In this work, Corbett-Detig et al. use proximity-ligation sequencing (Hi-C) of several Anopheles gambiae and A. coluzzii inversionbearing individuals to detect and map inversion breakpoints. They show that inversion breakpoints can be mapped precisely...
Keywords: Anopheles, Chromosomal Inversion, Hi-C
Abstract
Chromosomal inversions are fundamental drivers of genome evolution. In the main Afrotropical malaria vector species, belonging to the Anopheles gambiae species complex, inversions play an important role in local adaptation and have a rich history of cytological study. Despite the importance and ubiquity of some chromosomal inversions across the species complex, inversion breakpoints are often challenging to map molecularly due to the presence of large repetitive regions. Here, we develop an approach that uses Hi-C sequencing data to molecularly fine-map the breakpoints of inversions. We demonstrate that this approach is robust and likely to be widely applicable for both identification and fine-mapping inversion breakpoints in species whose inversions have heretofore been challenging to characterize. We apply our method to interrogate the previously unknown inversion breakpoints of 2Rbc and 2Rd in An. coluzzii. We found that inversion breakpoints occur in large repetitive regions, and, strikingly, among three inversions analyzed, two breakpoints appear to be reused in two separate inversions. These breakpoint-adjacent regions are strongly enriched for the presence of a 30 bp satellite repeat sequence. Because low frequency inversion breakpoints are not correlated with genomic regions containing this satellite, we suggest that interrupting this particular repeat may result in arrangements with higher relative fitness. Additionally, we use heterozygous individuals to quantitatively investigate the impacts of somatic pairing in the regions immediately surrounding inversion breakpoints. Finally, we discuss important considerations for possible applications of this approach for inversion breakpoint identification in a range of organisms.
CHROMOSOMAL inversions, reversals in the linear map order of chromosomes, are among the primary drivers of genome structure evolution across diverse species (Krimbas and Powell 1992; Hoffmann and Rieseberg 2008). Because they suppress recombination in heterozygous individuals, chromosomal inversions can maintain combinations of alleles that are more fit in similar contexts. Inversions are therefore theorized to be key contributors to local adaptation (Kirkpatrick and Barton 2006), speciation (Noor et al. 2001), and the maintenance of complex multigenic phenotypes (Lowry and Willis 2010; Joron et al. 2011). Owing to their myriad roles, uncovering the molecular and fitness consequences of inversions is a central goal for addressing numerous fundamental questions in evolutionary biology.
In the Anopheles gambiae species complex, inversions are known to play an important role in facilitating adaptation to a broad range of environments, and to affect behavioral traits that may enhance their efficiency for vectorial capacity (Coluzzi et al. 1979; Petrarca et al. 2000; Rocca et al. 2009; Cheng et al. 2012; Ayala et al. 2014, 2017). In particular, inversions affecting chromosome 2R are disproportionately common in the species’ genomes. Because this bias is evident in both common and rare inversions, it is thought to reflect a widespread mutational bias where inversions occur preferentially on this chromosome arm (Pombi et al. 2008). Furthermore, along 2R, specific cytological bands are strongly overrepresented for the presence or absence of inversion breakpoints, possibly consistent with mutational biases affecting the distribution of inversion breakpoints on short genomic scales as well (Coluzzi et al. 2002; Pombi et al. 2008). Uncovering the mutational patterns correlated with widespread chromosomal inversions in the An. gambiae species complex is key to understanding the ecological and evolutionary prospects for this group.
The precise identification and characterization of inversion breakpoints is a fundamental goal of evolutionary genomics. Breakpoint adjacent regions experience little or no recombination between arrangements and are particularly valuable for inferring the evolutionary histories of inversions (Wesley and Eanes 1994; Corbett-Detig and Hartl 2012), and provide ideal substrates for designing arrangement-specific PCR assays (e.g., Andolfatto et al. 1999; White et al. 2007; Lobo et al. 2010). Additionally, the genomic regions and specific structure of inversion breakpoints can yield key information about the molecular mechanisms underlying inversion formation, as well as the potential functional consequences of chromosomal inversions (Wesley and Eanes 1994; Puig et al. 2004; Sharakhov et al. 2006). Nonetheless, precisely mapping inversion breakpoints at the molecular level is not always straightforward due to the presence of repetitive elements and large-scale duplications that are sometimes found in breakpoint adjacent regions of the genome.
Extensive variation in inversion breakpoint structure impacts the prospects of successfully mapping them. Inversion breakpoints sometimes occur as simple “cut-and-paste” changes in unique sequences (e.g., Andolfatto et al. 1999; Ranz et al. 2007; Corbett-Detig et al. 2012), or they induce inverted duplications in breakpoint adjacent regions via a “staggered-break” in otherwise largely unique sequences (e.g., Sharakhov et al. 2006; Ranz et al. 2007). Nonetheless, it is perhaps more common for breakpoints to occur in, or to generate, structurally complex regions that include repetitive elements (Cáceres et al. 1999; Lobo et al. 2010; Aguado et al. 2014). The former type of inversion breakpoint is relatively easily mapped using standard short-insert Illumina sequencing, as long as breakpoints occur in sequences that are otherwise unique in the reference genome (e.g., Cridland and Thornton 2010; Corbett-Detig et al. 2012). The latter can be particularly challenging to identify, and often require the development of sophisticated molecular approaches (e.g., Aguado et al. 2014).
In the An. gambiae species complex, some important inversion breakpoints have proven to be a persistent challenge for accurate breakpoint detection and assembly. In particular, Lobo et al. (2010) used three Sanger assemblies (PEST, Pimperena, and Mali-NIH) together with directed BAC clone sequencing to accurately detect one of the breakpoints of 2Rb and one in 2Rbc, but were unable to identify the other breakpoints of either arrangement. The detected breakpoint contains a number of repetitive sequences and assembly gaps, suggesting that this has been an important impediment to sequence-based detection of inversion breakpoints for these species. As it is relevant through the manuscript, we note that 2Rc is virtually always associated with 2Rb in the An. gambiae species complex, so previous studies and ours necessarily rely on 2Rbc individuals to study 2Rc. More recently, Kingan et al. (2019) produced a de novo PacBio-based assembly of An. coluzzii. Despite high contiguity, we show here that their assembly fails to span important repetitive regions adjacent to known, and our predicted, inversion breakpoints. Thus, some of the inversion breakpoint adjacent regions in the An. gambiae species complex have been challenging to detect and assemble using an array of genome sequencing technologies.
Proximity-ligation sequencing, or Hi-C, has recently emerged as a powerful method of detecting chromosome structure variation (Harewood et al. 2017; Himmelbach et al. 2018). Briefly, this technology enables one to sequence short reads from DNA molecules that existed close together in the chromatin of living cells, but not necessarily adjacent to each other in the primary chromosome sequence (Lieberman-Aiden et al. 2009). Importantly, Hi-C often produces read pairs that span large distances along a chromosome. Consequently, the complexity of breakpoint adjacent sequences has little impact on the ability to detect chromosomal inversions, but it is not always possible to resolve the breakpoint structures at the sequence level. Despite strong interest and several recent applications, there are few straightforward and automated approaches for basepair resolution characterization of structural variation breakpoints using Hi-C data.
One complication for the successful application of proximity-ligation sequencing for identifying inversion breakpoints is the presence of somatic chromosome pairing which is prevalent in Dipterans, including An. gambiae and Drosophila melanogaster (Grell 1946). Somatic pairing occurs when homologous chromosomes are maintained in close physical proximity to each other in the interphase nucleus. This might be important for gene expression because enhancers on one paired chromosome may be able to initiate transcription of genes on the other, an effect known as transvection (Fukaya and Levine 2017). Inversion breakpoints interfere with the somatic pairing in heterozygotes, and may affect the pairing and allele proximity of breakpoint adjacent regions in heterokaryotypic individuals (Golic and Golic 1996). In particular, somatic pairing is expected to obscure the signal of physical proximity because paired chromosomes in heterokayotypes bring together genomic regions that are not adjacent along either chromosome’s two-dimensional genome sequence. Hi-C-based identification of inversion breakpoints is therefore likely to be impacted by somatic pairing and the application of proximity-ligation sequencing methods offers an opportunity to quantitatively investigate this phenomenon in a precise, high throughput format.
Here, we apply Hi-C proximity-ligation sequencing to identify inversion breakpoints of 2Rbc and 2Rd arrangements in An. coluzzii. We develop a simple approach for fine-mapping the positions of inversion breakpoints using Hi-C data, and we use this to discover that all breakpoints in the inversions we study here occur in regions that contained repetitive elements. Because Hi-C assays sequence proximity within chromatin, this method also enabled accurate estimation of the impact of somatic pairing on contact frequencies in the regions adjacent to chromosomal inversion breakpoints. Strikingly, between just three inversions (c, d, and b), there are only four unique breakpoint regions as two were reused twice. Three breakpoints contain large arrays of the same satellite repeat. Our results suggest that repetitive regions are an important contributor to inversion formation or retention in the An. gambiae species complex.
Materials and Methods
Anopheles gambiae s.l. colonies
We obtained adult or larval mosquitoes of the Pimperena (2Rb), Mali-NIH (2La, 2Rbc), and Ndokayo (2R+b;+c) colonies from BEI Resources (Anopheles program; https://www.beiresources.org/AnophelesProgram/Anopheles/WildStocks.aspx). In addition, carcasses of homokaryotypic and heterokaryotypic 2Rd carriers were selected by cytological analysis of ovarian polytene chromosomes (della Torre 1997) of half-gravid females from a 2Rd-polymorphic An. coluzzii Banfora M colony (Liverpool School of Tropical Medicine and Hygiene, LSTMH, UK), established from samples collected in 2014 from Banfora District, Burkina Faso, by LSTMH with support from the Centre National de Recherche et de Formation sur le Paludisme (CNRFP, Burkina Faso). Samples were kept at −80° until library preparation.
