Abstract
Antimicrobial resistance poses a threat in the treatment of infectious diseases in Bangladesh as well as in the world. Multidrug-resistant (MDR) Enterobacteriaceae, the most common cause of one such infectious disease, urinary tract infection (UTI), has contributed to the escalating problem of selecting empiric antibiotics against UTIs. The aim of this study was to investigate the presence of the efflux pump in MDR Escherichia coli isolates from UTI in the North-East region of Bangladesh, to isolate and characterize the AcrAB-TolC efflux pump genes of these locally isolated strains and to do mutation analysis of the efflux pump repressor AcrR gene to understand the AcrAB-TolC efflux pump mechanism. In the presence of omeprazole, an efflux pump inhibitor, every MDR E. coli isolate showed increased susceptibility to at least 1 of the 7 antibiotics investigated, indicating that efflux pump might be involved in their antibiotic resistance. Omeprazole decreased the minimum inhibitory concentration of every antibiotics being investigated by 2- to 8-fold. DNA and the deduced amino acid sequences of the polymerase chain reaction (PCR) products analyzed by bioinformatics tools revealed that the chromosomal AcrAB-TolC and AcrR genes were present in all MDR and antibiotic-susceptible E. coli isolates. However, the deduced amino acid sequences of the amplification refractory mutation system (ARMS) PCR product of the AcrR gene revealed that the substitution of arginine to cysteine at position 45 of AcrR was observed only in the MDR E. coli whose antibiotic susceptibility increased in the presence of omeprazole. Data reported herein support the notion that the increased antibiotic susceptibility of the MDR E. coli isolates in the presence of omeprazole might be due to efflux pump(s) inhibition and the AcrAB-TolC efflux pump might be a contributor to antibiotic resistance when the mutation of arginine to cysteine occurs at position 45 of AcrR.
Keywords: Multidrug resistant, MIC, UTI, efflux, AcrAB-TolC, mutation, AcrR
Introduction
Multidrug resistance of pathogenic microorganisms, for example, bacteria, is a global noteworthy concerning issue. Pathogens resistant to 3 or more antibiotics are called multidrug resistant (MDR).1,2 Infections by MDR pathogens are difficult to treat and have been associated with worse disease outcomes, and hence, impose a major threat.3,4 Recent studies suggest that antibiotic-resistant bacteria can be more virulent than their antimicrobial-susceptible counterparts.5 The antimicrobial resistance (AMR) in bacteria is emerging globally unprecedentedly and the resistant pattern differs based on geographical locations.6 There are a plethora of means by which humans have inadvertently accelerated the evolution of bacterial resistance. The over prescription of antibiotics by doctors for symptoms that in many cases may not be caused, unnecessary use of antimicrobials in agriculture and aquaculture and their dissemination in the environment are contributing to AMR worldwide. Recently, global warming and climate change are also supposed to contribute to AMR.7,8 Furthermore, in developing countries, such as Bangladesh, self-medication with antibiotics due to their easy availability in the pharmacy, selling pill-by-pill, misuse and abuse of antibiotics, and not fulfilling the prescribed antibiotic regimens once the patient feels better are the reasons of increasing AMR.2,9-11 Emerging AMR in developing countries is not necessarily regionally confined, as today’s globalized world allows for resistant bacteria as well as people to travel around the world.12 Therefore, molecular elucidation of multidrug resistance in pathogen of different geographical regions is required due to variance in antibiotic intake, agricultural use, and climate change in different areas of the world.
Susceptible bacteria can acquire resistance to an antimicrobial agent through mutation, degradation, or changing the structure of antibiotic, efflux pump, and conjugation and/or transformation of the plasmid.2,13,14 Efflux pump proteins, found in both Gram-positive and Gram-negative bacteria as well as in eukaryotic microorganisms,15 are transporter proteins involved in the extrusion of toxic substrates from within cells into the external environment. The impact of efflux mechanisms on AMR is large, and antibiotics can act as inducers and repressors of the expression of some efflux pump proteins.16 AcrAB-TolC, one of the efflux pumps, constitutively expressed in Escherichia coli, is composed of the outer membrane protein TolC, the inner membrane transporter AcrB, and the periplasmic adaptor protein AcrA.14,17 Overexpression of the AcrAB-TolC efflux pump is an intrinsic mechanism of multidrug resistance in Gram-negative bacteria.14,17,18 It can be due to the mutation in AcrR gene which is the repressor of the AcrAB operon system.19 Substrate profiles of the E. coli housekeeping efflux system AcrAB-TolC include chloramphenicol, fluoroquinolone, tetracycline, novobiocin, fusidic acid, nalidixic acid and β-lactam antibiotics, dyes, detergents, and most lipophilic antibiotics.16,20 The expression of AcrAB is regulated by the repressor AcrR, encoded by AcrR.19 It has been shown that a C133T transversion resulting in arginine to cysteine substitution at codon 45 of AcrR is responsible for overexpression of the AcrAB efflux system and results in AMR.21 Therefore, the antibiotic therapy can be made effective if (1) efflux pumps are inhibited, (2) the expression of efflux pumps is downregulated, or (3) the antibiotics are structurally redesigned, so that they are no longer suitable efflux substrates.22 One of the rational approaches toward confronting efflux of clinically relevant antibiotics is to discover or design potent efflux pump inhibitors. A number of known compounds have been reported as efflux pump inhibitors such as verapamil, biricodar, moxifloxacin, and omeprazole.14,19,23,24 Although several reports show the inhibitory effects of omeprazole on bacterial efflux pumps,23,25,26 very few studies have been done to show its inhibitory effects on efflux pumps of E. coli.
