Abstract
Comparative studies of quantitative and neutral genetic differentiation (QST-FST tests) provide means to detect adaptive population differentiation. However, QST-FST tests can be overly liberal if the markers used deflate FST below its expectation, or overly conservative if methodological biases lead to inflated FST estimates. We investigated how marker type and filtering criteria for marker selection influence QST-FST comparisons through their effects on FST using simulations and empirical data on over 18 000 in silico genotyped microsatellites and 3.8 million single-locus polymorphism (SNP) loci from four populations of nine-spined sticklebacks (Pungitius pungitius). Empirical and simulated data revealed that FST decreased with increasing marker variability, and was generally higher with SNPs than with microsatellites. The estimated baseline FST levels were also sensitive to filtering criteria for SNPs: both minor alleles and linkage disequilibrium (LD) pruning influenced FST estimation, as did marker ascertainment. However, in the case of stickleback data used here where QST is high, the choice of marker type, their genomic location, ascertainment and filtering made little difference to outcomes of QST-FST tests. Nevertheless, we recommend that QST-FST tests using microsatellites should discard the most variable loci, and those using SNPs should pay attention to marker ascertainment and properly account for LD before filtering SNPs. This may be especially important when level of quantitative trait differentiation is low and levels of neutral differentiation high.
Keywords: QST-FST, natural selection, microsatellite, quantitative genetics, Pungitius pungitius, single-locus polymorphisms
1. Introduction
Geographical and temporal differentiation in mean values of quantitative traits are of commonplace occurrence in animal and plant populations. Before such differentiation can be ascribed to adaptation, two premises need to be fulfilled. First, the observed differentiation needs to be genetically based rather than environmentally induced. Second, the observed differentiation has to have been caused by directional natural selection instead of neutral processes, such as gene flow or genetic drift. Comparative studies of quantitative trait and molecular marker differentiation (QST-FST tests) provide a way of identifying footprints of directional selection as a cause of population differentiation in quantitative traits (reviewed in [1–5]). The rationale behind QST-FST comparisons is simple: the level of genetically based population differentiation in quantitative traits (i.e. QST) is compared with that in neutral marker loci (i.e. FST). If QST > FST, this provides evidence for adaptive population differentiation, as only directional selection is expected to elevate divergence in quantitative traits above the neutral expectation. Similarly, if QST < FST, this would be indicative of uniform stabilizing selection that has prevented populations from diverging less than would be expected under random genetic drift alone. An outcome of QST ≈ FST would be inconclusive: the null-hypothesis of differentiation by genetic drift could not be rejected (e.g. [1]). While the majority of studies using the QST-FST (reviewed in [3,4,6]) or a related approach [7,8] to test for the adaptive nature of quantitative trait differentiation have found support for it, there is a concern that many of these studies might have tested QST against overly liberal (i.e. too low) FST estimates [9,10]. This concern stems from the fact that most QST-FST comparisons have used microsatellite markers for FST estimation. Since microsatellite loci are highly variable due to their high mutation rates [11,12], they may underestimate levels of neutral genetic differentiation [13–16], and hence, render QST-FST comparisons biased towards finding evidence for adaptive differentiation [9,10]. Recent advances in sequencing technology have started to replace microsatellite markers with single-locus polymorphisms (SNPs) as the tool of the trade-in population genetic investigations of non-model organisms [17–19]. Since SNP loci are (mostly) bi-allelic and experience, on average, lower mutation rates than microsatellite loci, they might provide a way to obtain less biased estimates of neutral baseline differentiation than microsatellite loci [9,20]. However, since per nucleotide mutation rates are highly variable, and may vary depending on their genomic location [21,22], SNP loci may not automatically yield less biased neutral baseline estimates of divergence (as measured by FST) than microsatellites. Similarly, not all SNPs are or behave as being neutral; they may be subject to various forms of selection depending on their genomic location and/or functionality [23–25]. For instance, hitchhiking under positive and background selection can increase allele frequency shifts between populations, especially in genomic regions of low recombination [26–28]. Bias in the estimation of the neutral baseline from SNP data could also arise from analytical methods involved in marker selection and FST estimation [29]. Specifically, different averaging methods to estimate FST across loci have been shown to yield different results between studies [29], which should be particularly concerning in the context of QST-FST comparisons. On the other hand, FST estimates can be affected by ascertainment bias (sensu [17]) when the selected markers are not representative of the variability observed in all sampled individuals. They can also be sensitive to how rare low-frequency alleles are collected and filtered from the data [30–33], and how each researcher decides to deal with markers that are in linkage disequilibrium (LD) with each other [34]. The latter issue may be particularly important in QST-FST studies where divergent populations differing in their LD patterns are compared: due to methodological constraints, typically, only a small subset of SNPs can be included and failure to account for LD in marker selection for one or more populations in the data has potential to bias FST estimates. To the best of our knowledge, there have not been any studies of these effects of SNPs filtering in the context of QST-FST comparisons, although the problem of defining neutral baseline differentiation level has been well recognized in related contexts (e.g. [25,35,36]).
The aim of this study was to explore how marker type and various filtering criteria could influence inferences from QST-FST comparisons through their effect on estimates of neutral baseline differentiation. To this end, we used empirical data on quantitative trait and microsatellite differentiation among four populations of nine-spined sticklebacks (Pungitius pungitius) used in an earlier QST-FST study [8]. These data were supplemented with information on variability in millions of SNP loci and over 18 000 in silico genotyped microsatellite loci to see how marker type and different filtering criteria influenced the neutral baseline differentiation level as reflected in FST. In particular, we were interested to evaluate the conjecture (cf. [10]) that QST-FST comparisons using highly variable microsatellite markers could be biased towards finding evidence for adaptive differentiation. Because the P. pungitius study system is known for its particularly high level of quantitative trait differentiation [8,37,38], we also simulated genomic datasets under varying levels of QST-FST divergence and evaluated the different marker types' ability to detect the signature of divergent selection under different evolutionary scenarios in this particular system.
2. Material and methods
2.1. Phenotype data
The phenotype data used here were adopted from an earlier study [8] which used a full-sib mating design comprised of 92 F1-generation nine-spined stickleback individuals from four populations (Baltic Sea, n = 21; White Sea, n = 24, Bynastjärnen, n = 22, Pyöreälampi, n = 25) and five families per population. Five morphological traits including standard length (M1), body depth (M2), head length (M3), pelvic girdle length (M4) and caudal peduncle length (M5), as well as two behavioural traits including aggressiveness (B1) and propensity for risk-taking (B2) of all 92 individuals were quantified as described in Herczeg et al. [37,38]. The study populations are highly differentiated in most of these traits, and the available evidence suggested that this differentiation has been driven by natural selection stemming from local adaptation to pond environments (Bynastjärnen & Pyöreälampi) lacking piscine predators [8,37–39].
2.2. Genetic data
An assembled and annotated reference genome (110× coverage) for P. pungitius from a Pyöreälampi individual [40] was used for obtaining the genetic data. SNP data and in silico microsatellite data were obtained by whole genome resequencing of 10 female individuals from each of the above-mentioned four populations. The processing and analysis of these are explained in detail below. The empirical microsatellite data for 12 loci were the same as that used by Karhunen et al. [8].
2.3. Variant calling
The 40 samples for this study were processed as a part of a larger sample set (n = 140) that included populations not used in this study. For each sample, the sequence reads were mapped to the reference genome with bwa (v. 0.7.12; [41]) using default options, duplicate reads were removed with SAMtools (v. 1.3.1; [42]) and those around indels were realigned with GenomeAnalysisToolkit's IndelRealigner (GATK; v. 3.4; [43]). Each sample was then called with GATK HaplotypeCaller using mode ‘discovery’, output format ‘gvcf’, and emit and call confidence of 3 and 10, respectively. The full sample set was jointly genotyped with GATK GenotypeGVCFs. From this, the 40 samples from the four study populations were extracted and their analysis continued in isolation.
2.4. SNP filtering
The full datasets consist of 3 806 181 SNPs that are variable within the four populations and have data for at least 80% of the individuals in each population. We applied different SNP filtering criteria to obtain the following three datasets: (i) SNPs in all genomic regions (ALL), (ii) the SNPs in non-coding regions excluding exons and repeat regions and their immediate neighbourhood (50 bp of flanking sequence) (NONCOD) and (iii) the SNPs in synonymous third codon position (C3SYNO). C3SYNO includes variable third codon positions that were inferred to contain synonymous substitutions by the R package VariantAnnotation (v. 1.18.1; [44]). From these, we removed sites with missing genotype data or monomorphic among the study samples and the three final datasets consisted of 1 702 105, 1 165 221 and 41 927 SNPs, respectively. To account for the possibility of ascertainment bias [17,29], we also filtered markers based on their variability in the marine populations. This is because FST can be seen as either a parameter of the evolutionary process, or a statistic derived from observed samples. As such, it is used to measure correlation between randomly drawn alleles from a single population relative to either the most recent common ancestral population or the combination of the two population samples [29]. Under the former definition, the markers included in the analysis should have been variable in the ancestral population; alternatively, markers can often be ascertained by their variability in an outgroup or maximally diverged modern populations. In our case, the marine populations are representative of the ancestral genetic variation and represent the geographical extremities among the study populations. Hence, variability-based marker ascertainment was done using the two marine populations and for each category, markers invariable in either of the marine populations were excluded.
