Abstract
Precise control of interferons (IFNs) is crucial to maintain immune homeostasis. Here, we demonstrated that homeodomain-interacting protein kinase 2 (HIPK2) was required for the production of type I IFNs in response to RNA virus infection. HIPK2 deficiency markedly impaired IFN production in macrophages after vesicular stomatitis virus (VSV) infection, and HIPK2-deficient mice were more susceptible to lethal VSV disease than were wild-type mice. After VSV infection, HIPK2 was cleaved by active caspases, which released a hyperactive, N-terminal fragment that translocated to the nucleus and further augmented antiviral responses. In part, HIPK2 interacted with ELF4 and promoted its phosphorylation at Ser369, which enabled Ifn-β transcription. In addition, HIPK2 production was stimulated by type I IFNs to further enhance antiviral immunity. These data suggest that the kinase activity and nuclear localization of HIPK2 are essential for the production of type I IFNs.
INTRODUCTION
Antiviral responses are triggered after viral recognition by host pattern recognition receptors (PRRs) of the innate immune system, including several Toll-like receptors (TLR3, TLR7/8, and TLR9) (1), RIG-I–like receptors (RLRs; RIG-I and MDA5) (2, 3), and cytosolic double-stranded DNA receptors (DAI, IFI16, DDX41, and cGAS) (4–7). RLRs recognize cytoplasmic viral RNA and recruit the adaptor protein MAVS (mitochondrial antiviral signaling protein, also called IPS-1, VISA, and Cardif) (8–10). MAVS localizes on mitochondria and peroxisomes and, after receiving danger signals from RLRs, recruits downstream molecules to form large aggregates, including TNF receptor–associated factor 2/3/6 and the kinases TANK-binding kinase 1 (TBK1) and IκB kinase α/β(11). The complex then triggers the activation of signaling events, leading to the transcription of nuclear factor κB (NF-κB) and interferon (IFN) regulatory factors (IRFs), such as IRF3 and IRF7 (12). These cytokines ultimately promote the transcription of IFNs, including type I IFNs (IFN-α and IFN-β), type II IFN (IFNg), and type III IFN (IFNλ) (13).
E26 transformation-specific (ETS)–related transcription factor Elf-4 (ELF4) is a transcription factor that belongs to the ETS family, which have an evolutionarily conserved ETS domain that binds to a consensus “GGA(A/T)” DNA sequence (14). ELF4 facilitates the transcription of type I IFNs and is critical for host defense (15). After virus infection, ELF4 is recruited by the adaptor proteins STING (stimulator of IFN genes protein) and MAVS, phosphorylated and activated by TBK1, and translocated into the nucleus (15). Through binding to ELF4-IRF or ELF4–NF-κB composite motifs in the type I IFN promoters, ELF4 cooperates with IRF3, IRF7, or p65 and initiates the transcription of type I IFNs. Molecular regulation of IFN gene expression is tightly controlled, and ETS proteins are a family of multifunctional transcription factors (16). There may be unknown mechanisms by which ELF4 participates in type I IFN production regulation.
Homeodomain-interacting protein kinase 2 (HIPK2) is a serine-threonine kinase that acts as a transcriptional regulator and corepressor for homeodomain transcription factors with pleiotropic functions (17). The protein is made up of multiple regions, an N-terminal kinase domain, a small ubiquitin-related modifier attachment site, a protein-protein interaction region, a homeobox-interacting domain, a speckle-retention signal (SRS) domain, and a C-terminal autoinhibitory domain (18). Two proline-glutamate-serine-threonine (PEST) motifs within the SRS domain are required for the subcellular localization of HIPK2 to nuclear bodies (17). In addition, cleavage of the C-terminal autoinhibitory domain, which contains a ubiquitination site, by caspases leads to full activation of HIPK2 (19). HIPK2 controls a wide spectrum of biological processes such as apoptosis, DNA damage response, angiogenesis, hypoxia, and cell proliferation and invasion by directly targeting p53, C-terminal binding protein, heterochromatin protein 1γ, vascular endothelial growth factor, and the RNA polymerase II–binding protein Che-1/AATF (18, 20–22). HIPK2 selectively phosphorylates p53 at Ser46, thus facilitating p53-dependent gene expression and subsequent apoptosis (23). p53 plays a dual role in innate antiviral immunity by both enforcing the type I IFN response and inducing cell apoptosis in response to viral infection (23). Loss of HIPK2 function has been implicated in several diseases, such as Alzheimer’s disease, cardiovascular disease, acute myeloid leukemia, and myelodysplastic syndrome (22, 24, 25). However, it remains unclear whether HIPK2 affects antiviral innate immunity.
In the present study, we established that HIPK2 is essential for the production of type I IFNs. Loss of HIPK2 reduced host defense to vesicular stomatitis virus (VSV) infection, which was dependent upon type I IFN production. Expression of HIPK2, which was stimulated by type I IFN itself, promoted the expression of IFN-β and IFN-stimulated genes (ISGs) in cooperation with IRF3 and IRF7. VSV infection induced caspase-dependent cleavage of HIPK2 and promoted HIPK2 nuclear translocation necessary for its antiviral activity. HIPK2 interacted with ELF4 and increased its phosphorylation at Ser369, an ELF4 site necessary for DNA binding and IFN-β transcription. These findings advance our understanding of the pathways that activate IFN expression critical for innate antiviral immunity.
RESULTS
HIPK2 is critical for antiviral immunity in vivo
ELF4 promotes innate immune responses and is critical for host defense (15). To further investigate how ELF4 is involved in antiviral responses, we performed a genome-wide RNA sequencing (RNA-seq) screen to identify ELF4-dependent genes during viral infection. We infected wild-type and Elf4−/− peritoneal macrophages with VSV, a commonly used virus for studying host-virus interactions and innate immunity, and then sequenced the RNAs from these cells. We selected the genes whose expression was substantially increased or decreased by VSV infection in Elf4−/− macrophages when compared to wild-type macrophages. Then, we analyzed this gene set in other datasets using Enrichr to reveal genes that were also induced by Kaposi’s sarcoma–associated herpesvirus and influenza H1N1 infection (26, 27). Among them, we found that HIPK2 was prominent (fig. S1A). HIPK2 is a conserved serine-threonine kinase that interacts with a variety of transcription factors and can function as both a corepressor and a coactivator (17) to direct essential cellular functions such as proliferation and apoptosis. The role of HIPK2 in antiviral host defense, however, has not been elucidated.
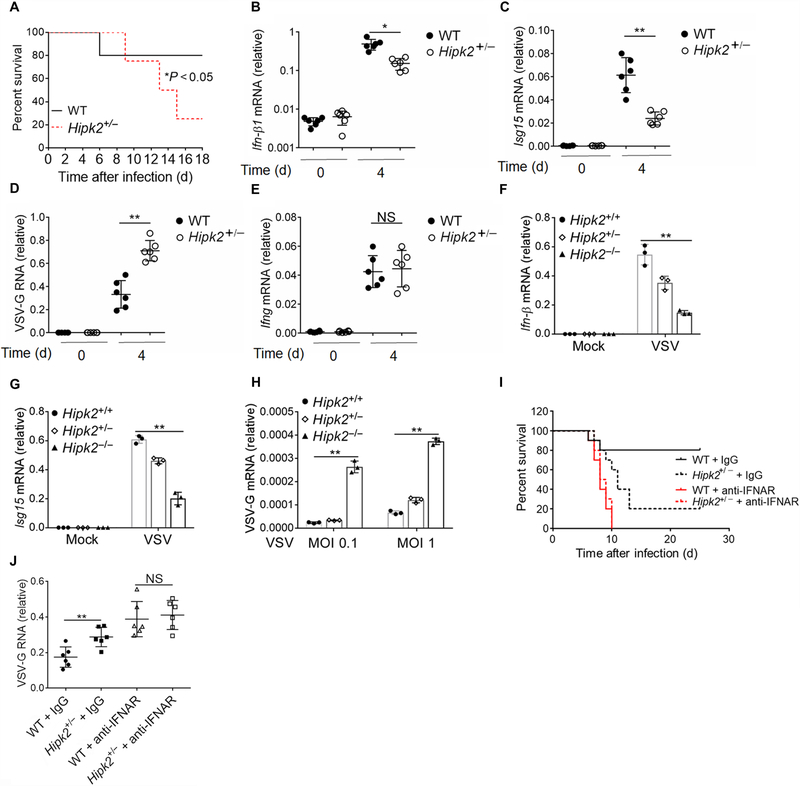
Hipk2−/− mice displayed an obvious defect in development (28). Their body size and body weight were much smaller than the age- and sex-matched wild-type mice (fig. S1, B and C). Because interpretations of any phenotypic results after infection can be misconstrued by the abnormalities of Hipk2−/− mice, we performed studies in Hipk2+/− mice, which grew normally (fig. S1, C and D). We confirmed that the abundance of Hipk2 was reduced in Hipk2+/− spleen and macrophages than in wild-type cells (fig. S1D). Hipk2+/− mice were highly susceptible to rapid, lethal infection with VSV (Fig. 1A). Because type I IFNs are primary antiviral effectors that are induced robustly after viral infection, we examined the expression of type I IFNs in the blood. We found that Hipk2+/− mice expressed less Ifn-β in response to VSV infection than wild-type mice (Fig. 1B). Consistent with this, the induction of Isg15 was impaired in Hipk2+/− mice (Fig. 1C). Hipk2+/− mice also exhibited markedly enhanced VSV replication in these animals compared with wild-type mice (Fig. 1D). In comparison, we found no defect in the induction of type II IFN (Ifng) in Hipk2+/− mice (Fig. 1E). In vitro, Hipk2−/− macrophages were more severely impaired than Hipk2+/− cells (Fig. 1, F and G). Consequently, quantitative real-time polymerase chain reaction (qRT-PCR) showed that Hipk2−/− cells lost the ability to effectively restrict VSV infection, in contrast to Hipk2+/− or wild-type macrophages (Fig. 1H). Moreover, we treated the mice with anti–IFN-α receptor (IFNAR) antibody and infected them with VSV. The wild-type and Hipk2+/− mice showed similar mortality and viremia after treatment of anti-IFNAR neutralizing antibody (Fig. 1, I and J). Collectively, these results suggest that HIPK2 is critical for the production of type I IFNs necessary for antiviral immunity.
