Abstract
Tomato (Solanum lycopersicum L.) belongs to the Solanaceae family and is the second most important fruit or vegetable crop next to potato (Solanum tuberosum L.). It is cultivated for fresh fruit and processed products. Tomatoes contain many health-promoting compounds including vitamins, carotenoids, and phenolic compounds. In addition to its economic and nutritional importance, tomatoes have become the model for the study of fleshy fruit development. Tomato is a climacteric fruit and dramatic metabolic changes occur during its fruit development. In this review, we provide an overview of our current understanding of tomato fruit metabolism. We begin by detailing the genetic and hormonal control of fruit development and ripening, after which we document the primary metabolism of tomato fruits, with a special focus on sugar, organic acid, and amino acid metabolism. Links between primary and secondary metabolic pathways are further highlighted by the importance of pigments, flavonoids, and volatiles for tomato fruit quality. Finally, as tomato plants are sensitive to several abiotic stresses, we briefly summarize the effects of adverse environmental conditions on tomato fruit metabolism and quality.
Keywords: abiotic stress, fruit set, fruit ripening, genetic control, hormonal control, primary metabolism, secondary metabolism, Solanum lycopersicum
Introduction
Tomato (Solanum lycopersicum L.) is the second most important fruit or vegetable crop next to potato (Solanum tuberosum L.), with approximately 182.3 million tons of tomato fruits produced on 4.85 million ha each year (FAOSTAT, 2019). Asia accounts for 61.1% of global tomato production, while Europe, America, and Africa produced 13.5%, 13.4%, and 11.8% of the total tomato yield, respectively. Tomato yields are highly variable, ranging from more than 508 tons per ha in the Netherlands to fewer than 1.5 tons per ha in Somalia in 2017 (FAOSTAT, 2019), with an average global yield of 376 tons per ha. Tomato consumption is concentrated in China, India, North Africa, the Middle East, the US, and Brazil with tomato consumption per capita, ranging from 61.9 to 198.9 kg per capita (FAOSTAT, 2019). Tomato is a member of the Solanaceae family, which includes several other economically important crops such as potato, pepper (Capsicum annuum L.), and eggplant (Solanum melongena L.), representing one of the most valuable plant families for vegetable and fruit crops.
Tomatoes contain many health-promoting compounds and are easily integrated as a nutritious part of a balanced diet (Martí et al., 2016). In addition to consuming the fresh fruits, consumers use tomatoes in processed products such as soups, juices, and sauces (Krauss et al., 2006; Li et al., 2018b). Over the last decade, consumers have become more aware of foods as a source of health benefits and their roles in prevention of several chronic diseases and dysfunctions (Pem and Jeewon, 2015). Although a wealth of functional foodstuffs have been created to fulfil these requirements, it is important to note that the consumption of “conventional foods” such as fruits and vegetables is more effective for this purpose (Viuda-Martos et al., 2014).
The nutritional importance of tomatoes is largely explained by their various health-promoting compounds, including vitamins, carotenoids, and phenolic compounds (Raiola et al., 2014; Liu et al., 2016; Martí et al., 2016; Li et al., 2018b). These bioactive compounds have a wide range of physiological properties, including anti-inflammatory, anti-allergenic, antimicrobial, vasodilatory, antithrombotic, cardio-protective, and antioxidant effects (Raiola et al., 2014). Tomatoes are rich in carotenoids, representing the main source of lycopene in the human diet (Viuda-Martos et al., 2014). Carotenoids and polyphenolic compounds contribute to the nutritional value of tomatoes and improve their functional attributes and sensory qualities, including taste, aroma, and texture (Raiola et al., 2014; Tohge and Fernie, 2015; Martí et al., 2016). Tomatoes also have the naturally occurring antioxidants Vitamins C and E (Agarwal and Rao, 2000; Martí et al., 2016) as well as large amounts of metabolites, such as sucrose, hexoses, citrate, malate, and ascorbic acid (Li et al., 2018b).
Tomato fruit quality and metabolite biosynthesis are affected by plant growing conditions (Diouf et al., 2018). Tomato production is challenged by several problems around the world, including the scarcity of water resources, soil salinization, and other abiotic stresses (Fahad et al., 2017; Gharbi et al., 2017; Zhou et al., 2019). In particular, in countries with a Mediterranean climate, including some regions in southern Europe and North and South America, tomato cultivation is increasingly confronted with limiting conditions such as drought and salinity, which ultimately reduce the competitiveness of tomato farmers in these areas. This, in turn, impacts the integrity of the ecosystem, contributing to the relocation (abandonment) of rural sectors.
In addition to its economic and nutritional importance, tomatoes have become the model for the study of fleshy fruit development (Karlova et al., 2014; Kim et al., 2018; Li et al., 2018b). The entire tomato genome has been sequenced, serving as a rich genomic resource, and both genetic and physical maps and molecular markers are available for this species (The Tomato Genome Consortium, 2012; Suresh et al., 2014; Zhao et al., 2019). Moreover, a range of well-characterized monogenic mutants, TILLING populations, wild tomato species, recombinant inbred lines and genome editing tools are available (Eshed and Zamir, 1994; Minoia et al., 2010; Pérez-Martín et al., 2017; Li et al., 2018b; Martín-Pizarro and Posé, 2018; Tomato Genetics Resource Center, 2019; Rothan et al., 2019). Several databases contain gene expression analysis data (Fei et al., 2006; Suresh et al., 2014; Zouine et al., 2017; Shinozaki et al., 2018b), while recent progress in tomato metabolomics has provided substantial information about the primary and specialized metabolism of this species and the pathways involved in molecular biosynthesis and turnover (Luo, 2015; Tieman et al., 2017; Zhu et al., 2018).
Dramatic metabolic changes occur during tomato fruit development (Carrari and Fernie, 2006). Tomato is a climacteric fruit, meaning it undergoes a surge in respiration and ethylene production at the onset of ripening (Li et al., 2019a). As ripening progresses, tomato fruits transit from partially photosynthetic to true heterotrophic tissues through the parallel differentiation of chloroplasts into chromoplasts and the dominance of carotenoids and lycopene in the cells of the ripe fruits (Carrari and Fernie, 2006). The ripening process has evolved to make fruit palatable to the organisms that consume them and disperse their seeds. In doing so, ripening activates pathways that generally influence the levels of pigments, sugars, acids, and aroma-associated volatiles to make the fruit more appealing, while simultaneously promoting tissue softening and degradation to permit easier seed release (Matas et al., 2009).
In this review, we provide an overview of our current understanding of tomato fruit metabolism. We begin by detailing the genetic and hormonal control of fruit development and ripening, after which we document the primary metabolism of tomato fruits, with a special focus on sugar, organic acid, and amino acid metabolism. Links between primary and secondary metabolic pathways are further highlighted by the importance of pigments, flavonoids, and volatiles for tomato fruit quality. Finally, as tomato plants are sensitive to several abiotic stresses, we briefly summarize the effects of adverse environmental conditions on tomato fruit metabolism and quality.
Genetic Regulation of the Development and Ripening of Tomato Fruit
Fruit Set and Early Fruit Development
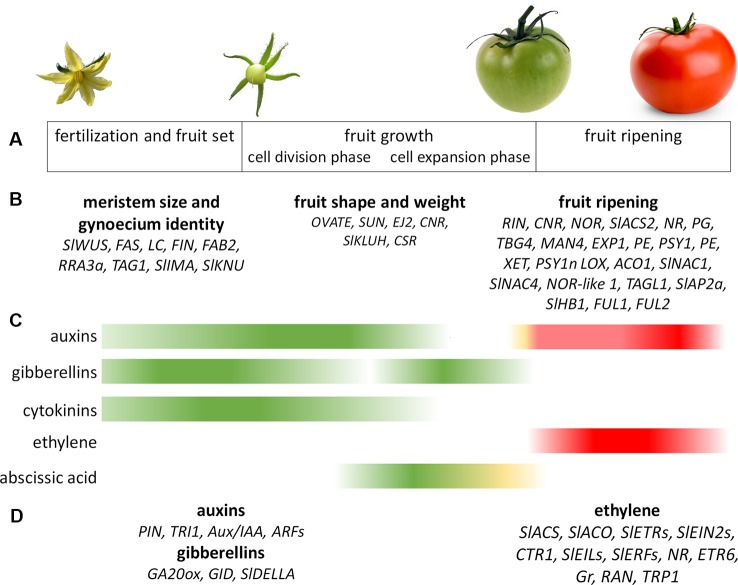
The genetic regulation of fruit development begins in the floral meristem (FM), where the architecture and organization of this tissue is determined, and continues until the later developmental stages before fruit ripening (Gillaspy et al., 1993) ( Figures 1A, B ). At the initial stage of tomato fruit development, the CLAVATA-WUSCHEL (CLV-WUS) feedback loop controls meristem activity and regulates FM size, which in turn determines the final number of carpels in flowers and, hence, seed locules in fruits (Rodríguez-Leal et al., 2017). The signaling peptide CLV3 directly interacts with leucine-rich repeat receptor kinases, such as CLV1 or CLV2, to activate a signaling cascade that negatively regulates the stem cell-promoting transcription factor WUS (Somssich et al., 2016). Loss-of-function mutations in any of the CLV genes will therefore cause stem cell over proliferation, resulting in the development of extra floral organs and larger fruits (Xu et al., 2015; Rodríguez-Leal et al., 2017); for example, the joint action of the natural mutations fasciated (fas) and locule number (lc) gave rise to large-fruited cultivars, in contrast to the bilocular fruits of tomato wild species and most small-fruited varieties (Tanksley, 2004; Barrero et al., 2006). The fas mutation is a 294-kb inversion disrupting the tomato CLV3 (SlCLV3) promoter (Xu et al., 2015), whereas lc is associated with two single-nucleotide polymorphisms in a putative CArG box regulatory element downstream of WUS (SlWUS) (Muños et al., 2011; van der Knaap et al., 2014). Furthermore, using forward genetics and CRISPR/Cas9 genome editing technology, Xu et al. (2015) identified the arabinosyltransferase genes FASCIATED INFLORESCENCE (FIN), FASCIATED AND BRANCHED2 (FAB2), and REDUCED RESIDUAL ARABINOSE 3a (RRA3a) as new components of the CLV-WUS pathway. The SlCLV3 peptide must therefore be fully arabinosylated to maintain meristem size since the loss of an arabinosyltransferase cascade causes floral and fruit fasciation.
Figure 1.
Genetic and hormonal control of tomato fruit development. (A) Main stages of tomato fruit development. (B) Genes involved in the control of tomato fruit development that are mentioned in this article. (C) Main hormones involved in tomato fruit development during fruit set and fruit growth (green) and fruit ripening (red). (D) Genes involved in the hormonal regulation of fruit development that are mentioned in this article. The Figure summarizes data collected by Gillaspy et al. (1993); Srivastava and Handa (2005); Karlova et al. (2014) and Obroucheva (2014).
