Abstract
Protein lysine fatty acylation is increasingly recognized as a prevalent and important protein post-translation modification. Recently, it has been shown that K-Ras4a, R-Ras2, and Rac1 are regulated by lysine fatty acylation. Here we investigated whether other members of the Ras superfamily could also be regulated by lysine fatty acylation. Several small GTPases exhibit hydroxylamine resistant fatty acylation, suggesting they may also have protein lysine fatty acylation. We further characterized one of these GTPases, RalB. We show that RalB has C-terminal lysine fatty acylation, with the predominant modification site being Lys200. The lysine acylation of RalB is regulated by SIRT2, a member of the sirtuin family of nicotinamide adenine dinucleotide (NAD+)-dependent protein lysine deacylases. Lysine fatty acylated RalB exhibited enhanced plasma membrane localization and recruited its known effectors Sec5 and Exo84, members of the exocyst complex, to the plasma membrane. RalB lysine fatty acylation did not affect the proliferation or anchorage-independent growth, but did affect the trans-well migration of A549 lung cancer cells. This study thus identified an additional function for protein lysine fatty acylation and the deacylase SIRT2.
Keywords: SIRT2, RalB, Lysine Fatty Acylation, cell migration, exocyst
Introduction:
Protein lipidation is a well-established mechanism that regulates various cellular homeostasis and human diseases 1,2. Lipidation has been shown to regulate membrane anchoring, protein stability, and protein-protein interactions 3. Much of our understanding of this class of modifications comes from the study of N-terminal myristoylation, cysteine palmitoylation (S-palmitoylation), cysteine prenylation, and modifications by Glycosylphosphatidylinositol (GPI) anchors 1–3. A similar, but not well studied modification is protein lysine fatty acylation, the modification of lysine residues with long-chain fatty acyl groups such as myristoyl or palmitoyl. Until 2017, only three proteins were identified to undergo lysine fatty acylation: TNF-α, IL-1α and lens integral membrane protein aquaporin-0 4–6. Recent work identified several other proteins that are modified by lysine fatty acylation and point to the prevalence and importance of this PTM 7–11. Interestingly, two bacterial toxins are also reported to fatty acylate lysine residues on host proteins 12,13.
Sirtuins, the nicotinamide adenine dinucleotide (NAD+)-dependent Class III of histone deacetylase (HDAC) proteins have attracted much research interest due to their connection to metabolism, cancer, and aging 14. Sirtuins were initially identified as protein lysine deacetylases. However, recently it was discovered that several HDACs, including Sirtuin 1 (SIRT1), SIRT2, SIRT3, SIRT6, SIRT7, HDAC8, and HDAC11, possess lysine defatty-acylation activity 9, 11, 15–20. These findings suggested that lysine fatty acylation is more prevalent than initially thought. Consistent with this hypothesis, it was recently reported that several Ras family small GTPases had protein lysine fatty acylation, and two of these proteins K-Ras4a and R-Ras2, are SIRT2 and SIRT6 defatty-acylation substrates, respectively7,8,12.
The Ras family of small GTPases are molecular switches; they are active when bound to guanosine triphosphate (GTP) and inactive when bound to guanosine-diphosphate (GDP) 21. There are over 150 members of the Ras superfamily that are classified into five subfamilies: Ras, Rho, Rab, Ran and Arf. The Ras subfamily has 36 members and includes the Rap, R-Ras, Ral and Rheb proteins 22, 23. Ras small GTPases are responsible for regulating a wide range of cellular functions such as cellular senescence, apoptosis, cell growth, autophagy, and are involved in numerous signaling pathways, making them key regulators of cancer 22. Thus understanding the regulation of small GTPases is important for the development of therapeutic strategies.
It is well established that protein lipidation, such as cysteine prenylation and palmitoylation serve as regulatory mechanisms for several small GTPases 7, 8, 24. Looking closely at the Ras small GTPases that were recently identified to have lysine fatty acylation, we saw that many of them shared common structural components in their C-terminal hypervariable region (HVR) 7, 8, 12, with multiple lysine residues in addition to the palmitoylated and prenylated cysteine residues. We hypothesized that protein lysine fatty acylation may be a regulatory mechanism for these small GTPases and investigated this possibility here.
RESULTS AND DISCUSSION:
Results: Several members of the Ras superfamily exhibit hydroxylamine resistant fatty acylation
To visualize the level of lysine fatty acylation on a specific protein, we used the biorthogonal palmitic acid analogue Alkyne-14 (Alk14) 9, 25. This probe is metabolically incorporated and subsequently labels fatty acylated proteins and allows in-gel fluorescence visualization of protein fatty acylation after conjugation of an azide-containing fluorescent dye with click chemistry (Figure 1A). This method labels both cysteine and lysine fatty acylation, but hydroxylamine can be used to remove S-palmitoylation and allows for visualization of lysine fatty acylation, which is resistant to hydroxylamine (Figure 1A). To test our hypothesis that other Ras superfamily GTPases also are regulated by lysine fatty acylation, we chose several small GTPases that contain multiple lysine residues, a palmitoylated cysteine, and/or a prenylated cysteine (characteristics that were seen with K-Ras4a, R-Ras2 and Rac1) (Figure 1B) 7, 8, 12, 24, 26, 27.
Figure 1.
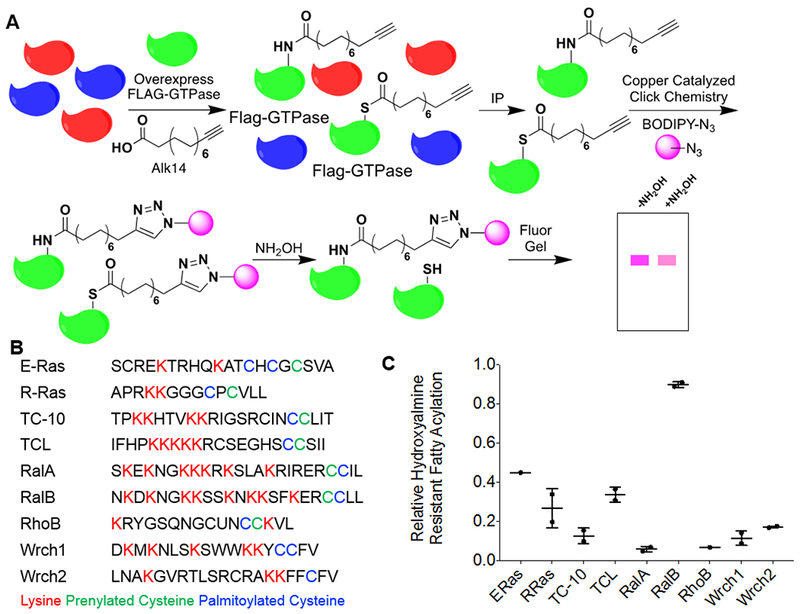
Several members of the Ras family small GTPases contain hydroxylamine-resistant fatty acylation. (A) The method used to detect lysine fatty acylation on proteins using the palmitic/myristic acid analogue, Alk14. N-terminal Flag-tagged small GTPases were overexpressed in HEK-293T cells. The cells were treated with 50 μM Alk14 (or the vehicle control DMSO) for 6 hours. After FLAG immunoprecipitation and on-bead click chemistry, fatty acylation levels were visualized using in-gel fluorescence with and without hydroxylamine treatment. (B) A list of related Ras family small GTPases and their respective c-terminal hypervariable region. Palmitoylated cysteine residues are highlighted in blue, prenylated cysteine residues are highlighted in green, and lysine residues that are potential sites for lysine fatty acylation are highlighted in red. (C) Quantification of the relative amount of hydroxylamine-resistant fatty acylation for each small GTPase. The fatty acylation level of each of the proteins listed was determined from two independent experiments (n=2). To quantify the relative hydroxylamine resistant fatty acylation, the fluorescence signal after hydroxylamine treatment, normalized to protein loading as determined by blue staining or a flag western blot, was divided by the fluorescence signal prior to hydroxylamine treatment, normalized to protein loading as determined by blue staining or a flag western blot.