Urinary tract infection (UTI) is recognized as the second most common infection in community practice.27 Globally, about 150 million people are diagnosed with UTI each year, costing the global economy in excess of US$6 billion.28 Recently, it is reported that about 60% of UTIs are caused by E. coli and ~90% of these E. coli isolates are MDR in the North-East region of Bangladesh.2 In 2000, only 7.1% MDR E. coli were reported in patients of the United States,29 whereas in 2011, 77% MDR E. coli were reported in UTI patients in Iran,30 indicating rapid and threatening emergence of antibiotic resistance in E. coli causing UTI. Although about 90% of the E. coli isolates causing UTI are MDR,2 the molecular reasons of multidrug resistance in these MDR E. coli are not yet elucidated. In this study, the efflux pump system has been assayed using omeprazole and ethidium bromide (EtBr). Because the sequence of AcrAB-TolC and mutation of AcrR in the E. coli isolates may differ due to the reasons aforementioned and contribute to AMR,2,6-11 the AcrA, AcrB, TolC, and AcrR genes have been isolated and sequenced. The present report shows that the omeprazole increases the susceptibility of MDR E. coli isolates against almost all the antibiotics being investigated, AcrAB-TolC are constitutive genes, and there is a correlation between antibiotic susceptibility and the mutation in AcrR in MDR E. coli isolates causing UTI in the North-East region of Bangladesh.
Materials and Methods
E. coli isolates and their maintenance
The MDR E. coli isolates were obtained from urine samples of UTI patients in North-East region of Bangladesh.2 The pure culture of E. coli isolates was routinely subcultured on nutrient agar (NA; 0.5% peptone, 0.15% yeast extract, 0.15% beef extract, 0.5% NaCl, 1.5% agar; pH 7.0) and preserved at −20°C in nutrient broth (NB; 0.5% peptone, 0.15% yeast extract, 0.15% beef extract, 0.5% NaCl; pH 7.0) with 15% glycerol.
Antibiogram and efflux assay of antibiotics with proton pump inhibitor
The presence of efflux pumps in antibiotic-resistant E. coli isolates was screened by the increase in antibiotic susceptibility in the presence of omeprazole, an H+/K+ proton pump inhibitor.14,23,25,26,31 The antibiotic disks of amoxicillin (AMX), azithromycin (AZM), ciprofloxacin (CIP), cefixime (CFM), ceftriaxone (CTR), gentamicin (GEN), and sulfamethoxazole (SXT) were added to the surface of culture on Mueller Hinton Agar (MHA; beef extract 0.2%, acid hydrolysate of casein 1.75%, starch 0.15%, Agar 1.7%; pH 7.3) of each plate supplemented with or without 100 µg/mL omeprazole. The zone of inhibition produced by the antibiotic disks was compared with the standard clear zone diameters of these disks against Enterobacteriaceae stated in the guidelines of Clinical and Laboratory Standards Institute (CLSI).32
The minimum inhibitory concentration (MIC) of each of the antibiotics aforementioned was determined in the presence or absence of 100 µg/mL omeprazole by the broth dilution method as described previously.33
EtBr efflux assay of E. coli isolates
Efflux of EtBr was tested by the EtBr cartwheel procedure.34 Bacterial isolates were cultured overnight with NB (0.5% peptone, 0.15% yeast extract, 0.15% beef extract, 0.5% NaCl; pH 7.0) in a shaker and the cell concentration was adjusted to 0.5 of a McFarland standard35 in the following day. The NA plates were divided by radial lines, forming a cartwheel pattern. The bacterial cultures were then swabbed (using sterilized cotton swab) on NA plates containing 1.5 µg/mL EtBr in the presence or absence of 100 µg/mL omeprazole, and incubated at 37°C for 16 h. The culture on the NA plates was examined under a UV transilluminator.
Isolation and sequencing of AcrA, AcrB, and TolC genes
Chromosomal DNA was extracted from overnight culture of E. coli isolates in NB with Favorgen genomic DNA purification kit in accordance with the manufacturer’s instruction (Favorgen Biotech. Corp., Taiwan). AcrA, AcrB, and TolC genes were amplified by polymerase chain reaction (PCR) using gene-specific primers (Table 1). The PCR reaction mixture for each gene was prepared in a total volume of 25 µL consisting of 12.5 µL of master mixture (GoTaq® Green Master Mix; Promega, USA), 1.5 µL of 50 µM forward and reverse primers (IDT, Singapore), 2.5 µL of genomic DNA (⩽250 ng) and 7 µL of sterile deionized water. Polymerase chain reaction conditions for amplification of AcrA were programmed to 1 cycle at 94°C for 1 min; 35 cycles at 94°C for 30 s, at 55°C for 45 s, and at 72°C for 2 min; and 1 cycle at 72°C for 10 min. For amplification of AcrB and TolC, the PCR cycle was programmed to 1 cycle at 94°C for 1 min; 35 cycles at 94°C for 30 s, at 65°C for 45 s, and 72°C for 2 min; and 1 cycle at 72°C for 10 min. The PCR products were purified and sequenced as described previously.37 The sequence was analyzed by a free computer program BioEdit 7.2.5. The similarities of the sequences were searched by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) search program, and nucleotide sequences were submitted to DNA Data Bank of Japan (DDBJ).
Table 1.
Primers used for the detection of AcrAB-TolC efflux pump genes and ARMS PCR for AcrR.
| Primer | Sequence (5′→3′) | Reference |
|---|---|---|
| AcrB-FP | GAAAGGCCAACAGCTTAAC | 36 |
| AcrB-RP | GAGCTGGAGTCAGGATCAAC | |
| TolC-FP | TGCTCCCCATTCTTATCGGC | |
| TolC-RP | GCTCTTGCTTGGCGTTGTAC | |
| AcrA-FP | CAATTTGAAATCGGACACTCG | This study |
| AcrA-RP | GGCATGTCTTAACGGCTCCT | |
| AcrR FP | CGACCGCCAGAGGCGTAAACCC | |
| Inner FP | CGCTGGGCGAGATTGCAAAAGCAGCTGGCGTTTCGT | |
| AcrR RP | ACGAAGCGTGGGGCACAGGAGA |
Abbreviation: ARMS PCR, amplification refractory mutation system polymerase chain reaction.
