Abstract
Purpose
Liquid biopsies, including circulating tumor DNA (ctDNA) and circulating tumor cells (CTCs), can be used to understand disease prognosis, tumor heterogeneity, and dynamic response to treatment in metastatic breast cancer (MBC). We explored a novel, 180-gene ctDNA panel and the association of this platform with CTCs and CTC clusters.
Methods
A total of 40 samples from 22 patients with MBC were included in the study. For the primary analysis, all patients had ctDNA sequencing using the PredicinePLUS™ platform. CTCs and CTC clusters were examined using the CellSearch™ System. Clinical and pathological variables were reported using descriptive analyses. Associations between CTC count and specific genomic alterations were tested using the Mann-Whitney U test.
Results
Of 43 sequenced patients, 40 (93%) had at least one detectable genomic alteration with a median of 6 (range 1–22). Fifty-seven different genes were altered, and the landscape of genomic alterations was representative of MBC, including the commonly encountered alterations TP53, PTEN, PIK3CA, ATM, BRCA1, CCND1, ESR1, and MYC. In patients with predominantly hormone-receptor-positive MBC, the number of CTCs was significantly associated with alterations in ESR1 (P < 0.005), GATA3 (P < 0.05), CDH1 (P < 0.0005), and CCND1 (P < 0.05) (Mann-Whitney U test). Thirty-six percent of patients had CTC clusters, which were associated with alterations in CDH1, CCND1, and BRCA1 (all P < 0.05, Mann-Whitney U test). In an independent validation cohort, CTC enumeration confirmed significant associations with ESR1 and GATA3, while CTC clusters were significantly associated with CDH1.
Conclusions
We report on a novel ctDNA platform that detected genomic alterations in the vast majority of tested patients, further indicating potential clinical utility for capturing disease heterogeneity and for disease monitoring. Detection of CTCs and CTC clusters was associated with particular genomic profiles.
Keywords: ctDNA, NGS, CTCs, CTC clusters, MBC
Introduction
Liquid biopsies have emerged as clinical tools for prognostication, molecular analysis, and detection of genomic alterations in blood [1]. The most well-studied components of liquid biopsies, circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA), give insight into the “liquid phase” of solid tumors by providing information on the spatial and temporal heterogeneity of metastatic breast cancer (MBC). Applications with potential clinical utility are being explored, including detection of minimal residual disease, dynamic treatment monitoring, and disease resistance [2, 3]. In MBC, prior work demonstrated that CTC detection and enumeration defined two subgroups of patients, stage IVindolent (< 5 CTCs per 7.5 mL of blood) and stage IVaggressive (≥ 5 CTCs) [4, 5]. This staging has been previously validated to stratify patients into two predefined cohorts with dramatically different prognostic outcomes, regardless of tumor type, site of disease, or line of therapy. Understanding the genomic changes that define these cohorts of patients is critical.
In comparison to CTCs, CTC clusters, which consist of aggregates of two or more cells, are encountered more rarely in the circulation. However, these cell groupings are associated with high metastatic potential, particular DNA methylation patterns, and potentially poor prognosis [6–8]. Initial studies have explored the cellular and genetic makeup of these cell groupings, but much remains unknown regarding how these clusters form and the genetic changes that contribute to the mechanisms of how these groupings metastasize in breast cancer [9].
In MBC, ctDNA genomic alterations have been studied to understand the genetic heterogeneity of tumor resistance. Specifically, markers of endocrine resistance emerge in response to the selective pressures of endocrine therapy with either tamoxifen or aromatase inhibitors as single agents or in combination with CDK4/6 inhibitors in MBC [10]. These events lead to clonal selection and emergence of specific genomic alterations, such as in RB1, PIK3CA driver mutations, and new ESR1 mutations. Furthermore, early changes in PIK3CA ctDNA can predict progression-free survival for patients treated with palbociclib and fulvestrant [11]. The impact of novel sequencing ctDNA panels in terms of longer DNA sequencing length and a greater number of genomic alterations on detection of resistance mutations in MBC is unknown.
