Abstract
Huntington's disease (HD) is a fatal hereditary neurodegenerative disorder, best known for its clinical triad of progressive motor impairment, cognitive deficits and psychiatric disturbances, is caused by CAG-repeat expansion in exon 1 of Huntingtin (HTT). However, in addition to the neurological disease, mutant HTT (mHTT), which is ubiquitously expressed in all tissues, impairs other organ systems. Not surprisingly, cardiovascular dysautonomia as well as the deterioration of circadian rhythms are among the earliest detectable pathophysiological changes in individuals with HD. Mitochondrial dysfunction in the brain and skeletal muscle in HD has been well documented, as the disease progresses. However, not much is known about mitochondrial abnormalities in the heart. In this study, we describe a role for Drp1/Fis1-mediated excessive mitochondrial fission and dysfunction, associated with lysosomal dysfunction in H9C2 expressing long polyglutamine repeat (Q73) and in human iPSC-derived cardiomyocytes transfected with Q77. Expression of long polyglutamine repeat led to reduced ATP production and mitochondrial fragmentation. We observed an increased accumulation of damaged mitochondria in the lysosome that was coupled with lysosomal dysfunction. Importantly, reducing Drp1/Fis1-mediated mitochondrial damage significantly improved mitochondrial function and cell survival. Finally, reducing Fis1-mediated Drp1 recruitment to the mitochondria, using the selective inhibitor of this interaction, P110, improved mitochondrial structure in the cardiac tissue of R6/2 mice. We suggest that drugs focusing on the central nervous system will not address mitochondrial function across all organs, and therefore will not be a sufficient strategy to treat or slow down HD disease progression.
Keywords: Drp1, Fis1, P110, Huntington’s disease, Bioenergetics, Mitochondria, Heart
1. Introduction
Huntington's disease (HD) is an autosomal-dominant inherited neurodegenerative disease, caused by an abnormal expansion of a CAG repeat located in the exon 1 of the gene encoding for the Huntingtin protein (HTT) and predominantly characterized by chorea, dyskinesia, and dystonia (1). The age of disease onset inversely correlates with CAG expansion length, and death occurs on average 10 to15 years after symptom onset (1, 2). Although there is no cure for HD, recent developments using anti-sense oligonucleotide (ASO) and adeno-associated virus (AAV)-mediated gene therapies to reduce HTT levels in the central nervous system (CNS) have demonstrated safety and efficacy in animal models and are progressing in clinical trials (3, 4). Since HTT expression is ubiquitous throughout the body, CNS-specific therapies may unmask peripheral HD-associated pathologies (5). In particular, skeletal muscle atrophy, cardiovascular diseases and heart failure have been identified as other leading causes of mortality in HD patients (6, 7).
Mitochondria are highly dynamic organelles, forming either elongated or fragmented structures through the processes of mitochondrial fusion and fission (8). This mitochondrial dynamic is essential for the maintenance of normal mitochondrial function, enabling mitochondrial quality control and mobility throughout the cell. Under physiological conditions, Drp1, a cytosolic large GTPase, is recruited to the outer mitochondrial membrane by mitochondrial fission factor (Mff), with yet to be fully characterized roles for mitochondrial dynamics proteins of 49 kDa and 51 kDa (Mid49/Mid51) (9, 10). However, under pathological stress, Drp1 predominantly interacts with fission 1 (Fis1), leading to excessive mitochondrial fragmentation, production of reactive oxygen species (ROS), and oxidative stress and loss of mitochondrial membrane potential (11). Under these conditions, selective inhibition of Drp1/Fis1 interaction, using P110, a rationally designed peptide inhibitor (11), provides a potential therapeutic intervention (12-14). In this study, we determined whether Drp1 hyperactivation through its interaction with Fis1 plays a role in the pathogenesis of polyglutamine induced cardiac mitochondrial dysfunction and whether inhibition of Drp1/Fis1-interaction can alleviate this cardiac pathology.
2. Materials and Methods
2.1. Peptides
Drp1/Fis1 interaction peptide inhibitor, P110, and control peptide (used for peptide delivery, TAT47-57) were synthesized by Ontores Biotechnologies (Hangzhou, China) (15). The purity of peptides was > 90% as measured by RP-HPLC chromatogram.
2.2. Peptide treatment in mouse model
All the experiments were done in accordance with protocols approved by the Institutional Animal Care and Use Committee of Stanford University and were performed based on the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Hemizygous R6/2 HD mice and their WT littermates were purchased from The Jackson Laboratory and shipped to us at 5-weeks of age.
2.3. Cell lines and transient expression of mutant proteins
H9C2 cardiomyocytes were cultured in either 75 cm2 flasks, 6-well or 24-well dishes, containing DMEM medium supplemented with 10% FBS, 4 mM glutamine, 100 U/ml penicillin, 10 μg/ml streptomycin and grown in a 5% CO2 atmosphere at 37 °C. H9C2 cardiomyocytes were seeded and allowed to attach before transfection with either green fluorescent protein (GFP)-Q23 (control) or Q73 (HD-like model), using the transfection reagent, according to the instructions provided by the manufacturer (Lipofectamine 2000 reagent, Invitrogen) and studied 48 h after transfection.
2.4. Culture and cardiac differentiation of human iPSCs.
A human iPSC line described previously (16) was cultured on Matrigel-coated plates (ES qualified, BD Biosciences, San Diego), using chemically defined E8 medium and culture conditions, as described (17, 18). Cells were passaged using Accutase (Global Cell Solutions) every four days. For expression of the Q23 and Q77 repeats, respective DNA sequences were subcloned into a pCDH-MSCV-GFP plasmid. Stable iPSC lines expressing the Q23- and Q77 repeat-GFP fusion proteins were generated by lentiviral transduction. Following lentiviral transduction, GFP-positive human iPSCs were sorted via fluorescent activated cell sorting (FACS) and expanded. For cardiac differentiation, human iPSCs stably expressing the Q23- and Q77-GFP fusion proteins were grown to 90% confluence. A small molecule-based monolayer method described in Ebert et al (19), was employed subsequently for differentiation of iPSCs into beating cardiomyocytes. Cardiomyocytes were used 30 days after differentiation, and their characterization has been previously described (19).
