Abstract
While the use of ATP-competitive tyrosine kinase inhibitors of oncoprotein BCR-ABL1 has enabled durable responses in patients with chronic myeloid leukemia (CML), issues of drug resistance and residual leukemic stem cells remain. To test whether the degradation of BCR-ABL1 kinase could offer improved response, we developed a series of proteolysis targeting chimera (PROTACs) that allosterically target BCR-ABL1 protein and recruit the E3 ligase Von Hippel Lindau (VHL), resulting in ubiquitination and subsequent degradation of the oncogenic fusion protein. In both human CML K562 cells and murine Ba/F3 cells expressing BCR-ABL1, lead compound GMB-475 induced rapid proteasomal degradation and inhibition of downstream biomarkers, such as STAT5, and showed increased sensitivity compared to diastereomeric controls lacking degradation activity. Notably, GMB-475 inhibited the proliferation of certain clinically relevant BCR-ABL1 kinase domain point mutants and further sensitized Ba/F3 BCR-ABL1 cells to inhibition by imatinib, while demonstrating no toxicity toward Ba/F3 parental cells. Reverse phase protein array analysis suggested additional differences in levels of phosphorylated SHP2, GAB2, and SHC associated with BCR-ABL1 degradation. Importantly, GMB-475 reduced viability and increased apoptosis in primary CML CD34+ cells, with no effect on healthy CD34+ cells at identical concentrations. GMB-475 degraded BCR-ABL1 and reduced cell viability in primary CML stem cells. Together, these findings suggest that combined BCR-ABL1 kinase inhibition and protein degradation may represent a strategy to address BCR-ABL1-dependent drug resistance, and warrants further investigation into the eradication of persistent leukemic stem cells, which rely on neither the presence nor the activity of the BCR-ABL1 protein for survival.
Keywords: PROTAC, kinase inhibitors, stem cells, allosteric
INTRODUCTION
Chronic myeloid leukemia (CML) is a myeloproliferative disorder characterized by the presence of the oncogenic fusion protein BCR-ABL1, the product of a translocation of chromosomes 9 and 22. This fusion results in a constitutively active BCR-ABL1 kinase that drives the overproduction and expansion of white blood cells in the bone marrow, and ultimately crowds out normal cells present in the bone marrow niche. Development of tyrosine kinase inhibitors (TKIs) specific to ABL1, such as imatinib, have drastically improved prognosis for patients by blocking BCR-ABL1 kinase activity without harming the normal cells (1).
Although in the vast majority of cases this disease is controlled by treatment with approved ABL1 TKIs, approximately 80% of patients must remain on treatment indefinitely, given that discontinuation results in relapse (2). This disease persistence can be attributed to residual leukemic stem cells (LSCs) which evade cell death even when BCR-ABL1 kinase activity is inhibited and drive relapse when TKI therapy is stopped. Previous studies have shown ABL1 TKIs effectively reduce kinase activity and downstream signaling of BCR-ABL1, yet induction of apoptosis is markedly lower in the CML stem cell fraction compared to progenitors (3). Others have investigated alternative signaling mechanisms independent of BCR-ABL1 modified in LSCs, such as persistent activation of the MEK signaling cascade (4). Furthermore, the BCR-ABL1 fusion protein is large and features multiple other domains beyond the tyrosine kinase domain with important biological roles. Some have proposed that the dependence of CML LSCs on BCR-ABL1 protein is scaffolding-dependent; for example, the autophosphorylated residue Y177 of BCR interacts with GRB2 to promote RAS and PI3K signaling or the less characterized interactions with PP2A and JAK2 signaling (5). The structure and leukemogenic signaling associated with specific regions of BCR-ABL1 also highlight involvement of non-kinase domains of the fusion protein. For example, the SH2 domain interacts with SHC and results in the recruitment of GRB2, which induces pro-cell survival mechanisms or SH3 domain interactions with signaling proteins regulating cell adhesion and migration (6). These non-kinase interactions of BCR-ABL1 can add to the recapitulation of disease despite kinase inhibition, suggesting a need to further eliminate BCR-ABL1 at the protein level. In this vein, our group and others have shown enhanced cell kill by employing compounds that interact with both BCR-ABL1 and proteasome degradation associated molecules (7-9).
The proteolysis targeting chimera (PROTAC) technology developed in the Crews lab (10-12) provides a unique toolkit for probing the non-kinase roles of proteins in a cellular context without resorting to genetic modification of the system. PROTACs are heterobifunctional molecules which recruit an E3 ligase to a target protein, resulting in ubiquitination and subsequent degradation of the target. The ability of PROTACs to induce degradation of the target is not limited to binding sites within the kinase domain, but have the potential to be implemented when kinase activity is not the singular role of the target protein (13). ABL1, one of the fusion partners in the BCR-ABL1 protein, contains an allosteric myristolation binding site, known to have a role in the autoinhibited state of the protein (14) and multiple compounds have been developed to target this region (15,16). To assess the additional treatment efficacy of both inhibiting the kinase and degrading the protein of BCR-ABL1, we further investigated the potential of a new series of PROTACs, utilizing an allosteric inhibitor (GNF-5) to degrade BCR-ABL1 protein and induce cell death in cell lines with relevant kinase domain mutants and primary CML LSCs.
MATERIALS AND METHODS
Cell lines
Ba/F3 murine cell lines (obtained from ATCC), either parental or with stable expression of BCR-ABL1 via pSRα vector backbone, were grown in R10 media consisting of RPMI (Invitrogen) supplemented with 10% FBS (Atlanta Biologicals), L-glutamine, penicillin/streptomycin (Invitrogen) and amphotericin B (HyClone). All BCR-ABL1 positive cell lines were confirmed by PCR to express the BCR-ABL1 p210 fusion; Ba/F3 were confirmed negative for BCR-ABL1 expression. In Ba/F3 parental cells, WEHI-3B-conditioned medium (15%) was included as a source of IL-3. The K562 cell line was purchased from ATCC and cultured in IMDM (Gibco) supplemented with 10% FBS and penicillin/streptomycin (Invitrogen). All cell lines were confirmed mycoplasma negative prior to experimental use. No additional cell line authentification was performed. None of the cell lines used in this study was cultured for longer than 6 months from initial purchase or characterization.
