Abstract
Longitudinal phenotypes have been increasingly available in genome-wide association studies (GWAS) and electronic health record-based studies for identification of genetic variants that influence complex traits over time. For longitudinal binary data, there remain significant challenges in gene mapping, including misspecification of the model for phenotype distribution due to ascertainment. Here, we propose L-BRAT (Longitudinal Binary-trait Retrospective Association Test), a retrospective, generalized estimating equation-based method for genetic association analysis of longitudinal binary outcomes. We also develop RGMMAT, a retrospective, generalized linear mixed model-based association test. Both tests are retrospective score approaches in which genotypes are treated as random conditional on phenotype and covariates. They allow both static and time-varying covariates to be included in the analysis. Through simulations, we illustrated that retrospective association tests are robust to ascertainment and other types of phenotype model misspecification, and gain power over previous association methods. We applied L-BRAT and RGMMAT to a genome-wide association analysis of repeated measures of cocaine use in a longitudinal cohort. Pathway analysis implicated association with opioid signaling and axonal guidance signaling pathways. Lastly, we replicated important pathways in an independent cocaine dependence case-control GWAS. Our results illustrate that L-BRAT is able to detect important loci and pathways in a genome scan and to provide insights into genetic architecture of cocaine use.
Keywords: ascertainment, genome-wide association studies, model misspecification, robustness, score test
GENOME-WIDE association studies (GWAS) have successfully discovered many disease susceptibility loci and provided insights into the genetic architecture of numerous human complex diseases and traits. In some genetic epidemiological studies, longitudinally collected phenotype data are available. This is the case for many electronic health record (EHR)-based studies. As many of these studies continue to follow enrolled subjects [e.g., the UK Biobank (UKB) and the Million Veteran Program (MVP)], longitudinal phenotypes will be increasingly available with the passage of time, providing new data resources that require appropriate analytical tools for optimal analysis. Standard association tests that consider one time point, or collapse repeated measurements into a single value such as an average, do not capture the trajectory of phenotypic traits over time, and may result in a loss of statistical power to detect genetic associations. In addition, the effects of time-varying covariates cannot be easily incorporated in such analyses. Recently, methodological developments for GWAS have proliferated to make full use of the available longitudinal data. For population cohorts, methods that account for dependence among observations from an individual include mixed effects models (Furlotte et al. 2012; Sikorska et al. 2013), generalized estimating equations (GEE) (Sitlani et al. 2015), growth mixture models (Das et al. 2011; Londono et al. 2013), and empirical Bayes models (Meirelles et al. 2013). Most of these approaches are prospective analyses and have been successfully applied to quantitative phenotypes.
As many diseases are rare, efficient designs, such as the case-control design, are commonly applied in epidemiological studies to recruit study subjects. Despite the enhanced efficiency in the study sample, nonrandom ascertainment can be a major source of model misspecification that may lead to inflated type I error and/or power loss in association analysis. The linear mixed model and the logistic mixed model do not perform well when the case-control ratio is unbalanced in large-scale genetic association studies (Zhou et al. 2018). Prospective analysis, in which a population-based model is used, ignores ascertainment bias and can result in compromised statistical inference. Furthermore, in the ascertained sample, the prospective approach conditional on the genotype and covariates may lose information when the joint distribution of the genotype and covariates carries additional information on whether the phenotype is associated with the genotype (Jiang et al. 2015). In this regard, several retrospective association methods have been proposed for analyzing ascertained population-based case-control studies (Hayeck et al. 2015; Jiang et al. 2016), family-based studies of continuous traits (Jakobsdottir and McPeek 2013), family-based case-control studies (Zhong et al. 2016; Hayeck et al. 2017), and family-based longitudinal quantitative traits (Wu and McPeek 2018). Compared to prospective tests, retrospective tests conditional on the phenotype and covariates are more robust to misspecification of the trait model (Jiang et al. 2015).
To generalize case-control sampling, outcome-dependent sampling designs have become popular for binary data in longitudinal cohort studies (Schildcrout and Heagerty 2008; Schildcrout et al. 2018a,b). However, association tests for longitudinally measured binary data are less well developed in GWAS. Here, we propose L-BRAT (Longitudinal Binary-trait Retrospective Association Test), a retrospective, GEE-based method for genetic association analysis of longitudinal binary outcomes. It requires specification of the mean of the outcome distribution and a working correlation matrix for repeated measurements. L-BRAT is a retrospective score approach in which genotypes are treated as random conditional on the phenotype and covariates. Thus, it is robust to ascertainment and trait model misspecification. It allows both static and time-varying covariates to be included in the analysis. We note that the Generalized linear Mixed Model Association Test (GMMAT), a recently proposed prospective test using the logistic mixed model to control for population structure and cryptic relatedness in case-control studies (Chen et al. 2016), can be adapted for repeated binary data. For comparison, we also develop RGMMAT, a retrospective, generalized linear mixed model (GLMM)-based association test for longitudinal binary traits.
We performed simulation studies to evaluate type I error and power of L-BRAT and RGMMAT, and compared them to the existing prospective methods. The results demonstrate that the retrospective association tests have better control of type I error when the phenotype model is misspecified, and are robust to various ascertainment schemes. Moreover, they are more powerful than the prospective tests. Finally, we applied L-BRAT and RGMMAT to a genome-wide association analysis of repeated measurements of cocaine use in a longitudinal cohort, the Veterans Aging Cohort Study (VACS), and replicated the results using data from an independent cocaine dependence case-control GWAS.
Materials and Methods
Suppose a binary trait is measured over time on a study population of individuals. We have their genome-wide measures of genetic variation. A set of covariates, static or dynamic, are also available. Let be the number of repeated measures on individual and be the total number of observations. For individual , let and be the -dimensional covariate vector, assumed to include an intercept, and the binary response at time , respectively. In this setting, individuals are permitted to have measurements at different time points and different number of observations. We let denote the outcome vector of length , and let denote the covariate matrix. Here, we focus on the problem of testing for association between a genetic variant and the longitudinal binary outcomes. Let denote the vector of genotypes for the individuals at the variant to be tested, where , 1, or 2 is the number of minor alleles of individual at the variant.
GEE model
We consider a GEE approach in which the mean of the outcome distribution, given the genotype and covariates, is specified as
| (1) |
where is a -dimensional vector of covariate effects and is a scalar parameter of interest representing the effect of the tested variant. Writing in a matrix form, we have the mean model
| (2) |
where is an matrix representing the measurement clustering structure, and its th entry is an indicator of the th entry of being a measurement on individual . Here, the vector is the vertically expanded genotype vector that maps the genotype data from the individual level to the measurement level. The covariance structure of is given by
| (3) |
where is an -dimensional diagonal matrix and is an correlation matrix. The covariance specification in Equation 3 ensures that the variance of the dichotomous response depends on its mean in a way that is consistent with the Bernoulli distribution. To apply the GEE method, a working correlation structure such as independent, exchangeable, and first-order autoregressive [AR(1)] must be specified. For a given within-cluster correlation matrix , which may depend on some parameter , the estimating equations for the unknown parameters are written as
Prospective GEE test
To detect association between the genetic variant and the phenotype, we consider a score approach to test against . The null estimate of , denoted by , is the solution to a system of estimating equations under the constraint , which can be computed iteratively between a Fisher scoring algorithm for and the method of moments for until convergence. Then, the score function for is
| (4) |
where , and are , and evaluated at .
In the GEE approach, the prospective score statistic for testing takes the form
| (5) |
where the null variance of is evaluated using a robust sandwich variance estimator, conditional on the genotype and covariates. Here , where and the sample covariance of , , is estimated by . Under the null hypothesis, the test statistic has an asymptotic distribution.
