Abstract
Genetic reporters such as the green fluorescent protein (GFP) can facilitate measurement of promoter activity and gene expression. However, animal autofluorescence limits the sensitivity of GFP and other fluorescent reporters in whole-animal settings like in the nematode Caenorhabditis elegans. Here, we present a highly sensitive Nanoluciferase (NanoLuc)-based method in a multiwell format to detect constitutive and inducible gene expression in C. elegans. We optimize detection of bioluminescent signals from NanoLuc in C. elegans and show that it can be detected at 400,000-fold over background in a population of 100 animals expressing intestinal NanoLuc driven by the vha-6 promoter. We can reliably detect signal in single vha-6p::Nanoluc-expressing worms from all developmental stages. Furthermore, we can detect signal from a 1/100 dilution of lysate from a single vha-6p::Nanoluc-expressing adult and from a single vha-6p::Nanoluc-expressing adult “hidden” in a pool of 5000 N2 wild-type animals. We also optimize various steps of this protocol, which involves a lysis step that can be performed in minutes. As a proof-of-concept, we used NanoLuc to monitor the promoter activity of the pals-5 stress/immune reporter and were able to measure 300- and 50-fold increased NanoLuc activity after proteasome blockade and infection with microsporidia, respectively. Altogether, these results indicate that NanoLuc provides a highly sensitive genetic reporter for rapidly monitoring whole-animal gene expression in C. elegans.
Keywords: Nanoluciferase, C. elegans intestine, intracellular pathogen response, genetically encoded reporter, luminescence
GENETICALLY encoded reporters are important tools to monitor gene expression and provide faster read-outs than other methods. For example, promoter-driven reporters are often used as proxies for assessing messenger RNA (mRNA) expression, and are faster than more direct measurements of mRNA expression, such as quantitative RT-PCR (qRT-PCR) or single-molecule fluorescence in situ hybridization. In Caenorhabditis elegans, genetic reporters have traditionally been introduced into the genome as multicopy arrays that contain 100–1000 copies (Mello et al. 1991). While multiple copies of a transgene can increase the reporter signal, a small RNA-mediated process called transgene silencing often reduces expression from these multicopy reporters in C. elegans (Minkina and Hunter 2018; De-Souza et al. 2019). Because of this phenomenon, measurement of reporter gene expression in C. elegans can be confounded by factors that regulate transgene silencing instead of regulating expression of the gene of interest itself. Generation of low-copy array strains (Schweinsberg and Grant 2013), or integration of reporters as single-copy transgenes via Mos1-mediated single-copy insertion (MosSCI) or clustered regularly interspaced short palindromic repeats/Cas9 (Frøkjaer-Jensen et al. 2008; Dokshin et al. 2018), can reduce or eliminate silencing, but has the disadvantage of producing lower signals than multicopy transgenes (Mendenhall et al. 2015). The most commonly used genetically encoded reporters include fluorophores like the green fluorescent protein (GFP). Unfortunately, weak-background fluorophores present in multicellular organisms can decrease the overall sensitivity of such fluorophore-based assays. In particular, gut granules of the C. elegans intestine are highly autofluorescent and can hamper the measurement of gene expression from this tissue (Teuscher and Ewald 2018). Thus, there is a need for genetically encoded reporters in C. elegans with better overall signals than fluorophores like GFP.
In contrast to fluorescent reporters, bioluminescent reporters generate de novo light without the need for external excitation through photons, and they are highly sensitive with a broad dynamic range (Thorne et al. 2010). The bioluminescent signal is generated through oxidation of a substrate (luciferin) by a luciferase enzyme and there are many luciferin/luciferase pairs. These reporters are commonly used in mammalian systems and less often in C. elegans. To date, the ATP-dependent firefly luciferase has been used in C. elegans to measure mitochondrial function, larval molting, feeding behavior, and circadian rhythm, and also to monitor viral infection (Lagido et al. 2015; Olmedo et al. 2015; Palikaras and Tavernarakis 2016; Gammon et al. 2017). While the ATP dependence of firefly luciferase is a means to monitor mitochondrial activity, the requirement for ATP hampers the ability of this luciferase to accurately investigate other processes like gene expression, because a lack of signal may simply reflect lowered ATP levels in the cell (Brock 2012). Therefore, the use of highly sensitive, ATP-independent luciferases would facilitate broader use of bioluminescent reporters in C. elegans.
In this study, we establish the ATP-independent Nanoluciferase (NanoLuc) as both a constitutive and inducible genetically encoded luciferase reporter in C. elegans. NanoLuc was developed by Promega (Madison, WI) (Hall et al. 2012), who optimized a subunit of the Oplophorus gracilirostris deep sea shrimp luciferase to generate a reporter with small size, high physical stability, and high brightness. Promega further recommends the use of the coelenterazine 2-furanylmethyl-deoxy-coelenterazine (furimazine) as a substrate for NanoLuc to achieve a high luminescent signal with increased half-life and decreased autoluminescence compared to other luciferase/luciferin pairs. The 19-kDa monomer NanoLuc exerts an ATP-independent glow-type blue signal (emission maximum 460 nm) with a half-life > 2 hr. Compared to firefly and Renilla luciferases, NanoLuc emits a ∼150-times-brighter signal, has better assay stability at higher temperatures, and operates at a wider pH range and in the presence of urea (Hall et al. 2012). NanoLuc has been used for several applications in non-C. elegans systems, like mammalian cells (England et al. 2016). Here, we used the MosSCI technique to generate single-copy reporter strains that have constitutive and inducible expression of NanoLuc in C. elegans. We used these strains to develop a sensitive and quantitative plate-based assay that has the ability to detect a bioluminescent signal in a fraction of a worm constitutively expressing NanoLuc, and to detect induction of NanoLuc being driven by an immune-/stress-regulated promoter.
Materials and Methods
C. elegans strains and culture
N2 was used as the wild-type strain. Transgenic reporter strains ERT412, ERT513, ERT529, and ERT729 are described in Table 1. All transgenic strains carry single-copy insertions on chromosome II introduced by the MosSCI method (Frøkjaer-Jensen et al. 2008) and were backcrossed at least three times before use. All C. elegans strains were maintained at 20° on Nematode Growth Media (NGM) plates seeded with Escherichia coli OP50-1 bacteria according to standard methods (Brenner 1974). Stocks of synchronized, starved first larval-stage (L1) animals were generated by bleaching gravid adults (Emmons et al. 1979).