Patient samples
All patient samples were collected following written informed consent and used in accordance with the Declaration of Helsinki. The OHSU Institutional Review Board approved all studies involving human specimens. Mononuclear cells (MNCs) were isolated from either peripheral blood, bone marrow or leukapheresis samples by Ficoll gradient. MNCs were then treated with ammonium-chloride-potassium (ACK) lysis buffer to remove any residual red blood cells. CD34+ cells were isolated via magnetic bead isolation (MACS, Miltenyi Biotec #130-046-703) and stored in liquid nitrogen storage in 90% FBS plus 10% DMSO for long term storage. For cell proliferation assays, samples were thawed and cultured in R10. For immunoblot assays, samples were thawed and cultured in IMDM with 40μg/mL low-density lipoprotein (LDL, Stem Cell), 20% FBS, and 100μM beta-mercaptaethanol (Sigma).
Flow cytometry
Samples sorted for CD38 were stained with CD34 PE-Cy™7 (BD Biosciences #348801) and BV421 Mouse Anti-Human CD38 (BD Biosciences #562444), washed in sterile PBS with 10% BSA, and sorted on a BD FACS Aria instrument. Sorted samples were recovered overnight in culture in IMDM with 40μg/mL low-density lipoprotein (LDL, Stem Cell), 20% FBS, and 100μM beta-mercaptaethanol (Sigma).
Immunoblot analysis
Samples were lysed in 1X Cell Signaling lysis buffer (#9803S) supplemented with PMSF and complete mini tablets (Sigma #11836153001). Lysates were quantified and separated on 4-15% Tris-glycine polyacrylamide gels. Gels were transferred and then blocked in TBS-Tween buffer with 5% BSA. The following primary antibodies were used: tABL, BD #554148 anti-mouse 1:400; c-ABL1, SantaCruz #24-11 anti-mouse 1:1000; B-tubulin, Millipore #05-66—MI anti-mouse 1:5000; pSTAT5, #CS9351S anti-rabbit 1:1000 pSTAT5, #CS4322 anti-rabbit 1:1000; pBCR-Abl, #CS2865 anti-rabbit 1:1000; pCRKL, #CS491 anti-rabbit 1:1000; pAKT, #CS4060 anti-rabbit 1:1000; pERK, #CS4695 anti-rabbit 1:1000; pSHP-2, Abcam #62322 anti-rabbit 1:1000; pGAB1, #CS3233 anti-rabbit 1:1000; pGAB2, #CS3882 anti-rabbit 1:1000; pSHC, #CS2434 anti-rabbit 1:1000; VHL, #CS68547 anti-rabbit 1:1000; pCRKL, #CS3181S anti-rabbit 1:1000. Following incubation with appropriate HRP-conjugated secondary antibodies, membranes were imaged on a BioRad ChemiDoc using BioRad Clarity Western ECL substrate and Thermo Super Signal West Femto Maximum Sensitivity substrate.
Cell viability assay
Cell lines and patient samples were exposed to dose ranges of single or combination agents and incubated for 3 days at 37°C, 5% CO2 and subjected to a CellTiter 96 AQueous One solution cell proliferation assay (Promega). IC50 values calculated and analyzed using Prism 6 software (GraphPad).
Apoptosis analysis
Patient samples were incubated ranging from 48 to 96 h, stained and analyzed according to the Guava Nexin Reagent analysis kit (Milipore #4500-0450) or ApoScreen Annexin V-FITC (Southern Biotech #10040-02) for flow cytometry analysis.
PROTAC synthesis
For chemical synthesis, see supporting information.
Reverse phase protein arrays (RPPA)
K562 cells were treated with DMSO, GMB-475 (5 μM) or GMB-651 (5 μM) for 8 h in duplicate, washed twice with PBS and lysed in RPPA lysis buffer (1% Triton X-100, 50 mM HEPES, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 10 mM Na pyrophosphate, 1 mM Na3VO4, 10% glycerol, containing freshly added protease and phosphatase inhibitors from Roche Applied Science). RPPA was performed in the MDACC CCSG core as described at http://www.mdanderson.org/education-and-research/resources-for-professionals/scientific-resources/core-facilities-and-services/functional-proteomics-rppa-core/index.html.
RESULTS
Development and synthesis of potent BCR-ABL1-specific PROTACs
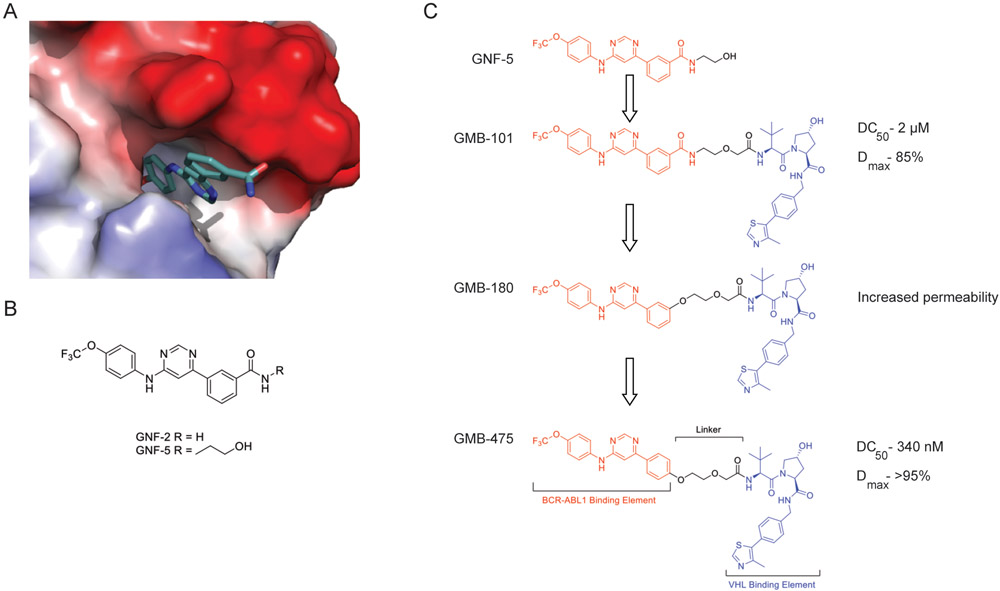
Previously, we and others have demonstrated the efficacy of synthesizing ATP-competitive compounds with both the capability to inhibit kinase activity and degrade the total protein. (7,17). Here we embarked on a program to develop BCR-ABL1 PROTACs derived from the allosteric GNF family of compounds (15,18). The GNF compounds bind at the membrane tethering myristate binding pocket present on ABL1/BCR-ABL1 (Fig. 1A/B) (15,18). Inspection of the crystal structure of the GNF-2/ABL1 complex (PDB ID:3K5V) revealed solvent exposed regions suitable for linker attachment and therefore PROTAC conversion. Initial synthesis focused on extension of the solubilizing group from GNF-5 into a linker and conjugation to the VHL ligand developed in our laboratory (19,20). This ligand addition gave us rapid access to a BCR-ABL1 PROTAC (GMB-101) with moderate activity (Fig. S1A).
