Abstract
We report that placental growth factor (PlGF) negatively affects the endothelial cell (EC) barrier function through a novel regulatory mechanism. The PlGF mAb promotes (but recombinant protein disrupts) EC barrier function, thus affecting the barrier-forming protein levels, membrane distribution, and EC monolayer impedance by the electrical cell-impedance sensing system, Western blot, and immunofluorescence staining. RNA sequencing–based transcriptome analysis identified the up-regulation of the pentose phosphate pathway (PPP) and the antioxidant defense protein by PlGF blockade. The PlGF and PlGF/VEGF dimers (but not VEGF-A) down-regulated the protein expression of glucose-6-phosphate dehydrogenase (G6PD) and peroxiredoxin (PRDX). G6PD inhibition and gene silencing (small interfering RNA) abolished the beneficial effects of PlGF inhibition on EC barrier function and PRDX3/6 protein expression. VEGF receptor (VEGFR)1 or VEGFR2 blockade prevented the inhibitory effect of PlGF on G6PD protein expression and EC barrier function. The PRDX6 played dual roles in EC barrier function through glutathione peroxidase and phospholipase A2 activity. In sum, PlGF negatively regulates EC barrier function through the activation of VEGFR1 and VEGFR2 and the suppression of the G6PD/PPP and the antioxidant pathways.—Huang, H., Lennikov, A., Saddala, M. S., Gozal, D., Grab, D. J., Khalyfa, A., Fan, L. Placental growth factor negatively regulates endothelial cell barrier function through suppression of glucose-6-phosphate dehydrogenase and antioxidant defense systems.
Keywords: diabetic retinopathy, G6PD, PlGF, retina, EC
Placental growth factor (PlGF), which was initially identified from a human placental cDNA library in 1991 (1), is a member of the expanded VEGF family and a homolog of VEGF-A. The PlGF and VEGF-A proteins have only 42 aa sequence identity. However, the 2 proteins share marked similarities at the level of their 3-dimensional structures, particularly a conserved cysteine knot motif. This facilitates their interaction and the formation of heterodimers, thereby activating the VEGF receptor (VEGFR)1 and VEGFR2 receptors and modulating vascular permeability and angiogenesis (2).
PlGF plays a pivotal role in pathologic angiogenesis and inflammation by stimulating endothelial cell (EC) migration and recruiting pericytes and inflammatory cells, such as microglia and macrophages (3–5). However, the genetic ablation of PlGF in mice results in healthy animals whose reproductive capacity is unaffected (6). Clinically, PlGF has been implicated in a variety of disorders associated with angiogenesis and ischemia. For example, reduced levels of PlGF caused by the excessive release of soluble VEGFR1 are a hallmark of preeclampsia (7). PlGF mediates therapeutic angiogenesis in myocardial infarction, diabetic wound healing, and limb ischemia (8, 9). It can promote cancer angiogenesis and the metastasis of cancer cells, thereby indicating a potential target for cancer therapy (10). Additionally, PlGF may play a role in the pathogenesis of proliferative diabetic retinopathy (DR) because of its increased expression in the vitreous of patients with diabetes. PlGF overexpression can lead to the characteristics of DR (11–13). Using a genetic mouse model of diabetes (Akita, PlGF−/− mouse), the study found that PlGF gene deletion leads to retinal protection against diabetic damage, such as the breakdown of blood-retinal barrier (BRB). In addition, PlGF gene ablation inhibits the insulin resistance pathway and increases the neuroprotective and antioxidant factors through retinal proteome analysis (14).
The cumulative evidence suggests that PlGF can act as a therapeutic target in the treatment of angiopathology disorders in humans, particularly retinal vascular diseases, such as neovascular or “wet” age-related macular degeneration, proliferative DR, and diabetic macular edema (DME) (15). For example, the intravitreal injection of aflibercept (Eylea), which blocks not only VEGF-A/B but also PlGF, has been recently approved for the treatment of wet age-related macular degeneration, proliferative DR, and DME, following the approval of the anti-VEGF agent ranibizumab, which targets only the VEGF-A isoforms (16). Further clinical investigation revealed that aflibercept exhibits superior efficacy to bevacizumab and ranibizumab for the improvement of visual acuity and the reduction of central subfield thickness over a 1-yr period. This is especially true for the subpopulation of patients with DME with visual acuity of 20/50 or worse (17). Two phase-II clinical trials to assess the safety and efficacy of an anti-PlGF mAb are currently underway.
Despite the emerging circumstantial evidence, additional studies are necessary to determine the direct involvement of PlGF in vascular EC barrier function, the extent to which targeting PlGF alone can promote EC barrier function, and the underlying mechanisms of PlGF activity. To clarify these questions, the effects of PlGF blockade by an mAb (PL5D11D4) and treatment by a recombinant human (rh)PlGF protein on the EC barrier function in primary cell cultures were investigated. The genes and signaling pathways that are regulated by PlGF blockade in human retinal ECs (HRECs) were identified through RNA sequencing (RNA-seq)-based transcriptome analysis. The membrane receptor signaling that mediates the effects of PlGF was characterized by the use of experimental rescue approaches through signal transduction blockade. In addition, a new mechanism through which PlGF regulates EC barrier function was discovered through the use of pharmacological inhibition and gene silencing.
MATERIALS AND METHODS
Primary retinal microvascular cell cultures
The primary bovine retinal ECs (BRECs) and calf retinal pericytes (CRPs) were isolated and cultured, as was previously described by Antonelli-Orlidge et al. (18). The primary HRECs (ACBR1 181) and human retinal pericytes (ACBR1 183) were purchased from Cell Systems (Kirkland, WA, USA) and cultured in accordance with the manufacturer’s instructions.
Cell treatments and small interfering RNA transfection
After HRECs or BRECs gained ∼80–90% confluence, the culture medium was replaced with fresh medium with the following desired treatment agents: d-glucose (25 mM), l-glucose (25 mM), mannitol (25 mM), anti-PlGF antibody (PL5D11D4), anti-VEGFR1 antibody (MF1; ImClone Systems, New York, NY, USA), anti-VEGFR2 antibody (DC101; ImClone Systems), rhPlGF protein (264-PGB-010/CF; R&D Systems, Minneapolis, MN, USA), rhVEGF/PlGF heterodimers (297-VP-005/CF; R&D Systems), VEGF-165 (293-VE-010/CF; R&D Systems), mouse IgG, peroxiredoxin (PRDX)6 inhibitor (MJ33 lithium salt, 1007476-63-2; Cayman Chemicals, Ann Arbor, MI, USA), and the glucose-6-phosphate dehydrogenase (G6PD) inhibitor dehydroepiandrosterone (DHEA; MilliporeSigma, Burlington, MA, USA). PRDX6 small interfering RNA (siRNA) (4390824; Thermo Fisher Scientific, Waltham, MA, USA), G6PD siRNA (AM16708; Thermo Fisher Scientific), and control siRNA (4390843; Thermo Fisher Scientific) were transfected into the HRECs with Lipofectamine 2000 or RNAi Max (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions.
Transendothelial electrical resistance measurement by an electrical cell-impedance sensing system
The primary HRECs or BRECs were seeded on an 8-well electrical cell-impedance sensing (ECIS) array and cultured as described above. Transendothelial electrical resistance (TEER) was monitored with the ECIS system (Applied BioPhysics, Troy, NY, USA) in real time. The changes in TEER were monitored automatically every 300 s at 4 kHz AC frequency and recorded with ECIS software. The embedded mathematical model of impedance change was used to calculate the TEER (Ω/cm2), a measure of the cell-to-cell barrier and cell-to-substratum function (19).
RNA-seq and bioinformatics analyses
The confluent HRECs were treated with the PBS control and PlGF antibody for 4 d. The collected HRECs were processed to isolate the total RNA through the use of the RNeasy Plus Mini Kit (Qiagen, Germantown, MD, USA) in accordance with the manufacturer’s instructions. After quality confirmation, RNA-seq was performed by Novogene (Beijing, China). Bioinformatics analysis was performed in the author’s laboratory in accordance with the previously described procedures (20). In brief, the raw data were used for the visualization of the read quality before and after preprocessing by the use of FastQC software (Babraham Bioinformatics, Cambridge, United Kingdom; https://www.bioinformatics.babraham.ac.uk/index.html). The data were processed for removal of adapters and ambiguity quality reads by Trimmomatic 0.36 tool (21). All of the data sets on the human genome reference sequence Genome Reference Consortium Human Build 38 (GRCh38) were mapped with TopHat 2.0.9 software (https://ccb.jhu.edu/software/tophat/index.shtml). The differentially expressed genes (DEGs) that satisfied significance expressed as a q value representing the false discovery rate–adjusted value of P < 0.05 were identified through Cufflinks 2.1.1 software (http://cole-trapnell-lab.github.io/cufflinks/install/). The Bioconductor tool with the CummeRbund package was used for the differential expression analysis in the assembled transcriptome. Gene ontology enrichment analysis and the Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/) functional annotation tool were used for the functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (https://www.genome.jp/kegg/).
Western blot and densitometric analysis
Western blot (WB) was performed, as was previously described, with some modifications (3, 22). HRECs grown in 6-well plates were washed with cold Dulbecco’s PBS 3 times, detached with a cell scraper, and collected by centrifugation. The harvested cell pellets were sonicated in cold RIPA buffer containing Fast protease inhibitors (S8830; MilliporeSigma). The protein concentration was determined with the DC Protein Assay Kit (Bio-Rad, Hercules, CA, USA) and/or Qubit 4 Fluorometer (Thermo Fisher Scientific).
