Abstract
Background
While the utility of parasite genotyping for malaria elimination has been extensively documented in low to moderate transmission settings, it has been less well-characterized in holoendemic regions. High malaria burden settings have received renewed attention acknowledging their critical role in malaria elimination. Defining the role for parasite genomics in driving these high burden settings towards elimination will enhance future control programme planning.
Methods
Amplicon deep sequencing was used to characterize parasite population genetic diversity at polymorphic Plasmodium falciparum loci, Pfama1 and Pfcsp, at two timepoints in June–July 2016 and January–March 2017 in a high transmission region along the international border between Luapula Province, Zambia and Haut-Katanga Province, the Democratic Republic of the Congo (DRC).
Results
High genetic diversity was observed across both seasons and in both countries. No evidence of population structure was observed between parasite populations on either side of the border, suggesting that this region may be one contiguous transmission zone. Despite a decline in parasite prevalence at the sampling locations in Haut-Katanga Province, no genetic signatures of a population bottleneck were detected, suggesting that larger declines in transmission may be required to reduce parasite genetic diversity. Analysing rare variants may be a suitable alternative approach for detecting epidemiologically important genetic signatures in highly diverse populations; however, the challenge is distinguishing true signals from potential artifacts introduced by small sample sizes.
Conclusions
Continuing to explore and document the utility of various parasite genotyping approaches for understanding malaria transmission in holoendemic settings will be valuable to future control and elimination programmes, empowering evidence-based selection of tools and methods to address pertinent questions, thus enabling more efficient resource allocation.
Keywords: Malaria, Genetic, Border, Diversity, Control, Amplicon deep sequencing, csp, ama1
Background
Significant progress has been made in reducing Plasmodium falciparum malaria transmission since the early 2000s, due in part to massive distributions of insecticide-treated bed nets (ITNs), increased coverage with indoor residual spraying (IRS), and the introduction of artemisinin-based combination therapy (ACT) [1, 2]. In fact, it is estimated that the global burden of malaria declined by 40% between 2000 and 2015, leading 35 countries to establish malaria elimination targets as of September 2015 [1, 3]. While the current arsenal of malaria control tools has been broadly effective, it has not been sufficient to reduce transmission everywhere [4]. Despite malaria control programmes, the ten countries with the highest malaria burden in Africa experienced increases in malaria cases between 2015 and 2017 [5, 6]. Acknowledging that the success of malaria control in these and other high burden regions is critical for the attainment of malaria elimination targets, the World Health Organization (WHO) renewed its focus on high transmission settings as a key component of malaria elimination programmes in the recent High Burden to High Impact (HBHI) response plan [6]. Achieving malaria control in regions where transmission has previously been refractory to interventions will require an enhanced understanding of the unique mechanisms that perpetuate transmission in different settings and data-driven approaches that target them [6].
Plasmodium falciparum molecular epidemiology has emerged as a tool to genetically track transmission patterns [7–10]. Population genetic diversity and multiplicity of infection (MOI) have been used to monitor changes in transmission intensity and evaluate whether programmes or interventions have altered transmission patterns [11–19]. Parasite genotyping may allow more refined discrimination of parasite locality of origin than travel surveys alone, possibly enabling gene flow between populations to be estimated [20–22] and source populations of on-going transmission to be identified [23]. Parasite genetic approaches have demonstrated their value in complementing routine epidemiological and entomological surveillance to inform control programmes in moderate to low transmission settings where elimination efforts have been concentrated [7]. With renewed attention to malaria control in high burden regions as a component of elimination planning, it is important to define the potential role of molecular epidemiology in such settings.
Zambia, a malaria-endemic country in southern Africa, aims to eliminate malaria from every district by 2021. As a country, Zambia has made great strides towards achieving this goal. In fact, P. falciparum prevalence by microscopy among children under 5 years old declined from 19.4% in 2015 to 9.1% in 2018 [24]. Unfortunately, the success in malaria control has been heterogeneous across the country. Luapula Province, located in northern Zambia along the international border with the Democratic Republic of the Congo (DRC), continues to experience holoendemic transmission despite use of control interventions for over a decade [4]. In Nchelenge District, located in Luapula Province, ITN distributions were carried out in 2007, 2011, and 2014, and IRS has been conducted annually, using pyrethroids between 2008 and 2010, carbamates between 2011 and 2012, and the organophosphate, pirimiphos-methyl (Actellic® 300CS), since 2014 [4, 25]. IRS with Actellic® led to a moderate decrease in P. falciparum prevalence by 25% in the sprayed regions within Nchelenge District; however, the effect was short-lived, lasting only for 6 months after spraying, and did not impact unsprayed areas [25]. Despite control measures, Luapula Province, including Nchelenge District, continues to experience the highest malaria prevalence among children under 5 years in the country at 30.4% by RDT [24, 26].
Across Lake Mweru from Nchelenge, situated along the international border, are two villages, Kilwa and Kashobwe in Haut-Katanga Province, the DRC. The most recent Demographic and Health Survey (DHS) from the DRC conducted in 2013–2014 reported that 23.1% of children ages 6 months to 5 years tested positive for malaria by microscopy in Haut-Katanga Province [27]. This Province has received fewer malaria control interventions historically compared to neighboring Nchelenge District in Zambia. Four cross-sectional surveys in Kilwa and Kashobwe conducted between August 2016 and July 2017 revealed that very few households (range: 0.2–2.5%) reported ever receiving IRS in these villages (Kobayashi, unpublished). During September and October 2016, a large-scale ITN distribution took place in Haut-Katanga Province. Epidemiological survey data revealed that following the ITN campaign, there was a significant increase in the proportion of individuals who reported sleeping under a bed net and a decline in parasite prevalence by microscopy in Kilwa and Kashobwe from 32% in 2016 to 18% in 2017 (Kobayashi, unpublished).
In high transmission settings including Nchelenge District, Zambia and Kilwa and Kashobwe, the DRC, individuals are typically infected with multiple genetically distinct parasites. Classic population genetic approaches that utilize neutrally-evolving SNPs or microsatellites across the genome inadequately address high polyclonality. Amplicon deep sequencing enables each parasite clone in an infection to be enumerated, but typically only at specific, high-diversity and non-neutral amplicons in the genome. Given the interest in molecular epidemiology for malaria control in high burden regions and the limitations of classic population genetic analysis in these settings, the performance of amplicon deep sequencing to enhance the understanding of malaria epidemiology in a key region for malaria elimination and control was evaluated. P. falciparum genetic diversity at two time points in June-July 2016 and January-March 2017 in Nchelenge District, Zambia and Kilwa and Kashobwe, the DRC is described. Genetic relatedness of parasites from northern Zambia and southern DRC were compared in order to assess whether this border region represents one contiguous transmission foci. The impact of declining parasite prevalence in Haut-Katanga Province on parasite population genetics was evaluated over the two time periods of the study. Finally, methods for detecting epidemiologically relevant genetic signals amidst high genetic diversity in a holoendemic transmission setting are discussed.
Methods
Sample collection and DNA extraction
At two timepoints, between June and July 2016 (dry season) and between January and March 2017 (rainy season), randomly selected households in Nchelenge District, Zambia and two villages, Kilwa and Kashobwe, in Haut-Katanga Province, the DRC were enrolled in a cross-sectional survey. Consenting individuals living within enrolled households participated in a demographic survey and had finger prick blood collected as dried blood spots (DBS) on filter paper (Whatman 903™ Protein Saver Card). DBS were stored with desiccant in sealed plastic bags and transported to the Johns Hopkins Bloomberg School of Public Health in Baltimore, Maryland, USA. DNA was extracted from the DBS using 20% Chelex and the concentration of P. falciparum specific DNA was quantified by qPCR targeting a fragment of the Pfldh gene using previously published primers [28] with SYBR green chemistry (Applied Biosystems/Thermo Fisher Scientific, Foster City, CA).