Fig. 1. HIPK2 is critical for antiviral immunity in vivo.
(A) Survival analysis of age-and sex-matched wild-type (WT) and Hipk2+/− mice inoculated with VSV. Data are from a total of eight mice per group from three experiments. d, days. (B to E) qRT-PCR analysis of (B) Ifn-β, (C) Isg15, (D) VSV glycoprotein (VSV-G), and (E) Ifng mRNA expression in blood cells from mice at the indicated times after infection with VSV. Data with means ± SEM of six mice per group from three experiments. NS, not significant. (F to H) qRT-PCR analysis of (F) Ifn-β, (G) Isg15, or (H) VSV-G mRNA expression in wild-type, Hipk2+/−, or Hipk2−/− peritoneal macrophages infected with VSV for 24 hours. MOI, multiplicity of infection. (I) Survival analysis of age-and sex-matched wild-type and Hipk2+/− mice treated with antibody against IFNAR and infected with VSV. Data are from a total of 10 mice per group from three experiments. IgG, immunoglobulin G. (J) Viral load in the blood after wild-type and Hipk2+/− mice were treated with antibody against IFNAR and infected with VSV. Data with means ± SEM are from three experiments. *P < 0.05 and **P < 0.01 by log-rank Mantel-Cox test (A and I) or an unpaired two-tailed nonparametric Mann-Whitney U test (B to E and J).
HIPK2 is an ISG
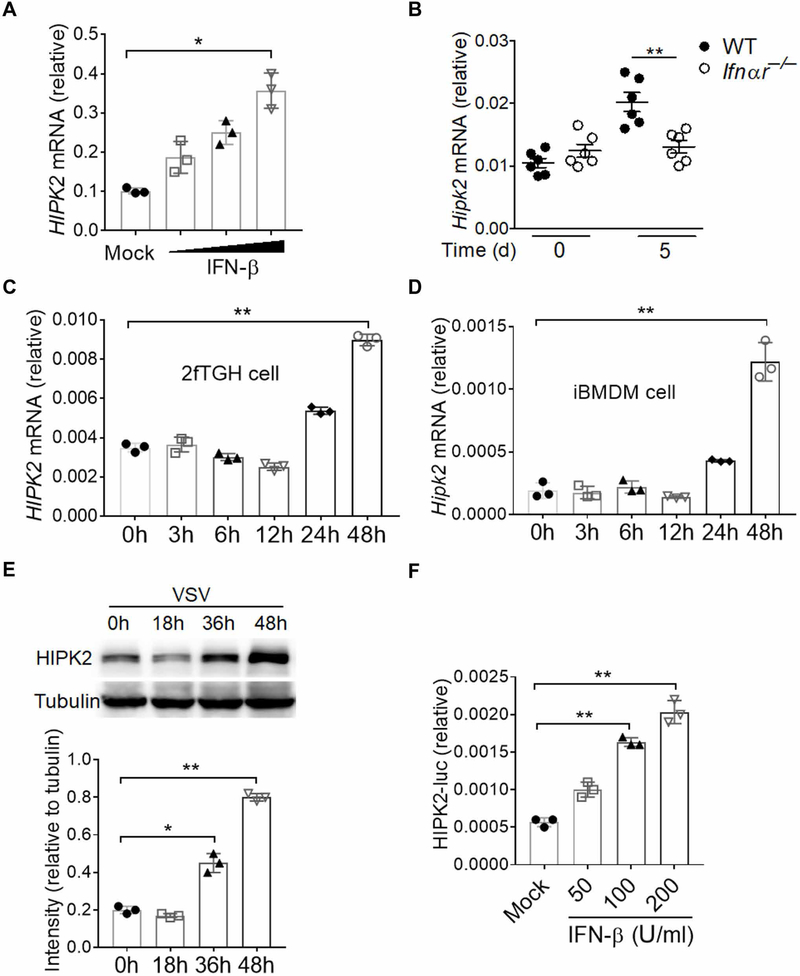
To sustain the expression of inflammation genes, type I IFNs can act in an autocrine loop by binding a heterodimeric complex of the IFNAR1 and IFNAR2 on the cell surface (29). IFNAR1/2 activation initiates a signaling cascade through the Janus kinase and signal transducer and activator of transcription pathway that induces the transcription of hundreds of ISGs (30, 31). Many antiviral effector molecules are ISGs, such as ELF4, STING, and DDX58 (15, 32, 33). We also observed that IFN treatment induced the expression of HIPK2, suggesting that HIPK2 may be an ISG (Fig. 2A). To determine whether these results extended to an additional RNA virus that commonly causes clinical disease, we infected mice with West Nile virus (WNV), a positive single-stranded RNA (ssRNA) virus that causes meningoencephalitis in humans (34). We found that HIPK2 mRNA was increased in the blood of wild-type mice but not in Ifnαr−/− mice that had been infected with WNV (Fig. 2B). We also found that HIPK2 expression was increased in 2fTGH cells and immortalized bone marrow–derived macrophages (iBMDMs) at later times after VSV infection (Fig. 2, C to E, and fig. S2A). These results indicate that viral infection augments HIPK2 expression. Furthermore, we constructed a HIPK2 promoter–driven reporter (fig. S2B) and found that the reporter was activated by type I IFN treatment of cells (Fig. 2F). Together, these results suggest that HIPK2 is an ISG stimulated by viral infection.
Fig. 2. HIPK2 is an ISG.
(A) qRT-PCR analysis of HIPK2 mRNA expression in 2fTGH cells treated with increasing amounts of human IFN-β for 24 hours. Data with means ± SEM are from three experiments. (B) qRT-PCR analysis of HIPK2 mRNA in whole blood from wild-type or Ifnαr−/− mice at the indicated times after infection with 200 plaque-forming units (PFU) of WNV. Data with means ± SEM are from three experiments. (C and D) qRT-PCR analysis of HIPK2 mRNA expression in 2fTGH cells (C) and iBMDMs (D) infected with VSV for indicated times. Data with means ± SEM are from three experiments. h, hours. (E) Western blot analysis of HIPK2 abundance in lysates of 2fTGH cells after infection with VSV for the indicated times. Blots (top) are representative of three experiments. Quantified band intensity values (bottom) with means ± SEM are from all experiments. (F) Luciferase assay analysis of HIPK2 promoter activity in HT1080 cells treated with increasing amounts of human IFN-β for 24 hours. Data with means ± SEM of three experiments. *P < 0.05 and **P < 0.01 by Student’s t test.
