Abstract
Background
This study was designed to investigate whether household cockroaches harbor cephalosporin-resistant enterobacteria that share resistance determinants with human inhabitants. From February through July 2016, whole cockroach homogenates and human fecal samples from 100 households were cultured for cephalosporin-resistant enterobacteria (CRe). The CRe were examined for plasmid-mediated AmpC, ESBL, and carbapenemase genes; antibiotic susceptibility patterns; and conjugative transfer of antibiotic resistance mechanisms. Clonal associations between CRe were determined by multi-locus sequence typing (MLST).
Results
Twenty CRe were recovered from whole cockroach homogenates from 15 households. The prevalence of households with cockroaches that harbored CRe, AmpC- (based on phenotype, with no identifiable blaAmpC genes), ESBL-, and carbapenemase-producers were 15, 4, 5%(2 blaCTX-M-15/TEM-1; 1 blaCTX-M-15/TEM-4; 1 blaTEM-24; 1 blaSHV-4) and 3%(2 blaNDM-1 genes and 1 blaOXA-48 gene), respectively. Overall, 20 CRe were recovered from 61 fecal samples of inhabitants from all 15 households that had cockroach samples positive for CRe. Of these, 5CRe (1 per household) were positive for ESBLs (blaTEM-24, blaTEM-14, blaCTX-M-15/TEM-4, blaSHV-3, blaCTX-M-15/TEM-1) and none carried AmpCs or carbapenemases. From 4% of households, the pair of cockroach and human CRe shared the same sequence type (ST), clonal complex (CC), antibiogram, and conjugable bla gene sequence (house 34, E. coli ST9/CC20-blaTEM-4; house 37, E. coli ST44/CC10-blaCTX-15/TEM-4; house 41, E. coli ST443/CC205-blaCTX-15/TEM-1; house 49, K. pneumoniae ST231/CC131-blaSHV-13).
Conclusion
The findings provide evidence that household cockroaches may carry CTX-M-15-, OXA-48- and NDM-1-producers, and share clonal relationship and beta-lactam resistance determinants with humans.
Keywords: Cockroach, ESBL, Carbapenemase, Multilocus sequence typing, Conjugation
Background
Production of extended-spectrum beta-lactamases (ESBL), Class C cephalosporinases and carbapenemases constitutes the primary antibiotic resistance mechanism in Enterobacteriaceae [1, 2]. Together, these beta-lactamases confer resistance to all available beta-lactam antibiotics, and are associated with significantly high levels of co-resistance to other classes of broad spectrum antimicrobials [1–6]. In several regions in Africa, the CTX-M-15 type ESBLs are becoming increasingly predominant [7, 8]. Occurrence of the recently described OXA-48-type carbapenemase, and widespread reports of blaNDM-1 genes across Africa, further compound the outlook of the antibiotic resistance problem [9–13]. Indeed, emergence and spread of such resistance determinants in bacteria is often related to abuse of beta-lactam antibiotics [14, 15]. However, distribution and persistence of the resistant pathogens may be hugely aided by poor sanitation [16–18]. Cockroaches, which widely colonize the environment, including human dwellings, may well act as vehicles for antibiotic-resistant bacteria.
Cockroaches are common in many households and are known to harbor an array of pathogens, some of which may be carriers of drug-resistance determinants including beta-lactamases [19–23].They often reside in household sewage pipe systems, which are a repository of diverse infectious microorganisms. However, only few studies have investigated AmpC, ESBL or carbapenemase resistance elements in bacteria from cockroaches [24]. Consequently, the vector potential of cockroaches in the dissemination of beta-lactamase-related drug resistance mechanisms is largely under-reported. This study was designed to determine whether cephalosporin resistant enterobacteria recovered from cockroaches in a household were identical to those that colonized human inhabitants from the same household □ with particular focus on clonal relationships, beta-lactam-resistance mechanisms, conjugability of resistance determinants and antibiogram.
Results
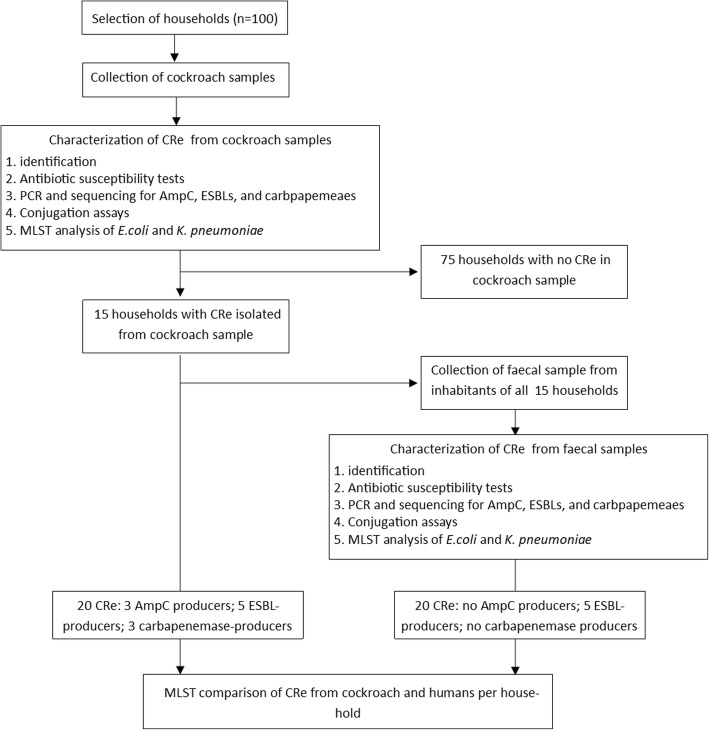
A flowchart of the study outcomes in shown in Fig. 1. All 100 insect homogenates yielded polymicrobial cultures after inoculation on SSI agar with 30 μg cefpodoxime disks. Cockroach samples from 15 households had homogenate culturesscreen positive for CRe. Ten of the samples grew one dominating CRe colony type and 5 had two morphologicaly distinct dominant CRe colonies with different antibiogram □ corresponding to 20 screen positive Enterobacteriaceae isolates (Table 1). The 20 isolates were each resistant to cefotaxime or ceftazidime, and were assigned a CRe phenotype. From the inhabitants of the 15 households that had cockroach samples positive for CRe, a total of 61 fecal samples were collected. The average number of inhabitants per household was 4 ± 1.3. Twenty (32.8%) of the 61 faecal samples were screen positive for CRe on SSI agar plate. The 20 fecal cultures each yielded only one dominant CRe colony type □ corresponding to 20 screen positive CRe (1 isolate per faecal sample) with different antibiogram. All 20 screen-positive CRe were resistant to cefotaxime or ceftazidime, and were also assigned a CRe phenotype. Ten and 5 households respectively had 1 and 2 inhabitants fecally colonized by CRe (Table 2).
Fig. 1.
