Abstract
Background
Implementation of phenotype algorithms requires phenotype engineers to interpret human-readable algorithms and translate the description (text and flowcharts) into computable phenotypes – a process that can be labor intensive and error prone. To address the critical need for reducing the implementation efforts, it is important to develop portable algorithms.
Methods
We conducted a retrospective analysis of phenotype algorithms developed in the Electronic Medical Records and Genomics (eMERGE) network and identified common customization tasks required for implementation. A novel scoring system was developed to quantify portability from three aspects: Knowledge conversion, clause Interpretation, and Programming (KIP). Tasks were grouped into twenty representative categories. Experienced phenotype engineers were asked to estimate the average time spent on each category and evaluate time saving enabled by a common data model (CDM), specifically the Observational Medical Outcomes Partnership (OMOP) model, for each category.
Results
A total of 485 distinct clauses (phenotype criteria) were identified from 55 phenotype algorithms, corresponding to 1153 customization tasks. In addition to 25 non-phenotype-specific tasks, 46 tasks are related to interpretation, 613 tasks are related to knowledge conversion, and 469 tasks are related to programming. A score between 0 and 2 (0 for easy, 1 for moderate, and 2 for difficult portability) is assigned for each aspect, yielding a total KIP score range of 0 to 6. The average clause-wise KIP score to reflect portability is 1.37±1.38. Specifically, the average knowledge (K) score is 0.64±0.66, interpretation (I) score is 0.33±0.55, and programming (P) score is 0.40±0.64. 5% of the categories can be completed within one hour (median). 70% of the categories take from days to months to complete. The OMOP model can assist with vocabulary mapping tasks.
Conclusion
This study presents firsthand knowledge of the substantial implementation efforts in phenotyping and introduces a novel metric (KIP) to measure portability of phenotype algorithms for quantifying such efforts across the eMERGE Network. Phenotype developers are encouraged to analyze and optimize the portability in regards to knowledge, interpretation and programming. CDMs can be used to improve the portability for some ‘knowledge-oriented’ tasks.
Keywords: Phenotyping, Electronic health records, Portability
Graphical Abstract
1. Background
Electronic health record (EHR) phenotyping refers to the identification of a cohort of patients with a specific clinical profile by developing an algorithm to query a wide spectrum of EHR data elements, such as diagnoses, laboratory tests, medications, procedures and derived concepts from clinical narratives using natural language processing (NLP) [1]. With domain experts’ input, phenotyping algorithms can be developed using knowledge engineering [2–6] methods of developing “rule-based” strategies or machine learning methods [6]. When a phenotype algorithm is developed, relevant features and the corresponding EHR data representations are identified and combined using logic to define cohort selection rules [3]. The resulting cohort can then be used for observational studies, clinical trial cohort identification, comparative effectiveness research and more recently, genome-wide and phenome-wide association studies for specific disease [7], syndrome [8] or complex trait [9].
The Electronic Medical Records and Genomics (eMERGE) Network (an NIH-funded national consortium that aims to conduct large scale and cross-site genomic studies by linking DNA biobanks to EHRs) leverages EHRs to develop phenotype algorithms across its participating sites in order to have larger sample size for its genomic research [2,10]. The process of developing and validating a phenotype algorithm can take a very long time, sometimes up to 6–10 months, depending on the complexity [11]. eMERGE has created the Phenotype KnowledgeBase website (PheKB, http://phekb.org) [11] to enable cross-site sharing of phenotype algorithms. Up to date, 90 phenotypes are under development or testing, 41 phenotypes have been validated, and 53 phenotypes have been finalized and are available to public access. A phenotype algorithm shared in the eMERGE network is represented by a narrative description, augmented by pseudocodes and flowcharts to illustrate the logical reasoning. Necessary codes for clinical concepts are embedded in the description or attached as an appendix. Additionally, each phenotype is tagged with the owner institution, owner phenotyping groups, development status and other metadata.
To implement a phenotype at another site, the first step is to understand the algorithm described in the narrative within the site-specific context of the data and clinical sublanguage [12] available. Then, phenotype engineers translate the site-dependent interpretation of the algorithm to machine executable code and deploy it within a site-specific clinical environment. Consequently, human intervention on both interpretation and implementation is required. Using a narrative description to promote the algorithm sharing, while assisting the interpretation, may also generate a labor-intensive and error-prone implementation process affecting efficiency. Given this, implementing a phenotype algorithm can also take several months. Developing and implementing phenotype algorithms consumes substantial resources from each eMERGE site. In addition, the current process does not scale well and the resulting phenotype algorithms often have limited portability.
In this paper, a “phenotype engineer” is defined as a person who develops or implements electronic phenotypes that mainly use knowledge engineering methods which transform and model problem-solving expertise on defining diseases, syndromes or complex traits from a knowledge source to computer programs such as SQL [13,14]. From our point of view, “phenotype engineers” are “knowledge engineers” who are specialized at electronic phenotyping. Knowledge engineers are different from software engineers considering that knowledge engineers focus on transforming domain knowledge to computer programs while software engineers focus on directing the life-cycle of software development [15] For example, software engineers focus on developing MedLEE for medical concept extraction from clinical narratives. Knowledge engineers focus on applying and customizing MedLEE to deliver a specific phenotyping solution.