As the flower develops, the gynoecium is initiated in the fourth whorl to terminate FM activity. The MADS box transcription factor AGAMOUS (AG) is required to form the carpel primordium (Yanofsky et al., 1990). Consequently, the downregulation of TOMATO AGAMOUS1 (TAG1), the tomato ortholog of Arabidopsis thaliana AG, gives rise to alterations in carpel development and determinacy by producing fruits that continue to develop in an indeterminate fashion (Pnueli et al., 1994; Pan et al., 2010; Gimenez et al., 2016). Furthermore, in Arabidopsis, AG turns off the stem cell maintenance program through the transcriptional repression of WUS via two different pathways: directly, by promoting the recruitment of Polycomb Group (PcG) proteins to methylate histone H3K27 at the WUS locus (Liu et al., 2011); and indirectly, by inducing the expression of a gene encoding the C2H2 zinc-finger protein KNUCKLES (KNU) (Sun et al., 2009). The induction of KNU expression by AG requires a time delay regulated by the epigenetic modification of histones at the KNU locus (Sun et al., 2014). Recently, Bollier et al. (2018) demonstrated that the AG-KNU-WUS pathway is conserved in Arabidopsis and tomato and regulates the timed termination of floral stem cell activity. In this context, the tomato mini zinc-finger protein INHIBITOR OF MERISTEM ACTIVITY (SlIMA) recruits SlKNU to form a transcriptional repressor complex together with TOPLESS and HISTONE DEACETYLASE19, which binds to the SlWUS locus to repress its transcription (Bollier et al., 2018). Additionally, it has been hypothesized that lc is a weak gain-of-function mutation that reduces or blocks the binding of TAG1 to the SlWUS 3′ regulatory region, which impairs the ability of TAG1 to repress SlWUS, resulting in the formation of larger fruits as a consequence of the development of extra carpels (van der Knaap et al., 2014).
The variation in tomato fruit morphology not only depends on CLV-WUS signaling pathway-related genes, but also on OVATE and SUN, which have a large effect on fruit shape ( Figure 1B ). The ovate null mutation gives rise to changes in cell division patterns during the earliest stages of gynoecium development, with more cells produced in the proximo-distal direction and fewer in the medio-lateral direction, causing the development of elongated fruits (Ku et al., 1999; Liu et al., 2002; Rodríguez et al., 2011). In contrast, the effect of SUN on fruit shape is most noticeable at flower anthesis, when it begins to increase cell division along the proximo-distal axis and cell elongation immediately after fertilization (Xiao et al., 2009; Wu et al., 2011; van der Knaap et al., 2014). Thus, a profound shift in the expression of genes involved in cell division, cell wall development, and patterning processes was observed in the elongating fruit tissues of the sun mutant (Clevenger et al., 2015). Moreover, the MADS box gene ENHANCER OF J2 (EJ2) also seems to be involved in determining fruit shape; ej2 knockout mutants develop slightly elongated fruits together with several pleiotropic effects, such as branched inflorescences and jointless pedicels (Soyk et al., 2017).
Among the fruit weight regulators, CELL NUMBER REGULATOR (CNR) was found to underlie the fw2.2 quantitative trait locus (QTL), acting early during the development of the gynoecium to increase ovary size (Frary et al., 2000; Guo and Simmons, 2011) and enlarge the placenta and columella fruit tissues (Cong et al., 2002; Gonzalo et al., 2009). SlKLUH is the causal gene for the fw3.2 QTL and encodes a CYP450 of the 78A class (Chakrabarti et al., 2013). One single-nucleotide polymorphism in the SlKLUH promoter leads to its enhanced expression in meristems and young flower bud tissues; however, the increased fruit weight of these mutant plants becomes evident only after fertilization. An increased number of cell layers in the pericarp gives rise to heavier fruits with a ripening delay, which has been hypothesized to be the result of the extension of the cell proliferation stage (Chakrabarti et al., 2013). Studies in Arabidopsis have suggested that KLUH is involved in generating a mobile growth-promoting signal, although its exact molecular and biochemical nature is yet to be deciphered (Anastasiou et al., 2007; Adamski et al., 2009). Cell expansion in the pericarp is responsible for the dramatic increase in fruit size from a 1- to 2-mm gynoecium to a 5- to 10-cm tomato fruit (Gillaspy et al., 1993; Xiao et al., 2009). The CELL SIZE REGULATOR (CSR) gene controls pericarp cell size and underlies the fw11.3 QTL (Huang and van der Knaap, 2011; Mu et al., 2017). CSR expression is restricted to fruits, starting about 5 days after pollination and decreasing at the onset of ripening. Along with the increased cell size, coexpression studies suggest that CSR is also involved in shoot development and phloem/xylem histogenesis; however, the molecular function of CSR in controlling these developmental processes remains unclear (Mu et al., 2017).
Fruit Ripening
At the end of fruit development, when seeds are mature and ready for dispersal, tomato fruits undergo ripening, a complex developmental program involving the coordinated regulation of numerous physiological and biochemical changes that determine flavor, color, texture, and aroma. These changes involve the up- or downregulation of numerous genes in various metabolic pathways (Alba et al., 2005; Fujisawa et al., 2011; Osorio et al., 2011). Multiple studies of the development and maturation of tomato fruits have facilitated the identification of specific genes that participate in ripening (Vrebalov et al., 2002; Manning et al., 2006; Giovannoni, 2007; Wang et al., 2009; Chung et al., 2010; Nashilevitz et al., 2010; Karlova et al., 2011; Pesaresi et al., 2014) ( Figure 1B ).
Tomatoes are classified as climacteric fruits, exhibiting a peak of respiration and ethylene production at the start of ripening (Alexander and Grierson, 2002). The biosynthesis and perception of ethylene are highly regulated, involving genes conserved in various plant taxa (Seymour et al., 2013). Some transcription factors modulate ethylene biosynthesis and signal transduction during fruit ripening, among which it is worth highlighting RIPENING INHIBITOR (RIN) (Vrebalov et al., 2002), COLORLESS NON-RIPENING (CNR) (Manning et al., 2006), and NON-RIPENING (NOR) (Yuan et al., 2016). RIN acts as the main regulator of fruit ripening, directly controlling the expression of target genes involved in a wide range of ripening-related events (Fujisawa et al., 2011; Qin et al., 2012). RIN encodes a SEPALLATA (SEP)-class MADS-box transcription factor (Vrebalov et al., 2002), which was previously considered to be an essential regulator of the induction of ripening (Vrebalov et al., 2002); however, its role in fruit ripening was recently reassessed following the publication of studies showing that RIN, although necessary to complete ripening, is not required for the initiation of this process (Ito et al., 2017). The rin mutant was found to be caused by the deletion of a genomic DNA fragment between RIN and MACROCALYX (MC), forming the chimeric gene RIN-MC (Vrebalov et al., 2002). MC affects inflorescence determinacy and sepal development (Vrebalov et al., 2002), and the rin mutant was found to be a gain-of-function mutant that produced a protein that actively represses ripening (Ito et al., 2008; Li et al., 2018a). RIN binds to the demethylated promoter regions of several genes, such as the ethylene biosynthesis genes SlACS2 (1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE 2), SlACS4, SlACO1 (ACC OXIDASE 1), the ethylene receptor NEVER RIPE (NR), and others whose products are involved in fruit softening and the transcriptional regulation of cell wall hydrolases [POLYGALACTURONASE (PG), β-GALACTOSIDASE4 (TBG4), ENDO-(1,4)-β- MANNANASE4 (MAN4), and α-EXPANSIN1 (EXP1)] (Klee and Tieman, 2002; Ito et al., 2008; Fujisawa et al., 2011; Martel et al., 2011; Shima et al., 2013; Ito et al., 2017).
RIN also positively stimulates the expression of CNR (Cardon et al., 1999; Manning et al., 2006). The cnr mutation is the result of a spontaneous epigenetic change that increases cytosine methylation in the promoter of a SQUAMOSA promoter-binding protein-encoding gene, which strongly decreases gene expression and produces colorless fruits with an altered pericarp texture (Manning et al., 2006). During ripening, the CNR promoter is progressively demethylated, but in cnr mutants, the promoter remains hypermethylated, preventing RIN from binding to it (Zhong et al., 2013). In addition, CNR was involved in the positive regulation of many ripening-related genes, including PG, PECTINESTERASE (PE), XYLOGLUCAN ENDOTRANSGLYCOSYLASE (XET), PHYTOENE SYNTHASE1 (PSY1), LIPOXYGENASE (LOX), and ACO1 (Eriksson et al., 2004).
The nor mutant exhibits abnormal ripening as a result of a 2-bp deletion in the NOR coding sequence, leading to the early termination of protein translation (Tigchelaar et al., 1973; Martel et al., 2011; Osorio et al., 2011). NOR encodes a NAC family transcription factor that regulates fruit ripening through a currently unclear mechanism, while mutations in this gene inhibit multiple metabolic processes and prolong fruit shelf life (Kumar et al., 2018). A study of the role of NOR and RIN in tomato fruit ripening confirmed that the nor mutation had a more global effect on ethylene/ripening-related gene expression than rin, suggesting that NOR might even act upstream of RIN in the transcriptional network controlling tomato fruit ripening (Osorio et al., 2011). In addition to NOR, three other NAC family genes, SlNAC1, SlNAC4, and NOR-like1, are known to be involved in the regulation of tomato fruit ripening (Ma et al., 2014; Zhu et al., 2014; Meng et al., 2016).
Other ripening factors, such as the MADS box TOMATO AGAMOUS-LIKE1 (TAGL1) (Vrebalov et al., 2002; Giménez et al., 2010), tomato APETALA2 (SlAP2a) (Karlova et al., 2011), and the tomato homeodomain leucine zipper homeobox protein SlHB1 (Lin et al., 2008), exercise their regulatory functions by interacting with RIN (Fujisawa et al., 2011; Qin et al., 2012; Seymour et al., 2013). TAGL1 (also referred to as ARLEQUIN in some publications), a PLENA lineage gene orthologous to Arabidopsis SHATTERPROOF1/2, controls many aspects of tomato fruit ripening (Vrebalov et al., 2009; Garceau et al., 2017), including the direct activation of the expression of the ethylene biosynthesis gene ACS2 (Itkin et al., 2009). Tomato fruits produced by TAGL1-silenced plants had defects in ripening without their floral organ specification being affected (Vrebalov et al., 2009; Giménez et al., 2010; Pan et al., 2010). Plants with reduced TAGL1 expression produced fruits with a narrow pericarp and reduced firmness at the breaker stage, which remained yellow and produced significantly less ethylene than the control fruits (Vrebalov et al., 2009). The MADS box proteins TAGL1 and two homologs of FRUITFULL (FUL1/TDR4 and FUL2/MBP7) function as coregulators of RIN (Leseberg et al., 2008; Itkin et al., 2009; Vrebalov et al., 2009; Giménez et al., 2010; Martel et al., 2011; Bemer et al., 2012; Shima et al., 2013; Wang et al., 2014). Fujisawa et al. (2014) demonstrated that RIN, TAGL1, and the FUL homologs form a DNA-binding complex, probably a tetramer, which is believed to regulate tomato fruit ripening. The RIN and CNR regulators have been shown to function upstream of SlAP2a and to positively regulate its expression (Karlova et al., 2014), whereas SlHB1 controls ethylene metabolism by binding to the regulatory regions of ACO1 (Lin et al., 2008). On the other hand, transcriptomic studies have shown that SlAP2a participates in the control of fruit ripening as a negative regulator of several processes involved in ethylene biosynthesis, and signaling pathways, as well as in the differentiation of chromoplasts (Chung et al., 2010; Karlova et al., 2011).
