Abstract
Heart failure continues to be a common and deadly sequela of myocardial infarction (MI). Despite strong evidence suggesting the importance of myocardial mechanics in cardiac remodelling, many MI studies still rely on two-dimensional analyses to estimate global left ventricular (LV) function. Here, we integrated four-dimensional ultrasound with three-dimensional strain mapping to longitudinally characterize LV mechanics within and around infarcts in order to study the post-MI remodelling process. To induce infarcts with varying severities, we separated 15 mice into three equal-sized groups: (i) sham, (ii) 30 min ischaemia–reperfusion, and (iii) permanent ligation of the left coronary artery. Four-dimensional ultrasound from a high-frequency small animal system was used to monitor changes in LV geometry, function and strain over 28 days. We reconstructed three-dimensional myocardial strain maps and showed that strain profiles at the infarct border followed a sigmoidal behaviour. We also identified that mice with mild remodelling had significantly higher strains in the infarcted myocardium than those with severe injury. Finally, we developed a new approach to non-invasively estimate infarct size from strain maps, which correlated well with histological results. Taken together, the presented work provides a thorough approach to quantify regional strain, an important component when assessing post-MI remodelling.
Keywords: infarction, ischaemia–reperfusion, left ventricular remodelling, murine, myocardial strain, ultrasound
1. Introduction
Coronary artery disease remains the leading cause of death in the USA, with over 1 million acute coronary events predicted to take place in 2019 [1]. Despite recent advances in percutaneous coronary intervention technologies, which have improved patient survival rates, heart failure continues to be a common long-term complication of acute myocardial infarction (MI) with high morbidity and mortality [2]. Cardiac remodelling post-MI encompasses a series of complex molecular, structural and functional changes in the left ventricle, driven by inflammatory, neurohormonal and mechanical factors [3,4]. Although the short-term effects of remodelling are vital in repairing the damaged myocardium, sustained imbalance between increased haemodynamic load, compromised myocardial mechanics and impaired cardiac function feeds a pathological response that results in left ventricular (LV) dilation and eventual heart failure [3,4]. Specifically, changes in the mechanical microenvironment regulate myofibroblast proliferation and subsequent collagenous scar formation at the infarct border zone, providing the heart with the structural rigidity necessary to minimize infarct expansion and prevent ventricular rupture [4,5]. The developing myocardial scar, although beneficial early in remodelling, reduces LV compliance over time, directly inhibiting LV pumping function [6]. Taken together, the time course, mechanical properties and size of the myocardial scar tissue are all critical components that determine the fate of the remodelling left ventricle.
Despite strong evidence supporting the importance of myocardial mechanics in remodelling post-MI [5–7], longitudinal assessment of regional LV mechanics proves to be challenging. The majority of in vivo infarction studies still rely on two-dimensional (2D) image analyses to estimate the global metrics of LV function such as ejection fraction and global longitudinal strain [8–10]. These metrics, while valuable in evaluating the overall impact of ischaemic injury on cardiac health, do not capture regional differences in myocardial contractility. Furthermore, strain measurements derived from 2D images are sensitive to through-plane motion caused by LV twisting during contraction [11]. Irrespective of these limitations, 2D maps highlighting regional strain differences still provide important spatial and temporal information regarding changes in LV contractility throughout remodelling [12,13].
Recent developments in non-invasive four-dimensional (4D) imaging techniques have made it possible for researchers to reconstruct volumetric maps of patient- or mouse-specific LV geometries throughout a cardiac cycle [14–16], opening the possibility for three-dimensional (3D) strain mapping of the heart. Indeed, several groups have quantified regional differences in 3D strain in both healthy [17,18] and ischaemic left ventricles [19–21], with results revealing significant strain reductions within infarcted tissue. However, these studies either evaluated strain at only sparse time points [19,20] or relied on contrast agents to quantify strain in the remodelling infarct [21]. The reported strain difference between the infarcted and remote myocardium suggests the presence of a strain gradient near infarct border zones that may play an important role in infarct expansion.
A thorough longitudinal study investigating changes in the spatial distribution of 3D myocardial strain in a murine model of acute MI has not yet been conducted. Here, we integrated high-resolution 4D ultrasound imaging [14] with 3D strain mapping [19] to monitor cardiac remodelling over 28 days. By employing two surgical mouse models to induce ischaemic damage with varying severities, we identified unique remodelling patterns that differed between ischaemia–reperfusion and permanent ligation models. By expanding ultrasound strain studies to three dimensions, we aim to provide further evidence that the mechanical behaviour of the left ventricle near infarct border zones contributes to infarct expansion and ventricular remodelling.
2. Material and methods
2.1. Coronary artery ligation
We randomly assigned 15 male, wild-type, C57BL/6J mice (age = 14 ± 1 weeks; weight = 27 ± 3 g; The Jackson Laboratory, Bar Harbor, ME) into three surgical groups: (i) sham (n = 5), (ii) ischaemia–reperfusion (I/R; n = 5), and (iii) permanent ligation (PL; n = 5). For surgery, each mouse was anaesthetized with 1–3% isoflurane and endotracheally intubated using a small-animal ventilator (SomnoSuite, Kent Scientific, Torrington, CT). Pressure-controlled ventilation supplied air to the lungs with a target inspiratory pressure between 16 and 18 cm H2O and a peak end-expiratory pressure between 3 and 5 cm H2O. We secured the mouse to a heated surgical stage and coupled a rectal temperature probe to a homeothermic control module to maintain body temperatures between 36°C and 37°C (RightTemp, Kent Scientific, Torrington, CT). We made a small incision in the third intercostal space of the left thorax and retracted the ribs to expose the left ventricle. The pericardium was dissected to visualize the left coronary artery (LCA). In the sham-operated controls, an 8–0 nylon suture was looped around the LCA without ligating the vessel. In the I/R group, we used PE-10 tubing in combination with a suture to temporarily ligate the LCA for 30 min before restoring blood flow to the ischaemic myocardium (reperfusion) as described previously [22]. In the PL group, the LCA was permanently ligated to induce an infarct [22]. At the end of the procedure, we sutured the incision site and recovered the mouse. All surgical procedures were performed aseptically, and buprenorphine (0.05 mg kg−1; intraperitoneal) was administered as an analgesic. All procedures were approved by the Purdue Animal Care and Use Committee.
2.2. Longitudinal ultrasound imaging
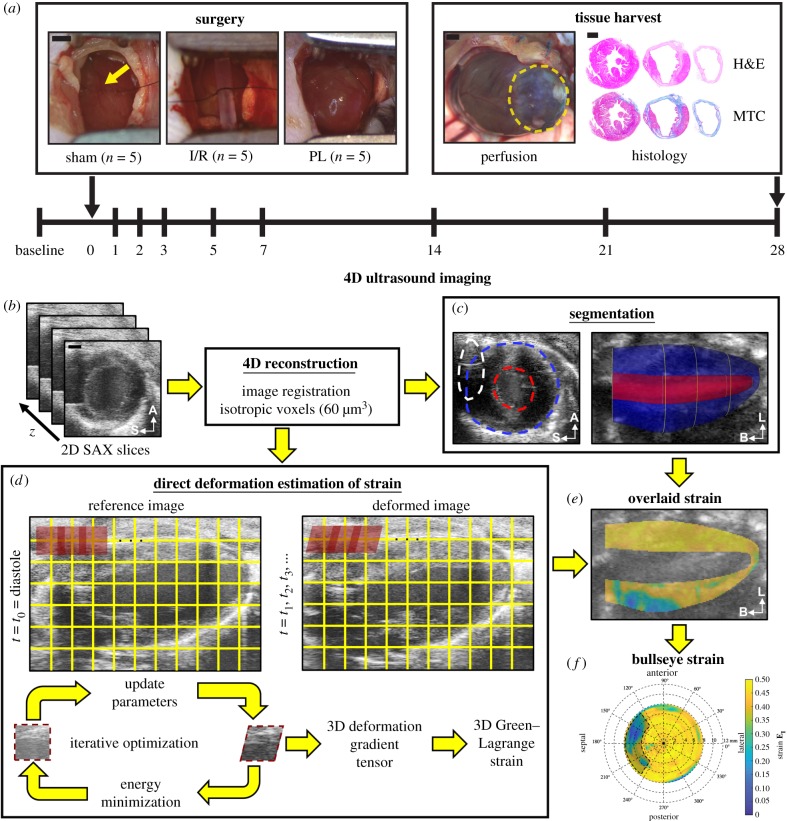
All ultrasound images were collected with a Vevo2100 small-animal ultrasound system (FUJIFILM VisualSonics Inc., Toronto, Canada) and a 40 MHz centre frequency linear array transducer (22–55 MHz; MS550D). Ultrasound images of the left ventricle were acquired at baseline and on days 1, 2, 3, 5, 7, 14, 21 and 28 post-surgery. Figure 1a summarizes the study design for the presented work. We acquired 4D ultrasound data as described previously [14]. Briefly, successive cardiac and respiratory-gated 2D cine loops were obtained at 1000 Hz in the short axis from the apex to the base of the heart by using a linearly translating 3D motor (step size = 0.2 mm; figure 1b). Respiratory waveforms obtained during imaging were used to ensure that ultrasound images were only acquired in between breaths to minimize breathing motion artefacts. Sequential 2D images were then spatially registered, temporally matched based on their relative time in the cardiac cycle and resampled to isotropic 60 µm voxels in Matlab (MathWorks Inc., Natick, MA). Additionally, we measured mitral valve inflow velocities from the four-chamber view of the heart with pulsed-wave Doppler.