Figure 1. Development of an allosteric BCR-ABL1 PROTAC.
(A) X-ray Crystal Structure of GNF-2 bound to the myristate pocket of ABL1 (PDB ID: 3K5V)(15). (B) Structures of GNF-2 and GNF-5. (C) Schematic of GNF-5 to PROTAC conversion and PROTAC optimization.
Next, we aimed to further optimize the PROTAC to both improve potency and cell permeability (Fig. 1C). Replacement of the aryl amide bond with an ether linkage (GMB-180) gave modest boost in degradation and improved cell permeability, evidenced by enhanced inhibition of downstream signaling. Finally, altering the vector where the VHL ligand extends from the myristate pocket by moving the aryl ether linkage from the meta to the para position further enhanced the activity, resulting in our lead compound (GMB-475) (Fig. S1B). The diastereomeric control compound (GMB-651), which has equal cell permeability but is unable to recruit VHL, was also prepared to allow a direct comparison with respect to inhibition alone (Fig. S2A-C) (21).
Validation of BCR-ABL1 allosteric PROTAC function in CML model systems
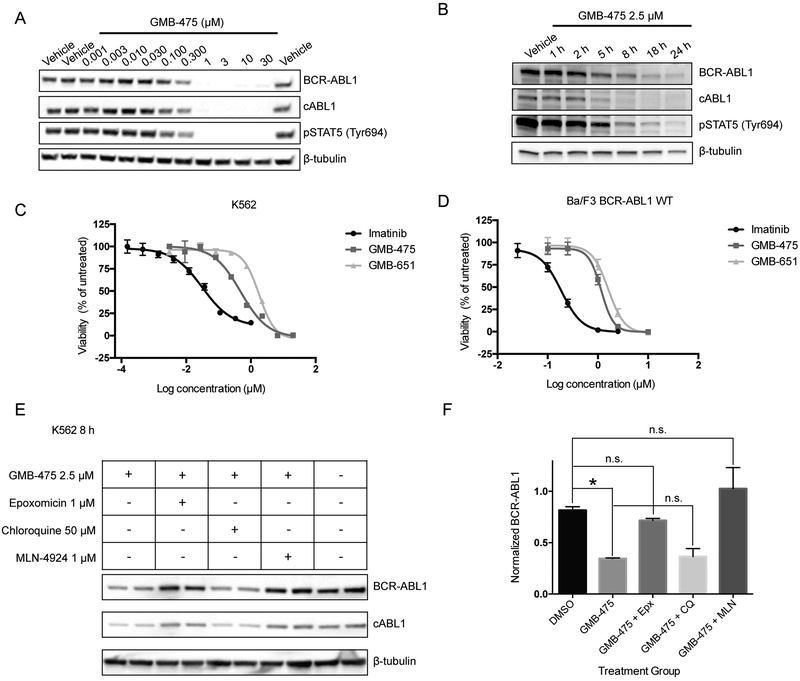
To further characterize the potency and efficacy of GMB-475, we sought to study its function in CML model systems: human K562 cells and murine BCR-ABL1 transformed Ba/F3 cells. GMB-475 induced the degradation of BCR-ABL1 and c-ABL1 in the context of both K562 (Fig. 2A) and Ba/F3 (Fig. 2B) cells with concomitant inhibition of downstream signaling via the STAT5 pathway, in a dose- and time-dependent fashion (human and murine VHL share >70% identity). In both cases, GMB-475 was capable of inhibiting cell proliferation with an IC50 of approximately 1 μM (Fig. 2C/D). Notably, neither GMB-475 nor GMB-651 displayed toxicity against parental Ba/F3 cells up to 10 μM, emphasizing the selectivity of these compounds (Fig. S2D and S3A). Furthermore by co-treatment with pharmacological modulators, we determined that the degradation of BCR-ABL1 progresses by a ubiquitination and proteasome-dependent mechanism and is not lysosome-dependent (Fig. 2E/F). Co-treatment of K562 cells with the proteasome inhibitor epoxomicin (22) and GMB-475 restored the levels of BCR-ABL1 and c-ABL1 compared to GMB-475 alone, while modulation of lysosomal pH with chloroquine had no effect. Additionally, inhibition of neddylation using MLN-4924 inhibited the degradation of BCR-ABL1 and ABL1, since VHL neddylation is required for its E3 ligase activity (Fig. 2E/F) (23).
Figure 2. PROTACs inhibit and degrade BCR-ABL1 via the proteasome in CML cell lines.
(A) Dose response of GMB-475 in K562 cells by immunoblot after 18 h. (B) Time course of degradation in Ba/F3 BCR-ABL1 cells.. (C) K562 sensitivity to PROTACs assessed by cell proliferation assay (D) Effect of PROTACs on Ba/F3 BCR-ABL1 cell proliferation. (E) Degradation mechanism interrogation by immunoblot in K562 cells following 8 h treatment. (F) Quantification of BCR-ABL1 protein levels, from panel (E), by densitometry.