Amplicon library preparation
Plasmodium falciparum positive DNA extracts were amplified at the Pfama1 and Pfcsp loci. Pfcsp amplicon generation in 2016 was previously described [29]. Pfama1 amplicons in 2016 were similarly generated. In brief, primers (forward primer: CCAACAAAACCTCTTATGTCACCA; reverse primer: TTAGGTTGATCCGAAGCACTCA) were fused with Illumina adapter sequences and were used to amplify a 454-basepair (bp) region of Pfama1 [30]. Pfama1 amplification reaction components included 9.25 μL of the extracted template DNA, 12.5 μL KAPA Hifi HotStart ReadyMix (Kapa Biosystems, Wilmington, Massachusetts), 0.25 μL of the 20 μM reverse primer, 1.0 μL of the 5 μM forward primer, and 2 μL of 25 μM magnesium chloride. Amplicon generation cycling conditions were: 95 °C for 5 min, then 32 cycles of 98 °C for 30 sec, 58 °C for 1 min, and 72 °C for 1 min, followed by 72 °C for 5 min, and 4 °C thereafter. Amplicon sizes were confirmed using TapeStation (Agilent 4200, Santa Clara) and purified using AMPure beads (Beckman Coulter, Brea, California). Unique Nextera (Illumina, San Diego, California) indexes were added to amplicons in a subsequent PCR reaction described by Illumina [30]. Indexed amplicons underwent an additional TapeStation verification and AMPure bead purification. Confirmed amplicons were quantified using PicoGreen (ThermoFisher Scientific, Waltham, Massachusetts), diluted, and combined in equimolar volumes into a single pool comprised of 96 samples for 300-bp paired-end sequencing on the Illumina MiSeq platform at the Johns Hopkins School of Medicine Sequencing and Synthesis Facility. Samples from 2017 were prepared for sequencing as described above with the following alterations: KAPA Pure Beads (Roche, Basel, Switzerland) were used for post-amplification clean-up instead of AMPure beads, Quant-iT (ThermoFisher Scientific, Waltham, Massachusetts) was used to quantity sample concentrations instead of Pico-Green, and samples were sequenced at the University of North Carolina School of Medicine High Throughput Genomic Sequencing Facility.
Bioinformatic processing
Bioinformatic processing was done for both sequencing runs at the same time using the same parameters. Forward and reverse read pairs were stitched, sorted by primer sequence, filtered for quality (any reads with > 75% of quality scores less than 20 were excluded), and collapsed into unique parasite haplotypes within an individual using SeekDeep (January, 9, 2019) [31]. Haplotypes abundant at less than 1% within an individual were excluded. Finally, samples with less than 200 sequencing reads, or less than the 10th percentile of all reads, were excluded from the final analysis. ArcGIS, version 10.6 (ESRI, Redlands, CA) was used to map the locations of the samples used in subsequent analysis [32].
Rarefaction
Sequencing was performed in two different facilities in 2016 and 2017. In order to ensure that potential differential read depth by sequencing facility did not bias these estimates of population diversity by year, the R package, RTK, was used to perform a rarefaction analysis [33]. Rarefaction is a process by which sequencing reads are re-sampled to a uniform, lower read depth equivalent to the lowest read coverage in the study. Diversity metrics can be computed for each re-sampling iteration to compare the raw data to the rarefied distribution and test whether read depth is responsible for higher diversity among samples with higher sequencing coverage. To test whether differential read depth biased the diversity metrics, 1000 rarefaction re-sampling replicates were performed for each amplicon, each time sampling raw sequencing reads to the read depth of the sample with the lowest read coverage in the study. For each replicate, the MOI was calculated for each individual using the rarefied sample and compared MOI estimates from the raw and rarefied data. MOI was determined by calculating number of unique Pfcsp or Pfama1 haplotypes for each individual.
Genetic diversity
Software DnaSP (version 6.11.01) was used to calculate the number of unique Pfama1 and Pfcsp haplotypes in each country, nucleotide diversity (π) within each country and among all samples, and the average number of nucleotide differences between samples from within and between countries. DnaSP was used to calculate haplotype diversity (Hd), which equals 1 in a population where every haplotype is unique and 0 in a population where every haplotype is identical. Genetic diversity metrics were compared across years and sampling locations.
Genetic distance, population structure, and differentiation by country
FASTA files containing sequences for each parasite haplotype were aligned using Geneious (version 9.1.5) (https://www.geneious.com). Geneious was used to calculate pairwise relatedness between parasite pairs as the proportion of identical single nucleotide polymorphisms (SNPs). Pairwise comparisons were then grouped into two categories depending on whether they occurred between two parasites isolated from individuals living in the same country or between two parasites within individuals residing in different countries. Mean within-country relatedness was compared between parasite pairs to the mean between-country relatedness between parasite pairs, both in an unstratified analysis as well as an analysis stratified by age. To detect signatures of population structure between parasites from Zambia and the DRC, the R package, adegenet [34] was used to conduct a discriminant analysis of principal components (DAPC) [35]. DAPC seeks to detect the linear axes which explain the most between-group variability in the data, unlike classical principal components analysis (PCA) which identifies the linear axes that explain the most variability overall [35]. DAPC was performed using country of origin as group priors. Templeton, Crandall, and Sing (TCS) haplotype networks [36] were constructed for each amplicon and visualized using tcsBU [37]. Finally, software DnaSP was used to calculate FST and test for population differentiation between P. falciparum isolates from Zambia and the DRC at each of the Pfama1 and Pfcsp loci.
Genetic diversity following ITN scale-up and parasite prevalence decline
Parasite population genetic diversity indicators were compared before and after the scale-up of ITNs in both the DRC and Zambia to determine whether the parasite population was genetically bottlenecked by the decline in transmission. MOI was calculated as the higher of the number of unique Pfama1 or Pfcsp haplotypes for each individual and compared MOI estimates from 2016 and 2017 in Zambia and the DRC. Population bottlenecks in small populations have been associated with a loss of rare alleles [38]. In fact, such recently bottlenecked populations have been shown to exhibit a characteristic mode-shift in their allele frequency distributions, such that rare alleles cease to be the most common allele type following a bottleneck [38]. Allele frequency distributions were calculated using segregating loci to detect evidence of a mode-shift resulting in a loss of rare alleles. In addition to considering the loss of rare alleles as an indicator of a recent population bottleneck, it was determined whether there was loss of rare haplotypes between 2016 and 2017. Rare haplotypes were defined in two ways: first, rare haplotypes were considered to be those present at 2% or less in the population. Next, rare haplotypes were considered as those occurring just once in a population, hereafter referred to as “singletons.” The proportion of Pfama1 and Pfcsp haplotypes that were rare in 2016 and 2017 in both countries were compared by each of these two definitions.
Population structure from rare variants
In high transmission settings, it is challenging to identify epidemiologically relevant signals in genetic data given the high background genetic diversity. Particularly for genes like Pfama1 and Pfcsp which are thought to be under balancing selection, genetically similar variants found in different populations could reflect frequency-dependent selection rather than true population panmixia. Studies using human genomes have revealed that rare variants tend to be more geographically informative [39]. In fact, analysing rare variants in the human exome has been shown to more accurately detect signatures of recent population structuring events [40]. Given that high genetic diversity among genes under balancing selective pressure could lead to the false conclusion that populations are panmictic, it was assessed whether analysing rare variants alone led to detection of population structure between countries. For both Pfama1 and Pfcsp, 100 binning thresholds ranging from the minimum to the maximum frequency of haplotypes across all samples were randomly selected. For each of the 100 binning thresholds, the data was subset to include only rare haplotypes present at less than or equal to the binning threshold. Using the subset data, FST between Zambia and DRC was calculated using the R package hierfstat [41]. To verify that the inferences using rare variants were not solely driven by the smaller sample size from selecting rare haplotypes the analysis was repeated for both amplicons 1000 times, each time randomly permuting the country of origin associated with each rare haplotype.
