Abstract
Outcomes for patients with chronic myeloid leukemia (CML) have substantially improved due to advances in drug development and rational treatment intervention strategies. Despite these significant advances there are still unanswered questions on patient management regarding how to more reliably predict treatment failure at the time of diagnosis and how to select frontline tyrosine kinase inhibitor (TKI) therapy for optimal outcome. The BCR-ABL1 transcript level at diagnosis has no established prognostic impact and cannot guide frontline TKI selection. BCR-ABL1 mutations are detected in approximately 50% of TKI resistant patients but are rarely responsible for primary resistance. Other resistance mechanisms are largely uncharacterized and there are no other routine molecular testing strategies to facilitate the evaluation and further stratification of TKI resistance. Advances in next-generation sequencing technology has aided the management of a growing number of other malignancies, enabling the incorporation of somatic mutation profiles in diagnosis, classification and prognostication. A largely unexplored area in CML research is whether expanded genomic analysis at diagnosis, resistance and disease transformation can enhance patient management decisions, as has occurred for other cancers. The aim of this article is to review publications that reported mutated cancer-associated genes in CML patients at various disease phases. We discuss the frequency and type of such variants at initial diagnosis and at the time of treatment failure and transformation. Current limitations in the evaluation of mutants and recommendations for future reporting are outlined. The collective evaluation of mutational studies over more than a decade suggests a limited set of cancer-associated genes are indeed recurrently mutated in CML and some at a relatively high frequency. Genomic studies have the potential to lay the foundation for improved diagnostic risk classification according to clinical and genomic risk, and to enable more precise early identification of TKI resistance.
Introduction
Chronic myeloid leukemia (CML) is a clonal disorder of hematopoietic stem cells resulting from the BCR-ABL1 rearrangement. The BCR-ABL1 fusion protein is a target of tyrosine kinase inhibitor (TKI) therapy. Advances over the last two decades in drug development, patient management guidelines and the introduction of standardized molecular monitoring based on the BCR-ABL1 quantitative PCR technique have dramatically improved patient outcomes. Molecular monitoring is now recommended to routinely assess TKI response and to guide disease management.[1,2] BCR-ABL1 kinase domain mutation analysis is the primary molecular monitoring tool for the assessment of TKI resistance.
Recent advances in next-generation sequencing (NGS) technology have allowed investigation of genetic abnormalities in small subpopulations of malignant clones and initiated the era of precision medicine for the management of cancer.[3] Somatic mutation profiles are now incorporated at diagnosis, risk classification and prognostication in diverse subtypes of myeloid neoplasms, including acute myeloid leukemia (AML), myelodysplastic syndrome and myeloproliferative neoplasms. In particular, genetic factors and the associated biology identified by NGS technology in AML have provided critical insights on mutational landscapes and clonal evolution in myeloid leukemia, and have driven drug development. In AML, the mainstay of prognostic stratification is based on mutation profiles including FLT3, NPM1, CEBPA and gene fusions.[4] The presence of mutant FLT3, ASXL1, RUNX1 or TP53 indicates adverse prognosis, while the presence of a bi-allelic CEBPA mutation aligns with favorable outcomes in AML patients. Accordingly, a mutation profile-based risk stratification system can guide treatment selection for leukemia patients.
In CML, risk scores, such as Sokal,[5] EUTOS[6] and ELTS,[7] are based on clinical parameters at diagnosis and can stratify patients according to their long-term prognosis. Historically considered a genetically uniform disease because of the universal presence of the BCR-ABL1 rearrangement, a largely unexplored area in CML research is the risk conferred by additional genomic lesions at diagnosis. A recent study did indeed demonstrate that genomic profiling of the BIM deletion polymorphism in Asian populations can be used to complement clinical risk scores.[8] However, a comprehensive and systematic evaluation of the mutational landscape at CML diagnosis, TKI resistance and at blast crisis (BC) transformation is lacking. Whole-exome sequencing of tumor/normal pairs provides an unbiased discovery tool, with the potential for unearthing a broad range of somatic coding variants of potential clinical relevance. However, studies using this strategy are limited and most studies examined a small set of genes or regions that were selected based on their known association with hematologic cancer.
The aim of this review is to analyze the current evidence in the published literature of mutated genes in adult patients with CML, and to collate the data and evaluate for mutant recurrence and potential significance at diagnosis, TKI resistance or transformation. We discuss whether clonal hematopoiesis[9-11] could complicate the identification and interpretation of leukemia-specific mutant gene detection, outline criteria for assignment of clinical relevance for somatic mutations and suggest the minimum core set of genes for targeting in future studies. The available data would suggest that incorporation of genomic factors into risk calculations will likely enhance the management of CML, as has occurred with other hematologic malignancies,[4,12,13] and is worthy of further exploration.
Mutational landscape at CML diagnosis
One of the first reports of molecular heterogeneity at chronic phase (CP) diagnosis was a case report published in 2005.[14] The patient failed interferon and imatinib therapy and progressed to accelerated phase (AP) within 1 year. The emergence of trisomy 21 prompted Sanger sequencing of the RUNX1 gene, which revealed a known loss-of-function variant within the runt domain. A BCR-ABL1 kinase domain mutation was also detected at imatinib resistance and at AP. To our knowledge, this was the first report of a concurrent BCR-ABL1 mutation and a cancer-associated gene. Importantly, sequencing at diagnosis only revealed the RUNX1 variant. The report suggested that additional screening at diagnosis of some defined genes could be complementary to clinical scoring systems to aid therapeutic decisions.
Genes of clinical relevance were not defined in 2005 and the standard technology of the time, Sanger sequencing, precluded a large scale search for mutated genes. RUNX1 was a good candidate for mutation screening. In 1988 a chromosomal translocation at the RUNX1 locus was described at CML diagnosis in a patient with ensuing BC 15 months after diagnosis[15] and a RUNX1 runt domain mutation was first described in 1999 at BC.[16] By the early 2000’s RUNX1 was one of the most frequently mutated genes in leukemia.[17] It is essential for normal hematopoiesis, functioning as a master regulator of myeloid and lymphoid differentiation.[18] Its function is deregulated by translocation, mutation and amplification.[19] However, RUNX1 was not generally selected as a candidate gene for sequencing at CML diagnosis in the early studies.
In 2011 Roche-Lestienne et al. selected four genes for DNA sequencing in 91 patients at CML diagnosis and at other timepoints.[20] The genes were TET2, ASXL1, IDH1 and IDH2, and were selected because they were somatically mutated in patients with various BCR-ABL1-negative hematologic malignancies. One of the 91 patients had TET2 mutations at diagnosis and this patient transformed to MBC at 4 months. No mutations were detected in IDH1 or IDH2 at diagnosis, which is consistent with another study that detected no IDH1/2 mutations among 50 patients at the time of CP CML diagnosis.[21] Similarly, an NGS study of 49 patients at CP diagnosis only detected an IDH1 variant (R132H) in one patient with lymphoid BC (LBC) at 6 months of imatinib.[22] These studies suggest that IDH1/2 variants are rare at diagnosis. In contrast, the Roche-Lestienne study found ASXL1 frameshift and nonsense variants in 8/91 patients (8.8%) at diagnosis. The prognostic significance of mutated ASXL1 was not clear from this study. Of the 8 patients with mutated ASXL1, 3 achieved a major molecular response (MMR) and the other 5 had primary or acquired imatinib resistance. Three of the five patients with resistance had both ASXL1 and BCR-ABL1 kinase domain mutations. Overall, this initial study of only 4 genes found variants in 9.9% of patients at the time of diagnosis in CP.
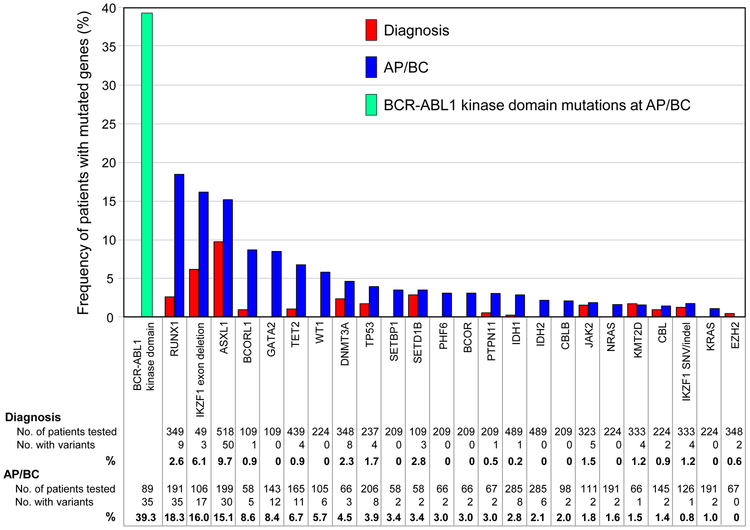
The clinical relevance of mutated ASXL1 at the time of CML diagnosis has not been clarified with subsequent genomic studies. Multiple studies reported patients at diagnosis with mutated cancer-associated genes.[14,20-32] The genes are as defined by the Catalogue of Somatic Mutations in Cancer (COSMIC) Cancer Gene Census.[33] Many of the studies did not detail whether the patients were in CP at diagnosis or how the patients were selected for testing. Therefore, the results at this stage cannot be considered indicative of the actual frequency of cancer-associated genes at CP diagnosis. Most samples were sequenced using Sanger sequencing or an NGS gene panel for specific genes known to be associated with hematologic cancer. Whole-exome, whole-genome or whole-transcriptome sequencing (RNA-Seq) was also performed (Table 1). Figure 1 summarizes the number of patients sequenced at diagnosis for specific cancer-associated genes and the frequency of patients with mutations.[14,20-32] Only genes mutated in more than one patient at diagnosis or AP/BC are listed. The analysis is focused on single nucleotide variants (SNVs), small insertions and deletions (indels) and IKZF1 exon deletions. Seven genes were recurrently mutated in more than one study: ASXL1, RUNX1, KMT2D, DNMT3A, JAK2, TP53 and TET2. Mutations in three of these genes, ASXL1, RUNX1 and TP53, confer an adverse risk for patients with AML.[4] ASXL1 was the most frequently sequenced gene (518 patients) and was also the most frequently mutated: 9.7% of all adult patients tested. The IDH1/2 hotspot mutation sites were sequenced in 489 patients at diagnosis. Only one patient had an IDH1 mutation.[22] This data then suggests that in CML mutated ASXL1 is relatively common at diagnosis, whereas IDH1/2 mutants are exceedingly rare.
