Abstract
Studies of colorectal cancer (CRC) originating through the conventional adenoma-carcinoma sequence have provided insight into the molecular mechanisms controlling its initiation and progression. Much less is known about the alternative “serrated” pathway, which has been associated with BRAF mutation and microsatellite instability. Recent transcriptomics approaches to classify human CRC revealed that mesenchymal/desmoplastic features combined with an immunosuppressive microenvironment are key determinants of CRC with the poorest prognosis. Importantly, these very aggressive CRCs harbor the characteristics of serrated tumors, suggesting that initiation through this alternative pathway determines how aggressive the CRC becomes. We review recent evidence on how serrated carcinogenesis contributes to the subtype of CRC with the poorest prognosis.
Keywords: serrated, colorectal cancer, mesenchymal, atypical PKC, microenvironment, immune checkpoint therapy
Two Pathways Leading to CRC
Colorectal cancer (CRC) is one of the leading causes of cancer-related deaths worldwide [1]. The development of sporadic CRC is thought to occur through precursor lesions that originate from either a “conventional” or an “alternative” (serrated) pathway [2, 3]. The precursor lesions of the conventional pathway are often referred to as conventional adenomas, which display tubular, tubulovillous, or villous adenoma histology [4]. In contrast, the serrated pathway is initiated by the formation of serrated adenomas or polyps, displaying so-called “saw-tooth” or stellate architecture of the crypt epithelium [5–7]. Although almost all sporadic CRCs were previously thought to develop through the conventional pathway, it is now recognized that this pathway only accounts for approximately 60–85% of CRCs and that most of the remaining 15–40% of CRCs occur through the alternative serrated pathway [6, 8–11]. The incidence of CRCs arising via the serrated pathway (hereafter referred to as “serrated CRCs”) is therefore potentially larger than the incidence of many other cancers, including gastric, oesophageal, and ovarian, indicating that this CRC subtype has become a significant public health problem [1]. Serrated adenoma/polyps are classified into sessile serrated adenomas or polyps (SSA/Ps), traditional serrated adenomas (TSAs), and hyperplastic polyps (HPs), all of which have the potential to progress to fatal cancers [7, 12]. SSA/Ps in particular have been reported to have a significant risk for malignant transformation [13, 14]. A recent improvement in colonoscopy tests has increased the detection rate of serrated polyps and will enable long-term follow-up of individual lesions. However, we are still far from a complete understanding of the biology and pathogenesis of serrated CRCs [7, 12]. Because relatively little attention has been paid to the mechanisms of serrated tumorigenesis, as compared with the tumorigenesis of conventional adenomas, reliable longitudinal observational data are sorely lacking for serrated lesions. Furthermore, the serrated morphology is preserved in only about one-third of CRCs arising via the serrated path, whereas the remaining two-thirds lose the serrated histological features when they have progressed to advanced cancer [12, 13, 15, 16]. These issues make it difficult to morphologically (i.e., endoscopically and histologically) distinguish whether a given CRC developed through the conventional or the serrated pathway.
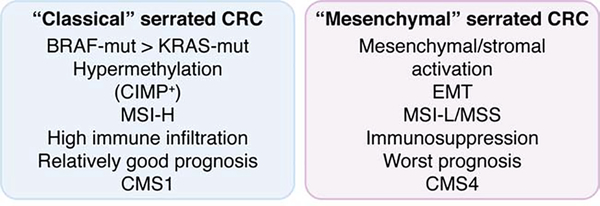
Recently, transcriptomics has been used to classify CRC into relevant subgroups. Transcriptomics provides a better way to identify the origin and characteristics of individual CRCs because it is not influenced by the final histological appearance of the tumors [17–24]. In addition, this type of analysis allows the tumor phenotype to be comprehensively determined in a way that also includes the contribution of the tumor microenvironment, which has not been studied in sufficient detail in CRC, especially for the serrated type. Molecular markers and gene expression profiles have suggested that the serrated pathway gives rise to at least two CRC subgroups: (1) an inflammatory subtype with a BRAF mutation and microsatellite instability (MSI) (hereafter referred to as “classical serrated” CRCs); and (2) an immunosuppressive subtype with stromal/mesenchymal characteristics (referred to as “mesenchymal serrated” CRCs) [19, 20, 25, 26] (Figure 1). Integration of the traditional morphological approach and the recent transcriptomic strategy allows for better characterization of serrated lesions as well as a fuller understanding of their etiopathogenesis.
Figure 1. Molecular characterization of two types of serrated CRC: classical and mesenchymal.
Typical molecular features of the two types (“Classical (left)” vs “Mesenchymal (right)”) of serrated CRC proposed in this review based on the previously reported evidence. CIMP, CpG island methylation phenotype; CRC, colorectal cancer; MSI-H, microsatellite instability-high; MSI-H, microsatellite instability-low; MSS, microsatellite stable.
A major clinical gap remains between the detection of serrated precursor lesions and the development of therapies to treat them. Understanding the pathogenesis, including the role of the immunological environment, of both types of serrated CRCs is essential for developing more efficacious therapies. Emerging evidence suggests that immunotherapy could be a viable option for patients with various types of cancers. Unfortunately, most CRCs, except those in the high MSI (MSI-H) category, respond poorly to this type of treatment [27–31]. This is particularly important because, although the classical serrated CRCs have an MSI-H phenotype, the mesenchymal serrated CRCs are considered microsatellite stable (MSS) [19, 20, 26]. Therefore, it is imperative to establish a therapeutic consensus on whether and how immunotherapy should be applied to the treatment of these two types of serrated CRCs.
Experimental animal models, beyond the most popular xenografts, that recapitulate at least the most salient features of the human disease in immunocompetent settings, are powerful tools for genetic and preclinical cancer research. They provide an amenable system for elucidating the molecular mechanisms and pathobiology of specific tumor subtypes and for developing potential treatments. Substantial efforts have been made to generate mouse models for classical serrated CRCs, especially by introducing the oncogenic BRAF mutation into mouse intestinal epithelial cells (IECs) in combination with genetic knockout of tumor suppressor genes. This approach has led to the generation of valuable information on the mechanisms of initiation and progression of serrated tumorigenesis [32, 33]. However, the role of the tumor stroma and the mechanisms underlying the appearance of mesenchymal serrated CRCs have not been addressed in these mouse models until recently [34]. The goal of this perspective article is to review the most recent literature on serrated tumorigenesis with special focus on the existing mouse models for both classical and mesenchymal serrated CRCs. We also discuss the role of the tumor microenvironment in the pathogenesis of this disease and its clinical and therapeutic implications.