Summary of study protocols and outcomes
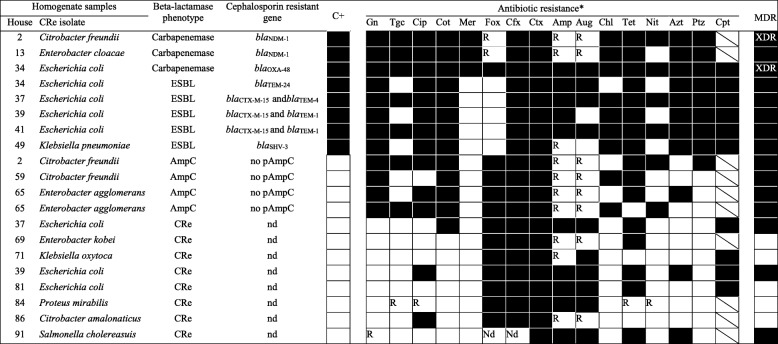
Table 1.
Antibiogram of CRe recovered from whole insect homogenates
*As per column headings, dark cells indicate “yes”; white cells indicate “no” C+, successfully transferred the beta-lactamase encoding genotype to J53 E.coli recipient via conjugation;CRerefers to cephalosporin resistant Enterobacteriaceae with no detectable phenotype for AmpC, ESBL or carbapenemases; Gn, gentamicin; Tgc, tigecycline; Cip, ciprofloxacin; Cot, cotrimoxazole; Mer, meropem; Fox, cefoxitin; Cfx, cefuroxime; Ctx, cefotaxime; Amp, ampicillin; Aug, aumentin; Chl, chloramphenicol; Tet, tetracycline; Nit, nitrofurantoin; Azt, aztreonam; Aug, amoxicillin/clavulanic acid, Ptz, piperacillin/tazobactam; Cpt, ceftaroline (approved for only E.coli, K. pneumoniae, K oxytoca. For cells with diagonal lines, Cpt is not approved for testing organism); pAmpC, plasmid mediated AmpC gene; MDR, multidrug resistanct; XDR, extensively drug resistant; R, not reported (test organisms intrinsically resistant to antibiotic); nd, no detectable plasmid AmpC, ESBL or carbapenemase gene
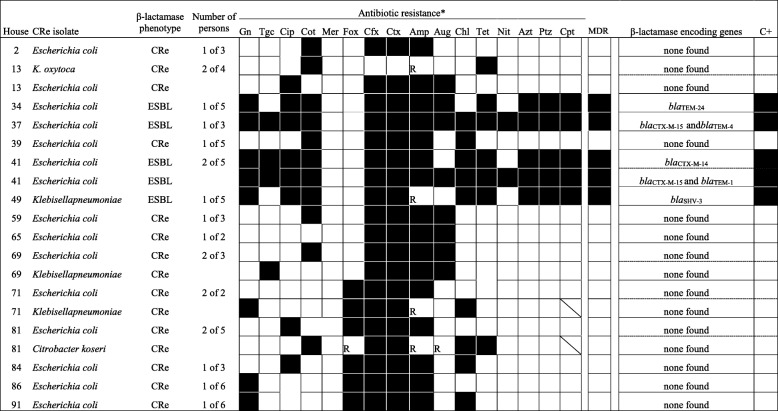
Table 2.
Antibiogram of CRe recovered from human inhabitants in households with cockroach CRe
*As per column headings, dark cells indicate “yes”; white cells indicate “no” C+, successfully transferred the beta-lactamase encoding genotype to J53 E.coli recipient via conjugation; CRe refers to cephalosporin resistant Enterobacteriaceae with no detectable phenotype for AmpC, ESBL or carbapenemases; Gn, gentamicin; Tgc, tigecycline; Cip, ciprofloxacin; Cot, cotrimoxazole; Mer, meropem; Fox, cefoxitin; Cfx, cefuroxime; Ctx, cefotaxime; Amp, ampicillin; Aug, aumentin; Chl, chloramphenicol; Tet, tetracycline; Nit, nitrofurantoin; Azt, aztreonam; Aug, amoxicillin/clavulanic acid, Ptz, piperacillin/tazobactam; Cpt, ceftaroline (approved for only E.coli, K. pneumoniae, K oxytoca. For cells with diagonal lines, Cpt is not approved for testing organism); pAmpC, plasmid mediated AmpC gene; MDR, multidrug resistanct; XDR, extensively drug resistant; R, not reported (test organisms intrinsically resistant to antibiotic); nd, no detectable plasmid AmpC, ESBL or carbapenemase gene
Characterization of CRe
Cockroach samples
Of the 20 CRe, 12 isolates (from 10 samples) expressed ESBL (n = 5), AmpC (n = 4) or were resistant to meropenem (n = 3) (Table 1). The AmpC-producers were Enterobacter freundii (n = 2) and Enterobacter agglomerans (n = 2). Isolates with ESBL phenotype were 4 E. coli and 1 K. pneumoniae. Two of the 3 meropenem resistant isolates (E. freundii and C. cloacae) were found to be class B metallo-beta-lactamase (MBL)-producers. The third meropenem resistant isolate (E. coli) was non-susceptible to temocillin (30 μg), and was deemed presumptively positive for OXA-48 like carbapenemase (Table 1). None of the CRe isolates was positive for any combination of the three phenotypes. Of the 5 ESBL-producers, PCR and nucleotide sequencing identified blaCTX-M-15/TEM-1 in 2 E. coli isolates (Table 1). The remaining 3 harbored either blaCTX-M-15/TEM-4, blaTEM-24 or blaSHV-3 ESBL genes. None of the AmpC-producers had an identifiable plasmid-mediated AmpC gene. The 2 MBL-positive isolates each carried blaNDM-1 gene. The meropenem resistant E. coli with non-susceptibility to temocillin carried blaOXA-48 gene. Thus, the overall prevalence of households with cockroaches that harbored CRe, ESBL-, AmpC- and carbapenemase-producers were respectively 15, 5, 4 and 3%. Antibiogram of the 20 CRe revealed differences in resistance patterns between AmpC-, ESBL-, or carbapenemase-positive isolates and those 8 CRe negative for any of the 3 phenotypes (Table 1). Multi drug-resistant (MDR) phenotype was indicated in all isolates with AmpC, ESBL or carbapenemase phenotype. Two of the 3 carbapenemase-positive CRe were XDR. Amongst the 8 CRe negative for the three phenotypes, 3 showed MDR phenotype.
Human faecal samples
Of the 20 CRe, none was positive for AmpC or carbapenemase phenotype. Five CRe (1 per household) expressed ESBLs (4 E. coli and 1 K. pneumoniae). When the ESBL-producing faecal isolates were subjected to PCR and sequencing, the K. pneumoniae carried blaSHV-3. The 4 E. coli separately harbored blaTEM-24, blaTEM-14, blaCTX-M-15/TEM-4, and blaCTX-M-15/TEM-1. The antibiotic susceptibility profile of all 20 human CRe mirrored that observed for cockroach isolates □ with obvious differences in resistance pattern between ESBL- and non-ESBL producers (Table 2).