Figure 1.
Study design. (a) Fifteen adult male mice were randomly assigned into three surgical groups: (i) sham-operated controls, (ii) 30 min ischaemia–reperfusion (I/R), and (iii) permanent ligation (PL) of the left coronary artery (yellow arrow). A Vevo2100 ultrasound system was used to acquire 4D ultrasound data and flow information of the left ventricle at baseline and on days 1, 2, 3, 5, 7, 14, 21 and 28 post-surgery. At the end of the study, the heart was stained with haematoxylin–eosin (H&E) and Masson's trichrome (MTC). Yellow dashed outlines highlight the infarcted myocardium. (b) We reconstructed 4D ultrasound data from ECG and respiratory-gated 2D short-axis ultrasound images of the left ventricle. (c) Three-dimensional endocardial (red), epicardial (blue) and sternal artefact (white) boundaries were segmented at end-diastole and peak-systole. (d) Maximum principal 3D Green–Lagrange strain (EI) was calculated using a direct deformation estimation technique. (e) Strain was then localized within the myocardium using segmented boundaries and presented as bullseye maps (f). A, anterior; S, septal; L, lateral; B, base. Scale bar: 1 mm. (Online version in colour.)
2.3. Ultrasound image analysis
2.3.1. Segmentation of left ventricular boundaries
Reconstructed 4D ultrasound data were matched spatially with a custom Matlab script by using anatomical landmarks such as the sternum, apex and heart valves. The reoriented 4D data were then loaded into SimVascular for segmentation [23]. First, we created a centreline path from the aortic valve to the apex of the left ventricle and manually segmented the endocardial and epicardial boundaries. Two-dimensional segmentations were created at least every 1 mm apart, with smaller spacing for regions showing significant changes in geometry. This process was performed at both end-diastole and peak-systole. We also segmented sternal shadowing artefacts to identify regions where strain could not be reliably calculated. Finally, 3D surface models of the endocardial, epicardial and sternal artefact boundaries at both end-diastole and peak-systole were rendered with uniform meshing (figure 1c) and exported as STL files for further Matlab analysis.
2.3.2. Assessment of global cardiac function
The 3D surface models were converted to solid, volumetric meshes and spatially registered to the 4D ultrasound data. We calculated end-diastolic volume (EDV) and peak-systolic volume (PSV) by multiplying the number of voxels in the endocardial solid mesh at the corresponding time point by the isotropic voxel dimensions of the 4D data. These volumes were then used to evaluate global metrics of LV systolic function including stroke volume (SV), ejection fraction (EF) and cardiac output (CO), which were calculated as follows (equations (2.1)–(2.3)):
| 2.1 |
| 2.2 |
| 2.3 |
LV diastolic function was also assessed using transmitral flow velocity waveforms obtained from pulsed-wave Doppler. E- and A-wave peak velocities from five different cardiac cycles were measured, and their corresponding averages were used to calculate the E/A ratio. We used the E/A ratio to identify whether blood flow into the left ventricle was primarily driven by passive filling (pressure gradient caused by LV relaxation; E-wave) or active filling (atrial contraction; A-wave).
2.3.3. Estimation of three-dimensional maximum principal Green–Lagrange strain
We implemented a direct deformation estimation (DDE) algorithm in Matlab to estimate the 3D deformation gradient tensor as described previously (figure 1d; [19]). Briefly, we defined a rectangular coordinate grid spaced 5 pixels apart on the 4D ultrasound data. At each grid intersection, an 11 × 11 × 11 investigation region was assigned. Using the image at end-diastole as a reference template, we iteratively optimized a warping function that best mapped the affine transformation of this region from the template image to a deformed image at the next time point. The warping function was optimized by best matching voxel intensities between the template and deformed image. The warping function was designed to be analogous to the 3D deformation gradient tensor, F3D, such that we could directly estimate F3D during voxel intensity mapping. This process was repeated until F3D was determined at each grid intersection (i,j,k) across all time points in the cardiac cycle. We then calculated the 3D Green–Lagrange (GL) strain tensor, E3D, as shown in equation (2.4), where I is the second-order identity tensor,
| 2.4 |
Finally, the maximum principal component of the 3D GL strain tensor was calculated and superimposed onto the 4D ultrasound data. Three-dimensional interpolation was performed to approximate strain values in regions between the coordinate grid points.
2.3.4. In vivo strain comparison with Vic2D
In one animal, we compared strain values calculated from the 3D-DDE technique with those measured from Vic2D, a commercially available digital image correlation software that has been previously used to quantify tissue strains from clinical images [24,25]. Briefly, representative short-axis slices of the left ventricle were obtained from the isotropic 4D data at baseline and on days 1, 7 and 28 post-PL of the LCA. Myocardial strains were evaluated with Vic2D by manually selecting a region of interest around the LV wall (ring geometry; 21 × 21 investigation window, step = 1 pixel). The end-diastolic image was used as the reference configuration. The top 10% of the maximum principal strains in the cardiac cycle were then averaged and compared between the Vic2D and 3D-DDE methods along the anterior and posterior walls of the left ventricle.
2.3.5. Bullseye mapping of myocardial strain
We created volumetric meshes of the myocardial wall by subtracting the rendered endocardial volumes from the epicardial volumes in order to visualize strain within the myocardium (figure 1e). The mid-surface of the myocardium was obtained by calculating the midpoints between paired endocardial and epicardial boundary points located in a plane normal to the centreline of the left ventricle. We defined paired endocardial and epicardial boundary points as points aligned radially from the centreline of the left ventricle. Myocardial strain values between paired endocardial and epicardial boundary points were averaged together to create a representative mean strain metric. The mid-surface of the myocardium was then unwrapped to polar coordinates, relative to the apex of the left ventricle, to represent the averaged strain values as a bullseye map in accordance with the American Heart Association's 17-segment model (figure 1f, [26]).
2.3.6. Non-invasive estimation of infarct size
Two approaches to non-invasively estimate infarct size were performed. First, we used myocardial wall thickness at peak-systole as a criterion for determining infarct size. Wall thickness was calculated by measuring the distance between paired endocardial and epicardial boundary points along the left ventricle. Regions with thickness values smaller than 0.5 mm were defined to be infarcted as used by others [27]. We then quantified infarct size as the percentage of the myocardium with systolic thickness values below 0.5 mm. Infarct size was reported as a per cent of LV size to take into account ventricular dilation. This infarct sizing method was only applicable for mice with transmural infarcts (PL) and not for subepicardial infarcts (I/R).
