Abstract
Extracellular β-amyloid (Aβ) plaque deposits and inflammatory immune activation are thought to alter various aspects of tissue microstructure, such as extracellular free water, fractional anisotropy and diffusivity, as well as the density and geometric arrangement of axonal processes. Quantifying these microstructural changes in Alzheimer’s disease and related neurodegenerative dementias could serve to monitor or predict disease course. In the present study we used high-field diffusion magnetic resonance imaging (dMRI) to investigate the effects of Aβ and inflammatory interleukin-6 (IL6), alone or in combination, on in vivo tissue microstructure in the TgCRND8 mouse model of Alzheimer’s-type Aβ deposition. TgCRND8 and non-transgenic (nTg) mice expressing brain-targeted IL6 or enhanced glial fibrillary protein (EGFP controls) were scanned at 8 months of age using a 2-shell, 54-gradient direction dMRI sequence at 11.1 Tesla. Images were processed using the diffusion tensor imaging (DTI) model or the neurite orientation dispersion and density imaging (NODDI) model. DTI and NODDI processing in TgCRND8 mice revealed a microstructure pattern in white matter (WM) and hippocampus consistent with radial and longitudinal diffusivity deficits along with an increase in density and geometric complexity of axonal and dendritic processes. This included reduced FA, mean, axial and radial diffusivity, and increased orientation dispersion (ODI) and intracellular volume fraction (ICVF) measured in WM and hippocampus. IL6 produced a ‘protective-like’ effect on WM FA in TgCRND8 mice, observed as an increased FA that counteracted a reduction in FA observed with endogenous Aβ production and accumulation. In addition, we found that ICVF and ODI had an inverse relationship with the functional connectome clustering coefficient. The relationship between NODDI and graph theory metrics suggests that currently unknown microstructure alterations in WM and hippocampus are associated with diminished functional network organization in the brain.
Keywords: Alzheimer’s disease, diffusion MRI, functional connectivity, connectomic, clustering, NODDI, free water, DTI, inflammation, interleukin 6
INTRODUCTION
Excess production and oligomerization of the β-amyloid (Aβ) protein, the main constituent of amyloid plaques, substantially modifies neuronal morphology (Biscaro et al., 2009; Spires and Hyman, 2004). Importantly, Aβ alters neuronal homeostasis and synaptic plasticity by modifying the extracellular environment of neuronal soma, axons, and synapses (Mueggler et al., 2004; Sykova et al., 2005). Aβ is implicated in the formation of dystrophic neurites (Sadleir et al., 2016; Zhang et al., 2009), deficits in synaptic plasticity and excitability (Cummings et al., 2015; Varga et al., 2015), cell membrane protein aggregation (Shrivastava et al., 2017), altered microstructure of major fiber tracts (Mayo et al., 2017; O’Dwyer et al., 2011; Teipel et al., 2014) and impaired activity in resting state networks (Hedden et al., 2009; Jones et al., 2016). In addition, Aβ is immunogenic (Dalgediene et al., 2013), leading to the increased activated microglia in the brain (Mirzaei et al., 2016; Serriere et al., 2015). Positron emission tomography (PET) studies using microglia- or Aβ-specific radiotracers provide evidence of an overlapping presence of activated microglia and amyloid plaques in Alzheimer’s disease (Kreisl et al., 2013; Liu et al., 2015; Mirzaei et al., 2016; Parbo et al., 2017; Schuitemaker et al., 2013). A longitudinal study identified a transient elevation in microglia early in mild cognitive impairment (MCI) which peaked later in Alzheimer’s disease (Fan et al., 2017). Aged transgenic mice expressing familial Alzheimer’s mutations to amyloid precursor protein and presenilin-1 with deletion of exon-9 (APP/PS1dE9) had higher in vivo cortical and hippocampal accumulation of an activated microglial-targeted radiotracer, 18kD translocator protein (TSPO), than aged wildtype or young Tg mice (Liu et al., 2015). The above reports suggest that accrual of Aβ plaques and concomitant reactive microgliosis could underpin pathologic mechanisms in Alzheimer’s, perhaps involving deleterious changes to brain parenchymal microstructure. In this study, we asked the question whether we can utilize high field diffusion magnetic resonance imaging (dMRI) to quantitatively assess the effects of Aβ accumulation and altered neuroimmune activity on tissue microstructure in a mouse model of Alzheimer’s disease. An additional objective was to determine the association between dMRI-based microstructure indices and functional activity measured by resting state functional MRI (fMRI) (Brown et al., 2018).
Diffusion tensor imaging (DTI) offers a powerful approach to generate metrics that describe the movement of water in brain tissue compartments. Advanced methods for the modeling of tissue diffusivity beyond standard DTI processing schemes may provide additional insight on the effects of pathogenic Aβ and immune mechanisms in Alzheimer’s disease. For example, increased radial diffusivity was associated with worsening of dementia symptoms in dMRI scans processed using a two-compartment model of diffusion known as the free water imaging method (Hoy et al., 2017; Ji et al., 2017; Maier-Hein et al., 2015). Also, a multi-compartment diffusion model termed Neurite Orientation Dispersion and Density Imaging (NODDI) revealed significant changes in NODDI metrics in young onset Alzheimer’s disease (Parker et al., 2018; Slattery et al., 2017), and correlations with cortical tau immunoreactivity in the rTg4510 mouse tauopathy model of frontal temporal dementia (Colgan et al., 2016). While the free water method (Pasternak et al., 2009) provides quantitative maps that have been associated with neuroinflammatory and neurodegenerative mechanisms (Ofori et al., 2015; Pasternak et al., 2012; Pasternak et al., 2015), the NODDI model can provide quantitative maps associated with axonal and dendritic dispersion and density while also modeling an isotropic compartment corresponding with unrestricted free water (Colgan et al., 2016; Grussu et al., 2017; Sepehrband et al., 2015; Zhang et al., 2012). Despite the advances in dMRI processing, preclinical high field imaging studies in mouse models are still needed to clarify how proteinopathies and immune cellular mechanisms involved in Alzheimer’s disease related to changes in brain microstructure, and in turn their impact on functional network activity.
In the present study we investigated the effects of Aβ on dMRI-derived NODDI metrics in transgenic (Tg)CRND8 mice with early onset amyloidosis (Chishti et al., 2001). To mimic the effects of chronic microgliosis and astrogliosis, we additionally used adeno-associated viral vector (AAV)-mediated brain-targeted expression of an inflammatory cytokine, interleukin (IL)-6. IL6 expression results in widespread gliosis in the CNS in the absence of overt amyloidogenic or neurodegenerative pathology (Chakrabarty et al., 2010b), and was thus considered ideal for testing the individual or combined effects of Aβ and neuroinflammation on in vivo tissue microstructure in mouse brain. NODDI processing in Aβ-expressing TgCRND8 mice revealed a microstructure pattern consistent with radial and longitudinal diffusivity deficits and also suggests the possibility of an increase in geometrically destabilized axonal bundles or misoriented dendritic processes in white matter (WM) and hippocampus. Interestingly, we discovered a novel relationship between NODDI metrics that may model the density and orientation dispersion of ‘neurites’ and a measure of functional network clustering that was similarly affected by Aβ and IL6.
MATERIALS AND METHODS
Subjects
Mice were housed in sex- and aged-matched groups of 3–4 in conventional cages (29 × 18 × 13 cm) at 20–26°C (lights on from 0700–1900 hours) with food and water ad libitum. TgCRND8 mice were maintained in-house by breeding amyloid precursor protein (APP) transgenic (Tg) males (carrying the wild-type retinal degeneration gene) with C57B6/C3H F1 females (Envigo) (Janus et al., 2000). TgCRND8 mice have early onset expression of human mutant APP (Swedish APP KM670/671NL and Indiana APP V717F), which increases human APP 5-times above endogenous murine APP. These mice have aggressive cognitive impairment, Aβ plaque deposits and increased inflammation at 3–4 months (mo.) of age, synaptic deficits and some synaptic and neuronal loss in hippocampus by 6 m.o. (Chishti et al., 2001). In this study, we used 8 mo. mice (non-transgenic -nTg- and TgCRND8), an age at which levels of forebrain amyloidosis are high. All groups were sex and age matched. All procedures received prior approval from the Institutional Animal Care and Use Committee of the University of Florida and follow all applicable NIH guidelines.
Adeno-associated viral vector (AAV) preparation and brain delivery
Recombinant adeno-associated virus subtype 2/1 (rAAV2/1) for intracerebroventricular (ICV) injections expressing murine IL6 (m-IL6) or enhanced green fluorescent protein (EGFP) under the control of the cytomegalovirus enhancer/chicken β actin promoter were generated as described previously (Chakrabarty et al., 2010b). Briefly, rAAV vectors expressing IL6 under the control of the cytomegalovirus enhancer/chicken beta actin (CBA) promoter, a WPRE, and the bovine growth hormone polyA were generated by plasmid transfection with helper plasmids in HEK293T cells. Forty-eight hours after transfection, cells were harvested and lysed in the presence of 0.5% Sodium Deoxycholate and 50U/ml Benzonase (Sigma) by freeze thawing and the virus isolated using a discontinuous Iodixanol gradient and affinity purified on a HiTrap HQ column (Amersham). The genomic titer of each virus was determined by quantitative polymerase chain reaction (qPCR). TgCRND8 and nTg mice were injected with 2 μl of rAAV (~2E10 viral genomes) into cerebral ventricles using a 10 μl Hamilton syringe with a 30 g needle on day postnatal day 0 (P0) as described before (Chakrabarty et al., 2010b; Chakrabarty et al., 2015). Animals were euthanized and brains harvested within 1–2 days of imaging.
Experimental groups
The following experimental groups were included in this study: nTg-Controls expressing AAV-EGFP (n = 14), nTg-IL6 expressing IL6 (n = 14), TgCRND8-Controls expressing AAVEGFP (n = 10) and TgCRND8-IL6 expressing IL6 (n = 10). All 4 groups were sex balanced. Statistical analysis showed no sex differences, thus data for male and female mice were grouped. One dMRI scan in the nTg-IL6 group was considered to be of poor quality and was removed (leaving 13 subjects in this dMRI group).
MRI acquisition
Mouse brain images were acquired on an 11.1 Tesla MRI scanner (Magnex Scientific Ltd., Oxford, UK) with a Resonance Research Inc. gradient set (RRI BFG-113/60, maximum gradient strength of 1500 mT/m at 150 Amps and a 130 μs risetime; RRI, Billerica, MA) and controlled by a Bruker Paravision 6.01 console (Bruker BioSpin, Billerica, MA) as previously reported (Sahara et al., 2014). An in-house 2.0 × 3.5 cm quadrature surface transmit/receive coil tuned to 470.7MHz (1H resonance) was used for B1 excitation and signal detection (Advanced Magnetic Resonance Imaging and Spectroscopy Facility, Gainesville, FL). Anesthesia was induced under 1.5–2.0% isoflurane (0.1–0.15 L/min) delivered in medical grade air (70%N2/30% O2) and levels were then kept at 1% through functional neuroimaging and then increased to 1.5% during diffusion MRI to maintain a slower breathing rate during the latter acquisition. Mice were placed on a custom plastic bed with a respiratory pad underneath the abdomen. Respiratory rates were monitored continuously, and body temperature was maintained at 37–38°C using a warm water recirculation system (SA Instruments, Inc., New York). For each mouse, a high resolution T2 weighted anatomical was first acquired in 10 minutes, followed by a resting state fMRI scan in 6 minutes, and finally a diffusion scan in 1 hour.
A T2-weighted Turbo Rapid Acquisition with Refocused Echoes (RARE) sequence was used for acquiring anatomical scans with the following parameters: echo time (TE) = 41 ms, repetition time (TR) = 4 seconds, RARE factor = 16, number of averages = 12, field of view (FOV) 15 mm × 11 mm and 0.7 mm thick slice, and 256 × 256 × 17 coronal (axial) slices covering the entire brain from the rostral-most extent of the prefrontal cortical region and caudally to the upper brainstem/cerebellum.