Combination treatment with ATP-competitive BCR-ABL1 TKIs
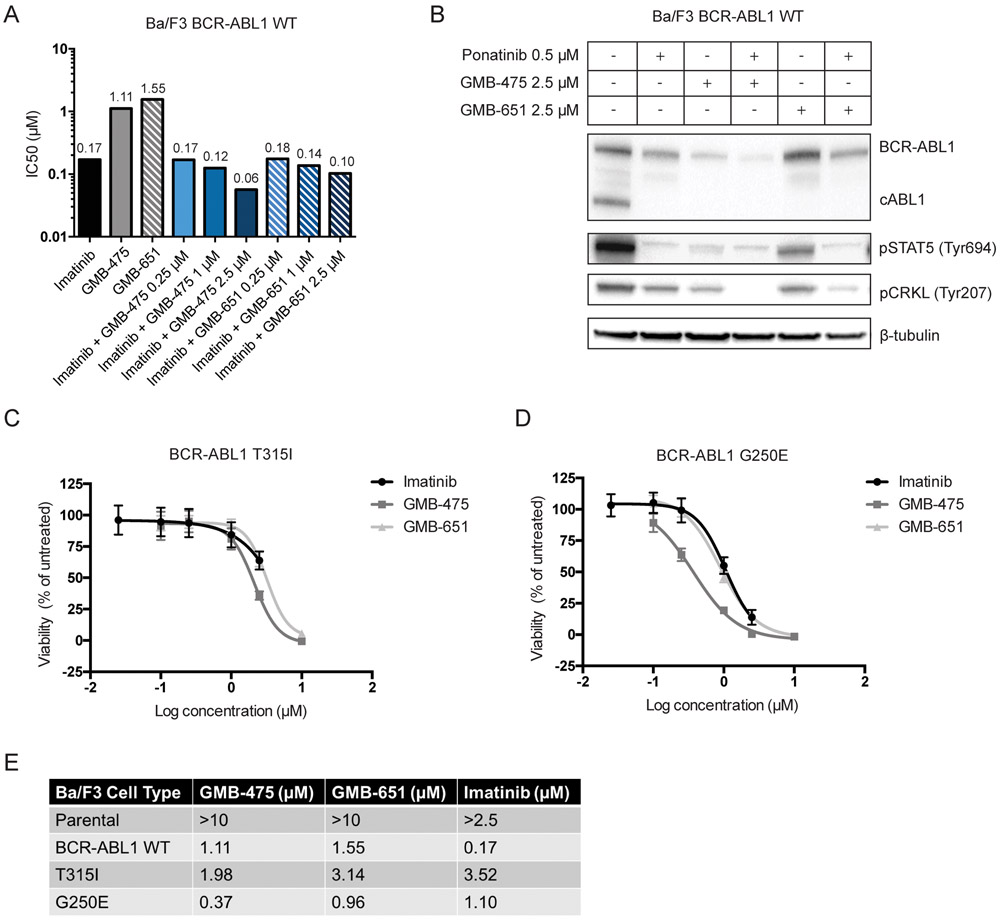
Since PROTACs and imatinib bind at orthogonal sites on the protein, we sought to characterize the effects of co-treatment on cell proliferation. We performed dose response titrations with BCR-ABL1 transformed Ba/F3 cells for imatinib, GMB-475 and GMB-651 and found IC50 values of 0.17 μM, 1.11 μM and 1.55 μM respectively (Fig. 3A/S3A). Furthermore, we determined the IC50 of imatinib in the presence of increasing concentrations of GMB-475 or GMB-651 (Fig. 3A). Co-treatment with 2.5 μM GMB-475 reduced the IC50 of imatinib almost 3-fold, likely due to degradation reducing the BCR-ABL1 protein present, suggesting a lower dose of imatinib can entirely abrogate signaling. In comparison, the co-treatment of the diastereomer GMB-651 slightly reduced the IC50 value for imatinib, demonstrating that co-treatment with an active degrader is advantageous over co-treatment with the equivalent allosteric inhibitor. Furthermore, we examined the effect of co-treatment with ponatinib (24), a potent BCR-ABL1 inhibitor effective against imatinib-resistant kinase domain point mutations such as T315I, with our PROTACs by immunoblot blot (Fig. 3B). Dual treatment with ponatinib and the active degrader was able to fully inhibit phosphorylation of CRKL in Ba/F3 BCR-ABL1 wild-type cells.
Figure 3. GMB-475-mediated degradation enhances efficacy of ATP-competitive TKIs and retains potency against imatinib resistant point mutations.
(A) IC50 values for single agents and combinations in Ba/F3 BCR-ABL1 cells. (B) Immunoblot of overnight co-treatment with ponatinib in Ba/F3 BCR-ABL1 cells. (C and D) Effects of imatinib and PROTACs on cell proliferation in Ba/F3 cells expressing mutant BCR-ABL1.
The Ba/F3 system also allowed us to explore the ability of GMB-475 to inhibit the proliferation of clinically relevant imatinib-resistant BCR-ABL1 point mutants (Fig. 3C-D, Table 1). As shown above, GMB-475 demonstrated a slight advantage over GMB-651 against Ba/F3 cells expressing wild-type BCR-ABL1 but neither were as potent as imatinib (Table 1). Introduction of a T315I mutation significantly reduced the ability of imatinib to inhibit cell proliferation by more than 20-fold but had little effect on the IC50 for GMB-475 (Fig. 3C). Interestingly, cells bearing a G250E mutation in BCR-ABL1 were particularly susceptible to GMB-475 displaying enhanced antiproliferative activity (Figs. 3D). At the level of degradation, GMB-475 treatment only modestly degraded the BCR-ABL1 T315I protein while thoroughly degrading the G250E mutant (Figs. S3B-D). When combined with ponatinib, GMB-475 afforded a slight additive reduction in BCR-ABL1 protein levels and downstream signaling in Ba/F3 BCR-ABL1 T315I cells (Fig. S3B). Interestingly, ponatinib stabilizes the inactive conformation of BCR-ABL1 upon binding, which appears to enhance the ability of GMB-475 to induce degradation in the wild-type cells (Fig. 3B). Co-treatment of ponatinib and GMB-651 showed little additional effect beyond the level of inhibition of kinase activity of ponatinib alone (Fig. S3A-C). Together, these data suggest that PROTAC treatment could be used in concert with traditional inhibitors to reduce the dose of inhibitor required and therefore potentially reduce side effects of ATP mimics.
Table 1 -.
Summary of IC50 values for PROTAC compounds and imatinib in Ba/F3 cell lines.
