How do boundaries between cells of different types form? Such boundaries are critical for metazoan development, but even in microorganisms, boundaries between cells of different types form in communities such as colonies...
Keywords: Rlm1, Rim101, Ime1, Slt2, cell autonomy, cell–cell signaling
Abstract
Under conditions in which budding yeast form colonies and then undergo meiosis/sporulation, the resulting colonies are organized such that a sharply defined layer of meiotic cells overlays a layer of unsporulated cells termed “feeder cells.” This differentiation pattern requires activation of both the Rlm1/cell-wall integrity pathway and the Rim101/alkaline-response pathway. In the current study, we analyzed the connection between these two signaling pathways in regulating colony development by determining expression patterns and cell-autonomy relationships. We present evidence that two parallel cell-nonautonomous positive-feedback loops are active in colony patterning, an Rlm1-Slt2 loop active in feeder cells and an Rim101-Ime1 loop active in meiotic cells. The Rlm1-Slt2 loop is expressed first and subsequently activates the Rim101-Ime1 loop through a cell-nonautonomous mechanism. Once activated, each feedback loop activates the cell fate specific to its colony region. At the same time, cell-autonomous mechanisms inhibit ectopic fates within these regions. In addition, once the second loop is active, it represses the first loop through a cell-nonautonomous mechanism. Linked cell-nonautonomous positive-feedback loops, by amplifying small differences in microenvironments, may be a general mechanism for pattern formation in yeast and other organisms.
WHILE pattern formation during metazoan development has been studied extensively (reviewed in Dahmann et al. 2011; Perrimon et al. 2012; Sjöqvist and Andersson 2017), much less is known about pattern formation in microorganisms. Communities of microorganisms, e.g., colonies and biofilms, are not homogeneous or random structures; instead, they are organized to contain sharply defined regions of cell types. These patterned communities, which are found in both bacteria and eukaryotic microorganisms, share the properties of requiring chemical signals between cells (Shank and Kolter 2011; Claessen et al. 2014; Du et al. 2015) and providing biological function not possible for single microbial cells (reviewed in Allocati et al. 2015; van Gestel et al. 2015; Fischbach and Segre 2016; Honigberg 2016).
In the model genetic eukaryotic microorganism, Saccharomyces cerevisiae, several types of community organization have been observed (reviewed in Honigberg 2011; Vachova and Palkova 2018). For example, a diploid yeast colony is partitioned into sharply defined layers of sporulating (meiotic) and nonsporulating cells. Specifically, after colony growth ceases, a thin layer of cells near the center of the colony induce Ime1 (reviewed in Neiman 2011; Honigberg 2016), a master regulator that activates sporulation in this layer (Piccirillo et al. 2010). Over time, this thin sporulation layer expands to eventually include the top half of the colony, whereas the cells underlying the sporulation layer never sporulate (Piccirillo et al. 2010). This colony sporulation pattern is observed in a range of S. cerevisiae laboratory strain backgrounds as well as in S. cerevisiae and S. paradoxus strains newly isolated from the wild. Indeed, in these wild strains, this sporulation pattern can be observed on a range of carbon and nitrogen sources (Piccirillo and Honigberg 2010). Through a range of conditions, colony sporulation patterning is characterized by the sharp boundary between a top layer of sporulating cells and a bottom layer with essentially no sporulation. The cells of the underlying unsporulated layer in colonies are termed feeder cells. Feeder cells are more permeable than undifferentiated cells, and, probably as a result of this permeability, provide signals and/or nutrients that stimulate sporulation in the overlying layer (Piccirillo et al. 2015).
Both the cell-wall integrity (CWI) MAPK pathway and the alkaline response (AR) pathway have been implicated in the dual-layer colony pattern described above. The CWI pathway activates cell-wall remodeling (reviewed in Levin 2011; Sanz et al. 2017), and a target of this pathway, the Rlm1 transcription factor, is activated in colonies specifically in the feeder-cell layer (Piccirillo et al. 2015). Indeed, examination of sections from embedded colonies indicates a feeder layer does not form in rlm1Δ colonies, sporulation is less efficient, and the residual spores are distributed throughout colonies rather than residing only in the upper layer. Because Rlm1 is activated in the lower colony layer, but required for sporulation in the upper layer, Rlm1 must activate sporulation through a cell-nonautonomous mechanism. In other words, Rlm1 expressed in one cell layer activates sporulation in an overlying cell layer through cell-to-cell signaling.
The rlm1Δ colony sporulation-pattern described above contrasts with the pattern observed in rim101Δ colonies. Rim101 is essential for activation of the AR pathway (reviewed in Maeda 2012; Serra-Cardona et al. 2015). In rim101Δ colonies, sporulation initiates in the same narrow central band of cells as in the wild type, but in the mutant colonies, this band fails to expand over time (Piccirillo et al. 2010). In addition, whereas the CWI pathway is only required for efficient sporulation in colonies (Piccirillo et al. 2015), the AR pathway is required for efficient induction of IME1, and, hence, sporulation, in both colonies or cultures (Li and Mitchell 1997; Piccirillo et al. 2010). To explain the rim101Δ colony sporulation pattern, we proposed that Rim101 forms a nonautonomous positive-feedback loop with IME1 (Piccirillo et al. 2010). In this model, IME1 expression leads to increased respiration, and, hence, raises the environmental pH (Hayashi et al. 1998; Piccirillo et al. 2010); in turn, this alkaline environment activates the Rim101 pathway in neighboring cells, and, hence, induces IME1 in these cells (Piccirillo et al. 2010).
The CWI and AR pathways respond to different cellular stresses, and act in parallel to regulate cell-wall synthesis (Castrejon et al. 2006). However, the relationship between these pathways in controlling colony sporulation is unknown. In the current study, we determined the interaction between these pathways through epistasis, cell autonomy, and colony expression/localization assays. Our results indicate that linked cell-nonautonomous positive-feedback loops coupled with cell autonomous repression of alternative fates underlies pattern formation.
Materials and Methods
Strains and plasmids
All strains used in the study (Supplemental Material, Table S1) were constructed in a modified W303 strain background (SH777) and are prototrophs. Deletion mutants were constructed by designing PCR fragments to replace >90% of the ORF with the URA3, TRP1, or LEU2 gene as described (Gray and Honigberg 2001; Gray et al. 2005). The RIM20-GFP allele was constructed by integrating GFP-TRP1 at the C-terminus of the RIM20 ORF as described (Boysen and Mitchell 2006). All newly constructed genomic alleles were verified by diagnostic PCR with primers flanking the targeted region (Gray and Honigberg 2001; Gray et al. 2005). The rim101Δ rlm1Δ prIME1-mCherry mutant was constructed by crossing single mutants, sporulating the heterozygous diploid, and dissecting tetrads. Plasmids used in the study were YCp50(URA3)-UASRlm1-LacZ (Jung et al. 2002) and YEp353(URA3)-prZPS1-LacZ (Frey et al. 2011).
Media and growth conditions
Except as noted below, all colonies in this study were grown as spot colonies inoculated with 0.5 μl of a 2 × 108 cells/ml suspension in water on Sp2% medium (2% potassium acetate, 0.5% yeast extract, 2% agar, pH 7.0) or the same medium containing 40 μg/ml X-gal to monitor LacZ expression (Piccirillo et al. 2015). A total of 20 colonies were inoculated per plate in a circular arrangement, with all colonies equally spaced and ∼0.5 cm from the edge of the plate. For chimeric colonies specifically, inoculations contained equal numbers of signal and response strains.
High frequencies of Rim20-GFP foci were achieved by growing colonies on Sp6% media (6% potassium acetate, 0.5% yeast extract, 2% agar, pH 7.0). High expression levels of ZPS1-LacZ (as well as prIME1-LacZ and UASRlm1-LacZ) were achieved by growing colonies on MPX medium (0.5% sodium pyruvate, 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 40 μg/ml X-gal, 2% agar, pH 7.0). Other media used in this study have been described previously (Rose et al. 1990; Piccirillo and Honigberg 2010).
Colony expression assays
Expression levels of promoter-fusion alleles (LacZ, GFP, and mCherry) were determined from images of the colony surface using Image J as described, and, hence, are expressed in arbitrary units (Piccirillo et al. 2010, 2015). When necessary, differences in illumination across plates was minimized by background subtraction based on pixels adjoining colonies. For the chimeric colony assay (CCA), to ensure that the ratio of signal cells to response cells in the colony was ∼1:1, and to adjust expression/activity levels for small variations in this ratio, we resuspended sample colonies in sorbitol solutions at the same osmolarity after growth was complete (2 days). These resuspended cells were plated and the signal cell:response cell ratio determined by replica-plating on X-gal medium (Piccirillo et al. 2015). Three biological replicas of these samples were assayed, and the mean fraction of response cells to total cells in the colony was used to standardize the overall signal. As a second control, to ensure that response activity remained in the linear range of this assay, we verified that activity/response cell was not significantly different in pure colonies of the response strain relative to chimeric colonies containing an equal mixture of this response strain and an otherwise isogenic strain lacking the response gene (i.e., P > 0.1).