Hormonal Regulation of the Development and Ripening of Tomato Fruit
Fruit Set and Early Fruit Development
Fruit set and fruit development are complex processes that require the coordination of different phytohormones (McAtee et al., 2013; Shinozaki et al., 2018b; Li et al., 2019b) ( Figures 1C, D ). From flower initiation to fertilization, the morphogenesis and growth of carpels and ovules require the spatial and temporal biosynthesis and action of auxins, cytokinins (CKs), and gibberellins (GAs) (Azzi et al., 2015). Shortly before anthesis, when the ovary has reached its mature size, abscisic acid (ABA) and ethylene work to stop growth within the ovary to maintain a temporally protected and dormant state (Gillaspy et al., 1993; Azzi et al., 2015). After the successful pollination and fertilization of the ovules, ovary growth resumes and the fruit and seeds develop concomitantly (Azzi et al., 2015). These changes are associated with a decrease in ABA and ethylene concentrations and an increase in auxin, GAs, and CKs (de Jong et al., 2009; McAtee et al., 2013; Shinozaki et al., 2015; Shinozaki et al., 2018a). GAs produced by pollen may increase auxin production in the ovary, which in turn may act as a signal for fruit set and the subsequent activation of cell division (Gillaspy et al., 1993; de Jong et al., 2009). Active fruit growth involving pericarp cell division and elongation is promoted by the biosynthesis of auxin in the developing seeds and GAs in the pericarp (Obroucheva, 2014). Auxins and GAs appear to be the predominant hormones required for tomato fruit initiation in response to fertilization, since the exogenous application of both hormones leads to fruit initiation and parthenocarpic development (de Jong et al., 2009). CKs, ethylene, ABA, brassinosteroids, and polyamines (PAs) have also been shown to play a role in fruit formation, but this is currently less well documented (Srivastava and Handa, 2005; McAtee et al., 2013; Azzi et al., 2015; Shinozaki et al., 2015; Liu et al., 2018; Shinozaki et al., 2018a).
In tomato, early fruit development is governed by the allocation of auxin to tissues and cells, which initiates signal transduction pathways (Azzi et al., 2015). The PIN-FORMED (PIN) auxin efflux transport proteins were shown to be involved in fruit set and early tomato fruit development (Mounet et al., 2012; Pattison and Catalá, 2012). Silencing SlPIN4 resulted in the production of small parthenocarpic fruits exhibiting precocious development (Mounet et al., 2012). The auxin signaling pathway involves an auxin receptor called TRANSPORT INHIBITOR RESPONSE1 (TIR1) (Azzi et al., 2015). In the presence of auxin, TIR1 recruits the transcriptional repressors AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) and triggers their degradation by the 26S proteasome (Azzi et al., 2015), releasing the Aux/IAA-bound auxin response factors (ARFs) and initiating the auxin response through auxin-responsive element-mediated gene transcription (Azzi et al., 2015). In tomato, the misexpression of TIR1 and specific members of the Aux/IAA and ARF gene family alters the normal flower-to-fruit transition and results in parthenocarpic fruit production (de Jong et al., 2009; Ren et al., 2011; Mounet et al., 2012; Azzi et al., 2015). However, Aux/IAA and ARF genes may have opposing functions to TIR regarding fruit set; the transcript abundance of SlIAA9 and SlARF7 decreased in SlTIR1-overexpressing plants, which resulted in the formation of seedless fruit (Ren and Wang, 2016; Goldental-Cohen et al., 2017). The silencing of the Aux/IAA transcriptional repressor SlIAA17 resulted in larger fruits with thicker pericarp tissues, a phenotype caused by enhanced cell expansion (Su et al., 2014). Ren and Wang (2016) showed that SlTIR was regulated by GAs, auxins, ABA, and ethylene, suggesting that TIR may be a key mediator of the crosstalk between auxin and other phytohormones. The SlARF7/SllAA9 complex also mediates crosstalk between auxin and GA pathways to regulate fruit initiation through their interaction with the GA-signaling repressor SlDELLA (Hu et al., 2018). SlARF7/SllAA9 complex and SlDELLA antagonistically regulate genes involved in auxin and GA metabolism while they additively coregulate genes involved in fruit growth (Hu et al., 2018).
Indeed, auxins do not act alone to trigger fruit development and fruit set; these processes are partly mediated by GAs, as part of a complex hormonal cross-talk with auxin (de Jong et al., 2009; McAtee et al., 2013; Azzi et al., 2015). Pollination triggers the upregulation of transcripts encoding GA 20-oxidases (GA20ox), which biosynthesize active GA1 and GA4 (Azzi et al., 2015). It was suggested that the expression of more than one GA20ox gene is required to control fruit set in tomato because the silencing of individual GA20ox genes did not strongly affect fruit set or development (Xiao et al., 2006; Olimpieri et al., 2011; Azzi et al., 2015). Despite this, the heterologous overexpression of citrus CgGA20ox1 in tomato resulted in an elevated GA4 content and parthenocarpic fruit development, demonstrating the influence of GA and GA20ox activity on fruit set and development (García-Hurtado et al., 2012). The GA signal transduction pathway requires the recognition of GA by its receptor, GA INSENSITIVE DWARF1 (GID1) (Azzi et al., 2015). The GID1-GA complex interacts with the nuclear repressor DELLA to target it for ubiquitin-dependent proteolytic degradation by the 26S proteasome (Azzi et al., 2015). This removes the repression of the GA-responsive genes, which are then able to initiate GA signal transduction. Consistent with this, the silencing of the SlDELLA gene in tomato resulted in small, facultative parthenocarpic fruits with an elongated shape (Martí et al., 2007). The procera (pro) mutant, which carries a point mutation in the GRAS region of SlDELLA, has also very strong parthenocarpic capacity and shows enhanced growth of preanthesis ovaries (Jones, 1987; Carrera et al., 2012; Shinozaki et al., 2018c). The parthenocarpic capacity of pro is mainly associated with changes in the expression of genes involved in GA and auxin pathways (Carrera et al., 2012). A new SlDELLA mutant containing a single nucleotide substitution, procera2 (pro2), has been recently identified and shows a potential for high fruit yield in both optimal and unfavorable growing conditions due to its facultative parthenocarpic capacity (Shinozaki et al., 2018c). Parthenocarpy is indeed an attractive trait for fruit production (Shinozaki et al., 2018c).
As mentioned previously, other phytohormones are involved in fruit set and growth. A number of ABA-deficient mutants have provided valuable insights into the role of ABA in fruit growth (Azzi et al., 2015). Phenotypic characterization of the ABA biosynthesis not/flc double mutant showed that its small fruits had considerably reduced ABA levels and smaller cell sizes, especially within the pericarp (Nitsch et al., 2012). It was suggested that ABA stimulates fruit growth by restricting the level of ethylene in normal fruits (Azzi et al., 2015), which may indeed induce fruit set as tomato plants treated with the ethylene action inhibitor 1-methylcyclopropene (1-MCP) produce parthenocarpic fruits (Shinozaki et al., 2015). In the same way, tomato plants carrying either of two allelic mutations in ETHYLENE RECEPTOR1 (Sletr1-1 or Sletr1-2) were insensitive to ethylene, resulting in parthenocarpy (Shinozaki et al., 2015; Shinozaki et al., 2018a). Ethylene is involved in the senescence of unpollinated ovaries and prevents fruit set by downregulating GA accumulation, acting downstream of auxin and upstream of GA in the control of fruit set (Shinozaki et al., 2018a). Exogenous CK application induces parthenocarpic fruits (Matsuo et al., 2012; Ding et al., 2013), suggesting a role for CKs during tomato fruit initiation. Cytokinins induce parthenocarpy in tomato partially through modulation of GA and auxin metabolisms (Ding et al., 2013). Moreover, transcriptomic and metabolomic studies showed that although CKs mainly control cell division during tomato fruit development, they also play a critical role in fruit-set and early growth of tomato fruits (Mariotti et al., 2011; Matsuo et al., 2012). A key role for PAs during fruit set was also suggested, with tomato genes encoding enzymes involved in PA biosynthesis, such as arginine/ornithine decarboxylase (ADC/ODC) and spermine synthase (SPMS), suggested to be particularly important during the process of fruit setting (Liu et al., 2018).
Fruit Ripening
Fruit ripening has been widely studied in tomato, with ethylene known to play a key role in this process (Osorio et al., 2013; Seymour et al., 2013; Liu et al., 2015; Borghesi et al., 2016; Shinozaki et al., 2018b; Li et al., 2019a) ( Figures 1C, D ). Two systems of ethylene biosynthesis have been proposed in climacteric fruits (McMurchie et al., 1972): System 1 is responsible for producing basal ethylene levels during fruit growth and is ethylene autoinhibitory, while system 2 operates during climacteric ripening and is autocatalytic (Liu et al., 2015). At the onset of ripening, an increase in ethylene is observed in mature green tomatoes, resulting in an eventual 100- to 300-fold increase in the ethylene concentration during fruit ripening (Karlova et al., 2014; Li et al., 2019a). Ethylene initiates a cascade of changes, which culminate in the transformation of the hard, unpalatable green tomato into an attractive, brightly colored succulent and nutritious fruit (Giovannoni, 2004; Li et al., 2019a).
Ethylene signaling can be regulated at several levels, including ethylene biosynthesis and its perception (Karlova et al., 2014; Mata et al., 2018; Li et al., 2019a). Ethylene biosynthesis involves multiple aminocyclopropane-1-carboxylic acid (ACC) synthase and ACC oxidase enzymes and genes (Osorio et al., 2013; Karlova et al., 2014; Kou et al., 2016; Li et al., 2019a). Fourteen putative ACS genes and six ACO genes have been identified in the tomato genome (Liu et al., 2015). Among them, it has been proposed that SlACS2, SlACS4, SlACO1, SlACO2, and SlACO4 play important roles in ethylene production during tomato fruit maturation (Cara and Giovannoni, 2008; Liu et al., 2015). Some transcription factors are known to act upstream of the ethylene biosynthesis genes to regulate fruit ripening, including RIN, SlHB-1, and the NAC transcription factors SNAC4 and SNAC9 (Liu et al., 2015; Kou et al., 2016).