Potential Molecular Evolution of Serrated CRCs
The molecular events that occur during the initiation and progression of intestinal tumors through the serrated pathway have not been fully characterized yet, in contrast to our detailed understanding of the mechanisms controlling the conventional pathway. The conventional pathway predominately initiates with the inactivation of the APC tumor suppressor, resulting in adenomas that need to undergo alterations in additional genes such as KRAS, TP53, and SMAD4 in order to progress to CRC (the adenoma-carcinoma sequence) [2, 4]. In contrast, the analyses of the mutational landscape of serrated lesions (including SSA/Ps, TSAs, and HPs) have identified an activating mutation in BRAF as a key gene alteration in the serrated pathway; this mutation results in the constitutive stimulation of the MAPK signaling cascade [7, 11, 35–37]. This oncogenic event initially results in the dysregulation of cell proliferation, differentiation, and survival that ultimately gives rise to the serrated lesions [35–38]. A hotspot mutation in codon 15 of BRAF that results in a Val600Glu amino acid change (BRAFV600E) is the most commonly identified mutation in serrated tumors. These mutated lesions develop into serrated precursors (microvesicular HPs and SSA/Ps) that are associated to another common molecular event in this pathway, the hypermethylation of the CpG island promoter regions (the so-called CpG island methylation phenotype; CIMP-H), which results in the epigenetic silencing of a number of tumor suppressor genes such as p16INK4a (encoded by CDKN2A) and MLH1 [36–40]. MLH1 is a mismatch repair (MMR) gene whose silencing leads to the development of CIMP-H/MSI-H CRCs [6, 39, 41]. The precise mechanism linking BRAF mutation and the CIMP-H and MSI-H phenotypes has been an open question in the field. It was not clear whether BRAF mutations may directly induce CIMP or whether CIMP may generate a cellular context that favors the survival and growth of cells with BRAF mutations. A more recent study using long-term culture of colon-derived organoids provided compelling evidence that aging-driven changes in DNA methylation, similar to those found in human patients of proximal CRC, create an epigenetic landscape permissive of transformation driven by BRAF mutation [42]. Interestingly, tumors developing in patients with Lynch syndrome (also called as “hereditary nonpolyposis colorectal cancer (HNPCC)”) that harbors a germline mutation in MMR genes show mixed morphology, including conventional adenomatous, sessile serrated and hyperplastic polyps even though these tumors are MSI-H like sporadic CIMP-H/MSI-H CRCs with BRAF mutation [43]. While polyps are more prevalent in patients with Lynch syndrome than in the general population, the detection rate of serrated lesions in Lynch syndrome individuals is comparable with a control population [44, 45]. These observations suggest that the serrated tumorigenesis seems not to depend on MSI-H phenotype itself but rather on somatic driver mutations in BRAF. However, given that BRAF mutation is observed in only around 10% of all CRCs, whereas 15–40% of CRCs develop through the serrated pathway, alterations other than the BRAF mutation must contribute to the development of the remaining serrated CRC cases. The other known driver in serrated tumorigenesis is the oncogenic mutation of KRAS (typically codon 12/13), that, like the BRAF mutation, also results in the constitutive activation of the MAPK signaling cascade [46]. Serrated polyps emerging from the KRAS mutant pathway evolve into carcinomas that are characterized by low levels of CIMP. In contrast to serrated tumors driven by BRAF mutation, the MLH1 gene is intact in KRAS-mutant cancers, and they are MSS in most cases [11, 46]. Inactivating mutations of tumor suppressor genes such as TP53, rather than aberrant methylation of their promoters, seem to be the main drivers for the evolution of KRAS-mutant serrated cancers [11, 47]. Although serrated cancers harboring a KRAS mutation may account for only approximately 5% of all CRCs, it is difficult to make a precise estimate because this type of oncogenic alteration, unlike that of BRAF, is also observed in about 50% of CRCs arising via the conventional CRC pathway [11, 47]. Furthermore, KRAS is altered much less frequently in serrated lesions than BRAF, and it seems unlikely that KRAS mutation alone accounts for all of the serrated-origin CRCs that do not have mutations in BRAF. A detailed description and discussion on the BRAF and KRAS mutations, as well as alterations in CIMP and MSI characteristics observed in serrated tumors, have been recently reviewed [7]. In any case, it should be borne in mind that these “typical” molecular characteristics (i.e., BRAF or KRAS mutation, CIMP-H, and MSI-H) are not present in all serrated lesions [7]. Therefore, the identification of other molecular driving events in serrated tumorigenesis is urgently needed. Indeed, the detailed transcriptomic analyses of human CRCs and the generation of new, more physiologically relevant, experimental mouse models, especially for the mesenchymal type, can provide a more complete understanding of the mechanisms controlling the initiation and progression of serrated tumorigenesis.
Transcriptomic Subtyping of Conventional and Serrated CRCs
Changes in gene expression are intimately linked to the cellular phenotype, common disease patterns, and clinical features of cancer. A number of recent studies have used these patterns to establish criteria for the classification of CRCs into three to six biologically homogeneous subgroups [18–20, 23]. De Sousa et al. proposed the CCS (colon cancer subtype) classification, which includes three subgroups: CCS1 represents chromosomal instability (CIN), CCS2 is MSI-H/CIMP-H, and CCS3 is mesenchymal and shows the worst prognosis and drug resistance [20]. Whereas the transcriptomic profile of CCS1 CRC is related to that of the conventional-type adenoma, the poor-prognosis CCS3 CRC is enriched with gene signatures also identified in serrated adenomas, suggesting that the serrated lesion is a potential precursor of this poor-prognosis CRC subtype [20]. Importantly, this particular subgroup displays high expression of genes related to the epithelial-mesenchymal transition (EMT) and activation of matrix remodeling, consistent with a desmoplastic phenotype [20]. A more recent study revealed that the CRCs with low expression levels of both atypical protein kinase Cs (aPKCs: PKCζ and PKCλ/ι; Figure 2 and Box 1) were strongly enriched for the gene expression signatures of serrated tumors, EMT, and stromal activation, when compared with CRCs with high expression levels of both aPKCs [34]. Consistent with this, CCS3 CRCs display lower expression of aPKCs than CCS1 or CCS2 CRCs [34]. Therefore, the aPKCs emerge as new players in the most aggressive form of CRC that have features of EMT and serrated gene expression signatures [34]. Further new genetic and transcriptomics classifications of CRC resulted in the generation of a more comprehensive consensus molecular subtype (CMS) classification that was intended to integrate six independent gene expression-based subtyping systems [24]. Two subgroups in the CMS classification, CMS1 and CMS4, correspond to serrated phenotypes. The CMS1 subgroup is characterized by MSI-H, CIMP, BRAFV600E mutation, and high infiltration of immune cells, whereas the CMS4 subgroup is characterized by signatures indicative of EMT and a stromal-enriched immune microenvironment and is the subgroup with the poorest prognosis [24]. Fessler et al. proposed SSA/Ps as the potential precursors for both CMS1 and CMS4 CRCs [25, 26]. Notably, they found that, whereas administration of TGFβ promotes cell death in organoids generated from conventional adenomas, the same treatment induces the mesenchymal phenotype in organoids harboring the BRAFV600E mutation [25]. The hyperproduction of TGFβ by a hyperactivated stroma in the tumor microenvironment has been proposed to be key to skewing the serrated precursors from the high-immune-infiltration CMS1 type to the poor-prognosis mesenchymal CMS4 type [25, 26]. These are very important observations because, although CMS1 CRCs harbor the common molecular features of serrated lesions as described in the previous section, all of the recent gene expression studies point to an association of mesenchymal CMS4 CRCs with serrated tumorigenesis [18–20, 23, 24]. Given that CMS4 CRCs are enriched in neither BRAF nor KRAS mutations and are neither MSI-H nor CIMP-H, we posited that there must be additional, still-unidentified molecular events that confer serrated phenotypes to this CRC subtype [24].
