Abstract
Background
The metabolic consequences of preoperative carbohydrate load in breast cancer patients are not known. The present explorative study investigated the systemic and tumor metabolic changes after preoperative per-oral carbohydrate load and their influence on tumor characteristics and survival.
Methods
The study setting was on university hospital level with primary and secondary care functions in south-west Norway. Serum and tumor tissue were sampled from a population-based cohort of 60 patients with operable breast cancer who were randomized to either per-oral carbohydrate load (preOp™; n = 25) or standard pre-operative fasting (n = 35) before surgery. Magnetic resonance (MR) metabolomics was performed on serum samples from all patients and high-resolution magic angle spinning (HR-MAS) MR analysis on 13 tumor samples available from the fasting group and 16 tumor samples from the carbohydrate group.
Results
Fourteen of 28 metabolites were differently expressed between fasting and carbohydrate groups. Partial least squares discriminant analysis showed a significant difference in the metabolic profile between the fasting and carbohydrate groups, compatible with the endocrine effects of insulin (i.e., increased serum-lactate and pyruvate and decreased ketone bodies and amino acids in the carbohydrate group). Among ER-positive tumors (n = 18), glutathione was significantly elevated in the carbohydrate group compared to the fasting group (p = 0.002), with a positive correlation between preoperative S-insulin levels and the glutathione content in tumors (r = 0.680; p = 0.002). In all tumors (n = 29), glutamate was increased in tumors with high proliferation (t-test; p = 0.009), independent of intervention group. Moreover, there was a positive correlation between tumor size and proliferation markers in the carbohydrate group only. Patients with ER-positive / T2 tumors and high tumor glutathione (≥1.09), high S-lactate (≥56.9), and high S-pyruvate (≥12.5) had inferior clinical outcomes regarding relapse-free survival, breast cancer-specific survival, and overall survival. Moreover, Integrated Pathway Analysis (IPA) in serum revealed activation of five major anabolic metabolic networks contributing to proliferation and growth.
Conclusions
Preoperative carbohydrate load increases systemic levels of lactate and pyruvate and tumor levels of glutathione and glutamate in ER-positive patients. These biological changes may contribute to the inferior clinical outcomes observed in luminal T2 breast cancer patients.
Trial of registration
ClinicalTrials.gov; NCT03886389. Retrospectively registered March 22, 2019.
Keywords: Breast cancer, Carbohydrate load, Proliferation, Insulin, Insulin c-peptide, S-lactate, S-pyruvate, Tumor glutamate, Tumor glutathione, Fasting state, Ketonic bodies, Clinical outcome
Background
Breast cancer is the most common female malignancy and one of the most frequent causes of death among women in the Western world [1]. Breast cancer incidence has more than doubled in the last 50 years, probably due to increased estrogen exposure and a change towards high levels of alimentary carbohydrates and fat [2, 3]. Even though breast cancer originates locally in the breast, circulating tumor cells (CTCs) may spread to the systemic circulation before and during surgery [4] and establish distant micrometastases [5]. These CTCs must thrive and survive attacks from the innate and adaptive immune system. Thus, tumor cells have to establish a favorable metabolism that can produce energy, protection mechanisms, and the necessary biomass to survive the journey from the breast tumor to remote locations, including transformation into dormancy [6]. The luminal breast cancer subtype, which express estrogen receptor (ER) and/or progesterone receptor (PR) in the tumor cells, comprise the largest subgroup, accounting for approximately 75% of all breast cancers. Endocrine resistance in this subtype can creates micrometastases that escape anti-estrogen therapy and can hibernate for many years before they become clinically overt [7]. The molecular features underlying these cellular characteristics are driven by hallmarks of cancer [8], including changes in cellular energetics and metabolism, followed by a vast number of necessary metabolic modifications to strengthen the metabolic needs of breast cancer cells [9]. A well-known cellular characteristic of tumor cells is increased glucose consumption and glycolysis towards lactate despite the presence of oxygen, a feature called ‘the Warburg effect’ [10, 11]. This metabolic switch includes the production of ribose for DNA synthesis and allowing amino acids to be a source for ATP production [12]. Furthermore, the Warburg effect extends to increased choline metabolism for cell membrane synthesis and increased amino acid turnover for protein synthesis [10, 13].
Even though much is known about metabolism in breast cancer cells [14], little is known about the influence of carbohydrate loading in the early recovery after surgery (ERAS) program [15] on peri-operative metabolism in the systemic circulation and locally in the breast tumor. We recently conducted a randomized controlled trial (RCT) in which operable breast cancer patients were treated with either two oral loads of enriched carbohydrate solution or a standard fasting procedure comprising free drinking of tap water before surgery [16]. In this study, luminal breast cancer patients, who received oral pre-operative carbohydrates, had a higher tumor proliferation and an adverse survival. The goal of the present paper, using the same patients, was to further explore the metabolic differences in serum and the tumor. Based on our previous findings, we hypothesize that the metabolic changes after carbohydrate loading will correlate with proliferation and outcome in patients with ER positive tumors. Also, we also wanted to study whether such metabolic alterations correlate with other tumor characteristics or translate into differences in clinical outcome.
Methods
Ethics statement
This paper is an explorative study based upon a recently published randomized controlled trial (RCT) approved by the Regional Ethics Committee in Western Norway (#2015/1445) and was retrospectively registered at Clinicaltrials.gov (NCT03886389).
Patients
Details on these patients have been described previously [16]. In short, between 12 May 2009 and 23 June 2010 a population-based cohort of 61 operable breast cancer patients (Stage I and II) were randomized into an intervention group receiving preoperative per-oral carbohydrate loading (n = 26) or a control group (n = 35) receiving the standard preoperative fasting protocol.
The patients in the carbohydrate group drank 200 mL pre-Op™ (Nutricia, the Netherlands). This non-carbonated carbohydrate enriched drink contained 100 kCal per bottle containing 4.2 g (2.1%) glucose and 20 g (10%) polysaccharides. A loading dose of two bottles pre-Op™ were given 18 h before surgery (i.e. the evening before surgery) and another 2 bottles were administered 2–4 h before surgery (i.e. the morning of the operation day). In contrast, the control group practiced the standard fasting procedure with free intake of tap water 12–14 h before surgery. From this cohort, patients with available fresh frozen tissue and serum samples were included in the present study (Fig. 1). The patient characteristics are given in Table 1.
Fig. 1.
Flowchart of study participants
Table 1.
Clinical variables in the randomized groups
| Carbohydrate group (N=26) | Fasting group (N=35) | Carbohydrate group with tissue (n=16) | Fasting group with tissue (n=13) | |
| Age | ||||
| <55 | 12 (46%) | 16 (46%) | 9 (56%) | 7 (53%) |
| ≥ 55 | 14 (54%) | 19 (54%) | 7 (44%) | 6 (46%) |
| Lymph Node status | ||||
| Negative | 19 (70%) | 25 (71%) | 11 (69%) | 9 (69%) |
| Positive | 8 (30%) | 10 (29%) | 5 (31%) | 4 (31%) |
| Tumor size (pT) | ||||
| pT1 (<2cm) | 16 (61%) | 30 (85%)a | 7 (44%) | 9 (69%) |
| pT2 (≥2cm) | 10 (39%) | 5 (14%) | 9 (57%) | 4 (31%) |
| Grade | ||||
| 1 | 4 (15%) | 7 (20%) | 2 (13%) | 2 (15%) |
| 2 | 10 (37%) | 20 (57%) | 4 (25%) | 7 (53%) |
| 3 | 13 (48%) | 8 (23%) | 10 (63%) | 4 (31%) |
| ER status | ||||
| Positive | 21 (81%) | 29 (83%) | 11 (69%) | 9 (69%) |
| Negative | 5 (19%) | 6 (17%) | 5 (31%) | 4 (31%) |
| PR statusa | ||||
| Positive | 13 (50%) | 28 (80%)b | 7 (44%) | 11 (85%) |
| Negative | 13 (50%) | 7 (20%) | 9 (56%) | 2 (15%) |
| HER2 status | ||||
| Negative | 23 (88%) | 34 (97%) | 13 (81%) | 12 (92%) |
| Positive | 3 (12%) | 1 ( 3%) | 3 (19%) | 1 (8%) |
| MAI | ||||
| <10 | 14 (56%) | 27 (77%) | 6 (38%) | 10 (77%) |
| ≥ 10 | 11 (44%) | 8 (23%) | 10 (62%) | 3 (23%) |
| PPH3 | ||||
| <13 | 14 (56%) | 21 (60%) | 7 (44%) | 6 (46%) |
| ≥ 13 | 12 (44%) | 14 (40%) | 9 (56%) | 7 (54%) |
| Ki67 | ||||
| ≥ 15 | 17 (65%) | 17 (50%) | 3 (19%) | 5 (42%) |
| <15 | 9 (35%) | 17 (50%) | 13 (81%) | 7 (58%) |
| ≥ 30 | 12 (46%) | 10 (29%) | 6 (38%) | 8 (67%) |
| <30 | 14 (54%) | 24 (71%) | 10 (62%) | 4 (33%) |
| TILs | ||||
| <10% | 24 (92%) | 31 (89%) | 15 (94%) | 13 (100%) |
| ≥10% | 2 (8%) | 4 (11%) | 1 (6%) | 0 (0%) |
| End of follow-up status | ||||
| No distant metastasis | 22 (85%) | 33 (94%) | 11 (67%) | 11 (85%) |
| Distant metastasis | 4 (15%) | 2 (6%) | 3 (20%) | 1 (7%) |
aSignificantly different between fasting and carbohydrate group (Fisher’s exact test)
bp=0.052 in tissue subset
Blood sampling
Blood samples were drawn immediately before surgery. In total three serum gel tubes and one EDTA plasma tube were drawn in this study. One serum gel tube and one EDTA plasma tube were delivered within an hour to the department of medical biochemistry for standard analysis. For metabolomics analyses, two serum gel tubes were centrifuged within one hour at 4 °C, 2500 x g in 10 min. After centrifugation, the serum of the two tubes were mixed and a minimum of 1.1 mL serum were sent for analyses in Haukeland University Hospital, Bergen, Norway, the rest of the serum were stored in 1 mL cryotubes at − 80 °C in the biobank at Stavanger University Hospital, Stavanger, Norway.
Tumor tissue sampling
Immediately after removal of the surgical specimen from the systemic circulation, it was transported to the Department of Pathology for further sampling. To avoid necrotic areas, cancerous tissue from the invasive front of the tumor (i.e. tumor periphery) was immediately snap-frozen in liquid nitrogen and stored at − 80 °C until assayed for tissue metabolomics. Before HR-MAS analysis, tissues from all of the patients were analyzed consecutively for histopathology and immunohistochemistry as described preciously [16].
Serum hormone and protein analyses
Serum was transported to the Hormone Laboratory, Haukeland University Hospital, Bergen, Norway. Insulin, insulin c-peptide, insulin growth factor 1 (IGF-1), and insulin growth factor binding protein 3 (IGFBP-3) were measured by the IMMULITE 2000 two-site chemiluminescent immunometric assay (Siemens Medical Solutions Diagnostics).
Serum metabolomics analyses
A separate aliquot of serum was transported to the MR Core Facility at NTNU, Trondheim, Norway for metabolomics analyses. Thawed samples (100 μL) were mixed with bacteriostatic buffer (100 μL; pH 7.4, 0.075 mM Na2HPO4, 5 mM NaN3, 5 mM TSP), transferred to 3-mm NMR tubes, and stored at 5 °C until analysis (< 15 h). The MR analysis was performed using a Bruker Avance III Ultrashielded Plus 600 MHz spectrometer (Bruker Biospin GmbH, Germany) equipped with a 5 mm QCI Cryoprobe with integrated, cooled pre-amplifiers for 1H, 2H, and 13C. Experiments were fully automated using the SampleJet™ in combination with Icon-NMR in TopSpin 3.1 software (Bruker Biospin). One-dimensional 1H Nuclear Overhauser effect spectroscopy (NOESY) and Carr–Purcell–Meiboom–Gill (CPMG) spectra with water presaturation were acquired at 310.15 K. The spectra were Fourier transformed to 128 K after 0.3 Hz exponential line broadening and automatically phased and baseline-corrected. Spectra were further processed in Matlab 2013b (The Mathworks Inc., Natick, MA, USA). The CPMG spectral region between 0.1 and 4.2 ppm was selected for further processing. Chemical shifts were referenced to the left alanine peak at 1.47 ppm. Metabolites were identified based on previous assignment [17, 18]. Twenty-eight metabolites were identified as measurable and their areas calculated by integrating the area under the signal curve.
Breast tumor tissue metabolomics analyses
In the 29 patients with available tissue, the tumors were larger (45% vs. 9% pT2/3/4, p = 0.003), had a higher histological grade (52% vs. 18% grade 3, p = 0.022), were more often ER-negative (35% vs. 3%, p = 0.002), and had higher proliferation (59% vs. 27% PPH3-positive, p = 0.002) than those without tissue. Thus, we had a selection bias of larger, non-luminal and a more proliferative tumors into the present study compared to the original study [16]. Tissue was transported on dry ice to the MR Core Facility at NTNU, Trondheim, Norway, for metabolomics analyses. Tissue samples were prepared frozen on a metal plate bathed in liquid nitrogen to minimize tissue degradation. Biopsies (11.0 ± 2.3 mg) were cut to fit 30 μL disposable inserts (Bruker Biospin Corp, USA) filled with 3 μL D2O containing 25 mM formate. The insert containing the frozen sample was placed in a 4-mm diameter zirconium rotor (Bruker, Biospin GmbH, Germany) and kept at − 20 °C until analysis (< 8 h). Spin-echo spectra were acquired on a Bruker Avance DRX600 spectrometer with a 1H/13C magic angle spinning (MAS) probe with gradient (Bruker Biospin GmbH, Germany) using the following parameters: 5 KHz spin rate, 5 °C probe temperature, 5-min temperature acclimatization before shimming and spectral acquisition, CPMG pulse sequence (cpmgpr1d; Bruker) with 4 s water suppression prior to a 90° excitation pulse, total echo time 77 ms, 256 scans, and spectral width 20 ppm. Spectra were Fourier transformed into 64 K following 0.3 Hz line broadening. Phase correction was performed automatically for each spectrum using TopSpin 3.1.