Prior to the electrophoretic transfer to 0.45-μm-pore-size nitrocellulose membranes, 30–50 μg total protein/lane was separated by SDS-PAGE (4–20% polyacrylamide gel). The membranes were blocked with 5% nonfat milk (Bio-Rad) at room temperature for 1 h and then incubated overnight at 4°C with the following primary antibodies: anti–zonula occludens-1 (ZO-1; 1:1000, 402200; Thermo Fisher Scientific), anti–occludin-1 (1:500, 33-1500; Thermo Fisher Scientific), anti–claudin-5 (1:500, 1873694; Thermo Fisher Scientific), anti–vascular endothelial (VE)-cadherin (1:1000, 5012896; Thermo Fisher Scientific), anti–β-catenin (1:1000, MAB 1329-200; R&D Systems), anti-G6PD (1:500, MA5-15918; Thermo Fisher Scientific), anti-transketolase (TKT, 1:500, AV48540-50UG; MilliporeSigma), anti-transaldolase (TALDO) 1 (1:500, PA5-27614; Thermo Fisher Scientific), anti-PRDX3 (1:500, WH0010935M1-100UG; MilliporeSigma), anti-PRDX6 (1:500, WH10009588M1-100UG; MilliporeSigma, or PA5-24632; Thermo Fisher Scientific), anti-VEGF-PlGF heterodimer antibody (1:1000, MAB297-SP; R&D Systems), and anti–β-actin (1:1000, PA5-16914; Cell Signaling Technology, Danvers, MA, USA).
After being washed with PBS with Tween (PBS-T) buffer, the blots were incubated with horseradish peroxidase–conjugated secondary antibody (1:2000; Cell Signaling Technology) for 1 h at room temperature. Signals were developed with ECL with a SuperSignal West Pico Kit (Thermo Fisher Scientific) and detected with an ImageQuant LAS 500 (GE Healthcare, Waukesha, WI, USA). Densitometry analysis was performed through the use of ImageJ (National Institutes of Health, Bethesda, MD, USA).
The densitometric analysis of WBs was performed with ImageJ software. All the quantification results were averaged from 3 protein blots and expressed as the mean ratio of the target protein and β-actin ± sd unless otherwise specified.
Dot immunoblotting assay
Modification of the Dot immunoblotting assay (23, 24) was used to validate the affinity and specificity of PL5DLLD4 antibody with human PlGF. rhPlGF protein (200 ng; 264-PGB-010/CF; R&D Systems), VEGF-165 (200 ng, 293-VE-010/CF; R&D Systems), and 500 ng bovine serum albumin (BSA) (MilliporeSigma) were deposited in the volume of 2 µl on dry nitrocellulose membrane (Bio-Rad) and immobilized by 15 min incubation at room temperature The membranes were blocked with 5% nonfat milk (Bio-Rad) in PBS at room temperature for 1 h and then incubated overnight at 4°C with mouse PL5DLLD4 antibody 0.5 µg/ml. The membrane was washed 3 times for 5 min with PBS-T (0.05% Triton) and incubated with horseradish peroxidase–conjugated secondary antibody goat anti-mouse IgG (172-1011, 1:1000; Bio-Rad) for 1 h at room temperature. Following additional washing, signals were developed with ECL using a Super Signal West Pico Kit (Thermo Fisher Scientific) and detected with ImageQuant LAS 500 (GE Healthcare).
Immunocytofluorescence analysis
HRECs and BRECs were seeded into the Millicell EZ Slide (MilliporeSigma) with the same experimental conditions as those for the TEER and WB experiments. At the experimental end point, the samples were mildly fixed in 4% paraformaldehyde (VVR Life Science, Radnor, PA, USA) for 10 min, permeabilized by incubation in 0.05% Triton X-100 for 10 min, and blocked with 10% normal goat serum for 1 h at room temperature. The samples were then incubated with anti–claudin-5 (1:100), anti–occludin-1 (1:100), anti–ZO-1 (1:100), and anti-PRDX6 (1:100) antibodies. After PBS-T washing, they were then visualized by goat anti-rabbit IgG (H+L), cyanine5 (A10523, 1:1000; Thermo Fisher Scientific), goat anti-rabbit IgG (H+L), Alexa Fluor 488 (A-11034, 1:1000; Thermo Fisher Scientific), goat anti-mouse IgG (H+L), and Cyanine5 (A10524, 1:1000; Thermo Fisher Scientific). The cell nuclei were visualized by incubation with DAPI 1:5000 (MilliporeSigma). The slides were mounted with a ProLong Diamond Antifade Reagent (Thermo Fisher Scientific) and imaged with an LSM 700 Inverted Laser Confocal Microscope (Carl Zeiss GmbH, Oberkochen, Germany).
Cytotoxicity assay of DHEA in HREC and BREC cultures
HRECs and BRECs were cultured in 24-well plates with 25 µM of DHEA in 1 µl of DMSO or DMSO 1 µl/ml for 4 d. Triton X-100 0.02% (MilliporeSigma) was added to positive control wells of the HREC and BRECs 15 min before the assay started. The fluorescent ReadyProbes Cell Viability Imaging Kit (blue/green) (R37609; Thermo Fisher Scientific) was used to evaluate the cell death in fluorescent images; active components of the kit were added to the cell culture medium with the dilution 1:100 and, following 10 min incubation, fluorescent images were acquired using Evos Fl Fluorescent Microscope (Thermo Fisher Scientific). Lactate dehydrogenase (LDH) assay (Thermo Fisher Scientific) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Thermo Fisher Scientific) were used to quantitively evaluate the DHEA toxicity; both kits were used according to the instruction manual, and the resulting absorbances were read at 490 nm (LDH assay) and 570 nm (MTT assay), respectively, using 800 TS Absorbance Reader (BioTek Instruments, Winooski, VT, USA). Absorbances at 650 nm were used as a reference wavelength.
Statistical analysis
All of the values were expressed as means ± sd for the respective groups. Statistical analysis was performed with Prism 8 software (GraphPad Software, La Jolla, CA, USA). ANOVA or a linear mixed model was used for the statistical comparisons of multiple groups (25). The P values for the comparison of the treatments were adjusted for multiple comparisons with Dunnett’s test. The nonparametric Mann-Whitney U test was performed to determine the significance level between the 2 groups. Statistical significance was set at P < 0.05.
The TEER or resistance data were obtained from the ECIS experiments; the values were averaged from 4 duplicates, and the error bars indicate sd. The statistical analysis was applied to all data points following the treatment (not including the culture proliferation). Therefore, the statistical significance answered the question of whether there is a difference over the whole course of a treatment between 2 groups (not a particular time point).
RESULTS
PlGF mAb specificity and affinity with human and bovine PlGF protein
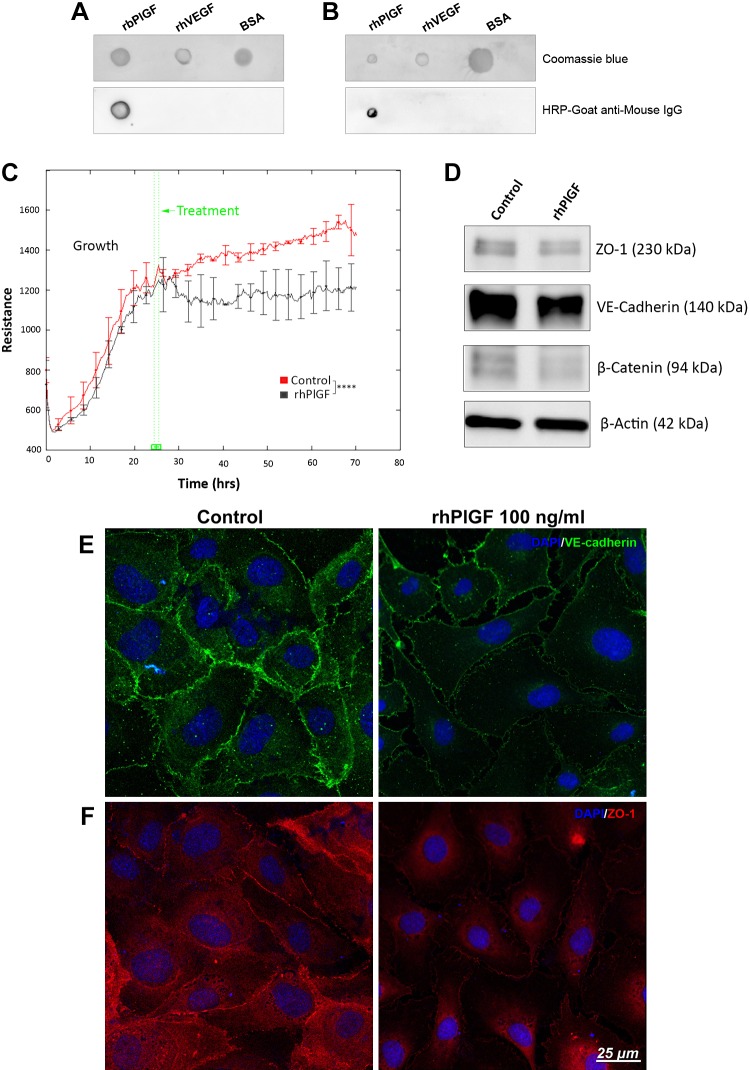
As the PlGF mAb (mAb: PL5D11D4) was generated to target mouse PlGF-2, we evaluated this antibody’s specificity and affinity with recombinant human and bovine PlGF proteins (rhPlGF and rbPlGF). Dot immunoassay results demonstrated strong affinity and specificity of this PlGF mAb with the rbPlGF (Fig. 1A) and rhPlGF (Fig. 1B), visualized by presence of PL5D11D4 antibody signal within the spot generated by immobilized rhPlGF and rbPlGF, with no binding with rhVEGF or BSA indicated by lack of signal within control spots.
