Abstract
Polyunsaturated fatty acids (PUFAs) play important roles in health and disease. PUFA levels are influenced by nutrition and genetic factors. The relationship between PUFA composition in red blood cells (RBCs) and genetic variations involved in PUFA metabolism has not been investigated in children with obesity. This study evaluated the association between several genetic variations and PUFA levels in RBCs in children with obesity. One hundred ninety-six children with obesity (101 females, 95 males) were evaluated using anthropometric measurements, dietary intakes, plasma and RBC PUFA quantification, blood biochemistry, and 55 single nucleotide polymorphisms within 14 genes. phosphatidylethanolamine N-methyltransferase (PEMT) rs1109859 and methylenetetrahydrofolate reductase gene (MTHFR) rs4846052 genotypes were associated with PUFA levels in RBCs. PUFA intake did not influence the RBC eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) levels. Higher RBC DHA and EPA levels were observed for PEMT rs1109859 GG and GA genotypes versus the AA genotype. Higher levels of RBC DHA, EPA, arachidonic acid (ARA), and linoleic acid (LA) and were observed for MTHFR rs4846052 TT genotype versus TC and CC genotypes. Genetic variations in PEMT rs1109859 and MTHFR rs4846052 were associated with different PUFA levels in RBC membranes and are estimators for PUFA species in RBCs. Further research is needed to establish whether these genotype-specific alterations are specific to overweight children.
Keywords: polyunsaturated fatty acids, PEMT, MTHFR, children, obesity, red blood cells
1. Introduction
Within the research devoted to obesity, nutrition studies are at the forefront of the efforts aimed at combating this major epidemic. Among these, considerable efforts are involved in finding how the quality, quantity, and types of fats are contributing to both the obesity onset and progression, or to its reduction. This study addresses the potential role of genetic variations in the modulation of polyunsaturated fatty acid (PUFA) levels in children with obesity. PUFA levels are influenced by both nutrition and genetic factors, separately [1,2], or by diet–gene interactions [3]. PUFAs are obtained from diet and synthesized endogenously from their precursor molecules (e.g., linoleic acid for omega-6, and alpha-linolenic acid for omega-3 species). Several genes may influence PUFAs’ status in the body. In PUFAs’ endogenous synthesis, the fatty acid desaturase (FADS) gene cluster (controlling the desaturation) and genes controlling the elongation (ELOVL genes) [4] are involved; variations in these genes impact PUFA levels in humans, including children [5]. Additionally, genetic variations in genes involved in the synthesis of phospholipids and one-carbon metabolism may further impact PUFAs’ status. Phosphatidylcholine (PtdCho) is the most common phospholipid in cell membranes. PtdCho is synthesized through distinct pathways, either through the Cytidine 5’-diphosphocholine (CDP-choline) pathway or by the conversion of phosphatidylethanolamine to PtdCho by phosphatidylethanolamine N-methyltransferase (PEMT) [6], which influences the abundance of certain PUFAs in phospholipids structure [2]. The methylenetetrahydrofolate reductase gene (MTHFR), involved in one-carbon metabolism, may also impact omega-3 PUFA levels [7,8], probably by influencing S-Adenosyl methionine synthesis, which in its turn influences PUFAs’ phospholipid composition.
The fatty acid composition of red blood cells (RBCs) reflects the fatty acid composition in other organs [9]. Therefore, the fatty acid levels in RBC membranes could be potentially relevant biomarkers for the evaluation of PUFAs’ status in the human body, and could potentially improve the assessment of PUFA homeostasis, adding relevant information about the roles PUFA might have in relation to obesity-associated metabolic disorders such as dyslipidemia [10], and also reveal other relationships with dietary intakes and genetic variations.
The PUFA status of RBC membranes has been scarcely studied in the context of childhood obesity and genetic variability. Considering the importance of the fatty acid composition in cell membranes, this study focused on the association between several single nucleotide polymorphisms (SNPs) and PUFA content in the RBCs of children with obesity.
2. Materials and Methods
2.1. Participants and Samples
Two hundred children (95 males, 105 females) aged 7–18 years, with obesity defined by body mass index (BMI) > +2 SD over the World Health Organization (WHO) 2007 reference [11], were evaluated at the 2nd Pediatric Clinic of Clinical Emergency County Hospital Timisoara, Romania. Exclusion criteria were diagnosis of any type of cancer or medical history of cancer; any psychiatric disorder; blood coagulation disorders; endocrine-induced obesity (Cushing syndrome, hypothyroidism, growth hormone deficit); hypothalamus-induced obesity (Babinski–Fröhlich syndrome); genetic syndromes (Prader–Willi, achondroplasia, Bardet–Biedl, Fanconi, Turner, etc.), and personal history for convulsive disorders, nephrotic syndrome, or asthma with corticoid treatment. Four individuals (females) were excluded due to incomplete assessment data.
Participants and their parents or legal guardians were informed about the aims and methods of the study. Informed consent was obtained verbally from the participating children and in writing from their parents or legal guardians. The study was approved by the Ethics Committee of the “Victor Babes” University of Medicine and Pharmacy (6/20.06.2016), Timisoara, Romania, and conducted in accordance with the Declaration of Helsinki. The study was registered at ClinicalTrials.gov (NCT02837367).
Blood samples were collected after overnight fasting (at least 6 h) in EDTA sterile vacutainers.
2.2. Anthropometric Measurements
Anthropometric measurements were performed in light clothing, without shoes, in the morning, following international guidelines as previously described [12]. Weight and height were measured using an electronic scale with a stadiometer. Measurements for height and weight were recorded to the nearest 0.5 cm and 0.5 kg, respectively. BMI was calculated as kg/m2. Standardized BMI-for-age z-scores (zBMI) were calculated according to the WHO guidelines in order to account for the age and gender of children [11].
2.3. Food Intake
Food intake was evaluated using 5-pass 24 h dietary recalls as previously described [13]. Briefly, the recalls were administered four times to each participant if older than 13 years of age, or to both a parent and the child if the participant was younger. The declared amounts for each day investigated (foods and drinks) were converted to energy and macro- and micronutrient intakes using a web-application (Nutritio, Bucharest, Romania, https://nutritioapp.com) based on the U.S. Department of Agriculture (USDA) Food and Nutrient Database for Dietary Studies, with appropriate adaptations for local foods.
2.4. Hematological and Biochemical Tests
Complete blood count was assessed using flux cytometry and cytochemistry by ISO 15189-accredited medical laboratory, acting as an external partner.
Total plasma concentrations of aspartate aminotransferase (AST), alanine aminotransferase (ALT), C reactive protein (CRP), total cholesterol, high-density lipoprotein (HDL) cholesterol, triglycerides, and glucose were performed on an Ortho Clinical Vitros 350 Chemistry System (Ortho Clinical Diagnostics Inc, Raritan, NJ, USA), using its standardized reagents, following the manufacturer’s protocols. Homocysteine and insulin were measured by ELISA method on an Epoch Microplate Spectrophotometer (BioTek Instruments Inc., Winooski, VT, USA). The kits used for insulin were acquired from Fortress Diagnostics (Antrim, United Kingdom), and for homocysteine (Axis Homocysteine Enzyme Immunoassay) from IBL International GMBH, Hamburg, Germany. The analyzer was calibrated and maintained according to the manufacturer’s instructions.