Spectra were preprocessed in Matlab 2013b as follows [19]. The spectral region between 1.4–4.70 ppm, which contained the majority of the metabolite signals, was selected for further processing. Chemical shifts were referenced to the creatine peak at 3.03 ppm. The spectra were baseline-corrected using asymmetric least squares [20] with parameters λ = 1e7 and p = 0.0001, setting the lowest point in each spectrum to zero. Lipid peaks at 4.34–4.27, 4.19–4.14, 2.90–2.7, 2.31–2.18, 2.11–1.92, and 1.68–1.5, and ethanol at 3.67–3.62, were excluded. The resulting spectra were normalized to the total area to correct for differences in sample size and tumor cell content. Metabolite peak assignment was based on previous identification [21]. Twenty metabolites were identified as measurable, and the area under the signal curve in the preprocessed spectra was used to calculate their relative intensities. The metabolite integrals were log10 transformed to satisfy prerequisite assumptions of normality.
Endpoints
Proliferation differences between the carbohydrate and fasting groups were evaluated by Ki67 (< 15% or ≥ 15 and < 30% or ≥ 30%), mitotic activity index (MAI; < 10 or ≥ 10), and PPH3 (< 13 or ≥ 13). The metabolic response to preoperative oral carbohydrate loading was evaluated in serum (preoperative) by 1H NMR and in tumor tissue by HR-MAS MRS.
Univariate analysis
Metabolite differences between groups were assessed by student T-tests. Correlations between continuous variables were assessed by Pearson correlation. Categorical variables were compared by Chi square tests. P-values were considered significant when p < 0.05. When multiple variables were compared, the resulting p-value tables were corrected for multiple testing by the Benjamini-Hochberg method [22].
Multivariate analyses (serum and tissue)
Multivariate analyses were performed in R V.3.5 [23] using the package PLS [24] and MetaboAnalyst [25]. Metabolite values were auto-scaled (mean-centered and divided by variance) before multivariate analysis. Principal component analysis (PCA) was performed to evaluate the data sets for outliers. Partial least squares discriminant analysis (PLS-DA) was performed to explore differences in serum and tissue metabolic profiles between categories: carbohydrate loading vs fasting. Partial least squares (PLS) was used to find correlations between the tissue metabolic profile and variables (MAI, PPH3, Ki67, serum (S)-glucose, S-insulin, S-insulin c-peptide, S-IGFR, S-IGFPB3, S-estradiol). Metabolites were evaluated by Variable Importance in Projection (VIP) score. The VIP score is a measure of how important each variable was for creating the discrimination model. It is calculated as a weighted sum of squares of the PLS loadings, where the weights are based on the amount of y-variance explained in each dimension [26]. PLS and PLS-DA classification parameters were evaluated by ‘leave-one-out’ cross validation due to the limited sample numbers. Permutation testing was carried out as an additional model validation; sample classes or responses were shuffled, and the model rebuilt with the same numbers of latent variables as the original model. One thousand permutations were performed, and models were considered significant if the final accuracy (of classification models) or R2 (of regression models) were > 95% of the permuted accuracy values (p < 0.05).
Thresholds in survival analyses
Relapse-free survival (RFS) was defined as the time from surgery until a relapse from any site. Breast cancer-specific survival (BCSS) was defined as the time from surgery until death from breast cancer, whereas overall survival (OS) was until death from any cause. Receiver-operator characteristic (ROC) analysis identified optimal thresholds for the various continuous metabolite variables using relapse ‘Yes/No’ as the categorical variable (Table 8 in Appendix). The cut-off values obtained in RFS analysis were also used in the BCSS and OS analyses. In ER-negative patients, none of the explanatory variables with ROC-derived thresholds were significant for analysis of RFS, BCSS, or OS. Therefore, further analyses were limited to ER-positive patients. The ROC-obtained thresholds were confirmed with the minimal p-value/maximal Wald-value in a Cox model. In the multivariabel Cox analyses the ‘Forward Wald’ method was primarily used. In cases of an unstable model, a stepwise backward analysis was performed.
Table 8.
ROC – analysis with ‘Relapse / No relapse’ as dichotomous variable in ER+ patients
| Test variable | AUC | 95% CI |
Sensitivity (%) |
Specificity (%) |
P | Threshold |
| S-lactate | 0.769 | 0.609 – 0.929 | 57 | 88 | 0.024 | 56.9 |
| S-pyruvate | 0.765 | 0.541 – 0.989 | 71 | 86 | 0.026 | 12.5 |
| Tumor-Glutathione | 0.711 | 0.485 – 0.938 | 100 | 66 | 0.260 | 1.09 |
| S-preoperative Insulin | 0.724 | 0.554– 0.896 | 86 | 67 | 0.059 | 18.3 I.U./L |
| S-preoperative insulin c-peptide | 0.735 | 0.566 – 0.903 | 86 | 67 | 0.049 | 1.22 nM |
Metabolite set enrichment analysis and ingenuity pathway analysis (IPA)
Serum metabolite levels were uploaded to the Enrichment module of MetaboAnalyst to explore the pathways affected by the carbohydrate intervention. Pathway-associated metabolite sets with sets containing at least two metabolites were used. Pathways with p-values ≤0.05 (after FDR correction) were interpreted as significant. Serum metabolites with significantly different expression (p = 0.05) and their corresponding fold changes were imported into the Ingenuity Pathway Analysis (IPA) software (Ingenuity, Redwood City, USA) to explore which biological and molecular functions these metabolites were involved in and how these and their direct and indirect target molecules were connected, using the network function in IPA. Additionally, we examined if there were a direct or indirect connection between the top network and seven microRNAs related to tamoxifen resistance from our previous paper [27], using the grow function with a moderate or experimentally observed confidence level.
Results
Systemic metabolism
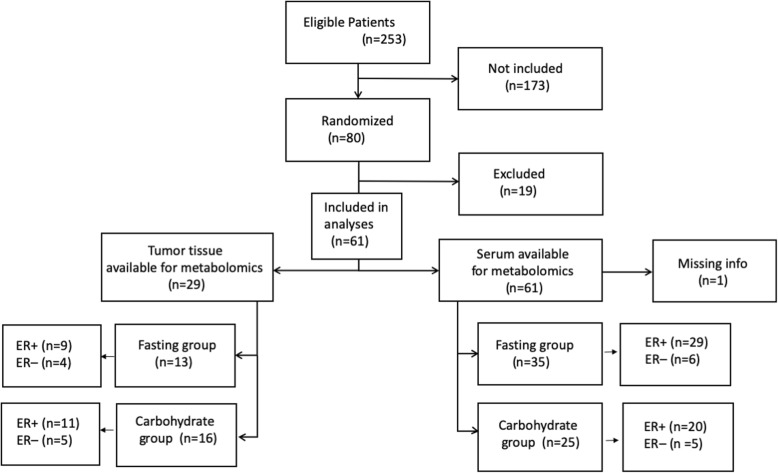
The results of the quantification of serum metabolites in the carbohydrate and fasting groups are given in Table 2. Fourteen out of 28 metabolites were significantly altered between the groups. PLS-DA revealed a significant difference in metabolic profiles between the two groups.; (one component, classification accuracy = 0.85; p < 0.001; Fig. 2a). The main increased markers were increased serum (S) lactate and S-pyruvate in the carbohydrate group (p < 0.0001; Fig. 2a and b). Among the patients in the fasting group, the levels of ketone bodies, such as S-acetate, S-acetoacetate, and S-3-hydroxybutyrate, were increased (Table 2). In addition, we observed increased S-N-acetylated groups, S-leucine, S-valine and S-isoleucine in the fasting group (all p < 0.05; Fig. 2b). We found positive correlations between tumor size and S-lactate (r = 0.344; p = 0.016) and tumor size and S-pyruvate (r = 0.370; p = 0.009).
Table 2.
Serum metabolites with p-values from t-tests, fasting group versus carbohydrate (CH), for all patients and for the ER positive subset
| Metabolite | p-valuea | Fold change | p-valuea ER+ | Fold change ER+ |
| 3-Hydroxybutyrate | 0.010 | -1.06 | 0.010 | -1.07 |
| Acetate | <0.001 | -1.22 | <0.001 | -1.21 |
| Acetoacetate | <0.001 | -1.25 | <0.001 | -1.20 |
| Acetone | 0.250 | -1.18 | 0.508 | -1.11 |
| Alanine | 0.692 | 1.01 | 0.544 | -1.02 |
| Asparagine | 0.237 | -1.05 | 0.376 | -1.04 |
| Citrate | 0.503 | 1.03 | 0.726 | 1.01 |
| Creatine | 0.905 | -1.01 | 0.704 | -1.02 |
| Creatinine | 0.066 | -1.06 | 0.039 | -1.07 |
| Dimethylsulfone | 0.319 | -1.09 | 0.154 | -1.15 |
| Glucose | 0.969 | 1.00 | 0.972 | 1.00 |
| Glutamine | 0.005 | -1.06 | 0.013 | -1.07 |
| Glycerol | 0.065 | -1.05 | 0.054 | -1.06 |
| Glycoprotein | 0.243 | -1.06 | 0.408 | -1.05 |
| Isoleucine | <0.001 | -1.26 | 0.001 | -1.22 |
| Isopropyl alcohol | 0.009 | -1.12 | 0.038 | -1.10 |
| Lactate | <0.001 | 1.36 | <0.001 | 1.26 |
| Leucine | <0.001 | -1.20 | 0.002 | -1.17 |
| Lysine | <0.001 | -1.12 | <0.001 | -1.11 |
| Methanol | 0.495 | -1.04 | 0.511 | -1.04 |
| Methionine | 0.052 | -1.11 | 0.062 | -1.11 |
| N-acetylated groups | <0.001 | -1.15 | <0.001 | -1.15 |
| Phenylalanine | <0.001 | -1.12 | <0.001 | -1.13 |
| Proline | 0.298 | -1.03 | 0.236 | -1.03 |
| Propylene Glycol | <0.001 | -1.13 | 0.004 | -1.10 |
| Pyruvate | <0.001 | 1.27 | <0.001 | 1.23 |
| Threonine | 0.035 | -1.07 | 0.016 | -1.08 |
| Valine | <0.001 | -1.31 | <0.001 | -1.29 |
Abbreviations: ER+ Estrogen Receptor positive
aSignificant at p ≤ 0.016 after Benjamini-Hochberg correction for multiple testing
Fig. 2.
Partial Least Square Discriminant Analysis (PLS-DA) in serum. a Scores plot showing serum samples from the fasting group (green) and carbohydrate group (red). The carbohydrate and fasting groups have significantly different metabolic profiles as evidenced by permutation testing. b Variable Importance in Projection (VIP) scores showing the top 14 metabolites contributing to differences between the groups. The right column indicates increased (red) or decreased (green) metabolite in the indicated group
In the carbohydrate group, there was a positive linear correlation between proliferation (Ki-67) and tumor size (r = 0.782, p = 0.038). When Ki-67, PPH3 and MAI were included in a forward and backward stepwise linear regression MAI was the only independent factor explaining increment in tumor size with a Beta = 0.530 (95%CI, 0.201 to 0.875) P = 0.009. In the fasting group, there was no correlation between tumor size and proliferation.’
Serum glucose and insulin responses
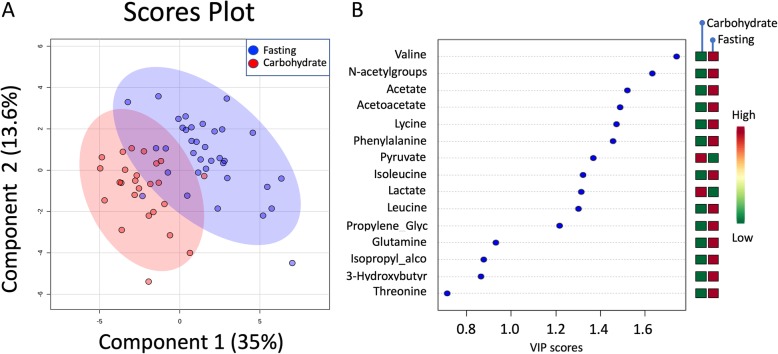
The mean fasting glucose and insulin values at admission were 5.4 mmol/L (95% CI 5.1 to 10.0) and 9.4 mIU (95% CI 6.8 to 32.5), respectively (normal ranges: glucose, 4.0 to 6.0 mmol/L; insulin, 6.0 to 27.0 mIU; c-peptide, 0.3 to 2.4 nmol/L). In the carbohydrate group, the mean preoperative insulin value was 35.6 mIU (26.7 to 106 mIU), compared to 9.1 (8.6 to 22 mIU) in the fasting group (student’s t-test p < 0.001). For C-peptide, the mean values in the carbohydrate and fasting groups were 2.10 nmol/L and 0.76 nmol/L, respectively (p < 0.001). We found significant univariate correlations between the serum concentrations of preoperative insulin (Table 3), Insulin C peptide (Table 9 in Appendix) IGFBP3 (Table 10 in Appendix), but not to IGF1 (Table 11 in Appendix). Multivariate analysis with leave-one-out cross-validation showed significant correlations between the serum metabolic profile and insulin (Cross-validated (CV) (R2 = 0.33, p < 0.001; Fig. 3a+b), Insulin C-peptide (CV R2 = 0.35, p < 0.001; Fig. 3c+d), IGFBP3 (CV R2 = 0.11, p < 0.001; Fig. 3e+f), but not IGF-1. For both insulin and insulin C-peptide, the most important metabolites for predictions were increased S-glucose, S-lactate and decreased S-Leucine. For IGFBP3, the most important metabolites were increased S-Acetone, S-Glycoprotein, and S-Leucine. We also found positive correlations between S-lactate and the preoperative increase in S-insulin and S-insulin / c-peptide (r = 0.57; p < 0.001 and r = 0.61; p < 0.0001), and between S-pyruvate and the increase in preoperative S-insulin and S-insulin c-peptide (r = 0.54; p < 0.001 and r = 0.60; p < 0.001).