L-BRAT retrospective test
In what follows, we introduce a new GEE-based association testing method, L-BRAT. The L-BRAT test statistic is also based on the score function in Equation 4. In contrast to the prospective GEE score test, L-BRAT takes a retrospective approach in which the variance of is assessed using a retrospective model of the genotype given the phenotype and covariates. An advantage of the retrospective approach is that the analysis is less dependent on the correct specification of the phenotype model. We assume that, under the null hypothesis of no association between the genetic variant and the phenotype, the quasi-likelihood model of conditional on and is
| (6) |
where is the minor allele frequency (MAF) of the tested variant, is a vector of length with every element equal to 1, is an unknown variance parameter, and is an genetic relationship matrix (GRM) representing the overall genetic similarity between individuals due to population structure. Because , which is the first column of that encodes an intercept, and , the -dimensional vector of transformed null phenotypic residuals, is orthogonal to the column space of , then the null mean model of in Equation 6 ensures that
where is the individual-level transformed phenotypic residual vector of length .
In model (6), the GRM can be obtained using genome-wide data, given by
where is the total number of genotyped variants, is the genotype vector at the th variant, and is the estimated MAF. For example, , the sample MAF at the th variant. For the variant of interest, let be its sample MAF. Under Hardy-Weinberg equilibrium, the variance of the genotype is estimated by . Or we can use a more robust variance estimator (Jakobsdottir and McPeek 2013) given by
| (7) |
where . Finally, the L-BRAT test statistic can be defined as
| (8) |
Under regularity conditions, L-BRAT asymptotically follows a distribution under the null hypothesis.
Generalized linear mixed model
The GMMAT test was originally designed to use multiple random effects in logistic mixed models to account for complex sampling designs in case-control studies (Chen et al. 2016). To extend the GMMAT method for case-control analysis to repeated binary data, we consider the following logistic mixed model:
| (9) |
where is the probability of a binary response at time for individual , conditional on his/her genotype, covariates, and random effects and , and are the same as defined in model (1), is the individual random effect, and is the individual-specific time-dependent random effect. The two random effects were used to capture the correlation among repeated measures in gene-based association test for longitudinal traits (Wang et al. 2017). Here, values are assumed to be independent and . The vector of time-dependent random effects has a multivariate normal distribution, , where an AR(1) structure is assumed for the correlation matrix , in which is the unknown parameter. The binary responses are assumed to be independent, given the random effects and . In model (9), population structure in the longitudinal data setting can be controlled for by including another random effect to account for genetic relationships (Chen et al. 2016; Wu and McPeek 2018), or including top principal components (PCs) of the genotype data as additional covariates.
GMMAT test
To construct a score test for the null hypothesis vs. the alternative , we use the penalized quasi-likelihood method (Breslow and Clayton 1993) to fit the null logistic mixed model and obtain the null estimates of and , denoted by and (Chen et al. 2016). Similarly, the best linear unbiased predictor (BLUP) of random effects, and , can be obtained. Then, the resulting score function for is
| (10) |
where is a vector of fitted values under .
In GMMAT, the null variance of the score is evaluated prospectively (Chen et al. 2016), i.e., , where , and . Here and are and evaluated at , where is the same as defined in Equation 3 and is a block diagonal matrix. The GMMAT test statistic can be written as
| (11) |
RGMMAT retrospective test
Like L-BRAT, we can construct a retrospective test to assess the significance of the GLMM score function of Equation 10, which we call RGMMAT, based on the quasi-likelihood model of in Equation 6. Thus, we define the RGMMAT statistic by
| (12) |
where is the -dimensional vector of phenotypic residuals at the individual level by summing over all time points for an individual, and the phenotypic residuals are obtained by fitting the null logistic mixed model. Both the GMMAT and RGMMAT test statistics are assumed to have asymptotic null distributions.
Simulation studies
We performed simulation studies to evaluate the type I error and power of the two retrospective tests, and compared them to the prospective GEE and GMMAT methods. We also assessed sensitivity of L-BRAT and RGMMAT in the presence of model misspecification and ascertainment. In the simulations, we considered two different trait models and three different ascertainment schemes. Because both the L-BRAT and GEE methods require specification of a working correlation matrix, we implemented three working correlation structures: (1) independent, (2) AR(1), and (3) a mixture of exchangeable and AR(1).
To generate genotypes, we first simulated 10,000 chromosomes over a 1 Mb region using a coalescent model that mimics the linkage disequilibrium (LD) and recombination rates of the European population (Schaffner et al. 2005). We then randomly selected 1000 noncausal single nucleotide polymorphisms (SNPs) with MAF > 0.05. In addition, we generated two causal SNPs that were assumed to influence the trait value with epistasis. In the type I error simulations, we tested association at the 1000 noncausal SNPs. In each simulation setting, we generated 1000 sets of phenotypes at five time points. Putting these together, 106 replicates were used for the type I error evaluation. In the power simulations, we tested the first of the two causal SNPs, and empirical power was assessed using 1000 simulation replicates. In all tests considered, the genotypes at the untested causal SNP(s) were assumed to be unobserved.
Trait models
We simulated two types of binary trait models at five time points, in which the two unlinked causal SNPs were assumed to act on the phenotype epistatically. The first type is a logistic mixed model, given by
where is a continuous, time-varying covariate generated independently from a standard normal distribution, is a binary, time-invariant covariate taking values 0 or 1 with a probability of 0.5, and are the genotypes of individual at the two causal SNPs, is a scalar parameter encoding the effect of the causal SNPs, is an indicator function that takes value 1 when individual has at least one copy of the minor allele at both the causal SNPs, and are the individual-level time-independent and time-dependent random effects, respectively. Here we assume and , where is a correlation matrix specified by the AR(1) structure with a correlation coefficient . The two causal SNPs are assumed to be unlinked with MAFs 0.1 and 0.5, respectively. The variance components are set to and .
The second type of trait model we considered is a liability threshold model in which an underlying continuous liability determines the outcome value based on a threshold. Specifically, the phenotype is given by
where is the underlying liability for individual at time , and represents independent noise, with . All other parameters are the same as those in the logistic mixed model.
In both models, we included a time effect and assumed that the mean of the outcome increases with time. The effect of the causal SNPs was set to in the type I error simulations. For the power evaluation, we considered a range of values for , where we set , 0.32, 0.34, 0.36, and 0.38. At the given parameter values, the prevalence of the event of interest ranges from 12.8 to 27.7% over time. The proportion of the phenotypic variance explained by the two causal SNPs ranges from 0.69 to 1.10% in the logistic mixed model, and from 0.49 to 0.78% in the liability threshold model.
Sampling designs
We considered three different sampling designs. In the “random” sampling scheme, the sample contains 2000 individuals that were randomly selected from the population regardless of their phenotypes. Thus, ascertainment is population based. In the “baseline” sampling scheme, we sampled 1000 case subjects and 1000 control subjects according to their outcome value at baseline only. In the “sum” sampling scheme, individuals were stratified into three strata (1, 2, and 3) based on a total count that sums each subject’s response over time, where samples in stratum 1 never experienced the event of interest, i.e., , samples in stratum 2 sometimes experienced the event, i.e., , and samples in stratum 3 always experienced the event, i.e., . Following the outcome-dependent sampling design for longitudinal binary data (Schildcrout et al. 2018b), we selected 100, 1800, and 100 individuals from the three strata respectively to oversample subjects who have response variation over the course of the study.
Cocaine use data from VACS
We illustrated the utility of our proposed methods by analyzing a GWAS dataset of cocaine use from VACS (Justice et al. 2006). VACS is a multi-center, longitudinal observational study of HIV infected and uninfected veterans whose primary objective is to understand the risk of alcohol and other substance abuse in individuals with HIV infection. Our use of the VACS data were approved by the Yale Human Research Protection Program and the Institutional Review Board of the Veterans Affairs Connecticut Healthcare System. We analyzed longitudinal cocaine use in patient surveys collected at six clinic visits on 2470 participants. Among them, 69.8% are African Americans (AAs), 19.3% are European Americans (EAs), and 10.9% are of other races. We considered the responses at each visit as zero if individuals had never tried cocaine or had not used cocaine in the last year, and as one if individuals had used cocaine in the last year. The proportion of case subjects at each visit ranges from 13.7% to 24.3% , and the missing rate at each visit ranges from 3.0 to 44.2%.