A homeostatic model assessment for insulin resistance (HOMA-IR) was calculated using the following formula: fasting insulin (mIU/L) × fasting glucose (mmol/L)/22.5.
2.5. Fatty Acids Quantification
The blood samples were subjected to centrifugation at 1000× g for 10 min at room temperature. After the plasma was transferred into another tube, the RBCs were washed twice with PBS. If not immediately processed, the samples were stored at −80 °C. The samples (plasma and RBCs) were processed and analyzed using a previously described protocol [14]. Three different measurements were obtained for each sample: plasma free fatty acids (FFA), plasma conjugated fatty acids, and RBC membrane fatty acids. The results were expressed in µmol/L. The conjugated fatty acid levels in plasma were estimated by subtracting the FFA plasma levels from total fatty acids levels.
2.6. Preparation of Libraries for Next-Generation Sequencing
Genomic DNA was isolated from whole blood using the MagCore® Extractor System and MagCore® Genomic DNA Whole Blood Kit (RBC Bioscience, New Taipei City, Taiwan), following the manufacturer’s protocol. Genotyping was performed on a MiSeq sequencer (Illumina, San Diego, CA, USA) using a custom-made hotspot sequencing kit for 55 SNPs within 14 genes selected as previously being associated with increased lipids, non-alcoholic fatty liver, or cardiovascular disease [15].
Amplicon sequencing libraries were prepared from 20 ng of DNA per sample according to the AmpliSeq protocol (Illumina Inc, San Diego, CA, USA). Libraries were generated with dual indices (19 PCR cycles) followed by normalization and pooling. The pooled libraries were paired-end (2 × 150) sequenced on a micro flow cell with V2 chemistry on a MiSeq instrument (Illumina Inc, San Diego, CA, USA).
2.7. Analysis of Genetic Variants
After demultiplexing and the generation of FASTQ files, sequence alignment to the reference genome and sequence quality filtering were performed using the Illumina MiSeq Reporter v2.6 platform. The sequences were aligned with Burrows-Wheeler Aligner (BWA)and variant calling was performed with Genome Analysis Toolkit (GATK) using the human reference sequence hg19/GRCh37. Variant calling was performed on the variant call format (VCF) output files by evaluating the coverage (the number of times that a targeted variant is read during the sequencing) and the quality score (Q-score; the estimated probability of the base call being wrong). VCF files were further subjected to annotation using ANNOVAR [16] with the dataset dbNSFP 35a [17].
2.8. Statistical Analyses
Data were analyzed using IBM-SPSS version 25 (IBM, Armonk, New York, USA). The Shapiro–Wilk test was used to determine if the data set had normal distribution. The Levene test was used for assessing homogeneity of variance. The Mann–Whitney U test was used to evaluate the significance of differences between females and males in all variables studied. Correlations were evaluated for statistical significance using the Spearman’s test.
The Kruskal–Wallis test for independent samples was performed using PUFA measurements in plasma and RBCs (alpha-linolenic acid (ALA), arachidonic acid (ARA), docosahexaenoic acid (DHA), eicosapentaenoic acid (EPA), and linoleic acid (LA)) as the dependent variables, and with the 55 SNPs as predictors. Where the tests gave significant results, the Mann–Whitney U test was used for the supplementary evaluation of differences in PUFA levels between the three genotypes for the SNPs found to be significantly associated with fatty acid measurements. Boxplot graphics were used to display the distribution of DHA, LA, ARA, and EPA between PEMT and MTHFR genotypes, respectively.
Univariate analyses of variance models were performed for evaluating joint variability of genotypes, gender, hemoglobin, hematocrit, and PUFA dietary intakes.
3. Results
Sixty-five percent (128/196) of children in the cohort lived in an urban environment, while the rest lived in rural areas. Descriptive statistics for anthropometric data, biochemical analysis, PUFA measurements in RBC membranes, and dietary intakes of the 196 participants are presented in Table 1. As most of the variables were non-normally distributed, median and interquartile range (IQR) are presented. Significant differences were found between genders for hemoglobin, zBMI, and RBC levels of ALA and LA; all were higher in males.
Table 1.
Descriptive statistics for anthropometric data, biochemical analysis, PUFA measurements in RBCs, and dietary intakes. Mann–Whitney U test was used to analyze gender differences (test is significant at p value ≤ 0.05).
| Variables | All n = 196 | Females n = 101 | Males n = 95 | p Value | |||
|---|---|---|---|---|---|---|---|
| Median | IQR | Median | IQR | Median | IQR | ||
| Anthropometric data | |||||||
| Age (years) | 12 | 5 | 12.00 | 5.0 | 12.00 | 4.75 | 0.787 |
| zBMI | 3.13 | 1.20 | 2.83 | 1.21 | 3.44 | 1.37 | 0.001 |
| Hematological and biochemical analysis | |||||||
| Hemoglobin (g/dL) | 13.60 | 1.20 | 13.50 | 1.10 | 13.70 | 1.47 | 0.034 |
| Hematocrit (%) | 40.20 | 2.85 | 39.90 | 2.80 | 40.25 | 3.40 | 0.152 |
| HOMA-IR | 3.95 | 2.95 | 4.04 | 3.04 | 3.94 | 3.09 | 0.470 |
| TC (mg/dL) | 174.00 | 54.00 | 166.00 | 52.00 | 182.50 | 55.50 | 0.109 |
| TG (mg/dL) | 129.00 | 86.50 | 125.00 | 88.00 | 133.00 | 99.75 | 0.237 |
| HDL (mg/dL) | 46.00 | 18.00 | 43.00 | 19.00 | 48.00 | 6.75 | 0.085 |
| AST (U/L) | 29.00 | 14.00 | 29.00 | 14.00 | 29.50 | 15.50 | 0.127 |
| ALT (U/L) | 32.00 | 15.00 | 31.00 | 16.00 | 33.50 | 14.75 | 0.167 |
| CRP (mg/dL) | 4.70 | 7.90 | 4.80 | 8.70 | 4.75 | 7.72 | 0.315 |
| Homocysteine (µmoL/L) | 14.78 | 8.78 | 14.16 | 8.08 | 15.33 | 9.28 | 0.398 |
| PUFA measurements in RBCs | |||||||
| ALA RBC (µmol/L) | 1.41 | 1.00 | 1.28 | 1.00 | 1.46 | 1.1 | 0.038 |
| ARA RBC (µmol/L) | 354.22 | 168.25 | 338.64 | 175.45 | 368.18 | 152.23 | 0.142 |
| DHA RBC (µmol/L) | 160.13 | 116.52 | 150.06 | 117.12 | 175.11 | 113.39 | 0.309 |
| EPA RBC (µmol/L) | 2.86 | 2.12 | 2.64 | 2.34 | 2.92 | 1.72 | 0.115 |
| LA RBC (µmol/L) | 209.49 | 135.82 | 193.86 | 119.7 | 239.89 | 140.5 | 0.029 |
| Selected daily nutrient intakes evaluated in 24 h dietary recalls | |||||||
| Kilocalories | 1201.59 | 494.72 | 1201.49 | 440.11 | 1205.77 | 580.32 | 0.889 |
| Protein (g) | 66.60 | 22.11 | 66.63 | 23.00 | 66.48 | 23.80 | 0.832 |
| Lipids (g) | 44.59 | 25.08 | 44.17 | 22.99 | 45.67 | 28.42 | 0.781 |
| Carbohydrates (g) | 135.93 | 58.15 | 138.52 | 51.27 | 129.93 | 61.29 | 0.266 |
| Water (g) | 21.96 | 829.49 | 2146.43 | 749.58 | 2280.80 | 912.78 | 0.207 |
| Cholesterol (mg) | 260.54 | 157.94 | 255.68 | 129.83 | 274.79 | 158.47 | 0.853 |
| ALA (mg) | 77.16 | 217.21 | 57.29 | 196.85 | 87 | 272.44 | 0.280 |
| DHA (mg) | 10.63 | 24 | 10.16 | 19.47 | 10.95 | 24.74 | 0.547 |
| EPA (mg) | 3.4 | 5.99 | 3.09 | 5.81 | 4.00 | 6.20 | 0.971 |
| LA (mg) | 943.08 | 2306.07 | 953.40 | 2072.57 | 943.08 | 2334.76 | 0.527 |
Legend: PUFA—polyunsaturated fatty acid, IQR—interquartile range (IQR = Q3–Q1), zBMI—standardized body mass index (BMI) to account for age and gender, HOMA-IR—homeostatic model assessment for insulin resistance, TC—total cholesterol, TG—triglycerides, HDL—high density cholesterol, g—grams, mg—milligrams, µmol/L—micromoles/liter, AST—aspartate aminotransferase, ALT—alanine aminotransferase, CRP—C reactive protein, HDL chol—high-density lipoprotein cholesterol, ALA—alfa-linolenic acid, ARA—arachidonic acid, DHA—docosahexaenoic acid, EPA—eicosapentaenoic acid, LA—linoleic acid, RBC—red blood cell membrane.
