Abstract
Objective
We explored the role and mechanism of miR-214 involvement in the progression of diffuse large B-cell lymphoma (DLBCL).
Methods
The expression levels of miR-214 and PD-L1 in human DLBCL cell lines and in tissue samples from patients with DLBCL were determined using quantitative RT-PCR. The dual-luciferase reporter assay was employed to determine the correlation between the expressions of miR-214 and PD-L1. Cell viability, invasiveness and apoptosis were respectively examined in cells of the DLBCL line OCI-Ly3 using CCK-8, transwell and flow cytometry assays. The expression level of PD-L1 was determined via immunoblotting. Inflammatory cytokine secretion was determined via enzyme-linked immune sorbent assay (ELISA).
Results
miR-214 was downregulated and PD-L1 was upregulated in DLBCL tissues and cell lines in comparison to normal adjacent tissues or normal B-cell. This indicates a negative correlation in the expression levels. Overexpression of miR-214 inhibited cell viability and invasion and induced apoptosis of OCI-Ly3 cells. Moreover, miR-214 was shown to target PD-L1 mRNA by binding to its 3′-untranslated region (UTR). Knockdown of PD-L1 attenuated the malignant phenotype of OCI-Ly3 cells. Overexpression of miR-214 inhibited tumor growth by targeting PD-L1 in vivo.
Conclusion
By targeting PD-L1, miR-214 regulates the progression of DLBCL in vitro and in vivo.
Keywords: Diffuse large B-cell lymphoma, miR-214, PD-L1, Proliferation, Invasion, T cells
Background
Diffuse large B-cell lymphoma (DLBCL) is a highly heterogeneous malignant lymphoma [1, 2]. Despite the current greater understanding of DLBCL pathological subtypes and the effectiveness of rituximab-based chemoimmunotherapy, 35–40% of patients show diminished treatment efficacy due to the rapid emergence of drug resistance [3]. To improve survival rates, there is an urgent need for an in-depth understanding of DLBCL pathogenesis and the mechanisms that lead to drug resistance.
Previous studies found that microRNAs (miRNAs) participate in the regulation of cancer cells’ malignant biological behavior [4, 5]. For example, miR-214 has been confirmed to act as a tumor suppressor gene in the development of various malignant tumor types, including colon [6], breast [7], ovarian [8], non-small cell lung [9] and gastric cancer [10], by decreasing cell proliferation and invasion and increasing apoptosis rates. Furthermore, miR-214 is downregulated in DLBCL tissues [11]. In addition, elevated miR-214 was associated with a superior outcome for this cancer type [12]. This led us to speculate that overexpression of miR-214 may inhibit the malignant behavior of DLBCL cells by targeting a downstream gene, but the mechanism details are not clear. The role of miR-214 in DLBCL progression is the focus of this study.
Programmed death ligand-1 (PD-L1) is a commonly found negative immunoregulatory protein [13]. It has great significance in the avoidance harmful autoimmune reactions [3]. Many studies have found that PD-L1 expression significantly increases in various malignant tumor tissues and cell lines, inducing the increase of immune cell apoptosis, which is an important regulatory mechanism for tumor immunosuppression [14, 15]. PD-L1 is widely distributed in various organs, circulatory systems and tumor tissues, with its distribution mainly modulated by the molecular microenvironment (IFN-γ, miRNAs, etc.) in which the cells are located [16, 17].
Several studies have confirmed that PD-L1 targeting underlies the involvement of multiple miRNAs in regulating the development of malignant tumor types, including gastric cancer [18], melanoma [19] and DLBCL [20]. He et al. reported that overexpressed miR-195 targets PD-L1 to attenuate DLBCL progression by decreasing the immune escape of DLBCL cells [21]. However, further information on the role of PD-L1 in the malignant behavior of DLBCL cells is needed.
This study investigated how the miR-214–PD-L1 axis is involved in the regulation of tumor growth and the function of T cells in vitro and in vivo. In addition, a new therapeutic biomarker for DLBCL treatment in the clinic is assessed.
Materials and methods
Tissue specimen
DLBCL tissues and paired normal adjacent tissues were collected from patients (n = 15) at Liaocheng People’s Hospital. The clinical specimens were immediately frozen at − 80 °C. None of the patients had received chemotherapy or radiotherapy prior to surgery. All the patients signed informed consent. Approval for the study was obtained from the Ethical Committee of Liaocheng People’s Hospital and all procedures complied with the guidelines and principles of the Declaration of Helsinki.
Cell culture
Human DLBCL cell lines (OCI-Ly3, SU-DHL-2 and OCI-Ly10), a normal B-cell line (NBC) and HEK-293 T cells were purchased from the Shanghai Institute for Biological Sciences of the Chinese Academy of Sciences. Cells were cultured according to the manufacturer’s instructions in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 1% penicillin–streptomycin and 10% fetal bovine serum (FBS; Thermo Fisher Scientific) at 37 °C in a 5% CO2 atmosphere.
T cell and OCI-Ly3 cell co-culture
T cells were obtained from the peripheral blood of healthy donors and DLBCL patients. A total of 2 × 105 T cells/ml were seeded in 96-well plates cultured in 5% CO2 at 37 °C. T-cell Activation (Thermo Fisher Scientific) was added according to the manufacturer’s protocol. OCI-Ly3 cells were co-cultured with the activated T cells at a ratio of 9:1 for 24 h before transfection with miR-214 inhibitor and PD-L1 siRNA or with PD-L1 siRNA alone.
Cell transfection
OCI-Ly3 cells were seeded in 6-well plates with 2 × 105 cells per well and then incubated for 24 h. The miR-214 mimic and inhibitor, PD-L1 siRNA (si-PD-L1), and control were transfected into OCI-Ly3 cells with Lipofectamine 3000 reagent and Opti-MEM medium (Invitrogen) according to the manufacturer’s protocols. The PD-L1 siRNA, miR-214 mimic and inhibitor, and control (blank plasmid) were purchased from Tolo Biotech. The sequences were miR-214 mimic, 5′-ACAGCAGGCACAGACAGGCAGU-3′; miR-214 inhibitor, 5′-ACUGCCUGUCUGUGCCUGCUGU-3′; and control, 5′-UUGUACUACACAAAAGUACUG-3′.