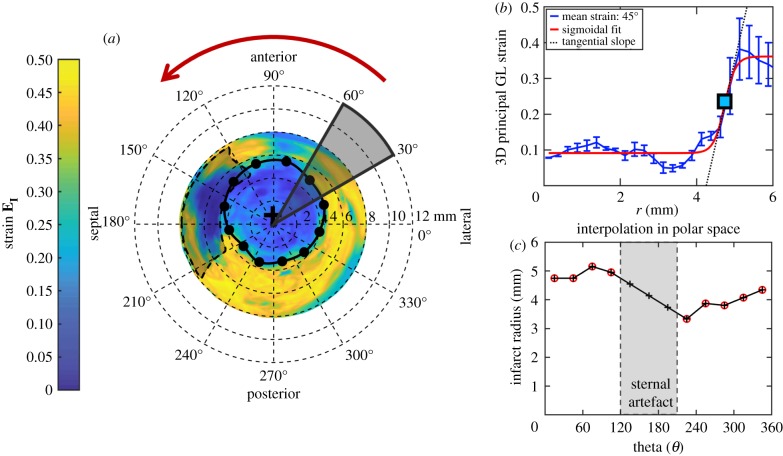
In the second approach, strain profiles were used to estimate infarct size. We first plotted bullseye maps of principal 3D GL strain throughout the cardiac cycle. The maximum strain values at each spatial position were then extracted across all time points to construct a representative bullseye map. This step was implemented to account for dyssynchrony in LV contractile patterns in mice with ischaemic injury. An initial estimate for infarct centre was obtained from the centre of the wall-thinned myocardium in the PL group. In the I/R group, which did not exhibit significant wall thinning, we manually identified the centre of low-strain regions to identify the infarct centre. Strain profiles were then plotted radially from the infarct centre, and a sigmoidal fit was implemented across every 30° region (figure 2a,b). The location of the inflection point was determined to be the boundary of the infarct zone, and the spatial strain gradient at the infarct boundary was calculated from the slope of the linear portion of the sigmoidal curve fit. In regions with significant sternal artefacts where the inflection points could not be identified, infarct boundaries were approximated by interpolating adjacent infarct boundaries in polar coordinates (figure 2c). Infarct size was then reported as an area percentage of the myocardium that fell within the strain-estimated infarct boundary.
Figure 2.
Strain estimation of infarct size. (a) We extracted myocardial strain profiles radially from the infarct centre (black crosshair) and performed sigmoidal fitting on strain profiles averaged across every 30° region. (b) The spatial positions of the inflection points (square box and black dots) are defined as the infarct border and unwrapped from the infarct centre to estimate infarct boundaries in regions with sternal artefacts (c). Infarct size is defined as the percentage of the myocardium that lies within the strain-estimated infarct boundary. (Online version in colour.)
2.4. Histological analysis
2.4.1. Tissue preparation for staining
At the end of the study, we euthanized the mice and perfused 30 mM KCl solution to arrest the heart in diastole. Harvested hearts were then sliced into three or four uniform segments in the short axis and fixed in 4% paraformaldehyde for 7 days at 4°C before being sent for histology. Briefly, cardiac segments were embedded in paraffin, thinly sectioned (5 µm) and stained with haematoxylin–eosin (H&E) and Masson's trichrome (MTC). MTC stain was used to differentiate muscle fibres (red) from collagen-rich scars (blue). We imaged stained tissues in segments at 10× magnification with a Leica ICC50 W stereomicroscope (Leica Microsystems Inc., Buffalo Grove, IL) and quantified collagen content and infarct size using ImageJ [28].
2.4.2. Collagen quantification and infarct sizing
We stitched adjacent cardiac images from a representative slice using MosaicJ [29] and removed image background from the rendered image. MTC images were then separated into their red–green–blue channels to isolate red pixels corresponding to muscle fibres from blue pixels representative of collagen-rich scars. Per cent collagen was then calculated as follows:
| 2.5 |
We calculated infarct size from MTC images using a midline length approach [30]. The LV myocardial midline was traced in ImageJ by identifying the midpoints between the endocardial and epicardial boundaries. The midline circumference corresponds to the total midline length. The midline infarct length was measured as the midline arc length in regions where the collagen scar encompassed more than 20% of the myocardial thickness. This 20% threshold was used to represent infarct size in left ventricles with subepicardial infarcts in the I/R group. Infarct size (IS) was then calculated by dividing the sum of the midline infarct lengths, linfarct, by the sum of the total midline length, ltotal, from all cross-sectional slices of the left ventricle, n,
| 2.6 |
2.5. Statistical analysis
All data were reported as mean ± standard deviation and tested for normality using the Shapiro–Wilk test. We implemented a log transformation on non-normal and heteroscedastic data before running statistical tests. A repeated measures analysis of variance (ANOVA) with the post hoc Tukey's test was performed to study the effects of surgery on cardiac function at the different time points. Similarly, we ran a two-way ANOVA with the post hoc Tukey's test to study the effects of spatial position and surgery on both maximum principal 3D GL strain and collagen content. Lastly, linear regression analyses comparing the different methods of infarct sizing were conducted. All statistical analyses were performed using GraphPad Prism v. 8.1.1 (GraphPad Software, San Diego, CA) with p < 0.05 representing statistical significance.
3. Results
3.1. Longitudinal assessment of cardiac function
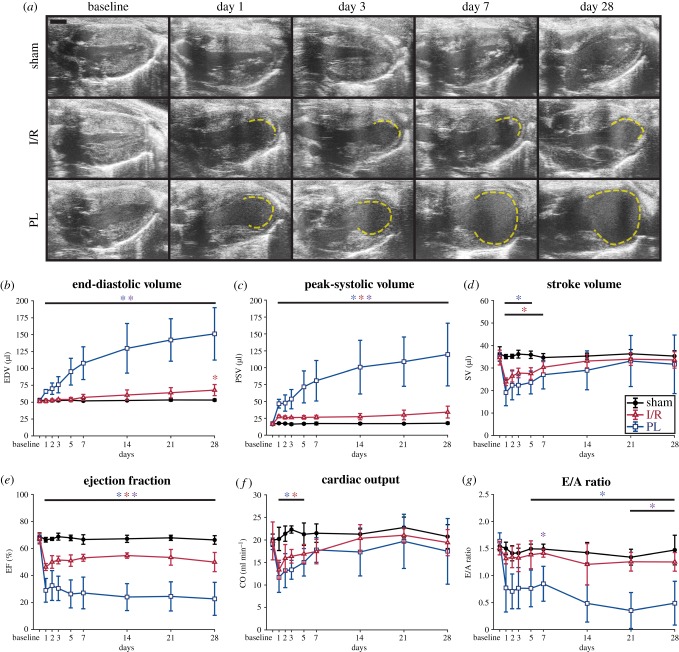
Long-axis ultrasound images of the left ventricles from three representative mice are presented at peak-systole in figure 3a, highlighting geometrical differences between surgical groups. A video of LV motion throughout the cardiac cycle is provided in the electronic supplementary material, figure S1. Figure 3a shows that LV geometry was preserved in the sham group, while myocardial damage was evident in the I/R and PL groups as early as day 1. Akinetic regions, marked in dashed yellow lines, indicated that ischaemic injury was primarily localized to the apex of these left ventricles. A closer inspection revealed that mice in the PL group experienced significant wall thinning and chamber dilation by day 7 post-surgery. We did not observe this trend in mice in the I/R group.
Figure 3.
LV remodelling post-infarction. (a) Representative long-axis ultrasound images of mouse left ventricles taken at peak-systole. Akinetic myocardial walls, indicative of ischaemic damage, are outlined in dashed yellow lines. Global metrics of LV function (b–g) showed that mice in both I/R and PL groups exhibited significant reductions in LV contractile function post-surgery, but significant dilation and diastolic dysfunction were only consistently measured in the PL group. Taken together, mice in the PL group experienced greater degrees of cardiac remodelling than those in the I/R group. Data are shown as mean ± standard deviation (*p < 0.05). I/R, ischaemia–reperfusion; PL, permanent ligation. Blue asterisks, PL versus sham; red asterisks, I/R versus sham; purple asterisks, PL versus I/R. Scale bar: 1 mm. (Online version in colour.)