Table 1. Transgenic reporter strains used in this study.
| Strain | Genotype |
|---|---|
| ERT412 | unc-119(ed3) III; jySi20[vha-6p::strepII::3XFLAG::GFP::let-858 3′ UTR; cb-unc-119(+)] II |
| ERT513 | unc-119(ed3) III; jySi35[vha-6p::Nanoluc::unc-54 3′ UTR; unc-119 (+)] II |
| ERT529 | unc-119(ed3) III; jySi40[vha-6p::Nanoluc::3XFLAG::unc-54 3′ UTR; unc-119 (+)] II |
| ERT729 | unc-119(ed3) III; jySi44[pals-5p::Nanoluc::unc-54 3′ UTR; unc-119 (+)] II |
Fluorescence microscopy
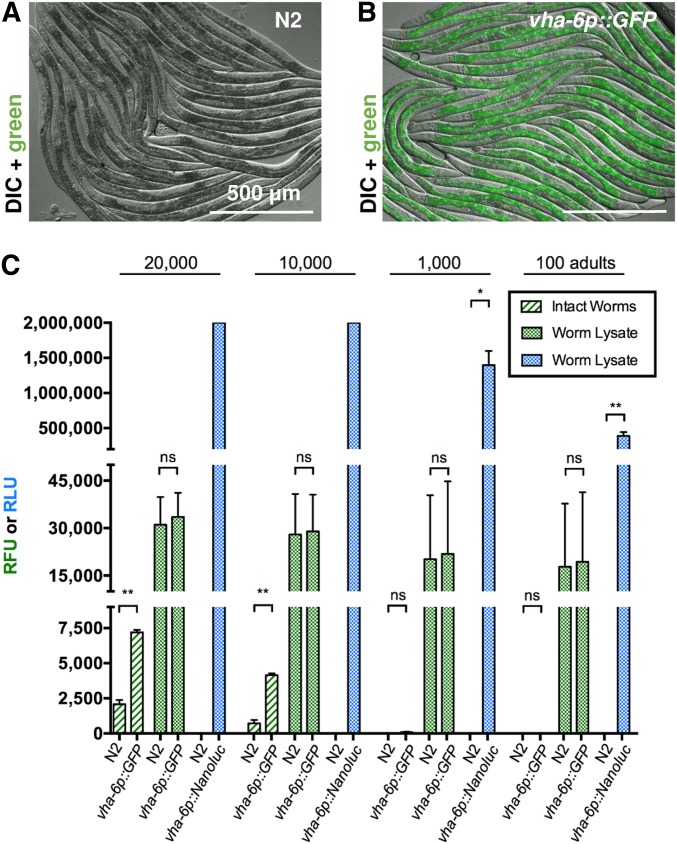
Worms were anesthetized with 10 mM levamisole in M9 buffer and mounted on 5% agarose pads for imaging. Images were captured with a Zeiss ([Carl Zeiss], Thornwood, NY) Axio Imager M1 using similar exposure times for differential interference contrast (DIC) and GFP measurements for both images in Figure 1, A and B.
Figure 1.
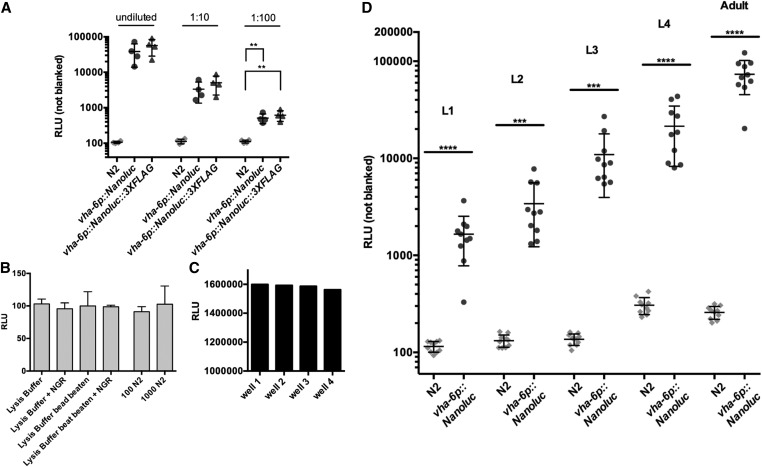
NanoLuc sensitivity exceeds GFP sensitivity on a plate reader. Fluorescence microscopy images show strong intestinal expression of GFP in vha-6p::GFP transgenic C. elegans (B), but not in N2 adults (A). Images obtained by DIC microscopy with GFP fluorescence overlay. (C) Fluorescent signals (RFU, green) or luminescent signals (RLU, blue) measured on a plate reader from intact young adult worms, and worm lysates of either N2, vha-6p::GFP, or vha-6p::Nanoluc animals. RFU was measured with 485-nm excitation and 520-nm emission filters using 10 flashes per well and cycle. RLU was measured for 1 sec, 10 min after addition of furimazine (Nano-Glo reagent, Promega) without filters. n = 2–4 trials for each condition. Error bars are SD; ** P < 0.01 and * P < 0.05. NanoLuc, Nanoluciferase; ns, not significant; RFU, relative fluorescent units; RLU, relative luminescent units.
GFP measurement on plate reader
For both N2 and ERT412, synchronized L1s were grown to the adult stage at 20°, and GFP was measured in either intact animals or in lysate from disrupted animals. In both cases, animals were first washed with M9 with 0.1% Tween 20 (M9-T) to remove excess OP50-1. For measurement of intact animals, 100, 1000, 10,000, or 20,000 young adults in 200 μl M9-T were transferred to a black 96-well assay plate with a clear bottom. To prepare worm lysates, 100, 1000, 10,000, or 20,000 young adults in 210 μl M9-T with protease inhibitor (Roche, cOmplete mini, EDTA-free) were vortexed on a Disruptor Genie vortexer (Scientific Industries) in microfuge tubes with 15–20 silicon carbide beads (#11079110sc; BioSpec Products, 1-mm diameter) for 5 min at 4°. These vortexed samples were spun at 20,000 × g for 5 min, and then 200 μl of the worm lysate was transferred to a black, clear-bottomed 96-well assay plate (#3603; Costar). Fluorescent signals in intact and lysed worms were measured on a NOVOstar plate reader (BMG Labtech), detecting 10 flashes per well and cycle using a 485-nm excitation and a 520-nm emission filter. M9-T was used as a blank for measurements of intact animals, and M9-T vortexed with silicon carbide beads was used as a blank for measurements of lysates.
NanoLuc assay in C. elegans
A step-by-step protocol can be found at: dx.doi.org/10.17504/protocols.io.7mhhk36.
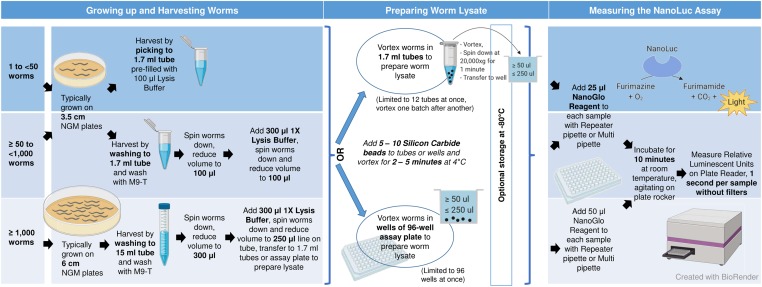
Growing up and harvesting worms:
For all NanoLuc Assays, synchronized L1s were grown at 20° on NGM plates seeded with E. coli OP50-1. L1s were harvested after 4 hr, L2s after 20 hr, L3s after 29 hr, L4s after 44 hr, and young adults 52 hr after plating. Worms were harvested by washing off NGM plates with M9-T. If not indicated otherwise, all spins to settle worms were performed at 2,000 × g for 1 min.