Real time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from clinical specimens and cell lines using Trizol reagent (QIAGEN) according to the manufacturer’s instructions. For mRNA detection, RNA samples were reverse-transcribed into cDNA using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa). Quantitative real-time PCR was performed with GoTaq qPCR Master Mix (TaKaRa) using the CFX96 Sequence Detection System (Bio-Rad). For miRNA detection, RNA samples were reverse transcribed using a Mir-X miRNA First-Strand Synthesis kit (TaKaRa) and real-time PCR was done with an Applied Biosystems 7300 Real-Time PCR system. The primer sequences are given in Table 1. U6 and GAPDH were respectively used as the endogenous reference for miRNA and mRNA. Real-time PCR was performed in triplicate.
Table 1.
Name and sequences of the primers
| Name | Primer sequences |
|---|---|
| miR-214 |
F: 5′ – CAATACTGACAGCAGGCACA – 3′ R: 5′ – TATGGTTGTTCACGACTCCTTAC – 3′ |
| U6 |
F: 5′ – CTCGCTTCGGCAGCACA – 3′ R: 5′ – AACGCTTCACGAATTTGCGT – 3′ |
| PD-L1 |
F: 5′ – GGTGAGGATGGTTCTACACAG – 3′ R: 5′ – GAGAACTGCATGAGGTTGC – 3′ |
| GAPDH |
F: 5′ – GGAGCGAGATCCCTCCAAAAT – 3′ R: 5′ – GGCTGTTGTCATACTTCTCATGG – 3’ |
F: Forward primer; R: Reverse primer
CCK-8 assay
OCI-Ly3 cells were cultured under standard conditions and transfected with different vectors until the cell confluence reached about 70%. The cells were then collected and seeded in the 96-well plates at a density of 1 × 104 cells per well. A CCK-8 kit (Sigma) was used to assess cell proliferation according to the manufacturer’s protocol. Briefly, the CCK-8 solution (10 μl per well) was added to the wells and incubated with the cells for 2 h. The optical density (OD) values were determined at a wavelength of 450 nm and used to assess cell proliferation abilities.
Flow cytometry analysis
DLBCL and healthy tissue samples were washed with phosphate-buffered saline (PBS), centrifuged at 800×g for 6 min, suspended in ice-cold 70% ethanol/PBS, centrifuged at 800×g for another 6 min, and suspended with PBS. An Annexin V-FITC/Propidium Iodide (PI) Apoptosis Detection Kit (Thermo Fisher Scientific) was used to determine the cell apoptosis ratio according to the manufacturer’s instructions. Briefly, the OCI-Ly3 cells were collected and suspended using 1× Annexin V Binding Buffer. After that, the Annexin V and PI staining solution was incubated with the cell suspensions at room temperature for 25 min without light. A BD LSR II Flow Cytometer (BD Biosciences) was used to determine the rate of apoptosis.
Transwell invasion assay
A total of 2 × 105 OCI-Ly3 cells/ml were plated in 200 μl of serum-free medium in the upper layer of a Corning Transwell chamber that was coated with Matrigel (BD Biosciences), while 800 μl medium supplemented with 10% FBS was added to the bottom chamber. After 24 h of incubation, the cells that had invaded were fixed with 4% paraformaldehyde (PA), stained with 0.1% crystal violet for 10 min and rinsed three times with PBS. For quantification, 5 randomly selected fields were analyzed.
Elisa
The expressions of IL-10, IFN-γ and TNF-α were determined using enzyme-linked immune sorbent assay (ELISA). The cytokines secreted from T cells in a co-culture T cell–OCI-Ly3 cell system were analyzed in triplicate using the IL-10, IFN-γ and TNF-α ELISA kits (R&D Systems) according to the manufacturer’s instructions.
Western blot
Total proteins were extracted from tissue samples and cells using RIPA lysis buffer (Beyotime Biotechnology) according to the manufacturer’s protocol. The BCA kit (Beyotime Biotechnology) was used to quantify protein concentrations. The target proteins were then separated via electrophoresis with 10% SDS-polyacrylamide gel (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). The membranes were incubated for 1 h at 37 °C with 5% skim milk diluted with TBS containing 0.1% Tween-20, then were incubated overnight at 4 °C with the primary rabbit antibodies against human PD-L1 (11,000, #13684, Cell Signaling Technology) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:2500, ab9485, Abcam). Horseradish peroxidase-linked goat anti-rabbit IgG (12,000, ab205718, Abcam) was incubated with the membranes for 1 h at room temperature. An ECL Western blot detection kit (Bio-Rad) was employed to determine the optical density of the protein bands to evaluate the expression levels of the proteins.
Dual-luciferase reporter gene assay
The PD-L1 fragment containing miR-214 binding sites was synthesized to generate wild-type (PD-L1-WT) or mutant-type PD-L1 (PD-L1-MUT). The PD-L1-WT and PD-L1-MUT fragments were subcloned into the Renilla luciferase gene Pgl3-Luciferase reporter vectors (Promega) to generate the pGL3-PD-L1 (WT) and pGL3-PD-L1 (MUT) vectors, respectively. After that, the vectors were co-transfected with miR-214 mimics or control mimics into HEK-293 T cells for 24 h. Finally, the cells were lysed using a Dual-Luciferase Assay Kit (Promega), and the luciferase activities were evaluated using the luminescence plate reader (Molecular Devices).
Nude mouse model
The animal experiments were approved by the Ethical Committee of Liaocheng People’s Hospital. A total of 20 female BALB/c nude mice (4~5 weeks old) were randomly separated into two groups of 10 mice. The miR-214 mimic or control mimics was transfected into OCI-Ly3 cells and cultured in serum-free DMEM for 24 h. These OCI-Ly3 cells (1 × 107) were subcutaneously inoculated into the mice when they were between 6 and 7 weeks old. After 4 weeks, all the mice were killed and the tumor tissues were collected for further experiments. Immunohistochemistry staining was used to observe the histomorphology and examine the expression of Ki-67 as described in an earlier study [22].
