Abstract
Three-prime Repair Exonuclease (TREX1) degrades ssDNA and dsDNA. TREX1 localizes to the perinuclear space in cells and degrades cytosolic DNA to prevent aberrant nucleic acid sensing and immune activation in humans and mice. Mutations in the TREX1 gene cause a spectrum of human autoimmune diseases including Aicardi-Goutières syndrome, familial chilblain lupus, retinal vasculopathy with cerebral leukodystrophy, and are associated with systemic lupus erythematosus. More than 60 disease-causing TREX1 variants have been identified including dominant and recessive, missense, and frameshift mutations that map to the catalytic core region and to the C-terminal cell localization region. The TREX1-disease causing mutations affect exonuclease activity at varied levels. In this chapter, we describe methods to purify variant recombinant TREX1 enzymes and measure the exonuclease activity using ssDNA and dsDNA substrates. The relationships between TREX1 activities, types of TREX1 mutations, and TREX1-associated autoimmune diseases are considered.
1. Introduction
The closely related TREX1 and TREX2 single-open-reading-frame genes encode the 314-amino-acid TREX1 and the 236-amino-acid TREX2 deoxyribonucleases located on chromosomes 3p21.2–21.3 and Xq28, respectively (Mazur & Perrino, 1999). The TREX1 catalytic core (amino acids 1–242) is about 40% identical to TREX2 (amino acids 1–236) and TREX1 contains an extended C-terminal region of 76 amino acids, not present in TREX2. These closely related nonprocessive 3′ → 5′ exonucleases degrade ssDNA and dsDNA, removing mononucleotides from DNA 3′termini (Mazur & Perrino, 2001b; Perrino, Miller, & Ealey, 1994), and modified DNA 3′ termini are poor substrates for these enzymes (Harrigan, Fan, Momand, et al., 2007; Perrino et al., 1994). TREX1 and TREX2 are Mg2+ ion-dependent nucleases belonging to the DnaQ-like subfamily (DEDD-h) characterized by the presence of four conserved carboxylate residues and a histidine positioned in the active site. TREX1 and TREX2 are the only mammalian deoxyribonuclease members of the DEDD-h subfamily, and degrade deoxynucleotide polymers at rates >1000-fold higher than ribonucleotide polymers (Mazur & Perrino, 1999).
TREX1 and TREX2 are functional dimers, indicating that the dimeric architecture is critical in their biological functions. Structural studies reveal the homodimeric nature of TREX1 and TREX2, and distinct structural elements, that point to different biological roles for these proteins (de Silva, Choudhury, Bailey, et al., 2007; Perrino, Harvey, McMillin, & Hollis, 2005). Between the TREX homodimers’ catalytic domains (TREX1 amino acids 1–242; TREX2 amino acids 1–236) exists a twofold axis of symmetry perpendicular to the central β-strands that connect the individual β-sheets of each protomer at the interface to form an extended β-sheet through the dimer core. Due to the dimers’ symmetries, active sites are positioned at opposite outer edges providing open access for the DNA. The side chains of the four conserved carboxylate residues in the active sites coordinate two Mg2+ ions, and the histidine is positioned to deprotonate a water producing a nucleophile to attack the scissile phosphodiester bond and effect hydrolysis. TREX1 and TREX2 contain a flexible loop of unique structure adjacent to the active site. TREX2 contains three conserved arginine residues on this loop that contribute to tight DNA binding (Perrino et al., 2005). The corresponding loop in TREX1 has only a single conserved arginine residue with additional residues outside the loop region contributing to DNA binding. TREX1 contains a unique polyproline II helix (PPII) formed by a non-repetitive proline rich region that contains six prolines within an 8-amino acid stretch. The corresponding region in TREX2 is a β-hairpin. TREX1 contains a unique C-terminal region of amino acids 243–314 that is responsible for its perinuclear ER localization and oligosaccharyltransferase (OST) complex interaction (de Silva et al., 2007; Hasan, Fermaintt, Gao, et al., 2015; Kucej, Fermaintt, Yang, Irizarry-Caro, & Yan, 2017; Mazur & Perrino, 1999, 2001a, 2001b; Orebaugh, Fye, Harvey, et al., 2013).
TREX1 mutations cause a spectrum of autoimmune diseases including Aicardi-Goutières syndrome (AGS), familial chilblain lupus (FCL), retinal vasculopathy with cerebral leukodystrophy (RVCL), and are associated with systemic lupus erythematosus (SLE). The TREX1-mediated disease spectrum is recapitulated in the mouse models with specific TREX1 mutations. The aggressive immune-activating phenotype of the TREX1 null mouse most closely mimics AGS (Morita, Stamp, Robins, et al., 2004), the spontaneous lupus-like inflammation in the TREX1 D18N mouse is most similar to FCL (Grieves, Fye, Harvey, et al., 2015), and the knock-in mouse carrying human TREX1 frame-shift mutations truncating the TREX1 C-terminus reflect the unique serologic autoimmune-like phenotypes caused by dysregulation of the ER oligosaccharyltransferase (OST) complex (Sakai, Miyazaki, Shin, et al., 2017). TREX1 is ubiquitously expressed with higher levels measured in some lymphoid tissues (Grieves et al., 2015; Mazur & Perrino, 2001a), and degrades cytosolic damaged DNA (Yang, Lindahl, & Barnes, 2007), retroelements (Stetson, Ko, Heidmann, & Medzhitov, 2008), and unprocessed erythroblast DNA (Rego, Harvey, Simpson, et al., 2018) to prevent inappropriate immune activation. TREX2 mutations have not been identified in humans, but TREX2 knockout mice exhibit increased chemically- (Parra, Manils, Castellana, et al., 2009) and UVB-induced skin tumorigenesis phenotypes consistent with the high level TREX2 expression detected specifically in keratinocytes (Ablasser, Hemmerling, Schmid-Burgk, et al., 2014; Manils, Gómez, Salla-Martret, et al., 2015; Rice, Rodero, & Crow, 2015).
The diverse immune-activating phenotypes caused by TREX1 mutations reflect the various types and positions of mutations within the catalytic and the C-terminal regions (Rice et al., 2015). In attempts to understand the link between TREX1 mutation and human disease we have developed strategies to measure TREX1 exonuclease activity focusing on the N-terminal catalytic domain. In this chapter, we describe assays to measure exonuclease activity of recombinant TREX1 using ssDNA and dsDNA substrates in vitro. These assays can be used with recombinant TREX1 dimeric enzymes (AA 1–242) of wild-type and mutant variant homo- and heterodimers providing insight into TREX1 catalytic mechanism. While the focus here is the TREX1 catalytic core, the high structural and catalytic similarities to TREX2 make these assays relevant and applicable to both enzymes.