Figure 1.
Recombinant PlGF has specific affinity with its mAb (PL5D11D4) and disrupts HREC barrier function. A, B) Dot immunoassay demonstrated the specific affinity of PlGF antibody (PL5D11D4) with rbPlGF (A) and rhPlGF (B). rhVEGF and BSA acted as negative controls. HRP, horseradish peroxidase. C) HRECs were cultured and grown with endothelial growth medium 2 (EGM-2) (with growth factors such as VEGF-A). After the cells grew to confluence, the culture medium was changed to endothelial cell growth basal medium 2 (EBM-2) (without growth factors), and PBS control and rhPlGF protein (100 ng/ml) were added to the culture medium and incubated for at least 2 d. TEER was monitored by an ECIS system at an AC frequency of 4 KHz in real time. TEER curves of HRECs that were treated with rhPlGF protein and PBS control. Error bars represent sd out of 4 duplicate samples. The experiments were repeated at least 3 times. D) WB result for VE-cadherin, β-catenin, and ZO-1. β-Actin was used as the protein loading control. E, F) Immunofluorescence staining results of tight junction ZO-1 and adhesion protein VE-cadherin. ****P < 0.0001.
PlGF acts as a negative regulator of EC barrier function
To investigate the role of PlGF as a negative regulator of EC barrier function, the rhPlGF protein and PlGF mAb were used to treat the HRECs. In comparison with the PBS control, the rhPlGF protein reduced HREC resistance (Fig. 1C) and the expression of the barrier function proteins (ZO-1, VE-cadherin, and β-catenin) (Fig. 1D). Immunofluorescent analysis indicated a strong presence of VE-cadherin (Fig. 1E) at the edges of the HREC membranes and cellular interaction sites, whereas treatment with rhPlGF indicated the reduced expression of VE-cadherin as well as the decreased intensity of HREC cellular processes interaction. These data were further supported by ZO-1 immunostaining (Fig. 1F), whereupon rhPlGF treatment ZO-1 was predominantly detected in the cytoplasm with the markedly reduced presence at cellular borders.
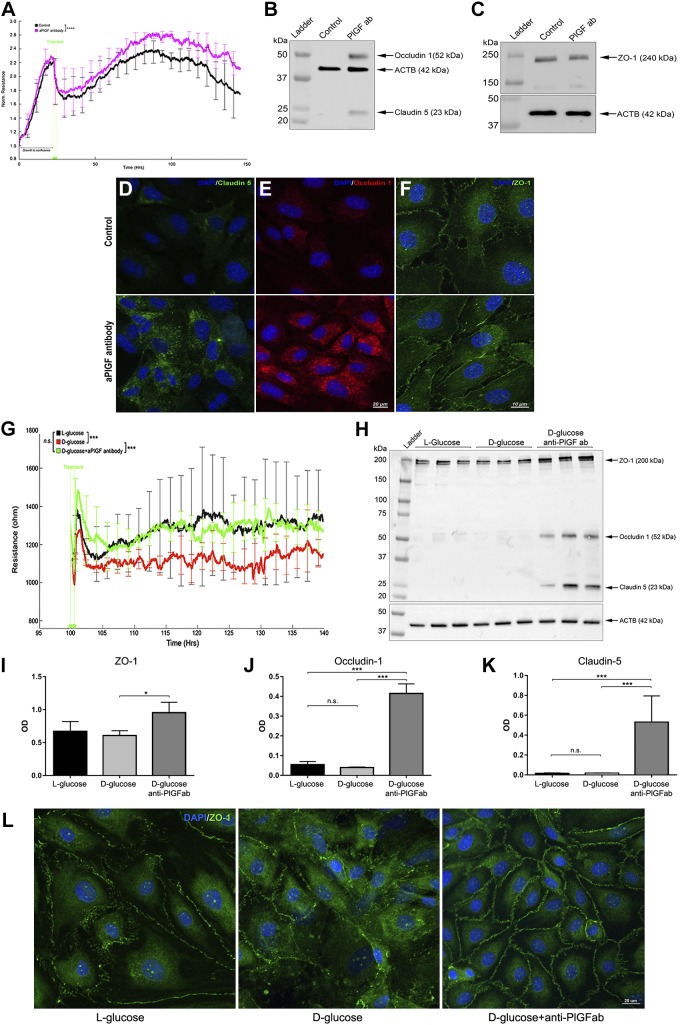
PlGF antibody (PL5D11D4) treatment promoted HREC resistance (Fig. 2A), up-regulated the tight junction proteins (occludin-1, claudin-5, and ZO-1) (Fig. 2B, C), and increased their cell membrane integration (Fig. 2D–F). Thus, the loss- and gain-of-function findings robustly supported the proposition that PlGF acts as a negative regulator of retinal EC barrier function.
Figure 2.
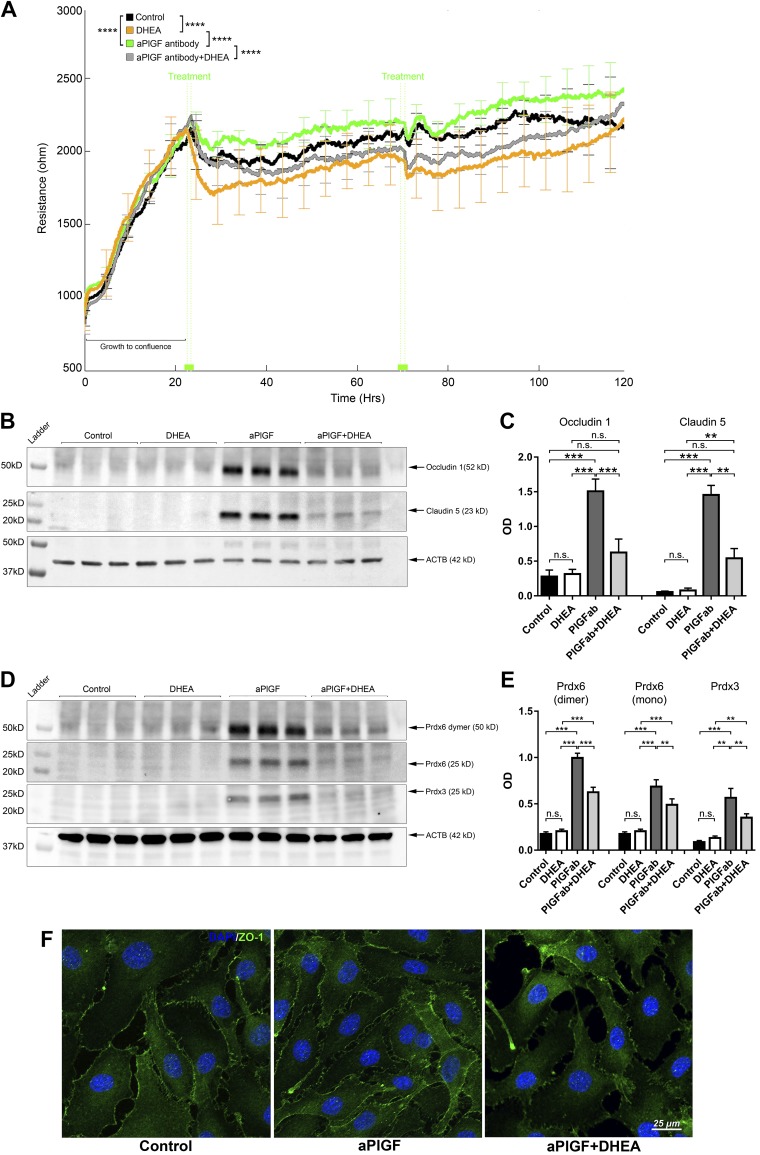
PlGF antibody promotes EC barrier function and prevents barrier impairment by high glucose (HG). Confluent HRECs were treated with the PlGF antibody (100 µg/ml) and PBS control. TEER was monitored by the ECIS system in real time. A) TEER curves of HRECs treated with PlGF antibody (purple) and PBS control (black). Error bars represent sd out of 4 duplicate samples. The experiments were repeated 3 times. B, C) WB results of the 3 tight junction proteins occludin-1, claudin-5, and ZO-1. D–F) Immunofluorescence staining of the 3 tight junction proteins. Note the increased protein levels of occludin-1 and claudin-5 and cell membrane localization of ZO-1 compared with those of the control. G) TEER curves of HRECs under normal glucose (NG; 25 l-glucose as osmotic control), HG (25 mM d-glucose), and HG + αPlGF antibody (100 µg/ml). Compared with the NG control (black), the HG + IgG control (red) induced a significant reduction of TEER starting at ∼5 d (100 h). The TEER values for the HG + αPGF antibody (green) were similar to those for the control (no significant difference). Resistance values were expressed as means ± sd. The experiments were repeated 3 times. H–K) WB and densitometry quantification results for ZO-1, occludin-1, and claudin-5 under the conditions of NG, HG, and HG + PlGF antibody. The HG (7 d) caused a reduction of ZO-1 protein expression levels, which were prevented by the PlGF antibody. The occludin-1 and claudin-5 proteins were not detected under either the NG control or HG conditions but were obviously up-regulated under the HG + anti-PlGF antibody: β-actin (ACTB) was used as the protein loading control. L) Immunofluorescence staining results of ZO-1 under NG (25 mM l-glucose as control), HG (25 mM d-glucose + IgG), and HG (25 mM d-glucose) + anti-PlGF antibodies. Compared with the NG control, HG caused ZO-1 protein disintegration in the plasma membrane; however, this was prevented by the αPGF antibody treatment. N.s., not significant; OD, optical density. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The role of PlGF blockade in preventing retinal EC barrier dysfunction by high glucose was investigated to explore the potential application of targeting PlGF in DR. Starting at ∼5 d of culture in high-glucose conditions, HREC resistance was significantly reduced (P < 0.01). In contrast, PlGF blockade prevented HREC resistance reduction by high-glucose environment (Fig. 2G). Concomitantly, PlGF blockade up-regulated the protein levels of the tight junction proteins (ZO-1, occludin-1, and claudin-5) (Fig. 2H–K) and the cell membrane distribution of ZO-1 protein under high glucose (Fig. 2L). PlGF blockade in the BRECs had similar effects as the HRECs under high-glucose conditions (Supplemental Fig. S1).
