Abstract
Background
Hepatocellular carcinoma (HCC) is the most common primary liver cancer in the world, with a high degree of malignancy and recurrence. The influence of the ceRNA network in tumor on the biological function of liver cancer is very important, It has been reported that many lncRNA play a key role in liver cancer development. In our study, integrated data analysis revealed potential eight novel lncRNA biomarkers in hepatocellular carcinoma.
Methods
Transcriptome data and clinical data were downloaded from the The Cancer Genome Atlas (TCGA) data portal. Weighted gene co-expression network analysis was performed to identify the expression pattern of genes in liver cancer. Then, the ceRNA network was constructed using transcriptome data.
Results
The integrated analysis of miRNA and RNAseq in the database show eight novel lncRNAs that may be involved in important biological pathways, including TNM and disease development in liver cancer. We performed function enrichment analysis of mRNAs affected by these lncRNAs.
Conclusions
By identifying the ceRNA network and the lncRNAs that affect liver cancer, we showed that eight novel lncRNAs play an important role in the development and progress of liver cancer.
Keywords: lncRNA, Micrornas, Diagnostic biomarkers, HCC
Introduction
Liver cancer, which is most common among male patients, is the high leading cause of death in men worldwide (Wang et al., 2015). In the United States, there are as many as 42,030 new cases and 31,780 deaths related to liver cancer every year, based on the latest statistics record (Siegel, Miller & Jemal, 2019). The degree of malignancy of cancer can be determined by study of the histology of the tumor, and patients can be divided into three classes, low, medium, and high according to the degree of malignancy. Clinically, primary liver carcinoma is considered to be one of the most common malignant tumors, and about 90% of these tumors are hepatocellular carcinoma (HCC). In cases of patients diagnosed with HCC, 50.7% of them achieve a 5-year survival rate, while the median survival time is 60 months (Lee et al., 2006). The prognosis of HCC patients is related to the patient’s disease stage. Currently, the tumor-node-metastasis (TNM) pathological staging standard developed by the American Joint Committee on Cancer (AJCC) is the most commonly used malignant tumor staging system worldwide. However, the relationship between long non-coding RNAs (lncRNAs) and tumor staging has raised concern among researchers (Chen et al., 2015; Ou et al., 2018; Yao et al., 2018), suggesting that many lncRNAs play progressive key roles in HCC development (Chen et al., 2016a). For example, the MALAT-1 gene is upregulated in HCC and also correlates with prognostics and recurrence (Guerrieri, 2015; Lai et al., 2012); the overexpression of the HULC gene may reduce the mir-372 gene, while at the same time may promote reprogramming during tumorigenesis (Du et al., 2012); and DANCR induces stemness features and could serve as a potential prognostic marker and therapeutic target for HCC (Yuan et al., 2016). Previous studies have mainly focused on the single biomarker use of miRNAs in HCC.
The weighted gene co-expression network analysis (WGCNA) is a popular bioinformatic method used in the construction of gene networks and the detection of gene modules and their phenotypic traits (Langfelder & Horvath, 2008; Yin et al., 2018). In this study, we identified eight novel lncRNAs correlated with TNM using WGCNA. Additionally, functional enrichment analysis shows that these eight novel lncRNAs play an important role in the regulation of gene expression that affect development and progression of liver cancer.
Materials and Methods
Data preprocessing and differential gene selection
The clinical data, RNAseq data and microRNA data of LIHC were downloaded from the The Cancer Genome Atlas (TCGA) database (Akbani et al., 2014) (Table S1). There were 49 pairs of microRNA samples and 50 pairs of RNAseq samples in total. The variation between the RNA and microRNAs data was calculated using EdgeR package (Reimers & Carey, 2006; Varet et al., 2016). Only microRNAs recorded with first 10 and last 10 FC values were selected for subsequent analysis (Li et al., 2018; Shao & Li, 2019). Because these biggest expression changed miRNA may have real major function in HCC. For RNAseq data, the EdgeR package was used to calculate the differential correlationship, and the threshold value for FDR was set at <0.01, — logFC —>1 (Table S2). Clinical data were used to calculate the correlation matrix of clinical information for integrated analysis. We used the “heatmap.2” function in the “gplots” package to create the heatmap.
Determination of ceRNA
Using microRNAs and differential expression genes as input data for target prediction, the RNA22 program was used to predict binding sites of microRNAs based on their sequence characteristics (Loher & Rigoutsos, 2012). Based on these ceRNA interactions, we obtained all the mRNA-miRNA pairs with sharing the number of common miRNAs. Because the number of the ceRNA pairs is obey hypergeometric distribution. We estimated their statistical significance by a hypergeometric test. The potential top 20 different expression miRNA with hypergeometric test in ceRNA network P value less than 0.05 was obtained (Li et al., 2018; Shao & Li, 2019). Specific formulas, such as the differential expression matrix of lncRNA, were used to get the Pearson correlation coefficient (PCC) (Table S3). Based on ceRNA’s mechanism, the expression matrix (EM) for lncRNA to bind between themselves resulted in a PCC of EM >0. Co-expression is one of the features of ceRNA network on account of their interactions. The final ceRNA pair was obtained by intersecting the ceRNA threshold with cutoff p-value <0.05. Cytoscape v3.0 was used to construct the ceRNA network. The overall Kaplan–Meier (KM) survival analysis in each subtype was performed using GEPIA2 database (Tang, 2017).