PUFA species in RBC membranes did not correlate with total RBC number, hemoglobin, hematocrit, estimated macronutrients, ALA, ARA, DHA, EPA, and LA dietary intakes, nor with zBMI (stratification by gender did not show significance, data not shown). PUFAs from RBCs did not correlate with age, except for EPA (−0.218, p = 0.002). Environment (urban versus rural) did not correlate with the PUFA levels. Mann–Whitney U test showed that gender is significantly associated with ALA and LA levels in RBC (p values of 0.038 and 0.029, respectively). The zBMI levels positively correlated with AST (0.161, p = 0.023), ALT (0.232, p = 0.001), and with age (0.441, p ≤ 0.001).
Descriptive statistics for the measurement of fatty acids in RBC membranes and plasma are presented as median and IQR in Table A1. DHA in RBC membranes correlated with plasma free DHA (0.236, p = 0.001) and with plasma conjugated DHA (0.220, p = 0.002). EPA from RBCs correlated with plasma free EPA (0.375, p ≤ 0.001) and with plasma conjugated EPA (0.433, p ≤ 0.001). ALA, ARA, and LA did not correlate between measurements in plasma and RBC membranes.
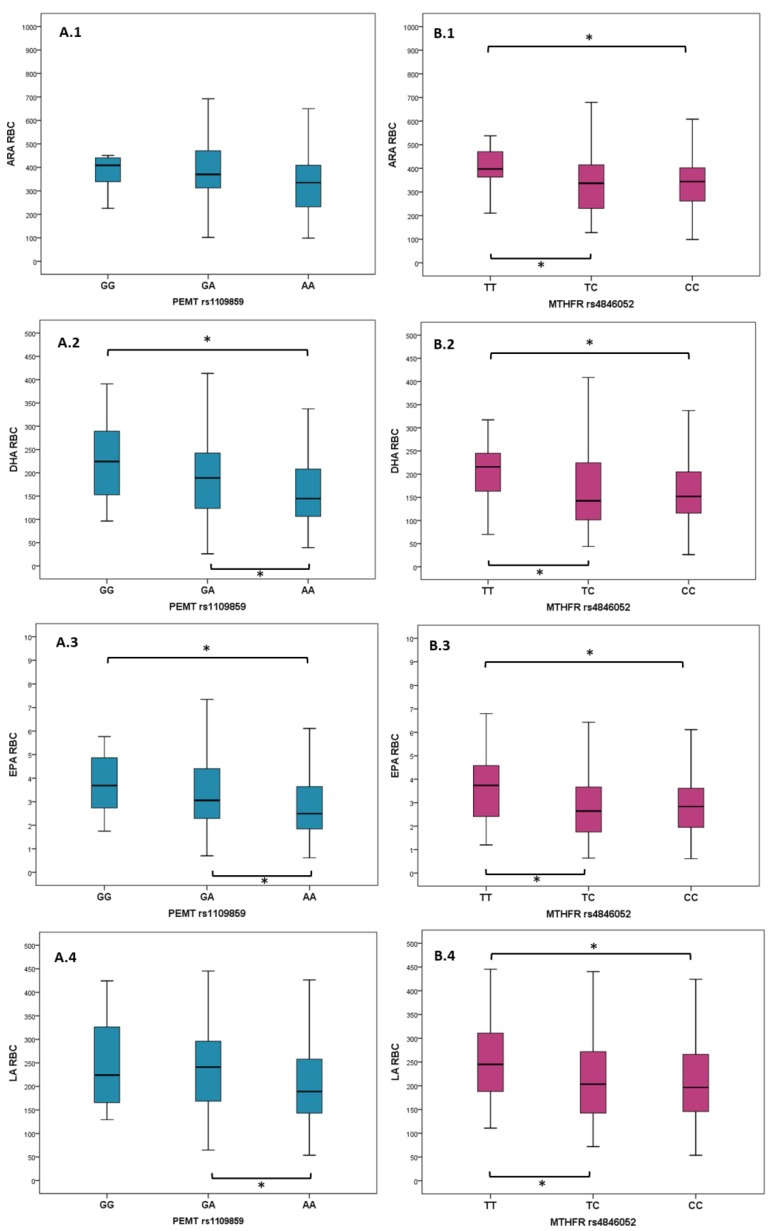
Using the independent-samples Kruskal–Wallis test, PEMT rs1109859 and MTHFR rs4846052 genotypes were found to significantly associate with fatty acid levels (except for ALA) measured in RBC membranes, but not in plasma (data not presented). The rest of the SNPs did not show significant association with RBC PUFAs (frequencies of all 55 SNPs are shown in Table A2). ARA, DHA, EPA, and LA levels in RBCs were significantly different between the patient’s groups identified based on their PEMT rs1109859 genotype. ARA, DHA, and EPA levels were significantly different between the patient’s groups identified based on their MTHFR rs4846052 genotype (Table 2). Figure 1 displays the distribution of ARA, EPA, DHA, and LA levels between genotypes and differences that are statistically significant between groups (Mann–Whitney U test). The p values are presented in Table A3).
Table 2.