All samples were genotyped on the Illumina OmniExpress BeadChip. After data cleaning, there are 2458 individuals available for genotype imputation. IMPUTE2 (Howie et al. 2009) was used for imputation using the 1000 Genomes Phase 3 data as a reference panel. We excluded subjects who did not meet either of the following criteria: (1) completeness (i.e., proportion of successfully imputed SNPs) > 95% and (2) empirical self-kinship < 0.525 (i.e., empirical inbreeding coefficient < 0.05). Based on the above criteria, 2231 individuals were retained in the analysis, with 2114 males and 117 females, of whom 1557 are AAs, 431 are EAs, and 243 are of other races. There are 1433 individuals who had never used cocaine during the study period, 639 individuals who sometimes used cocaine, i.e., exhibited response variation, and 159 individuals who had used cocaine at least once every year over the course of the study.
We performed a GWAS with longitudinally measured cocaine use in the entire VACS sample. SNPs that satisfied all of the following quality-control conditions were included in the analysis: (1) call rate > 95%, (2) Hardy-Weinberg statistic P-value > 10−6, and (3) MAF > 1%. All together we analyzed 10,215,072 SNPs using L-BRAT, RGMMAT, and the prospective GEE and GMMAT tests. Sex, age at baseline, HIV status, and time were included as covariates in the analysis. Because the VACS samples include AAs, EAs, and other races, the top 10 PCs were included as covariates in the analysis to control for population structure. In addition, we analyzed the data separately in each population, adjusted for the top 10 PCs obtained within the group, and then combined the results from the three groups by meta-analysis using the optimal weights for score statistics that have essentially the same power as the inverse variance weighting (Zhou et al. 2011).
To compare the performance of longitudinal association analysis with that of univariate analysis on the summary metrics of cocaine use in VACS, we considered two alternative cocaine phenotypes: baseline and trajectories. CARAT (Jiang et al. 2016), a case-control retrospective association test, was used to test for association with cocaine use at baseline, adjusted for sex, age at baseline, and HIV status. Longitudinal cocaine use trajectories were obtained using a growth mixture model that clusters longitudinal data into discrete growth trajectory curves (Muthén 2004). We fit a logistic model with a polynomial function of time. The number of groups was chosen based on the Bayesian information criterion (BIC). Once each individual was assigned to the trajectory with the highest probability of membership, we then performed association tests with the ordered cocaine use trajectory groups using a cumulative logit model. Sex, age at baseline, HIV status, and the top 10 PCs were included as covariates in the analysis.
Pathway and enrichment analyses
Pathway analysis was conducted on the association results for longitudinally measured cocaine use using the Ingenuity Pathway Analysis (IPA) software. The top SNPs with a P-value were annotated and evaluated to identify an overrepresentation of genes within defined canonical pathways based on information from multiple sources. The Ingenuity database contains information from manually reviewed literature and large public databases. The list of the top SNPs was mapped to the reference set in the Ingenuity knowledge. Then, Fisher’s exact test was used to determine whether the SNP list belongs to a gene set of a functional annotation more than expected by chance. Both the unadjusted P-value and adjusted P-value using the Benjamini-Hochberg method were reported. Pathways with the adjusted P-value <0.05 were considered to be significant. Enrichment analysis was also performed to assess whether the top association signals identified from the VACS data are more likely to regulate brain gene expression. Fisher’s exact test was used to test whether the associated SNPs with cocaine use is overrepresented in the brain expression quantitative trait loci (eQTLs) reported from the Genotype-Tissue Expression (GTEx) project (GTEx Consortium 2013, 2017).
Replication data
We used an independent cocaine dependence case-control GWAS from the Yale-Penn study (Gelernter et al. 2014) to replicate the top findings in VACS. The summary statistics from the Yale-Penn cocaine dependence GWAS were obtained. Pathway analysis using IPA was applied to the summary statistics of Yale-Penn on the top SNP list identified from VACS. The Fisher’s exact test P-values were calculated for each pathway to evaluate if there were more associated SNPs than would be expected by chance.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. An R package implementing the proposed methods is available at https://github.com/ZWang-Lab/LBRAT. Additional data analysis results of cocaine use from VACS are presented as supporting information including 1 table and 2 figures. Supplemental material available at figshare: https://doi.org/10.25386/genetics.9936881.
Results
Type I error assessment
To assess type I error, we simulated phenotype data at five time points under two trait models and three sampling designs, and tested for association at unlinked and unassociated SNPs. We compared the proportion of simulations in which the test statistic exceeded the th quantile of the distribution to the nominal type I error level , for , 0.01, 0.001, and 0.0001. Table 1 gives the empirical type I error of the L-BRAT, RGMMAT, GEE, and GMMAT tests, based on 106 replicates. For the GEE-based methods, three working correlation structures were considered: (1) independent, (2) AR(1), and (3) a mixture of exchangeable and AR(1). In all simulations, the type I error of the two retrospective tests, L-BRAT and RGMMAT, exhibited no inflation at any of the nominal levels considered. In contrast, the prospective GEE tests, regardless of the choice of working correlation, had inflated type I error at most of the nominal levels in all settings. This is likely due to the fact that the asymptotic distribution of robust sandwich variance estimators used in GEE are not well calibrated. The inflated type I error was also reported in longitudinal GWAS with quantitative traits using GEE (Sitlani et al. 2015). In GMMAT, the type I error was much lower than the nominal level when , 0.01, 0.001, and 0.0001. These results demonstrate that the two retrospective tests, L-BRAT and RGMMAT, are robust to trait model misspecification and ascertainment, whereas GEE has type I error inflation and GMMAT is overly conservative. Overall, the choice of the working correlation matrix does not have much impact on the type I error of the L-BRAT method.