| Ba/F3 Cell Type | GMB-475 (μm) | GMB-651 (μm) | Imatinib (μm) |
|---|---|---|---|
| Parental | >10 | >10 | >2.5 |
| BCR-ABL1 WT | 1.11 | 1.55 | 0.17 |
| T315I | 1.98 | 3.14 | 3.52 |
| G250E | 0.37 | 0.96 | 1.10 |
Non-kinase roles of BCR-ABL1 assessed by protein array analysis in K562 cells
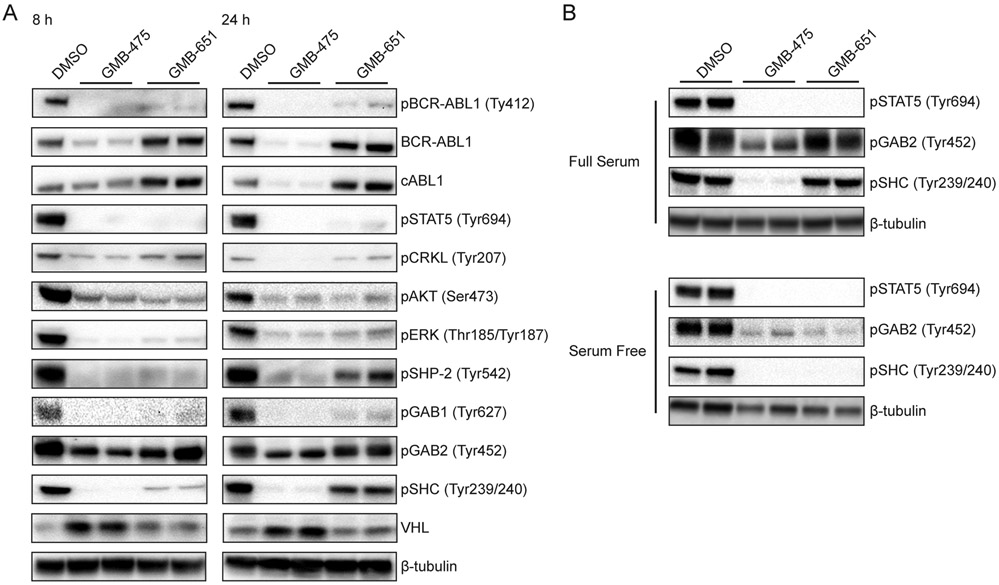
To address potential non-kinase roles of BCR-ABL1 which may be contributing to the additional effect of the PROTAC, we employed a functional proteomic approach to compare degradation to allosteric inhibition. Using reverse phase protein arrays (RPPA) (25), we analyzed changes in protein levels and post-translational modifications in a rapid and efficient manner. K562 cells were treated with 5 μM of either GMB-475 or GMB-651 for 8 h to probe the acute changes in protein states which occur on the degradation/inhibition of BCR-ABL1 (Table S1). As a proof of principle, treatment with GMB-475, but not GMB-651, showed a decrease in total ABL1 protein by RPPA (Table S1 and Fig. S2C). Immunoblot analysis was used to validate selected target proteins at 8 h and 24 h (Fig. 4A). Both GMB-475 and GMB-651 were able to inhibit the kinase activity of BCR-ABL1 as verified by both pBCR-ABL1 (Tyr412) and pSTAT5 (Tyr694) (Fig. 4A). Interestingly, differences between degradation and inhibition alone became evident at 8 h and were further accentuated after 24 h of treatment. For example, downstream signaling of BCR-ABL1 via the pCRKL and pERK pathways was inhibited to a greater extent with the active PROTAC compared to the inhibitor only control (Fig. 4A), indicating the advantages of degradation on suppression of oncogenic signaling.
Figure 4. Combined inhibition and degradation of BCR-ABL1 by GMB-475 reduces scaffolding of downstream interactors.
(A) Immunoblot analysis of downstream signaling and scaffolding proteins in K562 cells treated with 2.5 μM PROTAC or diastereomer. (B) Differences in pSTAT5, pGAB2, and pSHC between stimulated and unstimulated K562 cells.
However, the most extreme differences observed were for pSHP-2, pGAB2, pSHC and VHL. The increase in VHL protein can be explained by the stabilizing effect the binding of a ligand imparts to the VHL protein itself, as exemplified by the increase in the intensity of the VHL band in K562 cells treated with the VHL ligand alone (See Fig. S4A/B). GAB2, SHC and SHP-2 are all in the canonical network of BCR-ABL1 and together contribute to the activation of the MAPK signaling cascade (26). Phosphorylation of Y177 of BCR-ABL1 yields a docking site for GRB2 which in turn recruits GAB2 and/or SHC . Once phosphorylated, GAB2 recruits and activates SHP-2. Inhibition of BCR-ABL1 had no effect on the phosphorylation state of GAB2 but degradation of BCR-ABL1 reduced the level of GAB2 phosphorylation (Figs. 4 and S4C). To further investigate this phenomenon, we repeated the experiment in the presence and absence of serum. Under full serum (10% FBS) conditions, both GMB-475 and GMB-651 were able to inhibit the kinase activity of BCR-ABL1, as measured by loss of pSTAT5 signal, but only GMB-475 was able to reduce phosphorylation of GAB2 and SHC (Fig. 4B). Conversely, under serum-free conditions, both GMB-475 and GMB-651 were able to inhibit phosphorylation of GAB2 and SHC, as well as STAT5 (Fig. 4B). This suggests a scaffolding role for BCR-ABL1 in signaling via this pathway (Fig. S4D). Under serum-free conditions, only the constitutively active BCR-ABL1 kinase domain is able to (auto)-phosphorylate Y177, a key docking site, and thus both degrader (GMB-475) and inhibitor (GMB-651) are able to block signaling.
PROTAC efficacy in primary CML patient samples
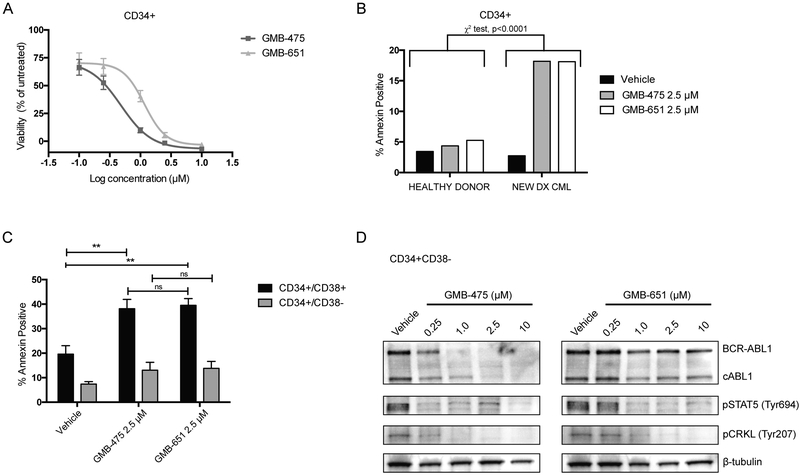
To further explore the scaffolding roles of BCR-ABL1 and the pro-survival effects in CML stem and progenitor cells, in the context of targeted protein degradation, we evaluated our compounds in experiments utilizing primary CML patient samples (Fig. S5A/B). Initially, we assessed the anti-proliferative activity of GMB-475 and GMB-651 in CD34+ cells from newly diagnosed CML patients (patient 1 and 2, Table S2; Fig. 5A/S5C, respectively). PROTACs, as well as the positive control (imatinib) (Fig. S5D), inhibited in vitro proliferation and induced apoptosis in primary CML CD34+ patient cells but had no effect on healthy donor CD34+ cells (Fig. 5B/S5E, p<0.0001, Chi-square; Fig. S5F, p<0.0001, One-way ANOVA; patients 4 and 3, respectively). Notably, GMB-475 was greater than 2-fold more potent at inhibiting proliferation than GMB-651. We further sorted the CD34+ cells from patient 1 into CD34+CD38+ (progenitor cells) and CD34+CD38− (stem cells) and evaluated the ability to induce apoptosis in these populations (Figs. 5C/S5B). Treatment of these cells with either GMB-475 or GMB-651 induced apoptosis in the progenitor cells (p=0.0068 and p=0.0064 respectively; 2-way ANOVA) and to a lesser extent in the stem cells, with no appreciable difference between the PROTAC and the diastereomer control, possibly due to the saturating doses. Finally, we confirmed by immunoblot that GMB-475, but not GMB-651, was indeed able to induce degradation of both BCR-ABL1 and ABL1 in primary patient LSCs (Fig. 5D).
