Abstract
First appearing in 2011 in Northern Europe, Schmallenberg virus (SBV), an Orthobunyavirus of the Simbu serogroup, is associated with clinical disease mainly in ruminants such as cattle, sheep and goats. The clinical signs are characterized by abortion and congenital deformities in newborns. The virus is transmitted by Culicoides midges of the Obsoletus complex. SBV infection induces a solid protective immunity that persists for at least 4 or 6 years in sheep and cattle, respectively. SBV infection can be diagnosed directly by real-time RT-qPCR and virus isolation or indirectly by serological assays. Three vaccines are commercially available in Europe. This article provides a comprehensive literature review on this emerging disease regarding pathogenesis, transmission, diagnosis, control and prevention. This review also highlights that although much has been learned since SBV’s first emergence, there are still areas that require further study to devise better mitigation strategies.
Keywords: Schmallenberg virus, Culicoides, ruminants
1. Discovery and Genomic Structure of the Virus
In the fall of 2011, a new cattle disease was reported in Germany and The Netherlands associated with a drop in milk production, hyperthermia and diarrhea. Using a metagenomic approach on blood samples collected from clinically sick dairy cows, the causative agent was identified as a novel RNA virus [1]. Consequently, the agent was named Schmallenberg virus (SBV) after the locality in Germany where the outbreak occurred.
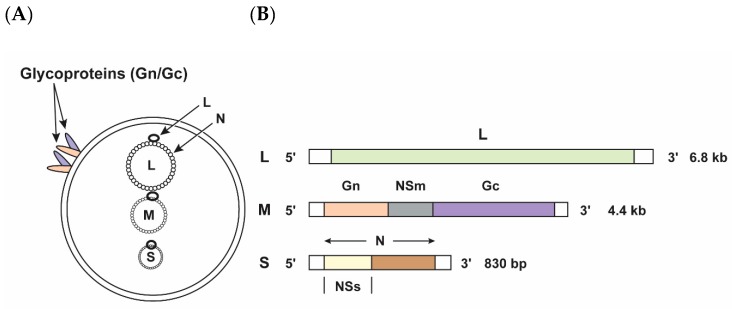
The newly discovered SBV has sequence similarities to other viruses in the Peribunyaviridae family, genus Orthobunyavirus, such as Akabane, Aino and Shamonda. Further sequence analysis also revealed high similarities with Sathuperi, and Douglas viruses [2]. SBV is an enveloped, single-stranded, negative-sense RNA virus with three genomic segments: L (large), M (medium) and S (small) segments (Figure 1A) [3]. The RNA-dependent RNA polymerase (RdRp) is encoded by the L segment, whereas the M segment encodes a polyprotein that is further cleaved into the envelope glycoproteins Gn and Gc and the non-structural protein NSm (Figure 1B). Overlapping open reading frames (ORFs) of the S segment encode the nucleoprotein N and the non-structural protein NSs (Figure 1B) [1,4]. Like other viruses in the order Bunyavirales, the NSs of SBV has been shown to be a major virulence factor that downregulates host-cell mRNA synthesis and type I interferon production in mammalian cells, thereby enhancing viral replication [4,5]. SBV is transmitted by culicoides midges [1].
Figure 1.
Schmallenberg virus (SBV) virion and genome organization. (A) The SBV large (L), medium (M), and small (S) segments are present in the virion and encapsulated with an envelope lipid bilayer containing the surface glycoproteins Gn and Gc. (B) Schematic representation of the SBV segments and the SBV coding strategy: L-segment: L, RNA-dependent RNA polymerase protein; M-segment: Gn and Gc, glycoproteins Gn and Gc; NSm, non-structural protein; S-segment: N, nucleoprotein; NSs, non-structural protein.
2. Epidemiology
Various virulent arboviruses, such as West Nile virus, Rift Valley fever virus, Chikungunya virus, Zika virus, and Bluetongue virus (BTV), have emerged and caused epidemics in humans and/or animals in North and South America, the Arabian Peninsula or Europe. Their emergence has been partially attributed to the phenomenon of climate change [6]. In the fall of 2011, Schmallenberg virus appeared in Northern Europe, where also BTV serotype 8 (BTV-8) first appeared in 2006 [1,7]. Schmallenberg virus infection manifested itself as congenital defects in newborn calves, lambs and kids, as well as an arthrogryposis and hydranencephaly syndrome [8]. SBV quickly spread to the rest of the continent, with wind playing an important role in the transmission of the virus, as infected midges are easily carried on air currents [9]. The rate of spread of SBV is estimated to be in the range of 0.9 to 1.5 km/day [10]. Despite a high seroprevalence of up to 98% reported in outbreak regions in 2011 and 2012 [11], new infections occurred again in the summer of 2012 in the same regions. Both serological and genome detection methods revealed the re-emergence of SBV infection in Belgium in 2012 [12]. It became evident that the cold winter season could not eradicate the virus since new cases were also observed in ruminants in Germany in June 2012 [13]. In late 2014, SBV outbreaks in cattle and sheep were once again detected in The Netherlands; and in 2016, an increase in SBV-specific antibody titers and prevalence in heifers was noted in The Netherlands [14]. Additionally, the detection of SBV genomic sequences in aborted calves in Belgium in 2015 was a further indication of SBV circulation in the area at the time [15]. Although there were several years of limited to no circulation of the virus in the UK and France, outbreaks of the disease occurred again in 2016 and 2017 [16,17].
The original source of SBV is still unclear; however, there are reports of SBV cross-reactive antibodies to other Simbu serogroup viruses found in African cattle, prior to and after the initial European outbreak [18,19,20]. A report from Jordan in 2013 indicated detection of antibodies against Aino virus, another Simbu serogroup virus, in ruminants on farms where similar clinical symptoms associated with SBV infections were observed [21]. Turkey reported detecting SBV RNA in aborted cattle and sheep fetuses a year after the initial outbreak in Europe, suggesting a spread of the virus from Northern Europe [22]. Furthermore, blood samples collected before 2011 were found to be positive for SBV antibodies by ELISA [23]. Since the SBV ELISA has low specificity, the possibility of potentially detecting cross-reactive antibodies induced by other Simbu serogroup viruses cannot be ruled out. Remarkably, there have been multiple reports of fetal malformation in ruminants from the Mediterranean region, suggesting a possible circulation of Simbu serogroup viruses in this region [21,22,23,24].