Hi-C library preparation and sequencing
To extract nuclei, we placed five adult mosquitoes into a dounce homogenizer, and used 5–10 strokes of the pestle to homogenize the contents. In preliminary analyses, we found that this substantially improved the quality of resulting libraries and reproducibility. We next produced Hi-C libraries as described in Lazar et al. (2018). Specifically, the resulting homogenate was transferred to a 1.5 ml microcentrifuge tube, spun down, and resuspended in cold PBS before adding paraformaldehyde (EMS Catalog 15714) to a final concentration of 1%. Following a 15-min incubation at room temperature, crosslinked nuclei and cells were washed twice by alternatively centrifuging and resuspending in cold PBS. A final spin was performed before addition of a hypertonic buffer (50 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA) and SDS to a final concentration of 1%. Samples were subsequently vortexed until visibly homogenous to extract crosslinked chromatin.
Crosslinked chromatin samples were combined with SPRI beads in 18% PEG 8000 and allowed to bind for 10 min at room temperature. Bead-bound samples were washed three times before digesting with DpnII (20 unit, Catalog R0543S; NEB) for 1 hr at 37° in a Benchmark Multi-Therm thermal shaker (Catalog H5000-HC; Benchmark). After washing the digestion products twice, Biotin-11-dCTP (Catalog CC-6002-1; ChemCyte) was incorporated by DNA Polymerase I, Klenow Fragment (10 unit, Catalog M0210L; NEB) for 30 min at 25°. Following two additional washes, biotinylated blunt ends were ligated with T4 DNA Ligase (4000 unit, Catalog M0202T; NEB) overnight at 16°. We digested proteins to release proximity-ligated DNA in an 8% SDS solution with Proteinase K (Catalog 19133; Qiagen) for 15 min at 55° and then 45 min at 68°.
We used SPRI beads in 18% PEG 8000 to purify DNA from the resulting supernatant before samples were split into two replicates and sonicated to an average length of 350 bp using a Diagenode Bioruptor NGS platform. Library Preparation for Illumina Sequencing End Preparation and Adaptor Ligation reactions were carried out using the NEBNext Ultra II DNA Library Prep Kit for Illumina (Catalog E7645S; NEB) on each sonicated sample, following the manufacturer’s recommendation but using custom Y-adaptors in place of the NEB-preferred hairpin loop variant. Adaptor ligation products underwent SPRI bead purification before biotinylated molecules were captured by room temperature incubation with Dynabeads MyOne Streptavidin C1 beads (Catalog 65002; ThermoFisher) for 30 min with shaking. Streptavidin-bound samples were washed thoroughly before indexing PCR with KAPA HiFi HotStart ReadyMix (Catalog KK2602; KAPA) and unique forward and reverse indexing adaptors. Library molecules were purified from the resulting supernatant and simultaneously size-selected using SPRI beads. DNA yield was quantified by Qubit 2.0 fluorometer using a High Sensitivity dsDNA kit (Catalog Q32854; ThermoFisher), and the average length of each library was determined via TapeStation 2200 using a D1000 screentape (Catalog 5067- 5582; Agilent).
Read mapping and filtering
This library preparation procedure uses a blunt-end repair and therefore causes junctions between fragments to be demarcated with two intact copies of the enzyme’s recognition sequence. In this case “GATC.” We therefore searched all reads for the characteristic “GATCGATC” sequence, and replaced that sequence plus any remaining sequence on the read with a single GATC, which must have been present in the sequence.
We mapped trimmed short-read data to the AgamP4 An. gambiae reference genome using BWA v0.7.17 using the mem alignment function. We filtered all reads with a mapping quality of <30, and removed all reads whose pairs did not successfully map to the reference genome. Additionally, we removed read pairs that map within 1 kb of each other in an attempt to remove self-ligated molecules. We note that, because the distance between read pairs is based on mapping to a reference genome, if self-ligated molecules spanned an inversion breakpoint (e.g., this is possible if a breakpoint occurs in largely unique sequence), they would not necessarily be removed. However, including those read pairs would likely improve the performance of this approach despite being a self-ligated molecule. Regardless, such occurrences should have no impact here because our breakpoints appear to localize to large repetitive blocks of sequence.
Because our filtering strategy will remove reads that map to repetitive regions of the genome, this precludes any attempt to accurately map breakpoints, and, more generally, to characterize the intervening repeat content. However, the ambiguity associated with short-read mapping positions within repetitive elements is typically too challenging for most genomic analyses. We suggest that, if identifying the specific breakpoint positions within repetitive elements is desirable, an alternative approach should be used. For example, an ultralong read strategy potentially coupled with a sequence capture approach may be preferable because unique repeat cluster adjacent sequences could be used to “anchor” long reads and map into repetitive content.
Inversion detection
To evaluate the performance of the coarse grid search detection procedure that we described here, we began by evaluating the impact of a proposed inversion breakpoint in arrangements that we know from cytological evidence are consistent with the arrangement of the reference genome. That is, we proposed a grid of pairwise inversion breakpoint positions, computationally “reversed” that genomic region within the read pair mapping coordinates, and recomputed the total distance spanned by all read pairs after this. We then recorded the minimum of this ratio across the full grid search.
Simulating inversions
To evaluate the specificity of the coarse grid search detection procedure for detecting putative inversion breakpoints, and for determining the precision of the fine-mapping procedure, we computationally inverted a portion of the reference genome by transposing read pair mapping positions of our proximity ligation data to be consistent with the rearranged reference genome. We selected the distal inversion breakpoints at random from a uniform (1, chromosome length-inversion size) distribution, for a range of inversion sizes (0.5, 1, 2.5, 5, and 10 Mb). The proximal breakpoint position would then be equivalent to the distal breakpoint plus the inversion size. We performed 100 replicates for each size and recorded the minimum ratio of total distances spanned by read pairs for inverted and “reference” (computationally inverted) mapping positions. All grid searches were performed with a distance between adjacent points of 250 kb unless otherwise stated. A script to perform this function “uninvert.py” is available from the github repository associated with this project.
Fine-scale breakpoint position estimation
We estimated 2Rc and 2Rd inversion breakpoint positions as a two-parameter optimization task. Specifically, we seek to minimize the total distance spanned by all read pairs surrounding inversion breakpoints by inputting possible breakpoints positions, “reversing” the inverted region, and recomputing the distance spanned by all read pairs. We implemented this procedure in python (Supplemental Material, File S1), and used the scipy optimize() function to implement a Nelder-Mead (Nelder and Mead 1965) two parameter optimization procedure. We evaluated the accuracy of our approach by comparing our estimated breakpoint positions for 2La and 2Rb, which have been identified previously (Sharakhov et al. 2006; Lobo et al. 2010). We also estimated the accuracy of our fine-mapping procedure by running this approach on our simulated inversion samples (see above) and performing 100 bootstraps on each sample.
To investigate the robustness of our method to decreasing read depths, we randomly subsampling read pairs and re-estimated inversion breakpoints at increasingly small read depths for 2Rb. We selected this inversion for our analysis because its breakpoints are known to be situated in repetitive regions, and it is therefore representative of the types of challenges we seek to resolve with this method.
Bootstrap confidence intervals
To obtain mapping position confidence intervals, we applied a nonparametric bootstrap procedure. This entails sampling read pairs with replacement from the subset that were used to map inversion breakpoints initially, and repeating our breakpoint estimation procedure 1000 times. We validated this approach by evaluating concordance between estimated confidence intervals and known inversion breakpoints 2La and 2Rb on both the full dataset and subsampled sets containing smaller subsets of our total read data that we used to evaluate the impact of decreased sequencing depths. Functionality to perform these procedures are included in our script, which is available from the github page associated with this manuscript.
Permutation tests
We tested for an enrichment for large blocks of repetitive sequences adjacent to the breakpoints of inversions in 2Rbc and 2Rd arrangements using a permutation test framework. Specifically, we drew positions for the four breakpoints randomly from all positions on 2R. We then computed the proportion of sites annotated as repetitive or assembly gaps within surrounding 40 kb windows, and asked if the mean repetitive/gap sequence content equalled, or exceeded, the amount we obtained from the true breakpoint positions. We then recorded the proportion of replicates that satisfied these criteria.
We also used a permutation test to ask if the breakpoint colocalization among separate inversions could be expected by chance. Here, we assume that all inversion breakpoint are sampled independently from the chromosome arm. We constructed large contiguous blocks of repetitive sequence by merging adjacent coordinates for repeats >100 bp and that occurred with 50 bp of another annotated repeat element of >100 bp. We did this because repeat clusters in close proximity often have different specific annotations, but are abbutting or nearly abutting each other, and, therefore, represent a single large repetitive region. To accommodate our uncertainty with the exact breakpoint positions within large continuous repetitive blocks of sequence, we recorded two breakpoints as colocalized when they coincide to within the same block of repetitive sequence or within 5 kb in coordinate space if we did not draw a position within one of these large repetitive regions. We then asked if each replicate permutation produced two or more colocalized breakpoints, and recorded the proportion of such tests. We performed each permutation procedure 10,000 times.