A portable phenotype is one that can be implemented faithfully [16] and easily at a different site while maintaining a similar performance [17]. In other words, the portability of an algorithm can be measured by the performance achieved at an implementation site combined with the time and efforts cost during the process of implementation. Studies have developed extensive methods to improve phenotype portability on performance consistency [18,19]. However, it is hard to measure and achieve the portability of phenotype algorithms in respect of implementation efforts due to the heterogeneity of EHR systems, data representations and clinical documentation patterns among different sites. Various strategies have been explored to achieve portability, including the development of the NQF Quality Data Model [20], the Phenotype Execution and Modeling Architecture [21,22], and algorithm design patterns [23]. However, measuring portability and recognizing hurdles for algorithm portability have yet to be analyzed systematically.
In this study, we focus on quantifying the implementation work or the customization tasks (referred to as “task” hereafter) involved to make a phenotype algorithm portable. The task is defined as what work needs to be done by the implementation site before executing the computable phenotype (e.g., a database SQL query, SAS/R implementation, KNIME application, ATLAS query). Furthermore, we assess whether a common data model (CDM), such as the Observational Medical Outcomes Partnership (OMOP) CDM, can alleviate the implementation burden. Finally, we will discuss best practices in designing portable phenotyping algorithms.
2. Methods
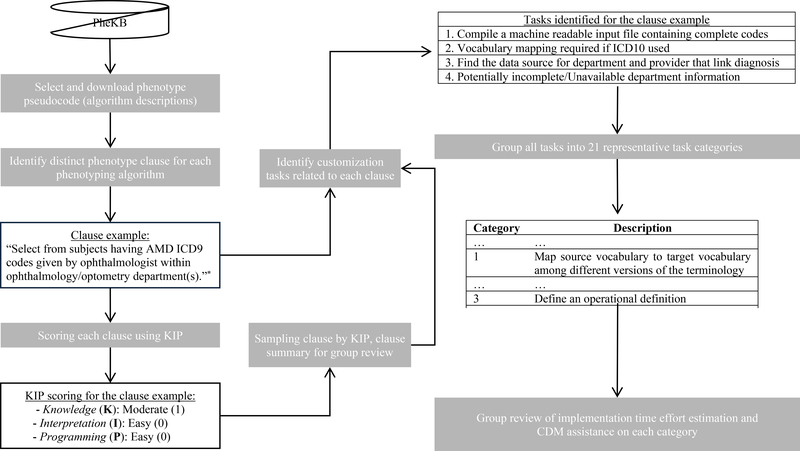
The workflow of this study is shown in Fig. 1. Overall, we disassembled 55 phenotype algorithms obtained from PheKB, identified the customization tasks from phenotyping engineers’ implementation experiences and reached agreement through iterative review and group consensus, and then grouped them into representative categories. A category is a conceptual grouping of customization tasks that are considered similar by relating to the same facet of the phenotype implementation. Experienced engineers from the participating eMERGE sites were then surveyed to estimate the time efforts for each identified category. A novel scoring system was created to quantify the portability of a phenotype algorithm. We will describe each step in detail next.
Fig. 1.
Overview of the study (*clause example is from https://phekb.org/phenotype/age-related-macular-degeneration)
2.1. Phenotype and clause selection
For this study, we choose phenotypes that have “final” or “validated” status, were developed and validated by eMERGE sites, and have been as well implemented by at least 5 of the 10 Phase III (current phase) eMERGE sites. These choices provide a reasonable proxy that sufficient firsthand implementation experiences from multiple participant sites would be available.
Fifty-five qualified phenotypes were downloaded from PheKB on February 7, 2019. One of the authors (NS) reviewed all phenotype algorithms and identified distinct clauses from the narrative definition of the phenotype algorithms. Each clause (Fig. 2 provides an example [7]) is generally one criterion for defining the phenotype or one instruction corresponding to a specific decision or action node in the algorithm and may contain one or more sentences. Figure workflows, chart review instructions, software installation instructions and data dictionaries from the phenotype algorithms were not included in the analysis.
Fig. 2.
Example of clause selected from an algorithm
All clauses were then reviewed and grouped into multiple clause summaries (see example in Fig. 2). The clause summary is defined as a brief statement in general terms to describe the goal of a phenotyping rule and is used for sampling clauses in the following tasks group review process. Details of clause summaries and corresponding examples can be found in Appendix 1. At the same time, relevant data types used in each clause were also identified (e.g., diagnosis, visit, pathology report).
2.2. Task identification and Knowledge-Interpretation-Programming (KIP) scoring
As not all variables defined by the algorithm development site are ready-to-extract from the clinical data warehouse of each implementing site, customization tasks can potentially affect the portability of the algorithm. Tasks may include getting input from domain experts and data curators, exploring data for understanding specific medical events, and generating middle-step products for extracting phenotype relevant variables. Using the set of curated clauses, two of the authors (NS and CL) identified all likely tasks involved in a specific clause, based on their collective experience in phenotype implementation. The task is from the implementation site’s perspective and reflects the actual work that two authors have conducted for converting phenotype algorithms to executable phenotypes. Overall, the task is defined as the customization that the implementation site needs to do for achieving the specific goal defined in the clause.