Results
Amplicon generation and sequencing
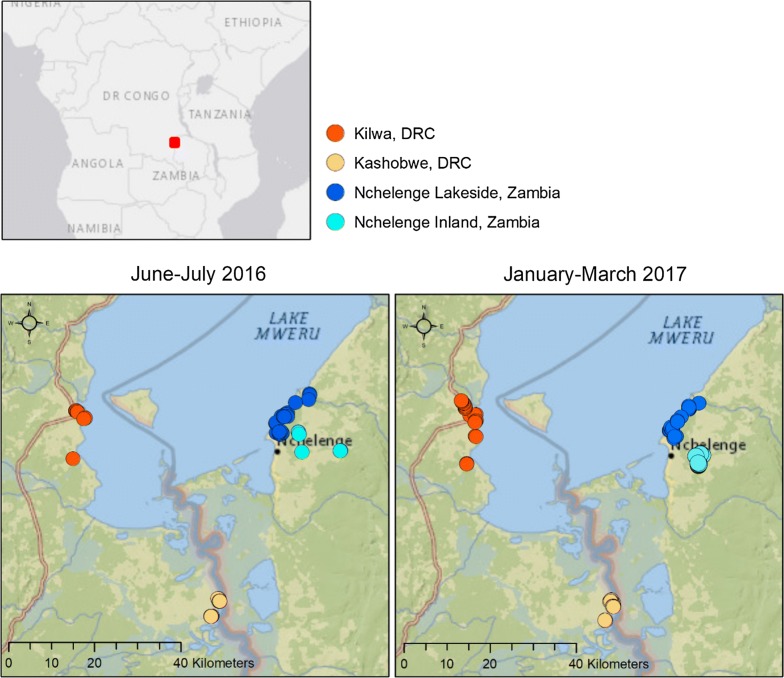
One hundred seventy-five samples underwent amplicon generation at both the Pfama1 and Pfcsp loci. Of those 175 samples, the 172 that yielded successful amplicons for Pfama1 and Pfcsp loci were sequenced. Among the 172 samples sequenced, 160 (DRC 2016 = 38, DRC 2017 = 47, Zambia 2016 = 27, Zambia 2017 = 48) resulted in sequencing reads passing quality filtering for Pfama1 and 144 (DRC 2016 = 30, DRC 2017 = 46, Zambia 2016 = 20, Zambia 2017 = 48) for Pfcsp. In total, 554 P. falciparum sequences representing 117 unique haplotypes were characterized at the Pfama1 locus and 592 sequences representing 83 unique haplotypes at the Pfcsp locus (Table 1). The locations of the 2016 and 2017 sequenced samples are shown in Fig. 1.
Table 1.
Genetic diversity by country and year
| DRC (2016 and 2017) | DRC (2016) | DRC (2017) | Zambia (2016 and 2017) | Zambia (2016) | Zambia (2017) | Overall | |
|---|---|---|---|---|---|---|---|
| Pfama1 (454 bp) | |||||||
| # Sequences | 301 | 131 | 170 | 255 | 78 | 176 | 554 |
| # Segregating sites (S) | 38 | 38 | 34 | 38 | 38 | 37 | 38 |
| Nucleotide diversity (π) | 0.024 | 0.025 | 0.024 | 0.024 | 0.024 | 0.024 | 0.024 |
| Nucleotide diversity (π): non-synonymous sites | 0.029 | 0.030 | 0.029 | 0.029 | 0.030 | 0.030 | 0.029 |
| # Unique haplotypes | 82 | 53 | 62 | 87 | 46 | 72 | 117 |
| Haplotype diversity (Hd) | 0.975 | 0.976 | 0.974 | 0.978 | 0.978 | 0.978 | 0.977 |
| Average # nucleotide differences (K) | 10.995 | 11.135 | 10.916 | 10.951 | 10.755 | 11.044 | 10.977 |
| Pfcsp (253 bp) | |||||||
| # Sequences | 303 | 106 | 197 | 288 | 66 | 222 | 592 |
| # Segregating sites (S) | 33 | 29 | 32 | 30 | 25 | 30 | 35 |
| Nucleotide diversity (π) | 0.024 | 0.024 | 0.024 | 0.025 | 0.025 | 0.026 | 0.025 |
| Nucleotide diversity (π): non-synonymous sites | 0.030 | 0.030 | 0.030 | 0.032 | 0.031 | 0.032 | 0.031 |
| # Unique haplotypes | 62 | 43 | 55 | 62 | 31 | 56 | 83 |
| Haplotype diversity (Hd) | 0.966 | 0.967 | 0.967 | 0.966 | 0.961 | 0.968 | 0.966 |
| Average # nucleotide differences (K) | 6.052 | 6.101 | 6.033 | 6.418 | 6.231 | 6.483 | 6.214 |
DnaSP (version 6.10.01) was used to calculate the number of segregating sites (S), nucleotide diversity (π) among all loci, nucleotide diversity (π) among non-synonymous sites, the number of unique DNA haplotypes, haplotype diversity (Hd), and the average number of nucleotide differences between P. falciparum haplotype for each of the populations
Fig. 1.
Locations of sequenced samples in 2016 (left) and 2017 (right). Samples came from two villages in Haut-Katanga Province, DRC, Kilwa (organge dots) and Kashobwe (yellow dots) and at two sites within Nchelenge District Zambia, along Lake Mweru (dark blue dots) and inland (aqua dots). Basemap Imagery Sources: National Geographic, Esri, DeLorme, HERE, UNEP-WCMC, USGS, NASA, ESA, METI, NRCAN, GEBCO, NOAA, iPC
Rarefaction analysis
Differential read coverage was observed between the Pfcsp amplicons from 2016 and 2017 sequencing runs. In 2016, amplicons were supported by an average of 52,600 reads for Pfama1 and 639 for Pfcsp. In 2017, samples were supported by an average of 41,813 reads for Pfama1 and 47,134 reads for Pfcsp. To test whether the lower read coverage in 2016 for Pfcsp could bias the estimates of genetic diversity, rarefaction analysis was performed to a read depth of 200 (the lowest read depth among the samples). Additional file 1: Figure S1 displays collector’s curves for each amplicon starting from either the raw data or the rarefied data. Collector’s curves show the increase in the number of unique haplotypes observed as more randomly selected samples are considered. For both amplicons, the collector’s curves are identical starting from either the raw data or the rarefied data and performing 1000 replicates of the collector’s curve analysis. This suggests that the raw data contains no more diversity than that which is captured in the rarefied subsample. Further, MOI estimates for both amplicons are nearly identical regardless of whether the estimate was derived from the raw or rarefied data (Pfama1 regression R2 = 0.99; Pfcsp regression R2 = 0.99) (Additional file 1: Figure S2), suggesting that the raw and rarefied datasets are equivalent in the diversity they explain. The total genetic diversity present among all sequencing reads was captured even when 200 reads per sample were used. Upon this demonstration that the lower read depth observed in 2016 did not bias the diversity estimates, remaining analyses were conducted using the unrarefied data.
Genetic distance, population structure, and differentiation by country
Genetic diversity was high in both Zambia and the DRC across both time points in 2016 and 2017. Among the samples, high Hd (Table 1) was observed across both countries and timepoints, highlighting the high degree of genetic diversity in these high transmission settings. Diversity, as measured by Hd and nucleotide diversity, remained high across both time points, with no significant differences by country or time (Table 1).