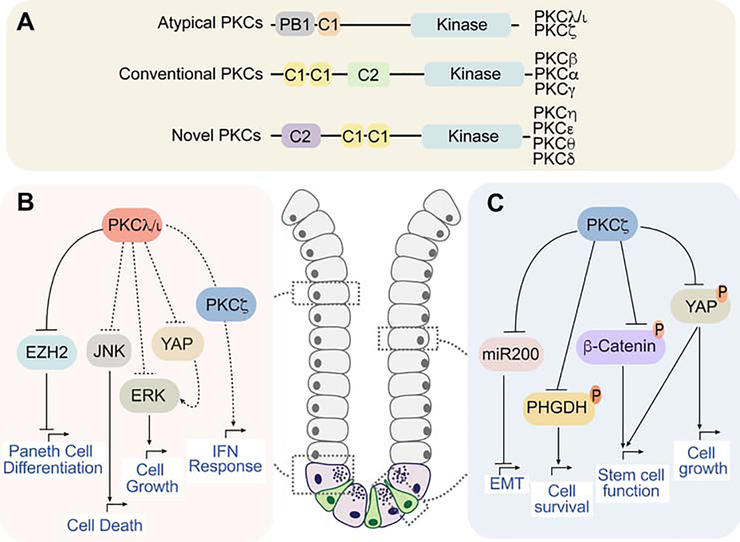
Figure 2. The roles of the aPKCs is intestinal homeostasis and cancer.
(A) The atypical protein kinases C (aPKCs) are part of the PKC family. Classification of the PKCs by subfamilies (atypical, conventional, novel). The protein names of the different members are indicated. Schematic showing structural domain organization characteristic of each PKC subfamily. The aPKCs contain a unique PB1 (Phox and Bem 1) and they do not harbor a C2 domain, which binds calcium. The C1 domain, which is critical for diacylglycerol binding, is not functional in the aPKCs. </p/> (B and C) Schematic representation of the functional role of the aPKCs in intestinal homeostasis and cancer. PKCλ/ι and PKCζ are both expressed in intestinal epithelial cells (grey cells). PKCλ/ι is highly expressed in Paneth cells (violet cells in the crypt) and PKCζ in intestinal stem cells (green cells in the crypt). (B) PKCλ/ι regulates Paneth cell differentiation through inhibition of EZH2 stability and intestinal cell death via JNK. PKCλ/ι is a tumor suppressor in CRC through the inhibition of ERK and YAP. It also regulates the IFN response. (C) PKCζ maintains intestinal stemness and is a versatile tumor suppressor by inhibiting miR200, PHGDH, β-Catenin and YAP to limit EMT, cell survival, stem cell function and cell growth, respectively.
Box 1. Role of the Atypical Protein Kinase Cs in intestinal homeostasis and cancer.
The atypical protein kinase Cs (aPKCs) constitute a subfamily of kinases that, together with the conventional or classical (cPKCs) and the novel (nPKCs), are part of the extended PKC family [79] (Fig. 2A). The aPKCs include two isoforms (PKCζ and PKCλ/ι), which are insensitive to Ca2+ and diacylglycerol due to the lack of functional C2 and C1 domains [79]. In contrast, both aPKCs harbor a unique protein-protein interaction domain (Phox and Bem 1; PB1), which is critical for their binding to other PB1-containing proteins such as the polarity regulator PAR6 or the autophagy and signaling adaptor p62 [79, 80]. Both aPKCs are closely related at the sequence level, but the use of in vivo knock-out (KO) models selectively for each aPKC has been instrumental in investigating the specific roles of these kinases in intestinal homeostasis and cancer.
PKCλ/ι is widely expressed in epithelial cells of small intestine and colon, especially in the crypt base, where its levels are remarkably high in Paneth cells [81]. Notably, conditional KO of PKCλ/ι in intestinal epithelial cells (lECs) resulted in the loss of mature Paneth cells, increased IEC death, inflammation, and dysbiosis [81] (Fig. 2B). PKCλ/ι is required for Paneth cell homeostasis through the control of the stability of EZH2, an epigenetic repressor of ATOH1 and GFI1, two master regulators of Paneth cell differentiation [81]. Importantly, PKCλ/ι expression is decreased in the intestine of Crohn’s disease and ulcerative colitis patients, consistent with its role in maintaining the intestinal barrier [81, 82]. Although the loss of PKCλ/ι in lECs is not sufficient by itself to drive tumorigenesis, it cooperates with the deficiency of other tumor suppressors such as APC or PKCζ to promote intestinal cancer by generating an inflammatory environment conducive to cancer [34, 81]. Furthermore, analysis of CRC patients revealed that low levels of PRKCI (gene encoding PKCλ/ι) correlated with worse patient survival, consistent with its role as a tumor suppressor in CRC [81].
PKCζ (encoded by the PRKCZ gene) is a well-established tumor suppressor in colorectal cancer (CRC) [79] (Fig. 2C), and consequently is downregulated in human CRC, as compared to normal colon tissue, and more significantly in metastasis, being a predictive factor of poor prognosis [83]. Consistently, an inactivating mutation in PKCζ (S514F) was identified in human CRCs [84, 85]. Importantly, the in vivo deletion of Prkcz in mice with inactivated adenomatous polyposis coli (Apc) increased intestinal tumorigenesis [83]. Mechanistically, PKCζ loss in intestinal cancer cells reprograms their metabolism under nutrient stress conditions to support tumor growth through the upregulation of phosphoglycerate dehydrogenase (PHGDH) [83]. Also, PKCζ KO in intestinal stem cells (ISCs) resulted in enhanced tumorigenic capacity of the stem cell population, and higher intestinal regeneration after irradiation [86]. PKCζ-deficient ISCs cope with nutrient deprivation-induced stress through the control of YAP and WNT/β-catenin signaling pathways [86]. On top of being a metabolic tumor suppressor in CRC cells, PKCζ, by reducing miR200 levels, is also critical for CRC liver metastasis, and for the regulation of sternness and the epithelial-mesenchymal transition of cancer cells [87].
Mouse Models of “Classical Serrated” CRC
Several mouse models have been generated to recapitulate the serrated pathway in vivo. Although few of them seem to succeed in fully mimicking the phenotype and behavior of human serrated lesions, especially in terms of tumor microenvironment, these mouse models provide great insights into the molecular events and pathophysiological features of the disease. On the basis of the mutation analyses of human serrated lesions described above, activation of the MAPK cascade has been considered to be the common driver for the initiation of serrated lesions in most cases. In support of this notion, Bennecke et al., (2010) aberrantly activated oncogenic KRAS (KRASG12D) in the mouse intestinal epithelium using the Villin-cre system and demonstrated that it results in the development of serrated hyperplasia in the colon, although the concomitant induction of cellular senescence prevented full transformation of these hyperplastic lesions [48]. Importantly, the abrogation of senescence by the inactivation of p16INK4a led to the formation of TSAs that further progressed to advanced carcinomas with metastases [48]. These p16Ink4a-deficient, KRASG12D-driven mouse serrated tumors were CIMP-negative and MSI-L/MSS, which closely resembles the molecular characteristics of human serrated polyps and carcinomas harboring mutant KRAS [7, 11, 46]. This study also demonstrated a significant infiltration of immune cells positive for CD3 and/or CD45 along the lamina propria and surrounding the adenomatous epithelia of the double p16Ink4a/KRASG12D-mutant serrated tumors, suggesting that this model potentially has an inflammatory phenotype [48].