Table 1. Empirical type I error of L-BRAT, RGMMAT, GEE, and GMMAT, based on 106 replicates.
| Analysis type | Test | Nominal level | Logistic mixed model | Liability threshold model | ||||
|---|---|---|---|---|---|---|---|---|
| Random | Baseline | Sum | Random | Baseline | Sum | |||
| Prospective | GEE(ind) | 0.05 | 5.38 × 10−2 | 5.08 × 10−2 | 5.27 × 10−2 | 5.36 × 10−2 | 5.19 × 10−2 | 5.38 × 10−2 |
| 0.01 | 1.18 × 10−2 | 1.04 × 10−2 | 1.13 × 10−2 | 1.17 × 10−2 | 1.07 × 10−2 | 1.17 × 10−2 | ||
| 0.001 | 1.32 × 10−3 | 1.16 × 10−3 | 1.23 × 10−3 | 1.37 × 10−3 | 1.14 × 10−3 | 1.37 × 10−3 | ||
| 0.0001 | 1.67 × 10−4 | 1.28 × 10−4 | 1.43 × 10−4 | 1.34 × 10−4 | 1.36 × 10−4 | 1.76 × 10−4 | ||
| GEE(AR1) | 0.05 | 5.36 × 10−2 | 5.02 × 10−2 | 5.26 × 10−2 | 5.34 × 10−2 | 5.17 × 10−2 | 5.37 × 10−2 | |
| 0.01 | 1.16 × 10−2 | 1.04 × 10−2 | 1.12 × 10−2 | 1.16 × 10−2 | 1.06 × 10−2 | 1.17 × 10−2 | ||
| 0.001 | 1.31 × 10−3 | 1.13 × 10−3 | 1.21 × 10−3 | 1.36 × 10−3 | 1.14 × 10−3 | 1.36 × 10−3 | ||
| 0.0001 | 1.73 × 10−4 | 1.19 × 10−4 | 1.37 × 10−4 | 1.32 × 10−4 | 1.35 × 10−4 | 1.78 × 10−4 | ||
| GEE(mix) | 0.05 | 5.34 × 10−2 | 5.07 × 10−2 | 5.26 × 10−2 | 5.34 × 10−2 | 5.19 × 10−2 | 5.37 × 10−2 | |
| 0.01 | 1.17 × 10−2 | 1.04 × 10−2 | 1.13 × 10−2 | 1.16 × 10−2 | 1.07 × 10−2 | 1.17 × 10−2 | ||
| 0.001 | 1.29 × 10−3 | 1.17 × 10−3 | 1.22 × 10−3 | 1.38 × 10−3 | 1.14 × 10−3 | 1.36 × 10−3 | ||
| 0.0001 | 1.70 × 10−4 | 1.29 × 10−4 | 1.37 × 10−4 | 1.31 × 10−4 | 1.30 × 10−4 | 1.70 × 10−4 | ||
| GMMAT | 0.05 | 3.89 × 10−2 | 3.53 × 10−2 | 4.76 × 10−2 | 4.80 × 10−2 | 4.89 × 10−2 | 4.91 × 10−2 | |
| 0.01 | 6.07 × 10−3 | 5.24 × 10−3 | 9.08 × 10−3 | 9.29 × 10−3 | 9.51 × 10−3 | 9.33 × 10−3 | ||
| 0.001 | 4.29 × 10−4 | 3.74 × 10−4 | 7.84 × 10−4 | 8.63 × 10−4 | 8.96 × 10−4 | 8.33 × 10−4 | ||
| 0.0001 | 2.20 × 10−5 | 2.20 × 10−5 | 6.80 × 10−5 | 6.30 × 10−5 | 9.10 × 10−5 | 8.80 × 10−5 | ||
| Retrospective | L-BRAT(ind) | 0.05 | 4.93 × 10−2 | 4.91 × 10−2 | 4.98 × 10−2 | 5.01 × 10−2 | 4.99 × 10−2 | 4.98 × 10−2 |
| 0.01 | 9.45 × 10−3 | 9.60 × 10−3 | 9.84 × 10−3 | 9.90 × 10−3 | 9.75 × 10−3 | 9.55 × 10−3 | ||
| 0.001 | 8.30 × 10−4 | 9.78 × 10−4 | 9.24 × 10−4 | 9.55 × 10−4 | 9.45 × 10−4 | 8.78 × 10−4 | ||
| 0.0001 | 7.20 × 10−5 | 9.50 × 10−5 | 8.20 × 10−5 | 8.20 × 10−5 | 9.40 × 10−5 | 9.20 × 10−5 | ||
| L-BRAT(AR1) | 0.05 | 4.93 × 10−2 | 4.88 × 10−2 | 4.97 × 10−2 | 4.99 × 10−2 | 4.98 × 10−2 | 4.97 × 10−2 | |
| 0.01 | 9.48 × 10−3 | 9.72 × 10−3 | 9.78 × 10−3 | 9.84 × 10−3 | 9.76 × 10−3 | 9.55 × 10−3 | ||
| 0.001 | 8.26 × 10−4 | 9.62 × 10−4 | 9.22 × 10−4 | 9.17 × 10−4 | 9.47 × 10−4 | 8.48 × 10−4 | ||
| 0.0001 | 8.80 × 10−5 | 9.60 × 10−5 | 8.20 × 10−5 | 7.10 × 10−5 | 1.02 × 10−4 | 8.90 × 10−5 | ||
| L-BRAT(mix) | 0.05 | 4.93 × 10−2 | 4.91 × 10−2 | 4.99 × 10−2 | 5.01 × 10−2 | 4.98 × 10−2 | 4.98 × 10−2 | |
| 0.01 | 9.57 × 10−3 | 9.61 × 10−3 | 9.86 × 10−3 | 9.88 × 10−3 | 9.79 × 10−3 | 9.54 × 10−3 | ||
| 0.001 | 8.35 × 10−4 | 9.86 × 10−4 | 9.26 × 10−4 | 9.57 × 10−4 | 9.37 × 10−4 | 8.78 × 10−4 | ||
| 0.0001 | 8.20 × 10−5 | 1.01 × 10−4 | 8.60 × 10−5 | 7.40 × 10−5 | 9.70 × 10−5 | 8.90 × 10−5 | ||
| RGMMAT | 0.05 | 4.72 × 10−2 | 4.91 × 10−2 | 4.98 × 10−2 | 4.93 × 10−2 | 4.99 × 10−2 | 4.98 × 10−2 | |
| 0.01 | 8.76 × 10−3 | 9.64 × 10−3 | 9.85 × 10−3 | 9.63 × 10−3 | 9.78 × 10−3 | 9.55 × 10−3 | ||
| 0.001 | 7.20 × 10−4 | 9.52 × 10−4 | 9.09 × 10−4 | 9.12 × 10−4 | 9.43 × 10−4 | 8.75 × 10−4 | ||
| 0.0001 | 6.80 × 10−5 | 8.90 × 10−5 | 8.20 × 10−5 | 7.70 × 10−5 | 9.10 × 10−5 | 9.30 × 10−5 | ||
Rates that are significantly larger than the nominal levels are in bold. Texts in the brackets following test statistics denote the working correlation structure. Specifically, L-BRAT(ind) and GEE(ind) denote the L-BRAT and GEE tests with an independent working correlation; L-BRAT(AR1) and GEE(AR1) denote the L-BRAT and GEE tests with an AR(1) working correlation; L-BRAT(mix) and GEE(mix) denote the L-BRAT and GEE tests with a mixture of exchangeable and AR(1) working correlation structure.
Power comparison
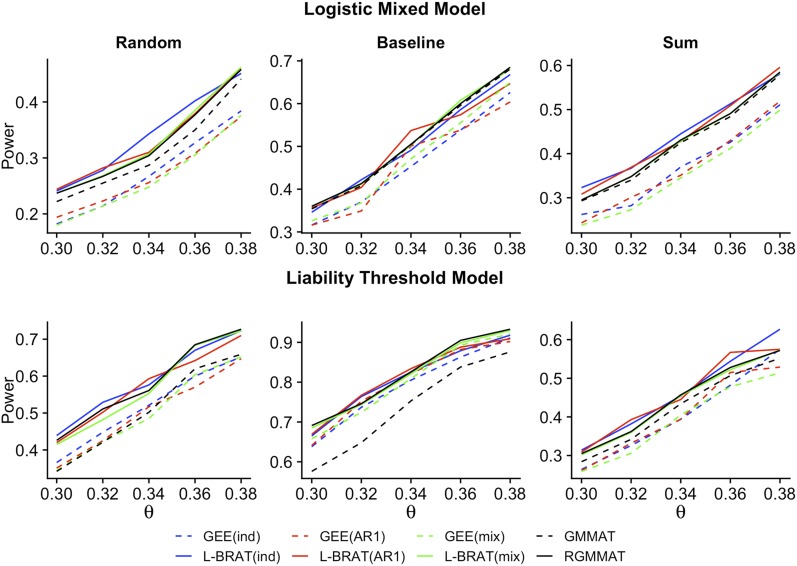
To compare the power of the methods, we simulated phenotype data at five time points under two types of trait models and three sampling designs. In each type of trait model, we considered five effect sizes at the two causal SNPs, and tested association between the trait and the first causal SNP. Empirical power was calculated at the significance level 10−3, based on 1000 simulated replicates. Figure 1 demonstrates the power results for each method. In all the simulation settings, the retrospective tests consistently had higher power than the prospective tests. The L-BRAT association tests under three different working correlation structures had similar power. The RGMMAT method also achieved high power. In contrast, the prospective GEE methods had the lowest power in all settings except under the baseline sampling and the liability threshold model, in which GMMAT performed the worst in power. Overall, we found that the baseline sampling scheme generated the highest power under different trait models, while the sum sampling scheme had a power gain over the random sampling scheme under the logistic mixed model, but was less powerful under the liability threshold model. These results suggest that L-BRAT and RGMMAT outperform the prospective tests, and the power of L-BRAT is not sensitive to the choice of the working correlation structure.
Figure 1.
Empirical power of L-BRAT, RGMMAT, GEE, and GMMAT. Power is based on 1000 simulated replicates at five time points with α = 10−3. In the upper panel, the trait is simulated by the logistic mixed model; in the lower panel, it is by the liability threshold model. Power results are demonstrated in samples of 2000 individuals according to three different ascertainment schemes: random, baseline, and sum. This figure appears in color in the electronic version of this article.
Analysis of cocaine use data from VACS
Genome-wide association testing for longitudinal cocaine use was performed on 10,215,072 SNPs in a total of the 2231 VACS samples including AAs, EAs, and other races, using L-BRAT, RGMMAT, GEE, and GMMAT, with adjustment for sex, age at baseline, HIV status, and time. To control for population structure, the top 10 PCs that explained 89.4% of the total genetic variation were included as covariates in the analysis. We considered two working correlation structures: independent and AR(1). For the L-BRAT and RGMMAT methods, the GRM was calculated using the LD pruned SNPs with MAF > 0.05.