We initially screened these small GTPases to determine how much of their fatty acylation was hydroxylamine resistant (Figure 1C, Figure S1). Interestingly, among the nine proteins tested, four of them (E-Ras, R-Ras, TCL, and RalB) contained >20% hydroxylamine-resistant fatty acylation, while only two of them (RalA and RhoB) contained <10% hydroxylamine-resistant fatty acylation. To ensure the hydroxylamine treatment was sufficient, we used a TNFα 4KR mutant as a positive control (Figure S1). The TNFα 4KR mutant only has S-palmitoylation, and therefore should exhibit no hydroxylamine resistant signal, which was what we saw 9. These results suggested lysine fatty acylation might be prevalent among Ras small GTPases.
RalB, but not RalA, has lysine fatty acylation under basal conditions
While many of the proteins exhibited hydroxylamine resistant fatty acylation, for further validation and functional studies, we decided to focus on the RAS like proto-oncogene A and B (RalA and RalB). RalA and RalB share 80% of their amino acid sequence, and most of their sequence divergence is found in their C-terminal hyper variable region (Figure 1B) 28. Despite their high sequence similarity, RalA and RalB often have different, or even opposing roles in cancer 28–31. The screening results indicated that RalB showed high level of hydroxylamine resistant fatty acylation while RalA had very little (Figure 1C). We therefore thought that lysine fatty acylation could be a factor to differentiate the function of these two small GTPases.
To further determine if either of the Ral GTPases, RalA or RalB, had lysine fatty acylation we ectopically overexpressed N-terminally Flag-tagged RalA (Flag-RalA) and RalB (Flag-RalB) in HEK293T cells, and labeled the proteins using the Alk14 method described above. RalA had higher levels of S-palmitoylation, but under basal conditions most of its fatty acylation signal could be removed by hydroxylamine, while RalB had labeling that was ~90% retained after hydroxylamine treatment (Figure 2A, 2B) 7. This suggested that under these cell culturing conditions, RalB, but not RalA, may have lysine fatty acylation.
Figure 2.
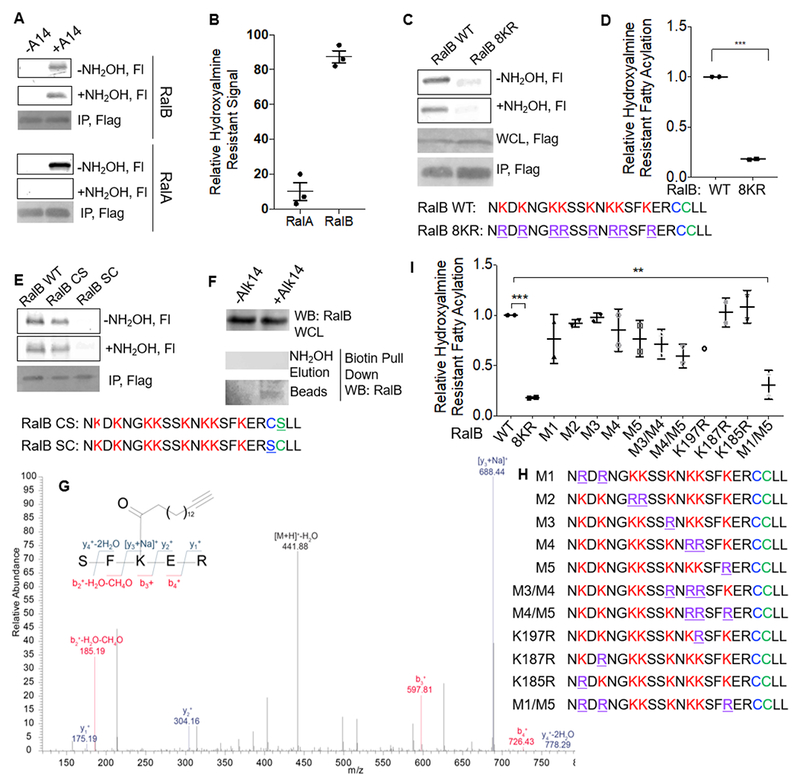
RalB contains Lysine Fatty Acylation. (A) Alk14 labeling of RalA and RalB with and without hydroxylamine treatment in HEK 293T Cells. The image shown is representative of three additional independent experiments to the data presented in Figure 1. (B) Quantification of the relative level of hydroxylamine-resistant fatty acylation of RalA and RalB shown in A for three additional independent experiments. Quantification was done as described in Figure 1. (C) Alk14 labeling of RalB WT and the 8KR mutant in HEK293T cells. The data shown is representative of two independent experiments. (D) Quantification of fluorescence labeling of RalB WT and the 8KR mutant shown in (C). For quantification, the total florescence signal for the 8KR mutant was normalized to its protein level as determined by a flag western blot or Coomassie blue staining and was then divided by the normalized florescence signal for WT RalB. The normalized (fluorescence signal/protein loading) for RalB WT was set to 1. (E) Alk14 labeling before and after hydroxylamine treatment of RalB WT, the non-prenylated (RalB SC) and non-S-palmitoylated (RalB CS) RalB mutants in HEK293T cells. The image shown is representative of three independent experiments. (F) Fatty acylation of endogenous RalB in HEK293T cells. (G) Tandem MS/MS spectra of a doubly charged Alk14 modified peptide on RalB. (H) C-terminal sequences of RalB lysine to arginine mutants generated. (I) Relative hydroxylamine-resistant fatty acylation levels of the various RalB lysine to arginine mutants in HEK293T cells. Statistical significance was calculated using Prism 7 software using an ordinary one-way ANOVA. ** p < 0.01, *** p < 0.001. (p=0.0008 for the difference between RalB-WT and RalB-8KR, and p=0.0034 for the difference between RalB-WT and the RalB-M1/M5 mutant.) Fl, fluorescence; WB, western blot.