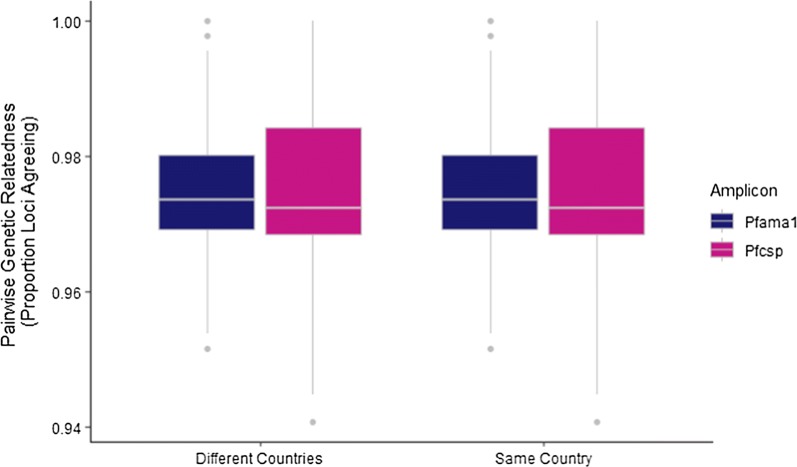
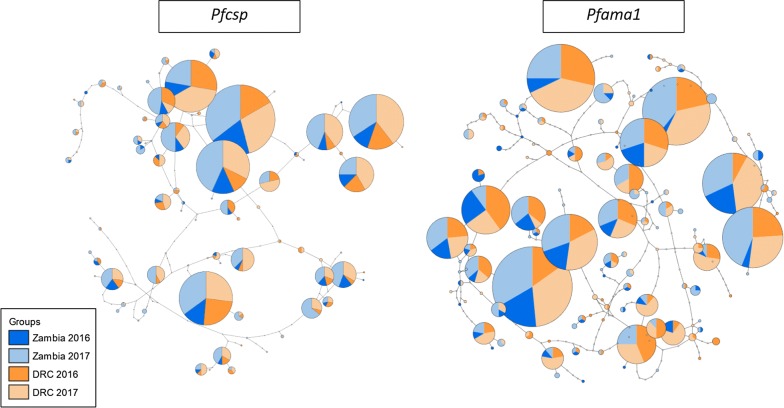
Further, after calculating pairwise genetic relatedness between all pairs of parasites, there was no significant difference in relatedness comparing parasites from within the same country to parasites from different countries for either Pfama1 or Pfcsp (Fig. 2). Age stratified analysis was similar to the unstratified analysis (Additional file 1: Figure S3). In genetically differentiated populations, within-country relatedness is expected to be higher than between-country relatedness, which was not demonstrated here. Similarly, the most common haplotypes in the study were shared at similar frequencies in both Zambia and the DRC (Fig. 3). DAPC analysis failed to identify a linear axis which could reliably discriminate isolates from Zambia and the DRC at either Pfama1 or Pfcsp (Additional file 1: Figure S6A, C), suggesting that these populations are not genetically distinct. Further, FST between the countries was found to be 0.00205 for Pfama1 and 0.00023 for Pfcsp, suggesting no evidence of population structure between countries. Finally, DnaSP detected no statistically significant population differentiation between parasites from Zambia and the DRC at either the Pfama1 (p = 0.10) or Pfcsp (p = 0.15) loci. Together, these observations are consistent with the hypothesis that parasites from Zambia and the DRC represent a single, highly genetically diverse population.
Fig. 2.
Pairwise genetic relatedness (the proportion of loci that match) is plotted for all pairs of parasites from different countries (left) or from the same country (right). Pfama1 comparisons are shown in blue and Pfcsp comparisons are shown in pink
Fig. 3.
TCS haplotype networks for Pfcsp (left) and Pfama1 (right). Each circle represents a unique haplotype; circles are scaled according to the frequency each haplotype was observed and colored by the proportion of sequences per haplotype originating from Zambia (blue) or the DRC (orange). Darker shades indicate that samples were collected in 2016, and lighter shades indicate that samples were collected in 2017. The number of mutations that differ between haplotypes is indicated by the number of notches in the lines connecting circles
Population bottleneck analysis
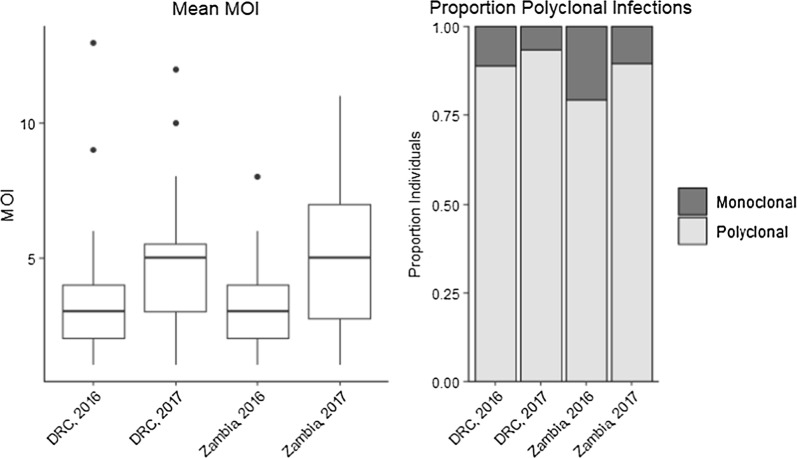
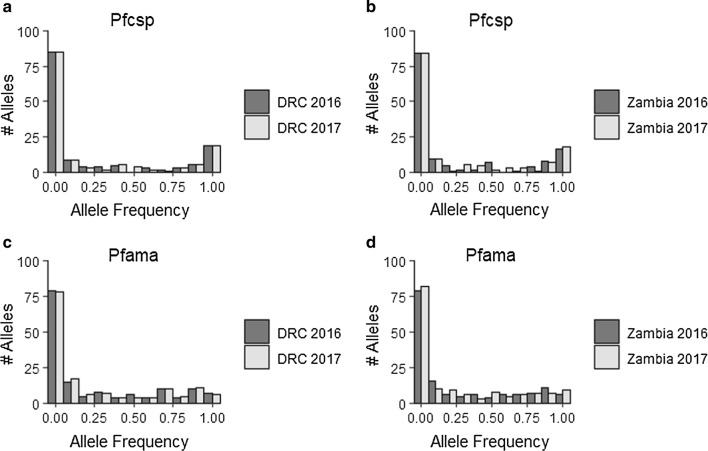
Multiple studies have reported declines in MOI that accompany decreases in transmission (12, 14, 17), but consistent with the other findings of this study, MOI did not decrease among individuals from the DRC in 2017 following the ITN distribution (DRC 2016: MOI = 3.78; DRC 2017: MOI = 4.64) (Fig. 4). Declines in parasite prevalence may result in a population bottleneck when comparing isolates from before (n = 131 Pfama1 isolates and n = 106 Pfcsp isolates in the DRC 2016) and after (n = 170 Pfama1 sequences and n = 197 Pfcsp sequences in the DRC 2017) the ITN distribution. There was no evidence of an allele frequency mode-shift indicative of a population bottleneck comparing the DRC isolates from 2016 and 2017. Similarly, a mode-shift was not detected from the haplotype frequency distributions either (Additional file 1: Figure S4). In fact, the proportion of haplotypes classified as rare by multiple thresholds was similar across countries and timepoints (Additional file 1: Figure S5) (Fig. 5).
Fig. 4.
MOI was determined to be the higher of the total number of unique haplotype present within an individual at either the Pfcsp or Pfama1 loci. Individuals were considered monoclonal if their MOI was estimated to be 1 and polyclonal if their MOI was > 1
Fig. 5.