These findings reflected longitudinal changes in global cardiac function (figure 3b–g). Although all cardiac parameters remained unaffected in the sham group over the course of 28 days, we identified detrimental changes in LV function in the I/R and PL groups. The left ventricles of mice in the PL group progressively dilated post-surgery until they reached EDVs close to triple those of the sham group at day 28 (EDVSham = 53 ± 2 µl versus EDVPL = 151 ± 39 µl, p < 0.01). Interestingly, minimal dilation was observed in the I/R group when compared with the sham group (EDVI/R = 68 ± 8 µl, p = 0.02). Reductions in LV contractile function due to ischaemic injury were detected from day 1 as increases in PSVs that either remained stable in the I/R group or increased proportionally to LV dilation in the PL group (PSVSham = 18 ± 2 µl versus PSVI/R = 34 ± 9 µl, p < 0.01; versus PSVPL = 119 ± 46 µl, p < 0.01). These resulted in immediate and significant decreases in EFs that remained depressed throughout the study (EFSham = 66 ± 3% versus EFI/R = 50 ± 7%, p < 0.01; versus EFPL = 23 ± 12%, p < 0.01). Surprisingly, we noticed transient reductions in SV and CO 7 days post-surgery before both returned to baseline values. In addition to compromised systolic function, we also observed significant diastolic dysfunction in the PL group that was not seen in the I/R group (E/ASham = 1.5 ± 0.3 versus E/AI/R = 1.3 ± 0.2, p = 0.59; versus E/APL = 0.5 ± 0.4, p = 0.04). Taken together, these results revealed that mice exposed to I/R injuries exhibited smaller degrees of LV remodelling than those subjected to permanent LCA ligation.
3.2. Spatial distribution of three-dimensional myocardial strain
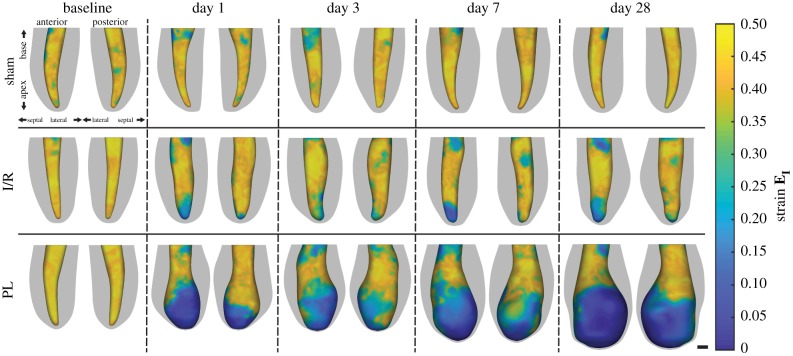
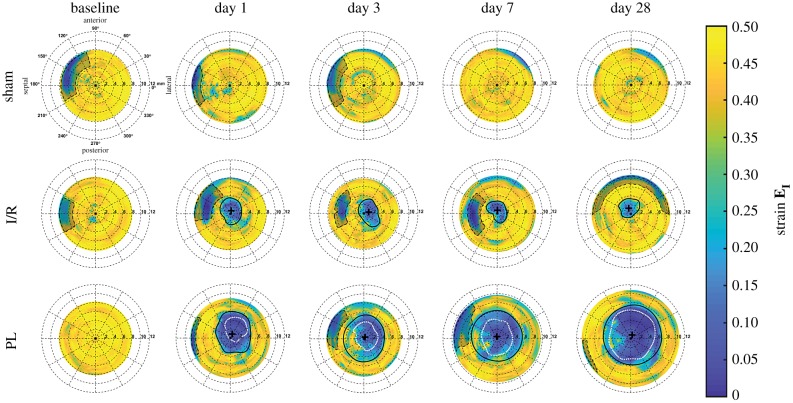
Longitudinal changes in peak-systolic LV geometries and endocardial wall strains from three representative mice are shown in figure 4. Maximum principal 3D GL strain (EI) of the endocardial wall was visualized along the anterior and posterior walls of the left ventricle to highlight regional differences in strain, with yellow and blue regions corresponding to areas of high and low strains, respectively. Bullseye plots mapping the peak myocardial strain profiles of the unwrapped LV surface are shown in figure 5, with video representations of day 28 left ventricles portrayed throughout the cardiac cycle in electronic supplementary material, figure S2. Sternal shadowing artefacts, commonly found near the base of the LV and marked as dashed black lines, artificially lowered strain values in these regions and were excluded from our analysis. Taken together, figures 4 and 5 illustrate that regions of low strain near the LV apex remained localized in the I/R group but continued to expand proportionally to chamber dilation in the PL group. Furthermore, LV wall thinning was only observed in the PL group, and wall-thinned boundaries continued to expand throughout remodelling, approaching infarct boundaries estimated from the strain profile inflection points.
Figure 4.
Longitudinal 3D representations of peak-systolic LV boundaries with maximum principal 3D Green–Lagrange myocardial strains (EI) overlaid onto the endocardial wall. Epicardial boundaries are shown in grey. Dark blue areas highlight akinetic regions of the myocardium. Scale bar: 1 mm. (Online version in colour.)
Figure 5.
Longitudinal bullseye maps of the maximum principal 3D Green–Lagrange strain (EI) within the myocardium. Strain-estimated infarct boundaries are outlined in solid black lines with infarct centres marked as black crosshairs. Wall-thinned infarct boundaries measured from segmentation are outlined as white dotted lines, while sternal artefacts are outlined as black dashed lines. (Online version in colour.)
Representative day 28 long-axis ultrasound images, 3D surface strains and bullseye strain maps of the remodelled left ventricles for all mice in the I/R and PL groups are included as electronic supplementary material, figures S3 and S4. Day 28 strains along the entire thickness of the LV wall are also provided for one representative mouse in each surgical group in electronic supplementary material, figure S5. These supplemental figures highlight substantial heterogeneity in LV remodelling across mice both within and between groups. However, clear patterns are present, and we noticed that most mice exhibited asymmetrical infarcts skewed towards the anterior wall. Lastly, a comparison of maximum principal strain values between the 3D-DDE and Vic2D methods is summarized in electronic supplementary material, figure S6 for a representative left ventricle with an asymmetrical anterior infarct. Although both techniques successfully captured strain reductions along the infarcted anterior wall when compared with the contractile posterior wall, the 3D-DDE algorithm more appropriately tracked changes in LV boundaries throughout a representative heartbeat, as strain values returned to 0 at the end of the cardiac cycle.
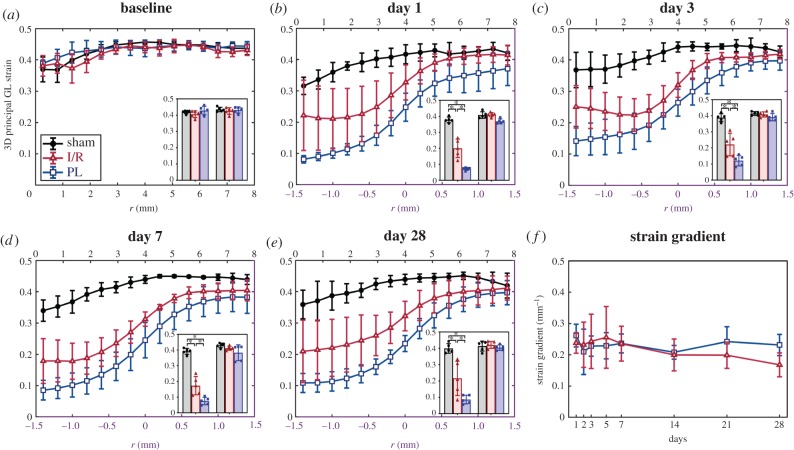
Interestingly, we identified significant differences in maximum principal 3D GL strain values within the infarcted myocardium between mice in the I/R and PL groups. Strain profiles averaged across all five mice in each surgical group are shown in figure 6a–e. Throughout the 28 days following surgery, we consistently observed a sigmoidal strain profile at the interface between infarcted and remote myocardium. We also detected significantly higher strain values within the infarcted myocardium of mice in the I/R group than in those in the PL group (day 28: EInfarct,I/R = 0.22 ± 0.10 versus EInfarct,PL = 0.09 ± 0.03, p = 0.01), while sham-operated mice maintained healthy strain values in the LV apex (EApex,Sham = 0.40 ± 0.03). Conversely, no differences in strain were seen in the remote myocardium between the three groups (EBase,Sham = 0.41 ± 0.03 versus ERemote,I/R = 0.42 ± 0.02, p = 0.97; versus ERemote,PL = 0.40 ± 0.02, p = 0.95). We observed no differences in the spatial strain gradient at the infarct boundary between the I/R and PL groups across all time points (figure 6f).