Detection of mutation in AcrR by ARMS PCR
Tri primer amplification refractory mutation system polymerase chain reaction (ARMS PCR)38 was done by using 1 set of gene-specific primer and 1 mutation-specific primer (Table 1) for determining the mutation in AcrR gene. The amplification efficiency of each pair of primers: (1) AcrR FP and AcrR RP and (2) inner FP and AcrR RP was tested. The PCR reaction mixtures were prepared as aforementioned concentration. The PCR condition was programmed to 1 cycle at 94°C for 1 min; 35 cycles at 94°C for 30 s, at 62°C for 45 s and at 72°C for 1 min; 1 cycle at 72°C for 10 min. For ARMS PCR, 40 µL reaction mixture consisting of 20 µL of master mixture (GoTaq® Green Master Mix; Promega, USA), 1 µL of AcrR FP (1 µM), 1 µL of inner FP (1 µM) and 2 µL AcrR RP (2 µM; IDT, Singapore), 3.5 µL of genomic DNA (⩽250 ng), and 12.5 µL of sterile deionized water was prepared. The ARMS PCR was programmed to 1 cycle at 94°C for 2 min; 35 cycles at 94°C for 30 s, at 62°C for 30 s, and at 72°C for 1 min, and final extension at 72°C for 2 min.
The PCR amplicons were subjected to electrophoresis and visualized in 1% agarose. Polymerase chain reaction products were purified from agarose gel and sequenced as described previously.39 The sequences were analyzed as aforementioned.
Results and Discussion
About 90% of the E. coli isolates causing UTI in the North-East region of Bangladesh were reported as MDR, and the multidrug resistance in E. coli might be either plasmid or chromosomal DNA mediated or the both.2,13 In this study, the efflux pump system has been discussed as one of the reasons for multidrug resistance in the MDR E. coli isolates causing UTIs and omeprazole may be an efflux pump inhibitor.
Antibiotic susceptibility in the presence of omeprazole
The efflux systems play important role in antibiotic resistance in MDR bacteria,40 and omeprazole is reported as an efflux pump inhibitor.23,31 To investigate the presence of efflux pump, we randomly selected 18 MDR E. coli isolates causing UTI.2 Their susceptibility to AMX, AZM, CFM, CIP, CTR, GEN, and SXT was assayed in the presence and absence of omeprazole (Figure 1 and Table 2). In the presence of omeprazole, the diameter of zone of inhibition by at least 1 of the 7 antibiotics increased against all MDR E. coli isolates (Table 2). The result indicated that the omeprazole, an efflux pump inhibitor,23 increased the antibiotic susceptibility, supporting the notion of the presence of efflux pump in the MDR E. coli isolates. The percentage of the MDR E. coli isolates that showed efflux activity to AMX, AZM, CFM, CIP, CTR, and SXT was about 22, 44, 72, 66, 38, and 66, respectively. About 77% of the MDR E. coli isolates showed efflux activity against 3 or more antibiotics. This result further indicated that efflux activity might be one of the mechanisms of multidrug resistance.
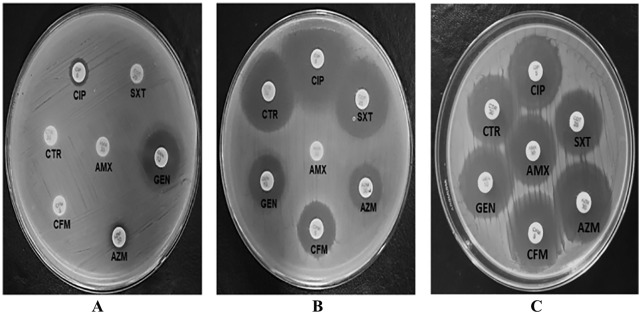
Figure 1.
Representative antibiogram of Escherichia coli isolate 15 in the absence (A) and presence (B) of omeprazole, and E. coli ATCC 25922 in the absence of omeprazole (C).
Table 2.
Increase in zone of inhibition of Escherichia coli isolates to different antibiotic disks in the presence of omeprazole.a
| Isolates | AMX |
AZM |
CFM |
CIP |
CTR |
GEN |
SXT |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | O | C | O | C | O | C | O | C | O | C | O | C | O | |
| 02 | NZ | NZ | NZ | NZ | NZ | NZ | NZ | NZ | NZ | NZ | 14 | 14 | 13 | 13 |
| 06 | NZ | NZ | 16 | 22 | 16 | 20 | 22 | 26 | 20 | 27 | 18 | 25 | 16 | 21 |
| 08 | NZ | NZ | 22 | 24 | 16 | 17 | 23 | 27 | 19 | 21 | 20 | 24 | 18 | 24 |
| 11 | NZ | NZ | 14 | 20 | NZ | 22 | NZ | 22 | NZ | 22 | 14 | 20 | 16 | 19 |
| 14 | NZ | 15 | 14 | 20 | NZ | 22 | NZ | 27 | NZ | 17 | 14 | 16 | 18 | 27 |
| 15 | NZ | NZ | 9 | 15 | NZ | 19 | 10 | 31 | NZ | 29 | 18 | 18 | NZ | 25 |
| 16 | NZ | NZ | 26 | 30 | 14 | 19 | 25 | 27 | 25 | 28 | 20 | 26 | 23 | 25 |
| 17 | NZ | NZ | 22 | 25 | NZ | NZ | NZ | 25 | NZ | NZ | 14 | 16 | 22 | 26 |
| 18 | NZ | 18 | NZ | 19 | NZ | NZ | NZ | NZ | NZ | NZ | NZ | 16 | 18 | 25 |
| 19 | NZ | NZ | 18 | 22 | NZ | NZ | 30 | 32 | 24 | 26 | 14 | 17 | 21 | 27 |
| 20 | NZ | NZ | 15 | 18 | 22 | 26 | 18 | 22 | 22 | 29 | 16 | 22 | 16 | 22 |
| 22 | NZ | NZ | 22 | 26 | 18 | 22 | NZ | 19 | 25 | 27 | 34 | 41 | 12 | 15 |
| 26 | NZ | NZ | 23 | 25 | 18 | 27 | 25 | 26 | 26 | 27 | 18 | 25 | 19 | 24 |
| 27 | NZ | NZ | 20 | 22 | NZ | NZ | NZ | 16 | NZ | NZ | 18 | 27 | NZ | NZ |
| 29 | NZ | NZ | 18 | 20 | NZ | 15 | NZ | 18 | NZ | NZ | 15 | 17 | NZ | 14 |
| 30 | NZ | 22 | 17 | 19 | NZ | 18 | NZ | 17 | 24 | 25 | 14 | 16 | 17 | 25 |
| 31 | NZ | NZ | 14 | 17 | 18 | 20 | 22 | 24 | 19 | 24 | 17 | 20 | 23 | 27 |
| 32 | NZ | 16 | 19 | 21 | NZ | 19 | NZ | NZ | NZ | 22 | 15 | 18 | 11 | 15 |
Abbreviations: AMX, amoxicillin; AZM, azithromycin; C, absence of omeprazole; CFM, cefixime; CIP, ciprofloxacin; CTR, ceftriaxone; GEN, gentamicin; NZ, no zone of inhibition; O, presence of omeprazole; SXT, sulfamethoxazole.