WGCNA model related computing
Weighted gene co-expression network analysis (WGCNA), a bioinformatic method used to find correlation patterns among genes, was utilized in this research. WGCNA assumes that gene expression networks is scale-free, it uses a ‘soft’ threshold to determine the weights of the edges connecting genes and merge individual genes to a module. An appropriate soft threshold will make the co-expression network closer to a scale-free network. Then we constructed a signed weighted co-expression network using WGCNA based on the gene expression value across the TCGA samples. We obtained 60 co-expression modules according to the correlations of fpkm value among samples. Each module is represented by an value belongs to the ’eigengene’. This value is identified from the principal component analysis (PCA) of all the gene expression value in the module. Then, we find the relationship between modules and the trait, Eventually, unsupervised WGCNA identified major lncRNAs expression modules with different degrees of correlation to TNM staging using WGCNA R software (Langfelder & Horvath, 2008).
Prediction of interrelationship between lncRNA-related mRNAs
Using RNAseq data from TCGA, the correlation prediction of the lncRNA-lncRNA network and lncRNA-mRNA network was constructed. The cutoff of PCC was 0.7. We retained the intersection with the lncRNA using a cutoff of the top 20% degree. Ultimately, we found eight potentially important lncRNAs with high degrees of connection in the ceRNA network and high correlation at the expression level.
Gene ontology and KEGG enrichment analysis
Gene ontology (GO) annotation analysis was performed using DAVID software (Huang da, Sherman & Lempicki, 2009a). Gene functions for these important indirectly regulated mRNA genes elucidate that this mRNA regulated by lncRNA may have some key biological functions (Huang da, Sherman & Lempicki, 2009a; Huang da, Sherman & Lempicki, 2009b). String database was used for protein-protein analysis (Szklarczyk et al., 2017).
Results
Analysis of differential miRNAs and differential lncRNAs
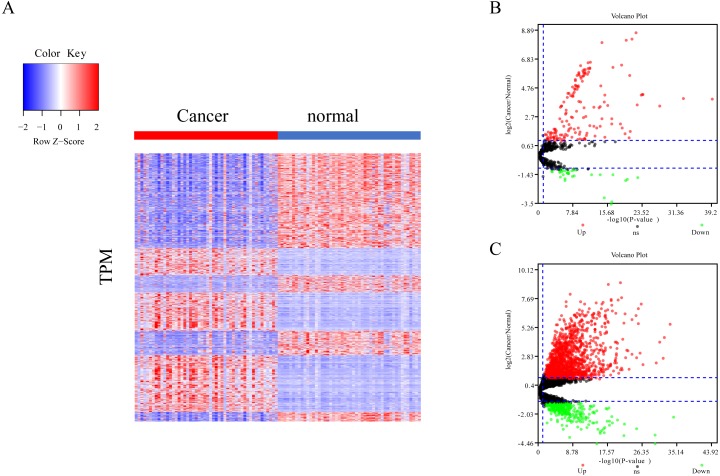
The differential expression of miRNAs and lncRNAs between normal samples and cancer samples was calculated separately using the EdgeR package (Chen et al., 2017; Law et al., 2016; Maza, 2016). There were 1962 significant differential expression genes in lncRNAs, and 310 differentially expressed miRNA genes found between healthy and cancer-treated samples. Almost half of the mRNAs are upregulated and half downregulated (Fig. 1A). Most of the gene expression for lncRNAs and miRNAs was upregulated in cancer-treated samples (Figs. 1B–1C).
Figure 1. Different expression of mRNA, miRNA and lncRNA levels between tumor and normal samples.
(A) Heatmap showing 2,763 different expression of mRNA between tumor and normal samples in LIHC. (B) Volcano map for 310 different expression miRNA. Dots in red and green indicate high and low expression of miRNA in cancer, respectively. (C) Volcano map for 1,962 different LncRNA. Dots in red and green indicate high and low expression of lncRNA in cancer, respectively.