Comparing rare and common breakpoints
To compare the frequencies with which rare inversion breakpoints intersect cytological bands that contain the large satellite arrays we discovered adjacent to common inversion breakpoints, we relied on the dataset compiled in Pombi et al. (2008). Then, for the three cytological bands that we can confidently assign as containing these satellite arrays, i.e., because we mapped cytologically known inversion breakpoints to those regions, we calculated the number of rare and common inversions whose breakpoints intersect those bands. We compared the ratios of breakpoints within-bands to outside using Fisher’s exact test.
Computing structural concordance and enrichment
As a means of quantifying the impact of somatic pairing on the breakpoint-adjacent contact map, We also estimated the relative enrichment in the second and fourth quadrants surrounding each breakpoint. To do this, we begin with the observation that the physical distance along both the inverted and standard chromosomes separating these two regions are actually the same for each arrangement. Therefore, to normalize the number of links mapping to this region, and to reduce the impacts of variance among library preparations, we obtained the proportion of links that span a similar distance (defined here as the proportion of total read pairs of the same length ± 250 kb) across the rest of the genome in otherwise colinear regions. We then used this proportion to normalize the observed number of read pairs mapping into each quadrant in the two homozygous arrangements and the heterozygote. Finally, we computed the ratio of the normalized read pair mapping proportions relative to the standard arrangement homozygote for the 2Rd/2R+d and 2Rd/2Rd libraries.
Comparison to a long-read assembly
We accessed the long-read assembly of An. coluzzi presented in Kingan et al. (2019), ASM413651v2, to determine if breakpoint adjacent regions are more completely assembled in this alternative genome assembly. We aligned all contigs in the “primary” assembly to the AgamP4 genome assembly using minimap2 (Li 2018), and filtered alignments to require a minimum mapping quality of 40 and a minimum alignment length of 1 kb to consider any contig as breakpoint adjacent. We then filtered this set of contigs to recover the set that are adjacent (defined here as mapping within 50 kb) to predicted and known breakpoint regions for each of the chromosomal inversions on 2R that we included in our study.
Data availability
All sequence data produced in this work are available from the sequence read archive under project accession number PRJNA564850. Software to perform the breakpoint mapping and uncertainty quantification procedures described herein is available from github, www.github.com/russcd/proximity_ligation_inversion_mapping. Supplemental material available at figshare: https://doi.org/10.25386/genetics.10006487.
Results and Discussion
Stocks and sequencing results
We obtained samples for homokaryotypic carriers of 2Rb, 2Rbc, and 2Rd arrangements (see Materials and Methods). For each arrangement, we produced Hi-C libraries for pools of five whole adult mosquitos or 15–25 larvae following the library preparation protocol in Lazar et al. (2018). We sequenced each library on a fraction of a Hiseq 4000 lane and obtained 12 million read pairs on average per sample. In each library, 23.5–36.8% of all read pairs mapped at distances of 1 kb or greater. Ultimately, we obtained relatively modest read depths (7.47× on average), but, because of the long distances spanned between read pairs in Hi-C libraries, this corresponds to exceptionally high clone coverage (37,547× on average per sample per site, Figure S1 and Table S1).
Overview of mapping approach and terminology
The nature of Hi-C data itself suggests a simple approach for mapping inversion breakpoint positions. We note that there are several methods for detection of structural variants from Hi-C (e.g., Harewood et al. 2017; Himmelbach et al. 2018). Nonetheless, to our knowledge, none of these have been validated for automated fine-mapping specific locations of structural rearrangements. The primary reason is that, by using a read binning strategy, previous automated approaches have placed a lower bound limit on breakpoint resolution, and, when specific breakpoint sites were identified later, these approaches required manual curation.
We therefore developed, validated, and applied a simple alternative method of mapping inversion breakpoints that does not require read binning. The key insight is that, although Hi-C links often span long distances, the vast majority are still relatively short (Lieberman-Aiden et al. 2009). Therefore, if a sample has an inversion relative to the reference genome, this will artificially increase the apparent distances spanned by read pairs, particularly in the regions surrounding inversion breakpoints. More specifically, here, we consider the breakpoint positions of an inversion to define the axes within a Cartesian graph, i.e., the first breakpoint defines a vertical axis, and the second a horizontal axis. Because inversions create new proximities between genomic regions in the upper right (quadrant one), and in the lower left (quadrant three), but leave proximity unchanged for quadrants two and four, in the absence of other factors, we expect to see a significant excess of read pairs whose mapping positions place them in quadrants one and three. However, somatic pairing within heterokaryotypic individuals has the potential to interact with inverted arrangements to redistribute links in quadrants two and four as well (Figure 1). Importantly, even read pairs that map relatively distantly from the inversion breakpoints contain some information (albeit quite imprecise) about the locations of the breakpoints.
Figure 1.
Cartoon of the locations of read pairs sampled along the genome in three possible genomic arrangements. (A) standard (reference), (B) inversion homozygote, and (C) inversion heterozygote paired in a classic “inversion loop” structure. In each, the direction of sequence along the standard arrangement is shown with bold arrowheads. (Blue) read pairs linking the same chromosome, (Light Green) read pairs linking sister chromosomes within regions that are consistent with pairs between collinear chromosomes, (Dark Green) read pairs linking sites that are in close proximity on the inverted, but not on the standard arrangement chromosome, and (Violet) read pairs linking regions that are not adjacent in the sequence of either standard or inverted chromosomes, but are proximal as a consequence of somatic pairing in heterokaryotypes. (D–F) Expected read pair mapping positions for Hi-C reads derived for each arrangement as in (A–C) and mapped onto a standard arrangement coordinate system. The colors are the same as in (A–C). The vertical and horizontal dashed lines represent positions for the distal and proximal inversion breakpoints, respectively, and generate the Cartesian graph system that we refer to throughout. Note that the lines subdivide the possible mapping positions of Hi-C links based on the type of pairing that generates them.
This expectation for read mapping positions within inversion homozygotes outlined above suggests a simple approach for estimating inversion positions from the mapping positions of Hi-C short-read data. Specifically, we seek to minimize the distance spanned by read pairs by transposing the mapping positions along a chromosome as defined by proposed breakpoint sites. In other words, our approach is based on “uninverting” the read pairs for a given proposed set of breakpoints, and recomputing the total distance spanned by the set of read pairs (see uninvert.py in the github repository associated with this project). The optimum for the minimization function should then present a good estimate for inversion breakpoint positions. Importantly, this method should be able to leverage information even from read pairs distributed at large distances away from inversion breakpoints.
Detection and broad-scale mapping
Although in our application in this work, we focus on samples known to contain karyotypically defined inversions, in many cases the chromosomal arrangement for a newly sequenced sample may not be known. Additionally, as it relies on a type of local optimization, our proposed fine-mapping approach (detailed below) requires an estimate of the breakpoint starting position. This may not be readily available even if the sample is known to contain inversions relative to a reference genome. We therefore sought to resolve these challenges by beginning with a coarse grid search across the range of pairs of possible inversion breakpoint positions along a chromosome. That is, we computed the expected total distance spanned by all read pairs along a chromosome before and after “uninverting” a proposed pair of breakpoint positions. We suggest that it is often convenient to express the values obtained from this procedure as a ratio (uninverted total distance spanned:unmodified total distance spanned) to partially account for differences in sequencing efforts among experiments and for simplicity of interpretation.
We expect that, when an inversion is not present in a sample relative to the reference genome, proposed breakpoints will tend to increase the apparent total distance spanned by proximity ligation read pairs. This should be particularly evident when proposed inversions are quite large, and should have a much smaller effect when the proposed breakpoints are relatively nearby. However, in the absence of misassemlies or other naturally occurring structural variants, the ratio of the total distance spanned should never be substantially <1. Alternatively, when a single inversion is present that distinguishes the sample and the reference genome, the region immediately surrounding the coordinate pair defined by the true inversion breakpoints will produce a significantly smaller total distance spanned by all read pairs. In evaluating these expectations across the genome for samples from the Ndokayo sub-colony (+a;+b; Cheng et al. 2018), and for samples known to be homozygous for an inversion on chromosome arms 2L or 2R, we find the expected pattern. More specifically, the ratio of the total distance spanned by read pairs after accounting for the inversion relative to their span without an inversion is substantially <1 in all cases (0.83–0.98, Figure 2), and is strongly correlated with the inversion length, but moderately significant due to a relatively small sample size (Spearman’s Rho = 1, P = 0.0833). Furthermore, the minimum ratio observed was always the closest grid point to the true inversion breakpoints for known inversions and the closest point to our fine-mapping estimates (see below). This suggests that a coarse grid search approach might be a useful way to identify candidate chromosomal inversions and for identification of a starting position for fine-scale position estimate optimization.
Figure 2.
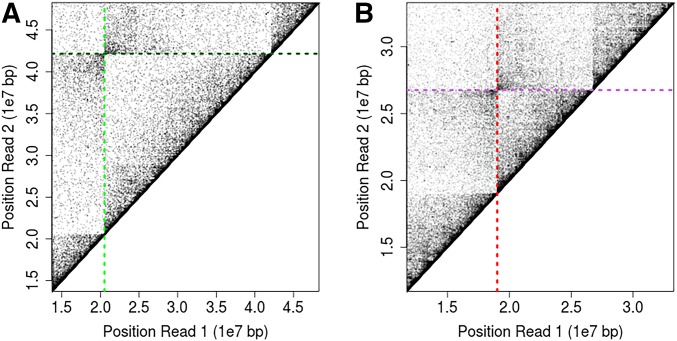
The proximity ligation read pair mapping positions for a sample homozygous for 2La (A), and the resulting proximity map after computationally reversing the inverted region based on its known coordinates (Sharakhov et al. 2006) (B). Ratio of the total distance spanned by all read pairs for a proposed inversion whose breakpoints intersect at the plotted position relative to the total distance spanned by standard arrangement reference mapping with no proposed inversion for the same 2La homozygote as above (C). The same ratio plotted across 2L for the same sample after computationally reversing the inverted region (D).