Figure 5. GMB-475 reduces cell viability, induces apoptosis, and degrades BCR-ABL1 in primary CML patient stem/progenitor cells.
(A) Cell viability dose response curves for CD34+ cells (patient 1) treated with PROTAC or diastereomer. (B) Annexin V staining healthy donor or CML primary CD34+ cells (patient 4, Chi-square test, ****p<0.0001). (C) Annexin V staining in sorted progenitor (CD34+/CD38+) and stem (CD34+/CD38−) CML cells (patient 1) by Guava Nexin assay (One-way ANOVA, **p<0.01; error bars represent SEM for at least 3 biological replicates). (D) BCR-ABL1 degradation in CML CD34+/CD38− cells (patient 1) treated overnight with GMB-475 or diastereomer.
DISCUSSION
While the activity of the BCR-ABL1 tyrosine kinase is essential to CML pathogenesis and the justification for, and basis of, the successful implementation of molecularly targeted small-molecule therapies, clinical responses to ABL1 TKIs run a spectrum from deep and durable molecular remission in most patients to overt drug resistance and disease progression in others. These differences are attributable to several known (e.g. resistant BCR-ABL1 kinase domain mutations, cellular drug transporter expression levels, drug intolerance) and not yet well characterized (e.g. primary, BCR-ABL1 kinase-independent resistance) mechanisms. Furthermore, clinical studies involving different approved ABL1 TKIs have identified a consensus association between rapid achievement of deep molecular response, such as a major molecular response or greater, improved overall and progression-free survival (27). To that end, opportunities to further improve outcomes for CML patients will need to focus on strategies that more extensively deplete the resistant and/or persistent leukemic cells through combined targeting approaches.
We and others have previously explored the development of PROTACs which bind and target the BCR-ABL1 protein for degradation by employing ATP-competitive ligands as recruiting elements (7,28). However, these compounds were unable to induce complete degradation of BCR-ABL1 and likely suffered from issues of selectivity, similar to those observed with other orthosteric kinase ligand-based degraders (29,30). As a potential strategy to circumvent this issue, our present PROTAC series was derived from the molecular scaffold of GNF-5(15), the first reported small-molecule allosteric inhibitor of ABL1, which very selectively binds to the myristoyl pocket of the kinase. Our lead PROTAC, GMB-475, which links this scaffold to the VHL ligand, achieved dramatic degradation of BCR-ABL1 protein in cell lines in a time- and concentration-dependent manner. Interestingly, previous attempts to develop VHL-recruiting BCR-ABL1 PROTACs, when employing active site recruiting elements, proved unsuccessful (7). This observation lends further credence to the consideration of favorable protein-protein interactions crucial for successful PROTAC development (30). It is also conceivable that the BCR-ABL1/PROTAC/CRBN trimer is functional at the ATP binding site while the BCR-ABL1/PROTAC/VHL trimer is functional only at the allosteric site (7,30).
Previous comparisons between PROTACs and inactive diastereomers have highlighted advantages of degradation versus inhibition alone and provide evidence for their use against hematological malignancies (21,31). In the present study, we observed modestly greater inhibition of cell proliferation for the degrader compared to the non-degrading control, suggesting that, much of the oncogenic signaling in native CML cells is critically dependent upon the tyrosine kinase activity of BCR-ABL1, hence the success of ATP competitive ligands. This finding is consistent with previous studies using critical tyrosine-mutated and kinase dead BCR-ABL1 mutant constructs (32,33). We also observed that our compound does not possess as potent anti-proliferative effects as imatinib, which is to be expected given the reduced affinity/efficacy of the GNF-5 scaffold compared to imatinib (15,18). Future efforts to develop more potent allosteric BCR-ABL1 PROTACs are ongoing, aiming to close the gap between imatinib and the molecules employed in the current study.
One of the intriguing possibilities of utilizing an allosteric PROTAC is the potential for synergy with an orthosteric inhibitor. Previous efforts to target the myristoyl pocket of ABL1 kinase have demonstrated the ability to inhibit many, but not all, of the problematic point mutations that reduce binding affinity of ATP-site TKIs, and this is enhanced by simultaneous inhibition at both sites on the kinase (15,16). We observed that, when combined with imatinib, GMB-475 demonstrated greater inhibition of Ba/F3 BCR-ABL1 cells compared to the non-degrader control. Combining GMB-475 with low concentrations of the third-generation ATP-site ABL1 TKI ponatinib also showed increased degradation compared GMB-475 alone. GMB-475 demonstrated varying degrees of retained sensitivity to imatinib-resistant BCR-ABL1 kinase domain mutants, suggesting that even partially limited target engagement is sufficient to induce degradation, which highlights the power of this event driven pharmacology model (34,35).
While clinically approved ABL1 TKIs have highlighted the importance of the downstream signaling activated by the tyrosine kinase activity of BCR-ABL1, selective BCR-ABL1-targeted PROTACs are uniquely able to facilitate interrogation of non-kinase-dependent scaffolding roles of this oncogene. Comparing GMB-475 and the inactive diastereomer, we found that, while both compounds inhibited BCR-ABL1 kinase activity, degradation of BCR-ABL1 uniquely resulted in decreased pSHC, pSHP2, and pGAB2 levels. Notably, this behavior phenocopies the mutation of a key autophosphorylation site (Y177) on the BCR portion of the fusion protein (36,37). While Y177 is normally auto-phosphorylated by the kinase domain of BCR-ABL1, under serum-stimulated conditions another kinase (likely HCK (38)) appears to phosphorylate Y177 on BCR-ABL1, allowing it to continue to act as a scaffold. Degradation of BCR-ABL1 prevents this scaffolding function, thus ameliorating signaling via GAB2, SHP-2 and SHC. This scaffolding role also partially explains the enhanced anti-proliferative activity of GMB-475 compared to GMB-651 in both model systems (K562, Ba/F3) and primary CML patient samples. Apparently, destruction of the protein, rather than inhibition of its kinase domain, has a more potent and sustained inhibition of downstream signaling, at least in part, due to the loss of the Y177 docking domain.
