Abstract
The prevalence of renal and cardiovascular disease (CVD) in patients with systemic lupus erythematosus (SLE) is higher than in general populations. Recently, a causal role of gut microbiota on the development of immune responses in SLE has been described. Probiotic consumption changes the composition of gut microbiota, preventing SLE progression. The aim of this review is to explore the role of the gut microbiota in the development of renal and cardiovascular disease in SLE and how probiotics could be a therapeutic option. Despite strong evidence on the beneficial effects of probiotics in the development of autoimmunity and nephritis in SLE, only a few studies described the protective effects of Lactobacillus in important risk factors for CVD, such as endothelial dysfunction and hypertension in mice. The preventive effects of probiotics in renal and CVD in humans have not been established yet.
Keywords: immune response, gut microbiota, hypertension, endothelial dysfunction, nephritis
1. Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by abnormally functioning B lymphocytes [1], which promote an exacerbated production of autoantibodies that trigger the formation and deposition of immune complexes that damage many organs and tissues [2]. While the causes are not known, it is widely considered that SLE is the consequence of the effects of environmental factors in genetically predisposed individuals, leading to the disruption of self-tolerance and to the activation/increase in innate immune cells and autoreactive lymphocytes [3].
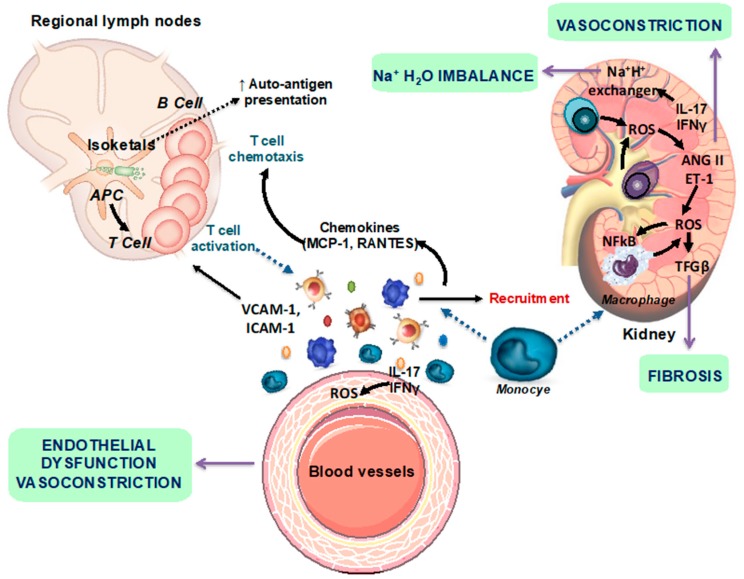
SLE is associated with renal and cardiovascular disease (CVD) [4]. In particular, hypertension is thought to be the greatest risk factor for cardiac events in lupus populations [5]. In fact, numbers ranging from 33% to 74% of SLE patients have been described to present hypertension [6]. Nevertheless, there has been little exploration of the pathophysiological mechanisms that promote SLE hypertension [7]. Several studies using female NZB/WF1 mice, a spontaneous model of SLE that mimics human disease and develops hypertension, have demonstrated that multiple factors contribute to the pathogenesis of hypertension, including inflammatory cytokines, tumor necrosis factor (TNF)-α, and oxidative stress. These mediators, which contribute to local inflammation and the subsequent renal and vascular dysfunction [8,9,10], are likely downstream of the initial immune system dysregulation [11]. Hypertension is associated with the infiltration of immune cells into the adventitia and periadventitial fat, as well as the activation of T cells that release proinflammatory cytokines such as interleukin (IL)-17a, interferon (IFN)-γ, and TNF-α [12,13]. However, T, but not B, cells play a key role in the modulation of blood pressure in response to angiotensin II-mediated hypertension [12]. Hypertension in female NZB/WF1 mice is associated with low plasma renin and is not sensitive to salt. In this mice model of SLE, treatment with anti-CD20 antibody, which lowered the percentage of B cells in the spleen and the quantity of anti-double-stranded (anti-dsDNA) antibodies in plasma, prevented the development of hypertension [14]. This study highlights the importance of B cells in the progression of SLE hypertension. However, the exact role of hyperactive T and B lymphocytes, both central to the development of autoimmune disorders, in the pathogenesis of hypertension remains unclear. Several studies suggest that immune cells, oxidative stress and inflammation are linked in a self-perpetuating cycle, which significantly contributes to the renal damage and vascular disease associated with hypertension in SLE [8]. T cell activation occurs in secondary lymphoid organs and locally within target organs, specifically the kidney and blood vessels. Neoantigen presentation is enhanced by oxidative stress in the antigen-presenting cells through isoketal production that enhance inflammatory responses. The effector mechanisms evoked by cytokines include fibrosis, vasoconstriction and Na+/H+ imbalance (Figure 1).
Figure 1.
Scheme representing the autoimmune mechanisms involved in the development of renal and vascular changes. AngII, angiotensin II; ET-1, endothelin-1; IFN-γ, interferon-γ; IL-17a, interleukin 17a; NF-kB, nuclear factor-kB; ROS, reactive oxygen species; Treg, T regulatory (modified from [10]).
However, whereas traditional risk factors associated with atherosclerosis, including hypertension, are present in lupus patients, they do not fully explain the high rate of ischemic events so far reported [15]. This implies that other factors inherent to disease itself contribute to the cardiovascular burden seen in these patients, such as, clinical, genetic, and immunological contributors. Disease duration, chronic organ damage and disease activity are important clinical factors for CVD development in the setting of SLE. Genetic data on lupus-related atherosclerosis are rather scarce. The presence of the rs10181656(G) signal transducer and activator of transcription factor 4 (STAT-4) allele or the prevalence of the minor A allele and the AA genotype of the rs12583006 B-cell activating factor (BAFF) variant conferred increased susceptibility for arterial events and ischemic cerebrovascular disease in the context of SLE. The imbalance between endothelial damage, as a result of several insults (deposition of oxLDL, autoantibodies, type I interferons, neutrophil extracellular traps), and atheroprotective mechanisms, seems to be a central event for immunological contributors to atherosclerosis in the setting of lupus [16].
Nowadays, the treatment and management of SLE is primarily based on non-steroidal anti-inflammatory drugs, glucocorticoids, hydroxychloroquine, and immunosuppressive agents [17]. Progress in the treatment of SLE has resulted in a significant improvement in prognosis. Nonetheless, SLE management is challenging because of the adverse effects of conventional therapies and the occurrence of refractory disease. In fact, corticosteroids and azathioprine therapies have been associated with an increased rate of CVD in lupus patients. Therefore, there is an imperative need for new treatment strategies that would allow us to treat renal and cardiovascular disorders in lupus patients without compromising their health state.
In this way, different studies have demonstrated that gut microbiome dysbiosis has been associated with autoimmune diseases such as type 1 diabetes, inflammatory bowel disease, rheumatoid arthritis, and multiple sclerosis. However, little is known on the role of gut microbiota in SLE in both animal models and humans [18,19,20]. Thus, the inclusion of probiotic supplementation in cardiovascular risk management should be considered. In fact, supplementation for a long period with multispecies probiotic mixtures exerts a favorable and dose-dependent effect on liver function and lipid profile in the rat model and may also have a favorable influence on cardiovascular impairments [21].
Furthermore, manipulation of the gut microbiota may lead to the development of novel therapies in SLE patients.
Specifically, we review the changes in the microbiota associated with SLE and discuss current knowledge on the impact of probiotics with immune-modulatory properties in the development of renal and cardiovascular disease on animal models and on human lupus patients as novel therapies.