Statistical analysis
All data are presented as the means ± standard deviation. The data were analyzed using SPSS 22.0 software (IBM). Spearman correlation analysis was performed to analyze the correlation between miR-214 and PD-L1 in DLBCL tissues using Graphpad Prism Version 8.0.2. p < 0.05 was considered statistically significant.
Results
MiR-214 is downregulated in DLBCL tissues and cell lines
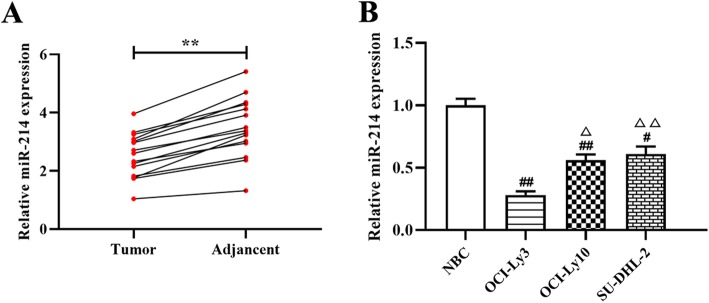
To explore the relationship between miR-214 and DLBCL development, quantitative RT-PCR was used to determine the expression level of miR-214 in DLBCL tissues (n = 15) and adjacent normal tissues (n = 15). As shown in Table 2, low miR-214 expression was positively associated with tumor size (p < 0.05), clinical stage (p < 0.05) and IPI scores (p < 0.05). The results also showed that the expression of miR-214 in DLBCL tissues was significantly lower than in the normal adjacent tissues (p < 0.01, Fig. 1a). Moreover, miR-214 was markedly downregulated in DLBCL cell lines compared with normal B-cell lines (NBC; p < 0.01, Fig. 1b), especially when comparing OCI-Ly3 cells (p < 0.01, Fig. 1b). Those results indicate that low expression of miR-214 may be related to the DLBCL progression. Based on these findings, OCI-Ly3 cells were chosen for subsequent experiments.
Table 2.
The clinicopathological features of patients with DLBCL
| Characteristic | Total number of patients | Expression of miR-214 | P value | |
|---|---|---|---|---|
| 15 | High (N = 7) | aLow (N = 8) | ||
| Age (years) | 0.447 | |||
| ≥ 55 | 8 (53.33%) | 3 | 5 | |
| < 55 | 7 (46.67%) | 4 | 3 | |
| Gender | 0.833 | |||
| Male | 9 (60.00%) | 4 | 5 | |
| Female | 6 (40.00%) | 3 | 3 | |
| Tumor size (cm) | 0.020 | |||
| ≥ 3 | 9 (60.00%) | 2 | 7 | |
| < 3 | 6 (40.00%) | 5 | 1 | |
| Clinical stage | 0.036 | |||
| I - II | 5 (33.33%) | 4 | 1 | |
| III - IV | 10 (66.67%) | 2 | 7 | |
| bLDH | 0.782 | |||
| High (≥ 300) | 8 (53.33%) | 4 | 4 | |
| Low (< 300) | 7 (46.67%) | 3 | 4 | |
| cIPI | 0.013 | |||
| Low (0–2) | 4 (26.67%) | 4 | 0 | |
| High (≥ 3) | 11 (73.33%) | 3 | 8 | |
aThe median of relative miR-214 expression level is 2.53, so the number of low miR-214 expression is 8 (< 2.53). bLDH: Lactate dehydrogenase; c IPI: International prognostic index
Fig. 1.
The expression of miR-214 in DLBCL tissues and cell lines. a and b—Quantitative RT-PCR was used to determine the expression levels of miR-214 in DLBCL tissues (a) and cell lines (b). **p < 0.01, compared with the adjacent normal tissues; #p < 0.05, ##p < 0.01, compared with the normal B-cell line (NBC); △p < 0.05, △△p < 0.01, compared with the OCI-Ly3 cells
Overexpression of miR-214 attenuates the malignant phenotype of OCI-Ly3 cells
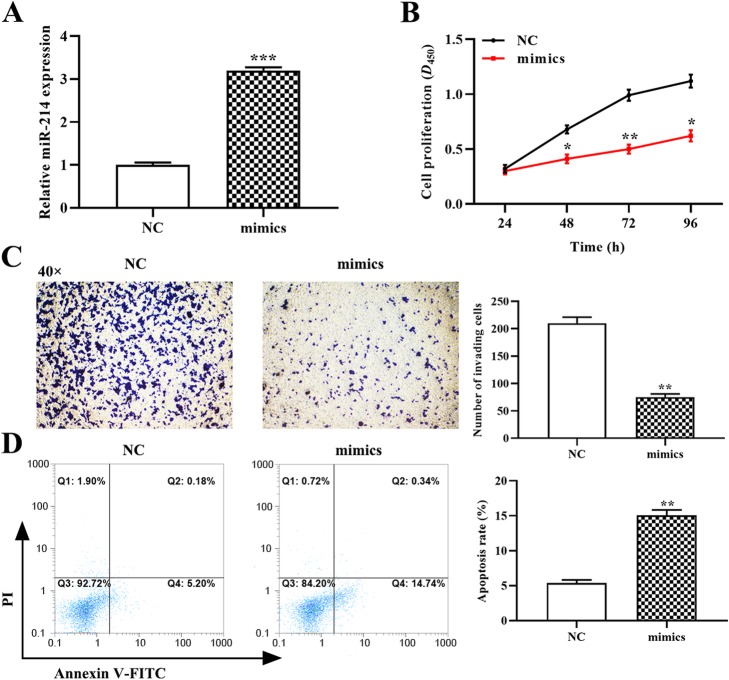
Based on the downregulation of miR-214 in DLBCL tissues and cell lines, we attempted to explore the effect of miR-214 on OCI-Ly3 cell proliferation, invasion and apoptosis. OCI-Ly3 cells were transfected with the miR-214 mimic to assess the gain-of-function of miR-214. The expression of miR-214 was significantly enhanced in the miR-214 mimic group compared with the control group (p < 0.001, Fig. 2a), confirming successful transfection and enhancement of miR-214 expression.