2. Considerations when purifying TREX1 enzymes
2.1. Preparation of truncated enzyme
Attempts to overexpress in bacteria and purify the complete TREX1 1–314 amino acid enzyme have not been successful, presumably due to the hydrophobic properties of residues 287–314. Thus, in order to test exonuclease activities we overexpress the truncated TREX1 N-terminal 1–242 amino acids, which contains the TREX1 catalytic domain and displays potent exonuclease activity. In contrast, the full-length TREX2 1–236 amino acid enzyme is readily overexpressed in bacteria and purified using column chromatography. Hereafter, mention of TREX1 is about the truncated form of the enzyme.
2.2. Preparation of heterodimers
The dimeric structure of TREX1 is unusual for 3′ → 5′ exonucleases. The recombinant TREX1 enzyme is a kinetically stable dimer that does not dissociate and re-equilibrate at measurable rates (Orebaugh, Fye, Harvey, Hollis, & Perrino, 2011). This unique property of TREX1 dimers provides opportunity to study the activities of purified heterodimers or compound heterodimers of variant TREX1 forms.
3. Purifying recombinant TREX1 enzymes
The TREX1 expression and purification procedure used in our lab generates large quantities of highly purified TREX1 enzyme for the assays described here. The materials and protocol described are applicable to TREX1 homodimer and heterodimer purification. The materials or procedural steps required specifically for heterodimer purification are indicated in the materials and protocol sections of this chapter. Two different TREX1 gene-encoding plasmids are listed, each of which should encode a different TREX1 protomer. For homodimers, the TREX1 gene is expressed as a fusion with maltose binding protein (MBP) in pLM303x (Lee-Kirsch, Chowdhury, Harvey, et al., 2007), with a rhinovirus 3C protease (PreScission Protease) recognition site between the MBP and TREX1 genes. For heterodimers, the pLM303x plasmid is co-expressed with a second plasmid containing a distinct TREX1 gene construct prepared as a fusion of the His-tagged NusA with TREX1 in pCDFDuet-1 (Novagen). Plasmids used for heterodimer expression and purification can encode either a wild-type and mutant TREX1 gene, or two different mutant TREX1 genes. Proteins are then purified via affinity chromatography. Common techniques such as gel preparation, electrophoresis, vector cloning, are mentioned, but details not provided. Methods of preparation for some materials not purchased directly from commercial sources will be discussed in more detail to ensure replication of our procedures. Enzyme concentration methods are not necessary for assays and will not be discussed here. The following equipment and materials are used in this assay.
3.1. Equipment
FPLC or low-pressure chromatography system at 4°C with UV detection
Six 1 L culture flasks
Temperature-controlled shaking incubator
SDS-PAGE system
Spectrophotometer UV–vis
- Temperature-controlled centrifuge(s) and adapters:
- Six 1 L samples at 4000 rpm/4°C for 20 min
- Four 40 mL samples at 15,000 rpm/4°C for 20 min
- 1 mL samples at >16,000 G/20°C for 1 min
- French Press or equivalent pressurized lysis instrument
- Ex. Emulsiflex (Avestin)
Thermometer
UV–vis gel imager
Magnetic stirrer
3.2. Materials
pLM303x-TREX1 plasmid (Lee-Kirsch et al., 2007)
- * pCDFDuet-1-TREX1 (Novagen)
- *Only needed for heterodimer purification.
Escherichia coli BL21(DE3) Rosetta 2 cells (Novagen)
LB media
PreScission Protease (GE Biosciences)
Kanamycin
Chloramphenicol
- *Streptomycin
- *Only needed for heterodimer purification.
Ice bath for five 1 L flasks
Isopropyl β-D-1-thiogalactopyranoside (IPTG)
Complete Protease Inhibitor Tablets (Roche; Sigma-Aldrich 50564890001)
Lysozyme
- Amylose A Buffer
- 20 mM Tris base-HCl (pH 7.5)
- 200 mM NaCl
- 1 mM EDTA
- Filter through 0.2 μm nitrocellulose filter.
- Amylose B Buffer
- Amylose A Buffer
- 10 mM maltose
Bacterial transformation materials and reagents
SDS-PAGE materials and reagents (ex. Molecular Weight Standard, SDS loading buffer, gels, gel stain)
Dialysis tubing of 6–8 kDa MWCO
- *Ni-NTA Agarose resin (Qiagen)
- *Only needed for heterodimer purification.
- *Wash Buffer
- *Only needed for heterodimer purification
- 50 mM NaH2PO4-HCl (pH 7.5)
- 300 mM NaCl
- 10 mM imidazole
- Filter with 0.2-μm filter.
- *Elution Buffer
- *Only needed for heterodimer purification.
- 50 mM NaH2PO4-HCl (pH 7.5)
- 300 mM NaCl
- 250 mM imidazole
- Filter with 0.2-μm filter.
- Phosphate Cellulose (P-cell) Resin (de Silva et al., 2007)
- Cellulose
- Dibasic sodium phosphate
- Sodium azide.
- P-cell A Buffer
- 50 mM Tris base-HCl (pH 7.5)
- 1 mM EDTA
- 10% (v/v) glycerol
- Filter through 0.2-μm filter.
- P-cell B Buffer
- 50 mM Tris base-HCl (pH 7.5)
- 1 mM EDTA
- 10% (v/v) glycerol
- 1 M NaCl
- Filter through 0.2-μm filter.
4. Measuring TREX1 activity on ssDNA
TREX1 ssDNA exonuclease activity is measured by quantifying the removal of mononucleotides from the 3′ termini of mixed or homogeneous sequence ssDNA oligonucleotides (Mazur & Perrino, 2001b). Reaction products are separated on urea-polyacrylamide gels that provide single nucleotide resolution in order to quantify the individual DNA band intensities and calculate activities. A modification of the standard assay has been used to measure Michaelis-Menten kinetics for precise Kcat and Km value determinations. Necessary deviations from the standard protocol are indicated in the corresponding protocol section of this chapter. In our standard ssDNA assay we measure the degradation of a FAM-labeled 30-mer oligonucleotide visualized on a denaturing urea-polyacrylamide sequencing-type gel. The amount of dNMP excised in a reaction is quantified by scanning the gel using a large gel imager to capture the FAM fluorescence present within each oligomer at each band position within a lane. The fraction of oligomer at each position is multiplied by the number of dNMPs excised from the 30-mer to generate the oligomer, and by the total fmol of 30-mer in the starting reaction. The sum of these values yields the total fmol of dNMP excised, which is used to calculate the rate of the enzyme (Fye, Orebaugh, Coffin, Hollis, & Perrino, 2011; Orebaugh et al., 2011; Perrino et al., 2005; Perrino, Krol, Harvey, et al., 2004). The following equipment and materials are used in this assay.