Another mouse model of serrated tumorigenesis was generated by the introduction of oncogenic BRAFV600E into the intestinal epithelium using the Ah-creER system, which produced tumors that faithfully recapitulate the characteristics of human serrated neoplasia positive for mutant BRAF, including serrated CRCs [32]. This model shows how the expression of BRAFV600E results in the formation of hyperplastic crypts and a serrated epithelium [32]. However, like the KRASG12D-driven serrated mouse model, BRAFV600E also gave rise to a senescent program controlled by the upregulation of p16Ink4a. Further experiments demonstrated that in this BRAF-driven mouse model, loss of p16Ink4a through age-dependent epigenetic suppression or genetic mutation is necessary for tumor progression from benign hyperplastic lesions to advanced cancer [32]. Notably, another study also reported that expression of oncogenic BRAF in the mouse intestinal epithelium drives serrated tumorigenesis; this model also used the Villin-cre system, but the mutant oncogene was BRAFV637E instead of BRAFV600E [33]. BRAFV637E in mouse exon 18 is in the orthologous position of the human BRAFV600E mutation, which affects exon 15. The study of the Villin-cre;BRAFV637E system allowed the long-term follow-up of intestinal tumorigenesis that was difficult in Ah-creER;BRAFV600E mice due to early lethality. Mice with the BRAFV637E mutation in the intestinal epithelium spontaneously develop benign serrated lesions that progress to more malignant adenomas, dysplasia, and finally adenocarcinomas after a long period [32]. The authors also found that all the BRAFV637E-induced HPs were MSS or MSI-low, whereas the proportion that were MSI-H increased considerably when tumors become more advanced [33]. CpG island methylation in p16Ink4a was observed in a subset of BRAFV637E-induced serrated adenomas with dysplasia and carcinomas [33]. This BRAFV637E mutation model was also used by other investigators who proposed that DNA methylation arises slowly in direct response to prolonged oncogenic BRAF signaling in the serrated polyps [49]. These results should be reconciled with those of Tao et al. (2019) [42] suggesting that the appearance of the CIMP phenotype over time allows transformation induced by mutant BRAF by suppressing senescence. Furthermore, an additional colon organoid-based model has been generated using colonic cells of transgenic BRAFV600E mice modified by CRISPR-Cas9 to incorporate five additional genetic alterations that occurred frequently in human CRCs with BRAFV600E These additional mutations included: CDKN2A, RNF43, ZNRF3, TGFΒR2 and MLH1. Interestingly, the injection of organoids incorporating all these mutations on top of BRAFV600E into the colon of immunodeficient mice led to tumors with a striking resemblance histologically and at the transcriptional level to human MSI serrated tumors [50].
Mouse Models of Mesenchymal Serrated CRC
In contrast to the models of classical serrated CRCs described above, there have been very few mouse models of non-classical or mesenchymal serrated CRCs. Inactivation of CDX2 combined with BRAFV600E expression using CDX2P-creERT2, which is expressed in terminal ileum and colon, promotes serrated benign and invasive tumors that are enriched with the EMT gene signature [51]. Balbinot et al. (2018) demonstrated that the mosaic knockout of CDX2 in the mouse intestinal epithelium using the Ah-creER system resulted in the development of tumors that share several stromal and immune properties with human serrated CRCs with mesenchymal activation [52]. These include high expression of extracellular matrix molecules, the myeloid chemokine CCL2, components of the complement pathway, angiogenic factors, and immunosuppressive molecules [52]. These characteristics suggest that this is a relevant animal model to investigate the complex modifications of the microenvironment leading to the neoplastic conversion of premalignant lesions into aggressive, desmoplastic, immunosuppressed CRCs. Furthermore, reduced expression of CDX2 associates with poor prognosis in CRC patients, and the down-regulation of CDX2 expression was observed in serrated CRCs with a mesenchymal phenotype, further suggesting the human relevance of this mouse model [51, 52]. However, CDX2 loss alone is not sufficient to drive serrated CRC; instead, mosaic loss of CDX2 combined with APC deficiency is required for tumors to develop [52]. As a result, tumors in this mouse model displayed mixed structure characterized by juxtaposition of areas resembling gastric-type metaplastic lesions and areas like conventional adenomatous polyps [52]. Therefore, careful consideration is needed to adequately interpret the phenotypes of these mixed tumors.
Importantly, recent data demonstrated that the simultaneous inactivation in the mouse intestinal epithelium of the only two aPKCs, PKCλ/ι and PKCζ, results in the rapid development of serrated hyperplasia, SSA/Ps, dysplasia, and adenocarcinomas in small intestines and proximal colon without any other induced oncogene or insult [34]. Strikingly, the intestinal adenocarcinomas in the aPKC-deficient mice (DKOIEC) displayed a highly invasive phenotype associated with stromal/mesenchymal activation [33]. Tumors in the aPKC-deficient mice are characterized by the MSS phenotype and histology showing poor differentiation and/or signet ring cell cancer. They therefore resemble human BRAFMUTANT/MSS CRCs, which are also associated with poorly differentiated, mucinous, or signet ring cell morphology and poor prognosis [12, 34]. Consistent with being MSS, DKOIEC tumors are also CIMP-L. In addition, SSA/Ps developed in DKOIEC mice show increased expression of p53, p21 and p16, which are all suppressed in invasive carcinoma, similarly to BRAFMUTANT tumors [33, 34].
Of potential great relevance for human disease, the serrated tumors in aPKC-deficient mice have a strongly immunosuppressed phenotype, as indicated by the immune exclusion of CD8+ T cells from the tumor and their accumulation in the stromal periphery, accompanied by increased infiltration of immunosuppressing cells such as Treg cells and myeloid-derived suppressor cells (MDSCs) (Figures 3 and 4) [34]. A striking aspect of this mouse model is the activation of the MAPK cascade through a novel YAP/EGFR-driven pathway. The activation is dependent on inflammation and takes place without mutations in BRAF or KRAS [34]. Interestingly, this oncogenic signaling pathway is activated not only in the DKOIEC mice but also in the single IEC-specific PKCλ/ι knockout (LKOIEC) mice [34], which do not develop tumors in the absence of additional mutations. This observation raises the question: If both DKOIEC and LKOIEC mice activate the same oncogenic signaling pathways, why are serrated intestinal tumors not seen in the LKOIEC mice as they are in the DKOIEC mice? The immune status provides the explanation to this conundrum. That is, whereas tumors in the DKOIEC mice present with strong immunosuppression, the intestinal tissue in the LKOIEC mice displays strong CD8+ T cell-dependent immunosurveillance, which is driven by the interferon (IFN) response and prevents tumor initiation [34]. Therefore, the contribution of PKCΖ deficiency to the phenotype of the DKOIEC mouse intestinal tissue is to switch the intestinal epithelium from the immunosurveillance triggered by the lack of PKCλ/ι to an immunosuppressive phenotype [34]. This is of relevance for human serrated CRC because human SSA/Ps express significantly less aPKC than normal mucosa or conventional-type adenoma [34].
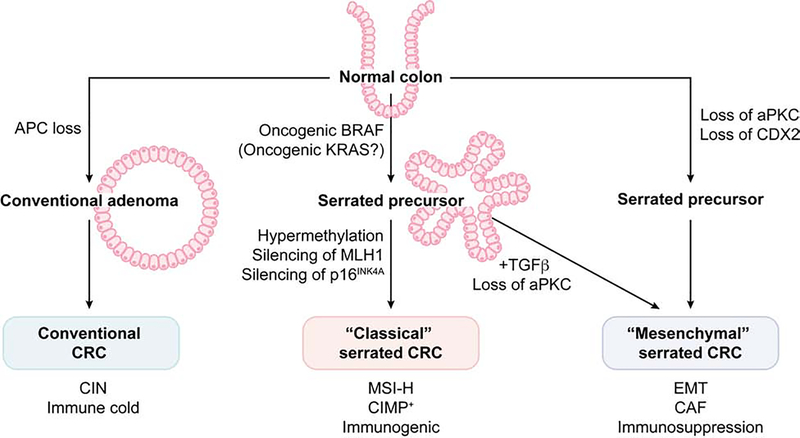
Figure 3. A simplified model of the molecular evolution of each colorectal cancer subtype.