Sensitivity and specificity of inversion detection
We next sought to more directly evaluate the specificity and sensitivity of our proposed approach for detecting chromosomal inversions. Owing to the biological complexity, there is currently no means of simulating realistic proximity ligation sequencing data, we therefore relied on the fact that our samples have karyotypically characterized arrangements, and that many match the reference genome. In all pairwise positions considered in the coarse-grid search described above, we found a range of minimum ratios of 0.99993–1 for all grid points across each chromosome arm in all samples that were known to be of the reference arrangement. We therefore conclude that, for our samples, and for this reference genome, the false positive rate of this method should be easily minimized using a simple thresholding approach, as the minimum value is quite close to the expectation for this ratio of 1.
To evaluate the sensitivity of our method, we introduced inversions of sizes (10, 5, 2.5, 1, and 0.5 Mb) into the reference genome by converting mapping positions of the underlying read data (i.e., using uninvert.py from our github repository associated with this project). Note that this approach faithfully preserves the underlying biological complexity of the proximity ligation data, although it may not reflect the distribution of biologically possible inversion breakpoint positions. For all sizes of simulated inversion ≥1 Mb, the distribution of the minimum read-length ratios observed does not overlap the ratio we obtained from univerted chromosome arms. However, for the smallest size class considered, 0.5 Mb, the distribution of the minimum observed ratio does slightly overlap that for univerted genomes, suggesting that this approach may not accurately identify relatively short chromosomal inversions (Table S2).
Fine-mapping approach and validation
Another fundamental challenge that we seek to resolve is to fine-map inversion breakpoints using the same basic approach. That is, we should be able to find the minimum of the function that defines the total distance spanned by read pairs for a set of read coordinates. We therefore sought to optimize the joint breakpoint position estimates using a Nelder-Mead direct search downhill simplex algorithm to minimize the distance spanned by univerted read pairs (Nelder and Mead 1965). Our implementation is available from the github page associated with this manuscript (github.com/russcd/proximity_ligation_inversion_mapping) and contains additional helpful information for practical considerations associated with running this software (see also Materials and Methods). As we suggested above, this approach requires an estimate of inversion breakpoint position. Throughout this work, we provide our program with an estimate obtained using the coarse grid search procedure, but we found that the starting position has little impact so long as the positions are within ∼1 Mb.
Validation with known inversion breakpoints
We validated this method using two previously mapped chromosomal inversions (2La and 2Rb). Both have been successfully characterized previously. Inversion 2La is fixed within the An. coluzzii Mali-NIH colony, the same colony that we used to identify the breakpoints of 2Rbc (Sharakhov et al. 2006; Lobo et al. 2010), and 2Rb is fixed within the An. gambiae Pimperena colony (Table S1). We therefore applied our method to these inversion breakpoints first, and we obtained strong concordance between the known inversion breakpoint position and our predicted mapping positions (Figure 3 and Table S3). This suggests that this approach can accurately fine-map inversion breakpoints despite relatively modest sequencing read depths (White et al. 2007; Lobo et al. 2010).
Figure 3.
Validation of the fine-mapping Hi-C sequencing approach on An. gambiae inversions with predicted breakpoint positions shown. Mapping positions of Hi-C read pairs and predicted breakpoint positions show as vertical and horizontal lines for 2La (A) and 2Rb (B) inversions.
We additionally sought to validate our method across a broader range of possible breakpoint positions. To do this, we applied our fine-mapping procedure to the simulated inversions described above. As before we caution that this approach will retain the realistic structure of chromatin contact data, but not necessarily a realistic distribution of inversion breakpoint positions. After applying our method, we find that breakpoint position estimates are consistently quite close to the true breakpoint positions, and that the error in estimated populations is negatively correlated with the size of the inversion considered (Table 1). We therefore conclude that this method can yield accurate fine-scale breakpoint estimates for the sizes of inversions we considered here.
Table 1. Error in breakpoint position estimates and confidence intervals for simulated chromosomal inversions.
| Size (Mb) | Error in Position | Proportion in 95% CI | CI Width |
|---|---|---|---|
| 10 | 207.6 | 0.93 | 942.1 |
| 5 | 198 | 0.89 | 1059.4 |
| 2.5 | 255.2 | 0.95 | 1165.3 |
| 1 | 335 | 0.94 | 1301 |
| 0.5 | 342.7 | 0.96 | 1489.4 |
Robustness to lower read depths
We next sought to evaluate the robustness of our fine-mapping approach to a more modest sequencing effort. To do this, we subsampled the read pairs used to estimate inversion breakpoint positions focusing on inversion 2Rb (Lobo et al. 2010). We selected 2Rb because it is known to have repeat rich complex regions at both breakpoints, and is therefore an exemplar of the types of cases where we expect this approach to be the most often applied. Despite light read coverage in many replicate subsampled sets (as low as ∼0.2× mean read depth), we find that our method is able to consistently and accurately identify inversion breakpoint positions (Figure S1). This suggests that our approach can be applied even with relatively modest read depths and importantly that this method will be applicable even for extremely large genomes, which could be cost prohibitive to sequence deeply using Hi-C or long-read technologies.
Breakpoint position confidence intervals
In addition to a single point estimate for the breakpoint position, it may be desirable to quantify uncertainty in breakpoint position estimates by constructing mapping confidence intervals. We therefore implemented a nonparametric bootstrapping approach, wherein we randomly resampled read pairs with replacement for each breakpoint estimate and repeated the optimization procedure. Across 1000 bootstrap replicates for each inversion, 2La and 2Rb, the known breakpoint position was within the 95% confidence interval of estimated breakpoint positions. In the case of 2La, confidence intervals are fairly narrow, ∼5 and 2 kb for proximal and distal breakpoints, respectively. These sizes are slightly larger than the mean, but well within the distribution of confidence interval widths that we obtained when fine-mapping the breakpoints of 10 Mb simulated inversions. Presumably because the 2Rb breakpoint adjacent regions contain substantial repeat content, the breakpoint mapping confidence intervals are quite large and spans essentially from the edges of the large repeat region (Figure 4 and Table S3).
Figure 4.
Breakpoint positions of inversions 2Rbc and 2Rd in An. coluzzii and schematic of breakpoint adjacent sequences. The breakpoint mapping positions for individuals carrying 2Rb and 2Rc (A) and 2Rd (B), with predicted breakpoints indicated for 2Rb (red and violet), 2Rc (violet and blue), and 2Rd (blue and cyan). Each breakpoint-containing region is denoted with a single color. A schematic of chromosome arm 2R (C) with positions of inversions indicated and a breakpoint structure schematic of approximately 40 kb surrounding each breakpoint. These schematics include satellite repeat sequences (blue), assembly gaps (black), and other repeats (red). Repeat annotations are from vectorbase.org, based on the AgamP4 genome assembly and exclude all repeats of <100 bp in length. Breakpoint confidence intervals are plotted below on the same scale as the repeat structure for each inversion. When two inversions share an approximate breakpoint region, confidence intervals obtained from applying our mapping approach to each inversion separately are shown. For that reason, there are two confidence intervals for the two reused breakpoints (violet and blue).
In simulated datasets (see above), the width of the confidence intervals depend somewhat on the inversion lengths where longer inversions tend to have larger confidence intervals than shorter inversions. Nonetheless, ∼95% of the simulated inversions included the true breakpoint positions within 95% bootstrap confidence intervals for all size classes considered (Table 1). Additionally, we find that the confidence interval width is strongly correlated with the number of mapped reads within 1 kb of the true breakpoint position (P < < 0.05 for all inversion sizes considered, Spearman’s rank correlation). When we aggregate data across all simulated inversion sizes, we find a positive correlation between the probability that the 95% confidence interval contains the true breakpoint position and the number of informative read pairs that map within 1 kb (P = 0.02008, Wilcox test). However, no test is significant for individual inversion sizes, possibly due to smaller sample size (P > 0.05 for all sizes considered). These results therefore suggest that this approach can provide a reasonable approximation of the uncertainty associated with breakpoint positions; however, breakpoint estimates should be scrutinized, particularly when the number of informative read pairs in the surrounding genomic region is low.
Breakpoint structures of 2Rb, 2Rc, and 2Rd
We applied our approach to map the breakpoints of 2Rb, 2Rc, and 2Rd in An. coluzzii and characterized the sequences surrounding each breakpoint. For both inversions, we found that all breakpoints localized to large annotated repeat clusters containing both transposable elements and satellite repeat sequences in the standard arrangement AgamP4 reference assembly (www.vectorbase.org; Giraldo-Calderón et al. 2015). These regions are also often flanked by assembly gaps, suggesting that they have presented a persistent challenge for comprehensive genome sequencing and annotation. Because short-read data cannot be accurately mapped within highly repetitive regions, we note that breakpoint estimates cannot be more accurate than localizing inversion breakpoints to within a specific repeat/gap cluster. This result is reflected by our broad confidence intervals, which span the majority of repetitive regions for each of the inversion breakpoints that we evaluated here (Figure 4). Especially when a repeat cluster is relatively large, few or no reads will map uniquely within the repetitive region. Therefore, breakpoint estimates will only be accurate to within the repetitive region identified but the breakpoints cannot precisely localized within the repeat cluster.