The allele frequency distribution is plotted for each population (a, c DRC; b, d Zambia; light grey bars: 2016 samples; dark grey bars: 2017 samples). Frequencies were calculated considering loci which were found to be segregating sites in the total dataset (Pfcsp: n = 35; Pfama1: n = 38)
Population structure from rare variants
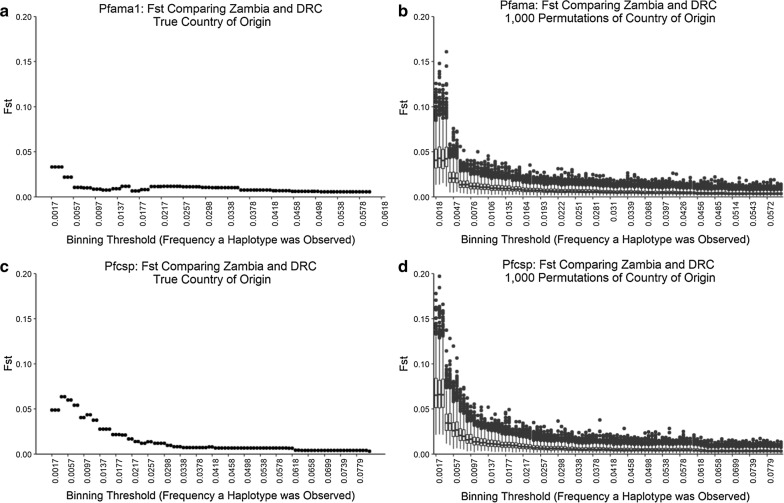
Using only rare variants, a possible signature was observed of low to moderate population structuring (FST = 0.06) between Pfcsp sequences from Zambia and the DRC (Fig. 6c). The FST estimates for the binning thresholds that included the rarest Pfama1 samples were low (FST = 0.025) and not indicative of population differentiation (Fig. 6a). However, although FST values were possibly indicative of genetic differentiation between Zambia and the DRC using rare Pfcsp isolates, the possibility that this observation was due to the small sample size remaining after subsetting rare Pfcsp haplotypes cannot be excluded. In fact, the FST value calculated from rare Pfcsp variants was not significantly different from the distribution of FST values obtained from randomly permuting the country of origin among rare haplotypes 1000 times and estimating FST (Fig. 6b, d). Further, DAPC using only rare variants resulted in improved population separation between Zambia and the DRC, particularly for Pfcsp (Additional file 1: Figure S6B, D). This was true when rare haplotypes were defined as singletons (Additional file 1: Figure S6) or as those present at 2% or less in the population (Additional file 1: Figure S7).
Fig. 6.
a, b For each of 100 randomly selected binning threshold (X-axis) ranging from the minimum to the maximum haplotype frequencies for each amplicon (a Pfama1, b Pfcsp), we classified parasites as rare or not, subset the data to include only rare parasites, and calculated FST (Y-axis) between Zambia and the DRC using only the subset data. c, d To test whether the reduced sample size was driving patterns in FST we randomly permuted the country of origin for each sample 1000 times. For each permutation, we subset the data to include only rare samples based on each binning threshold, and calculated FST comparing Zambia and the DRC. Boxplots show the range of FST estimates across the 1000 permutation replicates for Pfama1 (c) and Pfcsp (d)
Discussion
The utility of parasite genotyping to enhance malaria epidemiology has been well-demonstrated in low to moderate transmission settings. Although parasite genotyping has been useful in assessing the prevalence of drug resistance mutations in high transmission settings [42], it remains to be proven as a tool for evaluating control interventions or enhancing the understanding of transmission epidemiology in this context. In a holoendemic transmission region along the international border between Luapula Province, Zambia and Haut-Katanga Province, the DRC, high genetic Pfama1 and Pfcsp diversity was observed across two different seasons, indicating that these loci are diverse and unstructured in high transmission settings. Given the benefit of using amplicon deep sequencing in such regions with high polyclonality, this observation highlights the limitations of utilizing parasite population genetic analysis to understand transmission epidemiology in high-burden areas. Although multiple studies in moderate to low transmission settings reported decreases in genetic diversity and MOI following decreased transmission [11–19], no signatures of a parasite population bottleneck were detected in this study despite an ITN distribution campaign between sampling timepoints that reduced the parasite prevalence by microscopy in Kilwa and Kashobwe, the DRC using amplicon deep sequencing of two highly diverse antigens. This reflects the fact that transmission remains high in this region even after the observed decrease in parasite prevalence, and the parasite population remains sufficiently large and, therefore, shielded from a genetic bottleneck. Further, the choice of non-neutral genetic loci, which enabled haplotypic characterization of polyclonal infections, may have hindered the ability of this study to detect changes in population genetic diversity, underscoring the current limitations of implementing molecular epidemiologic approaches in high-burden transmission settings. It is also possible that genetic signatures of a population bottleneck may take longer than 6 months to become apparent, and were missed in this study. A much larger decline in transmission is likely required to bottleneck the parasite population. Further, although parasite genotyping has been touted as a tool for monitoring changes in transmission intensity and evaluating control interventions [7, 8], these utilities may be limited to moderate and low transmission settings, and may be less useful in high transmission regions, where high polyclonality necessitates analysis of non-neutral markers.
No evidence of population structure was detected comparing Pfama1 and Pfcsp isolates between Zambia and the DRC. While clear signatures of population differentiation are easily interpretable, it is more challenging to attribute cause for observations with no discernable population structure. A lack of population structure could either reflect the true underlying biology of an admixed population or may be an artifact of the use of genetic markers that are under selection and alone not ideal for assessing population structure on a small spatial scale in high transmission zones [43, 44]. Although the same Pfcsp amplicon that was examined in this study revealed parasite population genetic structure on a continental scale [20], additional research is merited to assess the utility of these Pfama1 and Pfcsp amplicons to detect population structure on smaller geographic scales. While these data suggest P. falciparum parasites from Nchelenge, Zambia and Kilwa and Kashobwe, the DRC, exist as a single panmictic population, increasing either the number of neutral SNPs characterized or the number of isolates sequenced could reveal finer scale population structuring.
It is typical to analyse neutral, unlinked SNPs in population genomic analysis. This study characterizes two highly variable P. falciparum genes, Pfama1 and Pfcsp which are known to be under balancing selective pressure [45]. If balancing selection were to occur independently at geographically separated sites, then the isolation by distance signal could be attenuated, which would lead to the inability to correctly identify population structure when it truly exists. Further, since Pfama1 and Pfcsp are under balancing selection, changes in their diversity do not necessarily reflect changes in transmission. Finally, the SNPs within each of these two amplicons are in linkage disequilibrium in the P. falciparum genome. It is possible that the use of non-neutral, linked loci biased these analyses such that true population differentiation was not detected between Nchelenge District, Zambia and Haut-Katanga Province, the DRC or failed to detect genetic signatures of a population bottleneck. However, in regions where most infections are comprised of multiple, genetically distinct parasite clones, amplicon deep sequencing is perhaps the most cost-effective method capable of preserving parasite haplotypes, bypassing the need to invoke potentially biased haplotype reconstruction methods or discarding polyclonal infections prior to analysis. In P. falciparum genetics, it has been common practice to exclusively analyse monoclonal infections [11, 46, 47] or disregard loci where two or more alleles are present in polyclonal infections [48, 49]. While such practices may be appropriate in some settings where MOI is low, they are not an option in high burden regions like Luapula and Haut-Katanga Provinces, where restricting an analysis to monoclonal infections would require discarding close to 80% of the data (Pringle, unpublished). As methods for handling polyclonal genetic data continue to improve, it may eventually be possible to select unlinked, neutral loci for additional analyses to assess parasite population structure in border regions and detect signatures of population bottlenecks in moderate to high transmission settings.