Figure 6.
Myocardial strain profiles of the remodelling left ventricles averaged across mice in each group and plotted at (a) baseline and (b–e) on days 1, 3, 7 and 28 post-surgery. Strain profiles in non-ischaemic mice (baseline and sham) are plotted radially away from the LV apex (black x-axis). In ischaemic mice, strain profiles are plotted only in regions near infarct boundaries (r = 0; purple x-axis). Strain values averaged within the infarcted and remote myocardium are shown as the left and right inset bar graphs, respectively. (f) Strain gradients, calculated within the linear component of the sigmoidal fit from the strain profiles, did not show significant changes between surgical groups over the 28 day period. Data are shown as mean ± standard deviation (*p < 0.05). (Online version in colour.)
3.3. Histological analysis of collagen content and infarct size
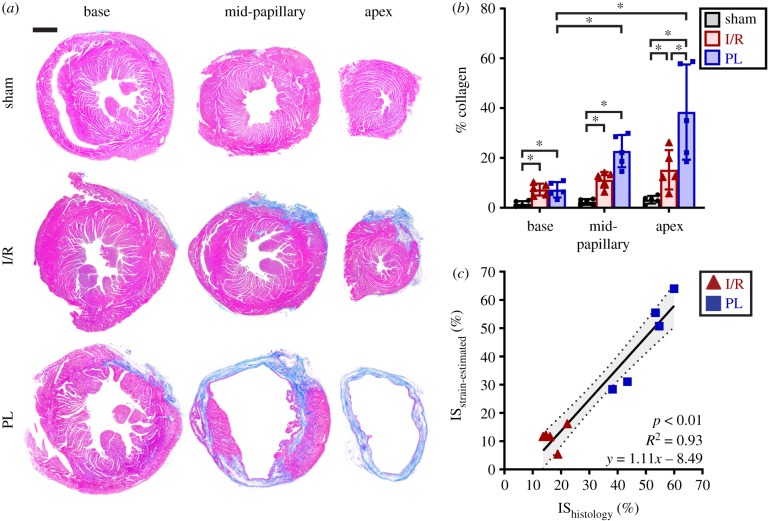
Representative histology images of mouse left ventricles stained with MTC revealed varying distributions of collagen-rich, fibrotic (blue) tissues between surgical groups (figure 7a,b). The absence of fibrosis within the myocardium of sham-operated mice demonstrated that sham surgeries contributed little to no myocardial damage. Significant scarring, however, was observed in both the I/R and PL groups (per cent collagen: shamApex = 3 ± 2% versus I/RApex = 15 ± 8%, p < 0.01; versus PLApex = 38 ± 17%, p < 0.01). While mice in the PL group developed transmural infarcts, as shown by the presence of collagen spanning the entire thickness of the myocardium, mice in the I/R group interestingly only developed subepicardial scarring. We also noticed an increase in interstitial collagen percentage towards the infarcted apex (per cent collagen: basePL = 7 ± 3% versus mid-papillaryPL = 23 ± 6%, p < 0.01; versus apexPL = 38 ± 17%, p < 0.01). Finally, we compared infarct sizes estimated from 3D strain maps with those calculated from histology and discovered a strong correlation between the two approaches (figure 7c; R2 = 0.93, p < 0.01).
Figure 7.
Histological analysis of collagen content and infarct size. (a) Histology images of mouse left ventricles, obtained 28 days post-surgery and stained with MTC, revealed subepicardial scarring in the I/R group and transmural infarcts in the PL group. Muscle fibres are stained red while collagen-rich scars are stained blue. (b) Bar graphs showing per cent collagen highlighted spatial variations in collagen content relative to infarct location. (c) Strain-estimated infarct size strongly correlated to infarct size measured from the histological midline length approach. Data are shown as mean ± standard deviation (*p < 0.05). Scale bar: 1 mm. (Online version in colour.)
3.4. Correlation of infarct size
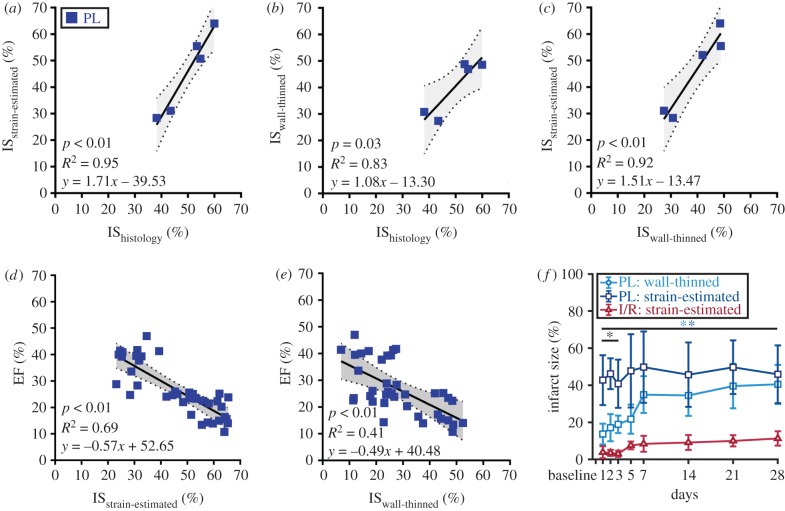
Correlation plots comparing infarct sizes estimated from three different approaches are summarized in figure 8a–c. Although we found a strong positive correlation between all methods (R2 > 0.80, p < 0.05), infarct sizes evaluated from histological staining at day 28 were better correlated with strain-estimated infarct sizes (R2 = 0.95) than those measured from wall thinning (R2 = 0.83). Similarly, a stronger negative correlation between strain-estimated infarct size and EF (R2 = 0.69; figure 8d) was observed, while infarct size approximated from wall-thinned regions exhibited only moderate correlation with EF (R2 = 0.41; figure 8e). Interestingly, we identified significant differences in measured infarct size between the strain-estimated and wall-thinned approaches between days 1 and 3 post-surgery (p < 0.05) which appeared to converge throughout cardiac remodelling (figure 8f).
Figure 8.
Correlation of varying infarct sizing techniques with cardiac function. (a–c) Correlation plots comparing three different infarct sizing methodologies showed that the proposed strain-estimation technique best correlated with gold standard histological estimation of infarct size. Correlation of (d) strain-estimated and (e) wall-thinned infarct size with EF. Linear regression lines are shown as solid black lines with 95% confidence intervals shaded in grey. (f) Line graphs summarizing infarct size growth throughout cardiac remodelling. Data are shown as mean ± standard deviation (*p < 0.05). Light blue asterisks, wall-thinned PL versus strain-estimated I/R; dark blue asterisks, strain-estimated PL versus strain-estimated I/R; black asterisks, wall-thinned PL versus strain-estimated PL. (Online version in colour.)
4. Discussion
We have demonstrated in two mouse models of MI that DDE, in conjunction with 4D ultrasound, provides regional in vivo estimates of 3D myocardial strain. Unlike 2D techniques, regional strain mapping not only helps explain the observed decrease in global LV function post-injury, but also reveals the importance of strain profiles in driving infarct expansion. Specifically, mice exhibiting higher strain values within infarcted tissue experienced smaller degrees of LV remodelling. Furthermore, our initial myocardial 3D maximum principal strain maps predicted final infarct size four weeks after ischaemic injury. Taken together, these strain data help characterize the role that mechanical strain plays in LV remodelling post-infarction.