Here, we explore the interplay between CTCs, CTC clusters, and ctDNA genomic alterations in a cohort of patients with predominantly hormone-receptor-positive (HR+) MBC. For CTC detection and enumeration, we utilized CellSearch™, and for ctDNA detection, we used the PredicinePLUS™ 180-gene panel with independent validation that was performed using Guardant360. These studies explore the potential utility of these platforms and how to integrate this information with other liquid-biopsy-derived biomarkers. We demonstrate the analytical validity of this novel ctDNA platform and the association of CTCs and CTC clusters with particular mutational profiles.
Methods
Patient selection and study design
The Institutional Review Board (IRB) at the Robert H. Lurie Comprehensive Cancer Center at the Northwestern University Feinberg School of Medicine approved the study. Informed consent from patients was waived per the IRB. The study was performed in concordance with the Health Insurance Portability and Accountability Act. Forty-nine samples were initially evaluated. Six samples had cfDNA yield less than 5 ng and, therefore, did not pass quality control, based on having less than 90% of regions with greater than 3000X coverage. In total, 40 of 43 (93%) passed next-generation sequencing (NGS) quality control for sequencing. As a result, the final cohort consisted of 40 samples from 22 patients with MBC with all samples collected between 2016 and 2017. Sequencing was performed in two batches in March and June 2018.
A cohort of patients with MBC with CTCs and CTC clusters were identified for ctDNA sequencing. CTCs were obtained under a prospective Investigator Initiated Trial (IIT) (NU16B06) at the Robert H. Lurie Comprehensive Cancer Center at Northwestern University (Chicago, IL, USA). CTC collection was performed at baseline prior to initiation of the next line of treatment.
CTC detection and enumeration
CTC analysis was performed using the CellSearch™ System (Menarini Silicon Biosystems, PA, USA). Approximately 10 mL of whole blood was collected into CellSave Stabilizing Tubes and processed via Celltracks Autoprep. Immunomagnetic sorting was used to characterize epithelial cell adhesion molecule (EpCAM)-positive, pan-cytokeratin (CK)-positive, DAPI-positive, and CD45-negative cells. Cells were reviewed via the Celltracks Analyzer II. CTC clusters were characterized as an aggregation of at least two cells with a distinct nuclei and an intact cytoplasm membrane.
ctDNA sequencing
Patients with 10 mL of whole blood specimens had ctDNA sequencing from plasma performed using the PredicinePLUS™ platform (Predicine, Inc., Hayward, CA, USA). The platform includes a 180-gene panel with 565 kb of sequencing coverage including single nucleotide variants (SNVs), copy number variants (CNVs), and a total of 88 fusion genes (Additional file 1: Table S1). cfDNA yield was measured using Bioanalyzer 2100, and sequencing was performed using an Illumina platform with an in-house proprietary bioinformatics pipeline to align ctDNA sequences and to determine genomic alterations. Assay sensitivity was set at a minimum of 0.25% mutant allele frequency (MAF) for all genomic regions and 0.1% MAF for hotspot variants. Targeted sequencing coverage was greater than 20,000X.
Based on a minimum cfDNA input of 15 nanograms (ng), sensitivity for SNVs, CNVs, and gene fusions were reported as 94.4%, 95.0%, and 83.3%, respectively, with positive predictive values (PPV) of 99.7%, 100%, and 100%, respectively. Of note, NGS of these samples was performed for research purposes only. Patients received standard, guideline-based, systemic treatments. For independent validation, NGS was performed on plasma samples in a subset of patients (N = 14) with concurrent commercial Guardant360 testing (Guardant Health, Redwood City, CA) [12]. In addition, 84 patients with Guardant360 testing and CTC evaluation were analyzed to confirm associations between CTCs and particular genomic alterations. CTC and ctDNA analyses were linked to a deidentified clinical database.
Statistical analysis
Clinical and pathological variables were reported using descriptive analyses. Associations between CTC count and specific genomic alterations were tested using the Mann-Whitney U test. All analyses were performed using STATA (StataCorp (2015) Stata Statistical Software: Release 14.2 College Station, TX: StataCorp, LP).