It has been shown previously that while ABL1 TKIs effectively inhibit BCR-ABL1 kinase activity in CML stem and progenitor cells, the stem population is preferentially less susceptible to apoptosis induction (3). Importantly, we confirmed that GMB-475 effectively inhibited BCR-ABL1 kinase activity and degraded BCR-ABL1 protein in the context of isolated CML stem (CD34+CD38−) and progenitor (CD34+CD38+) cells, albeit at higher concentrations than imatinib. While we observed significant induction of apoptosis to GMB-475 in the progenitors, only a minor induction of apoptosis was observed in CML stem cells, adding to the growing body of evidence that CML stem cells are not dependent on BCR-ABL1 kinase activity for survival (3,5,39-41). Additionally, the lack of differential between GMB-475 and GMB-651 in this population suggests that they are not dependent on potential non-kinase scaffolding roles associated with the presence of the BCR-ABL1 protein. While it has been postulated that the scaffolding roles of BCR-ABL1, including that of Y177, may be responsible for the survival of BCR-ABL1-positive LSCs (37,39), our results support a more limited dependence upon BCR-ABL1 in general in this population. Previous studies have also suggested the persistence of CML stem cells on treatment is likely attributable to several potential mechanisms, including quiescence, alternative survival signaling pathways, and protective signals from the bone marrow microenvironment niche (42). Our ongoing efforts to develop more potent allosteric BCR-ABL1 PROTACs will allow for further exploration of this phenomenon in vivo.
In conclusion, this study further demonstrates the power of the PROTAC technology both as a potential therapeutic approach and as a tool to probe fundamental biology. We demonstrate for the first time the power of allosteric PROTACs in concert with orthosteric inhibitors against native and mutant BCR-ABL1, providing an opportunity for reducing dose-dependent side effects and managing acquired resistance. In total, our findings highlight the potential benefits of combined inhibition and degradation of BCR-ABL1 in the context of drug resistance and inducing deeper responses, while underscoring the importance of auxiliary pathways activated independent of the oncogenic BCR-ABL1 fusion protein for future targeting efforts of residual CML stem cells and resistant patients who lack BCR-ABL1 mutations (5).
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Small molecule induced degradation of BCR-ABL1 in chronic myeloid leukemia provides an advantage over inhibition and provides insights into CML stem cell biology.
ACKNOWLEDGEMENTS
G.M.B is a Fellow of The Leukemia & Lymphoma Society. G.M.B and C.M.C gratefully acknowledge The Leukemia & Lymphoma Society for their support (MCG-11257-17). D.P.B. is supported by an National Cancer Institute (NCI) Predoctoral to Postdoctoral Fellow Transition award (F99 CA212229-02). C.M.C. gratefully acknowledges the US National Institutes of Health (NIH) for their support (R35CA197589). C.M.C. is founder, consultant, and shareholder in Arvinas, Inc, which supports research in his lab. B.J.D. is an Investigator for the Howard Hughes Medical Institute and is supported in part by the Leukemia & Lymphoma Society and the NIH/NCI (R01 CA065823).
Footnotes
Financial support and conflicts of interest:
B.J.D. potential competing interests -- SAB: Aileron Therapeutics, ALLCRON, Cepheid, Gilead Sciences, Vivid Biosciences, Celgene & Baxalta (inactive); SAB & Stock: Aptose Biosciences, Blueprint Medicines, Beta Cat, GRAIL, Third Coast Therapeutics, CTI BioPharma (inactive); Scientific Founder & Stock: MolecularMD; Board of Directors & Stock: Amgen; Board of Directors: Burroughs Wellcome Fund, CureOne; Joint Steering Committee: Beat AML LLS; Clinical Trial Funding: Novartis, Bristol-Myers Squibb, Pfizer; Royalties from Patent 6958335 (Novartis exclusive license) and OHSU and Dana-Farber Cancer Institute (one Merck exclusive license).
C.M.C. is founder, consultant, and shareholder in Arvinas, Inc, which supports research in his lab.
REFERENCES
- 1.Druker BJ. Perspectives on the development of imatinib and the future of cancer research. Nature Medicine 2009;15:1149. [DOI] [PubMed] [Google Scholar]
- 2.Mahon F-X, Réa D, Guilhot J, Guilhot F, Huguet F, Nicolini F, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. The Lancet Oncology 2010;11(11):1029–35. [DOI] [PubMed] [Google Scholar]
- 3.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. The Journal of Clinical Investigation 2011;121(1):396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma L, Shan Y, Bai R, Xue L, Eide CA, Ou J, et al. A therapeutically targetable mechanism of BCR-ABL–independent imatinib resistance in chronic myeloid leukemia. Science Translational Medicine 2014;6(252):252ra121–252ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ichim CV. Kinase-Independent Mechanisms of Resistance of Leukemia Stem Cells to Tyrosine Kinase Inhibitors. STEM CELLS Translational Medicine 2014;3(4):405–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren R Mechanisms of BCR–ABL in the pathogenesis of chronic myelogenous leukaemia. Nature Reviews Cancer 2005;5:172. [DOI] [PubMed] [Google Scholar]
- 7.Lai AC, Toure M, Hellerschmied D, Salami J, Jaime-Figueroa S, Ko E, et al. Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL. Angew Chem Int Ed Engl 2016;55(2):807–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ru Y, Wang Q, Liu X, Zhang M, Zhong D, Ye M, et al. The chimeric ubiquitin ligase SH2-U-box inhibits the growth of imatinib-sensitive and resistant CML by targeting the native and T315I-mutant BCR-ABL. Scientific Reports 2016;6:28352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimokawa K, Shibata N, Sameshima T, Miyamoto N, Ujikawa O, Nara H, et al. Targeting the Allosteric Site of Oncoprotein BCR-ABL as an Alternative Strategy for Effective Target Protein Degradation. ACS Medicinal Chemistry Letters 2017;8(10):1042–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burslem GM, Crews CM. Small-Molecule Modulation of Protein Homeostasis. Chemical Reviews 2017;117(17):11269–301. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proceedings of the National Academy of Sciences 2001;98(15):8554–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bondeson DP, Mares A, Smith IED, Ko E, Campos S, Miah AH, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol 2015;11(8):611–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cromm PM, Samarasinghe KTG, Hines J, Crews CM. Addressing Kinase-Independent Functions of Fak via PROTAC-Mediated Degradation. Journal of the American Chemical Society 2018;140(49):17019–26. [DOI] [PubMed] [Google Scholar]