To further support that the hydroxylamine resistant signal on RalB comes from lysine fatty acylation we mutated all eight lysine residues (K185, K187, K190, K191, K194, K196, K197, and K200) in the HVR of RalB to arginine to generate the RalB 8KR mutant. The RalB 8KR mutant showed approximately 20% of labeling compared to RalB WT, and exhibited essentially no hydroxylamine resistant signal (Figure 2C, 2D). This supports that the hydroxylamine resistant fluorescence signal detected on WT RalB is from lysine fatty acylation. Additionally, to show that the hydroxylamine resistant signal was not a result of hydroxylamine resistant S-palmitoylation, we mutated the palmitoylated cysteine to serine to generate the RalB C205S mutant. This mutant still contained fatty acylation that was hydroxylamine resistant (Figure 2E). However, removal of the prenylated cysteine (the C204S mutant) prevented RalB from having any fatty acylation signal (Figure 2E). The lack of labeling on the non-prenylated construct was not surprising, as prenylation has been established to be important for the proper processing and S-palmitoylation of proteins 26, 32, 33.
To assess whether endogenous RalB also has lysine fatty acylation, we treated cells with vehicle control (DMSO) or Alk14 and enriched Alk14-labeled proteins by attaching biotin-azide with click chemistry and subsequent streptavidin immunoprecipitation. S-palmitoylated proteins were removed from the streptavidin beads by hydroxylamine treatment, leaving proteins with lysine fatty acylation on the streptavidin beads, which were then detected by western blot. Indeed, we could detect RalB from the boiled streptavidin beads suggesting that endogenous RalB, like overexpressed RalB, has lysine fatty acylation (Figure 2F). We previously showed that RalA does not have endogenous lysine fatty acylation using basal conditions using the same method 7.
We further confirmed RalB has lysine fatty acylation by mass spectrometry (MS). After immunoprecipitation of Alk14-labeled Flag-RalB and trypsin digestion, we detected by tandem MS/MS a doubly charged Alk14-modified peptide in which RalB Lys200 was modified by Alk-14 (Figure 2G). To confirm the mass spectrometry results, we generated several different RalB lysine to arginine mutants (Figure 2H). We saw that the RalB M1 (K185,187R) and the RalB M5 (K200R) mutants both exhibited a slight decrease in fatty acylation levels. Further combinations suggested that the RalB M1M5 mutant exhibited a significant decrease in fatty acylation (Figure 2I, Figure S2). The redundancy of the lysine fatty acylation is not surprising, as it is similar to what was observed for R-Ras2, KRas4a, and Rac1 7, 8, 12. It is likely that if the predominant site of fatty acylation is mutated, other nearby lysine residues would have increased fatty acylation to compensate.
SIRT2 Regulates RalB Lysine Fatty Acylation
We next wanted to identify the mechanism by which RalB lysine fatty acylation is regulated. Over the past few years, it has been established that several HDACs, particularly sirtuins, possess lysine defatty-acylation activity 9, 11, 15–17. To see if RalB is a sirtuin target, we treated purified Alk14 labeled Flag-RalB from HEK293T cells, and examined the lysine fatty acylation levels of RalB after in vitro treatment with SIRT1, SIRT2, SIRT3, SIRT6, SIRT7, and HDAC8 (Figure 3A, Figure S3A–E). We saw that only SIRT2 was able to remove RalB lysine fatty acylation in an NAD-dependent manner. Using a previously established 32P-NAD assay, we detected the 2’-O-fatty acyl adenosine diphosphate ribose (ADPR) product in the SIRT2-catalyzed deacylation reaction, further supporting that SIRT2 was removing fatty acyl groups from a lysine residue of RalB (Figure 3B).
Figure 3.
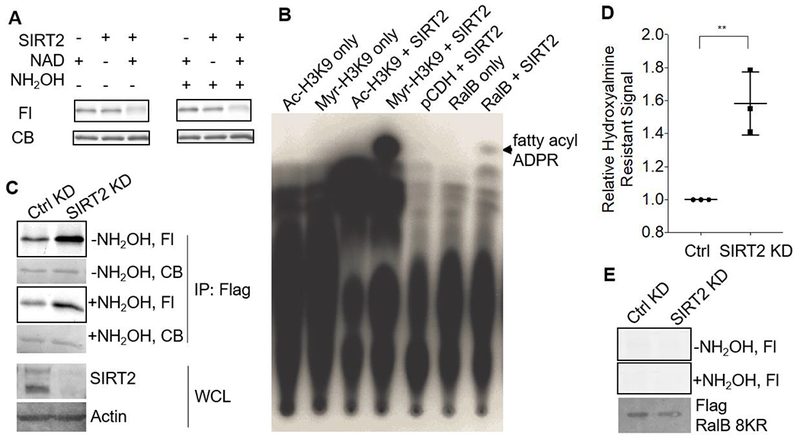
RalB is a SIRT2 defatty-acylation substrate. (A) SIRT2 decreased RalB hydroxylamine-resistant fatty acylation in vitro in an NAD-dependent manner. Representative images from three independent experiments are shown. (B) 32P-NAD assay with RalB and SIRT2. The formation of the 32P-labeled fatty acyl-ADPR was detected, showing that RalB contained lysine fatty acylation. A representative image from two independent experiments is shown. (C) Alk14 labeling of WT RalB in control and SIRT2 KD HEK 293T cells. Representative images from three independent experiments is shown. (D) Quantification of the relative hydroxylamine-resistant fatty acylation levels in SIRT2 WT and KD HEK293T cells. Fluorescence signal was normalized to the protein level based on blue staining. The normalized hydroxylamine resistant fatty acylation from SIRT2 KD cells was divided by the normalized hydroxylamine resistant fatty acylation signal from SIRT2 WT cells. The signal from SIRT2 WT cells was set to one. Statistical significance was calculated using Prism7 software with an unpaired student’s t-test. ** p < 0.01 (p=0.006) (E) Alk14 labeling of the RalB 8KR mutant in control and SIRT2 KD HEK 293T cells. The image shown is representative of two independent experiments. CB, Coomassie Blue staining.
We next wanted to see if RalB was a SIRT2 target in cells. To evaluate this, we looked at the fatty acylation levels of RalB in SIRT2 control and knockdown (KD) cells. In SIRT2 KD cells, RalB had significantly more hydroxylamine resistant fatty acylation (Figure 3C and 3D). In contrast, the RalB 8KR mutant did not have SIRT2-regulated lysine fatty acylation (Figure 3E).
SIRT2-Regulated RalB Lysine Fatty Acylation Affects Cancer Cell Migration
RalB has been shown to play a role in cell proliferation, anchorage independent growth and cell migration 34–36. We therefore sought to understand if lysine fatty acylation was important for the role of RalB in these physiological processes. We first investigated if lysine fatty acylation could promote cell proliferation. As it has previously been shown that RalB is important for A549 cell growth, to test if lysine fatty acylation was important for this, we transiently expressed pCDH control vector, Flag-RalB WT, and Flag-RalB 8KR and transiently knocked down SIRT2 in A549 cells (Supplemental Figure S4A). If RalB lysine fatty acylation was important for cell proliferation, we expect that SIRT2 KD should affect RalB WT expressing cells, but not RalB 8KR expressing cells. We monitored the cell proliferation and observed that lysine fatty acylation of RalB did not affect A549 cell proliferation (Supplemental Figure S4A).