Results
Patient characteristics
Patient characteristics for the 22 MBC patients included in the cohort are included in Table 1. All patients were female. The sample consisted of 17 HR+, HER2-negative patients, 1 HR-negative, HER2-positive, 1 HR+, HER2-positive, and 3 triple-negative breast cancer patients. There were 18 patients with invasive ductal carcinoma (IDC) and 4 patients with invasive lobular carcinoma (ILC). Median prior to lines of treatment in the metastatic setting was 2 [range 0–7]. In total, 81.8% of patients had bone involvement and 68.2% had visceral disease (Additional file 1: Table S2).
Table 1.
Patient characteristics
| Cohort | |
| Number of patients | 22 |
| Number of collections | 40 |
| Sex | |
| Female | 22 (100%) |
| Pathology | |
| IDC | 18 (81.8%) |
| ILC | 4 (18.2%) |
| Histologic subtype | |
| HR+, HER2− | 17 (77.3%) |
| HR−, HER2+ | 1 (4.5%) |
| HR+, HER2+ | 1 (4.5%) |
| TNBC | 3 (13.6%) |
| Clinical subtype | |
| IBC | 6 (27.3%) |
| Non-IBC | 16 (72.7%) |
| Prior therapies in metastatic setting | 2* [0–7] |
| Sites of disease | |
| Bone | 18 (81.8%) |
| Visceral | 15 (68.2%) |
| CTC clusters | |
| Yes | 8 (36.4%) |
| No | 14 (63.6%) |
| Total blood draws with clusters | 14 (35.0%) |
IDC invasive ductal carcinoma, ILC invasive lobular carcinoma, HR hormone receptor, TNCB triple-negative breast cancer, IBC inflammatory breast cancer
*The median
Platform characteristics
Forty samples from 22 patients were included in the final analyses [range 1–4 samples per patient]. Of 43 initial samples, 40 passed NGS quality control (40/43, 93%) and had detectable genomic alterations (Table 2). Mean, median, and range for number of genomic alterations were 6.7, 6.0, and 1–22, respectively. cfDNA yield ranged from 7.8 to 272.5 ng. MAF of detectable mutations ranged from 0.11 to 68.6%.
Table 2.
Characteristics of the PredicinePLUS™ platform and detected alterations
| Cohort | |
| Total cases | 49 |
| Cases included in final analyses | 40 |
| Regions analyzed | 180 genes |
| Panel size | 565 kb |
| Samples with detectable alterations | 40/43 (93%) |
| Number of genomic alterations | |
| Mean | 6.7 |
| Median | 6.0 |
| Range | 1–22 |
| Number of genes with detected alterations | 57 |
| Variant allele frequency of detected alterations | 0.11–68.6% |
| Commonly detected SNV/indels | TP53, PTEN, PIK3CA, ATM, ESR1 |
| Commonly detected copy number amplifications | MYC, CCND1, PIK3CA |
| Commonly detected copy number losses | BRCA1, CDKN2A, ATM |
SNV single nucleotide variant, Indels insertion-deletion mutations
ctDNA genomic alterations