2. Gut Microbiota and SLE
The mammalian microbiome consists of a unique set of microorganisms (i.e., bacteria, archaea, fungi, and viruses) associated with various niches in and on the body. The gut microbiota is dominated chiefly by Firmicutes and Bacteroidetes, and, to a lesser extent, by Actinobacteria, Proteobacteria, Synergistetes, Verrucomicrobia and Fusobacteria. However, gut microbiota constantly adapts to lifestyle modifications, such as diet, hormones and exercise [22]. In recent years, several parameters of health and disease have been found linked to shifts in the human gut microbiome.
2.1. Dysbiosis in SLE Patients
Human healthy gut microbiota is difficult to identify taking into account its interindividual variability and plasticity. However, ecological parameters of microbial stability, such as richness and diversity, are often used as indicators of gut health, since they are inversely associated with chronic diseases [23]. Lupus has been associated with several changes in gut microbiota (Table 1), which could be correlated with the manifestations of the pathology. However, those alterations are not fully understood yet, because there are variances among humans from different countries [24]. Contradictory results linking SLE to shifts in ecological parameters, such as richness and diversity, have been described. In fact, He et al. [24] showed significantly lower alpha diversity (Phylogenetic Diversity (PD) whole tree and observed species) in SLE patients, without differences in Shannon or Simpson. According to this, Li et al. [25] described a reduction in diversity metrics as Chao and observed species in SLE patients. In contrast, Hevia et al. [18] did not find significant differences in any alpha diversity measures (Chao, PD whole tree, observed species, Shannon, and Simpson indexes). Maybe these discrepancies could be explained by the influence of sex, age, progression of the disease and genetics background.
Table 1.
Gut microbiota shifts in systemic lupus erythematosus (SLE) patients.
| Patients | Ratio F/B | α-Diversity | Phylum | Family | Genus | Species | Reference |
|---|---|---|---|---|---|---|---|
| Women C 49.2 ± 10.7 years 20 patients |
↓F/B | No change | ↓Firmicutes ↓Tenericutes ↑Bacteroidetes |
↓Lachnospiraceae ↓Ruminococcaceae |
[18] | ||
| Women A 46.0 ± 1.8 years 35 patients |
↓F/B | ↓PD_whole_ tree ↓Observed species |
↓Firmicutes ↑Bacteroidetes ↑Actinobacteria ↑Proteobacteria ↑Fusobacteria |
↑Bacteroidaceae ↑Prevotellaceae ↑Rikenellaceae |
↓Pseudobutyvibrio ↓Dialister ↓Bifidobacterium ↑Rhodococcus ↑Eggerthella ↑Klebsiella ↑Prevotella ↑Flavonifractor ↑Eubacterium |
[24] | |
| Women A 37.46 ± 14.17 years 40 patients |
↓F/B | ↓Chao Richness ↓PD_whole_ tree ↓Observed species |
↓Tenericutes | ↑Streptococcaceae ↑Lactobacillaceae ↑Megasphaera |
↓Mollicutes ↓RF39 ↓Faecalobacteriu, ↓Cryptophyta ↓Roseburia |
↑Streptococcus anginosus ↑Lactobacillus mucosae ↑Veinella dispar |
[25] |
| Women 3 AA (42.33 ± 13.39 years), 7 C (49.42 ± 8.51 years) Men 3 C (33 ± 6.57 years) 1 AA (29) 14 patients |
Not change | ↑Proteobacteria | ↑Blautia | [26] | |||
| Women 10 C (38.3 ± 4.32 years) 13A (38.3 ± 4.32 years) 16 AA (46.69 ± 4.33 years) 19 WH (44.84 ± 3.5 years) 3 BH (43 ± 9.57 years) 61 patients |
↓Chao Richness | ↓Ruminococcaceae | ↑Blautia | ↓Ruminococcus gnavus ↓Bacteroides uniformis |
[27] |
C, Caucasian; AA, Afro-American; A, Asian; WH, White Hispanic; BH, Black Hispanic; Age (means ± SD); F/B, Firmicutes/Bacteroidetes. ↓Reduction, ↑Increase.
When the microbiota has been more deeply analyzed, the authors have found certain differences that could explain its role in SLE. At the phylum level, there seems to be a clear increase in Bacteroidetes, Actinobacteria and Proteobacteria and shrinking of Firmicutes in SLE patients, with these changes stable being between patients of different countries [24]. Recently, the reduction in Tenericutes and a rise in Fusobacteria in SLE has been described [25]. Overall, these changes are characterized by an intestinal dysbiosis associated with an alteration in the Firmicutes/Bacteroidetes (F/B) ratio (increase in Bacteroidetes and reduction in Firmicutes), although some authors have not found significant differences in this parameter between SLE and healthy patients [26]. At the family level, SLE patients from different studies presented varied results. He et al. [24] has described how SLE patients from Spain showed a depletion of Lachnospiraceae and Ruminococcaceae and an enrichment of Bacteroidaceae and Prevotellaceae but only Prevotellaceae showed significant increases in SLE patients from China [24]. Furthermore, Rikenellaceae, Streptococcaceae, Lactobacillaceae and Megasphaera might increase in SLE. These changes at the family and phylum levels are joined to changes at the genera level. At the genera level, we can highlight alterations in Pseudobutyvibrio, Dialister, Lactobacillus, Bifidobacterium, Mollicutes, RF39, Faecalobacterium, Cryptophyta and Roseburia, which are depleted in SLE patients. On the other hand, the genera Rhodococcus, Eggerthella, Klebsiella, Prevotella, Eubacterium, Flavonifractor and Blautia are enriched in this pathology [24,25,26]. The analysis of intestinal microbiota from patients with the pathology in the remissive stage found an increase in the Bifidobacterium genus [25]. At the species level, certain species were elevated in the gut microbiota of SLE [25], specifically, Streptococcus anginosus, Lactobacillus mucosae, and Veinella dispar. These changes appeared accompanied by a reduction in Ruminococcus gnavus and Bacteroides uniformis [27].
It might be an important milestone on the subject that Li et al. [25] found a positive correlation between the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) and the amount of Streptococcus, Campylobacter and S. anginosus in the fecal sample of SLE patients.
2.2. Dysbiosis in SLE Mice
Studying the microbiota in animal models, research has found some differences with humans and even between the different animal models (Table 2). Focusing on animal genetic models, changes have been described in the composition of gut microbiota during the course of the pathology; finding higher diversity in NZB/WF1, MRL/Mp-Faslpr (lpr) and SNF1 and toll-like receptor (TLR)-7.1 when the SLE had already set and even in the early stage of SLE [26,28]. Recently, no significant changes between female NZB/WF1 mice (33 weeks old) and age-matched control mice were observed regarding microbial richness, diversity, and evenness [29]. Regarding the F/B ratio, consensus has not been reached because some authors have found this parameter reduced [20] but others have not found any changes in NZB/WF1 or lpr mice [26,29].
Table 2.