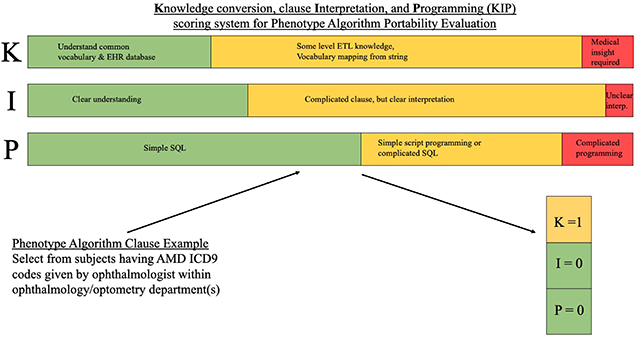
To the best of our knowledge, no previous work has defined a quantitative measure for portability to date, we developed a scoring system to quantify the portability of each clause based on the tasks involved. Our scoring system considers three aspects of an implementation: Knowledge conversion (K), clause Interpretation (I) and Programming (P). Table 1 provides the definitions and examples of the KIP scoring system. In general, knowledge conversion defines how much knowledge of the data or clinical knowledge related to this phenotype that a phenotype engineer is required to know before finishing this task. If the process to acquire the knowledge is more difficult and time consuming, then the portability is lower. For example, since consulting a domain expert often takes more time than checking a mapping table between two vocabularies, the knowledge conversion difficulty can be higher for defining a medical concept based on local clinical environment than a simple vocabulary mapping. Clause interpretation mainly refers to the consistency of phenotype engineers’ understanding of the algorithm according to the algorithm developers’ intent. Programming is defined by the level of computational complexity required for the implementation. If a task involves more advanced skills, the implementation may be more difficult, and therefore result in a lower portability. For example, a “GROPU BY” SQL query to count a diagnosis event is, in general, simpler than implementing a complicated name entity recognition (NER) system for specific events from unstructured data.
Table 1.
Definition of the KIP (knowledge, interpretation and programming) scoring system for evaluating algorithm portability
| Aspect | Score | Definition | Example |
|---|---|---|---|
| Knowledge conversion | 0 | Require basic understanding of common vocabulary (ICD, CPT, RxNorm, LOINC) and EHR database (e.g., diagnosis, lab test, procedure, medication, visit) | “...with one or more code of kidney transplant. (ICD9, ICD10 and CPT4 codes are provided)...” |
| 1 | Require some level of ETL knowledge for understanding EHR database underlying structure, or require vocabulary mapping from strings | “...with at least one T1DM medications order. (Drug generic name provided in the table)...” | |
| 2 | Medical insight is required from both domain experts and engineers to define a medical concept | “...has continuous enrollment for 5 or more years …” | |
| Clause interpretation | 0 | Very clear understanding | “…with diabetes family history…” |
| 1 | Complicated or long clause, but clear interpretation | “…T2DM Dx by physician >=2 …” | |
| 2 | Unclear interpretation | “…Control excl: Any cause of hypo- or hyper-thyroidism…” | |
| Programming | 0 | Simple SQL implementation only | “…one or more existing diagnosis of chronic kidney disease or other kidney disease…” |
| 1 | Simple script programming (e.g., keywords search) or complicated SQL | “…identify all radiology reports with keyword ‘pneumonia’…” | |
| 2 | Complicated programming (e.g., software configuration, post script programming, NegEx implementation) | “…search for a MedLEE attribute of “certainty” with any of the following values…” |
Two authors (NS and CL) worked independently to identify customization tasks and assign a KIP score for each clause (scaled with 0 as easy, 1 as moderate, and 2 as difficult for each aspect K, I, and P). Through iterative discussion and review, they developed a consensus set of customization tasks, and also arrived at a consensus for KIP score on each clause. Due to the iterative and collaborative nature of this process, the agreement rate was not calculated. To assess the potential bias introduced by two reviewers, a subsample (28%) of the clauses (Appendix 2) whose KIP total score is more than 0 was randomly selected proportionally from clause summary and KIP total score groups. The subsample was subsequently reviewed by 9 experienced phenotype engineers from eight eMERGE sites. To assess the consensus on any revision of identified tasks among reviewers, the overall agreement rate was calculated as the ratio of the number of sampled clauses where identified tasks are agreed by all reviewers over the number of total sampled clauses. To assess the consensus on the completeness of identified tasks among reviewers, the completeness agreement rate was calculated as the ratio of the number of sample clauses where identified tasks are deemed as complete (specifically, no extra task is identified) by all reviewers over the number of total sampled clauses.
2.3. Estimate time efforts on customization task categories and common data model assistance for portable phenotyping
By incorporating the feedback, recommendations, and comments provided by all the phenotyping engineers participating in this collaborative study, two authors (NS and CL) finalized the tasks and grouped them into categories by subjective assessment. For example, the task of “mapping International Classification of Diseases 9 (ICD9) to ICD10 codes” belongs to the category of “map source vocabulary to target vocabulary among different versions of the terminology” (Fig. 1). This bottom-up approach facilitated the recognition of comprehensive considerations for algorithm portability from the detailed customized tasks identified by phenotype engineers.
One of the key measurements of portability of a phenotype algorithm is the implementation time [22]. Here the implementation time is not just the execution time for the program. Instead, it consists of the time required for humans to interpret the algorithm, consult the domain experts, make decisions for development, translate the free-text description to computable codes, and deploy the final algorithm product. In this study, 17 phenotype engineers from eight eMERGE sites with at least two-years of experience in phenotyping algorithm implementation were asked to estimate the time needed to implement each task involved in each category aforementioned. Among them, 8 respondents claim their role as investigator, 7 respondents claim as informatician or programmer, 1 respondent claims as physician and 1 respondent claims as data analyst. Given the anonymous nature of the second survey, we are not able to calculate the overlap between the two sets of respondents. We evaluated the time consumption for 20 task categories, collapsed them into five levels: (1) 0–1 hour, (2) < 1 hour - 1 day, (3) < 1 day - 1 week, (4) 1 week - 1 month, and (5) > 1 month. In addition, the variance of reported time consumption was also assessed.