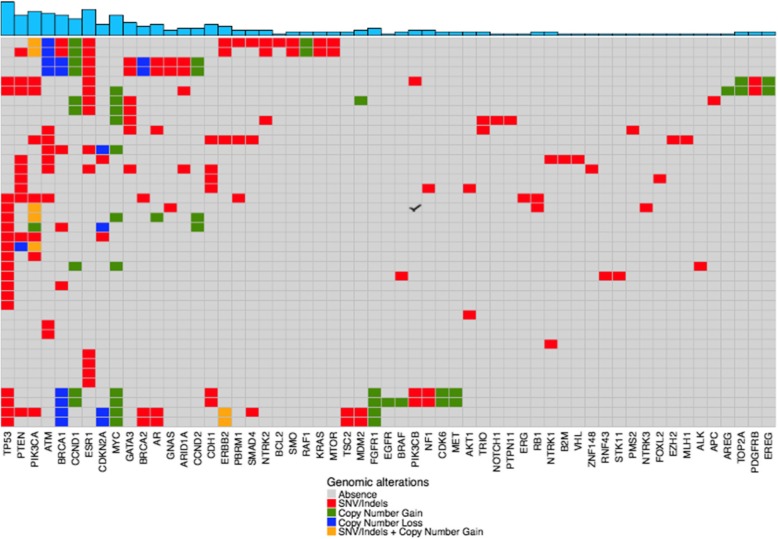
The landscape of genomic alterations in the cohort is included in Fig. 1. Identified alterations were representative of commonly encountered tissue and blood-based NGS alterations including the following: TP53, PTEN, PIK3CA, ATM, BRCA1, CCND1, ESR1, and MYC. In total, 267 genomic alterations from 57 different genes were reported. Copy number gains were encountered in 16 genes and copy number losses in 5 genes. MAF of common variants are shown in Additional file 1: Figure S1. For patients with concurrent PredicinePLUS™ and Guardant360 testing (N = 14), high concordance was observed in orthogonal samples across representative genomic alterations (ESR1 92.9%, PIK3CA 100%, MYC copy number variations 71.4%) (Gerratana L, Zhang Z, Shah A, Davis A, Zhan Y, Qiang W, Finkelman B, Flaum L, Behdad A, Gradishar WJ et al: Analytical and clinical performance of a novel next generation sequencing-based (NGS) circulating tumor DNA (ctDNA) platform for the evaluation of samples from metastatic breast cancer (MBC). In. Under Review; 2019).
Fig. 1.
Landscape of genomic alterations. Shown is a heatmap of all detected genomic alterations. Top blue panel indicates the total number of alterations detected for each gene. The colors below indicate the specific types of genomic alterations including SNV/indels (red), copy number gain (green), copy number loss (blue), and SNV/indel + copy number gain (yellow). Each row indicates a sample (N = 40) and each column represents a gene
Association of CTC enumeration with ctDNA alterations
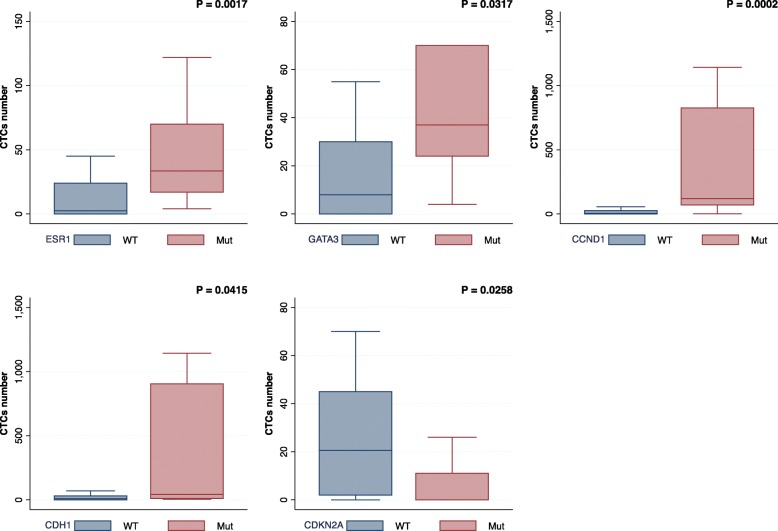
Patients were categorized into stage IVindolent (< 5 CTCs per 7.5 mL of blood) and stage IVaggressive (≥5 CTCs) disease based on CTC enumeration. In total, there were 8 patients (36%) with stage IVindolent and 14 patients (64%) with stage IVaggressive at baseline CTC draw in the cohort. Samples with CDKN2A alterations had a significantly lower number of CTCs (P < 0.05, Mann-Whitney U test) (Fig. 2). In contrast, samples with a higher number of CTCs were significantly associated with alterations in ESR1 (P < 0.005), GATA3 (P < 0.05), CDH1 (P < 0.0005), and CCND1 (P < 0.05) with stage IVaggressive disease associated with ESR1 mutations (Mann-Whitney U test). In independent validation using Guardant360 testing of 84 patients, number of CTCs was confirmed to have significant associations with ESR1 (P < 0.005) and GATA3 (P < 0.05), as well as copy number changes in MYC (P < 0.05). Characteristics of this cohort are included in Additional file 1: Table S3.