Functional images were collected using a single-shot spin-echo echo planar imaging (EPI) sequence with the following parameters: TE = 15 ms, TR = 2 seconds, 180 repetitions, 15 × 11 mm in plane, 14 slices with 0.9 mm thickness per slice, data matrix = 64 × 48. For all EPI sequences, a fat saturation pulse was used to suppress chemical shift artifact and phase correction parameters were acquired during receiver gain adjustment for Nyquist ghost correction. No stimuli were presented during functional scanning and respiratory rates, isoflurane concentration, body temperature, lighting and room conditions were kept constant across subjects.
Diffusion weighted scans were acquired using a 4-shot, 2-shell spin echo planar diffusion imaging (DTI EPI) sequence in Bruker Paravision, with TR = 4 seconds, TE = 19 ms, number of averages = 4, gradient duration δ = 3 ms, diffusion time Δ = 8 ms, 54 images with 3 different diffusion weightings, two b=0, 6 directions with b=600 s/mm2, and 46 directions with b=2000 s/mm2. A navigator signal was used by the Bruker reconstruction software to improve signal stability in the 4-shot EPI. The signal-to-noise ratio of B0 images were determined to range from 25–45 across subjects, which is above the lower limit where eigenvalues may diverge from typical values for a given tissue (Bastin et al., 1998). Image saturation bands were placed on either side and below the brain to suppress non-brain signal during image acquisition. Diffusion images had the same FOV and slice thickness as the anatomical scan but with a lower resolution data matrix size of 128 × 96 and 17 slices (resolution: 0.117 mm × 0.117 mm × 0.7 mm) in the same space as anatomical scans. This allowed careful manual outlining of regions to be analyzed by using T2 scans and diffusion maps (see below).
Brain region of interest (ROI) segmentation for dMRI scans
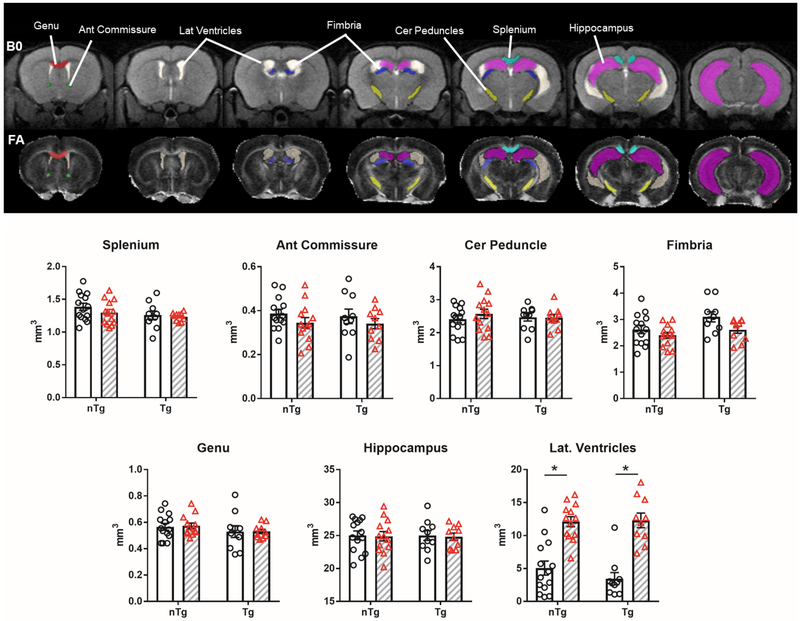
ITKSNAP (Yushkevich et al., 2006) was used to manually segment hippocampus and WM areas such as anterior commissure, fimbria, genu and splenium of the corpus callosum, and cerebral peduncle (Figure 1, top panel). We selected these axonal pathways because of their roles in carrying information between nuclei in striatum and forebrain, prefrontal cortical areas, sensorimotor, temporal, parietal and retrosplenial cortical structures, and the hippocampal-septal system. The T2 high resolution anatomical and the fractional anisotropy (FA) maps were used as guides to determine boundaries between tissue types (e.g., white matter, grey matter, and cerebrospinal fluid filled ventricles). In preliminary analyses of control mouse brain dMRI scans, we observed distinctive values for DTI and NODDI metrics in these different sub-compartments that suggests minimal overlap (see Supplemental Figure 1).
Figure 1.
IL6 increased lateral ventricle volume in transgenic CRND8 mice (TgCRND8) and non-transgenic (nTg) controls. No volumetric differences were observed in white matter (WM) and hippocampus. Top) Segmented ROIs are overlaid on representative B0 and FA maps and ROI names are indicated on the B0 map. Color-filled ROI are: anterior commissure (green), genu (red), ventricles (beige), fimbria (purple). Cerebral peduncle (yellow), splenium (light blue), hippocampus (fuscia). Bottom) WM and hippocampus volumes (y-axis is mm3). Clear bars (and clear circles) are control groups and grey hashed bars (and red triangles) are IL6 treated mice. *significant difference between control and IL6 (Tukey’s post hoc test, p<0.05). All data represented as bar graphs are mean ± standard error.
Diffusion MRI processing
Diffusion MRI scans were processed using tools available on FMRIB software library – FSL (Smith et al., 2004), as previously reported for mouse dMRI scans obtained at 11.1 Tesla (Sahara et al., 2014). In 17 out of the 48 subjects, individual diffusion images were removed from the large shell image series due to low image quality. The gradient b-vector files were corrected by removing gradient directions for the removed images. In the 17 edited scans, a median of 8 images were removed (range: 3–15). However, all 17 large shell scans retained over 30 directions. No images were removed from the smaller shell image series. Eddy correction was used for adjusting slight movements during image acquisition and gradient files rotated according to the motion correction vectors. After eddy correction, tensor element reconstruction and estimates of 1st, 2nd and 3rd eigenvectors and eigenvalues (λ1, λ2, λ3, where λ1 is axial diffusivity and the average of λ2 and λ3 provides radial diffusivity values) was performed using weighted least squares regression on DTIFIT in FSL (Behrens et al., 2003). This last step generated independent images of FA, mean, axial and radial diffusivities (MD, AD, and RD, respectively).
NODDI processing
We analyzed dMRI scans following processing with the NODDI model (Zhang et al., 2012). This was implemented within the Accelerated Microstructure Imaging via Convex Optimization (AMICO) framework (Daducci et al., 2015), which accelerates the model fitting procedure in standard Linux PC’s running Python 2.7. NODDI incorporates a multi-compartment model of the diffusion signal. The model includes an isotropic fraction of free unrestricted and hindered water (e.g., cerebral spinal fluid) and an anisotropic model that includes an intracellular fraction with zero radius infinite cylinders (high diffusion coefficients along the length of the principal axis only) and an extracellular fraction with non-zero diffusion perpendicular to the principal axis. The latter adds morphological information to the tissue model of the extracellular environment surrounding axons and dendrites (occupied by neurons and non-neuronal cell bodies) (Zhang et al., 2012). The fitting of the intracellular volume fraction (i.e., zero-radius cylinders or ‘sticks’) involves the use of a Bingham-Watson series used in statistical shape analysis (Maier-Hein et al., 2015; Tariq et al., 2016) and the results of the fitting process provides an index of the degree of orientation dispersion of so-called neurites. For the parameter fitting procedure, we assumed a value of in vivo intrinsic diffusivity of 1.7 μm2/ms and isotropic diffusivity of 3.0 μm2/ms (Grussu et al., 2017). The whole brain model-fitting generates maps of an intracellular volume fraction (ICVF: relative concentration of zero-radius cylinders modeling ‘neurites’), orientation dispersion (ODI: 0 for no dispersion as in highly organized parallel fiber bundles to a maximum of 1 for the highest degree of dispersion, as in cerebral cortical grey matter), and the isotropic free water fraction (ISO: 0 low isotropy to a maximum isotropy of 1) (Grussu et al., 2017).
Resting state fMRI processing
Binary masks outlining mouse brain boundaries were automatically generated on MATLAB using Pulsed Coupled Neural Networks (PCNN3D) on high resolution T2 Turbo-RARE anatomical scans (Chou et al., 2011). N4 bias field correction (Tustison et al., 2010) was used to remove B1 field inhomogeneities and improve anatomical image quality prior to alignment to a segmented mouse brain template (Moore et al., 2016). The binary ‘brain-only’ masks were multiplied by T2 anatomical scans to null voxels outside the brain. The cropped T2 anatomical brain images were aligned to a mouse brain template using the FSL program FLIRT (Jenkinson et al., 2002) (Supplemental Figure 2). Linear registration matrices for each subject were saved and used to transform functional datasets to template space for analysis (Supplemental Figure 2). Resting state processing steps included: (1) removal of time series spikes, (2) slice timing correction, (3) motion and linear drift correction, (4) subject-to-atlas registration, (5) regression of white matter and CSF (ventricle) signals, (6) bandpass filtering (0.01–0.1Hz), and (7) spatial blurring (0.3 mm FWHM). For step 4 (subject-to-atlas registration), a binary mask was created for the functional MRI scan and was used for skull stripping (as described above for the T2 anatomical scans). Functional scans were then linearly registered to their corresponding T2 scans (12 affine parameter registration) and then warped to the T2 anatomical scans using a nonlinear symmetric normalization (SyN) in Advanced Normalization Tools (ANTs) (Avants et al., 2008; Grandjean et al., 2017). The linear registration matrices moving the anatomical-to-atlas space were then applied to functional scans (Supplemental Figure 2).
Average BOLD signals from each region of interest (ROI) were extracted based on their atlas location and used in voxel-wise cross correlation to generate Pearson correlation maps and 4005 pairwise correlation coefficients to be imported to MATLAB as edge weights for network analysis. All correlation r values are z transformed prior to statistical analyses. For seed-based resting state fMRI analysis of cortical and memory structures, connectivity maps of somatosensory cortex, hippocampus, and entorhinal cortex were generated for each subject (Supplemental Figure 3) and analyzed using a multivariate modeling approach in Analysis of Functional Neuroimages (AFNI 3dMVM)(Cox, 1996). All group statistical maps for main effects and full factorial interactions were FDR corrected (Chen et al., 2014).
Network analyses
Functional network graphs were analyzed using Brain Connectivity Toolbox (Rubinov and Sporns, 2010). Graphs with 4,005 entries were organized in MATLAB [graph size = n(n-1)/2, where n is the number of nodes represented in the graph, or 90 ROI]. The z-scores were thresholded to equalize density prior to analyses (multiple node density thresholds from 2–20% are applied). Edge weights were normalized between 0 to 1. Node strength (sum of edge weights), clustering coefficient (the degree to which nodes cluster together in groups), average path length (the potential for communication between pairs of structures), modularity (quantifies the degree to which the network may be subdivided into such clearly delineated groups or communities), and small worldness (the degree to which mouse functional brain networks under study deviate from randomly connected networks) were calculated for undirected binarized graphs and statistically compared between groups (Boccaletti et al., 2006; Humphries and Gurney, 2008; Newman and Girvan, 2004; Newman, 2003; Saramaki et al., 2007). Brain networks were visualized using BrainNet Viewer (Xia et al., 2013). The 3D networks were generated with undirected edges weights ≥ 0.15. In these 3D mouse brain functional connectomes, the node size and color were scaled by the node strength, and edges were scaled by z-scores.
Preparation of brain tissue for immunohistochemistry and ELISA
Following euthanasia, the right hemisphere for each mouse brain was fixed in 10% normal buffered formalin and the left hemisphere was snap frozen. Formalin fixed brains were paraffin embedded and analyzed using the following antibodies according to immunohistochemistry protocols described previously (Chakrabarty, 2010; Chakrabarty, 2015): Glial Fibrillary Acidic Protein (GFAP, Cell Signaling, 1:1000) and ionized calcium binding adaptor molecule 1 (Iba1, Wako, 1:1000). Immunohistochemically stained sections were captured using the Scanscope XT image scanner (Aperio) and analyzed using the ImageScope program. Brightness and contrast alterations were applied identically on captured images using Adobe Photoshop CS3. Snap-frozen forebrains (without olfactory bulb and cerebellum) were homogenized in RIPA Buffer (Boston Bioproducts) and the homogenate was centrifuged at 100,000 × g for 1 h at 4°C. Protein concentration in the supernatant was determined using BCA Protein Assay kit (Pierce). mIL-6 levels were evaluated using sandwich capture ELISA assays using the RIPA soluble mouse forebrain lysates as per manufacturer’s instructions (BD OptiEIA, BD Biosciences).