4.1. Equipment
Large sequencing-style polyacrylamide gel electrophoresis system
- System for drying samples in vacuo
- Ex. SpeedVac Plus SC110 A and Refrigerated Vapor Trap RVT400 (Savant)
- Large gel imager capable of imaging at excitation/emission of 495/520 nm
- Ex. Typhoon FLA 9500 (GE Healthcare Life Sciences)
4.2. Materials
23% urea polyacrylamide gel (large, sequencing-style)
Reagents/buffers for polyacrylamide gel electrophoresis
- “Rxn mix”
- 50 mM Tris base-HCl (pH 7.5)
- 12.5 mM MgCl2
- 5 mM DTT
- We recommend pre-making aliquots of this mix and storing at −20°C until use, or making the mix fresh immediately preceding the assay. Aliquots stored up to 6 months at −20°C have no detectable effect on enzyme activity compared to fresh mix.
5′-FAM-labeled 30-mer oligonucleotide
Bovine serum albumin
TREX1 enzyme(s)
- Sample Loading Buffer (SLB)
- 4% (v/v) 500 mM EDTA
- 10% (v/v) Bromophenol Blue
- 10% (v/v) xylene cyanol
- 76% (v/v) deionized formamide
Cold, 100% ethanol
5. Measuring TREX1 activity on dsDNA with fluorescence
A fluorescence-based assay has been developed to measure TREX1 activities using dsDNA. A single nick is first introduced into the supercoiled plasmid pMYC using the restriction endonuclease nicking enzyme Nt. BbvCI. Reactions are prepared containing TREX1 and the nicked dsDNA plasmid, incubated, and samples are removed and quenched at various times in 15 × SYBR green dye solution. Samples are then excited, and emission intensity measured on a microplate reader. The fluorescent signal is directly related to the amount of remaining dsDNA. The rate of decreased fluorescent signal is used to quantify TREX1 activity. Some TREX1 mutant enzymes competitively bind to the nicked-dsDNA substrate, inhibiting TREX1 wild-type enzyme activity that is added in the same reaction. This competitive binding is not observed using ssDNA substrates (Lehtinen, Harvey, Mulcahy, Hollis, & Perrino, 2008). Thus, a variation of this dsDNA assay can be used with TREX1 mutants and wild-type enzyme to assess competitive DNA binding activity. The necessary deviations from the protocol are indicated in the protocols section of the chapter. Common techniques are mentioned, but not discussed in detail. The following equipment and materials are used for the assay.
5.1. Equipment
Microplate reader with 495/520 nm excitation/emission
5.2. Materials
- “Rxn mix”
- Defined in Section 4.2.
Bovine serum albumin
TREX1 enzyme(s)
- Black, 384-well plate
- Ex. Corning #3573.
SYBR green dye (ThermoFisher), 10,000 × concentrate
- Nicked plasmid of about 10 kb
- Ex. pMYC nicked with Nt.BbvCI (NEB)
6. Visualizing TREX1 activity on dsDNA with agarose gel electrophoresis
TREX1 degradation of dsDNA can be visualized using agarose gels. Reactions are prepared and performed as described for the fluorescence assay, then quenched by addition of ethanol, dried in vacuo, and resuspended in gel loading solution. Products are separated and visualized by electrophoresis through agarose gels prepared with GelRed dsDNA stain. All variations of the fluorescence-based assay can be applied to the agarose gel-based assay. Common techniques used for this assay are mentioned but not detailed. The following equipment and materials are used in this assay.
6.1. Equipment
Agarose gel electrophoresis system
- System for drying samples in vacuo
- Ex. SpeedVac Plus SC110 A and Refrigerated Vapor Trap RVT400 (Savant)
- UV–vis gel imaging system
- Ex. ChemiDoc (Bio-Rad)
6.2. Materials
- “Rx mix”
- Defined in Section 4.2.
Bovine serum albumin
TREX1 enzyme(s)
Plasmid (~10 kb)
- Nicked plasmid (~10 kb)
- Ex. pMYC nicked with Nt.BbvCI (NEB)
Agarose gel electrophoresis buffers/reagents
GelRed dsDNA stain (Biotium #41003)
- Loading dye
- Ex. 6 × Gel Loading Dye Blue (NEB)
Cold, 100% ethanol
7. Protocols
7.1. Preparation of phosphate cellulose resin
Add 100 g cellulose to a 2 L beaker.
Suspend in 0.5 N NaOH to 1.5 L volume, stir, and let sit at room temperature for 5 min.
Decant the top layer of fluid, add deionized water to 1.5 L volume, and let cellulose sit again until it settles. Repeat this wash phase 2 more times.
Suspend washed cellulose in 0.6 N HCl to 1.5 L volume, then let sit at room temperature for 5 min.
Repeat step 3.
Resuspend slurry to 1.5 L volume with deionized H2O.
Adjust pH of slurry to 7.0 using dibasic sodium phosphate.
Let the slurry sit until it packs, then decant to a remaining 1 L volume.
Resuspend the slurry, add 0.1% sodium azide, and transfer to bottle for long term storage. Final product should be a ~50% slurry.
7.2. Overexpression and affinity chromatography purification of TREX1
- Protein Induction
- Transform Rosetta II cells with pLM303x-TREX1 plasmid, and plate on selective media using kanamycin (Kan) and chloramphenicol (CP).
- *If performing a heterodimer purification, co-transform cells with pCDFDuet-1-TREX1 and pLM303x-TREX1 plasmids, and include Kan, CP, and streptomycin (SP) in selective media and subsequent culturing broths.
- Using a colony from the transformation plate, inoculate a ~200 mL LB/Kan/CP broth. Incubate this starter culture overnight at 37°C/180 rpm.
- Determine the A600 of the starter culture. Then, dilute the starter culture into five 1 L broths such that their A600~0.1.
- Let the new cultures incubate at 37°C/180 rpm, periodically measuring A600 of the cultures, until A600 = 0.5–0.8. This should take ~2 h.
- Induce each 1 L culture with 0.5 mM IPTG (final concentration).
- Place the cultures back in incubation at 37°C/180 rpm for 10 min. Then, retrieve the cultures and cool them in ice baths until their temperature reaches 20°C as measured using a thermometer. After cooling, place the cultures into incubation at 16°C/180 rpm to incubate overnight (15–20 h).