RalB has been shown to modulate colony formation of HCT116 cells in soft agar 34, 36. To evaluate if lysine fatty acylation was important for the role RalB plays in anchorage independent growth, we generated stable RalB KD HCT116 cells, and subsequently overexpressed RalB WT or 8KR with and without SIRT2 KD. Lysine fatty acylation did not significantly affect anchorage independent growth (Supplemental Figure S4B).
To evaluate if RalB lysine fatty acylation could promote cell migration, we generated stable pCDH (control), Flag-RalB WT, and Flag-RalB 8KR overexpressing A549 cells. We then transiently knocked down SIRT2 in these cell lines and examined cell migration. Expression of RalB WT or 8KR had very little effect on cell migration in control KD cells. Interestingly, when SIRT2 levels were decreased, overexpression of RalB WT, but not RalB 8KR, dramatically increased cell migration (Figure 4A and 4B). This result suggests that RalB lysine fatty acylation promotes cell migration in A549 cells in a SIRT2 dependent manner.
Figure 4.

RalB lysine fatty acylation promotes A549 cell migration. (A) Images of the stained Boyden chambers after 24 hour cell migration. For the experiment cells were starved of serum for 12hr, prior to being counted and seeded in Boyden chambers. Experiments were done in at least biological duplicates and images shown are from one representative experiment. Images were taken on a Motic AE21 microscope, using a 20X objective. Motic Images Plus 2.0 software was used to obtain the images. (B) Quantification for experiments shown in A. Quantification was done by measuring the absorbance of the crystal violet stain of migrated cells and normalizing to the starting cell number as determined by cell titer blue. (C) Western blots to confirm SIRT2 KD, and Flag-RalB WT and Flag-RalB 8KR overexpression in A549 cells.
RalB Lysine Fatty Acylation Promotes RalB Plasma Membrane Localization
Protein lipidation such as S-palmitoylation and cysteine prenylation are known to be important for protein membrane localization 3. Like other small GTPases, RalB cysteine prenylation and S-palmitoylation are also important for its membrane localization 26, 32. Previously it was reported that lysine fatty acylation could regulate the plasma membrane localization of K-Ras4a and R-Ras2 7, 8. We therefore hypothesized that lysine fatty acylation could also promote RalB plasma membrane localization. To test this hypothesis, we ectopically expressed Flag-RalB WT and Flag-RalB 8KR constructs in HEK239T cells and compared the intracellular immunofluorescence signal to the signal observed for the whole cell. We saw that the RalB 8KR mutant had a larger fraction of its fluorescence signal from the intracellular portion than RalB WT. This suggests that lysine fatty acylation may be important for plasma membrane localization of RalB (Figure 5A, B).
Figure 5.
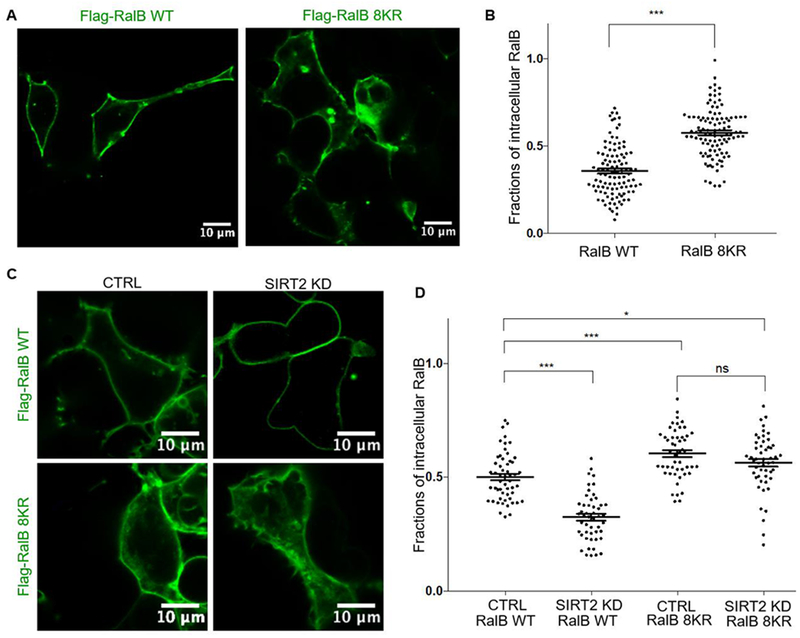
RalB lysine fatty acylation promotes membrane localization (A) Immunofluorescence to evaluate the subcellular localization of Flag-RalB WT and Flag-RalB 8KR in HEK293T cells. (B) Quantification of the intracellular RalB signal over the whole cell fluorescence signal. Statistical significance determined by a two-tailed unpaired student’s t-test, *** p < 0.001 (C) Immunofluorescence to evaluate the subcellular localization of Flag-RalB WT and Flag-RalB 8KR in Ctrl and SIRT2 KD HEK293T cells to evaluate the subcellular localization. (D) Quantification of the intracellular RalB signal over the whole cell fluorescence signal. Statistical significance determined using one-way Anova, * p < 0.05, ** p < 0.01, *** p < 0.001.
To rule out that this was an artifact from mutating multiple lysine residues, which form the polybasic region on the C-terminal hypervariable region of RalB, we compared the localization of RalB in SIRT2 Control and KD HEK293T cells. We saw that SIRT2 KD promoted membrane localization of RalB WT, but not the lysine fatty acylation deficient RalB 8KR mutant (Figure 5C, D). The data provide further support that RalB lysine fatty acylation is important for the plasma membrane localization of RalB.
RalB Lysine Fatty Acylation Promotes RalB GTP binding and Promotes Membrane Localization of the Exocyst Components Exo84 and Sec5
When bound to GTP, RalB can associate with its effector proteins such as Exo84, RalBP1, and Sec5 31. We first evaluated if RalB fatty acylation affects its activation (GTP binding). We compared the GTP binding of RalB-WT and RalB-8KR using a well-established GTP binding assay using the Sec5 Ral binding domain (RBD). The Sec5 RBD selective binds to active, or GTP-bound Ral. We saw that RalB WT bound more GTP than the 8KR mutant, suggesting that fatty acylation promotes the GTP binding of RalB (Figure 6A, B).
Figure 6.