Same-cell localization studies
The fraction of cells in a prIME1-mCherry RIM20-GFP colony that formed Rim20-GFP foci and/or expressed prIME1-mCherry was determined by suspending the spot colony in 20 μl of sorbitol, placing 2 μl of the suspension on a microscope slide, covering this suspension with 4 μl mounting media (80% glycerol, 1 mg/ml p-phenylene diamine), and visualizing cells using conventional fluorescent microscopy. The fraction of cells in a prIME1-mCherry UASRlm1-LacZ colony that expressed one or both alleles was determined by suspending spot colonies in 20 μl 0.1% SDS, 1 mM fluorescein di-β-d-galactopyranoside (FDG) in a PCR tube and incubating cells in a thermocycler for 10 cycles of 40 sec at 20° alternating with 60 sec at 30°, followed by 1 hr at 30°. Cell suspensions were then harvested, washed 4× with 200 μl water, and visualized by fluorescent microscopy as above.
Cell and colony imaging and analysis
Images of individual cells in Figure 1 were captured by confocal microscopy, red and green images merged, and Z-projections assembled in ImageJ (NIH). To visualize spatial expression patterns in RIM20-GFP prIME1-mCherry colonies, colonies were covered with agar and cleaved vertically through the center with a razor blade as described (Piccirillo et al. 2015). Colony hemispheres were then trimmed into blocks, flipped 90° on to a coverslip with the exposed side down, confocal images collected, Z-projections assembled, and red and green images merged as above. Sections were aligned using an ImageJ plug-in (Thevenaz et al. 1998). The distribution of fluorescent cells in the colony was quantified by superimposing a stack of 10 equal-size adjacent rectangles on the colony image and then manually determining the frequency of fluorescent cells in each rectangle. Images were adjusted for brightness and contrast.
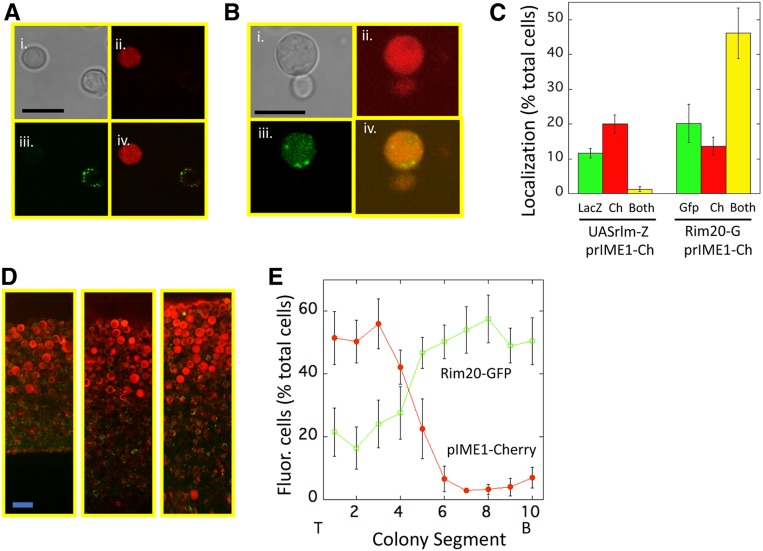
Figure 1.
Colocalization of IME1 expression, Rlm1 activity and Rim101 activity within colonies. (A) Cells from a 3-day colony of UASRlm1-LacZ prIME1-mCherry strain (SH5904) visualized for (i) bright field, (ii) mCherry (iii), FDG metabolite, (iv) merge of FDG and mCherry images. As a control for possible interference of mCherry on the FDG signal, a control strain lacking the prIME1-mCherry allele (SH5408) yielded approximately the same frequency of FDG+ cells (13.2 ± 0.9, n = 3) as did the experimental strain (11.7 ± 1.0, n = 3) (B) cells from a 3-day colony of RIM20-GFP prIME1-mCherry strain (SH5252) visualized for (i) bright field, (ii) mCherry, (iii) GFP, (iv) merge of GFP, and mCherry images. (C) (Left) Cells resuspended from 4 day prIME1-mCherry UASRlm1-LacZ colonies (SH5904) scored by fluorescence microscopy for expression of UASRlm1-LacZ (“LacZ,” green bar) visualized by FDG (see Materials and Methods), prIME1-mCherry (“Ch,” red bar), or both fluorophores (“Both,” yellow bar) (n = 3). (Right) Cells resuspended from prIME1-mCherry RIM20-GFP (Rim20-G) colonies (SH6054) scored for expression of Rim20-GFP foci (“Gfp,” green bar), prIME1-mCherry (“Ch,” red bar) or both fluorophores (“Both,” yellow bar), n = 3. (D) Representative regions from three independent colonies of RIM20-GFP prIME1-mCherry/IME1 strain (SH6054). (E) Distribution of cells (SH6054) expressing prIME1-mCherry (red filled symbols) and forming Rim20-GFP foci (green open circles) from colony images (n = 5). Graph shows fraction of cells that display these responses in 10 equal sized regions from top (T, left) to bottom (B, right) of colony. Bars, A and B = 10 μm, D = 20 μm.
Sporulation, viability, and pH assays
To determine sporulation efficiency, colonies were resuspended and examined by light microscopy for the fraction of cells that had formed asci. To measure viability of diploid cells in colonies, colonies were resuspended from Sp2% plates, diluted in 0.5 M sorbitol, and plated on His− Lys− synthetic medium containing 0.5 M sorbitol (Lee and Honigberg 1996). To monitor changes in pH, spot colonies were grown on Sp2% medium containing + 20 mg/liter phenol red for 4 days, an image of the top surface of a colony captured, and the red intensity of this image quantified as above.
Statistics and reproducibility
Except where noted in the figure legends, all quantitative data in the study are expressed as the mean of four biological replicate experiments with error bars representing the SEM. P-values are from unpaired Student’s t-tests. All experiments were replicated on at least two separate dates, and except as noted in Table S1 all experiments on mutants were performed with at least two independently derived isolates of each mutant. Experiments based on scoring cell populations by microscopy were performed blind when possible, and at least 250 cells examined for each sample.
Data availability
All strains used in this study are described in Table S1 and are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.9978863.
Results
Localization of Rim101 activity within colonies
To investigate the connection between Rlm1 and Rim101 in regulating colony patterning, we asked whether the Rim101 pathway was activated in meiotic cells and/or feeder cells within colonies. As a first experiment, we used fluorescence microscopy to verify a previous conclusion that the population of meiotic cells is distinct from the population of feeder cells (Piccirillo et al. 2015). The former population was visualized as expressing prIME1-mCherry, whereas the latter population was visualized as expressing UASRlm1-LacZ. For this purpose, spot colonies of a prIME1-mCherry UASRlm1-LacZ strain were incubated for 3–4 days, then colonies resuspended, incubated with the lactose analog, FDG, and examined by fluorescence microscopy (see Materials and Methods). As expected from the previous results, these colonies yielded many cells expressing one or the other allele but few, if any, cells expressing both alleles (Figure 1, A and C, left).
Next, we examined whether cells in colonies activated for the Rim101-pathway were also activated for IME1 expression. Rim101 activity was detected by the formation of Rim20-GFP foci (Boysen and Mitchell 2006). To identify cells activated for one or both pathways, we resuspended RIM20-GFP prIME1-mCherry colonies and examined them by fluorescence microscopy for Rim20-GFP foci and/or prIME1-mCherry expression. We found that these colonies contained many cells that had activated both pathways, as well as cells with only one or the other pathway active (Figure 1, B and C, right). Thus, meiosis was initiated in a subset of the cells with the Rim101 pathway activated.
We were not able to determine whether some cells activate both Rlm1 and Rim101 pathways due to interference between LacZ and Rim20-GFP detection. However, to address this point, we identified the location of cells with Rim20-GFP foci (Rim101 activity) within colonies. For this purpose, RIM20-GFP prIME1-mCherry colonies were embedded in agar, cleaved vertically, and the exposed plane of the colony examined by confocal microscopy (see Materials and Methods). As expected from earlier studies (Piccirillo et al. 2015), prIME1-mCherry was expressed specifically in the top layer of the colony. In contrast, we observed Rim20-GFP foci in both the prIME1-mCherry top layer and in the underlying layer (Figure 1, D and E, separate red and green images in Figure S1). Indeed, quantification of multiple colony sections demonstrated that at this stage of colony development, cells displaying Rim20-GFP foci were more prevalent in the underlying layer. Thus, the results presented in Figure 1, C–E demonstrate that the Rim101 pathway is active in both the meiotic layer and the feeder cell layer of colonies.
Epistasis experiments indicate that Rim101 and Rlm1 pathways regulate sporulation through distinct pathways
Since both the Rlm1 pathway, which is activated only in the lower (feeder cell) layer, and the Rim101 pathway, which is activated throughout the colony, induce IME1 transcription, we investigated the relationship of these two pathways in regulating IME1 using epistasis (double mutant) analysis. In particular, we compared prIME1-LacZ expression in wild-type, rim101Δ, rlm1Δ, and rim101Δ rlm1Δ colonies by growing these colonies on medium containing X-gal and then imaging the top surface of these colonies for the level of blue X-gal metabolite produced (see Materials and Methods). We found that either single mutant displayed ∼2–4-fold diminished expression of prIME1-LacZ relative to the wild type, and the double mutant was decreased ∼10-fold relative to the wild type (Figure 2A). If Rim101 and Rlm1 acted only in the same linear pathway to activate IME1 transcription, the double deletion mutant would not be more defective than either single mutant. Thus, our results are consistent with Rlm1 and Rim101 activating sporulation through distinct pathways. For clarity, conclusions from this and all subsequent experiments are diagrammed in Figure S2.