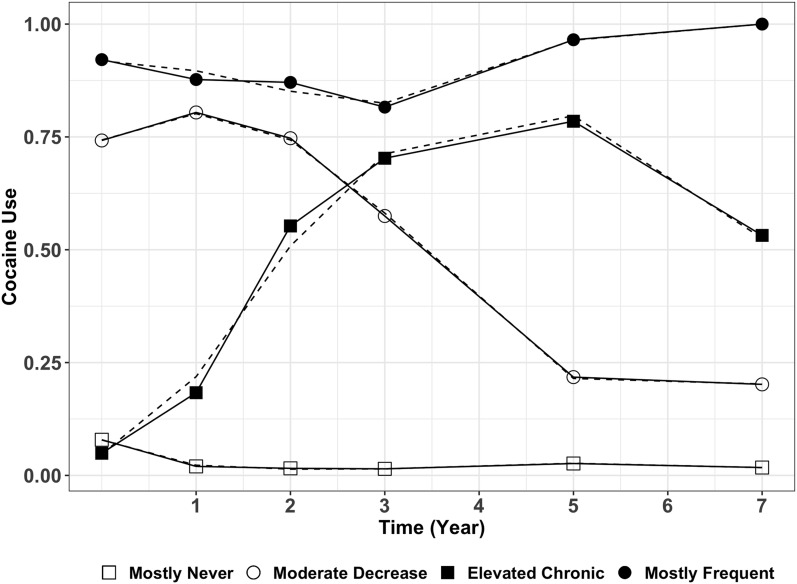
For comparison, we created two alternative summary characterizations of cocaine use: baseline and trajectories. Figure 2 shows the four cocaine use trajectory groups identified in the VACS sample. They were labeled as mostly never (0, ), moderate decrease (1, ), elevated chronic (2, ), and mostly frequent (3, ). We used CARAT for the analysis of cocaine use at baseline, adjusted for sex, age at baseline, and HIV status. Cumulative logit model was used to test for association between the four ordered cocaine use trajectory groups and each of the SNPs, with adjustment for sex, age at baseline, HIV status, and the top 10 PCs.
Figure 2.
Group-based cocaine use trajectories in VACS. Dashed lines represent the estimated trajectories, solid lines represent the observed mean cocaine use for each trajectory group. Time is the number of years since the baseline visit.
None of the retrospective tests exhibited evidence of inflation in the quantile-quantile (Q-Q) plot (Supplemental Material, Figure S1). The genomic control inflation factors were 0.993 and 0.991 for the L-BRAT genome scan under the independent and AR(1) working correlation, respectively, and 0.984 for the RGMMAT analysis. The prospective GEE tests showed some evidence of deflation in the Q-Q plot. The genomic control factors were 0.938 and 0.937 for the GEE tests under the independent and AR(1) working correlation. The most conservative test was GMMAT, with a genomic control factor 0.802.
Table 2 reports the results for SNPs for which at least one of the longitudinal tests gives a P-value . Among them, the L-BRAT tests produced the smallest P-values, RGMMAT and the trajectory-based analysis had comparable results, while GEE, GMMAT, and CARAT generated much larger P-values. The Manhattan plot of the smallest P-value from these tests in the VACS cocaine use data are shown in Figure S2. Among the top SNPs listed in Table 2, there are two SNPs, rs551879660 and rs150191017 ( and ), located at 3p12 and 13q12, respectively. Each of these SNPs was reported to have MAF < 1% in the 1000 Genomes (MAF = 0.68% and 0.98%, respectively). The MAFs of the two SNPs were 1.2% and 1.1% in the entire VACS sample, respectively, and were slightly higher in the AA sample (MAF = 1.6% and 1.5%, respectively). Although both SNPs have MAF > 1%, given the small sample size of VACS, there is limited information on them. SNP rs150191017 is located 31.5 kb from the gene AL161616.2, which was reported to be associated with venlafaxine treatment response in a generalized anxiety disorder GWAS (Jung et al. 2017). A cluster of five SNPs in complete LD (r2 = 1), rs76386683, rs114386843, rs186274502, rs376616438, and rs187855416, located at 9q33, showed association with longitudinal cocaine use . They are near OR1L4, an olfactory receptor gene that was reported to be associated with major depressive disorder (Wong et al. 2017). A cluster of olfactory receptor genes between OR3A1 and OR3A2 that belong to the olfactory receptor gene family were identified in a recent GWAS of cocaine dependence and related traits (Gelernter et al. 2014). The other three SNPs, rs188222191, rs1014278, and rs75132056, are located at 5q21 (, and , respectively), close to the gene EFNA5, which was identified in several GWAS to be associated with bipolar disorder and schizophrenia (Wang et al. 2010). There was also evidence of association with rs114629793 . This SNP is in an intron of the gene encoding PSD3, located at 8p22. Recently, two schizophrenia GWAS have identified association between PSD3 and schizophrenia (Goes et al. 2015; Li et al. 2017b), and one study has shown that PSD3 is associated with paliperidone treatment response in schizophrenic patients (Li et al. 2017a). Gene network analysis revealed that PSD3 is one of the differentially expressed hub genes that involve dysfunction of brain reward circuitry in cocaine use disorder (Ribeiro et al. 2017).
Table 2. SNPs with P-value < 2 × 10−7 in at least one of the longitudinal tests in the entire VACS sample.
| Chr | Gene Region | SNP | Position | MAF | GEE (ind) | GEE (AR1) | GMMAT | L-BRAT (ind) | L-BRAT (AR1) | RGMMAT | CARATa(BL) | CLb(traj) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | NIPA2P2 | rs551879660 | 75,146,492 | 0.012 | 1.87 × 10−4 | 7.14 × 10−4 | 9.07 × 10−4 | 2.00 × 10−8 | 3.19 × 10−6 | 4.13 × 10−5 | 5.78 × 10−4 | 3.35 × 10−5 |
| 5 | EFNA5 | rs188222191 | 105,411,547 | 0.042 | 6.86 × 10−6 | 1.65 × 10−5 | 8.87 × 10−5 | 1.28 × 10−7 | 4.17 × 10−7 | 2.69 × 10−6 | 8.95 × 10−5 | 2.72 × 10−5 |
| rs1014278 | 105,471,506 | 0.057 | 1.02 × 10−5 | 1.10 × 10−5 | 1.24 × 10−4 | 1.50 × 10−7 | 1.43 × 10−7 | 4.88 × 10−6 | 5.94 × 10−5 | 3.00 × 10−5 | ||
| rs75132056 | 105,480,442 | 0.05 | 1.05 × 10−5 | 2.42 × 10−5 | 1.89 × 10−4 | 8.92 × 10−8 | 2.89 × 10−7 | 8.55 × 10−6 | 2.59 × 10−4 | 2.31 × 10−5 | ||
| 8 | PSD3 | rs114629793 | 18,403,754 | 0.012 | 3.12 × 10−4 | 4.73 × 10−4 | 1.44 × 10−4 | 8.65 × 10−8 | 3.60 × 10−7 | 2.82 × 10−6 | 5.12 × 10−4 | 3.06 × 10−6 |
| 9 | OR1L4 | rs76386683 | 125,467,023 | 0.012 | 1.48 × 10−4 | 9.15 × 10−5 | 2.86 × 10−4 | 1.03 × 10−6 | 1.93 × 10−7 | 5.92 × 10−6 | 4.80 × 10−4 | 3.30 × 10−6 |
| rs114386843 | 125,469,425 | 0.012 | 1.47 × 10−4 | 9.05 × 10−5 | 2.82 × 10−4 | 1.01 × 10−6 | 1.88 × 10−7 | 5.78 × 10−6 | 4.75 × 10−4 | 3.22 × 10−6 | ||
| rs186274502 | 125,471,416 | 0.012 | 1.47 × 10−4 | 9.05 × 10−5 | 2.82 × 10−4 | 1.01 × 10−6 | 1.88 × 10−7 | 5.78 × 10−6 | 4.75 × 10−4 | 3.22 × 10−6 | ||
| rs376616438 | 125,472,267 | 0.012 | 1.44 × 10−4 | 8.95 × 10−5 | 2.77 × 10−4 | 9.79 × 10−7 | 1.85 × 10−7 | 5.62 × 10−6 | 4.79 × 10−4 | 3.20 × 10−6 | ||
| rs187855416 | 125,474,459 | 0.012 | 1.44 × 10−4 | 8.95 × 10−5 | 2.77 × 10−4 | 9.79 × 10−7 | 1.85 × 10−7 | 5.62 × 10−6 | 4.79 × 10−4 | 3.20 × 10−6 | ||
| 11 | AP000851.1 | rs139780693 | 102,509,700 | 0.03 | 2.60 × 10−5 | 1.04 × 10−5 | 2.78 × 10−4 | 5.83 × 10−7 | 1.26 × 10−7 | 1.35 × 10−5 | 1.06 × 10−4 | 2.00 × 10−6 |
| 13 | AL161616.2 | rs150191017 | 31,962,649 | 0.011 | 4.26 × 10−5 | 9.72 × 10−5 | 7.32 × 10−5 | 3.77 × 10−8 | 3.09 × 10−7 | 7.87 × 10−7 | 3.74 × 10−4 | 5.48 × 10−7 |
The smallest P-value among all tests at the given SNPs are in bold.