Table 1.
Studies that sequenced one or more genes of samples collected at the time of diagnosis or at AP/BC in the TKI era
| Study* | Disease stage |
No. of patients |
Sequencing Method |
No. of genes |
Genes targeted in studies that included <10 genes |
|---|---|---|---|---|---|
| Corm et al. 2005 [14] | Diagnosis | 1 | Sanger | 1 | RUNX1 |
| Corm et al. 2005 [14] | BC | 1 | Sanger | 1 | RUNX1 |
| Zhang et al. 2008 [42] | AP/BC | 85 | Sanger | 13 | |
| Mullighan et al. 2008 [43] | AP/BC | 22 | SNP Array / PCR | IKZF1 deletion (PCR) | |
| Piccaluga et al. 2009 [44] | BC | 1 | Sanger | 1 | NPM1 |
| Boultwood et al. 2010 [45] | BC | 21 | Sanger | 3 | ASXL1 (21 patients), TP53, IKZF1 (2 patients) |
| Roche-Lestienne et al. 2011 [20] | Diagnosis | 91 | Sanger | 4 | ASXL1, TET2, IDH1, IDH2 |
| Roche-Lestienne et al. 2011 [20] | AP/BC | 20 | Sanger | 4 | ASXL1, TET2, IDH1, IDH2 |
| Soverini et al. 2011 [21] | Diagnosis | 1 | RNA-Seq | ||
| Soverini et al. 2011 [21] | BC | 1 | RNA-Seq | ||
| Soverini et al. 2011 [21] | Diagnosis | 49 | Sanger | 2 | IDH1, IDH2 |
| Soverini et al. 2011 [21] | BC | 119 | Sanger | 2 |
IDH1, IDH2 (119 patients) JAK2, IKZF1 exon deletion (4 patients) |
| Grossmann et al. 2011 [47] | BC | 39 | Targeted NGS panel / Sanger / PCR | 12 | |
| Makishima et al. 2011 [46] | BC | 40 | Sanger | 7 | ASXL1, TET2, IDH1, IDH2, JAK2, CBL, CBLB |
| Menezes et al. 2013 [23] | Diagnosis | 1 | Whole-exome | ||
| Menezes et al. 2013 [23] | BC | 1 | Whole-exome | ||
| Menezes et al. 2013 [23] | Diagnosis | 13 | Sanger | 2 | ASXL1, TP53 |
| Menezes et al. 2013 [23] | BC | 13 | Sanger | 2 | ASXL1, TP53 |
| Konoplev et al. 2013 [48] | BC | 5 | Sanger | 1 | NPM1 |
| Watkins et al. 2013 [24] | Diagnosis | 33 | Sanger | 1 | NPM1 |
| Watkins et al. 2013 [24] | BC | 14 | Sanger | 1 | NPM1 |
| Schmidt et al. 2014 [25] | Diagnosis | 15 | Targeted NGS panel | 25 | |
| Huang et al. 2014 [49] | BC | 1 | Whole-exome | ||
| Mitani et al. 2016 [26] | Diagnosis | 15 | Whole-exome | ||
| Valikhani et al. 2017 [27] | Diagnosis | 66 | Sanger | 2 | ASXL1, JAK2 |
| Mologni et al. 2017 [28] | Diagnosis | 19 | Whole-exome | ||
| Togasaki et al. 2017 [29] | Diagnosis | 24 | Whole-exome | ||
| Sloma et al. 2017 [30] | Diagnosis | 1 | Whole-genome | ||
| Sloma et al. 2017 [30] | BC | 1 | Whole-genome | ||
| Kim Blood et al. 2017 [50] | BC | 8 | Whole-exome | ||
| Kim Blood et al. 2017 [31] | Diagnosis | 100 | Targeted NGS panel | 92 | |
| Kim Blood et al. 2017 [31] | AP/BC | 8 | Targeted NGS panel | ||
| Sklarz et al. 2018 [51] | BC | 1 | Targeted NGS panel | 50 | |
| Branford et al. 2018 [22] | Diagnosis | 49 | Whole-exome / RNA-Seq | ||
| Branford et al. 2018 [22] | BC | 39 | Whole-exome / RNA-Seq | ||
| Magistroni et al. 2019 [61] | BC | 8 | Whole-exome | ||
| Nteliopoulos et al. 2019 [32] | Diagnosis | 124 | Targeted NGS panel | 71 |
Studies were included if the disease stage was clearly defined as diagnosis or AP/BC. Patients defined as advanced phase at diagnosis were not included in the diagnosis group. The sensitivity of mutant detection and the criteria for selection of patients for sequencing varied between the studies.
Figure 1. Frequency of mutated cancer genes at diagnosis and AP/BC.
The data from 15 studies of patients at diagnosis and 20 studies at AP/BC are reported where cancer genes were mutated in more than one patient at diagnosis and/or BC.[14,20-32,42-51] Some variants were not included, such as the ASXL1 E1102D variant, which is reported in the population databases at a frequency suggesting it represents the germline and is not pathogenic. Other variants reported in PAX5, TP53 and TET2 were also not included for the same reason. Four other genes associated with hematologic malignancy were mutated in one patient each at BC: U2AF1, XPO1, NPMI and SETD2. The number of samples sequenced for individual genes is highly variable and ranges from 49 for IKZF1 exon deletions to 518 for ASXL1 exon 12 variants at diagnosis. BCR-ABL1 kinase domain mutation status was reported in few studies. Therefore, the confidence for the mutation frequency for some genes is low compared with other genes. Only genes listed in the COSMIC Cancer Gene Census are included.
Does clonal hematopoiesis have prognostic value at CML diagnosis?
In 1981, Fialkow et al. proposed that the Philadelphia chromosome (Ph) may not always initiate CML and suggested a multistep process.[34] It is now known that normal hematopoietic stem cells acquire certain somatic mutations as a part of the aging process, a phenomenon referred to as clonal hematopoiesis of indeterminate potential (CHIP) or age-related clonal hematopoiesis.[9-11] Mutations involved in CHIP include DNMT3A, TET2, ASXL1, TP53, IDH1, IDH2, SF3B1, SRSF2 and U2AF1. These clonal acquired events that occur with normal aging slightly increase the risk of hematologic cancer and are associated with an increased risk of all-cause mortality in the general population. They are detected in <1% of people under 50, but in >10% over 65,[9] which can complicate the evaluation of hematologic cancer.
Schmidt et al. reported sequencing data focusing on 29 CML patients with a 25 gene panel that explored Ph-negative and Ph-positive cell subsets.[25] Some patients had the same gene mutation present in both Ph-negative and Ph-positive cells, including one patient carrying a DNMT3A mutation in both cell subsets. This suggests that some mutations precede the BCR-ABL1 rearrangement.
Kim et al. recently reported CML patients carrying mutations in CHIP-associated genes, including DNMT3A, TET2, ASXL1, BCOR and CREBBP, without any significant change in the variant allele frequency following TKI therapy.[31] This finding suggests that cells carrying a CHIP mutation but without BCR-ABL1 rearrangement can expand after successful TKI therapy and can contribute to hematopoiesis. It also demonstrates the potential for selection of small clones that may contribute to the development of other malignancies in the future. One could speculate whether this is related to the development of Ph-negative AML observed in some CML patients with chromosomal abnormalities in Ph-negative metaphases.
Mutations associated with CHIP in the non-leukemic cells are evident by persistent detection in remission. A recent study in AML demonstrated that the detection of the CHIP-associated mutations DNMT3A, TET2 and ASXL1 in remission samples was not prognostic for relapse, whereas low-level detection of other mutations in remission was significantly associated with relapse.[35] The impact of mutations at diagnosis likely varies depending on whether they occur in the Ph-positive or Ph-negative clone. Sequencing a corresponding sample at diagnosis that was not derived from the leukemic clone will be needed to clarify the interpretation of mutations.
In our review of the literature, ASXL1 was the most frequently mutated gene at diagnosis and is also one of the most frequently mutated genes associated with CHIP. There is no strong evidence for an age-related mechanism of acquisition of mutated ASXL1 in CML. The median age at diagnosis for patients with ASXL1 exon 12 mutation was reported in two small series to be 55 years (range 44-62, n=8)[31] and 47 years (range 37-82, n=9).[22] Only 3 of these patients were >60 years of age. Furthermore, a study by Ernst et al. in children and young adults (median age 14 years) found ASXL1 mutations at CML diagnosis in 6 of 21 patients with no other gene mutations detected.[36] Clonal analysis in one patient demonstrated the ASXL1 mutation was acquired after BCR-ABL1 acquisition.