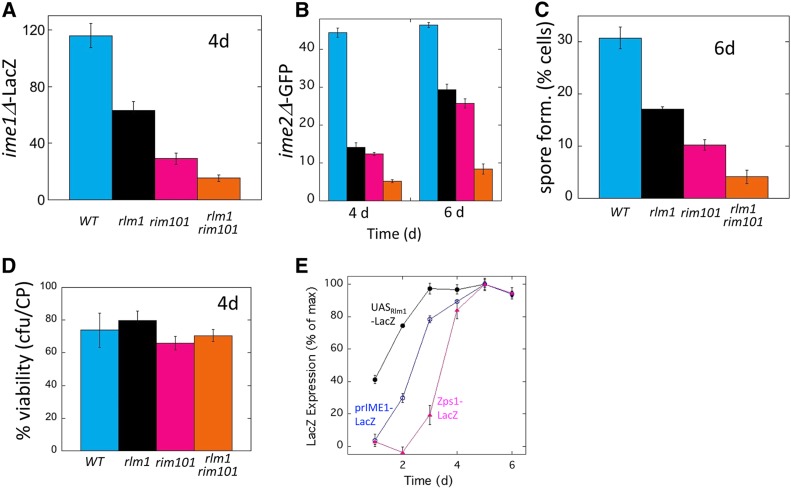
Figure 2.
Rlm1 and Rim101 have distinct roles in activating sporulation: (A) prIME1-LacZ expression in wild-type (SH3830), rlm1Δ (SH4800) and rim101Δ (SH5312), and rim101Δ rlm1Δ (SH5729) 4-day colonies at the indicated times relative to a LacZ− control strain (SH3972). Bars 3 and 4 are significantly different (P = 0.023). (B) ime2Δ-GFP expression in wild-type (SH4805), rlm1Δ (SH5509), rim101Δ (SH5545), and rim101Δ rlm1Δ (SH5513) colonies at the indicated times. SH3883 was used as a GFP− control strain. (C) Spore formation in wild-type (WT, SH3881), rlm1Δ (SH4708), rim101Δ (SH4377), and rim101Δ rlm1Δ (SH5550) 6-day colonies. Bars 3 and 4 are significantly different (P = 0.009). (D) Cell viability in 4-day colonies of same strains as (C). (E) Timing of UASRlm1-LacZ expression (SH5186), prIME1-LacZ expression (SH5900), and Zps1-LacZ expression (SH5830) in colonies incubated on MP medium for the times indicated. Expression at each time is shown as a percentage of the maximum level of expression of that gene during the time course and is standardized to a LacZ− strain (SH3881).
To confirm the above results, we used the same rim101Δ and rlm1Δ single and double mutants to measure transcriptional induction of a second meiotic gene, IME2 (Figure 2B), and to measure the formation of spores, which is the final event in the sporulation program (Figure 2C). Note that each assay monitors a separate level of control on sporulation. For example, IME2, which encodes a Ser/Thr kinase required through both early and middle stages of meiosis, is regulated by IME1 as well as through IME1-independent pathways (Mitchell et al. 1990). Similarly, spore formation requires IME1, IME2, and additional pathways (Kassir et al. 2003; Honigberg 2004). Nevertheless, we found that measuring either prIME2-GFP expression or spore formation yielded similar results to measuring prIME1-LacZ (Figure 2, A–C).The differences between strains described above could in theory reflect differential cell viabilities. To test this possibility, we measured cell viability in the above wild-type and mutant strains. We found that all four strains maintained comparable and relatively high cell viabilities during the course of the experiment (Figure 2D). Thus, measurement of IME1 expression, IME2 expression, spore formation, and cell viability all point to the conclusion that the Rim101 and Rlm1/CWI pathways activate sporulation through distinct pathways. As described below, although these pathways are distinct, they are not independent.
Rim101 is activated after Rlm1 and IME1
To further investigate the relationship between Rlm1 and Rim101 pathways in regulating IME1, we compared the timing of activation of these pathways to the timing of IME1 transcription. Rim101 pathway activity in this experiment was detected using the ZPS1-LacZ response allele, which requires both RIM101 and alkaline pH for expression (Lamb et al. 2001; Frey et al. 2011). We compared the timing of LacZ expression in whole colonies in three strains that differed only in the reporter-fusion allele—either ZPS1-LacZ, prIME1-LacZ, or UASRlm1-LacZ. In this experiment, all three strains were ime1Δ to eliminate downstream effects of sporulation on gene expression, and expression of each gene is represented as a percentage of its maximum expression. As shown previously (Piccirillo et al. 2015), UASRlm1-LacZ was expressed in colonies 1–2 days earlier than IME1. Furthermore, we found that ZPS1-LacZ was induced much later in colony development than UASRlm1-LacZ, and slightly later than prIME1-LacZ (Figure 2E). This expression pattern is consistent with Rlm1 and Rim101 activating IME1 through distinct pathways, with the Rlm1/CWI pathway acting earlier.
Cell autonomy tests of mutant alleles
The gradual Rim101-dependent expansion of an initially narrow central layer of sporulated cells to eventually include the entire top half of the colony led to the hypothesis that Ime1 activates Rim101 through a nonautonomous Rim101-Ime1 positive feedback loop, i.e., Ime1 is required to produce a signal that activates Rim101 in neighboring cells (see Introduction). To test this idea directly, we used the CCA (Piccirillo et al. 2010, 2015). This assay, which is used throughout the current study, utilizes “chimeric” spot colonies, which are colonies inoculated with an equal number of cells of two different genotypes.
Chimeric colonics always contain an approximately equal mixture of a “response strain” and a “signal strain” (Figure 3A). The response strain carries a “response gene” that allows the activity of a gene product to be monitored. For example, in the CCA diagrammed in Figure 3A, the response gene is RIM20-GFP. In contrast, the signal strain lacks this response gene, but instead contains alternative alleles of a “signal gene.” For example, in the CCA diagrammed in Figure 3A, the signal gene is either IME1 or ime1Δ. In a standard CCA, the response strain is also deleted for this signal gene, so any signal dependent on this gene can only originate from the signal strain.
Figure 3.
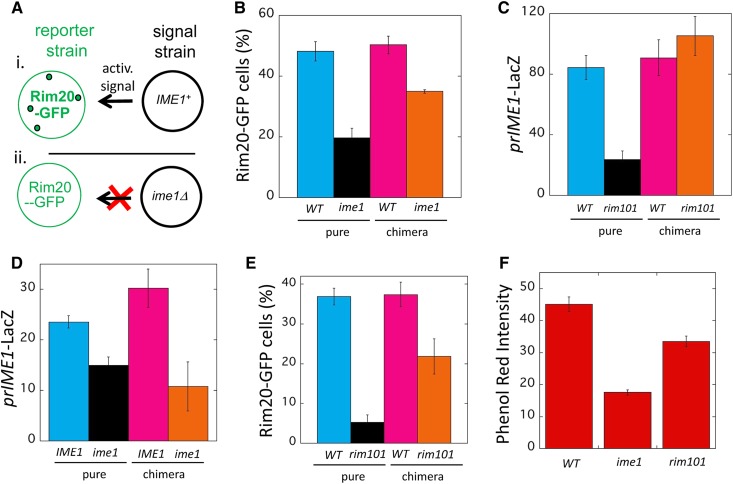
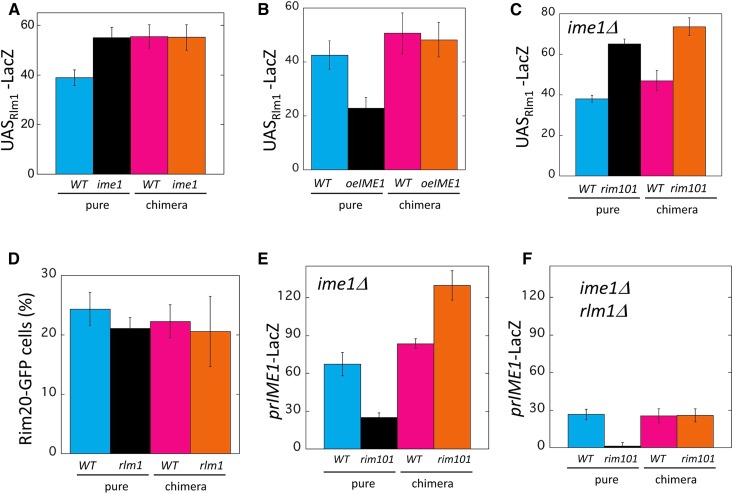
Nonautonomous Rim101-Ime1 positive feedback loop. (A) Diagram of chimeric colony assay: the activity of a response allele is compared between the chimeric colonies diagrammed in (i and ii). Each chimeric colony is a 50:50 mixture of a response strain (left side) and signal strain (right side). Note that the RIM20-GFP response strain is identical in both chimeric colonies, and the signal strain is either IME1 (top right) or ime1Δ (bottom right). If the chimeric colony diagrammed in (i) forms Rim20-GFP foci (indicated by green dots in diagram) to higher levels than the chimeric colony diagrammed in (ii), then Ime1 must activate Rim20 foci formation in neighboring cells, i.e., IME1 is required for cell-nonautonomous activation of Rim20 foci. (B) Results of chimeric colony assay (CCA) diagrammed in (A) measuring effect of ime1Δ on Rim20-GFP foci formation. Pure colonies of RIM20-GFP strains that were either IME1 (WT) (blue, SH6055) or ime1Δ (black, SH5252) and chimeric colonies containing an ime1Δ RIM20-GFP response strain mixed with either a WT (magenta, SH3826) or an ime1Δ (orange, SH3830) signal strain. Y-axis represents the fraction of cells in the colony that display Rim20-GFP foci. (C) CCA measuring effect of rim101Δ on prIME1-LacZ expression. Pure colonies of prIME1-LacZ strains that were either RIM101 (WT) (blue, SH3827) or rim101Δ (black, SH6034) and chimeric colonies containing a RIM101 prIME1-LacZ response strain mixed with either a WT (magenta, SH3881) or a rim101Δ (orange, SH4376) signal strain. (D) Autochimeric colony assay measuring effect of ime1Δ on expression of prIME1-LacZ. Pure colonies of a prIME1-LacZ strain that were either IME1 (WT) (blue, SH5868) or ime1Δ (black, SH5900) and chimeric colonies containing an ime1Δ prIME1-LacZ response strain mixed with either a WT (magenta, SH3882) or an ime1Δ (orange, SH5225) signal strain. Bars 3 and 4 are significantly different (P = 0.02). (E) Autochimeric whole colony expression assay measuring effect of rim101Δ on RIM20-GFP foci formation measured as in (C). Pure colonies of RIM20-GFP strains that were either RIM101 (WT) (blue, SH5252) or rim101Δ (black, SH5839) and chimeric colonies containing a RIM101 RIM20-GFP response strain mixed with either a WT (magenta, SH3826) or rim101Δ (orange, SH6035) signal strain. Bars 3 and 4 are significantly different (P = 0.03). (F) The relative pH in wild-type (SH3881), ime1Δ (SH3972) and rim101Δ (SH4375) colonies grown on medium containing phenol red was measured in arbitrary units. These units were based on the red intensity in colony surface images quantified as in Materials and Methods.