HIPK2 promotes type I IFN production
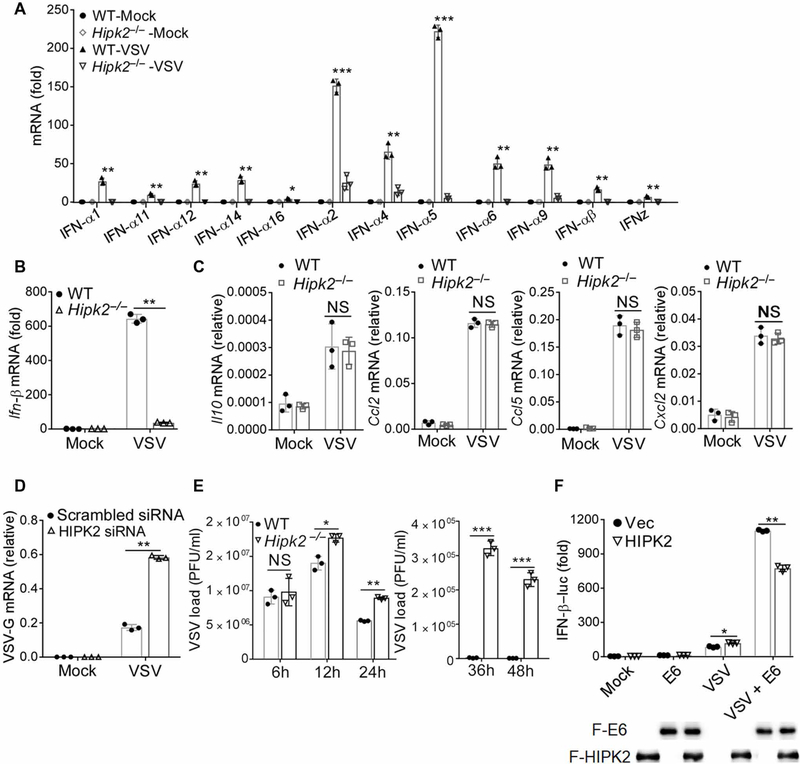
To gain further insight into the role of HIPK2 in antiviral immunity, we performed a genome-wide RNA-seq analysis on VSV-infected wild-type and Hipk2−/− iBMDMs (fig. S3, A and B). Consistent with the in vitro macrophage experiments, RNA-seq analysis showed that HIPK2 was required for type I IFN expression during VSV infection, including IFN-β and IFN-α (fig. S3A). In line with this, the qRT-PCR results showed that the induction of type I IFNs was significantly impaired in HIPK2-deficient cells during viral infection (Fig. 3, A and B). However, there was no substantial difference in the expression of interleukins or chemokines because of the loss of HIPK2 (Fig. 3C and fig. S3A). To further confirm that HIPK2 promotes the expression of type I IFNs, we overexpressed HIPK2 in human emborynic kidney– 293 (HEK293) cells that transiently expressed luciferase under the control of either the IFN-β or the IFN-α promoter. When these cells were infected with VSV, HIPK2 markedly increased the activation of these promoters (fig. S3, C and D). Sendai virus (SeV), which is another ssRNA virus (35), also stimulated HIPK2-dependent IFN-α promoter activation (fig. S3D). Consistently, knockdown of HIPK2 reduced IFN-β promoter activation after viral infection or double-stranded RNA analog poly(I:C) treatment (fig. S3, E and F) and increased VSV replication (Fig. 3D). When we detected VSV production over time in wild-type and Hipk2−/− iBMDMs, we found no difference in viral loads at the early times after infection (Fig. 3E). These data imply that HIPK2 is required for an essential antiviral activity instead of VSV replication. The viral proteins US11 of Herpes simplex virus 1 and E6 of human papillomavirus inhibit the activity of HIPK2 (36, 37). When we co-overexpressed US11 or E6 with HIPK2 after VSV infection, we found that the activation of IFN-β promoter was partially blocked (Fig. 3F and fig. S3G). Thus, our data further suggested that HIPK2 augments the expression of type I IFNs.
Fig. 3. HIPK2 promotes the expression of type I IFNs.
(A and B) qRT-PCR analysis of Ifnαs (A) and Ifn-β (B) mRNA expression 24 hours after VSV infection of wild-type and Hipk2−/− iBMDMs. Data with means ± SEM are from three experiments. (C) qRT-PCR analysis of Il10, Ccl2, Ccl5, and Cxcl2 mRNA expression in wild-type or Hipk2−/− iBMDMs after VSV infection for 6 hours. Data with means ± SEM are from three experiments. (D) qRT-PCR analysis of VSV-G mRNA expression in HEK293 cells transfected with scrambled control or HIPK2 small interfering RNA (siRNA) and infected with VSV for 24 hours. Data with means ± SEM are from three experiments. (E) Plaque assay analysis of viral production by wild-type or Hipk2−/− iBMDMs infected with VSV for indicated times. Data with means ± SEM are from three experiments. (F) Luciferase assay analysis of IFN-β promoter activity in HEK293 cells transfected with the indicated expression plasmids and then infected with VSV for 24 hours. Data (top) with means ± SEM are from three experiments. Blots confirming expression of the indicated proteins (lower) are representative of all experiments. *P < 0.05, **P < 0.01, and ***P < 0.001, by Student’s t test
HIPK2 is involved in antiviral immune signaling
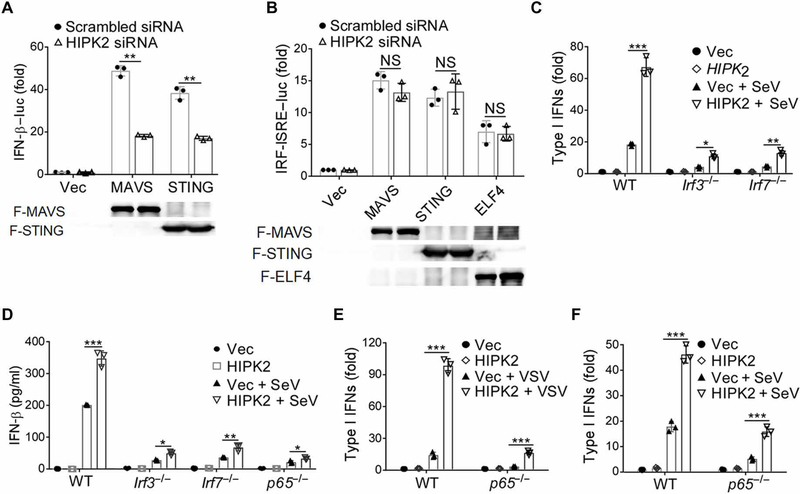
MAVS and STING are critical adaptor proteins of innate immune PRRs (12, 38). They link TLR activation and intracellular nucleic acid detection to TBK1, a key downstream kinase (13) that activates the transcription factors IRF3, IRF7, ELF4, and NF-κB involved in the transcription of type I IFNs (15, 39). When we overexpressed MAVS, STING, TBK1, or ELF4 in HEK293 cells, we found that increasing HIPK2 abundance enhanced IFN-β promoter activation (fig. S4, A and B). Moreover, knockdown of HIPK2 reduced MAVS- and STING-driven induction of IFN-β luciferase (Fig. 4A). These data suggest that HIPK2 may promote IFN-β expression downstream of these adaptor proteins.
Fig. 4. HIPK2 is involved in antiviral immune signaling.
(A and B) Luciferase assay analysis of IFN-β (A) or IRF-mediated ISRE (B) promoter activity in HEK293 cells transfected with scrambled control or HIPK2 siRNA and the indicated expression plasmids for 24 hours. Data (top) with means ± SEM are from three experiments. Blots confirming expression of the indicated proteins (lower) are representative of all experiments. (C) Bioassay of type I IFN production by wild-type, Irf3−/−, or Irf7−/− iBMDMs transfected with Vec control or HIPK2 expression plasmids and infected with SeV for 24 hours. Data with means ± SEM are from three experiments. (D) Enzyme-linked immunosorbent assay (ELISA) analysis of type I IFNs secreted from wild-type, Irf3−/−, Irf7−/−, and p65−/− iBMDMs transfected with Vec control or HIPK2 expression plasmids and infected with SeV for 6 hours. Data with means ± SEM are from three experiments. (E and F) Bioassay of IFN production by wild-type or p65−/− iBMDMs transfected with Vec or HIPK2 expression plasmids and infected with VSV (E) or SeV (F) for 6 hours. Data with means ± SEM are from three experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 by Student’s t test.
IRF3 and IRF7 are part of the transcriptional complexes that bind to specific nucleotide sequences called IFN-stimulated response elements (ISREs) in the IFN promoter area (40). When we examined whether HIPK2 promotes the activity of ISRE-luc, we found that HIPK2 had no effect on ISG factor 3 (ISGF3)–independent ISRE-luc activity stimulated by MAVS overexpression or VSV infection (fig. S4, C and D). Knockdown of HIPK2 also did not influence IRF-ISRE luc activity after overexpression of MAVS or STING (Fig. 4B). These results suggested that HIPK2 may exert its effects by stimulating IFN-β promoter regions outside of IRF-dependent ISRE sites. In addition, loss of HIPK2 did not reduce expression of IFN-inducible genes that harbor ISRE elements (ISGF3 dependent) after IFN-β treatment (fig. S4E). Together, these data suggest that HIPK2 was not directly involved in ISRE-mediated IFN-α/β responses. However, HIPK2-mediated production of type I IFNs after viral infection of macrophages required both IRF3 and IRF7 (Fig. 4, C and D, and fig. S4F). Although, in IRF3- or IRF7-deficient cells, expression of type I IFN was severely impaired, overexpression of HIPK2 still increased IFN production. These data suggest that HIPK2 cooperated with IRF3 and IRF7 to facilitate type I IFN production, although additional factors might be involved in this process.
The canonical NF-κB pathway, which activates the p50 and p65 subunits of NF-κB, also promotes the cellular response to IFN (41, 42). We observed that loss of p65 reduced the expression of type I IFNs stimulated after VSV infection, although, similar to loss of IRF3 or IRF7 when HIPK2 was overexpressed, IFNs were still produced (Fig. 4E and fig. S4F). Infection of wild-type and p65−/− macrophages that overexpressed HIPK2 with SeV also showed similar results (Fig. 4, D and F). These data suggest that HIPK2 may promote type I IFN production independently of canonical NF-κB activation.