2.5. Effect of LD on FST
Filtering of SNP dataset based on the degree of linkage between loci is a common pruning method in population genomics studies. For QST-FST comparison, no clear guidelines are available on whether SNPs in LD should or should not be included in the data used for FST estimation. A general assumption is that unlinked markers should be preferred as they provide a closer estimation of what would be expected under neutrality. For practical and computational reasons, LD pruning should at least be performed on the basis that some QST-FST methods—particularly MCMC-based approach—cannot handle large numbers of markers. Consequently, we investigated the effect of LD pruning on the estimation of FST by using genome-wide sets of unlinked SNPs to estimate the mean and variance of FST. Two different approaches were used to extract a subset of unlinked SNPs from the full SNP datasets. First, a thinning approach was applied to select roughly 2000 SNPs located at ca 150–200 kb from each other, and the rest of the SNPs were discarded. The thinning approach was carried out with VCFtools [45]. Second, we used the more sophisticated LD pruning approach of Zheng et al. [46] implemented in the R package SNPrelate. This approach divides each chromosome first into multiple 500 kb non-overlapping sliding windows. Within each sliding window, a single SNP was firstly randomly included into the active set, and this was followed by including another SNP into the active set if its correlation (as defined by the LD composite measure; [47,48]) with the SNPs in the active set was lower than a defined threshold. This procedure was repeated for all the SNPs in the same sliding window. A sequence of LD-values ranging from 0.05 to 0.95 (the pruned data have low LD when the threshold is small) were used to evaluate how the FST estimates were affected by the changes of LD. To see how the thinning and sliding window approaches influenced the baseline FST levels as estimated from the data, we evaluated FST as a function of standardized (to the total number of markers in the data) marker number included into the estimation. If the pruning is effective, we would expect no relationship between FST and number of markers included into the estimation. However, if the pruning is ineffective and linked markers become included into the data, we expect to see that the FST varies as a function of number of markers included. Comparison of LD between pond and marine populations was done by calculating the mean LD (defined as squared composite measure; [47,48]) between all the SNP pairs within each non-overlapping 500 kb sliding window across the genome.
2.6. In silico microsatellite analysis
The in silico microsatellite discovery and genotyping was performed with Tandem repeats finder (TRF; v. 4.09; [49]) and RepeatSeq (v. 0.8.2; [50]), respectively. Dinucleotide repeats were identified in the P. pungitius reference genome using TRF with options ‘2 7 7 80 10 20 2 -f -d -m -h’. The parameter values are as recommended by the software developers with the exception of the last two, minimum score (20) and maximum period size (2). From the output, repeat loci were chosen with the following criteria: period and consensus pattern of 2, length of 10–20 copies, alignment score of 100 and distance of at least 500 bp to the previous locus. The selected loci were genotyped in the 40 study samples using RepeatSeq. The output was processed using bcftools [42] and custom scripts, and the resulting data analysed using the R package diveRsity (v. 1.9.89; [51]). After removing loci with more than 20% missing data in any population, the resulting dataset contained 18 824 microsatellite loci.
2.7. Estimation of local recombination rate
Since variation in recombination rate along chromosomes can influence patterns of polymorphism and hence FST through background and/or hitchhiking selection (e.g. [28,52]), we explored how the FST in our data was influenced by variation in recombination rate. To this end, the local recombination rate was estimated using a sex-averaged genetic map (an improved [40] version of that reported in [53]). First, non-monotonic regions were manually removed and outlier markers discarded by performing loess regression (α = 0.2, degree = 1), interpolating genetic distance from physical positions and removing any points that lie further than 2 cM from the line. Using the remaining regions and markers, we estimated the local recombination rate by performing loess regression on a fixed number of markers (α adjusted to include 250 markers, degree = 2) and predicting the change on genetic distance for a pair of adjacent physical positions. We could estimate the local recombination rate for 54.7% of the genome (electronic supplementary material, figure S1). Although a small fraction of the genome still got negative rate estimates, a great majority of sites had inferred recombination rates between 0 and 20 cM Mb−1 (mean = 6.6). Using the inferred rate, the genome was split into two roughly equally large subsets of regions with low (1–7 cM Mb−1) and high (7–15 cM Mb−1) local recombination rates, discarding the loci with either no rate estimate or an estimate from the extreme ends of the rate distribution (electronic supplementary material, figure S2a). The microsatellite loci were further divided into those with few (1–4), intermediate (5–8) and many (9–21) alleles, the two extremes approximately corresponding to the first and fourth quartile of the distribution (electronic supplementary material, figure S2b). FST was separately estimated for the different subsets of SNP dataset NONCOD and the microsatellite data. Since the impact of background selection was expected to be weak in small pond populations subjected to strong drift, the impact of recombination rate on FST was assessed only in the two large marine populations (LEV & HEL).
2.8. Outlier analyses
Since loci linked to selected sites are expected to show elevated FST [36], we used two approaches to identify possible outliers and evaluate their impact on baseline FST: BayeScan [54] and OutFLANK [33]. In both methods, the outliers were detected while controlling for the false discovery rate with a liberal p = 0.1 threshold.
2.9. FST estimation and comparison
We investigated whether different estimation methods would yield critically different FST values as expected from previous studies [29]. First, we used the Weir & Cockerham [55] theta estimator of FST. This estimator can be efficiently obtained even from genome-wide datasets, and it was used for estimation of both single and multilocus FST. This estimator was used to (i) calculate the pairwise FST values for each individual marker and produce empirical FST distributions for different SNP (using VCFtools; [45]) and microsatellite datasets (R package diveRsity; [51]), and (ii) evaluate how different ways of filtering and pruning the SNP data can affect the estimation of the neutral baseline FST (using R package SNPrelate; [46]). We then combined the estimates across loci using either the ‘ratio of average' (ROA) or the ‘average of ratio' (AOR) [29].
2.10. QST-FST comparisons
We used two approaches to conduct the QST-FST comparisons: the Bayesian Driftsel [7] and the frequentist QstFstComp approach [56,57]. One advantage of the former over the latter approach is that it has been shown to be more powerful in detecting signatures of selection even when the number of populations is low (see [58] for a detailed statistical demonstration). However, since the standard QST-FST comparisons continue to be widely used, and because they might be more intuitive in providing direct comparison of the quantities of FST and QST, QstFstComp tests were included for comparison. Driftsel provides two test statistics, referred to as S and H statistics. S statistic accounts for patterns of relatedness among populations, as well as ancestral genetic correlations among the traits of interest [58]. S-values close to zero are indicative of stabilizing selection; those close to one indicate directional selection; and values close to 0.5 are consistent with evolution due to drift [58]. In addition to the factors accounted for with the S statistic, the H statistic allows the environment to be accounted for by including similarity in the distribution of population means and habitat parameters [8]. In other words, the H statistics allows one to test whether population means from similar habitats are more similar than expected by random genetic drift. Following the testing criteria proposed in Karhunen et al. [8], S or H > 0.95 implies that a quantitative trait has evolved under divergent selection at the 95% credibility level, whereas S or H < 0.05 would imply stabilizing selection at the same credibility level. The default non-informative priors were used in the Driftsel analyses. 15 000 Markov chain Monte Carlo (MCMC) samples of the posterior distribution were simulated, by considering the first 5000 as a burn-in, and the remaining were stored in every 10th iteration, so that eventually 1000 MCMC samples were used for calculating S and H statistics. We calculated both statistics for all datasets, using the binary habitat type (pond versus marine) as the input data for the distance matrix of the environmental covariates required by Driftsel [8]. The frequentist QstFstComp constructs a null distribution of the difference between QST and FST using a parametric simulation approach. The sampling error of the FST was evaluated by random sampling (i.e. bootstrapping) of the given loci, and the expected distribution of the QST under neutrality was simulated on the basis of the mean FST. The observed QST-FST quantity was compared with 1000 samples simulated from the QST-FST null distribution for a statistical test of neutrality, from where uncertainty quantities (p-values and confidence intervals) were derived. If QST-FST quantity is positive, and p < 0.05, the quantitative trait in question is inferred to have been subject to divergent selection. The QstFstComp was not applicable to the microsatellite loci with more than 10 alleles due to software limitation. Since both methods, and in particular the MCMC-based Driftsel approach, are computationally very demanding, we restricted all the analyses to 2000 loci. For SNP analyses, SNP loci obtained with the thinning approach (see above) were derived from the ALL, NONCOD and C3SYNO datasets. Likewise, loci for in silico microsatellite analyses were obtained with the thinning approach, and the analyses were conducted separately for all loci irrespectively of their variability, only loci with a low number of alleles, and only loci with a high number of alleles. Finally, we investigated the effect of marker ascertainment (cf. SNP filtering section above and [29]) on QST-FST comparisons. For each genomic dataset (i.e. ALL, NONCOD and C3SYNO) and each set of in silico microsatellites, we ran Driftsel using full datasets and using loci ascertained based on their variability in the marine populations. QstFstComp was also run for each genomic dataset using unascertained and ascertained SNP markers.