Weighted gene co-expression network analysis of lncRNA
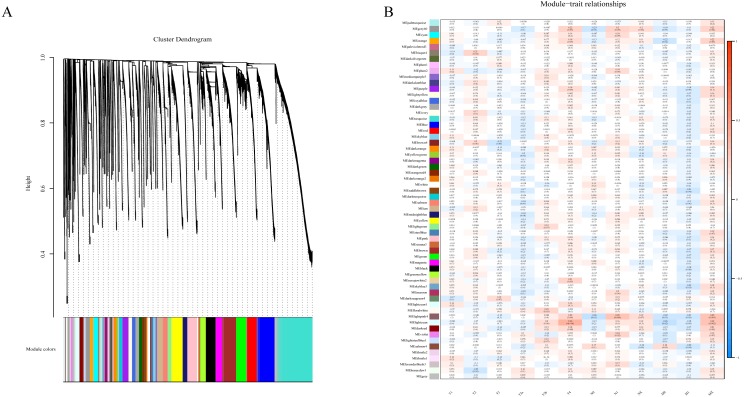
To determine if any of the identified coexpression modules were associated with TNM stage, we calculated the PCC between the MEs and TNM stage. All lncRNAs were merged to 60 modules according the degree of coexpression across the data set in WGCNA. As in the previous study, we assigned each coexpression module an arbitrary color for reference (Di et al., 2019; Zhussupbekova et al., 2016). The hierarchical clustering dendrogram of the eigengenes shows the module size (the number of genes per module) and relationships among these modules (Fig. 2A). Most modules had minimal relationships with each other.
Figure 2. Results of Weight Gene Co-expression Network Analysis (WGCNA).
(A) shows the clustering dendrogram of co-expression lncRNA based on topological overlap. (B) Module-TNM stage correlative analysis. Each row corresponds to a module eigengene, each column corresponds to a TNM stage. Heatmap block with p-values and correlation coefficient. The red box in the figure shows the module with higher correlation coefficient in the three stages of TNM. The blue box in the figure shows the module with negative correlation coefficient in the three stages of TNM.
Comparison of module-characteristic eigengenes showed TNM was best correlated with the module MElightpink4 (p = 4E–04) and MElightcyan (P < 0.006), composed of 125 lncRNAs (Fig. 2B). The PCC values ranged from −1 to +1 depending on the power of the relationship. A positive value indicated that the lncRNA within a particular co-expression module increased as the TNM increased, whereas the opposite occurred if the sign of the PCC was negative. We learned that the correlationship between the module and TNM stage with PCC value was accompanied by the corresponding P-value in brackets. These modules included genes that were co-expressed in a particular TNM stage can be used to represent the TNM stage of HCC development (Fig. 2B). These gene may be the risk factors and therapeutic targets in the treatment of HCC.
ceRNA network of lncRNA reveals potential biomarkers in liver cancer
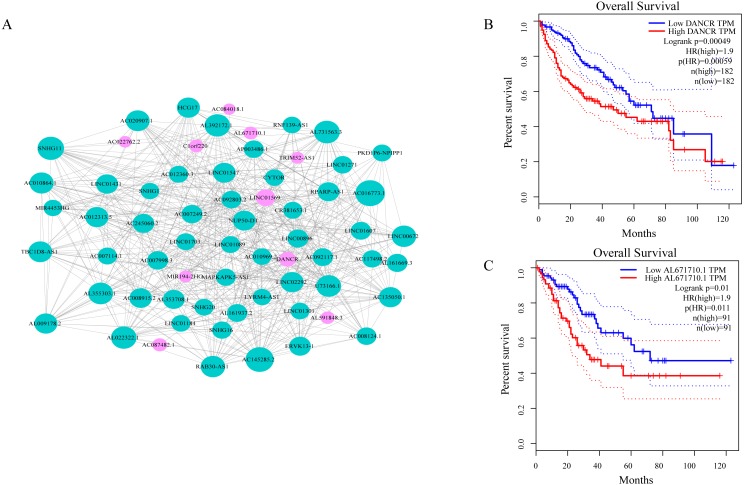
As shown in Fig. 3A, ceRNA network was constructed by high degree lncRNAs. The topological characteristics of ceRNA network were analyzed with degree, betweenness, and closeness (Table S4). Many genes in ceRNA network are with high degree, betweenness, or closeness like AC016773.1, AC145285.2, LINC01569 and DANCR, and so on. This implied ceRNA network may regulate many gene expression through these lncRNAs to have effect on progression of liver cancer. To verify the function of these lncRNAs, Kaplan–Meier survival analysis was perform ed for expression level of these lncRNAs. The results of the survival analysis presented in Figs. 3B and 3C show patients with high expression level of DANCR or AL671710.1 have poor prognosis. It is a validation of our analysis and two of the lncRNAs may affect prognosis.
Figure 3. CeRNA network in LIHC and survival curves.
(A) Red dots represent overlaping lncRNA selected from co-expression analysis and RNA22 which can identify microRNA binding sites. The bigger the dots, the higher the degree value, the more important nodes in the network. (B) Survival curve of patients with high expression of DANCR and low expression of DANCR. Survival curve of patients with high expression of AL671710.1 and low expression of AL671710.1.
