsx long noncoding RNAs suggests shared functions of tandem repeat domains. RNA 25: 1004–1019. 10.1261/rna.069815.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar, B., M. Rivas-Astroza, T. C. Nguyen, W. Chen, Z. Yan et al., 2017 Systematic mapping of RNA-chromatin interactions in vivo. Curr. Biol. 27: 602–609 (erratum: Curr. Biol. 27: 610–612). doi: 10.1016/j.cub.2017.01.011 10.1016/j.cub.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh V., Liu L., Adjeroh D., and Zhou X., 2015. RPI-pred: predicting NcRNA-protein interaction using sequence and structural information. Nucleic Acids Res. 43: 1370–1379. 10.1093/nar/gkv020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R., Yokobayashi S., Stadler M. B., Otte A. P., van Lohuizen M. et al. , 2008. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev. Cell 15: 668–679. 10.1016/j.devcel.2008.08.015 [DOI] [PubMed] [Google Scholar]
- Trendel J., Schwarzl T., Horos R., Prakash A., Bateman A. et al. , 2019. The human RNA-binding proteome and its dynamics during translational arrest. Cell 176: 391–403.e19. 10.1016/j.cell.2018.11.004 [DOI] [PubMed] [Google Scholar]
- Tsai M. C., Manor O., Wan Y., Mosammaparast N., Wang J. K. et al. , 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J., Jensen K., Mele A., and Darnell R. B., 2005. CLIP: a method for identifying protein–RNA interaction sites in living cells. Methods 37: 376–386. 10.1016/j.ymeth.2005.07.018 [DOI] [PubMed] [Google Scholar]
- Ulitsky I., Shkumatava A., Jan C. H., Sive H., and Bartel D. P., 2011. Conserved function of LincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550. 10.1016/j.cell.2011.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium , 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43: D204–D212. 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.-Q., Wang Y., Xiong Y., Chen X.-C., Ma M.-L., 2016. Sirt1 AS LncRNA interacts with its MRNA to inhibit muscle formation by attenuating function of MiR-34a. Scientific Rep. 6: 21865. . 10.1038/srep21865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Mager J., Chen Y., Schneider E., Cross J. C. et al. , 2001. Imprinted X inactivation maintained by a mouse polycomb group gene. Nat. Genet. 28: 371–375. 10.1038/ng574 [DOI] [PubMed] [Google Scholar]
- Wang K. C., Yang Y. W., Liu B., Sanyal A., Corces-Zimmerman R. et al. , 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124. 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Goodrich K. J., Gooding A. R., Naeem H., Archer S. et al. , 2017. Targeting of Polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell 65: 1056–1067.e5. 10.1016/j.molcel.2017.02.003 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xu Z., Jiang J., Xu C., Kang J. et al. , 2013. Endogenous MiRNA sponge LincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 25: 69–80. 10.1016/j.devcel.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Weng R., Lu C., Liu X., Li G., Lan Y. et al. , 2018. Long noncoding RNA-1604 orchestrates neural differentiation through the MiR-200c/ZEB axis. Stem Cells 36: 325–336. 10.1002/stem.2749 [DOI] [PubMed] [Google Scholar]
- Wheeler T. J., and Eddy S. R., 2013. Nhmmer: DNA homology search with profile HMMs. Bioinformatics 29: 2487–2489. 10.1093/bioinformatics/btt403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzi M., Casas Vila N., Paszkowski-Rogacz M., Ding L., Noack S. et al. , 2018. The long noncoding RNA LncR492 inhibits neural differentiation of murine embryonic stem cells. PLoS One 13: e0191682 10.1371/journal.pone.0191682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A., Rasmussen T. P., and Jaenisch R., 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30: 167–174. 10.1038/ng820 [DOI] [PubMed] [Google Scholar]
- Xue Z., Hennelly S., Doyle B., Gulati A. A., Novikova I. V. et al. , 2016. A G-rich motif in the LncRNA Braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol. Cell 64: 37–50. . 10.1016/j.molcel.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Deng X., Ma W., Berletch J. B., Rabaia N. et al. , 2015. The LncRNA firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16: 52 10.1186/s13059-015-0618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. H., Li J. H., Shao P., Zhou H., Chen Y. Q. et al. , 2011. StarBase: a database for exploring MicroRNA–MRNA interaction maps from argonaute CLIP-seq and degradome-seq data. Nucleic Acids Res. 39: D202–D209. 10.1093/nar/gkq1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. W., Flynn R. A., Chen Y., Qu K., Wan B. et al. , 2014. Essential role of LncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife 3: e02046 10.7554/eLife.02046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y., Zhao Y., Li C., Zhang L., Huang H. et al. , 2017. RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic Acids Res. 45: D115–D118. 10.1093/nar/gkw1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Yan P., Lu J., Song G., Zhu Y. et al. , 2015. Opposing roles for the LncRNA Haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16: 504–516. 10.1016/j.stem.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Yoon J.-H., Abdelmohsen K., and Gorospe M., 2013. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 425: 3723–3730. 10.1016/j.jmb.2012.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T. L., Matsuda T., and Cepko C. L., 2005. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 15: 501–512. 10.1016/j.cub.2005.02.027 [DOI] [PubMed] [Google Scholar]
- Yu C.-Y., and Kuo H.-C., 2016. The trans-spliced long noncoding RNA tsRMST impedes human embryonic stem cell differentiation through WNT5A-mediated inhibition of the epithelial-to-mesenchymal transition. Stem Cells 34: 2052–2062. 10.1002/stem.2386 [DOI] [PubMed] [Google Scholar]
- Zarnegar B. J., Flynn R. A., Shen Y., Do B. T., Chang H. Y. et al. , 2016. IrCLIP platform for efficient characterization of protein-RNA interactions. Nat. Methods 13: 489–492. 10.1038/nmeth.3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wu D., Chen L., Li X., Yang J. et al. , 2014. RAID: a comprehensive resource for human RNA-associated (RNA-RNA/RNA-Protein) interaction. RNA 20: 989–993. 10.1261/rna.044776.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Dai Q.-S., Zhu S.-C., Han Y.-H., Han H.-L. et al. , 2016. AK048794 maintains the mouse embryonic stem cell pluripotency by functioning as an MiRNA sponge for MiR-592. Biochem. J. 473: 3639–3654. 10.1042/BCJ20160540 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Xu G., Yang Y. T., Xu Z., Chen X. et al. , 2018. POSTAR2: deciphering the post-transcriptional regulatory logics. Nucleic Acids Res. 47: D203–D211. 10.1093/nar/gky830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żylicz, Jakub Jan, Bousard Aurélie, Žumer Kristina, Dossin Francois, Mohammad Eusra, Teixeira da Rocha Simão, Schwalb Björn, et al. 2019. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 176 (1–2): 182–197.e23. 10.1016/j.cell.2018.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]