Despite the use of non-neutral, linked SNPs, the data suggesting a contiguous P. falciparum population are consistent with whole genome sequencing analyses from Anopheles funestus mosquitoes that did not detect population structure of vectors between Nchelenge, Zambia and Haut-Katanga Province, the DRC ([50], Lee, unpublished). These data supporting a single and large primary vector population suggest a possible mechanism that might drive the regular genetic crossing and lack of population differentiation among P. falciparum isolates from across the country border. The observation of a contiguous parasite population across the border between Zambia and DRC suggest that collaborative malaria control efforts targeting both regions together may enhance intervention success. Border regions of a country frequently experience higher malaria transmission than non-border regions and often harbor the final transmission foci prior to elimination [51]. The observation that Luapula Province, Zambia and Haut-Katanga Province, the DRC together comprise a contiguous high transmission foci along an international border highlights the importance of expanding existing regional partnerships [52] like Elimination 8 (E8) in southern Africa that can facilitate the coordination of elimination efforts across multiple nations. A study which looked at how frequently the Global Fund funded malaria projects aiming to establish multi-national control efforts [53] found that these proposals are rarely funded, and that there is little guidance for what makes these projects successful. Developing new strategies to guide, fund, and support regional initiatives that encourage international cooperation towards malaria elimination may enhance current and future efforts. As efforts to eliminate malaria across the globe continue to expand, addressing the unique challenge of controlling border malaria is essential.
Conclusions
Achieving Zambia’s malaria elimination target date of 2021 will require substantially reducing the burden of malaria in the holoendemic transmission region in Luapula Province. Although parasite genotyping may be valuable in addressing specific questions, like the prevalence of drug resistance mutations, it is challenging to use parasite genotyping to draw inferences regarding transmission epidemiology in high burden regions characterized by high genetic diversity using current tools. This study explored whether analysing rare haplotypes enhanced the ability to elucidate transmission patterns in a holoendemic setting. While restricting the analysis to rare variants did lead to the detection of a possible genetic signature of population structure, it is unclear whether this signal is real, or merely an artifact from the reduced sample size. Incorporating rare variant analytical approaches in P. falciparum population genetic analysis may be beneficial but should be interpreted with caution when sample sizes are significantly reduced. Continued decreases in the cost of whole genome sequencing, improved computational methods for phasing sequencing reads from polyclonal data, and genetic distance metrics that account for polyclonality and high background diversity may lead to an enhanced value of parasite genotyping in high burden regions.
Supplementary information
Additional file 1: Fig. S1. Top: Pfama1; bottom: Pfcsp. Distributions across 1000 replicates of collector’s curve analysis show the number of unique haplotypes (Y-axis) found among a randomly selected group of samples of increasing size (X-axis). Curves on the left were generated using the raw (unrarefied) dataset, while curves on the right were generated from the dataset rarefied to a depth of 200 reads per sample. Fig. S2. For each amplicon (top: Pfama1, bottom: Pfcsp), we performed 1000 re-sampling replicates of rarefaction. For each replicate we estimated MOI in each individual using the rarefied data. The distribution of MOI estimates across all rarefaction re-sampling replicates are plttoed along the Y-axis. The X-axis shows the MOI estimate from the raw data. The red dashed line is the Y = X line, or what would be expected if there was no difference between the estimate using rarefied data and the true estimate. Fig. S3. Pairwise genetic relatedness (the proportion of loci which are identical between two parasites) is plotted for all pairs of parasites from different countries or from the same country for each amplicon, Pfama1 (left) and Pfcsp (right). Comparisons between two parasites from individuals both under 5 years old are shown in pink. Comparisons between two parasites from individuals both 5 years or older are shown in blue. Comparisons between parasites from an individual 5 years or older and an individual under 5 years are shown in yellow. Fig. S4. Haplotype frequency distributions are plotted for the haplotypes present in each population (left: DRC right: Zambia; light grey bars: 2016 samples; dark grey bars: 2017 samples). Fig. S5. Left: plots the proportion of haplotypes within the four country-year “populations” considered to be rare. Here rare haplotypes are those represented at 2% or less in the “population.” Right: plots the proportion of haplotypes within the four country-year “populations” that were observed only once (singletons). Fig. S6. Dicriminatory analysis of principal components (DAPC) was performed using R package, adgenet. DAPC performs linear discriminat analysis on principal components in order to maximize separation of a priori groups. A, B: Pfama1; C, D: Pfcsp. A, C: DAPC performed using all sequences regardless of population frequency shows no linear function that can classify the principal components of the parasite seqeunces reliably by country. B, D: DAPC using only rare haplotypes (singletons) results in more refined population discrimination for Pfcsp. Fig. S7. Dicriminatory analysis of principal components (DAPC) was performed using R package, adgenet. DAPC performs linear discriminat analysis on principal components in order to maximize separation of a priori groups. A, B: Pfama1; C, D: Pfcsp. A, C: DAPC performed using all sequences regardless of population frequency shows no linear function that can classify the principal components of the parasite seqeunces reliably by country. B, D: DAPC using only rare haplotypes (2% or less frequency) results in more refined population discrimination for Pfcsp.
Acknowledgements
We gratefully acknowledge Anne Jedlicka and Amanda Dziedzic in the Johns Hopkins Bloomberg School of Public Health Genomic Analysis and Sequencing Core for assistance with amplicon library preparation for sequencing. We are appreciative of the communities of Luapula Province, Zambia and Haut-Katanga Province, the DRC who participated in this research.
Abbreviations
- DRC
Democratic Republic of the Congo
- IRS
indoor residual spray
- ITN
insecticide-treated bed net
- ACTs
artemisinin-combination therapies
- HBHI
high burden to high impact
- WHO
World Health Organization
- RDT
rapid diagnostic test
- DHS
Demographic and Health Survey
- DBS
dried blood spots
- Pfama1
Plasmodium falciparum apical membrane antigen 1
- Pfcsp
Plasmodium falciparum circumsporozoite protein
- qPCR
quantitative polymerase chain reaction
- SNP
single nucleotide polymorphism
- DAPC
discriminatory analysis of principal components
- TCS
Templeton, Crandall, and Sing
- MOI
multiplicity of infection
Authors’ contributions
JCP, GC, DEN, WJM, SM, JJJ conceived of and designed the research project. TB, TK, MC, MM collected samples. JCP, MG performed molecular analyses and sample processing. JCP, GC, SB, AW conducted bioinformatic, genetic, and statistical analyses. The manuscript was written by JCP with input from all authors. All authors read and approved the final manuscript.
Funding
This research was funded in part by the National Institutes of Health (5U19AI089680 to the Southern and Central African International Centers of Excellence in Malaria Research; 5T32AI007417 to JCP as a Molecular and Cellular Basis of Infectious Diseases Training Grant Trainee in the Department of Molecular Microbiology and Immunology and the Johns Hopkins Bloomberg School of Public Health). JCP received a post-doctoral research fellowship award from the Johns Hopkins Malaria Research Institute and Bloomberg Philanthropies.
Availability of data and materials
Pfcsp sequences from 2016 were previously deposited to GenBank (accession numbers: MG715504-MG715555) (29). Pfama1 sequences from 2016 and 2017 as well as Pfcsp sequences from 2017 also were deposited to GenBank (accession numbers MN044107- MN044259).
Ethics approval and consent to participate
This research underwent ethical review and was approved by the Institutional Review Board at the Johns Hopkins Bloomberg School of Public Health (Baltimore, MD USA), the Ethics Review Committee of the Tropical Diseases Research Centre (Ndola, Zambia), and Le Comité d’éthique de l’Université Protestante au Congo (Kinshasa, the Democratic Republic of the Congo). All adult participants gave informed consent. All child participants gave assent and had parental consent for participation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Julia C. Pringle, Email: jpringl3@jhu.edu
Douglas E. Norris, Email: douglas.norris@jhu.edu
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12936-019-3023-4.
