Abstract
Multiple human proteins have been shown to both support and restrict viral replication, and confirmation of virus-associated changes in the expression of these genes is relevant for future therapeutic efforts. In this study a well-characterized panel of 49 individuals either infected with HIV-1 or uninfected was compiled and analyzed for the effect of HIV infection status, viral load, and antiretroviral treatment on specific gene expression. mRNA was extracted and reverse transcribed from purified CD4+ cells, and quantitative real-time PCR was utilized to scrutinize differences in the expression of four host genes that have been demonstrated to either stimulate (HSP90 and LEDGF/p75) or restrict (p21/WAF1 and APOBEC3G) proviral integration. HIV infection status was associated with slight to moderate alterations in the expression of all four genes. After adjusting for age, mRNA expression levels of HSP90, LEDGF/p75 and APOBEC3G were found to all be decreased in infected patients compared to healthy controls by 1.43-, 1.26-, and 4.71-fold, respectively, while p21/WAF1 expression was increased 2.35-fold. Furthermore, individuals receiving raltegravir exhibited a 1.28-fold reduction in LEDGF/p75 compared to those on non-raltegravir antiretroviral treatment. Identification of these and similar HIV-induced changes in gene expression may be valuable for delineating the extent of host cell molecular mechanisms stimulating viral replication.
Keywords: HIV, HSP90, LEDGF/p75, p21/WAF1, APOBEC3G, gene expression
INTRODUCTION
HIV-1 persists as a global epidemic with no known cure. Over 30 antiretroviral drugs targeting various stages of the viral life cycle have been developed to-date [Broder, 2010], but the emergence of drug-resistant viral strains has been a constant, insurmountable problem facing patients, pharmaceutical scientists, and clinicians alike since the initial prescription of AZT almost three decades ago [Cozzi-Lepri et al., 2007; Serrao et al., 2009]. The initial phase of HIV infection is characterized by high levels of circulating HIV RNA and a rapid decline in CD4+ T lymphocyte count [Cadogan and Dalgleish, 2008; McMichael et al., 2010]. Following infection there is a state of chronic immune activation, characterized by high T cell turnover, apoptosis, and cell death. Activation of CD4+ T cells is essential for HIV to establish a productive infection [Zack et al., 1990; Bukrinsky et al., 1991; Zhou et al., 2005], and a stable state of latent infection is thought to develop mainly as a subset of long-lived activated cells reverts back to the resting state [Siliciano and Greene, 2011]. Even intense pressure from HAART has been unable to eradicate the virus from stable latent reservoirs [Siliciano et al., 2003]. Productive proviral integration by HIV-1 integrase (IN) is a prerequisite for latency to occur, and multiple host proteins have been discovered to assist or restrict this enzymatic process [Al-Mawsawi and Neamati, 2007]. Of note, APOBEC3G and p21/WAF1 are host restriction factors also shown to bind and inhibit IN [Luo et al., 2007; Zhang et al., 2007]. On the other hand, HSP90 has been shown to stimulate both HIV-1 transcription and integration [O’Keeffe et al., 2000; Vozzolo et al., 2010], and small-molecule HSP90 inhibitors have shown antiviral effects and a reduction in integration efficiency [Vozzolo et al., 2010; Roesch et al., 2012]. Similarly, LEDGF/p75 is a host cofactor of integration that has been elegantly demonstrated to tether IN to the host chromosome and promote integration into transcriptional active genomic regions [Cherepanov et al., 2003; Shun et al., 2007]. Discoveries such as this have demonstrated that understanding the host antiviral response and the full extent of HIV’s manipulation of host cell machinery is crucial for therapeutic innovation.
In this cross-sectional study, a well-characterized panel of 49 individuals uninfected or infected with HIV-1 was compiled and analyzed for the effect of various parameters on the mRNA expression levels of HSP90, LEDGF/p75, p21/WAF1, and APOBEC3G. Specific parameters that were scrutinized included subject sex, ethnicity, HIV transmission status (behavioral or perinatal), HIV infection and exposure status, viral load, and antiretroviral treatment. While sex, ethnicity, exposure status, and transmission status were not associated with age-adjusted mRNA expression changes, subtle to significant differences in the expression of each gene in subjects infected with HIV compared to uninfected individuals were apparent. Additionally, individuals infected with HIV that were treated with raltegravir exhibited a reduction in LEDGF/p75 compared to those on a non-raltegravir regimen.
METHODS
Study Design
Subjects were selected from a Natural History, Long-term Follow-up and Pathogenesis Study of HIV Infection at the Maternal, Child, Adolescent Center for Infectious Diseases and Virology at the Los Angeles County, and the University of Southern California Medical Center in Los Angeles, CA. Informed consent was obtained for each patient in this IRB-approved study. At each routine clinic visit, a blood specimen was stored in the MCA repository for future research. Additional specimens from uninfected subjects were provided by individuals volunteering for another IRB approved sub-study of the Natural History Study.
