Abstract
Background
Plasma cells (PCs) are terminally differentiated B-lymphocytes producing antibodies. In coeliac disease (CeD) there is increased density of PCs in the small-intestinal lesion. Many of these PCs produce disease-specific autoantibodies targeting transglutaminase 2 (TG2).
Objective
The plasmacytosis of CeD motivated us to study the transcriptional programme of PCs from coeliac gut lesions.
Methods
RNA-seq was performed on the PCs of CeD patients and disease controls, being specific or non-specific for TG2.
Results
Being antibody-producing cells, 67% of the PCs’ transcript was aligned to immunoglobulin genes. Strikingly, genes encoding ligands and receptors of chemokines and cytokines were abundant. Higher transcript levels of genes associated with cell activation and immune responses were observed in PCs of CeD patients compared to controls. TG2-specific compared to non-TG2 specific PCs expressed increased levels of CXCR3, CXCL10 and interleukin-15; factors that have been implicated in the pathogenesis of CeD yet with production attributed to other cells than PCs. The presence of transcripts of HLA class II and T-cell co-stimulatory molecules suggests that PCs may serve as antigen-presenting cells for CD4 + helper T cells.
Conclusions
Our findings shed new light on the biology of intestinal PCs, implicating functions that go beyond the production of immunoglobulins.
Keywords: Plasma cells, coeliac disease, RNA-seq, gene ontology, plasmacytosis
Key summary
Current knowledge
Plasma cells (PCs) of immunoglobulin M (IgM) and immunoglobulin A (IgA) isotypes are found in mucosal tissues.
Patients with active coeliac disease (CeD) have increased numbers of PCs in the small intestine.
PCs in CeD produce disease-specific antibodies towards deamidated gluten peptides and the enzyme transglutaminase 2.
Key findings of this study
Mucosal PCs express genes of immune mediators and receptors.
Interleukin-15 as well as CXCR3 and CXCL10 that have a role in CeD are expressed by PCs. The production of these factors was attributed to cells others than PCs.
PCs may have a central role in immune function and regulation and hence, may be an attractive target for inflammatory intestinal disorders.
Introduction
Plasma cells (PCs) are highly specialised antibody-producing cells, mostly localised in special survival niches in the bone marrow and at mucosal tissues. In coeliac disease (CeD), a gluten-mediated enteropathy characterised by villous atrophy and malabsorption, and a two- to three-fold increased density of PCs in the lamina propria are the hallmarks of active disease.1,2 A substantial proportion of the PCs in CeD gut lesions is specific for the disease-associated antigens transglutaminase 2 (TG2) and deamidated gluten peptides.3,4 A strict avoidance of gluten from the diet normally results in the recovery of the villus structures and normalisation of PC density.
Little is known about the contribution of PCs to the generation of coeliac lesions and their immunological functions. Recent reports, however, have indicated that PCs exert functions that go beyond antibody production. Distinct functions were attributed to PCs based on their cytokine profile.5 Malignant plasma cells were demonstrated to produce cytokines and chemokines, and to activate cluster of differentiation (CD)4 + helper T cells.6 Expression of HLA-DR, HLA-DQ, and the co-stimulatory molecules CD40, CD80 and CD86 by PCs were also reported.6-10 Such functions would be particularly notable in conditions where PCs are prevalent, for example in CeD, where production of inflammatory mediators may have significant effects due to their vast numbers.
To understand whether and how PCs may contribute to intestinal inflammation, we characterised the transcriptional profiles of the small-intestinal PCs of disease controls and patients with active CeD. We demonstrate that gut PCs express transcripts of genes that encode for inflammatory mediators and receptors, co-stimulatory molecules and HLA class II. Our findings suggest that PCs have the ability to respond to external stimuli via their surface receptors and thus interact with other immune cells, in particular, CD4 + T cells.
Methods
Study material
Seven CeD patients with active disease and four disease controls were enrolled in this study (Table 1). All subjects visited the gastroenterology unit for clinical assessment. CeD was diagnosed according to the guidelines of the British Society of Gastroenterology.11 The disease control subjects were examined due to irritable bowel syndrome and histological assessment of routine biopsies revealed no signs of inflammation. Single cell suspensions were prepared from small-intestinal biopsies as described.12
Table 1.