Nonetheless, it is striking that each inversion breakpoint appears to be situated within large repetitive regions. In fact, the probability of selecting four regions at random along chromosome arm 2R with the same average rate of repetitive sequence annotation per basepair is small (P < 1e−4, Permutation Test see Materials and Methods), indicating that inversion breakpoint adjacent regions are strongly enriched for the presence of large blocks of repetitive sequences and assembly gaps.
It is also noteworthy that two repetitive regions appear to be reused to the level of large-scale repeat clusters between just these three inversions. This is even more surprising considering that 2Ru, another common inversions of this species complex, likely shares the same breakpoint as 2Rc and 2Rd share (Love et al. 2019). In light of the term’s long history in inversion biology, we clarify that in our terminology, “reuse” refers only to the recurrent breakage of the same large repeat cluster. Recurrent breaks within the same approximate repeat cluster is an extremely improbable event by chance (P < 1e−4, Permutation Test; see Materials and Methods). Specifically, we find reused breakpoints in the 2Rbc arrangement between the proximal breakpoint of 2Rb and the distal breakpoint of 2Rc, and between the proximal breakpoint of 2Rc and the distal breakpoint of 2Rd (Figure 4 and Table S3).
Previous work supports the shared breakpoint positions of 2Rc and 2Rd, which are cytologically indistinguishable, but not those of 2Rb and 2Rc within 2Rbc arrangement chromosomes, because of the presence of a thin band between the two breakpoints (Figure S2). This suggests that a relatively large genomic segment lies between them (Coluzzi et al. 2002). Nevertheless, it needs to be highlighted that, although the inclusion of the thin band between these inversion breakpoints was considered the most probable interpretation during the creation of the polytene chromosome map, cytological interpretation leaves some elements of uncertainty. Given the demonstrated accuracy of our mapping approach, one possible explanation for this discrepancy is that the breakpoints are close in reference coordinates but that the reference has a large gap, possibly due to the presence of large intervening collapsed repeat sequences. It is also possible that the cytological bands differ in appearance between arrangements possibly as a result of structural rearrangements (as in Semeshin et al. 2008).
Repeat sequences in breakpoint adjacent regions
We next sought to determine if specific sequences might be disproportionately represented in inversion breakpoint adjacent regions. Such sequences could yield clues into the mechanisms underlying inversion breakpoint formation or inversion retention in natural populations. Lobo et al. (2010), previously reported that the telomere proximal breakpoint of 2Rb contains a large array of an ∼30 bp satellite repeat sequence (Figure 4). It is therefore particularly noteworthy that we find the same satellite sequences, [(TTTTGCGATTGTCGCAAAAACTTYTGCGAC)n where Y indicates a C/T to accommodate the most common variant we observe] in large arrays at the shared 2Rc/2Rd breakpoint, and at the centromere proximal 2Rd breakpoint region (Figure 4). In fact, of the four arrays of this satellite across chromosome arm 2R, three intersect breakpoint-adjacent regions considered in this work (P < 1e−4, permutation test). These observations therefore strongly suggest that there is a mechanistic relationship between this specific satellite sequence and inversion breakpoint formation or retention in natural populations within the An. gambiae species complex.
More generally, of the other inversions whose breakpoints have been characterized in this species complex, 2La and 2Rj, neither has a breakpoint that is associated with this same satellite repeat sequence (Sharakhov et al. 2006; Coulibaly et al. 2007). Nonetheless, both of these inversions contain noncoding repeat elements at, or immediately adjacent to, their breakpoints. Collectively, the emerging pattern implicates repetitive elements as a consistent feature of inversion breakpoints in this species complex.
Possible evolutionary causes of breakpoint reuse in the An. gambiae species complex
If regions that contain this satellite sequence are simply more prone to breakage due to higher intrinsic fragility, breakpoint reuse could be expected as a consequence of neutral mutational biases (Krimbas and Powell 1992; Cáceres et al. 1997). Consistent with this idea, we note that the satellite sequence that we identified encodes at least two potential hairpin sequences (TTTTGCGATTGTCGCAAAAACTTYTGCGAC and TTTTGCGATTGTCGCAAAAACTTYTGCGAC, where the pairs of complementary sequences that could form hairpins are underlined). Additionally, the multi-copy nature of these satellite arrays could generate larger-scale secondary structures possibly with erroneous pairing between DNA strands. Extensive repeat arrays and hairpin sequences are known to result in chromosomal instability and increased rates of double-strand breaks (Lobachev et al. 2007). That breakpoints colocalize with specific, presumably unstable, repetitive satellite arrays suggests that a mutational bias might be an important contributor to variation in the fine-scale distribution of inversion breakpoint positions.
If a mutational bias associated with the shared satellite repeat element is the major force driving recurrent breakpoint formation that we report here, we expect that the breakpoints of low frequency (rare) inversions would intersect the cytological bands that contain these repetitive arrays at similar rates as those of high frequency (common) inversions that we described above. Pombi et al. (2008) compiled an extensive list of the cytologically mapped breakpoints of all of the known rare and common inversions in this species. We therefore compared the distributions of rare and common inversion breakpoints with respect to the proportion that intersect cytological bands containing these satellite repeat sequences. Cytological bands tend to encompass broader regions than each array, and the fine-scale positions might still differ substantially. This test is therefore conservative because a rare inversion breakpoint might intersect the same cytological band, but not intersect the satellite.
In performing this analysis, we found that, of the 12 common inversion breakpoints on 2R, five breakpoints are located within one of these three cytological bands. Conversely only 3 breakpoints of 134 total rare inversion breakpoints on chromosome arm 2R intersect the same cytological bands. This is substantially less than would be expected if the common and rare inversion breakpoints were equally likely to occur proximally to these satellite sequence (P = 0.0002, Fisher’s exact test). Even more intriguing, these arrays are the only annotated satellites on the autosomal chromosomes in the AgamP4 assembly that exceed 1 kb in length. This suggests that the rare inversion breakpoints are not generally associated with other repeat arrays, although it is possible that other satellites are less well assembled. Therefore we conclude that a biased mutagenic process might contribute to the observed colocalization of common inversion breakpoints and satellite arrays, but additional factors likely influence the fine-scale distribution of common inversion breakpoints.
Breakpoint colocalization with satellite arrays among common inversions might instead result if natural selection favors breakpoints that occur within or adjacent to these sequences. For example, breakpoints that occur in satellite arrays may be less disruptive to normal function than the average inversion breakpoint. Therefore the subset of inversions that reach high frequencies in natural populations would tend to have breakpoints that colocalize with repeat arrays or other “safe” genomic regions. However, it is not immediately obvious why this particular satellite should be so permissive to novel breakpoints given the moderate rates of noncoding repetitive sequence across the genome outside of these arrays.
Another explanation could be if these genomic regions constitute pairing sites whose disruption decreases recombination in heterokayotypic individuals. Because inversions are generally thought to be favorable when they reduce recombination among favorable combinations of alleles, natural selection for decreased recombination might favor recurrent breaks in those regions (Corbett-Detig 2016). Consistent with this, satellite DNA is often involved in chromosome pairing and stabilization of meiosis (Palomeque and Lorite 2008). Circumstantial evidence also supports this idea as the distribution of relatively evenly spaced satellite arrays in small numbers across each of the autosomes is qualitatively consistent with the distribution of sensitive sites in the D. melanogaster genome (Roberts 1970, 1972; Hawley 1980; Coyne et al. 1993; Sherizen et al. 2005). However, at present, it is not known if pairing-sensitive sites exist within the An. gambiae genome, therefore this hypothesis should be regarded as speculative.
Finally, it is also possible that these genomic regions harbor genes that have recurrently contributed to ecological differentiation during the evolution of Anopheles species. If maintaining linkage among these gene complexes is favored by natural selection, inversions that reach high frequency would be expected to contain breakpoints in these regions. In support of this hypothesis, it has previously been observed that this region on 2R is frequently rearranged during Anopheles evolution. Furthermore, this region contributes to adaptive differentiation associated with oviposition site—a fundamental characteristic of these species—and other ecologically important traits (Coluzzi et al. 2002; Ayala et al. 2014, 2017). However, while this might produce a biased distribution of breakpoint positions along a chromosome on broad scales, it is not obvious why selection to maintain large-scale linkage disequilibrium across large genomic intervals should in itself select for inversion breakpoints that intersect a specific satellite array.
It is likely that a combination of factors drive the biased distribution of inversions and breakpoint reuse in the An. gambiae species complex as none of the possible causes above precludes the contributions of another. Although a mutational bias unto itself is not consistent with these data, this effect in combination with higher relative fitness might drive breakpoint reuse and colocalization with these specific satellite sequence arrays.
Repeat content is unlikely to explain the abundance of inversions on 2R
We next asked if a biased distribution of repetitive elements across the genome might explain the extreme excess of chromosomal inversions on arm 2R in this species complex. Although chromosome arm 2R exhibits the highest rate of inversion polymorphism in natural populations (Holt et al. 2002; Pombi et al. 2008; Xia et al. 2010), the other chromosome arms have similar per basepair rates of annotated repetitive elements. In fact, chromosome arm-2R has a relatively low rate of annotated repetitive elements (5.3% of sites on 2R are annotated as repeats >1 kb, vs. 5.3–6.3% across 2L, 3L, and 3R). Similarly, 2R does not contain an excess of satellite repeat elements (0.09% of sites on 2R are annotated as satellites, vs. 0.09–1.8% on 2L, 3L, and 3R). Furthermore, there is no excess of the specific satellite sequence that is shared among three of the four breakpoints addressed in this work. There are four such satellite arrays on 2R, whereas there are between two and five arrays on the other autosomes. We therefore conclude that a simple excess of repetitive sequences generally, and these satellites specifically, on 2R is unlikely to be the major driver of disproportionate inversion accumulation on this chromosome arm. However, increased breaks at these satellite sequences might still contribute to the formation or retention of inversions on 2R in combination with additional factors.