Table 3.
Serum metabolite values correlated to insulin (Pearson’s correlation) for the total study population, and the carbohydrate and fasting groups separately
| Metabolite | R (All) | P (All)* | R (CH) | P (CH) | R (F) | P (F) |
| Lactate | 0.57 | <0.001 | 0.31 | 0.136 | 0.70 | <0.001 |
| pyruvate | 0.54 | <0.001 | 0.26 | 0.203 | 0.54 | 0.001 |
| Acetate | -0.53 | <0.001 | -0.40 | 0.046 | -0.22 | 0.212 |
| N.acetylgroups | -0.41 | 0.001 | -0.06 | 0.788 | 0.10 | 0.576 |
| Acetoacetate | -0.34 | 0.008 | -0.04 | 0.847 | 0.21 | 0.221 |
| Valine | -0.31 | 0.016 | 0.31 | 0.137 | 0.28 | 0.105 |
| Lysine | -0.29 | 0.027 | 0.01 | 0.947 | 0.43 | 0.010 |
| Citrate | 0.28 | 0.029 | 0.27 | 0.192 | 0.50 | 0.002 |
| Isoleucine | -0.28 | 0.030 | -0.03 | 0.881 | 0.36 | 0.035 |
| Glucose | 0.26 | 0.043 | 0.40 | 0.047 | -0.09 | 0.622 |
| Propylene_Glycol | -0.24 | 0.062 | 0.07 | 0.748 | 0.21 | 0.219 |
| Creatine | -0.23 | 0.075 | -0.39 | 0.054 | -0.18 | 0.292 |
| Leucine | -0.23 | 0.079 | 0.17 | 0.425 | 0.25 | 0.149 |
| Phenylalanine | -0.19 | 0.149 | 0.43 | 0.033 | 0.24 | 0.163 |
| Glycerol | -0.19 | 0.152 | -0.12 | 0.555 | 0.11 | 0.541 |
| Alanine | 0.15 | 0.262 | 0.18 | 0.386 | 0.30 | 0.076 |
| Isopropyl alcohol | -0.12 | 0.344 | 0.20 | 0.331 | 0.12 | 0.500 |
| 3-Hydroxybutyrate | -0.10 | 0.442 | 0.12 | 0.562 | 0.36 | 0.035 |
| Methanol | -0.10 | 0.457 | -0.06 | 0.778 | -0.05 | 0.761 |
| Glutamine | -0.09 | 0.506 | 0.30 | 0.150 | 0.06 | 0.737 |
| Creatinine | -0.08 | 0.543 | 0.23 | 0.263 | -0.05 | 0.755 |
| Threonine | -0.08 | 0.567 | 0.10 | 0.627 | 0.32 | 0.062 |
| Acetone | 0.04 | 0.780 | 0.15 | 0.489 | 0.39 | 0.019 |
| Proline | -0.04 | 0.789 | 0.11 | 0.593 | -0.03 | 0.880 |
| Glycoproteins | 0.02 | 0.873 | 0.06 | 0.787 | 0.49 | 0.003 |
| Asparagine | -0.01 | 0.923 | -0.05 | 0.818 | 0.47 | 0.005 |
| Methionine | 0.01 | 0.941 | 0.37 | 0.067 | 0.08 | 0.653 |
| Dimethylsulfone | 0.00 | 0.997 | 0.17 | 0.404 | 0.05 | 0.777 |
Abbreviations: CH Carbohydrate group, F Fasting group, P Pearson’s correlation p-value, R Pearson’s correlation R value.
*Significant at p ≤ 0.043 after Benjamini-Hochberg correction
** Significant at p ≤ 0.03 after Benjamini-Hochberg correction
*** Significant at p ≤ 0.035 after Benjamini-Hochberg correction
Table 9.
Metabolites correlated to serum insulin C peptide (Pearson’s correlation) for all patients, carbohydrate group, and fasting groups
| Metabolite | R All | P Alla | R CH | P CHa | R F | P Fb |
| Lactate | 0.611 | <0.001 | 0.401 | 0.047 | 0.577 | <0.001 |
| Pyruvate | 0.596 | <0.001 | 0.395 | 0.051 | 0.431 | 0.010 |
| Acetate | -0.513 | <0.001 | -0.344 | 0.092 | -0.092 | 0.598 |
| N-acetylgroups | -0.398 | 0.002 | -0.092 | 0.663 | 0.397 | 0.018 |
| Valine | -0.366 | 0.004 | 0.189 | 0.366 | 0.385 | 0.023 |
| Acetoacetate | -0.352 | 0.006 | -0.058 | 0.781 | 0.348 | 0.041 |
| Isoleucine | -0.333 | 0.009 | -0.166 | 0.428 | 0.488 | 0.003 |
| Lysine | -0.301 | 0.020 | -0.064 | 0.763 | 0.646 | <0.001 |
| Propylene Glycol | -0.260 | 0.045 | 0.044 | 0.833 | 0.305 | 0.075 |
| Citrate | 0.244 | 0.060 | 0.262 | 0.205 | 0.348 | 0.041 |
| Leucine | -0.242 | 0.063 | 0.125 | 0.551 | 0.416 | 0.013 |
| Glucose | 0.233 | 0.074 | 0.345 | 0.092 | 0.187 | 0.282 |
| Creatine | -0.209 | 0.110 | -0.444 | 0.026 | 0.005 | 0.978 |
| Phenylalanine | -0.206 | 0.115 | 0.357 | 0.080 | 0.503 | 0.002 |
| Methanol | -0.185 | 0.157 | -0.162 | 0.438 | -0.269 | 0.118 |
| Glycerol | -0.152 | 0.246 | -0.111 | 0.596 | 0.323 | 0.059 |
| Glutamine | -0.152 | 0.248 | 0.216 | 0.300 | 0.058 | 0.740 |
| Alanine | 0.150 | 0.252 | 0.207 | 0.320 | 0.303 | 0.077 |
| Isopropyl alcohol | -0.134 | 0.307 | 0.185 | 0.375 | 0.222 | 0.199 |
| Threonine | -0.108 | 0.409 | 0.000 | 1.000 | 0.462 | 0.005 |
| 3-Hydroxybutyrate | -0.098 | 0.455 | 0.153 | 0.464 | 0.439 | 0.008 |
| Creatinine | -0.077 | 0.557 | 0.176 | 0.399 | 0.158 | 0.365 |
| Dimethylsulfone | 0.076 | 0.565 | 0.264 | 0.202 | 0.332 | 0.051 |
| Acetone | 0.027 | 0.835 | 0.078 | 0.710 | 0.530 | 0.001 |
| Proline | -0.014 | 0.913 | 0.154 | 0.462 | 0.049 | 0.778 |
| Asparagine | -0.012 | 0.927 | -0.126 | 0.547 | 0.624 | <0.001 |
| Glycoproteins | 0.012 | 0.928 | 0.003 | 0.989 | 0.606 | <0.001 |
| Methionine | 0.010 | 0.938 | 0.412 | 0.041 | 0.138 | 0.430 |
aSignificant at p ≤ 0.045 after Benjamini-Hochberg correction
bSignificant at p ≤ 0.041 after Benjamini-Hochberg correction
Table 10.
Metabolites correlated to serum Insulin Growth Factor Binding Protein 3 (IGFBP3) (Pearson’s correlation) for all patients, carbohydrate group, and fasting groups
| Metabolite | R (All) | P (All)* | R (CH) | P (CH)* | R (F) | P (F)* |
| Isoleucine | 0.424 | 0.001 | 0.414 | 0.040 | 0.351 | 0.039 |
| Glycoproteins | 0.410 | 0.001 | 0.224 | 0.282 | 0.478 | 0.004 |
| Asparagine | 0.401 | 0.001 | 0.364 | 0.073 | 0.399 | 0.018 |
| Leucine | 0.393 | 0.002 | 0.107 | 0.612 | 0.440 | 0.008 |
| Acetone | 0.383 | 0.003 | 0.298 | 0.148 | 0.397 | 0.018 |
| Lysine | 0.378 | 0.003 | -0.017 | 0.937 | 0.459 | 0.006 |
| N.acetylgroups | 0.342 | 0.007 | -0.241 | 0.247 | 0.484 | 0.003 |
| Phenylalanine | 0.322 | 0.012 | 0.114 | 0.586 | 0.314 | 0.066 |
| Propylene-Glycol | 0.321 | 0.012 | 0.044 | 0.833 | 0.349 | 0.040 |
| Isopropyl-alcohol | 0.319 | 0.013 | -0.079 | 0.706 | 0.434 | 0.009 |
| Alanine | 0.310 | 0.016 | 0.100 | 0.635 | 0.413 | 0.014 |
| Acetoacetate | 0.243 | 0.062 | 0.036 | 0.866 | 0.192 | 0.268 |
| Threonine | 0.206 | 0.114 | -0.250 | 0.228 | 0.395 | 0.019 |
| Valine | 0.196 | 0.134 | 0.050 | 0.814 | 0.082 | 0.640 |
| Acetate | 0.180 | 0.169 | 0.079 | 0.707 | 0.066 | 0.707 |
| Lactate | -0.166 | 0.205 | -0.347 | 0.090 | 0.207 | 0.232 |
| pyruvate | -0.164 | 0.211 | -0.287 | 0.164 | 0.131 | 0.453 |
| Methionine | -0.158 | 0.228 | -0.484 | 0.014 | -0.066 | 0.704 |
| Glycerol | 0.137 | 0.296 | -0.384 | 0.058 | 0.360 | 0.034 |
| Proline | -0.124 | 0.345 | -0.358 | 0.079 | 0.004 | 0.982 |
| Creatine | 0.103 | 0.435 | 0.175 | 0.401 | 0.051 | 0.773 |
| Creatinine | 0.084 | 0.522 | -0.189 | 0.365 | 0.146 | 0.403 |
| Methanol | 0.079 | 0.549 | 0.288 | 0.162 | -0.110 | 0.528 |
| Glutamine | 0.075 | 0.569 | -0.176 | 0.399 | 0.123 | 0.481 |
| Glucose | 0.055 | 0.678 | 0.007 | 0.972 | 0.186 | 0.285 |
| Citrate | 0.031 | 0.812 | -0.052 | 0.804 | 0.134 | 0.442 |
| Dimethylsulfone | -0.005 | 0.967 | -0.255 | 0.219 | 0.096 | 0.584 |
| 3-Hydroxybutyrate | -0.001 | 0.993 | -0.320 | 0.119 | 0.046 | 0.794 |
Abbreviations: CH Carbohydrate group, F Fasting group, P Pearson’s correlation p-value, R Pearson’s correlation R value.
*Significant at p ≤ 0.01 after Benjamini-Hochberg correction for multiple testing
**Significant at p ≤ 0.037 after Benjamini-Hochberg correction for multiple testing
***Significant at p ≤ 0.04 after Benjamini-Hochberg correction for multiple testing
Table 11.