Gut microbiota shifts in different lupus animal models.
| Ratio F/B | α-Diversity | Phylum | Family | Genus | Species | Reference | |
|---|---|---|---|---|---|---|---|
| NZB/WF1 | ↓F/Bor no change | ↑α-diversity or no change | ↓Firmicutes ↑Bacteroidetes |
Pre-SLE ↓Bifidobacterium ↑Lactobacillus ↓Lactobacillus High severity ↓Anerostipes |
[26] [28] [29] [30] |
||
| MRL/lpr | ↓F/Bor no change | ↑α-diversityor no change | ↓Firmicutes ↑Bacteroidetes |
↓Lactobacillaceae ↑Rikenellaceae ↑Desulfovibrionacea ↑Ruminococcaceae ↑Lachnospiraceae ↑Streptococcaceae |
↓Lactobacillus ↓Bifidobacterium ↑Tenericutes ↑Mollicutes ↑Butyrivibrio ↑Roseburia |
[19] [26] [28] [30] |
|
| SNF1 | ↓F/Bor no change | ↑α-diversity | ↓Firmicutes ↑Bacteroidetes |
↑Rikenellaceae ↑Lachnospiraceae |
Pre-SLE ↓Lactobacillus SLE ↑Lactobacillus ↑Clostridium ↑Dehalobacterium ↑Oscillospira ↑Dorea ↑Bilophila |
[1] [26] |
|
| TLR-7.1 | ↓F/B | ↑α-diversity | ↓Firmicutes ↑Bacteroidetes |
↓Clostridaceae ↑Coriobacteriaceae ↑Rikenallecea |
↓Turicibacter ↓Bifidobacterium ↓Coprobacillus ↓Anaerostipes ↑Prevotella ↑Desulfovibrio |
↑Lactobacillus reuteri | [31] |
F/B, Firmicutes/Bacteroidetes; SLE, systemic lupus erythematosus. ↓Reduction, ↑Increase.
Broadly, in the NZB/WF1, lpr and SNF1 and TLR-7.1 models, at the phylum level, there are changes present that are similar to those in human patients, namely elevated Bacteroidetes and reduced Firmicutes. Nevertheless, the main changes happen within the sublevel categories as family and genera. At the family level in the lpr mouse model, a decrease in Lactobacillaceae and a higher abundance of Rikenellaceae, Desulfovibrionaceae, Ruminococcaceae, Lachnospiraceae, or Streptococcaceae has been found [19,28]. These alterations are usually associated with changes in several genera such as Tenericutes, Mollicutes, Butyrivibrio, and Roseburia, which were enhanced in that model. Conversely, other genera were reduced, such as Lactobacillus or Bifidobacterium [28]. Furthermore, it has been found to modulate the genus Anaerostipes, which was negatively correlated with lupus activity [26]. Anaerostipes is able to maintain the gut health state due to the capacity to produce butyrate.
Furthermore, of these changes in the lupus-prone mouse model SNF1, authors have described some similar alterations described in other animal models such as the abundance of Rikenellaceae or Lachnospiraceae [1]. However, others have found differences in this model between pre-disease and disease stages, showing elevated Lactobacillus, which is usually decreased in other animal models and in humans. Other genera such as Clostridium, Dehalobacterium, Oscillospira, Dorea and Bilophila may also be elevated [26].
Another main animal model of SLE is NZB/WF1, in which the microbiota suffers alterations similar to lpr and humans at the phylum, family and genus levels. NZB/WF1 mice displayed a higher abundance of Lactobacilli in the gut microbiota, which may be associated with more severe clinical signs, especially the impairment of systemic autoimmunity and renal functions [26]. In lpr mice, some bacteria from the genus Anerostipes such as Akkermansia muciniphila have been significantly decreased from pre-disease to disease stage and also bacteria from the genus Lactobacillus or Bifidobacterium appear reduced [32]. Recently, in hypertensive female NZB/WF1, we found that the gut microbiota of these mice had a significantly higher abundance of Pedobacter, Lactobacillus, and Prevotella than the age-matched control group, without changes in other genera such as Bifidobacterium [29].
In recent years, a new model has been described, the TLR-7.1 model, and the microbiota has been analyzed by Zegarra-Ruiz et al. [30]. The alterations found are similar to those found in the other animal models. The microbiota is characterized by an increase in the families Coriobacteriaceae and Rikenallecea, and a reduction in Clostridaceae. At the genus level, the tise in Prevotella and Desulfovibrio, and the decrease in the abundance of Turicibacter, Bifidobacterium, Coprobacillus and Anaerostipes stand out. Finally, they found an increase in the species Lactobacillus reuteri, which was also found in mesenteric lymph nodes (MLNs), spleen and liver, linking its translocation to the evolution of SLE.
2.3. Gut Dysbiosis in Lupus Is Linked to Leaky Gut, Changes in Immune Cell Populations, and Cardiovascular Complications
Besides these changes, in gut microbiota from patients and animal models of SLE an alteration in the epithelium of the intestinal barrier characterized by an impairment in junction proteins like occludin, zonulin-1 and claudin and an increase in intestinal permeability, measured through fluorescein isothiocyanate (commonly referred to as FITC)-dextran, takes place [19]. Researchers are linking these alterations with the course of SLE because some bacteria and their structural components or products from bacterial metabolism might be able to cross the intestinal epithelium and reach blood and even several organs.
Recently, Manfredo-Vieira et al. [31] have shown how Enterococcus gallinarum is able to translocate from bowel to liver and activate the production of anti-dsDNA antibodies through TLR-7/8 activation in genetically predisposed hosts ((NZW × BXSB) F1 mice, and SLE patients). In fact, antibiotic treatment prevented mortality in this animal model, suppressed the growth of E. gallinarum in tissues, and eliminated pathogenic autoantibodies and T cells. In this fashion, Katz-Agranov et al. [32] found a negative correlation between gut levels of Synergistetes and plasma anti-dsDNA antibody titers and IL-6 levels. IL-6 can promote an increase in both the differentiation of T-helper (Th) 17 and in the production of IL-17a, which is an important driver of autoimmunity in SLE [33,34]. In relation to this, in in vitro studies, the fecal microbiota of SLE patients was a stronger inducer of Th17 [35].
Besides the modulation of anti-dsDNA antibody levels, bacteria could modulate the immune system in other ways, inducing beneficial or harmful effects in the course of SLE. Other possible mechanisms to modulate the immune system could be induced by the components of bacteria like lipopolysaccharide (LPS), which is derived from the wall of Gram-negative bacteria belonging to Bacteroidetes (this phylum is elevated in the gut microbiota of SLE mice), and thus, through TLR-4 activation, inducing the production of proinflammatory cytokines such as TNF-α, IL-6 and type I interferons (IFN-α, IFN-γ) that are elevated in SLE patients and also in animal models of SLE [36]. IFN-α could also be induced through TLR-7, a high-risk locus for excessive activation of RNA sensing, which is overexpressed in SLE [31]. This high level of IFN-α is able to induce endothelial dysfunction in SLE patients [37]. Inflammatory cytokines interact with important blood pressure regulatory systems, such as the renin–angiotensin system and the sympathetic nervous system [38]. The role of IL-6 has not specifically been examined in SLE hypertension. TNF-α has also been shown to be elevated in the serum of SLE patients and can correlate with disease activity [39]. Etanercept, a clinically available recombinant TNF-α receptor that reduces the biological activity of TNF-α, reduces mean arterial pressure in a female mouse model of SLE, suggesting that TNF-α mechanistically contributes to the development of hypertension [40].
TLR4 activation also contributes to increased blood pressure and low-grade vascular inflammation displayed by spontaneously hypertensive rats [41]. In fact, TLR4−/− mice demonstrated full blood pressure protection against chronic endothelial nitric oxide synthase (eNOS) blockade-induced hypertension [42]. Bacterial LPS stimulates and increases the expression of TLR4 in the vasculature, which resulted in increased NADPH oxidase-dependent superoxide production, inflammation, and endothelial dysfunction [43,44]. Thus, enhanced TLR4 activation might be linked to the development and maintenance of hypertension in SLE. In fact, elevated plasma LPS levels have been previously described in both SLE patients [45,46,47] and hypertensive SLE mice [29].