2.5. Cell health assays
Cell viability was determined by measuring the activity of lactate dehydrogenase (LDH) released into the culture media using a Cytotoxicity detection kit (LDH; Roche) according to the manufacturer’s instructions. Briefly, at the end of the incubation, media supernatants were collected and centrifuged at 1000 × g to remove any residual cells and cell debris. The reaction mixture was added to the cell-free supernatant, and color development was measured spectrophotometrically at 492 nm, with a reference wavelength at 610 nm. Caspase-3 and −9 activities were assessed using a caspase-3 or −9 colorimetric assay kit (Abcam, Cambridge, MA, USA) according to the manufacturer's protocol. Briefly, the cells were harvested and resuspended in lysis buffer. Following incubation on ice for 30 min, cell lysates were centrifuged at 11,000 g for 15 min at 4°C, and the protein concentration in the supernatants was measured using the Bradford method. The supernatants were incubated with reaction buffer containing 2 mmol/L Ac-DEVD-AFC for caspase-3 and LEHD-AFC for caspase-9 (Abcam) in a caspase assay buffer at 37°C with 10 mmol/L DTT for 30 min. Caspase activity was determined by measuring the absorbance at 405 nm. Aconitase activity was measured using an Aconitase Assay Kit (Sigma, MAK051). To determine the aconitase activity of harvested cells, the manufacturer’s protocol was followed without the addition of the activating solution. To determine mitochondrial ROS production, cells were treated with 5 μM MitoSOX™ Red mitochondrial superoxide indicator (Invitrogen) for 10 min at 37°C according to the manufacturer’s protocol, and fluorescence was analyzed with excitation/emission at 510/580 nm using SpectraMax M2e (Molecular devices). Cells were incubated with tetramethylrhodamine methyl ester (TMRM, Invitrogen) in HBSS (Hank's balanced salt solution) for 30 min at 37°C, as per the manufacture's protocol, and the fluorescence was analyzed using SpectraMax M2e (Molecular devices, using excitation at 360 nm and emission at 460 nm). Cyto-ID Green Detection Reagent Kit (Enzo Life Sciences) detects autophagosomes and autolysosomes as recommended by manufacturer. In brief, cells were stained with Cyto-ID Green for 30 minutes at 37°C and fluorescence levels were measured using SpectraMax M2e (Molecular devices, using excitation at 480 nm and emission at 530 nm).
2.6. Seahorse analysis
Oxygen consumption rate (OCR) was measured with an XFe24 Extracellular Flux Analyzer (Seahorse Bioscience). Cells were seeded in an XF 24-well cell culture microplate (Seahorse Bioscience) in 250 μl of DMEM and incubated for 24 h at 37°C under a 5% CO2 atmosphere. The growth medium was replaced with 575 μl of pre-warmed bicarbonate-free DMEM, pH 7.4, and cells were incubated at 37°C for 1 h before starting the assay. After baseline measurements of OCR and ECAR, OCR was measured after sequentially adding to each well 75 μl of oligomycin, 75 μl of FCCP, 75 μl of rotenone and 75 μl of antimycin A to reach working concentrations of (1 μM, 2 μM and 0.5 μM respectively). OCR and ECAR values were normalized to protein content measured by sulforhodamine (SRB) assay as described before (13).
2.7. Immunofluorescence analysis
Immunocytochemistry were performed as described previously (14). Cells seeded at 12,000 , were cultured on 8-well chamber slides for 24 hours after 48-hour transfection, washed with cold PBS, fixed in 4% formaldehyde, and permeabilized with 0.1% Triton X-100. After incubation with 2% normal goat serum (to block nonspecific staining), fixed cells were incubated overnight at 4°C with anti-TOM20 primary antibody (1:500; Santa Cruz, USA). Cells were washed with PBS and incubated for 60 minutes with FITC-conjugated goat anti-rabbit IgG (1:500 dilution). The cells were then washed gently with PBS and counterstained with Hoechst 33342 (1:10,000 dilution, Molecular Probes) to visualize nuclei. The coverslips were mounted with Slowfade-antifade reagent (Invitrogen). Parameters of mitochondrial morphology were further quantified with Fiji (ImageJ) as described before (20). Images were acquired using an All-in-One Fluorescence Microscope BZ-X700 (Keyence) and analyzed using ImageJ, as described (21).
2.8. Lysate preparation and isolation of mitochondria-enriched fraction and lysosomal fraction
Cells were washed with cold phosphate-buffered saline (PBS) at pH 7.4 and scraped off using mannitol–sucrose (MS) buffer containing 210 mM mannitol, 70 mM sucrose, 5 mM MOPS (3-(N-morpholino) propanesulfonic acid), 1 mM EDTA, and protease inhibitor cocktail, pH 7.4. The collected cells were passed through a 27-gauge ½-inch needle for lysis, followed by centrifugation at 800 × g to pellet nuclei. The post-nuclear and cell debris supernatant was further centrifuged at 10,000 × g for 20 min to collect a mitochondria-enriched fraction, as previously described (14). Lysosomal fractions were extracted from cell homogenates by differential centrifugation followed by density centrifugation according to the manufacturer's protocol (LYSISO1; Sigma-Aldrich) and as previously described (22). The integrity of the lysosomal membrane was monitored by analyzing the presence of lysosomal enzyme, acid phosphatase, in the cytosol. Phosphatase activity was assayed with the help of the acid phosphatase assay kit (CS0740; Sigma), according to the manufacturer’s instructions. The quantification of the lysosomal pH was performed using LysoSensor™ Yellow/Blue DND-160 (L7545; Invitrogen) according to the manufacturer’s instructions.