CDMs are generally recommended for assisting portable implementation. CDMs organize EHR data according to a common standard and can dramatically speed phenotype implementation at the cost of having to populate that data model [24]. Due to the resources required to populate structured and unstructured EHR data into a CDM, current CDMs are not fully functional for phenotyping implementation. Therefore, the same 17 phenotyping engineers reported their opinion whether or not each task category can be assisted by the CDM. Since the OMOP CDM from Observational Health Data Sciences and Informatics (OHDSI) has been piloted in the eMERGE network for phenotyping implementation, the OMOP CDM was chosen for this study. Appendix 3 provides the survey instrument.
3. Results
3.1. Landscape of phenotype algorithms
Fifty-five phenotypes were selected, and their corresponding pseudocode were downloaded from PheKB. Table 2 shows a summarization of data types used among the phenotype algorithms (details can be found in Appendix 4). These types include structured and unstructured EHR data, as well as non-EHR data (such as diagnosis from cancer registry data, meta information of the cohort). As unstructured data, notes can refer to general clinical notes or specific relevant clinical notes (e.g., progress notes, observation reports). Different types of reports are used in different phenotypes, such as ECG, lab report, pathology report, radiology report. The most frequently used data type is ICD-coded diagnosis, which is used in 52 phenotype algorithms.
Table 2.
Prevalence of data types used in the 55 phenotype algorithms
| Data type | Count of distinct phenotype |
|---|---|
| EHR | |
| Diagnosis | 52 |
| Procedure | 31 |
| Rx / Medical device | 31 |
| Provider / Specialty / Department | 31 |
| Demo | 26 |
| Lab | 24 |
| Visit | 19 |
| Note | 16 |
| Report | 13 |
| Problem list | 9 |
| Encounter | 8 |
| Allergy | 1 |
| Death | 1 |
| Family History | 1 |
| non-EHR (e.g., cancer registry, meta information of the cohort) | 14 |
3.2. KIP score for clauses and agreement on identified tasks
485 distinct clauses with corresponding 1153 customization tasks were identified. 0–2 is defined as the range from easy to difficult for portability for each aspect yielding the total score range of 0–6. The average clause-wise KIP total score 1.37±1.38 (mean ± standard deviation). The K (Knowledge conversion) score is 0.64±0.66, I (clause Interpretation) score is 0.33±0.55, and P (Programming) score is 0.40±0.64. Among 1153 customization tasks, 46 tasks are related to interpretation, 613 tasks are related to knowledge conversion, and 469 tasks are related to programming (Table 3).
Table 3.
The task category list by grouping relevant customization tasks
| Cat. ID | Customization Task Category | Customization Task Example | Estimated time consumption (median) | Score | Count |
|---|---|---|---|---|---|
| Knowledge aspect | |||||
| C1 | Map source vocabulary to target vocabulary (among different terminologies or same terminology different versions) | Vocabulary mapping to get non-ingredient RxCUI from ingredient RxCUI since medication may also be coded using brand drug codes Vocabulary mapping to get ICD10 procedure codes from provided ICD9 procedure codes considering the data contains ICD10 coded data |
<1day | K 0 | 185 |
| C2 | Map free text to target vocabulary | Vocabulary mapping to get RxCUI from provided medication names | <1week | K 1 | 88 |
| C3 | Define an operational definition of a specific EHR data element | Find “provider specialty” that links to procedure to check eye exam from ophthalmology department | <1week | K 1 | 138 |
| C4 | Define an operational definition of a non-EHR data element event | Define “Continuous enrollment/contact” for implementation | <1day - <1week | K 2 | 20 |
| C5 | Locate the data source for a group of data | Identify where to find “carotid imaging study” | <1day | K 1 | 89 |
| C6 | Retrieve data attribute representation and contextual knowledge through exploring structured data | Find lab unit, categorical range for urine protein tests | <1hour | K 1 | 61 |
| C7 | Retrieve data attribute representation and contextual knowledge through exploring unstructured data | Explore radiology reports to validate the local use of the “intravenous contrast” keywords provided and their occurrence prevalence | <1week | K 2 | 18 |
| C8 | Acquire knowledge of unstructured clinical data from domain expert and through programming | Find relevant “note types” and “service groups” which clinical notes may contain PAD information | <1week and <1month | K 2 | 14 |
| Interpretation aspect | |||||
| C9 | Understanding phenotype algorithm pseudocode clause | Understand if “ever” from “Taking ARBs (angiotensin receptor blockers) ever” means both structured and unstructured medication lists should be used | <1day | I 0–2 | 46 |
| Programming aspect | |||||
| C10 | Compile machine readable input file | Compile ICD codes or code groups provided in a pdf pseudocode appendix to a machine-readable file | <1day | P 0 | 258 |
| C11 | Pre-processing data by simple programming | Find if relevant pathology reports exist from clinical data warehouse programmatically | <1week | P 1 | 11 |
| C12 | Search keywords from unstructured data | Find at least 2 unique DSM-IV social interaction terms from notes | <1week | P 1 | 56 |