Ethylene perception is mediated through ethylene receptors encoded by ETHYLENE RESPONSE (ETR) genes, which activate a signal transduction cascade through the release of the block on ETHYLENE INSENSITIVE2 (EIN2) exerted by CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) (Karlova et al., 2014; Liu et al., 2015; Mata et al., 2018; Li et al., 2019a). Seven ETR genes and four CTR1 homologs have been identified in tomato thus far, all of which control ethylene sensitivity by balancing the turnover of the components of the ethylene signaling pathway, combining positive and negative feedback (Liu et al., 2015; Mata et al., 2018). This release then activates the EIN3/EIN3-like (EIL) primary transcription factor genes, resulting in the expression of secondary transcription factor genes encoding the ethylene response factors (ERFs) (Karlova et al., 2014; Liu et al., 2015; Mata et al., 2018). The final result of this signaling pathway is the transcriptional regulation of the target genes by the EILs or ERFs (Karlova et al., 2014). Some of the ERF genes have been characterized in tomato, including SlERF1, SlERF.B3, and SlERF6 (Li et al., 2007; Liu et al., 2013; Karlova et al., 2014), but many of their functions and ethylene-responsive target genes remain unknown (Li et al., 2019a). Six EIL genes have been identified in tomato, although SlEIL5 and SlEIL6 may not be involved in tomato ripening (Liu et al., 2015). Several genes that regulate tomato ripening through the transduction of ethylene signals have been identified (Karlova et al., 2014), including the ethylene receptor genes NR, ETR6, and GREEN-RIPE (Gr) (Yen et al., 1995; Barry and Giovannoni, 2006; Kevany et al., 2007). Two other proteins, RESPONSE TO ANTAGONIST1 (RAN1) and TETRATRICOPEPTIDE REPEAT1 (TRP1), also play important roles at the receptor levels (Liu et al., 2015).
Ripening is also influenced by the balance of other hormones, including ABA, auxin, and the brassinosteroids (Seymour et al., 2013; Karlova et al., 2014; Liu et al., 2015; Shinozaki et al., 2018b; Li et al., 2019a; Shin et al., 2019). ABA is known to promote ripening, whereas auxin seems to have an antagonistic effect (Liu et al., 2015). ABA is a key intermediate regulator of tomato fruit ripening, and its levels change according to fruit development stages (Zhang et al., 2009; Borghesi et al., 2016). In tomato, the suppression of the gene that catalyzes the first step in ABA biosynthesis [9-cis-epoxy carotenoid dioxygenase (NCED1)] results in the downregulation of some ripening-related cell wall genes, such as those encoding polygalacturonase and pectin methylesterase, promoting an increase in firmness and a longer shelf life (Sun et al., 2012). ABA interacts with ethylene signaling; the expression of genes involved in ethylene biosynthesis are induced by exogenous ABA (Liu et al., 2015).
Low levels of auxins are also required at the onset of ripening, and auxin signaling declines at this stage (Gillaspy et al., 1993; Karlova et al., 2014; Shin et al., 2019); however, it seems that the ratio between indole acetic acid (IAA) and its conjugated forms is more important than the level of free IAA for the regulation of tomato ripening (Karlova et al., 2014). Indeed, the decrease of free IAA at the onset of ripening is associated with an increase in its conjugated form, IAA-Asp (Buta and Spaulding, 1994; Karlova et al., 2014). SlSAUR69 is involved in the decrease of auxin levels and/or signaling in the pericarp tissue at the onset of fruit ripening via the repression of polar auxin transport (Shin et al., 2019). ARF genes are also involved in fruit ripening; the downregulation of SlARF4 or SlARF2 resulted in fruits with dramatic ripening defects (Jones et al., 2002; Karlova et al., 2014; Hao et al., 2015). Auxin–ethylene interactions are crucial for the fruit ripening process, although the molecular basis of the regulatory network is still relatively unclear (Li et al., 2017; Shin et al., 2019). An antagonistic effect between auxin and ethylene has been observed during the ripening of tomatoes (Li et al., 2017), with ethylene inhibiting auxin transport, metabolism, and signaling processes, while auxin represses the expression of genes involved in ethylene biosynthesis and signaling (Chaabouni et al., 2009; Liu et al., 2015; Li et al., 2016a; Li et al., 2017). Moreover, both auxin and ethylene differentially regulate CK metabolism and signaling processes during tomato ripening (Li et al., 2017).
Brassinosteroids might also be involved in tomato ripening, as exogenous applications of this hormone can promote ripening and ethylene production in tomatoes (Karlova et al., 2014). PAs are also actively involved in climacteric fruit ripening (Liu et al., 2018); for example, putrescine levels progressively increase during fruit maturation and peak in ripe tomatoes, while spermine and spermidine levels decrease gradually until the fruits are fully ripe (Tsaniklidis et al., 2016; Liu et al., 2018). Moreover, although the expression levels of SPMS, ADC, and ODC were minimal during the fruit ripening process, the SPDS genes may play an important role during tomato fruit ripening (Liu et al., 2018).
Phytohormones also play a key role in the regulation of tomato fruit metabolism and quality (Van Meulebroek et al., 2015; Cruz et al., 2018; Li et al., 2019b). The hormones discussed above all contribute to the metabolism of tomato fruits, although ABA and ethylene play the most important roles (Li et al., 2019b). ABA had a greater effect on the regulation of the primary metabolism, while ethylene plays an important role in the transition of primary to secondary metabolism in tomatoes (Li et al., 2019b). Regarding secondary metabolism, ethylene and auxins were described as the most important regulators of carotenoid biosynthesis during tomato fruit ripening (Van Meulebroek et al., 2015; Cruz et al., 2018).
Primary Metabolism in Tomato Fruit
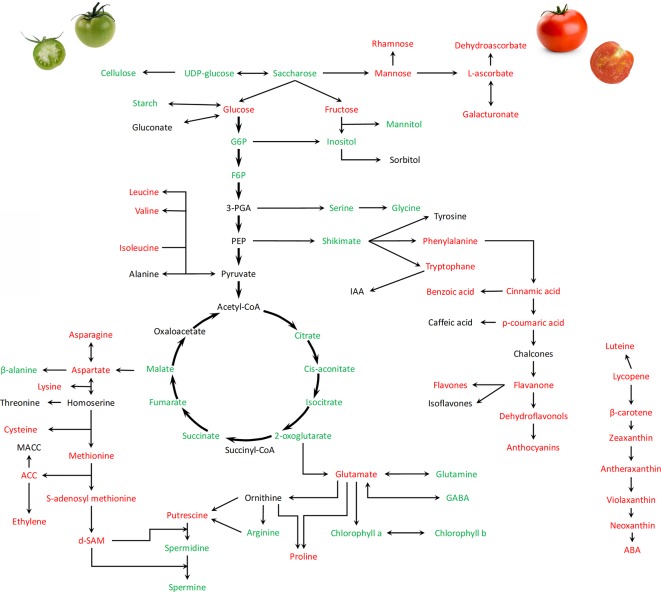
Development of the tomato fleshy fruit occurs in three distinct phases : i) cell division phase occurs in the early days following fertilization until 10 DAA ii) cell expansion (from 10 DAA to 40 DAA) and iii) fruit ripening and maturation ( Figure 1A ). During this evolution, tomato fruits follows a transition from partially photosynthetic to complete heterotrophic metabolism. Typical morphophysiological steps are considered and include immature, mature green, breaker, pink and red ripe fruits. Although the fruit ripening is an important step determining the fruit quality and nutritional values, recent works provided evidences that the early fruit development also assumes key roles for acquisition of quality traits, including the accumulation of sugars and organic acids (Carrari and Fernie, 2006; Beauvoit et al., 2014; Biais et al., 2014; Bauchet et al., 2017). Postgenomic approaches including analyses of fruit transcriptomes, proteomes, and metabolomes as well as multilevel studies integrating enzyme profiling generated a large set of useful data improving our knowledge on the regulation of metabolites turnover during tomato fruit development (Mounet et al., 2009; Centeno et al., 2011; Van de Poel et al., 2012; Van Meulebroek et al., 2015). Hierarchical clustering performed by Biais et al. (2014) revealed tight associations between enzyme activities and developmental phase and concluded that metabolites are more sensitive to growth conditions than enzyme activities. A global overview of the main recorded changes in metabolites recorded during fruit transition from green to red mature fruits is provided in Figure 2 .
Figure 2.
Global overview of metabolic changes occurring during the transition from green expanding fruit to ripening processes (from 30 DAA to 60 DAA) in tomato fruit. Names of metabolites in red, green and black indicate increase, decrease or no changes, respectively. Metabolites are analyzed mainly in pericarps. The Figure summarizes data collected by Carrari and Fernie (2006); Gilbert (2009); Mounet et al. (2009); Centeno et al. (2011); Beauvoit et al. (2014); Biais et al. (2014), Van Meulebroek et al. (2015), Van de Poel et al. (2012), and Zhao et al. (2018).
Carbohydrate Metabolism
Immature Green Fruit Photosynthesis
Sugars are closely related to fruit yield and quality. In tomato fruits, sugars provide sweetness and are important for the generation of turgor pressure to promote cell expansion (Kanayama, 2017). Sugars also act as signal molecules controlling fruit development and metabolism. Green fruits remain able to perform photosynthesis which can produce up to 20% of the fruit photosynthetates, the remaining part being imported by source leaves (Pesaresi et al., 2014). The light harvesting electron transfer and CO2 fixation proteins are conserved in their active state in green fruit tissues (Matas et al., 2011). Fruit chloroplasts contain sufficient amounts of plastocyanin, ferredoxins, Rieske proteins, cytochrome f and cytochrome b559 and ribulose-1,5-biphosphate carboxylase activity is detected in the fruits (Hetherington et al., 1998). The triose phosphate and glucose phosphate transporters are active in the tomato chloroplasts. Unexpectedly, genes associated with photosynthesis are highly expressed in the locule which is in fact the main site of respiration (Lemaire-Chamley et al., 2005).
Nevertheless, the importance of green fruit photosynthesis is still a matter of debate. According to Carrara et al. (2001), tomato fruits do not show signs of CO2 fixation, even if photochemical activity is detectable and an effective electron transport observed. Xu et al. (1997) reported that a small fruit (fresh weight lower than 10 g) is able to perform a gross photosynthesis equivalent to a 3-cm2 leaf blade but that this activity rapidly decreases thereafter: in heavier fruits, gross photosynthesis decreases to negligible values. These authors even assume that the aim of the photosynthetic process in maturing fruit is mainly to delete CO2 produced by respiration rather than contributing to photosynthate production. Kahlau and Bock (2008) showed that RNA, translation and protein accumulation downregulation was observed for all plastid-encoded photosynthesis genes already in the green fruit. Hetherington et al. (1998) however demonstrated that all truss tissues, including fruits, are quite active photosynthetically. These authors interestingly demonstrated that the relative contribution of the fruit versus the leaf photosynthesis for fruit photosynthate accumulation tend to narrow under low light intensities.