2.9. RNA isolation and gene expression analysis
RNA isolation was performed using GenElute™ Mammalian Total RNA Miniprep Kit, according to manufacturer’s protocols. RNA concentration was measured using a Nanodrop (ND −1000; NanoDrop Technologies, Rockland, DE, USA). cDNA synthesis was performed using the High-Capacity cDNA Reverse Transcription Kit (4368814; Invitrogen) according to manufacturer’s instructions, with a minimal input of 200 ng total RNA. Quantitative PCR (qPCR) was performed using the 7300 Real Time PCR system (Applied Biosystems, Foster City, USA) using the equivalent cDNA amount of 1-2 ng total RNA used in cDNA synthesis. SYBR green master-mix (Applied Biosystems) and a 2 pmol/ml mix of forward and reverse primer sequences were used for 40 cycles of target gene amplification. The primers used in this study are in Supplementary Data Table 1.
2.10. Western blot
Protein concentrations were determined using Bradford assay (Thermo Fisher Scientific). Proteins were resuspended in Laemmli buffer containing 2-mercaptoethanol, loaded on SDS–PAGE, and transferred on to nitrocellulose membrane, 0.45 μm (Bio-Rad), as before (14). Membranes were probed with the indicated antibody and then visualized by ECL (0.225 mM p-coumaric acid; Sigma), 1.25 mM 3-aminophthalhydrazide (Luminol; Fluka) in 1 M Tris pH 8.5 (GE Healthcare, NA931 (anti-mouse), NA934 (anti-rabbit)). Blots were quantitated using ImageJ. Quantification was performed on samples from independent cultures for each condition. The antibodies used in this study are in Supplementary Data Table 2.
2.11. Transmission electron microscopy
Hearts from wild-type mice and HD R6/2 transgenic mice were fixed (after excision) with a solution containing 2% (v/v) glutaraldehyde and 4% formaldehyde in 0.1 M sodium cacodylate, pH 7.4, for 15 min at room temperature. The fixed samples were then treated at the Stanford Electron Microscopy Facility for imaging, as previously described (14). Images were acquired using a JEOL1400 transmission electron microscope.
2.12. Statistical analysis
No statistical methods were used to predetermine sample size. Results were expressed as mean ± s.d. Statistical analyses were performed using GraphPad Prism 6. All experimental analysis was performed by personnel blinded to the conditions and the genotypes.
3. Results
3.1. Drp1/Fis1-mediated hyperactivation causes mitochondrial fragmentation in cardiac cells expressing expanded polyglutamine repeats
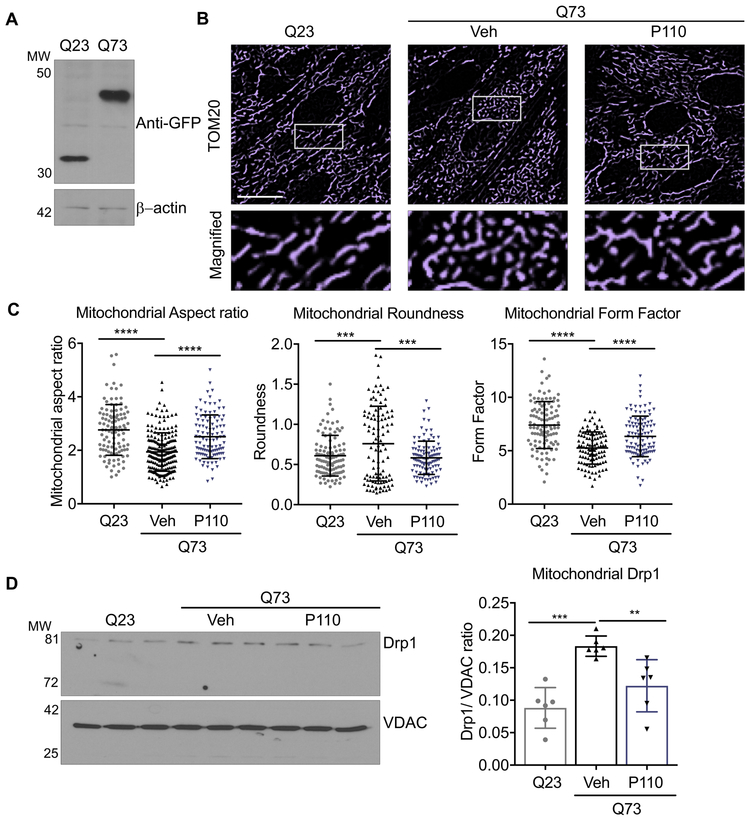
Exon 1 of the huntingtin gene followed by long polyglutamine repeats is sufficient to form intracellular aggregates and cellular toxicity. Therefore, its expression in cells provides a model to study HD. Here, we transiently transfected H9C2 cardiomyocytes with EGFP-tagged exon 1 of huntingtin with 23 or 73 polyglutamine repeats (Q23 or Q73) (Fig. 1A) for 48 hours. Expression of Q73 but not Q23 led to increased mitochondrial structural defects, with an increased number of small, rounded mitochondria and loss of mitochondrial interconnected network (Fig. 1B). Using computer-assisted morphometric analyses, we quantified the changes in mitochondrial structure, measuring mitochondrial aspect ratio (AR), roundness and form factor (FF) (Fig. 1C). We then determined if these mitochondrial structural defects were mediated by Drp1/Fis1 interaction, using P110, a heptapeptide conjugated to TAT47–57 (TAT, for intracellular delivery) that selectively inhibits the interaction between Drp1 and Fis1, one of its adaptor proteins on mitochondria . P110 treatment (1 μM for 24 hours) significantly improved mitochondrial structural defects as represented across the three measured parameters (Fig. 1C). Drp1 association with the mitochondria is a hallmark of activated fission machinery, and the levels of mitochondrial Drp1 upon Q73 expression in these cells, which increased by 2-folds, was significantly reduced following P110 treatment (Fig. 1D).
Figure 1. Q73 expression in H9C2 cells induces mitochondrial damage mediated through Drp1 hyperactivation.