PlGF inhibition up-regulates G6PD and PRDX
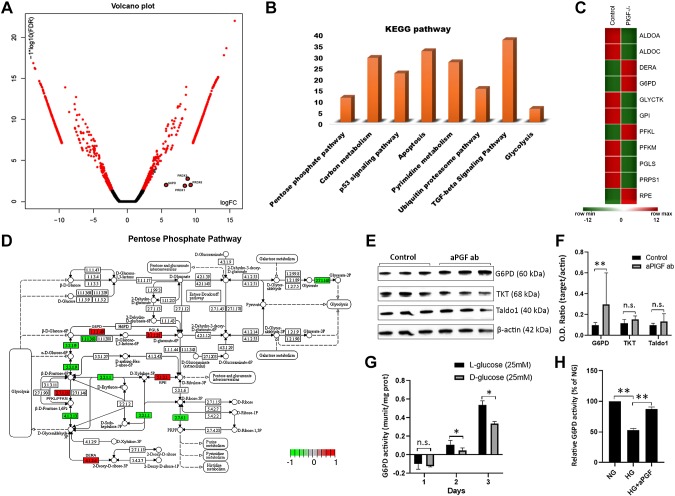
RNA-seq-based transcriptome analysis was performed to identify the genes and pathways that are regulated by PlGF inhibition in HRECs. Of the 53,808 identified gene transcripts, 3275 genes were found to be differentially expressed between the PBS control and PlGF antibody treatment. The pathway-enriched analysis identified the pentose phosphate pathway (PPP) and antioxidant defense pathways as being up-regulated by PlGF inhibition (Fig. 3A–D and Supplemental Table S1). G6PD, the first and rate-limiting enzyme of PPP, was up-regulated at transcript levels in the RNA-seq data set. The up-regulated antioxidant defense genes included PRDX (PRDX1, 3, and 6). The up-regulation of G6PD, PRDX3, and PRDX6 at the protein level by PlGF inhibition was further validated by WBs (Figs. 3E, F and 4). This also demonstrated that the protein levels of 2 other critical nonoxidative PPP enzymes (namely, TKT and TALDO1) were not reduced by PlGF inhibition (Fig. 3E, F). The transcriptome data analysis revealed that several nonoxidative PPP enzymes were down-regulated by PlGF inhibition (Supplemental Table S1). These results suggest that the targeting of PlGF might not necessarily activate the nonoxidative phase of the PPP. It would only increase the flux of glucose into the oxidative phase PPP (oxPPP) to generate NADPH. Additionally, PlGF inhibition could prevent the G6PD enzyme activity impairment by high glucose in retinal EC. This result suggests the potential implication in DR (Fig. 3G, H).
Figure 3.
PlGF inhibition up-regulates the PPP revealed by RNA-seq analysis and prevents G6PD activity impairment caused by high glucose in HRECs. After the confluent HRECs were treated with PBS control and PlGF antibody for 4 d, the cells were isolated for RNA isolation, sequencing, and bioinformatics analyses. Six replicates for each condition (control vs. antibody) were used for experimentation. The HRECs that were treated with l-glucose (25 mM), d-glucose (25 mM), and the PlGF antibody were collected for the G6PD enzyme activity assay. The assay experiments were repeated 3 times. A) Volcano plot represented by significantly up-regulated and down-regulated genes on the basis of the logFC and log10 [false discovery rate (FDR)]. Note that G6PD, PRDX1, PRDX3, and PRDX6 were marked in the plot. B) DEGs were classified in accordance with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The PPP, TGF-β, and other metabolic pathways were identified. C) Significantly up-regulated and down-regulated PPP genes in the HRECs treated with PBS control and PlGF antibody represented as a heat map (red color shows up-regulated genes, and green color shows down-regulated genes). D) PPP genes highlighted in red color (up-regulated) and green color (down-regulated) and represented in KEGG pathway. The DEGs, such as 1.1.1.49 (G6PD), 1.1.1363 (G6PD), 5.3.1.9 [glucose-6-phosphate isomerase (GPI)], 2.7.1.11 [phosphofructokinase liver/muscle type (PFKL/PFKM)], 4.1.2.13 [aldolase, fructose-bisphosphate A/C (ALDOA/ALDOC)], 4.1.2.4 [deoxyribose-phosphate aldolase (DERA)], 3.1.1.31 [6-phosphogluconolactonase (PGLS)], 5.1.3.1 [ribulose-phosphate 3-epimerase (RPE)], 2.7.2.1 [phosphoribosylpyrophosphate synthetase 1 (PRPS1)], and 2.7.1.165 [glycerate kinase (GLYCTK)], are involved in the PPP. E) WB results of G6PD, TKT, and TALDO in HRECs: β-actin was used as the protein loading control. F) Densitometry analysis of protein bands. O.D., optical density. G) G6PD enzyme activity under high glucose (HG; 25 mM d-glucose) and normal glucose (NG; 25 mM l-glucose for osmotic control) for 1, 2, and 3 d. H) G6PD enzyme activity of HRECs treated with NG control, HG, and HG + PlGF antibodies. The results were averaged from 3 independent experiments and expressed as a percentage relative to the NG condition. N.s., not significant. *P < 0.05, **P < 0.01.
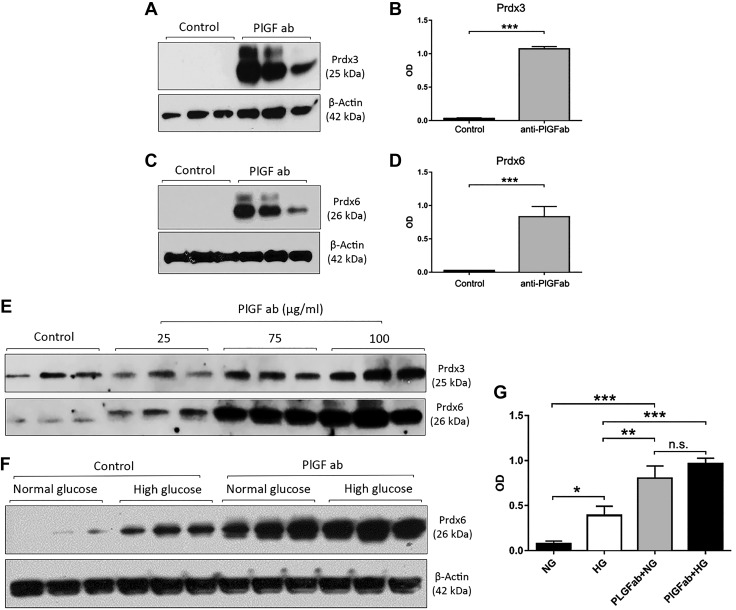
Figure 4.
PlGF inhibition up-regulates antioxidant Prdx3 and Prdx6 protein expression levels in HRECs. The confluent HRECs were treated with the PlGF antibody for 4 d. WB analysis was performed with the antibodies against Prdx3 and Prdx6. A–D) Densitometric analysis was performed from 3 protein blots with ImageJ software. Prdx3 (A, B) and Prdx6 (C, D) protein levels were significantly up-regulated by PlGF antibody in normal glucose condition: β-actin was used as the protein loading control, and each condition was in triplicate. E) Protein levels of Prdx3 (upper) and Prdx6 (lower) up-regulated by PlGF inhibition on the basis of antibody concentration (25, 75, and 100 µg/ml). F, G) Protein expression levels of Prdx6 under the 4 conditions: IgG control + normal glucose, IgG control + high glucose, PlGF antibody + normal glucose, and PlGF antibody + high glucose. Note that both the high-glucose and the PlGF antibody up-regulated Prdx6 protein expression. HG, high glucose; NG, normal glucose; n.s., not significant; OD, optical density. *P < 0.05, **P < 0.01, ***P < 0.001.