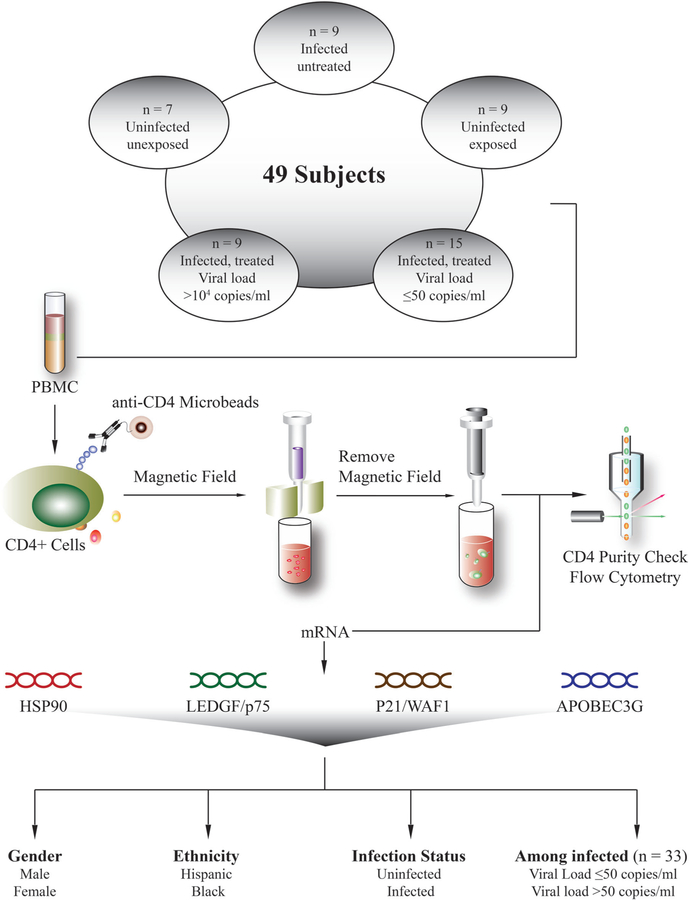
Based on the availability of repository specimens collected between 2008 and 2011, patients fitting desired experimental criteria were selected and organized into groups with careful definition of clinical and virologic parameters. The most recently collected specimen available for each subject was selected to be used to measure gene expression. Subjects were chosen based on their HIV infection status, HIV exposure status, HIV viral load, and antiretroviral treatment status. The goal was to select approximately 10 subjects in three groups of patients infected with HIV based on their HIV viral load and treatment status, and two groups of uninfected patients based on their HIV exposure status. Among the patients infected with HIV, the following were selected: (i) 15 patients who received antiretroviral treatment and had an undetectable viral load (<50 copies/ml) for at least 2 years (infected/treated/non-viremic); (ii) nine subjects who also received antiretroviral treatment but who had a high viral load at the time of specimen collection of >10,000 copies/ml (infected/treated/high viral load); and (iii) nine patients who were not receiving treatment at the time of the specimen (infected/untreated). Among the uninfected individuals, nine were exposed perinatally to HIV but uninfected, and seven were unexposed. Other variables collected in the Natural History Study included antiretroviral drug regimen, gender, age at the time of the specimen collection, sex, ethnicity, perinatal versus behavioral mode of HIV transmission, race/ethnicity, and CD4 count at the time of the specimen collection.
Isolation of CD4+ T Cells
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by density gradient centrifugation. The anticoagulant used was either EDTA or acid citrate dextrose. The cells were then cryopreserved in 90% fetal bovine serum with 10% DMSO and stored in liquid nitrogen until needed. All cells were cryopreserved between November, 2007 and October, 2011. Aliquots of frozen PBMCs were rapidly thawed and washed twice using 37°C water-bath warmed RPMI-1640 supplemented with 10% FBS. Cells were resuspended in PBS containing 2% FBS, and then aliquots were stained with trypan blue and evaluated for cell viability and recoverability. The CD4+ fraction of the total PBMCs was magnetically labeled with CD4 Microbeads (Miltenyi Biotec, Cambridge, MA). CD4+ cells were separated from the cell suspension in a column within a magnetic field. The column was then removed from the magnetic field, and the purified cells were eluted as the positively selected cell fraction and immediately processed for RNA isolation. Sample purity was assessed by flow cytometry, as described below.
Quantitative Real-Time PCR
To determine the gene expression levels of HSP90, LEDGF/p75, p21/WAF1, and APOBEC3G in CD4+ cells, real-time PCR was carried out on cDNA reverse transcribed (iScript™ cDNA Synthesis Kit, Bio-Rad, Hercules, CA) from RNA isolated from 1 to 8 × 106 purified CD4+ cells (Aurum™ Total RNA Mini Kit, Bio-Rad). One microgram of cDNA was then used as a template for real-time PCR in a 7900HT Fast Real-Time PCR System (ABI, Foster City, CA). The amplification reactions were carried out in triplicate using SsoAdvanced™ SYBR® Green Supermix (Bio-Rad) in 25 μl reaction volumes. The following primers were used: HSP90 forward, 5ʹ-ATTTGTCCCACGACGTGCT-CCTT-3ʹ; HSP90 reverse, 5ʹ-ATCCTCCGAGTCTACCACCCCTCTAAT-3ʹ; LEDGF/p75 forward, 5ʹ-AGGCAGGAGTAGTGACAACAGCA-3ʹ; LEDGF/p75 reverse, 5ʹ-TGTCACTCTCTGAAGGACAGGGCT-3ʹ; p21/WAF1 forward, 5ʹ-TCGACTTTGTCACCGAGACACCACT-3ʹ; p21/WAF1 reverse, 5ʹ-TGACAGGTCCACATGGTCTTCCTCT-3ʹ; APOBEC3G forward, 5ʹ-ACCAGAAGCTTGGAGCAGAAAGTGA-3ʹ; APOBEC3G reverse, 5ʹ-ACGGTATTCCGACGAGAAAGGATGGGT-3ʹ; Beta actin forward, 5ʹ-AGCCATGTACGTTGCTATCCAGGCT-3ʹ; Beta actin reverse, 5ʹ-TCGGTGAGGATCTTCATGAGGTAGTCAGT-3ʹ. Beta actin was chosen as the reference/house-keeping gene for this study based upon a previous report showing that this gene is the most stably expressed in PBMCs [Chege et al., 2010]. Primer efficiencies were calculated using standard curves generated from 1:5 serial dilutions of an arbitrary sample cDNA, using the calculation E = 10(−1/slope). All primer sets were validated to generate reproducible PCR efficiencies between 90% and 100%. PCR conditions consisted of a denaturation step at 95°C for 30 sec, followed by 40 cycles of denaturation and annealing/extension at 95°C for 5 sec and 55°C for 30 sec, respectively, followed by dissociation curve analysis programmed by the ABI PCR System.
