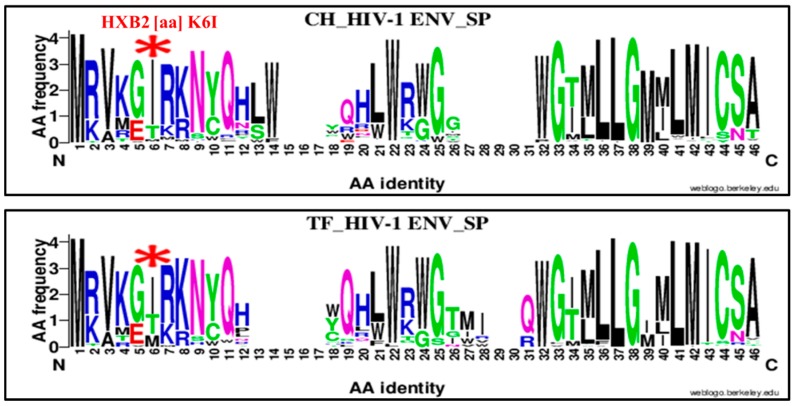
Figure 6.
Genetic signature identified under the HIV-1 envelope signal peptide (SP) associate to clade B HIV-1 chronic compared to TF viruses using WebLogo. The X axis represents sequences and amino acids (AA) identities composing the SP (direction N to C). The X axis represents amino acids (AA) composing the LLP-1 sequence (direction N to C). The Y axis represents the normalized AA frequency identified at each position of the SP sequence for each category of infection. The top line box represents chronic (CH) viruses envelope SP sequence (N = 105) and the bottom line for TF viruses (N = 98). As indicated for Weblogo analysis, the overall height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of symbols within the stack reflects the relative frequency of the corresponding amino or nucleic acid at that position. The isoleucine (I) amino acid signature was localized at position six of alignment (HXB2 position K6I) and identified by a red asterisk.