The interactions between lncRNAs and mRNAs
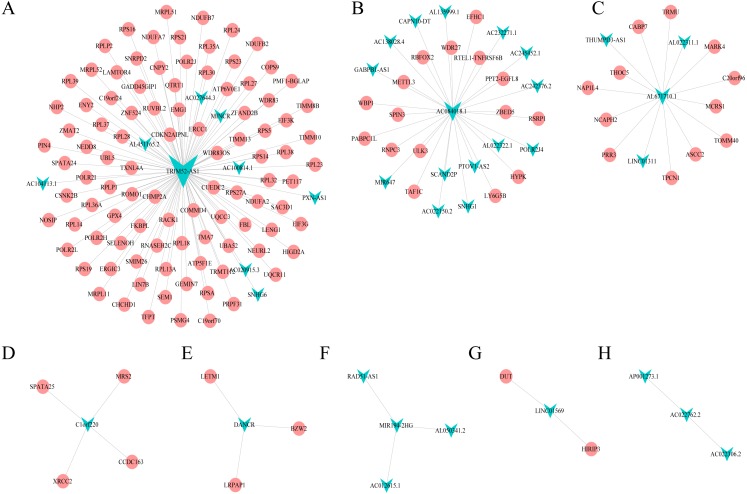
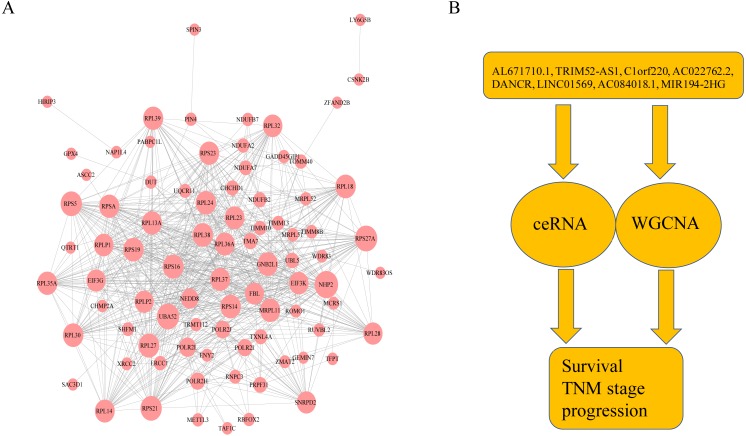
Combining WGCNA with the results of ceRNA prediction analysis, lncRNA was combined with correlation prediction to find eight important lncRNAs including AL671710.1, TRIM52-AS1, C1orf220, AC022762.2, DANCR, LINC01569, AC084018.1 and MIR194-2HG. Among these lncRNAs, downregulation of TRIM52-AS1 play key role in renal cell carcinoma (Liu et al., 2016). DANCR increases stemness features of hepatocellular carcinoma (Yuan et al., 2016). DANCR is also associated with various cancer (Lu et al., 2018a; Lu et al., 2018b; Mao et al., 2017; Sha et al., 2017; Xu et al., 2018; Yuan et al., 2016). However, there is little known about the function of other six lncRNAs in cancer. There were 124 genes targeted by these eight lncRNAs (Figs. 4A–4H), including EIF3 which functions during the initiation phase of translation. TRIM52-AS1 may influence cancer behavior and function through interactions with regulator EIF3. EIF3 plays a key role in human diseases (Gomes-Duarte et al., 2018; Valasek et al., 2017). Also, there were 30 genes targeted by lncRNA ac084018.1, including m6A reader methyltransferase like 3 (METTL3). As reported in previous research, METTL3 promotes liver cancer progression through YTHDF2 (Balacco & Soller, 2019; Berlivet et al., 2019; Chen et al., 2018; Weng et al., 2018). DANCR and AL671710.1 also have crucial roles through certain key genes (Figs. 4C, 4E).
Figure 4. Interaction of eight lncRNA and target mRNA.
The green arrows represent the lncRNA, and the pale red dots represent the mRNA. It can be seen that the eight important lncRNA AL6717.1, TRIM52-AS1, C1220C1, DANCR, LINC01569, AC084018.1 participated in many regulation of mRNA.
GO and KEGG pathway enrichment analyses of lncRNA-targeted gene
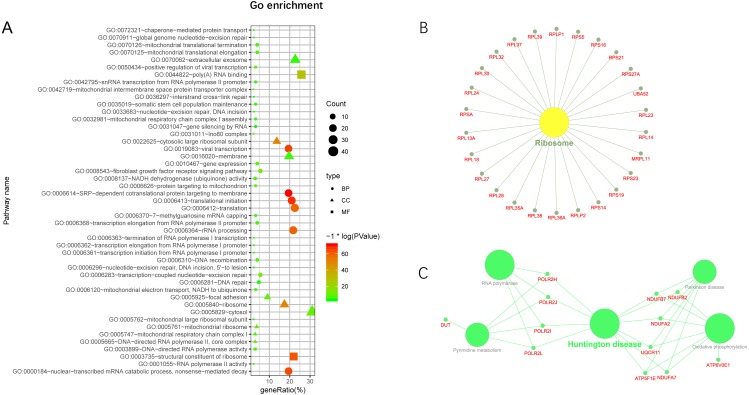
Additionally, we performed GO and KEGG enrichment analysis of the mRNAs in the network (Figs. 5A–5C). We analysed target genes of the lncRNA based on their enrichment scores for associated GO terms and KEGG pathways using David tools (Huang da, Sherman & Lempicki, 2009a; Huang da, Sherman & Lempicki, 2009b) . The GO and KEGG enrichment analysis concerning the target genes of lncRNAs indicated that the top regulated pathways of lncRNAs were Huntington’s disease, RNA polymerase and Pyrimidine metabolism, and the top regulated functions of lncRNAs were SRP-dependent co-translational protein targeting to membrane, translational initiation, viral transcription, nuclear-transcribed mRNA catabolic process, and nonsense-mediated decay. Further-more, those essential mRNA may interact with each other and function in HCC (Fig. 6A). We can conclude that those lncRNAs affect HCC through the functions and pathways listed above (the flow chart was depicted in Fig. 6B).