Metabolites correlated to serum Insulin Growth Factor 1 (IGF1) (Pearson’s correlation) for all patients, carbohydrate group, and fasting groups
| Metabolite | R (All) | P (All)a | R (CH) | P (CH)a | R (F) | P (F)a |
| Methionine | -0.318 | 0.013 | -0.591 | 0.002 | -0.135 | 0.438 |
| Isopropyl_alcohol | -0.314 | 0.015 | -0.318 | 0.121 | -0.330 | 0.052 |
| Creatinine | -0.302 | 0.019 | -0.381 | 0.061 | -0.265 | 0.124 |
| Proline | -0.274 | 0.034 | -0.465 | 0.019 | -0.095 | 0.585 |
| Valine | -0.251 | 0.053 | -0.122 | 0.563 | -0.406 | 0.015 |
| Propylene_Glycol | -0.230 | 0.077 | -0.159 | 0.446 | -0.291 | 0.089 |
| Acetoacetate | -0.230 | 0.077 | -0.168 | 0.421 | -0.308 | 0.072 |
| Methanol | -0.218 | 0.094 | -0.001 | 0.996 | -0.400 | 0.017 |
| Acetone | -0.184 | 0.159 | -0.061 | 0.772 | -0.247 | 0.152 |
| pyruvate | -0.173 | 0.186 | -0.271 | 0.191 | -0.179 | 0.303 |
| Leucine | -0.160 | 0.222 | -0.090 | 0.670 | -0.208 | 0.231 |
| 3.Hydroxybutyrate | -0.157 | 0.230 | -0.231 | 0.266 | -0.114 | 0.515 |
| Dimethylsulfone | 0.155 | 0.238 | 0.032 | 0.880 | 0.245 | 0.155 |
| Threonine | -0.152 | 0.248 | -0.397 | 0.050 | 0.013 | 0.941 |
| Lactate | -0.139 | 0.289 | -0.323 | 0.115 | -0.026 | 0.882 |
| N.acetylgroups | -0.131 | 0.320 | -0.385 | 0.057 | -0.038 | 0.830 |
| Glycerol | -0.125 | 0.342 | -0.430 | 0.032 | 0.075 | 0.668 |
| Lysine | -0.118 | 0.370 | -0.169 | 0.418 | -0.104 | 0.551 |
| Glutamine | 0.100 | 0.447 | -0.053 | 0.801 | 0.257 | 0.136 |
| Isoleucine | -0.096 | 0.468 | 0.116 | 0.582 | -0.225 | 0.193 |
| Acetate | -0.092 | 0.483 | 0.031 | 0.883 | -0.148 | 0.395 |
| Creatine | -0.087 | 0.507 | -0.002 | 0.994 | -0.154 | 0.378 |
| Glycoproteins | -0.081 | 0.539 | -0.059 | 0.779 | -0.088 | 0.614 |
| Citrate | 0.063 | 0.631 | 0.115 | 0.584 | 0.014 | 0.936 |
| Alanine | -0.048 | 0.717 | -0.070 | 0.738 | -0.048 | 0.783 |
| Glucose | 0.017 | 0.899 | -0.008 | 0.969 | 0.079 | 0.652 |
| Phenylalanine | 0.011 | 0.931 | 0.253 | 0.222 | -0.080 | 0.648 |
| Asparagine | -0.006 | 0.962 | 0.393 | 0.052 | -0.163 | 0.349 |
Abbreviations: CH Carbohydrate group. F Fasting group, P Pearson’s correlation p-value, R Pearson’s correlation R value.
aSignificant at p ≤ 0.002 after Benjamini-Hochberg correction for multiple testing
Fig. 3.
Correlation between serum metabolic profile and serum insulin, insulin C-peptide, and IGFBP3. Samples from carbohydrate-fed patients are shown in red, while samples from fasting patients are shown in blue. Metabolites are colored according to their variable importance in projection (VIP) score and labeled when VIP≥1. a Measured insulin vs. predicted insulin levels based on metabolic profile (cross-validated measurements). b Metabolites versus regression coefficient for insulin. Increased S-glucose, S-lactate, and decreased S-Leucine are important to prediction of serum insulin from the metabolic profile. c Measured insulin C peptide vs. predicted insulin C-peptide levels. d Regression weight plot showing metabolites versus the regression coefficient for insulin C-peptide. Increased S-Glucose, S-Lactate, and decreased S-Leucine are important to prediction of serum insulin C-peptide from the metabolic profile. e Measured Insulin Growth Factor Binding Protein 3 (IGFBP3) vs. predicted IGFBP3 based on metabolic profile. f Regression weight plot showing metabolites versus the regression coefficient for IGFBP3. Increased S-Acetone, S-Glycoproteins, and S-Leucine are important to prediction of serum IGFBP3 from the metabolic profile
Tumor metabolism
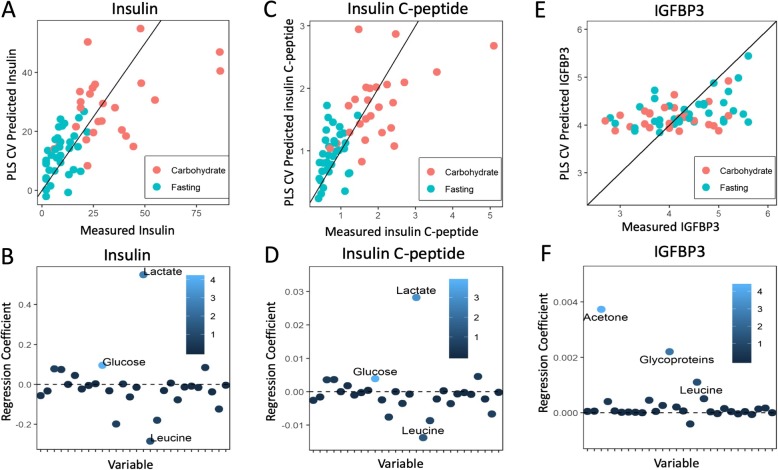
Metabolites included in the analysis are presented in Table 4. PLS-DA did not result in a significant model discriminating between fasting and carbohydrate-fed patients, and no metabolites were significantly different in univariate testing when all tumors were analyzed (Fig. 4a). However, for ER-positive tumors (n = 18), glutathione was significantly elevated in the carbohydrate group compared to the fasting group (p = 0.002; Fig. 4b), even after adjusting for tumor size. In the ROC analysis, we found an area under the curve (AUC) of 0.894 (95%CI = 0.687–1.000, p = 0.0015) for glutathione in discriminating between fasting and carbohydrate-fed patients with ER-positive tumors (Fig. 4c). The difference was also significant in the ER-positive tumors with low proliferation (MAI < 10; n = 7). Moreover, we found a positive correlation between preoperative S-insulin levels and the glutathione content in tumor tissue (r = 0.680; p = 0.002). Furthermore, we observed a higher level of tissue glutamate in tumors with a high proliferation as measured by Ki67</≥ 15% (p = 0.004). This association remained significant when adjusted for intervention group using a general linear model with intervention status as fixed factor, Ki67</≥ 15% as random factor, and tissue Glutamate as dependent variable (p = 0.009). Also, choline (p = 0.002) and phosphoetanolamine (p = 0.019) were increased in T2 tumors compared to T1 tumors.
Table 4.
Tumor metabolites with fold changes and t-test p-values in carbohydrate vs fasting groups
| Tumor metabolite | P (All)a | FC (All) | P (ER+)b | FC (ER+) |
| Acetate | 0.844 | -1.030 | 0.620 | -1.095 |
| Alanine | 0.322 | 1.038 | 0.163 | 1.067 |
| Ascorbate | 0.300 | -1.099 | 0.991 | -1.001 |
| Aspartate | 0.385 | 1.100 | 0.545 | 1.088 |
| Choline | 0.136 | 1.056 | 0.547 | 1.027 |
| Creatine | 0.418 | -1.062 | 0.558 | -1.051 |
| Glucose | 0.495 | -1.151 | 0.500 | -1.201 |
| Glutamate | 0.172 | 1.047 | 0.146 | 1.055 |
| Glutamine | 0.955 | 1.003 | 0.816 | -1.015 |
| Glutathione | 0.006 | 1.082 | 0.002 | 1.103 |
| Glycerophosphocholine | 0.712 | -1.018 | 0.762 | -1.018 |
| Glycine | 0.186 | 1.063 | 0.162 | 1.090 |
| Lactate | 0.862 | 1.006 | 0.922 | 1.004 |
| Leucine | 1.000 | 1.000 | 0.947 | -1.004 |
| Myoinositol | 0.445 | -1.038 | 0.768 | -1.018 |
| Phosphocholine | 0.517 | 1.027 | 0.291 | 1.051 |
| Phosphoethanolamine | 0.211 | 1.050 | 0.544 | 1.031 |
| Scylloinositol | 0.926 | -1.007 | 0.565 | 1.060 |
| Succinate | 0.788 | 1.022 | 0.503 | 1.067 |
| Taurine | 0.982 | 1.001 | 0.902 | 1.004 |
Abbreviations: ER+ Estrogen receptor positive, FC Fold change, P T-test p-value
aSignificant at p ≤ 0.001 after Benjamini-Hochberg correction
bSignificant at p ≤0.030 after Benjamini-Hochberg correction
Fig. 4.
a Principal Component Analysis (PCA) of tumor metabolites. No grouping of fasting vs carbohydrate groups observed. b Glutathione levels in ER positive tumors. c ROC curve for classification into carbohydrate or fasting group by glutathione concentration in ER-positive tumors. AUC= 0.894; 95%CI=0.0.687-1.000, P=0.002
Survival analysis
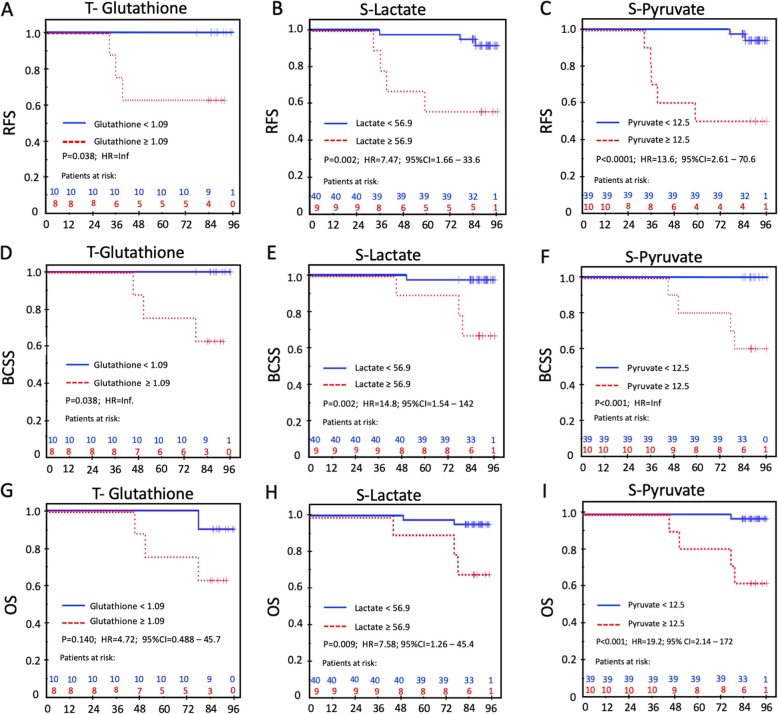
First, we used S-lactate, S-pyruvate, and tissue (T) glutathione as continuous variables in a univariate Cox model for RFS, BCSS and OS. Both S-pyruvate and S-lactate, but not T-glutathione reached significance with a hazard ratio (HR) for RFS of 1.53 (95% CI, 1.11 to 2.11; p = 0.009) and 1.08 (95% CI, 1.01 to 1.17; p = 0.029), respectively. For BCSS the HR for the continuous variables of S-pyruvate and S-Lactate were 1.85 (95%CI, 1.15 to 2.97; p = 0.011) and 1.13 (95%CI, 1.01 1.26; p = 0.028) respectively. The corresponding observations for OS were 1.63 (95%CI, 1.11 to 2.40; p = 0.014) for lactate and 1.10 (95%CI, 1.002 to 1.20; p = 0.045) for pyruvate. Thereafter, the following independent variables were dichotomized according to the optimal ROC-derived thresholds: S-lactate, S-pyruvate, preoperative S-insulin, preoperative S-insulin-c-peptide, and tissue glutathione. In addition, the well-established prognostic factors tumor size, nodal status, histological grade, MAI 10, Ki-67-30 and PPH3–13 were deemed clinically relevant and included as explanatory variables in the multivariable analyses. The results of the univariate RFS, BCSS, and OS analyses are given in Tables 5, 6 and 7, respectively. Patients with a high glutathione content in the tumor (≥1.09) had a 37% risk of experiencing a relapse and 37% risk of dying of breast cancer compared to no relapses and no deaths in patients with a low glutathione content in the tumor (both comparisons: p = 0.038; HR = Inf.; Fig. 5a and d). Patients with high S-lactate (≥56.9) had RFS of 71% compared to 97% for those with lower S-lactate (p = 0.002, HR = 7.47; 95% CI 1.66–33.6; Fig. 5b). Patients with S-pyruvate ≥12.5 had an adverse RFS of 50% compared to 95% for the patients with S-pyruvate < 12.5 (p < 0.0001; HR = 13.6; 95% CI 2.61–70.6; Fig. 5c). The same pattern was observed in the BCSS and OS analyses for these three prognostic variables (Fig. 5e-i). Notably, only one contralateral relapse occurred in the fasting group – all others were in the carbohydrate group. Even though the relapses were restricted to patients with T2 tumors, tumor category was not an independent prognostic factor in the multivariable analyses. In the multivariable analysis for RFS, S-pyruvate was the only factor left in the final model (HR = 12.8; 95% CI, 2.47 to 66.8), and only S-lactate remained in the final multivariable model for BCSS (HR = 14.8; 95% CI 1.54 to 142). Furthermore, S-pyruvate was the sole factor to reach significance in the multivariable model of the OS analysis (HR = 18.2; 95% CI 2.03 to 164).
Table 5.