⩾4 mm increase in zone of inhibition in the presence of omeprazole compared with that in the absence of omeprazole was considered as the increase in antibiotic susceptibility.
The efflux pump inhibition by omeprazole was further investigated through determining the MIC of each antibiotic against the E. coli isolates as well as the E. coli ATCC 25922. Omeprazole treatment reduced the MIC of almost all antibiotics being investigated for the MDR E. coli isolates by 2-fold to 8-fold (Table 3). Minimum inhibitory concentration of the same antibiotic varied up to 2-fold based on different E. coli isolates. However, the MIC of the same antibiotic for the E. coli ATCC 25922 was at least 2-fold to 4-fold less than that found against the E. coli isolates treated with omeprazole. Nevertheless, the MIC of the E. coli ATCC did not further change in the presence of omeprazole. This result indicated the presence of active efflux pumps in the E. coli isolates but not in the E. coli ATCC 25922, which was susceptible to all the antibiotics being investigated (Figure 1C). In addition, this result suggests that omeprazole can act as an efflux pump inhibitor in agreement with other studies.23,25,26 Molecular docking shows that omeprazole has significant interaction to NorA, an efflux pump in Staphylococcus aureus with low binding energy.24 However, further study is necessary to clarify which efflux pump(s) in E. coli isolates is inhibited by omeprazole.
Table 3.
MIC of MDR Escherichia coli isolates in the absence and presence of omeprazole.a
| Antibiotics | MIC of antibiotics against E. coli isolates |
MIC reduced in the presence of omeprazole (fold) | MIC of antibiotics against E. coli ATCC 25922 (µg/µL) | |
|---|---|---|---|---|
| in the absence of omeprazole (µg/µL) | in the presence of omeprazole (µg/µL) | |||
| AMX | 4-8 | 2-4 | 2 | 0.5 |
| AZM | 4-8 | 1-2 | 4 | 0.5 |
| CFM | 2-4 | 0.25-0.5 | 8 | 0.125 |
| CIP | 4-8 | 0.5-1 | 8 | 0.5 |
| CTR | 2-4 | 0.5-1 | 4 | 0.5 |
| GEN | 1-2 | 0.125-0.25 | 8 | 0.0625 |
| SXT | 4-8 | 2-4 | 2 | 2 |
Abbreviations: AMX, amoxicillin; AZM, azithromycin; CFM, cefixime; CIP, ciprofloxacin; CTR, ceftriaxone; GEN, gentamicin; MIC, minimum inhibitory concentration; MDR, multidrug resistant; SXT, sulfamethoxazole.
Every experiment was repeated at least 3 times.
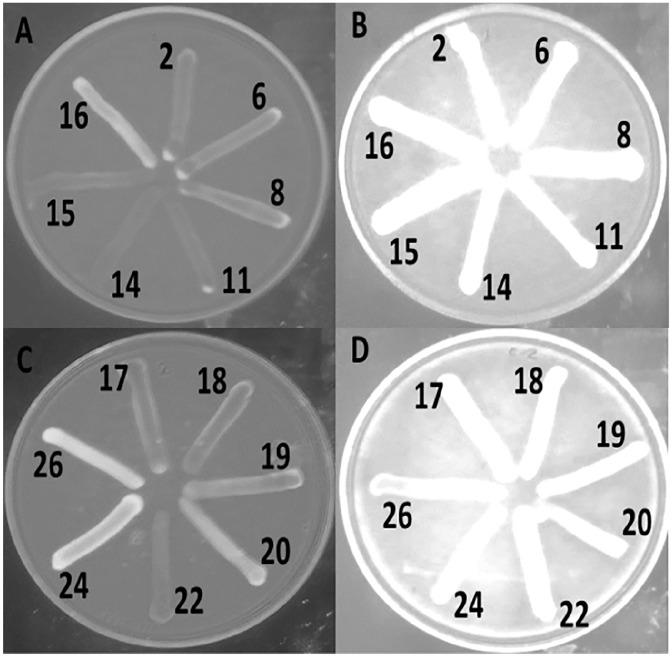
The presence of efflux pump was further tested by using EtBr. In the absence of omeprazole, many of the MDR E. coli isolates grown in the media containing EtBr failed to produce the characteristic intense orange fluorescence, indicating the possible efflux of EtBr from the cell (Figure 2A and C). However, when these strains were grown in the presence of omeprazole, strong fluorescence was observed indicating possible inhibition of the efflux process by omeprazole (Figure 2B and D). In the presence of omeprazole, EtBr could not come out from more than 70% of the bacterial isolates, and therefore, retained in the cell. Consequently, strong fluorescence was observed under a UV transilluminator and the antibiotic susceptibility increased (Table 4). This result supports the correlation between the efflux pump and the AMR of the E. coli isolates. The efflux system might be largely responsible for intrinsic resistance of some E. coli isolates to antibiotics.23 Therefore, antibiotics combined with efflux system blockers may increase the susceptibility of bacteria to a given antibiotic to a clinical relevant level.41,42 However, although a few bacterial isolates showed strong fluorescence under a UV transilluminator, they moderately increase susceptibility to antibiotics in the presence of omeprazole.