Information concerning the study participants, number of sorted PCs and RNA concentration and integrity.
| Subject | Age | Gender | %PCs (from total) | %PCs (from parent cells) | % IgA PCs | % TG2-PCs | DGP-PCs | Number of sorted non-TG2-PCs | Number of sorted TG2-PCs | RNA concentration non-TG2-PCs | RNA concentration TG2-PCs | RIN non-TG2-PCs | RIN TG2-PCs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD1283 | 48 | Female | 1.94% | 30.5% | 36.7% | 6.73% | 0.56% | 96,000 | 5000 | 4374 pg/µl | 178 pg/µl | 7.6 | 7.7 |
| CD1287 | 26 | Female | 1.45% | 34.8% | 58.7% | 7.90% | 0.52% | 29,000 | 5000 | 1076 pg/µl | 71 pg/µl | 7.4 | 8.6 |
| CD1291 | 50 | Female | 2.84% | 39.9% | 55.6% | 7.77% | 0.35% | 80,000 | 10,000 | 4519 pg/µl | 254 pg/µl | 8.5 | 9.5 |
| CD1292 | 58 | Female | 1.92% | 26.1% | 65.9% | 2.17% | 0.058% | 60,000 | 2900 | 3901 pg/µl | 71 pg/µl | 8.5 | 8.6 |
| CD1516 | 39 | Female | 6.09% | 54.5% | 67.6% | 4.21% | 1.02% | 48,000 | 3500 | 280 pg/µl | 211 pg/µl | 8.8 | 9.8 |
| CD1518 | 44 | Female | 2.86% | 23.1% | 72.1% | 5.39% | 0.41% | 25,000 | 2900 | 177 pg/µl | 132 pg/µl | 9.3 | 9.1 |
| CD1521 | 44 | Female | 1.24% | 11.4% | 63.5% | 8.28% | 0.44% | 15,000 | 2300 | 166 pg/µl | 240 pg/µl | 10 | 9 |
| CD1492 | 43 | Male | 1.57% | 33.7% | 75.2% | 0.056% | 0.039% | 30,000 | – | 609 pg/µl | – | 9.8 | – |
| CD1493 | 59 | Male | 0.93% | 26.4% | 68.8% | 0.009% | 0.028% | 24,500 | – | 722 pg/µl | – | 9.9 | – |
| CD1494 | 23 | Female | 0.94% | 22.7% | 41.8% | 0.14% | 0% | 16,500 | – | 645 pg/µl | – | 9.9 | – |
| CD1495 | 53 | Female | 2.12% | 39.4% | 44.0% | 0.012% | 0.012% | 42,000 | – | 588 pg/µl | – | 9.6 | – |
CD1492, CD1493, CD1494 and CD1495 are disease controls (DC); ‘–’ = no TG2-specific PCs were found in these individuals. PCs: plasma cells; TG2: transglutaminase 2; IgA: immunoglobulin A; DGP: deamidated gluten peptides; RIN: RNA integrity number.
All other methodological procedures are available as supplementary information.
Results
RNA-seq libraries
RNA-seq was performed on sorted intestinal IgA PCs from disease controls (hereafter referred to as DC-PCs), and the PCs of CeD patients, being specific or not specific for the autoantigen TG2 (hereafter TG2-PCs and non-TG2-PCs, respectively). The library sizes ranged from 36–67 million read pairs per sample. Around 78–82% of the total reads were mapped to the known transcripts annotated in GENCODE GRCh38.p7. In line with previous observations13 61–67% of the mappable