A fruit specific antisense inhibition of the chloroplastic fructose 1,6-biphosphatase (FBPase) led to an obvious decrease in final weight of ripe fruits (Obiadalla-Ali et al., 2004) while, conversely, tomato lines with a fruit specific reduction in the expression of glutamate-1-semialdehyde aminotransferase (GSA) and thus a lower level of chlorophyll and photosynthetic rate, remained unaffected in terms of fruit weight (Lytovchenko et al., 2011). Ntagkas et al. (2019) recently demonstrated that phosynthetically active fruits able to respond to light may trigger ascorbate synthesis while non-photosynthetic red maturing fruits are unable to produce this antioxidant in response to light.
Auxin plays an important role for determining final fruit stage through the control of cell division and cell expansion. Auxin-responsive factors (ARF) can either activate or repress transcription of auxin-responsive genes. Combined metabolomics and transcriptomic studies of plants deficient in the expression of the tomato Aux/IAA transcription factor IAA9 suggest a role for photosynthesis in the initiation of fruit development (Wang et al., 2009). Downregulation of SlARF4 enhanced fruit firmness and increased chlorophyll content in green fruits in relation to an increased number of chloroplasts (Guillon et al., 2008). SlARF4 also has a direct impact on fruit sugar metabolism: the SlARF4 underexpression tomato lines accumulated more starch at early stages of fruit development associated with an improved photochemical efficiency (Sagar et al., 2013). Moreover, SlARF4 is highly expressed in the pericarp tissues of immature fruits and undergoes decline at the onset of ripening. Down-regulated tomatoes also present a higher starch content than the wild type in developing fruits which is directly related to up-regulation of several genes and enzyme activities involved in starch biosynthesis (Sagar et al., 2013).
Plastid numbers and chlorophyll content in fruits are positively correlated with photosynthesis and photosynthate accumulation and both are influenced by numerous environmental and genetic factors. In tomato fruits, the GOLDEN2-LIKE (GLK) transcription factor induces the expression of numerous genes related to chloroplast differentiation and photosynthesis (Powell et al., 2012). The genome of S. lycopersicum possesses two copies of this gene: SlGLK1 is predominantly expressed in the leaves while SlGLK2 is expressed in the fruits, especially in the area of pedicel junction (Nguyen et al., 2014). A latitudinal gradient of SlGLK2 expression induces a typical uneven coloration in ripe fruit SlGLK2 is preferentially expressed in the shoulder of the fruit (Sagar et al., 2013). Sl-GLK2 belongs to the GARP subfamily of the myb transcription factor and is encoded by the UNIFORM (U) gene (Powell et al., 2012). The u mutation has been widely selected in modern tomato varieties which consequently exhibit a uniform ripening attractive to consumers and suitable for industrial processing. This mutant contains less sugar and chloroplasts present a lower number of thylakoid grana. According to Nadakuduti et al. (2014), some class I KNOTTED1-LIKE HOMEOBOX gene (TKN2 and TKN4) also influence chloroplast development in tomato fruits and act upstream of SlGLK2. A dominant gain-of-function mutation of TKN2 induces ectopic fruit chloroplast development that resembles SlGLK2 overexpression. More recently, Lupi et al. (2019) demonstrated that SlGLK2 expression is partly regulated by a phytochrome-mediated light perception. Auxin appears as a negative regulator of SlGLK2 expression and SlGLK2 enhances cytokinin responsiveness. This study also demonstrated that SlGLK2 enhances tocopherol and total soluble solid through amylase stimulation, so that selection of the u mutation in commercial varieties probably inadvertently compromise ripe fruit quality.
Sugar Unloading in Fruits
Sugar unloading in tomato fruit is a controlled process and its pattern is not constant during the fruit development. In green developing fruits, sugar is mainly unloaded via the symplasm. Numerous plasmodesmata and cell connections are present at this stage (Ruan and Patrick, 1995) but then are progressively lost. During this early phase of development, only a small amount of sucrose is unloaded by the apoplastic invertase and transported into the fruit cells by hexose transporters (Nguyen-Quoc and Foyer, 2001; Beckles et al., 2012). Although it has been demonstrated that sucrose unloads in tomato pericarp until 35 DAA, a precocious role for apoplastic invertase has however been postulated on the basis of kinetics properties explaining a moderate QTL for Brix index (Fridman et al., 2004).
Sugar Metabolism At the Cell Division Stage
In growing fruits, sucrose represents less than 1% DW while fructose and glucose are the main accumulated soluble sugars (25 and 22% DW; Gilbert, 2009). Glucose and fructose content strongly increased during early fruit developmental phase. Most studies until recent year have focused on the ripe stage but omics analysis need to be conducted throughout fruit development since several interactions may occur between the different stages (Kanayama, 2017). In green fruits, hexose phosphates are mainly used for starch synthesis until 13 DPA. Starch accumulation in pericarp and columella tissues at this early stage is a key factor determining the final soluble solid content of mature fruits (Carrari and Fernie, 2006).
The sink strength of a developing fruit depends on both sink activity and sink size, the latter being a function of both the number and the size of the fruit cells. According to Kataoka et al. (2009), gibberellic acid just after anthesis can promote an increased sink size of individual pericarp cell through the activation of vacuolar acid invertase and neutral invertase. During the cell division phase, a high rate of mitotic activity is observed and the final cell number is determined at the end of this period. It is, at least partly, influenced by endoreduplication processes, seed number and hormonal cues. During cell division, enzymes involved in glycolysis (especially gluokinase and fructokinase) are activated. According to Biais et al. (2014), Glc-6-P is accumulating during this phase, and maintenance of a low ATP to ADP ratio and high hexose-P results in high flux through glycolysis. Pyruvate kinase and tricarboxylic acid cycle enzymes also exhibit high activities, indicating that ATP production as a priority. Beauvoit et al. (2014) postulated that the close match of the catalytic capacity to flux needs may be partly due to protein neosynthesis occuring during the early cell division phase, although protein-protein interactions and post-translational modifications may modulate enzyme V max even if enzyme content remains constant.
At the end of the cell division phase, most soluble sugars accumulated in the vacuoles, together with malic and citric acid (Carrari and Fernie, 2006; Centeno et al., 2011). The osmotic potential of the vacuole consequently dropped to about ‑0.6 MPa and triggers water inflow in the dividing cells. Cytosolic sucrose synthase (SuSy) is involved in sucrose cleavage at the cell division stage. According to Nguyen-Quoc and Foyer (2001) cell vacuoles at this stage accumulate high concentration of hexose (up to 100 µmol g‑1 FW) and contain equal amounts of glucose and fructose. This implies that soluble sugars must be transported to the vacuoles by specific transporters. Vacuolar proton ATPase (V-ATPase) and vacuolar proton pyrophosphatase (V-PPase) generate the electrochemical gradient to transport sugar to the vacuolar compartment (Kanayama, 2017). Amemiya et al. (2006) showed that fruit specific V-ATPase suppression in antisense-transgenic tomato reduces fruit growth and seed formation. It is noteworthy that the highest expression of the V-PPase gene was observed during the cell division stage and not during latter stages of fruit development (Kanayama, 2017). Sucrose loading into the vacuole by the sucrose antiport–transporter is an efficient component of vacuolar storage and there is no requirement for sucrose hydrolysis to allow vacuolar loading or unloading. The fact that regulation of sugar transporters may be influenced by endogenous sugar levels through kinases provide an additional level of complexity regarding carbohydrate subcellular distribution at the end of the cell dividing phase (Lecourieux et al., 2010).
Beside Susy, acid invertase (AI; EC 3.2.1.26) may also be involved in sucrose cleavage and this implies that sucrolytic activity occurs within the vacuole and not only in the cytosol. According to Beauvoit et al. (2014), AI even assumes most of the sucrose cleavage in dividing cells while cytosolic neutral invertase (NI) and SuSy are mainly involved during the following cell expansion phase.
Although sucrose-phosphate-synthase (SPS : 2.4.1.14) activity remains low throughout the fruit development, it may significantly contribute to sucrose re-synthesis in the cytosol, inducing a « futile » cycle between sucrose and hexose characterized by a continuous sugar exchange between cytosol and vacuoles (sucrose influx and hexose efflux) (Nguyen-Quoc and Foyer, 2001). The extent of such resynthesis however remains limited from a quantitative point of view and never exceeds 10% of the cleavage (Beauvoit et al., 2014). Seeds may somewhat control the expression of genes coding for UGPase and SPS : Rounis et al. (2015) found drastic differences in transcript accumulation and enzyme activities of both UGPase and SPS between seeded and parthenocarpic fruits but only minor differences were recorded for sugar levels.
Sugar Metabolism During the Cell Expansion Phase
Mounet et al. (2009) explored transcriptional and metabolic changes in expanding fruit tissues (12–35 DAA) using multivariate analysis and gene-metabolites correlation networks. These authors demonstrated that cell expansion during fruit development proceeds differently in mesocarp and locular tissues which clearly differ in their metabolic composition. Mesocarp represent approximatively 50% (w/v) of the fruit fresh weight and its quantitative importance remains stable throughout fruit development while the locular tissues strongly develop reaching 23% (w/v) of the fruit fresh weight at the mature green stage. Some soluble sugars (mainly Suc and UDP-Glc) are most abundant in locular tissues at the end of the cell expansion phase while others such as hexoses mainly accumulate in the mesocarp. Beside differences in terms of distribution, discrepancies may also result from the mode of expression of enzyme activities or metabolites concentration. Biais et al. (2014) indeed estimated that expressing enzyme activities per protein content minimizes the influence of vacuolar expansion comparatively to an expression on a fresh weight basis.
The cell expansion phase itself is commonly divided in two distinct steps corresponding to « early » and « late » expansion. During the early cell expansion, enzymes involved in the middle part of the glycolysis (NAD-GAPDH, P6K, enolase [EC 4.2.1.11)] are activated in a coordinated way. The main enzyme controlling starch synthesis (ADP-Glc pyrophoshorylase) is also activated in order to produce ADP-Glc for starch synthesis. Sucrose synthase activity presented its highest value during this phase and could be involved in providing UDP-Glc for cell wall cellulose synthesis. Cell expansion is mainly driven by the hexose content. Hexose accumulation in the vacuole is responsible for at least 50% of the fruit osmotic potential during the time course of cell expansion. The crucial role of hexose in cell expansion may thus explain the small fruit size produced by shaded plants.