Figure 5. GO and KEGG pathway enrichment analysis of target gene.
(A) GO enrichment of mRNA interact with lncRNA. Points of different shapes represent BP, CC and MF of GO, and the size of points represents the number of gene enriched in the GO function; the number of different colors represents P value. (B) The KEGG pathway is enriched in ribosome and in Huntington’s disease.
Figure 6. The protein–protein interactions of RNA and conclusion summary.
(A) The PPI network of RNA with degree bigger than 10 in string database. The larger the dots in the graph, the more interaction between the RNA corresponding proteins and other proteins. (B) Conclusion summary.
Discussion
Hepatocellular carcinoma (HCC) is one of the primary causes of cancer-related death worldwide (Balogh et al., 2016). Many genes influence HCC, TP53 tumor-suppressor gene, p16INK4A and Rb-associated with various risk factors have been largely reported (Bae et al., 2016; Buendia, 2000; Nishida & Fukuda, 2001; Peng et al., 2013; Wang et al., 2017; Zhang et al., 2014). Some even take part in the cancer biography progress through lncRNA (Su et al., 2017). Long non-coding RNAs (lncRNAs) which are transcribed but do not encode proteins, play key roles in HCC development (Abbastabar et al., 2018). These include MALAT-1 and also NEAT-2, which regulates splicing factors mostly situated in nuclear speckles. In addition, MALAT-1 is a biomarker in various cancers including HCC (Lai et al., 2012; Wang et al., 2016). LncRNA GAS5 is a biomarker and have potential applications in HCC therapy (Fang et al., 2019). Using WGCNA and hypergeometric test analysis, we found eight lncRNAs with important functions: AL671710.1, TRIM52-AS1, C1orf220, AC022762.2, DANCR, LINC01569, AC084018.1 and MIR194-2HG. TRIM52-AS1 is one of the eight lncRNAs that has been reported as a function of a tumor suppressor (Liu et al., 2016; Zhang et al., 2017).Targeted by MYC, DANCR promotes cancer (Chen et al., 2016b; Dhanasekaran et al., 2017; Huang, Deng & Zhou, 2013; Kron et al., 2012; Lu et al., 2018b). Taken together, we concluded that these lncRNAs may function as a potential tumor regulator in HCC.
Additionally, some lncRNAs were associated with the TNM stage in HCC tissues (Abbastabar et al., 2018). The American Joint Committee on Cancer (AJCC) stratifies patients using a Tumor-Node-Metastasis (TNM) classification, representing a group of models useful in the assessment of tumor extension (Selcuk, 2017; Tellapuri et al., 2018). Among several staging systems, the TNM system was one of the most widely accepted, and had a higher prognostic competency than the other systems (Prognosis Evaluation in Patients with Hepatocellular Carcinoma after Hepatectomy, Comparison of BCLC and Hangzhou Criteria Staging Systems). In our work, we found two lncRNAs modules associated with the TNM stage. Those lncRNAs may function as the biomarker of node size and metastasis status in HCC. Systematic analysis of transcriptomics data reveal those novel potential therapeutic target may be involved in cancer-related pathway in liver cancer. Our study has limitations, the specific mechanism of these lncRNAs remains unexplored.
Conclusions
In summary, our results demonstrated that lncRNAs AL671710.1, TRIM52-AS1, C1orf220, AC022762.2, DANCR, LINC01569, AC084018.1, and MIR194-2HG play an essential role in the HCC stage, and their targeted mRNA have key functions in HCC. Those lncRNAs might be a novel prognostic biomarker for HCC.
Supplemental Information
Funding Statement
This work was supported by National Natural Science Foundation of China (No. 81660399 and 81860423), Yunnan Provincial Clinical Center of Hepato-biliary-pancreatic Diseases [no specific number], the Doctor Newcomer Award of Yunnan Province in 2017 [no specific number], and Ph.D. Student Innovation Fund of Kunming Medical University (No. 2019D004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Sanqi An, Email: ansq@mail2.sysu.edu.cn, ansanqi2016@gmail.com.
Lin Wang, Email: wanglinfey@126.com.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Ren-chao Zou performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Zhi-tian Shi performed the experiments, analyzed the data, prepared figures and/or tables, approved the final draft.
Shu-feng Xiao analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, approved the final draft.
Yang Ke analyzed the data, prepared figures and/or tables, approved the final draft.
Hao-ran Tang analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Tian-gen Wu analyzed the data, prepared figures and/or tables, approved the final draft, review and editing.