Statistical analyses
For dMRI, manual segmentation masks were used to import to MATLAB the average values for FA, MD, AD, RD (all diffusivities in units of mm2/sec), ODI, ICVF, and ISO for 7 ROI’s. Diffusion metrics were analyzed in MATLAB (Natick, MA) using a two-factor analysis of variance (ANOVA: Strain [2 levels: nTg and Tg] × Treatment [2 levels: control and IL6]). Tukey’s multiple comparison test was used to assess specific differences between group means. Unless otherwise noted, Tukey’s post hoc test results are reported in figure legends, whereas full factorial ANOVA results are provided in the results sections.
For fMRI, mouse brain atlas-based segmentation masks were used to import to MATLAB the average values for small world coefficient, global node strength, clustering coefficient, modularity, and mean path length. For these measures, we used a two-factor ANOVA (Strain × Treatment) and Tukey’s posthoc test. Statistical analyses of correlation coefficient values per ROI pairs were not carried out on exported values, as these were analyzed on somatosensory, hippocampal and entorhinal cortex maps using ANOVA computations on AFNI 3dMVM (see above). Main effects of strain and treatments and their interactions were statistically assessed.
As an additional step to supplement seed-based and network analyses, we conducted analyses using probabilistic independent component analysis (ICA) in FSL MELODIC (multivariate exploratory linear decomposition into independent components) for 48 resting state fMRI mouse brain scans (Beckmann and Smith, 2004). Prior to ICA, post-processed and atlas-aligned functional scans (see above) were cropped using manually segmented masks. This provided optimal co-registration of fMRI scans with each subject anatomical and with the mouse brain template(Ma et al., 2014). In MELODIC, the following steps were carried out: voxel-wise de-meaning, variance normalization, pre-whitening, and data were then projected into a 12, 20 or 30-dimensional subspace using PCA, and then ICA estimation was conducted in concatenated maps. In initial assessments, ICA maps for the 20-dimensional subspace (20 predetermined components) were observed to adequately separate distinct bilateral components that were consistent with previously reported ICA results for mouse brain (Grandjean et al., 2017; Sforazzini et al., 2014; Shah et al., 2016). Mixture model fitting, rescaling and thresholding was carried out on z-transformed ICA maps (alternative hypothesis test set at p > 0.5). ICA maps likely representing noise or not deemed to include well-defined regions were removed and the rest were considered of interest. The latter were assembled for visualization using slices summary and overlay scripts in FSL, using a lower z threshold of 2.5. Using the Glm tool in FSL a general linear model design matrix was generated to test for main effects of strain, treatment, and strain × treatment interactions, and used in dual regression analysis and randomise permutation testing in FSL to test for differences between groups (Nickerson et al., 2017).
RESULTS
IL6 treatment increased lateral ventricle volume
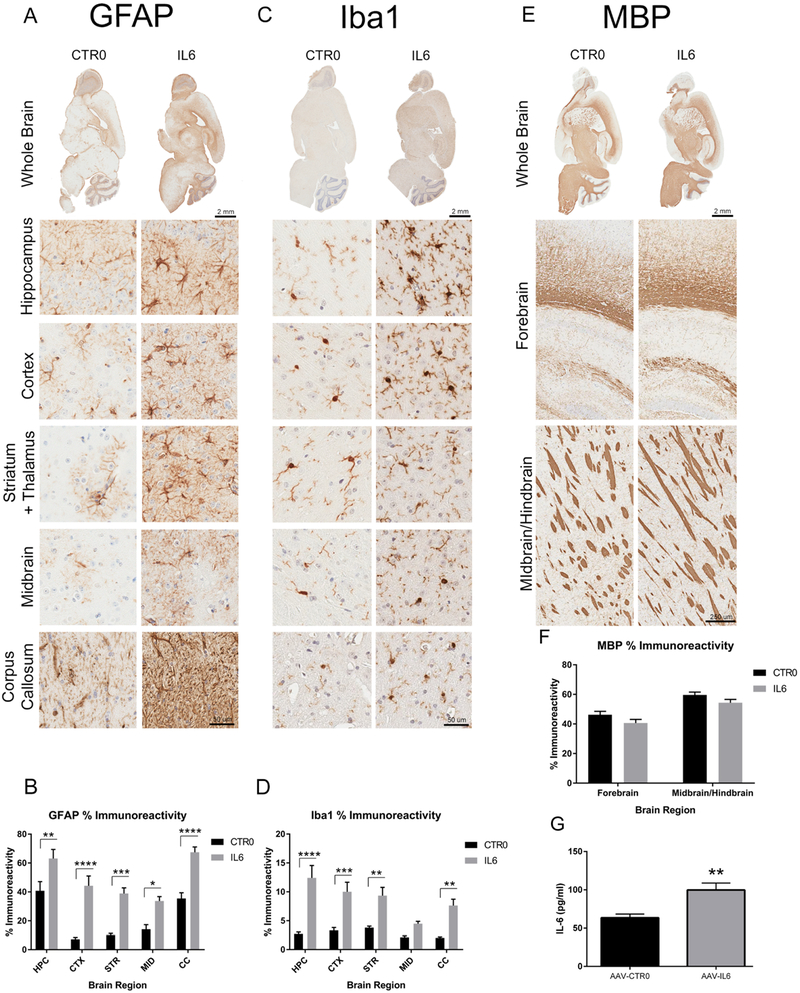
To understand how inflammatory conditioning alters brain connectivity in an Alzheimer’s disease mouse model, we delivered AAV-IL6 into the cerebral ventricles of neonatal TgCRND8 mice. We previously demonstrated the utility of similar paradigms to examine pathological alterations in Alzheimer’s disease mouse models (Chakrabarty, 2010; Chakrabarty, 2015). Mice were aged to 8 months, imaged and euthanized to confirm the histopathological effects of intracranial expression of IL6. We observed robust upregulation of astrogliosis (GFAP immunohistochemistry) and microgliosis (Iba-1 immunohistochemistry) in all areas of the brain examined, e.g., forebrain (cortex and hippocampus), basal ganglia (striatum) and midbrain (Figure 2A–D). However, IL-6 did not cause any significant effects on myelin basic protein staining (Figure 2E–F). IL-6 protein levels were also upregulated in the whole brain lysates (p<0.01; Figure 2G).
Figure 2.
Brain expression of IL6 resulted in widespread gliosis. A) Analysis of astrocytosis (GFAP, A-B), microgliosis (Iba-1, C-D) and IL6 protein (G) in nTg mice demonstrate that IL6 expression leads to dramatic upregulation of immune response throughout the brain. E-F) Show myelin basic protein (MBP) is not altered by IL6. HPC, hippocampus; CTX, cortex; STR, Striatum; MID; midbrain. B-D. One-way ANOVA, *p<0.05; **p<0.01, ***p<0.005, n=6; E. Student’s t test, **p<0.01, n=4.
To determine if there were any morphological differences in WM and hippocampus as a result of IL6 or Aβ, we analyzed ROI volumes (Figure 1). Thus, the number of voxels contained within each ROI mask was used as a proxy for ROI size. No differences in the size of the different WM ROIs was observed between nTg and Tg or control and IL6 treatment groups. However, we discovered that elevated brain IL6 significantly increased the size of the segmented lateral ventricles (main effect IL6 treatment: F1,43 = 63.6 p < 0.0001). The enlargement of lateral ventricles was observed in nTg and Tg mice given IL6 treatment.
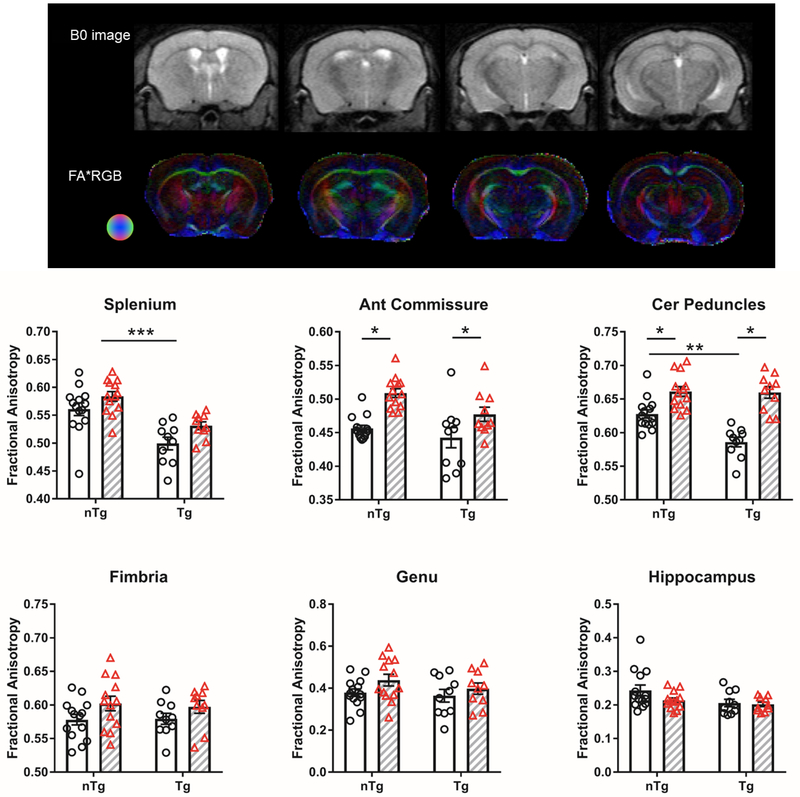
Opposite effects of Aβ and IL6 on white matter FA values
Figure 3 summarizes the results for FA as a function of strain (nTg vs. Tg) and IL6 treatment (IL6 vs. control). Representative mouse brain B0 and FA maps in Figure 3 illustrate WM structures such as the corpus callosum, fimbria, cerebral peduncle, and their directionality. In splenium, anterior commissure, and cerebral peduncle we observed a main effect of Aβ on FA (splenium: main effect strain F1,43 = 31.5 p < 0.0001; anterior commissure: main effect of strain F1,43 = 6.2 p = 0.02; cerebral peduncle: strain × treatment interaction F1,43 = 7.6 p = 0.008; main effect strain F1,43 = 8.8 p = 0.005). In the splenium and cerebral peduncles, we observed a reduced FA in TgCRND8 mice with increased Aβ (p<0.05, Tukey’s test). In splenium, fimbria, and cerebral peduncle we observed a main effect of IL6 on FA (splenium: main effect treatment F1,43 = 7.3 p = 0.009; anterior commissure: main effect of treatment F1,43 = 22.8 p < 0.0001; fimbria: main effect treatment F1,43 = 10.3 p = 0.03; cerebral peduncle: main effect treatment F1,43 = 53.8 p < 0.0001). In the anterior commissure and cerebral peduncles IL6 increased FA in both nTg and Tg mice (p < 0.05, Tukey’s test). No significant effects of IL6 or Aβ were observed in genu and hippocampus. To sum, Aβ was observed in WM structures to reduce FA while IL6 treatment increased FA (in both Tg and nTg mice).
Figure 3.
Aβ reduces and IL6 increased fractional anisotropy (FA) in WM regions. Top panel shows representative B0 and FA color maps. Diffusion direction indicated by colored sphere. Bottom graphs show clear bars (and clear circles) corresponding to control groups and grey hashed bars (and red triangles) are IL6-treated. *significant difference between control and IL6 mice; **significant difference between Tg and nTg mice (Tukey’s post hoc test, p<0.05). All data are mean ± standard error.