Pathways depicted are based largely on evidence from mouse models. The conventional pathway is initiated by inactivation of the tumor suppressor APC in normal colonic epithelium which results in the formation of conventional-type adenoma, further followed by the additional sequential mutations of oncogenes or tumor suppressor genes to progress to adenocarcinoma (adenoma-carcinoma sequence). This type of CRCs generally displays chromosomal instability (CIN) with immune cold tumor microenvironment (TME). Oncogenic mutation in BRAF (less frequently in KRAS) leads to the development of serrated precursors such as microvesicular HPs and SSA/Ps. Hypermethylation in the CpG island promoter regions (CpG island methylation phenotype; CIMP) of tumor suppressor genes such as p16INK4a results in the silencing of these genes and allow a complete malignant transformation of these serrated lesions (classical serrated pathway). Epigenetic silencing of MLH1 in these tumor cells leads to the development of MSI-H CRCs with enhanced tumor mutational burden, which invokes a strong immune response in TME. Simultaneous loss of both PKCλ/ι and PKCζ results in the rapid development of serrated benign to invasive carcinoma lesions. These tumors display the highly activated mesenchyme and EMT/CAF signatures accompanied by a highly immunosuppressive TME. Loss of CDX2 in combination with alteration in other tumor suppressors (such as APC), also display the mesenchymal/stromal tumor phenotype. The loss of aPKCs or the hyperproduction of TGFβ in the TME has been proposed to skew the serrated precursors from the classical to the mesenchymal subtype.
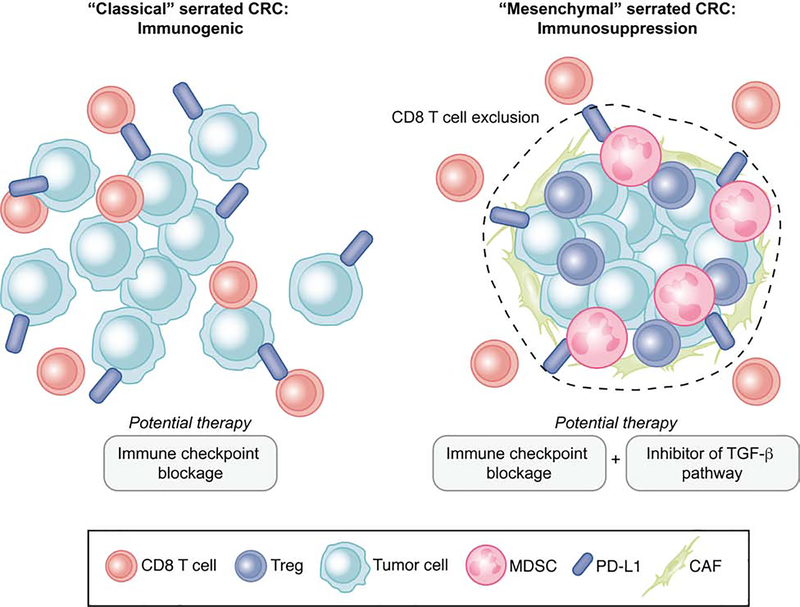
Figure 4. Immune backgrounds and potential therapeutic strategy for classical and mesenchymal serrated CRC.
(A) Tumor microenvironment (TME) of classical serrated CRC. This type of serrated tumor displays dense CD8+ T cell infiltrate (immunogenic), which is counterbalanced by the expression of checkpoint inhibitors, such as programmed cell death protein 1 ligand 1 (PD-L1). For this type of serrated CRC, the immune checkpoint inhibition therapy (ICI) inhibits the PD-1/PD-L1 interaction to reactivate the CD8 T cells, leading to tumor cell killing.
(B) In the TME of mesenchymal serrated CRC, tumor cells interact with stromal cells including cancer-associated fibroblasts (CAFs) and suppressive immune cells, such as myeloid-derived suppressor cells (MDSCs) and regulatory T (Treg) cells, to exclude cytotoxic CD8+ T cells. ICI combined with TGFβ pathway inhibitors is proposed to suppress tumorigenesis in this serrated CRC subtype.
Collectively, these data suggest that low levels of aPKC contribute to the development of serrated CRCs in the absence of BRAF or KRAS mutations [34]. This notion is further supported by data from The Cancer Genome Atlas (TCGA) that the CRC subgroup with low aPKC expression and wild-type BRAF and KRAS is enriched in a serrated gene signature [34, 53]. Furthermore, low aPKC expression associates with EMT and immunosuppressive gene expression signatures in human CRC samples irrespective of the presence of mutations in KRAS or BRAF. This highlights that the stromal and inflammatory response induced by aPKC deficiency confers aggressive mesenchymal phenotypes to CRC even in tumors harboring mutations in the conventional APC pathway [34]. Thus, the simultaneous deficiency of both aPKCs in the IECs results in aggressive serrated CRC in mice, and the reduced expression of aPKCs in human CRCs is associated with the transcriptional features of serrated tumorigenesis, stromal/mesenchymal characteristics, and poor prognosis found in the CCS3 and CMS4 patient groups. Together, these discoveries support the use of this new mouse model to investigate the mechanisms underlying the generation of the activated and immunosuppressed stroma in CRC. As discussed below, this is of potentially great therapeutic significance for the treatment of these types of tumors with immune checkpoint inhibition therapy (ICI).
Therapeutic Implications
The pathway for classical serrated CRCs seems to have been well recapitulated in the model combining BRAF mutation with deletion of the tumor suppressors p16Ink4a and p53 [32, 33, 49, 51]. Insights from these models revealed the pathobiology of initiation and development of the classical serrated tumors and have provided some therapeutic implications. In humans, there is a paradoxical situation in terms of the effectiveness of targeting the BRAF mutation in different contexts. Thus, inhibition of the BRAFV600E oncoprotein by the small molecule vemurafenib, which is highly effective in the treatment of melanoma, however, is quite ineffective in BRAFV600E-positive CRCs [54–56]. Several potential pathways have been identified as a cause of this unresponsiveness, at least in cell lines, including the activation of phosphoinositide 3-kinase (PI3K)/AKT or the reactivation of an EGFR-mediated MAPK pathway [57–59]. Rad et al. (2013) demonstrated in a high-throughput screen that murine BRAF-mutant tumors and human BRAF-mutant CRC cell lines respond similarly to many compounds [33]. They identified and validated small molecules that overcome the resistance to BRAF-inhibitor therapy in selected cell lines or across the whole cell panel (e.g., MEK and combinatorial BRAF/PI3K inhibition). This is important because several MEK, PI3K, and BRAF inhibitors are in late-stage clinical development [60–63], and these results provide a rationale for their clinical evaluation in BRAF-mutant CRCs. In addition, recent use of ICI offers the promise of harnessing the body’s own immune system to combat tumor initiation and progression in several types of cancers [31]. Unfortunately, the efficacy of ICI is very low in MSS CRCs with only approximately 10% of these patients showing a clinical benefit [64, 65]. Since MSS cancers account for 85% of all CRCs, ICI is currently limited to patients with MSI-H/deficient-MMR tumors. Although ICI has shown beneficial effects on human MSI-H CRCs [28, 31, 66], to our knowledge, the BRAF-mutant mouse models have not been used to investigate the potential effects and related mechanisms of action of ICI in MSI-H CRCs, which is one of the most salient features of this mouse model.