Flow Cytometry
To determine the purity of isolated CD4+ T cells, purified cell samples were stained with PE-Cy7-labeled antibody to CD3 and APC-labeled antibody to CD4 (BD Biosciences, San Jose, CA), and then analyzed by six-color flow cytometry. The purity of sorted CD4+ T cells was identified according to the percentages of CD3+/CD4+ cells among total PBMCs. For each subject 200,000 live-sorted cells were stained for 30 min with the anti-antibody cocktail at 4°C in the dark in BD Falcon round bottom tubes. After completion of the staining, cells were washed with PBS and resuspended in an appropriate volume of cold PBS. Acquisition of flow cytometry data was performed on a FACSCanto A flow cytometer (Becton Dickinson, Franklin Lakes, NJ) utilizing FACSDiva Software Version 6.2. Electronic compensation was performed with BD CompBeads (Becton Dickinson) stained separately with individual mAbs. Analyses were performed with FlowJo Version 9.4 software (Tree Star, Ashland, OR). The gating strategy and background levels of staining were determined by utilizing Fluorescence Minus One with Isotype controls.
Statistical Analysis
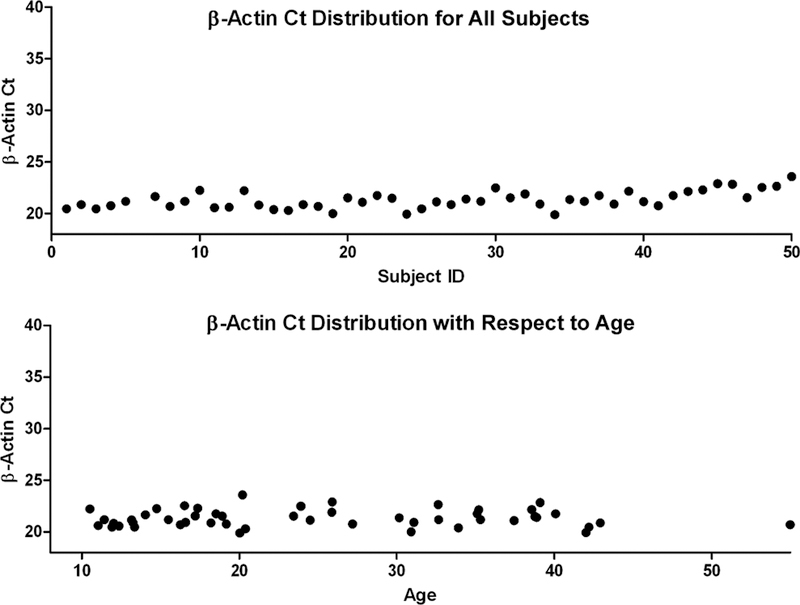
Descriptive statistics for demographic and clinical characteristics regarding age at the time of specimen collection, sex, ethnicity (Hispanic, African American, or Asian), CD4 counts (cells/µl), CD4 percentages, raltegravir/protease inhibitors treatment status, viral RNA level (copies/ml), and perinatal/behavioral exposure status are reported in Table I. It was investigated whether the mRNA expression of HSP90, LEDGF/p75, p21/WAF1, and APOBEC3G normalized to β-Actin varied significantly due to the following parameters: sex (male vs. female), ethnicity (Hispanic vs. Non-Hispanic), infection status (uninfected vs. infected), raltegravir/protease inhibitors treatment status (raltegravir/protease inhibitors vs. non-raltegravir/non-protease inhibitors), and viral load level among total infected [viremic (≥100 copies/ml in this study) vs. non-viremic subjects (<50 copies/ml)] and also specifically among infected/untreated subjects (viral load between 100 copies/ml and 1,000 copies/ml vs. viral load ≥1,000 copies/ml). The expression of genes HSP90, LEDGF/p75, p21/WAF1, and APOBEC3G were measured in cycles to threshold (Ct), which represented the cycle number at which the amount of DNA reached an arbitrarily placed threshold level. The normalized ΔCt values were obtained by subtracting the Ct numbers of HSP90, LEDGF/p75, p21/WAF1, and APOBEC3G from that of the reference gene, β-Actin. Scatter plots were constructed for inspection of the Ct distribution of β-Actin for all the participants (Fig. 2). Fold change, the expression ratio, was then calculated using the following method:
where ΔΔCt was the difference of the average ΔCt values between two comparison groups.
TABLE I.