reads were mapped to immunoglobulin (Ig) genes. Following filtration of Ig-mapped reads, the libraries varied from 12–25 million reads. Based on a conservative definition,14 around 45% of the total known protein-coding genes (>10,000 genes) were considered to be ‘active’ in each of the Ig-filtered libraries (Supplementary Table 1). To examine the purity of sorted PCs we searched for genes that were specific for T cells, monocyte, dendritic cells, eosinophils and stromal cells; all were absent from the data (Figure 1).
Figure 1.
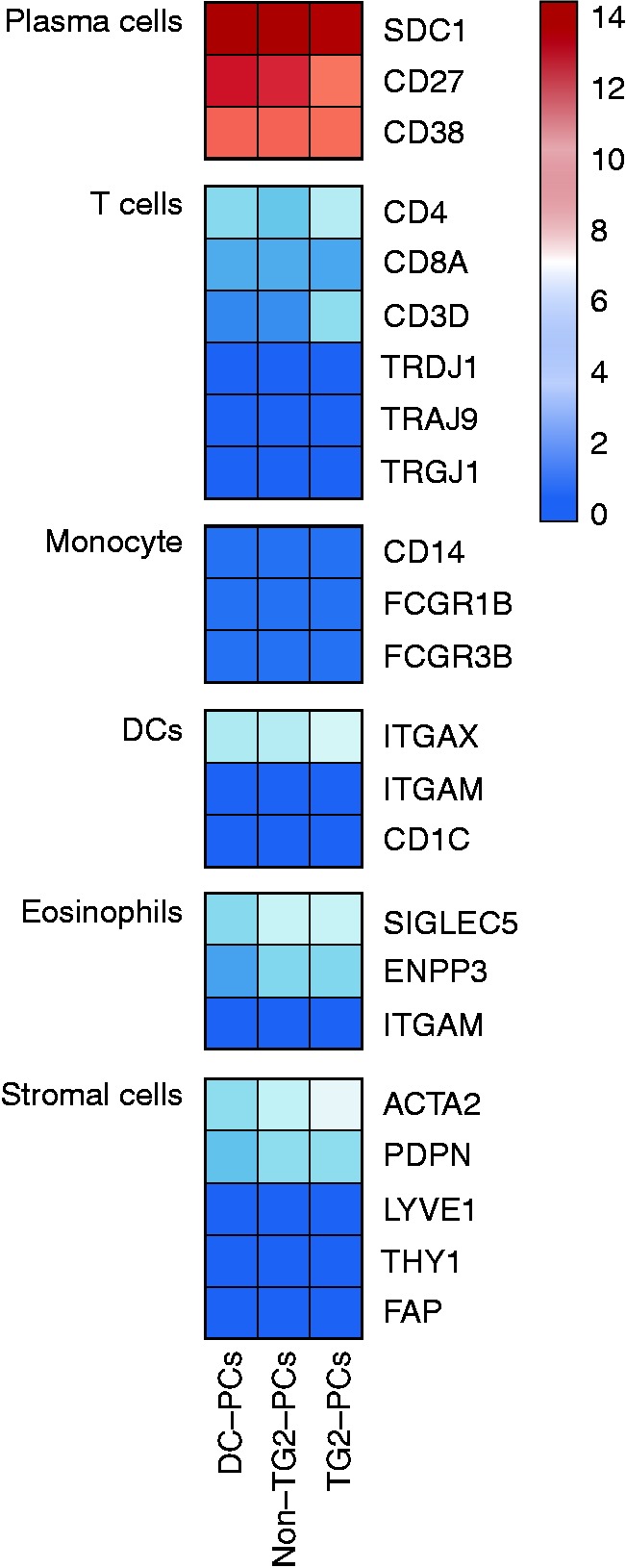
Purity test for PC libraries.
PC libraries were tested for expression of possible contaminating cells. The expression levels of cellular markers of T cells, monocytes, dendritic cells, eosinophils and stromal cells were tested and further compared with the PC expressed gene. The colour scale shows the normalised expression counts on a log2 scale (regularised logarithmic values reported by DESeq2).
Transcriptional profiles of intestinal IgA PCs
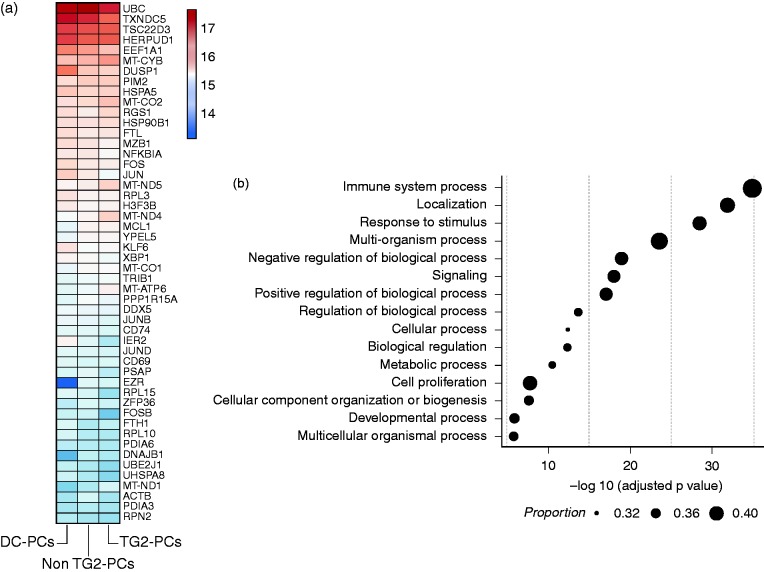
To survey the transcriptional programme of intestinal IgA PCs (other than Ig-encoding transcripts), we looked into the biological functions ascribed to the most abundantly expressed genes (n = 3000) in (i) DC-PCs, (ii) TG2-PCs and (iii) non-TG2-PCs. Notably, each of the PC groups expressed the same set of top 3000 genes, including known PC transcripts such as Blimp-1, syndecan-1, XBP1, IRF4, BCMA, TACI, CD27 and CD38. B-cell specific genes that are repressed in PCs such as CD20, PAX5 and BACH2 were not observed. The 50 most abundantly expressed genes across the three groups are depicted in Figure 2a and Supplementary Figure 1.
Figure 2.
Transcript profile of intestinal IgA PCs following exclusion of immunoglobulin transcripts.
(a) The top 50 expressed genes (mean geometric rank) in small-intestinal IgA PCs of disease controls (DC-PCs) and CeD patients that are either non-specific or specific for TG2 (non-TG2-PCs and TG2-PCs, respectively). The colour scale shows the normalised expression counts on a log2 scale (regularised logarithmic values reported by DESeq2); (b) enriched gene ontology (GO) categories within the top 3000 genes in PCs. On the x-axis, the negative logarithm of the adjusted p-value of the enriched category is shown. The size of the points represent the proportion of genes in the category (in the database) overlapping with the list of the top 3000 abundant genes.