Insect vectors such as mosquitoes and biting flies (Culicoides spp.) are often responsible for the transmission of bunyaviruses associated with human and animal diseases in Asia and Africa. Immediately after the discovery of SBV, it became evident that Culicoides spp. play a role in its transmission [25]. SBV genomic sequences were detected in biting flies, namely the Culicoides obsoletus species group of the Ceratopogonidae family, and it was shown that multiple Culicoides spp. (Culicoides dewulfi, Culicoides chiopterus, Culicoides punctatus, etc) were positive for SBV genomic markers. Culicoides caught as early as summer and autumn of 2011 in Belgium [26], Italy [27], The Netherlands [28], and Denmark [25] were found positive for SBV genomic markers. Interestingly, Culicoides sonorensis, a vector for BTV, was also shown to support the replication and dissemination of SBV under laboratory conditions [29]. The question of how the vector-borne SBV persists over winter has not been solved yet; however, one field study has demonstrated that the potential mechanism is transovarial transmission in the Culicoides vector [30].
Vertical transmission of SBV from infected dam to fetus occurs during the first and early-second trimester of gestation and results in abortion, stillbirth and birth of malformed newborns [31,32]. Although experimentally infected animals shed SBV RNA in feces, oral and nasal fluids [33], direct transmission of SBV from infected ruminants to naïve animals by contact or oro-nasal/feco-oral routes has not been reported [33]. Both, oral inoculation of cattle and nasal inoculation of sheep failed to produce viremia in the animals [33]. Interestingly, SBV was detected in semen from infected bulls [34]; however, transmission of SBV from infected bulls to dams either through natural mating or artificial insemination has not been extensively studied yet [34]. In one study, viral RNA was isolated from blood samples of cattle experimentally injected with SBV-RNA-positive semen [34]. The presence of SBV RNA in amniotic fluid and fetal tissues [35,36] was suggested in a previous review on Orthobunyaviruses as one possibility the virus may persist over winter [37].
Multiple domestic and wild animal species have been shown to be susceptible to SBV infection under natural and experimental conditions. This was determined through direct and/or indirect detection of SBV in animals with clinical manifestations or subclinical infections [14]. Importantly, overt clinical manifestations of SBV infection have been seen exclusively in domestic ruminants [14], whereas only indirect serological evidence of SBV infection has been reported for wild ruminants (e.g., alpaca, buffalo, deer, chamois, mouflon, bison), zoo animals (e.g., kudu, zebra, oryx), and some other mammalian species (e.g., horse, wild boar) [38,39,40,41,42]. Interestingly, virological and serological evidence of SBV infection has been also reported in dogs [43]. Experimental infection of piglets with SBV resulted only in seroconversion, and no RT-PCR positivity was detected, suggesting the inability of the virus to efficiently replicate in this host species [44].
3. Clinical and Pathological Findings
Infection of adult ruminants with SBV usually results in non-specific clinical signs. In cattle, SBV infection often manifests as a mild and transient disease, with anorexia, hyperthermia, and in some animals with diarrhea and reduced milk yield (up to 50%) [1,45]. In contrast, SBV infection in adult sheep and goats is mostly subclinical. Acute clinical cases of SBV are not common, but there are a few reports of clinical disease in adult animals (6% cattle, 3% sheep and 1% goats) [46]. Clinical signs of diarrhea and reduced milk yield have been reported at least once in goats [47]. Even though the causal relationship has not been clearly established, there have been reports of fever, diarrhea and reduced milk yield in sheep [46]. Under experimental conditions, in sheep and cattle, SBV infection exhibits a short viremic period of 5–7 days, which starts at day 2 or 3 post infection (pi) and peaks around day 4 pi [47]. The clinical outcomes of abortion, stillbirth, and malformed newborns associated with SBV infection in cattle and sheep are similar to those observed for other Simbu serogroup viruses such as Akabane and Aino [8,48]. A relationship between herd immunity and the birth of congenitally malformed newborns has been observed during SBV infection. A decline in herd immunity is followed by an increase in seroprevalence against SBV [45,49]. Common musculoskeletal deformities observed in fetuses during transplacental infection include arthrogryposis, lordosis, scoliosis, torticollis and brachygnathia inferior [37,50]. Sacral spina bifida and cleft palate are also observed in stillborn lambs. Meanwhile, musculoskeletal defects are not uniform in the case of twin gestation, in which case, one twin may present malformations, whereas the other is born healthy without any malformation or clinical signs [37]. A recent study revealed that experimental in utero infection of bovine fetuses resulted in a very low incidence rate of fetal abortion/malformation; only one abortion and one malformed fetus out of 36 experimentally in utero SBV-infected pregnant heifers were reported when the animals were infected between 60 and 150 days of gestation (first and second trimester) [51].
At necropsy, hydranencephaly, porencephaly, lissencephaly, hydrocephalus, cerebellar and cerebral hypoplasia and micromyelia are commonly observed in the central nervous system (CNS) of SBV-infected young ruminants. Among the microscopic lesions, glial nodules mainly in the mesencephalon and hippocampus of lambs and goats, lymphohistiocytic meningoencephalomyelitis, as well as neuronal degeneration and necrosis mainly in the brain stem of calves are observed [52]. The musculoskeletal defects manifested as arthrogryposis in fetuses are most likely due to lesions in the spinal cord [3]. Hence, these musculoskeletal lesions in SBV-infected aborted fetuses or neonates led to the description of the arthrogryposis and hydranencephaly syndrome (AG-HE syndrome) [14].