Univariate analysis of Relapse Free Survival in ER+ patients
| Variable | Events / At risk | % Survival | P | HR | 95% CI |
| Fasting / Carbohydrate | |||||
| Fasting | 1/29 | 97 | |||
| Carbohydrate | 6/21 | 71 | 0.012 | 9.34 | 1.12 –77.7 |
| S-Pyruvatea | |||||
| < 12.5 | 2/39 | 95 | |||
| ≥ 12.5 | 5/10 | 50 | <0.0001 | 13.59 | 2.61– 70.6 |
| S-Lactatea | |||||
| < 56.9 | 3/40 | 93 | |||
| ≥ 56.9 | 4/9 | 56 | 0.002 | 7.47 | 1.66 – 33.6 |
| S-Preoperative Insulin | |||||
| < 18.3 I.U. | 1/29 | 97 | |||
| ≥ 18.3 I.U. | 6/21 | 71 | 0.012 | 9.34 | 1.12 – 77.7 |
| S-Preoperative C-peptide | |||||
| < 1.22 nM | 1/29 | 97 | |||
| ≥1.22 nM | 6/21 | 71 | 0.011 | 9.51 | 1.14-79.0 |
|
Tumor Glutathione | |||||
| < 1.09 | 0/10 | 100 | |||
| ≥ 1.09 | 3/8 | 63 | 0.038 | Inf. | |
| Tumor size | |||||
| T1 | 3/40 | 93 | |||
| T2 | 4/10 | 60 | 0.003 | 7.09 | 1.57-31.9 |
| Nodal status | |||||
| N0 | 3/33 | 91 | |||
| N+ | 4/17 | 73 | 0.160 | 2.80 | 0.625-12.6 |
| Grade | |||||
| 1 | 0/11 | 100 | |||
| 2+3 | 7/39 | 82 | 0.136 | 31.1 | 0.019 – 50547 |
| MAIa | |||||
| <10 | 4/39 | 90 | |||
| ≥10 | 3/10 | 70 | 0.092 | 3.38 | 0.751–15.2 |
| Ki67a | |||||
| <30% | 3/37 | 92 | |||
| ≥30% | 4/12 | 67 | 0.023 | 4.84 | 1.08 – 21.8 |
| PPH3 | |||||
| <13 | 3/35 | 91 | |||
| ≥13 | 4/15 | 73 | 0.116 | 3.13 | 0.699-14.0 |
aMissing information on one patient in the ER+ group leading to n=49 patients analyzed for this variable
Table 6.
Univariate analysis of Breast Cancer Specific Survival in ER+ patients
| Variable |
Events / At risk |
% Survival | P | HR | 95% CI |
| Fasting /Carbohydrate | |||||
| Fasting | 0/29 | 100 | |||
| Carbohydrate | 4/21 | 81 | 0.015 | Inf. | |
| S-Pyruvate | |||||
| < 12.5 | 0/40 | 100 | |||
| ≥ 12.5 | 4/10 | 60 | <0. 0001 | Inf. | |
| S-Lactatea | |||||
| < 56.9 | 1/40 | 98 | |||
| ≥ 56.9 | 3/9 | 67 | 0.002 | 14.8 | 1.53-142 |
| S-Preoperative Insulin | |||||
| < 18.3 I.U. | 0/29 | 100 | |||
| ≥ 18.3 I.U. | 4/21 | 81 | 0.015 | Inf. | |
| S-Preoperative C-peptide | |||||
| < 1.22 nM | 0/29 | 100 | |||
| ≥1.22 nM | 4/21 | 81 | 0.015 | 103 | 0.025-429676 |
|
Tumor Glutathione | |||||
| < 1.09 | 0/10 | 100 | |||
| ≥ 1.09 | 3/8 | 63 | 0.038 | Inf. | |
| Tumor size | |||||
| T1 | 0/40 | 100 | |||
| T2 | 4/10 | 60 | <0.0001 | Inf. | |
| Nodal status | |||||
| N0 | 1/33 | 97 | |||
| N+ | 3/17 | 82 | 0.080 | 5.92 | 0.615 – 56.9 |
| Grade | |||||
| 1 | 0/11 | 100 | |||
| 2+3 | 4/39 | 90 | 0.277 | 30.1 | Inf. |
| MAIa | |||||
| <10 | 2/39 | 95 | |||
| ≥10 | 2/10 | 80 | 0.124 | 4.12 | 0.580- 29.3 |
| Ki67a | |||||
| <30% | 1/37 | 97 | |||
| ≥30% | 3/12 | 75 | 0.014 | 9.91 | 1.03-95.3 |
| PPH3 | |||||
| <13 | 2/35 | 94 | |||
| ≥13 | 2/15 | 87 | 0.399 | 2.27 | 0.320 – 16.1 |
aMissing information on one patient in the ER+ group leading to n=49 patients analyzed for this variable
Table 7.
Univariate analysis of Overall Survival in ER+ patients
| Variable | Events /At risk | % survival | P | HR | 95% CI |
| Carbo/Faste | |||||
| Faste | 1/29 | 97 | |||
| Carbohydrate | 4/21 | 81 | 0.068 | 6.02 | 0.675–53.8 |
| S-Pyruvata | |||||
| < 12.5 | 1/39 | 97 | |||
| ≥ 12.5 | 4/10 | 60 | <0.0001 | 19.2 | 2.14–172 |
| S-Lactatea | |||||
| < 56.9 | 2/40 | 95 | |||
| ≥ 56.9 | 3/9 | 67 | 0.009 | 7.58 | 1.26–45.4 |
| S-Preop Insulin | |||||
| < 18.3 I.U. | 1/29 | 97 | |||
| ≥ 18.3 I.U. | 4/21 | 81 | 0.068 | 6.016 | 0.672–53.9 |
| S-Preoperative C-peptide | |||||
| < 1.22 nM | 1/29 | 97 | |||
| ≥1.22 nM | 4/21 | 81 | 0.068 | 6.02 | 0.672–53.9 |
| Tissue Glutathione | |||||
| ≤1.0855 | 1/10 | 90 | |||
| >1.0855 | 3/8 | 63 | 0.140 | 4.72 | 0.488–45.7 |
| Tumor size | |||||
| T1 | 1/40 | 98 | |||
| T2 | 4/10 | 60 | < 0.0001 | 19.2 | 2.20 –176 |
| Nodal status | |||||
| N0 | 2/33 | 94 | |||
| N+ | 3/17 | 82 | 0.205 | 3.01 | 0.502–18.0 |
| Grade | |||||
| 1 | 0/11 | 100 | |||
| 2+3 | 5/39 | 87 | 0.222 | 30.2 | 0.004–223736 |
| MAIa | |||||
| <10 | 3/39 | 92 | |||
| ≥10 | 2/10 | 80 | 0.235 | 2.83 | 0.471–16.9 |
| Ki67a | |||||
| <30% | 2/37 | 95 | |||
| ≥30% | 3/12 | 75 | 0.049 | 5.040 | 0.842–30.2 |
| PPH3 | |||||
| <13 | 3/35 | 91 | |||
| ≥13 | 2/15 | 87 | 0.641 | 1.53 | 0.255–9.13 |
aMissing information on one patient in the ER+ group leading to n=49 patients analyzed for this variable
Fig. 5.
Survival analyses for Tumor-Glutathione, Serum-lactate and Serum-pyruvate. a-c Relapse Free Survival (RFS); d-f Breast Cancer Survival (BCSS); g-i Overall Survival (OS)
Pathway analyses
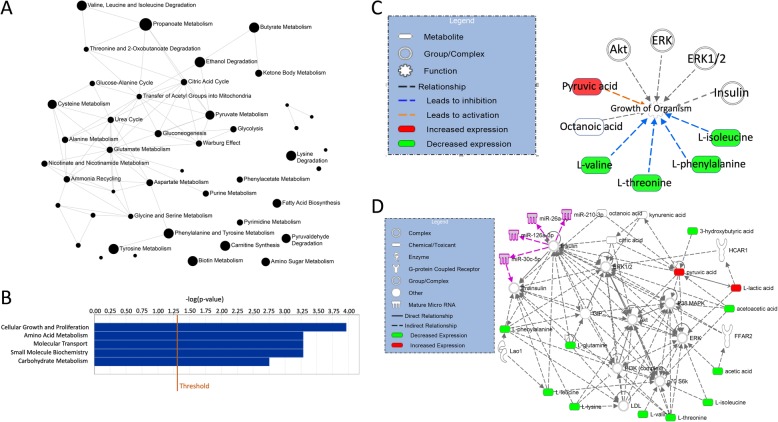
In the Pathway analyses, MetaboAnalyst and IPA showed complimentary information. Quantitative metabolite set enrichment analysis (MSEA) identified biologically meaningful patterns in serum metabolite concentration changes (Fig. 6a and Table 12 in Appendix). Significantly enriched pathways included energy associated metabolic pathways (amino sugar metabolism and pyruvate metabolism which links to glutamate metabolism, the citric acid cycle, gluconeogenesis and the Warburg effect). IPA showed the main functions of the involved metabolites as cellular growth and proliferation, molecular transport, small molecule biochemistry, carbohydrate metabolism and amino acid metabolism (Fig. 6b). Interestingly, the metabolites showed a pattern congruent with growth of organism (Fig. 6c) with metabolites increased in carbohydrate-fed patients activating growth pathways, and downregulation of metabolites acting as inhibitors of growth. Finally, four (miR-26a-5p, miR-30c-5p, miR126-3p and miR-210-3p) out of the seven microRNAs found to be involved in resistance to tamoxifen in our previous review [27] could indirectly be associated with the metabolic network through insulin signaling pathways (Fig. 6d). The same metabolic pathways were evident when only ER positive patients were considered.
Fig. 6.
Pathway analyses in serum metabolites. a Metabolite Set Enrichment Analysis of serum metabolism. Significantly enriched pathways are annotated in the pathway network. The circle size denotes significance of the pathway, and lines denote at least 25% shared metabolites in the pathways. b Ingenuity pathway analysis (IPA) bar chart showing the top 5 functions enriched in the dataset. c IPA pathway network showing the metabolites connected to four microRNAs found to be involved in tamoxifen resistance. Metabolites in green are downregulated in carbohydrate-fed patients, while metabolites in red are upregulated. MicroRNAs are colored purple. d IPA Function plot showing metabolites involved in organismal growth. Orange arrows indicate activation, while blue arrows indicate inhibition
Table 12.
Results from Quantitative Metabolite Set Enrichment Analysis
| Metabolic pathway | Total Cmpd | Hits |
Statistic Q (Expected 1.613) |
Raw p | FDR |
| Amino Sugar Metabolism | 33 | 3 | 24.50 | 0.000 | 0.000 |
| Propanoate Metabolism | 42 | 1 | 44.09 | 0.000 | 0.000 |
| Valine, Leucine and Isoleucine Degradation | 60 | 4 | 31.60 | 0.000 | 0.000 |
| Pyruvate Metabolism | 48 | 3 | 28.66 | 0.000 | 0.000 |
| Phenylalanine and Tyrosine Metabolism | 28 | 2 | 31.54 | 0.000 | 0.000 |
| Fatty Acid Biosynthesis | 35 | 3 | 25.58 | 0.000 | 0.000 |
| Aspartate Metabolism | 35 | 3 | 16.23 | 0.000 | 0.000 |
| Ethanol Degradation | 19 | 1 | 33.64 | 0.000 | 0.000 |
| Tyrosine Metabolism | 72 | 1 | 32.22 | 0.000 | 0.000 |
| Butyrate Metabolism | 19 | 1 | 32.22 | 0.000 | 0.000 |
| Lysine Degradation | 30 | 1 | 31.52 | 0.000 | 0.000 |
| Biotin Metabolism | 8 | 1 | 31.52 | 0.000 | 0.000 |
| Carnitine Synthesis | 22 | 1 | 31.52 | 0.000 | 0.000 |
| Ammonia Recycling | 32 | 3 | 14.09 | 0.000 | 0.000 |
| Warburg Effect | 58 | 5 | 13.16 | 0.000 | 0.000 |
| Cysteine Metabolism | 26 | 1 | 27.23 | 0.000 | 0.000 |
| Pyruvaldehyde Degradation | 10 | 1 | 27.23 | 0.000 | 0.000 |
| Urea Cycle | 29 | 3 | 13.38 | 0.000 | 0.000 |
| Glutamate Metabolism | 49 | 3 | 13.38 | 0.000 | 0.000 |
| Gluconeogenesis | 35 | 3 | 17.45 | 0.000 | 0.000 |
| Ketone Body Metabolism | 13 | 2 | 17.24 | 0.000 | 0.000 |
| Glycolysis | 25 | 2 | 13.61 | 0.000 | 0.000 |
| Citric Acid Cycle | 32 | 2 | 14.00 | 0.000 | 0.001 |
| Glycine and Serine Metabolism | 59 | 5 | 8.26 | 0.000 | 0.001 |
| Alanine Metabolism | 17 | 2 | 13.75 | 0.000 | 0.001 |
| Transfer of Acetyl Groups into Mitochondria | 22 | 3 | 9.34 | 0.001 | 0.001 |
| Glucose-Alanine Cycle | 13 | 3 | 9.17 | 0.001 | 0.002 |
| Pyrimidine Metabolism | 59 | 1 | 12.64 | 0.005 | 0.008 |
| Nicotinate and Nicotinamide Metabolism | 37 | 1 | 12.64 | 0.005 | 0.008 |
| Purine Metabolism | 74 | 1 | 12.64 | 0.005 | 0.008 |
| Phenylacetate Metabolism | 9 | 1 | 12.64 | 0.005 | 0.008 |
| Threonine and 2-Oxobutanoate Degradation | 20 | 1 | 7.42 | 0.035 | 0.048 |
| Methionine Metabolism | 43 | 1 | 6.38 | 0.052 | 0.065 |
| Betaine Metabolism | 21 | 1 | 6.38 | 0.052 | 0.065 |
| Spermidine and Spermine Biosynthesis | 18 | 1 | 6.38 | 0.052 | 0.065 |
| Glycerolipid Metabolism | 25 | 1 | 5.74 | 0.065 | 0.080 |
| Galactose Metabolism | 38 | 2 | 2.87 | 0.184 | 0.219 |
| Arginine and Proline Metabolism | 53 | 2 | 0.94 | 0.575 | 0.665 |
| Glutathione Metabolism | 21 | 1 | 0.27 | 0.692 | 0.743 |
| Selenoamino Acid Metabolism | 28 | 1 | 0.27 | 0.692 | 0.743 |
| Tryptophan Metabolism | 60 | 1 | 0.27 | 0.692 | 0.743 |
| Sphingolipid Metabolism | 40 | 1 | 0.00 | 0.969 | 0.969 |
| Lactose Synthesis | 20 | 1 | 0.00 | 0.969 | 0.969 |
| Lactose Degradation | 9 | 1 | 0.00 | 0.969 | 0.969 |
Discussion
We present the first study to examine the effect of per-oral preoperative carbohydrate load on perioperative metabolism in operable breast cancer patients. Among the 15 different serum metabolites that distinguished fasting from the per-oral carbohydrate load, we observed increased systemic lactate and pyruvate, decreased ketone bodies, increased glycerol, and reduced amino acids in the patients who received the carbohydrate load. Moreover, we found highly significant positive correlations between S-insulin and S-lactate and S-pyruvate. Thus, changes in these 15 key metabolites are consistent with increased glycolysis, increased ketolytic activity, reduced lipolysis, and reduced proteolysis, which are exactly the same metabolic modifications seen after carbohydrate challenge in healthy persons [28]. Being able to capture these well-known metabolic effects of insulin increases the reliability of our model to detect other changes that may follow a carbohydrate load.