Fig. 2.
The impact of miR-214 on the proliferation, invasion and apoptosis of OCI-Ly3 cells. (a) The relative expression of miR-214 in cells transfected with an miR-214 mimic was determined using quantitative RT-PCR. (b) The proliferation of OCI-Ly3 cells was determined using the CCK-8 assay. (c) The invasion ability of OCI-Ly3 cells was assessed using a Transwell assay (magnification, × 40). (d) The rate of OCI-Ly3 cell apoptosis was measured using flow cytometry. *p < 0.05, **p < 0.01, ***p < 0.001, compared with the negative control (NC) group
Next, we investigated the impact of miR-214 upregulation on the proliferation and invasion of OCI-Ly3 cells using the CCK-8 and transwell assays. Overexpression of miR-214 significantly inhibited OCI-Ly3 cell viability compared with the negative control group (p < 0.05, Fig. 2b). Upregulated miR-214 also significantly suppressed the invasion capacity of OCI-Ly3 cells as compared to the negative control group (p < 0.01, Fig. 2c). Furthermore, Annexin V-FITC/PI double staining results showed that the increased expression of miR-214 contributed to inducing apoptosis of OCI-Ly3 cells (p < 0.01, Fig. 2d). These results strongly imply that overexpression of miR-214 suppresses cell proliferation and invasion and promotes apoptosis of OCI-Ly3 cells.
MiR-214 negatively regulates the expression of PD-L1
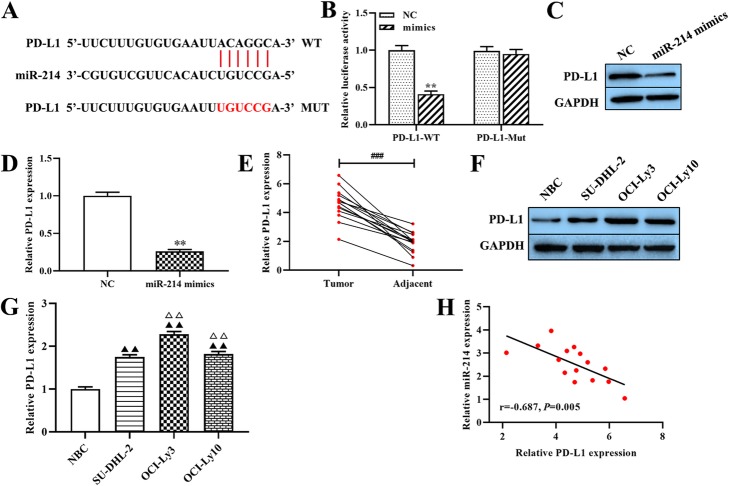
The starBase database analysis revealed that miR-214 may target at PD-L1 directly (Fig. 3a). The dual-luciferase reporter gene assay result showed that co-transfection of miR-214 mimics and PD-L1-WT significantly decreased the luciferase activity (p < 0.01, Fig. 3b), but co-transfection of miR-214 mimics and PD-L1-MUT did not affect luciferase activity. Moreover, overexpression of miR-214 significantly decreased the expression levels of PD-L1 protein in OCI-Ly3 cells compared with the levels for the NC group (p < 0.01; Fig. 3c and d). Additionally, the expression of PD-L1 was markedly higher in DLBCL tissues than in the adjacent normal tissues (p < 0.001, Fig. 3e). The same as the result was obtained for PD-L1 protein expression in the DLBCL cell line compared to the normal B cell line (p < 0.01, Fig. 3f and g). Furthermore, Spearman’s correlation analysis revealed a marked negative correlation between the expressions of miR-214 and PD-L1 in DLBCL tissues (r = − 0.687, p < 0.01, Fig. 3h). These results show that PD-L1 is a target of miR-214 and that it has a lower expression in OCI-Ly3 cells.
Fig. 3.
The regulatory relationship between miR-214 and PD-L1. (a) The bioinformatics analysis showed that miR-214 has a binding site with PD-L1. (b) The dual-luciferase reporter gene assay was used to verify the targeted relationship between miR-214 and PD-L1. (c and d) The expression of PD-L1 protein was determined using western blot. (e) Quantitative RT-PCR was used to determine the expression of PD-L1 in NSCLC tissues and adjacent tissues. (f and g) The expressions of PD-L1 in DLBCL cell lines were determined using quantitative RT-PCR. (h) The expression relationship between miR-214 and PD-L1 was evaluated using Spearman’s correlation analysis. **p < 0.01, compared with the NC group; ###p < 0.001, compared with the adjacent tissues; ▲p < 0.05, ▲▲p < 0.01, compared with the NBC group; △△p < 0.01, compared with the OCI-Ly3 cells
MiR-214 targets PD-L1 and attenuates the malignant phenotype of OCI-Ly3 cells
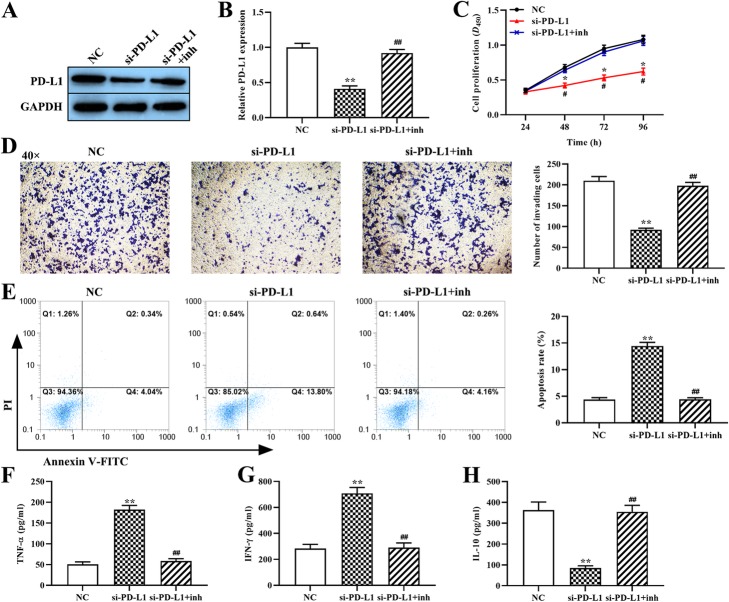
We attempted to determine whether upregulation of miR-214 inhibits cell proliferation and invasion and induces apoptosis of OCI-Ly3 cells by targeting PD-L1. Crucially, western blot analysis results showed that PD-L1 knockdown decreased the levels of PD-L1 protein (p < 0.01, Fig. 4a, b), while co-transfection with miR-214 inhibitor restored them. Moreover, the CCK-8 and Transwell assays showed that knockdown of PD-L1 significantly decreased the proliferation (p < 0.05, Fig. 4c) and invasion (p < 0.01, Fig. 4d) of OCI-Ly3 cells compared with the negative control group. Furthermore, compared with the control group, PD-L1 knockdown increased the percentage of apoptotic OCI-Ly3 cells (p < 0.01, Fig. 4e). However, the effect of silencing PD-L1 on the behavior of OCI-Ly3 cells was reversed by co-transferction with the miR-214 inhibitor. These results suggest that miR-214 negative regulates PD-L1 to inhibit the proliferation and invasion and induce the apoptosis of OCI-Ly3 cells in vitro.