Participant Demographics
| Parameter | Infected/untreated/ detectable |
Infected/ART/high viral load |
Infected/ART/ non-viremic |
Uninfected |
|---|---|---|---|---|
| Number participants | 9 | 9 | 15 | 16 |
| Median age, years (range) | 30 (17–55) | 24 (13–40) | 35 (17–43) | 14 (11–34) |
| Sex, % female | 56 | 78 | 80 | 69 |
| Ethnicity, % Hispanic | 56 | 78 | 93 | 88 |
| Median CD4+ count (range) | 541 (338–942) | 299 (23–713) | 774 (405–976) | — |
| Median CD4% (range) | 23 (6.8–42) | 21.1 (1.1–29.8) | 32.6 (18–45.9) | 43a (33.3–53.8) |
| Median viral RNA copies (range) | 2,505 (129–1,880,000) | 73,258 (11,776–172,136) | <50 | — |
| Number perinatally infected | 0 | 5 | 3 | — |
| Number behaviorally infected | 9 | 4 | 12 | — |
Among 12 patients with CD4% data.
Fig. 2.
Scatter plots representing β-Actin Ct distribution for each subject and each subject respect to age in this study’s participant cohort.
Having a positive fold-change suggested that a certain gene was upregulated, and a negative fold-change suggested the opposite. The non-parametric Wilcoxon rank-sum test was performed to determine whether the expression difference was statistically significant. The associations between ethnicity, gender, and HIV infection status were modeled by linear regressions adjusted for age. The normality of the residuals of all final models was confirmed by histograms of residuals and statistical tests of normality. The FDR approach was used to adjust for multiple testing [Benjamini et al., 2001].
RESULTS
Demographic and Clinical Characteristics of Study Subjects
The demographic and clinical characteristics of this study’s 49 participants are shown in Table I. The infected/untreated and treated/non-viremic subjects were older with a median age of 30 and 35 respectively, compared to uninfected individuals with a median age of 14. Within the infected group, 25 participants were infected behaviorally and 8 perinatally. The infected/treated/non-viremic individuals had the highest median CD4 count (range: 405–976 cells/µl) and initial (pre-purification) CD4 percentage (range: 18–45.9%), while those in the infected/treated/high viral load group had the lowest CD4 count (range: 23–713 cells/µl) and initial CD4 percentage (range: 1.1–29.8%). The infected/untreated group included patients with low viral loads (consistently <1,000 copies/ml for 3–16 years), as well as those with much higher viral loads. This group displayed a large viral load range from 129 to 1,880,000 copies/ml. The infected/treated/high viral load group had viral loads ranging from 11,776 to 172,136 copies/ml, as they were chosen to have viral loads greater than 10,000 copies/ml. This study’s design, illustrated in Figure 1, consisted of purification of participants’ CD4+ cells from total PBMCs and comparison of HSP90, LEDGF/p75, p21/WAF1, and APOBEC3G gene expression with respect to sex, ethnicity, infection status, viremic status, raltegravir/protease inhibitor treatment, and viral load.
Fig. 1.
Scheme detailing subject panel construction, sample processing, and gene expression analysis involved in this study.
Real-Time PCR Analysis of Host Gene Expression in CD4+ T Cells from Infected and Non-Infected Participants
To make conclusions about the significance of the effect of certain parameters on the expression of HIV-1 integration-modulating host genes, the normalized expression (ΔCt) of different participant stratifications were compared, and the fold-change in gene expression was calculated using the 2−ΔΔCt method. The significance of difference in gene expression was tested by the non-parametric Wilcoxon rank-sum test using 0.05 P-value cut-off for significance (see Tables II and III). Due to a high association between age and HIV status among participants of this study, ΔCt values were modeled by way of linear regressions adjusted for age as a continuous variable, in which gene expression was analyzed by Wald test (Table II). As shown in Figure 2, β-Actin Ct values remained relatively stable with respect to age in this cohort, giving confidence in calculated variance for mRNA levels of each of the genes of interest. No significant difference in the expression of any of the four genes with respect to sex, ethnicity, HIV exposure status or mode of transmission was observed. In addition, viral load level (100–1,000 copies/ml vs.≥1,000 copies/ml) among specifically the infected/untreated was not associated with expression differences for any of the four genes. In linear regression models adjusted for age, mRNA expression levels of HSP90, LEDGF/p75, and APOBEC3G were all slightly to moderately decreased in infected patients with respect to uninfected individuals by 1.43-fold (P = 0.01), 1.26-fold (P = 0.001), and 4.71-fold (P = 0.003), respectively. Alternatively, p21/WAF1 expression was increased in these patients by 2.35-fold (P = 0.03).
TABLE II.
Analysis of Change in Gene Expression Relative to β-Actin Adjusting for Age
| HSP90 vs. β-Actin |
LEDGF/p75a vs. β-Actin |
P21 vs. β-Actin |
APOBEC3G vs. β-Actin |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | Total | No. (%) | Fold change |
P-value | Fold change |
P-value | Fold change |
P-value | Fold change |
P-value |
| Gender | 49 | |||||||||
| Female | 35 (71.4) | Ref | Ref | Ref | Ref | |||||
| Male | 14 (28.6) | 1.05 | 0.68 | 1.02 | 0.69 | −1.14 | 0.70 | 1.73 | 0.23 | |
| Ethnicity | 49 | |||||||||
| Hispanic | 40 (83.3) | Ref | Ref | Ref | Ref | |||||
| Not Hispanic | 9 (18.4) | 1.06 | 0.68 | 1.04 | 0.61 | −1.18 | 0.67 | 1.45 | 0.48 | |
| Infection status | 49 | |||||||||
| Uninfected | 16 (32.7) | Ref | Ref | Ref | Ref | |||||
| Infected | 33 (67.3) | −1.43 | 0.01 | −1.26 | 0.001 | 2.35 | 0.03 | −4.71 | 0.003 | |
Note: The fold changes were obtained from the 2−ΔΔCt method in multiplicative effect. Ref, Reference group.