2.11. Simulation study
A simulation study was conducted to evaluate the ability of Driftsel and QstFstComp to detect signatures of divergent selection with different marker types and varying levels of QST-FST divergence. Because our empirical data show a high level of quantitative trait differentiation, the rationale here was to simulate different levels of QST-FST divergence to evaluate the effect of marker types on QST-FST inference in a more general context. To do so, a population genomic dataset of four sub-populations corresponding to our four study populations (Helsinki [HEL], White Sea [LEV], Bynastjärnen [BYN] and Pyöreälampi [PYO]) was first simulated using the software fastsimcoal2 [59,60]. Simulations were based on the most likely demographic history of real nine-spined stickleback populations (electronic supplementary material, figure S3 and see [61–63]): first, a population split was modelled between the two marine (LEV and HEL) sub-populations from an ancestral population 5000 and 4000 generations ago, respectively. Then the pond populations (BYN and PYO, respectively) split from HEL and LEV, 600 generations before present and experienced bottleneck for 300 subsequent generations. Each population consisted of 200 individuals. Like in the real stickleback genome, we simulated 20 chromosomes (sex chromosomes were not simulated). Each chromosome contained four LD blocks: one comprising 250 SNPs, and the other three 25 microsatellites each. The recombination rate was set to be 10−5 between loci within each of the LD blocks. In the three microsatellite LD blocks, mutation rates were specified to be 5 × 10−6 (low mutation rate; Mi_l) 5 × 10−5 (medium mutation rate; Mi_m) and 5 × 10−4 (high mutation rate; Mi_h), with 10, 22 and 64 alleles, respectively. We then randomly picked 10 individuals from each of the four sub-populations, and used both SNPs and microsatellites as neutral markers to estimate FST. To simulate the quantitative traits, we randomly picked 20 individuals from each of the four sub-population as founders, and then used a full-sib design to simulate 10 full-sib families (five offspring per family) within each of the sub-populations which evolved through 30 generations using a simulation procedure similar to that used in Karhunen et al. [7].
This simulation scheme was used to model different divergence scenarios corresponding to varying intensity of selection. Specifically, different evolutionary forces over the 30 generations were applied as follows: in scenario (i), the populations bred completely randomly without any selection (neutral scenario; QST = FST). In scenarios (ii) to (iv), populations were subjected to increasing levels of directional selection (QST > FST) by multiplying the neutral pattern by a factor of 1.5, 2 or 4, respectively, thus generating weak (ii), moderate (iii) and strong selection (iv). All the genetic and environmental parameters needed in simulations were specified in the same way as in Karhunen et al. [7]. Due to the high computational cost of MCMC-based approach (approx. 24 h for each run), the whole simulation procedure was replicated 10 times, and the averaged performances of both Driftsel and QstFstComp on the replicated datasets were recorded.
3. Results
3.1. FST comparisons: effects of marker type, genomic location and averaging approach
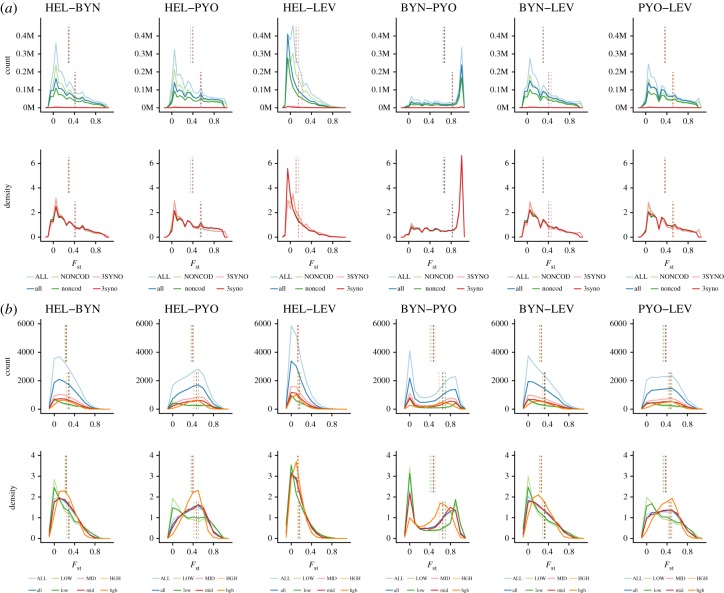
The estimated neutral baseline for the three SNP datasets based on the Weir & Cockerham [55] theta estimator of FST was around 0.51, with very narrow credibility intervals (electronic supplementary material, table S1). Hence, the mean and distribution of FST values were not significantly affected whether estimated using all, non-genic or genic loci (electronic supplementary material, table S1; figure 1). However, all of these values were significantly higher than the FST = 0.35 estimated from the 12 microsatellite loci used by Karhunen et al. [8]. While the FST = 0.37 estimated from the in silico genotyped microsatellite loci was similar to that of Karhunen et al.'s [8] estimate, comparison of estimates calculated for the different sets of microsatellites differing in their variability (see electronic supplementary material, figure S4 for details of variability in these loci) revealed that the FST estimates declined with increasing variability (electronic supplementary material, table S2; figure 1). The effect of marker variability on FST was particularly clear from the distribution of FST values: microsatellites with few alleles (1–4) were skewed towards extreme FST values and looked most similar to equivalent distributions for SNPs (figure 1); those with intermediate numbers of alleles (5–8) had the broadest distribution of FST and most strongly differed between the population pairs, whereas the ones with many alleles (9–21) were least diverged across the different population pairs (figure 1).
Figure 1.
Distribution of per locus pairwise FST. Values of per locus FST were estimated between all pairs of the four populations based on: (a) 3 806 181 SNP loci and (b) 18 824 microsatellite loci. Colours represent for each marker type the different datasets used to estimate FST estimate: in (a) all SNP loci (‘ALL' dataset; blue), non-coding/non-repeat sites (‘NONCOD' dataset; green) and codon third position synonymous changes (‘C3SYNO' dataset; red); light colours (in capitals) indicate full datasets whereas dark colours (in lowercase) show the distributions for loci ascertained by variability in marine populations. Absolute counts of sites (top) and density plots (bottom) are shown. Loci with minor allele frequency below 0.025 were discarded. In (b), FST for full microsatellite data (‘ALL' dataset; blue) and three subsets of data containing either low (1–4; ‘LOW'), moderate (5–8; ‘MID’) or high (9–21; ‘HIGH') number of alleles per locus. In all plots, dashed vertical lines indicate the mean value for the corresponding category: the upper and lower halves show the average of ratios and the ratio of averages, respectively.
We further compared two approaches for combining FST estimates across loci: average of ratios (AOR) and ratio of averages (ROA). Overall, ROA gave consistently higher estimates of FST than AOR (figure 1). The relative difference between the two approaches differed between comparisons and data types, but was the smallest for the two least diverged populations, FIN-HEL and RUS-LEV. While the three SNP datasets produced identical FST estimates under both averaging approaches, the relative order of marker categories in the microsatellites dataset changed drastically: the FST estimates based on highly variable microsatellites were clearly the lowest when using ROA, whereas they were one of the highest when using AOR (figure 1b).
3.2. FST comparisons: effects of variability-based ascertainment and minor allele filtering
We compared the FST estimates from the full data with those estimated from loci ascertained using the two marine populations, FIN-HEL and RUS-LEV. The effects of ascertainment differed between population comparisons, datasets and averaging approaches (figure 1). Notably, the greatest impact among SNP-based estimates was in the comparison of populations used for the ascertainment, whereas the estimates for the two pond populations—neither included in the ascertainment—were virtually unchanged (figure 1a). Among microsatellites, variability-based ascertainment only affected estimates based on the least variable category (figure 1b).