At this stage the role that CHIP may play in CML is unknown. Furthermore, it is not known if the BCR-ABL1 fusion occurs more frequently in a clone bearing a CHIP mutation than a clone without a CHIP mutation.
Mutational landscape at advanced phase CML
The mechanisms underlying transformation of CML to BC are not well understood. Single genetic abnormalities, such as BCR-ABL1, can be very successfully targeted, but molecular heterogeneity presents a more difficult therapeutic situation, contributing to the poor response to therapy for patients with BC. It has been proposed that unfaithful DNA repair in BCR-ABL1 expressing cells leads to an accumulation of mutations. Up to 60-80% of patients in BC have additional clonal chromosomal abnormalities,[37] many of which are known to be associated with a worse prognosis.[38] However, the extent that mutations in hematologic cancer genes contribute to transformation is unknown. A greater understanding of these molecular events may lead to improved therapeutic approaches for BC and possibly multi-agent therapy to combat specific mutations.
Sequencing of patients in the advanced phase of CML in search of mutated genes has been ongoing for almost 2 decades, either as single gene candidates or more recently, in an unbiased fashion using whole-exome or genome sequencing or RNA-Seq. In the early 1990s, sequencing studies revealed a high rate of point or frameshift mutations in TP53 at the time of BC: 13/65 patients (20%).[39-41] In the TKI era a number of studies have revealed mutations in other genes at varying frequencies in advanced phase patients (Figure 1).[14,20-23,30,31,42-51] Once again, the analysis is focused on SNVs/indels in cancer-associated genes[33] and IKZF1 exon deletions. BCR-ABL1 kinase domain mutation status was only reported for 89 patients and were detected in 39%. RUNX1 was most frequently mutated (18.3%) and only 2 other genes were mutated at a frequency >15%: IKZF1 (exon deletions, 16.0%) and ASXL1 (15.1%). The BC phenotype was not reported for most patients. However, IKZF1 deletions are associated with a lymphoid phenotype.[43] Thirteen genes were mutated in more than one study: RUNX1, IKZF1, ASXL1, GATA2, TET2, TP53, SETBP1, PTPN11, IDH1, IDH2, NRAS, JAK2 and CBL.
These genomic studies consistently found a lower frequency of mutated TP53 (3.9% overall) than was reported in the pre-TKI studies (20%).[39-41] WT1 was among the most frequently mutated genes in one study,[47] but has been reported at a lower frequency in other studies. An early targeted gene study of variants associated with BC identified a recurrent gain-of-function GATA2 variant (L359V) in 11% of patients at BC.[42] However, to our knowledge, this variant has not been reported in other studies that included GATA2 in the variant screen, although the number of patients studied is small. Mutant BCOR/BCORL1 was reported in one published study at a relatively high frequency at BC (15%) using whole-exome sequencing and RNA-Seq,[22] and is reported in other mutation screens in abstract form.[52,53]
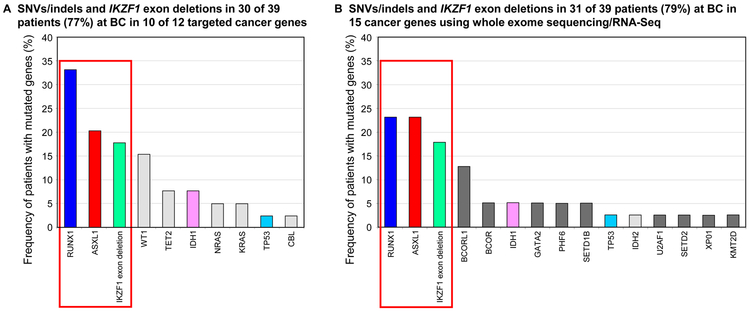
The studies to date suggest that a relatively small number of clinically relevant genes may be recurrently mutated in advanced phase CML. The spectrum of mutated cancer genes can be directly compared for two studies that each included 39 patients in BC. The first study by Grossman et al. targeted 12 genes, which included an analysis of IKZF1 exon deletions.[47] These 12 genes overlapped those of the second study by Branford et al. which used whole-exome and RNA-Seq.[22] Copy number variation, genomic breakpoint detection and atypical RNA splicing identified IKZF1 deletions in the study by Branford et al. Figure 2 highlights that SNVs/indels in RUNX1 and ASXL1, plus IKZF1 exon deletions, were the most frequent variants in both studies. Mutations in one or more of these 3 genes were detected in 23/39 patients (59%) in the targeted gene study[47] and in 21/39 patients (54%) in the unbiased mutation screen of the second study.[22] The frequency of mutations in RUNX1, IKZF1 and ASXL1 is comparable or higher than the frequency of BCR-ABL1 kinase domain mutations.[54-56] Interestingly, both studies reported a very high frequency of co-occurrence of mutated cancer genes and BCR-ABL1 kinase domain mutations: 11/13 patients (85%)[47] and 17/19 patients (89%)[22] with kinase domain mutations.
Figure 2. Comparison of variants in two studies of patients at BC.
(a) Mutant genes at BC reported in the study of Grossman et al. [47]. (b) Mutant genes at BC reported in the study of Branford et al. [22]. The most frequently mutated genes were common to both studies: RUNX1, ASXL1 and IKZF1 exon deletions. These were detected in >50% of patients in both studies. Genes mutated in both studies are indicated by matching colors. Light grey shading indicates genes that were sequenced in both studies but were only detected in one of the studies. Dark grey shading indicates the genes that were only sequenced in one study.
The comparison between these two studies is revealing. By sequencing only 12 genes, Grossmann et al. found that 10 of the genes were mutated in 30/39 patients (77%),[47] whereas sequencing of all genes by Branford et al. found the same type of mutations (SNVs/indels and IKZF1 deletions) in 15 cancer genes in 31/39 patients (79%).[22] Only 5 of the 10 cancer genes mutated in the targeted gene study were also mutated in the study of Branford et al. Figure 2. This suggests a core set of cancer genes account for most of the SNVs/indels at BC and a variety of other genes are mutated at low frequency.
An important consideration when searching for mutated genes associated with disease transformation is that a screen for only SNVs/indels and IKZF1 deletions does not detect a major class of mutation, which is gene fusion. Multiple gene fusions have been described in advanced phase CML and fusion partners include RUNX1,[57] MLL (KMT2A)[58] and CBFB.[59] RNA-Seq analysis for the 39 patients at BC in the study by Branford et al. revealed a high frequency of gene fusions.[22] Ten of the 39 patients (26%) had 11 fusions at BC where a partner gene was a known hematologic cancer gene, including MLL, RUNX1, IKZF1, CBFB, MECOM and PAX5. Some of the fusions were novel and/or cytogenetically cryptic, including cryptic MLL fusions. In 15% of the 39 patients, the fusion was the sole additional mutated cancer gene at BC. This analysis further demonstrates the important role of mutated RUNX1 in CML transformation. Using whole-exome sequencing and RNA-Seq, novel RUNX1 exon deletion and fusion, in addition to SNVs and indels, were detected at BC and at CML diagnosis in some patients with poor outcome.[22] Overall, the combined sequencing strategy revealed mutated cancer genes in 37/39 patients (95%) and the types of variants included SNVs/indels, focal deletions and fusions.[22] The majority of these patients were treated with imatinib and only two patients had BCR-ABL1 kinase domain mutations as the sole detectable mutation.
Dynamics of mutation profiles following TKI therapy in CML
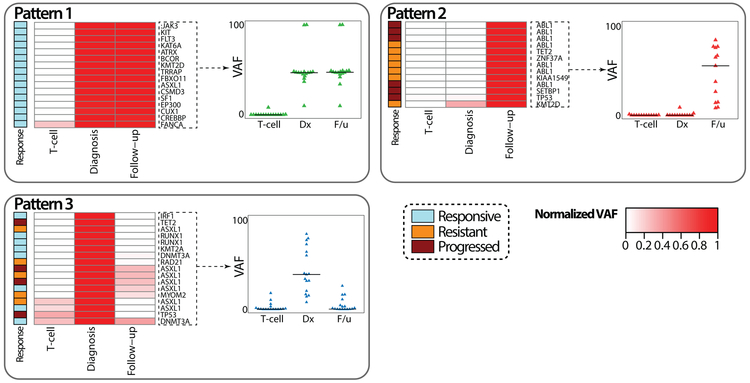
Most of the studies focusing on mutation profiles in CML have investigated a single time point: at diagnosis, resistance or transformation. Accordingly, they generally lack an evaluation of the serial changes of mutation profiles throughout the disease course. However, two studies have evaluated the dynamics of somatic mutations over time.[22,31] Kim et al. studied 100 patients at diagnosis pre-TKI and at 6-12 months after initiation of TKI therapy. The T-cell fraction was also tested as a representative of the hematopoietic stem cell fraction. This study suggested five patterns of mutation profile dynamics with TKI therapy. Pattern 1 included ASXL1, BCOR, CREBBP1 and several genes involved in transcription (Figure 3). The mutations were not cleared at all with TKI therapy despite a significant reduction of BCR-ABL1 transcript level, suggesting that these variants exist independent of the Ph-positive clone. Pattern 2 included ABL1, TP53, KMT2D, and TET2. These were strongly associated with TKI resistance or disease progression to advanced disease. Patterns 3 to 5 included somatic mutation of epigenetic pathway genes in the Ph-positive clone. There was a linear relationship between the reduction of mutant allele burden and BCR-ABL1 transcript level reduction in Pattern 3, which implies these mutations were acquired in the Ph-positive clone. Longitudinal follow-up of patients in the study of Branford et al. also demonstrated different patterns of mutation acquisition, retainment and clearance.[22] Clonal competition between mutant genes was evident over time. Interestingly, mutations in cancer-associated genes predated the acquisition of BCR-ABL1 kinase domain mutations in 62% of patients.