Conjugation assays
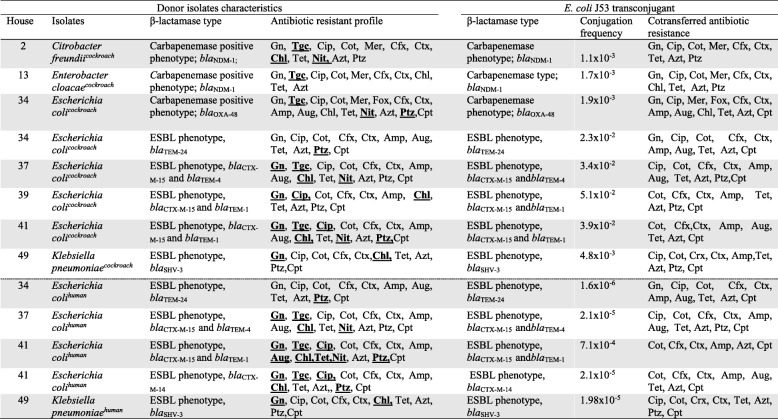
All CRe from cockroach and human samples were subjected to conjugation experiments. Successful conjugation events were demonstrated only in ESBL- or carbapenemase-producing isolates (Table 3). For these isolates, PCR amplification and sequencing of the ESBL or carbapenemase genes from the E. coli J53 transconjugants showed the same bla gene types previously identified in the donor isolates. The ESBL genes in human isolates transferred at slower conjugation frequencies (range: 1.1 × 10− 5-1.9 × 10− 4) compared to those from cockroach isolates (range: 2.3 × 10− 3-4.8 × 10− 2). For cockroach isolates, carbapenemase genes appeared to conjugate with lower frequencies (range: 1.1 × 10− 3-1.9 × 10− 3) compared to the ESBL genes (range: 2.3 × 10− 3-4.8 × 10− 2). Resistance to non-β-lactam antimicrobials was co-transferred in some cases (Table 3). The most frequently co-transferred phenotypes were resistance to cotrimoxazole, tetracycline and ciprofloxacin. Tigecycline and nitrofurantoin resistance did not transfer to recipients despite repeated attempts. All CRe isolates negative for ESBL or carbapenemase could not transfer their cephalosporin resistance to E. colij53 recipients. Similarly, none of the AmpC-producing isolates transferred the phenotype. In both cases, there was no further susceptibility testing to other antibiotics after the CRe phenotype was found to be absent in recipients.
Table 3.
Conjugation characteristics of ESBL- and carbapenemase-producing CRe isolated from cockroaches and humans
*Highlighted antibiotics were not cotransferred to transconjugants; Gn, gentamicin; Tgc, tigecycline; Cip, ciprofloxacin; Cot, cotrimoxazole; Mer, meropem; Fox, cefoxitin; Cfx, cefuroxime;Ctx, cefotaxime; Amp, ampicillin; Aug, aumentin; Chl, chloramphenicol; Tet, tetracycline; Nit, nitrofurantoin; Azt, aztreonam; Aug, amoxicillin/clavulanic acid, Ptz, piperacillin/tazobactam, Cpt, ceftaroline (approved for only E.coli, K. pneumoniae, K oxytoca)
Phylogenetic analysis
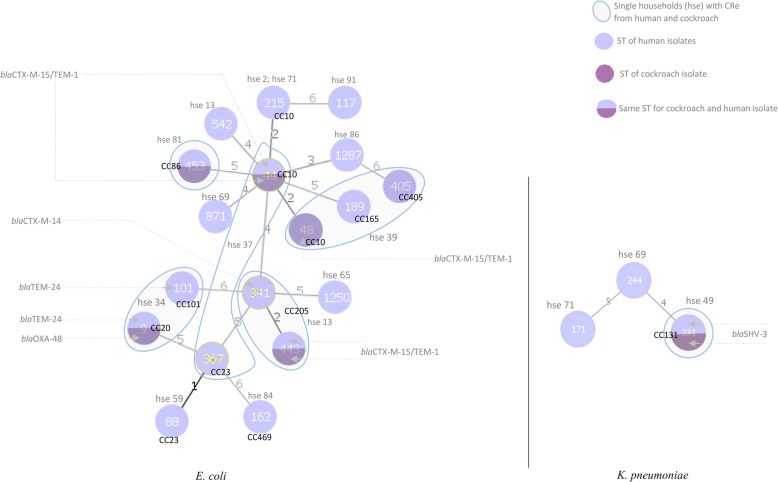
Our focus was to determine whether CRe cultured from household cockroaches were identical to those which colonized human inhabitants. There were 20 cockroach CRe belonging to 15 households with 20 human CRe. The MLST was considered if CRe from cockroach and human samples per household belonged to the same bacterial species, or were E. coli and K. pneumoniae. Eighteen human CRe (15 E. coli and 3 K. pneumoniae) and 9 cockroach CRe (8 E. coli and 1 K. pneumoniae) from the 15 households were thus included. Of the 15 households, 6 had human and cockroach CRe of the same bacterial species (E. coli or K. pneumoniae) (Table 4). From 4 of the 6 households, the pair of human and cockroach CRe shared the same ST and blaESBL gene sequence (house 34, E. coli ST9-blaTEM-4; house 37, E. coli ST44-blaCTX-15/TEM-4; house 41, E. coli ST443-blaCTX-15/TEM-1; house 49, K. pneumoniae ST231-blaSHV-13). The pairs also had the same antibiogram; and successfully transferred their ESBL phenotype and genotype by conjugation to E. coli J53 recipients (Table 3). In 1 of the 6 households (house 81), the pair of human and cockroach CRe belonged to the same E. coli ST453but differed in antibiogram and had no identifiable ESBL, AmpC or carbapenemase genes. The pair did not transfer their cephalosporin resistance phenotypes by conjugation to E. coli J53 recipients. In another of the 6 households (house 47), a human inhabitant was colonized by E. coli ST341-blaCTX-M-14 which was different from the corresponding cockroach CRe (E. coli ST443 with blaCTX-15/TEM-1 ESBL gene). Both isolates however belonged to the same clonal complex CC205. Generally, we observed low intra-species similarity regardless of beta-lactamase gene sequences (Fig. 2). Four E. coli STs, belonging to different clonal complexes, were found in only cockroach isolates (ST48/CC10, ST101/CC101, ST367/CC23, ST405/CC405). Similarly, 5 E. coli singleton STs (ST117, ST542, ST871, ST1250, ST1287) and 5 E. coli STs with associated clonal complexes (ST88/CC23, ST162/CC469, ST189/CC165, ST215/CC10, ST341/CC205) and were found in human isolates only. The E. coli ST215/CC10 was identified in 2 human isolates from separate households. The following STs with associated clonal complexes were detected in both human and cockroach isolates: E. coli; ST9/CC20, ST44/CC10, ST443/CC205, ST453/CC86 and K. pneumonia; ST231/CC86). The K. pneumoniae STs identified from only human CRe were ST171 and ST244 singletons.
Table 4.