4. Immunity
Infections of Schmallenberg virus in naïve populations spread quickly but result in solid protective immunity. This was evident when, within a two-month period, many cattle herds in Germany became infected [53]. Studies indicate that in the majority of SBV-infected cattle and sheep, anti-SBV antibodies last for at least 38 and 48 months, respectively [14]. A recent report indicates that among 17 naturally infected cattle studied over a period of 6 years, three animals became seronegative, while the other 14 animals still had measurable anti-SBV antibodies after six years [53]. Most calves and lambs born to SBV-infected cows or ewes were shown to be protected from SBV infection for at least the first six and four months, respectively [53,54]. Under experimental conditions, anti-SBV antibodies are detected between 14 and 21 days post-infection in cows and sheep [33,45,49]. Interestingly, the role of CD8+ T lymphocytes seems insignificant during SBV infection as these cells are not detected after initial infection, implying that virus clearance is most likely independent of T-cell-mediated cytotoxicity. However, CD8+ T-cells may be involved in the protective immune response against the virus during secondary exposures. In addition, between 3–7 days post-infection, the number of CD4+ T helper cells decrease and SBV genomic markers are not detectable in peripheral blood leukocytes [33].
5. Diagnosis
In adult animals, SBV mostly causes subclinical infections; however, occasionally, especially in cattle, clinical signs of fever, diarrhea and a reduction in milk yield are not uncommon [1]. In transplacental infections, congenital CNS and musculoskeletal malformations, such as the AG-HE syndrome, aplasia or hypoplasia of the cerebrum or cerebellum and hydranencephaly are observed [33,45,49]. Virological and/or serological diagnosis is needed to confirm suspicion of SBV infection because of similarities between clinical features of SBV and other ruminant virus infections [33].
SBV can be isolated in multiple insect and mammalian cell types such as: BHK-21 (baby hamster kidney), Vero (African green monkey) and KC (Culicoides sonorensis) cells [1]. Only mammalian cells are permissive to SBV infection with cytopathic effects. However, virus isolation is not always possible from clinical specimens including blood samples, due to the low viral load in most samples. Real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR) targeting the L and S segments of the SBV genome allows sensitive and specific detection of SBV RNA, which is indicative of SBV infection [55]. The presence of SBV RNA in various organs has been tested by RT-qPCR, but only some of them were determined as positive [31,56]. SBV genomic sequences are readily detected in the cerebrum, spinal cord, placental fluid and umbilical cord of malformed lambs or calves [31]. The brain stem is also an appropriate sample for the detection of SBV RNA [57].
Detecting anti-SBV antibodies in serum represents an indirect method for the diagnosis of SBV infections. Both, the virus neutralization test (VNT) and enzyme-linked immunosorbent assay (ELISA) have been developed as techniques for serological diagnosis. The former method is time-consuming; hence its use is restricted as a confirmatory test. On the other hand, the ELISA is a rapid, less expensive and high-throughput test. However, interpretation of test results in areas where other Simbu serogroup viruses are circulating could give false-positive results due to potential antibody cross-reactivity and, therefore, caution should be exercised in interpreting these results [58]. However, a unique advantage of the ELISA test is that it can be used to detect anti-SBV antibodies in milk [59]. On the other hand, discrepancies in specificity and sensitivity between VNT and ELISA have been reported [60]. ELISA tests seem to exhibit lower specificity and sensitivity than VNTs [60]. Anti-SBV antibodies can also be detected by indirect immunofluorescence assay [61]; however, this method is not often used as a routine detection technique. Immunohistochemistry and in situ hybridization are techniques which are employed for the detection of SBV proteins and genomic RNA in paraffin-embedded tissue sections, respectively [62].
6. Surveillance and Vaccination
Surveillance of the dynamics of competent vectors and vector infection rates seems to be the optimal strategy for predicting future SBV outbreaks. Furthermore, vaccination of replacement stocks and control of insect populations are the two most important methods for the prevention of SBV outbreaks [14], as vaccination in particular helps reduce SBV infection in ruminants [61]. Farmers in some countries, however, are unwilling to vaccinate their animals against SBV, claiming vector surveillance to be a more effective prevention strategy than vaccination. The re-emergence of SBV in Germany and The Netherlands in late 2014, and more recently in Belgium, France and the UK, is an indication that SBV is able to infect and disseminate in cattle and sheep flocks in the face of declining immunity [17,63,64]. SBV re-emerging events should be a reminder for farmers to regularly implement SBV vaccination of their animals.
7. Inactivated Vaccines
Inactivated SBV vaccines have been developed shortly after the initial isolation and characterization of the virus in Europe by various companies [65,66]. These vaccines effectively prevent viremia and clinical disease, including the prevention of fetal malformation and premature birth or stillbirth [67,68]. Three commercial vaccines, namely Zulvac SBV (Zoetis), Bovilis SBV (MSD Animal Health) and SBVvax (Merial) have been in use in Europe to protect sheep and cattle from SBV infection (see Table 1). In an experimental study using cattle and sheep, the onset of inactivated vaccine-induced immunity was demonstrated as early as 2 weeks after vaccination [65,66]. Furthermore, the efficacy of these vaccines appeared to be dependent on the production cell line and the virus titer in the vaccine [66].
Table 1.
Summary of SBV Vaccines and SBV Vaccine Candidates.