Figure 2.
Efflux of EtBr from some Escherichia coli isolates in the absence (A and C) and presence (B and D) of omeprazole.
EtBr indicates ethidium bromide.
Table 4.
Omeprazole-mediated efflux inhibition of EtBr and increase in antibiotic susceptibility.
| Escherichia coli isolates | Name of resistant antibiotics | Increase in antibiotic susceptibility in the presence of omeprazole | EtBr fluorescence in the presence of omeprazole |
|---|---|---|---|
| 02 | CTR, CIP, AMX, CFM, AZM, NA, F | No | Moderate |
| 06 | AMX, NA, F | Yes | Strong |
| 08 | AMX, NA, F | Yes | Strong |
| 11 | CTR, CIP, NA, AMX, CFM, F | Yes | Strong |
| 14 | CTR, CIP, AMX, CFM, NA, F | Yes | Strong |
| 15 | CTR, CIP, AMX, CFM, AZM, SXT, NA | Yes | Strong |
| 16 | AMX, CFM, NA | Moderate | Strong |
| 17 | CTR, CIP, CFM, NA, AMX | Yes | Strong |
| 18 | CTR, CIP, AMX, NA, CFM, GEN, AZM | Moderate | Strong |
| 19 | AMX, NA, CFM | Yes | Strong |
| 20 | AMX, NA | Yes | Strong |
| 22 | CIP, AMX, NA | Moderate | Strong |
| 26 | AMX, NA, F | Yes | Strong |
| 27 | CTR, CIP, NA, AMX, CFM, SXT | Moderate | Strong |
| 29 | CTR, CIP, AMX, NA, CFM, SXT, F | Moderate | Strong |
| 30 | CIP, AMX, CFM, NA, F | Yes | Strong |
| 31 | AMX, NA, F | Yes | Strong |
| 32 | CTR, CIP, AMX, CFM, NA | Moderate | Moderate |
Abbreviations: AMX, amoxicillin; AZM, azithromycin; CFM, cefixime; CIP, ciprofloxacin; CTR, ceftriaxone; EtBr, ethidium bromide; GEN, gentamicin; SXT, sulfamethoxazole.
Molecular detection and in silico characterization of AcrA, AcrB, and TolC genes in E. coli isolates
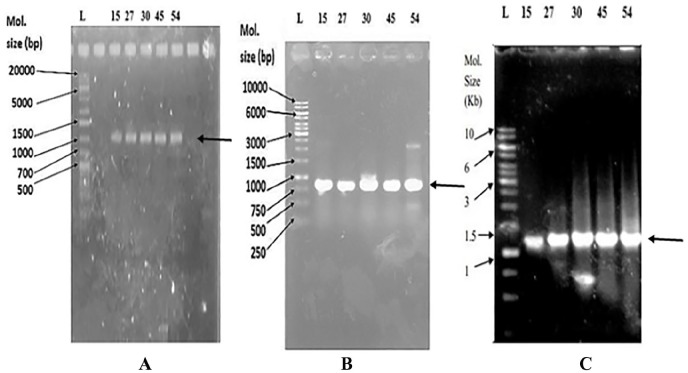
The association of the AcrAB-TolC efflux pump with antibiotic resistance is reported in bacteria.43 As more than 70% of the E. coli isolates have been assayed to have the efflux pump, an attempt was made to isolate and identify AcrA, AcrB, and TolC genes in their chromosomal DNA. In the presence of omeprazole, the MDR E. coli isolates that showed increased susceptibility to antibiotics and those that showed no increased susceptibility were selected for molecular detection of AcrA, AcrB, and TolC through PCR and DNA sequencing. The presence of AcrA, AcrB, and TolC genes with the size of 1194, 761, and 1170 bp, respectively, were found in both groups of E. coli isolates (Figure 3). The similarity search with the BLAST revealed that AcrA, AcrB, and TolC of E. coli isolates were 99%, 99%, and 95% identical, respectively, to those of E. coli. The nucleotide sequences of AcrA, AcrB, and TolC had been deposited to DDBJ under the accession number LC385717, LC385718, and LC271678, respectively. The amino acid sequences of AcrA, AcrB, and TolC were deduced by GENETYX-SV/RC version 7.0 program (Genetyx Corp., Japan) for further structural proteomics analysis. The physicochemical properties of these 3 deduced proteins investigated by ProtParam tools (https://web.expasy.org/protparam/) of the ExPASy server revealed that the theoretical pI of TolC is basic in nature and that of AcrA and AcrB is slightly acidic (Table 5). The extinction coefficient that is necessary during protein purification is different for AcrA, AcrB, and TolC. AcrAB proteins are much stable than TolC, and their aliphatic index indicated them as thermostable proteins (Table 5). The value of GRAVY (grand average of hydropathicity) calculated using the hydropathy value from Kyte and Doolittle44 is low for AcrA and TolC, indicating the possibility of their better interaction with water than that of AcrB. The pI and the hydropathy values are important for subcellular localization of channel forming membrane proteins. Recently, the in situ structure and assembly of the AcrAB-TolC efflux pump have been reported.17 Therefore, it will be interesting if molecular docking of omeprazole and other efflux pump inhibitors14,24 is done to predict the inhibitor profiles of the AcrAB-TolC efflux pump.
Figure 3.
PCR products of AcrA (A), AcrB (B), and TolC (C) genes in 1% agarose gel.
L indicates the DNA ladder; PCR, polymerase chain reaction. Escherichia coli isolate numbers are indicated above the image of the gel with PCR products shown by an arrow.
Table 5.
Physicochemical properties of AcrA, AcrB, and TolC proteins of the Escherichia coli isolates.
| Parameters | Values |
||
|---|---|---|---|
| AcrA (397 aa) | AcrB (253 aa) | TolC (389 aa) | |
| Molecular weight (Da) | 42,283.78 | 27,093.00 | 42,602.59 |
| Theoretical pI | 6.06 | 5.54 | 10.05 |
| Extinction coefficients (M−1 cm−1) | 0.447 | 0.275 | 1.205 |
| The estimated half-life (E. coli, in vivo) | >10 h | 3 min | >10 h |
| Instability index | 29.46 | 24.35 | 43.51 |
| Aliphatic index | 91.44 | 119.49 | 69.07 |
| Grand average of hydropathicity (GRAVY) | −0.266 | 0.690 | −0.587 |
Abbreviation: aa, amino acids.