MLST analysis of CRe from cockroach and human inhabitants per household
| Household code | Isolates | MLST | ||
|---|---|---|---|---|
| ST | CC | |||
| 2 | E. coli human | 215 | 10 | |
| 2 | C. freundiicockroach | – | ||
| 2 | C. freundii cockroach | – | ||
| 13 | K. oxytoca human | – | ||
| 13 | E. coli human | 542 | singleton | |
| 13 | E. cloacae cockroach | – | ||
| 34 | E. coli human | 9 | 20 | |
| 34 | E. coli cockroach | 9 | ||
| 34 | E. coli cockroach | 101 | 101 | |
| 37 | E. coli human | 44 | 10 | |
| 37 | E. coli cockroach | 44 | ||
| 37 | E. coli cockroach | 367 | 23 | |
| 39 | E. coli human | 189 | 165 | |
| 39 | E. coli cockroach | 48 | 10 | |
| 39 | E. coli cockroach | 405 | 405 | |
| 41 | E. coli human | 341 | 205 | |
| 41 | E. coli human | 443 | ||
| 41 | E. coli cockroach | 443 | ||
| 49 | K. pneumoniaehuman | 231 | 131 | |
| 49 | K. pneumoniae cockroach | 231 | ||
| 59 | E. coli human | 88 | 23 | |
| 59 | C. freundii cockroach | – | ||
| 65 | E. coli human | 1250 | singleton | |
| 65 | E. agglomeranscockroach | – | ||
| 65 | E. agglomeranscockoach. | – | ||
| 69 | E. coli human | 871 | singleton | |
| 69 | K. pneumoniae human | 244 | singleton | |
| 69 | E. kobei cockroach | – | ||
| 71 | E. coli human | 215 | 10 | |
| 71 | K. pneumoniae human | 171 | singleton | |
| 71 | K. oxytoca cockroach | – | ||
| 81 | E. coli human | 453 | 86 | |
| 81 | C. koseri human | – | ||
| 81 | E. coli cockroach | 453 | 86 | |
| 84 | E. coli human | 162 | 469 | |
| 84 | P. mirabilis cockroach | – | ||
| 86 | E. coli human | 1287 | singleton | |
| 86 | C. amalonaticus cockroach | – | ||
| 91 | E. coli human | 117 | singleton | |
| 91 | S. cholaeraesuis cockroach | – | ||
*C+, conjugation tests; dark cells within the C+ column show that the isolate successfully transferred their β-lactamase phenotype and genotype to J53 E.coli recipient via conjugation; white cells within the C+ column indicate unsuccessful conjugation of β-lactamase phenotype and genotype to J53 E.coli recipient via conjugation; MLST, multilocus sequence typing; ST, Sequence Type; CC, clonal complex
Fig. 2.
Minimum spanning tree based on MLST allelic profiles of cephalosporin resistant E. coli and K. pneumoniaefrom human and cockroach samples. Each circle represents an identified MLST sequence type (ST). The circle in red (lower half) represents an ST identified in the cockroach CRe. The circle in blue (the upper half) represents an ST identified in the human CRe. The numbers on the connecting lines illustrate the number of differing alleles. The clonal complexes (CC) if present are indicated for the STs. The pAmpC, ESBL or carbapenemase genes if present are indicated for the CRe from each household
Discussion
This study investigated whether within individual households, cockroaches and humans may share bacterial isolates of the same clone, and also antibiotic resistance determinants of public health concern. Generally, interpretation of this data is driven by findings from other reports that have indicated direct transfer of ESBL-producing bacteria to humans through close contact with animal sources [25–27]. A number of reports implicate insects, including cockroaches, in the carriage and spread of antibiotic resistant bacteria [28–32]. This is the first published description of blaNDM-1 and blaOXA-48 carbapenemases in enterobacteria recovered from household cockroaches.
Four observations from this study merit special attention. First, evidence is presented to demonstrate that household cockroaches may carry drug-resistant isolates, including multidrug-resistant blaCTX-M-15-, extensively drug-resistant blaOXA-48- and extensively drug-resistant blaNDM-1-producing enterobacteria. In human dwellings, cockroaches may move about freely but are usually found breeding in bathrooms, toilets and cupboards for storing food [28, 33]. Outside the home, they are commonly observed in drains and around damaged septic tanks. Their preferred living quarters, coupled with the fact that they are omnivorous, brings them in frequent contact with stored foods as well as sewage and different kinds of biological wastes [28, 34]. The isolation of these bacteria in household cockroaches is alarming and of public health concern, given that the strains could very quickly disseminate and initiate a pandemic spread of clones for which effective antibiotics may not be readily available. The ease of transmission of blaNDM-1 genes has already been described [2, 35–37]. Interestingly, although the identified blaNDM-1 and blaOXA-48 genes were conjugable, no household inhabitant was fecally colonized with a carbapenem resistant isolate. In Ghana, there are no published data on carbapenemases, but it is the experience that carbapenem resistance is low [6, 38, 39].
Second, about 4% of households had cockroach and human CRe that shared the same sequence type and clonal complex, antibiogram, blaESBL gene sequence, and successfully transferred their ESBL phenotypes and genotypes by conjugation. These findings suggest insect-mediated transmission by clonal spread. The presence of cephalosporin-resistant Salmonella choleraesuis in household cockroach, which is a pig typhoid strain, is a strong indication of the extent to which cockroaches could carry infectious contamination from diverse sources to humans. Cockroaches are known to be a common pest of confined pigs [40, 41] and have been shown to acquire and harbour bacterial pathogens from pigs [42]. Acquisition of bacterial pathogens by insects, from farm or domestic animals, is documented [43–45]. In the study site, as in many neighborhoods in rural and urban Africa, domestic and/or farm animals, including dogs, chicken and goats, may roam freely. The relatively high fecundity of cockroaches, coupled with the fact that mature insects may live for up to 2 years or more in association with humans [46], highlight household cockroaches as constituting a formidable ‘revolving door’ through which ESBL-producing enterobacteria may disseminate to human contacts.
Third, the successful inter-genus conjugation events reported in this study suggest that, at least for the non-E. coli isolates used as donors, the plasmids involved are likely to be conjugative plasmids with broad host range. This hints at the potential for these resistance determinants to spread freely across bacterial species in the environment, including the possibility of rescue of susceptible bacteria during antibacterial stress [47].
Last, we did not detect AmpCs, ESBLs, or carbapenemases in 8 of 12 cockroach CRe and 15 of 20 human CRe with resistance phenotypes to 3rd-generation cephalosporins. The results show a clear separation, in which the level of resistance to various antibiotics was higher among strains expressing ESBL, AmpC or carbapenemase than those negative for the enzymes. Similarly, resistance mechanisms to several other non-beta-lactam antibiotics were cojugated in parralel with ESBL and carbapenemase genotypes. The CRe isolates without ESBL, AmpC or carbapenemase were susceptible to several antibiotics tested. None of these CRe also successfully transferred their cephalosporin resistance phenotype by conjugation. The results may suggest that this CRe cluster of isolates harbour chromosomal-borne 3rd-generation cephalosporin-resistant determinants with limited functional spectrum compared to AmpCs, ESBLs or carbapenemases [1–6]. The findings highlight from the clinical point of view that, if we can succeed at curtailing the spread of ESBL-, AmpC-, or carbapenemase producing enterobacteria, we may somewhat succeed at reducing the prevalence of antimicrobial resistance among enterobacteria.