PUFA levels in RBC membranes grouped by PEMT and MTHFR genetic variation. The Kruskal–Wallis test was used to analyze differences between genotypes. (Test is significant at p value ≤ 0.05).
| PEMT rs1109859 | GG, n = 12 (6.1%) | GA, n = 68 (34.7%) | AA, n = 116 (59.2%) | p Value | |||
| Median | IQR | Median | IQR | Median | IQR | ||
| ALA RBC (µmol/L) | 1.65 | 1.46 | 1.54 | 1.11 | 1.34 | 0.99 | 0.090 |
| ARA RBC (µmol/L) | 408.72 | 112.81 | 370.48 | 162.76 | 366.92 | 178.73 | 0.007 |
| DHA RBC (µmol/L) | 224.10 | 151.89 | 190.15 | 120.4 | 144.62 | 102.81 | 0.040 |
| EPA RBC (µmol/L) | 3.69 | 2.51 | 3.09 | 2.27 | 2.50 | 1.83 | 0.017 |
| LA RBC (µmol/L) | 224.02 | 191.77 | 243.88 | 129.83 | 188.88 | 116 | 0.022 |
| MTHFR rs4846052 | TT, n = 33 (16.8%) | TC, n = 97 (49.5%) | CC, n = 66 (33.7%) | p Value | |||
| Median | IQR | Median | IQR | Median | IQR | ||
| ALA RBC (µmol/L) | 1.54 | 0.99 | 1.34 | 0.92 | 1.35 | 1.32 | 0.061 |
| ARA RBC (µmol/L) | 397.38 | 133.61 | 337.05 | 186.2 | 344.86 | 148.14 | 0.006 |
| DHA RBC (µmol/L) | 215.69 | 87.64 | 142.47 | 1338.86 | 153.82 | 91.28 | 0.015 |
| EPA RBC (µmol/L) | 3.74 | 2.33 | 2.67 | 1.92 | 2.83 | 1.77 | 0.066 |
| LA RBC (µmol/L) | 244.93 | 140.51 | 199.15 | 130.24 | 199.41 | 134.80 | 0.044 |
Legend 2: IQR—interquartile range, ALA—alfa-linolenic acid, ARA—arachidonic acid, DHA—docosahexaenoic acid, EPA—eicosapentaenoic acid, LA—linoleic acid, RBC—red blood cell membrane; MTHFR—methylenetetrahydrofolate reductase; PEMT—phosphatidylethanolamine N-methyltransferase.
Figure 1.
Distribution of PUFA species grouped by PEMT rs1109859 (A) and MTHFR rs4846052 genotypes (B). Mann–Whitney U test was used to assess statistical significance between groups. Significant differences are marked with *, and the brackets indicate the two groups for which these differences were identified. Horizontal lines, within each boxplot, indicate minimum, first quartile (Q1), median, third quartile (Q3), and maximum. Legend: ARA—arachidonic acid, DHA—docosahexaenoic acid, EPA—eicosapentaenoic acid, LA—linoleic acid, RBC—red blood cell membrane; MTHFR—methylenetetrahydrofolate reductase; PEMT—phosphatidylethanolamine N-methyltransferase.
Univariate analysis of variance controlled for the dietary intakes and indicated that DHA, EPA, and LA dietary intakes did not influence the DHA, EPA, and LA levels in RBC membranes. Univariate analysis of variance also indicated that gender influenced ALA and LA levels in RBCs, while hematocrit influenced ARA levels. Complete results for univariate analysis of variance models are shown in Table A4.
4. Discussion
This study analyzed omega-6 and omega-3 fatty acid levels in plasma and in the RBC membranes of children with obesity in relation to their food intake and to 55 SNPs from 13 genes associated with lipid metabolism.
Significant differences were identified between females and males regarding standardized body mass index for age (zBMI), with males presenting a higher degree of obesity. This is a previously described feature of the Romanian population [18,19] and in other countries [20]. However, the zBMI did not suggest an association with PUFAs measured in plasma or RBC membranes. The difference in hemoglobin levels between females and males, lower in females, has been already described in other studies, and it is probably explained by menstrual cycle blood loss [21]. Although this difference is significant, the hemoglobin and hematocrit did not correlate with the PUFAs measured in RBC membranes. However, hematocrit, when added in the model with PEMT rs1109859, was found to influence the ARA level in RBCs. The differences between males and females in ALA and LA, measured in RBCs, were not reported previously and need to be re-evaluated in larger studies.
Aminotransferases directly correlated with zBMI in the present study. This finding was previously reported by others and could be considered as a surrogate marker for nonalcoholic fatty liver disease and associated with metabolic syndrome in children [22]. Additionally, zBMI correlated directly with the age of children in our cohort. In other cohorts, higher adiposity was associated with older age of children in national and international studies [20,23,24].
The negative correlation of EPA with age and weight in children was previously shown in another cohort and was associated with response to supplementation, EPA increasing less in those with higher BMIs [25].
The correlations between EPA and DHA measurements from plasma and RBCs in children with obesity have not been previously investigated and need to be re-evaluated in other larger studies that should include control (lean) groups.
PUFA levels measured in plasma were not associated with any of the investigated genotypes, possibly because the plasma reflects a short-term metabolic status for PUFAs [26]. RBCs, on the other hand, contain fatty acids esterified in phospholipids, which are structural constituents of cell walls, and therefore their fatty acid composition is stable over a longer period, considering that RBCs have a 100-day life-span [27].
Although the RBC PUFA levels in the present study were not influenced by estimated intakes, they associated significantly with two SNPs in the PEMT and MTHFR genes, suggesting that, in children with obesity, genetic variability could be predictive for PUFA composition in RBCs. GG and GA genotypes, when compared to the AA genotype for the PEMT rs1109859, were associated with higher levels of DHA and EPA in RBCs. This is in agreement with previous findings [28], suggesting that the PtdCho synthetized via the PEMT pathway contains mainly PUFAs (mainly ARA and DHA) while the CDP-choline pathway forms PtdCho containing medium-chain and saturated fatty acids. Another study also found similar evidence, indicating that DHA composition from plasma PtdCho may be a marker for PEMT activity [2]. Our results strengthen further the hypothesis that fatty acid composition of PtdCho derived from the PEMT pathway is different from that obtained through the CDP–choline pathway.
PUFA levels in RBCs were also associated with genetic variations for the MTHFR gene (rs4846052). To the best of our knowledge, there have been no other previously published reports assessing this association. However, there is evidence that 5-methyltetrahydrofolate (5-mTHF) supplementation can modify the phospholipid fatty acid pattern [29] and can be explained by the fact that 5-mTHF influences methionine bioavailability that is necessary for S-adenosylmethionine synthesis. S-adenosylmethionine is involved in several cellular transmethylation reactions, including phosphatidylethanolamine methylation [6]. Although the mechanism still needs to be further clarified, our results suggest that the MTHFR rs4846052 genotype influences PUFA levels in the RBC membranes, with the TT genotype being associated with higher levels of PUFAs in the RBC membranes compared to that of TC and TT genotypes. However, it is unknown if higher levels of PUFAs in the RBC membranes represent a protective phenotype.
This study examined, for the first time, the association between PEMT and MTHFR polymorphisms and fatty acid concentrations in RBC membranes in children with obesity. The study indicated that two SNPs analyzed (located on chromosomes 1 and 17, respectively) could have an important role on the composition of PUFAs in RBCs, possibly due to alterations in PUFA metabolism. Therefore, it is possible that such genetic variations could also contribute to a better understanding of whether PUFA intakes are dependent on such genotypes, and subsequently refine PUFA recommended intakes in children with obesity who may have different requirements as a result of these genetic variations. PEMT and MTHFR genes contribute, within the methylation pathways, to the regulation of methionine and PUFA homeostasis via the de novo choline synthesis and its incorporation into PtdCho [6]. As the fatty acid composition of PtdCho in cell membranes, especially EPA and DHA distribution, has a protective role against the development of Alzheimer’s disease [30], obesity-induced metabolic disorders, and cardiovascular diseases in mice [31], this remains a significant point for further research.