- Cell Lysis
- Retrieve induced cultures and centrifuge them at 4000 rpm/4°C for 20 min (JLA-8.1000 rotor).
- Decant the supernatants, and transfer the pellets by spatula to a beaker containing 120 mL Amylose A Buffer, three protease inhibitor tablets, and 120 mg lysozyme. Cover the beaker and place it a on a magnetic stirrer at 4°C until the pellet is resuspended. This usually takes about 20 min.
- Keeping sample and lysate on ice at all times, lyse the cell suspension by pressure homogenization at 15,000–18,000 psi. Two rounds of lysis are recommended. Do not cycle the sample through the homogenizer continuously. Allow the outgoing lysate to cool on ice until all sample is finished, then reapply lysate for second lysis.
- Centrifuge the lysate for 20 min at 15,000 rpm/4°C (JA-25.50 rotor).
- Retrieve the supernatant by decanting, and place it at 4°C until further use.
- Amylose Column Purification
- Prepare a 15 mL amylose column in a 4°C chromatography system. Retrieve the supernatant sample from 4°C and place it on ice. Then, using a flow rate of 1.5 mL/min, perform the purification as follows making sure to record A280 throughout:
- Equilibrate column with 75 mL Amylose A Buffer.
- Apply the sample to the column.
- Wash column with 120 mL Amylose A Buffer, or until A280 returns to baseline.
- Elute from column with Amylose B Buffer, and collect protein-containing eluent as a single sample.
- Add glycerol to eluted sample to a final concentration of 10% (v/v).
- *If performing a heterodimer purification, skip the glycerol addition. Glycerol will be added after elution from the Ni-NTA column.
- Add PreScission Protease (final concentration 7.5 μg/mL) to the eluted sample, and mix gently.
- *If performing a heterodimer purification, skip the protease addition. Protease will be added after the Ni-NTA column purification.
- Dialyze the protein at 4°C overnight in 1 L of P-cell A Buffer containing 50 mM NaCl, using 6–8 kDa MWCO dialysis tubing.
- *If purifying heterodimer, protein should instead be dialyzed in 1L of Wash Buffer.
- **Ni-NTA Column Purification (**Skip step 4 procedures contained here if purifying homodimer.)
- Prepare a 15 mL Ni-NTA Agarose resin column in a 4° C chromatography system. Retrieve the dialyzed sample from 4°C and place it on ice. Then, using a flow rate of 1.5 mL/min, perform the purification as follows making sure to record A280 throughout:
- Equilibrate column with 75 mL Wash Buffer.
- Apply the sample to the column.
- Wash column with 120 mL Wash Buffer, or until A280 reaches baseline.
- Elute column with Elution Buffer, and collect protein-containing eluent in a single sample.
- Add glycerol to the eluted sample to a final concentration of 10% (v/v).
- Add PreScission Protease (final concentration 7.5 μg/mL) to the eluted sample, and mix gently.
- Dialyze the protein at 4°C overnight in 1 L of P-cell A Buffer containing 50 mM NaCl, using 6–8 kDa MWCO dialysis tubing.
- Phosphocellulose Column Purification
- Prepare a 15 mL phosphocellulose column in a 4°C chromatography system. Retrieve the dialyzed sample from 4°C and place it on ice. Then, using a flow rate of 1.5 mL/min, perform the purification as follows making sure to record A280 throughout:
- Equilibrate column with 75 mL P-cell A Buffer.
- Apply sample to column.
- Wash column with 75 mL P-cell A Buffer.
- Elute column with 200 mL gradient of 0 ≤ 100% P-cell B Buffer. Collect eluent in 2 mL fractions. The elution peak should appear at ~15% P-cell B Buffer.
- Combine the eluent fractions containing the TREX1 enzyme. This should be indicated on the chromatogram by a single large A280 peak during the elution phase. We recommend confirming the protein content of the fractions with SDS-PAGE.
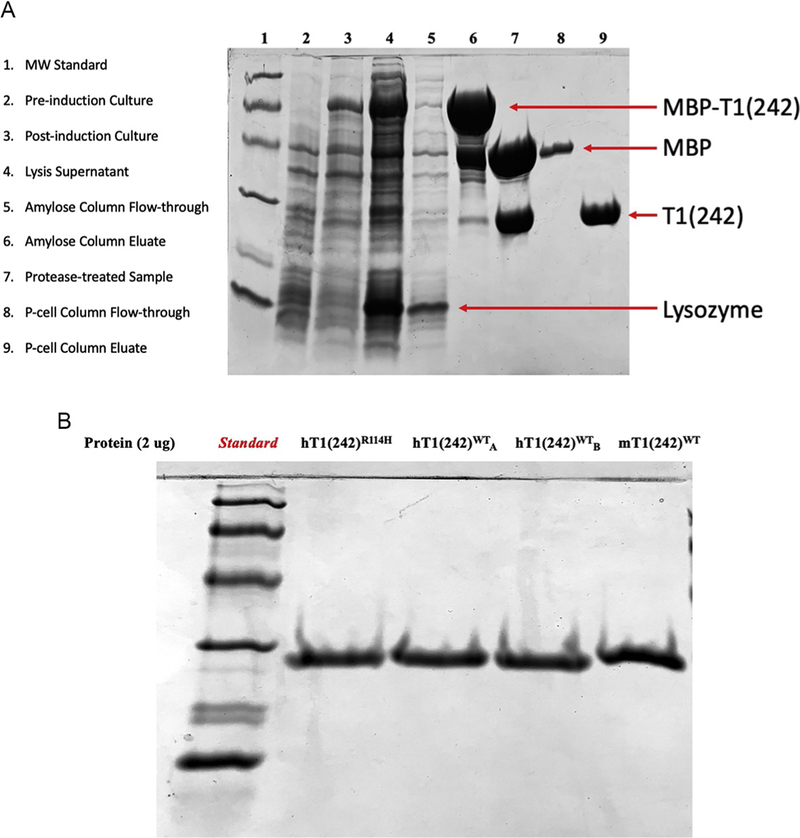
Fig. 1A illustrates the expected protein content during various stages of purification.
The concentration of purified enzyme is determined by absorbance (A280) and confirmed by subjecting equivalent-mass samples of multiple different TREX1 preparations to SDS-PAGE and visualizing polypeptides by Coomassie blue dye staining. Gels are imaged, and ImageJ software is used to determine the densitometric profile of the TREX1 electropherogram. The intensity profile for each lane is baseline corrected and the area under the TREX1 peak determined to calculate the percent purity and final concentration of each preparation. Example data of this step using various enzymes is illustrated in Fig. 1B.