References
- 1.Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526:207–211. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cibulskis RE, Alonso P, Aponte J, Aregawi M, Barrette A, Bergeron L, et al. Malaria: global progress 2000–2015 and future challenges. Infect Dis Poverty. 2016;5:61. doi: 10.1186/s40249-016-0151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newby G, Bennett A, Larson E, Cotter C, Shretta R, Phillips AA, et al. The path to eradication: a progress report on the malaria-eliminating countries. Lancet. 2016;387:1775–1784. doi: 10.1016/S0140-6736(16)00230-0. [DOI] [PubMed] [Google Scholar]
- 4.Mukonka VM, Chanda E, Haque U, Kamuliwo M, Mushinge G, Chileshe J, et al. High burden of malaria following scale-up of control interventions in Nchelenge District, Luapula Province, Zambia. Malar J. 2014;13:153. doi: 10.1186/1475-2875-13-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO . World malaria report 2018. Geneva: World Health Organization; 2018. [Google Scholar]
- 6.World Health Organization & Roll Back Malaria Partnership to End Malaria . High burden to high impact: a targeted malaria response. Geneva: World Health Organization; 2018. [Google Scholar]
- 7.Auburn S, Barry AE. Dissecting malaria biology and epidemiology using population genetics and genomics. Int J Parasitol. 2017;47:77–85. doi: 10.1016/j.ijpara.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Neafsey DE, Volkman SK. Malaria genomics in the era of eradication. Cold Spring Harb Perspect Med. 2017;7:a025544. doi: 10.1101/cshperspect.a025544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Roch KG, Chung DWD, Ponts N. Genomics and integrated systems biology in Plasmodium falciparum: a path to malaria control and eradication. Parasite Immunol. 2012;34:50–60. doi: 10.1111/j.1365-3024.2011.01340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pringle JC, Tessema S, Wesolowski A, Chen A, Murphy M, Carpi G, et al. Genetic evidence of focal Plasmodium falciparum transmission in a pre-elimination setting in Southern Province, Zambia. J Infect Dis. 2019;219:1254–1263. doi: 10.1093/infdis/jiy640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels R, Chang HH, Séne PD, Park DC, Neafsey DE, Schaffner SF, et al. Genetic surveillance detects both clonal and epidemic transmission of malaria following enhanced intervention in Senegal. PLoS ONE. 2013;8:e60780. doi: 10.1371/journal.pone.0060780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniels RF, Schaffner SF, Wenger EA, Proctor JL, Chang H-H, Wong W, et al. Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proc Natl Acad Sci USA. 2015;112:7067–7072. doi: 10.1073/pnas.1505691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ofosu-Okyere A, Mackinnon MJ, Sowa MP, Koram KA, Nkrumah F, Osei YD, et al. Novel Plasmodium falciparum clones and rising clone multiplicities are associated with the increase in malaria morbidity in Ghanaian children during the transition into the high transmission season. Parasitology. 2001;123:113–123. doi: 10.1017/S0031182001008162. [DOI] [PubMed] [Google Scholar]
- 14.Gatei W, Kariuki S, Hawley W, ter Kuile F, Terlouw D, Phillips-Howard P, et al. Effects of transmission reduction by insecticide-treated bed nets (ITNs) on parasite genetics population structure: I. The genetic diversity of Plasmodium falciparum parasites by microsatellite markers in western Kenya. Malar J. 2010;9:353. doi: 10.1186/1475-2875-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escalante AA, Ferreira MU, Vinetz JM, Volkman SK, Cui L, Gamboa D, et al. Malaria molecular epidemiology: lessons from the International Centers of Excellence for Malaria Research Network. Am J Trop Med Hyg. 2015;93:79–86. doi: 10.4269/ajtmh.15-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baliraine FN, Afrane YA, Amenya DA, Bonizzoni M, Vardo-Zalik AM, Menge DM, et al. A cohort study of Plasmodium falciparum infection dynamics in Western Kenya Highlands. BMC Infect Dis. 2010;10:283. doi: 10.1186/1471-2334-10-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibara-Okabande R, Koukouikila-Koussounda F, Ndounga M, Vouvoungui J, Malonga V, Casimiro PN, et al. Reduction of multiplicity of infections but no change in msp2 genetic diversity in Plasmodium falciparum isolates from Congolese children after introduction of artemisinin-combination therapy. Malar J. 2012;11:410. doi: 10.1186/1475-2875-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck HP, Felger I, Huber W, Steiger S, Smith T, Weiss N, et al. Analysis of multiple Plasmodium falciparum infections in Tanzanian children during the phase III trial of the malaria vaccine SPf66. J Infect Dis. 1997;175:921–926. doi: 10.1086/513991. [DOI] [PubMed] [Google Scholar]
- 19.Buchholz U, Kobbe R, Danquah I, Zanger P, Reither K, Abruquah HH, et al. Multiplicity of Plasmodium falciparum infection following intermittent preventive treatment in infants. Malar J. 2010;9:244. doi: 10.1186/1475-2875-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo E, Lam N, Hemming-Schroeder E, Nguyen J, Zhou G, Lee M-C, et al. Frequent spread of Plasmodium vivax malaria maintains high genetic diversity at the Myanmar-China border, without distance and landscape barriers. J Infect Dis. 2017;216:1254–1263. doi: 10.1093/infdis/jix106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Hamidhi S, Mahdy MAK, Idris MA, Bin Dajem SM, Al-Sheikh AAH, Al-Qahtani A, et al. The prospect of malaria elimination in the Arabian Peninsula: a population genetic approach. Infect Genet Evol. 2014;27:25–31. doi: 10.1016/j.meegid.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 22.Tessema S, Wesolowski A, Chen A, Murphy M, Wilheim J, Mupiri A-R, et al. Using parasite genetic and human mobility data to infer local and cross-border malaria connectivity in Southern Africa. Elife. 2019;8:e43510. doi: 10.7554/eLife.43510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo E, Zhou G, Oo W, Lee M-C, Baum E, Felgner PL, et al. Molecular inference of sources and spreading patterns of Plasmodium falciparum malaria parasites in internally displaced persons settlements in Myanmar-China border area. Infect Genet Evol. 2015;33:189–196. doi: 10.1016/j.meegid.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Republic of Zambia Ministry of Health . Zambia malaria indicator survey 2018. Lusaka: Republic of Zambia Ministry of Health; 2018. [Google Scholar]
- 25.Hast MA, Chaponda M, Muleba M, Kabuya J-B, Lupiya J, Kobayashi T, et al. The impact of three years of targeted IRS with pirimiphos-methyl on malaria parasite prevalence in a high-transmission area of northern Zambia. Am J Epidemiol. 2019 doi: 10.1093/aje/kwz107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moss WJ, Dorsey G, Mueller I, Laufer MK, Krogstad DJ, Vinetz JM, et al. Malaria epidemiology and control within the International Centers of Excellence for Malaria Research. Am J Trop Med Hyg. 2015;93:5–15. doi: 10.4269/ajtmh.15-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banque Mondiale. République Démocratique du Congo Deuxième Enquête Démographique et de Santé (EDS-RDC II 2013–2014). Le Fonds mondial de lutte contre le SIDA, la tuberculose et le paludisme. Septembre 2014.
- 28.Parr JB, Belson C, Patel JC, Hoffman IF, Kamthunzi P, Martinson F, et al. Estimation of Plasmodium falciparum transmission intensity in Lilongwe, Malawi, by microscopy, rapid diagnostic testing, and nucleic acid detection. Am J Trop Med Hyg. 2016;95:373–377. doi: 10.4269/ajtmh.16-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pringle JC, Carpi G, Almagro-Garcia J, Zhu SJ, Kobayashi T, Mulenga M, et al. RTS, S/AS01 malaria vaccine mismatch observed among Plasmodium falciparum isolates from southern and central Africa and globally. Sci Rep. 2018;8:6622. doi: 10.1038/s41598-018-24585-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Illumina. 16S Metagenomic sequencing library preparation. 2013;(B):1–28.