In addition, some bacteria produce short-chain fatty acids (SCFAs), which are able to activate free fatty acid receptors with beneficial [48] or deleterious effects in the pathology. Clostridium and Lachnospiraceae are both producers of butyrate, which through the activation of G-protein-coupled receptor (GPR)-109a promotes differentiation of regulatory T cells in the colon, spleen, and lymph nodes and also IL-18, hence being capable of suppressing inflammation [49]. On the other hand, acetate and propionate seem elevated in stool samples of SLE patients, but Rodríguez-Carrio et al. [50] did not find differences in butyrate levels. These alterations in the production of bacterial by-products could be derived from changes in some metabolic pathways. The glycan degradation pathways are slightly overrepresented in the microbiota from SLE patients, likely due to the higher abundance of Bacteroidetes in these patients [51]. The treatment with vancomycin showed downregulated LPS biosynthesis [52].
Currently, researchers are seeking how to modulate the gut microbiota to improve this disease prognosis. According to this, certain bacteria could be correlated with the stages of remission in the course of the pathology or even with modifications carried out in some treatments. Katz-Agranov et al. [32] proved treatment with retinol decreased Erysipelotrichaceae and increased Lachnospiraceae and Rikenellaceae, restoring some changes observed in lupus. The treatment with dexamethasone or prednisone [53] (glucocorticoids widely used in patients with SLE) was able to increase the alpha diversity (Shannon index), which may lead to a more stable community [26]. The prednisone treatment was able to reduce the phyla Proteobacteria and Deferribacteres. The modulation of the gut microbiota induced several changes at the genus level, characterized by an increase in Prevotella and Anaerostipes and a decrease in Rikenella, Mucispirillum, Oscillospira, and Bilophila [53]. Antibiotic treatment with vancomycin was able to remove Clostridiales and Bacteroidales, while increasing Lactobacilli such as L. rhamnosus and L. reuteri [19].
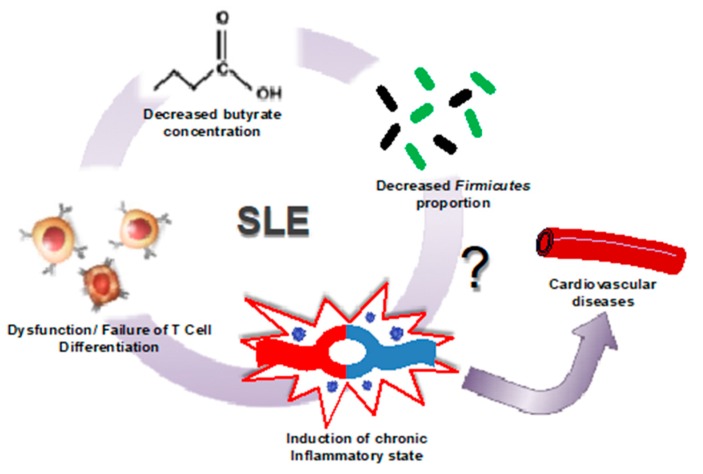
In summary, the evidence is suggesting that the renal and cardiovascular complications associated with SLE are a consequence of an immune response aggravated by a high multitude of antigen-presenting bacteria such as Lachnospiraceae, or Bacteroidetes, and a decrease in Firmicutes abundance within the gut microbiome, resulting in lower butyrate levels, T cell dysfunction, and the onset of chronic inflammation and cardiovascular risk (Figure 2) [54]. However, it is not clear whether intestinal dysbiosis in SLE is cause or consequence [35]. Current evidence suggests that intestinal microbes could be involved in the initiation and amplification of autoimmune diseases, such as SLE. Despite recent progress in understanding how these microbes influence the pathophysiology of lupus, studies are still limited. Recently, an association between gut microbiota, free fatty acid serum pool, and biomarkers of endothelial activation in lupus patients has been stablished, thus emphasizing the systemic effect of the gut microbiota in this condition [50]. However, there is no direct link between gut dysbiosis and endothelial dysfunction and hypertension in SLE. It is possible that reducing Th17 polarization by increasing butyrate-producing bacteria, or decreasing endotoxaemia by improving gut integrity, or reducing autoantibodies production by both reducing commensal bacteria containing an RNA-binding autoantigen, Ro60 structural homologs [55], or by preventing translocation of specific bacteria, such as Enterococcus gallinarum [31], or Lactobacillus reuteri [30] to secondary lymph organs and liver, could prevent cardiovascular complications in SLE.
Figure 2.
Microbiome shift in SLE induces cardiovascular risk. In patients with SLE, it has been observed that a decrease in the population of butyrate-producing Firmicutes leads to a decrease in butyrate production. This decreased production of butyrate leadsto the failure of T cells to properly differentiate in lupus patients, resulting in the induction of the chronic inflammatory state representative of this disease (modified from [44]).
3. SLE and Probiotics
Due to the aforementioned discoveries, and especially because of the depletion of Lactobacillus and Bifidobacterium in the gut microbiota present in the lpr SLE models, researchers thought that treating lupus patients with bacterial supplementation (Table 3), such as different strains of Lactobacillus that had already shown some effects in other autoimmune diseases [56,57], could help ameliorate the disease symptomatology. In fact, Mu et al. [19] showed that supplementation with Lactobacillus spp. in lpr mice displayed a striking effect mitigating lupus nephritis and prolonging survival. These effects are linked to a reduced plasma anti-dsDNA levels induced by probiotic consumption.
Table 3.
Probiotic effects in different lupus animal models.
| Probiotic | Model | Observed Effects | Reference |
|---|---|---|---|
| Lactobacillus delbrueckii subsp. lactis PTCC 1743 | Pristane-induced murine model | ↓Th17 ↓IL-17a ↓Th1 ↓IFN-γ | [61] |
| Lactobacillus rhamnosus ATCC 9595 | Pristane-induced murine model | ↓RORγ ↓Th17 ↓Th1 ↓IFN-γ | [61] |
| Ruminococcus obeum DSM25238 | In vitro | ↓Th17/Th1 ratio | [35] |
| Blautia coccoides DSM935 | In vitro | ↓Th17/Th1 ratio | [35] |
| Lactobacillus reuteri GMNL 263 | NZB/W F1 | ↑FoxP3 ↑Treg ↓TLR-4 ↓TLR-5 ↓TLR-7 ↓TLR-9 | [62] |
| ↓IL-1β ↓TNF-α ↓IL-6 | [63] | ||
| Bifidobacterium bifidum LMG13195 | In vitro | ↓T lymphocytes activation | [35] |
| Lactobacillus fermentum CECT5716 | NZB/W F1 | ↓B and T lymphocytes↓IL-17a ↓IFN-γ ↓TNF-α ↓IL-21 | [29] |
| Lactobacillus reuteri GMNL 89 | NZB/W F1 | ↓TLR-4 ↓TLR-5 ↓TLR-7 ↓TLR-9 | [62] |
| ↓IL-1β ↓TNF-α ↓IL-6 | [48] | ||
| Lactobacillus paracasei GMNL 32 | NZB/W F1 | ↓TLR-4 ↓TLR-5 ↓TLR-7 ↓TLR-9 | [62] |
| ↓IL-1β ↓TNF-α ↓IL-6 | [63] |
FoxP3, forkhead box P3; IFN, interferon; IL, interleukin; RORγ, RAR-related orphan receptor gamma; Th, T-helper; TLR, toll-like receptor; TNF-α, tumor necrosis factor alpha; Treg, T regulatory. ↓Reduction, ↑Increase.