| Type of Vaccine | Host Species Evaluated | DIVA Compatibility | References | ||
|---|---|---|---|---|---|
| Mice | Cattle | Sheep | |||
| Inactivated vaccines: | |||||
| Binary ethylenimine inactivated | NO | YES | YES | NO | [65,66] |
| Bovilis SBV (MSD Animal Health) | NO | YES | YES | NO | [67] |
| Zulvac SBV (Zoetis) | NO | YES | YES | NO | [75] |
| SBVvax (Merial) | NO | YES | YES | NO | [68] |
| Genetically modified live virus vaccines: | |||||
| Recombinant NSm and/or NSs deletion mutants | YES | YES | YES | YES | [69] |
| DNA vaccines: | |||||
| SBV Gc (N-terminal) | YES | NO | NO | YES | [72] |
| SBV Nucleoprotein | YES | NO | NO | YES | [73] |
| SBV Gn (ectodomain), SBV Gc (ectodomain 1 and 2) | YES | NO | NO | YES | [73] |
| Virus-vectored vaccines: | |||||
| Recombinant Equine Herpes Virus 1, Gc (N-terminal) | NO | YES | NO | YES | [74] |
| Modified Vaccinia Virus Ankara, Gc (N-terminal) | YES | NO | YES | YES | [74] |
| Recombinant subunit Vaccines: | |||||
| Baculovirus-expressed Gc or Gc/Gn | NO | YES | NO | YES | [71] |
| Gc (N-terminal), HEK cells | YES | YES | NO | YES | [72] |
| Gc + Gn linked ectodomains, HEK cells | YES | YES | NO | YES | [72] |
| Gc (N-terminal) of SBV and Akabane, HEK cells | YES | YES | NO | YES | [72] |
8. Modified-Live and Subunit Vaccines
Recombinant modified-live vaccines with NSm and/or NSs deletions in the M and S segments of SBV have been developed and conferred a high level of protection from infection [69]. Although these vaccines are DIVA (differentiate infected from vaccinated animals) compatible, safety concerns related to reversion to virulence may prevent their wider use. It was reported that serial passages of a NSs-deleted, attenuated virus in cell culture was able to restore its virulence when a single mutation was acquired in the Gc protein [70]. Using baculovirus produced Gc or Gc and Gn as a subunit vaccine generated a low level of neutralizing antibody response but did not protect against SBV challenge [71]. Another subunit vaccine based on the SBV Gc amino terminal domain (aa. 468–702), expressed in human embryonic kidney (HEK) cells; however, provided partial protection in cattle (three out of four animals were protected after challenge) [72]. Meanwhile, another subunit vaccine construct, which contained the entire ectodomains of Gc and Gn in a covalently linked fashion, conferred only low protective efficacy in cattle (one out of four animals protected after challenge) [72]. In contrast, full protection was obtained when an antigen containing the covalently linked Gc amino-terminal domains of both SBV and Akabane viruses was used [72]. Meanwhile, a DNA vaccine encoding the SBV Gc amino-terminal domain and the SBV nucleoprotein reduced viremia and protected animals against weight loss [73]. Vaccination with recombinant Equine Herpes Virus 1 or Modified Vaccinia Virus Ankara expressing the SBV Gc amino-terminal domain resulted in partial (two out of four animals) or full protection in cattle, respectively [74] (see Table 1). In general, in order to prevent fetal infection with SBV, vaccination should be targeted to breeding animals.
9. Potential for Re-Emergence of SBV
Like other viruses in the Simbu serogroup such as Akabane and Aino, SBV re-emerges when herd immunity declines and favorable conditions for the vector population occur [37]. Aino and Akabane virus-based epidemics occur every 3–6 years in Japan, as more naïve animals become available [50,76]. Similarly, in Australia, Akabane virus outbreaks occur every 10–15 years, due to the temporary change in the vector populations and the availability of naïve susceptible animals [77]. In 2015, the re-emergence of another arbovirus, BTV serotype 8, which shares the same vector species as SBV, occurred in Europe after several years of absence of clinical disease [78]. The fact that only 20% of newborn animals carried anti-SBV antibodies in 188 herds examined during a cross-sectional study in Belgium in 2012, demonstrates a significant loss of herd immunity compared to 2011 [79]. The mean herd seroprevalence in calves declined from 65.7% and to 20.6% [79], which represents a recipe for another SBV outbreak in this country. Between 2014 and 2015, no anti-SBV antibodies were detected in ruminants in the UK and Ireland, suggesting an absence of SBV circulation in the ruminant and vector population in these countries [80,81]. In contrast, from 2016 to 2018, virus circulation at low levels was detected in many countries in Europe, such as Belgium [15], UK [17] and France [16]. These epidemiological data re-enforce the idea of a possible resurgence of SBV infections when favorable conditions for its occurrence such as high proportion of naïve susceptible animals and increased vector populations exist [82].
10. Conclusions
Schmallenberg virus, a newly emerged arbovirus, was first reported in Central and Northwestern Europe in the fall of 2011. The initial source of introduction has not been identified to date. In addition, the mechanism by which the virus is maintained during the winter period when the vector population is low or absent remains unknown. Therefore, future studies should address the possibility of SBV introduction/re-introduction through infected midges imported from tropical or endemic regions where serological, molecular and vector prevalence of SBV has been reported. It is equally important to examine whether persistently infected, clinically healthy offspring born to infected domestic and wild ruminants exist and act as a constant source of SBV infection. Additional investigations of the competence of other arthropod vectors in the transmission of SBV, as well as for other viruses of the Simbu serogroup, are needed. Furthermore, studies on the cross-reactivity of current diagnostic tests and cross-protection between viruses from the Simbu serogroup need to be conducted. The major lesson learned from the emergence of SBV, is that maintaining vigilance and expertise is of great importance to be able to respond rapidly to new threats to animal and public health.
Author Contributions
W.C.W. and J.A.R. conceived the review; A.D.E. wrote the manuscript; J.A.R., B.F., and W.C.W. provided critical comments and edited the manuscript.
Funding
This work was funded by the Kansas Bioscience Authority grant #586, the USDA-ARS, NP-103, CRIS #3020-32000-009-00D, and the National Bio and Agro-defense Facility (NBAF) Transition Fund from the State of Kansas.
Conflicts of Interest
All authors declare no conflicts of interest.