Enzyme activity is confirmed using one of the assays described in this chapter.
Store the enzyme at −80°C. We have found that the enzyme can be thawed up to three times without detectable loss of activity.
Fig. 1.
Purification of hTREX1WT/WT. (A) SDS-PAGE samples from critical phases in the enzyme purification process. Bands corresponding to various key proteins are identified using 12% polyacrylamide gels. (B) SDS-PAGE gel results demonstrating equal-mass quantities of TREX1 enzymes using 12% polyacrylamide gels. All non-standard lanes contain ~2 μg of protein. Two lanes contain the same human TREX1 wild-type enzyme from different preparations (labeled subscripts “A” and “B”). Note, the TREX1 murine enzyme migrates slightly more slowly relative to the TREX1 human enzyme.
7.3. Measuring TREX1 activity on ssDNA
- This procedure is performed in two phases described below. All steps should be performed on ice unless otherwise stated.
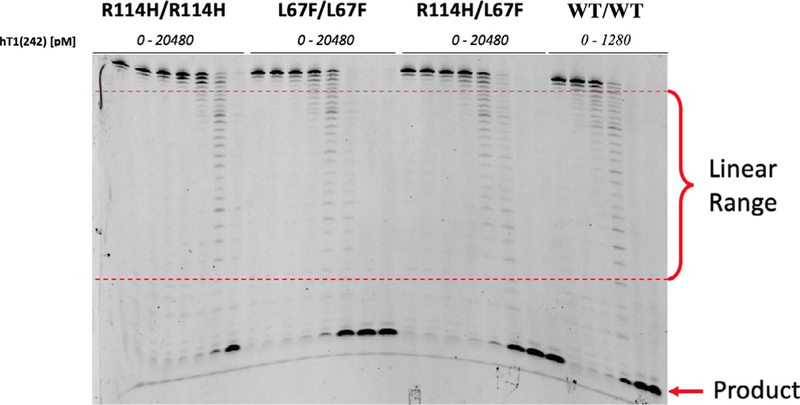
- Phase I: Identify the concentration range for each TREX1 enzyme that is within the linear range of the assay. This is determined by the generation of DNA bands migrating to positions on the gel that are visibly degraded, but do not migrate to positions near the bottom of the gel where background bands are evident as indicated in the no-enzyme control lanes. Multiple experiments might be required to identify appropriate TREX1 concentrations in the linear range of the assay. It is recommended to begin with broad concentration ranges and to narrow the selected concentrations identifying three different enzyme concentrations within the linear range of the assay. Data demonstrating the identification of linear range concentrations for hTREX1WT/WT and several hTREX1 mutants is shown in Fig. 2.
- Phase II: TREX1 activity is determined using the identified three enzyme concentrations in the assay linear range with triplicate reactions for each concentration. Band intensities for each reaction are quantified by densitometry and used to calculate the initial velocities of each reaction. Initial velocities of reaction triplicates are averaged to calculate activity rates for each enzyme tested, and rates are compared among enzyme variants (if applicable).
- Prepare a volume of the following “Master Mix” equal to the product of 30 μL and the number of reactions to be performed. Make sure to account for an extra no-enzyme control reaction.
- 44.4% Rxn mix
- 22.2% 250 nM FAM-labeled 30-mer oligonucleotide
- 33.3% H2O
- For an experiment consisting of 10 reactions you would need 300 μL of Master Mix composed of 133.3 μL Rx mix, 66.7 μL of 30-mer, and 100 μL of deionized water.
Obtain a microcentrifuge tube for each reaction to be performed, and add 27 μL of Master Mix to each tube. Leave the tubes at room temperature.
For each enzyme, prepare aliquots of each concentration to be tested at 10 × the desired reaction concentration. This should be done by dilution of stock enzyme into 1 mg/mL BSA. If the inhibitive properties of a mutant enzyme on wild-type enzyme are being tested, include wild-type enzyme in the prepared mutant enzyme aliquots at 10 × the desired reaction concentration.
Add 3 μL of each enzyme aliquot to its respective reaction tube to initiate the reactions. Vortex quickly after enzyme addition to mix. For the no-enzyme control reaction, add 3 μL of 1 mg/mL BSA. Incubate reactions at room temperature.
After a 20 min incubation at room temperature, quench each reaction with 90 μL of cold, 100% ethanol. Quenched samples may be stored at −20°C overnight or until the protocol can be continued.
Evaporate the quenched reactions in vacuo at low/no heat setting.
Resuspend the evaporated samples in 8 μL each of SLB.
Make sure at this point you have a 23% urea polyacrylamide gel prepared with VERY clean casting plates. The cleanliness of the gel image will be critical for quantification later.
Load 8 μL of each sample onto a 23% urea polyacrylamide gel, and electrophorese the gel with constant power at 55 W/3000 V for ~3.5 h. These electrophoresis conditions may vary depending on the gel apparatus and electrophoresis system. In our gel system, optimal band separation with minimal “smiling” of the bands occurs at ~60% progression of the dye front down our gels.
After electrophoresis, separate the two plates of the gel cast, making certain the gel completely adheres to the bottom plate. Cover the gel in clear plastic wrap (ex. all-purpose polyvinyl-chloride wrap), making sure that there are no wrinkles or bubbles in the wrap over the gel or excess wrap accumulating on the back of the casting plate. Be wary of touching the gel, wrap, or plate in areas other than the edges, as this can affect image quality. We recommend wiping the front and back of the finished gel/plate with 70% ethanol and letting it dry in a vertical position for a few minutes before imaging to obtain cleaner gels.
Image the prepared gel at excitation/emission of 495/520 nm.
Quantify and record band intensities for every reaction (lane) on the gel image by densitometry. These values will be used for activity quantification, to be discussed later in this chapter.
Fig. 2.
Phase I gel results for TREX1 ssDNA exonuclease assay. Degradation of a FAM-labeled 30-mer oligonucleotide substrate [50 nM] by varying concentrations of hTREX1 enzymes visualized on a 23% urea polyacrylamide gel. Banding consistent with the linear range of the assay is identified as “Linear Range.” Banding that corresponds to accumulating product is labeled as “Product.”