Some enzymes exhibit a high activity during the late elongation phase and culminate at the green mature stage. This is the case for phosphoglucoisomerase (PGI; EC 5.3.1.9), ATP-phosphofructokinase (PFK; EC 2.7.1.11) and for UDP-Glc pyrophosphorylase (UGPase; EC 2.7.7.9). These enzymes are involved in recycling of hexose-P issued from starch degradation (Carrari et al., 2007). Starch accumulation in the fruit occurs during the early expansion phase while net starch degradation occurs during the late cell expansion phase. Nevertheless, all enzymes required for starch synthesis and degradation are present in the fruits at all developmental stages and there is a continuous starch synthesis and breakdown in tomato fruits. The most important enzyme for starch degradation in fruit is starch phosphorylase which produces G1P while amylase activity remains rather low (Yelle et al., 1991). Beside regulation of ADP-Glc pyrophosphorylase, the concentration of hexose phosphate in the amyloplasts and the rate of hexose phosphate exchange between cytosol and amyloplast constitute major control points to regulate the balance between starch synthesis and starch degradation (Nguyen-Quoc and Foyer, 2001; Centeno et al., 2011).
SlARF4 represses the expression of SlAGPase gene (Sagar et al., 2013). Other transcription factors play key roles in the regulation of gene expression during cell expansion phase. Mounet et al. (2009) reported important roles for zinc finger proteins, MYB, bZIP, an ERF and a NAC transcription factors. The homeobox-Leu zipper protein HAT22 appears to be implicated in the complex regulation of the metabolic shift occurring between fruit early development and subsequent ripening. Some transcription factors assume important roles in the mesocarp while others are more specifically acting in the locular tissues (Lemaire-Chamley et al., 2005; Mintz-Oron et al., 2008; Mounet et al., 2009). Sugar signaling during the cell expansion phase may involve direct sugar-binding: hexokinase is acting as sugar sensor with dual independent functions in hexose phosphorylation and glucose sensing. Sugar signaling may also involve upstream open reading frame as reported for the sucrose-induced repression of translation in which the translation of the normal ORF of a bZIP transcription factor is repressed by sucrose (Kanayama, 2017). Sagor et al. (2016) expressed a tomato homolog of the bZIP gene lacking the uORF in fruit using a ripe fruit specific E8 promoter and strongly increased the fruit sugar concentration in the transgenic lines.
Both cell division and cell expansion phases imply the regulation of the cell wall metabolism, which also directly influences the fruit firmness and texture. Cell wall polysaccharides largely derive from sugar and sugar phosphates, and in tomato fleshy fruits mainly formed by unlignified parenchyma cells, pectic and hemicellulose polysaccharides account for nearly 95% of the cell wall. Regulation of the cell wall-related enzymes are however mainly studied in relation to the ripening phase of tomato fruit development.
Sugar Metabolism During Repining Phase and Putative Interest of Wild-Related Tomato Species
Ripening phase involves both catabolism and accumulation of key metabolites. During ripening, fruit weight still slightly increases and hexoses exhibit their highest concentration. Total protein content also increases and enzymes involved in TCA cycle and glycolysis strongly increased while glucokinase and fructokinase activities decreased. Degradation of starch hence becomes the main source of hexose-P used as substrate for respiration (Carrari and Fernie, 2006; Beckles et al., 2012; Biais et al., 2014). Sucrose-phosphate-synthase activity, which remains low during the previous expanding phases, significantly increased at the beginning of ripening phase (Biais et al., 2014). Accumulation of sucrose, however, remains limited since invertase activities also increased during ripening in the cultivated tomato species S. lycopersicum (Yelle et al., 1991). According to Bastías et al. (2011), ABA which increases before ethylene at the early beginning of maturation phase may be involved in stimulating the expression of genes coding acid vacuolar invertase. The ABA-responsive element binding factor SlAREB1 is indeed present in the fruit pericarp at the end of the mature green stage (Yáñez et al., 2009) and plays an important role for up-regulation of genes involved in sugar metabolism during ripening. During the breaker stage, chlorophyll content strongly declines and the dedifferentiation of chloroplasts in chromoplasts occur under the control of anterograd and retrograd mechanisms leading to the breakdown of starch granules and lysis of thylakoid membrane (Pesaresi et al., 2014). Cell walls are then degraded as a consequence of activation of rhamnogalacturonase and β-galactosidase which depolymerize branched pectins resistant to attack by endo-polygalacturonase (Carrari and Fernie, 2006). Pectin methylesterase catalyses de-esterification of pectin and are encoded by three genes, one being fruit specific and involved in shelf-life of tomato upon storage at room temperature. Fruit softening is also determined by cellulase (endo-β-1,4 glucanases) and by xyloglucan endotransglucosylase (Jiang et al., 2019).
Fructose is sweeter than other sugars and metabolic engineering was therefore specifically performed using fructokinase targets to increase fructose content in commercial tomato fruits (Odanaka et al., 2002; Kanayama, 2017). According to Schaffer et al. (1998), the trait of high fructose to glucose is independently inherited from that of sucrose accumulation. Numerous wild species differ from domesticated tomato cultivars and contain high TSS (Total Soluble Solid, a convenient proxy for sugar content) (more than 10% against 4–6% for S. lycopersicum). These wild species often present an increased import of sugar from source leaves, especially during the latter stage of development. Some of them (Solanum chmielewskii, Solanum peruvianum, Solanum neorickiim, and Solanum habrochaites) store large amounts of sucrose and present constitutively low invertase activities (Miron and Schaffer, 1991). Others (Solanum cheesmanii, Solanum pennellii, and Solanum pimpinelifolium) accumulate mainly glucose and fructose in relation to a high apoplastic invertase in the columella which increases the sugar gradient with the phloem (Beckles et al., 2012). Introgression lines thus constitute convenient tools to investigate the control of sugar content (Eshed and Zamir, 1994; Gur and Zamir, 2004). The line IL8-3 contains a single short segment from S. pennellii in the S. lycopersicum background. This promising line contains a high level of sugar resulting from an increased hexose content, probably as a consequence of a high activity of ADP-glucose pyrophosphorylase leading to accumulation of starch during the middle part of development, followed by an active starch remobilization during ripening (Ikeda et al., 2016).
Beside structural enzymes involved in sugar metabolism in fruits, sugar transporters also appear to play a key role in soluble sugar profile (Schroeder et al., 2013). This is especially the case for members of the SWEET gene family: the expression pattern of those genes frequently coincides with sugar accumulation pattern in tomato fruit (Feng et al., 2015). Two interacting chromosomal regions introgressed from the inedible S. habrochaites present an almost 3-fold epistatic increase in the fructose to glucose ratio in mature fruits (Levin et al., 2000). More recently, Shammai et al. (2018) reported that introgressions of the FgrH allele from S. habrochaites into cultivated tomato increased the fructose to glucose ratio of the ripe fruit. These authors clearly demonstrated that the SlFgr gene encodes a plasma membrane-localized glucose efflux transporter of the SWEET family. Its overexpression in transgenic tomato plants strongly reduced glucose concentration and increased fructose:glucose ratio. Interestingly, no clear impact of the Fgr gene overexpression on the expression of sugar metabolizing genes was recorded and the relationship between glucose efflux and fructose increase still remains an open question.
Organic Acid Metabolism
Organic acid content in fruits is one of the most important properties from a commercial point of view and have a strong influence on the sensorial qualities of the product. Acid taste in tomato is attributed to citric and malic acid which constitute together more than 90% of the total pool of organic acid in harvestable fruits (Bastías et al., 2011). High sugar content and relatively high acid content are required for a favorable taste. High level of acids with low level of sugar will produce a tart tomato, while high levels of sugars and low acids will result in a bland taste (Davies and Hobson, 1981).
The cell division phase is characterized by very high rates of organic acids accumulation (from 2 to 5 nmol min‑1 g‑1 FW) between 4 and 15 DAA according to Beauvoit et al. (2014). It is consequently tempting to speculate that such a high level of accumulation contribute with soluble sugars to decrease the cell water potential allowing water uptake. However, beside this osmotic function, organic acids are also of paramount importance at the cellular level for various biochemical pathways. According to Carrari and Fernie (2006), manipulation of central organic acids is a promising approach to improve tomato fruit yield.
During the cell expansion phase, clear differences were recorded between locular and mesocarp tissues since most organic acids were more abundant in the former than in the latter, and this is especially the case for citrate and malate (Mounet et al., 2009). According to this study, among genes related to organic acid metabolism, 13 were differentially expressed in the two types of tissues. In both tissues, however, organic acid concentration increased between 20 and 35 DAA, mainly in locular tissues and this was correlated with an increased expression of gene coding for aconitase, a key enzyme involved in TCA cycle.
At the ripening stage, tomato climacteric fruits strongly increase ethylene synthesis and respiration, although both subsequently decreased during post-climacteric storage (Van de Poel et al., 2012). Increasing respiration implies hastening of the TCA cycle. Before the ethylene burst, a transient increase in ABA may induce an accumulation of citric and malic enzymes. At the beginning of the ripening phase, fruit preferentially accumulates citrate through stimulation of citrate synthase and the expression of a gene encoding mitochondrial citrate synthase is upregulated by SlAREB1 (Bastías et al., 2011).
Centeno et al. (2011) experimentally decreased the activities of mitochondrial malate dehydrogenase or fumarase via targeted fruit-specific antisense approach in tomato. These authors demonstrated that the line containing higher concentration of malate exhibited a lower starch accumulation during the cell expansion phase and lower soluble sugars at harvest. Although modification of organic acid content in the mitochondria could be relevant from modification in the TCA cycle, it has to be mentioned that mitochondrial pool represents only a small portion of the total cellular organic acid. According to Centeno et al. (2011), correlation between malate and starch concentration could be related to an altered redox status of the AGPase protein allowing an allosteric enhancement of its maximal catalytic activity.
During the ripening stage, phosphoenolpyruvate carboxykinase (PEPCK; which was almost undetectable in green fruits) is suspected to act in the dissimilation of malate/citrate to provide sugar through neoglucogenesis. This hypothesis was confirmed by Huang et al. (2015) who analyzed the effect of an excessive PEPCK in transgenic lines overexpression SlPEPCK by either the constitutive CaMV35S or the fruit-specific E8 promoter. Soluble sugars increased while malate content decreased in both lines, confirming the participation of gluconeogenesis in sugar/acid metabolism during fruit ripening. Similarly, Schouten et al. (2016) recently confirmed that an important part of malate is converted to hexose
Amino Acids Metabolism
Total concentration of free amino acids in tomato fruits varies between 2.0 and 2.5% on a dry weight basis. The most quantitatively important are Glu, Asp and GABA (γ-aminobutiric acid) (Sorrequieta et al., 2010; Snowden et al., 2015). GABA is a four carbon non-protein amino acid which assumes important functional properties in reducing blood pressure in the human body (Zhao et al., 2018). It is also an important metabolite in plants and control cytosolic pH under acid load via the GABA shunt pathway. It is present at high concentration at the green mature stage but then progressively declines during ripening processes (Klee and Giovannoni, 2011). Threonine also declines during ripening and could be metabolized to pyruvate involved with glyceraldehyde 3-phosphate in the synthesis off isopentenyl pyrophosphate acting as a precursor of carotenoids. Most of the other free amino acids increased during ripening while the protein content decreased in relation to an increment in exopeptidase activity and non-specific protease activity pattern (Sorrequieta et al., 2010).