| C13 | Search keywords with modifier from unstructured data | Identify non-negated diverticulosis terms (e.g., diverticulitis, diverticula) from relevant radiology reports | <1week | P 2 | 16 |
| C14 | Extract information from unstructured data using advanced NLP implementation | Extract heart rate from ECG report | <1month | P 2 | 16 |
| C15 | Extract information from NLP tool processed documents | Search heart disease concepts (UMLS CUIs) from MedLEE parsed ECG report | <1day | P 1 | 11 |
| C16 | Configure, install and execute NLP tools | Install cTAKES for parsing clinical notes | <1week | P 2 | 23 |
| C17 | Populate NLP search results for SQL query | To exclude patient with cancer using ICD-9 and keywords, keywords search results from unstructured data need to be imported to the database | <1week | P 1 | 37 |
| C18 | Implement complex SQL query | Implement extrapolating height at serum creatinine measurement time from its pre- and post- height measurement based on a formula | <1day and <1week | P 1 | 41 |
| Other (not specific to the phenotype) | |||||
| C19 | Check the availability and completeness of the needed data element | Potentially unavailable information of medication administration route, which is required for glaucoma phenotyping | <1week | 22 | |
| C20 | Implement another existing phenotype | Use existing eMERGE T2DM algorithm to check if a patient has type 2 diabetes | <1week | 3 | |
| Total (1153) | |||||
The overall agreement rate is 59% (51/86) and the completeness agreement rate is 87% (75/86). For the surveyed 293 tasks, 35 tasks were suggested revisions and 11 extra tasks were identified. All suggested changes were not only revised on the sampled clauses but also integrated to the complete clauses. As expected, the disagreement is mainly from discrepancies in the source EHR data and implementation methods. For example, target vocabularies for medications or laboratory tests can be different terminologies (e.g., local legacy codes) instead of RxNorm and LOINC. Different data format of required variable may cause different methods for conducting tasks, for example an unstructured problem list may require NLP effort comparing with the code search from a structured problem list. Different NLP tools are used in different institutions for solving same phenotyping tasks.
3.3. Task categories
The task categories resulting from the assessment are summarized in Table 3. Prior to implementing a phenotype algorithm, experienced phenotype engineers will familiarize themselves with the documentation to have an accurate understanding of the algorithm developer’s intent. As part of this process, clarifying communication may be necessary, requiring further communication, which is described by the 46 tasks in Category 9. Engineers then usually work together with domain experts, informaticians (who sometimes play an engineer’s role) and data curators to define and identify all algorithm-defined variable representations from the site-dependent clinical data warehouse. The knowledge required can be vocabulary-related (Category 1–2), operational definition of events (Category 3–4), where to retrieve specific variables (Category 5), and how the variable is represented in the data (Category 6–8). Finally, phenotype engineers translate the algorithm logic into site-compatible executable programs, which include pre-processing data (Category 10–11), extracting relevant data from non-structured EHR data (Category 12–17) using NLP techniques, and writing SQL query for structured EHR data retrieval (Category 18). Since not all variables are available from each institution, data availability (Category 19) can affect the portability for specific institutions at the very beginning. Some algorithms may contain multiple algorithms within them, thus, the task to retrieve individual variables can often become a full implementation process in and of themselves (Category 20).
In Table 3, estimated time efforts resulting from the survey and scoring from the KIP definition (Table 1) are also summarized for each category. As we can see, most categories’ estimated time efforts are positively correlated with the KIP score, which justified our initial KIP score assignment.
3.4. Time effort and CDM usefulness
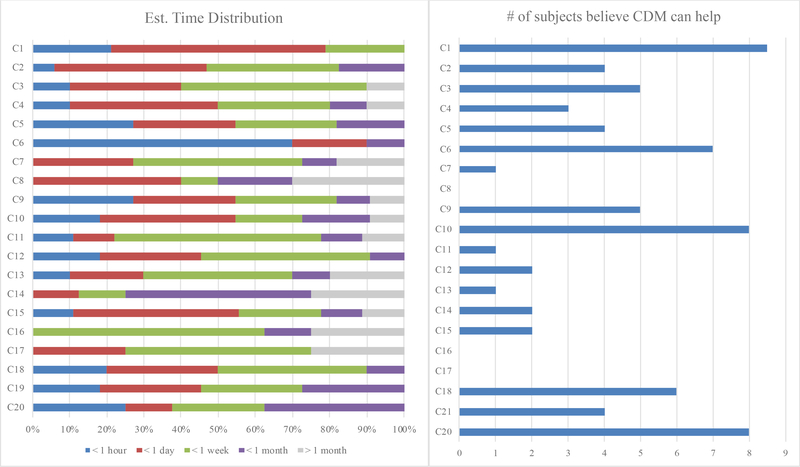
Experienced phenotype engineers reported time estimation and the perceived usefulness of a CDM for the 20 task categories. Fig. 3 summarizes the distribution of the estimated time consumed for different tasks and whether a CDM can be used to reduce the time needed. In general, implementation of a phenotype is not an easy task. Most tasks (19/20, 95%) take more than one hour (median value) to finish, the majority of the tasks (14/20, 70%) take more than one day to finish, and a small proportion of tasks (2/20, 1%) can take more than one week to finish. The most time-consuming task are often NLP-related tasks (i.e., Category 12, 13, 14, and 16) or tasks requiring external domain expertise (Category 8). The variance across different researchers and sites are large. The distribution of the estimated time consumption can even range from less than one hour to more than one month for some tasks (7/20, 35%), which may indicate the portability is unevenly distributed across different sites and researchers.