Fig. 2.
Genomic alterations associated with CTCs. Genomic alterations in ESR1, GATA3, CCND1, and CDH1 were significantly associated with higher number of CTCs (P < 0.05, Mann-Whitney U test). In contrast, alterations in CDKN2A were more commonly observed in samples with low CTC count (P < 0.05, Mann-Whitney U test). In the validation cohort, significant associations were confirmed for ESR1 (P < 0.005) and GATA3 (P < 0.05)
Association of CTC clusters with ctDNA alterations
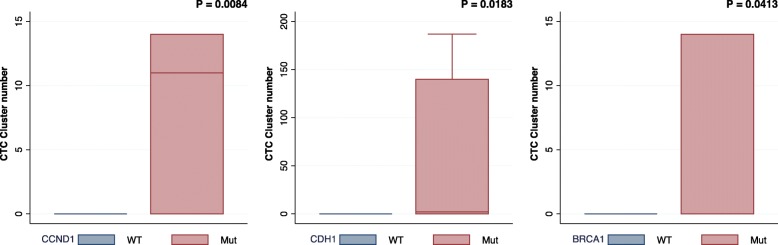
Eight patients (36%) were found to have CTC clusters in at least one blood collection in the cohort. Furthermore, 12/40 (30%) of total blood draws contained CTC clusters. All patients with CTC clusters had stage IVaggressive disease. CTC clusters were significantly associated with somatic genomic alterations in CDH1, CCND1, and BRCA1 (all P < 0.05, Mann-Whitney U test) (Figs. 3 and 4). In the validation cohort, CTC clusters were significantly associated with CDH1 (P < 0.005).
Fig. 3.
Genomic alterations in patients with CTC clusters. Genomic alterations in CCND1, CDH1, and BRCA1 were significantly associated with the number of CTC clusters (P < 0.05, Mann-Whitney U test). In the validation cohort, a significant association was confirmed for CDH1 (P < 0.005)
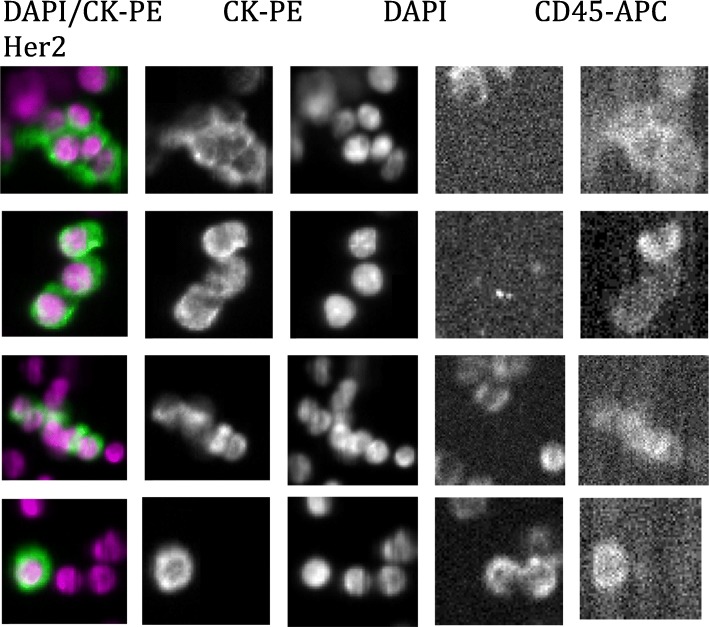
Fig. 4.