It may be considered that 18 h is too short to expect effect of the carbohydrate load on tumor cell proliferation and metabolism. However, in vitro studies show that glucose fed MCF-7 cells increase their proliferation after 12–24 h [3] .Others found the same pattern in three different breast cancer cell lines [29]. As the cell lines lack the in vivo endocrine response to glucose the increased proliferation was based on GTP-ase driven phosphorylation of EGFR with increased activity and longevity of this receptor as a consequence. Also, animals fed with a diet containing increased glucose show an increased epithelial mesenchymal transition (EMT) [30].
The increased S-lactate and S-pyruvate in the carbohydrate patients stems primarily from two sources. Firstly, lactate is the product of glycolysis, especially in muscle cells, and is transported to the liver for conversion back to glucose, known as the Cori cycle [31]. The intended effect of preOP is to contra act and reduce insulin resistance that follows surgical stress [32]. This stressor leads to reduced mitochondrial ATP production and lactate formation [33, 34]. In healthy individuals, an oral glucose tolerance test (OGTT) showed a negative correlation between differences in S-glucose concentrations and differences in S-lactate levels (i.e. a rise in S-glucose leads to a reduction in S-lactate) [35]. Moreover, during 180 min after an OGTT among non-insulin dependent diabetic mellitus (NIDDM) patients there was no significant alteration in S-lactate levels [36]. Thus, it is unlikely that preOp itself creates a systemic lactate production. Therefore, S-lactate in our patients may come from excretion of intracellular lactate and pyruvate produced in the breast cancer cells. Consequently, lactate and pyruvate in the present study are probably translocated into the systemic circulation via mono carboxylate transporter type 4 (MCT-4), which is a known part of the Warburg effect [13]. Despite the fact that systemic metabolite concentrations are functional read outs of the numerous homeostatic reactions in the body, which will blur the contribution from the cancer cell metabolism to the serum levels [18], our present observation of positive correlation between larger tumor size and increasing S-lactate is supported by Hui S et al. [37].. Also, the positive correlation between proliferation and tumor size solely occurs in the carbohydrate group this suggests that carbohydrate exposure to larger tumors (i.e.T2 tumors) increases both proliferation and S-lactate. Thus, this indicates that lactate from the Warburg effect in the tumor cells may have a substantial contribution to the systemic lactate and pyruvate levels. This observation also adheres to the lack of correlation between intra tumor lactate/pyruvate and fasting/carbohydrate status in the present study, as the former are probably excreted from the cells into the systemic environment.
Moreover, tumor cells not only produce lactate for excretion through MCT-4, they are also able to take up systemic circulating lactate and pyruvate via the MCT-1 transporters [38]. Regardless of the source, systemic lactate and pyruvate will certainly benefit the free CTCs shed from the tumor during surgery that are on their way to distant tissue to form micrometastases [39] but may also benefit the preoperatively established occult micrometastases [40, 41]. Lactate and pyruvate are the most preferred substrates for lactate/pyruvate dehydrogenase (LDH/PDH), ensuring a 1:1 ratio between lactate and pyruvate when equilibrium is reached. Thus, LDH provides substrate for both the production of ATP via the tricarboxylic acid (TCA) cycle [37] and also increased gluconeogenesis for the production of ribose for nucleotide synthesis via the pentose phosphate pathway (PPP) [38]. Notably, increased levels of serum LDH [42] and increased expression of LDH in breast cancer tissue [43] and lung tumors [44] are associated with an inferior prognosis.
In line with other studies [45], we observed a positive correlation between higher proliferation and increased glutamate content in tumor tissue. Glutamate is a metabolic product of glutaminolysis, which drives membrane trafficking to promote breast cancer cell invasiveness [46]. In addition, the expression of glutaminase genes GLS and GLS2 correlates with increased tumor growth rates [47]. Many tumors become glutamine-dependent, as it serves as a direct route into the TCA cycle at the alpha-ketoglutaric acid level with consequential ATP production. Together with glycine and cysteine, glutamate is a precursor to the tripeptide glutathione, which is an antioxidant molecule that serves to ‘buffer’ superoxide insults encountered in the tumor microenvironment [45]. Glutathione is the major thiol-containing endogenous antioxidant and serves as a redox buffer against various sources of oxidative stress. In tumors, maintaining a supply of glutathione is critical for cellular survival because it allows cells to resist the oxidative stress associated with rapid metabolism, DNA-damaging agents, and inflammation, among others [48, 49]. Glucose metabolism and biosynthesis of glutathione are often modulated by the PI3K/Akt pathway, which is often dysregulated in breast cancer tumors [50, 51]. Importantly, one of the effects of targeting the PI3K/Akt-pathway upstream [52] and downstream [19] is reduced glutathione content in tumor cells. In the PPP-pathway, NAD+ and NADP are converted into NADH and NADPH, respectively, which contribute to maintaining glutathione (GSSG) in the reduced state (GSH) [53]. Thus, the PPP-pathway in the Warburg effect secures a high intracellular level of glutathione, which is regarded as the most important cellular protection system against attack from reactive oxygen species (ROS) in both dividing and hibernating luminal cells [10], and also in cancer stem cells [54]. Thus, preoperative carbohydrate loading seems to create a doubly favorable environment that will probably serve the CTCs liberated during surgery [39] more than the already established micrometastases [55]. First, CTCs have a surplus of cellular fuel via lactate and pyruvate available systemically. Second, they benefit from an increased level of intracellular protection systems against ROS via increased tumor glutathione. Both effects will increase the probability of CTCs thriving and surviving as micrometastases, which then may erupt as clinical relapse years later, compatible with the tumor biology of luminal breast cancers. However, our observed clinical endpoint between 3 to 7 years must be regarded as ‘early relapses’ when coming to luminal cancers [56]. Thus, we need a much longer follow up to capture the late recurrences in order to get the correct picture of the clinical outcome of the present study.
Several attempts have been made to reverse the above-mentioned metabolic pathways for treatment purposes. The first attempt was to reverse the Warburg effect with the polyphenol resveratrol, which blocks PDH/LDH. In colon cancer cells, resveratrol inhibits proliferation, gluconeogenesis, and PPP [57]. By blocking PDH, resveratrol promotes mitochondrial electron transport chain overload with increased ROS production, ultimately resulting in apoptosis [58]. Secondly, a ketogenic diet has been shown to be effective in preclinical studies [59]. A ketogenic diet produces a large amount of intracellular ketone bodies that have a direct cytotoxic effect. Furthermore, the ketogenic state inhibits insulin/IGF signaling and downstream signaling pathways, such as PI3K/Akt/mTOR [60]. Interestingly, in the present study, the patients in the fasting group reached a ketogenic state with increased ketone bodies, which may have created an unfavorable environment for the cancer cells in the tumor and for the liberated CTCs. This is in line with a recent RCT of using ketogenic diet as adjuvant treatment in one of the study arms. They observed a better overall survival in the group that received ketogenic diet [61]. Others have recently shown a profound effect of ketogenic diet in a xenografted breast cancer mouse model with increased ketone bodies and increased aminoacidic [62], which is in line with our observations. The authors hypothesize that the anti-cancer effect may be mediated through immunological mechanisms [62]. Thus, use of a ketogenic diet as adjuvants to conventional therapy is rooted in several studies [63].
Likewise, physical activity is known to prevent and improve survival in several cancer forms and is thus recommended as a measure to both prevent and treat breast cancer [64, 65]. One of the mechanisms behind these observation is a change in the estrogen metabolism after 180 min exercise pr. week. They found an increased 2 hydroxy-estrone level known to antagonize the estradiol action [66] This observation is important for both in the preventive setting as breast cancer risk is correlated to total life exposure of estrogens [67]. Also, changes in diet affect the cancer incidence [68], and also prognosis in breast cancer patients [69].
A combination of calorie restriction and physical exercise in postmenopausal women did also reduce insulin levels [70]. In our patients, we found that metabolic changes after the carbohydrate load affected the ER-positive breast cancer patients. Thus, ketogenic diet combined with physical exercise would probably be beneficial for our patients as this approach will affect both the ER and insulin signaling pathways.
Interestingly, intermittent fasting (i.e. caloric restriction for 16–48 h [71] has been proven to affect the metabolism and disease process in a beneficial manner. Notably, intermittent fasting in animal studies have demonstrated reduction of tumor size [72]. In humans, intermittent fasting improves insulin sensitivity and thus reduces insulin and IGF-1 related signaling in over weighted individuals [72, 73]. Preclinical studies show that intermittent fasting more than 2 days is as effective as chemotherapy to reduce cancer load [74]. Thus, the ketones derived from intermittent fasting decreases cancer cell viability by attacking several hallmarks of cancer [75].
The IPA-analyses confirmed that the systemic response to the carbohydrate load converge towards pathways involved in proliferation and growth of the organism. Moreover, other pathways related to the Warburg effect were also involved. Thus, peroral preoperative carbohydrate load shifts the systemic metabolism towards a very fortunate and beneficial environment for CTCs liberated from the tumor under the operation. Interestingly, four out of seven microRNAs related to endocrine resistance [27] also regulate the same metabolic pathways through insulin signaling pathways, which are known to be involved in endocrine resistance with reduced effect of tamoxifen and aromatase inhibitors. Thus, it seems plausible to introduce metformin early on as adjuvant treatment to regain the endocrine sensitivity. Intriguingly, circulating microRNAs from the tumor in exosomes [76] can perform cell-independent microRNA biogenesis and promote tumorigenesis away from the primary tumor [77]. Thus, we may speculate that one of the steps in the metastatic process is to control the systemic metabolic pathways to ensure a beneficial environment and survival of the liberated cancer cells [54]. Moreover, increased cellular uptake of glucose via the Warburg effect [10] favor differentiating glycosylation of intracellular proteins included paucimannosylation [78]. Intriguingly, the metastatic Epithelial-Mesenchymal-Transition (EMT) process is regulated through glycosylation of key regulator proteins, that are frequently modulated via the insulin /IGF signaling [79]. Thus, glycosylation opens up a connection between the glucose/insulin signaling and increased survival of CTCs trough enhancement of the EMT-processes.
Taken together, this explorative study indicates that the carbohydrate loading state and fasting state have opposite systemic and micro-environmental effects, which may explain why the relapses in the present study were skewed towards the carbohydrate group, with an inferior RFS, BCSS, and OS in patients with high tissue glutathione, high S-lactate, and high S-pyruvate. The favorable macro- and micro-environmental changes for the tumor that come from carbohydrate loading reflect the Warburg effect, which serves the CTCs and micrometastases more than the patient [80]. In luminal cancers, the Warburg pathway enzyme PFKFB4 acts as a molecular fulcrum that couples sugar metabolism to transcriptional activation by stimulating the ER co-activator SRC-3 to promote aggressive metastatic tumors [81].
The present study has several weak points. First, it is a post hoc explorative analysis of an RCT. Therefore, the various analyses are not sufficiently powered regarding the various endpoints. In addition, tissue samples were not available for all patients, which reduces the number of patients in the various analyses. Thus, this creates a greater risk of a type II error than a type I error. Furthermore, the tissue analyses were skewed towards patients with larger tumors. This could introduce systematic error in the analysis. However, tumor size was not included in the final Cox models in any survival analysis, indicating that this error was not strong enough to blur the effects of the metabolites. Also, including diet recalls and demographic data of the patients would have strengthened the study. Detecting the well-known endocrine metabolic fingerprint of insulin strengthens the method and the reliability of the various findings in this study. However, the study is too small to conclude on preoperative preparation guidelines; fasting or carbohydrate loading. Moreover, the pilot nature of the present study calls for validation in a larger study with a long-term follow-up. Introducing a ketogenic diet as a third study arm may test out whether ketone bodies could wipe out the liberated CTCs and thus improve survival.