Preparing worm lysates:
To prepare worm lysates of 100, 1000, 10,000, or 20,000 N2 and ERT513 young adults for Figure 1, worms were harvested and washed with M9-T. After centrifugation, the total volume including the pellet was reduced to 300 μl by removing the supernatant to the 300-μl label on the tube. Then, 300 μl of 1× lysis buffer with protease inhibitor (50 mM HEPES pH 7.4, 1 mM EGTA, 1 mM MgCl2, 100 mM KCl, 10% glycerol, 0.05% NP40, 0.5 mM DTT, and protease inhibitor: cOmplete Cat# 11836170001; Sigma [Sigma Chemical], St. Louis, MO) were added. After spinning down the worm pellet, the supernatant was reduced to 250 μl. The samples were then vortexed on a Disruptor Genie vortexer (Scientific Industries) with 15–20 silicon carbide beads in microfuge tubes for 5 min at 4°. The ground samples were spun down at 20,000 × g for 5 min and 200 μl of the worm lysate was transferred to a black, clear-bottomed 96-well assay plate. Nano-Glo reagent (#N1110; Promega, Nano-Glo Luciferase Assay System) was prepared as per the manufacturer’s instructions (50 volumes Nano-Glo Buffer and one volume Nano-Glo substrate) and 50 μl were added to each sample using a repeater pipette. Worm lysate and Nano-Glo reagent were briefly resuspended by pipetting up and down with a multichannel pipette.
To prepare worm lysates of 100 and 5000 N2, ERT513, or ERT529 worms for Figure 3, Figure 5, and Figure 6, animals were harvested and washed with M9-T. Each worm pellet was spun down and the supernatant was removed to the 100-μl mark on the tube. Next, 300 μl of 1× lysis buffer with protease inhibitor were added and, after spinning the samples down, the volume was reduced to 100 μl. Then, the samples were vortexed on a Disruptor Genie vortexer with ∼10–15 silicon carbide beads for 4 min at 4°. The samples were spun down at 20,000 × g for 1 min and 50 μl of worm lysate were transferred to black, clear-bottomed 96-well assay plates. To each sample, 25 μl of freshly prepared Nano-Glo reagent were added.
Figure 3.
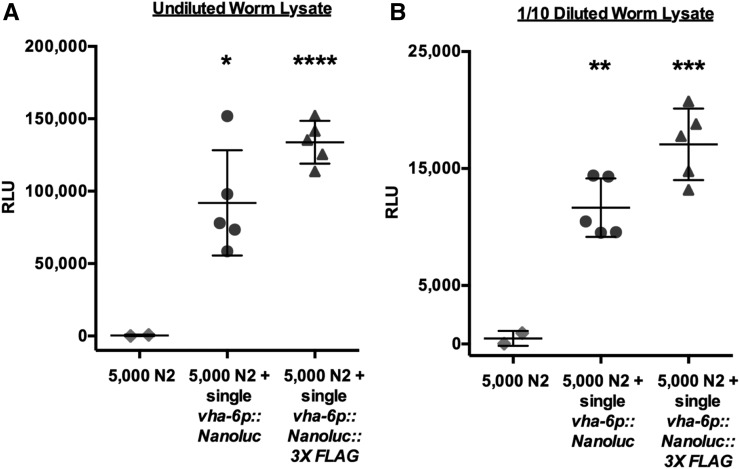
NanoLuc is a highly sensitive genetic reporter in the C. elegans intestine. (A) Bioluminescent signal measured in the lysate of 5000 N2 adults mixed with a single N2 adult, or a single adult expressing vha-6p::Nanoluc or vha-6p::Nanoluc::3XFLAG. (B) Signal of 1/10 dilution of the samples measured for Figure 3A. Error bars are SD; **** P < 0.0001, *** P < 0.001, ** P < 0.01, and * P < 0.05. NanoLuc, Nanoluciferase; RLU, relative luminescent units.
Figure 5.
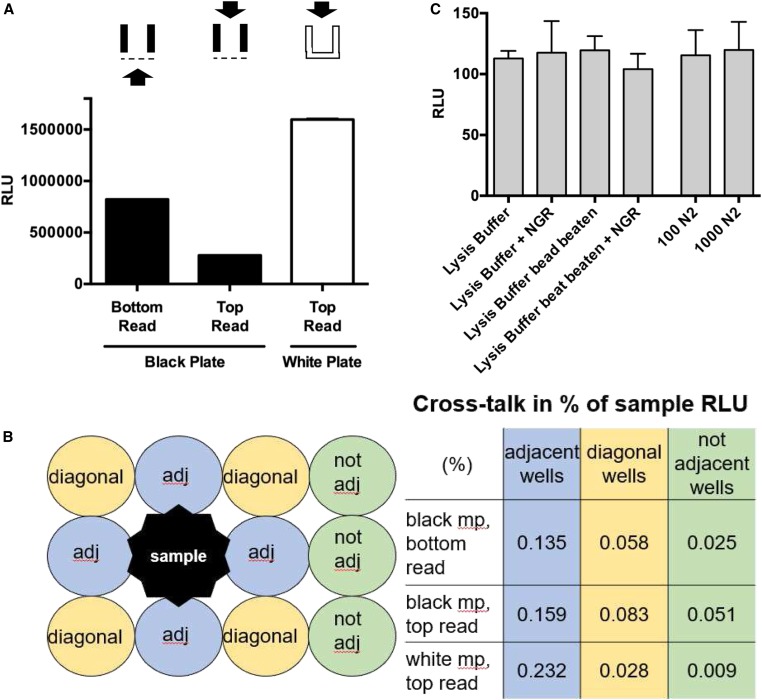
White 96-well assay plates increase detectable signal with minimal cross talk and background signal. (A) Bioluminescent signals from lysate aliquots of 50 vha-6p::Nanoluc adults measured in either black or white 96-well assay plates. Black plates had clear bottoms and white plates were completely opaque. Signal was detected from either the bottom or the top of wells as indicated. Error bars are SD (n = 2). (B) Signals from lysate of 50 vha-6p::Nanoluc adults measured in adj, diagonal, or not adj wells on 96-well assay plates. Cross talk expressed as percentage of signal in sample well (1.5 million RLU). (C) Background signal of white 96-well assay plate determined with buffers, reagents, and N2 lysate. Bead-beating in microfuge tubes with silicon carbide beads performed for 5 min at 4°. Error bars are SD (n = 3). adj, adjacent; mp, multiplate; NGR, Nano-Glo reagent; RLU, relative luminescent units.