The AcrB and TolC are localized on inner and outer parts of the membrane, respectively, and AcrA protein performs as an adapter of AcrB-TolC on the periplasmic region.45-47 Both AcrB and TolC comprise membrane domain as well as transmembrane region.48,49 The similar result was found in this study by computational analysis of AcrA, AcrB, and TolC with Pfam (https://pfam.xfam.org/) and TMpred (https://embnet.vital-it.ch/software/TMPRED_form.html). Three transmembrane helices were predicted in AcrB, 1 in TolC but not in AcrA (Table 6). Domain and transmembrane analyses indicate that AcrB is a member of integral membrane protein and cooperates with a fusion membrane protein AcrA and an outer membrane channel TolC. Two domains found by Pfam analysis may make the TolC unique as the outer membrane efflux protein through which antibiotic may be expelled.
Table 6.
Transmembrane regions of AcrA, AcrB, and TolC of the Escherichia coli isolates.
| Proteins | From | To | Length | Orientation |
|---|---|---|---|---|
| AcrA | − | − | − | − |
| AcrB | 119 | 137 | 19 | i-o |
| 145 | 163 | 19 | o-i | |
| 213 | 235 | 23 | i-o | |
| TolC | 114 | 138 | 25 | o-i |
Abbreviations: i-o, inner to outer; o-i, outer to inner.
Detection of mutation in AcrR
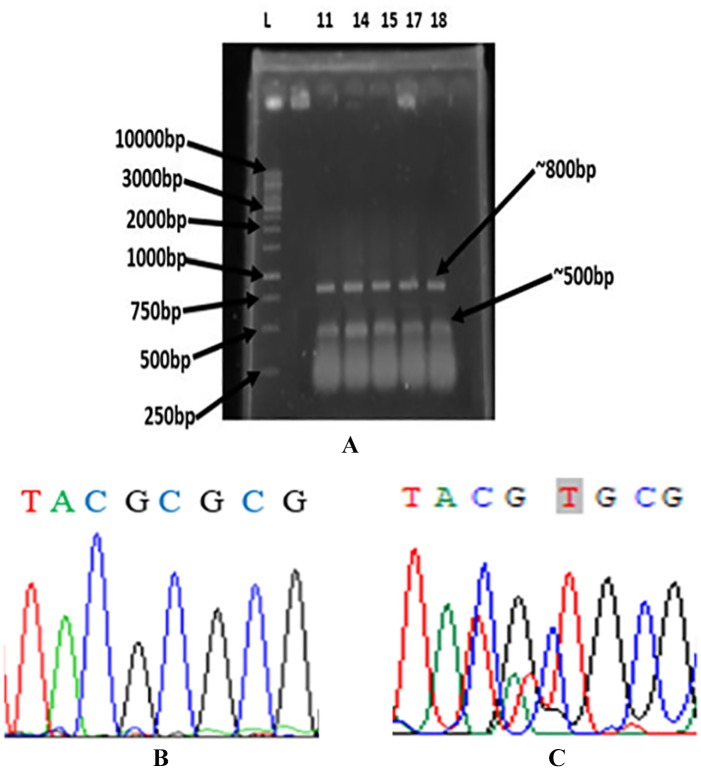
The ARMS-PCR assay used 3 primers in a single PCR reaction tube. Two outside primers designed to amplify a common fragment of 805 bp flanking the mutation site served as an internal amplification control. The inner primer was specific for the mutation at position 133 of the gene and the amplicon was 530-bp long (Figure 4A). The both amplicons were subjected to DNA sequencing and the result of sequencing confirmed the substitution of cytosine by thymine at position 133, which resulted a cysteine instead of an arginine at amino acid position 45 of AcrR (Figure 4B and C). The substitution of arginine to cysteine at position 45 of AcrR was observed only in the E. coli isolates that were assayed to have efflux pump (Figure 2, Tables 2 and 4). However, no substitution of arginine to cysteine at position 45 of AcrR was observed in the E. coli isolates showing no EtBr fluorescence and no increased susceptibility to antibiotics in the presence of omeprazole. Similarly, this mutation was not observed in AcrR of E. coli ATCC 25922 that was susceptible to all antibiotics investigated.2 These results suggested that the substitution of arginine to cysteine at position 45 of AcrR might be responsible for activation of the AcrAB-TolC efflux system.
Figure 4.
Agarose gel electrophoretogram and sequence output of ARMS PCR product for AcrR. All representative 5 isolates showed positive result for the presence of mutation (A). Normal AcrR has cytosine (B)19 and mutant AcrR sequence has thymine (C) at position 133.
L indicates the DNA ladder; ARMS PCR, amplification refractory mutation system polymerase chain reaction. Escherichia coli isolate numbers are indicated above the image of the gel with PCR products ~800 and 500 bp.
Conclusions
This study revealed that omeprazole increased the antibiotic susceptibility of the MDR E. coli isolates causing UTI in the North-East region of Bangladesh. AcrAB-TolC, one of the efflux pumps, has been partially characterized in the MDR E coli isolates, which might be important in future molecular docking study to predict and evaluate the inhibitor profiles of the AcrAB-TolC efflux pump. The efflux pump might be active only when substitution of arginine to cysteine occurs at position 45 of AcrR.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author contributions: NC and SS equally contributed to experiments, data analysis, and preparing the draft manuscript. AP performed experiments for TolC. MKB and TR performed experiments for screening efflux pump and antibiotic susceptibility. MJA and KI participated in manuscript revision. AKA conceived and designed the experiments, analyzed and interpreted the data, and wrote the paper. All the authors have approved the final article.