HIPK2 activates the transcription of type I IFNs by interacting with ELF4
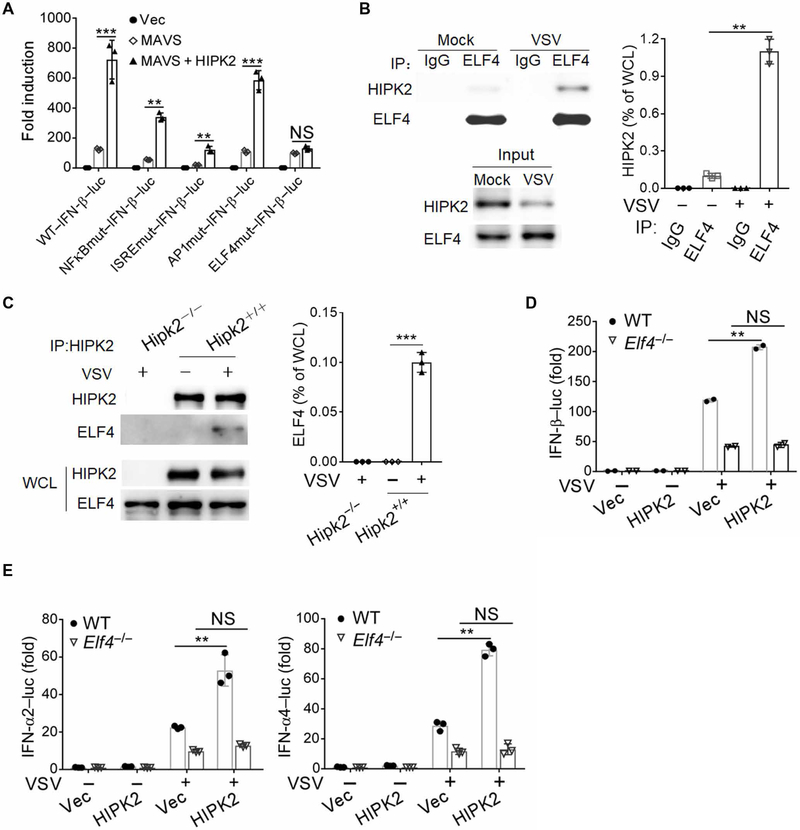
The collective data suggested that HIPK2 activates type I IFN production but may not directly target ISRE elements in the IFN-β promoter or require NF-κB p65 activity. To further elucidate how HIPK2 exerts its effects, we used promoter activity reporter assays to define the regions of the IFN-κ promoter that were required. The IFN-β promoter contains several transcription factor binding sites as follows: NF-κB, ISRE, AP1 (activator protein 1), YY1/2, and ELF4 binding sites (15, 43, 44). When we mutated specific sites in the IFN-β promoter, we found that HIPK2-driven IFN-β promoter activity stimulated by overexpression of MAVS in HEK293 cells required an intact ELF4 binding site (Fig. 5A). Coimmunoprecipitation indicated that HIPK2 interacted with ELF4 (Fig. 5, B and C). In agreement with these observations, HIPK2 enhanced promoter activation of IFN-β, IFN-α2, and IFN-α4 in wild-type but not in Elf4−/− HEK293 cells (Fig. 5, D and E, and fig. S4F). These results suggest that HIPK2 augments type I IFN production at least, in part, through binding to ELF4.
Fig. 5. HIPK2 activates the transcription of IFNs by interacting with ELF4.
(A) Luciferase assay analysis of wild-type and mutant IFN-β promoter activity in HEK293 cells transfected with the indicated expression plasmids for 24 hours. Data with means ± SEM are from three experiments. (B) Co-immunoprecipitation analysis of ELF4 interaction with HIPK2 in lysates of HEK293 cells infected with VSV for 6 hours. Blots (left) are representative of three experiments. Quantified HIPK2 band intensity values (right) normalized to whole-cell lysate (WCL) are pooled from all experiments. IP, immunoprecipitation. (C) Coimmunoprecipitation analysis of HIPK2 interaction with ELF4 in lysates of wild-type and Hipk2−/− iBMDMs infected with VSV for 6 hours. Blots (left) are representative of 3 experiments. Quantified HIPK2 band intensity values (right) normalized to WCL are pooled from all experiments. (D and E) Luciferase assay analysis of IFN-β (D) or IFN-α2–luc and IFN-α4–luc (E) promoter activity in wild-type or ELF4−/− HEK293 cells transfected with Vec control or HIPK2 expression plasmids and infected with VSV for 24 hours. Data with means ± SEM are from three experiments. **P < 0.01 and ***P < 0.001 by Student’s t test.
HIPK2 translocates to nucleus and is cleaved after virus infection
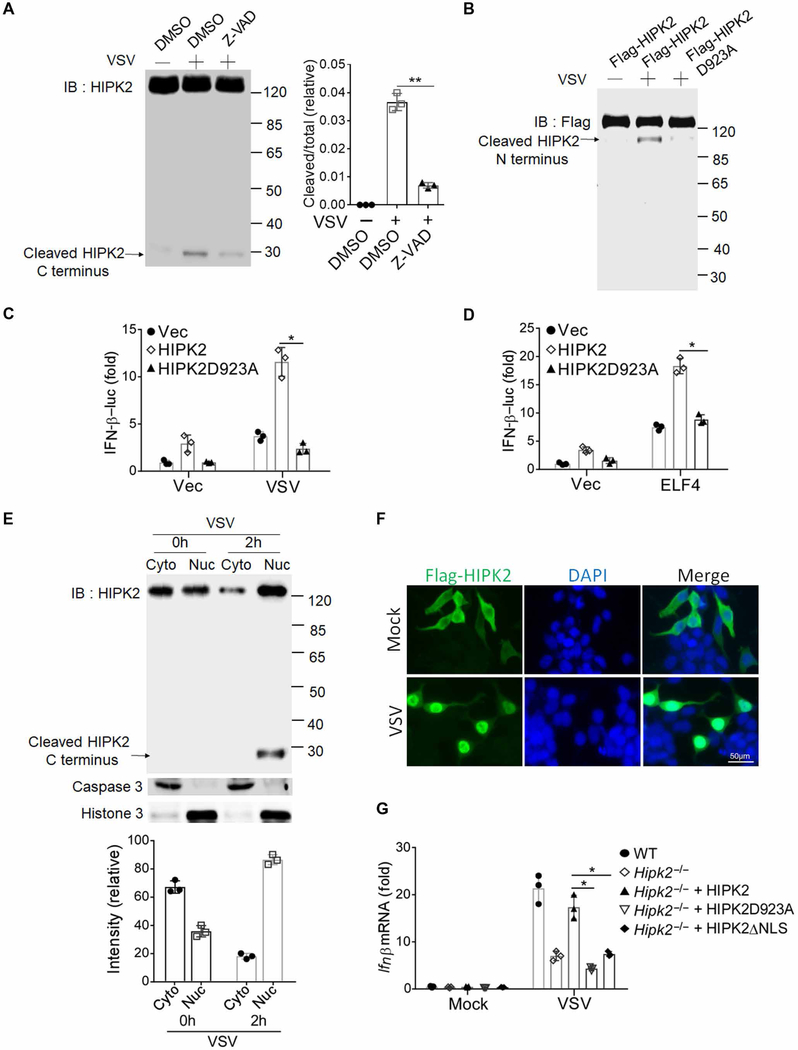
Caspase-6 cleavage of HIPK2 at Asp923 removes an autoinhibitory domain in the C terminus of HIPK2, which augments HIPK2 activity and rapidly amplifies the apoptotic response (19). When we investigated whether viral infection stimulated HIPK2 cleavage by Western blot, we found that VSV infection stimulated cleavage of HIPK2, which was blocked by the pan-caspase inhibitor Z-VAD (Fig. 6A). Furthermore, when we infected HEK293 cells that overexpressed wild-type HIPK2 or the HIPK2 D923A mutant that was N-terminal Flag-tagged with VSV, we found that mutation of this site abrogated HIPK2 cleavage after viral infection (Fig. 6B). In addition, in cells that expressed this mutated HIPK2, VSV infection did not augment IFN-β promoter activation (Fig. 6, C and D). Overexpression of the N-terminal fragment of HIPK2 (HIPK2-NT) exhibited higher antiviral activity than the full-length HIPK2 (fig. S5, A to C). These data suggest that cleavage of HIPK2 was required for its effects on IFN production.
Fig. 6. HIPK2 translocates to nucleus and is cleaved after virus infection.
(A) Western blot analysis of HIPK2 cleavage in lysates of HEK293 cells pretreated with dimethyl sulfoxide (DMSO) or caspase inhibitor Z-VAD for 2 hours and infected with VSV for 2 hours. Blots (left) are representative of three experiments. Quantified band intensity values (right) are pooled from all experiments. IB, immunoblotting. (B) Western blot analysis of HIPK2 cleavage in lysates of HEK293 cells transfected with wild-type or D923A mutant HIPK2 and infected with VSV for 2 hours. Blots are representative of three experiments. (C and D) Luciferase assay analysis of IFN-β promoter activity in HEK293 cells transfected with the indicated expression plasmids 24 hours after infection with VSV (C) or transfection with ELF4 (D). Data with means ± SEM are from three experiments. (E) Western blot analysis of the indicated proteins in nuclear and cytoplasmic fractionations of lysates from MEF cells infected with VSV for 2 hours. Blots (top) are representative of three experiments. Quantified HIPK2 band intensity values (right) are pooled from all experiments. (F) Confocal microscopy analysis of HIPK2 localization in Hela cells transfected with Flag-HIPK2 and infected with VSV for 4 hours. Images are representative of three experiments. DAPI, 4′,6-diamidino-2-phenylindole. (G) qRT-PCR analysis of IFN-β mRNA expression in wild-type or Hipk2−/− MEFs reconstituted with full-length HIPK2, HIPK2-D923A, or HIPK2△NLS (deletion of nuclear localization sequence) and infected with VSV for 24 hours. Data with means ± SEM are from three experiments. *P < 0.05 and **P < 0.01 by Student’s t test.