Fig. 4.
The effect of the miR-214–PD-L1 axis on the malignant behavior of OCI-Ly3 cells and on the cytokine secretion from T cells. (a and b) The expression of PD-L1 protein was determined using western blot. (c) The proliferation of OCI-Ly3 cells was determined using the CCK-8 assay. (d) The invasion ability of OCI-Ly3 cells was assessed using the Transwell assay (magnification, × 40). (e) The rate of OCI-Ly3 cell apoptosis was measured using flow cytometry. (f through h) The expression levels of TNF-α, IFN-γ and IL-10 were measured using ELISA. si-PD-L1: PD-L1 siRNA; si-PD-L1 + inh: PD-L1 siRNA + miR-214 inhibitor. *p < 0.05, **p < 0.01, compared with the NC group; #p < 0.05, ##p < 0.01, compared with the si-PD-L1 group
MiR-214 targets PD-L1 to modulate cytokine secretion from T cells
To further determine the impact of the miR-214–PD-L1 axis on inflammatory cytokine secretion, we set up a co-culture system of OCI-Ly3 cells and T cells. The ELISA results show that knockdown of PD-L1 significantly increased the levels of IFN-γ and TNF-α compared with the control group (p < 0.01, Fig. 4f and g), but decreased those of IL-10 (p < 0.01, Fig. 4h). However, there was no significant difference between the cells co-transfected with miR-214 inhibitor and PD-L1 siRNA and those in the negative control group. This indicates that miR-214 targets PD-L1 to regulate the function of T cells and to further mediate the immune response of tumor cells.
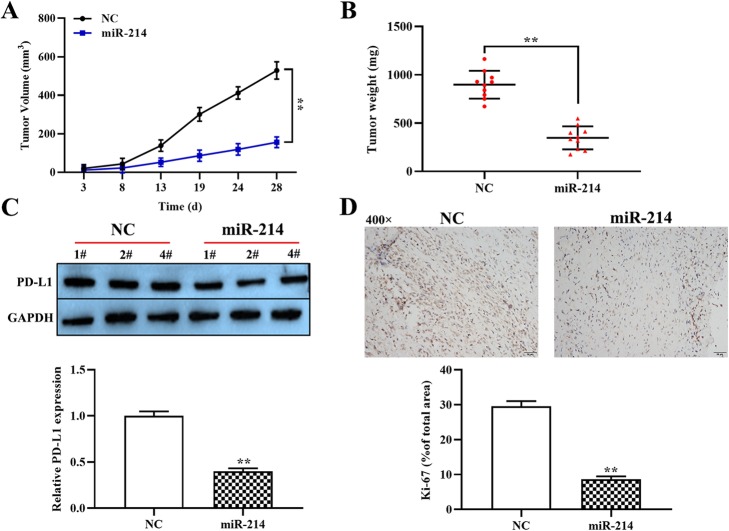
Upregulation of miR-214 suppresses DLBCL development in vivo
Having determined the impact of miR-214 on the proliferation, invasion and apoptosis of OCI-Ly3 cells, we attempted to examine the effect of miR-214 on the tumor growth of DLBCL in vivo. In a DLBCL mouse model with overexpression of miR-214, the tumor volume and weight were noticeably lower than in the negative control group (p < 0.01, Fig. 5a and b). Moreover, as in the cell lines, upregulation of miR-214 markedly decreased the expression of PD-L1 protein (p < 0.01, Fig. 5c). Immunohistochemistry results demonstrate that elevated miR-214 decreased the expression of Ki-67 in xenograft tumor tissues (p < 0.01, Fig. 5d). Our findings suggest that overexpression of miR-214 may restrict DLBCL progression by targeting PD-L1 in vivo.
Fig. 5.
Effect of miR-214 overexpression on the progression of DLBCL in vivo. (a) The tumor volume curve of nude mice treated with miR-214 or NC was analyzed. (b) The tumor weight was measured in the transfected miR-214 or NC group. (c) The expression of PD-L1 protein in the tumor tissues was measured using western blot. (d) The expression of Ki-67 was determined in tumor tissues using immunohistochemistry (magnification, × 400). *p < 0.05, **p < 0.01, compared with the NC group
Discussion
This study focused on the molecular biomarkers in the development and progression of DLBCL. We found that miR-214 is significantly downregulated in DLBCL tissues and cell lines. We also established that it inhibits the proliferation and invasion and promotes the apoptosis of OCI-Ly3 cells by targeting PD-L1. Furthermore, in a co-culture system of OCI-Ly3 cells and T cells, knockdown of PD-L1 was found to increase the levels of IFN-γ and TNF-α, but to decrease the level of IL-10. This effect was reversed by knockdown of miR-214. Therefore, miR-214 plays an important role in regulating DLBCL progression and may be used as a target in DLBCL treatment.