Our data should be interpreted considering some potential limitations. We accept the possibility that the cockroach isolates reported in this study may be of transient colonization. However, it is notable that the cockroach CRe were the only dominant colonies in culture□ suggesting stable colonization rather than the consequence of sudden external contamination. The data on the complimentary sampling of humans included only inhabitants of households where CRe was cultured from cockroach samples. Due to the small sample size and provincial household concentration, our data is not meant to be representative of the human prevalence of AmpC-, ESBL-, or carbapenemase-producing bacteria in the study site or similar locations in Ghana. Suffice it to say that faecal carriage of beta-lactamase-producing enterobacteria in the Ghanaian community setting is the focus of another manuscript under consideration for publication elsewhere. Nevertheless, it is noteworthy that a profiling of antibiotic resistance patterns in Accra, Ghana indicated high incidence of cephalosporin resistance in antibiotic resistant bacteria [48]. It is the experience in Ghana that resistance to commonly used antibiotics in primary health care is high, and the prevalence is rising. Although antibiotic use is a known risk factor for the emergence of antibiotic resistance, we did not collect data on antibiotic use by study participants for this report. Information on antibiotic therapy would have better helped to delineate the faecal carriage patterns of household inhabitants with the endemic beta-lactam resistance in the community, their interrelated factors for spread, and the correlating role of household cockroaches as important reservoir of resistant genes. Notwithstanding the shortcomings, our findings highlight the importance of cockroaches as a potential reservoir of epidemiologically significant multidrug resistant pathogens of public health concern.
Conclusion
We report the alarming colonization of household cockroaches with multidrug resistant CTX-.
M-15 ESBL-producers, and extensively drug resistant NDM-1 and OXA-48 carbapenemase-positive Enterobacteriacaea. Our findings highlight cockroaches as insects of public health concern, and call for regulations on their control, especially in healthcare settings.
Methods
Study design and setting
Between February and July 2016, cockroaches and human fecal samples were collected from 100 households in Ashaley Botwe, an urban municipality in Accra, Ghana. The municipality has a population of approximately 78,215, with most households occupied by an average of 5.6 persons [49]. The major source of water is pipe borne, and most of the households have proper biological waste disposal systems with flush toilets and standardized septic tanks. Bathwater and liquid wastes from kitchens may, however, be observed running freely in open drains. Households included in the study were at least 150 m from each other. Hundred households were selected by systematic random sampling using the Kish method [50] which statistically allowed for equal chances of selecting any household in the community. From each household, live indoor cockroaches were collected. At the same time, all inhabitants per household were requested to self-collect and submit stool samples. The participants provided written, informed consent. The study received ethical approval from the Ethics and Protocol Review Committee of the School of Biomedical and Allied Health Sciences, University of Ghana, with approval identification number: SBAHS-MD./10,512,194/aa/5a/2016–2017.
Sample collection and processing
Households were provided with cockroach collection kits (sterile 100 mL containers with screw caps, sterile zipper bags, sterile gloves, sterile entomological forceps) and guidelines for collection to avoid any human contamination. A member from each household was trained to capture the cockroaches. Only live cockroaches found indoors were collected for this study. From each household, four cockroaches regardless of species were requested and pooled as one sample. Each cockroach sample was soaked in 40 mL Brain Heart Infusion broth (Sigma, UK), vortexed vigorously for approximately 5 min, and ground with a sterile rod. The triturate was again vortexed vigorously for 6 min to obtain a whole insect homogenate. A loop-full (approximately 10 μL) of suspension from each sample was inoculated onto SSI agar plate (SSI, Diagnostica, Denmark) with 30 μg cefpodoxime disks (MAST, UK) and incubated overnight at 37 °C. For stool samples, 1 g of each specimen was suspended in 10 mL of sterile 0.9% physiological saline and vigorously vortexed. 1 mL of the suspension was cultured on SSI agar plateswith 30 μg cefpodoxime disks as described above. From each culture plate, distinct morphological phenotypes of enterobacteria growing within an inhibition zone of ≤21 mm for cefpodoxime were considered as screen positive for third-generation cephalosporin resistance. Each distinct morphological phenotype was identified to the species level using biochemical test kits Minibact-E® (SSI, Diagnostica, Denmark) according to the manufacturer’s guidelines. Subsequently, four colonies of each species were tested for confirmation of CRe status by Kirby-Bauer disk diffusion using cefotaxime (30 μg) and ceftazidime (30 μg) antibiotic disks, per guidelines of the Clinical and Laboratory Standards Institute (CLSI) [51]. Isolates resistant to cefotaxime or ceftazidime were confirmed as third-generation cephalosporin resistant (CRe).
Susceptibility test and assays for ESBL, AmpC and carbapenemase
Confirmed CRe isolates were tested for susceptibility to the following antibiotic disks (MAST, UK) according to CLSI guidelines [51]: ampicillin (10 μg), augmentin (10/260 μg); meropenem (10 μg), tetracycline (30 μg), chloramphenicol (30 μg), cotrimoxazole (100/240 μg), gentamicin (10 μg), ciprofloxacin (5 μg), nitrofurantoin (100 μg); piperacillin/tazobactam (10/30 μg), cefoxitin (30 μg), ceftaroline (30 μg), and tigecycline (30 μg).Klebsiella pneumoniae ATCC 700603 and Escherichia coli ATCC 25922 were used as quality control strains. Ceftaroline breakpoint was interpreted with European Committee for Antibiotic Susceptibility Testing (EUCAST) guidelines [52]. Phenotypic detection of ESBL production was by the combination disk method with cefotaxime (30 μg) or ceftazidime (30 μg), alone or in combination with clavulanic acid (10 μg) [53]. AmpC expression was suspected in isolates with reduced susceptibility (inhibition zone < 20 mm) to cefoxitin (30 μg). AmpC confirmation was by susceptibility testing to cefotaxime (30 μg) or ceftazidime (30 μg) antibiotic disks, with or without boronic acid (250 μg) as per the manufacturer’s guidelines (Rosco Diagnostica, Taastrup, Denmark). Isolates with an increase of ≥5 mm in zone diameter, due to boronic acid, were considered AmpC positive. Isolates with inhibition zone of < 21 mm to meropenem(10 μg) were considered carbapenem resistant [53]. Carbapenem resistant isolates were confirmed for class A and B carbapenemases using boronic acid (600 μg) and EDTA (750 μg) effects, respectively, on meropenem (10 μg). Strains with boronic acid or EDTA effect of ≥5 mm increase in zone diameter were considered positive for class A or B carbapenemase phenotype, respectively. Carbapenem resistant strains that were not susceptible to temocillin (30 μg) were considered positive for class D carbapenemase. Carbapenem resistant strains were also subjected to the Modified Hodges Test [53].
Conjugation
All CRe isolates were included in the conjugation assay. Conjugations were done using sodium azide-resistant E. coli J53 as recipient [54]. None of the CRe isolates showed resistance to sodium azide. The donors were cultured in Luria-Bertani (LB) (MAST, UK) broth with cefotaxime (8 μg/mL) overnight. The recipient was also cultured overnight in LB broth but with no antibiotic. Subsequently, 1 mL aliquots of each donor and the recipient were separately transferred into fresh 10 mL LB broth and incubated for 2 h at 37 °C. For each donor, 100 μL of culture was mixed with an equal volume of the recipient and the mixture was incubated for 6 h at 37 °C. Selection for transconjugants was on MacConkey agar supplemented with sodium azide (150 mg/L) and cefotaxime (8 mg/L) or cefoxitin (32 mg/L) or meropenem (2 mg/L). Transconjugants were confirmed for AmpC, ESBL or carbapenemase phenotype as previously described.