CARAT applied to cocaine use at baseline.
Cumulative logit model applied to the four ordered cocaine use trajectory group.
We further performed separate analyses by population group. Table S1 gives the results in the 1557 AA samples. All the top 12 SNPs listed in Table 2 had a P-value in at least one of the longitudinal tests in AAs. L-BRAT consistently gave the smallest P-values among all the longitudinal tests. The results from the three groups (AAs, EAs, and other races) were combined by meta-analysis. The meta-analysis P-values were of the same order of magnitude as that obtained from the entire sample adjusted for population structure for each longitudinal test (Table 3). All the top 12 SNPs listed in Table 2 had a meta-analysis P-value in at least one of the longitudinal tests. Among them, the L-BRAT test with either an independent or AR(1) working correlation gave the smallest meta-analysis P-values.
Table 3. Meta-analysis results of the top 12 SNPs from Table 2 in the VACS data.
| Chr | Gene Region | SNP | Position | GEE (ind) | GEE (AR1) | GMMAT | L-BRAT (ind) | L-BRAT (AR1) | RGMMAT |
|---|---|---|---|---|---|---|---|---|---|
| 3 | NIPA2P2 | rs551879660 | 75,146,492 | 1.81 × 10−4 | 5.86 × 10−4 | 8.98 × 10−4 | 5.26 × 10−8 | 6.41 × 10−6 | 6.49 × 10−5 |
| 5 | EFNA5 | rs188222191 | 105,411,547 | 7.57 × 10−6 | 1.28 × 10−5 | 1.80 × 10−4 | 2.55 × 10−7 | 5.52 × 10−7 | 1.10 × 10−5 |
| rs1014278 | 105,471,506 | 1.26 × 10−5 | 8.44 × 10−6 | 3.15 × 10−4 | 1.03 × 10−6 | 5.59 × 10−7 | 2.44 × 10−5 | ||
| rs75132056 | 105,480,442 | 1.31 × 10−5 | 2.00 × 10−5 | 4.24 × 10−4 | 7.31 × 10−7 | 1.27 × 10−6 | 3.56 × 10−5 | ||
| 8 | PSD3 | rs114629793 | 18,403,754 | 2.92 × 10−4 | 4.31 × 10−4 | 1.66 × 10−4 | 1.79 × 10−7 | 7.98 × 10−7 | 6.83 × 10−6 |
| 9 | OR1L4 | rs76386683 | 125,467,023 | 1.44 × 10−4 | 8.78 × 10−5 | 3.75 × 10−4 | 2.32 × 10−6 | 5.12 × 10−7 | 1.46 × 10−5 |
| rs114386843 | 125,469,425 | 1.42 × 10−4 | 8.62 × 10−5 | 3.68 × 10−4 | 2.25 × 10−6 | 4.97 × 10−7 | 1.41 × 10−5 | ||
| rs186274502 | 125,471,416 | 1.42 × 10−4 | 8.62 × 10−5 | 3.68 × 10−4 | 2.25 × 10−6 | 4.97 × 10−7 | 1.41 × 10−5 | ||
| rs376616438 | 125,472,267 | 1.39 × 10−4 | 8.51 × 10−5 | 3.60 × 10−4 | 2.18 × 10−6 | 4.86 × 10−7 | 1.37 × 10−5 | ||
| rs187855416 | 125,474,459 | 1.39 × 10−4 | 8.51 × 10−5 | 3.60 × 10−4 | 2.18 × 10−6 | 4.86 × 10−7 | 1.37 × 10−5 | ||
| 11 | AP000851.1 | rs139780693 | 102,509,700 | 1.15 × 10−5 | 4.16 × 10−6 | 1.07 × 10−4 | 4.04 × 10−7 | 6.05 × 10−8 | 4.41 × 10−6 |
| 13 | AL161616.2 | rs150191017 | 31,962,649 | 3.55 × 10−5 | 6.77 × 10−5 | 1.26 × 10−4 | 6.68 × 10−8 | 5.80 × 10−7 | 3.12 × 10−6 |
The smallest P-value among all tests at the given SNPs are in bold.
Pathway and enrichment analysis results
We then performed pathway analysis on the top SNPs for which at least one of the longitudinal tests had a P-value using IPA. We identified two significant canonical pathways that belong to the neurotransmitters and nervous system signaling. The first one is the opioid signaling pathway (, adjusted P = 0.010), which plays an important role in opioid tolerance and dependence. Studies have shown that chronic administration of cocaine and opioids are associated with changes in dopamine transporters and opioid receptors in various brain regions (Le Merrer et al. 2009; Soderman and Unterwald 2009). The second significant pathway is the axonal guidance signaling pathway (, adjusted P = 0.012), which is critical for neural development. The mRNA expression levels of axon guidance molecules have been found to be altered in some brain regions of cocaine-treated rats, which may contribute to drug abuse-associated cognitive impairment (Bahi and Dreyer 2005; Jassen et al. 2006). Each of the two pathways remained significant when we performed pathway analysis, using the same P-value cutoff value to select top SNPs, based on the L-BRAT results generated under the independence and AR(1) working correlation, respectively. In contrast, only the opioid signaling pathway was significant based on the results from the GEE analysis using the independent working correlation, and only the axonal guidance signaling pathway was significant based on the RGMMAT results, whereas neither of them remained significant based on the GMMAT results and that from the GEE analysis with an AR(1) working correlation. These results demonstrate that L-BRAT provides more informative association results to help identify biological relevant pathways.
Lastly, we performed an enrichment analysis to see whether the top SNPs in our analysis are more likely to regulate brain gene expression. We considered the cis-eQTLs reported in 13 human brain regions from the GTEx project (GTEx Consortium 2013, 2017), including amygdala, anterior cingulate cortex, caudate, cerebellar hemisphere, cerebellum, cortex, frontal cortex, hippocampus, hypothalamus, nucleus accumbens, putamen, spinal cord, and substantia nigra. Fisher’s exact test was used to assess the enrichment of eQTLs (FDR < 0.05) in the top 2778 SNPs for which at least one of the longitudinal tests had a P-value in the VACS sample. Among the 13 brain regions, amygdala is the only region in which eQTLs showed significant enrichment in our top SNP list (odds ratio = 2.06, ).
Replication of top findings
We used an independent cocaine dependence case-control GWAS from the Yale-Penn study (Gelernter et al. 2014) to replicate the top findings from our longitudinal analysis results in VACS. Note that the lifetime cocaine dependence diagnosis was made using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) (Pierucci-Lagha et al. 2005), which is different from the outcome used in VACS, and there were no longitudinal phenotype measures in Yale-Penn. Nevertheless, we performed pathway analysis using the SNP summary statistics of Yale-Penn to replicate the two pathways identified in the VACS sample. Among the top 2778 SNPs for which at least one of the longitudinal tests had a P-value , we were able to retrieve 2602 SNP summary statistics from Yale-Penn. Pathway analysis was conducted on the top 84 SNPs that had a P-value < 0.05. Although none of the top 12 SNPs in Table 2 had a P-value < 0.05 in the Yale-Penn AA sample, each of the two pathways remained significant: the opioid signaling pathway (, adjusted ) and the axonal guidance signaling pathway (, adjusted ).