4.1. Advantages of direct three-dimensional strain estimation
A significant advantage of the 3D-DDE technique is its ability to capture regional strain differences along the entire thickness of the myocardium (electronic supplementary material, figure S5). Most preclinical [8,10,13] and clinical [11,17] ultrasound studies to date rely on commercially available ultrasound software packages to estimate 2D myocardial strain (ε) as the change in length of a segment, ΔL, divided by its original length, L0,
| 4.1 |
The observed change in length, in either the circumferential, longitudinal or radial direction, is measured by tracking the endocardial and epicardial LV boundaries throughout the entire cardiac cycle using speckle-tracking algorithms. Since a simple change in length is used to approximate strain within large subregions, this approach cannot resolve regional strain differences within the myocardium. This linear approximation also does not consider the nonlinear components of strain and is only appropriate when estimating small deformations less than 5% [31]. This is not the case in many soft tissues, such as the heart, where large deformations are observed in vivo. Finally, as the heart undergoes twisting during systolic contraction, 2D techniques are negatively impacted by through-plane motion [11]. By directly estimating the 3D deformation gradient tensor from small image subregions within 4D ultrasound data, these issues can be mitigated.
Another advantage of the presented technique lies in its ability to yield reproducible measures of 3D strain. Unlike existing techniques, which often rely on displacement regularization prior to strain estimation, the DDE method estimates the 3D deformation gradient tensor directly during voxel intensity mapping as reported previously [19]. This results in a noise-insensitive algorithm that provides a more accurate and precise strain-field estimation when compared with displacement-based methods, as supported by in silico validation [19]. We demonstrate the reproducibility of our 3D strain measurements in electronic supplementary material, figure S7, which highlights similarities in the bullseye strain maps of all 15 healthy mice imaged at baseline. In all cases, we found high strain values ranging between 0.40 and 0.45 throughout the LV myocardium, except in regions with prominent sternal artefacts. This suggests that, across animals, we are consistently obtaining reproducible values of strain. Additionally, the fact that we observed (i) a consistent sigmoidal behaviour between the infarct and remote regions with similar strain values in these regions (figure 6) and (ii) reported a consistent strain-estimated infarct size for each animal at the same location (figures 5 and 8f) throughout disease progression further demonstrates the reproducibility of the technique. Taken together, these data suggest that, if sternal artefacts are minimized or avoided during image acquisition, 3D myocardial strain in remodelling left ventricles can be reliably quantified.
4.2. Three-dimensional strain map reveals myocardial tissue heterogeneity
Through our 3D approach, we can identify regional variations in strain values and profiles that compare well to previously published results. Many 2D ultrasound studies have reported significant decreases in global myocardial strains in mice subjected to infarction, with the remote myocardium exhibiting significantly higher strains than the infarcted tissue [8,10,12]. In the radial direction, where the largest deformation is observed [32], strain values range between 25% and 40% in the healthy myocardium but drop to less than 15% within the infarct [8,10]. Our 3D strain results are consistent with these findings (figure 6a–e). Furthermore, a short-axis comparison of maximum principal strains between the presented 3D-DDE technique and Vic2D yielded similar ranges of strain values (electronic supplementary material, figure S6).
Although LV kinematics in the remote and infarct zones have been widely studied, the interface between these regions remains to be fully characterized as previous work has only described intermediate strain values in this vulnerable border zone [12,13]. Unsurprisingly, given the original ultrasound data, heterogeneity in border zone strain patterns can be identified in the reconstructed 3D strain maps (figures 4 and 5; electronic supplementary material, figures S2–S5). These strain patterns are correlated with complex, non-uniform deposition of collagen along the LV wall, clearly visualized from histological staining of the mid-papillary level of the left ventricle in the PL group (figure 7a). Indeed, collagen fibre orientations are remarkably heterogeneous in the healing myocardial scar and likely influence the mechanical properties of the infarct border zone [33,34]. Taken together, capturing strain heterogeneity within the infarct border zone early in remodelling may provide important insights into the role of strain in infarct expansion and LV remodelling.
4.3. Correlation between strain profiles and left ventricular remodelling severity
Strain profiles near infarct border zones exhibit a unique sigmoidal behaviour (figure 6a–e), probably caused by a spatial decrease in collagen content away from the infarct (figure 7a,b). Interestingly, throughout the 28 days post-infarction, we found significantly higher strain values within the infarcted myocardium of mice in the I/R group than in those in the PL group (figure 6b–e). A sustained increase in strain within the infarct zone may suggest either a higher percentage of viable cardiomyocytes or an improved scar contractile function attributed to the mechanoregulation of myofibroblast activity [5,35]. Within the damaged myocardium, these elevated strains may be a unique characteristic of small infarcts, but further investigation is necessary to determine their role on infarct expansion during early remodelling. Indeed, we measured significant improvements in both systolic and diastolic LV functions (figure 3b–g) as well as significantly smaller final infarct sizes (figure 7c) in the I/R group than in PL group. Although direct regional strain comparisons between mice subjected to I/R and PL surgeries have yet to be conducted within a single study, our results closely match findings from existing ultrasound studies, which report a significant increase in infarct size and worse remodelling outcomes with prolonged ischaemic durations [36,37].
4.4. Strain profiles provide early estimates of infarct size
A key discovery from this study is the propensity for wall-thinned myocardial regions at early stages to expand towards the strain-estimated infarct boundaries (figures 5 and 8f). Wall thinning is generally accepted to be the final product of infarct healing; although the majority of wall thinning in murine models occurs within the first week, this gradual process may continue to take place up to a month post-infarction [6,38]. Additionally, wall thinning is often used to monitor infarct expansion in vivo [6], is primarily responsible for LV rupture [38] and directly impacts both systolic and diastolic function [5]. Thus, the ability to predict early the extent of wall thinning non-invasively may provide critical insights into LV remodelling and progression to heart failure.
Our data suggest that the damaged area with compromised 3D maximum principal strain values is predictive of final infarct size as early as day 1. As the strain-estimated infarct size in the PL group remained unchanged throughout ventricular remodelling, rapid expansion of wall-thinned regions was prevalent within the first week before converging with the strain-estimated infarct border (figure 8f). Other previous work similarly reported that changes in 2D myocardial principal strain also precede LV wall thinning in a genetic mouse model of dilated cardiomyopathy [39]. A potential explanation for this observation may be related to the creation of a stiff provisional matrix in the ischaemic region early in remodelling, which facilitates the gradual formation of collagen-rich scars. As non-viable cardiomyocytes are resorbed, a provisional granulation tissue rich in fibrin, laminin and glycosaminoglycans is quickly formed to provide the left ventricle with temporary structural support [6]. In the presence of a stiff extracellular matrix (ECM), transforming growth factor beta (TGF-β) is released from the latent-associated peptide complex as a result of increased mechanical resistance to cell tension [40]. TGF-β then promotes the differentiation of cardiac fibroblasts to myofibroblasts, which gradually replace the provisional structure with collagen-rich infarct tissue [6,40]. Thus, the presence of necrotic cardiomyocytes and a stiff provisional structure are likely detected as an immediate decrease in strain values 1 day after infarction (figures 4 and 5). As these regions eventually become collagen-rich scars, a process that take several days or weeks to fully develop, the initial changes in myocardial ECM may explain why early strain-estimated infarct size remains unchanged throughout disease progression and is predictive of final infarct size (figure 8f).
Our comparison of day 28 infarct sizes calculated from three different approaches revealed that gold standard histology infarct size is best correlated with strain-estimated infarct size (figure 8a–c). Furthermore, we observed a better correlation with EF for the strain-estimated infarct size than with the wall-thinned approach (R2 = 0.69 versus R2 = 0.41). These findings indicate that 3D principal strain profiles can be used to accurately predict final infarct size in rodents and may have similar utility in humans. This discovery is impactful because it presents a novel non-invasive method of estimating infarct size without the use of contrast agents or tissue collection. Previously, infarct size can only be reliably estimated using late-gadolinium-enhanced magnetic resonance imaging (MRI) [21] or ex vivo histological staining [30].