References
- 1.Hoffmann B., Scheuch M., Höper D., Jungblut R., Holsteg M., Schirrmeier H., Eschbaumer M., Goller K.V., Wernike K., Fischer M., et al. Novel Orthobunyavirus in Cattle, Europe, 2011. Emerg. Infect. Dis. 2012;18:469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goller K.V., Höper D., Schirrmeier H., Mettenleiter T.C., Beer M. Schmallenberg Virus as Possible Ancestor of Shamonda Virus. Emerg. Infect. Dis. 2012;18:1644. doi: 10.3201/eid1810.120835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elliott R.M. Orthobunyaviruses: Recent genetic and structural insights. Nat. Rev. Microbiol. 2014;12:673–685. doi: 10.1038/nrmicro3332. [DOI] [PubMed] [Google Scholar]
- 4.Varela M., Schnettler E., Caporale M., Murgia C., Barry G., McFarlane M., McGregor E., Piras I.M., Shaw A., Lamm C., et al. Schmallenberg virus pathogenesis, tropism and interaction with the innate immune system of the host. PLoS Pathog. 2013;9:e1003133. doi: 10.1371/journal.ppat.1003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott R.M., Blakqori G., van Knippenberg I.C., Koudriakova E., Li P., McLees A., Shi X., Szemiel A.M. Establishment of a reverse genetics system for Schmallenberg virus, a newly emerged orthobunyavirus in Europe. J. Gen. Virol. 2013;94:851–859. doi: 10.1099/vir.0.049981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein P.R. Chikungunya Fever resurgence and global warming. Am. J. Trop. Med. Hyg. 2007;76:403–404. doi: 10.4269/ajtmh.2007.76.403. [DOI] [PubMed] [Google Scholar]
- 7.Conraths F.J., Gethmann J.M., Staubach C., Mettenleiter T.C., Beer M., Hoffmann B. Epidemiology of bluetongue virus serotype 8, Germany. Emerg. Infect. Dis. 2009;15:433–435. doi: 10.3201/eid1503.081210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luttikholt S., Veldhuis A., van den Brom R., Moll L., Lievaart-Peterson K., Peperkamp K., van Schaik G., Vellema P. Risk factors for malformations and impact on reproductive performance and mortality rates of Schmallenberg virus in sheep flocks in the Netherlands. PLoS ONE. 2014;9:e100135. doi: 10.1371/journal.pone.0100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sedda L., Rogers D.J. The influence of the wind in the Schmallenberg virus outbreak in Europe. Sci. Rep. 2013;3:3361. doi: 10.1038/srep03361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balmer S., Vögtlin A., Thür B., Büchi M., Abril C., Houmard M., Danuser J., Schwermer H. Serosurveillance of Schmallenberg virus in Switzerland using bulk tank milk samples. Prev. Vet. Med. 2014;116:370–379. doi: 10.1016/j.prevetmed.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Elbers A.R.W., Elbers A.R., Stockhofe N., Stockhofe N., van der Poel W.H.M., van der Poel W.H. Schmallenberg virus antibodies in adult cows and maternal antibodies in calves. Emerg. Infect. Dis. 2014;20:901–902. doi: 10.3201/eid2005.130763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claine F., Coupeau D., Wiggers L., Muylkens B., Kirschvink N. Schmallenberg virus among female lambs, Belgium, 2012. Emerg. Infect. Dis. 2013;19:1115–1117. doi: 10.3201/eid1907.121768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conraths F.J., Kämer D., Teske K., Hoffmann B., Mettenleiter T.C., Beer M. Reemerging Schmallenberg virus infections, Germany, 2012. Emerg. Infect. Dis. 2013;19:513–514. doi: 10.3201/eid1903.121324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claine F., Coupeau D., Wiggers L., Muylkens B., Kirschvink N. Schmallenberg virus infection of ruminants: Challenges and opportunities for veterinarians. Vet. Med. 2015;6:261–272. doi: 10.2147/VMRR.S83594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delooz L., Saegerman C., Quinet C., Petitjean T., De Regge N., Cay B. Resurgence of Schmallenberg Virus in Belgium after 3 Years of Epidemiological Silence. Transbound. Emerg. Dis. 2017;64:1641–1642. doi: 10.1111/tbed.12552. [DOI] [PubMed] [Google Scholar]
- 16.Gache K., Zientara S., Collin E., Authié E., Dion F., Garin E., Zanella G., Calavas D. Spatial and temporal patterns of Schmallenberg virus in France in 2016. Vet. Rec. 2018;182:575. doi: 10.1136/vr.104769. [DOI] [PubMed] [Google Scholar]
- 17.Stokes J.E., Tarlinton R.E., Lovatt F., Baylis M., Carson A., Duncan J.S. Survey to determine the farm-level impact of Schmallenberg virus during the 2016–2017 United Kingdom lambing season. Vet. Rec. 2018;183:690. doi: 10.1136/vr.104866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molini U., Capobianco Dondona A., Hilbert R., Monaco F. Antibodies against Schmallenberg virus detected in cattle in the Otjozondjupa region, Namibia. J. S. Afr. Vet. Assoc. 2018;89:1–2. doi: 10.4102/jsava.v89i0.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oluwayelu D., Wernike K., Adebiyi A., Cadmus S., Beer M. Neutralizing antibodies against Simbu serogroup viruses in cattle and sheep, Nigeria, 2012–2014. BMC Vet. Res. 2018;14:277. doi: 10.1186/s12917-018-1605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sibhat B., Ayelet G., Gebremedhin E.Z., Skjerve E., Asmare K. Seroprevalence of Schmallenberg virus in dairy cattle in Ethiopia. Acta Trop. 2018;178:61–67. doi: 10.1016/j.actatropica.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 21.Abutarbush S.M., La Rocca A., Wernike K., Beer M., Al Zuraikat K., Al Sheyab O.M., Talafha A.Q., Steinbach F. Circulation of a Simbu Serogroup Virus, Causing Schmallenberg Virus-Like Clinical Signs in Northern Jordan. Transbound. Emerg. Dis. 2017;64:1095–1099. doi: 10.1111/tbed.12468. [DOI] [PubMed] [Google Scholar]
- 22.Yilmaz H., Hoffmann B., Turan N., Cizmecigil U.Y., Richt J.A., van der Poel W.H.M. Detection and partial sequencing of Schmallenberg virus in cattle and sheep in Turkey. Vector Borne Zoonotic Dis. 2014;14:223–225. doi: 10.1089/vbz.2013.1451. [DOI] [PubMed] [Google Scholar]
- 23.Azkur A.K., Albayrak H., Risvanli A., Pestil Z., Ozan E., Yılmaz O., Tonbak S., Cavunt A., Kadı H., Macun H.C., et al. Antibodies to Schmallenberg virus in domestic livestock in Turkey. Trop. Anim. Health Prod. 2013;45:1825–1828. doi: 10.1007/s11250-013-0415-2. [DOI] [PubMed] [Google Scholar]