Comparison to a long-read based assembly
Recently, Kingan et al. (2019) produced a de novo genome assembly for An. coluzzi using high coverage PacBio long-read sequence data. This colony bears the same arrangement as the AgamP4 reference genome. To determine if their approach could assemble across these large-scale repeats, and thereby reveal the molecular organization of the breakpoint-associated regions, we aligned the genome to the AgamP4 genome assembly and extracted the contigs that aligned adjacent to each large-scale repeat cluster. For all three putative breakpoints, we found large contigs (all >500 kb) that aligned collinear to the breakpoint adjacent regions in the An. gambiae genome assembly. However, we did not identify a scaffold that spanned any of the predicted breakpoints (Table S4), indicating that these genomic regions remain a persistent challenge for even the most advanced long-read sequence-based assembly methods. In fact, a single contig spans the length of the genomic segment between the breakpoints of 2Rc, and terminates on each end at the repeat clusters surrounding our predicted inversion breakpoints. Nonetheless, this does reinforce a key advantage of Hi-C-based inversion breakpoint detection. Specifically, chromatin conformation capture can span very large genomic distances, thereby mitigating the impacts of large repetitive regions that may be challenging or impossible to completely sequence.
Impact of somatic pairing on breakpoint identification
Whereas sister chromosomes in mammalian genomes maintain independent chromosome domains in somatic tissues, dipteran sister chromosomes are paired along their lengths in the vast majority of somatic cells (Metz 1916). Heterokaryotypy is therefore expected to impact our prospects for successfully mapping inversion breakpoints. Specifically, whereas a single inversion induces novel sequence proximity between lower left and upper right quadrants in paired-read mapping position coordinates, when an inverted chromosome is paired to a standard arrangement chromosome in a heterokaryotypic individual, DNA sequence in the other two quadrants will be brought into proximity as well due to somatic pairing (Figure 1). If the two chromosomes contact each other frequently, we expect strong enrichment for read pairs mapping to quadrants two and four in heterokaryotypic individuals. However, we caution that these expectations should be considered only rough approximations and the underlying chromatin structure can also impact the distribution of read pairs linking independent genomic regions.
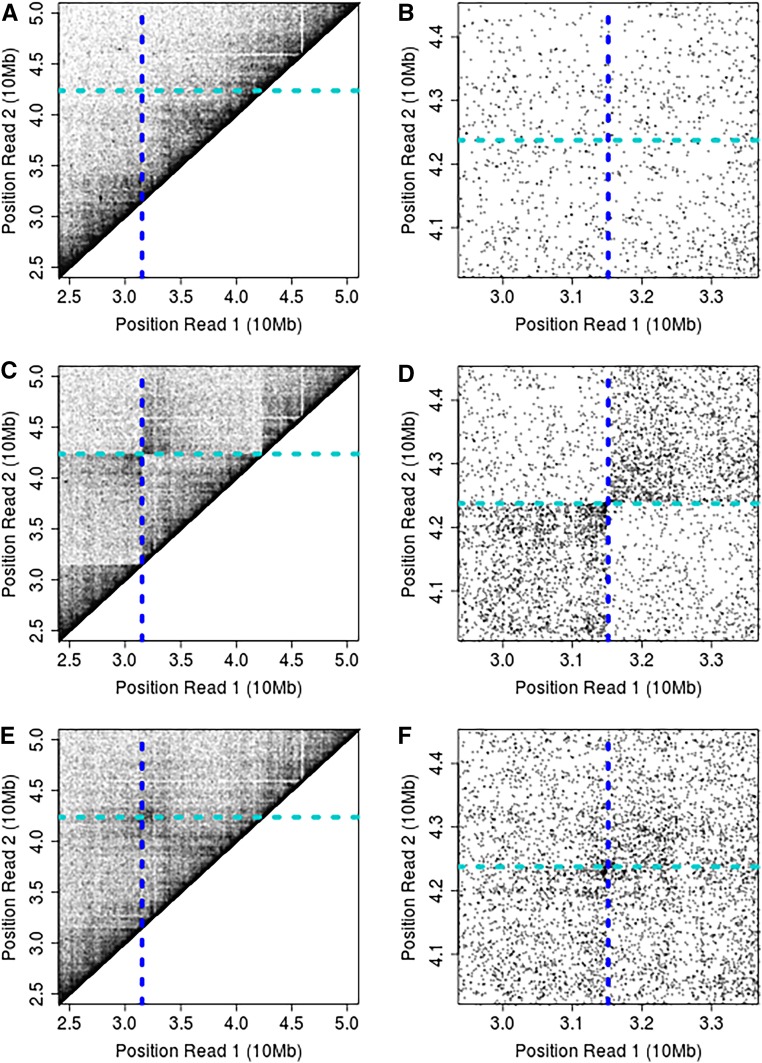
To investigate this phenomenon, we produced and sequenced an additional library from 2Rd/2R+d heterokaryotypic individuals. Whereas the homozygote library reveals a strong enrichment for Hi-C links in the lower left and upper right quadrants as expected, the heterokaryotype library is much less strongly delineated (Figure 5). When we attempt to bioinformatically map the breakpoints as described above, our method fails, presumably due to the challenges associated with somatic pairing.
Figure 5.
Impact of somatic pairing for 2Rd/2Rd+ heterokaryotypic An. coluzzii. For reference, the contact map of a standard arrangement, homokaryotypic individual (A and B). Inversion breakpoint predictions and contact map for inversion homokaryotypic individuals (C and D). Inversion breakpoint predictions and contact map for heterokaryotypic individuals (E and F). Figures on the left show ∼25 Mb on the centromere proximal portion of 2R (A, C, and E) and those on the right show the 2 Mb square region surrounding the predicted breakpoints of 2Rd (B, D, and F).
To attempt to map breakpoint positions in heterokaryotypes, we modified our mapping approach to accept only read pairs for which the first is within 5 Mb of the distal breakpoint, and the second is within 5 Mb of the proximal breakpoint. This has the effect of removing the “on-diagonal” reference consistent read pairs from impacting breakpoint estimates. In rerunning our mapping approach, the distal breakpoint estimated position is predicted at position 31,495,608, which is remarkably close to our estimate from homokaryotypic individuals, and within the same repetitive sequence block. However, the proximal breakpoint is predicted at position 42,550,800, which is ∼175 kb from the breakpoint we predicted from homokaryotypic individuals. This difference may reflect the challenges of the real chromatin domains, which alter the frequencies of links, and suggests that, whenever feasible, homokaryotypes should be used for mapping breakpoint positions when working with dipterans or other species that experience somatic pairing.
Breakpoint heterozygosity and somatic pairing
Despite the diffuse signal of association between sister chromosomes, there is still weak enrichment for the lower left and upper right quadrants (28 and 30% of read pairs respectively within a 4 Mb square centered on the breakpoint) in the 2Rd/2Rd+ heterokaryotype Hi-C mapping data (Figure 5). Read pairs mapping in these quadrants correspond to those that are physically proximal along the inverted chromosome, whereas read pairs mapping to the second and fourth quadrants are not physically proximal in either arrangement, but are brought into close proximity within chromatin presumably as a consequence of somatic pairing. This suggests that maternal and paternal chromosomes are almost equally likely to contact each other as to contact themselves in the region ∼2 Mb from inversion breakpoints and that inversion breakpoints present little barrier to somatic pairing despite different chromosome structures in intermediate to large distances away from breakpoint positions along the genome [similar to inversions in D. melanogaster (Golic and Golic 1996)]. Recent work using Hi-C to study somatic pairing in Drosophila embryos uncovered a highly regulated and “on-diagonal” structure of pairing between homologs (Erceg et al. 2019), our result appears approximately consistent, but suggest slightly more diffuse contacts between homologs in An. coluzzi. However, we caution that tissue types and specific molecular methods are different between studies, and more precise interspecific comparisons will require carefully controlled comparisons. Hi-C is therefore emerging as a powerful tool for investigating somatic pairing across many species.
To investigate the effect of somatic pairing on the relative contact frequencies more quantitatively, we examined the relative abundances of read pairs in quadrants two and four in homozygotes and heterozygotes. The genomic regions that contribute to the contacts in quadrants two and four are not physically proximal in any of the arrangements that we examined here, but as we describe above (Figure 1), pairing may induce proximity due the formation of inversion loop structures in heterozygotes. To approximately account for differences in the overall distribution of read pairs, we normalized the proportion that mapped in either quadrant by the proportion of all read pairs of similar length (see Materials and Methods). After doing this, we found no enrichment for read pairs mapping to quadrants two and four in the inversion homozygote at varying distances from the inversion breakpoints. Conversely, at close distances, there is a large enrichment for read pairs in quadrants two and four within inversion heterozygotes, and the effect decays with increasing distance from the breakpoints (Table S5). These results are therefore consistent with a model where there is an excess of interchromosomal contacts within regions that are brought into close physical proximity by the formation of inversion loops (or similar chromosomal conformations) during somatic pairing.