The potential activation of the tumor stroma and the type of immune response associated with the induction of CRC tumors in the BRAF-mutant mouse models are also still not well understood. The DKOIEC mouse model offers an excellent opportunity to address this issue due to the highly stromal and immunosuppressed characteristics of their serrated tumors [34]. That is, the DKO tumors are infiltrated by CD45+ cells that express PD-L1, which would account for the exclusion of the CD8+ T cells from the tumor [34]. However, treatment of these mice with anti-PD-LI therapy only resulted in the restoration of the CD8+ infiltration (and the concomitant reduction in the tumor burden) if they were young and the tumors had not yet become desmoplastic [34]. Treating mice with activated stromal tumors with an inhibitor of the TGFβ receptor (galunisertib) made these immune-resistant tumors susceptible to anti-PD-LI ICI [34]. This suggests that a potentially valid approach to sensitizing resistant serrated tumors to ICI is to target the tumor stroma (Figure 4). This is not just relevant for aPKC-deficient CRCs; it would also apply to other stromal and immunosuppressed models of intestinal carcinogenesis. Indeed, a similar response has been identified in a mouse model of metastasis using organoids harboring alterations in four genes: APC, KRAS, TP53, and TGFΒR2 or SMAD4 [67]. However, a significant difference between the endogenous DKOIEC tumor mouse model and the quadruple-mutant orthotopic metastasis model is that galunisertib alone was not sufficient to reduce the tumor burden of the DKOIEC mice, whereas in the quadruple-mutant model, it did reduced the tumor burden, albeit only when metastases were small [34, 67]. In aggregate, these two mouse models offer highly relevant preclinical systems to investigate the interplay between the immunological landscape and the stromal content and function in serrated and conventional CRCs. They will also inform potential combinatorial therapies aimed at reducing the resistance conferred by the stroma to ICI in CRC patients.
Concluding Remarks
There are several critical questions that arise from these studies. The first relates to the mechanisms whereby CRC cells drive immunosuppression. Is this mediated by the presence in the tumor microenvironment of high amounts of stroma-derived TGFβ, which is amply recognized as an immunosuppressive cytokine? This is unlikely to be a general cause of immunosuppression in CRCs because treatment of DKOIEC mice with galunisertib did not prevent immunosuppression, nor did it restore the infiltration of CD8+ T cells into the serrated tumors or reduce the levels of infiltrating PD-L1-expressing myeloid cells or Treg cells [34]. Interestingly, compelling evidence has been presented that the expression of mutant KRAS results in the inhibition of the IFN pathway in a mouse CRC model harboring mutation in APC, TP53, and KRAS in the intestinal epithelium (the iKAP model). Inhibition of the IFN pathway allows the expression of CXCL3, which serves as a chemoattractant for MDSCs to the tumor microenvironment [68]. Intriguingly, treatment of the iKAP mice with an inhibitor of CXCR2, the receptor for CXCL3, showed only modest efficacy and required anti-PD1 co-treatment to significantly extend their survival [68]. It is unclear why combined therapy is required in this model and might suggest that targeting the stroma like in the quadruple-mutant organoid-driven metastasis model and the DKOIEC mice might provide significant additional therapeutic benefit to the CXCR2 or anti-PD1 therapy in the iKAP system.
The second critical question that should be addressed is the cellular origin and evolution of serrated lesions. Although several potential cells of origin have been described, intestinal stem cells are believed to be a key contributor to tumor initiation in conventional type CRC by aberrant activation of the WNT/β-catenin pathway [69, 70]. However, whether this is a common cell of origin to the generation of serrated tumors that progress independently of canonical WNT activation, remains unexplored. In fact, stem cells have been shown to be lost in BRAF mutant models [71]. Considering the high cellular plasticity of the intestinal epithelium, especially under the influence of the tumor microenvironment, studies using new tracing technologies, in conjunction with single cell genomics, could shed light into a better understanding of the cell of origin in the heterogenous serrated CRCs.
Terminology for serrated lesions has changed drastically, and the best approach to differentiating between the serrated subtypes (i.e., HP, TSA, SSA/P) is still a matter of debate among endoscopists and pathologists [6, 7, 12]. Reliable longitudinal observational data for each subtype of serrated precursor will be vital for resolving this important issue, but the acquisition of such data has been hampered by the difficulties in detecting these lesions. In particular, SSA/Ps are often flat and are frequently located in the proximal (right side) colon, which makes it difficult to detect them until they have progressed to an advanced stage [13, 72]. For these reasons, serrated tumors have been proposed to be a major contributor to interval CRC (iCRC), which are CRCs diagnosed after screening or surveillance examination and before the next recommended colonoscopy [73, 74]. The prognosis of proximal (right-sided) CRC is worse than that of distal (left-sided) CRC, independent of the MSI and KRAS or BRAF mutation status [75]. Notably, proximal CRC is more resistant to anti-EGFR therapy even when BRAF and KRAS are not mutated, supporting the existence of an elusive underlying mechanism responsible for the development of serrated CRC that is independent of BRAF/KRAS mutations [76, 77]. Further improvements in detection methods and consensus on how to diagnose serrated lesions are essential for obtaining the evidence required to better characterize both serrated precursors and CRCs in terms of morphological appearance, molecular signature (classical or mesenchymal), and immune/stromal backgrounds.
Lastly, although gene expression-based stratification systems of CRCs patients, such as the CMS classification, provide great insight into the molecular mechanism of CRCs, the implementation of subtype information into the clinical practice is still at an early stage. A pending issue remains that approximately 13% of all CRCs represent either mixed or intermediate samples and cannot be properly assigned to any of the subtypes [24, 26]. Another caveat with this consensus effort is that there is not a clear integration of the serrated CRCs, which limits the robustness of these classifications. An improvement in sampling procedure and subtype assignment, together with a better understanding of the molecular drivers of serrated cancer, will definitively help discern how these tumors integrate into the global landscape of CRC. This will be important to more definitively link CMS4 tumors to poor prognosis serrated lesions. Combining longitudinal analysis of human CRCs of serrated origin with histological and molecular characterization along with a comparative analysis of the different mouse serrated models will advance our understanding of the cellular and molecular pathways driving serrated initial lesions into aggressive desmoplastic and immunosuppressed CRCs. This is an evolving field and likely better classification criteria should be generated to make this strategy valid for the clinic. In this regard, Vasaikar et al. 2019, combined the mRNA, protein, and MSI-based classifications, and recently proposed unified multi-omics subtypes (UMS) which stratify CRC patients into three subgroups, “MSI”, “CIN”, and “Mesenchymal”, corresponding to CMS1, CMS2, and CMS4 in the CMS system, respectively. Interestingly, the new UMS classification has eliminated the CMS3 subtype and assigned CMS3 tumors to other UMS categories due to the vague molecular boundary of the CMS3 subtype [78]. This suggests the invariable value of the remaining MSI, CIN, and mesenchymal phenotypes as distinct features of CRCs, and highlights the importance of integrating clinical and molecular features of tumors in order to obtain the comprehensive understanding that would to allow personalized therapy of both conventional and serrated CRCs.
Highlights.
A significant proportion of all colorectal cancers (CRC) develops through an alternative mechanism termed the “serrated” pathway.
Recent transcriptomic approaches have revealed that serrated precursor lesions give rise not only to BRAF-mutant/CIMP-H/MSI-H CRC but also to a mesenchymal-activated and immunosuppressive CRC subtype of poor prognosis.
The simultaneous loss of both atypical PKCs drives serrated colorectal cancer independently of BRAF and KRAS mutations.