LEDGF/p75 was transformed by the natural log in all models to better meet the residual normality criterion of linear regression.
The bold face highlights the significant p-values.
TABLE III.
Analyses for Infected Participants Only, Not Adjusted for Age
| HSP90 vs. β-Actin |
LEDGF/p75 vs. β-Actin |
P21 vs. β-Actin |
APOBEC3G vs. β-Actin |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | Total | No. (%) | Fold change |
P-value | Fold change |
P-value | Fold change |
P-value | Fold change |
P-value |
| Among infected | 33 | |||||||||
| Non-viremic | 15 (45.4) | Ref | Ref | Ref | Ref | |||||
| Viremic | 18 (54.6) | −1.48 | 0.002 | −1.15 | 0.05 | 1.03 | 0.94 | −1.71 | 0.34 | |
| Among treated | 21 | |||||||||
| Not on raltegravir | 17 (81.0) | Ref | Ref | Ref | Ref | |||||
| Raltegravir | 4 (19.0) | −1.39 | 0.19 | −1.28 | 0.04 | 1.12 | 0.85 | −3.54 | 0.16 | |
| Among non-raltegravir | 17 | |||||||||
| Not on protease inhibitors | 7 (41.2) | Ref | Ref | Ref | Ref | |||||
| On protease inhibitors | 10 (58.8) | −1.17 | 0.49 | −1.14 | 0.21 | −1.20 | 0.71 | −3.87 | 0.07 | |
| Among untreated | 9 | |||||||||
| Viral load 100–1,000 | 3 (33.3) | Ref | Ref | Ref | Ref | |||||
| Viral load >1,000 | 6 (66.7) | 1.00 | 1.00 | −1.16 | 0.36 | −2.07 | 0.36 | −4.39 | 0.28 | |
Note: The fold changes were obtained from the 2−ΔΔCt method and the P-values were obtained from Wilcoxon rank-sum test. Ref, Reference group.
HSP90 mRNA expression was 1.48-fold lower in infected/viremic patients than in infected/non-viremic, while expressions of LEDGF/p75, p21/WAF1, and APOBEC3G were similar in these two groups. Furthermore, patients on raltegravir exhibited a 1.28-fold reduction in LEDGF/p75 mRNA expression compared to those on non-raltegravir HAART (P = 0.04). HSP90, APOBEC3G, and p21/WAF1 expressions were unaffected by raltegravir in reference to non-raltegravir HAART. Also, among the treated patients who were not on a raltegravir regimen, no differences were observed in the expressions of all four genes in infected subjects receiving protease inhibitors compared to the patients receiving non-protease inhibitor treatment (Table III).
DISCUSSION
HIV-1 propagation depends on a delicate balance between cellular cofactors and restriction factors, and multiple host proteins have been demonstrated to facilitate or inhibit various stages of viral replication. Furthermore, HIV-1 disease progression [Wu et al., 2008; Vigneault et al., 2011] and response to HAART [Woelk et al., 2010] have been shown to be associated with certain host cell gene expression levels. Here, focus was placed on four host genes (HSP90, LEDGF/p75, p21/WAF1, and APOBEC3G) that have been described for their significant positive or negative effects specifically on proviral integration. Although all four of the genes studied here have been individually analyzed in infected patients before, there are some conflicting results [Jin et al., 2005; Cho et al., 2006; Ulenga et al., 2008; Reddy et al., 2010]. Further, this study represents the first time that these four genes have been scrutinized in the same, well-characterized participant cohort.
HSP90 is a molecular chaperone that is one of the most abundant proteins in the cell and assists in proper protein folding and stabilization [Csermely et al., 1998]. HSP90 has been well documented to stabilize cellular proteins involved in tumor growth [Kim et al., 2009] but has received relatively recent attention as being important for HIV-1 proviral integration [Vozzolo et al., 2010]. HSP90 protein production has previously been shown to be decreased in infected patients [Yohannes et al., 2011], and a minor but significant reduction in HSP90 mRNA levels was observed in this study, in infected individuals. LEDGF/p75 is a known coactivator of transcription [Ge et al., 1998; Fatma et al., 2001; Singh et al., 2001] and normally protects cells from thermal and oxidative stresses [Sharma et al., 2000; Singh et al., 2000]. LEDGF/p75 has been designated a cofactor of viral replication for its role in tethering IN to host cell chromatin [Vanegas et al., 2005; Ciuffi and Bushman, 2006; Hombrouck et al., 2007; Meehan et al., 2009] and targeting integration into transcriptionally active regions [Ciuffi et al., 2005, 2006 and Shun et al., 2007]. Although LEDGF/p75 expression was similar in viremic and non-viremic HIV patients, its expression mirrored that of HSP90 with respect to HIV infection status in general, with a 1.26-fold reduction compared to healthy subjects. This observation correlates directly with a similar cross-sectional study, which reported previously decreased LEDGF/p75 in infected individuals [Mous et al., 2012].