Representative images of a patient with CTC clusters. Shown are representative images of a patient with CTC clusters with nuclear (DAPI) and cytokeratin (CK-PE) staining. CD45 stains for non-CTC leukocytes. HER2/neu staining further distinguished CTCs from leukocytes in this patient. This sample was associated with the following ctDNA genomic alterations: CDH1, TP53, NF1, PIK3CB, BRCA1, CCND1, CDK6, FGFR1, MET, and MYC
Case vignette
We present a case illustrating the potential clinical utility for serial blood monitoring (Additional file 1: Figure S2). The patient was a 67-year-old female with a history of localized HR+ HER2-negative breast cancer treated with lumpectomy and radiation who declined endocrine therapy. About 2 years later, she developed right upper quadrant pain and was found to have metastatic disease to the liver, which was biopsy confirmed HR+ HER2-negative breast adenocarcinoma. She was initiated on single-agent anastrozole for approximately 2 years and subsequently developed bone metastasis in her scapula and lumbar spine. She was changed to fulvestrant (declined palbociclib) and zoledronic acid with progression of disease 4 months later in her liver and bones. At this time, CTC evaluation demonstrated 45 CTCs of which 5 were HER2 positive with 2 CTC clusters present (timepoint 1). ctDNA NGS revealed an ERBB2 (HER2) S310F mutation. Treatment was initiated with trastuzumab and capecitabine. Serial CTC collection 3 months later revealed 0 CTCs (timepoint 2). Eighteen months later, after 22 cycles of trastuzumab, ctDNA revealed increasing clonal heterogeneity with 9 alterations present in blood including 5 different HER2 mutations and 9 CTCs (1 HER2 positive) (timepoint 3). Subsequent imaging confirmed progression of disease in the liver and lymph nodes.
Discussion
The clinical potential for liquid biopsies, including CTCs and ctDNA, for prognostication and disease monitoring is expanding. We analyzed a novel, sensitive, 180-gene, 565-kb sequencing platform to analyze and report on the landscape of alterations and association of genomic changes with a cohort of predominantly HR+ MBC characterized by CTC enumeration and CTC clusters. The study demonstrated that using the sequencing length of this platform, 100% of samples that passed quality control and 40 of 43 that underwent sequencing (93%) had at least one somatic alteration detected. This suggests a clear potential for using this technique to detect and monitor dynamic changes in the blood of patients with MBC. In total, 57 genes with genomic alterations were detected including SNVs, indels, and CNVs. Longer sequencing panels may also aid in identifying novel resistance mutations in blood and better represent tumor heterogeneity to eventually capture tumor mutational burden, non-invasively.
We observed that higher number of CTCs was associated with genomic alterations in ESR1, GATA3, CDH1, and CCND1, while lower number of CTCs was associated with CDKN2A alterations. In independent validation, these findings were confirmed for ESR1 and GATA3. This supports the relation between CTCs as a marker of poor prognosis and the ability to detect specific resistance mutations (e.g., ESR1) with implications for clinical practice. Prior work in our group has demonstrated that ESR1 mutations in single CTCs matched mutations observed in ctDNA, which demonstrates a mechanism to link CTCs with the release of ctDNA into the blood [13]. Furthermore, alterations in CDH1, CCND1, and BRCA1 were associated with a higher number of CTC clusters with the validation cohort confirming CDH1 as statistically significant. The CDH1 gene is involved in the production of epithelial cadherin (E-cadherin). Therefore, the gene plays a role in cell adhesion, chemical signaling, and cell movement, all of which may contribute to the metastatic process. Somatic variants in CDH1 were seen in both ILC and IDC patients in our cohort, although classically this mutation is associated with ILC histology [14]. Further studies are needed to explore this mutation as an indicator of metastatic potential.
Conclusions
In summary, this novel ctDNA sequencing platform identified genomic alterations in the vast majority of tested patients, reflecting the genomic heterogeneity of patients with predominantly HR+ MBC. This suggests a clear clinical potential for disease monitoring using this platform given the frequency of genomic alterations encountered in our sample. Additional analyses enabled us to characterize particular genomic alterations and different biology based on CTC enumeration and the identification of CTC clusters. Limitations of this study include the relatively small sample size and that some patients with more than one ctDNA sample could have biased the analyses of specific ctDNA alterations. However, these data are consistent with previous experiences using different NGS platforms and were validated in an independent cohort in our study [15]. These findings, therefore, further demonstrate the potential of combining CTCs and ctDNA for comprehensive liquid biopsy analysis to accurately represent genomic heterogeneity and to detect resistance mutations in a non-invasive manner with implications for clinical management of patients with MBC.