Gene ontology (GO) categorisation of the top 3000 most highly expressed genes revealed that the core transcriptional programme of intestinal IgA PCs extends beyond antibody production and secretion. Specifically, genes related to immunological pathways, stimulus response, cellular processes, signalling, and cell proliferation were enriched (Figure 2b). Several GO terms related to the protein machinery (i.e. antibody production) of PCs were also enriched. Functions related to RNA and protein anabolism/catabolism, ribosome biogenesis, protein folding, biosynthesis of macromolecule and protein localisation were prominent.
Dissection of PC transcripts encoding for immunological mediators and receptors
The GO analysis demonstrated that immunological pathways are in the core of the transcriptional programmes of gut IgA PCs. Hence, the transcription patterns of genes related to (i) cytokines and cytokine receptors, (ii) chemokines and their receptors, (iii) antigen presentation and (iv) co-stimulatory molecules were closely examined.
(i) Cytokines and cytokine receptors: IL-16, IL-15 and TGFB1 were highly abundant, whereas IL-6 and IL-32 were expressed at lower levels (Figure 3a). Four TNF superfamily genes were expressed: TNFSF9 and TNFSF10 that induce T-cell activation and apoptosis, respectively; TNFSF13 (APRIL) and TNFSF13B (BAFF) that are essential for PCs development. IL-12A and IL-23A were also expressed; however, the transcript for their common p40 subunit was not detected.
Figure 3.
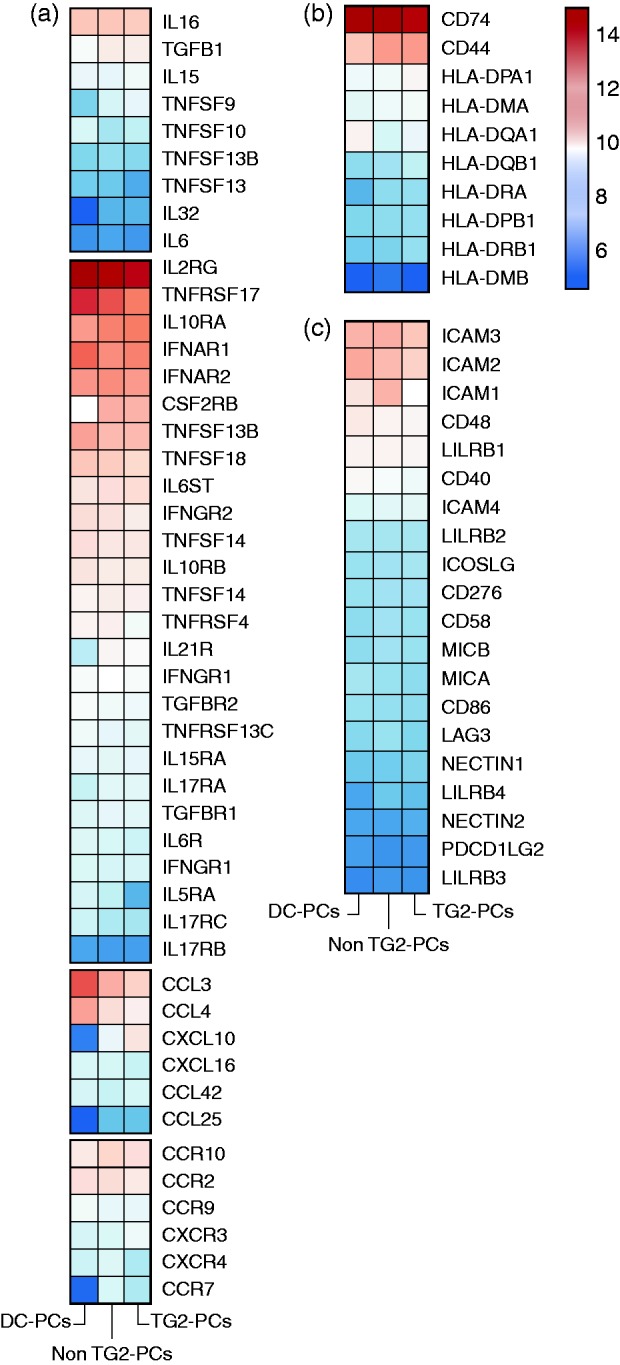
Immune-related mediators and receptor genes that are expressed in PCs.
The expression profile of the following immune-related genes in IgA PCs of disease controls and CeD patients: (a) cytokines, cytokine receptors as well as chemokines and chemokines receptors; (b) HLA class II genes and their associated and (c) co-stimulatory molecules. The colour scale shows the normalised expression counts on a log2 scale (regularised logarithmic values reported by DESeq2).
All subunits to form functional cytokine receptors of IL-5, IL-6, IL-10, IL-17A, IL-17F, IL-21 and IL-27 were expressed (Figure 3a), as well as the paired receptor genes for IFNα (IFNAR1/2), IFNγ (IFNGR1/2), IFNλ (IFNLR1 and IL10RB) and TGFBR1/R2. TACI, BAFF-R and BCMA (i.e. TNFRSF13B, TNFRSF13C and TNFRSF17, respectively) known to be essential for PCs differentiation and survival were abundant.