The upregulation of p21/WAF1 that was observed in this study in infected patients indicates that more than just stress-related host genes may play a role in this antiviral response. CD4+ T cells from elite controllers have consistently exhibited increased p21/WAF1 expression levels [Chen et al., 2011; Saez-Cirion et al., 2011], and induction of p53 and its target genes (including p21/WAF1) has been shown to occur in microglia, astrocytes, and oligodendrocytes as an adaptive response to HIV infection [Jayadev et al., 2007]. Furthermore, p21/WAF1 expression has been suggested to convey resistance to HIV-1 infection in hematopoietic stem cells through inactivation of integrase [Zhang et al., 2007]. Here, infected patients exhibited a 2.35-fold increase in p21/WAF1 mRNA expression relative to healthy individuals, though its expression was similar in viremic and non-viremic infected individuals. This data sheds light on the possibility that HIV-1 infection modulates host gene expression toward a potentially more extensive antiviral response than is currently understood, through decreasing production of host cofactors and increasing production of host restriction factors. It is, however, possible that the observed increase in p21/WAF1 expression is actually HIV-induced rather than host-induced, as p21/WAF1 is also an inhibitor of G1 and G2/M cell cycle phase transitions [Clark et al., 2003] and has been shown to be upregulated by HIV-1 Vpr as a means of stalling the cell cycle for maximizing viral transcription prior to host cell apoptosis [Goh et al., 1998]. This host gene has been clearly shown to exert a dual positive and negative effect on tumor growth [Stivala et al., 2012], and so further work will be required to clarify whether p21/WAF1 is exploited by the host cell as an antiviral defense or by HIV itself for counteracting this defense.
Alternatively, the mRNA expression of APOBEC3G, another potent restriction factor of HIV-1 replication, appears to be reduced by HIV infection in this patient cohort. APOBEC3G is a cytidine deaminase that decreases HIV-1 infectivity by inducing hypermutation of the viral genome [Sheehy et al., 2002; Wissing et al., 2010] and interfering with integration through IN-binding [Luo et al., 2007]. There has been some controversy in the literature as to whether APO-BEC3G is upregulated, downregulated, or unchanged in infected individuals compared to healthy controls. Two studies have reported that HIV-exposed seronegative individuals exhibit an increase in expression compared to (A) healthy controls [Vazquez-Perez et al., 2009] and (B) both healthy controls and infected individuals [Biasin et al., 2007]. Alternatively, there have been reports that APOBEC3G expression is unchanged in exposed seronegative individuals compared to healthy controls [Mous et al., 2011], and also that its expression is reduced in infected individuals compared to uninfected [Reddy et al., 2010; Mous et al., 2012]. The results of this study are in line with the latter studies, as a significant decrease in APOBEC3G expression was detected in infected individuals compared to uninfected. Furthermore, no difference was observed in expression levels between exposed seronegative subjects and healthy controls (data not shown). These findings suggest that HIV can circumvent this natural host restriction factor’s inhibitory action in its transcription during infection. The further study of possible molecular mechanisms involved in such inhibition may assist in developing novel strategies for new drug targets. Most HIV+ patients, and especially coinfected individuals, have skewed populations of T cell subsets characterized by elevated levels of T cell activation in their immune systems [Al-Harthi et al., 2006]. As memory CD4+ T cells have shown significantly different expression of certain genes than naïve CD4+ T cells, for example, expression of APOBEC3G and LEDGF/p75 [Mous et al., 2011], further work will be required to clarify how the homeostasis of CD4+ T cells subsets, other viral coinfections, and immune activation levels impact gene expression in CD4+ T cell during HIV infection.
In summary a well-characterized panel of infected and uninfected individuals was compiled in this study, and the expressions of four host genes were reported to positively or negatively affect the process of HIV-1 proviral integration were analyzed. The age-adjusted expression levels of HSP90, LEDGF/p75, and APOBEC3G were all downregulated, while p21/WAF1 mRNA levels were upregulated due to HIV infection. This work reveals that host genes exerting positive effects on replication may be manipulated by the host cell as an antiviral response, which is countered by manipulation of alternate genes by the virus itself. Elucidation of the complete mechanism underlying the battle between host and virus is relevant for future therapeutic efforts, and the genes scrutinized in this work could potentially serve as novel drug targets or biomarkers of disease progression.
ACKNOWLEDGMENTS
We thank Dr. Jiaao Xu, Chris Duymich, and Elizabeth Mcilvaine for excellent technical assistance.
Grant sponsor: NIH/NIAID; Grant numbers: R21; AI081610.; Grant sponsor: TL1 Training Grant from the Southern California Clinical Translational Science Institute (SC-CTSI) from the Neamati Lab and the Campbell Foundation; Grant sponsor: NIH R01 (Kovacs Lab); Grant number: AI052065-08.