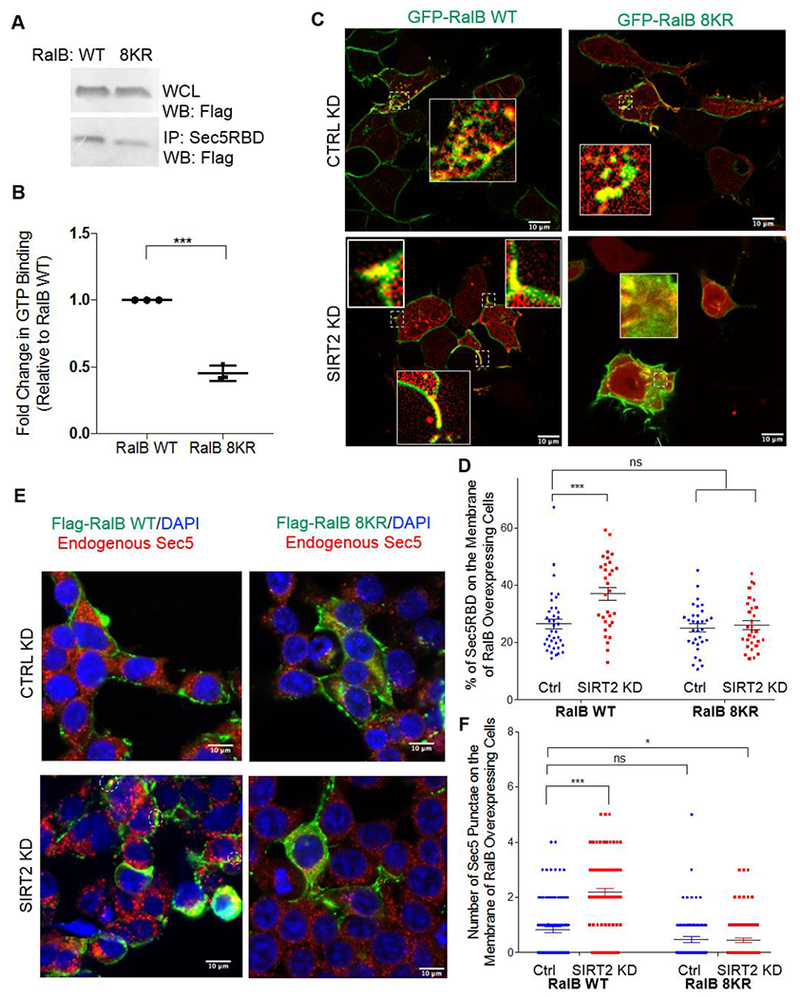
RalB lysine fatty acylation promotes RalB-GTP binding. (A) GTP binding of RalB WT and RalB 8KR determined using the Sec5-RBD after 30 min starvation in EBSS buffer. (B) Quantification of the GTP binding assay. Statistical significance was determined using a student’s unpaired t-test with Prism7 software. (C) Live cell imaging of HEK293T cells co-overexpressing GFP-RalB (WT and 8KR) and mCherry-Sec5RBD. (D) Quantification showing the percentage of the Sec5-RBD membrane localization in cells overexpressing RalB WT and RalB 8KR. Statistical significance determined by a one-way Anova using Prism7 software, *** p < 0.001, ns= not significant. (E) Immunofluorescence of CTRL and SIRT2 KD HEK293T cells overexpressing Flag-RalB WT and Flag-RalB 8KR and endogenous Sec5 (F) Quantification of the number of Sec5 puncta in cells overexpressing RalB. Statistical significance was determined by a one-way Anova using Prism7 software *** p < 0.001, ** p < 0.01, * p < 0.05, ns= not significant.
We next wanted to see where active RalB WT and RalB 8KR were located within the cell. We co-overexpressed GFP-tagged RalB WT or 8KR and mCherry-Sec5RBD (Figure 6C). Through live cell imaging, we observed that when RalB-WT was overexpressed in SIRT2 knockdown cells, there was a larger portion of the Sec5 RBD located on the plasma membrane compared to control knockdown cells (Figure 6C & D). RalB WT showed more co-localization with the Sec5RBD on the plasma membrane than the lysine fatty acylation deficient 8KR mutant in SIRT2 knockdown cells, with the RalB 8KR mutant showing predominately intracellular co-localization with the Sec5-RBD in both control and SIRT2 knockdown cells (Figure 6C & D).
When small GTPases are bound to GTP they have the ability to interact with their known effector proteins. Two of the most well established Ral effector proteins are members of the exocyst complex, Sec5 and Exo84 37–40. The exocyst complex has been implicated in cellular processes such as migration and exocytosis 41. RalA and RalB have been shown to interact with exocyst components Sec5 and Exo84, modulate the assembly of the exocyst as well as control its subcellular localization, and thus regulating trafficking, exocytosis and migration 39–43.
We hypothesized that the enhanced membrane localization of fatty acylated RalB could promote the membrane localization of the exocyst complex. To evaluate this, we looked at the localization of endogenous Sec5 in HEK293T cells overexpressing Flag-RalB WT or Flag-RalB 8KR. We saw that in a SIRT2 dependent manner, overexpression of lysine fatty acylated RalB promoted the localization of Sec5 punctae to the plasma membrane (Figure 6E, F). When we expressed the RalB G23V active mutant in HEK293T cells, we also observed that endogenous Exo84 was recruited more to the plasma membrane by the RalB G23V WT that contained lysine fatty acylation, than by the RalB G23V 8KR mutant that did not contain lysine fatty acylation (Supplemental Figure 5A). Furthermore, knockdown of SIRT2 also led to enhanced cell membrane localization of endogenous Sec5 in A549 cells (Supplemental Figure 5B, C). These findings suggest that lysine fatty acylation of RalB promotes the membrane localization of the exocyst complex, which may be important for the migration phenotype we observed (Figure 5A).
Discussion.
Despite being identified in the early 1990s, protein lysine fatty acylation has remained an under-recognized PTM. This is largely due to the fact that only a few proteins have been identified to have this modification. Our studies here provide important support for the prevalence and importance of lysine fatty acylation. The screening of nine GTPases suggested that many members in the Ras superfamily are likely regulated by lysine fatty acylation. It is possible that the abundance and importance of lysine fatty acylation is similar to cysteine palmitoylation that has been well studied.
In this study, we focused on the role lysine fatty acylation of RalB. RalB is predominately fatty acylated at Lys200. Interestingly only RalB, but not RalA, exhibited lysine fatty acylation under the conditions tested, despite their high sequence similarity. This observation highlights that there are specific requirements for lysine fatty acylation to occur. It is possible that the difference in the C-terminal sequence of RalA and RalB can partially explain why RalB but not RalA exhibits lysine fatty acylation under the conditions tested. We identified Lys200 as the predominate site of lysine fatty acylation on RalB, and RalA does not have this lysine residue. The RalA sequence actually has an arginine where RalB has a lysine. As arginine cannot be fatty acylated this can potentially explain our findings. Furthermore, it is already reported that RalA and RalB exhibit different subcellular localizations26, 28, 30. The difference in subcellular localization can also potentially explain our findings. Recently, two bacterial toxins that are reported to fatty acylate lysine residues12, 13. It is likely that mammalian cells have endogenous lysine fatty acyl transferases. Understanding what determines this interesting RalB over RalA specificity requires the identification of lysine fatty acyl transferases in the future.