Differential effects of Aβ and IL6 on MD, AD and RD metrics in white matter
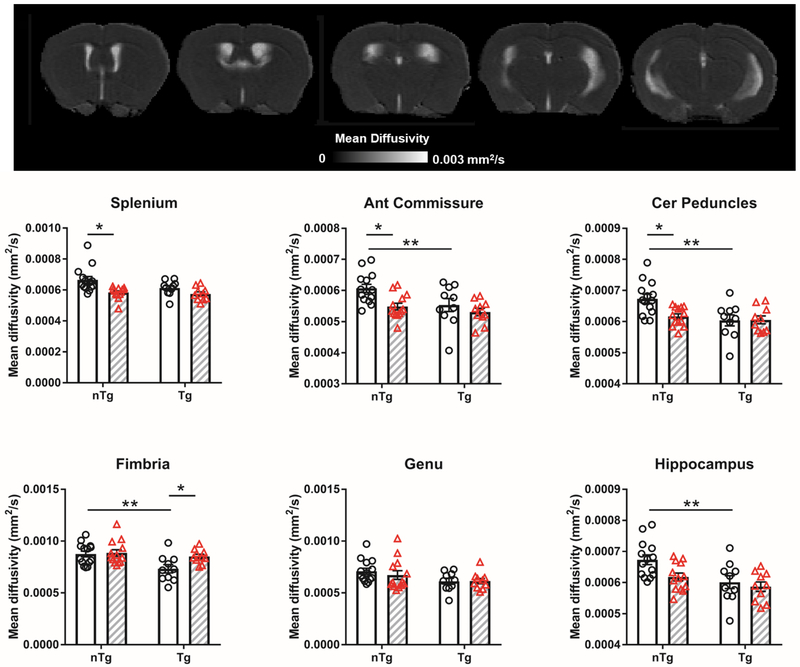
We observed that increased Aβ in Tg mice was associated with reduced MD in anterior commissure (Figure 4; main effect strain F1,43 = 6.1 p = 0.02), cerebral peduncle (main effect strain F1,43 = 8.8 p = 0.005; strain × treatment interaction F1,43 = 4.7 p = 0.04), fimbria (main effect strain F1,43 = 7.9 p = 0.007), genu (main effect strain F1,43 = 5.2 p = 0.03), and hippocampus (main effect strain F1,43 = 10.3 p = 0.003). Elevated IL6 expression reduced MD in splenium (main effect treatment F1,43 = 12.4 p = 0.001), anterior commissure (main effect treatment F1,43 = 8.0 p = 0.007), hippocampus (main effect strain F1,43 = 4.6 p = 0.04), and cerebral peduncle of nTg mice (main effect treatment F1,43 = 4.2 p = 0.04). IL6 increased MD in fimbria of Tg mice (main effect strain F1,43 = 4.2 p = 0.04).
Figure 4.
Aβ and IL6 reduced mean diffusivity (MD) in WM regions. Representative MD map shown in top panel. Clear bars (and clear circles) are control groups and grey hashed bars (and red triangles) are IL6 treated. *significant difference between control and IL6 mice; **significant difference between Tg and nTg mice (Tukey’s post hoc test, p<0.05). All data represented as bar graphs are mean ± standard error.
AD and RD were also assessed (Supplemental Figures 4–5). TgCRND8 mice had lower AD in splenium, anterior commissure, fimbria, hippocampus, and cerebral peduncle than nTg (splenium: main effect strain F1,43 = 12.2 p = 0.001; anterior commissure: main effect strain F1,43 = 14.6 p = 0.0004; fimbria: main effect strain F1,43 = 8.0 p = 0.007; hippocampus: main effect strain F1,43 = 9.8 p = 0.003; cerebral peduncle: main effect strain F1,43 = 30.0 p < 0.0001 and strain × treatment interaction F1,43 = 14.9 p = 0.0004). In cerebral peduncle and fimbria, IL6 treatment increased AD in Tg mice (cerebral peduncle: main effect treatment F1,43 = 6.9 p = 0.01; fimbria: main effect treatment F1,43 = 10.3 p = 0.003).
TgCRND8 mice had lower RD in genu and hippocampus than nTg mice (genu: main effect of strain F1,43 = 8.7 p = 0.005; hippocampus: main effect of strain F1,43 = 9.4 p = 0.004). IL6 reduced RD in splenium, anterior commissure, and cerebral peduncles, but only in TgCRND8 mice (splenium: main effect treatment F1,43 = 13.6 p = 0.0006; anterior commissure: main effect treatment F1,43 = 11.8 p = 0.001; cerebral peduncle: main effect treatment F1,43 = 17.8 p = 0.0001). To sum, in several WM structures both Aβ and IL6 treatment reduced MD, AD, and RD. In TgCRND8 mice, IL6 increased MD and AD but only in few WM areas.
Aβ and IL6 increase ICVF
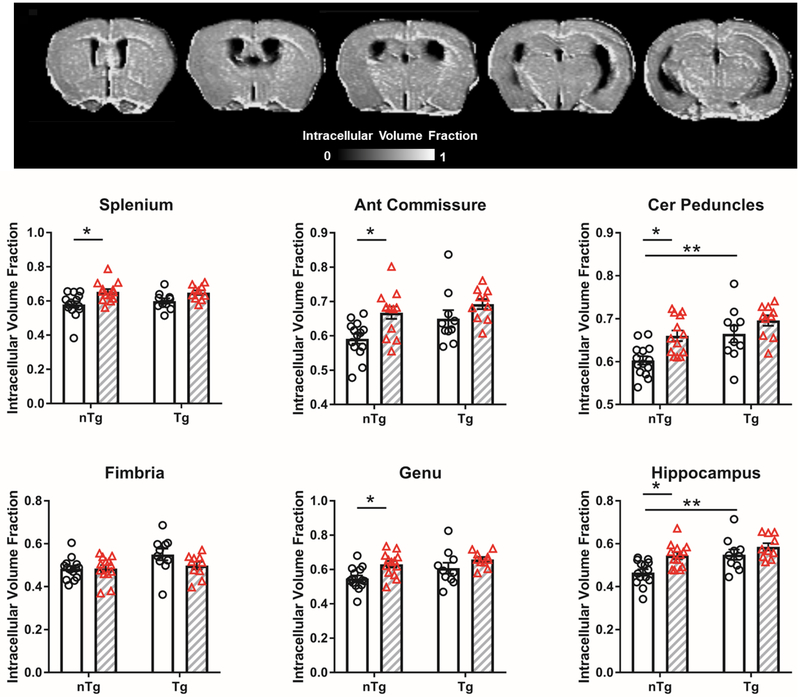
We conducted NODDI analysis to examine the whether Aβ and IL6 altered the fraction of tissue corresponding to axons or dendrites (Figure 5). Highest ICVF values were observed in large WM tracts such as the corpus callosum (which appear bright), and varying degrees of ICVF are observed across grey matter areas (which appear in various light grey tones). TgCRND8 mice had higher ICVF values in the cerebral peduncle and hippocampus (cerebral peduncle: main effect strain F1,43 = 13.4 p = 0.0007; hippocampus: main effect strain F1,43 = 11.8 p = 0.001). IL6 treatment increased ICVF values only in nTg mice in the splenium, anterior commissure, cerebral peduncle, genu and hippocampus (splenium: main effect treatment F1,43 = 12.9 p = 0.0008; anterior commissure: main effect treatment F1,43 = 10.6 p = 0.002; cerebral peduncle: main effect treatment F1,43 = 11.1 p = 0.002; genu: main effect treatment F1,43 = 9.9 p = 0.003; hippocampus: main effect treatment F1,43 = 10.3 p = 0.002).
Figure 5.
Aβ and IL6 increased intracellular volume fraction (ICVF) in WM regions and hippocampus. The effects of IL6 are observed only in nTg mice. Representative ICVF map shown. Clear bars (and clear circles) are control groups and grey hashed bars (and red triangles) are IL6 treated. *significant difference between control and IL6 mice; **significant difference between Tg and nTg mice (Tukey’s post hoc test, p<0.05). All data represented as bar graphs are mean ± standard error.
Aβ increases ODI, while effects of IL6 on ODI are strain-dependent
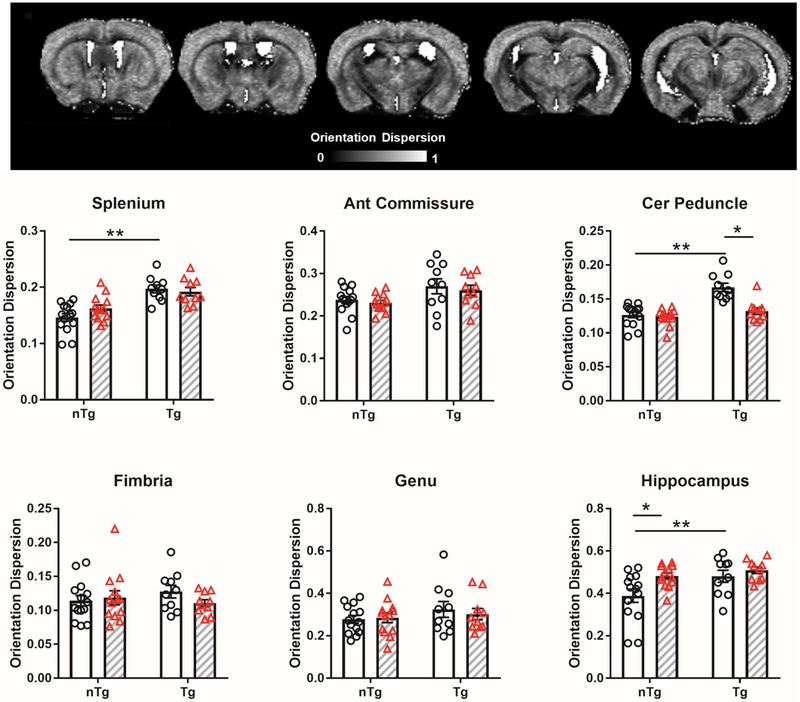
To further characterize spatial configuration of tissue microstructure, we performed analysis for ODI (Figure 6). Lower values of ODI are observed in large WM regions (which appear darker tones), such as splenium, corpus callosum, fimbria, and higher values are observed in grey matter areas (in various light grey tones). Consistent with an increase in ICVF (presumed increased density of modeled neurites), we observed that Aβ increased their degree of orientation dispersion (geometric complexity or misorientation of modeled neurites). Thus, TgCRND8 had increased ODI in splenium, cerebral peduncle and hippocampus (splenium: main effect strain F1,43 = 33.8 p < 0.0001; cerebral peduncle: main effect strain F1,43 = 28.3 p < 0.0001; hippocampus: main effect strain F1,43 = 5.8 p = 0.01). The effects of IL6 were not widespread in the assessed structures and were also strain/tissue type dependent. Thus, in the cerebral peduncle (WM) IL6 reduced ODI only in Tg mice (treatment × strain interaction F1,43 = 12.4 p = 0.001 and main effect treatment F1,43 = 17.0 p = 0.0002), whereas in the hippocampus (grey matter) IL6 increased ODI only in nTg mice (main effect treatment F1,43 = 6.1 p = 0.02).
Figure 6.
Aβ increased orientation dispersion index (ODI) in WM regions and hippocampus. The effects of IL6 differ in nTg and Tg mice. Representative ODI map shown. Clear bars (and clear circles) are control groups and grey hashed bars (and red triangles) are IL6 treated. *significant difference between control and IL6 mice; **significant difference between Tg and nTg mice (Tukey’s post hoc test, p<0.05). All data represented as bar graphs are mean ± standard error.