ORCID iDs: Nandan Chowdhury  https://orcid.org/0000-0003-0723-0071
https://orcid.org/0000-0003-0723-0071
Abul Kalam Azad  https://orcid.org/0000-0003-1918-3268
https://orcid.org/0000-0003-1918-3268
References
- 1. Uzodi AS, Lohse CM, Banerjee R. Risk factors for and outcomes of multidrug-resistant Escherichia coli infections in children. Infect Dis Ther. 2017;6:245-257. doi: 10.1007/s40121-017-0152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rabbee MF, Begum MK, Islam MJ, et al. Multidrug resistance phenotype and plasmid profiling of Escherichia coli isolates causing urinary tract infections in north east part of Bangladesh. Br Microbiol Res J. 2016;15:1-11. doi: 10.9734/bmrj/2016/27393. [DOI] [Google Scholar]
- 3. Martinez JL, Baquero F. Emergence and spread of antibiotic resistance: setting a parameter space. Ups J Med Sci. 2014;119:68-77. doi: 10.3109/03009734.2014.901444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. European Centre for Disease Control Prevention. Annual epidemiological report: antimicrobial resistance and healthcare-associated infections 2014. ECDC, Stock. 2015. https://ecdc.europa.eu/en/publications-data/antimicrobial-resistance-and-healthcare-associated-infections-annual. Accessed April 21, 2019.
- 5. Guillard T, Pons S, Roux D, Pier GB, Skurnik D. Antibiotic resistance and virulence: understanding the link and its consequences for prophylaxis and therapy. Bioessays. 2016;38:682-693. doi: 10.1002/bies.201500180. [DOI] [PubMed] [Google Scholar]
- 6. O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. The Review on Antimicrobial Resistance. Wellcometrust HM Gov. 2016. https://amr-review.org/sites/default/files/160518_Finalpaper_withcover.pdf. Accessed September 28, 2019.
- 7. Global warming could be fuelling the spread of drug resistant superbugs throughout Europe. Edinburgh Evening News. 2019. https://www.edinburghnews.scotsman.com/health/global-warming-could-be-fuelling-the-spread-of-drug-resistant-superbugs-throughout-europe-1-4907119. Accessed September 28, 2019.
- 8. Climate Change Could Worsen Antimicrobial Resistance Threat Scientists Predict. Newsweek 2019. https://www.newsweek.com/climate-change-could-worsen-antimicrobial-resistance-threat-scientists-1394346. Accessed September 28, 2019.
- 9. Planta MB. The role of poverty in antimicrobial resistance. J Am Board Fam Med. 2007;20:533-539. doi: 10.3122/jabfm.2007.06.070019. [DOI] [PubMed] [Google Scholar]
- 10. Wellington EM, Boxall AB, Cross P, et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis. 2013;13:155-165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 11. Blomberg B. Antibiotikaresistens i utviklingsland. Tidsskr Den Nor Legeforening. 2008;128:2462-2466. [PubMed] [Google Scholar]
- 12. Wright GD. Q&A: Antibiotic resistance: where does it come from and what can we do about it? BMC Biol. 2010;8:123. doi: 10.1186/1741-7007-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suhani S, Purkaystha A, Begum MK, Islam MJ, Azad AK. Plasmids for amoxicillin and ciprofloxacin resistance in Escherichia coli isolate causing urinary tract infection. Clin Microbiol Open Access. 2017;6:4. doi: 10.4172/2327-5073.1000284. [DOI] [Google Scholar]
- 14. Tegos GP, Haynes M, Strouse JJ, et al. Microbial efflux pump inhibition: tactics and strategies. Curr Pharm Des. 2011;17:1291-1302. doi: 10.2174/138161211795703726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Bambeke F, Balzi E, Tulkens PM. Antibiotic efflux pumps. Biochem Pharmacol. 2000;60:457-470. doi: 10.1016/S0006-2952(00)00291-4. [DOI] [PubMed] [Google Scholar]
- 16. Theuretzbacher U. Accelerating resistance, inadequate antibacterial drug pipelines and international responses. Int J Antimicrob Agents. 2012;39:295-299. doi: 10.1016/j.ijantimicag.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 17. Shi X, Chen M, Yu Z, et al. In situ structure and assembly of the multidrug efflux pump AcrAB-TolC. Nat Commun. 2019;10:2635. doi: 10.1038/s41467-019-10512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42-51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 19. Pourahmad Jaktaji R, Jazayeri N. Expression of acrA and acrB genes in Escherichia coli mutants with or without marR or acrR mutations. Iran J Basic Med Sci. 2013;16:1254-1258. doi:10.1016/j. ijantimicag.2011.12.006. [PMC free article] [PubMed] [Google Scholar]
- 20. Piddock LJV, White DG, Gensberg K, Pumbwe L, Griggs DJ. Evidence for an efflux pump mediating multiple antibiotic resistance in Salmonella enterica Serovar Typhimurium. Antimicrob Agents Chemother. 2000;44:3118-3121. doi: 10.1128/aac.44.11.3118-3121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Webber MA, Talukder A, Piddock LJ. Contribution of mutation at amino acid 45 of AcrR to acrB expression and ciprofloxacin resistance in clinical and veterinary Escherichia coli isolates. Antimicrob Agents Chemother. 2005;49:4390-4392. doi: 10.1128/AAC.49.10.4390-4392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kourtesi C, Ball AR, Huang YY, et al. Suppl 1: microbial efflux systems and inhibitors: approaches to drug discovery and the challenge of clinical implementation. Open Microbiol J. 2013;7:34-52. doi: 10.2174/1874285801307010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vidaillac C, Guillon J, Arpin C, et al. Synthesis of omeprazole analogues and evaluation of these as potential inhibitors of the multidrug efflux pump NorA of Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:831-838. doi: 10.1128/AAC.01306-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bhaskar BV, Babu TMC, Reddy NV, Rajendra W. Homology modeling, molecular dynamics, and virtual screening of NorA efflux pump inhibitors of Staphylococcus aureus. Drug Des Devel Ther. 2016;10:3237-3252. doi: 10.2147/DDDT.S113556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hashem RA, Yassin AS, Zedan HH, Amin MA. Fluoroquinolone resistant mechanisms in methicillin-resistant Staphylococcus aureus clinical isolates in Cairo, Egypt. J Infect Dev Ctries. 2013;7:796-803. doi: 10.3855/jidc.3105. [DOI] [PubMed] [Google Scholar]