Figure 6.
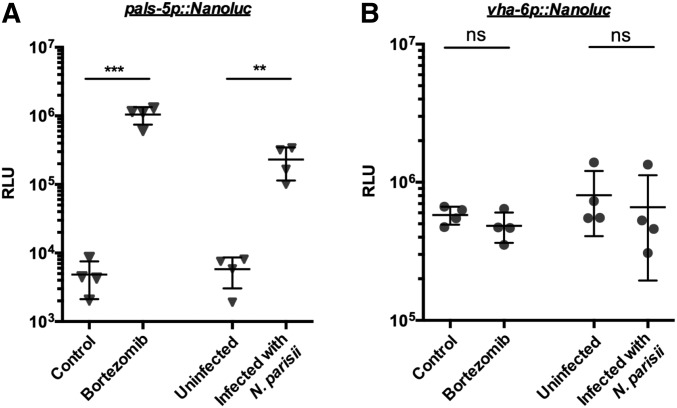
NanoLuc as an inducible genetic reporter in C. elegans. One hundred pals-5p::Nanoluc (A) and vha-6p::Nanoluc (B) L4s treated with 22 μM bortezomib, or infected with 166,000 N. parisii spores, for 5 or 4 hr, respectively. Bioluminescent signal measured in mock- vs. bortezomib-treated worms or uninfected vs. N. parisii-infected worms. One data point represents two independent measurements performed on the same day. Error bar is SD; *** P < 0.001 and ** P < 0.01. NanoLuc, Nanoluciferase; ns, not significant; RLU, relative luminescent units.
To prepare worm lysates of single worms for Figure 2 and Figure 4, an eyelash pick was used to transfer individual N2, ERT513, or ERT529 worms to microfuge tubes prefilled with 80 μl 1× lysis buffer. Next, 5–10 silicon carbide beads were added and samples were vortexed on a Disruptor Genie vortexer for 4 min at 4°. The samples were spun down at 20,000 × g for 1 min and 50 μl of worm lysate were transferred to black, clear-bottomed 96-well assay plates. To prepare dilutions, wells were prefilled with appropriate amounts of 1× lysis buffer and worm lysate was added to a total volume of 50 μl. To each sample, 25 μl of freshly prepared Nano-Glo reagent were added.
Figure 2.
NanoLuc signal can be detected throughout the C. elegans life cycle. (A) Bioluminescent signal measured in undiluted and diluted lysates of single vha-6p::Nanoluc or vha-6p::Nanoluc::3XFLAG adults, and N2 adults. Error bars are SD, ** P < 0.01. (B) Background signal of black 96-well assay plate determined with buffers, reagents, and N2 lysate. Bead-beating in microfuge tubes with silicon carbide beads performed for 5 min at 4°. Error bars are SD (n = 3). (C) Signals from worm lysate aliquots from the same sample measured in multiple wells of a black 96-well assay plate (wells B10, C10, D10, and E10). (D) Each dot is the signal from a single worm. Data show robust NanoLuc activity in lysates of vha-6p::Nanoluc adults throughout the life cycle, but not N2 adults. Error bars are SD; **** P < 0.0001 and *** P < 0.001. NanoLuc, Nanoluciferase; NGR, Nano-Glo reagent; RLU, relative luminescent units.
Figure 4.
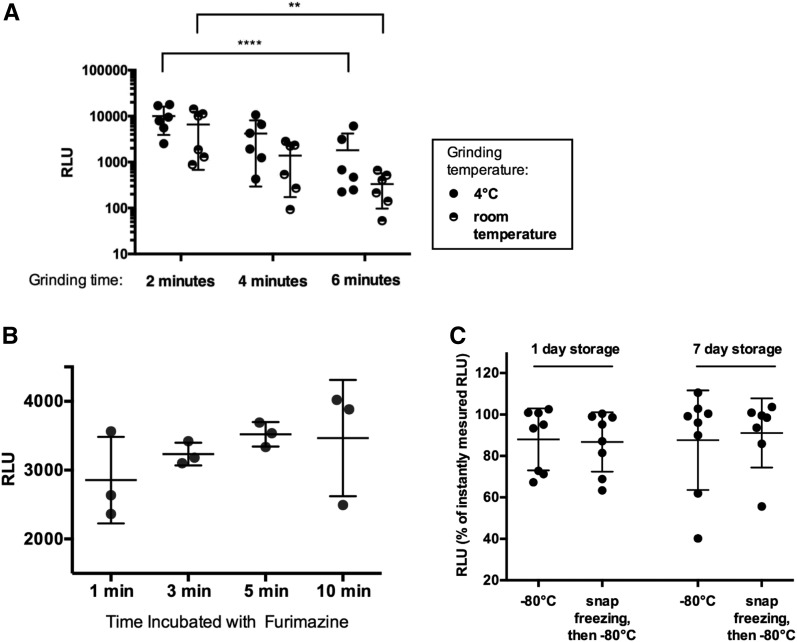
Optimizing grinding conditions, substrate incubation time, and sample storage for the NanoLuc assay. (A) Bioluminescent signal measured in lysates of single vha-6p::Nanoluc adults obtained by vortexing for 2, 4, and 6 min with silicon carbide beads at 4° or room temperature. Two-way ANOVA followed by Tukey’s post-test [grinding temperature: F (1,10) = 1.793; P = 0.21; grinding time: F (2,20) = 24.35; P < 0.0001]. ** P < 0.01 and *** P < 0.0001. (B) Bioluminescent signals measured in vha-6p::Nanoluc worm lysates immediately after harvesting, or storing for 1 or 7 days at −80°, with or without initial liquid nitrogen snap freezing. Measurements displayed as percentages of signals measured immediately after harvesting. (C) Bioluminescent signals measured in lysates of single vha-6p::Nanoluc adults incubated with furimazine for 1, 3, 5, and 10 min. (A–C) error bars are SD. NanoLuc, Nanoluciferase; RLU, relative luminescent units.
For storage experiments in Figure 4B, worm lysates were frozen at −80° in 1× lysis buffer. Worm lysate samples were either placed at −80° immediately after harvesting or first flash frozen in liquid N2 for a couple of seconds. To measure the bioluminescent signal, samples were thawed at room temperature and signal was obtained as previously described.
NanoLuc assay measurements:
The assay plate was agitated for 10 min at room temperature and luminescent signal was detected on a NOVOstar plate reader for 1 sec without filters. Unless indicated otherwise, the optimal gain across the plate was determined from the sample with the highest luminescent signal. For a blank, 1× lysis buffer vortexed with silicon carbide beads and Nano-Glo reagent were used.
Unless indicated otherwise, all assays were measured using black, clear-bottomed 96-well assay plates (#3603; Costar) and signals were read from the bottom. The reading signal from the top was tested in black plates and in opaque white plates (#3912; Costar) for Figure 5 and Supplemental Material, Figures S1 and S2.