Genotypic characterization
Gene amplification and sequencing were done for ESBL-, AmpC-, and carbapenemase-producing isolates as well as their transconjugants. For each isolate, 10 μL of pure culture on Mueller Hinton agar was suspended in 300 μL Milli-Q® water, heated for 10 min at 98 °C, and subsequently centrifuged for 5 min at 4 °C and 20,000×g. The supernatant DNA lysate was transferred into sterile 1.5 mL Eppendorf® tubes for storage at − 5 °C. See Additional file 1: Table S1 for amplification primers and conditions. For ESBLs, PCR was performed for blaTEM, blaSHV, blaCTX-M-1, blaCTX-M-2, blaCTX-M-9, blaOXA-2, blaOXA-10. Isolates with AmpC phenotypes were examined for 6 families of plasmid-mediated AmpC genes including MOX, CMY, DHA, ACC, EBC and FOX using the multiplex assay [55]. For carbapenemases, PCRs were designed for specific genes belonging to class A, B, and D carbapenemase phenotypes. A multiplex PCR assay was performed to differentiate5 genes (GES, KPC, SME, INI-NMC-A) for class A carbapenemase, and 6 genes (IMP, VIM, GIM, SPM, SIM, and NDM-1) for class B phenotypes [56]. Isolates with Class D carbapenemase phenotypes were examined for OXA-48 like genes. Additional internal primers (Additional file 1: Table S1) were used for sequencing CTX-M-1, CTX-M-9, SHV and TEM genes. Nucleotide and deduced protein sequences were compared with sequences in the NCBI database (http://www.ncbi.nlm.nih.gov/BLAST). TEM and SHV beta-lactamase sequences were compared to wild-type E. coli TEM-1 and SHV-1 at http://www.lahey.org/studies.
Phylogenetic analysis
Multilocus sequence typing (MLST) was conducted when cockroach and human CRe per household belonged to the same bacterial species, or were E. coli and K. pneumoniae. Isolates that satisfied the inclusion criteria were E. coli and Klebsiella pneumoniae. The previously described E. coli protocol [57] and the Institut Pasteur scheme for K. pneumoniae (http://bigsdb.pasteur.fr/klebsiella/) were used. Seven housekeeping genes were amplified and sequenced for each E. coli (adk, fumC, gyr, icd, mdh, purA and recA) and K. pneumoniae (gapA, infB, mdh, pgi, phoE, rpoB, tonB) isolates. Sequences of the seven house-keeping genes were analysed using the CodonCode Aligner software version 8.1 (Germany). The MLST sequence types (ST) and clonal complexes (CC) were assigned in accordance with the online platform PubMLST database (https://pubmlst.org/). Phylogenetic minimum spanning tree was constructed using the online programme PHYLOViZ [58].
Data analyses
Data were entered into a Microsoft Excel spreadsheet for analysis by proportions and percentages. Multidrug-resistant (MDR) isolates were those resistant to ≥1 agent in ≥3 antimicrobial categories (aminoglycosides, flouoroquinolones, penicillins, penicillins/β-lactamase inhibitors, antipseudomonal penicillins/ β-lactamase inhibitors, cephamycins, anti-MRSA cephalosporins, 1st and 2nd generation cephalosporins, 3rd and 4th generation cephalosporins, monobactams, carbapenems, polymixins, phosphonic acids, folate-pathway inhibitors, glycylcyclines, phenicols). Extensively drug-resistant (XDR) isolates were non-susceptible to ≥1 agent in all but ≤2 antimicrobial categories. Conjugation frequency per recipient was expressed by dividing the number of transconjugants by the initial number of recipients.
Supplementary information
Additional file 1: Table S1. Oligonucleotides and PCR conditions used for amplification (internal primers included for sequencing).
Acknowledgements
The authors are grateful to Bill Wickstead (Wickstead Lab, University of Nottingham School of Life Sciences, UK) who assisted with some reagents for the project. We thank the following for their laboratory support: Michael Olu-Taiwo (University of Ghana School of Biomedical and Allied Health Sciences, Department of Medical Laboratory Sciences); Mary Magdalene Osei (University of Ghana School of Biomedical and Allied Health Sciences, Medical Microbiology department); Mr. Thomas Dankwa (all of Korle-Bu Teaching Hospital Central Laboratory); and Mr. Seth Agyeman (Korle-Bu Teaching Hospital Central Laboratory).
Abbreviations
- AmpCs
Class C cephalosporins
- ATTC
American Type Culture Cells
- CC
Clonal complex
- CLSI
Clinical and Laboratory Standards Institute
- CRe
Cephalosporin resistant enterobacteria
- EDTA
Ethylene diamine tetraacetic acid
- ESBLs
Extended-spectrum beta-lactamases
- LB broth
Luria-Bertani Broth
- MBL
Metallo-beta-lactamases
- MLST
Multilocus Sequence Typing
- PCR
Polymerase Chain Reactions
- SSI
StatensSserum Agar
- TC
American Type culture Cells
Authors’ contributions
NON, RA, AKL conceptualized the study; participated in its design, laboratory work and coordination, and helped to draft the manuscript. HB, GAM, PFK participated in the study design, coordination, performed analysis, and drafted the manuscript. DOM participated in data interpretation and preparation of the manuscript. JA, SDK, JT, EB participated in the study design, laboratory work, and drafting the manuscript. All authors have read and approved the manuscript.
Funding
No specific funding was received for this study.
Availability of data and materials
The dataset for the current study is available from the corresponding author on reasonable request.
Ethics approval and consent to consent to participate
The study received ethical approval from the Ethics and Protocol Review Committee of the School of Biomedical and Allied Health Sciences, University of Ghana, with approval identification number: SBAHS-MD./10,512,194/aa/5a/2016–2017. All participants provided written informed consents. Patient data were treated with a high level of confidentiality. Computerized records of the survey were kept in electronic files. These documents were accessible to the lead investigator only.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests. The study findings were presented at ASM Microbe 2017.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Noah Obeng-Nkrumah and Appiah-Korang Labi contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12866-019-1629-x.