Nuclear fractionation experiments indicated that VSV infection increased HIPK2 nuclear translocation, including its cleaved form (Fig. 6E). By immunofluorescence, we also observed that VSV infection promotes nuclear accumulation of HIPK2 (Fig. 6F). When we deleted the nuclear localization sequence of HIPK2 (HIPK2△NLS) and overexpressed it in Hipk2−/− mouse embryonic fibroblast (MEF) cells, we found that only the full-length HIPK2 could rescue IFN-β production, but not HIPK2△NLS (Fig. 6G and fig. S5D). Moreover, VSV infection stimulated cleavage of wild-type HIPK2 but not HIPK2△NLS, which may suggest that cleavage could occur in the nucleus (fig. S5E). In contrast, cleavage did not prevent VSV-induced HIPK2 nuclear translocation because the HIPK2 D923A mutant was also found in nuclear fractions of transfected HEK293 cells after VSV infection (fig. S5F). These data indicate that viral infection stimulated nuclear localization of HIPK2, which was required for its cleavage necessary to promote IFN expression.
HIPK2 promotes phosphorylation of ELF4 at Ser369 and contributes to its binding to type I IFN promoters
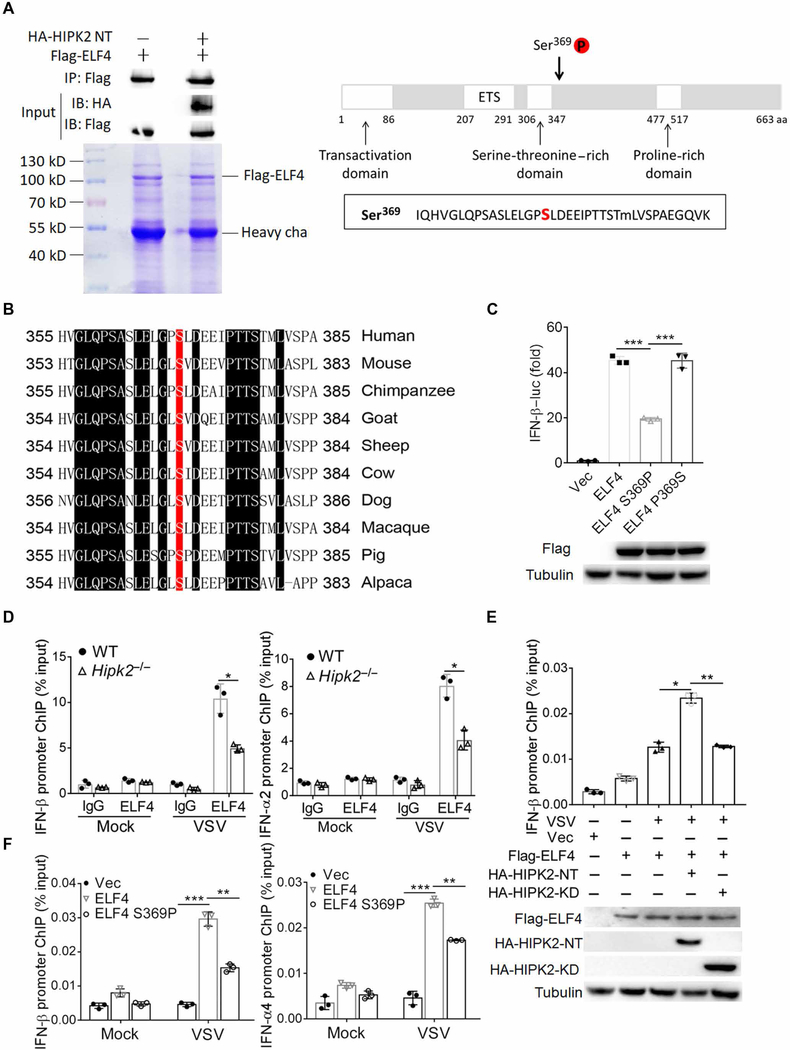
Because HIPK2 is a serine-threonine kinase, the above results raise a fundamental question as to the function of HIPK2—that is, is the kinase activity required for HIPK2-mediated type I IFN production? To address this, we constructed a kinase-defective mutant K221R and the N-terminal deletion HIPK2-KD, which lacks the kinase domain (192 to 520) (45). Wild-type HIPK2 enhanced VSV-induced IFN-β and IFN-α4 promoter activation, whereas the kinase-negative mutant K221R and HIPK2-KD had no effect, indicating that the enzymatic activity of HIPK2 is essential for its antiviral activity (fig. S6, A and B). To further examine whether HIPK2 could promote phosphorylation of ELF4, we immunoprecipitated ELF4 from lysate of HEK293 cells that overexpressed Flag-tagged ELF4 with or without the N-terminal fragment of HIPK2. After SDS–polyacrylamide gel electrophoresis (SDS-PAGE), ELF4 protein modification was analyzed by mass spectrometry. We found that when HIPK2-NT was coexpressed with ELF4, it was phosphorylated at Ser369 (Fig. 7A). Thus, HIKP2 can promote ELF4 phosphorylation at Ser369.
Fig. 7. HIPK2 phosphorylates ELF4 at Ser369and contributes to its binding to type I IFN promoters.
(A) Coimmunoprecipitation (top) and SDS-PAGE (bottom) analysis of lysates from HEK293 cells transfected with Flag-ELF4 and HA-HIPK2 plasmid. ELF4 bands were analyzed for protein modification by mass spectrometry (right). Data are representative of three experiments. aa, amino acid. (B) Alignment of ELF4 proteins from human and nonhuman species. (C) Luciferase assay analysis of IFN-β promoter activity in HEK293 cells transfected with IFN-β luc and the indicated expression plasmids after 24 hours. Data (top) with means ± SEM are from three experiments. Blots confirming expression of the indicated proteins (lower) are representative of all experiments. (D) ChIP analysis of IFN-β or IFN-α2 promoter DNA associated with ELF4 in wild-type or Hipk2−/− peritoneal macro-phages infected with VSV for 6 hours. Data with means ± SEM are from three experiments. (E) ChIP analysis of IFN-β promoter DNA associated with Flag-HIPK2 in HEK293 cells transfected with indicated plasmids and infected with VSV for 6 hours. Data (top) with means ± SEM are from three experiments. Blots confirming expression of the indicated proteins (lower) are representative of all experiments. (F) ChIP analysis of IFN-β or IFN-α4 promoter DNA associated with Flag-HIPK2 in ELF4−/− HEK293 cells transfected with indicated plasmids and infected with VSV for 6 hours. Data with means ± SEM are from three experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 by Student’s t test.
When we aligned protein sequences of ELF4 from human and several nonhuman species, we noted that the Ser369 residue is highly conserved (Fig. 7B). In X-linked hypogammaglobulinemia with growth hormone deficiency, a point mutation alters this residue from Ser to Pro, which could interfere with ELF4 interaction with other proteins or its ability to bind to DNA (46). To determine the importance of this residue in HIPK2-ELF4–mediated IFN production, we constructed the ELF4 mutant by replacing this serine with proline (S369P). This mutation impaired IFN-β promoter activation compared to wild-type ELF4 (Fig. 7C). Reversing this mutation (P369S) rescued expression of IFN-β (Fig. 7C). These data indicate that HIPK2 promotes ELF4 phosphorylation at a site that is critical for activating IFN-β expression.
ELF4 promotes type I IFN transcription by binding to IFN gene promoters through its conserved ETS domain (15). To determine how modification of ELF4 at Ser369 may alter association of ELF4 and type I IFN promoter DNA, we performed chromatin immunoprecipitation (ChIP) assay on VSV-infected wild-type and Hipk2−/− peritoneal macrophages. In the absence of HIPK2, less ELF4 bound to IFN-β and IFN-α2 promoters (Fig. 7D). In addition, the association between ELF4 and IFN-β promoter was significantly enhanced by overexpression of hyperactive HIPK2-NT but not the kinase-dead HIPK2 mutant (Fig. 7E). These data suggested that enzymatic activity of HIPK2 was required for ELF4-mediated IFN transcription initiation complex formation. We also reconstituted ELF4−/− cells with wild-type or S369P mutant ELF4, performed ChIP assay, and found that the S369P mutation reduced binding to the IFN-β promoter (Fig. 7F). Together, our results demonstrate that HIPK2 augments type I IFN expression by interacting with and promoting phosphorylation of ELF4, which was crucial for association between ELF4 and IFN promoters.