The basic idea of the CCA is to compare two chimeric colonies that contain the same response strain but alternative signal strains. We refer to the chimeric colony containing the wild-type signal strain as the “wild-type chimeric colony” and the one containing the mutant signal strain as a “mutant (i.e. mutXΔ) chimeric colony.” For example, in the CCA diagrammed in Figure 3A, if the fraction of cells forming Rim20-foci is higher in wild-type (IME1) chimeric colonies than in ime1Δ chimeric colonies, then IME1 must stimulate the Rim101 pathway through a nonautonomous mechanism. Finding a cell-nonautonomous effect of a signal allele on a response gene implies that the signal gene is required to produce a cell-to-cell signal, but it should be noted that other explanations are theoretically possible. For example, the signal allele might affect response gene activity by affecting metabolism, and hence concentration of nutrients in the medium.
Interpretation of CCA results involves two separate comparisons. The first of these is between the wild-type and mutant chimeric colonies as described above. The second comparison is between two pure (i.e., not chimeric) colonies. Both pure colonies contain the same response gene as the chimeric colonies, but they differ from one another (like the chimeric colonies) in having alternative signal alleles (wild type or mutXΔ). The pure colony comparison reveals the total effect (i.e., autonomous plus nonautonomous) of the mutant allele on the response gene. By subtracting the nonautonomous effect from the total effect, the autonomous effect can be estimated. This dual comparison is particularly necessary because, as shown later in this study, the same allele may have both autonomous and nonautonomous effects on a response, and these effects may act in either the same or opposite directions (i.e., activation and/or repression).
An important control for the CCA assay verifies that measured gene activity reflects activity/response cell and not variations in growth rates between signal and response strains (see Materials and Methods). Furthermore, a separate internal control comparison verifies that the biological activity being measured is in the linear range of the assay (see Materials and Methods).
Rim101 and IME1 form a cell-nonautonomous positive feedback loop
In the particular CCA described above, comparing wild-type to ime1Δ pure colonies for Rim20-GFP foci formation revealed higher levels of foci in the wild-type colony. Thus, the total effect of Ime1 is to activate the Rim101 pathway (Figure 3B, left two columns). Furthermore, comparing chimeric colonies revealed that the wild-type chimeric colony displayed significantly more cells with Rim20-GFP foci than did the ime1Δ chimeric colony (Figure 3B, right two columns). Indeed, the frequency of Rim20-GFP foci in IME1 colonies relative to ime1Δ colonies was approximately the same in chimeric colonies as in pure colonies. Thus, IME1 activates the Rim101 pathway largely through a nonautonomous mechanism consistent with the hypothesis of a nonautonomous positive-feedback loop (Figure 3A).
We next used the CCA to determine the converse regulatory relationship, i.e., to test the effect of rim101Δ on IME1 transcription. As expected, given that Rim101 activates of IME1 transcription (Figure 2A), in the pure colony comparison, RIM101 colonies displayed much higher prIME1-LacZ expression than did rim101Δ colonies (Figure 3C, bars 1 and 2). For the chimeric colony comparison, RIM101 colonies displayed approximately the same expression as rim101Δ colonies (Figure 3C, bars 3 and 4). Hence, even in the context of a cell-nonautonomous Rim101-Ime1 feedback loop, Rim101 activates IME1 through a largely cell autonomous mechanism. Thus, a cell-nonautonomous feedback loop can have some components that are cell autonomous.
Ime1 and Rim101 autochimeras confirm Rim101-Ime1 nonautonomous positive feedback loop
One prediction of a cell-nonautonomous Rim101-Ime1 positive feedback loop is that an ime1Δ allele will result in lower IME1 expression in neighboring cells. To test this prediction, we applied the CCA so that the response allele is prIME1-LacZ and the signal allele is either IME1 or ime1Δ (an “auto-chimeric colony”). In comparing pure colonies, we found significantly higher prIME1-LacZ expression in IME1 colonies than in ime1Δ colonies; this result is consistent with the Ime1 positive-feedback loop proposed previously (Shefer-Vaida et al. 1995; Moretto et al. 2018). In comparing chimeric colonies, we also found expression to be significant higher in IME1 colonies than ime1Δ colonies (Figure 3D). Thus, IME1 activates its own expression in colonies primarily through a cell-nonautonomous positive-feedback loop. That is, Ime1 is required for a signal that activates IME1 transcription in neighboring cells.
Similarly, we compared the frequency of cells with Rim20-GFP foci in auto-chimeric colonies containing either a RIM101 or a rim101Δ signal strain. In the pure colony comparison, many fewer cells displayed Rim20-GFP foci in rim101Δ colonies than in RIM101 colonies (Figure 3E, left two bars) as expected since Rim20-foci formation requires Rim101. In the chimeric colony comparison as well, RIM101 colonies yielded significantly higher frequencies of Rim20-GFP foci than rim101Δ colonies (Figure 3E, right two bars). Thus, both auto-chimeric colony experiments support the idea of an Rim101-Ime1 nonautonomous positive feedback loop.
Rim101-Ime1 feedback loop driven by extracellular alkaline pH
A nonautonomous feedback loop requires an extracellular signal, and one candidate signal for the Rim101-Ime1 loop is alkaline pH. For example, sporulating colonies increase the pH of their environment (Hayashi et al. 1998; Piccirillo et al. 2010; Zhao et al. 2018), and alkaline pH is the primary stimulus for the Rim101/AR pathway. To determine whether increased pH drives the Rim101-Ime1 positive-feedback loop, we performed two types of experiments. First, we compared colonies grown on pH 8.0 vs. pH 6.0 medium for both Rim20-GFP foci formation and prIME1-mCherry expression (Figure S3). We found that the frequency of cells that displayed either activity was increased at the higher pH. Second, we compared the pH of the medium surrounding wild-type rim101Δ or ime1Δ colonies using phenol red as a pH indicator. We found that the pH surrounding either mutant colony was significantly lower than wild-type colonies (Figure 3F). Thus, both IME1 expression and Rim101 activity were stimulated by extracellular pH, and both IME1 and RIM101 were required for the increase in extracellular pH as colonies develop. These results support the idea that alkaline pH is the extracellular signal driving the Rim101-Ime1 positive feedback loop.
Cell-nonautonomous positive feedback loop regulates Rlm1 in colonies
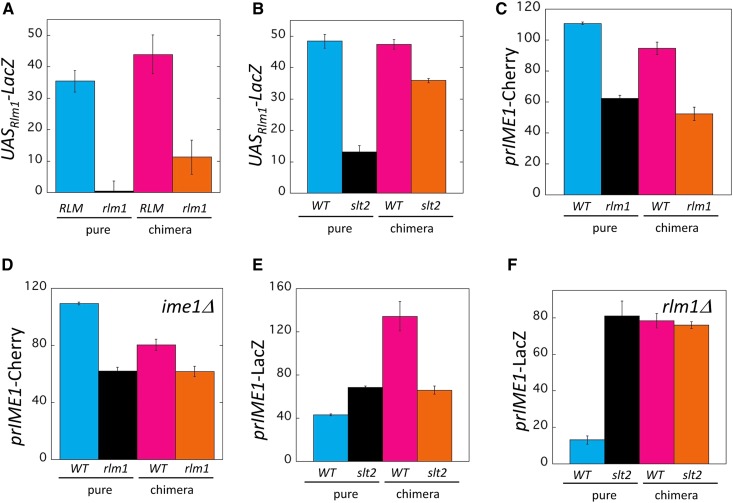
The discovery of the cell nonautonomous Rim101-Ime1 positive feedback loop in colonies led us to ask whether Rlm1, which is active in feeder cells, might also form a feedback loop. For this purpose, another autochimeric colony experiment was performed; in this case, with a UASRlm1-LacZ RLM1 response strain and either RLM1+ or rlm1Δ signal strains. In comparing pure colonies, UASRlm1-LacZ expression was much higher in the wild type colony than in the rlm1Δ colony (Figure 4A, left two bars), as expected given that Rlm1 activates transcription of this response gene. In comparing chimeric colonies, UASRlm1-LacZ expression was also higher in the wild-type colony than in the rlm1Δ colony (Figure 4A, right two bars). Thus, Rlm1 activates itself through a nonautonomous mechanism.