References
- 1.Rubin JE, Pitout JD. Extended-spectrum beta-lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Vet Microbiol. 2014;170:10–18. doi: 10.1016/j.vetmic.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 2.Bush K. Beta-lactamases. Ann N Y Acad Sci. 2013;1277:84–90. doi: 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- 3.Cornaglia Giuseppe, Giamarellou Helen, Rossolini Gian Maria. Metallo-β-lactamases: a last frontier for β-lactams? The Lancet Infectious Diseases. 2011;11(5):381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 4.Ćirković I, Pavlović L, Konstantinović N, Kostić K, Jovanović S, Djukić S. Phenotypic detection of beta-lactamases production in Enterobacteriaceae. Srp Arh Celok Lek. 2014;142:457–463. doi: 10.2298/SARH1408457C. [DOI] [PubMed] [Google Scholar]
- 5.Babic M, Hujer AM, Bonomo RA. What’s new in antibiotic resistance? Focus on beta-lactamases Drug Resist Updat. 2006;9:142–156. doi: 10.1016/j.drup.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Obeng-Nkrumah N, Twum-Danso K, Krogfelt KA, Newman MJ. High levels of extended-spectrum beta-lactamases in a major teaching hospital in Ghana: the need for regular monitoring and evaluation of antibiotic resistance. Am J Trop Med Hyg. 2013;89:960–964. doi: 10.4269/ajtmh.12-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agyekum Alex, Fajardo-Lubián Alicia, Ansong Daniel, Partridge Sally R., Agbenyega Tsiri, Iredell Jonathan R. blaCTX-M-15 carried by IncF-type plasmids is the dominant ESBL gene in Escherichia coli and Klebsiella pneumoniae at a hospital in Ghana. Diagnostic Microbiology and Infectious Disease. 2016;84(4):328–333. doi: 10.1016/j.diagmicrobio.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Hackman HK, Brown CA, Asmah RH, Badu CA, Amegashi G. Advances in life science and technology. Technology and Education (IISTE): Advances in Life Science and Technology. International Institute for Science; 2011. [Google Scholar]
- 9.Potron Anaïs, Poirel Laurent, Dortet Laurent, Nordmann Patrice. Characterisation of OXA-244, a chromosomally-encoded OXA-48-like β-lactamase from Escherichia coli. International Journal of Antimicrobial Agents. 2016;47(1):102–103. doi: 10.1016/j.ijantimicag.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Oueslati S, Nordmann P, Poirel L. Heterogeneous hydrolytic features for OXA-48-like ??-lactamases. J Antimicrob Chemother. 2014;70:1059–1063. doi: 10.1093/jac/dku524. [DOI] [PubMed] [Google Scholar]
- 11.Poirel L, Bonnin RA, Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother. 2012;56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasan B, Drobni P, Drobni M, Alam M, Olsen B. Dissemination of NDM-1. Lancet Infect Dis. 2012;12:99–100. doi: 10.1016/S1473-3099(11)70333-4. [DOI] [PubMed] [Google Scholar]
- 13.Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, Tripathi A, et al. Prevalence and molecular characterisation of New Delhi metallo-β- lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents. 2014;44:30–37. doi: 10.1016/j.ijantimicag.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Page MGP. Cephalosporins in clinical development. Expert Opin Investig Drugs. 2004;13:973–985. doi: 10.1517/13543784.13.8.973. [DOI] [PubMed] [Google Scholar]
- 15.Pfeifer Yvonne, Cullik Angela, Witte Wolfgang. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. International Journal of Medical Microbiology. 2010;300(6):371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez Jose Luis. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environmental Pollution. 2009;157(11):2893–2902. doi: 10.1016/j.envpol.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 18.Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. 2015;13:310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 19.Tetteh-Quarcoo PB, Donkor ES, Attah SK, Duedu KO, Afutu E, Boamah I, et al. Microbial carriage of cockroaches at a tertiary care hospital in ghana. Environmental health insights. 2013. pp. 59–66. doi:10.4137/EHI.S12820 [DOI] [PMC free article] [PubMed]
- 20.Salehzadeh A, Tavacol P, Mahjub H. Bacterial, fungal and parasitic contamination of cockroaches in public hospitals of Hamadan. Iran J Vector Borne Dis. 2007;44:105–110. [PubMed] [Google Scholar]
- 21.Fotedar R, Shriniwas UB, Verma A. Cockroaches ( Blattella germanica) as carriers of microorganisms of medical importance in hospitals. Epidemiol Infect. 1991;107:181–187. doi: 10.1017/S0950268800048809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassiri H, Kassiri A, Kazemi S. Investigation on American cockroaches medically important bacteria in Khorramshahr hospital. Iran Asian Pacific J Trop Dis. 2014;4:201–203. doi: 10.1016/S2222-1808(14)60505-3. [DOI] [Google Scholar]
- 23.Fakoorziba MR, Shahriari-Namadi M, Moemenbellah-Fard MD, Hatam GR, Azizi K, Amin M, et al. Antibiotics susceptibility patterns of bacteria isolated from American and German cockroaches as potential vectors of microbial pathogens in hospitals. Asian Pacific J Trop Dis. 2014;4:S790–S794. doi: 10.1016/S2222-1808(14)60728-3. [DOI] [Google Scholar]
- 24.Loucif L, Gacemi-Kirane D, Cherak Z, Chamlal N, Grainat N, Rolain JM. First report of German cockroaches (Blattella germanica) as reservoirs of CTX-M-15 extended-spectrum-β-lactamase- and OXA-48 carbapenemase-producing Enterobacteriaceae in Batna University hospital. Algeria Antimicrob Agents Chemother. 2016;60:6377–6380. doi: 10.1128/AAC.00871-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huijbers PMC, Graat EAM, Haenen APJ, van Santen MG, van Essen-Zandbergen A, Mevius DJ, et al. Extended-spectrum and AmpC β-lactamase-producing Escherichia coli in broilers and people living and/or working on broiler farms: prevalence, risk factors and molecular characteristics. J Antimicrob Chemother. 2014;69:2669–2675. doi: 10.1093/jac/dku178. [DOI] [PubMed] [Google Scholar]
- 26.Dierikx C, van der Goot J, Fabri T, van Essen-Zandbergen A, Smith H, Mevius D. Extended-spectrum-β-lactamase- and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother. 2013;68:60–67. doi: 10.1093/jac/dks349. [DOI] [PubMed] [Google Scholar]
- 27.Wu Guanghui, Day Michaela J., Mafura Muriel T., Nunez-Garcia Javier, Fenner Jackie J., Sharma Meenaxi, van Essen-Zandbergen Alieda, Rodríguez Irene, Dierikx Cindy, Kadlec Kristina, Schink Anne-Kathrin, Wain John, Helmuth Reiner, Guerra Beatriz, Schwarz Stefan, Threlfall John, Woodward Martin J., Woodford Neil, Coldham Nick, Mevius Dik. Comparative Analysis of ESBL-Positive Escherichia coli Isolates from Animals and Humans from the UK, The Netherlands and Germany. PLoS ONE. 2013;8(9):e75392. doi: 10.1371/journal.pone.0075392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cloarec A, Rivault C, Fontaine F, Le Guyader A. Cockroaches as carriers of bacteria in multifamily dwellings. Epidemiol Infect. 1992;109:483–490. doi: 10.1017/S0950268800050470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tachbele E, Erku W, Gebre-Michael T, Ashenafi M. Cockroach-associated food borne bacterial pathogens from some hospitals and restaurants in Addis Ababa, Ethiopia: distribution and antibiograms. J Rural Trop Public Health. 2006;5:34–41. doi: 10.3329/bmrcb.v44i1.36802. [DOI] [Google Scholar]
- 30.Fakoorziba MR, Eghbal F, Hassanzadeh J, Moemenbellah-Fard MD. Cockroaches (Periplaneta americana and Blattella germanica) as potential vectors of the pathogenic bacteria found in nosocomial infections. Ann Trop Med Parasitol. 2010;104(6):521–528. doi: 10.1179/136485910X12786389891326. [DOI] [PubMed] [Google Scholar]
- 31.Elgderi RM, Ghenghesh KS, Berbash N. Carriage by the German cockroach (Blattella germanica) of multiple-antibiotic-resistant bacteria that are potentially pathogenic to humans, in hospitals and households in Tripoli, Libya. Ann Trop Med Parasitol. 2006;100(1):55–62. doi: 10.1179/136485906X78463. [DOI] [PubMed] [Google Scholar]
- 32.Vazirianzadeh B, Dehghani R, Mehdinejad M, Sharififard M, Nasirabadi N. The first report of drug resistant Bacteria isolated from the Brown-banded cockroach, Supella longipalpa, in Ahvaz, South-western Iran. J Arthropod Borne Dis. 2014;8(1):53–59. [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson WH, Akers RC, Powell PK. German cockroaches in urban apartment buildings. Pest Control. 1980;48:18–20. [Google Scholar]
- 34.Cochran DG. Cockroaches, Their Biology, Distribution and Control. Switzerland World Health Organization. 1999;99.3. p. 83.