RalB lysine fatty acylation is regulated by SIRT2, one of the sirtuin family of NAD-dependent lysine deacylases. SIRT2 has been reported to regulate many biological pathways via different substrate proteins, including metabolic enzymes, histones, signaling proteins, and transcription factors 44–59. However, to date, almost all the known substrates (except K-Ras4a) of SIRT2 are deacetylation substrates, not defatty-acylation substrates. Here, we have identified the second SIRT2 defatty-acylation substrate. These findings show that SIRT2-regulated lysine fatty acylation can affect K-Ras4a-induced colony formation and RalB-mediated cell migration. Much still remains to be understood about the biological function of SIRT2. For example, SIRT2 inhibitors have shown promise in treating both cancer and neurodegenerative diseases, but the detailed molecular mechanisms still remain elusive 60–68. The identification of RalB as a defatty-acylation substrate may help to understand why SIRT2 inhibitors would be useful for treating cancer and neurodegeneration.
Our study identifies a novel regulatory mechanism for RalB lysine fatty acylation. We found that lysine fatty acylation is important for the GTP binding of RalB. This is similar to what was recently seen for Rac1, which is lysine fatty acylated by a Cholera effector protein RID 12. RalB has been known to regulate cell proliferation, anchorage-independent growth, cell migration, and cell adhesion. Interestingly, lysine fatty acylation does not significantly affect cell proliferation or anchorage-independent growth, but affects cell migration. Our finding that RalB lysine fatty acylation promotes Exo84 and Sec5 plasma membrane localization is consistent with its effect on cell migration. Lysine fatty acylation promotes RalB activation and localization to the cell migration front, which serves to recruit exocyst to the migration front, thus promoting cell migration.
In conjunction with recent findings that several other small GTPases also exhibit hydroxylamine resistant fatty acylation, this study suggests that lysine fatty acylation could be an additional mechanism which regulates many GTPases. As modulating the Ras family of GTPases can affect various cancer phenotypes such as cell migration, proliferation, or anchorage independent growth, elucidating the role of lysine fatty acylation as a regulatory mechanism may lead to new treatment strategies for human diseases 23, 29, 42, 69–74.
Material and Methods:
Detailed information for all methods used can be found in the supplemental information.
Supplementary Material
Acknowledgements:
We would like thank M. Linder for providing several of the GTPase plasmids. GFP-TCL was a gift from C. Der (Addgene plasmid # 23231; http://n2t.net/addgene:23231; RRID: Addgene_23231). The work is supported in part by NIH/NIDDK R01 DK107868. H.J. was an HHMI International Student Fellow. The work made use of the Cornell University NMR Facility, which is supported, in part, by the NSF through MRI award CHE-1531632. Imaging data was acquired through the Cornell University Biotechnology Resource Center, with NYSTEM (CO29155) and NIH (S10OD018516) funding for the shared Zeiss LSM880 confocal/multiphoton microscope.
Footnotes
Supporting Information: Supplemental figures and detailed methods used can be found in the supplemental information, which is available free of charge via the internet at http://pubs.acs.org.
Competing Interests: The authors declare no competing interests.
References:
- 1.Jiang H; Zhang X; Chen X; Aramsangtienchai P; Tong Z; Lin H, Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies. Chem Rev 2018, 118, 919–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nadolski MJ; Linder ME, Protein lipidation. FEBS J 2007, 274, 5202–5210. [DOI] [PubMed] [Google Scholar]
- 3.Resh MD, Covalent lipid modifications of proteins. Curr Biol 2013, 23, R431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schey KL; Gutierrez DB; Wang Z; Wei J; Grey AC, Novel fatty acid acylation of lens integral membrane protein aquaporin-0. Biochemistry 2010, 49, 9858–9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevenson FT; Bursten SL; Locksley RM; Lovett DH, Myristyl acylation of the tumor necrosis factor alpha precursor on specific lysine residues. J Exp Med 1992, 176, 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevenson FT; Bursten SL; Fanton C; Locksley RM; Lovett DH, The 31-kDa precursor of interleukin 1 alpha is myristoylated on specific lysines within the 16-kDa N-terminal propiece. Proc Natl Acad Sci U S A 1993, 90, 7245–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jing H; Zhang X; Wisner SA; Chen X; Spiegelman NA; Linder ME; Lin H, SIRT2 and lysine fatty acylation regulate the transforming activity of K-Ras4a. Elife 2017. pii: e32436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X; Spiegelman NA; Nelson OD; Jing H; Lin H, SIRT6 regulates Ras-related protein R-Ras2 by lysine defatty-acylation. Elife 2017. pii: e25158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H; Khan S; Wang Y; Charron G; He B; Sebastian C; Du J; Kim R; Ge E; Mostoslavsky R; Hang HC; Hao Q; Lin H, SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496, 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang H; Zhang X; Lin H , Lysine fatty acylation promotes lysosomal targeting of TNF-alpha. Sci Rep 2016, 6, 24371–24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao J; Sun L; Aramsangtienchai P; Spiegelman NA; Zhang X; Huang W; Seto E; Lin H, HDAC11 regulates type I interferon signaling through defatty-acylation of SHMT2. Proc Natl Acad Sci U S A 2019, 116, 5487–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y; Huang C; Yin L; Wan M; Wang X; Li L; Liu Y; Wang Z; Fu P; Zhang N; Chen S; Liu X; Shao F; Zhu Y, N(epsilon)-Fatty acylation of Rho GTPases by a MARTX toxin effector. Science 2017, 358, 528–531. [DOI] [PubMed] [Google Scholar]
- 13.Liu W; Zhou Y; Peng T; Zhou P; Ding X; Li Z; Zhong H; Xu Y; Chen S; Hang HC; Shao F, N(epsilon)-fatty acylation of multiple membrane-associated proteins by Shigella IcsB effector to modulate host function. Nat Microbiol 2018, 3, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haigis MC; Sinclair DA, Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 2010, 5, 253–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teng YB; Jing H; Aramsangtienchai P; He B; Khan S; Hu J; Lin H; Hao Q, Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci Rep 2015, 5, 8529–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aramsangtienchai P; Spiegelman NA; He B; Miller SP; Dai L; Zhao Y; Lin H, HDAC8 Catalyzes the Hydrolysis of Long Chain Fatty Acyl Lysine. ACS Chem Biol 2016, 11, 2685–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong Z; Wang M; Wang Y; Kim DD; Grenier JK; Cao J; Sadhukhan S; Hao Q; Lin H, SIRT7 Is an RNA-Activated Protein Lysine Deacylase. ACS Chem Biol 2017, 12, 300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno-Yruela C; Galleano I; Madsen AS; Olsen CA, Histone Deacetylase 11 Is an epsilon-N-Myristoyllysine Hydrolase. Cell Chem Biol 2018, 25, 849–856. [DOI] [PubMed] [Google Scholar]