Recent studies have established that miR-214 is downregulated in tumor tissues and cell lines, and that progression-free survival is shortened in patients with low expression of miR-214 [23]. MiR-214 participates in many cellular functions, including cell cycle control, DNA damage and repair, and gene transcription; but its aberrant expression affects cell migration, invasion and apoptosis of human malignant tumor cells by targeting downstream genes [24]. For example, overexpression of miR-214 inhibits proliferation and migration in hepatocellular carcinoma by targeting FOXM1 [25]. A low expression level of miR-214 is associated with lymph node metastasis, TNM stage and tumor size [26, 27]. Meanwhile, downregulation of miR-214 is associated with poor survival in leukemia [28, 29].
The miR-214 is not only significant for malignant tumor progression, but also plays an important role in regulating resistance to chemotherapy and radiotherapy in several tumors. For example, overexpression of miR-214 enhances the sensitivity of radiotherapy in colorectal cancer by decreasing ATG12-induced autophagy [30]. Elevated miR-214 reverses doxorubicin resistance in breast cancer by promoting cell apoptosis [31]. Our results demonstrate that overexpression of miR-214 significantly restricts the malignant behavior of OCI-Ly3 cells and decreases tumor growth in a xenograft mouse model.
PD-L1 monoclonal antibodies have recently been approved by the FDA for the United States, and they have been used in a variety of cancer therapies with good results [32, 33]. Importantly, PD-L1 acts as an immune checkpoint in cancer immunotherapy [34, 35]. Song et al. confirmed that the PD-1/PD-L1 pathway is an immune evasion mechanism associated with the progression of DLBCL [14]. Abnormal PD-L1 expression is used as a biomarker for early diagnosis and progression of several malignant tumors [36], such as lung cancer [37], thyroid cancer [38] and head and neck squamous cell carcinoma [39].
In addition, some studies found that miRNAs targets PD-L1 to regulate the proliferation, invasion and apoptosis of tumor cells and modulate the immune response. For example, miR-148a-3p overexpression inhibits the progression of colorectal cancer by targeting PD-L1 [40]. Repression of miR-940 promotes the proliferation and migration of gastric cancer by upregulating PD-L1 [41]. Overexpression of Epstein-Barr virus-encoded EBNA2 contributes to an increase in the immune escape of B-cell lymphomas through downregulation of the inhibitor effect of miR-34a on PD-L1 expression [42]. Here, we found that compared with the control group in a co-culture system of OCI-Ly3 cells and T cells, knockdown of PD-L1 significantly decreases the malignant behavior of OCI-Ly3 cells, increases the levels of IFN-γ and TNF-α, and decreases the level of IL-10.
In this study, we determined that overexpression of miR-214 could suppress the progression of DLBCL by targeting PD-L1 in vitro and in vivo. In addition, miR-214 targets PD-L1 to regulate the immune response of DLBCL by modulating the expressions of IL-10, IFN-γ and TNF-α. We hope these results will point the way to new molecular targets for the treatment of DLBCL and new biomarkers for its diagnosis and prognosis.
Acknowledgements
Not applicable.
Ethics approval and consent to participate (2,019,001 + 2018.05.20)
This study was approved by the Ethical Committee of Liaocheng People’s Hospital and complied with the guidelines and principles of the Declaration of Helsinki. All participants signed written informed consent. The nude mouse model was approved by the Ethical Committee of Liaocheng People’s Hospital.
Abbreviations
- CCK-8
Cell Counting Kit-8
- DLBCL
diffuse large B-cell lymphoma
- ELISA
enzyme-linked immune sorbent assay
- miR-214
microRNA-214
- MUT
mutant type
- PD-L1
programmed death ligand-1
- UTR
untranslated region
- WT
wild type
Authors’ contributions
ZY and SJR designed the study. ZY analyzed the data and reviewed the manuscript. SJR and ZX conducted the experiments. SJR wrote the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The datasets and material used and/or analyzed during this study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhao H, Tang DZ, Zhu J, Zhang X, Tang YD, Li ST. Diffuse large B cell lymphoma presented as trigeminal neuralgia: 2 cases reported and literature review. World Neurosurg. 2019;123:383–389. doi: 10.1016/j.wneu.2018.11.217. [DOI] [PubMed] [Google Scholar]
- 2.Moccia AA, Thieblemont C. Curing diffuse large B-cell lymphomas in elderly patients. Eur J Intern Med. 2018;58:14–21. doi: 10.1016/j.ejim.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Medeiros LJ, Young KH. Cancer immunotherapy in diffuse large B-cell lymphoma. Front Oncol. 2018;8:351. doi: 10.3389/fonc.2018.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larrabeiti-Etxebarria Ane, Lopez-Santillan Maria, Santos-Zorrozua Borja, Lopez-Lopez Elixabet, Garcia-Orad Africa. Systematic Review of the Potential of MicroRNAs in Diffuse Large B Cell Lymphoma. Cancers. 2019;11(2):144. doi: 10.3390/cancers11020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jardin F, Figeac M. MicroRNAs in lymphoma, from diagnosis to targeted therapy. Curr Opin Oncol. 2013;25(5):480–486. doi: 10.1097/CCO.0b013e328363def2. [DOI] [PubMed] [Google Scholar]