These microorganisms that in adequate amounts can provide a health benefit to the treated host receive the name of probiotics. Although certain limitations must be applied to this term according to the last panel of the International Scientific Association for Probiotics and Prebiotics (ISAPP), gathering only products that deliver live microorganisms with a suitable viable count of well-defined strains with a reasonable expectation of delivering benefits for the wellbeing of the host under this name [58]. Previous evidence showed that probiotic supplementation could be effective in the prevention and treatment of cardiovascular disease in obese postmenopausal women. In fact, supplementation with multispecies probiotic Ecologic® Barrier favorably modified both functional and biochemical markers of vascular dysfunction, reduced systolic blood pressure [59], and improved cardiometabolic parameters [60]. These protective effects are related to lower plasma LPS levels found in women with probiotic supplementation. However, whether probiotic consumption improves endothelial dysfunction in SLE patients is still unknown.
The physiological responses to different probiotics have proven to be consistently diverse. Typically, these microorganisms trigger immune shifts that effectively decrease the inflammatory response, thus, ameliorating the symptomatology of autoimmune diseases. Some of the most commonly observed mechanisms in the treatment of SLE are related to the modulation of Th17 and T regulatory (Treg) lymphocyte populations [61]. Nonetheless, other less studied mechanisms are also involved.
According to Mardani et al. [61], the administration of the probiotic Lactobacillus delbrueckii subsp. Lactis PTCC 1743 to a pristane-induced SLE mice model was able to improve the disease symptoms, decreasing Th17 populations and the expression of one of its main cytokines, IL-17a, well known as crucial elements in the development and maintenance of inflammation. In addition to this, another probiotic microorganism, Lactobacillus rhamnosus ATCC 9595, showed the capacity to modulate retinoic acid receptor-related orphan receptor gamma (RORγ), a transcription factor involved in the maturation of Th17 lymphocytes, which would explain the ability to decrease this lymphocytic population. Both probiotics were also able to reduce Th1 populations and its cytokine IFN-γ, which is regarded as one of the main mechanisms involved in the generation of the inflammatory response.
On the other hand, Lactobacillus reuteri GMNL 263 displayed a different pathway in the genetic murine model of SLE NZB/W F1 [62]. This microorganism can increase Treg lymphocyte expression and its transcription factor forkhead box P3 (FoxP3) levels. These cells are responsible for the regulation of the pro-inflammatory lymphocytes cited above, and with a marked anti-inflammatory character. Aside from this, levels in common pathogen-associated molecular pattern (or PAMPs) receptors such as TLR-4, TLR-5, TLR-7 and TLR-9 that mediate in the development of inflammation were also reduced in the liver with the probiotic treatment, as well as an increase in antioxidant activity. The aforementioned changes in TLRs and oxidative stress were also detected in similar experiments with probiotics Lactobacillus paracasei GMNL 32 (GMNL-32) and Lactobacillus reuteri GMNL 89, although the effects of Lactobacillus reuteri GMNL 263 on Treg expression were not present in those cases. Furthermore, in these three treatments, through the suppression of nuclear factor kB (NF-κB) and the mitogen-activated protein kinase signaling pathways, there was a reduction in hepatic pro-inflammatory cytokines IL-1β, TNF-α and IL-6 [63,64]. Interestingly, GMNL-32 treatment reduced left ventricular hypertrophy in this genetic model of lupus.
Furthermore, dendritic cells exposed to SLE microbiota enriched with Bifidobacterium bifidum LMG13195 tended to, when co-cultured with naïve T cells, lessen the activation of these lymphocytes when compared to normal SLE microbiota in vitro. In the same conditions but using Ruminococcus obeum DSM25238 and Blautia coccoides DSM935 instead of the Bifidobacterium strain, after recovering the T cells resulting from the co-incubation, decreased Th17/Th1 ratios were observed, changing the inflammation pattern when compared to normal SLE microbiota. However, when studying IFN-γ and Il-17a expression levels, no significant differences were observed [35].
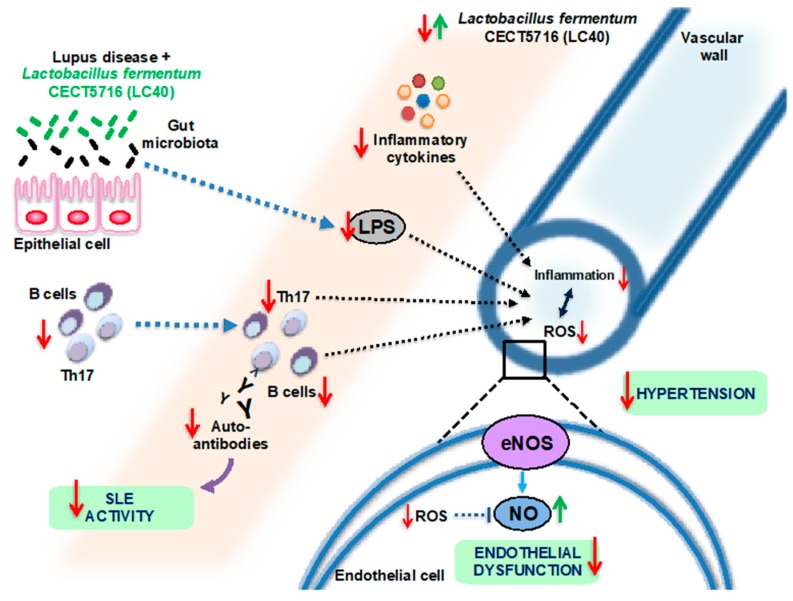
Female NZB/WF1 mice have several characteristics consistent with human SLE, including immune complex deposition in the glomerulus, dsDNA autoantibodies, albuminuria and, importantly, endothelial dysfunction and hypertension [65]. Chronic Lactobacillus fermentum CECT5716 (LC40) consumption in female NZB/WF1 mice was able to increase the Bifidobacterium count in the gut. LC40 reduced lupus disease activity and splenomegaly in SLE mice. It also improved gut barrier integrity, reducing LPS plasma levels, which in turn reduced immune activation, shown by a decrease in T and B cells in MLN and pro-inflammatory cytokines such as IL-17a, IFN-γ, TNF-α and IL-21 in plasma. As the probiotic prevented the development of the pro-inflammatory response, complications associated with SLE such as cardiac and renal hypertrophy were averted. Moreover, high blood pressure and impaired vascular endothelium-dependent vasodilation were prevented by L. fermentum as a result of reduced eNOS phosphorylation at the inhibitory site Thr495, and decreased NADPH oxidase activity, commonly considered as the main source of reactive oxygen species in the vascular system, which improved the bioavailability of nitric oxide [29] (Figure 3).
Figure 3.
Proposed mechanism by which Lactobacillus fermentum CECT5716 (LC40) administration could prevent SLE activity, endothelial dysfunction and hypertension in the NZB/WF1 model (based on data from publication no. [29]).
Therefore, adjustment of the SLE microbiome via dietary intervention has shown attenuation of SLE symptoms and, in one case, of its cardiovascular complications, marking this strategy as a possible novel therapeutic approach to this disease.
As seen with the examples above, despite having a clearer picture of the elements involved in the effect of probiotics in SLE, especially the role of Treg and Th17 modulation, a better understanding of the underlying mechanisms of probiotics is needed, which would not only allow for better targeting and selection of microorganisms but could also cast some light on the development and causes of SLE and its renal and cardiovascular complications.