7.4. Measuring TREX1 activity on dsDNA with fluorescence
- Prepare a volume of Master Mix equal to the product of the number of reactions and 140 μL. When determining the number of reactions, we recommend performing reactions in triplicate for each concentration of each enzyme tested. The Master Mix is prepared as follows:
- 44.4% Rx mix
- 55.6% 20 ng/μL nicked 10 kb plasmid
- For 10 reactions, you would need 1.4 mL of Master mix composed of 622 μL of Rx mix and 778 μL of nicked plasmid at 20 ng/μL.
Obtain microcentrifuge tubes for every reaction to be performed, and place 135 μL of Master Mix into each reaction tube.
Set up the 384-well microplate and add 20 μL of 15 × SYBR green (stock dissolved in H2O) to every well to be used. You will need seven wells per reaction, corresponding to the seven samples that will be taken for each of the time points used. Prepare this solution no > 1 h before reaction initiation, and make sure to cover the microplate throughout the experiment whenever possible to protect photosensitive contents.
For each enzyme to be tested, prepare aliquots for each of the intended enzyme concentrations at 10× the desired reaction concentration. This should be done via dilution of stock enzyme into 1 mg/mL BSA. If the inhibitive properties of a mutant enzyme on wild-type enzyme are being tested, include wild-type enzyme in the prepared mutant enzyme aliquots at 10× the desired reaction concentration.
Take an 18 μL sample of each reaction tube and quench it in its corresponding well in the microplate, then add 2 μL of 1 mg/mL BSA. This will act as the 0 min time point for the assay. Here, and at every sample quenching after, make sure to pipette-mix the well contents thoroughly to completely inactivate the TREX1. This is a major source of result variation if not done adequately.
Add 13 μL of the corresponding enzyme aliquot to each reaction tube, and vortex thoroughly to initiate the reactions.
Allow the reactions to incubate for 1 h at room temperature. Take 20 μL samples from each reaction tube at the 5, 10, 20, 30, 45, and 60-min time points and quench the samples in their intended wells on the microplate.
After reaction completion, read the fluorescence intensity of the plate wells at excitation/emission of 495/520 nm and record the values. These values will be used for activity quantification, to be discussed later in the chapter.
7.5. Visualizing TREX1 activity on dsDNA with agarose gel electrophoresis
- Prepare a volume of Master Mix equal to the product of the number of reactions and 140 μL. The Master Mix is prepared as follows:
- 44.4% Rx mix
- 55.6% 20 ng/μL nicked 10 kb plasmid
- For 10 reactions, you would need 1.4 mL of Master mix composed of 622 μL of Rx mix and 778 μL of nicked plasmid at 20 ng/μL.
Obtain microcentrifuge tubes for every reaction to be performed, and place 135 μL of Master Mix into each reaction tube.
Obtain five microcentrifuge tubes per planned reaction, and add 80 μL of cold 100% ethanol to each. These will be used to quench the samples from each reaction.
For each enzyme to be tested, prepare aliquots for each of the intended enzyme concentrations at 10 × the desired reaction concentration. This should be done via dilution of stock enzyme into 1 mg/mL BSA. If the inhibitive properties of a mutant enzyme on wild-type enzyme are being tested, include wild-type enzyme in the prepared mutant enzyme aliquots at 10 × the desired reaction concentration.
Take an 18 μL sample from each reaction tube and quench it in its corresponding ethanol tube, then add 2 μL of 1 mg/mL BSA. This will act as the 0 min time point for the assay.
Add 13 μL of the corresponding enzyme aliquot to each reaction tube, and vortex well to initiate the reactions.
Allow the reactions to incubate at room temperature. Take 20 μL samples from each reaction tube at the 5, 10, 20, and 40-min time points and quench the samples in their intended tubes.
Evaporate the quenched reactions in vacuo at low/no heat setting. This should take ~20 min.
Resuspend the evaporated samples in 11 μL of solution containing 80% 1 × TAE with 20% 5 × loading dye.
Load the samples onto a 0.8% agarose gel prepared with a GelRed dsDNA stain, and electrophorese the gel for 50 min at 110 V.
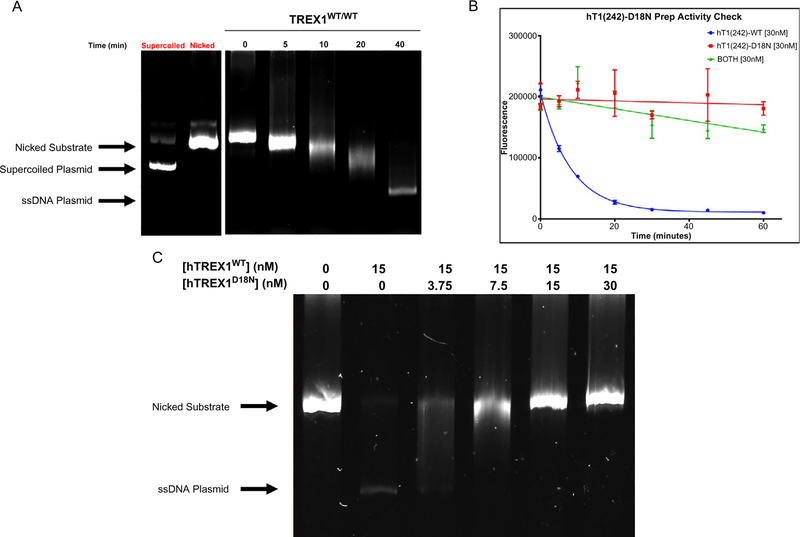
After electrophoresis, image the gel. Fig. 3A shows example data from this assay, and identifies the banding present.
Fig. 3.
Results of TREX1 dsDNA fluorescence and agarose gel assays. (A) Degradation of 10 ng/μL Nt.BbvCI-nicked pMYC substrate by 15 nM hTREX1WT/WT visualized on a 0.8% agarose gel. Bands corresponding to supercoiled and nicked plasmid are indicated. TREX1 excises dNMPs from the nicked polynucleotide of the dsDNA plasmid generating a single-stranded DNA polymer. The position of migration of the ssDNA polymer is identified as “ssDNA.” The gel lanes from left to right are: supercoiled (un-nicked) plasmid, nicked plasmid, reaction samples at the indicated times (min). (B) Degradation of 10 ng/μL Nt.BbvCI-nicked pMYC dsDNA by 30 nM hTREX1WT/WT and/or 30 nM hTREX1D18N/D18N, visualized via fluorescence. Fluorescence values for the indicated time points for each reaction indicated were plotted as a function of fluorescence vs time in Prism7 (GraphPad), and fitted with one-phase decay nonlinear regression. (C) Degradation of 5 ng/μL Nt.BbvCI-nicked pMYC substrate by 15 nM hTREX1WT/WT in the presence of increasing concentrations of hTREX1D18N, visualized on a 0.8% agarose gel. Bands corresponding to undegraded substrate are indicated.