- 24.Chaintoutis S.C., Kiossis E., Giadinis N.D., Brozos C.N., Sailleau C., Viarouge C., Breard E., Papanastassopoulou M., Zientara S., Papadopoulos O., et al. Evidence of Schmallenberg virus circulation in ruminants in Greece. Trop. Anim. Health Prod. 2014;46:251–255. doi: 10.1007/s11250-013-0449-5. [DOI] [PubMed] [Google Scholar]
- 25.Rasmussen L.D., Kristensen B., Kirkeby C., Rasmussen T.B., Belsham G.J., Bødker R., Bøtner A. Culicoids as vectors of Schmallenberg virus. Emerg. Infect. Dis. 2012;18:1204–1206. doi: 10.3201/eid1807.120385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Regge N., Deblauwe I., De Deken R., Vantieghem P., Madder M., Geysen D., Smeets F., Losson B., van den Berg T., Cay A.B. Detection of Schmallenberg virus in different Culicoides spp. by real-time RT-PCR. Transbound. Emerg. Dis. 2012;59:471–475. doi: 10.1111/tbed.12000. [DOI] [PubMed] [Google Scholar]
- 27.Goffredo M., Monaco F., Capelli G., Quaglia M., Federici V., Catalani M., Montarsi F., Polci A., Pinoni C., Calistri P., et al. Schmallenberg virus in Italy: A retrospective survey in Culicoides stored during the bluetongue Italian surveillance program. Prev. Vet. Med. 2013;111:230–236. doi: 10.1016/j.prevetmed.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 28.Elbers A.R.W., Elbers A.R., Meiswinkel R., Meiswinkel R., van Weezep E., van Weezep E., Sloet van Oldruitenborgh-Oosterbaan M.M., Sloet van Oldruitenborgh-Oosterbaan M.M., Kooi E.A., Kooi E.A. Schmallenberg virus in Culicoides spp. biting midges, the Netherlands, 2011. Emerg. Infect. Dis. 2013;19:106–109. doi: 10.3201/eid1901.121054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veronesi E., Henstock M., Gubbins S., Batten C., Manley R., Barber J., Hoffmann B., Beer M., Attoui H., Mertens P.P.C., et al. Implicating Culicoides biting midges as vectors of Schmallenberg virus using semi-quantitative RT-PCR. PLoS ONE. 2013;8:e57747. doi: 10.1371/journal.pone.0057747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kęsik-Maliszewska J., Larska M., Collins Á.B., Rola J. Post-Epidemic Distribution of Schmallenberg Virus in Culicoides Arbovirus Vectors in Poland. Viruses. 2019;11:447. doi: 10.3390/v11050447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bilk S., Schulze C., Fischer M., Beer M., Hlinak A., Hoffmann B. Organ distribution of Schmallenberg virus RNA in malformed newborns. Vet. Microbiol. 2012;159:236–238. doi: 10.1016/j.vetmic.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 32.Wernike K., Holsteg M., Schirrmeier H., Hoffmann B., Beer M. Natural Infection of Pregnant Cows with Schmallenberg Virus—A Follow-Up Study. PLoS ONE. 2014;9:e98223. doi: 10.1371/journal.pone.0098223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wernike K., Eschbaumer M., Schirrmeier H., Blohm U., Breithaupt A., Hoffmann B., Beer M. Oral exposure, reinfection and cellular immunity to Schmallenberg virus in cattle. Vet. Microbiol. 2013;165:155–159. doi: 10.1016/j.vetmic.2013.01.040. [DOI] [PubMed] [Google Scholar]
- 34.Schulz C., Wernike K., Beer M., Hoffmann B. Infectious Schmallenberg Virus from Bovine Semen, Germany. Emerg. Infect. Dis. 2014;20:338–339. doi: 10.3201/eid2002.131436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rapid spread and association of Schmallenberg virus with ruminant abortions and foetal death in Austria in 2012/2013. Prev. Vet. Med. 2014;116:350–359. doi: 10.1016/j.prevetmed.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Natural intrauterine infection with Schmallenberg virus in malformed newborn calves. Emerg. Infect. Dis. 2014;20:1327–1330. doi: 10.3201/eid2008.121890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Regge N. Akabane, Aino and Schmallenberg virus-where do we stand and what do we know about the role of domestic ruminant hosts and Culicoides vectors in virus transmission and overwintering? Curr. Opin. Virol. 2017;27:15–30. doi: 10.1016/j.coviro.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Larska M., Krzysiak M.K., Kęsik-Maliszewska J., Rola J. Cross-sectional study of Schmallenberg virus seroprevalence in wild ruminants in Poland at the end of the vector season of 2013. BMC Vet. Res. 2014;10:967. doi: 10.1186/s12917-014-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulz C., Beer M., Hoffmann B. Schmallenberg virus infection in South American camelids: Field and experimental investigations. Vet. Microbiol. 2015;180:171–179. doi: 10.1016/j.vetmic.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 40.García-Bocanegra I., Cano-Terriza D., Vidal G., Rosell R., Paniagua J., Jiménez-Ruiz S., Expósito C., Rivero-Juarez A., Arenas A., Pujols J. Monitoring of Schmallenberg virus in Spanish wild artiodactyls, 2006–2015. PLoS ONE. 2017;12:e0182212. doi: 10.1371/journal.pone.0182212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhai S.L., Lv D.H., Wen X.H., Zhu X.L., Yang Y.Q., Chen Q.L., Wei W.K. Preliminary serological evidence for Schmallenberg virus infection in China. Trop. Anim. Health Prod. 2018;50:449–453. doi: 10.1007/s11250-017-1433-2. [DOI] [PubMed] [Google Scholar]
- 42.Rasekh M., Sarani A., Hashemi S.H. Detection of Schmallenberg virus antibody in equine population of Northern and Northeast of Iran. Vet. World. 2018;11:30–33. doi: 10.14202/vetworld.2018.30-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sailleau C., Boogaerts C., Meyrueix A., Laloy E., Breard E., Viarouge C., Desprat A., Vitour D., Doceul V., Boucher C., et al. Schmallenberg virus infection in dogs, France, 2012. Emerg. Infect. Dis. 2013;19:1896–1898. doi: 10.3201/eid1911.130464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poskin A., Van Campe W., Mostin L., Cay B., De Regge N. Experimental Schmallenberg virus infection of pigs. Vet. Microbiol. 2014;170:398–402. doi: 10.1016/j.vetmic.