Supplementary information
Additional file 1 Table S1. PredicinePLUS™ 180-gene panel. Table S2. Treatment data and sites of disease. Table S3. Characteristics of Guardant360 validation cohort. Figure S1. Mutant allele frequency of 100 most common variants in the cohort. Figure S2. Case vignette demonstrating the potential clinical utility of serial liquid biopsy assessment.
Acknowledgements
n/a
Authors’ contributions
AAD, QZ, LG, and MC were responsible for the conception and design. AAD, QZ, LG, BSF, and MC were responsible for the acquisition and analysis; all authors were responsible for the interpretation of data and approval and editing of the final manuscript.
Authors’ information
n/a
Funding
Lynn Sage Cancer Research Foundation, Predicine, and REDCap support was funded in part by a Clinical and Translational Science Award (CTSA) grant from the National Institutes of Health UL1TR001422. The data interpretation and reporting included in the manuscript were performed independently from Predicine.
Availability of data and materials
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Informed consent was waived per the IRB.
Consent for publication
Consent for publication is covered under the OncoSET Registry (NUDB 16Z01) for inclusion of the case vignette.
Competing interests
Andrew Davis Travel expenses: Menarini Silicon Biosystems. Lorenzo Gerratana Travel expenses: Menarini Silicon Biosystems. Massimo Cristofanilli Honoraria: Pfizer, Merus, Novartis, CytoDyn. All other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13058-019-1229-6.
References
- 1.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 2.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 3.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 5.Cristofanilli M, J-Y P, Reuben JM. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): International expert consensus paper. Crit Rev Oncol Hematol. 2019;134:39–45. 10.1016/j.critrevonc.2018.12.004. [DOI] [PubMed]
- 6.Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, Krol I, Scheidmann MC, Beisel C, Stirnimann CU, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176(1–2):98–112. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Mu Z, Chervoneva I, Austin L, Ye Z, Rossi G, Palazzo JP, Sun C, Abu-Khalaf M, Myers RE, et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res Treat. 2017;161(1):83–94. doi: 10.1007/s10549-016-4026-2. [DOI] [PubMed] [Google Scholar]
- 8.Jansson S, Bendahl PO, Larsson AM, Aaltonen KE, Ryden L. Prognostic impact of circulating tumor cell apoptosis and clusters in serial blood samples from patients with metastatic breast cancer in a prospective observational cohort. BMC Cancer. 2016;16:433. doi: 10.1186/s12885-016-2406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158(5):1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, Andre F, Loibl S, Loi S, Garcia-Murillas I, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018;8(11):1390–1403. doi: 10.1158/2159-8290.CD-18-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, Liu Y, Bartlett CH, Koehler M, Cristofanilli M, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun. 2018;9(1):896. doi: 10.1038/s41467-018-03215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, Collisson EA, Divers SG, Hoon DS, Kopetz ES, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One. 2015;10(10):e0140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paolillo C, Mu Z, Rossi G, Schiewer MJ, Nguyen T, Austin L, Capoluongo E, Knudsen K, Cristofanilli M, Fortina P. Detection of activating estrogen receptor gene (ESR1) mutations in single circulating tumor cells. Clin Cancer Res. 2017;23(20):6086–6093. doi: 10.1158/1078-0432.CCR-17-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163(2):506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossi G, Mu Z, Rademaker AW, Austin LK, Strickland KS, Costa RLB, Nagy RJ, Zagonel V, Taxter TJ, Behdad A, et al. Cell-free DNA and circulating tumor cells: comprehensive liquid biopsy analysis in advanced breast cancer. Clin Cancer Res. 2018;24(3):560–568. doi: 10.1158/1078-0432.CCR-17-2092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Table S1. PredicinePLUS™ 180-gene panel. Table S2. Treatment data and sites of disease. Table S3. Characteristics of Guardant360 validation cohort. Figure S1. Mutant allele frequency of 100 most common variants in the cohort. Figure S2. Case vignette demonstrating the potential clinical utility of serial liquid biopsy assessment.
Data Availability Statement
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.