The absence of a lean control group is the main limitation of the study. Additionally, this study has not provided a longitudinal analysis for the status of fatty acids in children with different genotypes over time. The assessment of dietary intake using 24 h recalls also has limitations, mostly due to underreporting, as previously shown [32]. However, such recalls have been used widely and represent a validated method for dietary assessment [33,34,35]. Finally, one technical limitation consisted of ARA intakes not being available using the Nutritioapp at the time of this study.
5. Conclusions
Genetic variations in PEMT (rs1109859) and MTHFR (rs4846052) were associated with alterations in the content of PUFA species in RBC membranes. This finding suggests that the genetic status of PEMT and MTHFR genes may contribute to PUFA homeostasis and, therefore, could contribute to PUFA status in children with obesity. Further research is needed to establish whether these genotype-specific alterations are specific to overweight children.
Acknowledgments
This work was performed at the Centre of Genomic Medicine, “Victor Babes” University of Medicine and Pharmacy in Timisoara.
Appendix A
Table A1.
Descriptive statistics for fatty acids in plasma and RBC membranes.
| Fatty Acids (µmol/L) | Plasma Free | Plasma Conjugated | RBC | |||
|---|---|---|---|---|---|---|
| Median | IQR | Median | IQR | Median | IQR | |
| ALA | 3.72 | 4.27 | 16.47 | 15.42 | 1.41 | 1.02 |
| ARA | 7.15 | 4.31 | 375.51 | 156.61 | 354.22 | 168.25 |
| DHA | 6.13 | 4.52 | 271.70 | 159.49 | 160.13 | 116.52 |
| EPA | 0.24 | 0.14 | 7.23 | 5.39 | 2.86 | 2.12 |
| LA | 137.40 | 117.75 | 1208.13 | 462.86 | 209.49 | 135.82 |
Legend: ALA—alfa-linolenic acid, ARA—arachidonic acid, DHA—docosahexaenoic acid, EPA—eicosapentaenoic acid, LA—linoleic acid, RBC—red blood cell membrane, µmol/L—micromoles/liter, RBC—red blood cells, IQR—interquartile range.
Table A2.
The frequencies of 55 single nucleotide polymorphisms studied within 196 subjects.
| Gene | Rs Chr Position GRCh37 | AA/AB/BB | AA% | AB% | BB% |
|---|---|---|---|---|---|
| ABCB4 | rs1149222 chr7:87073775 | GG/GT/TT | 2.0 | 28.1 | 69.9 |
| rs2071645 chr7:87105276 | GG/GC/CC | 73.4 | 25.0 | 1.6 | |
| rs31672 chr7:87059699 | CC/CT/TT | 2.6 | 30.1 | 67.3 | |
| rs4148811 chr7:87101486 | TT/TG/GG | 73.5 | 24.5 | 2.0 | |
| rs9655950 chr7:87033561 | CC/CT/TT | 0.5 | 27.6 | 71.9 | |
| rs1202283 chr7:87082292 | GG/GA/AA | 16.0 | 49.5 | 34.5 | |
| APOC3 | rs2854117 chr11:116700142 | TT/TC/CC | 9.7 | 43.4 | 46.9 |
| CHDH | rs12676 chr3:53857803 | AA/AC/CC | 9.7 | 42.9 | 47.4 |
| rs2289209 chr3:53852835 | CC/CT/TT | 89.8 | 10.2 | 0.0 | |
| rs4563403 chr3:53850814 | CC/CT/TT | 78.6 | 21.4 | 0.0 | |
| rs4687591 chr3:53864407 | AA/AG/GG | 86.2 | 12.8 | 1.0 | |
| rs6807783 chr3:53859662 | GG/GC/CC | 65.3 | 31.1 | 3.6 | |
| rs7634578 chr3:53876728 | CC/CT/TT | 98.5 | 1.5 | 0.0 | |
| rs881883 chr3:53847805 | AA/AG/GG | 68.9 | 28.5 | 2.6 | |
| CHKB | rs1557502 chr22:51013998 | CC/CT/TT | 61.7 | 32.7 | 5.6 |
| rs1557503 chr22:51013072 | GG/GA/AA | 87.7 | 11.8 | 0.5 | |
| rs470117 chr22:51009953 | CC/CT/TT | 33.2 | 47.6 | 19.3 | |
| rs7238 chr22:51007488 | AA/AG/GG | 80.7 | 19.3 | 0.0 | |
| FADS2 | rs2526678chr1161623793 | GG/GA/AA | 82.1 | 17.9 | 0.0 |
| rs526126chr1161624885 | GG/GC/CC | 3.1 | 26.3 | 70.6 | |
| MTHFD1 | rs10135928 chr14:64866439 | TT/TC/CC | 95.4 | 4.6 | 0.0 |
| MTHFR | rs1801133 chr1:11856378 | GG/GA/AA | 43.4 | 43.4 | 13.3 |
| rs2066471 chr1:11860458 | CC/CT/TT | 68.4 | 29.6 | 2.0 | |
| rs4846048 chr1:11846252 | GG/GA/AA | 10.7 | 42.9 | 46.4 | |
| rs4846052 chr1:11857951 | TT/TC/CC | 16.9 | 49.7 | 33.3 | |
| rs7525338 chr1:11862332 | CC/CT/TT | 100.0 | 0.0 | 0.0 | |
| rs868014 chr1:11849447 | AA/AG/GG | 0.0 | 0.0 | 100.0 | |
| SCD | rs11557927 chr10:102121816 | TT/TG/GG | 88.8 | 10.7 | 0.5 |
| rs11599710 chr10:102105788 | GG/GA/AA | 89.3 | 10.2 | 0.5 | |
| rs12247426 chr10:102115327 | CC/CG/GG | 98.5 | 1.5 | 0.0 | |
| rs2167444 chr10:102124744 | TT/TA/AA | 74.5 | 22.4 | 3.1 | |
| rs7849 chr10:102122603 | TT/TC/CC | 69.6 | 25.8 | 4.6 | |
| SLC44A1 | rs10120572chr9108077756 | TT/TG/GG | 97.4 | 2.6 | 0.0 |
| rs10820799 chr9:108092216 | AA/AC/CC | 92.3 | 7.7 | 0.0 | |
| rs193008 chr9:108042806 | TT/TC/CC | 83.6 | 16.4 | 0.0 | |
| rs328006 chr9:108039808 | GG/GC/CC | 83.2 | 16.8 | 0.0 | |
| rs440290 chr9:107987290 | TT/TC/CC | 82.7 | 17.3 | 0.0 | |
| rs443094 chr9:108016685 | GG/GC/CC | 84.7 | 15.3 | 0.0 | |
| rs7018875 chr9:108077434 | CC/CA/AA | 95.4 | 4.6 | 0.0 | |
| STAT3 | rs9891119 chr17:40507980 | AA/AC/CC | 41.8 | 43.9 | 14.3 |
| PCYT1A | rs1580820 chr3:195966258 | GG/GA/AA | 0.5 | 20.9 | 78.6 |
| rs4898190 chrX:24607933 | AA/AC/CC | 1.5 | 6.7 | 91.8 | |
| PEMT | rs1109859chr1717424333 | GG/GA/AA | 6.2 | 34.9 | 59.0 |
| rs12103822 chr17:17418432 | CC/CG/GG | 99.5 | 0.5 | 0.0 | |
| rs16961845 chr17:17432456 | CC/CT/TT | 83.2 | 16.3 | 0.5 | |
| rs4244593 chr17:17420218 | TT/TG/GG | 20.0 | 51.8 | 28.2 | |
| rs4479310 chr17:17405504 | CC/CT/TT | 12.2 | 43.9 | 43.9 | |
| rs7214988 chr17:17491836 | CC/CG/GG | 83.2 | 16.3 | 0.5 | |
| rs7946 chr17:17409560 | CC/CT/TT | 13.3 | 44.1 | 42.6 | |
| rs8068641chr1717480187 | AA/AG/GG | 78.6 | 20.9 | 0.5 | |
| rs936108 chr17:17439793 | CC/CT/TT | 26.5 | 52.0 | 21.4 | |
| rs13342397 chr17:17460926 | TT/TC/CC | 79.5 | 19.9 | 0.6 | |
| rs6502603 chr17:17445680 | GG/GT/TT | 26.0 | 52.6 | 21.4 | |