The variance in link abundance across genomic regions, samples, and library preparations is unknown, but likely to be large. Therefore, more quantitative analyses into the impacts of somatic pairing and the resulting differences in the contact frequencies among arrangements will require a carefully designed and replicated experiment (as in, e.g., differential expression analysis). Additionally, as we produced sequencing libraries here from pools of individuals, we cannot be certain that all SNPs are in the same phase. Nonetheless, our results suggest that the resulting contact map in inversion heterozygotes is substantially more complex than simply the average of the two homozygote contact maps, and, more specifically, that somatic pairing reshapes contacts in part by introducing new contacts in the regions surrounding inversion breakpoints (Figure 5 and Table S5).
Recent work in Drosophila has found that chromosome structure in itself has little effect on gene expression patterns (Said et al. 2018). For clarity, we note that alleles linked to inversions can have substantial impacts on expression outside of the genome structure change itself as well (Huang et al. 2015; Fuller et al. 2016; Lavington and Kern 2017; Cheng et al. 2018; Said et al. 2018). The observation that genome structure has little impact might suggest that chromosomal conformations are modified within inversion homozygotes and heterozygotes to maintain chromatin regulatory architectures as in standard homozygous individuals. However, we observe that somatic pairing results in relatively widespread reshaping of contact frequencies and many novel interchromosomal contacts surrounding regions that are adjacent within somatically paired diploid genomes. This instead suggests that most genes’ overall expression levels are negligibly impacted by large-scale proximity across the genome. This does not preclude an effect on more subtle expression profiles that we and others have not assayed directly in the context of chromosomal inversions (e.g., specific expression timing or variance in expression across cells). Nonetheless, we speculate that compact regulatory architectures that are minimally impacted by the presence of inversions may help to explain the seeming excess of chromosomal inversions found in many dipteran populations.
Prospects for karyotyping field-collected specimens
It has long been possible to karyotype field-collected Anopheles females via cytological methods. However, this approach can be laborious and requires collection of half-gravid females (della Torre 1997). For genotyping high frequency, well-characterized arrangements, PCR-based genotyping assays will likely be a preferable option. Because of the strong haplotype structure imposed by chromosomal inversions, it is often straightforward to identify linked diagnostic variants even when precise breakpoints are not known (e.g., Love et al. 2019). These and related approaches may be much more straightforward, particularly when inversion breakpoints occur in challenging repeat-heavy regions. Alternatively, particularly if newly discovered arrangements are previously unknown but segregating at high frequency in sampled populations, our approach will likely provide good tradeoffs. In preliminary analyses, we have found that samples can be fixed in formaldehyde, shipped at room temperature, and processed in the laboratory later. The libraries we produce are often robust to extremely low input materials, so although in this work we pooled several samples, our ongoing efforts suggest that it may be feasible to produce these libraries from single field-collected individuals. We contend that genotyping inversions in heterokaryotypes will likely be successful for already characterized arrangements due to the strong enrichment of read pairs mapping around breakpoint regions. However, new arrangements might need to be locally high frequency—a not uncommon situation for localized Anopheles populations (Coluzzi et al. 2002)—such that a portion of samples are likely to be homozygous, a necessary prerequisite for this approach to accurately fine-map novel breakpoint positions due to concerns related to somatic pairing.
Considerations for successful inversion breakpoint detection
Many of the important considerations for the successful application of this approach are addressed above. Here, we focus on three considerations that we believe are the most pertinent to the success of this method. First, Hi-C libraries are made with a variety of restriction enzymes, including most common 6-cutters and 4-cutters restriction enzymes as we do in this work. It has also recently become feasible to cut the genome approximately at random using DNases (e.g., Ma et al. 2018). Because breakpoint resolution is limited in large part by the frequency that reads are sampled along the genome, DNase methods or four-cutters will likely be favorable because they should provide a more uniform coverage per requisite sequencing depth than enzymes that cut less frequently along the genome.
Second, because this approach leverages diffuse information from read pairs distributed across a chromosome, it should be applied when largely complete reference genomes are available. Additionally, this concern implies that this method will be most efficient for longer inversions. Hi-C links often span entire chromosomes, so there is unlikely to be a maximum size of detectable inversions, but there must exist a lower bound. For example, in the extreme case where no reads mapped within an inverted region, it would be indistinguishable from a collinear chromosome using our approach. What minimal inversion length is detectable will certainly depend on many factors. For example, in our analysis, we believe the lower limit for detection is ∼1/2 Mb of inversion length. Nonetheless, most inversions of interest in Anopheles and Drosophila populations are on the order of several Megabases in length, and this approach is likely to compare favorably to alternatives when breakpoints occur in repetitive regions.
Finally, our method might be confounded by the presence of other naturally occurring rearrangements in a sample. For example, translocations or very large gene duplications might also yield spurious evidence for a chromosomal inversion using the approach that we developed here. Although this does not appear to present a major challenge for our application in this work, many genome assemblies are less complete than An. gambiae, and this is likely to confound analyses where reference genome quality is low, or where the reference genome is highly divergent from the sample of interest. We therefore suggest that this approach is a useful starting point for inversion detection and characterization, but that each candidate should be carefully visually interrogated to be certain that the read pair mapping distribution is consistent with expectations for a chromosomal inversion (see above).
Conclusion
Accurate inversion breakpoint detection is central for evolutionary genomic inference and for developing molecular karyotyping diagnostics. Here, we have shown that Hi-C sequencing is a cost-effective means of accurately fine-mapping inversion breakpoints in members of the Anopheles species complex. Our results demonstrate that conventional binning approaches for analyzing Hi-C contact maps are not a prerequisite, and limitations imposed by these methods can therefore be avoided, even for samples with very modest sequencing depths. Importantly, Hi-C has virtually unlimited range, despite extensive repetitive sequences flanking the inversion breakpoints of interest in the An. gambiae species complex. Breakpoint identification reliant on Hi-C data and related approaches will therefore enable structural variation discovery across the An. gambiae species complex, as well as across life more generally.
Acknowledgments
We thank E. Dotson, Principal Investigator for MR4 Vector Activity, for kindly supplying Mali-NIH, Pimperena, and Nkdokayo mosquitoes, and H. Ranson (Liverpool School of Tropical Medicine and Hygiene; UK) for sharing the Banfora M colony. Support for this work came from the National Institutes of Health (NIH) (R01 AI125360 awarded to N.J.B. and R35 GM128932 to R.B.C.-D.) and from an Alfred P. Sloan Fellowship to R.B.C.-D. During this work J.M. and M.G. were supported by NIH training grant T32 HG008345-01.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.10006487.
Communicating editor: M. Lawniczak
Literature Cited
- Aguado C., Gayà-Vidal M., Villatoro S., Oliva M., Izquierdo D. et al. , 2014. Validation and genotyping of multiple human polymorphic inversions mediated by inverted repeats reveals a high degree of recurrence. PLoS Genet. 10: e1004208 10.1371/journal.pgen.1004208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Wall J. D., and Kreitman M., 1999. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics 153: 1297–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D., Ullastres A., and González J., 2014. Adaptation through chromosomal inversions in Anopheles. Front. Genet. 5: 129 10.3389/fgene.2014.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D., Acevedo P., Pombi M., Dia I., Boccolini D. et al. , 2017. Chromosome inversions and ecological plasticity in the main African malaria mosquitoes. Evolution 71: 686–701. 10.1111/evo.13176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M., Barbadilla A., and Ruiz A., 1997. Inversion length and breakpoint distribution in the Drosophila buzzatii species complex: is inversion length a selected trait? Evolution 51: 1149–1155. 10.1111/j.1558-5646.1997.tb03962.x [DOI] [PubMed] [Google Scholar]
- Cáceres M., Ranz J. M., Barbadilla A., Long M., and Ruiz A., 1999. Generation of a widespread Drosophila inversion by a transposable element. Science 285: 415–418. 10.1126/science.285.5426.415 [DOI] [PubMed] [Google Scholar]
- Cheng C., White B. J., Kamdem C., Mockaitis K., Costantini C. et al. , 2012. Ecological genomics of Anopheles gambiae along a latitudinal cline: a population-resequencing approach. Genetics 190: 1417–1432. 10.1534/genetics.111.137794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Tan J. C., Hahn M. W., and Besansky N. J., 2018. Systems genetic analysis of inversion polymorphisms in the malaria mosquito. Proc. Natl. Acad. Sci. USA 115: E7005–E7014. 10.1073/pnas.1806760115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi M., Sabatini A., Petrarca V., and Di Deco M. A., 1979. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans. R. Soc. Trop. Med. Hyg. 73: 483–497. 10.1016/0035-9203(79)90036-1 [DOI] [PubMed] [Google Scholar]
- Coluzzi M., Sabatini A., della Torre A., Di Deco M. A., and Petrarca V., 2002. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298: 1415–1418. 10.1126/science.1077769 [DOI] [PubMed] [Google Scholar]