- 31.Hathaway NJ, Parobek CM, Juliano JJ, Bailey JA. SeekDeep: single-base resolution de novo clustering for amplicon deep sequencing. Nucleic Acids Res. 2018;46:e21. doi: 10.1093/nar/gkx1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ESRI . ArcGIS Desktop: release 10.6. Redlands: Environmental Systems Research Institute; 2018. [Google Scholar]
- 33.Saary P, Forslund K, Bork P, Hildebrand F. RTK: efficient rarefaction analysis of large datasets. Bioinformatics. 2017;33:2594–2595. doi: 10.1093/bioinformatics/btx206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 35.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 37.Múrias dos Santos A, Cabezas MP, Tavares AI, Xavier R, Branco M. tcsBU: a tool to extend TCS network layout and visualization. Bioinformatics. 2016;32:627–628. doi: 10.1093/bioinformatics/btv636. [DOI] [PubMed] [Google Scholar]
- 38.Luikart G, Allendorf FW, Cornuet JM, Sherwin WB. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J Hered. 1998;89:238–247. doi: 10.1093/jhered/89.3.238. [DOI] [PubMed] [Google Scholar]
- 39.Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, et al. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci USA. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Connor TD, Fu W, Mychaleckyj JC, Logsdon B, Auer P, Carlson CS, et al. Rare variation facilitates inferences of fine-scale population structure in humans. Mol Biol Evol. 2015;32:653–660. doi: 10.1093/molbev/msu326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goudet J. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes. 2005;5:184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- 42.Aydemir O, Janko M, Hathaway NJ, Verity R, Mwandagalirwa MK, Tshefu AK, et al. Drug-resistance and population structure of Plasmodium falciparum across the Democratic Republic of Congo using high-throughput molecular inversion probes. J Infect Dis. 2018;218:946–955. doi: 10.1093/infdis/jiy223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnaud-Haond S, Alberto F, Teixeira S, Procaccini G, Serr EA, Duarte CM. Assessing genetic diversity in clonal organisms: low diversity or low resolution? Combining power and cost efficiency in selecting markers. J Hered. 2005;96:434–440. doi: 10.1093/jhered/esi043. [DOI] [PubMed] [Google Scholar]
- 44.Reusch TB. New markers- old questions: population genetics of seagrasses. Mar Ecol Prog Ser. 2001;211:261–274. doi: 10.3354/meps211261. [DOI] [Google Scholar]
- 45.Miller RH, Hathaway NJ, Kharabora O, Mwandagalirwa K, Tshefu A, Meshnick SR, et al. A deep sequencing approach to estimate Plasmodium falciparum complexity of infection (COI) and explore apical membrane antigen 1 diversity. Malar J. 2017;16:490. doi: 10.1186/s12936-017-2137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jalloh A, Jalloh M, Matsuoka H. T-cell epitope polymorphisms of the Plasmodium falciparum circumsporozoite protein among field isolates from Sierra Leone: age-dependent haplotype distribution? Malar J. 2009;8:120. doi: 10.1186/1475-2875-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chenet SM, Taylor JE, Blair S, Zuluaga L, Escalante AA. Longitudinal analysis of Plasmodium falciparum genetic variation in Turbo, Colombia: implications for malaria control and elimination. Malar J. 2015;14:363. doi: 10.1186/s12936-015-0887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice BL, Golden CD, Anjaranirina EJG, Botelho CM, Volkman SK, Hartl DL. Genetic evidence that the Makira region in northeastern Madagascar is a hotspot of malaria transmission. Malar J. 2016;15:596. doi: 10.1186/s12936-016-1644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ochola-Oyier LI, Okombo J, Wagatua N, Ochieng J, Tetteh KK, Fegan G, et al. Comparison of allele frequencies of Plasmodium falciparum merozoite antigens in malaria infections sampled in different years in a Kenyan population. Malar J. 2016;15:261. doi: 10.1186/s12936-016-1304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones CM, Lee Y, Kitchen A, Collier T, Pringle JC, Muleba M, et al. Complete Anopheles funestus mitogenomes reveal an ancient history of mitochondrial lineages and their distribution in southern and central Africa. Sci Rep. 2018;8:9054. doi: 10.1038/s41598-018-27092-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.WHO . Malaria Policy Advisory Committee Meeting Evidence review group on border malaria Summary of conclusions and recommendations. Geneva: World Health Organization; 2018. [Google Scholar]
- 52.Sturrock HJW, Roberts KW, Wegbreit J, Ohrt C, Gosling RD. Tackling imported malaria: an elimination endgame. Am J Trop Med Hyg. 2015;93:139–144. doi: 10.4269/ajtmh.14-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gueye C, Teng A, Kinyua K, Wafula F, Gosling R, McCoy D. Parasites and vectors carry no passport: how to fund cross-border and regional efforts to achieve malaria elimination. Malar J. 2012;11:344. doi: 10.1186/1475-2875-11-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Top: Pfama1; bottom: Pfcsp. Distributions across 1000 replicates of collector’s curve analysis show the number of unique haplotypes (Y-axis) found among a randomly selected group of samples of increasing size (X-axis). Curves on the left were generated using the raw (unrarefied) dataset, while curves on the right were generated from the dataset rarefied to a depth of 200 reads per sample. Fig. S2. For each amplicon (top: Pfama1, bottom: Pfcsp), we performed 1000 re-sampling replicates of rarefaction. For each replicate we estimated MOI in each individual using the rarefied data. The distribution of MOI estimates across all rarefaction re-sampling replicates are plttoed along the Y-axis. The X-axis shows the MOI estimate from the raw data. The red dashed line is the Y = X line, or what would be expected if there was no difference between the estimate using rarefied data and the true estimate. Fig. S3. Pairwise genetic relatedness (the proportion of loci which are identical between two parasites) is plotted for all pairs of parasites from different countries or from the same country for each amplicon, Pfama1 (left) and Pfcsp (right). Comparisons between two parasites from individuals both under 5 years old are shown in pink. Comparisons between two parasites from individuals both 5 years or older are shown in blue. Comparisons between parasites from an individual 5 years or older and an individual under 5 years are shown in yellow. Fig. S4. Haplotype frequency distributions are plotted for the haplotypes present in each population (left: DRC right: Zambia; light grey bars: 2016 samples; dark grey bars: 2017 samples). Fig. S5. Left: plots the proportion of haplotypes within the four country-year “populations” considered to be rare. Here rare haplotypes are those represented at 2% or less in the “population.” Right: plots the proportion of haplotypes within the four country-year “populations” that were observed only once (singletons). Fig. S6. Dicriminatory analysis of principal components (DAPC) was performed using R package, adgenet. DAPC performs linear discriminat analysis on principal components in order to maximize separation of a priori groups. A, B: Pfama1; C, D: Pfcsp. A, C: DAPC performed using all sequences regardless of population frequency shows no linear function that can classify the principal components of the parasite seqeunces reliably by country. B, D: DAPC using only rare haplotypes (singletons) results in more refined population discrimination for Pfcsp. Fig. S7. Dicriminatory analysis of principal components (DAPC) was performed using R package, adgenet. DAPC performs linear discriminat analysis on principal components in order to maximize separation of a priori groups. A, B: Pfama1; C, D: Pfcsp. A, C: DAPC performed using all sequences regardless of population frequency shows no linear function that can classify the principal components of the parasite seqeunces reliably by country. B, D: DAPC using only rare haplotypes (2% or less frequency) results in more refined population discrimination for Pfcsp.
Data Availability Statement
Pfcsp sequences from 2016 were previously deposited to GenBank (accession numbers: MG715504-MG715555) (29). Pfama1 sequences from 2016 and 2017 as well as Pfcsp sequences from 2017 also were deposited to GenBank (accession numbers MN044107- MN044259).