Figure 3. Dynamics of somatic mutation profiles following TKI therapy.
Five patterns of mutation dynamics are noted according to their longitudinal changes in mutation burden following TKI therapy. Pattern 1 does not show any significant changes but all the cases responded to TKI therapy optimally, implying independence of the clone carrying the mutation from the Ph-positive clone. Pattern 2 represents new acquisition of mutations, associated with treatment failure. Pattern 3 shows a linear relationship between the reduction rate of mutation allele burden and BCR-ABL1 transcript level reduction, implying that this mutation arose from the Ph-positive clone. Modified from Kim et al. [31].
These findings highlight that additional studies are needed to unravel the clinical relevance of mutant genes detected at diagnosis. Single-cell sequencing could also be important to define whether mutants represent single clones or multiple independent clones that can be selected, as occurs in AML.[60]
Emerging novel recurrently mutated genes
Broad mutation screens using NGS are revealing potentially novel mutated genes. However, a variant predicted to be damaging in a gene that is not associated with hematologic cancer requires careful assessment before assigning clinical relevance. Recurrence of damaging variants in individual genes that occur within or across genomic studies in patients with similar clinical phenotype provides a high level of evidence for causality.[3] For example, using whole-exome and RNA-Seq, frameshift/nonsense variants in the SETD1B gene were recently reported in 3 patients at CP diagnosis of CML. These patients were treated with first line imatinib and all transformed to BC.[22] Two other recent studies reported frameshift/nonsense variants in SETD1B at BC.[52,53] Interestingly, this histone methyltransferase was recently reclassified as a putative tumor suppressor gene in the COSMIC Cancer Gene Census,[33] with a colorectal and endometrial cancer association. SETD1B is categorized as a Tier 2 cancer gene, meaning it has a strong indication for a role in cancer, but the evidence is less extensive than other cancer genes. Another novel gene that was somatically mutated at CML BC across 3 whole-exome sequencing studies is UBE2A.[22,53,61] The gene is a ubiquitin-conjugating enzyme and mutated UBE2A has no reported association with cancer. SETD1B and UBE2A could have emerging roles as contributors to CML transformation. Mutation of these genes frequently co-occurred with other mutated genes.[22,53,61] Novel recurrent mutations should ideally be evaluated in vitro or in vivo using experimental models to better define the degree of their contribution to resistance.[61] Pooling datasets is a powerful means to strengthen phenotypic associations to identify less frequent, but potentially relevant genes. It also provides guidance for establishing genes for target in genomic studies.
How to assign clinical relevance to genes and variants for CML
The data presented in this review has focused on genes that have a known role in cancer and, in some cases, on the presumption that the variants were pathogenic. For some variants, sufficient information was supplied to review their frequency in population databases.[62,63] Some mutations reported in CML patients have a relatively high population frequency and have actually been described as probable non-pathogenic germline variants, such as the ASXL1 E1102D variant,[64] which is listed in population databases at a minor allele frequency of ~1%. Population databases do indeed contain somatic variants but their allele frequency is usually extremely low. The inclusion of a non-leukemic control for confirmation of the somatic status of variants aids interpretation. Types of controls include remission samples, buccal cells, saliva, skin biopsy, hair follicles, mature T cells, cultured mesenchymal stromal cells or nail clippings. However caution is advised in interpreting this data as saliva and skin biopsies may be contaminated with leukemic cells[22,65] and controls should be assessed for absence of the BCR-ABL1 fusion. A non-leukemic control should ideally be sourced at CML diagnosis for potential future use to assess the somatic status of variants.
Specific variants reported in general or region-specific population databases at an allele frequency ≥1% were excluded from the summary in Figure 1, which highlights the importance of interrogating multiple factors before assigning clinical relevance to variants. These include, but are not limited to, the impact of variants using in silico prediction tools,[66-71] conservation scores,[72] occurrence in important protein domains[73-75] and frequency in population databases. Listing in the COSMIC database[76] is valuable but does not confirm pathogenicity and clinical relevance.
A variant annotation pipeline is a valuable aid when assessing the relevance of variants. Table 2 is an example of an annotation pipeline where several factors were used to assess variants and to assign clinical relevance.[22] Seven variants in the cancer gene TP53 are shown but only 3/7 were deemed clinically relevant. One of the variants, p.Pro72Arg/c.215C>G, is listed 162 times in COSMIC but has a very high frequency in all population databases. It is a non-pathogenic germline variant. In contrast, a novel variant, p.Gly117Arg/c.349G>A, is not listed in COSMIC but meets multiple criteria for clinical relevance. Another important consideration is the type of variant in individual genes. The clinical relevance of a missense variant in a strong hematologic cancer gene such as MLL is questionable, whereas a gene fusion involving MLL is highly relevant.
Table 2.
Variant annotation aids the interpretation of clinical relevance
| Variant number |
Amino acid change | Chrom | Position, hg19 |
HGNC symbol |
Effect | Ensembl transcript ID |
COSMIC count, Version 86 |
COSMIC Cancer Gene Census |
gnomAD % |
1KG ALT Freq % |
UK10K ALT Freq % |
ExAC ALT % |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1* | p.P72R/c.215C>G | 17 | 7579472 | TP53 | missense | ENST00000269305 | 162 | TRUE | 66 | 54 | 75 | 66 | |
| 2 | p. V73G/C.218T>G | 17 | 7579469 | TP53 | missense | ENST00000269305 | TRUE | ||||||
| 3 | p.G117R/c.349G>A | 17 | 7579338 | TP53 | missense | ENST00000269305 | TRUE | ||||||
| 4 | p.S185N/c.554G>A | 17 | 7578376 | TP53 | missense | ENST00000269305 | 1 | TRUE | 0.0004 | 0.001 | |||
| 5 | p.N210D/c.628A>G | 17 | 7578221 | TP53 | missense | ENST00000269305 | 1 | TRUE | |||||
| 6 | p.R248Q/c.743G>A | 17 | 7577538 | TP53 | missense | ENST00000269305 | 1085 | TRUE | 0.002 | 0.005 | |||
| 7 | p.R283C/c.847C>T | 17 | 7577091 | TP53 | missense | ENST00000269305 | 22 | TRUE | 0.009 | 0.008 | |||
| Variant number |
Amino acid change | GERP | phyloP | CADD | SIFT | Poly Phen2 |
Mutation Taster |
FATHMM | InterPro | ALT allele Freq % |
Clinically relevant |
||
| 1* | p.P72R/c.215C>G | <2 | 1.355 | 0.4 | T | B | T | D | 49 | No | |||
| 2 | p.V73G/c.218T>G | <2 | <1 | 0.003 | T | B | T | D | 51 | No | |||
| 3 | p.G117R/c.349G>A | 4.75 | 2.63 | 32 | D | D | D | D | P53 DNA-binding domain | 32 | Yes | ||
| 4 | p.S185N/c.554G>A | <2 | <1 | 0.4 | T | B | T | D | P53 DNA-binding domain | 40 | No | ||
| 5 | p.N210D/c.628A>G | <2 | <1 | 9.6 | T | B | T | D | P53 DNA-binding domain | 20 | No | ||
| 6 | p.R248Q/c.743G>A | 3.65 | 1.305 | 34 | D | D | D | D | P53 DNA-binding domain | 48 | Yes | ||
| 7 | p.R283C/c.847C>T | 4.01 | 1.278 | 26 | D | B | D | D | P53 DNA-biriding domain | 27 | Yes | ||
Note this this variant is listed in COSMIC with a high count, but is also reported at a high frequency in population databases and is a non-pathogenic germline variant
Abbreviations. Population databases: gnomAD, The Genome Aggregation Database, Version r2.0.2; 1KG, The 1000 Genomes Project Phase 3 V5b; UK10K, The UK10K project Version 20160215; ExAC, Exome Aggregation Consortium Version 0.3.1.
Evolutionary conservation scores: GERP, Genomic Evolutionary Rate Profiling for SNVs. The higher the score, the more evolutionarily conserved the position is. Scores ≥2 were considered conserved; phyloP conservation score for SNVs. Scores ≥1 were considered conserved.
In silico prediction tools for SNVs: CADD, A Combined Annotation Dependent Depletion score Version 1.3. The higher the score, the more deleterious the variant. A score of 20 was used as a threshold for deleteriousness; SIFT, Sorting Intolerant from Tolerant. T= tolerated, D=damaging; Polyphen2, Polymorphism Phenotyping v2. B=benign, D=probably damaging; MutationTaster, T=tolerated, D=probably damaging; FATHMM, Functional Analysis through Hidden Markov Models. T=tolerated, D=damaging. The pipeline uses dbNSFP Version 2.6 for SIFT, Polyphen2, MutationTaster and FATHMM.