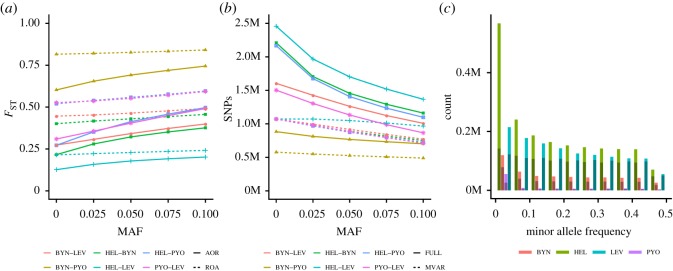
Another approach to reduce the impact of rare, novel variants is to filter the data using minor allele frequency (MAF). We tested the MAF filtering using SNP data and found that it had clear effects on FST estimates when using average of ratios (AOR) but only subtle effects when using ratio of averages (ROA; figure 2a). The effect on data ascertained by variability in marine populations was minimal (data not shown). This is consistent with the fact that the variability-based ascertainment itself heavily reduces the number of SNPs and the MAF filtering have only a minimal additional effect (figure 2b). Another notable effect of variability-based ascertainment was the changes in the shape of minor allele frequency: although the number of variable SNPs was reduced in all populations, the relative impact on numbers of low-frequency alleles was drastically different in the highly variable marine populations, especially on FIN-HEL, than in the small pond populations (figure 2c).
Figure 2.
Effect of minor allele frequency (MAF) cut-offs on mean FST estimated from the NONCOD SNP dataset. (a) Mean FST for different population pairs as a function of MAF, (b) number of SNPs retained, (c) site frequency spectrum for the four populations using the bin width of 0.05. In (a), solid and dashed lines indicate the two averaging approaches, average of ratios (AOR) and ratio of averages (ROA) whereas in (b) they indicate full data (FULL) and loci ascertained by variability (MVAR). In (c), dark shading shows the effect of ascertainment by variability in marine populations.
3.3. FST comparisons: effects of recombination rate
The effects of local recombination rate on FST between the two marine populations were statistically significant, but subtle. For SNPs, the average per-locus FST was 0.1265 and 0.1352 (t-test: p < 2.2 × 10−16) in regions of low and high recombination rate, respectively, while for microsatellite loci with few alleles, they were 0.1403 and 0.1540 (t-test: p < 0.05; electronic supplementary material, figure S2c). The differences were consistent but not significant for microsatellite loci with an intermediate number of alleles (mean FST 0.140 and 0.145, p = 0.15) while that for the most allele-rich loci were incongruent with the other categories of markers albeit not significantly so (0.1234 and 0.1186, p = 0.36; electronic supplementary material figure, S2c).
3.4. FST comparisons: effects of outliers
Outlier analyses did not detect any outliers either in the SNP nor microsatellite datasets, suggesting that outliers had little influence on mean FST estimates in our data (results not shown).
3.5. FST comparisons: effects of LD
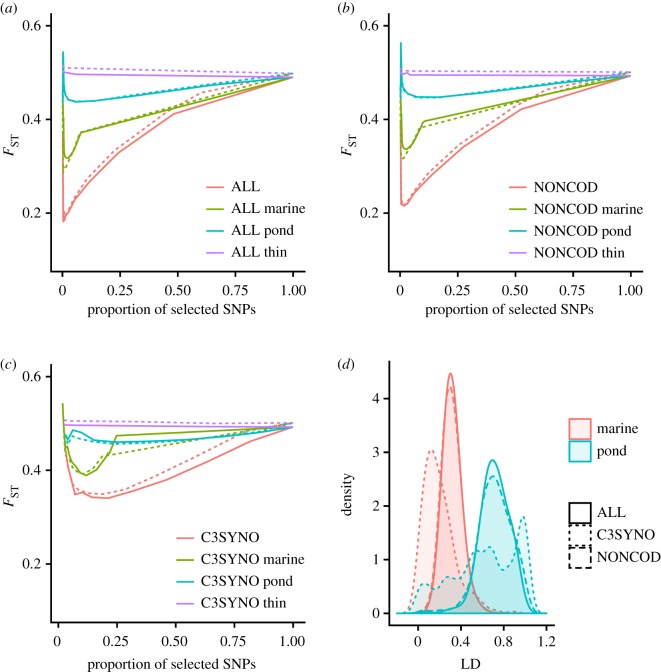
For all three SNP datasets, pruning of SNPs with the distance thinning approach had little effect on the mean FST: it remained constant irrespective of the number of SNPs sampled for estimation (figure 3a–c). The LD-based pruning yielded quite different results: when pooled data were used, the mean FST was heavily deflated for all three SNP datasets over much (0.1–0.7) of the parameter range (figure 3a–c). More specifically, this deflation was most marked at the lower end of this LD threshold range, generating a positive correlation between FST and the proportion of SNPs sampled (figure 3a–c). Only when the LD-threshold was set to be less than 0.1 or greater than 0.7 (corresponding to selection of less than 2000 or greater than 20 000 SNPs in C3SYNO), the sampled unlinked SNPs yielded FSTs similar to that in the full datasets (figure 3a–c). As we employed LD-based pruning separately for pond and marine populations, the FST values were more similar (but still somewhat deflated) to those obtained with the thinning approach (figure 3a–c). Although the extent of LD differs in different populations (figure 3d), this is unlikely to explain the FST deflation in the LD-based pruning.
Figure 3.
Effect of LD pruning on FST estimated from different datasets. Results are shown for (a) all SNPs (‘ALL' dataset), (b) non-coding/non-repeat sites (‘NONCOD' dataset) and (c) codon third position synonymous changes (‘C3SYNO' dataset). For each dataset, solid lines are colour coded to depict results obtained with the thinning approach (purple), or sliding window approach for all pooled populations (red), marine populations only (green) or pond populations only (blue). Dashed lines depict the same results after removing loci with MAF < 0.05. (d) The distribution of the average degree of linkage disequilibrium (LD) in marine and pond populations as estimated from non-overlapping 500 kb sliding windows across the genome.
3.6. QST-FST tests: effects of marker type and filtering
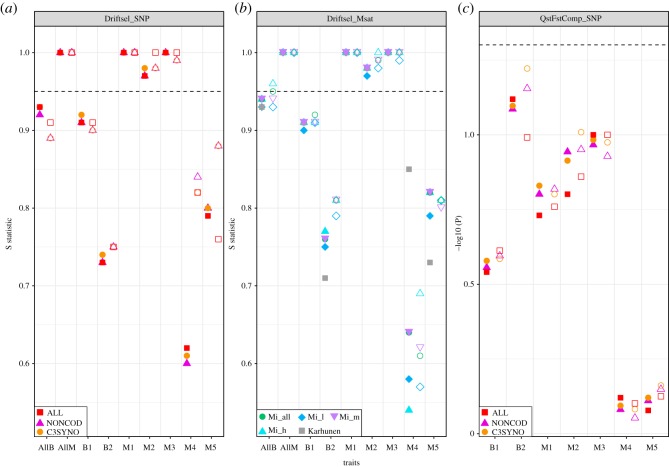
Irrespectively of the marker type or filtering criteria used, the Driftsel analyses detected significant signals of divergent natural selection for most of the traits while QstFstComp detected only very weak and non-significant signals of divergent selection (figure 4; electronic supplementary material, table S1–S7). In all analyses, Driftsel results were also fully consistent with earlier results based on 12 microsatellite loci ([8]; figure 4; electronic supplementary material, table S1, S5). Filtering of SNP data did not affect the outcomes: both Driftsel and QstFstComp yielded the same inference across datasets (i.e. ALL, NONCOD, C3SYNO; figure 4; electronic supplementary material, tables S1, S3, S5, S7). Marker type (i.e. microsatellites or SNP) did not influence the outcome of the Driftsel analyses: all traits displaying evidence for divergent natural selection with SNP markers did also with microsatellite markers (figure 4). Similarly, marker type did not influence the outcome of the QstFstComp analyses (figure 4; electronic supplementary material, table S4).
Figure 4.
Effect of marker ascertainment, marker type and filtering of SNP dataset on QST-FST comparisons. Results of Driftsel analyses using SNP and microsatellite markers (a,b, respectively) and QstFstComp using SNP markers (b) are shown. (b) The Driftsel results with the 12 microsatellites used in [8] (Karhunen; grey squares) as well as for different sets of in silico microsatellites: all (Mi_all; green circles), ones with low number of alleles (Mi_l; blue diamonds); with intermediate number of alleles (Mi_m; purple triangles), and with high number of alleles (Mi_h; cyan triangles). (a,c) The results of Driftsel and QstFstComp, respectively, using all SNPs (‘ALL' dataset, red squares); non-coding/non-repeat sites (‘NONCOD' dataset, pink triangles) and codon 3rd position synonymous changes (‘C3SYNO’ dataset, orange circles). Values of the S statistic (Driftsel) or (−log10) p-values (QstFstComp) are reported on the y-axis and dashed lines correspond to the significance threshold for each method. For all panels, filled and open shapes correspond to whether or not the full datasets (filled) or datasets with variability-based ascertained markers (open) were used.