The recorded increase is especially important for glutamate whose concentration may be as high as 10 mmol kg‑1 FW in mature fruits. Such an increase is partly due to stimulation of glutamate dehydrogenase (aminating reaction) and α-ketoglutarate-dependent γ-aminobutyrate transaminase. Cultivated S. lycopersicum has quite higher glutamate content than wild species (Schauer et al., 2005). Since glutamate is a direct precursor of chlorophyll, its accumulation in ripening fruit may be, at least partly, regarded as the consequence of downregulation of chlorophyll synthesis. Mature green fruit contain Fd-GOGAT putatively involved in glutamate synthesis but this enzyme was not detected in red mature fruits where glutamate accumulates (Sorrequieta et al., 2010). Considering the importance of glutamate in phloem sap, transfer of this amino acid from the source leaves to the maturing fruits could not be excluded. (Snowden et al., 2015) considered that GABA may be interconverted in Glu and Asp and provided evidences that these amino acids must be stored in the vacuoles. These authors identified SlCAT9 as a candidate protein for tonoplast transporter exporting GABA from the vacuole and importing Glu and Asp.
Aromatic amino acids also increase and are of special interest since they constitute precursor of flavor volatiles during the ripening process. Valine increased in relation to a stimulation of dihydroxy acid dehydratase (Mounet et al., 2009). MYB and bZIP transcription factors were shown to affect amino acid metabolism (Mounet et al., 2009). (Zhao et al., 2018) recently demonstrated that TAGL1, which play a major role in fruit development (see above), also directly influences fruit metabolism in relation to an increase in seven amino acids (tyrosine, glutamic acid, valine, phenylalanine, proline, leucine and isoleucine).
Secondary Metabolism in Tomato Fruit
Pigments and Flavonoids
The onset and progression of ripening in tomato is typically associated with changes in the external color of the pericarp, reflecting the accumulation of carotenoid and flavonoid pigments (Shinozaki et al., 2018b). Tomato fruits typically provide the principal dietary source of carotenoids in many Western diets (Carrari and Fernie, 2006). The characteristic red tomato color is a result of the accumulation of the carotenoid lycopene in both the fruit skin and pulp (Seymour et al., 2013; Borghesi et al., 2016; D’Ambrosio et al., 2018). During tomato ripening, the concentrations of carotenoids increase by between 10- and 14-fold, mainly due to the accumulation of lycopene (Fraser et al., 1994), which increases as the fruit matures (Tamasi et al., 2019). Alterations in the pigment accumulation patterns have also been observed in several spontaneously occurring tomato mutants (Carrari and Fernie, 2006); for example, the recessive mutant high pigment (hp) produces fruits with two times more carotenoids than wild-type fruits and increased levels of other antioxidants (Yen et al., 1997; Bino et al., 2005; Carrari and Fernie, 2006).
Carotenoid biosynthesis has been studied extensively in tomato, and major steps in the pathway have been identified (Seymour et al., 2013). Light signaling and plant hormones, particularly ethylene and auxins, have been identified as important regulators of carotenoid biosynthesis during tomato fruit ripening (Cruz et al., 2018). Almost all the enzymes acting in the carotenoid biosynthesis pathway have been cloned, and metabolic engineering approaches have been developed to enhance pigment quantity and quality (Carrari and Fernie, 2006; Alseekh et al., 2015; D’Ambrosio et al., 2018). The first committed step of carotenoid biosynthesis is the formation of phytoene, which is dependent on the catalytic activity of phytoene synthase. Phytoene then undergoes two desaturation reactions to form ζ-carotene, catalyzed by phytoene desaturase, which in turn is desaturated to neurosporene and finally lycopene. Lycopene is then either cyclized at both ends of the molecule by lycopene b-cyclase to form β-carotene, or cyclized at one end by lycopene b-cyclase and at the other by lycopene e-cyclase to form α-carotene. These cyclic carotenoids can then be converted to xanthophylls.
Tomatoes also accumulate semipolar metabolites, such as flavonoids, phenolic acids, and alkaloids, which are important health-promoting compounds (Bovy et al., 2007; Tohge and Fernie, 2015; Ballester et al., 2016; Tohge et al., 2017; Tamasi et al., 2019; Wang et al., 2019). To identify the genes responsible for their biosynthesis, QTL analyses were performed in different populations of introgression lines between S. lycopersicum and wild tomato species such as S. chmielewskii and S. pennellii (Alseekh et al., 2015; Ballester et al., 2016; Liu et al., 2016; Alseekh et al., 2017). The flavonoids represent a large family of low molecular weight polyphenolic secondary metabolites, which are grouped into several classes based on their aglycone structure (Bovy et al., 2007; Ballester et al., 2016). The main flavonoid classes are the flavones, flavonols, flavanones, flavanols, anthocyanidins, and isoflavones (Bovy et al., 2007; Tohge et al., 2017). More than 500 different forms of flavonoids are present in tomato, with the most major being the chalcone naringenin chalcone and various sugar conjugates of the flavonols quercetin and kaempferol, including rutin (Bovy et al., 2007; Ballester et al., 2016; Tamasi et al., 2019). In tomato fruits, the accumulation of flavonoids is restricted to the peel, with only traces found in the flesh, which comprises approximately 95% of the whole fruit (Schijlen et al., 2008; Bovy et al., 2010; Ballester et al., 2016). As a result, in a typical tomato cultivar such as Moneymaker, quercetin levels rarely go above 10 mg kg‑1 fresh weight (Bovy et al., 2010). Usually, cultivated tomatoes lack high levels of anthocyanins, while some wild tomato species (S. chilense and S. cheesmaniae) have much higher levels, giving a purple tone to the skin of certain organs (Seymour et al., 2013; Borghesi et al., 2016; Wang et al., 2019). The main phenylpropanoids found in tomato are chlorogenic and caffeic acids (Tamasi et al., 2019).
Flavonoids, along with other phenylpropanoids, are biosynthesized from phenylalanine. Three enzymes [phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumaroyl CoA ligase (4CL)] convert phenylalanine into 4-coumaroyl CoA, the activated intermediate for the various branches of phenylpropanoid metabolism (Zhang et al., 2015). Chalcone synthase (CHS) is the first enzyme involved in the phenylpropanoid/flavonoid pathway and converts 4-coumaroyl CoA into naringenin chalcone (Ballester et al., 2016; Tohge et al., 2017). Most of the biosynthetic genes involved in the flavonoid pathway and the transcription factors regulating them have been identified (Adato et al., 2009; Bovy et al., 2010; Ballester et al., 2016; Tohge et al., 2017; Li et al., 2018b; Wang et al., 2019). These insights have been used to develop genetic engineering strategies to increase the flavonoid contents of tomatoes, since this species accumulates limited amounts of phenolic antioxidants relative to its content of lipophilic antioxidants such as carotenoids (Carrari and Fernie, 2006; Bovy et al., 2007; Bovy et al., 2010; Zhang et al., 2015; Tohge et al., 2017; Wang et al., 2019).
Alkaloids are generally considered to be antinutritional factors in our diet (Friedman, 2002). Breeding efforts have focused on reducing their levels in foods, but some of these substances still remain in our daily diet (Friedman, 2002). More than 100 glycoalkaloids have been found to be present in the tomato clade in various tissues and accessions (Tohge and Fernie, 2015). The main alkaloids present in tomato are α-tomatine and dehydrotomatine, which are often concurrently analyzed as tomatine (Friedman, 2015; Ballester et al., 2016; Tamasi et al., 2019). Immature green tomatoes contain up to 500 mg of tomatine per kilogram of dry weight, while the levels in red tomatoes are much lower (up to about 5 mg kg‑1) (Friedman, 2015; Tamasi et al., 2019). The tomatine contents of cherry tomatoes (grape tomatoes, minitomatoes) are several fold greater than those of the larger standard tomato varieties (Friedman, 2015; Tamasi et al., 2019). In tomato fruits, the bitter-tasting α-tomatine is present at high levels in early developmental stages, but its levels decrease upon ripening due to its conversion into its acetyl glucosylated forms lycoperoside G and F or esculeoside A, which are not bitter (Tohge and Fernie, 2015; Ballester et al., 2016). Dehydrotomatine is 10 times less abundant than α-tomatine in immature fruits (Tamasi et al., 2019). Despite their negative impact on nutrition and their toxicity, glycoalkaloids found in Solanaceous plants, such as α-tomatine, and their hydrolysis products were shown to have anticancer properties (Friedman, 2015).
Volatiles
Volatile metabolites biosynthesized during tomato ripening are responsible for fruit flavor and aroma (Carrari and Fernie, 2006; Ballester et al., 2016; Shinozaki et al., 2018b). More than 400 volatiles have been detected in tomatoes, but a smaller set of 15 to 20 are made in sufficient quantities to have an impact on human perception (Baldwin et al., 2000; Mathieu et al., 2009; Zanor et al., 2009). These volatile compounds are generally derived from various precursors, including fatty acids, carotenoids, and amino acids (Tieman et al., 2006; Zanor et al., 2009; Bauchet et al., 2017). The principal contributors to the ripe tomato flavor are cis-3-hexanal, cis-3-hexanol, hexanal, 3-methylbutanal, 6-methyl-5-hepten-2-one, 1-pentan-3-one, trans-2-hexanal, methyl salicylate, 2-isobutylthiazole, and β-ionone (Carrari and Fernie, 2006). There are differences of many orders of magnitude between the abundance of the various volatile compounds, with concentrations ranging from several micrograms per gram of fresh weight for the most abundant, such as (Z)-3-hexenal or hexanal, to nanograms per gram and even lower levels detected for β-damascenone or β-ionone (Rambla et al., 2014; Tieman et al., 2017). The levels of almost any volatile compound also vary substantially between varieties and accessions (Rambla et al., 2014). Modern commercial varieties contain significantly lower amounts of many of the important flavor chemicals than older varieties since it was not the focus of breeding programs (Tohge and Fernie, 2015; Bauchet et al., 2017; Tieman et al., 2017). Volatiles display a variable pattern of heritability, suggesting a high sensitivity to environmental conditions (Bauchet et al., 2017). Moreover, not all volatile compounds confer positive taste attributes to tomato (Carrari and Fernie, 2006). An example is the identification of malodorous, a wild tomato species allele affecting tomato aroma that was selected against during domestication (Tadmor et al., 2002).