No effects of Aβ or IL6 on ISO index
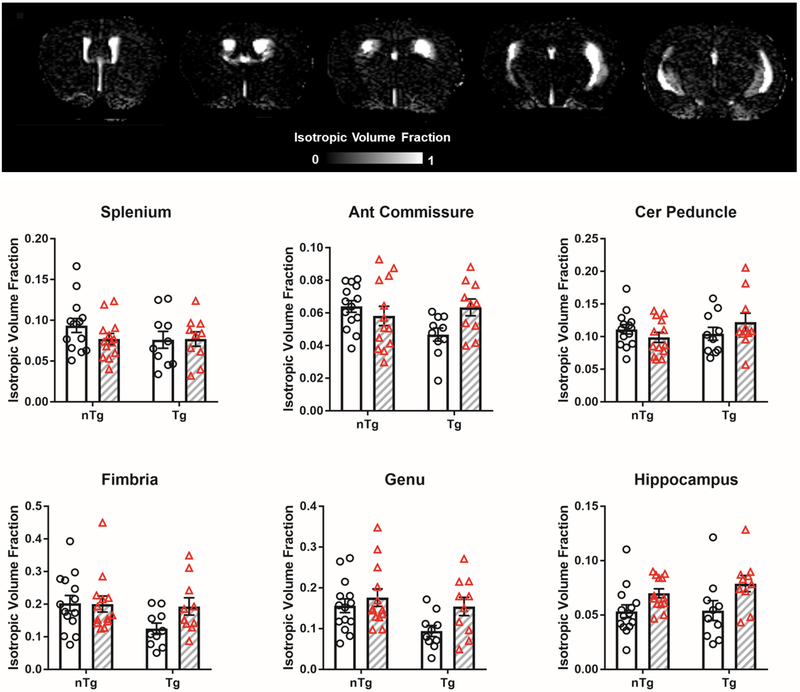
The ISO index is part of the NODDI model that considers freely diffusing, unrestricted/unhindered water, that would be found mostly in cerebral spinal fluid (CSF) containing structures in the brain. As shown in Figure 7, the greatest values of ISO are observed in ventricles. Darkest regions were found in grey matter and WM areas. Overall, we did not observe any effects of Aβ or IL6 on ISO values (Figure 7).
Figure 7.
No effect of Aβ or IL6 was observed on isotropic (ISO) volume fraction. Representative ISO map shown. Clear bars (and clear circles) are control groups and grey hashed bars (and red triangles) are IL6 treated. *significant difference between control and IL6 mice. All data represented as bar graphs are mean ± standard error.
Aβ and IL6 alters a novel relationship between functional network connectivity and microstructure measures
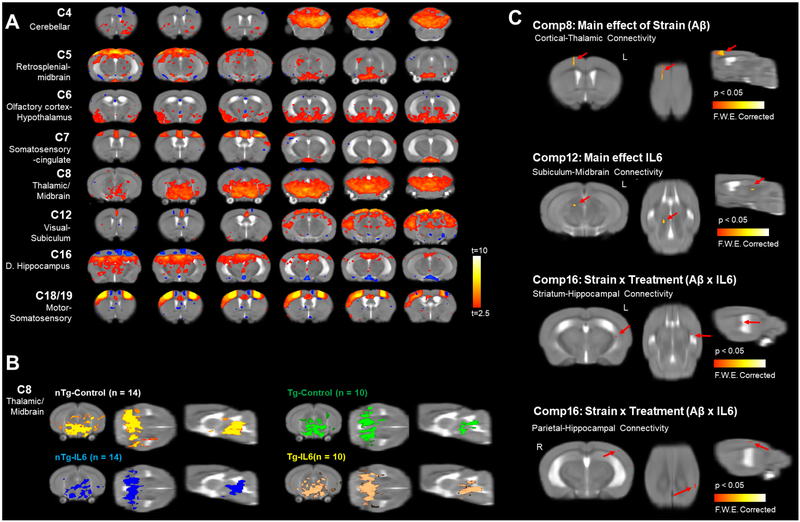
We carried out functional MRI scanning at 11.1 Tesla to investigate if in addition to microstructural changes, IL6 or Aβ expression may affect intrinsic brain activity. For seed-based analysis, we chose cortical and hippocampal areas serving spatial and multi-sensory memory functions (Supplemental Figure 3). Statistical analysis using AFNI 3dMVM did not reveal any significant differences between the groups using this a priori seed-selection approach (data not shown). To supplement the seed-based functional connectivity approach, we conducted model free probabilistic ICA. ICA produced 20 components of which 8 were deemed qualitatively consistent with published resting state functional MRI networks in mice (Grandjean et al., 2017; Sforazzini et al., 2014; Shah et al., 2016). Figure 8A shows 8 components that included cerebellar, visual cortical, olfactory, retrosplenial, somatosensory cortex, anterior cingulate, midbrain, hippocampal, subicular, and thalamic regions. Figure 8B shows a comparison of component-8 for a network connecting caudal thalamus and midbrain, which was similar in all 4 experimental groups. Permutation testing in FSL randomise revealed a significant main effect of Aβ on cortical-thalamic connectivity (component 8), and significant main effect of IL6 on subiculum-midbrain connectivity (component 12), a significant Aβ × IL6 interaction in striatumhippocampal connectivity (component 16) and parietal-hippocampal connectivity (component 16) (p < 0.05 familywise error corrected).
Figure 8.
Independent component analysis of resting state functional MRI revealed significant effects of Aβ and IL6 on hippocampal, thalamic, midbrain and striatal networks. A) Eight of 20 components were consistent with established ICA networks in mice. Shown here are 8, which include anterior cingulate, motor, somatosensory, retrosplenial, midbrain, thalamic, and hippocampal regions (t>2.3). B) Comparison of a thalamic/midbrain component in 4 experimental groups. C) Components that had significant main effects of Aβ, IL6, or Aβ × IL6 interactions following permutation tests (p<0.05 family-wise error corrected).
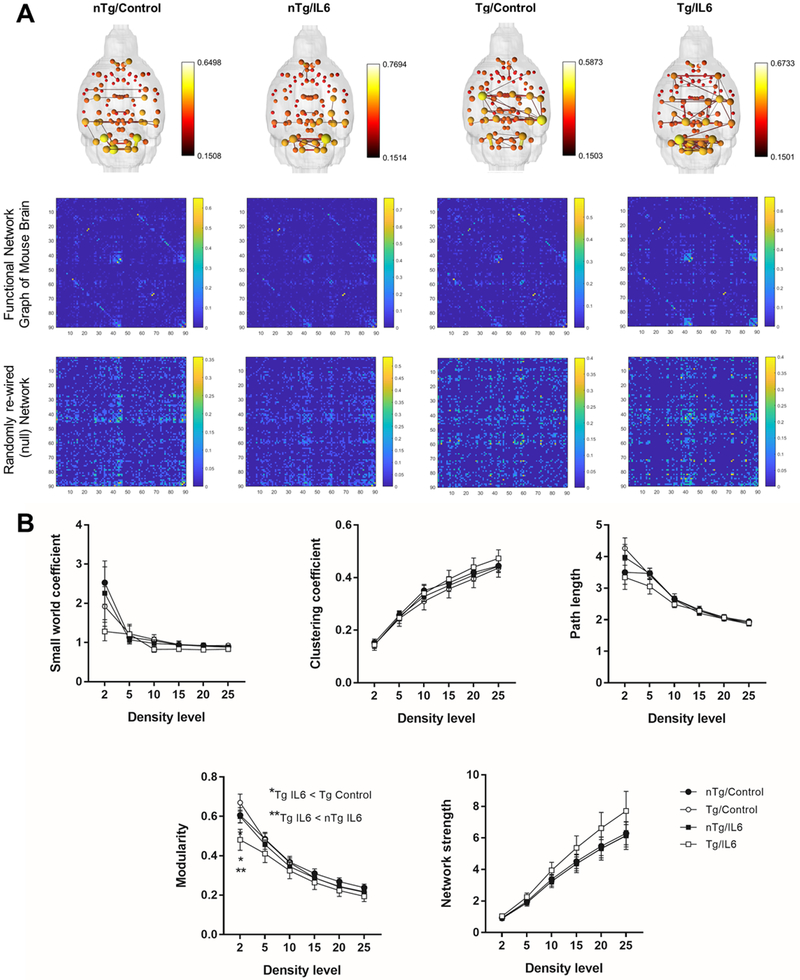
We additionally carried out graph theory analysis on a total of 4,005 pairwise comparisons between a total of 90 ROI’s (45 per mouse brain hemisphere) (Figure 9). Composite 3D functional connectome brains show similar node strengths and edge weights across the 4 experimental groups (network set at a density threshold to retain the top 2% of total edge weights) (Figure 9A). Composite matrices for 3D maps illustrate functional connectivity organizational patterns not observed after randomization of connections (null networks) (Figure 9A). TgCRND8 mice treated with IL6 had a lower modularity coefficient (Q) than Tg controls (F3,264 = 5.2 p = 0.002; p<0.05 Tukey’s test) and nTg IL6 mice (p<0.05 Tukey’s test). No additional differences were observed in clustering coefficient, small world coefficient, mean path length, or global node strength (Supplemental Figure 5).
Figure 9.
IL6 reduced modular organization but only in Tg mice. A) 3D functional connectome maps (density threshold set at 2%; map threshold at edge value of z=0.15). Below maps are composite graphs for each experimental group and their corresponding null networks. B) Connectomic metrics for graphs in A. Top row are functional connectome metrics and the bottom row show results for null networks. *significant difference between control and IL6 mice (Tukey’s post hoc test, p<0.05). All data are mean ± standard error.
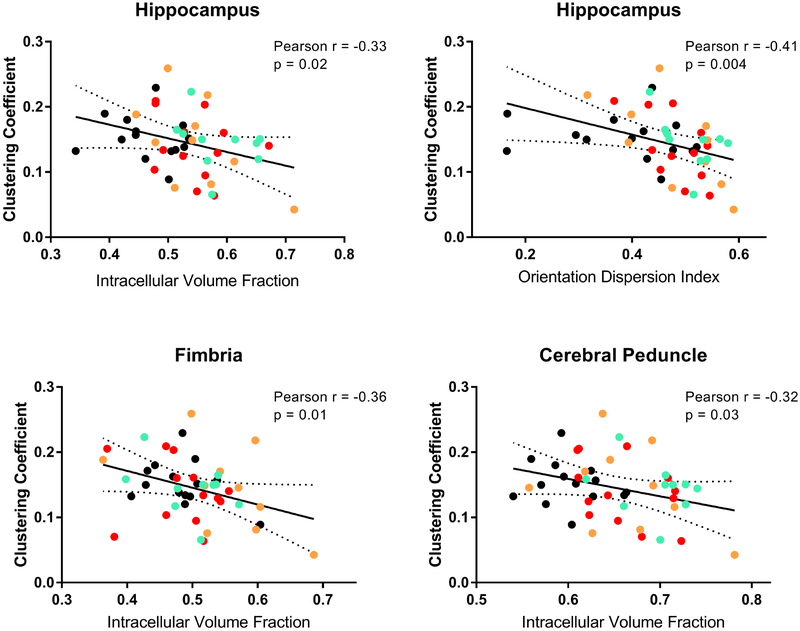
Finally, to investigate the relationship between functional network metrics and DTI and NODDI metrics we carried out correlation analyses. The first step involved normalization of dMRI metrics to their highest value and plotting these against modularity index, clustering coefficient, and node strength for each subject (Supplemental Figure 6). The results indicated that clustering coefficient correlated significantly NODDI metrics. Results from non-normalized NODDI data supported this correlation and indicated that clustering coefficient is altered by the degree of orientation dispersion and ‘neurite density’ (Figure 10). Low ODI or ICVF correlated with high clustering. We observed this relationship in the fimbria (ICVF: p = 0.01), hippocampus (ICVF: p = 0.04; ODI: p = 0.004), and cerebral peduncle (ICVF: p = 0.03) (Figure 10) but found no significant correlations in splenium, genu and anterior commissure (data not shown). Inspection of individual data points revealed that IL6-treated nTg or TgCNRD8 mice (orange and green data points in plots in Figure 10) tended to have higher ODI and ICVF values, which corresponded to lower clustering coefficient values than nTg control mice.