A. Expression of polyglutamine repeats (Q23 or Q73) in H9C2 cardiomyocytes. B. H9C2 cardiomyocytes were transfected with either Q23 or Q73 repeats and after 48 hours were treated with P110 (1μM once) or Veh, for an additional 24 h in serum free- galactose medium and stained with anti-TOM20 (a marker of mitochondria, 1:500 dilution). Scale bar: 20 μm. 5 independent experiments. C. Mitochondrial aspect, roundness and form factor were determined. D. Levels of mitochondrial Drp1 were determined by immunoblotting, quantified and presented as ratio vs VDAC, loading control, treated as in (B). (n=6/group). Data were evaluated by one-way ANOVA and Holm-Sidak's multiple comparisons test for multiple testing between each treatment group. **** p-value <0.0001; *** p-value <0.001; ** p-value <0.01. All graphs represent mean ± s.d.
3.2. Mitochondrial fragmentation in Q73-expressing H9C2 cells induces defects in mitochondrial integrity, autophagy and apoptosis.
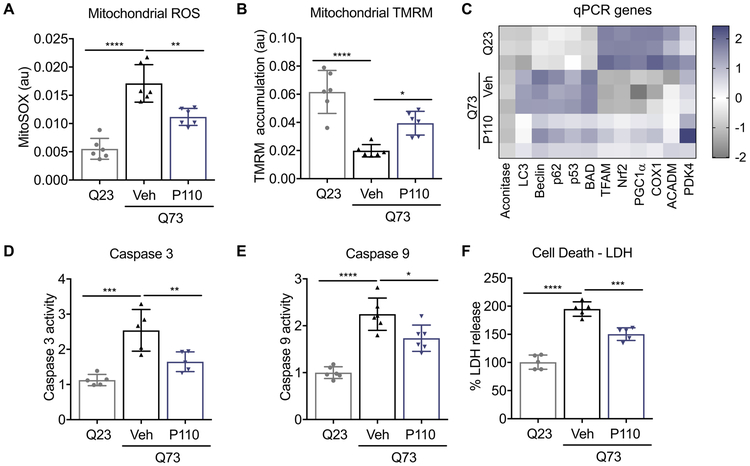
Q73 expression in H9C2 led to a marked increase in mitochondrial reactive oxygen species (mROS) as well as a significant reduction in mitochondrial membrane potential (Fig. 2A, B). P110 treatment (1 μM for 24 hrs) reduced mROS by 50% and improved mitochondrial membrane potential (Fig. 2A, B). Since mitochondrial integrity governs cell fate, we next measured the expression of genes associated with autophagy, mitochondrial biogenesis and cell death in cardiac cells (Fig. 2C). The expression of autophagy genes, LC3, Beclin and p62 as well as cell death genes p53 and BAD were significantly upregulated in Q73-expressing H9C2 cells as compared to Q23-H9C2. Furthermore, Q73 expression was sufficient to reduce the expression of critical mitochondrial associated genes, TFAM, Nrf2, PGC1-α and COX1, indicating a shift towards cellular stress. P110 treatment (1 μM) significantly reversed the overall gene expression in H9C2 cells expressing Q73, indicating a link between mitochondrial structure and cellular fate. Q73-induced gene upregulation of apoptosis-associated genes increased the activity of caspase 3 and caspase 9, which was also significantly reduced by P110 treatment (Fig. 2D, E). Finally, the expression of Q73 led to increased cellular toxicity (measured by increased LDH release to media), which was reduced upon P110 treatment (Fig. 2F).
Figure 2. Q73 expression in H9C2 cells induces defects in mitochondrial health, autophagy and apoptosis.
Mitochondrial health was evaluated using A. MitoSOX for mitochondrial ROS measurement and B. TMRM as a surrogate for mitochondrial membrane potential in H9C2 cardiomyocytes transfected with either Q23 or Q73 repeats for 48 hours and then treated with P110 (1μM once) or Veh, for an additional 24 h in serum free- galactose medium C. Heat map of RNA transcripts in these H9C2 cardiomyocytes, treated as in (A). D. Caspase 3 and E. Caspase 9 activities were measured using kits, in cells treated as in (A). LDH release as a surrogate for cell death in H9C2 cardiomyocytes transfected with either Q23 or Q73 repeats for 48 hours and then treated with P110 (1μM once) or Veh, for an additional 48 h in serum free- galactose medium. Data were evaluated by one-way ANOVA and Holm-Sidak's multiple comparisons test for multiple testing between each treatment group. **** p-value <0.0001; *** p-value <0.001; ** p-value <0.01; * p-value <0.05. All graphs represent mean ± s.d. (n= 5 or 6, as indicated in each panel).
3.3. Q73 expression in H9C2 cells leads to lysosomal dysfunction.
Previously, we demonstrated lysosomal defects in Q74-containing PC12 cells, resulting in accumulation of mitochondria in the lysosomal fraction. Similarly, we observed a significant increased accumulation of mitochondria in Q73-expressing H9C2 cells, as indicated by the presence of increased outer mitochondrial membrane proteins, mitofusin 2 (MFN2) and VDAC, in the lysosome-enriched membrane fractions (LAMP1) (Fig. 3A). Additionally, we also observed that the presence of expanded polyglutamine repeats in H9C2 significantly increased the amount of mitochondrial proteins, TOM20, and aconitase in the total lysates, possibly suggesting lack of mitochondrial elimination (Fig. 3B). Similar to observations in postmortem HD caudate samples (23), we also observed a significant decrease in aconitase activity (Fig. 3C). Since acidic pH is required for lysosomal activity, we used LysoSensor dye staining to evaluate whether Q73 expression affected lysosomal function in H9C2. Additionally, acid phosphatase activity assay was also performed as a secondary measure of lysosomal function. Q73 expression in H9C2 cells led to a marked decline in LysoSensor and acid phosphatase activities, indicating that Q73 expression causes lysosomal dysfunction, and this dysfunction was reduced with P110 treatment (Fig. 3D, E). Since lysosomal function and autophagic flux are intrinsically linked, the defects observed in lysosomal function translated into increased autophagosome accumulation indicative of a defect in autophagic flux (Fig. 3F). P110 treatment reduced this accumulation of autophagosomes, thereby improving the autophagic flux. Inhibition of autophagy and lysosomal machinery with 3-MA (10 mM) or chloroquine (20 μM) reduced autophagic flux and increased cell death in Q73 H9C2 cells, whereas rapamycin (100 nM) reduced cell death through an increase in autophagy (Fig 3G, H).