3.7. QST-FST tests: effects of marker ascertainment
Marker ascertainment did not influence the outcome of the QstFstComp analyses, regardless of the type of genomic dataset used (figure 4; electronic supplementary material, tables S3, S4, S7). The ascertainment did influence the outcome of the Driftsel analyses for microsatellite markers, however (figure 4). Specifically, Driftsel detected a significant signal of divergent selection on the multivariate behaviour phenotype (‘AllB'; figure 4; electronic supplementary material, table S6) when in silico microsatellites were used. However, this effect disappeared when using only in silico microsatellites with low to mid number of alleles (figure 4; electronic supplementary material, table S6). Moreover, the effect of marker ascertainment on the outcome of Driftsel was more pronounced when the H statistic [8] was computed (electronic supplementary material, figure S5). For this test, both NONCOD and C3SYNO ascertained SNP datasets failed to detect signal of divergent selection on behavioural traits (‘AllB' and ‘B1'; electronic supplementary material, figure S5) and morphological traits (‘M2' and ‘M3'; electronic supplementary material, figure S5). Similarly, the ALL SNP and Mi_h datasets failed to detect signals of selection for the first behavioural trait (‘B1'; electronic supplementary material, figure S5) when using ascertained markers. The ALL SNP dataset also failed to detect the signal of selection for the second morphological trait (‘M2'; electronic supplementary material, figure S5) when using ascertained markers.
3.8. Simulation study
FST values estimated by the Driftsel method were 0.49, 0.47, 0.43 and 0.31 for the SNP, Mi_l, Mi_m and Mi_h datasets, respectively, thus approaching the FST estimates obtained from the real stickleback data (electronic supplementary material, table S1 and S2). Under the scenarios (i) and (ii) (i.e. QST ≈ 0.45–0.68), the S statistics estimated from the Mi_h data were considerably higher than the estimates obtained with SNP or Mi_l datasets (electronic supplementary material, figure S6(a) and table S9), but in general, the averaged S statistics of all the datasets were under the significance threshold (0.95) to identify the divergent selection. In scenario (iii) (i.e. QST ≈ 0.85), the S statistic of Mi_h set started to go beyond the significance threshold, and had better power to identify the signature of selection compared with other datasets. In scenario (iv) with strong signature of selection (i.e. QST ≈ 0.99), the S statistic among all the datasets were consistent, and they all indicated strong selection signals (S ≈ 0.99). By contrast, the p-values of all the datasets by using the QstFstComp method did not reach the significance level even in scenario (iv) (electronic supplementary material, figure S6(b)), and the power to detect signature of selection appeared to be consistently low (electronic supplementary material, table S9).
4. Discussion
In accordance with earlier suggestions [9,10], our results indicate that microsatellite markers can indeed yield an overly liberal neutral baseline for QST-FST tests, and this problem increases with increasing marker variability. Yet this made little difference for the outcome of the QST-FST tests in the stickleback data: SNP and microsatellite data yielded the same evolutionary inference. Similarly, we found little evidence to suggest that the genomic location of SNP-markers would markedly influence the FST estimates. Filtering of the SNP data for minor alleles had some impact on the estimation of FST when using AOR, but the magnitude of this impact was low and varied from one population comparison to another. However, the ascertainment of the markers by variability in marine populations had a relatively large but inconsistent effect: it changed the FST estimates when comparing populations used in ascertainment, but had virtually no impact on the comparison of pond populations. Worryingly, the direction of change differed between comparisons from slight increase to significant decrease. If the results are generalizable to other systems involving a greater number of populations with less extreme divergences, the choice of populations used for marker ascertainment may have a relatively large impact on FST estimates, and thus, on the neutral baseline divergence estimation. Hence, our results highlight some challenges (and opportunities) which the earlier QST-FST studies based on low marker numbers did not meet. In the following, we will briefly discuss the implications of these findings for comparative studies of quantitative trait and molecular marker differentiation.
We compared two approaches for combining FST estimates across loci: average of ratios (AOR) and ratio of averages (ROA). Although AOR has been shown to be affected by rare alleles and underestimate the FST [29], it often seems the most natural way to compare subsets of genomic estimates (e.g. in sliding window analyses), and is probably still widely used [29]. Our analyses confirmed that ROA gives consistently higher estimates of FST than AOR. The relative difference between the two approaches differed between comparisons, but, more interestingly, the relative order FST estimates also changed within a comparison between different sets of markers: in our analysis of microsatellite data, the FST estimates based on highly variable loci were clearly the lowest with ROA but among the highest with AOR. As a side note, when doing the analyses, we learned that the proper averaging approach, ROA, has been made unnecessarily difficult as many softwares only output estimates of the sitewise ratio of variances. For example, the popular software VCFtools could be easily changed to output also the numerator and denominator of the FST estimates, and thus, allow computing ROA for arbitrary subsets of loci.
One of the major concerns in the application of QST-FST tests for adaptive differentiation has been the likely underestimation of neutral genetic differentiation when highly variable microsatellite markers are used [9,10]. Our results comparing FST values estimated using a very large number of microsatellite loci with differing levels of variability do indeed suggest, in accordance with earlier studies [13,14], that highly variable loci deflate FST estimates. However, to which degree this constitutes a concern for QST-FST studies depends also on the degree of differentiation in quantitative traits. In the case of the stickleback data analysed here, this made little difference for the outcome of the Driftsel analyses: although the FST values were lower for microsatellite than for SNP loci, all the tests that rejected the null hypothesis of differentiation by drift with microsatellites also did so with SNPs. However, this point should be treated cautiously because of the nature of our data. Very high degree of quantitative trait differentiation among marine and freshwater nine-spined sticklebacks are well documented and have been attributed to divergent natural selection for a suite of ecologically important traits [8,37]. Such a marked differentiation is not the norm in empirical QST-FST studies where the degree of differentiation can be more subtle (e.g. [64,65]). Hence, the observed insensitivity of the QST-FST tests towards biases in the neutral baseline FST should not be considered as a general rule. However, the particular case of the sticklebacks could be viewed as an exception confirming the rule: marker type and variability have the potential to bias the neutral baseline FST, and although they are less likely to affect inferences involving strong quantitative trait differentiation (i.e. high QST), they should be particularly acknowledged when degree of quantitative trait differentiation is at low to moderate levels. This view is reinforced by the effect of marker ascertainment on the outcome of our QST-FST tests. Although the observed effect was trait-specific and confined to highly variable microsatellites in Driftsel analyses, we found that QST-FST inference can be influenced by marker type and filtering criteria, even when QST is high.
One critical assumption underlying QST-FST analyses is that the markers used for estimation of FST are unlinked, as inclusion of tightly linked markers in FST estimation can lead to biases in neutral expectations. This assumption is not discussed much in the QST-FST literature, where the major methodological concerns have instead been in the estimation of QST (e.g. [66,67]), as well as in the effect of marker variability on FST. One reason for the lack of concern over possible effects of LD is that most QST-FST studies have used relatively few markers, and the likelihood of sampling tightly linked loci with few markers is low. With genome-scale data, accounting for LD can become a major concern, as linked markers do not provide independent information about the evolutionary history and demography of the study populations [34,68]. Using two different approaches to account for LD, we discovered that the LD-based sliding window approach [46], which is a commonly used tool in genome-wide association and population genetic studies (e.g. [69–72]), may lead to gross underestimation of FST. The reason for this appeared to be the strong structure among our study populations that created an impression of LD when the data were pooled. The pond populations had low nucleotide diversity (cf. figure 2c) and many loci showing variation in the marine populations were either fixed or nearly fixed in the ponds. Such differentiated loci showed high FST between the populations but also appeared to be in high LD with each other and were preferentially pruned by the sliding window approach. As such, a severe downward bias in FST ensued. In fact, LD pruning methods developed for populations with homogeneous LD structure are known to result in misleading outcomes when applied to data from structured populations [73]. Hence, care should be taken when pruning linked markers from multiple-population data, and as demonstrated here, a simple distance thinning approach can maintain the genetic structure of the original data and avoid downward bias in FST which could lead to overly liberal QST-FST tests. Moreover, our results suggest that the LD-based approach may work relatively well when the LD is estimated from one population or a set of closely related populations; in our case, thinning based on pond populations worked the best (figure 3a–c).
The question of how loci with low minor allele frequencies should be dealt with in population genomic investigations is contentious. Because such loci exhibit high sampling variance, and carry very little information about populations' demographic histories, it is commonly argued that they should be excluded from FST estimation (e.g. [31,33], but see [30]). We observed that FST was sensitive to decisions about minor allele filtering cut-off points: the higher the cut-off point, the higher the FST became. However, the magnitude of this effect varied from one population pair to another, apparently because of the fairly large differences in the frequency of minor alleles among populations. What may be useful to note is that the effect of minor allele filtering on FST may depend on how it is calculated [29]. The mean FSTs can be obtained by averaging locus-specific FST estimates, or by first calculating the average (within and between populations) variances of all the SNPs and then estimating the mean FST of those. Since the individual FSTs of the low-frequency variants are small, they weigh down the average FST as obtained with the first approach. However, since the low-frequency variants have very small allelic variances, their influence on the mean FST as estimated from the second approach (which is the one used by Driftsel and other QST-FST comparison methods) is small. Hence, given their relatively small contribution to FST, and in order to err on the conservative side, we believe that it would be justified to remove low-frequency alleles (e.g. MAF < 0.025) when estimating the FST baseline for QST-FST tests. Their inclusion would have only a minor impact and tend to make tests of adaptive hypotheses slightly more liberal.