Fig. 3.
Phenotype engineers reported time estimation and the usefulness of CDM for the 20 task categories (corresponding descriptions provided in Table 3). Left: The distribution of estimated time needed for each category. Different color represents different time consumption, and the bar length is proportional to the portion of the engineers reported the corresponding time consumption. Right: The number of engineers believing CDM’s assistance on task category implementation The phenotype engineer responses suggest a CDM specifically OMOP can reduce the time consumption for some tasks, especially for the vocabulary mapping part (Category 1–2) from the structured data. However, for the most time-consuming tasks such as those related to the NLP, respondents had less confidence that the OMOP would help reduce the time needed for implementation.
4. Discussion
By systematically reviewing 55 phenotype algorithms published within the eMERGE network, we identified the main sources for implementation inefficiency, and summarized the most common customization tasks required for implementation. With the high completeness agreement rate, we believe identified tasks are comprehensive and can reflect the implementation efforts from the majority of the eMERGE network. Consequently, this systematic and comprehensive analysis can provide insights for designing more portable phenotype algorithms.
The portability of a phenotype algorithm is related to replicability and efficiency. Replicability is almost always the first consideration while developing an algorithm, and in fact, all algorithms developed within the eMERGE network are validated by at least one secondary site to ensure replicability. Consequently, multi-site studies have established that the algorithms are portable with consistently good performance between sites [18,19,21]. However, inefficiency in implementation can be a practical barrier to reuse of an existing phenotype algorithm.
In our study, tasks such as compiling machine-readable input file, mapping a source vocabulary to a target vocabulary, identifying data source, and defining a medical event operational definition are identified. Some of those tasks can be easily avoided. For example, since the algorithms are usually described in a narrative format stored as a Microsoft Word or PDF document, the source vocabulary codes or the variable data dictionaries (e.g., medication names, keywords) embedded in free text are not easily to be retrieved by programming. Therefore, we encourage all algorithm authoring sites to share the vocabulary codes or variable data dictionaries with corresponding variable name as a plain text file to facilitate the programming at the implementation site. This step can potentially save some unnecessary file format transformation and programming difficulties.
Complicated SQL logic implementation is another time-consuming task. We believe the efforts can be largely reduced or even completely eliminated by directly sharing the code that implements the logic. We suggest that the authors divide any phenotype algorithms into two parts, data element extraction (and formatting) and logic implementation (or outcome prediction for machine-learning based phenotyping algorithm [6]) as described in Shang et al.’s study [25]. The logical separation has been explored initially by the Arden Syntax which demarcates health information system specific codes in curly braces to share task-specific knowledge implementations across institutions [26–28]. Once the extracted elements are provided in a standard format (such as the data dictionary usually required in the eMERGE studies), the logic code can be shared and executed easily across different sites by taking the standard format file as an input, which can save significant implementation time for the algorithm with complicated logical reasoning. Even better, implementation codes for cohort definition can be directly shared if a CDM is used. For example, implementation in a JSON/SQL format conducted through ATLAS [29] is shareable if OMOP is used as the source data model. For decreasing human efforts in cohort execution, Yuan et al. [30] have developed an NLP interface Criteria2Query to automatically parse eligibility criteria text into SQL queries on the OMOP model. However, the input criteria are trained mainly from ClinicalTrials.gov. Consequently, this solution should be further explored or evaluated for eMERGE phenotypes which have different writing patterns.
Many labor-intensive implementation tasks are introduced as a result of the knowledge gap between implementation sites and development sites, such as different data representations or different understandings of a specific medical concept in various clinical data warehouses. eMERGE has adopted some common clinical vocabularies for building phenotypes (e.g., ICD for diagnoses, RxNorm for drugs, LOINC for laboratory measures), which help reconcile common terminologies for representing structured data in phenotype development. Furthermore, we found that it is not rare for phenotype algorithms to leverage operational events (e.g., “continuous enrollment”) and organization-specific EHR elements (e.g., providers and departments). These administrative level data and definitions can be highly site-dependent, and thus make portable phenotypes difficult. We argue here that the algorithm authoring site should try to reduce the involvement of these administrative level data as much as possible (or define them with common knowledge). The highly site-dependent nature makes it a challenge to implement at other sites efficiently. In addition, more research is needed to understand if the accuracy increase found by including those criteria is justified given the lack of portability of those elements across the network.
Hripcsak et al. [24] have demonstrated that common data models can potentially bridge the knowledge gap to some extent, which is supported by our results. By adopting a CDM, both development sites and implementation sites put in efforts ahead of time to convert the localized knowledge to an agreed upon common ground. The initial operational overhead may be challenging and time consuming, especially when complicated data, such as unstructured clinical narratives, are considered. However, as phenotyping initiatives expand, the upfront investment in effort may be justified by the resulting improvement in phenotype implementation efficiency. If the CDM has not been adopted or only partially adopted due to difficulties such as incomplete NLP, the current common practice to bridge the knowledge gap is to put more burden on the implementation sites, that is to convert the knowledge (e.g., variable, concepts, events) in the algorithm to be compatible with local environment and knowledge. These efforts usually include hours of data exploration and days or even weeks of repeated consultation with domain experts. With algorithms shared to more and more implementation sites, the improvements in the efficiency of implementing a CDM compounds. Another strategy to consider is for the development site to collaborate with multiple external experts who are familiar with their local clinical practice and clinical data warehouses, and provide an algorithm that is more reflective of the broader data representation across these sites. However, this is not practical due to limited resources and time, but knowledge towards different data representation from different institutions can be curated in a collaborative matter for repeat use.