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 45.Wernike K., Eschbaumer M., Breithaupt A., Hoffmann B., Beer M. Schmallenberg virus challenge models in cattle: Infectious serum or culture-grown virus? Vet. Res. 2012;43:84. doi: 10.1186/1297-9716-43-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afonso A., Abrahantes J.C., Conraths F., Veldhuis A., Elbers A., Roberts H., Van der Stede Y., Méroc E., Gache K., Richardson J. The Schmallenberg virus epidemic in Europe-2011–2013. Prev. Vet. Med. 2014;116:391–403. doi: 10.1016/j.prevetmed.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 47.Helmer C., Eibach R., Tegtmeyer P.C., Humann-Ziehank E., Ganter M. Survey of Schmallenberg virus (SBV) infection in German goat flocks. Epidemiol. Infect. 2013;141:2335–2345. doi: 10.1017/S0950268813000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lievaart-Peterson K., Luttikholt S., Peperkamp K., Van den Brom R., Vellema P. Schmallenberg disease in sheep or goats: Past, present and future. Vet. Microbiol. 2015;181:147–153. doi: 10.1016/j.vetmic.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 49.Endalew A.D., Morozov I., Davis A.S., Gaudreault N.N., Wernike K., Bawa B., Ruder M.G., Drolet B.S., McVey D.S., Shivanna V., et al. Virological and Serological Responses of Sheep and Cattle to Experimental Schmallenberg Virus Infection. Vector Borne Zoonotic Dis. 2018;18:697–703. doi: 10.1089/vbz.2018.2297. [DOI] [PubMed] [Google Scholar]
- 50.Tsuda T., Yoshida K., Ohashi S., Yanase T., Yanase T., Sueyoshi M., Kamimura S., Misumi K., Hamana K., Sakamoto H., et al. Arthrogryposis, hydranencephaly and cerebellar hypoplasia syndrome in neonatal calves resulting from intrauterine infection with Aino virus. Vet. Res. 2004;35:531–538. doi: 10.1051/vetres:2004029. [DOI] [PubMed] [Google Scholar]
- 51.Herder V., Wohlsein P., Peters M., Hansmann F., Baumgartner W. Salient lesions in domestic ruminants infected with the emerging so-called Schmallenberg virus in Germany. Vet. Pathol. 2012;49:588–591. doi: 10.1177/0300985812447831. [DOI] [PubMed] [Google Scholar]
- 52.Doceul V., Lara E., Sailleau C., Belbis G., Richardson J., Breard E., Viarouge C., Dominguez M., Hendrikx P., Calavas D., et al. Epidemiology, molecular virology and diagnostics of Schmallenberg virus, an emerging orthobunyavirus in Europe. Vet. Res. 2013;44:31. doi: 10.1186/1297-9716-44-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wernike K., Holsteg M., Sasserath M., Beer M. Schmallenberg virus antibody development and decline in a naturally infected dairy cattle herd in Germany, 2011–2014. Vet. Microbiol. 2015;181:294–297. doi: 10.1016/j.vetmic.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 54.Claine F., Coupeau D., Wiggers L., Muylkens B., Kirschvink N. Modelling the evolution of Schmallenberg virus seroprevalence in a sheep flock after natural infection. Prev. Vet. Med. 2018;154:132–138. doi: 10.1016/j.prevetmed.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 55.Peperkamp N.H., Luttikholt S.J., Dijkman R., Vos J.H., Junker K., Greijdanus S., Roumen M.P., van Garderen E., Meertens N., van Maanen C., et al. Ovine and Bovine Congenital Abnormalities Associated with Intrauterine Infection with Schmallenberg Virus. Vet. Pathol. 2015;52:1057–1066. doi: 10.1177/0300985814560231. [DOI] [PubMed] [Google Scholar]
- 56.Van der Poel W.H.M. Diagnostics for Schmallenberg virus. Vet. Rec. 2012;171:294–295. doi: 10.1136/vr.e6278. [DOI] [PubMed] [Google Scholar]
- 57.De Regge N., van den Berg T., Georges L., Cay B. Diagnosis of Schmallenberg virus infection in malformed lambs and calves and first indications for virus clearance in the fetus. Vet. Microbiol. 2013;162:595–600. doi: 10.1016/j.vetmic.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 58.Breard E., Lara E., Comtet L., Viarouge C., Doceul V., Desprat A., Vitour D., Pozzi N., Cay A.B., De Regge N., et al. Validation of a commercially available indirect ELISA using a nucleocapside recombinant protein for detection of Schmallenberg virus antibodies. PLoS ONE. 2013;8:e53446. doi: 10.1371/journal.pone.0053446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Humphries D., Burr P. Schmallenberg virus milk antibody ELISA. Vet. Rec. 2012;171:511–512. doi: 10.1136/vr.e7739. [DOI] [PubMed] [Google Scholar]
- 60.Van der Poel W.H.M., Cay B., Zientara S., Steinbach F., Valarcher J.F., Botner A., Mars M.H., Hakze-van der Honing R., Schirrmeier H., Beer M. Limited interlaboratory comparison of Schmallenberg virus antibody detection in serum samples. Vet. Rec. 2014;174:380. doi: 10.1136/vr.102180. [DOI] [PubMed] [Google Scholar]
- 61.Tarlinton R., Daly J., Dunham S., Kydd J. The challenge of Schmallenberg virus emergence in Europe. Vet. J. 2012;194:10–18. doi: 10.1016/j.tvjl.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 62.Gerhauser I., Weigand M., Hahn K., Herder V., Wohlsein P., Habierski A., Varela M., Palmarini M., Baumgartner W. Lack of schmallenberg virus in ruminant brain tissues archived from 1961 to 2010 in Germany. J. Comp. Pathol. 2014;150:151–154. doi: 10.1016/j.jcpa.2013.11.210. [DOI] [PubMed] [Google Scholar]
- 63.Gache K., Touratier A., Bournez L., Zientara S., Bronner A., Dion F., Garin E., Calavas D. Detection of Schmallenberg virus in France since 2012. Vet. Rec. 2017;180:24. doi: 10.1136/vr.j38. [DOI] [PubMed] [Google Scholar]
- 64.Sohier C., Deblauwe I., Van Loo T., Hanon J.B., Cay A.B., De Regge N. Evidence of extensive renewed Schmallenberg virus circulation in Belgium during summer of 2016—Increase in arthrogryposis-hydranencephaly cases expected. Transbound. Emerg. Dis. 2017;64:1015–1019. doi: 10.1111/tbed.12655. [DOI] [PubMed] [Google Scholar]
- 65.Hechinger S., Wernike K., Beer M. Single immunization with an inactivated vaccine protects sheep from Schmallenberg virus infection. Vet. Res. 2014;45:79. doi: 10.1186/s13567-014-0079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wernike K., Nikolin V.M., Hechinger S., Hoffmann B., Beer M. Inactivated Schmallenberg virus prototype vaccines. Vaccine. 2013;31:3558–3563. doi: 10.1016/j.vaccine.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 67.Anonymous Merck Animal Health. [(accessed on 12 November 2019)]; Available online: http://wwwmerck-animal-healthcom/news/2013-5-21aspx.