REFERENCES
- Al-Harthi L, Voris J, Du W, Wright D, Nowicki M, Frederick T, Landay A, Kovacs A. 2006. Evaluating the impact of hepatitis C virus (HCV) on highly active antiretroviral therapy-mediated immune responses in HCV/HIV-coinfected women: Role of HCV on expression of primed/memory T cells. J Infect Dis 193:1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mawsawi LQ, Neamati N. 2007. Blocking interactions between HIV-1 integrase and cellular cofactors: An emerging antiretroviral strategy. Trends Pharmacol Sci 28:526–535. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. 2001. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284. [DOI] [PubMed] [Google Scholar]
- Biasin M, Piacentini L, Lo Caputo S, Kanari Y, Magri G, Trabattoni D, Naddeo V, Lopalco L, Clivio A, Cesana E, Fasano F, Bergamaschi C, Mazzotta F, Miyazawa M, Clerici M. 2007. Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G: A possible role in the resistance to HIV of HIV-exposed seronegative individuals. J Infect Dis 195:960–964. [DOI] [PubMed] [Google Scholar]
- Broder S 2010. The development of antiretroviral therapy and its impact on the HIV-1/AIDS pandemic. Antiviral Res 85: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky MI, Stanwick TL, Dempsey MP, Stevenson M. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadogan M, Dalgleish AG. 2008. HIV immunopathogenesis and strategies for intervention. Lancet Infect Dis 8:675–684. [DOI] [PubMed] [Google Scholar]
- Chege D, Chai Y, Huibner S, McKinnon L, Wachihi C, Kimani M, Jaoko W, Kimani J, Ball TB, Plummer FA, Kaul R, Rebbapragada A. 2010. Evaluation of a quantitative real-time PCR assay to measure HIV-specific mucosal CD8+ T cell responses in the cervix. PLoS ONE 5:e13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li C, Huang J, Cung T, Seiss K, Beamon J, Carrington MF, Porter LC, Burke PS, Yang Y, Ryan BJ, Liu R, Weiss RH, Pereyra F, Cress WD, Brass AL, Rosenberg ES, Walker BD, Yu XG, Lichterfeld M. 2011. CD4+ T cells from elite controllers resist HIV-1 infection by selective upregulation of p21. J Clin Invest 121:1549–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 278:372–381. [DOI] [PubMed] [Google Scholar]
- Cho SJ, Drechsler H, Burke RC, Arens MQ, Powderly W, Davidson NO. 2006. APOBEC3F and APOBEC3G mRNA levels do not correlate with human immunodeficiency virus type 1 plasma viremia or CD4+ T-cell count. J Virol 80:2069–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffi A, Bushman FD. 2006. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet 22:388–395. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 11:1287–1289. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Diamond TL, Hwang Y, Marshall HM, Bushman FD. 2006. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum Gene Ther 17:960–967. [DOI] [PubMed] [Google Scholar]
- Clark HF, Burke CJ, Volkin DB, Offit P, Ward RL, Bresee JS, Dennehy P, Gooch WM, Malacaman E, Matson D, Walter E, Watson B, Krah DL, Dallas MJ, Schodel F, Kaplan KM, Heaton P. 2003. Safety, immunogenicity and efficacy in healthy infants of G1 and G2 human reassortant rotavirus vaccine in a new stabilizer/buffer liquid formulation. Pediatr Infect Dis J 22:914–920. [DOI] [PubMed] [Google Scholar]
- Cozzi-Lepri A, Phillips AN, Ruiz L, Clotet B, Loveday C, Kjaer J, Mens H, Clumeck N, Viksna L, Antunes F, Machala L, Lundgren JD. 2007. Evolution of drug resistance in HIV-infected patients remaining on a virologically failing combination antiretroviral therapy regimen. AIDS 21:721–732. [DOI] [PubMed] [Google Scholar]
- Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. 1998. The 90-kDa molecular chaperone family: Structure, function, and clinical applications. A comprehensive review. Pharmacol Ther 79:129–168. [DOI] [PubMed] [Google Scholar]
- Fatma N, Singh DP, Shinohara T, Chylack LT Jr. 2001. Transcriptional regulation of the antioxidant protein 2 gene, a thiol-specific antioxidant, by lens epithelium-derived growth factor to protect cells from oxidative stress. J Biol Chem 276: 48899–48907. [DOI] [PubMed] [Google Scholar]
- Ge H, Si Y, Roeder RG. 1998. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J 17:6723–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WC, Rogel ME, Kinsey CM, Michael SF, Fultz PN, Nowak MA, Hahn BH, Emerman M. 1998. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: A mechanism for selection of Vpr in vivo. Nat Med 4:65–71. [DOI] [PubMed] [Google Scholar]
- Hombrouck A, De Rijck J, Hendrix J, Vandekerckhove L, Voet A, De Maeyer M, Witvrouw M, Engelborghs Y, Christ F, Gijsbers R, Debyser Z. 2007. Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog 3: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayadev S, Yun B, Nguyen H, Yokoo H, Morrison RS, Garden GA. 2007. The glial response to CNS HIV infection includes p53 activation and increased expression of p53 target genes. J Neuroimmune Pharmacol 2:359–370. [DOI] [PubMed] [Google Scholar]
- Jin X, Brooks A, Chen H, Bennett R, Reichman R, Smith H. 2005. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol 79:11513–11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Alarcon SV, Lee S, Lee MJ, Giaccone G, Neckers L, Trepel JB. 2009. Update on Hsp90 inhibitors in clinical trial. Curr Top Med Chem 9:1479–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol 81:7238–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. 2010. The immune response during acute HIV-1 infection: Clues for vaccine development. Nat Rev Immunol 10:11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan AM, Saenz DT, Morrison JH, Garcia-Rivera JA, Peretz M, Llano M, Poeschla EM. 2009. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog 5:e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mous K, Jennes W, Roo AD, Pintelon I, Kestens L, Ostade XV. 2011. Intracellular detection of differential APOBEC3G, TRI-M5alpha, and LEDGF/p75 protein expression in peripheral blood by flow cytometry. J Immunol Methods 372. [DOI] [PubMed] [Google Scholar]