- Corbett-Detig R. B., 2016. Selection on inversion breakpoints favors proximity to pairing sensitive sites in Drosophila melanogaster. Genetics 204: 259–265. 10.1534/genetics.116.190389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig R. B., and Hartl D. L., 2012. Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 8: e1003056 [corrigenda: PLoS Genet. 9 (2013)]. 10.1371/journal.pgen.1003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Cardeno C., and Langley C. H., 2012. Sequence-based detection and breakpoint assembly of polymorphic inversions. Genetics 192: 131–137. 10.1534/genetics.112.141622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly M. B., Lobo N. F., Fitzpatrick M. C., Kern M., Grushko O. et al. , 2007. Segmental duplication implicated in the genesis of inversion 2Rj of Anopheles gambiae. PLoS One 2: e849 10.1371/journal.pone.0000849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Meyers W., Crittenden A. P., and Sniegowski P., 1993. The fertility effects of pericentric inversions in Drosophila melanogaster. Genetics 134: 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland J. M., and Thornton K. R., 2010. Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol. Evol. 2: 83–101. 10.1093/gbe/evq001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- della Torre A., 1997. Polytene chromosome preparation from anopheline mosquitoes, pp. 329–336 in The Molecular Biology of Insect Disease Vectors, edited by Crampton J. M., Beard C. B., and Louis C.. Chapman & Hall, London. [Google Scholar]
- Erceg J., AlHaj Abed J., Goloborodko A., Lajoie B. R., Fudenberg G. et al. , 2019. The genome-wide multi-layered architecture of chromosome pairing in early Drosophila embryos. Nat. Commun. 10: 4486 10.1038/s41467-019-12211-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T., and Levine M., 2017. Transvection. Curr. Biol. 27: R1047–R1049. 10.1016/j.cub.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller Z. L., Haynes G. D., Richards S., and Schaeffer S. W., 2016. Genomics of natural populations: how differentially expressed genes shape the evolution of chromosomal inversions in Drosophila pseudoobscura. Genetics 204: 287–301. 10.1534/genetics.116.191429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderón G. I., Emrich S. J., MacCallum R. M., Maslen G., Dialynas E. et al. , 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43: D707–D713. 10.1093/nar/gku1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic M. M., and Golic K. G., 1996. A quantitative measure of the mitotic pairing of alleles in Drosophila melanogaster and the influence of structural heterozygosity. Genetics 143: 385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell M., 1946. Cytological studies in Culex I. somatic reduction divisions. Genetics 31: 60–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harewood L., Kishore K., Eldridge M. D., Wingett S., Pearson D. et al. , 2017. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol. 18: 125 10.1186/s13059-017-1253-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R. S., 1980. Chromosomal sites necessary for normal levels of meiotic recombination in Drosophila melanogaster. I. Evidence for and mapping of the sites. Genetics 94: 625–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A., Ruban A., Walde I., Šimková H., Doležel J. et al. , 2018. Discovery of multi-megabase polymorphic inversions by chromosome conformation capture sequencing in large-genome plant species. Plant J. 96: 1309–1316. 10.1111/tpj.14109 [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., and Rieseberg L. H., 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 39: 21–42. 10.1146/annurev.ecolsys.39.110707.173532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt R. A., Subramanian G. M., Halpern A., Sutton G. G., Charlab R. et al. , 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. 10.1126/science.1076181 [DOI] [PubMed] [Google Scholar]
- Huang W., Carbone M. A., Magwire M. M., Peiffer J. A., Lyman R. F. et al. , 2015. Genetic basis of transcriptome diversity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 112: E6010–E6019. 10.1073/pnas.1519159112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M., Frezal L., Jones R. T., Chamberlain N. L., Lee S. F. et al. , 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477: 203–206. 10.1038/nature10341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingan S., Heaton H., Cudini J., Lambert C., Baybayan P. et al. , 2019. A high-quality de novo genome assembly from a single mosquito using PacBio sequencing. Genes (Basel) 10: pii: E62. 10.3390/genes10010062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., and Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. 10.1534/genetics.105.047985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas C. B., and Powell J. R., 1992. Drosophila Inversion Polymorphism. CRC Press, Boca Raton, FL. [Google Scholar]
- Lavington E., and Kern A. D., 2017. The effect of common inversion polymorphisms In(2L)t and In(3R)Mo on patterns of transcriptional variation in Drosophila melanogaster. G3 (Bethesda) 7: 3659–3668. 10.1534/g3.117.1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar N. H., Nevonen K. A., O’Connell B., McCann C., O’Neill R. J. et al. , 2018. Epigenetic maintenance of topological domains in the highly rearranged gibbon genome. Genome Res. 28: 983–997. 10.1101/gr.233874.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T. et al. , 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K. S., Rattray A., and Narayanan V., 2007. Hairpin- and cruciform-mediated chromosome breakage: causes and consequences in eukaryotic cells. Front. Biosci. 12: 4208–4220. 10.2741/2381 [DOI] [PubMed] [Google Scholar]
- Lobo N. F., Sangaré D. M., Regier A. A., Reidenbach K. R., Bretz D. A. et al. , 2010. Breakpoint structure of the Anopheles gambiae 2Rb chromosomal inversion. Malar. J. 9: 293 10.1186/1475-2875-9-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love R. R., Redmond S. N., Pombi M., Caputo B., Petrarca V., et al. , 2019. In silico karyotyping of chromosomally polymorphic malaria mosquitoes in the Anopheles gambiae complex. G3 (Bethesda) 9: 3249–3262. 10.1534/g3.119.400445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B., and Willis J. H., 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8: pii: e1000500. 10.1371/journal.pbio.1000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Ay F., Lee C., Gulsoy G., Deng X. et al. , 2018. Using DNase Hi-C techniques to map global and local three-dimensional genome architecture at high resolution. Methods 142: 59–73. 10.1016/j.ymeth.2018.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz C. W., 1916. Chromosome studies on the Diptera. II. The paired association of chromosomes in the Diptera, and its significance. J. Exp. Zool. 21: 213–279. 10.1002/jez.1400210204 [DOI] [Google Scholar]
- Nelder J. A., and Mead R., 1965. A simplex method for function minimization. Comput. J. 7: 308–313. 10.1093/comjnl/7.4.308 [DOI] [Google Scholar]
- Noor M. A., Grams K. L., Bertucci L. A., and Reiland J., 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088. 10.1073/pnas.221274498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomeque T., and Lorite P., 2008. Satellite DNA in insects: a review. Heredity 100: 564–573. 10.1038/hdy.2008.24 [DOI] [PubMed] [Google Scholar]
- Petrarca V., Nugud A. D., Elkarim Ahmed M. A., Haridi A. M., Di Deco M. A. et al. , 2000. Cytogenetics of the Anopheles gambiae complex in Sudan, with special reference to An. arabiensis: relationships with East and West African populations. Med. Vet. Entomol. 14: 149–164. 10.1046/j.1365-2915.2000.00231.x [DOI] [PubMed] [Google Scholar]
- Pombi M., Caputo B., Simard F., Di Deco M. A., Coluzzi M. et al. , 2008. Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions. BMC Evol. Biol. 8: 309 10.1186/1471-2148-8-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M., Cáceres M., and Ruiz A., 2004. Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA. Proc. Natl. Acad. Sci. USA 101: 9013–9018. 10.1073/pnas.0403090101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz J. M., Maurin D., Chan Y. S., von Grotthuss M., Hillier L. W. et al. , 2007. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 5: e152 10.1371/journal.pbio.0050152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P. A., 1970. Screening for x-ray-induced crossover suppressors in Drosophila melanogaster: prevalence and effectiveness of translocations. Genetics 65: 429–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P. A., 1972. Differences in synaptic affinity of chromosome arms of Drosophila melanogaster revealed by differential sensitivity to translocation heterozygosity. Genetics 71: 401–415. [DOI] [PubMed] [Google Scholar]
- Rocca K. A. C., Gray E. M., Costantini C., and Besansky N. J., 2009. 2La chromosomal inversion enhances thermal tolerance of Anopheles gambiae larvae. Malar. J. 8: 147 10.1186/1475-2875-8-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said I., Byrne A., Serrano V., Cardeno C., Vollmers C. et al. , 2018. Linked genetic variation and not genome structure causes widespread differential expression associated with chromosomal inversions. Proc. Natl. Acad. Sci. USA 115: 5492–5497. 10.1073/pnas.1721275115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semeshin V. F., Demakov S. A., Shloma V. V., Vatolina T. Y., Gorchakov A. A. et al. , 2008. Interbands behave as decompacted autonomous units in Drosophila melanogaster polytene chromosomes. Genetica 132: 267–279. 10.1007/s10709-007-9170-5 [DOI] [PubMed] [Google Scholar]
- Sharakhov I. V., White B. J., Sharakhova M. V., Kayondo J., Lobo N. F. et al. , 2006. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc. Natl. Acad. Sci. USA 103: 6258–6262. 10.1073/pnas.0509683103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherizen D., Jang J. K., Bhagat R., Kato N., and McKim K. S., 2005. Meiotic recombination in Drosophila females depends on chromosome continuity between genetically defined boundaries. Genetics 169: 767–781. 10.1534/genetics.104.035824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley C. S., and Eanes W. F., 1994. Isolation and analysis of the breakpoint sequences of chromosome inversion In(3L)Payne in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 91: 3132–3136. 10.1073/pnas.91.8.3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. J., Santolamazza F., Kamau L., Pombi M., Grushko O. et al. , 2007. Molecular karyotyping of the 2La inversion in Anopheles gambiae. Am. J. Trop. Med. Hyg. 76: 334–339. 10.4269/ajtmh.2007.76.334 [DOI] [PubMed] [Google Scholar]
- Xia A., Sharakhova M. V., Leman S. C., Tu Z., Bailey J. A. et al. , 2010. Genome landscape and evolutionary plasticity of chromosomes in malaria mosquitoes. PLoS One 5: e10592 10.1371/journal.pone.0010592 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequence data produced in this work are available from the sequence read archive under project accession number PRJNA564850. Software to perform the breakpoint mapping and uncertainty quantification procedures described herein is available from github, www.github.com/russcd/proximity_ligation_inversion_mapping. Supplemental material available at figshare: https://doi.org/10.25386/genetics.10006487.