Figure 3. Q73 expression in H9C2 cells leads to lysosomal dysfunction.
A. Levels of Mfn2, VDAC and LAMP1 were determined by immunoblotting of lysosomal fraction from H9C2 cardiomyocytes transfected with either Q23 or Q73 repeats for 48 hours and then treated with P110 (1μM once) or Veh, for an additional 24 h in serum free- galactose medium. LAMP-1 was used as lysosomal marker, quantified and presented as ratio vs β-actin B. Levels of aconitase and TOM20 were determined by immunoblotting of total lysates, levels were quantified and presented as ratio vs actin (loading control), from cells treated as in A. C. Aconitase activity in H9C2 cardiomyocytes, treated as in A. D. Lysosomal function was measured using LysoSensor™ yellow/blue DND-160 in H9C2 cardiomyocytes, treated as in A. E. Acid phosphatase activity as a secondary readout for lysosomal function was measured in H9C2 cardiomyocytes, treated as in A. F. Autophagic flux was measured using Cyto-ID™ in H9C2 cardiomyocytes, treated as in A. G. Autophagic flux was measured using Cyto-ID™ in H9C2 cardiomyocytes transfected with either Q23 or Q73 repeats for 48 hours and then treated with 3-MA, chloroquine, rapamycin or Veh, for additional 24 h in serum free/galactose medium. H. LDH release was measured as a marker of cell death in H9C2 cardiomyocytes, treated as in G. Data were evaluated by one-way ANOVA and Holm-Sidak's multiple comparisons test for multiple testing between each treatment group. **** p-value <0.0001; *** p-value <0.001; ** p-value <0.01; * p-value <0.05. All graphs represent mean ± s.d. (n=4/5, as indicated in each panel.)
3.3. Mitochondrial bioenergetic failure correlates with lysosomal dysfunction in human iPSC-derived cardiomyocytes expressing expanded polyglutamine repeats.
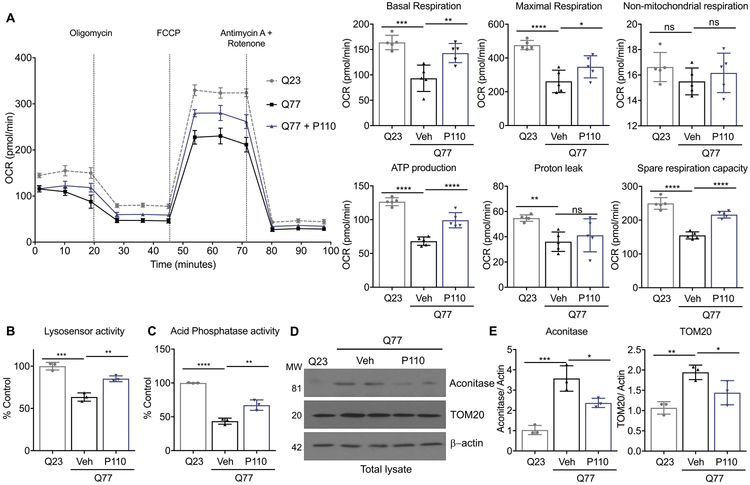
To maintain their acidic internal pH, lysosomes must actively concentrate H+ (protons). This is accomplished by a proton pump in the lysosomal membrane, which actively transports protons into the lysosome from the cytosol. This pumping requires expenditure of energy in the form of ATP hydrolysis, since it maintains approximately hundredfold higher H+ concentration inside the lysosome. Since loss of ATP affects lysosomal function (24), we measured bioenergetics in human iPSC-derived cardiomyocytes expressing the expanded polyglutamine repeats (Q77), using Seahorse XF24 extracellular flux analyzer and performed a baseline mitochondrial stress test (Fig. 4A). As expected, a significant decrease was observed in basal respiration, maximal mitochondrial respiration, and spare respiratory capacity, as assessed by subjecting cells to FCCP (carbonyl cyanide-4-(trifluoromethoxy)-phenylhydrazone) in Q77-expressing cells (Fig. 4A). Furthermore, baseline ATP production level was significantly lower in these cells. Importantly, blocking Drp1/Fis1 interaction with P110 (1μM, for 24 hrs) significantly improved these mitochondria dependent parameters. We also observed defects in lysosomal activities in Q77-expressing human iPSC-derived cardiomyocytes relative to Q23-expressing cells, which were remarkably recovered with P110 treatment, highlighting the interdependence of mitochondrial and lysosomal dysfunctions (Fig. 4B, C). Finally, mitochondrial accumulation, as measured by the levels of TOM20 and aconitase, were significantly reduced after P110 treatment in these cells (Fig. 4D, E).
Figure 4. Expanded poly-Q (Q77) expression in human iPSC-derived cardiomyocytes leads to bioenergetic failure and lysosomal dysfunction.
A. Metabolic health in human iPSC-derived cardiomyocytes stably expressing Q23 (control) or Q77 (as a HD model) 48 hours treated with P110 (1μM once24 h) or Veh, in serum free- galactose medium was measured with Seahorse extracellular flux analyzer. B. Lysosomal function was measured using LysoSensor™ Yellow/Blue DND-160 in iPSC-derived cardiomyocytes, treated as in A. C. Acid phosphatase activity as a secondary readout for lysosomal function was measured in iPSC-derived cardiomyocytes, treated as in A.D. Levels of aconitase (E) and TOM20 (F) were determined by immunoblotting in total lysates, quantified and presented as ratio vs actin, loading control, from cells treated as in A. Data were evaluated by one-way ANOVA and Holm-Sidak's multiple comparisons test for multiple testing between each treatment group. **** p-value <0.0001; *** p-value <0.001; ** p-value <0.01; * p-value <0.05. All graphs represent mean ± s.d. n=3-5, as indicated in each panel.