Comparing NanoLuc signal in intact and lysed worms for Supplemental Figure 1:
First, 10,000 pals-5p::GFP (not expressing NanoLuc, jyIs8) or vha-6p::Nanoluc (ERT513) adult worms were harvested into 15-ml tubes, and washed with M9-T to remove residual bacteria. After spinning the worm pellet down, the supernatant was reduced to the 1-ml marking on the tube, and 1 ml of 2× lysis buffer with protease inhibitor was added to generate a worm suspension with 5 worms/μl. To measure the signal in 100 intact worms, 20 μl of worm suspension and 30 μl of 2× lysis buffer were combined in wells of an opaque white 96-well plate. To measure the signal in 100 lysed worms, 20 μl of worm suspension, 40 μl of 2× lysis buffer, and five silicon carbide beads were combined in microfuge tubes and vortexed on a Disruptor Genie vortexer for 4 min at 4°. The samples were spun down at 20,000 × g for 1 min and 50 μl of worm lysate were transferred to an opaque white 96-well assay plate. To each sample, 50 μl of freshly prepared Nano-Glo reagent were added. The assay plate was agitated at room temperature, and luminescent signal was detected after 10 and 60 min on a NOVOstar plate reader for 1 sec without filters. Two measurements were performed at each time point, using either the optimal gain 2662 across all samples or the maximum gain 4095. For a blank, 2× lysis buffer vortexed with silicon carbide beads and Nano-Glo reagent was used.
Preparing worm lysates directly in 96-well assay plates for supplemental Figure 2:
First, 100 pals-5p::Nanoluc- or vha-6p::Nanoluc-expressing adult worms were harvested into 1.5 ml tubes, and washed with M9-T to remove residual bacteria, after which the supernatant was reduced to a volume of 100 μl. To prepare worm lysates, five silicon carbide beads were added and the samples were vortexed on a Disruptor Genie vortexer for 4 min at 4°. Then, the samples were spun down for 1 min at 20,000 × g, and 50 μl of the supernatant were transferred to an opaque, white 96-well assay plate. To prepare worm lysate directly inside a well of an assay plate, the 100-μl worm suspension was transferred into a well of an opaque, white 96-well assay plate. Five silicon carbide beads were added; the plate was sealed with sealing tape (Cat# MSB1001; Bio-Rad, Hercules, CA), fastened to a vortexer using an attachment (Cat# 504-0233-00 Model H301; Scientific Industries), and vortexed for 4 min at 4°. Then, the plate was spun down at maximum speed for 1 min, after which 50 μl of the supernatant were transferred to a fresh well. Luminescent signal was measured in lysate prepared in microfuge tubes or lysate (with or without silicon carbide beads) prepared in wells of an assay plate as described previously.
Testing inducible genetic reporter induction
One hundred starved vha-6p::Nanoluc and pals-5p::NanoLuc L1s were grown for 48 hr to the fourth larval state (L4) at 20°. Then, the animals were treated with bortezomib (or DMSO vehicle control) or infected with Nematocida parisii (ERTm1) (or M9 vehicle control). All biological replicates were prepared in duplicate. For bortezomib treatment, a 10-mM stock of bortezomib in DMSO was diluted with M9, then NGM plates were top-plated to reach a final concentration of 22 μM bortezomib in the plates. For mock treatment, DMSO and M9 were top-plated. Animals were incubated for 5 hr at 20° before harvesting and analysis. For infection with N. parisii (ERTm1), 166,000 spores in M9 and 50 μl OP50, or M9 and 50 μl OP50 only for mock treatment, were top-plated. Animals were incubated for 4 hr at 20° before harvesting and analysis. The worms were harvested and washed with M9-T, and the NanoLuc assay was performed as described in Preparing worm lysates for 100 worms and NanoLuc assay measurements.
Statistical analysis of expression data
When not indicated otherwise, statistical significance was determined with the parametric Student’s t-test for comparing two unpaired groups. The impacts of grinding temperature and time in Figure 4A were determined with a repeated measures two-way ANOVA followed by Tukey’s post-test.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.9898943.
Results
Comparisons of GFP and NanoLuc signals in transgenic C. elegans adults
With the goal of developing a sensitive, plate-based assay to measure reporter gene expression in C. elegans, we compared signals from transgenically expressed GFP or NanoLuc under similar conditions. First, we analyzed animals that contained an integrated, single-copy 3XFLAG-tagged GFP controlled by the vha-6 promoter (of note, the 3XFLAG tag serves the purpose of facilitating subsequent biochemical analysis). The vha-6 promoter is a commonly used promoter in C. elegans that drives strong, constitutive expression in the intestine (Oka et al. 2001). The strong intestinal GFP expression from these vha-6p::GFP::3XFLAG adult animals can be visualized with standard microscopy (Figure 1, A and B). We used a conventional plate reader (NOVOstar, BMG Labtech) to measure green fluorescent signals in 100–20,000 intact vha-6p::GFP::3XFLAG adults compared to nontransgenic wild-type N2 controls. The plate reader only detected a three- and sixfold-higher fluorescent signal in 20,000 and 10,000 vha-6p::GFP::3XFLAG animals compared to 20,000 and 10,000 N2 animals, respectively (Figure 1C). There was no significant signal over background when only 1000 or 100 animals were measured. In an attempt to increase signal, we lysed worms first and then measured the green fluorescent signal. Here, the plate reader did not detect any significant signal over background in lysates generated from 20,000, 10,000, 1000, or 100 vha-6p::GFP::3XFLAG adult animals.
Because of the poor signal from GFP measured on a plate reader, we explored a NanoLuc-based assay. First, we generated transgenic animals that contained a single-copy vha-6p::Nanoluc transgene. Using this strain, we found that luminescence from intact animals was detectable but highly variable, and did not correlate well with input amounts (Figure S1). Therefore, we lysed worms prior to performing the assay, to facilitate access of substrate to the NanoLuc enzyme and improve the signal. In contrast to the measurements from intact animals, we detected a robust and reproducible signal from lysates of NanoLuc-expressing animals. Impressively, we detected a 400,000-fold higher signal in lysate from 100 vha-6p::Nanoluc adult animals when compared to lysate from 100 N2 animals (Figure 1C). Lysates from 1000 vha-6p::Nanoluc adult animals had significantly higher signals, and lysates from 10,000 to 20,000 vha-6p::Nanoluc adult animals saturated the detection capability of the plate reader (Figure 1C). Because of the higher levels of bioluminescent signals obtained from lysates compared to intact animals, we used lysates for all subsequent analyses (see Figure 7 for assay schematic). Overall, these results demonstrate robust and reproducible signals in a plate reader-based assay using lysates from C. elegans expressing NanoLuc, which are much greater than the signals from GFP under similar conditions.
Figure 7.