- 14.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, et al. Structural Basis for the Autoinhibition of c-Abl Tyrosine Kinase. Cell 2003;112(6):859–71. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Adrián FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, et al. Targeting Bcr–Abl by combining allosteric with ATP-binding-site inhibitors. Nature 2010;463:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wylie AA, Schoepfer J, Jahnke W, Cowan-Jacob SW, Loo A, Furet P, et al. The allosteric inhibitor ABL001 enables dual targeting of BCR–ABL1. Nature 2017;543:733. [DOI] [PubMed] [Google Scholar]
- 17.Shibata N, Ohoka N, Hattori T, Naito M. Development of a Potent Protein Degrader against Oncogenic BCR-ABL Protein. Chemical and Pharmaceutical Bulletin 2019;67(3):165–72. [DOI] [PubMed] [Google Scholar]
- 18.Deng X, Okram B, Ding Q, Zhang J, Choi Y, Adrián FJ, et al. Expanding the Diversity of Allosteric Bcr-Abl Inhibitors. Journal of Medicinal Chemistry 2010;53(19):6934–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buckley DL, Gustafson JL, Van Molle I, Roth AG, Tae HS, Gareiss PC, et al. Small-Molecule Inhibitors of the Interaction between the E3 Ligase VHL and HIF1α. Angewandte Chemie International Edition 2012;51(46):11463–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buckley DL, Van Molle I, Gareiss PC, Tae HS, Michel J, Noblin DJ, et al. Targeting the von Hippel–Lindau E3 Ubiquitin Ligase Using Small Molecules To Disrupt the VHL/HIF-1α Interaction. Journal of the American Chemical Society 2012;134(10):4465–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burslem GM, Smith BE, Lai AC, Jaime-Figueroa S, McQuaid DC, Bondeson DP, et al. The Advantages of Targeted Protein Degradation Over Inhibition: An RTK Case Study. Cell Chemical Biology 2018;25(1):67–77.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim KB, Crews CM. From epoxomicin to carfilzomib: chemistry, biology, and medical outcomes. Natural Product Reports 2013;30(5):600–04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, et al. Substrate-Assisted Inhibition of Ubiquitin-like Protein-Activating Enzymes: The NEDD8 E1 Inhibitor MLN4924 Forms a NEDD8-AMP Mimetic In Situ. Molecular Cell 2010;37(1):102–11. [DOI] [PubMed] [Google Scholar]
- 24.O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a Pan-BCR-ABL Inhibitor for Chronic Myeloid Leukemia, Potently Inhibits the T315I Mutant and Overcomes Mutation-Based Resistance. Cancer Cell 2009;16(5):401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular Cancer Therapeutics 2006;5(10):2512–21. [DOI] [PubMed] [Google Scholar]
- 26.Cilloni D, Saglio G. Molecular Pathways: BCR-ABL. Clinical Cancer Research 2012;18(4):930–37. [DOI] [PubMed] [Google Scholar]
- 27.Branford S Molecular monitoring in chronic myeloid leukemia—how low can you go? ASH Education Program Book 2016;2016(1):156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibata N, Miyamoto N, Nagai K, Shimokawa K, Sameshima T, Ohoka N, et al. Development of protein degradation inducers of oncogenic BCR-ABL protein by conjugation of ABL kinase inhibitors and IAP ligands. Cancer Science 2017;108(8):1657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H-T, Dobrovolsky D, Paulk J, Yang G, Weisberg EL, Doctor ZM, et al. A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader. Cell Chemical Biology 2018;25(1):88–99.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa S, et al. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chemical Biology 2018;25(1):78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burslem GM, Song J, Chen X, Hines J, Crews CM. Enhancing Antiproliferative Activity and Selectivity of a FLT-3 Inhibitor by Proteolysis Targeting Chimera Conversion. Journal of the American Chemical Society 2018;140(48):16428–32. [DOI] [PubMed] [Google Scholar]
- 32.Smith KM, Yacobi R, Van Etten RA. Autoinhibition of Bcr-Abl through Its SH3 Domain. Molecular Cell 2003;12(1):27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Wu Y, Ma GZ, Lu D, Haataja L, Heisterkamp N, et al. Inhibition of Bcr serine kinase by tyrosine phosphorylation. Molecular and Cellular Biology 1996;16(3):998–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai AC, Crews CM. Induced protein degradation: an emerging drug discovery paradigm. Nature Reviews Drug Discovery 2016;16:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salami J, Alabi S, Willard RR, Vitale NJ, Wang J, Dong H, et al. Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Communications Biology 2018;1(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sattler M, Mohi MG, Pride YB, Quinnan LR, Malouf NA, Podar K, et al. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell 2002;1(5):479–92. [DOI] [PubMed] [Google Scholar]
- 37.Chu S, Li L, Singh H, Bhatia R. BCR-Tyrosine 177 Plays an Essential Role in Ras and Akt Activation and in Human Hematopoietic Progenitor Transformation in Chronic Myelogenous Leukemia. Cancer Research 2007;67(14):7045–53. [DOI] [PubMed] [Google Scholar]
- 38.Warmuth M, Bergmann M, Prieß A, Häuslmann K, Emmerich B, Hallek M. The Src Family Kinase Hck Interacts with Bcr-Abl by a Kinase-independent Mechanism and Phosphorylates the Grb2-binding Site of Bcr. Journal of Biological Chemistry 1997;272(52):33260–70. [DOI] [PubMed] [Google Scholar]
- 39.Hamilton A, Helgason GV, Schemionek M, Zhang B, Myssina S, Allan EK, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood 2012;119(6):1501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konig H, Holtz M, Modi H, Manley P, Holyoake TL, Forman SJ, et al. Enhanced BCR-ABL kinase inhibition does not result in increased inhibition of downstream signaling pathways or increased growth suppression in CML progenitors. Leukemia 2008;22:748. [DOI] [PubMed] [Google Scholar]
- 41.Zhang B, Ho Yin W, Huang Q, Maeda T, Lin A, Lee S-u, et al. Altered Microenvironmental Regulation of Leukemic and Normal Stem Cells in Chronic Myelogenous Leukemia. Cancer Cell 2012;21(4):577–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holyoake TL, Vetrie D. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood 2017;129(12):1595–606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.