(ii) Chemokines and chemokine receptors: Chemoattractant genes for T cells, monocytes and macrophages such as CCL3, CCL4, CCL4L2, CXCL10 and CXCL16 were found. mRNA for chemokine receptors responding in an inflammatory environment such as CCR2, CCR10 and CXCR3 were also detected (Figure 3a).
(iii) Antigen presentation: As expected, transcripts for HLA class I molecules were abundantly expressed. Remarkably, genes encoded to HLA class II molecules were also found consistently across all PCs samples (Figure 3b). The MHC class II antigen-associated invariant chain (CD74) was among the top 50 expressed genes.
(iv) Co-stimulatory molecules: Genes that aid cross-talk with T cells were expressed. Of particular note were TNFSF9 (4-1BBL) that interacts with 4-1BB; ICOSL, CD86; CD48 and CD58 that interact with CD2; CD40 that interacts with CD40L; and ICAM1/2/3/4 that interact with LFA1 on T cells. MIC-A and MIC-B that react with NKGTD and NKGTA that are expressed on NK cells, intraepithelial T cells and γδT cells were also detected (Figure 3c).
Supplementary Figure 2 depicts the individual variation of immunological mediators and receptors expressed by PCs.
PCs of CeD patients exhibit an increased inflammatory profile
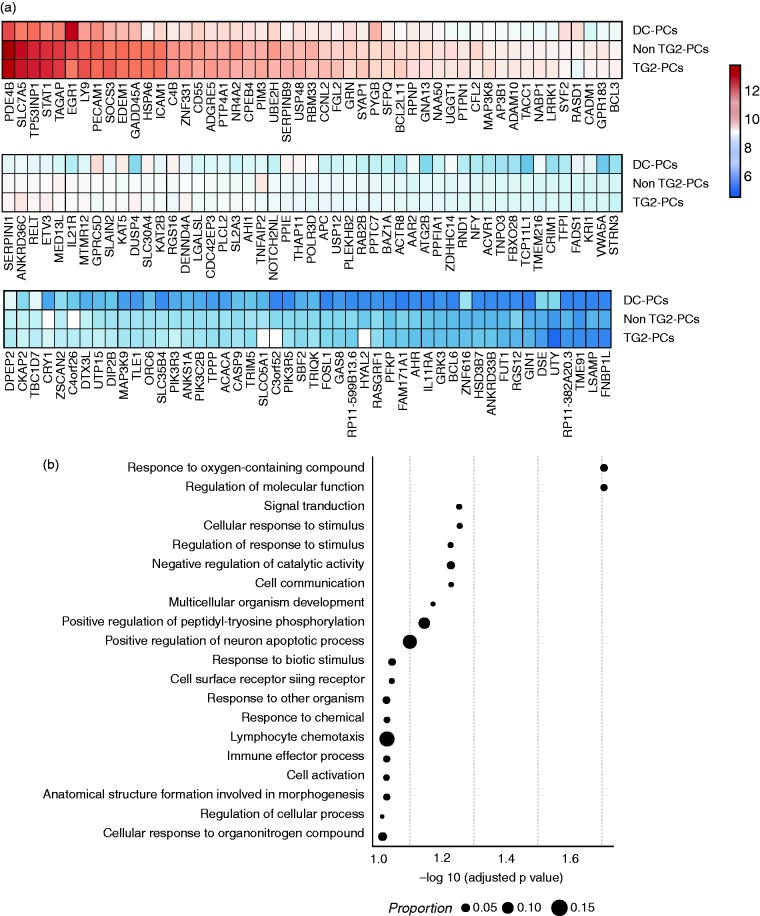
Next, we compared the transcriptional profiles (all expressed genes) of PCs of the disease control and CeD patients. Differential expression (DE) analysis was performed for DC-PCs vs. non-TG2-PCs and DC-PCs vs. TG2-PCs. A hundred and forty genes had increased activity in both non-TG2-PCs and TG2-PCs in comparison with DC-PCs (Figure 4a, Supplementary Figure 3 and Supplementary Table 2). GO categorisation of these 141 DE genes implied that PCs from patients were at higher activation levels relative to DC-PCs (Figure 4b). Such genes are known to participate in cellular processes such as (i) response to stimulus, (ii) cell activation, (iii) cell surface receptor signalling and others. Of particular note are genes that were grouped under ‘immune effector process’ and ‘lymphocyte chemotaxis’ (such as ADAM10, GPR183, CXCL10, and HSD3B7), which play a pivotal role in recruiting immune cells to an inflammatory site.
Figure 4.
Differentially expressed genes in CD vs. disease controls.