InterPro: Protein sequence analysis and classification. Provides protein domain information. hg19 version from the UCSC Genome Browser.
Clinically relevant variants were conserved variants with a population frequency ≤0.1%, CADD score ≥20 and predicted to be damaging by 3 of 4 prediction tools.
In order to prioritize genes and specific variants for inclusion in testing regimes, the evidence for clinical relevance should be examined. Evidence is enhanced by overlaying functional data, if available. For example, variants that occur at amino acid R132 of the IDH1 gene are detected in many cancer types and have been shown to generate an oncometabolite that blocks differentiation and contributes to a transformed phenotype (reviewed in [77]). The relevance of variants at other amino acids of IDH1 is unknown, even for those variants where in silico prediction models suggest pathogenicity.
A unified approach to the interpretation of variants in CML and the provision of essential minimum information for each reported variant will aid our understanding of their clinical relevance. The inclusion of variants in mutation lists that are likely benign or those listed in population databases at a significant frequency, risk obscuring the true clinical relevance of variants. Damaging variants with unknown clinical relevance are important since their relevance may become apparent with recurrence in other patients with a similar phenotype, as is emerging for SETD1B and UBE2A variants.[22,52,53,61] However, we suggest that these should be clearly classified as having unknown clinical relevance in the absence of strong supporting evidence.
The Association for Molecular Pathology, American Society of Clinical Oncology and the College of American Pathologists have recently published joint consensus standards and guidelines for the interpretation and reporting of sequence variants in cancer.[78] A framework for somatic variant interpretation is outlined and a four-tiered system to assign clinical relevance to variants is proposed. These guidelines are analogous to the published standards for the interpretation of variants associated with mendelian disorders.[79] We propose the new guidelines for the assignment of clinical relevance be considered for genomic studies in CML. They recognize that the classification of some variants will change with new knowledge. The inclusion of essential minimum information will aid comparison and validation across studies (Table 3). This information includes variant level data and clinical information.
Table 3.
Essential minimum information required for unambiguous identification of variants and for the evaluation of clinical relevance
| Genomic | Example |
|---|---|
| Version of the human genome assembly | hg19/GRCh37 or hg38/GRCh38 |
| Coordinates (chromosome and sequence location) | Chr17:7577538 |
| mRNA transcript reference | NM_000546 or ENST00000269305 |
| Sequence variant | TP53 p.R248Q, c.743G>A |
| Variant allele frequency | 48% |
| Annotation | |
| Version of in silico prediction models and other databases | CADD v1.3 |
| Threshold values for assignment of deleteriousness | CADD score ≥20 and predicted to be damaging in 3 of 4 prediction algorithms |
| Population database minor allele frequency threshold (for inclusion of variant as probably somatic) | >0.1% |
| Patient information | |
| Disease stage at time of testing | Diagnosis |
| Treatment | First line imatinib |
| Outcome | Blast crisis |
The clinical significance of mutated cancer genes, particularly at the time of CML diagnosis, awaits comprehensive assessment. If their risk is established through rigorous evaluation of the evidence, the adoption of the variant Tier classification system may be appropriate to aid clinician interpretation.[78] Briefly, Tier I variants are those with strong clinical significance: therapeutic; prognostic; and diagnostic. Tier II variants have potential clinical significance. Tier III have unknown clinical significance and Tier IV are benign or likely benign. BCR-ABL1 kinase domain mutations are clearly Tier I variants.
Which genes should be included in a targeted gene panel?
We propose that expanded clinical association studies are necessary to establish the risk conferred by mutated cancer genes at CML diagnosis and during therapy. For many clinical applications, targeted DNA sequencing using panels of specific disease associated genes is the most cost-effective method of choice for mutation screening. It also allows high coverage of targeted regions for improved sensitivity of variant detection. We suggest the genes outlined in Figure 1 be among those included in gene panels. However, few current gene panels include the newly emerging recurrent genes of potential clinical relevance identified from unbiased mutation screens, such as SETD1B and UBE2A, which will limit their clinical evaluation. The inclusion of other genes with well-defined clinical relevance in myeloid cancer and acute lymphoblastic leukemia is valid, for example, splicing factor genes, cohesin complex genes, CDKN2A and PAX5. Nevertheless, the current data suggest that a core set of genes will account for the majority of mutated genes in CML, as shown in Figures 1 and 2, and relevant clinical evaluation across studies is achievable without total concordance of the genes included for assessment. An evaluation of mutated IKZF1 using gene panels will necessitate appropriate assessment for focal deletions, such as copy number variation analysis.[80]
Users of pre-designed gene panels should be mindful of limitations. The TruSight Myeloid Sequencing Panel (Illumina, San Diego, California) targets 54 genes. Only ABL1 exons 4 to 6 are targeted. Therefore, it will likely underestimate the frequency of imatinib resistant mutations since some are not targeted, such as H396R, which is frequently detected.[54,56,81,82] Furthermore, individual technologies cannot encompass the entire biological complexity and the diverse mutation types that appear to contribute to response.[22] Integration of multiple technologies will increase the diagnostic yield, such as the incorporation of a gene fusion screen,[83] which will provide an opportunity to establish the role of additional gene fusions in treatment outcome.
Can cancer gene mutation status aid therapeutic decisions for CML?
Genomic studies have identified additional mutated cancer genes at diagnosis and transformation. However, important questions need to be answered before the true clinical relevance can be established. In particular, their utility at diagnosis for response prediction and therapy decisions. Furthermore, the clinical relevance of low-level mutated cancer genes at diagnosis is unknown and studies are needed to define the sensitivity required when testing diagnostic samples. Newer technologies, such as duplex sequencing and single-cell genotyping, will improve the detection of rare variants[84] and these could further define the mutational landscape and clonal heterogeneity.
Only one study to date using a screening panel of 92 genes has systematically assessed outcomes according to mutation profile at diagnosis.[31] Of 100 consecutively treated patients, those carrying mutated epigenetic regulation pathway genes had inferior complete cytogenetic response by 12 months (53% versus 79%, P=0.02), MMR by 2 years (35% versus 62%, P=0.04) and MR4.5 by 3 years (26% versus 47%, P=0.03). This initial study requires a substantially expanded patient base with well curated clinical outcome data and replication in multiple studies before appropriate recommendations can be made for the inclusion of targeted gene screens at diagnosis. Moreover, clinical risk scores at diagnosis may inform the selection of patients for screening. One study found an association between Sokal score and the detection of cancer gene variants at diagnosis, but the patients were highly selected according to outcome.[22] Another study used whole-exome sequencing and found an increased burden of somatic mutations in patients resistant to first-line imatinib.[28] The study used a variant ‘oncogenic potential’ scoring system and found a correlation with outcome. Further studies are necessary to evaluate whether genomic guided risk will enhance prognostication when combined with clinical risk. Furthermore, expanded evaluation is required to establish whether a potential poor outcome for imatinib treatment could be overcome with specific therapeutic interventions such as the use of more potent TKIs. This has been suggested in one study, but patients were selected based on their outcome.[32] NGS provides the opportunity for high throughput sequencing of multiple potentially relevant genes and comprehensive studies over the next few years should establish the specific mutations or combination of mutations at diagnosis that confer a worse prognosis. Of course, this is highly dependent on the uniform interpretation of variants and the appropriate assignment of their clinical relevance. Thus, at the present time, mutation status at diagnosis cannot be recommended to guide therapy decisions.
An unambiguous causal role of mutated cancer genes may emerge for patients with primary TKI resistance who fail early treatment milestone responses and for patients who relapse after an initial response. Using gene expression analysis, Radich et al. demonstrated that patients who relapsed after an initial response had a gene expression signature resembling that of advanced phase disease.[85] The current recommendation for patients who fail TKI therapy is to perform BCR-ABL1 kinase domain analysis.[1,2,86] BCR-ABL1 mutations are rarely responsible for primary resistance, but failure to meet early milestone responses is associated with their detection or an increased risk of their subsequent acquisition.[87] Supplementing BCR-ABL1 kinase domain mutation analysis with targeted gene sequencing may increase the diagnostic yield of true biological resistance in some cases. Currently, in the absence of a BCR-ABL1 mutation, nonadherence to therapy cannot always be excluded. However, exquisite sensitivity is achieved for BCR-ABL1 mutations since cells bearing the BCR-ABL1 fusion are isolated through PCR amplification.[88,89] This level of sensitivity may not be achievable for other genes where there is an admixture of non-leukemic cells. The relative ratio of non-mutated to mutated alleles will vary depending on treatment response. Therefore, for patients with acquired TKI resistance, interrogating other genes for causal mutations could proceed when the BCR-ABL1 level is above a critical threshold, unless the sequencing method is capable of high sensitivity. The sensitivity required for early detection of mutated cancer genes is not yet established.