- 26. Lo DY, Lee YJ, Wang JH, Kuo HC. Antimicrobial susceptibility and genetic characterisation of oxytetracycline-resistant Edwardsiella tarda isolated from diseased eels. Vet Rec. 2014;175:203. doi: 10.1136/vr.101580. [DOI] [PubMed] [Google Scholar]
- 27. Amin M, Mehdinejad M, Pourdangchi Z. Study of bacteria isolated from urinary tract infections and determination of their susceptibility to antibiotics. Jundishapur J Microbiol. 2009;2:118-123. [Google Scholar]
- 28. Gonzalez CM, Schaeffer AJ. Treatment of urinary tract infection: what’s old, what’s new, and what works. World J Urol. 1999;17:372-382. doi: 10.1007/s003450050163. [DOI] [PubMed] [Google Scholar]
- 29. Sahm DF, Thornsberry C, Mayfield DC, Jones ME, Karlowsky JA. Multidrug-resistant urinary tract isolates of Escherichia coli: prevalence and patient demographics in the United States in 2000. Antimicrob Agents Chemother. 2001;45:1402-1406. doi: 10.1128/AAC.45.5.1402-1406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Farshad S, Anvarinejad M, Tavana AM, et al. Molecular epidemiology of Escherichia coli strains isolated from children with community acquired urinary tract infections. African J Microbiol Res. 2011;5:4476-4483. doi: 10.5897/ajmr11.285. [DOI] [Google Scholar]
- 31. Mattsson JP, Vaananen K, Wallmark B, Lorentzon P. Omeprazole and bafilomycin, two proton pump inhibitors: differentiation of their effects on gastric, kidney and bone H+-translocating ATPases. Biochim Biophys Acta. 1991;1065:261-268. doi: 10.1016/0005-2736(91)90238-4. [DOI] [PubMed] [Google Scholar]
- 32. Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial DiskSusceptibility Tests; Approved Standard—Twelfth Edition 2015. https://clsi.org/media/1631/m02a12_sample.pdf.
- 33. Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163-175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 34. Martins M, Viveiros M, Couto I, et al. Identification of efflux pump-mediated multidrug-resistant bacteria by the ethidium bromide-agar cartwheel method. In Vivo. 2011;25:171-178. [PubMed] [Google Scholar]
- 35. McFarland J. The Nephelometer: an Instrument for Estimating the Number of Bacteria in Suspensions Used for Calculating the Opsonic Index and for Vaccines. JAMA J Am Med Assoc. 1907; XLIX:1176. doi: 10.1001/jama.1907.25320140022001f. [DOI] [Google Scholar]
- 36. Liu JH, Pan YS, Yuan L, Wu H, Hu GZ, Chen YX. Genetic variations in the active efflux pump genes acrA/B and tolC in different drug-induced strains of Escherichia coli CVCC 1547. Genet Mol Res. 2013;12:2829-2836. doi: 10.4238/2013.August.8.3. [DOI] [PubMed] [Google Scholar]
- 37. Iqbal A, Hakim A, Hossain MS, et al. Partial purification and characterization of serine protease produced through fermentation of organic municipal solid wastes by Serratia marcescens A3 and Pseudomonas putida A2. J Genet Eng Biotechnol. 2018;16:29-37. doi: 10.1016/j.jgeb.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramadan AR, Shawar SM, Alghamdi MA. Development and validation of a simple diagnostic method to detect gain and loss of function defects in fibroblast growth factor-23. Horm Res Paediatr. 2016;86:45-52. doi: 10.1159/000447113. [DOI] [PubMed] [Google Scholar]
- 39. Hakim A, Bhuiyan FR, Iqbal A, Emon TH, Ahmed J, Azad AK. Production and partial characterization of dehairing alkaline protease from Bacillus subtilis AKAL7 and Exiguobacterium indicum AKAL11 by using organic municipal solid wastes. Heliyon. 2018;4:e00646. doi: 10.1016/j.heliyon.2018.e00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Infect Control. 2006;34:s3-s10. doi: 10.1016/j.ajic.2006.05.219. [DOI] [PubMed] [Google Scholar]
- 41. Martins M, Dastidar SG, Fanning S, et al. Potential role of non-antibiotics (helper compounds) in the treatment of multidrug-resistant Gram-negative infections: mechanisms for their direct and indirect activities. Int J Antimicrob Agents. 2008;31:198-208. doi: 10.1016/j.ijantimicag.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 42. Bhardwaj AK, Mohanty P. Bacterial efflux pumps involved in multidrug resistance and their inhibitors: rejuvinating the antimicrobial chemotherapy. Recent Pat Antiinfect Drug Discov. 2012;7:73-89. doi: 10.2174/157489112799829710. [DOI] [PubMed] [Google Scholar]
- 43. Perez A, Poza M, Fernandez A, et al. Involvement of the AcrAB-TolC Efflux pump in the resistance, fitness, and virulence of Enterobacter cloacae. Antimicrob Agents Chemother. 2012;56:2084-2090. doi: 10.1128/AAC.05509-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105-132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 45. Elkins CA, Nikaido H. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of. J Bacteriol. 2002;184:6490-6498. doi: 10.1128/jb.184.23.6490-6499.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morona R, Manning PA, Reeves P. Identification and characterization of the TolC protein, an outer membrane protein from Escherichia coli. J Bacteriol. 1983;153:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ge Q, Yamada Y, Zgurskaya H. The C-terminal domain of AcrA is essential for the assembly and function of the multidrug efflux pump AcrAB-TolC. J Bacteriol. 2009;191:4365-4371. doi: 10.1128/JB.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Koronakis V, Li J, Koronakis E, Stauffer K. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol Microbiol. 1997;23:617-626. doi: 10.1046/j.1365-2958.1997.d01-1880.x. [DOI] [PubMed] [Google Scholar]
- 49. Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295-1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]