Genetically engineered mouse models that precisely recapitulate each subtype of serrated CRCs provide insights into their etiologies and the development of new therapies.
Outstanding Questions.
Which are the molecular mechanisms whereby colorectal cancer cells drive immunosuppression?
Which are the oncogenic signaling pathways driving the mesenchymal phenotype and desmoplasia in aggressive colorectal cancer?
Which is the molecular and cellular origin and evolution of serrated lesions?
How to improve detection and diagnosis of serrated lesions to inform better treatments of serrated colorectal cancer?
Acknowledgments
Research in the authors’ laboratories was supported by grants from the National Institutes of Health (R01CA192642 and R01CA218254 to M.T.D.-M.; R01DK108743, R01CA207177, and R01CA211794 to J.M.), Grants-in-Aid KAKENHI 19K16712 (to Y.N.) and the grants from Shimizu Foundation for Immunology and Neuroscience and Japan Foundation for Applied Enzymology (to Y.N.). We thank Naomi Ruff for editing this manuscript.
Glossary
- Sporadic colorectal cancer
colorectal cancers that develop without involvement of identifiable inherited gene in the carcinogenesis process
- Conventional pathway
the pathway that precursor lesions follow to develop into colorectal cancer that is predominately initiated by the inactivation of the Apc tumor-suppressor gene and results in a tubular adenoma histology
- Alternative pathway
the pathway that leads to colorectal cancer that is initiated by the formation of serrated adenomas or polyps and is mostly associated with the activation of the ERK cascade
- Sessile Serrated Adenomas or Polyps (SSA/Ps)
these are premalignant precursor lesions that develop through the alternative pathway to lead to serrated colorectal cancer. They have a flat morphology and therefore, they are difficult to detect. Histologically, they display a characteristic so-called saw-tooth or stellate architecture of the intestinal crypt
- CpG island methylation phenotype (CIMP)
tumor phenotype defined as hypermethylation of CpG islands in promoter regions of genes, which leads to epigenetic silencing of tumor suppressor genes
- Microsatellite instability (MSI)
tumor phenotype characterized by a near-diploid genome and instability in the form of insertions and deletions in microsatellite regions owing to a deficiency in DNA mismatch-repair genes. Microsatellite stable (MSS) tumors indicates a tumor subgroup without MSI
- Epithelial-mesenchymal transition (EMT)
physiological process in which cancer cells lose their characteristics, such as polarity and cell-cell adhesion, and gain mesenchymal and migratory features that allow them to disseminate to distant organs
- Immune checkpoint inhibition therapy (ICI)
treatments that use the immune system to fight cancer by targeting immune checkpoints, which are immune inhibitory molecules or pathways to keep T cells in standby mode. The most prominent examples are anti-programmed death receptor
Footnotes
Disclaimer Statement
J. Moscat, M.T. Diaz-Meco and Y. Nakanishi are inventors in a patent related to serrated colorectal cancer. J. Moscat and M.T. Diaz-Meco report receiving a commercial research grant from Halozyme Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Siegel RL et al. (2019) Cancer statistics, 2019. CA Cancer J Clin 69 (1), 7–34. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER and Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61 (5), 759–67. [DOI] [PubMed] [Google Scholar]
- 3.Jass JR (2001) Serrated route to colorectal cancer: back street or super highway? J Pathol 193 (3), 283–5. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B et al. (1988) Genetic alterations during colorectal-tumor development. N Engl J Med 319 (9), 525–32. [DOI] [PubMed] [Google Scholar]
- 5.Jass JR and Smith M (1992) Sialic acid and epithelial differentiation in colorectal polyps and cancer--a morphological, mucin and lectin histochemical study. Pathology 24 (4), 233–42. [DOI] [PubMed] [Google Scholar]
- 6.Snover DC (2011) Update on the serrated pathway to colorectal carcinoma. Hum Pathol 42 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- 7.IJspeert J et al. (2015) Serrated neoplasia-role in colorectal carcinogenesis and clinical implications. Nat Rev Gastroenterol Hepatol 12 (7), 401–9. [DOI] [PubMed] [Google Scholar]
- 8.Kane MF et al. (1997) Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 57 (5), 808–11. [PubMed] [Google Scholar]
- 9.Cunningham JM et al. (1998) Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res 58 (15), 3455–60. [PubMed] [Google Scholar]
- 10.Herman JG et al. (1998) Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A 95 (12), 6870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jass JR (2007) Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 50 (1), 113–30. [DOI] [PubMed] [Google Scholar]
- 12.Bettington M et al. (2013) The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology 62 (3), 367–86. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Solano J et al. (2010) Clinicopathologic study of 85 colorectal serrated adenocarcinomas: further insights into the full recognition of a new subset of colorectal carcinoma. Hum Pathol 41 (10), 1359–68. [DOI] [PubMed] [Google Scholar]
- 14.Rex DK et al. (2012) Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol 107 (9), 1315–29; quiz 1314, 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makinen MJ et al. (2001) Colorectal carcinoma associated with serrated adenoma--prevalence, histological features, and prognosis. J Pathol 193 (3), 286–94. [DOI] [PubMed] [Google Scholar]
- 16.Tuppurainen K et al. (2005) Morphology and microsatellite instability in sporadic serrated and non-serrated colorectal cancer. J Pathol 207 (3), 285–94. [DOI] [PubMed] [Google Scholar]
- 17.Schlicker A et al. (2012) Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genomics 5, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadanandam A et al. (2013) A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med 19 (5), 619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marisa L et al. (2013) Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 10 (5), e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Sousa EMF et al. (2013) Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med 19 (5), 614–8. [DOI] [PubMed] [Google Scholar]
- 21.Budinska E et al. (2013) Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol 231 (1), 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roepman P et al. (2014) Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer 134 (3), 552–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isella C et al. (2015) Stromal contribution to the colorectal cancer transcriptome. Nat Genet 47 (4), 312–9. [DOI] [PubMed] [Google Scholar]
- 24.Guinney J et al. (2015) The consensus molecular subtypes of colorectal cancer. Nat Med 21 (11), 1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fessler E et al. (2016) TGFβeta signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol Med 8 (7), 745–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fessler E and Medema JP (2016) Colorectal Cancer Subtypes: Developmental Origin and Microenvironmental Regulation. Trends Cancer 2 (9), 505–518. [DOI] [PubMed] [Google Scholar]
- 27.Robert C et al. (2011) Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med 364 (26), 2517–26. [DOI] [PubMed] [Google Scholar]
- 28.Topalian SL et al. (2012) Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 366 (26), 2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ansell SM et al. (2015) PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med 372 (4), 311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borghaei H et al. (2015) Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 373 (17), 1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le DT et al. (2015) PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372 (26), 2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carragher LA et al. (2010) V600EBraf induces gastrointestinal crypt senescence and promotes tumour progression through enhanced CpG methylation of p16INK4a. EMBO Mol Med 2 (11), 458–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rad R et al. (2013) A genetic progression model of Braf(V600E)-induced intestinal tumorigenesis reveals targets for therapeutic intervention. Cancer Cell 24 (1), 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakanishi Y et al. (2018) Simultaneous Loss of Both Atypical Protein Kinase C Genes in the Intestinal Epithelium Drives Serrated Intestinal Cancer by Impairing Immunosurveillance. Immunity 49 (6), 1132–1147 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L et al. (2003) BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res 63 (17), 5209–12. [PubMed] [Google Scholar]