- 68.Anonymous Merial. [(accessed on 12 November 2019)]; Available online: http://merialcom/en/press-releases/merial-receives-approval-for-new-vaccine-to-prevent-schmallenberg-disease-in-livestock/
- 69.Kraatz F., Wernike K., Hechinger S., König P., Granzow H., Reimann I., Beer M. Deletion Mutants of Schmallenberg Virus Are Avirulent and Protect from Virus Challenge. J. Virol. 2015;89:1825–1837. doi: 10.1128/JVI.02729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varela M., Pinto R.M., Caporale M., Piras I.M., Taggart A., Seehusen F., Hahn K., Janowicz A., de Souza W.M., Baumgärtner W., et al. Mutations in the Schmallenberg Virus Gc Glycoprotein Facilitate Cellular Protein Synthesis Shutoff and Restore Pathogenicity of NSs Deletion Mutants in Mice. J. Virol. 2016;90:5440–5450. doi: 10.1128/JVI.00424-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Endalew A., Faburay B., Trujillo J.D., Gaudreault N.N., Davis A.S., Shivanna V., Sunwoo S.Y., Ma W., Drolet B.S., McVey D.S., et al. Immunogenicity and Efficacy of Schmallenberg Virus Envelope Glycoprotein Subunit Vaccines. J. Vet. Sci. 2019 doi: 10.4142/jvs.2019.20.e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wernike K., Aebischer A., Roman-Sosa G., Beer M. The N-terminal domain of Schmallenberg virus envelope protein Gc is highly immunogenic and can provide protection from infection. Sci. Rep. 2017;7:42500. doi: 10.1038/srep42500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boshra H.Y., Charro D., Lorenzo G., Sánchez I., Lazaro B., Brun A., Abrescia N.G.A. DNA vaccination regimes against Schmallenberg virus infection in IFNAR-/- mice suggest two targets for immunization. Antivir. Res. 2017;141:107–115. doi: 10.1016/j.antiviral.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 74.Wernike K., Mundt A., Link E.K., Aebischer A., Schlotthauer F., Sutter G., Fux R., Beer M. N-terminal domain of Schmallenberg virus envelope protein Gc delivered by recombinant equine herpesvirus type 1 and modified vaccinia virus Ankara: Immunogenicity and protective efficacy in cattle. Vaccine. 2018;36:5116–5123. doi: 10.1016/j.vaccine.2018.07.047. [DOI] [PubMed] [Google Scholar]
- 75.Anonymous. Schmallenberg virus vaccine. Vet. Rec. 2015;177:321. [Google Scholar]
- 76.Kono R., Hirata M., Kaji M., Goto Y., Ikeda S., Yanase T., Kato T., Tanaka S., Tsutsui T., Imada T., et al. Bovine epizootic encephalomyelitis caused by Akabane virus in southern Japan. BMC Vet. Res. 2008;4:20. doi: 10.1186/1746-6148-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kirkland P.D. Akabane virus infection. Rev. Sci. Tech. 2015;34:403–410. doi: 10.20506/rst.34.2.2366. [DOI] [PubMed] [Google Scholar]
- 78.Sailleau C., Breard E., Viarouge C., Vitour D., Romey A., Garnier A., Fablet A., Lowenski S., Gorna K., Caignard G., et al. Re-Emergence of Bluetongue Virus Serotype 8 in France, 2015. Transbound. Emerg. Dis. 2017;64:998–1000. doi: 10.1111/tbed.12453. [DOI] [PubMed] [Google Scholar]
- 79.Meroc E., Poskin A., Van Loo H., Van Driessche E., Czaplicki G., Quinet C., Riocreux F., De Regge N., Caij B., van den Berg T., et al. Follow-up of the Schmallenberg Virus Seroprevalence in Belgian Cattle. Transbound. Emerg. Dis. 2015;62:e80–e84. doi: 10.1111/tbed.12202. [DOI] [PubMed] [Google Scholar]
- 80.Collins Á.B., Barrett D., Doherty M.L., Larska M., Mee J.F. Post-epidemic Schmallenberg virus circulation: Parallel bovine serological and Culicoides virological surveillance studies in Ireland. BMC Vet. Res. 2016;12:234. doi: 10.1186/s12917-016-0865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stokes J.E., Baylis M., Duncan J.S. A freedom from disease study: Schmallenberg virus in the south of England in 2015. Vet. Rec. 2016;179:435. doi: 10.1136/vr.103903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stavrou A., Daly J.M., Maddison B., Gough K., Tarlinton R. How is Europe positioned for a re-emergence of Schmallenberg virus? Vet. J. 2017;230:45–51. doi: 10.1016/j.tvjl.2017.04.009. [DOI] [PubMed] [Google Scholar]