Zhi-tang Guo prepared figures and/or tables, approved the final draft, validation.
Fan Ni analyzed the data, prepared figures and/or tables, approved the final draft, data curation.
Sanqi An conceived and designed the experiments, authored or reviewed drafts of the paper, approved the final draft.
Lin Wang conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplementary Files.
References
- Abbastabar et al. (2018).Abbastabar M, Sarfi M, Golestani A, Khalili E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI Journal. 2018;17:900–913. doi: 10.17179/excli2018-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbani et al. (2014).Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, Liu W, Yang JY, Yoshihara K, Li J, Ling S, Seviour EG, Ram PT, Minna JD, Diao L, Tong P, Heymach JV, Hill SM, Dondelinger F, Stadler N, Byers LA, Meric-Bernstam F, Weinstein JN, Broom BM, Verhaak RG, Liang H, Mukherjee S, Lu Y, Mills GB. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nature Communications. 2014;5 doi: 10.1038/ncomms4887. Article 3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae et al. (2016).Bae JS, Noh SJ, Kim KM, Jang KY, Park HS, Chung MJ, Park BH, Moon WS. PIN1 in hepatocellular carcinoma is associated with TP53 gene status. Oncology Reports. 2016;36:2405–2411. doi: 10.3892/or.2016.5001. [DOI] [PubMed] [Google Scholar]
- Balacco & Soller (2019).Balacco DL, Soller M. The m(6)A writer: rise of a machine for growing tasks. Biochemistry. 2019;58:363–378. doi: 10.1021/acs.biochem.8b01166. [DOI] [PubMed] [Google Scholar]
- Balogh et al. (2016).Balogh J, Victor 3rd D, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM, Monsour Jr HP. Hepatocellular carcinoma: a review. Journal of Hepatocellular Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlivet et al. (2019).Berlivet S, Scutenaire J, Deragon JM, Bousquet-Antonelli C. Readers of the m(6)A epitranscriptomic code. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2019;1862(3):329–342. doi: 10.1016/j.bbagrm.2018.12.008. [DOI] [PubMed] [Google Scholar]
- Buendia (2000).Buendia MA. Genetics of hepatocellular carcinoma. Seminars in Cancer Biology. 2000;10(3):185–200. doi: 10.1006/scbi.2000.0319. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2016a).Chen D, Sun Q, Cheng XF, Zhang LF, Song W, Zhou DK, Lin JJ, Wang WL. Genome-wide analysis of long noncoding RNA (lncRNA) expression in colorectal cancer tissues from patients with liver metastasis. Cancer Medicine. 2016a;5:1629–1639. doi: 10.1002/cam4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2018).Chen MN, Wei L, Law CT, Tsang FHC, Shen JL, Cheng CLH, Tsang LH, Ho DWH, Chiu DKC, Lee JMF, Wong CCL, Ng IOL, Wong CM. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2015).Chen T, Xie W, Xie L, Sun Y, Zhang Y, Shen Z, Sha N, Xu H, Wu Z, Hu H, Wu C. Expression of long noncoding RNA lncRNA-n336928 is correlated with tumor stage and grade and overall survival in bladder cancer. Biochemical and Biophysical Research Communications. 2015;468:666–670. doi: 10.1016/j.bbrc.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2016b).Chen YM, Lin CZ, Liu Y, Jiang Y. HMGB1 promotes HCC progression partly by downregulating p21 via ERK/c-Myc pathway and upregulating MMP-2. Tumor Biology. 2016b;37:4399–4408. doi: 10.1007/s13277-015-4049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2017).Chen Y, Pal B, Visvader JE, Smyth GK. Differential methylation analysis of reduced representation bisulfite sequencing experiments using edgeR. F1000Research. 2017;6 doi: 10.12688/f1000research.13196.2. Article 2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran et al. (2017).Dhanasekaran R, Baylot V, Mosley A, Felsher D. MYC is the master switch between tumor dormancy and relapse in Hepatocellular carcinoma (HCC) Hepatology. 2017;66:966a–966a. [Google Scholar]
- Di et al. (2019).Di Y, Chen D, Yu W, Yan L. Bladder cancer stage-associated hub genes revealed by WGCNA co-expression network analysis. Hereditas. 2019;156 doi: 10.1186/s41065-019-0083-y. Article 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du et al. (2012).Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. Journal of Biological Chemistry. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang et al. (2019).Fang P, Xiang L, Chen W, Li S, Huang S, Li J, Zhuge L, Jin L, Feng W, Chen Y, Pan C. LncRNA GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544/RUNX3. Innate Immunity. 2019;25:99–109. doi: 10.1177/1753425919827632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Duarte et al. (2018).Gomes-Duarte A, Lacerda R, Menezes J, Romao L. eIF3: a factor for human health and disease. RNA Biology. 2018;15(1):26–34. doi: 10.1080/15476286.2017.1391437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrieri (2015).Guerrieri F. Long non-coding RNAs era in liver cancer. World Journal of Hepatology. 2015;7:1971–1973. doi: 10.4254/wjh.v7.i16.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Deng & Zhou (2013).Huang ZS, Deng ZH, Zhou XH. Oxymatrine influence proliferation and expression of E2F1 and c-myc in HCC cell line Bel-7404. Journal of Gastroenterology and Hepatology. 2013;28:783–783. doi: 10.1111/jgh.12142. [DOI] [PubMed] [Google Scholar]