3.4. P110 treatment improves cardiac mitochondrial structure and promotes elimination of damaged mitochondria in hearts of R6/2 mice.
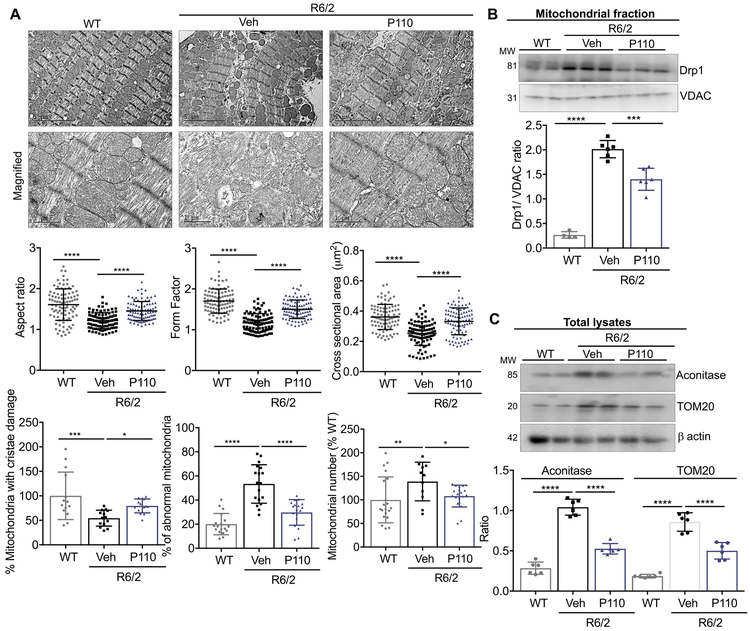
We next evaluated the mitochondrial ultrastructure in the hearts of 13-week-old R6/2 mice (an age correlating with severe HD symptoms; (15) (Fig. 5A). Healthy normal cardiac mitochondria generally present as elliptical, with uniform optical densities when observed under electron microscope. However, mitochondria in the hearts of R6/2 mice exhibited a more circular appearance, with a disruption of their uniform densities, and other ultrastructure differences. Using a computer-assisted analysis, we determined the mitochondrial aspect ratio, form factor, cross-sectional area and mitochondrial roundness in these images. All the measured parameters were significantly reduced in R6/2 mice as compared to these in hearts of WT-littermate mice indicative of damaged mitochondria (Fig. 5A). Furthermore, mitochondria were frequently abnormally shaped with damaged or lost cristae and the total number of mitochondria was elevated (**p < 0.01) in the hearts of R6/2 mice relative to those of WT littermates, suggesting defects in mitochondrial elimination. We previously described the beneficial effect of P110 treatment on neurodegeneration, behavior and life span of R6/2 mice (6). R6/2 mice treated with P110 inhibitor peptide or TAT (Veh; each at 3 mg/Kg/d), delivered by a subcutaneous osmotic pump for 8-weeks, as we described previously (20), markedly improved cardiac mitochondrial structure (Fig. 5A), reduced the increased mitochondrial Drp1 levels (Fig. 5B) and decreased mitochondrial accumulation, as evidenced by the levels of aconitase and TOM20 when comparing hearts of P110-treated R6/2 mice with vehicle-treated R6/2 mice (Fig. 5C).
Figure 5. Mitochondrial defects in hearts of R6/2 mice.
A. Electron microscopy analysis of mitochondria in heart from WT mice, R6/2 mice, and R6/2 mice treated with P110 for 8 weeks (3mg/Kg/day) as a continuous subcutaneous infusion). Normal mitochondria structure was dramatically reduced and mitochondria with damaged structure was higher in R6/2 mice, relative to hearts of littermate WT mice. Mitochondrial aspect ratio, form factor, cross sectional area, cristae damage, total number of damaged mitochondria and mitochondrial number were analyzed in each group by an observer blinded to the experimental conditions. B. Levels of mitochondrial Drp1 were determined by immunoblotting, quantified and presented as ratio vs VDAC, loading control in mitochondria enriched fractions from the heart. C. Levels of Aconitase and TOM20 were determined by immunoblotting in total heart lysates, quantified and presented as ratio vs actin, loading control. Data were evaluated by one-way ANOVA and Holm-Sidak's multiple comparisons test for multiple testing between each treatment group. **** p-value <0.0001; *** p-value <0.001; ** p-value <0.01; * p-value <0.05. All graphs represent mean ± s.d. (n=6/group).
4. Discussion
Many studies on HD and therapeutic interventions are focused solely on the brain pathology (25). However, it is becoming increasingly apparent that the heart is highly susceptible to the effects of mHTT (26-28). Like the brain, the heart is a highly metabolic organ. Unsurprisingly, HD patients exhibit a high rate of cardiac events, with heart failure being the second leading cause of death among HD patients (accounting for 20–30% of HD deaths) (6, 29, 30). However, not much is known about the cause of the cardiac dysfunction and the cells involved. In this study, we demonstrated that inhibition of Drp1/Fis1-mediated excessive mitochondrial fission and dysfunction using P110, a selective peptide inhibitor of Drp1/Fis1 interaction that we developed (6), reduced pathological mitochondrial fission in mouse and human cell models of HD as well as in an R6/2 HD mouse model.