Schematic of the C. elegans NanoLuc assay workflow. The number of worms needed for an assay depends on the strength of expression from the promoter in use, and can be measured in single animals or in populations of tens of thousands of animals. We routinely use 100 L4s to detect activation of a pals-5 stress/immune reporter. Worms are single-picked to 1.7-ml tubes prefilled with lysis buffer, or washed to 1.7-ml or 15-ml tubes with M9-T, and subsequently washed with M9-T to remove residual bacteria. The main goal is to harvest and lyse the worms in as small a volume as possible. After spinning the worms down, the supernatant is removed and then washed with 1× lysis buffer containing protease inhibitor. Lysate can be prepared either inside 1.7-ml tubes or directly in wells of a white, opaque 96-well assay plate. Worm lysate is prepared by adding 5–10 silicon carbide beads to the worm suspension in tubes or wells, then closing tubes or sealing the wells, and vortexing for 4 min at 4°. Optionally, worm lysate can be stored for several days at −80° after this step. The tubes or the assay plate are/is spun down at 20,000 × g for 1 min. If grinding in tubes, ≥ 50 µl of worm lysate are transferred to wells of a white, opaque 96-well assay plate. Next, 25–50 µl of Nano-Glo reagent (Promega) are added to the worm lysate and the plate is agitated for 10 min at room temperature on plate rocker. With a plate reader, relative luminescent units are measured by detecting luminescent signal for 1 sec per sample without filters. As a blank, lysis buffer is vortexed with silicon carbide beads and Nano-Glo reagent is added 10 min before measurement. The assay gain is set so that the highest signal does not saturate the detector capability of the photo multiplier tube. M9-T, M9 buffer with 0.1% Tween 20; NanoLuc, Nanoluciferase; PI, protease inhibitor.
Assessing the sensitivity of NanoLuc constitutively expressed in the intestine
To assess the sensitivity of the NanoLuc signal in C. elegans, we measured the reporter signal in lysates from single vha-6p::Nanoluc-expressing animals at different life stages (Figure 2). We detected a robust luminescent signal in single adults that was significantly higher than in the N2 background (Figure 2A). We also generated a vha-6p::Nanoluc::3XFLAG strain and found similar results as the vha-6p::Nanoluc strain, with robust signals detected in single adults (Figure 2A). Further, we detected strong signals in 10- and 100-fold dilutions of lysates made from single adult vha-6p::Nanoluc and vha-6p::Nanoluc::3XFLAG animals (Figure 2A). The background signals of lysis buffer, substrate, and lysed N2 were similar to the plate reader background determined with water, indicating very low background for this assay (Figure 2B). Importantly, we saw a very stable signal throughout different wells (B10, C10, D10, and E10) of a 96-well assay plate (Figure 2C). To assess NanoLuc signal sensitivity in younger animals, we measured the signal in all four larval stages (L1 through L4) and in young adults. We detected a robust signal in single worms from all of these life stages (Figure 2D). In summary, a significant signal over background can be detected throughout all C. elegans life stages from single animals constitutively expressing NanoLuc in the intestine (Figure 2D). Further, signal can be detected in as little as a 1/100 dilution of lysate from a single adult animal (Figure 2A).
Besides detecting low reporter expression, detecting rare events can be critical to the success of reporter-based assays. Therefore, we mimicked a rare event by mixing a single NanoLuc-expressing worm in a pool of 5000 N2s. Here, we detected ∼100,000- and 12,000-fold signals over the N2 background in undiluted worm lysate and 1-10 diluted worm lysate, respectively (Figure 3, A and B). Therefore, this NanoLuc assay should allow the detection of reporter expression coming from only a single animal in a large population of nonexpressing animals.
Optimizing lysate preparation conditions for the NanoLuc assay
Next, we tested several assay parameters for NanoLuc detection, including grinding conditions, substrate incubation time, and sample storage. The worm lysate described for the NanoLuc assays above was generated by vortexing worms in microfuge tubes with silicon carbide beads on a vortexer. We compared grinding times of 2, 4, and 6 min at 4° or at room temperature. We found that signal was decreased with longer grinding times (P < 0.0001 in two-way ANOVA followed by Tukey’s post-test, Figure 4A). Grinding at 4° compared to room temperature had no significant influence on NanoLuc signal (P = 0.21), although there was a trend toward a lower signal at room temperature. In cell culture studies, the bioluminescent NanoLuc signal is usually measured after 3 min of incubation of cell lysate with furimazine. Our data show that worm lysate can be incubated with substrate for 3–10 min before measurement (Figure 4B).
The workflow of many experiments can benefit from the option to store samples for several days prior to data collection. To evaluate if NanoLuc samples can be stored before measurement, we measured signals in vha-6p::Nanoluc worm lysates immediately after worm lysis, after 1 day of storage at −80°, and after 7 days of storage. We either froze down the lysate directly or after snap freezing in liquid nitrogen. With both storage methods, we were able to preserve the bioluminescent signal for up to 7 days without significant signal loss (Figure 4C). In summary, our analyses indicate that worm lysate should be prepared by grinding the worms for 2–4 min, then bioluminescence measured immediately or after storage at −80° for several days. The lysate can be incubated with substrate for up to 10 min before assessing the bioluminescent signal.
Testing multiwell plate assay parameters for the NanoLuc assay
Next, we optimized plate-reading parameters. While black assay plates are recommended for the detection of fluorescence because they minimize background, white assay plates are recommended for bioluminescence because of the increased signal possible due to the reflection that occurs in white plates together with the low background typical of bioluminescent assays (Judy Gibbs 2001; Wohlstadter et al. 2005). Another assay parameter to consider is whether the signal is detected from the top or the bottom of the plate. Top reading has been reported to minimize well-to-well cross talk compared to bottom reading, because the detector optics can function as a lid for the sample well and shield it from incoming light (Bjerke 2014). Therefore, we compared luminescent signals measured from the top of opaque white assay plates to bottom or top reading of the previously used black, clear-bottomed assay plates. We detected two-times-higher signals in aliquots of the same sample when measured in white plates from the top compared to when measured in black plates from the bottom (Figure 5A). This effect was not compensated by measuring in black plates from the top, as the signal was fourfold lower than when bottom reading in the same plates. Of note, both black and white plates show minimal well-to-well cross talk of < 1% of sample signals in wells adjacent and diagonal to a sample with a strong signal (1.5 million relative luminescent units, Figure 5B). These results indicate that white plates increase the detectable bioluminescent signal in C. elegans without increasing background and maintaining minimal cross talk (Figure 5C) .