- 35.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bush K. Bench-to-bedside review: the role of beta-lactamases in antibiotic-resistant gram-negative infections. Crit Care. 2010;14:224. doi: 10.1186/cc8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bush Karen. Carbapenemases: Partners in crime. Journal of Global Antimicrobial Resistance. 2013;1(1):7–16. doi: 10.1016/j.jgar.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Obeng-Nkrumah N, Labi A-K, Addison NO, Labi JEM, Awuah-Mensah G. Trends in paediatric and adult bloodstream infections at a Ghanaian referral hospital: a retrospective study. Ann Clin Microbiol Antimicrob. 2016;15:49. doi: 10.1186/s12941-016-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obeng-Nkrumah N, Labi A-K, Acquah ME, Donkor ES. Bloodstream infections in patients with malignancies: implications for antibiotic treatment in a Ghanaian tertiary setting. BMC Res Notes. 2015;8:742. doi: 10.1186/s13104-015-1701-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waldvogel MG, Moore CB, Nalyanya GW, String-ham SM, Watson DW, Schal C. Integrated cock-croach (Dictyoptera: Blattellidae) management in con-Þned swine production. In: Robinson WH, Rettich F, Rambo GW, editors. Proceedings of the3rd international conference of urban pests. Graflicke Zavody Hronov: Prague, Czech Republic; 1999. pp. 183–188. [Google Scholar]
- 41.Zurek L, Gore JC, Stringham SM, Wesley DW, Michael G. Boric acid dust as a component of an integrated cockroach management program in confined swine production. J Econ Entomol. 2003;96(4):1362–1366. doi: 10.1603/0022-0493-96.4.1362. [DOI] [PubMed] [Google Scholar]
- 42.Zurek L, Schal C. Evaluation of the German cockroach (Blattella germanica) as a vector for verotoxigenic Escherichia coli F18 in confined swine production. Verterinary Microbiology. 2004;101(4):263–267. doi: 10.1016/j.vetmic.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 43.Tian B, Fadhil, NH, Powell JE, Kwong WK, Moran NA. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio. 2012; 3(6). doi.org/10.1128/mBio.00377-12 [DOI] [PMC free article] [PubMed]
- 44.Allen HK, Cloud-Hansen KA, Wolinski JM, Guan C, Greene S, Lu S, Handelsman J. Resident microbiota of the gypsy moth midgut harbors antibiotic resistance determinants. DNA Cell Biol. 2009;28(3):109–117. doi: 10.1089/dna.2008.0812. [DOI] [PubMed] [Google Scholar]
- 45.Lowe CF, Romney MG. Bedbugs as vectors for drug-resistant bacteria. Emerg Infect Dis. 2011;17(6):1132–1134. doi: 10.3201/eid1706.101978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baumholtz MA, Parish LC, Witkowski JA, Nutting WB. The medical importance of cockroaches. Int J Dermatol. 1997;36(2):90–96. doi: 10.1046/j.1365-4362.1997.00077.x. [DOI] [PubMed] [Google Scholar]
- 47.Mattila S, Ruotsalainen P, Ojala V, Tuononen T, Hiltunen T, Jalasvuori M. Conjugative ESBL plasmids differ in their potential to rescue susceptible bacteria via horizontal gene transfer in lethal antibiotic concentrations. J Antibiotics. 2017;70:805–808. doi: 10.1038/ja.2017.41. [DOI] [PubMed] [Google Scholar]
- 48.Jibril M, Yaovi MGH, Line ET. Antimicrobial resistance among clinically relevant bacterial isolates in Accra: a retrospective study. BMC Research Notes. 2018; 11(254). doi.org/10.1186/s13104-018-3377-7 [DOI] [PMC free article] [PubMed]
- 49.Jupe A.J., Parker R.H., Takahashi H. Preface and acknowledgement. Geothermics. 1995;24(3):297. doi: 10.1016/0375-6505(95)90007-1. [DOI] [Google Scholar]
- 50.Sandelowski M. Combining qualitative and quantitative sampling, data collection, and analysis techniques in mixed-method studies. Res Nurs Health. 2000;23:246–255. doi: 10.1002/1098-240x(200006)23:3<246::aid-nur9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 51.CLSI. M02-A12 Performance Standards for Antimicrobial Disk. Clsi. 2015.
- 52.EUCAST. European Commitee for Antibiotic Susceptibilty testing. In: EUCAST [Internet]. 2017 [cited 7 Nov 2017]. Available: http://www.eucast.org/clinical_breakpoints/
- 53.CLSI . M100S performance standards for antimicrobial susceptibility testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- 54.Tamang MD, Nam HM, Jang GC, Kim SR, Chae MH, Jung SC, et al. Molecular characterization of extended-spectrum-β-lactamase- producing and plasmid-mediated AmpC β-lactamase-producing Escherichia coli isolated from stray dogs in South Korea. Antimicrob Agents Chemother. 2012;56:2705–2712. doi: 10.1128/AAC.05598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–2162. doi: 10.1128/JCM.40.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Azim A, Dwivedi M, Rao PB, Baronia AK, Singh RK, Prasad KN, et al. Epidemiology of bacterial colonization at intensive care unit admission with emphasis on extended-spectrum beta-lactamase- and metallo-beta-lactamase-producing Gram-negative bacteria--an Indian experience J Med Microbiol. 2010;59:955–960. doi: 10.1099/jmm.0.018085-0. [DOI] [PubMed] [Google Scholar]
- 57.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichiacoli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Francisco Alexandre P, Vaz Cátia, Monteiro Pedro T, Melo-Cristino José, Ramirez Mário, Carriço João A. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics. 2012;13(1):87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Oligonucleotides and PCR conditions used for amplification (internal primers included for sequencing).
Data Availability Statement
The dataset for the current study is available from the corresponding author on reasonable request.