DISCUSSION
Our results demonstrated that HIPK2 enhanced type I IFN expression, at least, in part, through interaction with ELF4. Loss of HIPK2 decreased IFN-β expression, which augmented VSV viral replication and rendered mice highly susceptible to VSV infection. HIPK2 interacted directly with ELF4 (Fig. 5, B and C), and ChIP assays indicated that HIPK2 was required for the interaction between ELF4 and the IFN promoter (Fig. 7D). Therefore, our data indicate that HIPK2 facilitates type I IFN induction in an ELF4-dependent fashion (fig. S7). NF-κB and IRF3/IRF7 are key transcription factors for antiviral cytokine production, including type I IFNs, and ELF4 cooperates with NF-κB and IRF3/IRF7 to activate antiviral gene transcription (15). In our study, although the enhancement of IFN by HIPK2 was impaired in p65−/−, Irf3−/−, or Irf7−/−cells (Fig. 4, C to F), we also observed that HIPK2 stimulated IFN production independently of these transcription factors. In contrast, our data indicate that HIPK2 has no function in the absence of ELF4 (Fig. 5, D and E). These data suggest that, although HIPK2 may cooperate with NF-κB and IRF3/IRF7 to initiate antiviral gene transcription, only its interaction with ELF4 is required. Consistent with this conclusion, IRF-dependent ISRE activation was intact in HIPK2 knockdown cells (Fig 4B). Similarly, ELF4 activated after viral infection is not required for IRF activation or phosphorylation and dimerization of IRF3 (15). Therefore, the HIPK2-ELF4 axis required for type I IFN induction may act in parallel with IRF3.
Our data indicate that HIPK2 promotes phosphorylation of ELF4 at Ser369 and that this residue is required for ELF4-mediated antiviral IFN production (Fig 7, A, C, and F). Phosphorylation at this site is distinct from TBK1-mediated phosphorylation of ELF4 at Ser331, which is essential for its nuclear translocation and indispensable for type I IFN induction (15). Thus, TBK1-mediated phosphorylation of ELF4 promotes its nuclear translocation, which may then be phosphorylated by active cleaved HIPK2 in the nucleus to stimulate ELF4 DNA binding. Phosphorylation of ELF4 at Ser369 also occurs downstream of ataxia-telangiectasia mutated kinase activation in response to DNA damage at its consensus kinase motif S/TQ sites Thr70, Ser369, and Ser472, which leads to ELF4 degradation (47). In our study, we found that N-terminal–activated HIPK2 fragment promotes phosphorylation of ELF4 at Ser369 but not Thr70 and Ser472 (Fig. 7A). Because we did not observe ELF4 degradation, it may be that these other two phosphorylation sites play a more important role in ELF4 degradation. We identified that Ser369 is required for the interaction between ELF4 and the IFN promoters (Fig. 7, C and F). Both loss of HIPK2 and the S369P mutation of ELF4 decreased ELF4 binding to type I IFN promoters, which is critical for virus-induced type I IFN expression (Fig. 7, D and F). However, it remains unclear whether HIPK2 directly phosphorylates ELF4. Improper IFN function is closely associated with many immunodeficiency diseases. Our results suggest a possible pathological mechanism of ELF4-related clinical disorders.
The activity of HIPK2 is promoted or inhibited by multiple posttranslational modifications that may occur in the context of viral infection. Sumoylated HIPK2 is directed to nuclear speckles (48). Ubiquitinoylation leads to rapid proteasome-dependent degradation of HIPK2 (49). In addition, caspase-dependent cleavage of HIPK2 removed autoregulatory domains and potentiates its apoptotic function (50). Viral infection can trigger apoptosis, which is one of the main host defense responses to eliminate invading pathogens, and it is intriguing to speculate that HIPK2 may also be involved in that process (51). We here found that HIPK2 was cleaved by caspase after viral infection (Fig. 6, A, B, and E), and subsequently, the N-terminal hyperactive form of HIPK2 was released to facilitate IFN-β induction (fig. S5, A to C). Our results are consistent with the literature on the importance of posttranslational modification in determining HIPK2 activity.
HIPK2 is known as a “caretaker” gene, which plays an important role in tumorigenesis. Its inactivation increases tumorigenicity, whereas its activation inhibits tumor growth. HIPK2 interacts with and phosphorylates the transcription factor p53 (39), facilitating p53-dependent gene expression and subsequent apoptosis to suppress tumor growth (51). In addition to apoptosis, p53 plays a role in innate antiviral immunity by directing transcriptional up-regulation of several innate immune genes, such as TLR3, IRF9, IRF5, and ISG15, influencing the type I IFN pathway (23). Similarly, DNA damage is also able to trigger the induction of type I IFNs (52). Although ELF4 inhibits the expression of p53 and modulates the cell cycle entry of hematopoietic stem cell (53), it may be interesting to explore whether the HIPK2-p53 axis could modulate the production of type I IFNs. In addition, via phosphorylation and degradation of β-catenin, HIPK2 acts as a negative regulator of the Wnt/β-catenin pathway (54). HIPK2 is also an important transcriptional cofactor that limits the bone morphogenetic protein signaling pathway and postnatal development of enteric dopaminergic neurons (55). Thus, HIPK2 may have direct and indirect roles in the regulation of IFN signaling that remain to be characterized.
Improper HIPK2 function has been implicated in the pathology of various diseases, such as cardiac dysfunction, Alzheimer’s disease, acute myeloid leukemia, and myelodysplastic syndrome (24, 52, 56). The promyelocytic leukemia (PML) protein is associated with acute PML (57), and HIPK2 is recruited to PML nuclear bodies in response to genotoxic stress (58). It is believed that HIPK2 plays a role in the process of autoimmune diseases. Type I IFNs are important modulators in the maintenance of immune homeostasis. Dysregulation of type I IFNs could result in inflammatory disease, autoimmune disease, and tumor. Therefore, further work could focus on clarifying the mechanism by which HIPK2 bridges the type I IFN expression and pathogenesis of related diseases.
Thus, HIPK2 is a multifunctional protein. Our results demonstrate that HIPK2 has an important role in immune homeostasis. Exploiting or enhancing HIPK2-mediated signaling may lead to the development of important new targets for the treatment of infectious diseases.
MATERIALS AND METHODS
Mice
All animals care and use adhered to the Guide for the Care and Use of Laboratory Animals of the Chinese Association for Laboratory Animal Science. All procedures of animal handling were approved by the Animal Care Committee of Peking University Health Science Center (permit number LA 2016240). The Hipk2+/− mice were a gift from E. J. Huang (University of California, San Francisco). Hipk2+/− mice were bred with Hipk2+/− mice to generate Hipk2−/− mice. Ifnαr−/− mice were a gift from R. Medzhitov (Yale University). Wild-type B6 mice were purchased from the Department of Laboratory Animal Science of Peking University Health Science Center, Beijing. Mice were kept and bred in pathogen-free conditions. All animal results were included, and no method of randomization was applied. Eight-week-old mice were infected with VSV with 1 × 107 PFU of viruses per mouse by intravenous injection. The body weight of the mice was monitored accordingly. For cytokine studies, blood was collected at days 0 and 4 after infection.
Cells
HEK293, HeLa, 2fTGH-ISRE (human fibrosarcoma cells expressing an ISRE), L929-ISRE (mouse fibrosarcoma cells expressing an ISRE), HT1080 (human fibrosarcoma cells), MEF, and Vero cells were purchased from American Type Culture Collection. iBMDM cells were provided by Z. Jiang (Peking University). Isolation of BMDMs and peritoneal macrophages was performed as described. Heparinized human blood was obtained with written informed consent from healthy donors. Primary macrophages were derived from peripheral blood monocytes and infected with WNV as described. Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin-streptomycin (100 U/ml). Cells were negative for mycoplasma.
CRISPR-Cas9 system
Elf4−/− HEK293 cells, Hipk2−/− MEF cells, and Hipk2−/−, Irf3−/−, Irf7−/−, p65−/− iBMDMs were generated by CRISPR-Cas9 knockout. Specific guide RNAs were ligated into the BsmB1 restriction site of the inducible lentiviral vector (lentiGuide-Puro). Lentivirus particles were produced by cotransfected HEK293 cells with guide RNA plasmids (2 μg), packaging plasmids pVSVg (800 ng, AddGene 8454), and psPAX2 (800 ng, AddGene 12260). At 24 hours after transfection, the medium was changed, and viral supernatant was collected at 48 to 72 hours. A total of 1 × 105 iBMDMs or MEF cells were infected with 2 ml of viral supernatant supplemented with polybrene (8 μg/ml) and incubated for 48 hours. Knockout cells were selected by puromycin (1 μg), and each monoclone was confirmed by sequencing. Cells were negative for mycoplasma. The specific guide RNA sequences are listed in table S1.
Viruses, antibodies, and reagents
VSV (Indiana strain) was gift from J. Rose (Yale University) and SeV (Cantell strain; VR-907) was purchased from the American Type Culture Collection. WNV (CT-2741) was isolated from naturally infected wild birds and was passaged in vitro once in Vero cells (African green monkey kidney cells). Cells were infected with indicated viruses at a multiplicity of infection of 0.1 to 1 PFU. The following antibodies were used: anti-HIPK2 (sc-100383, Santa Cruz Biotechnology), anti-ELF4 (AV38028, Sigma-Aldrich), anti-Flag (F3165, Sigma-Aldrich), anti-IFNAR (clone MAR1–5A3, Leinco Technologies), anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (G8795, Sigma-Aldrich), and anti–caspase 3 (sc-113427, Santa Cruz Biotechnology). Poly(I:C) (tlr-pic) and Z-VAD (HY16658) were from InvivoGen. Human IFN-β (114201) was from peripheral blood leukocyte IFN. HIPK2 antibody is a mouse monoclonal antibody raised against amino acid 9211065 in the C-terminal tail.