Not surprisingly, the most time-consuming tasks are NLP-related, with simple keyword search consuming less time than more complicated NLP tasks, i.e., using developed or customized components, pipeline, or tools to extract information. An NLP task using advanced tools is often inherently more complicated than simple keyword search, which can often be completed inside a database. The configuration and installation of NLP pipelines at an implementation site without previous experience of the tools can be as time consuming as the computational time to run them. Multiple frameworks and pipelines have been adopted across the eMERGE network, and different sites are likely to develop their own custom NLP pipelines. For example, Vanderbilt University Medical Center developed a combination of SecTag [31], MedEx [32], and the KnowledgeMap Concept Identifier [33], Mayo Clinic built an NLP pipeline around UIMA [34] based on cTAKES [35–37], Northwestern University has used KNIME [22,38] for distributing NLP code, and Columbia University utilized MedLEE [39] to process unstructured clinical notes. Compared to structured data, unstructured data currently lack the same common ground on clinical documentation template across the network, making the development of an implementation-portable algorithm using clinical narratives more challenging.
In addition to implementation of phenotype algorithms themselves, phenotype algorithms also include data dictionaries that specify what data (e.g., demographics, covariates) to include with the case/control designation. Several surveyed phenotype engineers reported a great amount of time was spent on extracting data compliant with specific data dictionaries, which is often required in the eMERGE network.
The ultimate goal of developing a portable phenotyping algorithm is to deliver an executable product-level software across different sites. Unfortunately, current state of phenotyping algorithm implementation is far away from this goal. As a beginning project of understanding phenotyping implementation efforts, this paper is to identify customization tasks in the implementation. We hope in the future when it is possible to deliver a product-level software for phenotype implementation, we can use support tools such as CASE (computer-aided software engineering) and metrics to ensure the development of a high-quality products.
There are a few limitations in this study. The first limitation of this study is lack of evaluation in complications when taking combination of clauses into consideration. Some clauses can be simple tasks if treated independently, but the logic can become complicated when many clauses are combined together. In addition, despite a subsample of our identified tasks and a team of reviewers for estimating time efforts for implementing task categories, we initially used only two reviewers, which may bias the overall task identification and summarization. In the future, we encourage more members from the phenotyping community or the broader biomedical informatics society to join and contribute to this discussion.
5. Conclusion
Recognizing the tasks makes phenotyping implementation efforts visible. The category of tasks presented in this study is a valuable lesson learned from the eMERGE network phenotyping practice. The time required to complete those tasks can vary from hours to months according to our results. Phenotype developers are encouraged to analyze and optimize the portability from the aspects of knowledge, interpretation and programming (KIP). The discussion on how to improve the portability under current phenotyping practice is intended as a starting point for a best practice in developing implementation-friendly algorithms.
Supplementary Material
Highlights.
We developed a novel method (Knowledge conversion, clause Interpretation, and Programming, KIP) to quantify the portability of phenotype algorithms
We uncovered substantial phenotype implementation efforts across the eMERGE network
We discussed the challenges of making electronic phenotype portable and opportunities in using the OMOP CDM to improve phenotype portability
Acknowledgments
The study was supported by the phase III of the eMERGE program funded by the NHGRI through the following grants: U01HG8657 (Kaiser Washington/University of Washington); U01HG8672 (Vanderbilt University Medical Center); U01HG8666 (Cincinnati Children’s Hospital Medical Center); U01HG6379 (Mayo Clinic); U01HG8680 (Columbia University); U01HG8684 (Children’s Hospital of Philadelphia); U01HG8673 (Northwestern University); U01HG8701 (Vanderbilt University Medical Center serving as the Coordinating Center); and U01HG8676 (Partners Healthcare/Broad Institute)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liao KP, Cai T, Savova GK, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 2015;350:h1885. doi: 10.1136/bmj.h1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011;4:13. doi: 10.1186/1755-8794-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conway M, Berg RL, Carrell D, et al. Analyzing the Heterogeneity and Complexity of Electronic Health Record Oriented Phenotyping Algorithms. AMIA Annu Symp Proc 2011;2011:274–83. [PMC free article] [PubMed] [Google Scholar]