Computation time
We implemented all four tests in an R software called LBRAT in which the robust variance estimator of Equation 7 was used in the two retrospective tests: L-BRAT and RGMMAT. The computational burden of the retrospective tests comes mainly from the eigendecomposition of the GRM in calculating the retrospective variance of the score functions. However, its impact on run time is minimal because the decomposition needs to be done only once per genome scan. When fitting the null models, the GLMM-based methods require extra time to obtain the estimates of random effects compared to the GEE-based methods. Once the null model is obtained, the transformed phenotypic residual vector, , in L-BRAT and the phenotypic residual vector, , in RGMMAT, need to be calculated just once per genome scan. Thus, the computational cost of the variance in the retrospective tests is much less than that in the prospective tests. We reported some example run times for analysis of simulated and real data. For a simulated dataset of phenotypes at five time points on 2000 individuals, the GEE-based methods took 0.9 sec while the GLMM-based methods took 37 sec to fit the null model. Overall, L-BRAT took 2.4 sec and GEE took 27.7 sec to analyze 1000 SNPs using a single processor on an Intel Xeon 2.6 GHz CPU machine. In the analysis of the VACS cocaine use data, L-BRAT and GEE took 1 sec, while RGMMAT and GMMAT took 2.5 min to fit the null model. Overall, L-BRAT, RGMMAT, GEE, and GMMAT took 0.8, 0.7, 24.8, and 26.2 hr, respectively, to analyze a total of 10,215,072 genome-wide SNPs on Intel Xeon 2.6 GHz CPU computing clusters with 22 nodes. These results demonstrate that L-BRAT and RGMMAT are computationally feasible for large-scale whole-genome association studies.
Discussion
Longitudinal data can be used in GWAS to improve power for identification of genetic variants and environmental factors that influence complex traits over time. In this study, we developed L-BRAT, a retrospective association testing method for longitudinal binary outcomes. L-BRAT is based on GEE, thus it requires assumptions on the mean but not the full distribution of the outcome. Correct specification of the covariance of repeated measurements within each individual is not required, instead, a working covariance matrix is assumed. The significance of the L-BRAT association test is assessed retrospectively by considering the conditional distribution of the genotype at the variant of interest, given phenotype and covariate information, under the null hypothesis of no association. Features of L-BRAT include the following: (1) it is computationally feasible for genetic studies with millions of variants, (2) it allows both static and time-varying covariates to be included in the analysis, (3) it allows different individuals to have measurements at different time points, and (4) it has correct type I error in the presence of ascertainment and trait model misspecification. For comparison, we also propose a retrospective, logistic mixed model-based association test, RGMMAT, which requires specification of the full distribution of the outcome. Random effects are used to model dependence among observations for an individual. Like L-BRAT, RGMMAT is a retrospective analysis in which genotypes are treated as random conditional on the phenotype and covariates. As a result, RGMMAT is also robust to misspecification of the model for the phenotype distribution.
Through simulation, we demonstrated that the type I error of L-BRAT was well calibrated under different trait models and ascertainment schemes, whereas the type I error of the prospective GEE method was inflated relative to nominal levels. In the GLMM-based methods, GMMAT, a prospective analysis, was overly conservative, whereas the retrospective version, RGMMAT, was able to maintain correct type I error. We further demonstrated that the two retrospective tests, L-BRAT and RGMMAT, provided higher power to detect association than the prospective GEE and GMMAT tests under all the trait models and ascertainment schemes considered in the simulations. The choice of the working correlation matrix in L-BRAT resulted in little loss of power. We applied L-BRAT and RGMMAT to longitudinal association analysis of cocaine use in the VACS data, where we identified six novel genes that are associated with cocaine use. Moreover, our pathway analysis identified two significant pathways associated with longitudinal cocaine use: the opioid signaling pathway and the axonal guidance signaling pathway. We were able to replicate both pathways in a cocaine dependence case-control GWAS from the Yale-Penn study. Lastly, we illustrated that the top SNPs identified by our methods are more likely to be the amygdala eQTLs in the GTEx data. The amygdala plays an important role in the processing of memory, decision-making, and emotional responses, and contributes to drug craving that leads to addiction and relapse (Hyman and Malenka 2001; Warlow et al. 2017). These findings verify that L-BRAT is able to detect important loci in a genome scan and to provide novel insights into the disease mechanism in relevant tissues. For repeated binary data, L-BRAT was more robust to trait model misspecification and ascertainment, and has comparable or higher power than RGMMAT in all simulation settings. In the real data analysis, L-BRAT generated smaller P-values on the top SNPs while the QQ plot of L-BRAT did not show any inflation of type I error. Therefore, we recommend L-BRAT when only one test is used for longitudinal binary data.
In this study, both the L-BRAT and RGMMAT methods were developed for population samples. When samples contain related individuals, we can extend L-BRAT and RGMMAT by including an extra variance component in the GEE model or an additional random effect in the GLMM model to account for genetic relationships. As a result, the GRM will appear in both the null model and the score test. The L-BRAT and RGMMAT methods are designed for single-variant association analysis of longitudinally measured binary outcomes. However, single-variant association tests in general have limited power to detect association for low-frequency or rare variants in sequencing studies. We have previously developed longitudinal burden test and sequence kernel association test, LBT and LSKAT, to analyze rare variants with longitudinal quantitative phenotypes (Wang et al. 2017). Both tests are based on a prospective approach. To extend L-BRAT and RGMMAT to rare variant analysis with longitudinal binary data, we could consider either a linear statistic or a quadratic statistic that combines the retrospective score test at each variant in a gene region. In addition, the genetic effect in L-BRAT and RGMMAT is assumed to be constant. We could consider an extension to allow for time-varying genetic effect so that the fluctuation of genetic contributions to the trait value over time is well calibrated.
Acknowledgments
The authors appreciate the support of the Veterans Aging Cohort Study and Yale Center for Genome Analysis. This work was supported by National Institutes of Health grants K01AA023321 and R21AA022870, and National Science Foundation grant DMS1916246. The authors also thank two anonymous reviewers for thoughtful and constructive comments to improve the manuscript.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.9936881.