Figure 4.
Nonautonomous Rlm1-Slt2 positive feedback loop regulates the Rim101-Ime1 feedback loop. (A) Autochimeric colony assay measuring effect of rlm1Δ on UASRlm1-LacZ expression. Pure colonies of UASRlm1-LacZ strains that were either RLM1(WT) (blue, SH5069) or rlm1Δ (black, SH5863) and chimeric colonies containing an RLM1 UASRlm1-LacZ response strain mixed with either a WT (magenta, SH3882) or an rlm1Δ (orange, SH4767) signal strain. (B) CCA measuring effect of slt2Δ on UASRlm1-LacZ expression. Pure colonies of UASRlm1-LacZ strains that were either SLT2 (WT) (blue, SH5069) or slt2Δ (black, SH6062) and chimeric colonies containing the SLT2 UASRlm1-LacZ response strain mixed with either a WT (magenta, SH3882) or slt2Δ (orange, SH5520) signal strain. Bars 3 and 4 are significantly different (P = 0.007, n = 3). (C) CCA measuring effect of rlm1Δ on prIME1-mCherry expression. Pure colonies of prIME1-mCherry strains that were either RLM1 (WT) (blue, SH4652) or rlm1Δ (black, SH5875) and chimeric colonies containing an rlm1Δ prIME1-mCherry response strain mixed with either a WT (magenta, SH3826) or an rlm1Δ (orange, SH4799) signal strain. (D) CCA measuring effect of rlm1Δ on prIME1-mCherry expression when IME1 is absent. Pure colonies of prIME1-mCherry/ime1Δ strains that were either RLM1 (WT) (blue, SH4414) or rlm1Δ (black, SH4967) strains and chimeric colonies containing an rlm1Δ prIME1-mCherry/ime1Δ response strain mixed with either a WT (magenta, SH3830) or an rlm1Δ (orange, SH4800) signal strain. All strains used were ime1 Δ.Bars 3 and 4 are significantly different (P = 0.04). (E) CCA measuring effect of slt2Δ on prIME1-LacZ expression. Pure colonies of prIME1-LacZ strains that are either SLT2 (WT) (blue, SH3826) or slt2Δ (black, SH6105) and chimeric colonies containing an slt2Δ prIME1-LacZ response strain mixed with either a WT (magenta, SH3882) or an slt2Δ (orange, SH5520) signal strain. (F) CCA measuring effect of slt2Δ on prIME1-LacZ expression when RLM1 is absent. Pure colonies of prIME1-LacZ strains that are either SLT2 (WT) (blue, SH5175) or slt2Δ (black, SH6104), and chimeric colonies containing an slt2Δ prIME1-LacZ response strain mixed with either a WT (magenta, SH4708) or an slt2Δ (orange, SH5495) signal strain. All strains used were rlm1Δ.
In cultures subject to cell-wall stress, Slt2 kinase (also termed Mpk1 kinase) phosphorylates and activates Rlm1. In turn, the Rlm1 transcription factor may activate SLT2 transcription (Dodou and Treisman 1997; Watanabe et al. 1997; Jung et al. 2002), forming an Rlm1-Slt2 positive feedback loop (García et al. 2016). These results suggest that Slt2 may be part of the nonautonomous mechanism by which Rlm1 activates itself. To test this idea, we performed CCA with a UASRlm1-LacZ response strain and either SLT2 or slt2Δ signal strains. As expected from the studies cited above, in pure colonies, the response gene was expressed to much lower levels in the slt2Δ mutant than in the wild type (Figure 4B, left two bars). Similarly, in chimeric colonies, UASRlm1-LacZ was expressed to significantly lower levels in the slt2Δ colony than in the wild-type colony (Figure 4B, right two bars). Thus, Slt2 activates Rlm1 in part through a nonautonomous mechanism; however, the total effect of Slt2 on UASRlm1-LacZ is much greater than its nonautonomous effect on this gene (Figure 4B, cf. right two bars to left two bars). Thus, even in the context of a cell-nonautonomous Rlm1-Slt2 positive feedback loop, Slt2 activates Rlm1 through largely a cell-autonomous mechanism.
Rlm1 activation of IME1 is amplified by Ime1 feedback loop
As described in the Introduction, we previously concluded that Rlm1 activates IME1 expression in colonies through a cell nonautonomous mechanism (Piccirillo et al. 2015). As an initial experiment, we confirmed this conclusion under the conditions of the current study. Consistent with the results of Figure 2A, in the pure colony comparison, rlm1Δ colonies displayed much lower prIME1-mCherry expression than wild-type colonies (Figure 4C, left two bars). In the chimeric colony comparison, prIME1-mCherry was expressed to higher levels in the wild-type colonies than in rlm1Δ colonies (Figure 4C, right two bars). Indeed, the ratio between wild-type and rlm1Δ colonies was very similar in the pure colony and chimeric colony comparison. Thus, as expected, under the growth conditions used in the current study, Rlm1 activates IME1 primarily through a cell nonautonomous mechanism. That is, Rlm1, which is active in the lower layer, is required to provide a signal that stimulates meiosis in the upper layer.
Once a signal gene is shown to nonautonomously regulate a response gene by an initial CCA, it is sometimes useful to determine if a second gene also acts in the same regulatory circuit. In the current study, this was accomplished by a performing a second CCA, which was identical to the initial one except with the second gene deleted in both signal strains, and comparing the results of the two CCAs. For example, to determine whether activation of IME1 transcription by Rlm1 requires the Ime1-dependent positive feedback loop, we repeated the CCA described above except that both signal strains (and the response strain) were ime1Δ. Pure colonies displayed similar results in the second CCA (Figure 4D) as in the initial CCA (Figure 4C). When wild-type and rlm1Δ chimeric colonies were compared in the second CCA, there was still a significantly higher prIME1-mCherry expression in the wild-type chimeric colony, as we had also seen in the initial CCA. However, this difference is much smaller in the second (ime1Δ) CCA than in the initial (IME1) CCA (Figure 4, C and D). Thus, the effect of Rlm1 on prIME1-mCherry expression is likely amplified by the Ime1-dependent feedback loop, but does not completely depend on this loop.
Slt2 represses IME1 through a cell autonomous mechanism
Given that Rlm1 activates IME1 transcription through a nonautonomous mechanism, we next asked whether the Slt2 component of the feedback loop also activates IME1 transcription. For this purpose, we performed the CCA using a prIME1-LacZ/IME1 response strain, and signal strains that were either SLT2 or slt2Δ (Figure 4E). Surprisingly, in the pure colony comparison, wild-type colonies yielded lower prIME1-LacZ expression than did slt2Δ colonies. In the chimeric colony comparison, wild-type colonies displayed higher prIME1-LacZ expression than slt2Δ colonies. Thus Slt2, like Rlm1, activates IME1 transcription through a cell nonautonomous mechanism, but, at the same time, Slt2 represses IME1 through a cell autonomous mechanism.
To explore the opposing roles of Slt2 on IME1 expression described above, we tested the role of Rlm1 in these roles. To this purpose, we performed a second CCA identical to the initial CCA described in the previous paragraph except that all strains were also rlm1Δ. In the pure colony comparison for this second CCA, prIME1-LacZ was expressed to lower levels in wild-type colonies than in slt2Δ colonies (Figure 4, E and F). In fact, the total effect of Slt2 on the response gene was much greater in this second CCA than in the initial CCA (Figure 4E). However, in the chimeric colony comparison for the second CCA, unlike in the initial CCA, prIME1-LacZ was expressed to approximately the same level in wild-type colonies as in slt2Δ colonies. Thus, Slt2 activates IME1 through an Rlm1-dependent cell nonautonomous mechanism, but it also represses IME1 through an Rlm1-independent cell-autonomous mechanism.
Effect of Rim101-Ime1 feedback loop on Rlm1 activity
Because the Rlm1-Slt2 feedback loop activates the Rim101-Ime1 feedback loop, we investigated other possible interactions between these two loops. First, we performed a CCA to determine the effect of IME1 on Rlm1 activity. In the pure colony comparison, ime1Δ colonies expressed the UASRlm1-LacZ response allele to significantly higher levels than wild-type colonies. In contrast, in the chimeric colony comparison, expression of UASRlm1-LacZ in ime1Δ colonies was approximately the same as in wild-type colonies (Figure 5A). Thus, Ime1 represses Rlm1 through a cell autonomous mechanism.
Figure 5.