We found that FST estimates for SNPs and less variable microsatellite loci were on average slightly, but significantly higher in regions of high than low recombination. While this effect was small, it is noteworthy since the difference was in the opposite direction than expected. Namely, background and hitchhiking selection are expected to have stronger effects on allele frequencies in areas of low than high recombination. This is because linkage between neutral and selected loci is tighter in the areas of low recombination [27,28,74]. In fact, several studies have documented negative correlations between population differentiation and recombination rate [75–82]. On the other hand, introgression between fish species was recently shown to be more common in regions of high recombination [83] and possibly the weaker linkage to negatively selected sites allow loci with smaller adaptive effects to diverge between populations. This would create correlation between genetic divergence and recombination rate, and could explain the slightly elevated FST values. Although the effect we observed was so subtle that it did not have practical consequences for our QST-FST comparisons, recombination rate and degree of population differentiation are often found to go hand-in-hand. Future QST-FST studies should thus pay particular attention to how the markers in test panels are distributed in respect to variation in recombination rate or its proxies, such as their location relative to centromeres (e.g. [82]).
Interestingly, we did not detect any outlier loci in our analyses, suggesting that their impact on baseline FST estimates, and hence on the outcome of our QST-FST tests, can be assumed to be negligible. However, the fact that no outliers were detected does not mean that they did not exist in the data: given the extremely strong impact of genetic drift on the genetic constitution of the pond populations, as for instance reflected in the generally very high FST estimates and lack of low-frequency alternative alleles, the outlier tests were probably ill-suited for detecting footprints of selection against such a highly divergent neutral background (e.g. [36]). However, even if some outliers might have become included in the data, it is likely that their impact is small given that a few high-FST loci make little difference for the overall FST when averaged over a large number of unlinked and presumably neutral loci.
QST-FST comparisons have a long history in evolutionary biology (reviewed in [4]), and recent years have seen several methodological refinements to the basic approach (e.g. [8,56–58,84]). Here, we employed two recently developed approaches and although our aim was not to provide formal performance comparisons between the two methods, we discovered that the Driftsel approach recovered consistently more evidence for selection in our data than the QstFstComp approach. This was expected, as the Driftsel approach has been shown to have more power to detect footprints of selection than other types of QST-FST tests, especially when the number of populations is small and the impact of random genetic drift is strong [58]. Our simulations further confirm the validity of this inference. Under a scenario of profound quantitative trait divergence (QST > FST), Driftsel was consistently able to detect the signal of selection, irrespective of the marker set used, whereas QstFstComp approach failed to do so. Under the scenario of intermediate differentiation, neither of the two approaches was able to pick up the signal of selection, albeit the Driftsel approach applied to highly variable markers showed tendency towards this. This observation underlines the validity of the concerns associated with the use of highly variable markers in QST-FST comparisons [9,10]. The generally poor performance of both Driftsel and QstFstComp approaches in detecting signatures of selection in our simulated datasets can be explained by two factors. First is the high level of neutral baseline differentiation used in the simulations, and the second is the small number of populations used. As shown earlier, the performance of both approaches increases with decreasing impact of drift and increasing number of populations included into analyses [58].
Another illuminating lesson from our comparisons was that increasing the number of microsatellite loci (from 12 to 2000) used for FST estimation did not influence the outcome of our QST-FST tests. However, given the many possible sources of error and bias in estimating neutral baseline differentiation, including as many judiciously filtered markers as possible, may be warranted. This also highlights the need for further developments in the current QST-FST testing tools. Although Driftsel is in principle capable of handling infinite number of loci, the run-times for individual tests quickly become prohibitive once the number of markers starts to exceed a few thousand. A nature of the stochastic sample-based MCMC algorithm is that the larger number of SNPs included, the slower the algorithm will converge to the target posterior distribution, and more iterations will be needed. Hence, fine-tuning of the underlying algorithm to meet the demands of modern-day marker-panels is needed. For example, a deterministic variational Bayes (VB) algorithm [85,86] might provide a faster alternative to analyse large-scale SNP data with the Driftsel model.
Finally, it may be also worth stressing that QST-FST tests are prone to biases stemming from the choice of study design used to obtain quantitative genetic parameters needed for the estimation of QST. Specifically, the use of full-sib design, such as used in the present study, yields estimates of additive genetic variance potentially confounded with dominance variance [87]. However, dominance variance tends to reduce, rather than inflate QST [88,89]. Thus, even if non-additive inheritance were at play in the differentiation among our stickleback populations, our results would most likely still be rather conservative in respect to the detection of signals of divergent selection.
5. Conclusion
In conclusion, our empirical investigation of genetic variation and differentiation based on large numbers of SNP and microsatellite loci shows that both under- and overestimation of the neutral baseline level of differentiation can occur depending on the filtering and ascertainment of marker data. The results highlight the fact that marker ascertainment, but sometimes also minor allele filtering, might influence the outcome of QST-FST tests through their effects of FST, especially if the degree of quantitative trait divergence is low. Similarly, while pruning of tightly linked markers from the SNP panels is needed to avoid biased neutral estimates of differentiation, standard filtering methods developed for data from unstructured populations may result in serious biases if populations are strongly differentiated. However, once founded on carefully assembled SNP-panels and sound quantitative genetic data, QST-FST comparisons continue to provide a useful framework for testing the adaptive basis of population differentiation in ecologically important quantitative traits. Our findings and considerations suggest that for QST-FST comparisons, SNP-markers are preferable over microsatellites, and if microsatellites are to be used, loci from the lower end of the allele number distribution should be preferred over those from the higher end.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank M.D. Edge and an anonymous reviewer for their comments that helped us to improve the manuscript. We thank J. De Faveri for comments and linguistic check (remaining errors introduced by us) of an earlier version of this manuscript. Thanks are also for all the people who helped in obtaining the data. Special thanks to S. Varadharajan, L. Nederbragt and K. Jacobsen for their generosity in allowing us tap into the unpublished nine-spined stickleback reference genome. We are also grateful to P. Momigliano and B. Guo for discussions relevant to FST outlier detection. Computational analyses were enabled by the resources of CSC—IT Center for Science, Finland.
Data accessibility
R scripts for the simulation study are attached as electronic supplementary material. SNP and in silico microsatellite data are available within the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.c2fqz6140 [90].
Authors' contributions
J.M. conceived the study; A.L. and Z.L. designed the methodology; all authors contributed to collecting of the data; A.L. processed the genomic data; A.L. and Z.L. analysed the data; J.M. and A.F. led the writing of the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
Competing interests
We have no competing interests.
Funding
We were supported by the Academy of Finland (grant nos 134728, 250435 and 265211 to J.M.).