8. Calculating TREX1 exonuclease activities
8.1. Quantifying TREX1 activity on ssDNA
All band intensities are determined in each lane (reaction) and summed (referred to hereafter as “lane intensity”). The lane intensity represents the total FAM-labeled substrate used in the reaction (for 50 nM in 30 μL, this would be 1.5 pmol of FAM-labeled oligonucleotide). The ratio of band intensity to lane intensity multiplied by the moles of FAM-labeled oligonucleotide yields the moles of n-mer oligonucleotide represented by each band in the lane. The highest molecular weight band is the starting 30-mer, and each band below represents oligonucleotide products of decreasing length by a single nucleotide. The amount of dNMPs excised in each reaction (lane) is determined by multiplying the moles of each length oligonucleotide by the number of nucleotides excised to produce the n-mer oligonucleotide associated with each band, and summing these values for all bands in the lane. The equation and variable definitions for calculating dNMPs excised in each reaction (lane) are detailed below.
If:
N = the band number of a given band in the chosen lane, such that N = 1 is the highest apparent molecular weight band in the chosen lane, N = 2 is the next highest, etc.
B = the band intensity value associated with the band identified by N.
L = the lane intensity of the chosen lane (sum of B for all N in chosen lane).
T = the total number of bands present in the chosen lane.
Then:
For the purposes of comparing TREX1 variant activities, we calculate TREX1 enzyme activity as the picomoles of dNMPs excised divided by seconds of incubation time divided by picomoles of enzyme. This is shown in the equation:
8.2. Quantifying TREX1 activity on dsDNA with fluorescence
Fluorescence values are obtained for each reaction sample and plotted as a function of time. The time-course reaction curve is generated and fit using single-phase decay nonlinear regression with Prism (GraphPad) as illustrated in Fig. 3B. Reaction rates are calculated using initial slopes of the fitted line and expressed as fluorescence units per second. A standard curve equating fluorescence units to DNA concentration can be generated if units of dNMPs per second are desired by including no-enzyme reactions and a range of dsDNA substrate concentrations in the experiment. In either case, the calculated reaction rates can be normalized to determine relative activities of TREX1 variant enzymes.
9. Exonuclease activities of TREX1 mutant enzymes
TREX1 mutations have been identified at positions throughout the gene with varying effects on ssDNA and dsDNA exonuclease activities and cell localization. Four nucleotides of ssDNA bind at the TREX1 active site through sequence independent hydrogen bonding and hydrophobic interactions (de Silva et al., 2007). The phosphate oxygens of the DNA 3′ nucleotide, along with the carboxylate oxygens of Asp18, Glu20, Asp130, and Asp200 and several water molecules contribute to the coordination of two divalent metal ions at the active site. The D18N, D18H, D200N, and D200H disease causing mutations at these active site residues abolish DNA exonuclease activity in TREX1 homodimers resulting from dysfunctional metal ion coordination (Lehtinen et al., 2008). However, heterodimers containing one of these active site mutant protomers with the wild-type protomer exhibit ~50% wild-type activity using the ssDNA assay and undetectable activity using the dsDNA assay. In addition, the heterodimers competitively inhibit TREX1 wild-type activity in the dsDNA assay (Lehtinen et al., 2008). DNA exonuclease assays support the idea that heterodimers bind DNA and position the 3′ nucleotide into the active site. The inability to perform chemistry of phosphodiester bond cleavage resulting from dysfunctional metal-ion coordination traps the TREX1 mutant enzyme onto the dsDNA in a nonproductive enzyme-DNA complex at the site of the nick and accounts for the inhibition of TREX1 WT DNA degradation activity detected in the presence of the Asp18 and Asp200 TREX1 mutant enzymes. Thus, the combination of TREX1 catalytic deficiency and DNA binding proficiency generates TREX1 mutant enzymes that fail to process nicks in dsDNA and block access to these sites by TREX1 WT enzyme helping to explain the dominant genetics exhibited by the TREX1 D18N, D18H, and D200N alleles (Rice et al., 2015) (see Fig. 3B and C) The TREX1 residues Arg-174 and Lys-175 positioned adjacent to the active sites act with the Arg-128 residues positioned in the catalytic core facilitate dsDNA melting and generate ssDNA for entry into the active sites. TREX1 mutations at R174 and K175 located in the flexible loop have varied effects on ssDNA and dsDNA exonuclease activities by altering TREX1 DNA binding (Fye et al., 2011).
The TREX1 R114H mutation is one of the most common autoimmune disease-causing mutations. DNA degradation properties of R114H have provided direct evidence that TREX1 residues in one protomer contribute to DNA degradation catalyzed in the opposing protomer and help to explain the dimeric TREX1 structure required for full catalytic competency (Orebaugh et al., 2011). The TREX1 Arg-114 residue is about 15 Å away from the active site positioned at the dimer interface where it hydrogen bonds with two backbone carbonyl oxygens of the opposing protomer where it contributes to the activity in the opposing protomer of the TREX1 dimer. The TREX1 R114H homodimer exhibits severely reduced enzymatic activity on ss- and dsDNA. The R114 H mutation causes recessive TREX1-associated autoimmune disorders (Rice et al., 2015).
The TREX1 flexible regions contribute to a nucleic acid-kinking mechanism used to unwind, melt, and reshape dsDNA and position the ssDNA 3′ termini required for catalysis into the active site (de Silva et al., 2007; Grieves et al., 2015). This DNA binding mechanism likely explains the varied effects on exonuclease activities using ssDNA and dsDNA assays exhibited by some TREX1 mutations such as R174 and K175 positioned on the flexible DNA-binding loop. Additionally, residues like Arg-62 in one TREX1 protomer provide necessary structural elements for full catalytic activity in the opposing TREX1 protomer (Fye, Coffin, Orebaugh, Hollis, & Perrino, 2014). These data provide compelling evidence for the required TREX1 dimeric structure for full catalytic function.
10. Summary
The nonprocessive 3′ →5′ exonuclease TREX1 degrades cytosolic DNA to prevent aberrant nucleic acid sensing. When TREX1 exonuclease activity is attenuated by mutation or altered cellular location autoimmune disease can result. In this chapter, we detailed methods to overexpress and purify recombinant TREX1 enzyme and to measure ssDNA and dsDNA exonuclease activities in vitro. The biochemical activities of TREX1 mutants determined using these assays have contributed to our understanding of TREX1-associated autoimmune disease. In addition, these methods help elucidate the mechanisms by which TREX1 mutation affects exonuclease activity and cytosolic nucleic acid sensing.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (AI116725), National Institute of General Medical Sciences (GM110734), Lupus Research Alliance, Wake Forest Innovations, Comprehensive Cancer Center of Wake Forest University National Cancer Institute (Center Support Grant, P30CA012197), and a NIH NRSA Predoctoral Fellowship (T32-GM095440).