Plasmids
Pcmv5-human ELF4 has been described previously (15). The full length HIPK2 was subcloned to Pcmv7.1 and Pcmv3xHA vectors. The truncated HIPK2-NT fragment (1923 aa), HIPK2△NLS (795828 aa), and all point mutants of HIPK2 were generated by QuikChange site-directed mutagenesis kits (Stratagene) using the construct encoding wild-type protein as the template. Each mutation was confirmed by sequencing. The plasmids coding human STING, MAVS, and TBK1 have been described previously (59). For reporter assays, pISRE-luc (Stratagene) was used. Other promoters were cloned into pGL3-Basic Vector (Promega) with the following promoter regions: −1 to −155 for mouse IFN-β, −1 to −957 for mouse IFN-α1, −1 to 855 for mouse IFN-α2, −1 to −476 for mouse IFN-α4, and −1 to −1205 for mouse IFN-α8. All constructs were verified by sequencing. Point mutants of IFN-β reporter were generated by QuikChange site-directed mutagenesis kits (200518, Stratagene) and the construct encoding wild-type IFN-β–luc as the template. Each mutation was confirmed by sequencing.
RNA interference
HIPK2 siRNA (SASI_Hs02_00325683) and scrambled siRNA (SASI_ Hs01_00224995) were from Sigma-Aldrich. For siRNA treatment, cells (30 to 50% confluent) were transfected with 20 nM siRNA using Lipofectamine 2000 for 24 hours.
Luciferase promoter reporter activity assays
HEK293 cells seeded on 24-well plates were transiently transfected with 50 ng of the luciferase reporter plasmid together with a total of 600 ng of various expression plasmids or empty control plasmids 24 hours before any treatment. As an internal control, 10 ng of pRL-TK was transfected simultaneously. After the indicated time or treatment, the luciferase activity in the total cell lysate was measured.
Type I IFN bioassay
Relative amounts of biologically active IFN in sera were quantified using a 2fGTH-ISRE (human derived) or L929-ISRE (mouse derived) cell line stably expressing an ISRE-luc reporter. Briefly, 200 μl of culture medium was incubated with confluent 2fGTH-ISRE or L929-ISRE cells (24-well plate) for 6 hours. Cells were lysed in passive lysis buffer and subjected to luciferase quantification (Promega). A serial dilution of human IFN-β was included as standards.
Enzyme-linked immunosorbent assay
The ELISA was conducted using the ELISA Kit for IFN-β (SEA222Mu, Cloud-Clone) according to the manufacturer’s protocol.
Western blotting
Immunoblotting was carried out by standard procedures. Briefly, samples eluted with 4× loading buffer were boiled at 100°C for 5 min and then loaded to gels (SDS-PAGE). After electrophoresis, proteins were transferred to a 0.22-um nitrocellulose membrane (PALL). Membranes were probed with antibodies using standard techniques and detected by enhanced chemiluminescence. The antibodies were diluted 1000 times for immunoblots. Western blotting data were quantified using ImageJ software. Normalized band intensity values were the ratio of bands intensity to corresponding internal reference. For Western blotting–involved cleavage, normalized band intensity values were the ratio of the abundance of the cleaved and uncleaved form to corresponding internal reference.
Coimmunoprecipitation
Cells were lysed in lysis buffer [0.5% Triton X-100, 20 mM Hepes (pH 7.4), 150 mM NaCl, 12.5 mM β-glycerolphosphate, 1.5 mM MgCl2, 2 mM EGTA, 10 mM NaF, 1 mM Na3VO4, and 2 mM dithiothreitol (DTT)] containing protease inhibitors. Lysates were centrifuged and incubated with indicated antibodies at 4°C overnight. The next day, prewashed protein A/G beads (Pierce) were added and incubated at 4°C for 4 hours. The beads were washed with cold phosphate-buffered saline (PBS) four times and eluted with DTT-containing SDS sample buffer by boiling for 10 min before immunoblotting.
Immunofluorescence microscopy
Treated Hela cells on coverslips were washed once with prewarmed PBS and fixed in 4% (w/v) paraformaldehyde for 15 min. After three washes in PBS, cells were permeabilized with 0.2% (v/v) Triton X-100 for 5 min. After three washes in PBS, cells were blocked in PBS containing 5% (w/v) bovine serum albumin (BSA) for 30 min and incubated with indicated antibodies in PBS containing 3% (w/v) BSA for 2 hours at 37°C. After three washes, cells were incubated with Alexa Fluor 488–conjugated secondary antibodies for 1 hour at 37°C and then with 4′,6-diamidino-2-phenylindole (Roche) for 15 min. The coverslips were washed extensively and mounted onto slides. Imaging of the cells was carried out using an Olympus BX51 microscope confocal system under a 40× objective.
Chromatin immunoprecipitation
ChIP was conducted using the Magna Chip A/G Chromatin immunoprecipitation Kit (Millipore). Ifn-β1 and Ifn-α2 promoter primers were described (15). ChIP samples were analyzed by qRT-PCR with the SYBR-Green Master Mix System (Transgene).
Nuclear-cytoplasmic fractionation
Nuclear-cytoplasmic fractionation was conducted using the NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Quantitative real-time PCR
Total RNA was isolated from cells using the RNeasy RNA Extraction Kit (Qiagen), and cDNA synthesis was performed using 1 ug of total RNA (iScript cDNA Synthesis kit). Expression of the indicated genes was analyzed by qRT-PCR amplified using SYBR Green (Transgene). Data shown are the relative abundance of the indicated mRNA normalized to Gapdh. The specific primers are listed in table S2.
Plaque assay
Viral titers from the cell culture medium were determined by plaque-forming assays, as previously described (15). Briefly, virus-containing medium was serially diluted and then added to confluent Vero cells. After 1-hour incubation, supernatants were removed, cells were washed with PBS, and culture medium containing 2% (w/v) methylcellulose was overlaid for 24 hours. Then, cells were fixed for 30 min with 0.5% (v/v) glutaraldehyde and then stained with 1% (w/v) crystal violet dissolved in 70% ethanol for 30 min. After washing twice with double-distilled water, plaques were counted, and average counts were multiplied by the dilution factor to determine the viral titer as PFU per milliliter.
RNA sequencing
Cells were infected with VSV for 6 hours, and total RNA was purified using the RNeasy Mini Kit (Qiagen no. 74104). The transcriptome library for sequencing was generated using the VAHTSTM mRNA-seq v2 Library Prep Kit for Illumina (Vazyme Biotech Co. Ltd., Nanjing, China) following the manufacturer’s recommendations. After clustering, the libraries were sequenced on Illumina HiSeq X Ten platform using (2 × 150 base pairs) paired-end module. The raw images were transformed into raw reads by base calling using CASAVA (http://www.illumina.com/support/documentation.ilmn). Then, raw reads in a fastq format were first processed using in-house perl scripts. Clean reads were obtained by removing reads with adapters, reads in which unknown bases were more than 5% and low-quality reads (the percentage of low-quality bases was more than 50% in a read; we defined the low-quality base to be the base whose sequencing quality was no more than 10). At the same time, Q20, Q30, and GC content of the clean data were calculated (Vazyme Biotech Co. Ltd., Nanjing, China). The original data of the RNA-seq were uploaded to the Gene Expression Omnibus (GEO) DataSets (GEO accession nos. GSE107174 and GSE120820).
Statistical analysis
RNA-seq data were analyzed using Enrichr (http://amp.pharm.mssm.edu/Enrichr) (26, 27). For all the bar graphs, data were expressed as means ± SEM. Prism 7 software (graphic software) was used for survival curves, charts, and statistical analyses.
Supplementary Material
Acknowledgments:
We thank E. J. Huang (University of California, San Francisco) for the Hipk2−/− mice. We thank R. Medzhitov (Yale University) for the Ifnαr−/− mice. We thank Z. Jiang (Peking University) for the iBMDMs. We thank J. Rose (Yale University) for the VSV virus.
Funding: This work was supported by grants from the National Natural Science Foundation of China (31570891) and the National Key Research and Development Program of China (2016YFA0500302).