References
- 1.Merilä J, Crnokrak P. 2001. Comparison of genetic differentiation at marker loci and quantitative traits. J. Evol. Biol. 14, 892–903. ( 10.1046/j.1420-9101.2001.00348.x) [DOI] [Google Scholar]
- 2.McKay JK, Latta RG. 2002. Adaptive population divergence: markers, QTL and traits. Trends Ecol. Evol. 17, 285–291. ( 10.1016/S0169-5347(02)02478-3) [DOI] [Google Scholar]
- 3.Leinonen T, McCairns RS, O'hara RB, Merilä J. 2013. QST–FST comparisons: evolutionary and ecological insights from genomic heterogeneity. Nat. Rev. Genet. 14, 179 ( 10.1038/nrg3395) [DOI] [PubMed] [Google Scholar]
- 4.Leinonen T, O'hara RB, Cano JM, Merila J. 2008. Comparative studies of quantitative trait and neutral marker divergence: a meta-analysis. J. Evol. Biol. 21, 1–17. ( 10.1111/j.1420-9101.2007.01445.x) [DOI] [PubMed] [Google Scholar]
- 5.Whitlock MC. 2008. Evolutionary inference from QST. Mol. Ecol. 17, 1885–1896. ( 10.1111/j.1365-294X.2008.03712.x) [DOI] [PubMed] [Google Scholar]
- 6.De Kort H, Vandepitte K, Honnay O. 2013. A meta-analysis of the effects of plant traits and geographical scale on the magnitude of adaptive differentiation as measured by the difference between QST and FST. Evol. Ecol. 27, 1081–1097. ( 10.1007/s10682-012-9624-9) [DOI] [Google Scholar]
- 7.Karhunen M, Merilä J, Leinonen T, Cano J, Ovaskainen OJ. 2013. driftsel: an R package for detecting signals of natural selection in quantitative traits. Mol. Ecol. Res. 13, 746–754. ( 10.1111/1755-0998.12111) [DOI] [PubMed] [Google Scholar]
- 8.Karhunen M, Ovaskainen O, Herczeg G, Merila J. 2014. Bringing habitat information into statistical tests of local adaptation in quantitative traits: a case study of nine-spined sticklebacks. Evolution 68, 559–568. ( 10.1111/evo.12268) [DOI] [PubMed] [Google Scholar]
- 9.Edelaar P, Björklund M. 2011. If FST does not measure neutral genetic differentiation, then comparing it with QST is misleading. Or is it? Mol. Ecol. 20, 1805–1812. ( 10.1111/j.1365-294X.2011.05051.x) [DOI] [PubMed] [Google Scholar]
- 10.Edelaar P, Burraco P, Gomez-Mestre I. 2011. Comparisons between QST and FST—how wrong have we been? Mol. Ecol. 20, 4830–4839. ( 10.1111/j.1365-294X.2011.05333.x) [DOI] [PubMed] [Google Scholar]
- 11.Li YC, Korol AB, Fahima T, Beiles A, Nevo E. 2002. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol. 11, 2453–2465. ( 10.1046/j.1365-294X.2002.01643.x) [DOI] [PubMed] [Google Scholar]
- 12.Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435 ( 10.1038/nrg1348) [DOI] [PubMed] [Google Scholar]
- 13.Hedrick PW. 2005. A standardized genetic differentiation measure. Evolution 59, 1633–1638. ( 10.1111/j.0014-3820.2005.tb01814.x) [DOI] [PubMed] [Google Scholar]
- 14.Heller R, Siegismund HR. 2009. Relationship between three measures of genetic differentiation GST, DEST and G′ST: how wrong have we been? Mol. Ecol. 18, 2080–2083. ( 10.1111/j.1365-294X.2009.04185.x) [DOI] [PubMed] [Google Scholar]
- 15.Meirmans PG, Hedrick PW. 2011. Assessing population structure: FST and related measures. Mole. Ecol. Resour. 11, 5–18. ( 10.1111/j.1755-0998.2010.02927.x) [DOI] [PubMed] [Google Scholar]
- 16.Wang J. 2015. Does GST underestimate genetic differentiation from marker data? Mol. Ecol. 24, 3546–3558. ( 10.1111/mec.13204) [DOI] [PubMed] [Google Scholar]
- 17.Morin PA, Luikart G, Wayne RK. 2004. SNPs in ecology, evolution and conservation. Trends Ecol. Evol. 19, 208–216. ( 10.1016/j.tree.2004.01.009) [DOI] [Google Scholar]
- 18.Pavey SA, Bernatchez L, Aubin-Horth N, Landry CR. 2012. What is needed for next-generation ecological and evolutionary genomics? Trends Ecol. Evol. 27, 673–678. ( 10.1016/j.tree.2012.07.014) [DOI] [PubMed] [Google Scholar]
- 19.Helyar SJ, et al. 2011. Application of SNPs for population genetics of nonmodel organisms: new opportunities and challenges. Mole. Ecol. Resour. 11(Suppl 1), 123–136. ( 10.1111/j.1755-0998.2010.02943.x) [DOI] [PubMed] [Google Scholar]
- 20.Whitlock MC. 2011. G′ST and D do not replace FST. Mol. Ecol. 20, 1083–1091. ( 10.1111/j.1365-294X.2010.04996.x) [DOI] [PubMed] [Google Scholar]
- 21.Baer CF, Miyamoto MM, Denver DR. 2007. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 8, 619–631. ( 10.1038/nrg2158) [DOI] [PubMed] [Google Scholar]
- 22.Hodgkinson A, Eyre-Walker A. 2011. Variation in the mutation rate across mammalian genomes. Nat. Rev. Genet. 12, 756–766. ( 10.1038/nrg3098) [DOI] [PubMed] [Google Scholar]
- 23.Nielsen R. 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39, 197–218. ( 10.1146/annurev.genet.39.073003.112420) [DOI] [PubMed] [Google Scholar]
- 24.Arbiza L, Zhong E, Keinan A. 2012. NRE: a tool for exploring neutral loci in the human genome. BMC Bioinf. 13, 301 ( 10.1186/1471-2105-13-301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schraiber JG, Akey JM. 2015. Methods and models for unravelling human evolutionary history. Nat. Rev. Genet. 16, 727–740. ( 10.1038/nrg4005) [DOI] [PubMed] [Google Scholar]
- 26.Maynard-Smith J, Haigh J. 1974. The hitch-hiking effect of a favorable gene. Genet. Res. 23, 23–35. ( 10.1017/S0016672300014634) [DOI] [PubMed] [Google Scholar]
- 27.Charlesworth B, Nordborg M, Charlesworth D. 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 70, 155–174. ( 10.1017/S0016672397002954) [DOI] [PubMed] [Google Scholar]
- 28.Nachman MW. 2002. Variation in recombination rate across the genome: evidence and implications. Curr. Opin. Genet. Dev. 12, 657–663. ( 10.1016/S0959-437x(02)00358-1) [DOI] [PubMed] [Google Scholar]
- 29.Bhatia G, Patterson N, Sankararaman S, Price AL. 2013. Estimating and interpreting FST: the impact of rare variants. Genome Res. 23, 1514–1521. ( 10.1101/gr.154831.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albrechtsen A, Nielsen FC, Nielsen R. 2010. Ascertainment biases in SNP chips affect measures of population divergence. Mol. Biol. Evol. 27, 2534–2547. ( 10.1093/molbev/msq148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roesti M, Salzburger W, Berner D. 2012. Uninformative polymorphisms bias genome scans for signatures of selection. BMC Evol. Biol. 12, Article number 94 ( 10.1186/1471-2148-12-94) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakobsson M, Edge MD, Rosenberg NA. 2013. The relationship between FST and the frequency of the most frequent allele. Genetics 193, 515–528. ( 10.1534/genetics.112.144758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitlock MC, Lotterhos KE. 2015. Reliable detection of loci responsible for local adaptation: inference of a null model through trimming the distribution of FST. Am. Nat. 186, S24–S36. ( 10.1086/682949) [DOI] [PubMed] [Google Scholar]
- 34.Williams LM, Oleksiak MF. 2011. Ecologically and evolutionarily important SNPs identified in natural populations. Mol. Biol. Evol. 28, 1817–1826. ( 10.1093/molbev/msr004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beaumont MA, Nichols RA. 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. B 263, 1619–1626. ( 10.1098/rspb.1996.0237) [DOI] [Google Scholar]
- 36.Hoban S, et al. 2016. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am. Nat. 188, 379–397. ( 10.1086/688018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herczeg G, Gonda A, Merila J. 2009. Evolution of gigantism in nine-spined sticklebacks. Evolution 63, 3190–3200. ( 10.1111/j.1558-5646.2009.00781.x) [DOI] [PubMed] [Google Scholar]
- 38.Herczeg G, Gonda A, Merila J. 2009. Predation mediated population divergence in complex behaviour of nine-spined stickleback (Pungitius pungitius). J. Evol. Biol. 22, 544–552. ( 10.1111/j.1420-9101.2008.01674.x) [DOI] [PubMed] [Google Scholar]
- 39.Merila J. 2013. Nine-spined stickleback (Pungitius pungitius): an emerging model for evolutionary biology research. Ann. N. Y. Acad. Sci. 1289, 18–35. ( 10.1111/nyas.12089) [DOI] [PubMed] [Google Scholar]
- 40.Varadharajan S, Rastas P, Löytynoja A, Matschiner M, Calboli FCF, Guo B, Nederbragt AJ, Jakobsen KS, Merilä J. 2019. Genome sequencing of the nine-spined stickleback (Pungitius pungitius) provides insights into chromosome evolution. bioRxiv. 741751 ( 10.1101/741751%JbioRxiv) [DOI] [PMC free article] [PubMed]
- 41.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. ( 10.1101/gr.107524.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obenchain V, Lawrence M, Carey V, Gogarten S, Shannon P, Morgan M. 2014. VariantAnnotation: a bioconductor package for exploration and annotation of genetic variants. Bioinformatics 30, 2076–2078. ( 10.1093/bioinformatics/btu168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27, 2156–2158. ( 10.1093/bioinformatics/btr330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328. ( 10.1093/bioinformatics/bts606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weir BS. 1979. Inferences about linkage disequilibrium. Biometrics 35, 235–254. ( 10.2307/2529947) [DOI] [PubMed] [Google Scholar]
- 48.Weir BS. 1996. Genetic data analysis II: methods for discrete population genetic data. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 49.Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. ( 10.1093/nar/27.2.573) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Highnam G, Franck C, Martin A, Stephens C, Puthige A, Mittelman D. 2013. Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles. Nucleic Acids Res. 41, e32 ( 10.1093/nar/gks981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keenan K, McGinnity P, Cross TF, Crozier WW, Prodohl PA. 2013. diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 4, 782–788. ( 10.1111/2041-210X.12067) [DOI] [Google Scholar]