- 6.Sun R, Liu Z, Han L, Yang Y, Wu F, Jiang Q, Zhang H, Ma R, Miao J, He K, Wang X, Zhou D, Huang C. miR-22 and miR-214 targeting BCL9L inhibit proliferation, metastasis, and epithelial-mesenchymal transition by down-regulating Wnt signaling in colon cancer. FASEB J. 2019;33(4):5411–5424. doi: 10.1096/fj.201801798RR. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, Tian Y, Li F, Zhao Z, Jiang X, Zhai C, Han X, Zhang L. Tumor-suppressing roles of miR-214 and miR-218 in breast cancer. Oncol Rep. 2016;35(6):3178–3184. doi: 10.3892/or.2016.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Zhou H, Ma L, Hou Y, Pan J, Sun C, Yang Y, Zhang J. MiR-214 suppressed ovarian cancer and negatively regulated semaphorin 4D. Tumour Biol. 2016;37(6):8239–8248. doi: 10.1007/s13277-015-4708-0. [DOI] [PubMed] [Google Scholar]
- 9.Dandan W, Jianliang C, Haiyan H, Hang M, Xuedong L. Long noncoding RNA MIR31HG is activated by SP1 and promotes cell migration and invasion by sponging miR-214 in NSCLC. Gene. 2019;692:223–230. doi: 10.1016/j.gene.2018.12.077. [DOI] [PubMed] [Google Scholar]
- 10.Wang R, Sun Y, Yu W, Yan Y, Qiao M, Jiang R, Guan W, Wang L. Downregulation of miRNA-214 in cancer-associated fibroblasts contributes to migration and invasion of gastric cancer cells through targeting FGF9 and inducing EMT. J Exp Clin Cancer Res. 2019;38(1):20. doi: 10.1186/s13046-018-0995-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer L, Hummel M, Korfel A, Lenze D, Joehrens K, Thiel E. Differential micro-RNA expression in primary CNS and nodal diffuse large B-cell lymphomas. Neuro-Oncology. 2011;13(10):1090–1098. doi: 10.1093/neuonc/nor107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim EL, Trinh DL, Scott DW, Chu A, Krzywinski M, Zhao Y, Robertson AG, Mungall AJ, Schein J, Boyle M, Mottok A, Ennishi D, Johnson NA, Steidl C, Connors JM, Morin RD, Gascoyne RD, Marra MA. Comprehensive miRNA sequence analysis reveals survival differences in diffuse large B-cell lymphoma patients. Genome Biol. 2015;16:18. doi: 10.1186/s13059-014-0568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signaling in cancer. Ann Oncol. 2016;27(3):409–416. doi: 10.1093/annonc/mdv615. [DOI] [PubMed] [Google Scholar]
- 14.Song Moo-Kon, Park Byeong-Bae, Uhm Jieun. Understanding Immune Evasion and Therapeutic Targeting Associated with PD-1/PD-L1 Pathway in Diffuse Large B-cell Lymphoma. International Journal of Molecular Sciences. 2019;20(6):1326. doi: 10.3390/ijms20061326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miao Y, Medeiros LJ, Xu-Monette ZY, Li J, Young KH. Dysregulation of cell survival in diffuse large B cell lymphoma: mechanisms and therapeutic targets. Front Oncol. 2019;9:107. doi: 10.3389/fonc.2019.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longo Vito, Brunetti Oronzo, Azzariti Amalia, Galetta Domenico, Nardulli Patrizia, Leonetti Francesco, Silvestris Nicola. Strategies to Improve Cancer Immune Checkpoint Inhibitors Efficacy, Other Than Abscopal Effect: A Systematic Review. Cancers. 2019;11(4):539. doi: 10.3390/cancers11040539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darragh LB, Oweida AJ, Karam SD. Overcoming resistance to combination radiation-immunotherapy: a focus on contributing pathways within the tumor microenvironment. Front Immunol. 2018;9:3154. doi: 10.3389/fimmu.2018.03154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X, Dong L, Wang K, Zou H, Zhao S, Wang Y, Wang G. MiR-21 participates in the PD-1/PD-L1 pathway-mediated imbalance of Th17/Treg cells in patients after gastric cancer resection. Ann Surg Oncol. 2019;26(3):884–893. doi: 10.1245/s10434-018-07117-6. [DOI] [PubMed] [Google Scholar]
- 19.Mastroianni J, Stickel N, Andrlova H, Hanke K, Melchinger W, Duquesne S, Schmidt D, Falk M, Andrieux G, Pfeifer D, Dierbach H, Schmitt-Graeff A, Meiss F, Boerries M, Zeiser R. miR-146a controls immune response in the melanoma microenvironment. Cancer Res. 2019;79(1):183–195. doi: 10.1158/0008-5472.CAN-18-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Qing-Ming, Lian Guang-Yu, Song Yuan, Huang Yan-Fang, Gong Yi. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sciences. 2019;231:116335. doi: 10.1016/j.lfs.2019.03.040. [DOI] [PubMed] [Google Scholar]
- 21.He B, Yan F, Wu C. Overexpressed miR-195 attenuated immune escape of diffuse large B-cell lymphoma by targeting PD-L1. Biomed Pharmacother. 2018;98:95–101. doi: 10.1016/j.biopha.2017.11.146. [DOI] [PubMed] [Google Scholar]
- 22.Han M, Gao H, Xie J, Yuan YP, Yuan Q, Gao MQ, Liu KL, Chen XH, Han YT, Han ZW. Hispidulin induces ER stress-mediated apoptosis in human hepatocellular carcinoma cells in vitro and in vivo by activating AMPK signaling pathway. Acta Pharmacol Sin. 2019;40:666–676. doi: 10.1038/s41401-018-0159-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Sharma T, Hamilton R, Mandal CC. miR-214: a potential biomarker and therapeutic for different cancers. Future Oncol. 2015;11(2):349–363. doi: 10.2217/fon.14.193. [DOI] [PubMed] [Google Scholar]
- 24.Penna E, Orso F, Taverna D. miR-214 as a key hub that controls cancer networks: small player, multiple functions. J Invest Dermatol. 2015;135(4):960–969. doi: 10.1038/jid.2014.479. [DOI] [PubMed] [Google Scholar]
- 25.Tian C, Wu H, Li C, Tian X, Sun Y, Liu E, Liao X, Song W. Downregulation of FoxM1 by miR-214 inhibits proliferation and migration in hepatocellular carcinoma. Gene Ther. 2018;25(4):312–319. doi: 10.1038/s41434-018-0029-4. [DOI] [PubMed] [Google Scholar]