Conclusion
Preoperative oral glucose loading increases systemic levels of lactate and pyruvate, and tumor levels of glutathione and glutamate in luminal breast cancer patients. In fasting patients, the proapoptotic ketone bodies are increased. These biological changes may contribute to the survival differences observed between these two study groups. Integrated Pathway Analysis (IPA) in serum revealed activation of five major anabolic metabolic networks contributing to proliferation and growth mainly through insulin signaling pathways.
Acknowledgements
In memory of our late and beloved co-author Bianca van Diermen Hidle, who all too early became a victim of cancer. We are very much in debt to her legacy, and for her excellent work on the quantitative pathology analysis — provided also for this paper.
Advices on the various statistical analyses given by Prof. Jan Terje Kvaløy at University of Stavanger, Stavanger, Norway are very much appreciated.
We would also like to thank the former department heads Dr. Ottar Bjerkeset and Dr. Kjell H. Kjellevold, who facilitated the study flow in the Department of Surgery and the Department of Pathology, respectively.
Abbreviations
- ATP
Adenosine triphosphate
- AUC
Area under the curve
- BCSS
Breast cancer specific survival
- CI
Confidence interval
- CPMG
Carr–Purcell–Meiboom–Gill spectra
- CTC
Circulating tumor cell
- EMT
Epithelial mesenchymal transition
- ER
Estrogen receptor
- ERAS
Enhanced recovery after surgery
- GSH
Glutathione, reduced form
- GSSG
Glutathione, oxidized form
- HER2
Human epithelial growth factor receptor 2
- HR
Hazard ratio
- HR-MAS-MR
High-resolution magic angle spinning - magnetic resonance
- IGF1
Insulin-like growth factor 1
- IGF1R
Insulin-like growth factor 1 receptor
- IGFBP3
Insulin-like growth factor 1 binding protein 3
- IHC
Immunohistochemistry
- IPA
Ingenuity Pathway Analysis
- IR
Insulin receptor
- LDH
Lactate dehydrogenase
- MAI
Mitotic activity index
- MCT-1
Monocarboxylate transporter 1
- MCT-4
Monocarboxylate transporter 4
- MR
Magnetic resonance
- MRI
Magnetic resonance imaging
- MSEA
Metabolite Set Enrichment Analysis
- NAD
Nicotinamide adenine dinucleotide
- NADP
Nicotinamide adenine dinucleotide phosphate
- NOESY
One-dimensional 1H Nuclear Overhauser effect spectroscopy
- NSD
Norwegian Center for Research Data
- OS
Overall survival
- PHD
Pyruvate dehydrogenase
- PLS
Partial least square
- PLS-DA
Partial least square discriminant analysis
- PPH3
Phosphorylated phosphohistone 3
- PPP
Pentose phosphate pathway
- PR
Progesterone receptor
- RCT
Randomized controlled trial
- RFS
Relapse free survival
- ROS
Reactive oxygen species
- S
Serum
- SRC-3
Steroid receptor co-activator 3
- TCA-cycle
Tri carboxyl acid cycle
Appendix
Authors’ contributions
THL included and operated on all of the patients, built the database, and contributed to statistical analyses and interpretation of data. MA contributed to analyses of the metabolite spectra of all serum and tissue samples and also to the IPA analyses. TFB provided all the metabolomic analyses in her MR-imaging lab and interpreted the results of the metabolic spectra. AEV obtained all of the blood samples and performed the laboratory analyses. IS contributed to the laboratory analyses. EG performed surgical pathological analysis with histological grading and morphological analysis of the tumor. NGE and SL provided detailed information of the micro RNA in the IPA analyses. LAA contributed with scientific support and advice. KJ performed the IPA analyses. EAMJ contributed to the concept of the study, assessed the pathological parameters and scorings, and participated in interpreting the data. HS contributed to the concept of the study, statistical analyses and interpretation of data. JPAB contributed to the concept of the study and the analysis and interpretation of the data. All co-authors contributed to writing the manuscript and gave their final approval of the last version to be published.
Authors’ information
N.A.
Funding
The present study was funded by Marathon Oil, the Folke Hermannsen Foundation, and the Inge Steenslands Foundation, Stavanger, Norway. The funding covered the cost of preOp™, blood chemistry, and hormone analysis. The funding bodies were not involved in the design of the study, data collection, analysis, interpretation of the data, or in writing the manuscript.
Availability of data and materials
The data that support the findings of this study are available from Stavanger Breast Cancer Research Group, but restrictions apply to the availability of these data, which were used under license for the current study and as such are not publicly available. However, data are available from the authors upon reasonable request and with permission from Stavanger Breast Cancer Research Group.
Ethics approval and consent to participate
The RCT, which this study is based upon, was approved by the Regional Ethics Committee (Accession number 2015/1445), Norwegian Centre for Research Data (#20984), and The Norwegian Biobank Registry (#2239). An informed consent form was signed by each patient. The trial was retrospectively registered at Clinicaltrials.gov (NCT03886389).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tone Hoel Lende and Marie Austdal shared First authorship.
Emiel A. M. Janssen, Håvard Søiland and Jan PA Baak are equal senior contributor.
Contributor Information
Tone Hoel Lende, Email: tonehl@yahoo.no.
Marie Austdal, Email: marie.austdal@sus.no.
Tone Frost Bathen, Email: tone.f.bathen@ntnu.no.
Anne Elin Varhaugvik, Email: anne.elin.varhaugvik@helse-mr.no.
Ivar Skaland, Email: skiv@sus.no.
Einar Gudlaugsson, Email: guei@sus.no.
Nina G. Egeland, Email: nina.gran.egeland@sus.no
Siri Lunde, Email: siri.lunde@sus.no.
Lars A. Akslen, Email: lars.akslen@uib.no
Kristin Jonsdottir, Email: kristin.jonsdottir@sus.no.
Emiel A. M. Janssen, Email: jaem@sus.no
Håvard Søiland, Email: hsoiland@gmail.com.
Jan P. A. Baak, Email: info@drjanbaak.com
References
- 1.Lundqvist A, Andersson E, Ahlberg I, Nilbert M, Gerdtham U. Socioeconomic inequalities in breast cancer incidence and mortality in Europe-a systematic review and meta-analysis. Eur J Pub Health. 2016;26(5):804–813. doi: 10.1093/eurpub/ckw070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yap Yoon-Sim, Lu Yen-Shen, Tamura Kenji, Lee Jeong Eon, Ko Eun Young, Park Yeon Hee, Cao A-Yong, Lin Ching-Hung, Toi Masakazu, Wu Jiong, Lee Soo-Chin. Insights Into Breast Cancer in the East vs the West. JAMA Oncology. 2019;5(10):1489. doi: 10.1001/jamaoncol.2019.0620. [DOI] [PubMed] [Google Scholar]
- 3.Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, Shi W, Jiang J, Yao PP, Zhu HP. Risk factors and preventions of breast Cancer. Int J Biol Sci. 2017;13(11):1387–1397. doi: 10.7150/ijbs.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baum M, Demicheli R, Hrushesky W, Retsky M. Does surgery unfavourably perturb the "natural history" of early breast cancer by accelerating the appearance of distant metastases? Eur J Cancer. 2005;41(4):508–515. doi: 10.1016/j.ejca.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 5.Pukazhendhi G, Gluck S. Circulating tumor cells in breast cancer. J Carcinog. 2014;13:8. doi: 10.4103/1477-3163.135578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price TT, Burness ML, Sivan A, Warner MJ, Cheng R, Lee CH, Olivere L, Comatas K, Magnani J, Kim Lyerly H, et al. Dormant breast cancer micrometastases reside in specific bone marrow niches that regulate their transit to and from bone. Sci Transl Med. 2016;8(340):340–373. doi: 10.1126/scitranslmed.aad4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wangchinda P, Ithimakin S. Factors that predict recurrence later than 5 years after initial treatment in operable breast cancer. World J Surg Oncol. 2016;14(1):223. doi: 10.1186/s12957-016-0988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Hua, Shen Jie, Moore Steven C., Ye Yuanqing, Wu Xifeng, Esteva Francisco J., Tripathy Debasish, Chow Wong-Ho. Breast cancer risk in relation to plasma metabolites among Hispanic and African American women. Breast Cancer Research and Treatment. 2019;176(3):687–696. doi: 10.1007/s10549-019-05165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warburg OPK, Negelein E. Ueber den stoffwechhsel der tumoren. Biochem Z. 1924;152(1):319–344. [Google Scholar]
- 12.Fernandez-de-Cossio-Diaz J, Vazquez A. Limits of aerobic metabolism in cancer cells. Sci Rep. 2017;7(1):13488. doi: 10.1038/s41598-017-14071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran Q, Lee H, Park J, Kim SH, Park J. Targeting Cancer metabolism - revisiting the Warburg effects. Toxicol Res. 2016;32(3):177–193. doi: 10.5487/TR.2016.32.3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart CD, Tenori L, Luchinat C, Di Leo A. Metabolomics in breast Cancer: current status and perspectives. Adv Exp Med Biol. 2016;882:217–234. doi: 10.1007/978-3-319-22909-6_9. [DOI] [PubMed] [Google Scholar]
- 15.Ljungqvist O. ERAS--enhanced recovery after surgery: moving evidence-based perioperative care to practice. JPEN J Parenter Enteral Nutr. 2014;38(5):559–566. doi: 10.1177/0148607114523451. [DOI] [PubMed] [Google Scholar]
- 16.Lende TH, Austdal M, Varhaugvik AE, Skaland I, Gudlaugsson E, Kvaløy JT, Akslen LA, Søiland H, Janssen EAM, Baak JPA. Influence of pre-operative oral carbohydrate loading vs. standard fasting procedure on tumor proliferation and clinical outcome in breast cancer patients — a randomized trial. BMC Cancer. 2019;19(1):1076. doi: 10.1186/s12885-019-6275-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Austdal M, Tangeras LH, Skrastad RB, Salvesen K, Austgulen R, Iversen AC, Bathen TF. First trimester urine and serum metabolomics for prediction of preeclampsia and gestational hypertension: a prospective screening study. Int J Mol Sci. 2015;16(9):21520–21538. doi: 10.3390/ijms160921520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giskeodegard GF, Madssen TS, Euceda LR, Tessem MB, Moestue SA, Bathen TF. NMR-based metabolomics of biofluids in cancer. NMR Biomed. 2018:e3927. 10.1002/nbm.3927. [DOI] [PubMed]
- 19.Euceda LR, Hill DK, Stokke E, Hatem R, El Botty R, Bièche I, Marangoni E, Bathen TF, Moestue SA. Metabolic response to Everolimus in patient-derived triple-negative breast Cancer Xenografts. J Proteome Res. 2017;16(5):1868–1879. doi: 10.1021/acs.jproteome.6b00918. [DOI] [PubMed] [Google Scholar]
- 20.Eilers PH. Parametric time warping. Anal Chem. 2004;76(2):404–411. doi: 10.1021/ac034800e. [DOI] [PubMed] [Google Scholar]
- 21.Sitter B, Sonnewald U, Spraul M, Fjosne HE, Gribbestad IS. High-resolution magic angle spinning MRS of breast cancer tissue. NMR Biomed. 2002;15(5):327–337. doi: 10.1002/nbm.775. [DOI] [PubMed] [Google Scholar]
- 22.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29(4):1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- 23.R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- 24.pls: Partial Least Squares and Principal Component Regression. R package version 2.7–1. https://cran.r-project.org/web/packages/pls/index.html
- 25.Xia J., Mandal R., Sinelnikov I. V., Broadhurst D., Wishart D. S. MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Research. 2012;40(W1):W127–W133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehmood TLK, Snipen L, Sæbø S. A review of variable selection methods in partial least squares regression. Chemom Intell Lab Syst. 2012;118:62–69. doi: 10.1016/j.chemolab.2012.07.010. [DOI] [Google Scholar]
- 27.Egeland NG, Lunde S, Jonsdottir K, Lende TH, Cronin-Fenton D, Gilje B, Janssen EA, Søiland H. The role of MicroRNAs as predictors of response to Tamoxifen treatment in breast Cancer patients. Int J Mol Sci. 2015;16(10):24243–24275. doi: 10.3390/ijms161024243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaham O, Wei R, Wang TJ, Ricciardi C, Lewis GD, Vasan RS, Carr SA, Thadhani R, Gerszten RE, Mootha VK. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou Y, Zhou M, Xie J, Chao P, Feng Q, Wu J. High glucose levels promote the proliferation of breast cancer cells through GTPases. Breast Cancer (Dove Med Press) 2017;9:429–436. doi: 10.2147/BCTT.S135665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang FY, Pai MH, Chiang EP. Consumption of high-fat diet induces tumor progression and epithelial-mesenchymal transition of colorectal cancer in a mouse xenograft model. J Nutr Biochem. 2012;23(10):1302–1313. doi: 10.1016/j.jnutbio.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Passarella S. Schurr a: l-lactate transport and metabolism in mitochondria of Hep G2 cells-the Cori cycle revisited. Front Oncol. 2018;8:120. doi: 10.3389/fonc.2018.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott MJ, Fawcett WJ. Oral carbohydrate preload drink for major surgery - the first steps from famine to feast. Anaesthesia. 2014;69(12):1308–1313. doi: 10.1111/anae.12921. [DOI] [PubMed] [Google Scholar]