4. Conclusions
This review opens new possibilities to the prevention SLE and its renal and cardiovascular disorders and the modulation of the gut microbiota through the administration of some probiotic strains. However, caution is advised when extrapolating these findings to humans because of the possible differences in the behavior of the animal and human gut microbiota. In fact, the relative abundance of Lactobacillales appears to be normal in SLE patients in remission [18], which might alter the possible applicability of the Lactobacillus treatment to clinical practice in humans. Although not a diet in itself, there is a common belief that probiotic-containing fermented foods modify the gut microbiota and confer host health benefits. However, fermented foods rarely contain adequate amounts of specific probiotic organisms. Moreover, there is limited evidence for the role of probiotics as modulators of the human gut microbiota, and recent data suggest that even supplemental quantities of probiotics exert limited effects on human gut ecology [66]. On the other hand, the composition of gut microbiota in a large population of hypertensive SLE patients is unknown.
The answer to these questions could clarify not only the mechanisms involved in the protective effects of specific probiotic bacteria but also their potential in human SLE treatment. Taking into account that the pathophysiological mechanism involved in disorder development in these patients may vary and that the mechanisms of improvement in SLE disease by specific probiotics are unknown, it is worth investigating whether we can choose a specific probiotic strain to gain benefits in a particular SLE patient. Taken together, preclinical and clinical evidence indicates that further research is needed to evaluate the safety profile before any of these probiotics can be marketed for the clinical treatment of hypertension, metabolic, and vascular complications in SLE. However, we suggest that beneficial bacteria, such as L. fermentum, capable of reducing dysbiosis, improving gut barrier function and reducing endotoxemia might prevent vascular complications in SLE patients. Unfortunately, when consulted for this review, clinicaltrials.gov does not yet include any trial on SLE patients with probiotics.
Author Contributions
Conceptualization, J.D. and M.T.; Preparing figures, M.T.; Writing—original draft, N.d.l.V., I.R.-V., M.T. and J.D.
Funding
This work was supported by grants from Comisión Interministerial de Ciencia y Tecnología, Ministerio de Economía y competitividad (MINECO) (SAF2014-55523-R, SAF2017-84894-R) and Junta de Andalucía (Proyecto de excelencia P12-CTS-2722, and CTS 164), with funds from the European Union, and by the Ministerio de Economia y Competitividad, Instituto de Salud Carlos III (CIBER-CV). I.R.-V. is a predoctoral fellow of MINECO; N.d.l.V. is a predoctoral fellow of Junta de Andalucía and Fondo Social Europeo “FEDER una manera de hacer Europa”, and M.T. is a postdoctoral fellow of Instituto de Salud Carlos III (Sara Borrell Program).
Conflicts of Interest
The authors declare no competing interests.
References
- 1.Johnson B.M., Gaudreau M.C., Al-Gadban M.M., Gudi R., Vasu C. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clin. Exp. Immunol. 2015;181:323–337. doi: 10.1111/cei.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanz I., Lee F.E. B cells as therapeutic targets in SLE. Nat. Rev. Rheumatol. 2010;6:326–337. doi: 10.1038/nrrheum.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Paglia G.M.C., Leone M.C., Lepri G., Vagelli R., Valentini E., Alunno A., Tani C. One year in review 2017: Systemic lupus erythematosus. Clin. Exp. Rheumatol. 2017;35:551–561. [PubMed] [Google Scholar]
- 4.Liu Y., Kaplan M.J. Cardiovascular disease in systemic lupus erythematosus: An update. Curr. Opin. Rheumatol. 2018;30:441–448. doi: 10.1097/BOR.0000000000000528. [DOI] [PubMed] [Google Scholar]
- 5.Bartels C.M., Buhr K.A., Goldberg J.W., Bell C.L., Visekruna M., Nekkanti S., Greenlee R.T. Mortality and cardiovascular burden of systemic lupus erythematosus in a US population-based cohort. J. Rheumatol. 2014;41:680–687. doi: 10.3899/jrheum.130874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frostegård J. Systemic lupus erythematosus and cardiovascular disease. Lupus. 2008;17:364–367. doi: 10.1177/0961203308089988. [DOI] [PubMed] [Google Scholar]
- 7.Ryan M.J. The pathophysiology of hypertension in systemic lupus erythematosus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;296:R1258–R1267. doi: 10.1152/ajpregu.90864.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J., Saleh M.A., Kirabo A., Itani H.A., Montaniel K.R., Xiao L., Chen W., Mernaugh R.L., Cai H., Bernstein K.E., et al. Immune activation caused by vascular oxidation promotes fibrosis and hypertension. J. Clin. Investig. 2016;126:50–67. doi: 10.1172/JCI80761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzik T.J., Korbut R., Adamek-Guzik T. Nitric oxide and superoxide in inflammation and immune regulation. J. Physiol. Pharmacol. 2003;54:469–487. [PubMed] [Google Scholar]
- 10.Small H.Y., Migliarino S., Czesnikiewicz-Guzik M., Guzik T.J. Hypertension: Focus on autoimmunity and oxidative stress. Free Radic. Biol. Med. 2018;125:104–115. doi: 10.1016/j.freeradbiomed.2018.05.085. [DOI] [PubMed] [Google Scholar]
- 11.Taylor E.B., Ryan M.J. Understanding mechanisms of hypertension in systemic lupus erythematosus. Ther. Adv. Cardiovasc. Dis. 2017;11:20–32. doi: 10.1177/1753944716637807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzik T.J., Hoch N.E., Brown K.A., McCann L.A., Rahman A., Dikalov S., Goronzy J., Weyand C., Harrison D.G. Role of the T cell in the genesis of angiotensin II induced hypertension and vascular dysfunction. J. Exp. Med. 2007;204:2449–2460. doi: 10.1084/jem.20070657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madhur M.S., Lob H.E., McCann L.A., Iwakura Y., Blinder Y., Guzik T.J., Harrison D.G. Interleukin 17 promotes angiotensin II-induced hypertension and vascular dysfunction. Hypertension. 2010;55:500–507. doi: 10.1161/HYPERTENSIONAHA.109.145094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathis K.W., Wallace K., Flynn E.R., Maric-Bilkan C., LaMarca B., Ryan M.J. Preventing autoimmunity protects against the development of hypertension and renal injury. Hypertension. 2014;64:792–800. doi: 10.1161/HYPERTENSIONAHA.114.04006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esdaile J.M., Abrahamowicz M., Grodzicky T., Li Y., Panaritis C., du Berger R., Côte R., Grover S.A., Fortin P.R., Clarke A.E., et al. Traditional Framingham risk factors fail to fully account for accelerated atherosclerosis in systemic lupus erythematosus. Arthritis Rheum. 2001;44:2331–2337. doi: 10.1002/1529-0131(200110)44:10<2331::AID-ART395>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 16.Giannelou M., Mavragani C.P. Cardiovascular disease in systemic lupus erythematosus: A comprehensive update. J. Autoimmun. 