Mitochondrial structure and function are intimately linked . Multiple studies, including ours, have demonstrated that reducing mitochondrial fragmentation is sufficient to restore mitochondrial functions (13-15, 31). Expression of expanded polyglutamine repeats in H9C2 cardiomyocytes resulted in a drastic increase in fragmented mitochondria, which was mediated through Drp1 association with Fis1 on the outer mitochondrial membrane; blocking this interaction with P110 significantly improved mitochondrial structure. Mitochondria are not only the main source of ATP; they also produce and are the target of ROS. Oxidative stress induces mitochondrial fragmentation and dysfunction (12, 32). We found that improvement in mitochondrial structure correlated with restoration of mitochondrial membrane potential, reducing mitochondrial ROS levels and increasing ATP production.
Alterations in mitochondrial function have been shown to impair communication between mitochondria and endoplasmic reticulum (ER). This crosstalk between the organelles has been identified as an important regulator of mitochondrial dynamics, lipid and calcium homeostasis, autophagy and apoptosis and disturbances in it, characterized by either an increase or a reduction of ER-mitochondria interactions, have been reported in several neurodegenerative and metabolic diseases (12, 33). Furthermore, there are many reports of mitochondrial mass increase under conditions of oxidative stress (34-36). These increases might be due to a defective autophagy and lysosomal pathways. In the present study, we observed an increase in mitochondrial mass in both Q73-expressing H9C2 cells as well as Q77-expressing human iPSC-derived cardiomyocytes. Furthermore, in contrast to the increase in mitochondrial mass, TFAM, Nrf2 and PGC-1α expression decreased, suggesting that enhanced mitochondrial biogenesis maybe not the primary cause of the mitochondrial accumulation. Since autophagy and lysosomal clearance requires ATP, this mitochondrial clearance impairment might be explained by the bioenergetic defects observed in these mouse and human cell models of HD. Importantly, by improving mitochondrial health through blocking excessive/pathological mitochondrial fragmentation with P110 treatment, we noted an improvement in lysosomal activities and thereby better clearance of mitochondria. Furthermore, we have previously demonstrated that blocking Drp1/Fis1-mediated mitochondrial fission reduces ER stress response (14). This, in turn, may reduce cellular stress, thus improving cell survival.
For effective mitochondrial clearance, mitochondrial fission is necessary. P110 inhibitory peptide is composed of a 7-amino acid peptide, representing a homology sequence between Drp1 and Fis1 (11). Importantly, P110 does not affect Drp1 interaction with any other mitochondrial adaptors of Drp1 nor with the mitochondrial fusion proteins, Mfn1 or Mfn2 (11, 14). P110 selectively inhibits excessive, but not basal, mitochondrial fission, thereby allowing normal mitophagy to occur (12-15) and a 5-month treatment of WT mice with P110 is safe and has no effect on mitochondrial structure or functions (24). Most importantly, in the R6/2 mouse model of HD, the mitochondrial structure in the heart was severely damaged and was associated with accumulation of damaged mitochondria as well as Drp1 hyperactivation. These detrimental modes of damage were corrected with sustained P110 treatment. Echocardiogram and other functional parameters to assess the benefit of P110 treatment in HD mice on myocardial functions have not yet been evaluated. Taken together, our study shows that expanded polyglutamine repeats cause mitochondrial and lysosomal dysfunctions in cultured mouse cardiac cell line and in human iPSc-derived cardiomyocytes as well as in an in vivo model of HD, which are greatly reduced by selective inhibition of mitochondrial fragmentation. Our study also indicates the need for therapeutic modality for HD that, in addition to targeting the CNS, will address the pathology in other organs, especially the heart.
Supplementary Material
Acknowledgements:
The authors thank Marie-Helene Disatnik her advice and John Perrino for technical support with EM. This work was supported, in part, by Takeda Pharmaceuticals’ Science Frontier Fund to DM-R. Although Takeda sponsored, in part, the cost of the project, experiments were designed and executed entirely at Stanford University and Takeda has no ownership on the findings.
Footnotes
Conflict of interest
Patents on P110 and its utility in HD, ALS and other neurodegenerative diseases have been filed by D.M-R and A.U.J, and were licensed to Mitoconix Bioscience, a company that DM-R founded and serves on its board. However, none of the work in her laboratory was carried out in collaboration with or with financial support from the company. AUJ advised the company, as part of technology transfer to the company, on his work related to Huntington's disease. The other authors declare that they have no conflict of interest.