PlGF blockade promotes EC barrier function through glucose-6-phosphate (PPP)
Because PlGF blockade up-regulates antioxidants and G6PD and promotes EC barrier function, it was hypothesized that PlGF inhibition would promote EC barrier function and up-regulate antioxidants PRDX3/6 through the activation of G6PD/PPP, the primary source of the reductant cofactor NADPH. Therefore, the inhibitor DHEA was used to inhibit G6PD activity. DHEA’s minimal effect on cell viability at the effective concentration of 25 µM was demonstrated by the results of fluorescent live/death assay and LDH assay (Supplemental Fig. S2); these data were further confirmed with MTT assay indicating low toxicity values of 3.8 ± 1.3% for HREC and 4.7 + 3.1% for BREC after 4 d of culture. Indeed, G6PD inhibition by DHEA (25 µM) abrogated the beneficial effects of PlGF blockade, including boosted HREC resistance (Fig. 5A), increased junction protein levels (Fig. 5B, C). Concomitantly, G6PD inhibition diminished the increased antioxidant protein levels (PRDX3 and PRDX6) by PlGF blockade (Fig. 5D, E). Additional immunofluorescence staining demonstrated that PlGF inhibition enhanced membrane integration of tight junction protein ZO-1 (Fig. 5F). Similar effects of PlGF blockade by DHEA were observed for the BRECs (Supplemental Fig. S3). G6PD gene knockdown by siRNA also reduced HREC resistance and the barrier proteins (Supplemental Fig. S4).
Figure 5.
G6PD inhibitor DHEA abrogates the effects of the PlGF antibody on HREC barrier function and PRDX3/6 protein expression. A) G6PD inhibitor DHEA abolished effects of PlGF antibody on HREC TEER, which was monitored by ECIS in real time. When the monolayer was formed, as indicated by the peak of the TEER values (mean ∼2200 Ω) at ∼24 h, treatment agents were added to the culture medium: PBS control, DHEA (25 µM), αPlGF (100 µg/ml), and DHEA + αPlGF. At ∼72 h, the same treatment schemes were reapplied. Four replicates were included for each treatment condition. Error bars represent sd out of 4 replicated samples. B, C) WB and densitometric quantification results for claudin-5 and occludin-1. DHEA abolished the effects of αPlGF on claudin-5 and occludin-1 protein expression. The treatment conditions were the same as those for the TEER experiments; however, the cells were grown on 6-well plates. For each sample, 50 µg proteins were loaded: β-actin (ACTB) was used as the protein loading control. D, E) WB and densitometric quantification results of PRDX3 and PRDX6. The G6PD inhibitor DHEA diminished the increased PRDX3 and PRDX6 protein expression by the PlGF antibody. F) Immunofluorescence staining of ZO-1. This illustrates increased ZO-1 allocation to the plasma membrane in the cell-cell interaction sites that were promoted by the PlGF antibody but prevented by the inhibitor DHEA. N.s., not significant. **P < 0.01, ***P < 0.001, ****P < 0.0001.
PlGF regulates G6PD, PRDX, and barrier function through VEGFR1 and R2 signaling
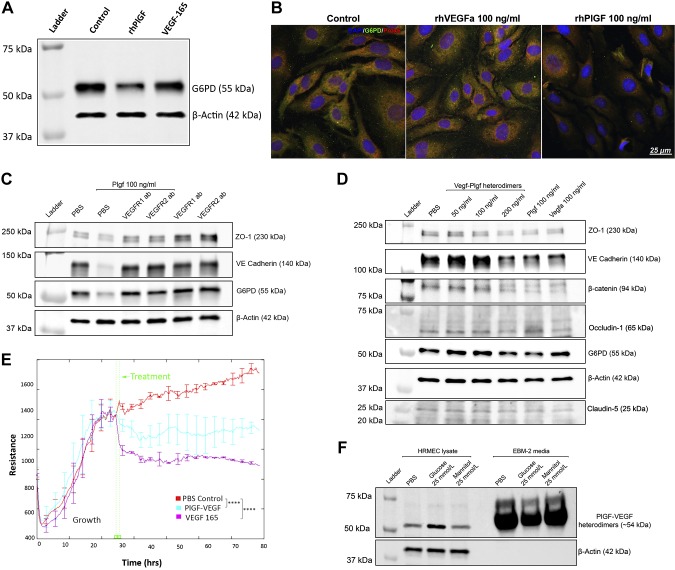
PlGF can bind and activate VEGFR1, and it also indirectly activates VEGFR2 through interaction with VEGF-A. Therefore, an evaluation was performed to determine the receptor signaling that mediates the effects of PlGF on G6PD, PRDX, and EC resistance. First, the effect of VEGF-165 (the most active VEGF-A isoform) on G6PD protein expression was examined. In comparison to the PBS control, the rhPlGF protein down-regulated G6PD protein expression; however, VEGF-165 did not (Fig. 6A). Immunofluorescence staining showed that the G6PD and PRDX6 were colocalized in the cytoplasm of the PBS control-treated and rhVEGF-treated cells and down-regulated in the PlGF antibody-treated samples (Fig. 6B and Supplemental Fig. S5). The results suggest that VEGF-A might not be involved in the regulation of G6PD and PRDX6 and that such functions are unique to PlGF.
Figure 6.
PlGF regulates G6PD, PRDX6, and EC barrier function through VEGFR1 and VEGFR2 signaling. A) WB results for G6PD. HRECs that were treated with rhPlGF and VEGF-165 protein (100 ng/ml each, for 2 d) were used for the WB assay: β-actin was used as the protein loading control. Note that rhPlGF, but not VEGF-165, down-regulated the protein level of G6PD compared with the PBS control. B) Double immunofluorescence labeling of G6PD and PRDX6. The HRECs that were treated with the PBS control, rhVEGF(a), and rhPlGF protein were fixed and stained with G6PD (green) and PRDX6 (red). The DAPI staining of the nucleus (blue) was used as the counterstaining. Note that the 2 proteins were colocalized in the PBS-treated and rhVEGF-treated cells (yellow); however, the staining signals were reduced in the rhPlGF-treated cells. The complete images of all 3 fluorescent staining channels are shown in Supplemental Fig. S5. C) WB results of ZO-1, VE-cadherin, and β-catenin. The confluent HRECs were treated with PBS, PBS + rhPlGF (100 ng/ml), rhPlGF + VEGFR1 antibodies (100 µg/ml), rhPlGF + VEGFR2 antibody (100 µg/ml), and VEGFR1 antibody alone or VEGFR2 antibody alone for 2 d. Note that rhPlGF protein down-regulated the protein levels of ZO-1, VE-cadherin, and G6PD, which were prevented by the VEGFR1 or VEGFR2 antibody. The antibody alone did not change the protein levels, which were equivalent to those of the PBS control. D) WB results of ZO-1, VE-cadherin, β-catenin, occludin-1, claudin-5, and G6PD. The confluent HRECs were treated with the PBS, PlGF/VEGF heterodimer (50, 100, or 200 ng/ml), PlGF (100 ng/ml), or VEGF-165 (100 ng/ml). Note that PlGF, the PlGF/VEGF heterodimer (200 ng/ml), and VEGF-165 down-regulated ZO-1, VE-cadherin, β-catenin, and claudin-5 (but not occludin-1); however, only PlGF and PlGF/VEGF down-regulated G6PD. E) TEER (or resistance) of HRECs. After the HRECs grew to confluence, the PBS control, PlGF/VEGF, or VEGF-165 was added to the medium, and the TEER was measured with ECIS in real time. The TEER values of the PlGF/VEGF heterodimer (blue) and VEGF-165 (purple) were significantly lower than those of the PBS control (red). Error bars represent sd out of 4 duplicate samples. F) WB results of PlGF/VEGF heterodimers in HREC lysate and culture medium. The confluent HRECs were treated with PBS, 25 mM d-glucose [high glucose (HG)], and 25 mM l-glucose [normal glucose (NG)]. Note that compared with the PBS controls, the HG up-regulated PlGF/VEGF in the cell lysates but decreased the levels in the culture medium. The β-actin was presented in the HREC lysate (but not in the culture medium) and used as the protein loading control. ****P < 0.0001.
Next, the role of the blockade of VEGFR1 and VEGFR2 signaling in the prevention of the down-regulation of G6PD and PRDX6 protein expression elicited by the rhPlGF protein was assessed. As is shown in Fig. 6C, the blockade of VEGFR1 or VEGFR2 prevented the protein degradation of G6PD by rhPlGF treatment, as well as the barrier function proteins ZO-1 and VE-cadherin. These results suggest that PlGF has a negative effect on G6PD and the barrier proteins, not only through VEGFR1 but also through interaction with VEGF-A, to activate VEGFR2 indirectly.
To confirm this finding, the ability of the PlGF/VEGF heterodimers, which can bind and activate both VEGFR1 and VEGFR2, to down-regulate the expression of G6PD and the EC barrier function proteins were investigated. The PlGF/VEGF heterodimers (200 ng/ml, but not 50 or 100 ng/ml) down-regulated the protein levels of G6PD and 4 barrier function proteins (ZO-1, VE-cadherin, claudin-5, and β-catenin), which are similar to the rhPlGF protein (100 ng/ml). VEGF-165 down-regulated the 3 barrier proteins, but not G6PD, thereby suggesting that the mechanism was different from that of PlGF or the PlGF/VEGF heterodimer. Interestingly, whereas PlGF treatment decreased ZO-1, VE-cadherin, claudin-5, and β-catenin, it did not show the effect on occludin-1 (Fig. 6D). Furthermore, the effect of the PlGF/VEGF heterodimer on EC resistance was examined. As is shown in Fig. 6E, PlGF/VEGF and VEGF-A led to a significant reduction of EC resistance in comparison to that in the PBS control. High glucose up-regulated the protein level of the PlGF/VEGF heterodimer in the cell lysates and correspondingly led to a decreased level in the supernatant. This was likely the result of its binding with its receptors in the cell membrane (Fig. 6F).