(a) The expression profile of the differentially expressed genes (n = 141 based on fold difference) in PCs from CeD patients (non-TG2- and TG2-PCs) and disease controls. The colour scale shows the normalised expression counts on a log2 scale (regularised logarithmic values reported by DESeq2); (b) a functional GO analysis was performed using the differentially expressed genes that were upregulated in both non-TG2- and TG2-PCs in comparison to DC-PCs. On the x-axis, the negative logarithm of the adjusted p-value of the enriched GO term is shown. The size of the points represents the proportion of genes in the term.
TG2-PCs exhibit increased expression of genes related to T-cell activation and chemotaxis
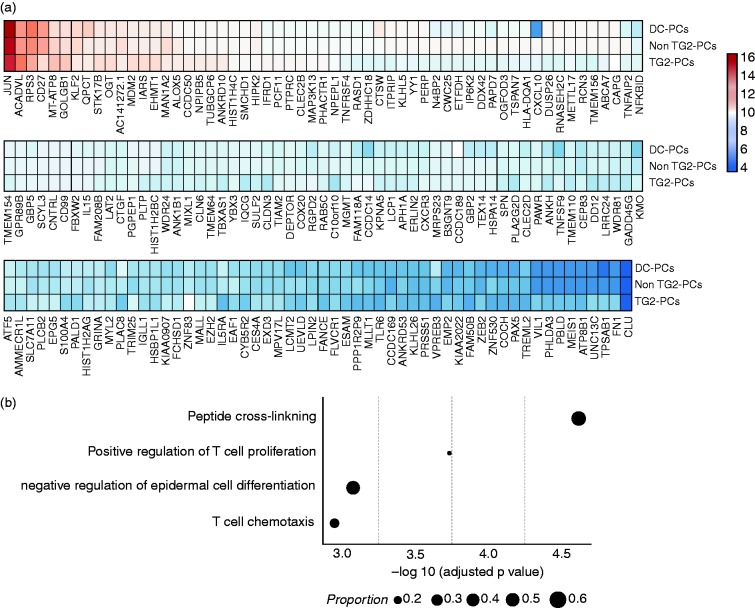
A hundred and fifty-one DE genes with increased expression were found in TG2-PCs in comparison with the non-TG2-PCs of CeD patients (Figure 5a, Supplementary Figure 4 and Supplementary Table 3). GO categorisation of those genes highlights a potential cross-talk between TG2-PCs and T cells (Figure 5b). Of particular interest are genes grouped under the terms ‘positive regulation of T-cell proliferation’ (ANXA1, IL15, TNFSF9) and ‘T-cell chemotaxis’ (CXCL10 and CXCR3).
Figure 5.
Differentially expressed genes in TG2-PCs vs. non-TG2-PCs.
(a) The expression profile of the top differentially expressed genes (n = 151) in TG2-PCs in comparison with non-TG2-PCs. The colour scale shows the normalised expression counts on a log2 scale (regularised logarithmic values reported by DESeq2); (b) a functional GO analysis was performed using the upregulated genes in TG2-PCs. On the x-axis, the negative logarithm of the adjusted p-value of the enriched GO term is shown. The size of the points represents the proportion of genes in the term.
Discussion
In this work, we analysed the transcriptomic profile of intestinal IgA PCs. Being highly specialised antibody-producing cells, PCs are not considered to be involved in immunoregulatory circuits. Our findings challenge this notion. We found expression of genes that play crucial roles in various immune system processes, thus suggesting that PCs have the ability to respond to stimuli and to interact with their surrounding environment. Interestingly, in the inflamed gut, the PCs showed higher transcript levels of inflammatory mediators and receptors.
The expression of gene encoding for cytokines, chemokines and their receptors by PCs suggests a cross-talk with other immune cells, primarily T cells. Of particular interest are the expressions of IL-15 and IL-16; the latter is the ligand for CD4 (and CD9). It attracts and activates CD4 + T cells,15-17 upregulates CD25 and CD122 and thus primes and facilitates their responsiveness to IL-2 and IL-15.18 Elevated levels of IL-15 have been reported in CeD19,20 as well as in inflammatory bowel disease.21 This cytokine supports the survival and proliferation of cytotoxic intraepithelial lymphocytes (IELs), which can directly damage epithelial cells and lead to tissue remodelling in CeD.22 IL-15 is therefore thought to have a central role in the pathogenesis of CeD, with epithelial cells and lamina propria dendritic cells being its main sources.23,24 Here, we demonstrated that intestinal PCs express high levels of IL-15 and IL-15RA transcripts. IL-15 functions chiefly via cell-to-cell contact (i.e. trans presentation) by which the membrane bound IL-15:IL-15Rα complex is presented to the interacting cell. Co-expression of IL-15 and IL-15RA suggests that PCs have the ability to activate CD4 + T cells, IELs and CTLs; all T-cell subsets being involved in gut inflammation and shaping of the coeliac lesion.