4.5. Limitations
One major limitation of this study is the impact of image quality on strain estimation. Since the 3D-DDE algorithm is a direct image-based approach, shadowing artefacts can affect the measured strain values. As mentioned previously, sternal shadowing artefacts commonly found near the base of the left ventricle resulted in underestimation of strains. Although we addressed this problem by removing these regions from our final strain analysis, care during data acquisition to minimize shadowing artefacts is needed. Another limitation is the computational time needed for the strain analysis. Owing to the large number of investigation regions and need to spatially resolve small differences between time points, the strain analysis requires 2–3 h to complete per dataset. It is important, however, to note that a trade-off exists between processing times and the desired spatial resolution of the analysed strain. In other words, if less refined strain maps are needed, the computational costs could be reduced dramatically. Lastly, surgical inductions of myocardial ischaemia in mice are not true reflections of the gradual series of events leading up to a heart attack in humans. Mice experience smaller increases in collagen content post-MI [38] and undergo substantially faster infarct healing than typically observed in patients [6], which may lead to species differences in the LV remodelling process. Beyond mice, however, similar strain mapping and profiling techniques could be applied to 4D ultrasound data acquired from other rodents, large animals and humans.
Although the present study focused on characterizing changes in 3D maximum principal strain, it is important to note that other metrics including the second and third principal strains, as well as principal strain direction, may provide additional insights into the remodelling process. A previous study using tagged MRI of the porcine left ventricle showed significant reductions in all three components of principal strains post-infarction [41]. Regional differences in principal strain directions were also detected; notably, maximum principal strain angles rotated away from the radial direction within the infarcted myocardium and its surrounding region. Future work will be needed to fully characterize the relationship between infarct expansion, principal strain directionality and other components of the 3D strain tensor.
5. Conclusion
In summary, we have demonstrated a novel and robust approach to non-invasively quantify 3D myocardial strain. By integrating 4D ultrasound with a 3D-DDE technique, we expanded existing 2D ultrasound strain studies to three-dimensions to better characterize the role of myocardial mechanics in disease progression. To the best of our knowledge, this study is the first demonstration of the use of 4D ultrasound to quantify 3D strain in order to characterize regional differences, instead of global changes, between two murine models with different infarct severities. By reconstructing 3D strain maps of the left ventricles, we were able to capture strain heterogeneity and characterize the sigmoidal strain profile at infarct border zones. We discovered that mice undergoing mild LV remodelling had significantly higher strain values within the infarcted tissue than those with severe remodelling, suggesting that a more contractile infarct scar may be a unique characteristic of small infarcts. Finally, we described a new method to non-invasively estimate and predict final infarct size, without the use of contrast agents, at an acute phase based on 3D strain maps. Taken together, the findings presented in this study highlight the importance of 3D strain when studying how the mechanical behaviour of the left ventricle near infarct border zones contributes to post-infarction remodelling. Future work will be needed to investigate if the presented technique can be used to better characterize the role of 3D strains in infarct expansion, infarct extension and cases of multiple infarcts.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
The authors thank Drs John Boyle, Guy Genin and Stavros Thomopoulos for their feedback on the strain code development. We would also like to thank Dr Kristiina Aasa, Stephen Buttars and Andrew Needles at FUJIFILM VisualSonics Inc. for their technical assistance with the Vevo2100 ultrasound system.
Ethics
The presented study was conducted in accordance with Purdue University's ethics guidelines regarding the use of animals in research. All surgical procedures have been approved by the Purdue Animal Care and Use Committee under protocol no. 1505001246.
Data accessibility
Additional data that support the findings of this study are available as part of the electronic supplementary material.
Authors' contributions
A.H.S. and C.J.G. conceptualized and designed the study. A.H.S. performed all surgical procedures and developed the Matlab codes for image analysis. A.H.S., A.K.Y. and A.D.C. were responsible for data acquisition, image analysis and histological analysis. A.H.S., S.E.B. and G.D.O. compared the 3D-DDE strain results with Vic2D. All authors discussed the results and contributed to the writing, editing and review of the manuscript. All authors gave final approval for publication.
Competing interests
The authors have no conflict of interest to report.
Funding
This work was funded by the Hugh W. and Edna M. Donnan Fellowship (A.H.S.), the American Heart Association through grant no. 14SDG18220010 (C.J.G.) and the Indiana Clinical and Translational Sciences Institute, funded in part by grant no. UL1TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award (C.J.G).
References
- 1.Benjamin EJ, et al. 2019. Heart disease and stroke statistics—2019 update: a report from the American Heart Association. Circulation 139, e56–e528. ( 10.1161/CIR.0000000000000659) [DOI] [PubMed] [Google Scholar]
- 2.Cahill TJ, Kharbanda RK.. 2017. Heart failure after myocardial infarction in the era of primary percutaneous coronary intervention: mechanisms, incidence and identification of patients at risk. World J. Cardiol. 9, 407–415. ( 10.4330/wjc.v9.i5.407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutton MG, Sharpe N. 2000. Left ventricular remodeling after myocardial infarction: pathophysiology and therapy. Circulation 101, 2981–2988. ( 10.1161/01.CIR.101.25.2981) [DOI] [PubMed] [Google Scholar]
- 4.Cohn JN, Ferrari R, Sharpe N.. 2000. Cardiac remodeling—concepts and clinical implications: a consensus paper from an international forum on cardiac remodeling. J. Am. Coll. Cardiol. 35, 569–582. ( 10.1016/s0735-1097(99)00630-0) [DOI] [PubMed] [Google Scholar]
- 5.van den Borne SW, Diez J, Blankesteijn WM, Verjans J, Hofstra L, Narula J.. 2010. Myocardial remodeling after infarction: the role of myofibroblasts. Nat. Rev. Cardiol. 7, 30–37. ( 10.1038/nrcardio.2009.199) [DOI] [PubMed] [Google Scholar]
- 6.Richardson WJ, Clarke SA, Quinn TA, Holmes JW. 2015. Physiological implications of myocardial scar structure. Compr. Physiol. 5, 1877–1909. ( 10.1002/cphy.c140067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Elia N, D'Hooge J, Marwick TH. 2015. Association between myocardial mechanics and ischemic LV remodeling. JACC Cardiovasc. Imaging 8, 1430–1443. ( 10.1016/j.jcmg.2015.10.005) [DOI] [PubMed] [Google Scholar]