Dual roles of PRDX in EC barrier function
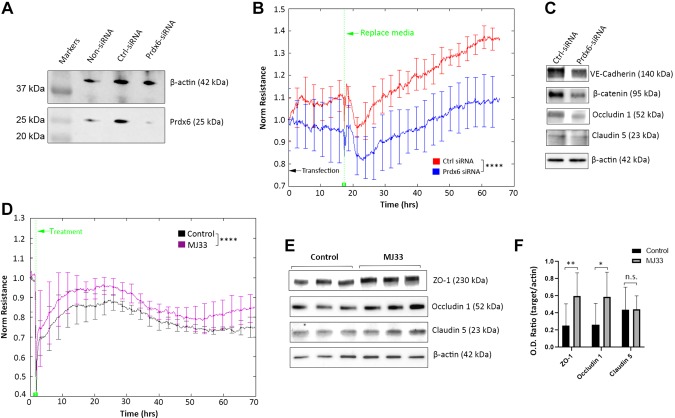
Finally, the role of PRDX6 in EC barrier function was investigated. The protein level of PRDX6 expression was up-regulated by both high glucose and PlGF inhibition (Fig. 4F, G). High glucose is harmful to EC barrier function; however, PlGF inhibition plays a beneficial role. Therefore, the increased level of PRDX6 by high glucose and PlGF inhibition makes its role in EC barrier function uncertain. PRDX6 siRNA was used to elucidate the role of PRDX6 in EC barrier function. The gene knockdown efficiency of PRDX6 siRNA was first validated by the verification of a dramatic decrease in its protein levels in the HRECs (Fig. 7A). PRDX6 knockdown disrupted EC barrier function, as indicated by the significant reduction of resistance of the PRDX6 siRNA treatment compared with that observed in the control siRNA (Fig. 7B). The protein levels of the barrier function proteins, including VE-cadherin, β-catenin, claudin-5, and occludin-1, were also down-regulated by PRDX6 siRNA (Fig. 7C). Because PRDX6 is a bifunctional enzyme that has the activities of glutathione peroxidase (GPX) and calcium-independent phospholipase A2 (PLA2) (26), the elucidation of the enzyme activity that contributes to EC barrier function was necessary. The PRDX6 inhibitor MJ133, a selective and reversible inhibitor of the calcium-independent PLA2 activity of PRDX6, was used to address this question.
Figure 7.
Dual roles of PRDX6 in EC barrier function. A) WB results of PRDX6 in HRECs under the 3 conditions of control siRNA and PRDX6 siRNA or mock transfection without siRNA: β-actin was used as the protein loading control. Note that PRDX6 siRNA effectively down-regulated its protein level compared with the mock (non-siRNA) and control siRNA. B) TEER (or resistance) of HRECs transfected with PRDX6 siRNA and control siRNA. The normalized resistance of control siRNA (red) and PRDX6 siRNA (blue) is shown. The normalized values were averaged from 4 duplicate samples and expressed as means ± sd. Note that the anti-PlGF antibody (100 µg/ml) was supplemented with culture medium to boost EC barrier function and junction formation. C) WB results of HRECs transfected with PRDX6 siRNA and control siRNA. PRDX6 siRNA down-regulated the tight junction and adhesion proteins, including VE-cadherin, β-catenin, occludin-1, and claudin-5. D) TEER (or resistance) of HRECs treated with PRDX6 phospholipase A2 inhibitor MJ33 (purple) and PBS control (black). Error bars represent sd out of 4 replicate samples (n = 4). The experiments were repeated 3 times. E, F) WB and densitometry quantification results of HRECs treated with MJ33. The protein levels of ZO-1 and occludin-1 (but not claudin-5) were significantly up-regulated by MJ33. Ctrl-siRNA, control siRNA; n.s., not significant; O.D., optical density. *P < 0.05, **P < 0.01, ****P < 0.0001.
Unexpectedly, PRDX6 PLA2 inhibition by MJ33 was found not to disrupt EC resistance; instead, it led to increased TEER in comparison to the result observed for the control (Fig. 7D). The expression levels of the tight junction proteins (i.e., the ZO-1, claudin-5, and occludin-1 proteins) were also increased by MJ33 (Fig. 7E, F). Therefore, PRDX6-GPX activity is advantageous to EC barrier function; however, PLA2 activity is detrimental.
DISCUSSION
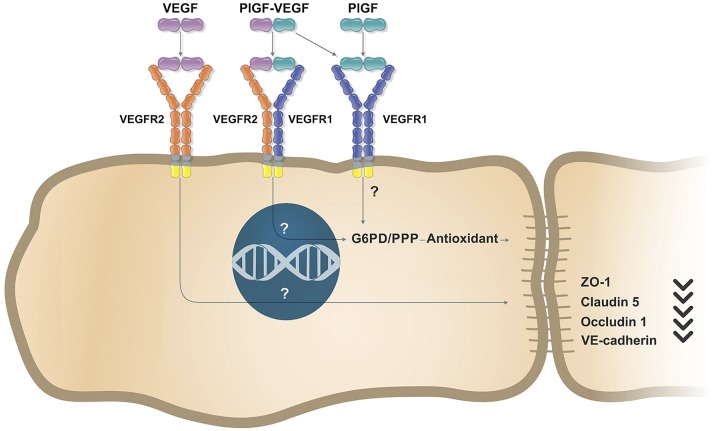
Although PlGF has been studied extensively for pathologic angiogenesis and inflammation, its role in vascular barrier function and permeability remains controversial. Through the use of the PlGF mAb (loss of function) and recombinant protein (gain of function) approaches, this study has unequivocally demonstrated that PlGF acts as a negative regulator of EC barrier function. It has also identified the novel mechanism whereby PlGF negatively regulates EC barrier function through G6PD (oxPPP) suppression and the antioxidant defense by the use of pharmacological inhibition (small inhibitor DHEA) and gene silencing (siRNA). In addition, the study has shown that PlGF regulates the G6PD (oxPPP)–antioxidant defense–EC barrier function pathway through the activation of VEGFR1 signaling and the interaction with VEGF-A (PlGF/VEGF dimer) to activate VEGFR2 signaling. The novel signaling pathway that regulates EC barrier function is illustrated in the schematic diagram in Fig. 8.
Figure 8.
Schematic diagram illustrating that PlGF signaling regulates EC barrier function through G6PD/PPP. PlGF/VEGFR1 or PlGF/VEGF-VEGFR1/R2 signaling regulates EC barrier function (e.g., tight junction and adherens junction proteins) through G6PD/PPP and antioxidant. The regulatory mechanism that VEGF-VEGFR2 signaling regulates EC barrier function or vascular permeability is different. The intracellular molecules that mediate the signaling transduction are unclear.
Growing evidence suggests that PlGF may act as a primary therapeutic target molecule in the treatment of DME. For example, the FDA-approved drug aflibercept (Eylea), which blocks not only VEGF-A/B but also PlGF, provides superior beneficial effects in the treatment of patients with DME to those of bevacizumab and ranibizumab, both of which exclusively target VEGF-A isoforms (17). VEGF-B has been predominantly characterized as performing nonvascular functions, such as antioxidant and insulin resistance (27, 28). Therefore, in patients with DME, it is plausible that PlGF could act as a primary target molecule in therapeutic interventions and contribute to the superior effect of aflibercept. However, several issues need to be resolved before a firm conclusion can be drawn. For example, is PlGF an active component of aflibercept in mediating the BRB function? If this is the case, does PlGF mediate the barrier function directly or indirectly through VEGF-A? The results of the present study unambiguously demonstrate that PlGF negatively affects EC barrier function through the use of its mAb and recombinant protein. These results, therefore, provide support for the argument that PlGF acts as a therapeutic target in treating DME in which BRB is impaired. In a recent study by Van Bergen et al. (29), PlGF neutralization was found to attenuate various DR features, including fibrosis, inflammation, and vascular leakage, in several animal models. In addition, 2 ongoing clinical trials were designed to apply the anti-PlGF antibody (THR-317) to the treatment of patients with DME. It is therefore appealing to determine the possible contribution of the PlGF/VEGFR1 (PlGF/VEGF-VEGFR1/R2)-G6PD (oxPPP)–antioxidant–EC barrier function pathway to the beneficial effect of anti-PlGF in animal models and humans.
As a member of the VEGF family and homolog to VEGF-A, PlGF has been mainly characterized as an angiogenetic factor, particularly in pathologic angiogenesis via interactions with VEGF-A and the activation of both VEGFR1 and VEGFR2, in addition to its proinflammatory role through its receptor VEGFR1. However, the role of PlGF in vascular permeability or barrier function has not been established as robustly as its roles in angiogenesis and inflammation; thus, this issue remains controversial under some circumstances. For example, Cai et al. (30) demonstrated that PlGF-1 (not PlGF-2) inhibited VEGF-induced vascular permeability through the stabilization of the tight junction and adherens proteins (e.g., VE-cadherin and claudin-5) 6 h after VEGF-A treatment.
The Behar-Cohen group showed that sustained PlGF overexpression disrupted the BRB (both inner and outer BRB) function and produced other DR characteristics in the cultures and the rats (12, 13). However, Deissler et al. (31) showed that PlGF failed to induce increased EC permeability with immortalized bovine ECs. Recent studies demonstrated the ability of PlGF genetic deletion to prevent BRB breakdown in DR that is associated with the up-regulation of antioxidant and neuroprotective proteins in the mouse retina (5, 14). The inconsistency of the PlGF function in vascular permeability or barrier function might be the result of the adoption of divergent experimental systems (i.e., different animal models and delivery methods) as well as the use of primary cell cultures vs. immortalized cell lines.