The current data suggest a role for expanded mutation sequencing at BC. Furthermore, there is emerging evidence for the co-occurrence of BCR-ABL1 kinase domain mutations and mutated cancer genes at BC.[22,47] These mutants may predate the acquisition of BCR-ABL1 mutations.[22] However, knowing the mutation status is primarily of clinical relevance if it opens opportunities for more effective therapy. In this setting rational combination therapy will likely be needed. Some mutants may be actionable currently or in the future. The most frequently mutated gene in advanced phase CML is the transcription factor RUNX1, which plays an indispensable role in hematopoiesis. It is required for efficient differentiation of both myeloid and lymphoid lineages and mutations were detected in both lineages at BC.[22] RUNX1 directed therapies are under development by inhibiting protein/protein interactions. Small-molecule targeting of CBFB, the binding co-factor of RUNX1, reduced RUNX1 binding to target genes and impacted cell survival and differentiation.[90]
ASXL1 is among the most frequently mutated genes in CML. It has a role in chromatin modification and regulates the expression of multiple genes. A recent study provided new evidence that ASXL1 exon 12 mutants confer a gain-of-function by abnormal binding to the bromodomain-containing BRD4.[91] ASXL1 mutants demonstrated hypersensitivity to recently developed bromodomain inhibitors, which may be a promising therapeutic approach. ASXL1 mutants have been predominantly described in MBC. Deletion involving the IKZF1 gene is a feature of lymphoblastic leukemia and exon deletion was commonly detected at BC CML.[22,43,47] IKZF1 is a tumor suppressor that encodes the lymphoid transcription factor IKAROS, which plays a crucial role in hematopoiesis. A mouse model of IKZF1 exon deletion demonstrated reduced responsiveness to dasatinib.[92] However, targeting the retinoid pathway augmented dasatinib activity. An inhibitor of the pro-oncogenic casein kinase II may restore IKZF1 tumor suppressor activity.[93]
Drugs that are already approved for clinical use in AML may have a role for a minority of CML patients with IDH1/2 mutations. These were detected at a markedly lower frequency in advanced phase CML compared with mutated RUNX1, ASXL1 and IKZF1. Nevertheless, the IDH2 inhibitor enasidenib achieved clinical responses in approximately 40% of patients with AML by promoting leukemic cell differentiation[94] and is approved by the FDA. Similarly, ivosidenib was FDA approved in 2018 as the first treatment of IDH1 mutated AML. Whether adding one of these agents to a TKI and/or other agents will improve outcome in this small subset of patients with CML with IDH1/2 mutations remains to be investigated.
Novel approaches may be beneficial for gene fusion driven advanced phase CML.[95] In a small cohort of 39 patients at BC, MLL gene fusions were detected at a frequency of 13%, including cytogenetically cryptic MLL fusions.[22] MLL fusions interact with different co-factors that are largely associated with epigenetic control, including the tumor suppressor Menin and the histone methyltransferase DOT1L. Inhibition of Menin and DOT1L enhanced induction of differentiation and cell death compared with targeting DOT1L alone in MLL rearranged leukemia.[96] The CBFB-MYH11 fusion has also been described in CML BC.[22,59] The fusion has strong binding affinity for RUNX1 and leads to deregulation of its transcription factor activity. Small-molecule inhibition of the protein-protein interaction between CBFB-MYH11 and RUNX1 showed efficacy against this fusion driven leukemia.[97] Targeting BCL2, although not a gene found mutated in CML but reported to be relevant for survival of leukemic progenitors, has been shown in preclinical studies to be synergistic with TKI and may prove a valuable clinical option in patients with a variety of additional mutations.[98]
Incorporation of non-genic factors into prognostic scores
We have focused on the potential of using gene mutation status to aid clinical decisions. However, non-genic factors may also provide prognostic information. These would include the non-coding regions of the genome, which are detectable by whole-genome sequencing. Furthermore, gene expression signatures have the potential to capture non-genic contributions, such as epigenetic factors, that could potentially contribute to worse prognosis.[85,99,100] In the future a combination of genic, non-genic and gene expression signatures may contribute to clinical risk scores.
Conclusion
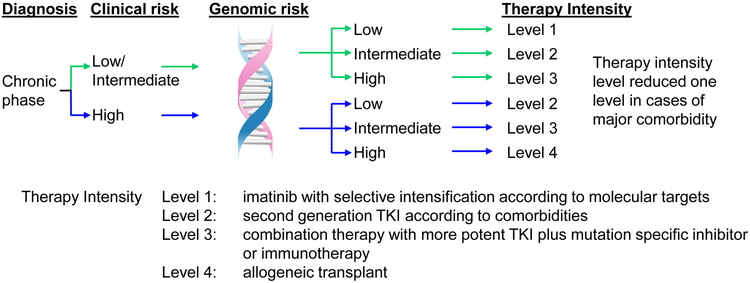
In all disease phases of CML, important questions remain to be answered; do specific mutants associate with treatment response? Does the presence of multiple mutants correlate with a poorer outcome? Do specific interactions occur between mutants, either by co-occurrence or mutual exclusivity? What is the impact of co-expression of mutants in the same or different clones? Is there an association between specific mutants and time to loss of response or transformation? Do specific mutants at diagnosis require more potent kinase inhibition to achieve an optimal response? Some mutations may indicate the need to target a second pathway in addition to BCR-ABL1 inhibition (Figure 4). As the field begins to address these important issues, we strongly believe that well-designed NGS studies offer a critical opportunity to lay the foundation for improved risk classification according to clinical and genomic risk.
Figure 4. Potential future model for an enhanced risk classification by incorporation of genomic factors.
Clinical risk assessed according to the EUTOS long-term survival score. Genomic risk may be classified as Low if no mutant genes of clinical relevance are detected, Intermediate if a mutation demonstrated to confer a moderate risk is detected, and High if specific mutations or multiple mutations are detected.
Acknowledgment
The authors thank Nicola Evans and the International CML Foundation (iCMLf) for facilitating meetings and discussion for preparation of the manuscript. SB receives support from the National Health and Medical Research Council of Australia (APP1104425). DK received research support from the Princess Margaret Cancer Foundation, Toronto, Canada. JFA is a NIHR Senior Investigator and acknowledges the support of the Imperial College NIHR Biomedical Research Centre. SM receives support from the Finnish Cancer Organizations and Gyllenberg Foundation. CC receives funding support from the National Research Foundation Singapore under Clinician Scientist Awards, NMRC/CSA/017/2010 (C.C.) and administered by the Singapore Ministry of Health’s National Medical Research Council (NMRC); by the Agency for Science, Technology, and Research (A*STAR) Biomedical Research Council Translational Clinical Research Partnership Grant, BMRC/13/1/96/681 (C.C.); and the Duke–National University of Singapore Signature Research Program funded by the A* STAR and the Ministry of Health, Singapore. BJD receives support from the Howard Hughes Medical Institute. TPH receives support from the National Health and Medical Research Council of Australia (APP1135949) and has the financial support of Cancer Council SA’s Beat Cancer Project on behalf of its donors and the State Government of South Australia through the Department of Health.
Footnotes
Conflict of Interest
SB: Member of the advisory board of Qiagen, Novartis and Bristol-Myers Squibb; Received honoraria from Qiagen, Novartis, Bristol-Myers Squibb and Cepheid. Research support from Novartis. CC: Honorarium from Novartis Oncology, Bristol-Myers Squibb, Korea Otsuka Pharmaceuticals, Chiltem International; Research funding from Bristol-Myers Squibb. BJD: Aileron Therapeutics, ALLCRON, Cepheid, Vivid Biosciences, Celgene, Gilead Sciences (inactive), Baxalta (inactive), Monojul (inactive); SAB & Stock: Aptose Biosciences, Blueprint Medicines, Beta Cat, Third Coast Therapeutics, GRAIL (inactive), CTI BioPharma (inactive); Scientific Founder: MolecularMD (inactive, acquired by ICON); Board of Directors & Stock: Amgen; Board of Directors: Burroughs Wellcome Fund, CureOne; Joint Steering Committee: Beat AML LLS; Clinical Trial Funding: Novartis, Bristol-Myers Squibb, Pfizer; Royalties from Patent 6958335 (Novartis exclusive license) and OHSU and Dana-Farber Cancer Institute (one Merck exclusive license). TPH: Holds a consultancy role and has received research funding and honoraria from Novartis, Bristol-Myers Squibb and Ariad. Other authors declare no conflicts of interest.