- 8.Bhan A, Sirker A, Zhang J, Protti A, Catibog N, Driver W, Botnar R, Monaghan MJ, Shah AM. 2014. High-frequency speckle tracking echocardiography in the assessment of left ventricular function and remodeling after murine myocardial infarction. Am. J. Physiol. Heart Circ. Physiol. 306, H1371–H1383. ( 10.1152/ajpheart.00553.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biere L, Donal E, Terrien G, Kervio G, Willoteaux S, Furber A, Prunier F. 2014. Longitudinal strain is a marker of microvascular obstruction and infarct size in patients with acute ST-segment elevation myocardial infarction. PLoS ONE 9, e86959 ( 10.1371/journal.pone.0086959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer M, Cheng S, Jain M, Ngoy S, Theodoropoulos C, Trujillo A, Lin FC, Liao R. 2011. Echocardiographic speckle-tracking based strain imaging for rapid cardiovascular phenotyping in mice. Circ. Res. 108, 908–916. ( 10.1161/CIRCRESAHA.110.239574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu VC, Takeuchi M, Otani K, Haruki N, Yoshitani H, Tamura M, Abe H, Lin FC, Otsuji Y. 2013. Effect of through-plane and twisting motion on left ventricular strain calculation: direct comparison between two-dimensional and three-dimensional speckle-tracking echocardiography. J. Am. Soc. Echocardiogr. 26, 1274–1281. ( 10.1016/j.echo.2013.07.006) [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Garson CD, Xu Y, Helm PA, Hossack JA, French BA. 2011. Serial ultrasound evaluation of intramyocardial strain after reperfused myocardial infarction reveals that remote zone dyssynchrony develops in concert with left ventricular remodeling. Ultrasound Med. Biol. 37, 1073–1086. ( 10.1016/j.ultrasmedbio.2011.04.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torres WM, Jacobs J, Doviak H, Barlow SC, Zile MR, Shazly T, Spinale FG. 2018. Regional and temporal changes in left ventricular strain and stiffness in a porcine model of myocardial infarction. Am. J. Physiol. Heart Circ. Physiol. 315, H958–H967. ( 10.1152/ajpheart.00279.2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soepriatna AH, Damen FW, Vlachos PP, Goergen CJ. 2018. Cardiac and respiratory-gated volumetric murine ultrasound. Int. J. Cardiovasc. Imaging 34, 713–724. ( 10.1007/s10554-017-1283-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damen FW, Berman AG, Soepriatna AH, Ellis JM, Buttars SD, Aasa KL, Goergen CJ. 2017. High-frequency 4-dimensional ultrasound (4DUS): a reliable method for assessing murine cardiac function. Tomography 3, 180–187. ( 10.18383/j.tom.2017.00016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grune J, et al. 2018. Evaluation of a commercial multi-dimensional echocardiography technique for ventricular volumetry in small animals. Cardiovasc. Ultrasound 16, 10 ( 10.1186/s12947-018-0128-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn HS, et al. 2017. The impact of preload on 3-dimensional deformation parameters: principal strain, twist and torsion. Cardiovasc. Ultrasound 15, 22 ( 10.1186/s12947-017-0111-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia JZ, Xia JY, Li G, Ma WY, Wang QQ. 2014. Left ventricular strain examination of different aged adults with 3D speckle tracking echocardiography. Echocardiography 31, 335–339. ( 10.1111/echo.12367) [DOI] [PubMed] [Google Scholar]
- 19.Boyle JJ, Soepriatna A, Damen F, Rowe RA, Pless RB, Kovacs A, Goergen CJ, Thomopoulos S, Genin GM. 2018. Regularization-free strain mapping in three dimensions, with application to cardiac ultrasound. J. Biomech. Eng. 141, 0110101-1-11 ( 10.1115/1.4041576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satriano A, et al. 2017. Clinical feasibility and validation of 3D principal strain analysis from cine MRI: comparison to 2D strain by MRI and 3D speckle tracking echocardiography. Int. J. Cardiovasc. Imaging 33, 1979–1992. ( 10.1007/s10554-017-1199-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young AA, French BA, Yang Z, Cowan BR, Gilson WD, Berr SS, Kramer CM, Epstein FH. 2006. Reperfused myocardial infarction in mice: 3D mapping of late gadolinium enhancement and strain. J. Cardiovasc. Magn. Reson. 8, 685–692. ( 10.1080/10976640600721767) [DOI] [PubMed] [Google Scholar]
- 22.Hashmi S, Al-Salam S. 2015. Acute myocardial infarction and myocardial ischemia-reperfusion injury: a comparison. Int. J. Clin. Exp. Pathol. 8, 8786–8796. [PMC free article] [PubMed] [Google Scholar]
- 23.Updegrove A, Wilson NM, Merkow J, Lan H, Marsden AL, Shadden SC. 2017. SimVascular: an open source pipeline for cardiovascular simulation. Ann. Biomed. Eng. 45, 525–541. ( 10.1007/s10439-016-1762-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chernak Slane L, Thelen DG. 2014. The use of 2D ultrasound elastography for measuring tendon motion and strain. J. Biomech. 47, 750–754. ( 10.1016/j.jbiomech.2013.11.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connell GD, Johannessen W, Vresilovic EJ, Elliott DM. 2007. Human internal disc strains in axial compression measured noninvasively using magnetic resonance imaging. Spine (Phila Pa 1976) 32, 2860–2868. ( 10.1097/BRS.0b013e31815b75fb) [DOI] [PubMed] [Google Scholar]
- 26.Cerqueira MD, et al. 2002. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation 105, 539–542. ( 10.1161/hc0402.102975) [DOI] [PubMed] [Google Scholar]
- 27.Diwan A, et al. 2007. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J. Clin. Invest. 117, 2825–2833. ( 10.1172/JCI32490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. ( 10.1038/nmeth.2089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thevenaz P, Unser M. 2007. User-friendly semiautomated assembly of accurate image mosaics in microscopy. Microsc. Res. Tech. 70, 135–146. ( 10.1002/jemt.20393) [DOI] [PubMed] [Google Scholar]
- 30.Nascimento DS, Valente M, Esteves T, de Pina Mde F, Guedes JG, Freire A, Quelhas P, Pinto-do OP. 2011. MIQuant--semi-automation of infarct size assessment in models of cardiac ischemic injury. PLoS ONE 6, e25045 ( 10.1371/journal.pone.0025045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphrey JD. 2002. Cardiac mechanics. In Cardiovascular solid mechanics: cells, tissues, and organs (eds Campbell J, Reagan B, Blum B, Rogers W, Humphrey R), pp. 599–729. New York, NY: Springer. [Google Scholar]
- 32.Geyer H, et al. 2010. Assessment of myocardial mechanics using speckle tracking echocardiography: fundamentals and clinical applications. J. Am. Soc. Echocardiogr. 23, 351–369; quiz 453–355 ( 10.1016/j.echo.2010.02.015) [DOI] [PubMed] [Google Scholar]
- 33.Richardson WJ, Holmes JW. 2016. Emergence of collagen orientation heterogeneity in healing infarcts and an agent-based model. Biophys. J. 110, 2266–2277. ( 10.1016/j.bpj.2016.04.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fomovsky GM, Rouillard AD, Holmes JW. 2012. Regional mechanics determine collagen fiber structure in healing myocardial infarcts. J. Mol. Cell. Cardiol. 52, 1083–1090. ( 10.1016/j.yjmcc.2012.02.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van De Water L, Varney S, Tomasek JJ. 2013. Mechanoregulation of the myofibroblast in wound contraction, scarring, and fibrosis: opportunities for new therapeutic intervention. Adv. Wound Care (New Rochelle) 2, 122–141. ( 10.1089/wound.2012.0393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preda MB, Burlacu A. 2010. Electrocardiography as a tool for validating myocardial ischemia-reperfusion procedures in mice. Comp. Med. 60, 443–447. [PMC free article] [PubMed] [Google Scholar]
- 37.Thibault H, Gomez L, Donal E, Pontier G, Scherrer-Crosbie M, Ovize M, Derumeaux G. 2007. Acute myocardial infarction in mice: assessment of transmurality by strain rate imaging. Am. J. Physiol. Heart Circ. Physiol. 293, H496-H502. ( 10.1152/ajpheart.00087.2007) [DOI] [PubMed] [Google Scholar]
- 38.Gao XM, White DA, Dart AM, Du XJ. 2012. Post-infarct cardiac rupture: recent insights on pathogenesis and therapeutic interventions. Pharmacol. Ther. 134, 156–179. ( 10.1016/j.pharmthera.2011.12.010) [DOI] [PubMed] [Google Scholar]
- 39.Hankiewicz JH, Goldspink PH, Buttrick PM, Lewandowski ED. 2008. Principal strain changes precede ventricular wall thinning during transition to heart failure in a mouse model of dilated cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 294, H330–H336. ( 10.1152/ajpheart.01109.2007) [DOI] [PubMed] [Google Scholar]
- 40.Wells RG. 2013. Tissue mechanics and fibrosis. Biochim. Biophys. Acta 1832, 884–890. ( 10.1016/j.bbadis.2013.02.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pilla JJ, Koomalsingh KJ, McGarvey JR, Witschey WR, Dougherty L, Gorman JH 3rd, Gorman RC. 2015. Regional myocardial three-dimensional principal strains during postinfarction remodeling. Ann. Thorac. Surg. 99, 770–778. ( 10.1016/j.athoracsur.2014.10.067) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional data that support the findings of this study are available as part of the electronic supplementary material.