CXCL9, CXCL10 and CXCL11 being ligands for CXCR3 attract cells to inflammatory sites. The expression of CXCL10 and CXCL11 has been shown to increase in duodenal biopsies of untreated CeD patients, and PCs and enterocytes were identified as CXCL10-producing cells.25 Our data support these findings and further demonstrate that autoreactive TG2-PCs express high levels of CXCL10 and CXCR3. Thus TG2-PCs and also non-TG2-PCs in CeD directly contribute to the recruitment of CXCR3-expressing cells such as Th1 cells, IELs, γδT cells, monocytes, PCs, and other immune cells that take part in ongoing gut inflammation. IL-15, IFNα, and IFNγ that are found at increased levels in the coeliac lesion, are known to induce the production of CXCL10.26 Both IFN receptors are expressed by PCs; it is thus conceivable that the elevated levels of CXCL10 expressed by PCs are due to the effect of these cytokines. Additionally, PCs express CCL3, CCL4, CCL4L2, CCL25 and CXCL16. Taken as a whole, the vast expression of chemokine genes suggests that PCs contribute to the recruitment of immune cells to gut inflammatory sites.
The expression of HLA class II molecules is thought to be silenced during PC differentiation. Several studies, however, reported various levels of surface expression of HLA class II molecules as well as co-stimulatory molecules such as CD86 on PCs.9,10 We have shown that PCs express HLA class II transcripts as well as HLA-DM and CD74. In line with Ellyard et al.,9 we found CD86 (but not CD80) to be expressed, thus suggesting that intestinal PCs can function as competent antigen-presenting cells (APCs) to CD4 + T cells. Intestinal IgM and IgA PCs maintain expression of Ig surface receptors.3,27 Being antigen-specific, PCs, via their B cell receptors (BCRs), can internalise antigen and thereafter load HLA class II molecules with peptides from the BCR-targeted antigen. Even though HLA class II expression on PCs may be low, the numbers of a specific peptide HLA II complex, might be sufficient to stimulate cognate activation of T cells. Intriguingly, by employing a monoclonal antibody specific for an HLA-DQ:gluten-peptide complex (i.e. HLA-DQ2.5:DQ2.5-glia-α1a), PCs were shown to be the main cells presenting a gluten epitope in untreated CeD patients.28 Functional testing of gut PCs as APCs for CD4 + T cells in general, and gluten-specific CeD CD4 + T cells in particular, should thus be a priority in future studies.
Here, we have shown that PCs express genes implicated in cellular and immunological functions that expand beyond their role as antibody-secreting cells. PCs have the potential to produce inflammatory cytokines and chemokines and respond to environmental cues via designated receptors. They may have an intimate cross-talk with CD4 + T cells that include chemotaxis, activation, and antigen presentation. Additional studies are needed to understand the function of PCs and their role in gut inflammation.
Supplemental Material
Supplemental material, Supplemental Material2 for Transcriptional profiling of human intestinal plasma cells reveals effector functions beyond antibody production by Omri Snir, Chakravarthi Kanduri, Knut E A Lundin, Geir Kjetil Sandve and Ludvig M Sollid in United European Gastroenterology Journal
Supplemental Material
Supplemental material, Supplemental Material1 for Transcriptional profiling of human intestinal plasma cells reveals effector functions beyond antibody production by Omri Snir, Chakravarthi Kanduri, Knut E A Lundin, Geir Kjetil Sandve and Ludvig M Sollid in United European Gastroenterology Journal
Acknowledgements
We thank the endoscopy unit at the Department of Gastroenterology at Oslo University Hospital-Rikshospitalet for samples collection; the study participants; Marie K. Johannesen and Carina Hinrichs for technical assistance; and Eivind Gard Lund for critical discussions.
Conflict of interest
The authors have declared that no conflict of interest exists.
Funding
This work was supported by grants from the Research Council of Norway through its Centres of Excellence funding scheme (project number 179573/V40), and Stiftelsen KG Jebsen (SKGJ MED-017).
Informed consent
Written informed consent was obtained from each patient.
Ethics approval
The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki, and was approved by the Regional Ethics Committee of South-Eastern Norway (2010/2720).