- Huang da, Sherman & Lempicki (2009a).Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da, Sherman & Lempicki (2009b).Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kron et al. (2012).Kron C, Wittke E, Mottar A, Bothwell P, Finger R, Bode BP. Amino acid transporters ASCT2 and LAT1 ubiquitously function in N-myc (+) epithelial and mesenchymal human HCC cells exhibiting a wide array of mTOR and glycolytic reliance for growth. Cancer Research. 2012;72(8):5148. doi: 10.1158/1538-7445.Am2012-5148. [DOI] [Google Scholar]
- Lai et al. (2012).Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Medical Oncology. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- Langfelder & Horvath (2008).Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law et al. (2016).Law CW, Alhamdoosh M, Su S, Dong X, Tian L, Smyth GK, Ritchie ME. RNA-seq analysis is easy as 1 − 2 − 3 with limma, Glimma and edgeR. F1000Research. 2016;5:1408. doi: 10.12688/f1000research.9005.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2006).Lee JG, Kang CM, Park JS, Kim KS, Yoon DS, Choi JS, Lee WJ, Kim BR. The actual five-year survival rate of hepatocellular carcinoma patients after curative resection. Yonsei Medical Journal. 2006;47(1):105–112. doi: 10.3349/ymj.2006.47.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2018).Li X, Li B, Ran P, Wang L. Identification of ceRNA network based on a RNA-seq shows prognostic lncRNA biomarkers in human lung adenocarcinoma. Oncology Letters. 2018;16:5697–5708. doi: 10.3892/ol.2018.9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2016).Liu Z, Yan HY, Xia SY, Zhang C, Xiu YC. Downregulation of long non-coding RNA TRIM52-AS1 functions as a tumor suppressor in renal cell carcinoma. Molecular Medicine Reports. 2016;13:3206–3212. doi: 10.3892/mmr.2016.4908. [DOI] [PubMed] [Google Scholar]
- Loher & Rigoutsos (2012).Loher P, Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics. 2012;28:3322–3323. doi: 10.1093/bioinformatics/bts615. [DOI] [PubMed] [Google Scholar]
- Lu et al. (2018a).Lu QC, Rui ZH, Guo ZL, Xie W, Shan S, Ren T. LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. Journal of Cellular and Molecular Medicine. 2018a;22(3):1527–1537. doi: 10.1111/jcmm.13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2018b).Lu Y, Hu Z, Mangala LS, Stine ZE, Hu X, Jiang D, Xiang Y, Zhang Y, Pradeep S, Rodriguez-Aguayo C, Lopez-Berestein G, DeMarzo AM, Sood AK, Zhang L, Dang CV. MYC targeted long noncoding RNA DANCR promotes cancer in part by reducing p21 levels. Cancer Research. 2018b;78:64–74. doi: 10.1158/0008-5472.CAN-17-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao et al. (2017).Mao Z, Li H, Du B, Cui K, Xing Y, Zhao X, Zai S. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Bioscience Reports. 2017;37(6) doi: 10.1042/BSR20171070. Article BSR20171070. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Maza (2016).Maza E. In Papyro comparison of TMM (edgeR), RLE (DESeq2), and MRN normalization methods for a simple two-conditions-without-replicates RNA-Seq experimental design. Frontiers in Genetics. 2016;7 doi: 10.3389/fgene.2016.00164. Article 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida & Fukuda (2001).Nishida N, Fukuda Y. Tumor suppressor RB gene and its related molecules in hepatocellular carcinoma. Nihon Rinsho. 2001;59(Suppl 6):134–137. [PubMed] [Google Scholar]
- Ou et al. (2018).Ou L, Wang D, Zhang H, Yu Q, Hua F. Decreased expression of miR-138-5p by lncRNA H19 in cervical cancer promotes tumor proliferation. Oncology Research. 2018;26:401–410. doi: 10.3727/096504017X15017209042610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng et al. (2013).Peng Q, Lao X, Chen Z, Lai H, Deng Y, Wang J, Mo C, Sui J, Wu J, Zhai L, Yang S, Qin X, Li S. TP53 and MDM2 gene polymorphisms, gene-gene interaction, and hepatocellular carcinoma risk: evidence from an updated meta-analysis. PLOS ONE. 2013;8:e82773. doi: 10.1371/journal.pone.0082773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers & Carey (2006).Reimers M, Carey VJ. Bioconductor: an open source framework for bioinformatics and computational biology. Methods in Enzymology. 2006;411:119–134. doi: 10.1016/S0076-6879(06)11008-3. [DOI] [PubMed] [Google Scholar]
- Selcuk (2017).Selcuk H. Prognostic factors and staging systems in hepatocellular carcinoma. Experimental and Clinical Transplantation. 2017;15(2):45–49. doi: 10.6002/ect.TOND16.L11. [DOI] [PubMed] [Google Scholar]