- 4.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013;20:e147–54. doi: 10.1136/amiajnl-2012-000896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei W-Q, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med 2015;7:41. doi: 10.1186/s13073-015-0166-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banda JM, Seneviratne M, Hernandez-Boussard T, et al. Advances in Electronic Phenotyping: From Rule-Based Definitions to Machine Learning Models. Annu Rev Biomed Data Sci 2018;1:53–68. doi: 10.1146/annurev-biodatasci-080917-013315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc 2012;19:212–8. doi: 10.1136/amiajnl-2011-000439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shim U, Kim H-N, Sung Y-A, et al. Pathway Analysis of Metabolic Syndrome Using a Genome-Wide Association Study of Korea Associated Resource (KARE) Cohorts. Genomics Inform 2014;12:195–202. doi: 10.5808/GI.2014.12.4.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kullo IJ, Ding K, Jouni H, et al. A genome-wide association study of red blood cell traits using the electronic medical record. PloS One 2010;5. doi: 10.1371/journal.pone.0013011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford DC, Crosslin DR, Tromp G, et al. eMERGEing progress in genomics—the first seven years. Front Genet 2014;5. doi: 10.3389/fgene.2014.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016;23:1046–52. doi: 10.1093/jamia/ocv202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patterson O, Hurdle JF. Document Clustering of Clinical Narratives: a Systematic Study of Clinical Sublanguages. AMIA Annu Symp Proc 2011;2011:1099–107. [PMC free article] [PubMed] [Google Scholar]
- 13.Studer R, Benjamins VR, Fensel D. Knowledge engineering: Principles and methods. Data Knowl Eng 1998;25:161–97. doi: 10.1016/S0169-023X(97)00056-6 [DOI] [Google Scholar]
- 14.Aylett RS, Doniat C. Supporting the Domain expert in planning domain construction. In: Proceedings of the AIPS’02 Workshop on Knowledge Engineering Tools and Techniques for AI Planning. Citeseer 2002. [Google Scholar]
- 15.Alonso F, Maté JL, Pazos J. Knowledge engineering versus software engineering. Data Knowl Eng 1990;5:79–91. doi: 10.1016/0169-023X(90)90005-X [DOI] [Google Scholar]
- 16.Hripcsak G, Albers DJ. High-fidelity phenotyping: richness and freedom from bias. J Am Med Inform Assoc doi: 10.1093/jamia/ocx110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richesson RL, Sun J, Pathak J, et al. Clinical phenotyping in selected national networks: demonstrating the need for high-throughput, portable, and computational methods. Artif Intell Med 2016;71:57–61. doi: 10.1016/j.artmed.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc 2012;19:e162–9. doi: 10.1136/amiajnl-2011-000583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denny JC, Crawford DC, Ritchie MD, et al. Variants Near FOXE1 Are Associated with Hypothyroidism and Other Thyroid Conditions: Using Electronic Medical Records for Genome- and Phenome-wide Studies. Am J Hum Genet 2011;89:529–42. doi: 10.1016/j.ajhg.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson WK, Rasmussen LV, Pacheco JA, et al. An evaluation of the NQF Quality Data Model for representing Electronic Health Record driven phenotyping algorithms. In: AMIA. Citeseer 2012. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.453.993&rep=rep1&type=pdf (accessed 1 Mar 2016). [PMC free article] [PubMed] [Google Scholar]
- 21.Mo H, Thompson WK, Rasmussen LV, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J Am Med Inform Assoc 2015;22:1220–30. doi: 10.1093/jamia/ocv112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacheco JA, Rasmussen LV, Kiefer RC, et al. A case study evaluating the portability of an executable computable phenotype algorithm across multiple institutions and electronic health record environments. J Am Med Inform Assoc 2018;25:1540–6. doi: 10.1093/jamia/ocy101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen LV, Thompson WK, Pacheco JA, et al. Design patterns for the development of electronic health record-driven phenotype extraction algorithms. J Biomed Inform 2014;51:280–6. doi: 10.1016/j.jbi.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hripcsak G, Shang N, Peissig PL, et al. Facilitating phenotype transfer using a common data model. J Biomed Inform Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang N, Weng C, Hripcsak G. A Method for Enhancing the Portability of Electronic Phenotyping Algorithms: An eMERGE Pilot Study. 2016. [Google Scholar]
- 26.Hripcsak G, Ludemann P, Pryor TA, et al. Rationale for the Arden Syntax. Comput Biomed Res 1994;27:291–324. doi: 10.1006/cbmr.1994.1023 [DOI] [PubMed] [Google Scholar]
- 27.Hripcsak G Writing Arden Syntax medical logic modules. Comput Biol Med 1994;24:331–63. doi: 10.1016/0010-4825(94)90002-7 [DOI] [PubMed] [Google Scholar]
- 28.Samwald M, Fehre K, de Bruin J, et al. The Arden Syntax standard for clinical decision support: Experiences and directions. J Biomed Inform 2012;45:711–8. doi: 10.1016/j.jbi.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 29.OHDSI. ATLAS. http://www.ohdsi.org/web/atlas (accessed 28 May 2019).
- 30.Yuan C, Ryan PB, Ta C, et al. Criteria2Query: a natural language interface to clinical databases for cohort definition. J Am Med Inform Assoc 2019;26:294–305. doi: 10.1093/jamia/ocy178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denny JC, Spickard A, Johnson KB, et al. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc JAMIA 2009;16:806–15. doi: 10.1197/jamia.M3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24. doi: 10.1197/jamia.M3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denny JC, Smithers JD, Miller RA, et al. “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc JAMIA 2003;10:351–62. doi: 10.1197/jamia.M1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng 2004;10:327–48. doi: 10.1017/S1351324904003523 [DOI] [Google Scholar]
- 35.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13. doi: 10.1136/jamia.2009.001560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kullo IJ, Fan J, Pathak J, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc 2010;17:568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Bielinski SJ, Sohn S, et al. An Information Extraction Framework for Cohort Identification Using Electronic Health Records. AMIA Summits Transl Sci Proc 2013;2013:149–53. [PMC free article] [PubMed] [Google Scholar]
- 38.Mo H, Pacheco JA, Rasmussen LV, et al. A Prototype for Executable and Portable Electronic Clinical Quality Measures Using the KNIME Analytics Platform. AMIA Summits Transl Sci Proc 2015;2015:127–31. [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc JAMIA 1994;1:161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.