Footnotes
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Schulz O, Diebold SS, Chen M, Näslund TI, Nolte MA, Alexopoulou L, Azuma Y-T, Flavell RA, Liljeström P, Reis e Sousa C, Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature 433, 887–892 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T, The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol 5, 730–737 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo Y-M, Gale M Jr., Akira S, Yonehara S, Kato A, Fujita T, Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol 175, 2851–2858 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T, DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG, IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol 11, 997–1004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu Y-J, The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol 12, 959–965 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun L, Wu J, Du F, Chen X, Chen ZJ, Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seth RB, Sun L, Ea C-K, Chen ZJ, Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S, Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med 203, 1795–1803 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L-G, Wang Y-Y, Han K-J, Li L-Y, Zhai Z, Shu H-B, VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Fang R, Jiang Q, Zhou X, Wang C, Guan Y, Tao J, Xi J, Feng J-M, Jiang Z, MAVS activates TBK1 and IKKε through TRAFs in NEMO dependent and independent manner. PLOS Pathog. 13, e1006720 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu Y-T, Grishin NV, Chen ZJ, Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Blach-Olszewska Z, Innate immunity: Cells, receptors, and signaling pathways. Arch. Immunol. Ther. Exp 53, 245–253 (2005). [PubMed] [Google Scholar]
- 14.Meadows SM, Myers CT, Krieg PA, Regulation of endothelial cell development by ETS transcription factors. Semin. Cell Dev. Biol 22, 976–984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.You F, Wang P, Yang L, Yang G, Zhao YO, Qian F, Walker W, Sutton R, Montgomery R, Lin R, Iwasaki A, Fikrig E, ELF4 is critical for induction of type I interferon and the host antiviral response. Nat. Immunol 14, 1237–1246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaikhibrahim Z, Wernert N, ETS transcription factors and prostate cancer: The role of the family prototype ETS-1 (review). Int. J. Oncol 40, 1748–1754 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Kim YH, Choi CY, Lee S-J, Conti MA, Kim Y, Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J. Biol. Chem 273, 25875–25879 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Feng Y, Zhou L, Sun X, Li Q, Homeodomain-interacting protein kinase 2 (HIPK2): A promising target for anti-cancer therapies. Oncotarget 8, 20452–20461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sombroek D, Hofmann TG, How cells switch HIPK2 on and off. Cell Death Differ. 16, 187–194 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH, Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 115, 177–186 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Rinaldo C, Prodosmo A, Siepi F, Soddu S, HIPK2: A multitalented partner for transcription factors in DNA damage response and development. Biochem. Cell Biol 85, 411–418 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Nardinocchi L, Puca R, Givol D, D’Orazi G, HIPK2—A therapeutical target to be (re)activated for tumor suppression: Role in p53 activation and HIF-1α inhibition. Cell Cycle 9, 1270–1275 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Rivas C, Aaronson SA, Munoz-Fontela C, Dual role of p53 in innate antiviral immunity. Viruses 2, 298–313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanni C, Nardinocchi L, Puca R, Stanga S, Uberti D, Memo M, Govoni S, D’Orazi G, Racchi M , Homeodomain interacting protein kinase 2: A target for Alzheimer’s beta amyloid leading to misfolded p53 and inappropriate cell survival. PLOS ONE 5, e10171 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang Y, Doan CN, Arnold TD, Lee S, Tang AA, Reichardt LF, Huang EJ, Transcriptional corepressors HIPK1 and HIPK2 control angiogenesis via TGF-β–TAK1– dependent mechanism. PLOS Biol. 11, e1001527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A, Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anzilotti S, Tornincasa M, Gerlini R, Conte A, Brancaccio P, Cuomo O, Bianco G, Fusco A, Annunziato L, Pignataro G, Pierantoni GM, Genetic ablation of homeodomain-interacting protein kinase 2 selectively induces apoptosis of cerebellar Purkinje cells during adulthood and generates an ataxic-like phenotype. Cell Death Dis. 6, e2004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lasfar A, Cook JR, Cohen Solal KA, Reuhl K, Kotenko SV, Langer JA, Laskin DL, Critical role of the endogenous interferon ligand-receptors in type I and type II interferons response. Immunology 142, 442–452 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider WM, Chevillotte MD, Rice CM, Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol 32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoggins JW, Rice CM, Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol 1, 519–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma F, Li B, Yu Y, Iyer SS, Sun M, Cheng G, Positive feedback regulation of type I interferon by the interferon-stimulated gene STING. EMBO Rep. 16, 202–212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu L, Wang W, Li Y, Zhou X, Yin Y, Wang Y, de Man RA, van der Laan LJW, Huang F, Kamar N, Peppelenbosch MP, Pan Q, RIG-I is a key antiviral interferon-stimulated gene against hepatitis E virus regardless of interferon production. Hepatology 65, 1823–1839 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Mostashari F, Bunning ML, Kitsutani PT, Singer DA, Nash D, Cooper MJ, Katz N, Liljebjelke KA, Biggerstaff BJ, Fine AD, Layton MC, Mullin SM, Johnson AJ, Martin DA, Hayes EB, Campbell GL, Epidemic West Nile encephalitis, New York, 1999: Results of a household-based seroepidemiological survey. Lancet 358, 261–264 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Faísca P, Desmecht D, Sendai virus, the mouse parainfluenza type 1: A longstanding pathogen that remains up-to-date. Res. Vet. Sci 82, 115–125 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Giraud S, Diaz-Latoud C, Hacot S, Textoris J, Bourette RP, Diaz J-J, US11 of herpes simplex virus type 1 interacts with HIPK2 and antagonizes HIPK2-induced cell growth arrest. J. Virol 78, 2984–2993 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muschik D, Braspenning-Wesch I, Stockfleth E, Rösl F, Hofmann TG, Nindl I, Cutaneous HPV23 E6 prevents p53 phosphorylation through interaction with HIPK2. PLOS ONE 6, e27655 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa H, Barber GN, STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.tenOever BR, Sharma S, Zou W, Sun Q, Grandvaux N, Julkunen I, Hemmi H, Yamamoto M, Akira S, Yeh W-C, Lin R, Hiscott J, Activation of TBK1 and IKKε kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol 78, 10636–10649 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrilenas KK, Ramlall V, Kurland J, Leung B, Harbaugh AG, Siggers T, DNA-binding landscape of IRF3, IRF5 and IRF7 dimers: Implications for dimer-specific gene regulation. Nucleic Acids Res. 46, 2509–2520 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oeckinghaus A, Ghosh S, The NF-kβ family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol 1, a000034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeffer LM, The role of nuclear factor κB in the interferon response. J. Interferon Cytokine Res. 31, 553–559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonnefoy E, Bandu M-T, Doly J, Specific binding of high-mobility-group I (HMGI) protein and histone H1 to the upstream AT-rich region of the murine beta interferon promoter: HMGI protein acts as a potential antirepressor of the promoter. Mol. Cell. Biol 19, 2803–2816 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klar M, Bode J, Enhanceosome formation over the beta interferon promoter underlies a remote-control mechanism mediated by YY1 and YY2. Mol. Cell. Biol 25, 10159–10170 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siepi F, Gatti V, Camerini S, Crescenzi M, Soddu S, HIPK2 catalytic activity and subcellular localization are regulated by activation-loop Y354 autophosphorylation. Biochim. Biophys. Acta 1833, 1443–1453 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart DM, Tian L, Notarangelo LD, Nelson DL, X-linked hypogammaglobulinemia and isolated growth hormone deficiency: An update. Immunol. Res 40, 262–270 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Sashida G, Bae N, Di Giandomenico S, Asai T, Gurvich N, Bazzoli E, Liu Y, Huang G, Zhao X, Menendez S, Nimer SD, The Mef/Elf4 transcription factor fine tunes the DNA damage response. Cancer Res. 71, 4857–4865 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim YH, Sung KS, Lee S-J, Kim Y-O, Choi CY, Kim Y, Desumoylation of homeodomain-interacting protein kinase 2 (HIPK2) through the cytoplasmic-nuclear shuttling of the SUMO-specific protease SENP1. FEBS Lett. 579, 6272–6278 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Choi DW, Seo Y-M, Kim E-A, Sung KS, Ahn JW, Park S-J, Lee S-R, Choi CY, Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J. Biol. Chem 283, 4682–4689 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Gresko E, Roscic A, Ritterhoff S, Vichalkovski A, del Sal G, Schmitz ML, Autoregulatory control of the p53 response by caspase-mediated processing of HIPK2. EMBO J. 25, 1883–1894 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, Piaggio G, Fanciulli M, Appella E, Soddu S, Homeodomaininteracting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol 4, 11–19 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Li T, Chen ZJ, The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med 215, 1287–1299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, Antipin J, Reva B, Koff A, Nimer SD, p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4, 37–48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim E-A, Kim JE, Sung KS, Choi DW, Lee BJ, Choi CY, Homeodomain-interacting protein kinase 2 (HIPK2) targets β-catenin for phosphorylation and proteasomal degradation. Biochem. Biophys. Res. Commun 394, 966–971 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Chalazonitis A, Tang AA, Shang Y, Pham TD, Hsieh I, Setlik W, Gershon MD, Huang EJ, Homeodomain interacting protein kinase 2 regulates postnatal development of enteric dopaminergic neurons and glia via BMP signaling. J. Neurosci 31, 13746–13757 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X-L, Arai Y, Harada H, Shima Y, Yoshida H, Rokudai S, Aikawa Y, Kimura A, Kitabayashi I, Mutations of the HIPK2 gene in acute myeloid leukemia and myelodysplastic syndrome impair AML1-and p53-mediated transcription. Oncogene 26, 7231–7239 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Brown NJM, Ramalho M, Pedersen EW, Moravcsik E, Solomon E, Grimwade DJ, PML nuclear bodies in the pathogenesis of acute promyelocytic leukemia: Active players or innocent bystanders? Front. Biosci. (Landmark Ed.) 14, 1684–1707 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Conrad E, Polonio-Vallon T, Meister M, Matt S, Bitomsky N, Herbel C, Liebl M, Greiner V, Kriznik B, Schumacher S, Krieghoff-Henning E, Hofmann TG, HIPK2 restricts SIRT1 activity upon severe DNA damage by a phosphorylation-controlled mechanism. Cell Death Differ. 23, 110–122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA, Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat. Med 10, 1366–1373 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.