References
- 1.Baccarani M, Deininger MW, Rosti G, Hochhaus A, Soverini S, Apperley JF, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122:872–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radich JP, Deininger M, Abboud CN, Altman JK, Berman E, Bhatia R, et al. Chronic Myeloid Leukemia, Version 1.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2018;16:1108–35. [DOI] [PubMed] [Google Scholar]
- 3.Ashley EA. Towards precision medicine. Nature Reviews Genetics. 2016;17:507–22. [DOI] [PubMed] [Google Scholar]
- 4.Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sokal JE, Cox EB, Baccarani M, Tura S, Gomez GA, Robertson JE, et al. Prognostic discrimination in "good-risk" chronic granulocytic leukemia. Blood. 1984;63:789–99. [PubMed] [Google Scholar]
- 6.Hasford J, Baccarani M, Hoffmann V, Guilhot J, Saussele S, Rosti G, et al. Predicting complete cytogenetic response and subsequent progression-free survival in 2060 patients with CML on imatinib treatment: the EUTOS score. Blood. 2011;118:686–92. [DOI] [PubMed] [Google Scholar]
- 7.Pfirrmann M, Baccarani M, Saussele S, Guilhot J, Cervantes F, Ossenkoppele G, et al. Prognosis of long-term survival considering disease-specific death in patients with chronic myeloid leukemia. Leukemia. 2016;30:48–56. [DOI] [PubMed] [Google Scholar]
- 8.Than H, Lye WK, Sng C, Allen JC Jr., Ong ST, Chuah C. BIM deletion polymorphism profiling complements prognostic values of risk scores in imatinib-treated Asian chronic myeloid leukemia patients. Leuk Lymphoma. 2019;60:234–37. [DOI] [PubMed] [Google Scholar]
- 9.Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shlush LI. Age-related clonal hematopoiesis. Blood. 2018;131:496–504. [DOI] [PubMed] [Google Scholar]
- 12.Nazha A, Narkhede M, Radivoyevitch T, Seastone DJ, Patel BJ, Gerds AT, et al. Incorporation of molecular data into the Revised International Prognostic Scoring System in treated patients with myelodysplastic syndromes. Leukemia. 2016;30:2214–20. [DOI] [PubMed] [Google Scholar]
- 13.Grinfeld J, Nangalia J, Baxter EJ, Wedge DC, Angelopoulos N, Cantrill R, et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N Engl J Med. 2018;379:1416–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corm S, Biggio V, Roche-Lestienne C, Lai JL, Yakoub-Agha I, Philippe N, et al. Coexistence of AML1/RUNX1 and BCR-ABL point mutations in an imatinib-resistant form of CML. Leukemia. 2005;19:1991–2. [DOI] [PubMed] [Google Scholar]
- 15.Coyle T, Najfeld V. Translocation (3;21) in Philadelphia chromosome-positive chronic myelogenous leukemia prior to the onset of blast crisis. Am J Hematol. 1988;27:56–9. [DOI] [PubMed] [Google Scholar]
- 16.Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, et al. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2alphaB gene associated with myeloblastic leukemias. Blood. 1999;93:1817–24. [PubMed] [Google Scholar]
- 17.Roumier C, Fenaux P, Lafage M, Imbert M, Eclache V, Preudhomme C. New mechanisms of AML1 gene alteration in hematological malignancies. Leukemia. 2003;17:9–16. [DOI] [PubMed] [Google Scholar]
- 18.Ichikawa M, Asai T, Saito T, Yamamoto G, Seo S, Yamazaki I, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nature Medicine. 2004;10:299–304. [DOI] [PubMed] [Google Scholar]
- 19.Mangan JK, Speck NA. RUNX1 mutations in clonal myeloid disorders: from conventional cytogenetics to next generation sequencing, a story 40 years in the making. Crit Rev Oncog. 2011;16:77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roche-Lestienne C, Marceau A, Labis E, Nibourel O, Coiteux V, Guilhot J, et al. Mutation analysis of TET2, IDH1, IDH2 and ASXL1 in chronic myeloid leukemia. Leukemia. 2011;25:1661–4. [DOI] [PubMed] [Google Scholar]
- 21.Soverini S, Score J, Iacobucci I, Poerio A, Lonetti A, Gnani A, et al. IDH2 somatic mutations in chronic myeloid leukemia patients in blast crisis. Leukemia. 2011;25:178–81. [DOI] [PubMed] [Google Scholar]
- 22.Branford S, Wang P, Yeung DT, Thomson D, Purins A, Wadham C, et al. Integrative genomic analysis reveals cancer-associated mutations at diagnosis of CML in patients with high-risk disease. Blood. 2018;132:948–61. [DOI] [PubMed] [Google Scholar]
- 23.Menezes J, Salgado RN, Acquadro F, Gomez-Lopez G, Carralero MC, Barroso A, et al. ASXL1, TP53 and IKZF3 mutations are present in the chronic phase and blast crisis of chronic myeloid leukemia. Blood Cancer J. 2013;3:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watkins DB, Hughes TP, White DL, D'Andrea RJ. NPM1 mutations occur rarely or not at all in chronic myeloid leukaemia patients in chronic phase or blast crisis. Leukemia. 2013;27:489–90. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt M, Rinke J, Schafer V, Schnittger S, Kohlmann A, Obstfelder E, et al. Molecular-defined clonal evolution in patients with chronic myeloid leukemia independent of the BCR-ABL status. Leukemia. 2014;28:2292–9. [DOI] [PubMed] [Google Scholar]
- 26.Mitani K, Nagata Y, Sasaki K, Yoshida K, Chiba K, Tanaka H, et al. Somatic mosaicism in chronic myeloid leukemia in remission. Blood. 2016;128:2863–66. [DOI] [PubMed] [Google Scholar]
- 27.Valikhani A, Poopak B, Ferdowsi S, Azizi Tabesh G, Ghaffari SH, Saraf Kazeruoni E, et al. ASXL1 and JAK2V617F gene mutation screening in Iranian patients with chronic myeloid leukemia. Asia Pac J Clin Oncol. 2017;13:e41–e47. [DOI] [PubMed] [Google Scholar]
- 28.Mologni L, Piazza R, Khandelwal P, Pirola A, Gambacorti-Passerini C. Somatic mutations identified at diagnosis by exome sequencing can predict response to imatinib in chronic phase chronic myeloid leukemia (CML) patients. Am J Hematol. 2017; 92: E623–E25. [DOI] [PubMed] [Google Scholar]
- 29.Togasaki E, Takeda J, Yoshida K, Shiozawa Y, Takeuchi M, Oshima M, et al. Frequent somatic mutations in epigenetic regulators in newly diagnosed chronic myeloid leukemia. Blood Cancer J. 2017;7:e559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sloma I, Mitjavila-Garcia MT, Feraud O, Griscelli F, Oudrhiri N, El Marsafy S, et al. Whole-genome analysis reveals unexpected dynamics of mutant subclone development in a patient with JAK2-V617F-positive chronic myeloid leukemia. Exp Hematol. 2017;53:48–58. [DOI] [PubMed] [Google Scholar]
- 31.Kim T, Tyndel MS, Kim HJ, Ahn JS, Choi SH, Park HJ, et al. Spectrum of somatic mutation dynamics in chronic myeloid leukemia following tyrosine kinase inhibitor therapy. Blood. 2017;129:38–47. [DOI] [PubMed] [Google Scholar]
- 32.Nteliopoulos G, Bazeos A, Claudiani C, Gerrard G, Curry C, Szydlo R, et al. Somatic variants in epigenetic modifiers can predict failure of response to imatinib but not to second generation tyrosine kinase inhibitors. Haematologica. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer. 2018;18:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fialkow P, Martin P, Najfeld V, Penfold G, Jacobson R, Hansen J. Evidence for a multistep pathogenesis of chronic myelogenous leukemia. Blood. 1981;58:158–63. [PubMed] [Google Scholar]
- 35.Jongen-Lavrencic M, Grob T, Hanekamp D, Kavelaars FG, Al Hinai A, Zeilemaker A, et al. Molecular Minimal Residual Disease in Acute Myeloid Leukemia. N Engl J Med. 2018;378:1189–99. [DOI] [PubMed] [Google Scholar]
- 36.Ernst T, Busch M, Rinke J, Ernst J, Haferlach C, Beck JF, et al. Frequent ASXL1 mutations in children and young adults with chronic myeloid leukemia. Leukemia. 2018;32:2046–49. [DOI] [PubMed] [Google Scholar]
- 37.Johansson B, Fioretos T, Mitelman F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002;107:76–94. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Cortes JE, Tang G, Khoury JD, Wang S, Bueso-Ramos CE, et al. Risk stratification of chromosomal abnormalities in chronic myelogenous leukemia in the era of tyrosine kinase inhibitor therapy. Blood. 2016;127:2742–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahuja H, Bar-Eli M, Arlin Z, Advani S, Allen SL, Goldman J, et al. The spectrum of molecular alterations in the evolution of chronic myelocytic leukemia. J Clin Invest. 1991;87:2042–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feinstein E, Cimino G, Gale RP, Alimena G, Berthier R, Kishi K, et al. p53 in chronic myelogenous leukemia in acute phase. Proc Natl Acad Sci U S A. 1991;88:6293–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakai H, Misawa S, Toguchida J, Yandell DW, Ishizaki K. Frequent p53 Gene Mutations in Blast Crisis of Chronic Myelogenous Leukemia, Especially in Myeloid Crisis Harboring Loss of a Chromosome 17p. Cancer Research. 1992;52:6588–93. [PubMed] [Google Scholar]
- 42.Zhang SJ, Ma LY, Huang QH, Li G, Gu BW, Gao XD, et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2008;105:2076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–14. [DOI] [PubMed] [Google Scholar]
- 44.Piccaluga PP, Sabattini E, Bacci F, Agostinelli C, Righi S, Salmi F, et al. Cytoplasmic mutated nucleophosmin (NPM1) in blast crisis of chronic myeloid leukaemia. Leukemia. 2009;23:1370–1. [DOI] [PubMed] [Google Scholar]
- 45.Boultwood J, Perry J, Zaman R, Fernandez-Santamaria C, Littlewood T, Kusec R, et al. High-density single nucleotide polymorphism array analysis and ASXL1 gene mutation screening in chronic myeloid leukemia during disease progression. Leukemia. 2010;24:1139–45. [DOI] [PubMed] [Google Scholar]