| PNPLA3 | rs2281135 chr22:44332570 rs738409 chr22:44324727 |
GG/GA/AA CC/CG/GG |
64.3 52.8 |
33.7 21.1 |
2.0 26.1 |
Legend: Gene name as per international nomenclature HUGO Gene Nomenclature Committee (HGNC); Chr—chromosome position reference genome GRCh37; AA—homozygous for one allele; AB—heterozygous; BB—homozygous for the other allele; ABCB4—ATP binding cassette subfamily B member 4; APOC3—apolipoprotein C3; CHDH—choline dehydrogenase; CHKB—choline/ethanolamine kinase beta; FADS2—fatty acid desaturase 2; MTHFD1—methylenetetrahydrofolate dehydrogenase; MTHFR—methylenetetrahydrofolate reductase; SCD—stearoyl-CoA desaturase; SLC44A1—solute carrier family 44 member 1; STAT3—signal transducer and activator of transcription 3; PCYT1A—phosphate cytidylyltransferase 1, choline, alpha; PCYT1B—phosphate cytidylyltransferase 1, choline, beta; PEMT—phosphatidylethanolamine N-methyltransferase; PNPLA3—patatin-like phospholipase domain containing 3.
Table A3.
The p values for the Mann–Whitney U test used to evaluate differences between PUFA measured in red blood cell membranes in regards to rs1109859 PEMT, and rs4846052 MTHFR genotypes. Test is significant at p ≤ 0.05.
| rs1109859 in PEMT gene | |||||
| ALA | ARA | DHA | EPA | LA | |
| GG/GA | 0.978 | 0.524 | 0.201 | 0.370 | 0.984 |
| GA/AA | 0.059 | 0.007 | 0.014 | 0.029 | 0.011 |
| GG/AA | 0.299 | 0.058 | 0.011 | 0.045 | 0.202 |
| rs4846052 in MTHFR gene | |||||
| ALA | ARA | DHA | EPA | LA | |
| TT/TC | 0.020 | 0.003 | 0.016 | 0.027 | 0.018 |
| TC/CC | 0.394 | 0.624 | 0.805 | 0.914 | 0.734 |
| TT/CC | 0.138 | 0.009 | 0.004 | 0.045 | 0.048 |
Legend: ALA—alfa-linolenic acid; ARA—arachidonic acid; DHA—docosahexaenoic acid; EPA—eicosapentaenoic acid; LA—linoleic acid; MTHFR—methylenetetrahydrofolate reductase; PEMT—phosphatidylethanolamine N-methyltransferase.
Table A4.
Univariate analysis of variance results. Models were created using the PUFA RBC levels as dependent variables; gender, genotypes rs1109859, and rs4846052 as fixed factors, and dietary intakes, hemoglobin, and hematocrit as covariates. Test is significant at p ≤ 0.05.
| Dependent Variable: ALA in RBC | |||||
| Source | Type III Sum of Squares | df | Mean Square | F | Sig. |
| Corrected Model | 12.449 a | 8 | 1.556 | 1.548 | 0.145 |
| Intercept | 6.563 | 1 | 6.563 | 6.528 | 0.012 |
| Gender | 4.011 | 1 | 4.011 | 3.989 | 0.048 |
| rs1109859 in PEMT gene | 4.900 | 2 | 2.450 | 2.437 | 0.091 |
| rs4846052 in MTHFR gene | 2.513 | 2 | 1.256 | 1.250 | 0.289 |
| Hemoglobin | 0.045 | 1 | 0.045 | 0.045 | 0.833 |
| Hematocrit | 0.356 | 1 | 0.356 | 0.354 | 0.553 |
| ALA intake | 0.275 | 1 | 0.275 | 0.273 | 0.602 |
| Error | 159.858 | 159 | 1.005 | ||
| Total | 656.808 | 168 | |||
| Corrected Total | 172.307 | 167 | |||
| a. R Squared = 0.072 (Adjusted R Squared = 0.026) | |||||
| Dependent Variable: ARA in RBC | |||||
| Source | Type III Sum of Squares | df | Mean Square | F | Sig. |
| Corrected Model | 552433.852 a | 7 | 78919.122 | 3.657 | 0.001 |
| Intercept | 284923.202 | 1 | 284923.202 | 13.204 | 0.000 |
| Gender | 38020.108 | 1 | 38020.108 | 1.762 | 0.186 |
| rs1109859 in PEMT gene | 290956.552 | 2 | 145478.276 | 6.742 | 0.001 |
| rs4846052 in MTHFR gene | 92203.136 | 2 | 46101.568 | 2.136 | 0.121 |
| Hemoglobin | 69691.519 | 1 | 69691.519 | 3.230 | 0.074 |
| Hematocrit | 95978.498 | 1 | 95978.498 | 4.448 | 0.036 |
| Error | 3927257.216 | 182 | 21578.336 | ||
| Total | 30801660.185 | 190 | |||
| Corrected Total | 4479691.068 | 189 | |||
| a. R Squared = 0.123 (Adjusted R Squared = 0.090) | |||||
| Dependent Variable: DHA in RBC | |||||
| Source | Type III Sum of Squares | df | Mean Square | F | Sig. |
| Corrected Model | 111644.265 a | 8 | 13955.533 | 1.831 | 0.075 |
| Intercept | 44569.962 | 1 | 44569.962 | 5.847 | 0.017 |
| Gender | 6232.763 | 1 | 6232.763 | 0.818 | 0.367 |
| rs1109859 in PEMT gene | 69225.385 | 2 | 34612.692 | 4.541 | 0.012 |
| rs4846052 in MTHFR gene | 33145.490 | 2 | 16572.745 | 2.174 | 0.117 |
| Hemoglobin | 2690.465 | 1 | 2690.465 | 0.353 | 0.553 |
| Hematocrit | 3791.682 | 1 | 3791.682 | 0.497 | 0.482 |
| DHA intake | 943.244 | 1 | 943.244 | 0.124 | 0.725 |
| Error | 1212070.616 | 159 | 7623.086 | ||
| Total | 7032444.484 | 168 | |||
| Corrected Total | 1323714.880 | 167 | |||
| a. R Squared = 0.084 (Adjusted R Squared = 0.038) | |||||
| Dependent Variable: EPA in RBC | |||||
| Source | Type III Sum of Squares | df | Mean Square | F | Sig. |
| Corrected Model | 33.140 a | 8 | 4.143 | 0.853 | 0.558 |
| Intercept | 19.043 | 1 | 19.043 | 3.921 | 0.049 |
| Gender | 2.858 | 1 | 2.858 | 0.588 | 0.444 |
| rs1109859 in PEMT gene | 21.406 | 2 | 10.703 | 2.204 | 0.114 |
| rs4846052 in MTHFR gene | 5.710 | 2 | 2.855 | 0.588 | 0.557 |
| Hemoglobin | 0.758 | 1 | 0.758 | 0.156 | 0.693 |
| Hematocrit | 1.599 | 1 | 1.599 | 0.329 | 0.567 |
| EPA intake | 1.324 | 1 | 1.324 | 0.272 | 0.602 |
| Error | 772.318 | 159 | 4.857 | ||
| Total | 2855.067 | 168 | |||
| Corrected Total | 805.458 | 167 | |||
| a. R Squared = 0.041 (Adjusted R Squared = −0.007) | |||||
| Dependent Variable: LA in RBC | |||||
| Source | Type III Sum of Squares | df | Mean Square | F | Sig. |
| Corrected Model | 220014.263 a | 8 | 27501.783 | 2.829 | 0.006 |
| Intercept | 142585.361 | 1 | 142585.361 | 14.665 | 0.000 |
| Gender | 86981.037 | 1 | 86981.037 | 8.946 | 0.003 |
| rs1109859 in PEMT gene | 64090.372 | 2 | 32045.186 | 3.296 | 0.040 |
| rs4846052 in MTHFR gene | 19491.620 | 2 | 9745.810 | 1.002 | 0.369 |