Both autonomous an nonautonomous mechanisms divide meiotic fate from feeder-cell fate. (A) Chimeric whole colony expression assay measuring effect of ime1Δ on expression of UASRlm1-LacZ. Pure colonies of UASRlm1-LacZ strains that are either IME1 (WT) (blue, SH5069) or ime1Δ (black, SH5410) and chimeric colonies containing an ime1Δ UASRlm1-LacZ response strain mixed with either a WT (magenta, SH3881) or an ime1Δ (orange, SH5484) signal strain. Bars 1 and 2 are significantly different (P = 0.01, n = 8). (B) Chimeric whole colony expression assay measuring effect of IME1 overexpression on UASRlm1-LacZ in an ime2Δ background. Pure colonies of UASRlm1-LacZ strains that are either IME1 (WT) (blue, SH5778) or contain a high-copy IME1 plasmid (oeIME1) (black, SH5799) and chimeric colonies containing a UASRlm1-LacZ response strain mixed with either a WT (magenta, SH5848) or an oeIME1 (orange, SH5850) signal strain. (C) CCA measuring effect of RIM101 on UASRlm1-LacZ expression when IME1 is absent. Pure colonies of UASRlm1-LacZ strains that were either RIM101 (WT) (blue, SH5186) or rim101Δ (black, SH5721) and chimeric colonies containing a rim101Δ UASRlm1-LacZ response strain mixed with either a RIM101 (magenta, SH3972) or a rim101Δ (orange, SH5865) signal strain. All strains were ime1Δ. Bars 3 and 4 are significantly different (P = 0.01). (D) CCA measuring effect of rlm1Δ on Rim20-GFP foci formation. Pure colonies of Rim20-GFP strains that were either RLM1 (WT) (blue, SH5252) or rlm1Δ (black, SH5768) and chimeric colonies containing a RIM20-GFP response strain mixed with either a WT (magenta, SH3830) or an rlm1Δ (orange, SH4800) signal strain. (E) CCA measuring effect of RIM101 on prIME1-LacZ expression in absence of IME1. Pure colonies of prIME1-LacZ strains that were either RIM101 (WT) (blue, SH3830) or rim101Δ (black, SH5312) and chimeric colonies containing a rim101Δ prIME1-LacZ response strain mixed with either WT (magenta, SH4414) or rim101Δ (orange, SH4563) signal strains. All strains used were ime1Δ. Bars 3 and 4 are significantly different (P = 0.03). (F) CCA measuring effect of RIM101 on prIME1-LacZ/expression when strains lack both IME1 and RLM1. Pure colonies of prIME1-LacZ strains that were either RIM101 (WT) (blue, SH4800) or rim101Δ (black, SH5729) and chimeric colonies containing a prIME1-LacZ response strain mixed with WT (magenta, SH4767) or rim101Δ (orange, SH5702) signal strains. All strains were ime1Δ and rlm1Δ.
As an independent test of the effect of IME1 on Rlm1 activity described above, we measured the effect of overexpressing IME1 using a high-copy plasmid (Honigberg and Lee 1998) on UASRlm1-LacZ expression (Figure 5B). All strains in these CCAs were ime2Δ, so they cannot initiate meiosis. In the pure colony comparison, overexpressing IME1 (oeIME1) inhibited UASRlm1-LacZ expression relative to the wild type. In the chimeric colony comparison, oeIME1 in the signal strain had no effect on UASRlm1-LacZ expression relative to the wild type. Thus, our results with both ime1Δ and oeIME1 colonies indicate that IME1 represses Rlm1, and that it does so through a cell autonomous mechanism.
Since Ime1 represses Rlm1 activity, we next performed a CCA to determine whether Rim101 also represses this activity. To eliminate the possibility that Rim101 regulates Rlm1 by activating IME1, these CCA experiments were performed in ime1Δ strains. In the pure colony comparison, rim101Δ colonies displayed increased UASRlm1-LacZ expression relative to RIM101 colonies. In the chimeric colony comparison, rim101Δ colonies also displayed increased UASRlm1-LacZ relative to wild-type colonies (Figure 5C). Thus, in contrast to the cell-autonomous repression of Rlm1 activity by IME1, Rim101 represses Rlm1 through a cell-nonautonomous mechanism.
The above result, along with the discovery that the Rim101 pathway is active in both colony layers (Figure 1A), and that this pathway is activated 2–3 days after Rlm1 (Figure 2E), suggests the hypothesis that colony alkalization activates Rim101 in feeder cells, and this activation inhibits Rlm1 activity in these cells. To test this possibility, we determined the effect of pH on UASRlm1-LacZ expression in RIM101 and rim101Δ strains. Both strains contain ime1Δ mutations so Ime1-dependent controls will be inactive. We found that, in RIM101 strains, UASRlm1-LacZ was expressed to significantly higher levels in medium buffered to pH 7.0 than in medium buffered to pH 9.0. In contrast in a rim101Δ strain, UASRlm1-LacZ was expressed at the same high level at either pH (Figure S4). These results are consistent with the idea that, as colonies mature and cells in the central region of the colony initiate meiosis, respiration in these cells results in increased pH not only in the upper layer of the colony, but also in the underlying feeder cells, activating Rim101, and, hence, repressing Rlm1 in this lower cell layer.
Rlm1 does not directly affect Rim101 activity
We next tested the reciprocal relationship to the one described above; namely, we asked whether Rlm1 regulates Rim101. For this purpose, we performed a CCA measuring the effect of rlm1Δ on Rim20-GFP foci formation. To eliminate IME1-dependent effects, this CCA used only ime1Δ strains. We found that in either the pure or chimeric colony comparisons, there was no significant effect of rlm1Δ on Rim20-GFP foci formation (Figure 5D). Thus, Rlm1 does not activate the Rim101 pathway independently of IME1.
Rim101 represses IME1 through an Rlm1-dependent mechanism
Interestingly, Rim101 both activates IME1 (Figure 3C) and represses the IME1-activator, Rlm1 (Figure 5C). To investigate these opposite effects, we repeated the initial rim101Δ prIME1-LacZ CCA as in Figure 3C except that strains in this second CCA were ime1Δ so that the Rim101-Ime1 feedback loop would be disabled. As expected from the initial CCA, in the pure colony comparison from the second CCA, prIME1-LacZ was expressed at higher levels in wild-type than in rim101Δ colonies (Figure 5E, left two bars). In contrast, unlike the initial CCA, for the chimeric colony comparison, prIME1-LacZ was expressed at lower levels in wild-type colonies than in rim101Δ colonies (Figure 5E, right two bars). To confirm this result, we also performed a CCA measuring the effect of rim101Δ on a second meiotic response gene, prIME2-GFP with all strains being ime2Δ, and similar results were obtained as with the IME1 response gene (Figure S5). These results indicate that in an imeΔ background, Rim101 activates the meiotic program autonomously while repressing this program nonautonomously.
How might Rim101 nonautonomously repress IME1? Our findings indicate that Rim101 represses Rlm1 and Rlm1 activates IME1; thus, Rim101 might repress IME1 by repressing Rlm1. To test this idea, we repeated the same CCA as in Figure 5E except with all strains being rlm1Δ as well as ime1Δ. In these latter chimeric colonies, autonomous activation of prIME1-LacZ by Rim101 was still observed, but nonautonomous repression of IME1 by Rim101 was not (Figure 5F). Thus, Rim101 activates IME1 through an Rlm1-independent autonomous mechanism while repressing this same gene through an Rlm1-dependent nonautonomous mechanism.
Discussion
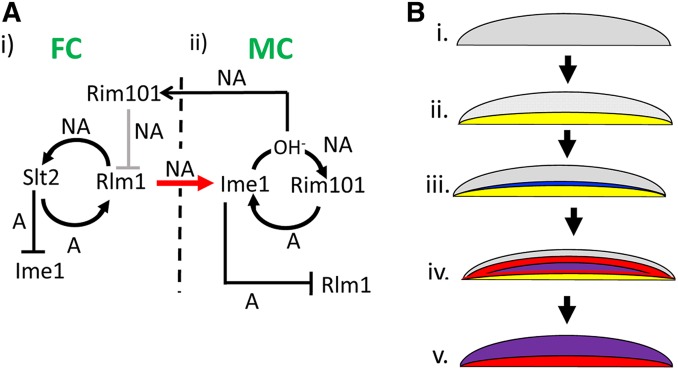
Many microbial communities are strikingly organized with respect to cell types, but the mechanisms underlying this organization remain mysterious. The results presented above, and in previous studies (Piccirillo et al. 2015, 2017), lead to a model for colony organization/pattern formation in yeast. These findings include localization of gene expression and identification of cell–cell signaling within colonies as cataloged diagrammatically in Figure S2; a model derived from these results is shown in Figure 6. By this model, colony pattern formation consists of five sequential steps. First, a cell nonautonomous positive feedback loop involving Rlm1 and Slt2 is activated in the bottom layer of cells (Figure 6A, i), causing them to differentiate into feeder cells (Figure 6B, i and ii, region of Rlm1 activity shown in yellow). Second, these feeder cells, which are more permeable than undifferentiated cells, produce a signal that triggers Ime1 expression in a narrow layer of cells overlying the feeder cell layer (Figure 6A, i and ii and Figure 6B, iii, Ime1 expression shown in blue). Once Ime1 is expressed, this initiates a second nonautonomous positive feedback loop involving Ime1 and Rim101 (Figure 6A, ii). Third, this second feedback loop allows the gradual expansion of the initial narrow layer of meiotic cells, driven by a gradual expansion of the region of increased pH, (Figure 6B, iv, Rim101 activation shown in red, and activation of both IME1 and Rim101 shown in purple). Fourth, through cell autonomous mechanisms, Slt2 inhibits IME1 expression in the lower layer (Figure 6A, i), whereas Ime1 inhibits Rlm1 activity in the top layer (Figure 6A, ii). Fifth, at late stages of colony development, the Ime1-Rim101 feedback loop becomes active throughout the top of the colony, and colony alkalization spreads to the feeder cell layer, activating Rim101, and, hence, repressing Rlm1 activity in this layer (Figure 6A, i and Figure 6B, v). Through these mechanisms, IME1 expression is limited to the top layer of the colony, whereas Rlm1 activity is transient and limited to the lower layer.