Demonstration of NanoLuc as a sensitive reporter for monitoring inducible gene expression
Finally, we investigated whether NanoLuc could be used as an inducible genetic reporter. Here, we generated animals with a single-copy NanoLuc driven by the pals-5 promoter. The pals-5 gene is used as a read-out for the intracellular pathogen response (IPR), which is a defense program induced by diverse, natural intracellular pathogens, as well as by proteotoxic stress (Reddy et al. 2019). Previous studies have used a multicopy pals-5::GFP reporter strain to monitor pals-5 induction in the intestine upon blockade of the proteasome or with intracellular infection by the microsporidian N. parisii (Bakowski et al. 2014). With the integrated, single-copy pals-5p::Nanoluc transgene, we detected a ∼300-fold increase upon treatment with the proteasome inhibitor bortezomib and a ∼50-fold increase in luminescent signal after infection with N. parisii in 100 L4 worms (Figure 6A). We controlled for changes in overall worm mass and treatment interaction with NanoLuc by measuring expression of the constitutively expressed vha-6p::Nanoluc in parallel. The luminescent signals in treated and untreated vha-6p::Nanoluc animals were not significantly different upon treatment (Figure 6B), indicating that bortezomib treatment and microsporidia infection did not simply increase the NanoLuc signal independent of the pals-5 promoter.
To streamline the NanoLuc assay workflow, we also tested whether signal could be measured directly from worm lysates prepared in a multiwell plate, to eliminate the need for a separate step where worms are first disrupted in microfuge tubes and then transferred into wells of an assay plate. Here, we tested two conditions: (1) we vortexed worms with silicon carbide beads in the wells and transferred one-half of this lysate away from the beads into fresh wells where it was measured, and (2) we directly measured signals from wells that contained both worm lysate and beads (Figure S2). In condition #1, we measured the same signals as from lysates that were prepared in microfuge tubes and transferred into wells to be measured. For condition #2, we found that the absolute signals measured were lower when beads were still present in the wells, but the fold increase upon bortezomib treatment was identical to that seen for measurements of lysates only. Therefore, in cases where there is strong induction such as with the pals-5p::NanoLuc reporter, the signal can conveniently be measured from worms disrupted in the wells and subsequently measured from the same wells. Overall, these results indicate that NanoLuc can be used effectively both as a constitutive as well as an inducible genetic reporter in C. elegans.
Discussion
Here, we describe a highly sensitive luminescence-based method to detect gene expression in C. elegans, using the luciferase NanoLuc. In a plate-reader setting, where intestinally expressed GFP could be detected only sixfold over background levels, we found that intestinally expressed NanoLuc could be detected at several million-fold over background (Figure 1C). We used a strain that expresses NanoLuc intestinally to detect signals in lysates of single worms at all life stages from L1s to adults. The signals were so robust that we could detect them confidently over background in worm lysate dilutions as small as 1/100 of a single NanoLuc-expressing adult animal. The optimization and testing that we performed of various assay conditions provide guidelines for grinding time and temperature, sample storage, and plate type. We also found that the assay can be further streamlined by grinding and measuring directly in a multiwell plate (see schematic in Figure 7).
Our initial motivation to translate this extremely bright genetic reporter from cell culture to the nematode C. elegans was to monitor rare transformation events in obligate intracellular pathogens of the C. elegans intestine from the Microsporidia phylum (Reinke and Troemel 2015). To date, there has been no successful genetic modification of any of the > 1400 species in the Microsporidia phylum, despite their widespread significance in the fields of agriculture, evolution, and pathogenesis (Vávra and Lukeš 2013; Munita and Arias 2016). The small size, high physical stability, and high brightness of NanoLuc should enable the detection of rare, single-microsporidia transformation events in the intestine of individual C. elegans animals within large populations being tested. Indeed, our analysis indicates that it is possible to easily detect a single NanoLuc-expressing worm in a population of thousands of non-NanoLuc-expressing worms.
In addition to measuring constitutive expression, we have demonstrated that NanoLuc could be used as a convenient tool to monitor inducible promoter activity, showing a 300- and 50-fold increase in expression from the pals-5 stress/immune reporter after proteasome blockade and infection with microsporidia, respectively. To assess if those treatments influence NanoLuc activity or animal size, we used vha-6 promoter-driven NanoLuc expression in treated and untreated animals as a control. In the future, dual luciferase assays that measure promoter-of-interest::Nanoluc and control-promoter::Firefly luciferase expression in the same animal could provide an internal control (#N1610; Promega, Nano-Glo Dual-Luciferase Reporter Assay System). However, an advantage of a NanoLuc-only system for monitoring gene expression is that it is not affected by changes in ATP levels, which regulate luminescence from firefly and other luciferases.
The advantages and disadvantages of a NanoLuc-based assay compared to other gene expression methods are summarized in Table 2. When compared to GFP, NanoLuc provides a much more sensitive and rapid method for plate-reader-based quantitation of gene expression. However, a major disadvantage of the NanoLuc assay is that it requires lysing the worms to generate a robust signal. This step is problematic for experiments where tissue-specific information and/or sample recovery is desirable. While it is not clear why lysis is required, a likely explanation is that the substrate furimazine cannot sufficiently penetrate the cuticle and tissues of C. elegans. Nonetheless, spatial resolution of NanoLuc-tagged organelles has been successfully monitored in live cells using microscopes that can detect bioluminescence (Ogoh et al. 2014). Additionally, Lagido and colleagues were able to image ubiquitous expression of firefly luciferase in live C. elegans (Lagido et al. 2008). We have detected luciferase activity 10 min after substrate addition to room temperature samples. Decrease of enzyme activity is not expected at this time (NanoLuc half-life > 2 hr; Hall et al. 2012), but a more likely confounder is lack of substrate in samples with a high NanoLuc content. We used 25–50 μl of Nano-Glo reagent for samples with 1000 worms.
Table 2. Comparisons between NanoLuc and other methods for measuring gene expression.
| NanoLuc | GFP | qRT-PCR | smFISH | |
|---|---|---|---|---|
| Sensitivity | High | Low | High | High |
| Time to unbiased quantitation | Minutes | Hours (depending on sample size) | Hours | Hours to days |
| Throughput | High | Medium | Medium | Low |
| Requires lysis (loses spatial information) | Yes | No | Yes | No |
| Input material required | Single worms | Single worms | Hundreds to thousands | Single worms |
| Readout | Promoter activity | Promoter activity | Endogenous mRNA | Endogenous mRNA |
| Strain development | Required | Required | Not required | Not required |
NanoLuc, Nanoluciferase; qRT-PCR, quantitative RT-PCR; smFISH, single-molecule fluorescence in situ hybridization.
Although it can be desirable to measure reporter gene expression in intact worms, the lysing procedure we describe only takes a few minutes, and thus is much faster than other endpoint assays for gene expression that lose spatial information, like qRT-PCR (Table 2). One excellent application for C. elegans expression of NanoLuc would be genetic screens where it is not necessary to recover live animals, such as in RNA interference-based screens. For these screens, NanoLuc would provide better sensitivity and quantitation than GFP, and would be more scalable than qRT-PCR. In addition, the NanoLuc assay requires less input material than assays like qRT-PCR, and in some cases it is preferable to determine promoter activity instead of overall mRNA levels (Table 2). This feature highlights the application that we have demonstrated in Figure 6, where a pals-5 promoter-driven NanoLuc strain provides a reporter for induction of the IPR stress/immune pathway. This use is analogous to luciferase-based methods for measuring the activation of stress and immune pathways in mammalian cells (Delhove et al. 2017), and may be generally useful for monitoring the activation of many different kinds of transcriptional responses in C. elegans.