- 19.Kutil Z; Novakova Z; Meleshin M; Mikesova J; Schutkowski M; Barinka C, Histone Deacetylase 11 Is a Fatty-Acid Deacylase. ACS Chem Biol 2018, 13, 685–693. [DOI] [PubMed] [Google Scholar]
- 20.Feldman JL; Baeza J; Denu JM, Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem 2013, 288, 31350–31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ten Klooster JP; Hordijk PL, Targeting and localized signalling by small GTPases. Biol Cell 2007, 99, 1–12. [DOI] [PubMed] [Google Scholar]
- 22.Colicelli J, Human RAS superfamily proteins and related GTPases. Sci STKE 2004, 13–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wennerberg K; Rossman KL; Der CJ, The Ras superfamily at a glance. J Cell Sci 2005, 118, 843–846. [DOI] [PubMed] [Google Scholar]
- 24.Aicart-Ramos C; Valero RA; Rodriguez-Crespo I, Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta 2011, 1808, 2981–2994. [DOI] [PubMed] [Google Scholar]
- 25.Gao X; Hannoush RN, A Decade of Click Chemistry in Protein Palmitoylation: Impact on Discovery and New Biology. Cell Chem Biol 2018, 25, 236–246. [DOI] [PubMed] [Google Scholar]
- 26.Gentry LR; Nishimura A; Cox AD; Martin TD; Tsygankov D; Nishida M; Elston TC; Der CJ, Divergent roles of CAAX motif-signaled posttranslational modifications in the regulation and subcellular localization of Ral GTPases. J Biol Chem 2015, 290, 22851–22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaelson D; Ali W; Chiu VK; Bergo M; Silletti J; Wright L; Young SG; Philips M, Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol Biol Cell 2005, 16, 1606–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentry LR; Martin TD; Reiner DJ; Der CJ, Ral small GTPase signaling and oncogenesis: More than just 15minutes of fame. Biochim Biophys Acta 2014, 1843, 2976–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan C; Theodorescu D, RAL GTPases: Biology and Potential as Therapeutic Targets in Cancer. Pharmacol Rev 2018, 70, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirakawa R; Horiuchi H, Ral GTPases: crucial mediators of exocytosis and tumourigenesis. J Biochem 2015, 157, 285–99. [DOI] [PubMed] [Google Scholar]
- 31.Neel NF; Martin TD; Stratford JK; Zand TP; Reiner DJ; Der CJ, The RalGEF-Ral Effector Signaling Network: The Road Less Traveled for Anti-Ras Drug Discovery. Genes Cancer 2011, 2, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falsetti SC; Wang DA; Peng H; Carrico D; Cox AD; Der CJ; Hamilton AD; Sebti SM, Geranylgeranyltransferase I inhibitors target RalB to inhibit anchorage-dependent growth and induce apoptosis and RalA to inhibit anchorage-independent growth. Mol Cell Biol 2007, 27, 8003–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimura A; Linder ME, Identification of a novel prenyl and palmitoyl modification at the CaaX motif of Cdc42 that regulates RhoGDI binding. Mol Cell Biol 2013, 33, 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin TD; Samuel JC; Routh ED; Der CJ; Yeh JJ, Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res 2011, 71, 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guin S; Ru Y; Wynes MW; Mishra R; Lu X; Owens C; Barn AE; Vasu VT; Hirsch FR; Kern JA; Theodorescu D, Contributions of KRAS and RAL in non-small-cell lung cancer growth and progression. J Thorac Oncol 2013, 8, 1492–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tecleab A; Zhang X; Sebti SM, Ral GTPase down-regulation stabilizes and reactivates p53 to inhibit malignant transformation. J Biol Chem 2014, 289, 31296–31309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cantor SB; Urano T; Feig LA, Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol 1995, 15, 4578–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda M; Ishida O; Hinoi T; Kishida S; Kikuchi A, Identification and characterization of a novel protein interacting with Ral-binding protein 1, a putative effector protein of Ral. J Biol Chem 1998, 273, 814–821. [DOI] [PubMed] [Google Scholar]
- 39.Moskalenko S; Henry DO; Rosse C; Mirey G; Camonis JH; White MA, The exocyst is a Ral effector complex. Nat Cell Biol 2002, 4, 66–72. [DOI] [PubMed] [Google Scholar]
- 40.Moskalenko S; Tong C; Rosse C; Mirey G; Formstecher E; Daviet L; Camonis J; White MA, Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem 2003, 278, 51743–51748. [DOI] [PubMed] [Google Scholar]
- 41.Liu J; Guo W, The exocyst complex in exocytosis and cell migration. Protoplasma 2012, 249, 587–597. [DOI] [PubMed] [Google Scholar]
- 42.Bodemann BO; Orvedahl A; Cheng T; Ram RR; Ou YH; Formstecher E; Maiti M; Hazelett CC; Wauson EM; Balakireva M; Camonis JH; Yeaman C; Levine B; White MA, RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 2011, 144, 253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camonis JH; White MA, Ral GTPases: corrupting the exocyst in cancer cells. Trends Cell Biol 2005, 15, 327–332. [DOI] [PubMed] [Google Scholar]
- 44.Cha Y; Han MJ; Cha HJ; Zoldan J; Burkart A; Jung JH; Jang Y; Kim CH; Jeong HC; Kim BG; Langer R; Kahn CR; Guarente L; Kim KS, Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c-SIRT2 axis. Nat Cell Biol 2017, 19, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dryden SC; Nahhas FA; Nowak JE; Goustin AS; Tainsky MA, Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol 2003, 23, 3173–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eskandarian HA; Impens F; Nahori MA; Soubigou G; Coppee JY; Cossart P; Hamon MA, A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science 2013, 341, 525–536. [DOI] [PubMed] [Google Scholar]
- 47.Fiskus W; Coothankandaswamy V; Chen J; Ma H; Ha K; Saenz DT; Krieger SS; Mill CP; Sun B; Huang P; Mumm JS; Melnick AM; Bhalla KN, SIRT2 Deacetylates and Inhibits the Peroxidase Activity of Peroxiredoxin-1 to Sensitize Breast Cancer Cells to Oxidant Stress-Inducing Agents. Cancer Res 2016, 76, 5467–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue T; Nakayama Y; Li Y; Matsumori H; Takahashi H; Kojima H; Wanibuchi H; Katoh M; Oshimura M, SIRT2 knockdown increases basal autophagy and prevents postslippage death by abnormally prolonging the mitotic arrest that is induced by microtubule inhibitors. FEBS J 2014, 281, 2623–2637. [DOI] [PubMed] [Google Scholar]