2017;82:1–12. doi: 10.1016/j.jaut.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Chan V.S., Tsang H.H., Tam R.C., Lu L., Lau C.S. B-cell-targeted therapies in systemic lupus erythematosus. Cell. Mol. Immunol. 2013;10:133–142. doi: 10.1038/cmi.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hevia A., Milani C., López P., Cuervo A., Arboleya S., Duranti S., Turroni F., González S., Suárez A., Gueimonde M., et al. Intestinal dysbiosis associated with systemic lupus erythematosus. MBio. 2014;5 doi: 10.1128/mBio.01548-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mu Q., Zhang H., Liao X., Lin K., Liu H., Edwards M.R., Ahmed S.A., Yuan R., Li L., Cecere T.E., et al. Control of lupus nephritis by changes of gut microbiota. Microbiome. 2017;5:73. doi: 10.1186/s40168-017-0300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B., Selmi C., Tang R., Gershwin M.E., Ma X. The microbiome and autoimmunity: A paradigm from the gut-liver axis. Cell. Mol. Immunol. 2018;15:595–609. doi: 10.1038/cmi.2018.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skrypnik K., Bogdański P., Łoniewski I., Reguła J., Suliburska J. Effect of probiotic supplementation on liver function and lipid status in rats. Acta Sci. Pol. Technol. Aliment. 2018;17:185–192. doi: 10.17306/J.AFS.0554. [DOI] [PubMed] [Google Scholar]
- 22.Gentile C.L., Weir T.L. The gut microbiota at the intersection of diet and human health. Science. 2018;362:776–780. doi: 10.1126/science.aau5812. [DOI] [PubMed] [Google Scholar]
- 23.Cotillard A., Kennedy S.P., Kong L.C., Prifti E., Pons N., Le Chatelier E., Almeida M., Quinquis B., Levenez F., Galleron N., et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 24.He Z., Shao T., Li H., Xie Z., Wen C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog. 2016;8:64. doi: 10.1186/s13099-016-0146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Wang H., Li X., Li H., Zhang Q., Zhou H., He Y., Li P., Fu C., Zhang X., et al. Disordered intestinal microbes are associated with the activity of Systemic Lupus Erythematosus. Clin. Sci. 2019;133:821–838. doi: 10.1042/CS20180841. [DOI] [PubMed] [Google Scholar]
- 26.Luo X.M., Edwards M.R., Mu Q., Yu Y., Vieson M.D., Reilly C.M., Ahmed S.A., Bankole A.A. Gut Microbiota in Human Systemic Lupus Erythematosus and a Mouse Model of Lupus. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02288-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azzouz D., Omarbekova A., Heguy A., Schwudke D., Gisch N., Rovin B.H., Caricchio R., Buyon J.P., Alekseyenko A.V., Silverman G.J. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 2019;78:947–956. doi: 10.1136/annrheumdis-2018-214856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Liao X., Sparks J.B., Luo X.M. Dynamics of gut microbiota in autoimmune lupus. Appl. Environ. Microbiol. 2014;80:7551–7560. doi: 10.1128/AEM.02676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toral M., Robles-Vera I., Romero M., de la Visitación N., Sánchez M., O’Valle F., Rodriguez-Nogales A., Gálvez J., Duarte J., Jiménez R. Lactobacillus fermentum CECT5716: A novel alternative for the prevention of vascular disorders in a mouse model of systemic lupus erythematosus. FASEB J. 2019;33:10005–10018. doi: 10.1096/fj.201900545RR. [DOI] [PubMed] [Google Scholar]
- 30.Zegarra-Ruiz D.F., El Beidaq A., Iñiguez A.J., Lubrano Di Ricco M., Manfredo Vieira S., Ruff W.E., Mubiru D., Fine R.L., Sterpka J., Greiling T.M., et al. A Diet-Sensitive Commensal Lactobacillus Strain Mediates TLR7-Dependent Systemic Autoimmunity. Cell Host Microbe. 2019;25:113–127. doi: 10.1016/j.chom.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manfredo Vieira S., Hiltensperger M., Kumar V., Zegarra-Ruiz D., Dehner C., Khan N., Costa F.R.C., Tiniakou E., Greiling T., Ruff W., et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359:1156–1161. doi: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katz-Agranov N., Zandman-Goddard G. The microbiome and systemic lupus erythematosus. Immunol. Res. 2017;65:432–437. doi: 10.1007/s12026-017-8906-2. [DOI] [PubMed] [Google Scholar]
- 33.Chen X.Q., Yu Y.C., Deng H.H., Sun J.Z., Dai Z., Wu Y.W., Yang M. Plasma IL-17A is increased in new-onset SLE patients and associated with disease activity. J. Clin. Immunol. 2010;30:221–225. doi: 10.1007/s10875-009-9365-x. [DOI] [PubMed] [Google Scholar]
- 34.Kimura A., Kishimoto T. IL-6: Regulator of Treg/Th17 balance. Eur. J. Immunol. 2010;40:1830–1835. doi: 10.1002/eji.201040391. [DOI] [PubMed] [Google Scholar]
- 35.López P., de Paz B., Rodríguez-Carrio J., Hevia A., Sánchez B., Margolles A., Suárez A. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci. Rep. 2016;6:24072. doi: 10.1038/srep24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu C.F., Peng W.M., Schlee M., Barchet W., Eis-Hübinger A.M., Kolanus W., Geyer M., Schmitt S., Steinhagen F., Oldenburg J., et al. SOCS1 and SOCS3 Target IRF7 Degradation to Suppress TLR7-Mediated Type I IFN Production of Human Plasmacytoid Dendritic Cells. J. Immunol. 2018;200:4024–4035. doi: 10.4049/jimmunol.1700510. [DOI] [PubMed] [Google Scholar]
- 37.Buie J.J., Renaud L.L., Muise-Helmericks R., Oates J.C. IFN-α Negatively Regulates the Expression of Endothelial Nitric Oxide Synthase and Nitric Oxide Production: Implications for Systemic Lupus Erythematosus. J. Immunol. 2017;199:1979–1988. doi: 10.4049/jimmunol.1600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison D.G., Guzik T.J., Lob H.E., Madhur M.S., Marvar P.J., Thabet S.R., Vinh A., Weyand C.M. Inflammation, immunity, and hypertension. Hypertension. 2011;57:132–140. doi: 10.1161/HYPERTENSIONAHA.110.163576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weckerle C.E., Mangale D., Franek B.S., Kelly J.A., Kumabe M., James J.A., Moser K.L., Harley J.B., Niewold T.B. Large-scale analysis of tumor necrosis factor α levels in systemic lupus erythematosus. Arthritis Rheum. 2012;64:2947–2952. doi: 10.1002/art.34483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venegas-Pont M., Manigrasso M.B., Grifoni S.C., LaMarca B.B., Maric C., Racusen L.C., Glover P.H., Jones A.V., Drummond H.A., Ryan M.J. Tumor necrosis factor-alpha antagonist etanercept decreases blood pressure and protects the kidney in a mouse model of systemic lupus erythematosus. Hypertension. 2010;56:643–649. doi: 10.1161/HYPERTENSIONAHA.110.157685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bomfim G.F., Dos Santos R.A., Oliveira M.A., Giachini F.R., Akamine E.H., Tostes R.C., Fortes Z.B., Webb R.C., Carvalho M.H. Toll-like receptor 4 contributes to blood pressure regulation and vascular contraction in spontaneously hypertensive rats. Clin. Sci. 2012;122:535–543. doi: 10.1042/CS20110523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sollinger D., Eißler R., Lorenz S., Strand S., Chmielewski S., Aoqui C., Schmaderer C., Bluyssen H., Zicha J., Witzke O., et al. Damage-associated molecular pattern activated Toll-like receptor 4 signalling modulates blood pressure in L-NAME-induced hypertension. Cardiovasc. Res. 2014;101:464–472. doi: 10.1093/cvr/cvt265. [DOI] [PubMed] [Google Scholar]