Acknowledgments
We acknowledge Robert Luallen for generating strain E.412; Kirthi Reddy for help in generating strain E.729; Spencer Gang, Vladimir Lazetic, Robert Luallen, and Crystal Chhan for comments on the manuscript; and Michael David’s laboratory for maintenance of the NOVOstar plate reader. This work was supported by the National Institutes of Health under grants R01 AG-052622 and R01 GM-114139 to E.R.T.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.9898943.
Communicating editor: H. Bülow
Literature Cited
- Bakowski M. A., Desjardins C. A., Smelkinson M. G., Dunbar T. A., Lopez-Moyado I. F. et al. , 2014. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 10: e1004200 (erratum: PLoS Pathog. 10: e1004371). 10.1371/journal.ppat.1004200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerke M., 2014 Getting the most from your plate-based assays. Available at: http://www.promega.co.uk/resources/pubhub/getting-the-most-from-your-plate-based-assays/.
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock M., 2012. Application of bioluminescence imaging for in vivo monitoring of fungal infections. Int. J. Microbiol. 2012: 956794 10.1155/2012/956794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhove J. M. K. M., Karda R., Hawkins K. E., FitzPatrick L. M., Waddington S. N. et al. , 2017. Bioluminescence monitoring of promoter activity in vitro and in vivo. Methods Mol. Biol. 1651: 49–64. 10.1007/978-1-4939-7223-4_5 [DOI] [PubMed] [Google Scholar]
- De-Souza E. A., Camara H., Salgueiro W. G., Moro R. P., Knittel T. L. et al. , 2019. RNA interference may result in unexpected phenotypes in Caenorhabditis elegans. Nucleic Acids Res. 47: 3957–3969. 10.1093/nar/gkz154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin G. A., Ghanta K. S., Piscopo K. M., and Mello C. C., 2018. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics 210: 781–787. 10.1534/genetics.118.301532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons S. W., Klass M. R., and Hirsh D., 1979. Analysis of the constancy of DNA sequences during development and evolution of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 76: 1333–1337. 10.1073/pnas.76.3.1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- England C. G., Ehlerding E. B., and Cai W., 2016. NanoLuc: a small luciferase is brightening up the field of bioluminescence. Bioconjug. Chem. 27: 1175–1187. 10.1021/acs.bioconjchem.6b00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M. et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon D. B., Ishidate T., Li L., Gu W., Silverman N. et al. , 2017. The antiviral RNA interference response provides resistance to lethal arbovirus infection and vertical transmission in Caenorhabditis elegans. Curr. Biol. 27: 795–806. 10.1016/j.cub.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J., 2001. Selecting the Detection System - Colorimetric, Fluorescent, Luminescent Methods. Life Sci. (5) pp. 14.
- Hall M. P., Unch J., Binkowski B. F., Valley M. P., Butler B. L. et al. , 2012. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 7: 1848–1857. 10.1021/cb3002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagido C., Pettitt J., Flett A., and Glover L. A., 2008. Bridging the phenotypic gap: real-time assessment of mitochondrial function and metabolism of the nematode Caenorhabditis elegans. BMC Physiol. 8: 7 10.1186/1472-6793-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagido C., McLaggan D., and Glover L. A., 2015. A screenable in vivo assay for mitochondrial modulators using transgenic bioluminescent Caenorhabditis elegans. J. Vis. Exp. 105: e53083 10.3791/53083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., and Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall A. R., Tedesco P. M., Sands B., Johnson T. E., and Brent R., 2015. Single cell quantification of reporter gene expression in live adult Caenorhabditis elegans reveals reproducible cell-specific expression patterns and underlying biological variation. PLoS One 10: e0124289 10.1371/journal.pone.0124289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkina O., and Hunter C. P., 2018. Intergenerational transmission of gene regulatory information in Caenorhabditis elegans. Trends Genet. 34: 54–64. 10.1016/j.tig.2017.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita J. M., and Arias C. A., 2016. Mechanisms of antibiotic resistance. Microbiol. Spectr. 4. Available at: doi:10.1128/microbiolspec.VMBF-0016-2015. 10.1128/microbiolspec.VMBF-0016-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogoh K., Akiyoshi R., May-Maw-Thet T., Sugiyama T., Dosaka S. et al. , 2014. Bioluminescence microscopy using a short focal-length imaging lens. J. Microsc. 253: 191–197. 10.1111/jmi.12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Toyomura T., Honjo K., Wada Y., and Futai M., 2001. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J. Biol. Chem. 276: 33079–33085. 10.1074/jbc.M101652200 [DOI] [PubMed] [Google Scholar]
- Olmedo M., Geibel M., Artal-Sanz M., and Merrow M., 2015. A high-throughput method for the analysis of larval developmental phenotypes in Caenorhabditis elegans. Genetics 201: 443–448. 10.1534/genetics.115.179242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K., and Tavernarakis N., 2016. Intracellular assessment of ATP levels in Caenorhabditis elegans. Bio Protoc. 6: e22048. 10.21769/BioProtoc.2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K. C., Dror T., Underwood R. S., Osman G. A., Elder C. R. et al. , 2019. Antagonistic paralogs control a switch between growth and pathogen resistance in C. elegans. PLoS Pathog. 15: e1007528 10.1371/journal.ppat.1007528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A. W., and Troemel E. R., 2015. The development of genetic modification techniques in intracellular parasites and potential applications to microsporidia. PLoS Pathog. 11: e1005283 10.1371/journal.ppat.1005283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg P. J., and Grant B. D., 2013. C. elegans gene transformation by microparticle bombardment (December 30, 2013), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.166.1, http://www.wormbook.org. 10.1895/WORMBOOK.1.166.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuscher A. C., and Ewald C. Y., 2018. Overcoming autofluorescence to assess GFP expression during normal physiology and aging in Caenorhabditis elegans. Bio Protoc. 8: e2940 10.21769/BioProtoc.2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne N., Inglese J., and Auld D. S., 2010. Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem. Biol. 17: 646–657. 10.1016/j.chembiol.2010.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vávra J., and Lukeš J., 2013. Microsporidia and ‘the art of living together.’. Adv. Parasitol. 82: 253–319. 10.1016/B978-0-12-407706-5.00004-6 [DOI] [PubMed] [Google Scholar]
- Wohlstadter J. N., E. Glezer, J. Wilbur, G. Sigal, K. Johnson, et al., 2005 Assay Plates, Reader Systems And Methods For Luminescence Test Measurements. US7842246B. United States Patent No. 6,977,722.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.9898943.