- Mous K, Jennes W, Camara M, Seydi M, Daneau G, Mboup S, Kestens L, Van Ostade X. 2012. Expression analysis of LEDGF/p75, APOBEC3G, TRIM5alpha, and tetherin in a Senegalese cohort of HIV-1-exposed seronegative individuals. PLoS ONE 7: e33934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keeffe B, Fong Y, Chen D, Zhou S, Zhou Q. 2000. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated tat stimulation of HIV-1 transcription. J Biol Chem 275:279–287. [DOI] [PubMed] [Google Scholar]
- Reddy K, Winkler CA, Werner L, Mlisana K, Abdool Karim SS, Ndung’u T. 2010. APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load. AIDS 24:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch F, Meziane O, Kula A, Nisole S, Porrot F, Anderson I, Mammano F, Fassati A, Marcello A, Benkirane M, Schwartz O. 2012. Hyperthermia stimulates HIV-1 replication. PLoS Pathog 8:e1002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Cirion A, Hamimi C, Bergamaschi A, David A, Versmisse P, Melard A, Boufassa F, Barre-Sinoussi F, Lambotte O, Rouzioux C, Pancino G. 2011. Restriction of HIV-1 replication in macrophages and CD4+ T cells from HIV controllers. Blood 118:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrao E, Odde S, Ramkumar K, Neamati N. 2009. Raltegravir, elvitegravir, and metoogravir: The birth of “me-too” HIV-1 integrase inhibitors. Retrovirology 6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Singh DP, Fatma N, Chylack LT Jr, Shinohara T. 2000. Activation of LEDGF gene by thermal-and oxidative-stresses. Biochem Biophys Res Commun 276:1320–1324. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. [DOI] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 21: 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9:727–728. [DOI] [PubMed] [Google Scholar]
- Siliciano RF, Greene WC. 2011. HIV latency. Cold Spring Harb Perspect Med 1:a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DP, Fatma N, Kimura A, Chylack LT Jr, Shinohara T. 2001. LEDGF binds to heat shock and stress-related element to activate the expression of stress-related genes. Biochem Biophys Res Commun 283:943–955. [DOI] [PubMed] [Google Scholar]
- Singh DP, Ohguro N, Kikuchi T, Sueno T, Reddy VN, Yuge K, Chylack LT Jr, Shinohara T. 2000. Lens epithelium-derived growth factor: Effects on growth and survival of lens epithelial cells, keratinocytes, and fibroblasts. Biochem Biophys Res Commun 267:373–381. [DOI] [PubMed] [Google Scholar]
- Stivala LA, Cazzalini O, Prosperi E. 2012. The cyclin-dependent kinase inhibitor p21CDKN1A as a target of anti-cancer drugs. Curr Cancer Drug Targets 12:85–96. [DOI] [PubMed] [Google Scholar]
- Ulenga NK, Sarr AD, Thakore-Meloni S, Sankale JL, Eisen G, Kanki PJ. 2008. Relationship between human immunodeficiency type 1 infection and expression of human APOBEC3G and APOBEC3F. J Infect Dis 198:486–492. [DOI] [PubMed] [Google Scholar]
- Vanegas M, Llano M, Delgado S, Thompson D, Peretz M, Poeschla E. 2005. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J Cell Sci 118:1733–1743. [DOI] [PubMed] [Google Scholar]
- Vazquez-Perez JA, Ormsby CE, Hernandez-Juan R, Torres KJ, Reyes-Teran G. 2009. APOBEC3G mRNA expression in exposed seronegative and early stage HIV infected individuals decreases with removal of exposure and with disease progression. Retrovirology 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneault F, Woods M, Buzon MJ, Li C, Pereyra F, Crosby SD, Rychert J, Church G, Martinez-Picado J, Rosenberg ES, Telenti A, Yu XG, Lichterfeld M. 2011. Transcriptional profiling of CD4 T cells identifies distinct subgroups of HIV-1 elite controllers. J Virol 85:3015–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vozzolo L, Loh B, Gane PJ, Tribak M, Zhou L, Anderson I, Nyakatura E, Jenner RG, Selwood D, Fassati A. 2010. Gyrase B inhibitor impairs HIV-1 replication by targeting Hsp90 and the capsid protein. J Biol Chem 285:39314–39328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Galloway NL, Greene WC. 2010. HIV-1 Vif versus the APOBEC3 cytidine deaminases: An intracellular duel between pathogen and host restriction factors. Mol Aspects Med 31:383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk CH, Beliakova-Bethell N, Goicoechea M, Zhao Y, Du P, Rought SE, Lozach J, Perez-Santiago J, Richman DD, Smith DM, Little SJ. 2010. Gene expression before HAART initiation predicts HIV-infected individuals at risk of poor CD4+ T-cell recovery. AIDS 24:217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Dwyer DE, Dyer WB, Yang YH, Wang B, Saksena NK. 2008. Transcriptional profiles in CD8+ T cells from HIV progressors on HAART are characterized by coordinated upregulation of oxidative phosphorylation enzymes and interferon responses. Virology 380:124–135. [DOI] [PubMed] [Google Scholar]
- Yohannes E, Ghosh SK, Jiang B, McCormick TS, Weinberg A, Hill E, Faddoul F, Chance MR. 2011. Proteomic signatures of human oral epithelial cells in HIV-infected subjects. PLoS ONE 6: e27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. 1990. HIV-1 entry into quiescent primary lymphocytes: Molecular analysis reveals a labile, latent viral structure. Cell 61:213–222. [DOI] [PubMed] [Google Scholar]
- Zhang J, Scadden DT, Crumpacker CS. 2007. Primitive hematopoietic cells resist HIV-1 infection via p21. J Clin Invest 117:473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang H, Siliciano JD, Siliciano RF. 2005. Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J Virol 79:2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]