- 49.Jing E; Gesta S; Kahn CR, SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab 2007, 6, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HS; Vassilopoulos A; Wang RH; Lahusen T; Xiao Z; Xu X; Li C; Veenstra TD; Li B; Yu H; Ji J; Wang XW; Park SH; Cha YI; Gius D; Deng CX, SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 2011, 20, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y; Matsumori H; Nakayama Y; Osaki M; Kojima H; Kurimasa A; Ito H; Mori S; Katoh M; Oshimura M; Inoue T, SIRT2 down-regulation in HeLa can induce p53 accumulation via p38 MAPK activation-dependent p300 decrease, eventually leading to apoptosis. Genes Cells 2011, 16, 34–45. [DOI] [PubMed] [Google Scholar]
- 52.Park SH; Ozden O; Liu G; Song HY; Zhu Y; Yan Y; Zou X; Kang HJ; Jiang H; Principe DR; Cha YI; Roh M; Vassilopoulos A; Gius D, SIRT2-Mediated Deacetylation and Tetramerization of Pyruvate Kinase Directs Glycolysis and Tumor Growth. Cancer Res 2016, 76, 3802–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rothgiesser KM; Erener S; Waibel S; Luscher B; Hottiger MO, SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci 2010, 123, 4251–4258. [DOI] [PubMed] [Google Scholar]
- 54.Sarikhani M; Maity S; Mishra S; Jain A; Tamta AK; Ravi V; Kondapalli MS; Desingu PA; Khan D; Kumar S; Rao S; Inbaraj M; Pandit AS; Sundaresan NR, SIRT2 deacetylase represses NFAT transcription factor to maintain cardiac homeostasis. J Biol Chem 2018, 293, 5281–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsusaka T; Guo T; Yagura T; Inoue T; Yokode M; Inagaki N; Kondoh H, Deacetylation of phosphoglycerate mutase in its distinct central region by SIRT2 down-regulates its enzymatic activity. Genes Cells 2014, 19, 766–777. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H; Park SH; Pantazides BG; Karpiuk O; Warren MD; Hardy CW; Duong DM; Park SJ; Kim HS; Vassilopoulos A; Seyfried NT; Johnsen SA; Gius D; Yu DS, SIRT2 directs the replication stress response through CDK9 deacetylation. Proc Natl Acad Sci U S A 2013, 110, 13546–13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pereira JM; Chevalier C; Chaze T; Gianetto Q; Impens F; Matondo M; Cossart P; Hamon MA, Infection Reveals a Modification of SIRT2 Critical for Chromatin Association. Cell Rep 2018, 23, 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang X; Park SH; Chang HC; Shapiro JS; Vassilopoulos A; Sawicki KT; Chen C; Shang M; Burridge PW; Epting CL; Wilsbacher LD; Jenkitkasemwong S; Knutson M; Gius D; Ardehali H, Sirtuin 2 regulates cellular iron homeostasis via deacetylation of transcription factor NRF2. J Clin Invest 2017, 127, 1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarikhani M; Mishra S; Desingu PA; Kotyada C; Wolfgeher D; Gupta MP; Singh M; Sundaresan NR, SIRT2 regulates oxidative stress-induced cell death through deacetylation of c-Jun NH2-terminal kinase. Cell Death Differ 2018, 25, 1638–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dai H; Sinclair DA; Ellis JL; Steegborn C, Sirtuin activators and inhibitors: Promises, achievements, and challenges. Pharmacol Ther 2018, 188, 140–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chopra V; Quinti L; Kim J; Vollor L; Narayanan KL; Edgerly C; Cipicchio PM; Lauver MA; Choi SH; Silverman RB; Ferrante RJ; Hersch S; Kazantsev AG, The sirtuin 2 inhibitor AK-7 is neuroprotective in Huntington’s disease mouse models. Cell Rep 2012, 2, 1492–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jing H; Hu J; He B; Negron Abril YL; Stupinski J; Weiser K; Carbonaro M; Chiang YL; Southard T; Giannakakou P; Weiss RS; Lin H, A SIRT2-Selective Inhibitor Promotes c-Myc Oncoprotein Degradation and Exhibits Broad Anticancer Activity. Cancer Cell 2016, 29, 767–768. [DOI] [PubMed] [Google Scholar]
- 63.Shah AA; Ito A; Nakata A; Yoshida M, Identification of a Selective SIRT2 Inhibitor and Its Anti-breast Cancer Activity. Biol Pharm Bull 2016, 39, 1739–1742. [DOI] [PubMed] [Google Scholar]
- 64.Outeiro TF; Kontopoulos E; Altmann SM; Kufareva I; Strathearn KE; Amore AM; Volk CB; Maxwell MM; Rochet JC; McLean PJ; Young AB; Abagyan R; Feany MB; Hyman BT; Kazantsev AG, Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 2007, 317, 516–519. [DOI] [PubMed] [Google Scholar]
- 65.Spiegelman NA; Price IR; Jing H; Wang M; Yang M; Cao J; Hong JY; Zhang X; Aramsangtienchai P; Sadhukhan S; Lin H, Direct Comparison of SIRT2 Inhibitors: Potency, Specificity, Activity-Dependent Inhibition, and On-Target Anticancer Activities. ChemMedChem 2018, 13, 1890–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozako T; Mellini P; Ohsugi T; Aikawa A; Uchida YI; Honda SI; Suzuki T, Novel small molecule SIRT2 inhibitors induce cell death in leukemic cell lines. BMC Cancer 2018, 18, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luthi-Carter R; Taylor DM; Pallos J; Lambert E; Amore A; Parker A; Moffitt H; Smith DL; Runne H; Gokce O; Kuhn A; Xiang Z; Maxwell MM; Reeves SA; Bates GP; Neri C; Thompson LM; Marsh JL; Kazantsev AG, SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc Natl Acad Sci U S A 2010, 107, 7927–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun S; Han X; Li X; Song Q; Lu M; Jia M; Ding J; Hu G, MicroRNA-212–5p Prevents Dopaminergic Neuron Death by Inhibiting SIRT2 in MPTP-Induced Mouse Model of Parkinson’s Disease. Front Mol Neurosci 2018, 11, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajalingam K; Schreck R; Rapp UR; Albert S, Ras oncogenes and their downstream targets. Biochim Biophys Acta 2007, 1773, 1177–1195. [DOI] [PubMed] [Google Scholar]
- 70.Bos JL, ras oncogenes in human cancer: a review. Cancer Res 1989, 49, 4682–4689. [PubMed] [Google Scholar]
- 71.Feig LA, Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol 2003, 13, 419–425. [DOI] [PubMed] [Google Scholar]
- 72.Thumkeo D; Watanabe S; Narumiya S, Physiological roles of Rho and Rho effectors in mammals. Eur J Cell Biol 2013, 92, 303–315. [DOI] [PubMed] [Google Scholar]
- 73.Maggio J; Gonzalez N; Cardama GA; Gomez DE, [Rho GTPases as molecular targets in cancer. Strategies and therapeutic opportunities]. Medicina (B Aires) 2017, 77, 497–504. [PubMed] [Google Scholar]
- 74.Cardama GA; Gonzalez N; Maggio J; Menna PL; Gomez DE, Rho GTPases as therapeutic targets in cancer (Review). Int J Oncol 2017, 51, 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