Communicating editor: E. Hauser
Literature Cited
- Bahi A., and Dreyer J.-L., 2005. Cocaine-induced expression changes of axon guidance molecules in the adult rat brain. Mol. Cell. Neurosci. 28: 275–291. 10.1016/j.mcn.2004.09.011 [DOI] [PubMed] [Google Scholar]
- Breslow N. E., and Clayton D. G., 1993. Approximate inference in generalized linear mixed models. J. Am. Stat. Assoc. 88: 9–25. [Google Scholar]
- Chen H., Wang C., Conomos M. P., Stilp A. M., Li Z. et al. , 2016. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am. J. Hum. Genet. 98: 653–666. 10.1016/j.ajhg.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K., Li J., Wang Z., Tong C., Fu G. et al. , 2011. A dynamic model for genome-wide association studies. Hum. Genet. 129: 629–639. 10.1007/s00439-011-0960-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlotte N. A., Eskin E., and Eyheramendy S., 2012. Genome-wide association mapping with longitudinal data. Genet. Epidemiol. 36: 463–471. 10.1002/gepi.21640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J., Sherva R., Koesterer R., Almasy L., Zhao H. et al. , 2014. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol. Psychiatry 19: 717–723. 10.1038/mp.2013.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goes F. S., McGrath J., Avramopoulos D., Wolyniec P., Pirooznia M. et al. , 2015. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 168: 649–659. 10.1002/ajmg.b.32349 [DOI] [PubMed] [Google Scholar]
- GTEx Consortium , 2013. The genotype-tissue expression (GTEx) project. Nat. Genet. 45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium , 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213 [corrigenda: Nature 553: 530 (2018)]. 10.1038/nature24277 [DOI] [PubMed] [Google Scholar]
- Hayeck T. J., Zaitlen N. A., Loh P.-R., Vilhjalmsson B., Pollack S. et al. , 2015. Mixed model with correction for case-control ascertainment increases association power. Am. J. Hum. Genet. 96: 720–730. 10.1016/j.ajhg.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayeck T. J., Loh P.-R., Pollack S., Gusev A., Patterson N. et al. , 2017. Mixed model association with family-biased case-control ascertainment. Am. J. Hum. Genet. 100: 31–39. 10.1016/j.ajhg.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B. N., Donnelly P., and Marchini J., 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5: e1000529 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman S. E., and Malenka R. C., 2001. Addiction and the brain: the neurobiology of compulsion and its persistence. Nat. Rev. Neurosci. 2: 695–703. 10.1038/35094560 [DOI] [PubMed] [Google Scholar]
- Jakobsdottir J., and McPeek M. S., 2013. MASTOR: mixed-model association mapping of quantitative traits in samples with related individuals. Am. J. Hum. Genet. 92: 652–666. 10.1016/j.ajhg.2013.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassen A. K., Yang H., Miller G. M., Calder E., and Madras B. K., 2006. Receptor regulation of gene expression of axon guidance molecules: implications for adaptation. Mol. Pharmacol. 70: 71–77. 10.1124/mol.105.021998 [DOI] [PubMed] [Google Scholar]
- Jiang D., Mbatchou J., and McPeek M. S., 2015. Retrospective association analysis of binary traits: overcoming some limitations of the additive polygenic model. Hum. Hered. 80: 187–195. 10.1159/000446957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Zhong S., and McPeek M. S., 2016. Retrospective binary-trait association test elucidates genetic architecture of Crohn disease. Am. J. Hum. Genet. 98: 243–255. 10.1016/j.ajhg.2015.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J., Tawa E. A., Muench C., Rosen A. D., Rickels K. et al. , 2017. Genome-wide association study of treatment response to venlafaxine XR in generalized anxiety disorder. Psychiatry Res. 254: 8–11. 10.1016/j.psychres.2017.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice A. C., Dombrowski E., Conigliaro J., Fultz S. L., Gibson D. et al. , 2006. Veterans aging cohort study (VACS): overview and description. Med. Care 44: S13–S24. 10.1097/01.mlr.0000223741.02074.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Merrer J., Becker J. A. J., Befort K., and Kieffer B. L., 2009. Reward processing by the opioid system in the brain. Physiol. Rev. 89: 1379–1412. 10.1152/physrev.00005.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wineinger N. E., Fu D.-J., Libiger O., Alphs L. et al. , 2017a Genome-wide association study of paliperidone efficacy. Pharmacogenet. Genomics 27: 7–18. 10.1097/FPC.0000000000000250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chen J., Yu H., He L., Xu Y. et al. , 2017b Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 49: 1576–1583. 10.1038/ng.3973 [DOI] [PubMed] [Google Scholar]
- Londono D., Chen K.-M., Musolf A., Wang R., Shen T. et al. , 2013. A novel method for analyzing genetic association with longitudinal phenotypes. Stat. Appl. Genet. Mol. Biol. 12: 241–261. 10.1515/sagmb-2012-0070 [DOI] [PubMed] [Google Scholar]
- Meirelles O. D., Ding J., Tanaka T., Sanna S., Yang H.-T. et al. , 2013. SHAVE: shrinkage estimator measured for multiple visits increases power in GWAS of quantitative traits. Eur. J. Hum. Genet. 21: 673–679 [corrigenda: Eur. J. Hum. Genet. 22: 154 (2014)]. 10.1038/ejhg.2012.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthén B., 2004. Latent variable analysis: growth mixture modeling and related techniques for longitudinal data, pp. 346–369 in The SAGE Handbook of Quantitative Methodology for the Social Sciences, edited by Kaplan D., Sage, Thousand Oaks, CA: 10.4135/9781412986311.n19 [DOI] [Google Scholar]
- Pierucci-Lagha A., Gelernter J., Feinn R., Cubells J. F., Pearson D. et al. , 2005. Diagnostic reliability of the semi-structured assessment for drug dependence and alcoholism (SSADDA). Drug Alcohol Depend. 80: 303–312. 10.1016/j.drugalcdep.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Ribeiro E. A., Scarpa J. R., Garamszegi S. P., Kasarskis A., Mash D. C. et al. , 2017. Gene network dysregulation in dorsolateral prefrontal cortex neurons of humans with cocaine use disorder. Sci. Rep. 7: 5412 10.1038/s41598-017-05720-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner S. F., Foo C., Gabriel S., Reich D., Daly M. J. et al. , 2005. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15: 1576–1583. 10.1101/gr.3709305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildcrout J. S., and Heagerty P. J., 2008. On outcome-dependent sampling designs for longitudinal binary response data with time-varying covariates. Biostatistics 9: 735–749. 10.1093/biostatistics/kxn006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildcrout J. S., Schisterman E. F., Aldrich M. C., and Rathouz P. J., 2018a Outcome-related, auxiliary variable sampling designs for longitudinal binary data. Epidemiology 29: 58–66. 10.1097/EDE.0000000000000765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildcrout J. S., Schisterman E. F., Mercaldo N. D., Rathouz P. J., and Heagerty P. J., 2018b Extending the case-control design to longitudinal data: stratified sampling based on repeated binary outcomes. Epidemiology 29: 67–75. 10.1097/EDE.0000000000000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorska K., Rivadeneira F., Groenen P. J. F., Hofman A., Uitterlinden A. G. et al. , 2013. Fast linear mixed model computations for genome-wide association studies with longitudinal data. Stat. Med. 32: 165–180. 10.1002/sim.5517 [DOI] [PubMed] [Google Scholar]
- Sitlani C. M., Rice K. M., Lumley T., McKnight B., Cupples L. A. et al. , 2015. Generalized estimating equations for genome-wide association studies using longitudinal phenotype data. Stat. Med. 34: 118–130. 10.1002/sim.6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderman A. R., and Unterwald E. M., 2009. Cocaine-induced mu opioid receptor occupancy within the striatum is mediated by dopamine D2 receptors. Brain Res. 1296: 63–71. 10.1016/j.brainres.2009.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.-S., Liu X.-F., and Aragam N., 2010. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr. Res. 124: 192–199. 10.1016/j.schres.2010.09.002 [DOI] [PubMed] [Google Scholar]
- Wang Z., Xu K., Zhang X., Wu X., and Wang Z., 2017. Longitudinal SNP-set association analysis of quantitative phenotypes. Genet. Epidemiol. 41: 81–93. 10.1002/gepi.22016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warlow S. M., Robinson M. J. F., and Berridge K. C., 2017. Optogenetic central amygdala stimulation intensifies and narrows motivation for cocaine. J. Neurosci. 37: 8330–8348. 10.1523/JNEUROSCI.3141-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M.-L., Arcos-Burgos M., Liu S., Vélez J. I., Yu C. et al. , 2017. The PHF21B gene is associated with major depression and modulates the stress response. Mol. Psychiatry 22: 1015–1025. 10.1038/mp.2016.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., and McPeek M. S., 2018. L-gator: genetic association testing for a longitudinally measured quantitative trait in samples with related individuals. Am. J. Hum. Genet. 102: 574–591. 10.1016/j.ajhg.2018.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Jiang D., and McPeek M. S., 2016. CERAMIC: case-control association testing in samples with related individuals, based on retrospective mixed model analysis with adjustment for covariates. PLoS Genet. 12: e1006329 10.1371/journal.pgen.1006329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Shi J., and Whittemore A. S., 2011. Optimal methods for meta-analysis of genome-wide association studies. Genet. Epidemiol. 35: 581–591. 10.1002/gepi.20603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Nielsen J. B., Fritsche L. G., Dey R., Gabrielsen M. E. et al. , 2018. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50: 1335–1341. 10.1038/s41588-018-0184-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. An R package implementing the proposed methods is available at https://github.com/ZWang-Lab/LBRAT. Additional data analysis results of cocaine use from VACS are presented as supporting information including 1 table and 2 figures. Supplemental material available at figshare: https://doi.org/10.25386/genetics.9936881.