- 36.Kambara T et al. (2004) BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 53 (8), 1137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koinuma K et al. (2004) Mutations of BRAF are associated with extensive hMLH1 promoter methylation in sporadic colorectal carcinomas. Int J Cancer 108 (2), 237–42. [DOI] [PubMed] [Google Scholar]
- 38.Minoo P et al. (2007) Role of BRAF-V600E in the serrated pathway of colorectal tumourigenesis. J Pathol 212 (2), 124–33. [DOI] [PubMed] [Google Scholar]
- 39.Toyota M et al. (1999) CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 96 (15), 8681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weisenberger DJ et al. (2006) CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38 (7), 787–93. [DOI] [PubMed] [Google Scholar]
- 41.Park SJ et al. (2003) Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol 162 (3), 815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao Y et al. (2019) Aging-like Spontaneous Epigenetic Silencing Facilitates Wnt Activation, Stemness, and Braf(V600E)-Induced Tumorigenesis. Cancer Cell 35 (2), 315–328 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch HT et al. (2015) Milestones of Lynch syndrome: 1895–2015. Nat Rev Cancer 15 (3), 181–94. [DOI] [PubMed] [Google Scholar]
- 44.Vleugels JLA et al. (2018) Endoscopic detection rate of sessile serrated lesions in Lynch syndrome patients is comparable with an age- and gender-matched control population: case-control study with expert pathology review. Gastrointest Endosc 87 (5), 1289–1296. [DOI] [PubMed] [Google Scholar]
- 45.Andersen SH et al. (2008) Sessile serrated polyps of the colorectum are rare in patients with Lynch syndrome and in familial colorectal cancer families. Fam Cancer 7 (2), 157–62. [DOI] [PubMed] [Google Scholar]
- 46.Stefanius K et al. (2011) Frequent mutations of KRAS in addition to BRAF in colorectal serrated adenocarcinoma. Histopathology 58 (5), 679–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang CS et al. (2011) The clinical significance of serrated polyps. Am J Gastroenterol 106 (2), 229–40; quiz 241. [DOI] [PubMed] [Google Scholar]
- 48.Bennecke M et al. (2010) Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell 18 (2), 135–46. [DOI] [PubMed] [Google Scholar]
- 49.Bond CE et al. (2018) Oncogenic BRAF mutation induces DNA methylation changes in a murine model for human serrated colorectal neoplasia. Epigenetics 13 (1), 40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lannagan TRM et al. (2019) Genetic editing of colonic organoids provides a molecularly distinct and orthotopic preclinical model of serrated carcinogenesis. Gut 68 (4), 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakamoto N et al. (2017) BRAF(V600E) cooperates with CDX2 inactivation to promote serrated colorectal tumorigenesis. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balbinot C et al. (2018) The Cdx2 homeobox gene suppresses intestinal tumorigenesis through non-cell-autonomous mechanisms. J Exp Med 215 (3), 911–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cancer Genome Atlas N (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487 (7407), 330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richman SD et al. (2009) KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 27 (35), 5931–7. [DOI] [PubMed] [Google Scholar]
- 55.Roth AD et al. (2010) Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60–00 trial. J Clin Oncol 28 (3), 466–74. [DOI] [PubMed] [Google Scholar]
- 56.Kopetz S et al. (2015) Phase II Pilot Study of Vemurafenib in Patients With Metastatic BRAF-Mutated Colorectal Cancer. J Clin Oncol 33 (34), 4032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prahallad A et al. (2012) Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483 (7387), 100–3. [DOI] [PubMed] [Google Scholar]
- 58.Corcoran RB et al. (2012) EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2 (3), 227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mao M et al. (2013) Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin Cancer Res 19 (3), 657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chappell WH et al. (2011) Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget 2 (3), 135–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flaherty KT et al. (2012) Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 367 (18), 1694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flaherty KT et al. (2012) Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 367 (2), 107–14. [DOI] [PubMed] [Google Scholar]
- 63.Rusconi P et al. (2012) RAS/RAF/MEK inhibitors in oncology. Curr Med Chem 19 (8), 1164–76. [DOI] [PubMed] [Google Scholar]
- 64.Le DT et al. (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357 (6349), 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Llosa NJ et al. (2019) Intratumoral Adaptive Immunosuppression and Type 17 Immunity in Mismatch Repair Proficient Colorectal Tumors. Clin Cancer Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Overman MJ et al. (2017) Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol 18 (9), 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tauriello DVF et al. (2018) TGFβeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554 (7693), 538–543. [DOI] [PubMed] [Google Scholar]
- 68.Liao W et al. (2019) KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell 35 (4), 559–572 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barker N (2014) Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 15 (1), 19–33. [DOI] [PubMed] [Google Scholar]
- 70.Medema JP and Vermeulen L (2011) Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature 474 (7351), 318–26. [DOI] [PubMed] [Google Scholar]
- 71.Tong K et al. (2017) Degree of Tissue Differentiation Dictates Susceptibility to BRAF-Driven Colorectal Cancer. Cell Rep 21 (13), 3833–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thibodeau SN et al. (1993) Microsatellite instability in cancer of the proximal colon. Science 260 (5109), 816–9. [DOI] [PubMed] [Google Scholar]
- 73.Shin SP (2017) Sessile Serrated Adenoma; the Hard-to-Catch Culprit of Interval Cancer. Clin Endosc 50 (3), 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee YM and Huh KC (2017) Clinical and Biological Features of Interval Colorectal Cancer. Clin Endosc 50 (3), 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee MS et al. (2017) Right Versus Left Colon Cancer Biology: Integrating the Consensus Molecular Subtypes. J Natl Compr Canc Netw 15 (3), 411–419. [DOI] [PubMed] [Google Scholar]
- 76.Missiaglia E et al. (2014) Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol 25 (10), 1995–2001. [DOI] [PubMed] [Google Scholar]
- 77.Moretto R et al. (2016) Location of Primary Tumor and Benefit From Anti-Epidermal Growth Factor Receptor Monoclonal Antibodies in Patients With RAS and BRAF Wild-Type Metastatic Colorectal Cancer. Oncologist 21 (8), 988–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vasaikar S et al. (2019) Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities. Cell 177 (4), 1035–1049 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reina-Campos M et al. (2019) The Dual Roles of the Atypical Protein Kinase Cs in Cancer. Cancer Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moscat J et al. (2016) p62 in Cancer: Signaling Adaptor Beyond Autophagy. Cell 167 (3), 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakanishi Y et al. (2016) Control of Paneth Cell Fate, Intestinal Inflammation, and Tumorigenesis by PKClambda/iota. Cell Rep 16 (12), 3297–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wald FA et al. (2011) Aberrant expression of the polarity complex atypical PKC and non-muscle myosin IIA in active and inactive inflammatory bowel disease. Virchows Arch 459 (3), 331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma L et al. (2013) Control of nutrient stress-induced metabolic reprogramming by PKCzeta in tumorigenesis. Cell 152 (3), 599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Galvez AS et al. (2009) Protein kinase Czeta represses the interleukin-6 promoter and impairs tumorigenesis in vivo. Mol Cell Biol 29 (1), 104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wood LD et al. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318 (5853), 1108–13. [DOI] [PubMed] [Google Scholar]
- 86.Liado V et al. (2015) Repression of Intestinal Stem Cell Function and Tumorigenesis through Direct Phosphorylation of beta-Catenin and Yap by PKCzeta. Cell Rep 10 (5), 740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shelton PM et al. (2018) The Secretion of miR-200s by a PKCzeta/ADAR2 Signaling Axis Promotes Liver Metastasis in Colorectal Cancer. Cell Rep 23 (4), 1178–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]