| Hemoglobin | 13805.541 | 1 | 13805.541 | 1.420 | 0.235 |
| Hematocrit | 29920.071 | 1 | 29920.071 | 3.077 | 0.081 |
| LA intake | 21692.633 | 1 | 21692.633 | 2.231 | 0.137 |
| Error | 1545925.144 | 159 | 9722.800 | ||
| Total | 10938772.094 | 168 | |||
| Corrected Total | 1765939.407 | 167 | |||
| a. R Squared = 0.125 (Adjusted R Squared = 0.081) | |||||
Legend: DHA—docosahexaenoic acid, EPA—eicosapentaenoic acid, LA—linoleic acid, RBC—red blood cells, MTHFR—methylenetetrahydrofolate reductase, PEMT—phosphatidylethanolamine N -methyltransferase.
Author Contributions
Conceptualization, V.S., A.C.-E., and M.D.N.; Data curation, D.-A.T., P.T., and C.G.Z.; Formal analysis, D.-A.T. and P.T.; Investigation, D.-A.T., P.T., A.M., and C.L.Ș.; Methodology, V.S., A.C.-E., N.A., and C.L.Ș.; Resources, C.P., I.V., and A.M.; Supervision, M.P. and M.D.N.; Validation, V.S. and A.C.-E.; Visualization, V.S., A.C.-E., and N.A.; Writing—original draft, V.S., and A.C.-E.; Writing—review and editing, N.A., M.P., and M.D.N.
Funding
This research was funded by the POC Project “Use of nutrigenomic models for the personalized treatment with medical foods in obese people” (NutriGen) 2016–2019, SMIS: 104852, 91/09.09.2016, ID P_37-684.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tocher D.R., Betancor M.B., Sprague M., Olsen R.E., Napier J.A. Omega-3 Long-Chain Polyunsaturated Fatty Acids, EPA and DHA: Bridging the Gap between Supply and Demand. Nutrients. 2019;11:89. doi: 10.3390/nu11010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Da Costa K.A., Sanders L.M., Fischer L.M., Zeisel S.H. Docosahexaenoic Acid in Plasma Phosphatidylcholine May Be a Potential Marker for in Vivo Phosphatidylethanolamine A/-Methyltransferase Activity in Humans. Am. J. Clin. Nutr. 2011 doi: 10.3945/ajcn.110.011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ainsworth H., Seeds M.C., Mathias R.A. Diet-Gene Interactions and PUFA Metabolism: A Potential Contributor to Health Disparities and Human Diseases. Nutrients. 2014;6:1993–2022. doi: 10.3390/nu6051993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sergeant S., Rahbar E., Chilton F.H. Gamma-Linolenic Acid, Dihommo-Gamma Linolenic, Eicosanoids and Inflammatory Processes. Eur. J. Pharmacol. 2016 doi: 10.1016/j.ejphar.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanjung C., Rzehak P., Sudoyo H., Mansyur M., Munasir Z., Immanuel S., Irawan R., Reischl E., Demmelmair H., Hadinegoro S.R., et al. The Association of Fatty Acid Desaturase Gene Polymorphisms on Long-Chain Polyunsaturated Fatty Acid Composition in Indonesian Infants. Am. J. Clin. Nutr. 2018;108:1135–1144. doi: 10.1093/ajcn/nqy166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niculescu M. Choline and Phosphatidylcholine. In: Caballero B., editor. Encyclopedia of Human Nutrition. 3rd ed. Elsevier; Amsterdam, The Netherlands: 2013. pp. 346–351. [Google Scholar]
- 7.Khot V., Kale A., Joshi A., Chavan-gautam P., Joshi S. Expression of Genes Encoding Enzymes Involved in the One Carbon Cycle in Rat Placenta Is Determined by Maternal Micronutrients (Folic Acid, Vitamin B12) and Omega-3 Fatty Acids. BioMed Res. Int. 2014;2014:613078. doi: 10.1155/2014/613078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kume A., Kurotani K., Sato M., Ejima Y., Pham N.M., Nanri A., Kuwahara K. Polyunsaturated Fatty Acids in Serum and Homocysteine Concentrations in Japanese Men and Women: A Cross-Sectional Study. Nutr. Metab. 2013;10:1. doi: 10.1186/1743-7075-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenton J.I., Gurzell E.A., Davidson E.A., Harris W.S. Red Blood Cell PUFAs Reflect the Phospholipid PUFA Composition of Major Organs. Prostaglandins Leukot. Essent. Fat. Acids. 2016 doi: 10.1016/j.plefa.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs S., Schiller K., Jansen E., Fritsche A., Weikert C., Di Giuseppe R., Boeing H., Schulze M.B., Kröger J. Association between Erythrocyte Membrane Fatty Acids and Biomarkers of Dyslipidemia in the EPIC-Potsdam Study. Eur. J. Clin. Nutr. 2014 doi: 10.1038/ejcn.2014.18. [DOI] [PubMed] [Google Scholar]
- 11.De Onis M., Onyango A.W., Borghi E., Siyam A., Nishida C., Siekmann J. Development of a WHO Growth Reference for School-Aged Children and Adolescents. Bull. World Health Organ. 2007;85:660–667. doi: 10.2471/BLT.07.043497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Onis M., Wijnhoven T.M.A., Onyango A.W. Worldwide Practices in Child Growth Monitoring. J. Pediatr. 2004 doi: 10.1016/j.jpeds.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 13.Șerban C.L., Sima A., Hogea C.M., Chiriță-Emandi A., Perva I.T., Vlad A., Albai A., Nicolae G., Putnoky S., Timar R., et al. Assessment of Nutritional Intakes in Individuals with Obesity under Medical Supervision. A Cross-Sectional Study. Int. J. Environ. Res. Public Health. 2019;16:3036. doi: 10.3390/ijerph16173036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serafim V., Tiugan D.-A., Andreescu N., Mihailescu A., Paul C., Velea I., Puiu M., Niculescu M. Development and Validation of a LC–MS/MS-Based Assay for Quantification of Free and Total Omega 3 and 6 Fatty Acids from Human Plasma. Molecules. 2019;24:360. doi: 10.3390/molecules24020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbin K.D., Abdelmalek M.F., Spencer M.D., da Costa K.-A., Galanko J.A., Sha W., Suzuki A., Guy C.D., Cardona D.M., Torquati A., et al. Genetic Signatures in Choline and 1-Carbon Metabolism Are Associated with the Severity of Hepatic Steatosis. FASEB J. 2013;27:1674–1689. doi: 10.1096/fj.12-219097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K., Li M., Hakonarson H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010;38:1–7. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., Wu C., Li C., Boerwinkle E. DbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chirita Emandi A., Puiu M., Gafencu M., Pienar C. Growth References for School Aged Children in Western Romania. Acta Endocrinol. 2012;8:133–152. doi: 10.4183/aeb.2012.133. [DOI] [Google Scholar]