Figure 6.
Model of colony patterning through linked Rim101-Ime1 and Rlm1-Slt2 nonautonomous positive feedback loops (A) Genetic circuitry of pattern formation. Arrows represent activation and T-bars represent repression; autonomous or nonautonomous regulation is represented by an A or NA respectively adjacent to the arrow or bar. (i) Positive feed-back loop in feeder cells (FCs) initiate colony development. FCs in the bottom layer of the colony activate a nonautonomous positive-feedback loop containing Rlm1 and Slt2. Rlm1 expressed in FCs also induces IME1 expressed in meiotic cells (MCs), through a cell nonautonomous mechanism (red arrow). (ii) Positive feed-back loop in MCs during colony development. Once IME1 is activated by Rlm1, a nonautonomous positive feedback loop involving both Ime1 and Rim101 is activated. In this regulatory circuit, Rim101 activates IME1 transcription by a cell autonomous mechanism, and IME1 induces sporulation, and, hence, alkalization of the environment. This alkalization feeds back to activate Rim101 in neighboring cells, i.e., by a cell-nonautonomous mechanism. In addition, Ime1 represses Rlm1 through a cell-autonomous mechanism, and Rim101 represses Rlm1 through a cell-nonautonomous mechanism. Thus, Rlm1 is initially induced by the Rlm1-Slt2 feedback loop and later repressed by Rim101. (B) Spatial pattern of gene expression in developing colonies. (i) Colonies cease growth but are initially undifferentiated (gray). (ii) Nonautonomous Rlm1-Slt2 positive feedback loop activates Rlm1(yellow) in the lower (feeder cell) layer of the colony. (iii) As colonies develop, feeder cells activate Ime1 (blue) in a narrow layer of cells overlying the feeder layer. (iv) Ime1 expression raises pH and activates Rim101 expression (red) in neighboring cells, a Rim101-Ime1 positive feedback loop expands the region of activity of both genes (purple). (v) This expanding region of Rim101-Ime1 activity eventually includes the top of the colony. At the same time, expansion of alkaline pH and Rim101 activity (red) to the lower colony layer represses Rlm1 activity in this region.
Remarkably, the above model displays three types of regulatory symmetry between upper and lower colony layers. The first type of symmetry is that cell-nonautonomous positive feedback loops act in both layers. Although positive feedback regulation is expected from prior studies of both Ime1 (Shefer-Vaida et al. 1995; Moretto et al. 2018) and Rlm1 (Dodou and Treisman 1997; Watanabe et al. 1997; Jung et al. 2002; García et al. 2016), it is striking that positive feedback in both layers is cell-nonautonomous. A second type symmetry is that within the cells in which a feedback loop is expressed, the components of this loop repress components of the other loop. A final type of symmetry is that each feedback loop regulates activity of the other loop through cross-layer cell-to-cell signals.
This symmetry suggests that nonautonomous positive feedback loops, coupled with autonomous repression of alternative fates, may be a general mechanism for pattern formation. In particular, cell nonautonomous feedback loops can amplify initially small differences between colony microenvironments. As an example, a gradient of nutrients diffusing upward in the colony or oxygen diffusing downward could result in very different microenvironments at the top and bottom of colonies, but it is a riddle how this gradual change in microenvironment would lead to sharp boundaries between regions of colonies containing different cell types, such as the horizontal boundary that forms between meiotic cells and feeder cells near the center of a colony (Piccirillo et al. 2010, 2015). Cell nonautonomous feedback loops driven by cell-cell signaling may act to reinforce initially small environmental differences along either side of a boundary. At the same time, cell autonomous repression mechanisms would inhibit cells in a particular region from adopting an ectopic fate.
The Rim101-Ime1 feedback loop may explain our earlier observation that, as colonies develop, the sporulating region expands from a narrow central band to include the entire top half of the colony. Based on our results, we propose that this expansion is driven by a cycle of intracellular and extracellular events. First, increased extracellular pH activates the Rim101/AR pathway. Second, this pathway induces IME1 in the same cell. Third, IME1, by activating sporulation, stimulates respiration, and, hence, further increases extracellular pH. Thus the sporulation wave likely reflects a wave of alkalization. At the same time, increased colony pH reverses the activation of Rlm1 in the lower cell layer.
The model presented in Figure 6 can be considered in the context of pattern formation during metazoan development. In both yeast and metazoan pattern formation, undifferentiated cells partition into sharply divided regions that adopt mutually exclusive differentiation fates. However, close homologs to highly conserved master regulators of metazoan pattern formation, such as Notch, Wnt, Hedgehog, and bone morphogenic protein, are absent from the fungal genome. Thus, pattern formation likely evolved independently in these two clades. However, some features of the mechanism of pattern formation are similar in yeast colonies as in animals. In particular, a common feature of embryonic patterning models is “lateral induction”; i.e., a differentiating cell induces neighboring cells to adapt the same fate (reviewed in Sjöqvist and Andersson 2017). Lateral induction may be analogous to the nonautonomous Rim101-Ime1 and Rlm1-Slt2 feedback loops in yeast colony pattern formation. Indeed, spreading of the meiotic cell fate from the central boundary to the top of the colony is reminiscent of the waves of differentiation that characterize some models of metazoan development (Sato et al. 2013; Tateya et al. 2013). In contrast to the parallels between metazoan lateral induction and nonautonomous positive feedback in yeast, another element of metazoan pattern formation—lateral inhibition—is absent or not yet recognized in colonies. In lateral inhibition, a differentiated cell inhibits its neighbors from adapting the same fate, allowing the establishment and maintenance of boundaries between cell types (Dahmann et al. 2011; Schweisguth and Corson 2019). In yeast colonies, rather than lateral inhibition, boundaries may instead be enforced by cell-autonomous repression mechanisms that prevent cells from adopting the wrong fate.
The current study illustrates two general caveats regarding genetic analysis in microorganisms. First, alleles may affect distinct subpopulations of a microbial community in different or even opposite ways. Indeed, even suspended cultures contain populations with considerable expression and phenotypic heterogeneity (Schwabe and Bruggeman 2014; Honigberg 2016; Gasch et al. 2017; Li et al. 2018; Nadal-Ribelles et al. 2019). Second, contrary to usual expectations, alleles may affect biological processes in single-cell organisms by cell-nonautonomous as well as cell-autonomous mechanisms. As demonstrated in the current study, both of these caveats can be addressed by a combination of cell autonomy assays and single-cell or community colocalization assays. In this respect, representing regulatory relationships by adding an “A” (autonomous) or “NA” (nonautonomous) above arrow or bar symbols and also incorporating spatial and/or subpopulation information in the regulatory diagram as in Figure 6 could prove generally useful.
In the CCA, as with any measurement of gene expression in any cell population, increased expression of the response gene could result from increased expression in the same subpopulation and/or equal expression in a larger subpopulation. For the response genes used in the current study, positive feedback regulation likely causes these genes to be either fully on or fully off in most cells. Thus, the difference in total response in colony mostly reflects the fraction of cells in the colony that express the gene. Indeed, for all CCA presented in this study, an internal control verifies that doubling the fraction of response cells in a colony doubles the total signal (see Materials and Methods).
Because the organization of microorganisms into communities is central to their biological function, these communities offer a unique opportunity to investigate fundamental mechanisms of pattern formation. However, it is important to point out that any cellular process could in principal be regulated by a combination of cell autonomous and nonautonomous mechanisms. Indeed, it is very likely that many more biological processes are regulated by cell-to-cell signals than currently suspected.
Acknowledgments
We thank David Eide (University of Wisconsin-Madison) for the pZPS1-LacZ plasmid. Research reported in this publication was supported by the National Institutes of General Medical Sciences of the National Institutes of Health under award number R15 GM-094770. We acknowledge the use of the confocal microscope in the University of Missouri-Kansas City (UMKC) School of Dentistry Confocal Microscopy Core. This facility is supported by the UMKC Office of Research Services, UMKC Center of Excellence in Dental and Musculoskeletal Tissues, and National Institutes of Health (NIH) grants S10RR027668 and S10OD021665. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.9978863.