- 33.Atkins RPVK, Constantin-Teodosiu D, Lobo DN, Greenhaff PL. Rates of skeletal muscle mitochondrial ATP production are reduced during elective abdominal surgery in humans. J Am Coll Surg. 2011;2013:S59. doi: 10.1016/j.jamcollsurg.2011.06.130. [DOI] [Google Scholar]
- 34.Varadan KKAR, Dimitru CT, Blackshaw E, Perkins AC, Greenhaff PL, Lobo DN. Gastrointestinal surgery mediated increases in gut permeability and expression of IL6 and PDK4 mRNAs in quadriceps muscle may underpin the post-operative increase in whole-body insulin resistance in humans. J Am Coll Surg. 2011;2013:S53. doi: 10.1016/j.jamcollsurg.2011.06.114. [DOI] [Google Scholar]
- 35.Prando R, Cheli V, Buzzo P, Melga P, Ansaldi E, Accoto S. Blood lactate behavior after glucose load in diabetes mellitus. Acta Diabetol Lat. 1988;25(3):247–256. doi: 10.1007/BF02624820. [DOI] [PubMed] [Google Scholar]
- 36.Moore MC, Davis SN, Mann SL, Cherrington AD. Acute fructose administration improves oral glucose tolerance in adults with type 2 diabetes. Diabetes Care. 2001;24(11):1882–1887. doi: 10.2337/diacare.24.11.1882. [DOI] [PubMed] [Google Scholar]
- 37.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Le Z, Yanxiang Guo J, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551(7678):115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, et al. Lactate metabolism in human lung tumors. Cell. 2017;171(2):358–371. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papavasiliou P, Fisher T, Kuhn J, Nemunaitis J, Lamont J. Circulating tumor cells in patients undergoing surgery for hepatic metastases from colorectal cancer. Proc (Baylor Univ Med Cent) 2010;23(1):11–14. doi: 10.1080/08998280.2010.11928572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, Schlimok G, Diel IJ, Gerber B, Gebauer G, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353(8):793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 41.de Boer M, van Dijck JA, Bult P, Borm GF, Tjan-Heijnen VC. Breast cancer prognosis and occult lymph node metastases, isolated tumor cells, and micrometastases. J Natl Cancer Inst. 2010;102(6):410–425. doi: 10.1093/jnci/djq008. [DOI] [PubMed] [Google Scholar]
- 42.Bidard FC, Hajage D, Bachelot T, Delaloge S, Brain E, Campone M, Cottu P, Beuzeboc P, Rolland E, Mathiot C, Pierga JY. Assessment of circulating tumor cells and serum markers for progression-free survival prediction in metastatic breast cancer: a prospective observational study. Breast Cancer Res. 2012;14(1):R29. doi: 10.1186/bcr3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu D, Wang D, Wu C, Zhang L, Mei Q, Hu G, Long G, Sun W. Prognostic significance of serum lactate dehydrogenase in patients with breast cancer: a meta-analysis. Cancer Manag Res. 2019;11:3611–3619. doi: 10.2147/CMAR.S199260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L, He Y, Ge G, Li L, Zhou P, Zhu Y, Tang H, Huang Y, Li W, Zhang L. Lactate dehydrogenase and creatine kinase as poor prognostic factors in lung cancer: a retrospective observational study. PLoS One. 2017;12(8):e0182168. doi: 10.1371/journal.pone.0182168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi JS, Yoon D, Koo JS, Kim S, Park VY, Kim EK, Kim S, Kim MJ. Magnetic resonance metabolic profiling of estrogen receptor-positive breast cancer: correlation with currently used molecular markers. Oncotarget. 2017;8(38):63405–63416. doi: 10.18632/oncotarget.18822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dornier E, Rabas N, Mitchell L, Novo D, Dhayade S, Marco S, Mackay G, Sumpton D, Pallares M, Nixon C, et al. Glutaminolysis drives membrane trafficking to promote invasiveness of breast cancer cells. Nat Commun. 2017;8(1):2255. doi: 10.1038/s41467-017-02101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knox WE, Horowitz ML, Friedell GH. The proportionality of glutaminase content to growth rate and morphology of rat neoplasms. Cancer Res. 1969;29(3):669–680. [PubMed] [Google Scholar]
- 48.Estrela JM, Ortega A, Obrador E. Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci. 2006;43(2):143–181. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- 49.Franco R, Cidlowski JA. Apoptosis and glutathione: beyond an antioxidant. Cell Death Differ. 2009;16(10):1303–1314. doi: 10.1038/cdd.2009.107. [DOI] [PubMed] [Google Scholar]
- 50.Lien EC, Lyssiotis CA, Juvekar A, Hu H, Asara JM, Cantley LC, Toker A. Glutathione biosynthesis is a metabolic vulnerability in PI (3) K/Akt-driven breast cancer. Nat Cell Biol. 2016;18(5):572–578. doi: 10.1038/ncb3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29(3):313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fack F, Espedal H, Keunen O, Golebiewska A, Obad N, Harter PN, Mittelbronn M, Bahr O, Weyerbrock A, Stuhr L, et al. Bevacizumab treatment induces metabolic adaptation toward anaerobic metabolism in glioblastomas. Acta Neuropathol. 2015;129(1):115–131. doi: 10.1007/s00401-014-1352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liberti MV, Locasale JW. The Warburg effect: how does it benefit Cancer cells? Trends Biochem Sci. 2016;41(3):211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee SY, Ju MK, Jeon HM, Lee YJ, Kim CH, Park HG, Han SI, Kang HS. Oncogenic metabolism acts as a prerequisite step for induction of Cancer metastasis and Cancer stem cell phenotype. Oxidative Med Cell Longev. 2018;2018:1027453. doi: 10.1155/2018/1027453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klauber-DeMore N, Van Zee KJ, Linkov I, Borgen PI, Gerald WL. Biological behavior of human breast cancer micrometastases. Clin Cancer Res. 2001;7(8):2434–2439. [PubMed] [Google Scholar]
- 56.Engstrom MJ, Opdahl S, Hagen AI, Romundstad PR, Akslen LA, Haugen OA, Vatten LJ, Bofin AM. Molecular subtypes, histopathological grade and survival in a historic cohort of breast cancer patients. Breast Cancer Res Treat. 2013;140(3):463–473. doi: 10.1007/s10549-013-2647-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saunier E, Antonio S, Regazzetti A, Auzeil N, Laprevote O, Shay JW, Coumoul X, Barouki R, Benelli C, Huc L, Bortoli S. Resveratrol reverses the Warburg effect by targeting the pyruvate dehydrogenase complex in colon cancer cells. Sci Rep. 2017;7(1):6945. doi: 10.1038/s41598-017-07006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blanquer-Rossello MD, Hernandez-Lopez R, Roca P, Oliver J, Valle A. Resveratrol induces mitochondrial respiration and apoptosis in SW620 colon cancer cells. Biochim Biophys Acta Gen Subj. 2017;1861(2):431–440. doi: 10.1016/j.bbagen.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 59.Weber DD, Aminazdeh-Gohari S, Kofler B. Ketogenic diet in cancer therapy. Aging (Albany NY) 2018;10(2):164–165. doi: 10.18632/aging.101382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan-Shalaby J. Ketogenic diets and Cancer: emerging evidence. Fed Pract. 2017;34(Suppl 1):37S–42S. [PMC free article] [PubMed] [Google Scholar]
- 61.Khodabakhshi A, Akbari ME, Mirzaei HR, Mehrad-Majd H, Kalamian M, Davoodi SH. Feasibility, safety, and beneficial effects of MCT-based Ketogenic diet for breast Cancer treatment: a randomized controlled trial study. Nutr Cancer. 2019:1–8. [Epub ahead of print]. [DOI] [PubMed]
- 62.Licha D, Vidali S, Aminzadeh-Gohari S, Alka O, Breitkreuz L, Kohlbacher O, Reischl RJ, Feichtinger RG, Kofler B, Huber CG. Untargeted metabolomics reveals molecular effects of Ketogenic diet on healthy and tumor Xenograft mouse models. Int J Mol Sci. 2019;20(16):E3873. doi: 10.3390/ijms20163873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paoli A, Rubini A, Volek JS, Grimaldi KA. Beyond weight loss: a review of the therapeutic uses of very-low-carbohydrate (ketogenic) diets. Eur J Clin Nutr. 2013;67(8):789–796. doi: 10.1038/ejcn.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McTiernan A, Friedenreich CM, Katzmarzyk PT, Powell KE, Macko R, Buchner D, Pescatello LS, Bloodgood B, Tennant B, Vaux-Bjerke A, et al. Physical activity in Cancer prevention and survival: a systematic review. Med Sci Sports Exerc. 2019;51(6):1252–1261. doi: 10.1249/MSS.0000000000001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Irwin ML, Smith AW, McTiernan A, Ballard-Barbash R, Cronin K, Gilliland FD, Baumgartner RN, Baumgartner KB, Bernstein L. Influence of pre- and postdiagnosis physical activity on mortality in breast cancer survivors: the health, eating, activity, and lifestyle study. J Clin Oncol. 2008;26(24):3958–3964. doi: 10.1200/JCO.2007.15.9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith AJ, Phipps WR, Thomas W, Schmitz KH, Kurzer MS. The effects of aerobic exercise on estrogen metabolism in healthy premenopausal women. Cancer Epidemiol Biomark Prev. 2013;22(5):756–764. doi: 10.1158/1055-9965.EPI-12-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yager JD, Davidson NE. Estrogen carcinogenesis in breast Cancer. N Engl J Med. 2006;354(3):270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 68.Donaldson MS. Nutrition and cancer: a review of the evidence for an anti-cancer diet. Nutr J. 2004;3:19. doi: 10.1186/1475-2891-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas F, Rome S, Mery F, Dawson E, Montagne J, Biro PA, Beckmann C, Renaud F, Poulin R, Raymond M, Ujvari B. Changes in diet associated with cancer: an evolutionary perspective. Evol Appl. 2017;10(7):651–657. doi: 10.1111/eva.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chlebowski RT, Pettinger M, Stefanick ML, Howard BV, Mossavar-Rahmani Y, McTiernan A. Insulin, physical activity, and caloric intake in postmenopausal women: breast cancer implications. J Clin Oncol. 2004;22(22):4507–4513. doi: 10.1200/JCO.2004.04.119. [DOI] [PubMed] [Google Scholar]
- 71.Mattson MP, Longo VD, Harvie M. Impact of intermittent fasting on health and disease processes. Ageing Res Rev. 2017;39:46–58. doi: 10.1016/j.arr.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harvie MN, Howell T. Could intermittent energy restriction and intermittent fasting reduce rates of Cancer in obese, overweight, and Normal-weight subjects? A summary of evidence. Adv Nutr. 2016;7(4):690–705. doi: 10.3945/an.115.011767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Longo VD, Panda S. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab. 2016;23(6):1048–1059. doi: 10.1016/j.cmet.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee C, Raffaghello L, Brandhorst S, Safdie FM, Bianchi G, Martin-Montalvo A, Pistoia V, Wei M, Hwang S, Merlino A, et al. Fasting cycles retard growth of tumors and sensitize a range of cancer cell types to chemotherapy. Sci Transl Med. 2012;4(124):ra127. doi: 10.1126/scitranslmed.3003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poff AM, Ari C, Arnold P, Seyfried TN, D'Agostino DP. Ketone supplementation decreases tumor cell viability and prolongs survival of mice with metastatic cancer. Int J Cancer. 2014;135(7):1711–1720. doi: 10.1002/ijc.28809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15(10):617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 77.Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chatterjee Sayantani, Lee Ling Y., Kawahara Rebeca, Abrahams Jodie L., Adamczyk Barbara, Anugraham Merrina, Ashwood Christopher, Sumer‐Bayraktar Zeynep, Briggs Matthew T., Chik Jenny H. L., Everest‐Dass Arun, Förster Sarah, Hinneburg Hannes, Leite Katia R. M., Loke Ian, Möginger Uwe, Moh Edward S. X., Nakano Miyako, Recuero Saulo, Sethi Manveen K., Srougi Miguel, Stavenhagen Kathrin, Venkatakrishnan Vignesh, Wongtrakul‐Kish Katherine, Diestel Simone, Hoffmann Peter, Karlsson Niclas G., Kolarich Daniel, Molloy Mark P., Muders Michael H., Oehler Martin K., Packer Nicolle H., Palmisano Giuseppe, Thaysen‐Andersen Morten. Protein Paucimannosylation Is an Enriched N ‐Glycosylation Signature of Human Cancers. PROTEOMICS. 2019;19(21-22):1900010. doi: 10.1002/pmic.201900010. [DOI] [PubMed] [Google Scholar]
- 79.de-Freitas-Junior JCM, Andrade-da-Costa J, Silva MC, Pinho SS. Glycans as Regulatory Elements of the Insulin/IGF System: Impact in Cancer Progression. Int J Mol Sci. 2017;18(9):E1921. doi: 10.3390/ijms18091921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu J. The Warburg metabolism fuels tumor metastasis. Cancer Metastasis Rev. 2019;38(1–2):157–164. doi: 10.1007/s10555-019-09794-5. [DOI] [PubMed] [Google Scholar]
- 81.Dasgupta S, Rajapakshe K, Zhu B, Nikolai BC, Yi P, Putluri N, Choi JM, Jung SY, Coarfa C, Westbrook TF, et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer. Nature. 2018;556(7700):249–254. doi: 10.1038/s41586-018-0018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from Stavanger Breast Cancer Research Group, but restrictions apply to the availability of these data, which were used under license for the current study and as such are not publicly available. However, data are available from the authors upon reasonable request and with permission from Stavanger Breast Cancer Research Group.