- 43.Liang C.F., Liu J.T., Wang Y., Xu A., Vanhoutte P.M. Toll-like receptor 4 mutation protects obese mice against endothelial dysfunction by decreasing NADPH oxidase isoforms 1 and 4. Arterioscler. Thromb. Vasc. Biol. 2013;33:777–784. doi: 10.1161/ATVBAHA.112.301087. [DOI] [PubMed] [Google Scholar]
- 44.Toral M., Gómez-Guzmán M., Jiménez R., Romero M., Sánchez M., Utrilla M.P., Garrido-Mesa N., Rodríguez-Cabezas M.E., Olivares M., Gálvez J., et al. The probiotic Lactobacillus coryniformis CECT5711 reduces the vascular pro-oxidant and pro-inflammatory status in obese mice. Clin. Sci. 2014;127:33–45. doi: 10.1042/CS20130339. [DOI] [PubMed] [Google Scholar]
- 45.Shi L., Zhang Z., Yu A.M., Wang W., Wei Z., Akhter E., Maurer K., Costa Reis P., Song L., Petri M., et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PLoS ONE. 2014;9:e93846. doi: 10.1371/journal.pone.0093846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Issara-Amphorn J., Surawut S., Worasilchai N., Thim-Uam A., Finkelman M., Chindamporn A., Palaga T., Hirankarn N., Pisitkun P., Leelahavanichkul A. The Synergy of Endotoxin and (1→3)-β-D-Glucan, from Gut Translocation, Worsens Sepsis Severity in a Lupus Model of Fc Gamma Receptor IIb-Deficient Mice. J. Innate Immun. 2018;10:189–201. doi: 10.1159/000486321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogunrinde E., Zhou Z., Luo Z., Alekseyenko A., Li Q.Z., Macedo D., Kamen D.L., Oates J.C., Gilkeson G.S., Jiang W. A Link Between Plasma Microbial Translocation, Microbiome, and Autoantibody Development in First-Degree Relatives of Systemic Lupus Erythematosus Patients. Arthritis Rheumatol. 2019 doi: 10.1002/art.40935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P., Liu H., Cross J.R., Pfeffer K., Coffer P.J., et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh N., Gurav A., Sivaprakasam S., Brady E., Padia R., Shi H., Thangaraju M., Prasad P.D., Manicassamy S., Munn D.H., et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodríguez-Carrio J., López P., Sánchez B., González S., Gueimonde M., Margolles A., de Los Reyes-Gavilán C.G., Suárez A. Intestinal Dysbiosis Is Associated with Altered Short-Chain Fatty Acids and Serum-Free Fatty Acids in Systemic Lupus Erythematosus. Front. Immunol. 2017;8:23. doi: 10.3389/fimmu.2017.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martens E.C., Lowe E.C., Chiang H., Pudlo N.A., Wu M., McNulty N.P., Abbott D.W., Henrissat B., Gilbert H.J., Bolam D.N., et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mu Q., Tavella V.J., Kirby J.L., Cecere T.E., Chung M., Lee J., Li S., Ahmed S.A., Eden K., Allen I.C., et al. Antibiotics ameliorate lupus-like symptoms in mice. Sci. Rep. 2017;7:13675. doi: 10.1038/s41598-017-14223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He Z., Kong X., Shao T., Zhang Y., Wen C. Alterations of the Gut Microbiota Associated with Promoting Efficacy of Prednisone by Bromofuranone in MRL/lpr Mice. Front. Microbiol. 2019;10:978. doi: 10.3389/fmicb.2019.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kasselman L.J., Vernice N.A., DeLeon J., Reiss A.B. The gut microbiome and elevated cardiovascular risk in obesity and autoimmunity. Atherosclerosis. 2018;271:203–213. doi: 10.1016/j.atherosclerosis.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 55.Greiling T.M., Dehner C., Chen X., Hughes K., Iñiguez A.J., Boccitto M., Ruiz D.Z., Renfroe S.C., Vieira S.M., Ruff W.E., et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aan2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schiffer C., Lalanne A.I., Cassard L., Mancardi D.A., Malbec O., Bruhns P., Dif F., Daëron M. A strain of Lactobacillus casei inhibits the effector phase of immune inflammation. J. Immunol. 2011;187:2646–2655. doi: 10.4049/jimmunol.1002415. [DOI] [PubMed] [Google Scholar]
- 57.Zamani B., Golkar H.R., Farshbaf S., Emadi-Baygi M., Tajabadi-Ebrahimi M., Jafari P., Akhavan R., Taghizadeh M., Memarzadeh M.R., Asemi Z. Clinical and metabolic response to probiotic supplementation in patients with rheumatoid arthritis: A randomized, double-blind, placebo-controlled trial. Int. J. Rheum. Dis. 2016;19:869–879. doi: 10.1111/1756-185X.12888. [DOI] [PubMed] [Google Scholar]
- 58.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 59.Szulińska M., Łoniewski I., Skrypnik K., Sobieska M., Korybalska K., Suliburska J., Bogdański P. Multispecies Probiotic Supplementation Favorably Affects Vascular Function and Reduces Arterial Stiffness in Obese Postmenopausal Women-A 12-Week Placebo-Controlled and Randomized Clinical Study. Nutrients. 2018;10:1672. doi: 10.3390/nu10111672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szulińska M., Łoniewski I., van Hemert S., Sobieska M., Bogdański P. Dose-Dependent Effects of Multispecies Probiotic Supplementation on the Lipopolysaccharide (LPS) Level and Cardiometabolic Profile in Obese Postmenopausal Women: A 12-Week Randomized Clinical Trial. Nutrients. 2018;10:773. doi: 10.3390/nu10060773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mardani F., Mahmoudi M., Esmaeili S.A., Khorasani S., Tabasi N., Rastin M. In vivo study: Th1-Th17 reduction in pristane-induced systemic lupus erythematosus mice after treatment with tolerogenic Lactobacillus probiotics. J. Cell. Physiol. 2018;234:642–649. doi: 10.1002/jcp.26819. [DOI] [PubMed] [Google Scholar]
- 62.Tzang B.S., Liu C.H., Hsu K.C., Chen Y.H., Huang C.Y., Hsu T.C. Effects of oral Lactobacillus administration on antioxidant activities and CD4 + CD25 + forkhead box P3 (FoxP3) + T cells in NZB/W F1 mice. Br. J. Nutr. 2017;118:333–342. doi: 10.1017/S0007114517002112. [DOI] [PubMed] [Google Scholar]
- 63.Hsu T.C., Huang C.Y., Liu C.H., Hsu K.C., Chen Y.H., Tzang B.S. Lactobacillus paracasei GMNL-32, Lactobacillus reuteri GMNL-89 and L. reuteri GMNL-263 ameliorate hepatic injuries in lupus-prone mice. Br. J. Nutr. 2017;117:1066–1074. doi: 10.1017/S0007114517001039. [DOI] [PubMed] [Google Scholar]
- 64.Hu W.S., Rajendran P., Tzang B.S., Yeh Y.L., Shen C.Y., Chen R.J., Ho T.J., Vijaya Padma V., Chen Y.H., Huang C.Y. Lactobacillus paracasei GMNL-32 exerts a therapeutic effect on cardiac abnormalities in NZB/W F1 mice. PLoS ONE. 2017;12:e0185098. doi: 10.1371/journal.pone.0185098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ryan M.J., McLemore G.R., Jr. Hypertension and impaired vascular function in a female mouse model of systemic lupus erythematosus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;292:R736–R742. doi: 10.1152/ajpregu.00168.2006. [DOI] [PubMed] [Google Scholar]
- 66.Zmora N., Zilberman-Schapira G., Suez J., Mor U., Dori-Bachash M., Bashiardes S., Kotler E., Zur M., Regev-Lehavi D., Brik R.B., et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell. 2018;174:1388–1405. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]