Interestingly the PlGF antibody treatment has increased occludin-1 expression among other barrier function proteins, whereas rhPlGF treatment decreased ZO-1, VE-cadherin, β-catenin, and claudin-5, but not occludin-1. VEGF-A treatment, as well as PlGF/VEGF heterodimers treatment, however, has decreased occludin-1 along with the rest of the barrier function proteins, hinting at a yet-unknown interaction or compensatory mechanism involving occludin-1 and VEGF and PlGF interaction.
The transcriptomic analysis comprehensively identified the genes that are regulated by PlGF in HRECs. Further functional classifications and pathway analyses revealed that the PPP, antioxidant defense system, TGF-β signaling pathway, adherent junction pathway, and other metabolic pathways were up-regulated by PlGF neutralization. The TGF-β signaling pathway regulates the interactions of ECs and pericytes that are critical for EC barrier maturation (32, 33). The adherent junction pathway is conducive to the establishment of tight EC-EC connections (34). Antioxidant systems, such as the PRDX family members (e.g., PRDX1, PRDX3, and PRDX6), can prevent EC barrier dysfunction by oxidative stress in diabetes (35). The PPP is one of the central glucose catabolic pathways that link glucose metabolism to ribose synthesis and NADPH production. Under physiologic conditions, the PPP is stringently regulated to use ∼5.5% glucose branched from the glycolysis pathway mainly through the control of G6PD activity, which is the first and rate-limiting PPP enzyme (36). G6PD activity can be regulated positively and negatively by a variety of factors. These include oncogene genes [e.g., PI3K and mammalian target of rapamycin complex 1 (mTORC1)], tumor suppression genes [e.g., p53, phosphatase and tensin homolog (PTEN), and AMPK] (37), and other factors, such as NF-κB, TP53-induced glycolysis regulatory phosphatase (TIGAR), and heat shock protein (HSP)27, that can affect the substrate of NADP+ and production of NADPH (38, 39).
The overactivation and underactivation of G6PD/PPP can have an adverse effect on cell growth, survival, and proliferation. For example, G6PD activity and PPP flux were increased in cancer cells to meet the demand for their rapid cell proliferation and mass production (37). However, G6PD activity was impaired in diabetes, and this resulted in high sensitivity to oxidative stress (40). Even G6PD deficiency per se can cause increased oxidative stress, NF-κB level, PKC activity, and elevated albuminuria. This is similar to the diabetic condition in mice (41). In patients with diabetes, PPP dysregulation and deficiency have been associated with DR (42, 43).
The results of the present study suggest that PlGF serves as a negative regulator of the G6PD/PPP in the endothelium. Therefore, its inhibition up-regulates G6PD protein expression and enzyme activity, thereby leading to enhanced EC barrier function. Because some critical enzymes of the nonoxidative branch of the PPP (e.g., TKT and TALDO1) were not affected by PlGF blockade, it is most likely that only the oxPPP, which is responsible for the production of NADPH, is involved in the pathway regulated by PlGF inhibition. Interestingly, a recent study by Quaegebeur et al. (44) found that increased oxPPP flux resulting from prolyl hydroxylase 1 (PHD1) deficiency protected the brain neurons in a murine stroke model against ischemic injury.
In summary, PlGF negatively regulates retinal vascular EC barrier function through suppression of the G6PD (oxPPP) and antioxidant defenses. PlGF mediates the negative effect on EC barrier function, G6PD (oxPPP), and antioxidant defenses through the activation of VEGFR1 signaling and interaction with VEGF-A (PlGF/VEGF dimer) to activate VEGFR2 signaling. PRDX6 plays dual roles in EC barrier function through its GPX and PLA2 enzyme activity. Further investigation should address the possible contribution of enhanced EC barrier function and increased G6PD (oxPPP) activity by the targeting of PlGF to the superior efficacy of aflibercept in the treatment of patients with DME. The possibility that the PlGF/VEGFR1 (PlGF/VEGF-VEGFR1/R2)-G6PD/oxPPP-antioxidant pathway might represent a universal mechanism that is applicable to other biologic systems should also be studied.
ACKNOWLEDGMENTS
The authors thank Dmitry Rumyancev (Belgorod, Russia) for graphical abstract artwork assets design. This work was supported by the U.S. National Institutes of Health, National Eye Institute (Grant R01 EY027824 to H.H.), and the Missouri University Start-Up Fund (to H.H.). The authors declare no conflicts of interest.
Glossary
- BRB
blood-retinal barrier
- BREC
bovine retinal EC
- BSA
bovine serum albumin
- DEG
differentially expressed gene
- DHEA
dehydroepiandrosterone
- DME
diabetic macular edema
- DR
diabetic retinopathy
- EC
endothelial cell
- ECIS
electrical cell-impedance sensing
- G6PD
glucose-6-phosphate dehydrogenase
- GPX
glutathione peroxidase
- HREC
human retinal EC
- LDH
lactate dehydrogenase
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- oxPPP
oxidative phase PPP
- PBS-T
PBS with Tween
- PLA2
phospholipase A2
- PlGF
placental growth factor
- PPP
pentose phosphate pathway
- PRDX
peroxiredoxin
- rb
recombinant bovine
- rh
recombinant human
- RNA-seq
RNA sequencing
- siRNA
small interfering RNA
- TALDO
transaldolase
- TEER
transendothelial electrical resistance
- TKT
transketolase
- VEGFR
VEGF receptor
- WB
Western blot, ZO-1, zonula occludens-1
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
H. Huang designed, performed, and analyzed experiments and wrote the manuscript; A. Lennikov and L. Fan performed and analyzed experiments, designed the figures, and edited the manuscript; M. Saddala performed the bioinformatics analyses for RNA-seq raw data; and D. Gozal, D. J. Grab, and A. Khalyfa consulted the project and edited the manuscript.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Maglione D., Guerriero V., Viglietto G., Delli-Bovi P., Persico M. G. (1991) Isolation of a human placenta cDNA coding for a protein related to the vascular permeability factor. Proc. Natl. Acad. Sci. USA 88, 9267–9271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmeliet P., Moons L., Luttun A., Vincenti V., Compernolle V., De Mol M., Wu Y., Bono F., Devy L., Beck H., Scholz D., Acker T., DiPalma T., Dewerchin M., Noel A., Stalmans I., Barra A., Blacher S., VandenDriessche T., Ponten A., Eriksson U., Plate K. H., Foidart J. M., Schaper W., Charnock-Jones D. S., Hicklin D. J., Herbert J. M., Collen D., Persico M. G. (2001) Synergism between vascular endothelial growth factor and placental growth factor contributes to angiogenesis and plasma extravasation in pathological conditions. Nat. Med. 7, 575–583 [DOI] [PubMed] [Google Scholar]
- 3.Huang H., Shen J., Vinores S. A. (2011) Blockade of VEGFR1 and 2 suppresses pathological angiogenesis and vascular leakage in the eye. PLoS One 6, e21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang H., Parlier R., Shen J. K., Lutty G. A., Vinores S. A. (2013) VEGF receptor blockade markedly reduces retinal microglia/macrophage infiltration into laser-induced CNV. PLoS One 8, e71808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H., He J., Johnson D., Wei Y., Liu Y., Wang S., Lutty G. A., Duh E. J., Semba R. D. (2015) Deletion of placental growth factor prevents diabetic retinopathy and is associated with Akt activation and HIF1α-VEGF pathway inhibition. Diabetes 64, 200–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmeliet P., Jain R. K. (2011) Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verlohren S., Herraiz I., Lapaire O., Schlembach D., Moertl M., Zeisler H., Calda P., Holzgreve W., Galindo A., Engels T., Denk B., Stepan H. (2012) The sFlt-1/PlGF ratio in different types of hypertensive pregnancy disorders and its prognostic potential in preeclamptic patients. Am. J. Obstet. Gynecol. 206, 58.e1-8. [DOI] [PubMed] [Google Scholar]
- 8.Cianfarani F., Zambruno G., Brogelli L., Sera F., Lacal P. M., Pesce M., Capogrossi M. C., Failla C. M., Napolitano M., Odorisio T. (2006) Placenta growth factor in diabetic wound healing: altered expression and therapeutic potential. Am. J. Pathol. 169, 1167–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luttun A., Tjwa M., Moons L., Wu Y., Angelillo-Scherrer A., Liao F., Nagy J. A., Hooper A., Priller J., De Klerck B., Compernolle V., Daci E., Bohlen P., Dewerchin M., Herbert J. M., Fava R., Matthys P., Carmeliet G., Collen D., Dvorak H. F., Hicklin D. J., Carmeliet P. (2002) Revascularization of ischemic tissues by PlGF treatment, and inhibition of tumor angiogenesis, arthritis and atherosclerosis by anti-Flt1. Nat. Med. 8, 831–840 [DOI] [PubMed] [Google Scholar]