- Sha et al. (2017).Sha S, Yuan D, Liu Y, Han B, Zhong N. Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biology Open. 2017;6:1310–1316. doi: 10.1242/bio.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao & Li (2019).Shao M, Li W. Transcriptional factor regulation network and competitive endogenous RNA (ceRNA) network determining response of esophageal squamous cell carcinomas to neoadjuvant chemoradiotherapy. PeerJ. 2019;7:e6668. doi: 10.7717/peerj.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, Miller & Jemal (2019).Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- Su et al. (2017).Su P, Wang F, Qi B, Wang T, Zhang S. P53 regulation-association long non-coding RNA (LncRNA PRAL) inhibits cell proliferation by regulation of P53 in human lung cancer. Medical Science Monitor. 2017;23:1751–1758. doi: 10.12659/MSM.900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk et al. (2017).Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, Von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellapuri et al. (2018).Tellapuri S, Sutphin PD, Beg MS, Singal AG, Kalva SP. Staging systems of hepatocellular carcinoma: a review. Indian Journal of Gastroenterology. 2018;37:481–491. doi: 10.1007/s12664-018-0915-0. [DOI] [PubMed] [Google Scholar]
- Valasek et al. (2017).Valasek LS, Zeman J, Wagner S, Beznoskova P, Pavlikova Z, Mohammad MP, Hronova V, Herrmannova A, Hashem Y, Gunisova S. Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Research. 2017;45:10948–10968. doi: 10.1093/nar/gkx805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet et al. (2016).Varet H, Brillet-Gueguen L, Coppee JY, Dillies MA. SARTools: a DESeq2- and EdgeR-Based R pipeline for comprehensive differential analysis of RNA-Seq data. PLOS ONE. 2016;11:e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2017).Wang H, Liu W, Liu L, Wu C, Wu W, Zheng J, Zhang M, Chen X, Zhou B, Gao Z, Huang J. Overexpression of centromere protein K (CENP-K) gene in hepatocellular carcinoma promote cell proliferation by activating AKT/TP53 signal pathway. Oncotarget. 2017;8:73994–74005. doi: 10.18632/oncotarget.18172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2015).Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, Yan X, Ye B, Li C, Xia P, Zhang G, Tian Y, Chen R, Fan Z. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2016).Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang B, Zhou M, Li X, Xiong W, Li G, Zeng Z, Yang T. The long noncoding RNA malat-1 is a novel biomarker in various cancers: a meta-analysis based on the GEO database and literature. Journal of Cancer. 2016;7(8):991–1001. doi: 10.7150/jca.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng et al. (2018).Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, Sheng Y, Wang Y, Wunderlich M, Zhang B, Dore LC, Su R, Deng X, Ferchen K, Li C, Sun M, Lu Z, Jiang X, Marcucci G, Mulloy JC, Yang J, Qian Z, Wei M, He C, Chen J. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–205. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu et al. (2018).Xu D, Yu J, Gao G, Lu G, Zhang Y, Ma P. LncRNA DANCR functions as a competing endogenous RNA to regulate RAB1A expression by sponging miR-634 in glioma. Bioscience Reports. 2018;38(1) doi: 10.1042/BSR20171664. Article BSR20171664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao et al. (2018).Yao J, Shen X, Li H, Xu J, Shao S, Huang JX, Lin M. LncRNA-ECM is overexpressed in esophageal squamous cell carcinoma and promotes tumor metastasis. Oncology Letters. 2018;16:3935–3942. doi: 10.3892/ol.2018.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin et al. (2018).Yin L, Cai Z, Zhu B, Xu C. Identification of key pathways and genes in the dynamic progression of HCC based on WGCNA. Gene. 2018;9(2) doi: 10.3390/genes9020092. Article 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan et al. (2016).Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, Fan J, Liu L, Sun SH, Zhou WP. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2014).Zhang JC, Gao B, Yu ZT, Liu XB, Lu J, Xie F, Luo HJ, Li HP. Promoter hypermethylation of p14 (ARF), RB, and INK4 gene family in hepatocellular carcinoma with hepatitis B virus infection. Tumour Biology. 2014;35:2795–2802. doi: 10.1007/s13277-013-1372-0. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2017).Zhang Y, Wu SS, Chen XH, Tang ZH, Yu YS, Zang GQ. Tripartite motif containing 52 (TRIM52) promotes cell proliferation in hepatitis B virus-associated hepatocellular carcinoma. Medical Science Monitor. 2017;23:5202–5210. doi: 10.12659/MSM.907242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhussupbekova et al. (2016).Zhussupbekova S, Sinha R, Kuo P, Lambert PF, Frazer IH, Tuong ZK. A mouse model of hyperproliferative human epithelium validated by keratin profiling shows an aberrant cytoskeletal response to injury. EBioMedicine. 2016;9:314–323. doi: 10.1016/j.ebiom.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplementary Files.