Figure 10.
Aβ and IL6 influenced the relationship between NODDI metrics and clustering coefficient. Values for intracellular volume fraction and orientation dispersion are negatively correlated with the clustering coefficient in fimbria, hippocampus, and cerebral peduncle. Shown are individual data points for nTg-controls (black circles), nTg-IL6 (red circles), Tg-controls (orange circles), and Tg-IL6 (green circles). Statistical p values indicate a significant non-zero slope (99% confidence intervals shown).
DISCUSSION
Using an Alzheimer’s disease mouse model, we carried out high field dMRI to investigate how amyloidosis with IL6-induced inflammatory conditioning alters DTI and NODDI metrics. Our data indicate that Aβ and IL6 differentially alter the various indices of tissue microstructure measured in WM using dMRI (e.g., fractional anisotropy, mean, axial and radial diffusivities, and NODDI indices of intracellular volume fraction and orientation dispersion). Aβ and IL6 had opposite effects on WM FA, the former reduced while the latter decreased FA. The reduction in FA associated with increased Aβ might be due to hindrance to diffusion in the presence of high levels of this protein. IL6 induced increases in FA could on the other hand be associated with some counteracting effect such as clearance of Aβ. Consistent with this notion of microstructural changes that may hinder water diffusivity, we observed reduced MD, AD, and RD in several WM areas as well as the hippocampus. Further, the effects of Aβ and IL6 on dMRI metrics were not additive, which argues against the possibility that Aβ and proinflammatory signaling through IL6 may constitute a double-hit in terms of exerting deleterious actions on tissue microstructure. Contrary to the effects of Aβ and IL6 on FA, their effects on NODDI metrics were similar in directionality. This finding could suggest that in the presence of Aβ or IL6 there is an increase in the density of axonal, dendritic, or perhaps non-neuronal processes, with a dispersed or geometrically disorganized arrangement in WM and hippocampus.
We show that TgCRND8 mice, which have early onset CNS amyloidosis (Chishti et al., 2001), had significant reductions in FA and MD values. This is consistent with previous dMRI research using TgCRND8 mice (Thiessen et al., 2010) and other APP mouse models (Mueggler et al., 2004; Sun et al., 2005), and our previous work using a frontal temporal dementia tauopathy model (Sahara et al., 2014). Reductions in these DTI metrics was associated with reduced AD (parallel diffusivity) and reduced RD (perpendicular diffusivity) (Sun et al., 2005). The overall reduction of white matter AD might be associated with hindered diffusion along axons, which could be related increased concentrations of Aβ, other proteins involved in its production, or deficits in axonal transport (Johnson et al., 2010; Kim et al.; Zhang et al., 2009). Reduced RD could be associated with decreased membrane permeability or atrophy-like changes in axons or dendritic processes (Adalbert et al., 2018; Zhang et al., 2017). Increased IL6 expression had mixed effects on tissue diffusivities and FA in nTg and TgCRND8 mice. The effect of IL6 was not synergistic with Aβ, and in the case of FA, IL6 appeared to counteract the effects of Aβ (Figure 3). This agrees with previous research suggesting protective actions of IL6 on Aβ burden in TgCRND8 mice (Chakrabarty et al., 2010b).
We also assessed NODDI metrics (Zhang et al., 2012) in the same animals processed for DTI. The ICVF has been shown to correlate with histological markers of axonal or dendritic processes (Grussu et al., 2017; Sepehrband et al., 2015), and changes in this NODDI metric has been associated with neurodegenerative changes (Colgan et al., 2016; Grussu et al., 2017; Parker et al., 2018). We observed that Aβ expression in TgCNRD8 mice was associated with increases in both ICVF and the orientation dispersion index (ODI) in WM and hippocampus. At first glance this finding may seem counterintuitive considering the above-described alterations in DTI metrics which might entail pathological mechanisms related to Aβ accumulation or IL6-induced neuroimmune activation. However, the results are consistent with previously reported negative correlations between FA and ODI (Zhang et al., 2012). Low FA values in WM correlate with high ODI, and this latter metric can show a positive correlation with ICVF (Zhang et al., 2012). From a physiological standpoint, the co-occurrence of pathological changes in or near axons/dendritic processes along with increased density of misoriented axonal/dendritic processes in WM and hippocampus seems plausible given previously reported increases in activated microglial cells along with amyloid plaque-associated structural changes to neurons in mouse models with increased Aβ expression.
These findings in the TgCRND8 mouse are in partial agreement with previous NODDI results using the rTg4510 tauopathy mouse model (Colgan et al., 2016). Colgan and colleagues reported increased MD and FA in hippocampus, reduced FA and increased MD in WM, and interestingly, reduced ODI and ICVF in hippocampus and increased ODI and reduced ICVF in WM. The rTg4510 mice were scanned at 8.5 mo when these mice are known to have extensive intracellular tangle pathology in cortex and hippocampus, with a reported loss of over 70% of pyramidal CA1 cells and dendritic spines (Santacruz et al., 2005). Further studies are needed to resolve differences in these two models representing the two major Alzheimer’s disease hallmarks, particularly changes that occur with aging and their direct association with neural activity and cognitive behavioral impairments. As with the effects of IL6 on DTI metrics, the effects of this inflammatory conditioning on NODDI measures varied. IL6 increased ICVF in the splenium, anterior commissure, cerebral peduncle, genu, and hippocampus, but only in nTg mice. Due the finding that Aβ and IL6 were independently observed to increase ICVF and ODI relative to nTg controls, we predict that Aβ proteinopathy and IL6 inflammation may be inducing similar microstructure changes to axons and dendritic processes in white matter and hippocampal areas.
As indicated in the introduction, Aβ and neuroinflammatory mechanisms overlap in the Alzheimer’s disease brain and their corresponding roles in pathological progression of dementia warrants investigation. Six-month old 5xFAD mice showed significant brain accumulation of activated microglia-specific protein tracer TSPO (Mirzaei et al., 2016). In PS2APP mice, there are significant increases in both Aβ and microglia levels from 5month to 16 months of age (Brendel et al., 2016). This is partially consistent with results in humans showing that in advanced Alzheimer’s disease there is a close relationship between amyloid burden with neuroinflammation (Kreisl et al., 2013). However, non-Alzheimer’s disease individuals with mild cognitive impairment (MCI) either fail to show significant microgliosis concomitant with regions of high amyloid burden (Kreisl et al., 2013), or only a subset of MCI subjects show overlapping increases in amyloid and neuroinflammation (Parbo et al., 2017). A longitudinal study identified a transient elevation in microglia early in MCI which then peaks again later in Alzheimer’s disease (Fan et al., 2017). Moreover, areas showing high plaque load do not necessarily show increased activated microglia (Serriere et al., 2015). It is possible that the functional and structural outcome of brain immune responses depend on the specific cytokine-initiated immune response, with some inflammatory mechanisms exacerbating neurodegenerative phenotypes and Aβ deposition while others serving protective roles (Chakrabarty et al., 2010a; Chakrabarty et al., 2011a; Chakrabarty et al., 2011b; Chakrabarty et al., 2010b; Chakrabarty et al., 2012). Comparisons of DTI and NODDI measures after treatment with these different cytokines could be important to understanding the link between dMRI metrics and brain-specific immunity.
Our results are for the most part in agreement with prior studies using APP mouse models. No changes in the apparent diffusion coefficient (ADC) were observed when comparing young 3 mo. to old 12–16 mo. TgCNRD8 mice (Thiessen et al., 2010), although reduced ADC was observed in grey matter areas of 25 mo. compared to 6 mo. APP23 mice (Mueggler et al., 2004). Reductions and increases in FA were measured in 23 mo. Tg2576 APP mice scanned at 11.7 Tesla, and this involved reduced MD, AD, and RD (Muller et al., 2013). In APP/PS1 mice, Qin et al observed increased FA, AD, and RD in cortex, dorsal striatum, thalamus, and in WM structures (Qin et al.), and their electron microscopy results indicated widespread swelling of axons, hypertrophic astrocytes and neuronal loss (Qin et al.). The latter results were highly consistent with those of Shu et al(Shu et al., 2013). In the triple transgenic (3xTgFAD) mouse model, reduced FA and AD was measured only in the hippocampus (Snow et al., 2017). Results similar to those obtained here were reported by Sun et al, who observed reduced relative anisotropy along with reduced AD and increased RD in the Tg2576 mouse model (Sun et al., 2005). Interestingly, opposite changes in hippocampal MD, AD and RD were observed between 6–8 mo. in the APP/PS1 mouse using a diffusion kurtosis image acquisition scheme (Praet et al., 2018). The varied results across studies illustrates difficulties inherent to in vivo dMRI of Alzheimer’s disease mouse models, which setting aside technical challenges, may be influenced by the dynamic nature of the cellular and microstructural changes occurring in response to APP associated mutations in the different mouse models and background genetics (Grandjean et al., 2016; Neuner et al., 2019). However, several caveats in the present study should be emphasized and will be important to consider in future studies. First, while we confirmed the effectiveness of the AAV-mediated IL6 expression and its effects on chronic levels of astrogliosis and microgliosis, a direct ‘ground truth’ histological correlates for IL6 or Aβ on NODDI or DTI metrics was not obtained here. This is an area that merits further investigation in order to improve the translational prospects of these dMRI processing schemes. Second, although care was taken in the manual delineation of ROI’s for dMRI processing, partial volume effects can always impact the results and voxels containing complex underlying tissue types might not accurately reflect the presence of pathology related to IL6 or Aβ. Third, the current study is cross-sectional and does not assess age-specific effects of IL6 and did not monitor overall progression of Aβ effects with age as in previous imaging study in the rTg4510 tauopathy mouse (Sahara et al., 2014).
Finally, we observed an inverse linear relationship between ICVF, ODI in WM and hippocampus and the functional connectomic clustering coefficient. High ICVF and ODI was negatively correlated with low clustering, which suggests that increasing the density and orientation dispersion of modeled neurites may diminish functional network organization in the brain. Implied in this relationship is that in addition to capturing neuronal fiber organization, NODDI metrics somehow measures components of microstructure that may describe or relate to neuronal activity and how neuronal communication is organized in the brain. Recently, it was reported that high ICVF values in auditory/temporal cortex in humans was associated with a decrease latency (faster processing) of neurophysiological processing of speech (Ocklenburg et al., 2018). Also recently, Brown and colleagues observed that decreased default mode network (DMN) deactivation during a working memory task was associated with Alzheimer’s disease pathology and cognitive decline (Brown et al., 2018). Interestingly, decline in WM integrity was reported to influence the reduced DMN deactivation, suggesting a relationship between microstructural differences between controls and Alzheimer’s disease subjects and network activity. Our results suggest a negative relationship potentially involving reduced activity or reduced modular organization in Alzheimer’s disease mice and with IL6 treatment, which is contrary to the positive relationship reported by Ocklenberg et al (Ocklenburg et al., 2018) and Brown et al (Brown et al., 2018). A possibility is that the increased ICVF and ODI in the present work, and its association with reduced clustering, might involve increased presence of neuroglial cells (as noted above) and not necessarily changes in the density of functionally active axons and dendritic processes. Moreover, the fact that both ICVF and ODI were increased may suggest atrophic microstructural differences in TgCRND8 and IL6 treated mice that can adversely impact neural processing. It will be interesting to resolve such interactions between these two different imaging modalities. The relationship between NODDI and graph theory metrics warrants further investigation in the development of novel biomarkers for Alzheimer’s disease and other neurodegenerative diseases.
Supplementary Material
Acknowledgements:
The authors thank the Advanced Magnetic Resonance Imaging and Spectroscopy (AMRIS) facility and National High Magnetic Field Laboratory (NHMFL) for their continued support (National Science Foundation Cooperative Agreement No. DMR-1157490 and the State of Florida).