- 10.Snuderl M., Batista A., Kirkpatrick N. D., Ruiz de Almodovar C., Riedemann L., Walsh E. C., Anolik R., Huang Y., Martin J. D., Kamoun W., Knevels E., Schmidt T., Farrar C. T., Vakoc B. J., Mohan N., Chung E., Roberge S., Peterson T., Bais C., Zhelyazkova B. H., Yip S., Hasselblatt M., Rossig C., Niemeyer E., Ferrara N., Klagsbrun M., Duda D. G., Fukumura D., Xu L., Carmeliet P., Jain R. K. (2013) Targeting placental growth factor/neuropilin 1 pathway inhibits growth and spread of medulloblastoma. Cell 152, 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al Kahtani E., Xu Z., Al Rashaed S., Wu L., Mahale A., Tian J., Abboud E. B., Ghazi N. G., Kozak I., Gupta V., Arevalo J. F., Duh E. J. (2017) Vitreous levels of placental growth factor correlate with activity of proliferative diabetic retinopathy and are not influenced by bevacizumab treatment. Eye (Lond.) 31, 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyamoto N., de Kozak Y., Jeanny J. C., Glotin A., Mascarelli F., Massin P., BenEzra D., Behar-Cohen F. (2007) Placental growth factor-1 and epithelial haemato-retinal barrier breakdown: potential implication in the pathogenesis of diabetic retinopathy. Diabetologia 50, 461–470 [DOI] [PubMed] [Google Scholar]
- 13.Kowalczuk L., Touchard E., Omri S., Jonet L., Klein C., Valamanes F., Berdugo M., Bigey P., Massin P., Jeanny J. C., Behar-Cohen F. (2011) Placental growth factor contributes to micro-vascular abnormalization and blood-retinal barrier breakdown in diabetic retinopathy. PLoS One 6, e17462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saddala M. S., Lennikov A., Grab D. J., Liu G. S., Tang S., Huang H. (2018) Proteomics reveals ablation of PlGF increases antioxidant and neuroprotective proteins in the diabetic mouse retina. Sci. Rep. 8, 16728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Bergen T., Etienne I., Cunningham F., Moons L., Schlingemann R. O., Feyen J. H. M., Stitt A. W. (2019) The role of placental growth factor (PlGF) and its receptor system in retinal vascular diseases. Prog. Retin. Eye Res. 69, 116–136 [DOI] [PubMed] [Google Scholar]
- 16.Nguyen Q. D., De Falco S., Behar-Cohen F., Lam W. C., Li X., Reichhart N., Ricci F., Pluim J., Li W. W. (2018) Placental growth factor and its potential role in diabetic retinopathy and other ocular neovascular diseases. Acta Ophthalmol. 96, e1–e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells J. A., Glassman A. R., Ayala A. R., Jampol L. M., Aiello L. P., Antoszyk A. N., Arnold-Bush B., Baker C. W., Bressler N. M., Browning D. J., Elman M. J., Ferris F. L., Friedman S. M., Melia M., Pieramici D. J., Sun J. K., Beck R. W.; Diabetic Retinopathy Clinical Research Network (2015) Aflibercept, bevacizumab, or ranibizumab for diabetic macular edema. N. Engl. J. Med. 372, 1193–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonelli-Orlidge A., Saunders K. B., Smith S. R., D’Amore P. A. (1989) An activated form of transforming growth factor beta is produced by cocultures of endothelial cells and pericytes. Proc. Natl. Acad. Sci. USA 86, 4544–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giaever I., Keese C. R. (1991) Micromotion of mammalian cells measured electrically. Proc. Natl. Acad. Sci. USA 88, 7896–7900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saddala M. S., Lennikov A., Mukwaya A., Fan L., Hu Z., Huang H. (2019) Transcriptome-wide analysis of differentially expressed chemokine receptors, SNPs, and SSRs in the age-related macular degeneration. Hum. Genomics 13, 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolger A. M., Lohse M., Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H., Van de Veire S., Dalal M., Parlier R., Semba R. D., Carmeliet P., Vinores S. A. (2011) Reduced retinal neovascularization, vascular permeability, and apoptosis in ischemic retinopathy in the absence of prolyl hydroxylase-1 due to the prevention of hyperoxia-induced vascular obliteration. Invest. Ophthalmol. Vis. Sci. 52, 7565–7573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rupprecht K. R., Nair R. K., Harwick L. C., Grote J., Beligere G. S., Rege S. D., Chen Y. Y., Lin Z., Fishpaugh J. R. (2010) Development of a dot-blot assay for screening monoclonal antibodies to low-molecular-mass drugs. Anal. Biochem. 407, 160–164 [DOI] [PubMed] [Google Scholar]
- 24.Sternberg J., Jeppesen P. (1983) Dot-blotting--a novel screening assay for antibodies in hybridoma cultures. J. Immunol. Methods 64, 39–43 [DOI] [PubMed] [Google Scholar]
- 25.Verbeke G., Molenberghs G. (2000) Linear Mixed Models for Longitudinal Data, Springer-Verlag, Inc, New York [Google Scholar]
- 26.Fisher A. B. (2018) The phospholipase A2 activity of peroxiredoxin 6. J. Lipid Res. 59, 1132–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arjunan P., Lin X., Tang Z., Du Y., Kumar A., Liu L., Yin X., Huang L., Chen W., Chen Q., Ye Z., Wang S., Kuang H., Zhou L., Xu K., Chen X., Zeng H., Lu W., Cao Y., Liu Y., Zhao C., Li X. (2018) VEGF-B is a potent antioxidant. Proc. Natl. Acad. Sci. USA 115, 10351–10356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagberg C. E., Mehlem A., Falkevall A., Muhl L., Fam B. C., Ortsäter H., Scotney P., Nyqvist D., Samén E., Lu L., Stone-Elander S., Proietto J., Andrikopoulos S., Sjöholm A., Nash A., Eriksson U. (2012) Targeting VEGF-B as a novel treatment for insulin resistance and type 2 diabetes. Nature 490, 426–430 [DOI] [PubMed] [Google Scholar]
- 29.Van Bergen T., Hu T. T., Etienne I., Reyns G. E., Moons L., Feyen J. H. M. (2017) Neutralization of placental growth factor as a novel treatment option in diabetic retinopathy. Exp. Eye Res. 165, 136–150 [DOI] [PubMed] [Google Scholar]
- 30.Cai J., Wu L., Qi X., Shaw L., Li Calzi S., Caballero S., Jiang W. G., Vinores S. A., Antonetti D., Ahmed A., Grant M. B., Boulton M. E. (2011) Placenta growth factor-1 exerts time-dependent stabilization of adherens junctions following VEGF-induced vascular permeability. PLoS One 6, e18076 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Deissler H. L., Deissler H., Lang G. K., Lang G. E. (2013) VEGF but not PlGF disturbs the barrier of retinal endothelial cells. Exp. Eye Res. 115, 162–171 [DOI] [PubMed] [Google Scholar]
- 32.Obermeier B., Daneman R., Ransohoff R. M. (2013) Development, maintenance and disruption of the blood-brain barrier. Nat. Med. 19, 1584–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Z., Nelson A. R., Betsholtz C., Zlokovic B. V. (2015) Establishment and dysfunction of the blood-brain barrier. Cell 163, 1064–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antonetti D. A., Klein R., Gardner T. W. (2012) Diabetic retinopathy. N. Engl. J. Med. 366, 1227–1239 [DOI] [PubMed] [Google Scholar]
- 35.He M., Siow R. C., Sugden D., Gao L., Cheng X., Mann G. E. (2011) Induction of HO-1 and redox signaling in endothelial cells by advanced glycation end products: a role for Nrf2 in vascular protection in diabetes. Nutr. Metab. Cardiovasc. Dis. 21, 277–285 [DOI] [PubMed] [Google Scholar]
- 36.Alving A. S., Carson P. E., Flanagan C. L., Ickes C. E. (1956) Enzymatic deficiency in primaquine-sensitive erythrocytes. Science 124, 484–485 [DOI] [PubMed] [Google Scholar]
- 37.Jiang P., Du W., Wu M. (2014) Regulation of the pentose phosphate pathway in cancer. Protein Cell 5, 592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bierhansl L., Conradi L. C., Treps L., Dewerchin M., Carmeliet P. (2017) Central role of metabolism in endothelial cell function and vascular disease. Physiology (Bethesda) 32, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cosentino C., Grieco D., Costanzo V. (2011) ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 30, 546–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z., Apse K., Pang J., Stanton R. C. (2000) High glucose inhibits glucose-6-phosphate dehydrogenase via cAMP in aortic endothelial cells. J. Biol. Chem. 275, 40042–40047 [DOI] [PubMed] [Google Scholar]
- 41.Xu Y., Zhang Z., Hu J., Stillman I. E., Leopold J. A., Handy D. E., Loscalzo J., Stanton R. C. (2010) Glucose-6-phosphate dehydrogenase-deficient mice have increased renal oxidative stress and increased albuminuria. FASEB J. 24, 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cappai G., Songini M., Doria A., Cavallerano J. D., Lorenzi M. (2011) Increased prevalence of proliferative retinopathy in patients with type 1 diabetes who are deficient in glucose-6-phosphate dehydrogenase. Diabetologia 54, 1539–1542 [DOI] [PubMed] [Google Scholar]
- 43.Chen L., Cheng C. Y., Choi H., Ikram M. K., Sabanayagam C., Tan G. S., Tian D., Zhang L., Venkatesan G., Tai E. S., Wang J. J., Mitchell P., Cheung C. M., Beuerman R. W., Zhou L., Chan E. C., Wong T. Y. (2016) Plasma metabonomic profiling of diabetic retinopathy. Diabetes 65, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 44.Quaegebeur A., Segura I., Schmieder R., Verdegem D., Decimo I., Bifari F., Dresselaers T., Eelen G., Ghosh D., Davidson S. M., Schoors S., Broekaert D., Cruys B., Govaerts K., De Legher C., Bouché A., Schoonjans L., Ramer M. S., Hung G., Bossaert G., Cleveland D. W., Himmelreich U., Voets T., Lemmens R., Bennett C. F., Robberecht W., De Bock K., Dewerchin M., Ghesquière B., Fendt S. M., Carmeliet P. (2016) Deletion or inhibition of the oxygen sensor PHD1 protects against ischemic stroke via reprogramming of neuronal metabolism. Cell Metab. 23, 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.