- 26.Liu F, Lou K, Zhao X, Zhang J, Chen W, Qian Y, Zhao Y, Zhu Y, Zhang Y. miR-214 regulates papillary thyroid carcinoma cell proliferation and metastasis by targeting PSMD10. Int J Mol Med. 2018;42(6):3027–3036. doi: 10.3892/ijmm.2018.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Z, Wang X, Huang C, Gao Y, Yang C, Zeng P, Chen Z. The FENDRR/miR-214-3P/TET2 axis affects cell malignant activity via RASSF1A methylation in gastric cancer. Am J Transl Res. 2018;10(10):3211–3223. [PMC free article] [PubMed] [Google Scholar]
- 28.He Z, Liao Z, Chen S, Li B, Yu Z, Luo G, Yang L, Zeng C, Li Y. Downregulated miR-17, miR-29c, miR-92a and miR-214 may be related to BCL11B overexpression in T cell acute lymphoblastic leukemia. Asia Pac J Clin Oncol. 2018;14(5):e259–e265. doi: 10.1111/ajco.12979. [DOI] [PubMed] [Google Scholar]
- 29.Dettori D, Orso F, Penna E, Baruffaldi D, Brundu S, Maione F, Turco E, Giraudo E, Taverna D. Therapeutic silencing of miR-214 inhibits tumor progression in multiple mouse models. Mol Ther. 2018;26(8):2008–2018. doi: 10.1016/j.ymthe.2018.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu JL, He GY, Lan XL, Zeng ZC, Guan J, Ding Y, Qian XL, Liao WT, Ding YQ, Liang L. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis. 2018;7(2):16. doi: 10.1038/s41389-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Su B, Gong C, Xi Q, Chao T. miR-214 promotes apoptosis and sensitizes breast cancer cells to doxorubicin by targeting the RFWD2-p53 cascade. Biochem Biophys Res Commun. 2016;478(1):337–342. doi: 10.1016/j.bbrc.2016.07.054. [DOI] [PubMed] [Google Scholar]
- 32.Kammerer-Jacquet Solène-Florence, Deleuze Antoine, Saout Judikaël, Mathieu Romain, Laguerre Brigitte, Verhoest Gregory, Dugay Frédéric, Belaud-Rotureau Marc-Antoine, Bensalah Karim, Rioux-Leclercq Nathalie. Targeting the PD-1/PD-L1 Pathway in Renal Cell Carcinoma. International Journal of Molecular Sciences. 2019;20(7):1692. doi: 10.3390/ijms20071692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenouvel D, Gonzalez-Moles MA, Talbaoui A, Ramos-Garcia P, Gonzalez-Ruiz L, Ruiz-Avila I, Gil-Montoya JA. An update of knowledge on PD-L1 in head and neck cancers: physiologic, prognostic and therapeutic perspectives. Oral Dis. 2019:1–16. [DOI] [PubMed]
- 34.Chen X, Song X, Li K, Zhang T. Fcgammar-binding is an important functional attribute for immune checkpoint antibodies in cancer immunotherapy. Front Immunol. 2019;10:292. doi: 10.3389/fimmu.2019.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozpiskin OM, Zhang L, Li JJ. Immune targets in the tumor microenvironment treated by radiotherapy. Theranostics. 2019;9(5):1215–1231. doi: 10.7150/thno.32648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maleki Vareki S, Garrigos C, Duran I. Biomarkers of response to PD-1/PD-L1 inhibition. Crit Rev Oncol Hematol. 2017;116:116–124. doi: 10.1016/j.critrevonc.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Bocanegra A, Fernandez-Hinojal G, Zuazo-Ibarra M, Arasanz H, Garcia-Granda MJ, Hernandez C, Ibanez M, Hernandez-Marin B, Martinez-Aguillo M, Lecumberri MJ, Fernandez de Lascoiti A, Teijeira L, Morilla I, Vera R, Escors D, Kochan G. PD-L1 expression in systemic immune cell populations as a potential predictive biomarker of responses to PD-L1/PD-1 blockade therapy in lung cancer. Int J Mol Sci. 2019;20(7):1631. doi: 10.3390/ijms20071631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn S, Kim TH, Kim SW, Ki CS, Jang HW, Kim JS, Kim JH, Choe JH, Shin JH, Hahn SY, Oh YL, Chung JH. Comprehensive screening for PD-L1 expression in thyroid cancer. Endocr Relat Cancer. 2017;24(2):97–106. doi: 10.1530/ERC-16-0421. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen JH, Lelkaitis G, Hakansson K, Vogelius IR, Johannesen HH, Fischer BM, Bentzen SM, Specht L, Kristensen CA, von Buchwald C, Wessel I, Friborg J. Intratumor heterogeneity of PD-L1 expression in head and neck squamous cell carcinoma. Br J Cancer. 2019;120(10):1003. doi: 10.1038/s41416-019-0449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashizawa M, Okayama H, Ishigame T, Thar Min AK, Saito K, Ujiie D, Murakami Y, Kikuchi T, Nakayama Y, Noda M, Tada T, Endo H, Fujita S, Sakamoto W, Saito M, Saze Z, Momma T, Ohki S, Mimura K, Kono K. miRNA-148a-3p regulates immunosuppression in DNA mismatch repair-deficient colorectal cancer by targeting PD-L1. Mol Cancer Res. 2019;17(6):1403–13. [DOI] [PubMed]
- 41.Fan Y, Che X, Hou K, Zhang M, Wen T, Qu X, Liu Y. MiR-940 promotes the proliferation and migration of gastric cancer cells through up-regulation of programmed death ligand-1 expression. Exp Cell Res. 2018;373(1–2):180–187. doi: 10.1016/j.yexcr.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 42.Anastasiadou E, Stroopinsky D, Alimperti S, Jiao AL, Pyzer AR, Cippitelli C, Pepe G, Severa M, Rosenblatt J, Etna MP, Rieger S, Kempkes B, Coccia EM, Sui SJH, Chen CS, Uccini S, Avigan D, Faggioni A, Trivedi P, Slack FJ. Epstein-Barr virus-encoded EBNA2 alters immune checkpoint PD-L1 expression by downregulating miR-34a in B-cell lymphomas. Leukemia. 2019;33(1):132–147. doi: 10.1038/s41375-018-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets and material used and/or analyzed during this study are available from the corresponding author on reasonable request.