- 46.Makishima H, Jankowska AM, McDevitt MA, O'Keefe C, Dujardin S, Cazzolli H, et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 2011;117:e198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grossmann V, Kohlmann A, Zenger M, Schindela S, Eder C, Weissmann S, et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011;25:557–60. [DOI] [PubMed] [Google Scholar]
- 48.Konoplev S, Yin CC, Kornblau SM, Kantarjian HM, Konopleva M, Andreeff M, et al. Molecular characterization of de novo Philadelphia chromosome-positive acute myeloid leukemia. Leuk Lymphoma. 2013;54:138–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Zheng J, Hu JD, Wu YA, Zheng XY, Liu TB, et al. Discovery of somatic mutations in the progression of chronic myeloid leukemia by whole-exome sequencing. Genet Mol Res. 2014;13:945–53. [DOI] [PubMed] [Google Scholar]
- 50.Kim T, Tyndel MS, Zhang Z, Ahn J, Choi S, Szardenings M, et al. Exome sequencing reveals DNMT3A and ASXL1 variants associate with progression of chronic myeloid leukemia after tyrosine kinase inhibitor therapy. Leuk Res. 2017;59:142–48. [DOI] [PubMed] [Google Scholar]
- 51.Sklarz LM, Wittke C, Krohn S, C GR-T, Junghanss C, Murua Escobar H, et al. Genetic Mutations in a Patient with Chronic Myeloid Leukemia Showing Blast Crisis 10 Years After Presentation. Anticancer Res. 2018;38:3961–66. [DOI] [PubMed] [Google Scholar]
- 52.Adnan Awad S, Kankainen M, Eldfors S, Yadav B, Kytölä S, Ellonen P, et al. Identification of progression-associated mutations in chronic myeloid leukemia (CML) by comparative genomic analysis (Abstract). Blood. 2017;130:250–50. [Google Scholar]
- 53.Ochi Y, Yoshida K, Huang Y-J, Kuo M-C, Shiozawa Y, Nannya Y, et al. Molecular profiling of blastic transformation in chronic myeloid leukemia (Abstract). Blood. 2018;132:1725–25.30337322 [Google Scholar]
- 54.Soverini S, Colarossi S, Gnani A, Rosti G, Castagnetti F, Poerio A, et al. Contribution of ABL kinase domain mutations to imatinib resistance in different subsets of Philadelphia-positive patients: By the GIMEMA Working Party on Chronic Myeloid Leukemia. Clin Cancer Res. 2006;12:7374–79. [DOI] [PubMed] [Google Scholar]
- 55.Cortes J, Rousselot P, Kim D-W, Ritchie E, Hamerschlak N, Coutre S, et al. Dasatinib induces complete hematologic and cytogenetic responses in patients with imatinib-resistant or -intolerant chronic myeloid leukemia in blast crisis. Blood. 2007; 109:3207–13. [DOI] [PubMed] [Google Scholar]
- 56.Branford S, Melo JV, Hughes TP. Selecting optimal second-line tyrosine kinase inhibitor therapy for chronic myeloid leukemia patients after imatinib failure: does the BCR-ABL mutation status really matter? Blood. 2009;114:5426–35. [DOI] [PubMed] [Google Scholar]
- 57.Roche-Lestienne C, Deluche L, Corm S, Tigaud I, Joha S, Philippe N, et al. RUNX1 DNA-binding mutations and RUNX1-PRDM16 cryptic fusions in BCR-ABL+ leukemias are frequently associated with secondary trisomy 21 and may contribute to clonal evolution and imatinib resistance. Blood. 2008;111:3735–41. [DOI] [PubMed] [Google Scholar]
- 58.Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. 2017;32:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salem A, Loghavi S, Tang G, Huh YO, Jabbour EJ, Kantarjian H, et al. Myeloid neoplasms with concurrent BCR-ABL1 and CBFB rearrangements: A series of 10 cases of a clinically aggressive neoplasm. Am J Hematol. 2017;92:520–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jan M, Majeti R. Clonal evolution of acute leukemia genomes. Oncogene. 2013;32:135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Magistroni V, Mauri M, D’Aliberti D, Mezzatesta C, Crespiatico I, Nava M, et al. De novo UBE2A mutations are recurrently acquired during chronic myeloid leukemia progression and interfere with myeloid differentiation pathways. Haematologica. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carbuccia N, Murati A, Trouplin V, Brecqueville M, Adelaide J, Rey J, et al. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23:2183–6. [DOI] [PubMed] [Google Scholar]
- 65.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2018;47:D886–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–2. [DOI] [PubMed] [Google Scholar]
- 70.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Piazza R, Ramazzotti D, Spinelli R, Pirola A, De Sano L, Ferrari P, et al. OncoScore: a novel, Internet-based tool to assess the oncogenic potential of genes. Scientific Reports. 2017;7:46290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lane L, Argoud-Puy G, Britan A, Cusin I, Duek PD, Evalet O, et al. neXtProt: a knowledge platform for human proteins. Nucleic Acids Res. 2012;40:D76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Losman JA, Kaelin WG Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kerkhof J, Schenkel LC, Reilly J, McRobbie S, Aref-Eshghi E, Stuart A, et al. Clinical Validation of Copy Number Variant Detection from Targeted Next-Generation Sequencing Panels. J Mol Diagn. 2017;19:905–20. [DOI] [PubMed] [Google Scholar]
- 81.Jabbour E, Kantaqian H, Jones D, Talpaz M, Bekele N, O'Brien S, et al. Frequency and clinical significance of BCR-ABL mutations in patients with chronic myeloid leukemia treated with imatinib mesylate. Leukemia. 2006;20:1767–73. [DOI] [PubMed] [Google Scholar]
- 82.Nicolini FE, Corm S, Le QH, Sorel N, Hayette S, Bories D, et al. Mutation status and clinical outcome of 89 imatinib mesylate-resistant chronic myelogenous leukemia patients: a retrospective analysis from the French intergroup of CML (Fi(varphi)-LMC GROUP). Leukemia. 2006;20:1061–6. [DOI] [PubMed] [Google Scholar]
- 83.Reeser JW, Martin D, Miya J, Kautto EA, Lyon E, Zhu E, et al. Validation of a targeted RNA sequencing assay for kinase fusion detection in solid tumors. J Mol Diagn. 2017;19:682–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018;19:269–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103:2794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Soverini S, Hochhaus A, Nicolini FE, Gruber F, Lange T, Saglio G, et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: recommendations from an expert panel on behalf of European LeukemiaNet. Blood. 2011;118:1208–15. [DOI] [PubMed] [Google Scholar]
- 87.Branford S, Rudzki Z, Walsh S, Parkinson I, Grigg A, Szer J, et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood. 2003;102:276–83. [DOI] [PubMed] [Google Scholar]
- 88.Parker WT, Lawrence RM, Ho M, Irwin DL, Scott HS, Hughes TP, et al. Sensitive detection of BCR-ABL1 mutations in patients with chronic myeloid leukemia after imatinib resistance is predictive of outcome during subsequent therapy. J Clin Oncol. 2011;29:4250–59. [DOI] [PubMed] [Google Scholar]
- 89.Soverini S, De Benedittis C, Machova Polakova K, Brouckova A, Horner D, Iacono M, et al. Unraveling the complexity of tyrosine kinase inhibitor-resistant populations by ultra-deep sequencing of the BCR-ABL kinase domain. Blood. 2013;122:1634–48. [DOI] [PubMed] [Google Scholar]
- 90.Illendula A, Gilmour J, Grembecka J, Tirumala VSS, Boulton A, Kuntimaddi A, et al. Small Molecule Inhibitor of CBFβ-RUNX Binding for RUNX Transcription Factor Driven Cancers. EBioMedicine. 2016;8:117–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang H, Kurtenbach S, Guo Y, Lohse I, Durante MA, Li J, et al. Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood. 2018;131:328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Churchman ML, Low J, Qu C, Paietta EM, Kasper LH, Chang Y, et al. Efficacy of Retinoids in IKZF1-Mutated BCR-ABL1 Acute Lymphoblastic Leukemia. Cancer Cell. 2015;28:343–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Song C, Gowda C, Pan X, Ding Y, Tong Y, Tan BH, et al. Targeting casein kinase II restores Ikaros tumor suppressor activity and demonstrates therapeutic efficacy in high-risk leukemia. Blood. 2015;126:1813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130:722–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pikman Y, Stegmaier K. Targeted therapy for fusion-driven high-risk acute leukemia. Blood. 2018;132:1241–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dafflon C, Craig VJ, Méreau H, Gräsel J, Schacher Engstler B, Hoffman G, et al. Complementary activities of DOT1L and Menin inhibitors in MLL-rearranged leukemia. Leukemia. 2016;31:1269–77. [DOI] [PubMed] [Google Scholar]
- 97.Illendula A, Pulikkan JA, Zong H, Grembecka J, Xue L, Sen S, et al. Chemical biology. A small-molecule inhibitor of the aberrant transcription factor CBFbeta-SMMHC delays leukemia in mice. Science. 2015;347:779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Carter BZ, Mak PY, Mu H, Zhou H, Mak DH, Schober W, et al. Combined targeting of BCL-2 and BCR-ABL tyrosine kinase eradicates chronic myeloid leukemia stem cells. Science translational medicine. 2016;8:117–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yong ASM, Szydlo RM, Goldman JM, Apperley JF, Melo JV. Molecular profiling of CD34+ cells identifies low expression of CD7, along with high expression of proteinase 3 or elastase, as predictors of longer survival in patients with CML. Blood. 2006;107:205–12. [DOI] [PubMed] [Google Scholar]
- 100.McWeeney SK, Pemberton LC, Loriaux MM, Vartanian K, Willis SG, Yochum G, et al. A gene expression signature of CD34+ cells to predict major cytogenetic response in chronic-phase chronic myeloid leukemia patients treated with imatinib. Blood. 2010;115:315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]