- 19.Chirita-Emandi A., Barbu C.G., Cinteza E.E., Chesaru B.I., Gafencu M., Mocanu V., Pascanu I.M., Tatar S.A., Balgradean M., Dobre M., et al. Overweight and Underweight Prevalence Trends in Children from Romania—Pooled Analysis of Cross-Sectional Studies between 2006 and 2015. Obes. Facts. 2016;9:206–220. doi: 10.1159/000444173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abarca-Gómez L., Abdeen Z.A., Hamid Z.A., Abu-Rmeileh N.M., Acosta-Cazares B., Acuin C., Adams R.J., Aekplakorn W., Afsana K., Aguilar-Salinas C.A., et al. Worldwide Trends in Body-Mass Index, Underweight, Overweight, and Obesity from 1975 to 2016: A Pooled Analysis of 2416 Population-Based Measurement Studies in 128·9 Million Children, Adolescents, and Adults. Lancet. 2017;390:2627–2642. doi: 10.1016/S0140-6736(17)32129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy W.G. The Sex Difference in Haemoglobin Levels in Adults—Mechanisms, Causes, and Consequences. Blood Rev. 2014;28:41–47. doi: 10.1016/j.blre.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Engelmann G., Hoffmann G.F., Grulich-Henn J., Teufel U. Alanine Aminotransferase Elevation in Obese Infants and Children: A Marker of Early Onset Non Alcoholic Fatty Liver Disease. Hepat. Mon. 2014;14 doi: 10.5812/hepatmon.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chirita Emandi A., Gafencu M., Pienar C. Impact of Increased Body Mass on Growth Patterns in School Children. Acta Endo. 2012;8:551–563. doi: 10.4183/aeb.2012.551. [DOI] [Google Scholar]
- 24.Svensson V., Jacobsson J.A., Fredriksson R., Danielsson P., Sobko T., Schiöth H.B., Marcus C. Associations between Severity of Obesity in Childhood and Adolescence, Obesity Onset and Parental BMI: A Longitudinal Cohort Study. Int. J. Obes. 2011;35:46–52. doi: 10.1038/ijo.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold L.E., Young A.S., Belury M.A., Cole R.M., Gracious B., Seidenfeld A.M., Wolfson H., Fristad M.A. Omega-3 Fatty Acid Plasma Levels Before and After Supplementation: Correlations with Mood and Clinical Outcomes in the Omega-3 and Therapy Studies. J. Child Adolesc. Psychopharmacol. 2017;27:223–233. doi: 10.1089/cap.2016.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann J., Skeaff M. Lipids. In: Mann J., Truswell A.S., editors. Essentials of Human Nutrition. Oxford University Press; New York, NY, USA: 2002. pp. 31–53. [Google Scholar]
- 27.Franco R.S. Critical Review the Measurement and Importance of Red Cell Survival. Am. J. Hematol. 2009;84:109–114. doi: 10.1002/ajh.21298. [DOI] [PubMed] [Google Scholar]
- 28.DeLong C.J., Shen Y.J., Thomas M.J., Cui Z. Molecular Distinction of Phosphatidylcholine Synthesis between the CDP-Choline Pathway and Phosphatidylethanolamine Methylation Pathway. J. Biol. Chem. 1999 doi: 10.1074/jbc.274.42.29683. [DOI] [PubMed] [Google Scholar]
- 29.Scambi C., Guarini P., De Franceschi L., Bambara L.M. Can 5-Methyltetrahydrofolate Modify the Phospholipid Fatty Acid Pattern in Cystic Fibrosis Pediatric Patients? J. Cyst. Fibros. 2006 doi: 10.1016/j.jcf.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Zhou M.M., Xue Y., Sun S.H., Wen M., Li Z.J., Xu J., Wang J.F., Yanagita T., Wang Y.M., Xue C.H. Effects of Different Fatty Acids Composition of Phosphatidylcholine on Brain Function of Dementia Mice Induced by Scopolamine. Lipids Health Dis. 2016 doi: 10.1186/s12944-016-0305-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X., Xue Y., Liu C., Lou Q., Wang J., Yanagita T., Xue C., Wang Y. Eicosapentaenoic Acid-Enriched Phospholipid Ameliorates Insulin Resistance and Lipid Metabolism in Diet-Induced-Obese Mice. Lipids Health Dis. 2013 doi: 10.1186/1476-511X-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livingstone M.B.E., Robson P.J., Wallace J.M.W. Issues in Dietary Intake Assessment of Children and Adolescents. Br. J. Nutr. 2004;92:S213–S222. doi: 10.1079/BJN20041169. [DOI] [PubMed] [Google Scholar]
- 33.Basch C.E., Shea S., Arliss R., Contento I.R., Rips J., Gutin B., Irigoyen M., Zybert P. Validation of Mothers’ Reports of Dietary Intake by Four to Seven Year-Old Children. Am. J. Public Health. 1990;80:1314–1317. doi: 10.2105/AJPH.80.11.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burrows T.L., Martin R.J., Collins C.E. A Systematic Review of the Validity of Dietary Assessment Methods in Children When Compared with the Method of Doubly Labeled Water. J. Am. Diet. Assoc. 2010;110:1501–1510. doi: 10.1016/j.jada.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 35.Kirkpatrick S.I., Gilsing A.M., Hobin E., Solbak N.M., Wallace A., Haines J., Mayhew A.J., Orr S.K., Raina P., Robson P.J., et al. Lessons from Studies to Evaluate an Online 24-Hour Recall for Use with Children and Adults in Canada. Nutrients. 2017;9:100. doi: 10.3390/nu9020100. [DOI] [PMC free article] [PubMed] [Google Scholar]