References
- 1.Baklien K, Brandtzaeg P, Fausa O. Immunoglobulins in jejunal mucosa and serum from patients with adult coeliac disease. Scand J Gastroenterol 1977; 12: 149–159. [PubMed] [Google Scholar]
- 2.Scott H, Ek J, Baklien K, et al. Immunoglobulin-producing cells in jejunal mucosa of children with coeliac disease on a gluten-free diet and after gluten challenge. Scand J Gastroenterol 1980; 15: 81–88. [DOI] [PubMed] [Google Scholar]
- 3.Di Niro R, Mesin L, Zheng NY, et al. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nat Med 2012; 18: 441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinsbo O, Henry Dunand CJ, Huang M, et al. Restricted VH/VL usage and limited mutations in gluten-specific IgA of coeliac disease lesion plasma cells. Nat Commun 2014; 5: 4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fillatreau S. Regulatory plasma cells. Curr Opin Pharmacol 2015; 23: 1–5. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, Floisand Y, Myklebust CV, et al. Autologous bone marrow Th cells can support multiple myeloma cell proliferation in vitro and in xenografted mice. Leukemia 2017; 31: 2114–2121. [DOI] [PubMed] [Google Scholar]
- 7.Yi Q, Dabadghao S, Osterborg A, et al. Myeloma bone marrow plasma cells: evidence for their capacity as antigen-presenting cells. Blood 1997; 90: 1960–1967. [PubMed] [Google Scholar]
- 8.Walz S, Stickel JS, Kowalewski DJ, et al. The antigenic landscape of multiple myeloma: mass spectrometry (re)defines targets for T-cell-based immunotherapy. Blood 2015; 126: 1203–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellyard JI, Avery DT, Phan TG, et al. Antigen-selected, immunoglobulin-secreting cells persist in human spleen and bone marrow. Blood 2004; 103: 3805–3812. [DOI] [PubMed] [Google Scholar]
- 10.Pelletier N, McHeyzer-Williams LJ, Wong KA, et al. Plasma cells negatively regulate the follicular helper T cell program. Nat Immunol 2010; 11: 1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludvigsson JF, Bai JC, Biagi F, et al. Diagnosis and management of adult coeliac disease: guidelines from the British Society of Gastroenterology. Gut 2014; 63: 1210–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snir O, Mesin L, Gidoni M, et al. Analysis of celiac disease autoreactive gut plasma cells and their corresponding memory compartment in peripheral blood using high-throughput sequencing. J Immunol 2015; 194: 5703–5712. [DOI] [PubMed] [Google Scholar]
- 13.Shi W, Liao Y, Willis SN, et al. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat Immunol 2015; 16: 663–673. [DOI] [PubMed] [Google Scholar]
- 14.Hebenstreit D, Fang M, Gu M, et al. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol 2011; 7: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi JC, Wang J, Mandadi S, et al. Human and mouse mast cells use the tetraspanin CD9 as an alternate interleukin-16 receptor. Blood 2006; 107: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Center DM, Kornfeld H and Cruikshank WW. Interleukin 16 and its function as a CD4 ligand. Immunol Today 1996; 17: 476–481. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Cruikshank WW, O'Loughlin T, et al. Identification of a CD4 domain required for interleukin-16 binding and lymphocyte activation. J Biol Chem 1999; 274: 23,387–23,395. [DOI] [PubMed] [Google Scholar]
- 18.Richmond J, Tuzova M, Cruikshank W, et al. Regulation of cellular processes by interleukin-16 in homeostasis and cancer. J Cell Physiol 2014; 229: 139–147. [DOI] [PubMed] [Google Scholar]
- 19.Maiuri L, Ciacci C, Ricciardelli I, et al. Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet 2003; 362: 30–37. [DOI] [PubMed] [Google Scholar]
- 20.Meresse B, Chen Z, Ciszewski C, et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity 2004; 21: 357–366. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Geboes K, Colpaert S, et al. IL-15 is highly expressed in inflammatory bowel disease and regulates local T cell-dependent cytokine production. J Immunol 2000; 164: 3608–3615. [DOI] [PubMed] [Google Scholar]
- 22.Abadie V, Discepolo V, Jabri B. Intraepithelial lymphocytes in celiac disease immunopathology. Semin Immunopathol 2012; 34: 551–566. [DOI] [PubMed] [Google Scholar]
- 23.Setty M, Discepolo V, Abadie V, et al. Distinct and synergistic contributions of epithelial stress and adaptive immunity to functions of intraepithelial killer cells and active celiac disease. Gastroenterology 2015; 149: 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jabri B, Abadie V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nature reviews Immunology 2015; 15: 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bondar C, Araya RE, Guzman L, et al. Role of CXCR3/CXCL10 axis in immune cell recruitment into the small intestine in celiac disease. PLoS One 2014; 9: e89068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 2011; 89: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Niro R, Mesin L, Raki M, et al. Rapid generation of rotavirus-specific human monoclonal antibodies from small-intestinal mucosa. J Immunol 2010; 185: 5377–5383. [DOI] [PubMed] [Google Scholar]
- 28.Hoydahl LS, Richter L, Frick R, et al. Plasma cells are the most abundant gluten peptide MHC-expressing cells in inflamed intestinal tissues from patients with celiac disease. Gastroenterology 2019; 156: 1428–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental Material2 for Transcriptional profiling of human intestinal plasma cells reveals effector functions beyond antibody production by Omri Snir, Chakravarthi Kanduri, Knut E A Lundin, Geir Kjetil Sandve and Ludvig M Sollid in United European Gastroenterology Journal
Supplemental material, Supplemental Material1 for Transcriptional profiling of human intestinal plasma cells reveals effector functions beyond antibody production by Omri Snir, Chakravarthi Kanduri, Knut E A Lundin, Geir Kjetil Sandve and Ludvig M Sollid in United European Gastroenterology Journal