Communicating editor: A. Mitchell
Literature Cited
- Allocati N., Masulli M., Di Ilio C., and De Laurenzi V., 2015. Die for the community: an overview of programmed cell death in bacteria. Cell Death Dis. 6: e1609 10.1038/cddis.2014.570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boysen J. H., and Mitchell A. P., 2006. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol. Biol. Cell 17: 1344–1353. 10.1091/mbc.e05-10-0949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrejon F., Gomez A., Sanz M., Duran A., and Roncero C., 2006. The RIM101 pathway contributes to yeast cell wall assembly and its function becomes essential in the absence of mitogen-activated protein kinase Slt2p. Eukaryot. Cell 5: 507–517. 10.1128/EC.5.3.507-517.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D., Rozen D. E., Kuipers O. P., Sogaard-Andersen L., and van Wezel G. P., 2014. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 12: 115–124. 10.1038/nrmicro3178 [DOI] [PubMed] [Google Scholar]
- Dahmann C., Oates A. C., and Brand M., 2011. Boundary formation and maintenance in tissue development. Nat. Rev. Genet. 12: 43–55. 10.1038/nrg2902 [DOI] [PubMed] [Google Scholar]
- Dodou E., and Treisman R., 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17: 1848–1859. 10.1128/MCB.17.4.1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q., Kawabe Y., Schilde C., Chen Z. H., and Schaap P., 2015. The evolution of aggregative multicellularity and cell-cell communication in the dictyostelia. J. Mol. Biol. 427: 3722–3733. 10.1016/j.jmb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach M. A., and Segre J. A., 2016. Signaling in host-associated microbial communities. Cell 164: 1288–1300. 10.1016/j.cell.2016.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A. G., Bird A. J., Evans-Galea M. V., Blankman E., Winge D. R. et al. , 2011. Zinc-regulated DNA binding of the yeast Zap1 zinc-responsive activator. PLoS One 6: e22535 10.1371/journal.pone.0022535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García R., Sanz A. B., Rodríguez-Peña J. M., Nombela C., and Arroyo J., 2016. Rlm1 mediates positive autoregulatory transcriptional feedback that is essential for Slt2-dependent gene expression. J. Cell Sci. 129: 1649–1660. 10.1242/jcs.180190 [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Yu F. B., Hose J., Escalante L. E., Place M. et al. , 2017. Single-cell RNA sequencing reveals intrinsic and extrinsic regulatory heterogeneity in yeast responding to stress. PLoS Biol. 15: e2004050 10.1371/journal.pbio.2004050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M., and Honigberg S. M., 2001. Effect of chromosomal locus, GC content and length of homology on PCR-mediated targeted gene replacement in Saccharomyces. Nucleic Acids Res. 29: 5156–5162. 10.1093/nar/29.24.5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M., Piccirillo S., and Honigberg S. M., 2005. Two-step method for constructing unmarked insertions, deletions and allele substitutions in the yeast genome. FEMS Microbiol. Lett. 248: 31–36. 10.1016/j.femsle.2005.05.018 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Ohkuni K., and Yamashita I., 1998. Control of division arrest and entry into meiosis by extracellular alkalization in Saccharomyces cerevisiae. Yeast 14: 905–913. [DOI] [PubMed] [Google Scholar]
- Honigberg S. M., 2004. Ime2p and Cdc28p: co-pilots driving meiotic development. J. Cell. Biochem. 92: 1025–1033. 10.1002/jcb.20131 [DOI] [PubMed] [Google Scholar]
- Honigberg S. M., 2011. Cell signals, cell contacts, and the organization of yeast communities. Eukaryot. Cell 10: 466–473. 10.1128/EC.00313-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg S. M., 2016. Similar environments but diverse fates: responses of budding yeast to nutrient deprivation. Microb. Cell 3: 302–328. 10.15698/mic2016.08.516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg S. M., and Lee R. H., 1998. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 4548–4555. 10.1128/MCB.18.8.4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung U. S., Sobering A. K., Romeo M. J., and Levin D. E., 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46: 781–789. 10.1046/j.1365-2958.2002.03198.x [DOI] [PubMed] [Google Scholar]
- Kassir Y., Adir N., Boger-Nadjar E., Raviv N. G., Rubin-Bejerano I. et al. , 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224: 111–171. 10.1016/S0074-7696(05)24004-4 [DOI] [PubMed] [Google Scholar]
- Lamb T. M., Xu W., Diamond A., and Mitchell A. P., 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276: 1850–1856 (erratum: J. Biol. Chem 276: 12476). 10.1074/jbc.M008381200 [DOI] [PubMed] [Google Scholar]
- Lee R. H., and Honigberg S. M., 1996. Nutritional regulation of late meiotic events in Saccharomyces cerevisiae through a pathway distinct from initiation. Mol. Cell. Biol. 16: 3222–3232. 10.1128/MCB.16.6.3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189: 1145–1175. 10.1534/genetics.111.128264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Giardina D. M., and Siegal M. L., 2018. Control of nongenetic heterogeneity in growth rate and stress tolerance of Saccharomyces cerevisiae by cyclic AMP-regulated transcription factors. PLoS Genet. 14: e1007744 10.1371/journal.pgen.1007744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., and Mitchell A. P., 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., 2012. The signaling mechanism of ambient pH sensing and adaptation in yeast and fungi. FEBS J. 279: 1407–1413. 10.1111/j.1742-4658.2012.08548.x [DOI] [PubMed] [Google Scholar]
- Mitchell A. P., Driscoll S. E., and Smith H. E., 1990. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 2104–2110. 10.1128/MCB.10.5.2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretto F., Wood N. E., Kelly G., Doncic A., and van Werven F. J., 2018. A regulatory circuit of two lncRNAs and a master regulator directs cell fate in yeast. Nat. Commun. 9: 780 10.1038/s41467-018-03213-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Ribelles M., Islam S., Wei W., Latorre P., Nguyen M. et al. , 2019. Sensitive high-throughput single-cell RNA-seq reveals within-clonal transcript correlations in yeast populations. Nat. Microbiol. 4: 683–692. 10.1038/s41564-018-0346-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M., 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189: 737–765. 10.1534/genetics.111.127126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N., Pitsouli C., and Shilo B. Z., 2012. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb. Perspect. Biol. 4: a005975 10.1101/cshperspect.a005975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., and Honigberg S. M., 2010. Sporulation patterning and invasive growth in wild and domesticated yeast colonies. Res. Microbiol. 161: 390–398. 10.1016/j.resmic.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., White M. G., Murphy J. C., Law D. J., and Honigberg S. M., 2010. The Rim101p/PacC pathway and alkaline pH regulate pattern formation in yeast colonies. Genetics 184: 707–716. 10.1534/genetics.109.113480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., Morales R., White M. G., Smith K., Kapros T. et al. , 2015. Cell differentiation and spatial organization in yeast colonies: role of cell-wall integrity pathway. Genetics 201: 1427–1438. 10.1534/genetics.115.180919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., Neog D., Spade D., Van Horn J. D., Tiede-Lewis L. M. et al. , 2017. Shrinking daughters: Rlm1-dependent G1/S checkpoint maintains Saccharomyces cerevisiae daughter cell size and viability. Genetics 206: 1923–1938. 10.1534/genetics.117.204206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., and Hieter P., 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sanz A. B., Garcia R., Rodriguez-Pena J. M., and Arroyo J., 2017. The CWI pathway: regulation of the transcriptional adaptive response to cell wall stress in yeast. J. Fungi (Basel) 4: PMC5872304. 10.3390/jof4010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Suzuki T., and Nakai Y., 2013. Waves of differentiation in the fly visual system. Dev. Biol. 380: 1–11. 10.1016/j.ydbio.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Schwabe A., and Bruggeman F. J., 2014. Single yeast cells vary in transcription activity not in delay time after a metabolic shift. Nat. Commun. 5: 4798 10.1038/ncomms5798 [DOI] [PubMed] [Google Scholar]
- Schweisguth F., and Corson F., 2019. Self-organization in pattern formation. Dev. Cell 49: 659–677. 10.1016/j.devcel.2019.05.019 [DOI] [PubMed] [Google Scholar]
- Serra-Cardona A., Canadell D., and Arino J., 2015. Coordinate responses to alkaline pH stress in budding yeast. Microb. Cell 2: 182–196. 10.15698/mic2015.06.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank E. A., and Kolter R., 2011. Extracellular signaling and multicellularity in Bacillus subtilis. Curr. Opin. Microbiol. 14: 741–747. 10.1016/j.mib.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer-Vaida M., Sherman A., Ashkenazi T., Robzyk K., and Kassir Y., 1995. Positive and negative feedback loops affect the transcription of IME1, a positive regulator of meiosis in Saccharomyces cerevisiae. Dev. Genet. 16: 219–228. 10.1002/dvg.1020160302 [DOI] [PubMed] [Google Scholar]
- Sjöqvist M., and Andersson E. R., 2017. Do as I say, Not(ch) as I do: lateral control of cell fate. Dev. Biol. 447: 58–70. 10.1016/j.ydbio.2017.09.032 [DOI] [PubMed] [Google Scholar]
- Tateya T., Imayoshi I., Tateya I., Hamaguchi K., Torii H. et al. , 2013. Hedgehog signaling regulates prosensory cell properties during the basal-to-apical wave of hair cell differentiation in the mammalian cochlea. Development 140: 3848–3857. 10.1242/dev.095398 [DOI] [PubMed] [Google Scholar]
- Thevenaz P., Ruttimann U. E., and Unser M., 1998. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7: 27–41. 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- Váchová L., and Palková Z., 2018. How structured yeast multicellular communities live, age and die? FEMS Yeast Res. 18 10.1093/femsyr/foy033 [DOI] [PubMed] [Google Scholar]
- van Gestel J., Vlamakis H., and Kolter R., 2015. Division of labor in biofilms: the ecology of cell differentiation. Microbiol. Spectr. 3: MB-0002-2014 10.1128/microbiolspec.MB-0002-2014 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Takaesu G., Hagiwara M., Irie K., and Matsumoto K., 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17: 2615–2623. 10.1128/MCB.17.5.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Wang Q., Liu C., Shang Y., Wen F. et al. , 2018. A role for the respiratory chain in regulating meiosis initiation in Saccharomyces cerevisiae. Genetics 208: 1181–1194. 10.1534/genetics.118.300689 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains used in this study are described in Table S1 and are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.9978863.