References
- 1.Damiano M, Galvan L, Deglon N, Brouillet E. Mitochondria in Huntington's disease. Biochimica et biophysica acta. 2010;1802(1):52–61. [DOI] [PubMed] [Google Scholar]
- 2.Jodeiri Farshbaf M, Ghaedi K. Huntington's Disease and Mitochondria. Neurotox Res. 2017;32(3):518–529. [DOI] [PubMed] [Google Scholar]
- 3.McColgan P, Tabrizi SJ. Huntington's disease: a clinical review. European journal of neurology. 2018;25(1):24–34. [DOI] [PubMed] [Google Scholar]
- 4.Travessa AM, Rodrigues FB, Mestre TA, Ferreira JJ. Fifteen Years of Clinical Trials in Huntington's Disease: A Very Low Clinical Drug Development Success Rate. J Huntingtons Dis. 2017;6(2):157–163. [DOI] [PubMed] [Google Scholar]
- 5.Saudou F, Humbert S. The Biology of Huntingtin. Neuron. 2016;89(5):910–926. [DOI] [PubMed] [Google Scholar]
- 6.Sorensen SA, Fenger K. Causes of death in patients with Huntington's disease and in unaffected first degree relatives. J Med Genet. 1992;29(12):911–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mihm MJ, Amann DM, Schanbacher BL, Altschuld RA, Bauer JA, Hoyt KR. Cardiac dysfunction in the R6/2 mouse model of Huntington's disease. Neurobiology of disease. 2007;25(2):297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Archer SL. Mitochondrial dynamics--mitochondrial fission and fusion in human diseases. N Engl J Med. 2013;369(23):2236–2251. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Human molecular genetics. 2009;18(R2):R169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharp WW, Archer SL. Mitochondrial dynamics in cardiovascular disease: fission and fusion foretell form and function. J Mol Med (Berl). 2015;93(3):225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi X, Qvit N, Su Y-C, Mochly-Rosen D. A novel Drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. Journal of cell science. 2013;126:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joshi AU, Kornfeld OS, Mochly-Rosen D. The entangled ER-mitochondrial axis as a potential therapeutic strategy in neurodegeneration: A tangled duo unchained. Cell Calcium. 2016;60(3):218–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi AU, Saw NL, Shamloo M, Mochly-Rosen D. Drp1/Fis1 interaction mediates mitochondrial dysfunction, bioenergetic failure and cognitive decline in Alzheimer's disease. Oncotarget. 2018;9(5):6128–6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi AU, Saw NL, Vogel H, Cunnigham AD, Shamloo M, Mochly-Rosen D. Inhibition of Drp1/Fis1 interaction slows progression of amyotrophic lateral sclerosis. EMBO Mol Med. 2018;10(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Disatnik MH, Joshi AU, Saw NL, Shamloo M, Leavitt BR, Qi X, Mochly-Rosen D. Potential biomarkers to follow the progression and treatment response of Huntington's disease. J Exp Med. 2016;213(12):2655–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun N, Panetta NJ, Gupta DM, Wilson KD, Lee A, Jia F, Hu S, Cherry AM, Robbins RC, Longaker MT, Wu JC. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(37):15720–15725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, Shibata T, Kunisada T, Takahashi M, Takahashi J, Saji H, Yamanaka S. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–412. [DOI] [PubMed] [Google Scholar]
- 18.Garbern JC, Lee RT. Cardiac stem cell therapy and the promise of heart regeneration. Cell stem cell. 2013;12(6):689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebert AD, Kodo K, Liang P, Wu H, Huber BC, Riegler J, Churko J, Lee J, de Almeida P, Lan F, Diecke S, Burridge PW, Gold JD, Mochly-Rosen D, Wu JC. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Science translational medicine. 2014;6(255):255ra130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valente AJ, Maddalena LA, Robb EL, Moradi F, Stuart JA. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119(3):315–326. [DOI] [PubMed] [Google Scholar]
- 21.Dagda RK, Cherra SJ 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. The Journal of biological chemistry. 2009;284(20):13843–13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Fang Y, Yang Y, Qin Y, Wu P, Wang T, Lai H, Meng L, Wang D, Zheng Z, Lu X, Zhang H, Gao Q, Zhou J, Ma D. Elaiophylin, a novel autophagy inhibitor, exerts antitumor activity as a single agent in ovarian cancer cells. Autophagy. 2015;11(10):1849–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabrizi SJ, Cleeter MW, Xuereb J, Taanman JW, Cooper JM, Schapira AH. Biochemical abnormalities and excitotoxicity in Huntington's disease brain. Ann Neurol. 1999;45(1):25–32. [DOI] [PubMed] [Google Scholar]
- 24.Demers-Lamarche J, Guillebaud G, Tlili M, Todkar K, Belanger N, Grondin M, Nguyen AP, Michel J, Germain M. Loss of Mitochondrial Function Impairs Lysosomes. The Journal of biological chemistry. 2016;291(19):10263–10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank S Treatment of Huntington's disease. Neurotherapeutics. 2014;11(1):153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cutler TS, Park S, Loh DH, Jordan MC, Yokota T, Roos KP, Ghiani CA, Colwell CS. Neurocardiovascular deficits in the Q175 mouse model of Huntington's disease. Physiol Rep. 2017;5(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellosta Diago E, Perez-Perez J, Santos Lasaosa S, Viloria Alebesque A, Martinez-Horta S, Kulisevsky J, Lopez Del Val J. Neurocardiovascular pathology in pre-manifest and early-stage Huntington's disease. European journal of neurology. 2018;25(7):956–962. [DOI] [PubMed] [Google Scholar]
- 28.Child DD, Lee JH, Pascua CJ, Chen YH, Mas Monteys A, Davidson BL. Cardiac mTORC1 Dysregulation Impacts Stress Adaptation and Survival in Huntington's Disease. Cell Rep. 2018;23(4):1020–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanska DJ, Lanska MJ, Lavine L, Schoenberg BS. Conditions associated with Huntington's disease at death. A case-control study. Arch Neurol. 1988;45(8):878–880. [DOI] [PubMed] [Google Scholar]
- 30.Mielcarek M Huntington's disease is a multi-system disorder. Rare Dis. 2015;3(1):e1058464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo X, Sun X, Hu D, Wang YJ, Fujioka H, Vyas R, Chakrapani S, Joshi AU, Luo Y, Mochly-Rosen D, Qi X. VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington's disease. Nat Commun. 2016;7:12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jezek J, Cooper KF, Strich R. Reactive Oxygen Species and Mitochondrial Dynamics: The Yin and Yang of Mitochondrial Dysfunction and Cancer Progression. Antioxidants (Basel). 2018;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi AU, Mochly-Rosen D. Mortal engines: Mitochondrial bioenergetics and dysfunction in neurodegenerative diseases. Pharmacol Res. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pohjoismaki JL, Boettger T, Liu Z, Goffart S, Szibor M, Braun T. Oxidative stress during mitochondrial biogenesis compromises mtDNA integrity in growing hearts and induces a global DNA repair response. Nucleic Acids Res. 2012;40(14):6595–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HC, Yin PH, Chi CW, Wei YH. Increase in mitochondrial mass in human fibroblasts under oxidative stress and during replicative cell senescence. J Biomed Sci. 2002;9(6 Pt 1):517–526. [DOI] [PubMed] [Google Scholar]
- 36.Crane JD, Abadi A, Hettinga BP, Ogborn DI, MacNeil LG, Steinberg GR, Tarnopolsky MA. Elevated mitochondrial oxidative stress impairs metabolic adaptations to exercise in skeletal muscle. PloS one. 2013;8(12):e81879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.