Funding: This work was supported by the National Institutes of Health grants P50AG047266, R01NS052318, R01NS075012, T32NS082168, P50NS091856, 1R01AG055798.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adalbert R, Milde S, Durrant C, Ando K, Stygelbout V, Yilmaz Z, Gould S, Brion JP, Coleman MP, 2018. Interaction between a MAPT variant causing frontotemporal dementia and mutant APP affects axonal transport. Neurobiol Aging 68, 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin ME, Armitage PA, Marshall I, 1998. A theoretical study of the effect of experimental noise on the measurement of anisotropy in diffusion imaging. Magn Reson Imaging 16, 773–785. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM, 2004. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging 23, 137–152. [DOI] [PubMed] [Google Scholar]
- Behrens TE, Woolrich MW, Jenkinson M, Johansen-Berg H, Nunes RG, Clare S, Matthews PM, Brady JM, Smith SM, 2003. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med 50, 1077–1088. [DOI] [PubMed] [Google Scholar]
- Biscaro B, Lindvall O, Hock C, Ekdahl CT, Nitsch RM, 2009. Abeta immunotherapy protects morphology and survival of adult-born neurons in doubly transgenic APP/PS1 mice. J Neurosci 29, 14108–14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletti S, Latora V, Moreno Y, Chavez M, Hwang DU, 2006. Complex networks: Structure and dynamics. Physics Reports 424, 175–308. [Google Scholar]
- Brendel M, Probst F, Jaworska A, Overhoff F, Korzhova V, Albert NL, Beck R, Lindner S, Gildehaus FJ, Baumann K, Bartenstein P, Kleinberger G, Haass C, Herms J, Rominger A, 2016. Glial Activation and Glucose Metabolism in a Transgenic Amyloid Mouse Model: A Triple-Tracer PET Study. J Nucl Med 57, 954–960. [DOI] [PubMed] [Google Scholar]
- Brown CA, Jiang Y, Smith CD, Gold BT, 2018. Age and Alzheimer’s pathology disrupt default mode network functioning via alterations in white matter microstructure but not hyperintensities. Cortex 104, 58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Ceballos-Diaz C, Beccard A, Janus C, Dickson D, Golde TE, Das P, 2010a. IFN-gamma promotes complement expression and attenuates amyloid plaque deposition in amyloid beta precursor protein transgenic mice. J Immunol 184, 5333–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Ceballos-Diaz C, Beccard A, Janus C, Dickson D, Golde TE, Das P, 2010b. IFN-gamma promotes complement expression and attenuates amyloid plaque deposition in amyloid beta precursor protein transgenic mice. Journal of immunology (Baltimore, Md : 1950) 184, 5333–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Ceballos-Diaz C, Lin WL, Beccard A, Jansen-West K, McFarland NR, Janus C, Dickson D, Das P, Golde TE, 2011a. Interferon-gamma induces progressive nigrostriatal degeneration and basal ganglia calcification. Nat Neurosci 14, 694–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Herring A, Ceballos-Diaz C, Das P, Golde TE, 2011b. Hippocampal expression of murine TNFalpha results in attenuation of amyloid deposition in vivo. Mol Neurodegener 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Jansen-West K, Beccard A, Ceballos-Diaz C, Levites Y, Verbeeck C, Zubair AC, Dickson D, Golde TE, Das P, 2010b. Massive gliosis induced by interleukin-6 suppresses Abeta deposition in vivo: evidence against inflammation as a driving force for amyloid deposition. FASEB J 24, 548–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Li A, Ceballos-Diaz C, Eddy JA, Funk CC, Moore B, DiNunno N, Rosario AM, Cruz PE, Verbeeck C, Sacino A, Nix S, Janus C, Price ND, Das P, Golde TE, 2015. IL-10 alters immunoproteostasis in APP mice, increasing plaque burden and worsening cognitive behavior. Neuron 85, 519–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Tianbai L, Herring A, Ceballos-Diaz C, Das P, Golde TE, 2012. Hippocampal expression of murine IL-4 results in exacerbation of amyloid deposition. Mol Neurodegener 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Adleman NE, Saad ZS, Leibenluft E, Cox RW, 2014. Applications of multivariate modeling to neuroimaging group analysis: a comprehensive alternative to univariate general linear model. Neuroimage 99, 571–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chishti MA, Yang DS, Janus C, Phinney AL, Horne P, Pearson J, Strome R, Zuker N, Loukides J, French J, Turner S, Lozza G, Grilli M, Kunicki S, Morissette C, Paquette J, Gervais F, Bergeron C, Fraser PE, Carlson GA, George-Hyslop PS, Westaway D, 2001. Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J Biol Chem 276, 21562–21570. [DOI] [PubMed] [Google Scholar]
- Chou N, Wu J, Bai Bingren J, Qiu A, Chuang KH, 2011. Robust automatic rodent brain extraction using 3-D pulse-coupled neural networks (PCNN). IEEE Trans Image Process 20, 2554–2564. [DOI] [PubMed] [Google Scholar]
- Colgan N, Siow B, O’Callaghan JM, Harrison IF, Wells JA, Holmes HE, Ismail O, Richardson S, Alexander DC, Collins EC, Fisher EM, Johnson R, Schwarz AJ, Ahmed Z, O’Neill MJ, Murray TK, Zhang H, Lythgoe MF, 2016. Application of neurite orientation dispersion and density imaging (NODDI) to a tau pathology model of Alzheimer’s disease. Neuroimage 125, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW, 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29, 162–173. [DOI] [PubMed] [Google Scholar]
- Cummings DM, Liu W, Portelius E, Bayram S, Yasvoina M, Ho SH, Smits H, Ali SS, Steinberg R, Pegasiou CM, James OT, Matarin M, Richardson JC, Zetterberg H, Blennow K, Hardy JA, Salih DA, Edwards FA, 2015. First effects of rising amyloid-beta in transgenic mouse brain: synaptic transmission and gene expression. Brain 138, 1992–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daducci A, Canales-Rodriguez EJ, Zhang H, Dyrby TB, Alexander DC, Thiran JP, 2015. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. Neuroimage 105, 32–44. [DOI] [PubMed] [Google Scholar]
- Dalgediene I, Lasickiene R, Budvytyte R, Valincius G, Morkuniene R, Borutaite V, Zvirbliene A, 2013. Immunogenic properties of amyloid beta oligomers. J Biomed Sci 20, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Brooks DJ, Okello A, Edison P, 2017. An early and late peak in microglial activation in Alzheimer’s disease trajectory. Brain 140, 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean J, Derungs R, Kulic L, Welt T, Henkelman M, Nitsch RM, Rudin M, 2016. Complex interplay between brain function and structure during cerebral amyloidosis in APP transgenic mouse strains revealed by multi-parametric MRI comparison. Neuroimage 134, 1–11. [DOI] [PubMed] [Google Scholar]
- Grandjean J, Zerbi V, Balsters JH, Wenderoth N, Rudin M, 2017. Structural Basis of Large-Scale Functional Connectivity in the Mouse. J Neurosci 37, 8092–8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grussu F, Schneider T, Tur C, Yates RL, Tachrount M, Ianus A, Yiannakas MC, Newcombe J, Zhang H, Alexander DC, DeLuca GC, Gandini Wheeler-Kingshott CAM, 2017. Neurite dispersion: a new marker of multiple sclerosis spinal cord pathology? Ann Clin Transl Neurol 4, 663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden T, Van Dijk KR, Becker JA, Mehta A, Sperling RA, Johnson KA, Buckner RL, 2009. Disruption of functional connectivity in clinically normal older adults harboring amyloid burden. J Neurosci 29, 12686–12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy AR, Ly M, Carlsson CM, Okonkwo OC, Zetterberg H, Blennow K, Sager MA, Asthana S, Johnson SC, Alexander AL, Bendlin BB, 2017. Microstructural white matter alterations in preclinical Alzheimer’s disease detected using free water elimination diffusion tensor imaging. PLoS One 12, e0173982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MD, Gurney K, 2008. Network ‘small-world-ness’: a quantitative method for determining canonical network equivalence. PLoS One 3, e0002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janus C, Pearson J, McLaurin J, Mathews PM, Jiang Y, Schmidt SD, Chishti MA, Horne P, Heslin D, French J, Mount HT, Nixon RA, Mercken M, Bergeron C, Fraser PE, St George-Hyslop P, Westaway D, 2000. A beta peptide immunization reduces behavioural impairment and plaques in a model of Alzheimer’s disease. Nature 408, 979–982. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S, 2002. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841. [DOI] [PubMed] [Google Scholar]
- Ji F, Pasternak O, Liu S, Loke YM, Choo BL, Hilal S, Xu X, Ikram MK, Venketasubramanian N, Chen CL, Zhou J, 2017. Distinct white matter microstructural abnormalities and extracellular water increases relate to cognitive impairment in Alzheimer’s disease with and without cerebrovascular disease. Alzheimers Res Ther 9, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VE, Stewart W, Smith DH, 2010. Traumatic brain injury and amyloid-beta pathology: a link to Alzheimer’s disease? Nat Rev Neurosci 11, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Knopman DS, Gunter JL, Graff-Radford J, Vemuri P, Boeve BF, Petersen RC, Weiner MW, Jack CR Jr., Alzheimer’s Disease Neuroimaging I, 2016. Cascading network failure across the Alzheimer’s disease spectrum. Brain 139, 547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Choi IY, Michaelis ML, Lee P, Quantitative in vivo measurement of early axonal transport deficits in a triple transgenic mouse model of Alzheimer’s disease using manganese-enhanced MRI. Neuroimage 56, 1286–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisl WC, Lyoo CH, McGwier M, Snow J, Jenko KJ, Kimura N, Corona W, Morse CL, Zoghbi SS, Pike VW, McMahon FJ, Turner RS, Innis RB, Biomarkers Consortium PETRPT, 2013. In vivo radioligand binding to translocator protein correlates with severity of Alzheimer’s disease. Brain 136, 2228–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Le KX, Park MA, Wang S, Belanger AP, Dubey S, Frost JL, Holton P, Reiser V, Jones PA, Trigg W, Di Carli MF, Lemere CA, 2015. In Vivo Detection of Age- and Disease-Related Increases in Neuroinflammation by 18F-GE180 TSPO MicroPET Imaging in Wild-Type and Alzheimer’s Transgenic Mice. J Neurosci 35, 15716–15730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Cardoso MJ, Modat M, Powell N, Wells J, Holmes H, Wiseman F, Tybulewicz V, Fisher E, Lythgoe MF, Ourselin S, 2014. Automatic structural parcellation of mouse brain MRI using multi-atlas label fusion. PLoS One 9, e86576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier-Hein KH, Westin CF, Shenton ME, Weiner MW, Raj A, Thomann P, Kikinis R, Stieltjes B, Pasternak O, 2015. Widespread white matter degeneration preceding the onset of dementia. Alzheimers Dement 11, 485–493 e482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo CD, Mazerolle EL, Ritchie L, Fisk JD, Gawryluk JR, Alzheimer’s Disease Neuroimaging I, 2017. Longitudinal changes in microstructural white matter metrics in Alzheimer’s disease. Neuroimage Clin 13, 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei N, Tang SP, Ashworth S, Coello C, Plisson C, Passchier J, Selvaraj V, Tyacke RJ, Nutt DJ, Sastre M, 2016. In vivo imaging of microglial activation by positron emission tomography with [(11)C]PBR28 in the 5XFAD model of Alzheimer’s disease. Glia 64, 993–1006. [DOI] [PubMed] [Google Scholar]