- 52.Charlesworth B. 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol. Biol. Evol. 15, 538–543. ( 10.1093/oxfordjournals.molbev.a025953) [DOI] [PubMed] [Google Scholar]
- 53.Rastas P, Calboli FCF, Guo BC, Shikano T, Merila J. 2016. Construction of ultradense linkage maps with lep-MAP2: stickleback F-2 recombinant crosses as an example. Genome Biol. Evol. 8, 78–93. ( 10.1093/gbe/evv250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foll M, Gaggiotti O. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180, 977–993. ( 10.1534/genetics.108.092221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population-structure. Evolution 38, 1358–1370. ( 10.1111/j.1558-5646.1984.tb05657.x) [DOI] [PubMed] [Google Scholar]
- 56.Gilbert KJ, Whitlock MC. 2015. QST–FST comparisons with unbalanced half-sib designs. Mole. Ecol. Resou. 15, 262–267. ( 10.1111/1755-0998.12303) [DOI] [PubMed] [Google Scholar]
- 57.Whitlock MC, Guillaume F. 2009. Testing for spatially divergent selection: comparing QST to FST. Genetics 183, 1055–1063. ( 10.1534/genetics.108.099812) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ovaskainen O, Karhunen M, Zheng C, Arias JMC, Merilä JJG. 2011. A new method to uncover signatures of divergent and stabilizing selection in quantitative traits. Genetics 189, 621–632. ( 10.1534/genetics.111.129387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Excoffier L, Foll M. 2011. fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics 27, 1332–1334. ( 10.1093/bioinformatics/btr124) [DOI] [PubMed] [Google Scholar]
- 60.Excoffier L, Dupanloup I, Huerta-Sanchez E, Sousa VC, Foll M. 2013. Robust demographic inference from genomic and SNP data. PLoS Genet. 9, e1003905 ( 10.1371/journal.pgen.1003905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shikano T, Shimada Y, Herczeg G, Merila J. 2010. History vs. habitat type: explaining the genetic structure of European nine-spined stickleback (Pungitius pungitius) populations. Mol. Ecol. 19, 1147–1161. ( 10.1111/j.1365-294X.2010.04553.x) [DOI] [PubMed] [Google Scholar]
- 62.Teacher AG. F., Shikano T, Karjalainen ME, Merila J. 2011. Phylogeography and genetic structuring of European nine-spined sticklebacks (Pungitius pungitius)—mitochondrial DNA evidence. PLoS ONE 6, e19476 ( 10.1371/journal.pone.0019476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bruneaux M, Johnston SE, Herczeg G, Merila J, Primmer CR, Vasemagi A. 2013. Molecular evolutionary and population genomic analysis of the nine-spined stickleback using a modified restriction-site-associated DNA tag approach. Mol. Ecol. 22, 565–582. ( 10.1111/j.1365-294X.2012.05749.x) [DOI] [PubMed] [Google Scholar]
- 64.Dubuc-Messier G, Caro SP, Perrier C, van Oers K, Reale D, Charmantier A. 2018. Gene flow does not prevent personality and morphological differentiation between two blue tit populations. J. Evol. Biol. 31, 1127–1137. ( 10.1111/jeb.13291) [DOI] [PubMed] [Google Scholar]
- 65.Blanckenhorn WU, et al. 2018. Life history traits, but not body size, vary systematically along latitudinal gradients on three continents in the widespread yellow dung fly. Ecography. 41, 2080–2091. ( 10.1111/ecog.03752) [DOI] [Google Scholar]
- 66.O'Hara RB, Merila J. 2005. Bias and precision in QST estimates: problems and some solutions. Genetics 171, 1331–1339. ( 10.1534/genetics.105.044545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lopez-Fanjul C, Fernandez A, Toro MA. 2007. The effect of dominance on the use of the QST-FST contrast to detect natural selection on quantitative traits. Genetics 176, 725–727. ( 10.1534/genetics.106.062901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kemppainen P, Knight CG, Sarma DK, Hlaing T, Prakash A, Maung YNM, Somboon P, Mahanta J, Walton C. 2015. Linkage disequilibrium network analysis (LDna) gives a global view of chromosomal inversions, local adaptation and geographic structure. Mole. Ecol. Resou. 15, 1031–1045. ( 10.1111/1755-0998.12369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crosslin DR, et al. 2014. Controlling for population structure and genotyping platform bias in the eMERGE multi-institutional biobank linked to electronic health records. Front. Genet. 5, 352 ( 10.3389/fgene.2014.00352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carbone M, et al. 2015. Combined genetic and genealogic studies uncover a large BAP1 cancer syndrome kindred tracing back nine generations to a common ancestor from the 1700s. PLoS Genet. 11, e1005633 ( 10.1371/journal.pgen.1005633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reed DH, Frankham R. 2001. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 55, 1095–1103. ( 10.1554/0014-3820(2001)055[1095:Hccama]2.0.Co;2) [DOI] [PubMed] [Google Scholar]
- 72.Fadista J, Manning AK, Florez JC, Groop L. 2016. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Europ. J. Hum. Genet. 24, 1202–1205. ( 10.1038/ejhg.2015.269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu L, Wu YH, Lonardi S, Jiang T. 2010. Efficient genome-wide TagSNP selection across populations via the linkage disequilibrium criterion. J. Comput. Biol. 17, 21–37. ( 10.1089/cmb.2007.0228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Corbett-Detig RB, Hartl DL, Sackton TB. 2015. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 13, e1002112 ( 10.1371/journal.pbio.1002112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Begun DJ, Aquadro CF. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356, 519–520. ( 10.1038/356519a0) [DOI] [PubMed] [Google Scholar]
- 76.Kraft T, Sall T, Magnusson-Rading I, Nilsson NO, Hallden C. 1998. Positive correlation between recombination rates and levels of genetic variation in natural populations of sea beet (Beta vulgaris subsp. maritima). Genetics 150, 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nachman MW. 2001. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17, 481–485. ( 10.1016/S0168-9525(01)02409-X) [DOI] [PubMed] [Google Scholar]
- 78.Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl Acad. Sci. USA 98, 9161–9166. ( 10.1073/pnas.151244298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takahashi A, Liu YH, Saitou N. 2004. Genetic variation versus recombination rate in a structured population of mice. Mol. Biol. Evol. 21, 404–409. ( 10.1093/molbev/msh030) [DOI] [PubMed] [Google Scholar]
- 80.Roselius K, Stephan W, Stadler T. 2005. The relationship of nucleotide polymorphism, recombination rate and selection in wild tomato species. Genetics 171, 753–763. ( 10.1534/genetics.105.043877) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGaugh SE, Heil CS. S., Manzano-Winkler B, Loewe L, Goldstein S, Himmel TL, Noor MA. F. 2012. Recombination modulates how selection affects linked sites in Drosophila. PLoS Biol. 10, e1001422 ( 10.1371/journal.pbio.1001422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roesti M. 2013. Recombination in the threespine stickleback genome—patterns and consequences. Mol. Ecol. 22, 5270–5270. ( 10.1111/mec.12322) [DOI] [PubMed] [Google Scholar]
- 83.Schumer M, et al. 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science 360, 656–659. ( 10.1126/science.aar3684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martin G, Chapuis E, Goudet J. 2008. Multivariate QST–FST comparisons: a neutrality test for the evolution of the G matrix in structured populations. Genetics 180, 2135–2149. ( 10.1534/genetics.107.080820) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jaakkola TS, Jordan MI. 2000. Bayesian parameter estimation via variational methods. Stat. Comput. 10, 25–37. ( 10.1023/A:1008932416310) [DOI] [Google Scholar]
- 86.Raj A, Stephens M, Pritchard JK. 2014. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197, 573 ( 10.1534/genetics.114.164350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lynch M, Walsh B. 1998. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer. [Google Scholar]
- 88.Goudet J, Martin G. 2007. Under neutrality, QST ≤ FST when there is dominance in an island model. Genetics 176, 1371–1374. ( 10.1534/genetics.106.067173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Santure AW, Wang J. 2009. The joint effects of selection and dominance on the QST–FST contrast. Genetics 181, 259–276. ( 10.1534/genetics.108.097998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Z, Löytynoja A, Fraimout A, Merilä J. 2019. Data from: Effects of marker type and filtering criteria on QST–FST comparisons Dryad Digital Repository ( 10.5061/dryad.c2fqz6140) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Li Z, Löytynoja A, Fraimout A, Merilä J. 2019. Data from: Effects of marker type and filtering criteria on QST–FST comparisons Dryad Digital Repository ( 10.5061/dryad.c2fqz6140) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
R scripts for the simulation study are attached as electronic supplementary material. SNP and in silico microsatellite data are available within the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.c2fqz6140 [90].