References
- Ablasser A, Hemmerling I, Schmid-Burgk JL, et al. (2014). TREX1 deficiency triggers cell-autonomous immunity in a CGAS-dependent manner. The Journal of Immunology, 192(12), 5993–5997. [DOI] [PubMed] [Google Scholar]
- Silva de U, Choudhury S, Bailey SL, et al. (2007). The crystal structure of TREX1 explains the 3′ nucleotide specificity and reveals a polyproline II helix for protein partnering. Journal of Biological Chemistry, 282(14), 10537–10543. [DOI] [PubMed] [Google Scholar]
- Fye JM, Coffin SR, Orebaugh CD, Hollis T, & Perrino FW. (2014). The Arg-62 residues of the TREX1 exonuclease act across the dimer interface contributing to catalysis in the opposing protomers. The Journal of Biological Chemistry, 289(16), 11556–11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fye JM, Orebaugh CD, Coffin SR, Hollis T, & Perrino FW. (2011). Dominant mutations of the TREX1 exonuclease gene in lupus and aicardi-goutières syndrome. The Journal of Biological Chemistry, 286(37), 32373–32382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieves JL, Fye JM, Harvey S, et al. (2015). Exonuclease TREX1 degrades double-stranded DNA to prevent spontaneous lupus-like inflammatory disease. Proceedings of the National Academy of Sciences of the United States of America, 112(16), 5117–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan JA, Fan J, Momand J, et al. (2007). WRN exonuclease activity is blocked by DNA termini harboring 3’ obstructive groups. Mechanisms of Ageing and Development, 128(3), 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M, Fermaintt CS, Gao N, et al. (2015). Cytosolic nuclease TREX1 regulates oligosaccharyltransferase activity independent of nuclease activity to suppress immune activation. Immunity, 43(3), 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucej M, Fermaintt CS, Yang K, Irizarry-Caro RA, & Yan N. (2017). Mitotic phosphorylation of TREX1 C terminus disrupts TREX1 regulation of the oligosaccharyltransferase complex. Cell Reports, 18(11), 2600–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Kirsch MA, Chowdhury D, Harvey S, et al. (2007). A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. Journal of Molecular Medicine, 85(5), 531–537. [DOI] [PubMed] [Google Scholar]
- Lehtinen DA, Harvey S, Mulcahy MJ, Hollis T, & Perrino FW. (2008). The TREX1 double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. The Journal of Biological Chemistry, 283(46), 31649–31656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manils J, Gómez D, Salla-Martret M, et al. (2015). Multifaceted role of TREX2 in the skin defense against UV-induced skin carcinogenesis. Oncotarget, 6(26), 22375–22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur DJ, & Perrino FW. (1999). Identification and expression of the TREX1 and TREX2 CDNA sequences encoding mammalian 3′ → 5′ exonucleases. Journal of Biological Chemistry, 274(28), 19655–19660. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, & Perrino FW. (2001a). Structure and expression of the TREX1 and TREX2 3′ → 5′ exonuclease genes. Journal of Biological Chemistry, 276(18), 14718–14727. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, & Perrino FW. (2001b). Excision of 3′ Termini by the Trex1 and TREX2 3′ → 5′ Exonucleases characterization of the recombinant proteins. Journal of Biological Chemistry, 276(20), 17022–17029. [DOI] [PubMed] [Google Scholar]
- Morita M, Stamp G, Robins P, et al. (2004). Gene-targeted mice lacking the trex1 (DNase III) 3′ → 5′ DNA exonuclease develop inflammatory myocarditis. Molecular and Cellular Biology, 24(15), 6719–6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orebaugh CD, Fye JM, Harvey S, Hollis T, & Perrino FW. (2011). The TREX1 exonuclease R114H mutation in aicardi-goutières syndrome and lupus reveals dimeric structure requirements for DNA degradation activity. The Journal of Biological Chemistry, 286(46), 40246–40254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orebaugh CD, Fye JM, Harvey S, et al. (2013). The TREX1 C-terminal region controls cellular localization through ubiquitination. The Journal of Biological Chemistry, 288(40), 28881–28892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra D, Manils J, Castellana B, et al. (2009). Increased susceptibility to skin carcinogenesis in TREX2 knockout mice. Cancer Research, 69(16), 6676–6684. [DOI] [PubMed] [Google Scholar]
- Perrino FW, Harvey S, McMillin S, & Hollis T. (2005). The human TREX2 3′ → 5′ -exonuclease structure suggests a mechanism for efficient nonprocessive DNA catalysis. Journal of Biological Chemistry, 280(15), 15212–15218. [DOI] [PubMed] [Google Scholar]
- Perrino FW, Krol A, Harvey S, et al. (2004). Sequence variants in the 3′ → 5′ deoxyribonuclease TREX2: identification in a genetic screen and effects on catalysis by the recombinant proteins. Advances in Enzyme Regulation, 44(1), 37–49. [DOI] [PubMed] [Google Scholar]
- Perrino FW, Miller H, & Ealey KA. (1994). Identification of a 3′ → 5′ -exonuclease that removes cytosine arabinoside monophosphate from 3’ termini of DNA. Journal of Biological Chemistry, 269(23), 16357–16363. [PubMed] [Google Scholar]
- Rego SL, Harvey S, Simpson SR, et al. (2018). TREX1 D18N mice fail to process erythroblast DNA resulting in inflammation and dysfunctional erythropoiesis. Autoimmunity, 51(7), 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Rodero MP, & Crow YJ. (2015). Human disease phenotypes associated with mutations in TREX1. Journal of Clinical Immunology, 35(3), 235–243. [DOI] [PubMed] [Google Scholar]
- Sakai T, Miyazaki T, Shin D-M, et al. (2017). DNase-active TREX1 frame-shift mutants induce serologic autoimmunity in mice. Journal of Autoimmunity, 81, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, & Medzhitov R. (2008). Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell, 134(4), 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-G, Lindahl T, & Barnes DE. (2007). Trex1 exonuclease degrades SsDNA to prevent chronic checkpoint activation and autoimmune disease. Cell, 131(5), 873–886. [DOI] [PubMed] [Google Scholar]