- Moore K, Madularu D, Iriah S, Yee JR, Kulkarni P, Darcq E, Kieffer BL, Ferris CF, 2016. BOLD Imaging in Awake Wild-Type and Mu-Opioid Receptor Knock-Out Mice Reveals On-Target Activation Maps in Response to Oxycodone. Front Neurosci 10, 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueggler T, Meyer-Luehmann M, Rausch M, Staufenbiel M, Jucker M, Rudin M, 2004. Restricted diffusion in the brain of transgenic mice with cerebral amyloidosis. Eur J Neurosci 20, 811–817. [DOI] [PubMed] [Google Scholar]
- Muller HP, Kassubek J, Vernikouskaya I, Ludolph AC, Stiller D, Rasche V, 2013. Diffusion tensor magnetic resonance imaging of the brain in APP transgenic mice: a cohort study. PLoS One 8, e67630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuner SM, Heuer SE, Huentelman MJ, O’Connell KMS, Kaczorowski CC, 2019. Harnessing Genetic Complexity to Enhance Translatability of Alzheimer’s Disease Mouse Models: A Path toward Precision Medicine. Neuron 101, 399–411 e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman ME, Girvan M, 2004. Finding and evaluating community structure in networks. Phys Rev E Stat Nonlin Soft Matter Phys 69, 026113. [DOI] [PubMed] [Google Scholar]
- Newman MEJ, 2003. The Structure and Function of Complex Networks. SIAM Review 45, 167–256. [Google Scholar]
- Nickerson LD, Smith SM, Ongur D, Beckmann CF, 2017. Using Dual Regression to Investigate Network Shape and Amplitude in Functional Connectivity Analyses. Front Neurosci 11, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dwyer L, Lamberton F, Bokde AL, Ewers M, Faluyi YO, Tanner C, Mazoyer B, O’Neill D, Bartley M, Collins DR, Coughlan T, Prvulovic D, Hampel H, 2011. Using diffusion tensor imaging and mixed-effects models to investigate primary and secondary white matter degeneration in Alzheimer’s disease and mild cognitive impairment. J Alzheimers Dis 26, 667–682. [DOI] [PubMed] [Google Scholar]
- Ocklenburg S, Friedrich P, Fraenz C, Schluter C, Beste C, Gunturkun O, Genc E, 2018. Neurite architecture of the planum temporale predicts neurophysiological processing of auditory speech. Sci Adv 4, eaar6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofori E, Pasternak O, Planetta PJ, Burciu R, Snyder A, Febo M, Golde TE, Okun MS, Vaillancourt DE, 2015. Increased free water in the substantia nigra of Parkinson’s disease: a single-site and multi-site study. Neurobiol Aging 36, 1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parbo P, Ismail R, Hansen KV, Amidi A, Marup FH, Gottrup H, Braendgaard H, Eriksson BO, Eskildsen SF, Lund TE, Tietze A, Edison P, Pavese N, Stokholm MG, Borghammer P, Hinz R, Aanerud J, Brooks DJ, 2017. Brain inflammation accompanies amyloid in the majority of mild cognitive impairment cases due to Alzheimer’s disease. Brain 140, 2002–2011. [DOI] [PubMed] [Google Scholar]
- Parker TD, Slattery CF, Zhang J, Nicholas JM, Paterson RW, Foulkes AJM, Malone IB, Thomas DL, Modat M, Cash DM, Crutch SJ, Alexander DC, Ourselin S, Fox NC, Zhang H, Schott JM, 2018. Cortical microstructure in young onset Alzheimer’s disease using neurite orientation dispersion and density imaging. Hum Brain Mapp 39, 3005–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak O, Sochen N, Gur Y, Intrator N, Assaf Y, 2009. Free water elimination and mapping from diffusion MRI. Magn Reson Med 62, 717–730. [DOI] [PubMed] [Google Scholar]
- Pasternak O, Westin CF, Bouix S, Seidman LJ, Goldstein JM, Woo TU, Petryshen TL, Mesholam-Gately RI, McCarley RW, Kikinis R, Shenton ME, Kubicki M, 2012. Excessive extracellular volume reveals a neurodegenerative pattern in schizophrenia onset. J Neurosci 32, 17365–17372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak O, Westin CF, Dahlben B, Bouix S, Kubicki M, 2015. The extent of diffusion MRI markers of neuroinflammation and white matter deterioration in chronic schizophrenia. Schizophr Res 161, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praet J, Manyakov NV, Muchene L, Mai Z, Terzopoulos V, de Backer S, Torremans A, Guns PJ, Van De Casteele T, Bottelbergs A, Van Broeck B, Sijbers J, Smeets D, Shkedy Z, Bijnens L, Pemberton DJ, Schmidt ME, Van der Linden A, Verhoye M, 2018. Diffusion kurtosis imaging allows the early detection and longitudinal follow-up of amyloid-beta-induced pathology. Alzheimers Res Ther 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YY, Li MW, Zhang S, Zhang Y, Zhao LY, Lei H, Oishi K, Zhu WZ, In vivo quantitative whole-brain diffusion tensor imaging analysis of APP/PS1 transgenic mice using voxel-based and atlas-based methods. Neuroradiology. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O, 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069. [DOI] [PubMed] [Google Scholar]
- Sadleir KR, Kandalepas PC, Buggia-Prevot V, Nicholson DA, Thinakaran G, Vassar R, 2016. Presynaptic dystrophic neurites surrounding amyloid plaques are sites of microtubule disruption, BACE1 elevation, and increased Abeta generation in Alzheimer’s disease. Acta Neuropathol 132, 235–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara N, Perez PD, Lin WL, Dickson DW, Ren Y, Zeng H, Lewis J, Febo M, 2014. Age-related decline in white matter integrity in a mouse model of tauopathy: an in vivo diffusion tensor magnetic resonance imaging study. Neurobiol Aging 35, 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, Guimaraes A, DeTure M, Ramsden M, McGowan E, Forster C, Yue M, Orne J, Janus C, Mariash A, Kuskowski M, Hyman B, Hutton M, Ashe KH, 2005. Tau suppression in a neurodegenerative mouse model improves memory function. Science 309, 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saramaki J, Kivel M, Onnela J-P, Kaski K, Kertesz J, 2007. Generalizations of the clustering coefficient to weighted complex networks. Physical Review E 75, 027105. [DOI] [PubMed] [Google Scholar]
- Schuitemaker A, Kropholler MA, Boellaard R, van der Flier WM, Kloet RW, van der Doef TF, Knol DL, Windhorst AD, Luurtsema G, Barkhof F, Jonker C, Lammertsma AA, Scheltens P, van Berckel BN, 2013. Microglial activation in Alzheimer’s disease: an (R)-[(1)(1)C]PK11195 positron emission tomography study. Neurobiol Aging 34, 128–136. [DOI] [PubMed] [Google Scholar]
- Sepehrband F, Clark KA, Ullmann JF, Kurniawan ND, Leanage G, Reutens DC, Yang Z, 2015. Brain tissue compartment density estimated using diffusion-weighted MRI yields tissue parameters consistent with histology. Hum Brain Mapp 36, 3687–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serriere S, Tauber C, Vercouillie J, Mothes C, Pruckner C, Guilloteau D, Kassiou M, Domene A, Garreau L, Page G, Chalon S, 2015. Amyloid load and translocator protein 18 kDa in APPswePS1-dE9 mice: a longitudinal study. Neurobiol Aging 36, 1639–1652. [DOI] [PubMed] [Google Scholar]
- Sforazzini F, Schwarz AJ, Galbusera A, Bifone A, Gozzi A, 2014. Distributed BOLD and CBV-weighted resting-state networks in the mouse brain. Neuroimage 87, 403–415. [DOI] [PubMed] [Google Scholar]
- Shah D, Deleye S, Verhoye M, Staelens S, Van der Linden A, 2016. Resting-state functional MRI and [18F]-FDG PET demonstrate differences in neuronal activity between commonly used mouse strains. Neuroimage 125, 571–577. [DOI] [PubMed] [Google Scholar]
- Shrivastava AN, Aperia A, Melki R, Triller A, 2017. Physico-Pathologic Mechanisms Involved in Neurodegeneration: Misfolded Protein-Plasma Membrane Interactions. Neuron 95, 33–50. [DOI] [PubMed] [Google Scholar]
- Shu X, Qin YY, Zhang S, Jiang JJ, Zhang Y, Zhao LY, Shan D, Zhu WZ, 2013. Voxel-based diffusion tensor imaging of an APP/PS1 mouse model of Alzheimer’s disease. Mol Neurobiol 48, 78–83. [DOI] [PubMed] [Google Scholar]
- Slattery CF, Zhang J, Paterson RW, Foulkes AJM, Carton A, Macpherson K, Mancini L, Thomas DL, Modat M, Toussaint N, Cash DM, Thornton JS, Henley SMD, Crutch SJ, Alexander DC, Ourselin S, Fox NC, Zhang H, Schott JM, 2017. ApoE influences regional white-matter axonal density loss in Alzheimer’s disease. Neurobiol Aging 57, 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM, 2004. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23 Suppl 1, S208–219. [DOI] [PubMed] [Google Scholar]
- Snow WM, Dale R, O’Brien-Moran Z, Buist R, Peirson D, Martin M, Albensi BC, 2017. In Vivo Detection of Gray Matter Neuropathology in the 3xTg Mouse Model of Alzheimer’s Disease with Diffusion Tensor Imaging. J Alzheimers Dis 58, 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spires TL, Hyman BT, 2004. Neuronal structure is altered by amyloid plaques. Rev Neurosci 15, 267–278. [DOI] [PubMed] [Google Scholar]
- Sun SW, Song SK, Harms MP, Lin SJ, Holtzman DM, Merchant KM, Kotyk JJ, 2005. Detection of age-dependent brain injury in a mouse model of brain amyloidosis associated with Alzheimer’s disease using magnetic resonance diffusion tensor imaging. Exp Neurol 191, 77–85. [DOI] [PubMed] [Google Scholar]
- Sykova E, Vorisek I, Antonova T, Mazel T, Meyer-Luehmann M, Jucker M, Hajek M, Ort M, Bures J, 2005. Changes in extracellular space size and geometry in APP23 transgenic mice: a model of Alzheimer’s disease. Proc Natl Acad Sci U S A 102, 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M, Schneider T, Alexander DC, Gandini Wheeler-Kingshott CA, Zhang H, 2016. Bingham-NODDI: Mapping anisotropic orientation dispersion of neurites using diffusion MRI. Neuroimage 133, 207–223. [DOI] [PubMed] [Google Scholar]
- Teipel SJ, Grothe MJ, Filippi M, Fellgiebel A, Dyrba M, Frisoni GB, Meindl T, Bokde AL, Hampel H, Kloppel S, Hauenstein K, group E.s., 2014. Fractional anisotropy changes in Alzheimer’s disease depend on the underlying fiber tract architecture: a multiparametric DTI study using joint independent component analysis. J Alzheimers Dis 41, 69–83. [DOI] [PubMed] [Google Scholar]
- Thiessen JD, Glazner KA, Nafez S, Schellenberg AE, Buist R, Martin M, Albensi BC, 2010. Histochemical visualization and diffusion MRI at 7 Tesla in the TgCRND8 transgenic model of Alzheimer’s disease. Brain Struct Funct 215, 29–36. [DOI] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, 2010. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29, 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga E, Juhasz G, Bozso Z, Penke B, Fulop L, Szegedi V, 2015. Amyloid-beta1–42 Disrupts Synaptic Plasticity by Altering Glutamate Recycling at the Synapse. J Alzheimers Dis 45, 449–456. [DOI] [PubMed] [Google Scholar]
- Xia M, Wang J, He Y, 2013. BrainNet Viewer: a network visualization tool for human brain connectomics. PLoS One 8, e68910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G, 2006. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–1128. [DOI] [PubMed] [Google Scholar]
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC, 2012. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chao FL, Luo YM, Xiao Q, Jiang L, Zhou CN, Zhang Y, Lv FL, He Q, Ma J, Tang Y, 2017. Exercise Prevents Cognitive Function Decline and Demyelination in the White Matter of APP/PS1 Transgenic AD Mice. Curr Alzheimer Res 14, 645–655. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Cai Y, Xiong K, Cai H, Luo XG, Feng JC, Clough RW, Struble RG, Patrylo PR, Yan XX, 2009. Beta-secretase-1 elevation in transgenic mouse models of Alzheimer’s disease is associated with synaptic/axonal pathology and amyloidogenesis: implications for neuritic plaque development. Eur J Neurosci 30, 2271–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.