QTL analyses, genome-wide association studies, and targeted metabolome quantifications were conducted in several cultivars and accessions of cultivated tomato, wild relatives, and inbred lines to identify tomato volatiles and their associated genetic loci (Saliba-Colombani et al., 2001; Tieman et al., 2006; Mathieu et al., 2009; Zanor et al., 2009; Alseekh et al., 2015; Ballester et al., 2016; Liu et al., 2016; Alseekh et al., 2017; Bauchet et al., 2017; Tieman et al., 2017). These studies revealed the complex and distinct regulation of metabolites in tomato subspecies (Rambla et al., 2014; Bauchet et al., 2017), demonstrating that there is ample genetic scope to improve the volatile composition of commercial varieties (Rambla et al., 2014).
As mentioned previously, several classes of volatiles exist in tomato. Volatiles derived from fatty acids constitute a class of compounds containing the most abundant volatiles produced in tomato fruits: the C6 volatiles 1-hexanol, (Z)-3-hexenal, (E)-2-hexenal, or hexanal, and the C5 volatile 1-penten-3-one (Rambla et al., 2014). These compounds are classified as green-leaf volatiles due to their characteristic fresh aroma of cut grass (Rambla et al., 2014). The production of these compounds increases as the fruit ripens (Klee, 2010). A second class are volatiles derived from amino acids. A significant number of volatile compounds considered important for the tomato aroma are derived from amino acids (Rambla et al., 2014). These volatiles can be grouped into two categories: phenolic and branched-chain compounds. Phenolic volatiles include a variety of compounds derived from the amino acid phenylalanine, while branched-chain volatiles have particularly low molecular weights and high volatility (Rambla et al., 2014). Additional classes are ester and terpenoid volatiles. Few esters are found in the volatile fraction of tomato (Rambla et al., 2014), while volatile terpenoids are among the most abundant volatiles in tomato vegetative tissues, but only a few of them, such as limonene, linalool, or α-terpineol, are present in the ripe fruit (Rambla et al., 2014), Volatile terpenoids can be classified into two groups, the monoterpenoids (C10) and sesquiterpenoids (C15), both of which are biosynthesized from the five-carbon precursors isopentenyl diphosphate and dimethylallyl diphosphate (Rambla et al., 2014). Carotenoid-derived volatiles are produced at low levels in ripe fruit but are important in our perception of tomato flavor due to their very low odor thresholds (Vogel et al., 2010; Rambla et al., 2014). This is particularly true for β-ionone or β-damascenone, which can be detected ortho-nasally at concentrations of 0.007 and 0.002 nL L‑1, respectively (Buttery et al., 1989). Volatile compounds first accumulate in a conjugated nonvolatile form, such as a glycoside, before being released during the ripening process (Rambla et al., 2014; Tohge and Fernie, 2015). The accumulation of the appropriate glycosidases in a separate subcellular location would allow the immediate liberation of high amounts of the aglycone when the enzyme and the conjugate glycosylated form come into contact with each other (Rambla et al., 2014).
Effects of Abiotic Stress on Tomato Fruit Metabolism
Tomato is one of the most cultivated vegetable species but its productivity is impaired by a wide range of abiotic stresses (Gerszberg and Hnatuszko-Konka, 2017). The presence of adverse environmental factors like extreme temperatures, salinity or drought affects tomato yield as a consequence of reduced fruit number and fruit size but it also affects fruit quality (Moretti et al., 2010; Li et al., 2012; Gerszberg and Hnatuszko-Konka, 2017). It has been shown that moderate stress conditions may improve fruit quality through higher concentration of flavor compounds (Zheng et al., 2013; Albert et al., 2016a; Albert et al., 2016b). In several studies, the concentrations of sugars, organic acids, vitamin C, phenolic compounds and carotenoids increased in tomato fruits in response to water deficit, salinity, or heat (Saito et al., 2009; Patanè et al., 2011; Zushi and Matsuzoe, 2015; Albert et al., 2016b; Flores et al., 2016; Marsic et al., 2018). However, increased CO2 levels increased fruit production but decreased fruit quality (Mamatha et al., 2014). Nevertheless, metabolic modifications in tomato fruits in response to abiotic stress may be cultivar-dependent (Sánchez-Rodríguez et al., 2012a; Sánchez‐Rodríguez et al., 2012b; Zushi and Matsuzoe, 2015; Albert et al., 2016b; Albert et al., 2016a; Flores et al., 2016; Marsic et al., 2018). Table 1 summarizes some recent studies regarding the impact of abiotic stress occurring during plant growth on primary and secondary metabolism of tomato fruits. Modification of fruit metabolism was mainly investigated in response to salinity and drought ( Table 1 ). The effect of salinity was investigated in various cultivars under hydroponic culture with NaCl concentrations varying between 0 to 100 mM and salinity overall increased the concentrations of sugars, organic acids, amino acids, pigments and antioxidants (Saito et al., 2009; Schnitzler and Krauss, 2010; Zushi and Matsuzoe, 2015; Flores et al., 2016; Marsic et al., 2018). The effect of drought was investigated under both greenhouse and field conditions. Most studies reported an increase in sugars and organic acids in response to drought in a wide range of tomato accessions (Patanè et al., 2011; Murshed et al., 2013; Zheng et al., 2013; Shao et al., 2014; Albert et al., 2016b; Albert et al., 2016a) while others reported less strong effects (Atkinson et al., 2011; Sánchez‐Rodríguez et al., 2012; Wei et al., 2018). The effect of drought on the concentration of secondary metabolites was more cultivar-dependent (Atkinson et al., 2011; Sánchez‐Rodríguez et al., 2012). In contrast to salinity and drought, heat mainly decreased the concentration of pigments and ascorbic acid in tomatoes (Li et al., 2012; Hernández et al., 2015) and increased CO2 levels decreased carotenoid, polyphenol and flavonoid concentrations but increased ascorbic acid concentration in tomatoes (Mamatha et al., 2014). All these compounds play an important role in the final nutritional and commercial quality of tomato and depend on genetic, environmental, agronomic and post-harvest factors (Flores et al., 2016). Several studies based on the influence of these factors on fruit composition have been carried out with the aim of increasing tomato quality (Flores et al., 2016).
Table 1.
Effect of abiotic stress occurring during plant growth on primary and secondary metabolite production in tomato fruits.
| Metabolites | Salinity | Drought | Heat | Cold | CO2 increase |
|---|---|---|---|---|---|
| Primary metabolites | |||||
| Sugars | |||||
| Soluble solid Content | ↑ | ↑ | = | ↓(↑) | |
| Total soluble Sugars | ↑ | ↑(=*) | ↑ | = | ↑ |
| Fructose | ↑ | ↑(↓, =*) | |||
| Glucose | ↑ | ↑(↓, =*) | |||
| Saccharose | ↑ | (=*) | |||
| Organic acids | |||||
| Citric acid | ↑(=*) | ↑(=*) | = | ↓ | |
| Malic acid | ↑(=*) | ↑(=*) | = | ↓ | |
| Glutamic acid | ↑(=*) | ||||
| Quinic acid | ↑(=*) | ||||
| Amino acids | |||||
| Arginine | ↑* | ||||
| Histidine | ↑* | ||||
| Isoleucine | ↑* | ||||
| Threonine | ↑ | ||||
| Serine | ↑ | ||||
| Proline | ↑ | ||||
| Phenylalanine | ↑ | ||||
| Secondary metabolites | |||||
| Pigments | |||||
| Carotenoids | ↑(=) | ↑(=*) | ↓ | ↓ | |
| Lycopene | ↑*(=) | ↑(↓*) | ↓ | ↓ | |
| β-carotenoid | ↑ | ↑(↓*) | ↓= | ||
| Antioxydants | |||||
| Total | ↑ | ||||
| Polyphenols | ↑* | ↑(=*) | ↓ | ||
| Flavonoids | ↑* | ↑(=,↓*) | ↓ | ||
| Ascorbic acid | ↑(=*) | ↑(=*) | ↓ | ↑= | |
| References | (Saito et al., 2009; Schnitzler and Krauss, 2010; Zushi and Matsuzoe, 2015; Flores et al., 2016; Marsic et al., 2018) | (Atkinson et al., 2011; Patanè et al., 2011; Sánchez‐Rodríguez et al., 2012; Murshed et al., 2013; Zheng et al., 2013; Shao et al., 2014; Albert et al., 2016a; Albert et al., 2016b; Wei et al., 2018) | (Li et al., 2012; Hernández et al., 2015) | (Kläring et al., 2015) | (Moretti et al., 2010; Mamatha et al., 2014; Wei et al., 2018) |
↑ : Increase; ↓: Decrease; =: No modification; *Cultivar-dependent; () Effect observed only in one study or few cultivars.
In addition to the environmental conditions to which plants are subjected during their growth, post-harvest conditions may also affect fruit quality and metabolism. The impact of low temperature storage on tomato quality has been extensively investigated (Sevillano et al., 2009; Luengwilai et al., 2012; Cruz-Mendívil et al., 2015; Wang et al., 2015; Raffo et al., 2018; Zhang et al., 2019). Among others, early harvesting and cold storage negatively affect tomato flavor and decrease the levels of aroma compounds (Wang et al., 2015; Raffo et al., 2018). Indeed, metabolomics data showed that 7 amino acids, 27 organic acids, 16 of sugars and 22 other compounds had a significantly different content in cold-stored tomatoes and transcriptomics data showed 1735 differentially expressed genes due to cold storage (Zhang et al., 2019). Some pre-treatments have been proposed to improve tomato fruit resistance to cold stress such as ozone exposition, high CO2 treatment, UV-C hormesis, oxalic acid application and heat treatment (Moretti et al., 2010; Luengwilai et al., 2012; Mattos et al., 2014; Charles et al., 2015; Cruz-Mendívil et al., 2015; Li et al., 2016b; Sangwanangkul et al., 2017; Raffo et al., 2018). These treatments provide protection from chilling in part by altering levels of fruit metabolites (Luengwilai et al., 2012; Wang et al., 2015; Sangwanangkul et al., 2017).
Conclusions
In this review, we focused on the tomato fruit development and metabolism. Tomato has long been the model for the study of fleshy fruits and the emergence of “omics” approaches (phenomics, genomics, transcriptomics, proteomics, and metabolomics) has largely contributed to improve our understanding of the genetic, hormonal and metabolic networks that govern tomato fruit development and metabolism. Tomatoes are climacteric fruits with high level of health-promoting compounds. As important as yield improvement and stress resistance, enhancement of tomato fruit quality has gained extensive attention. Improvement of tomato flavor and quality is a challenge for the coming years. The sequencing of tomato genome and genome-wide association studies provide genetic insights into the genetic control of tomato flavor and gives a roadmap for flavor improvement. Moreover, several techniques can now be exploited for breeding superior tomato varieties in the context of current changing climatic conditions.
Author Contributions
MQ and SL designed the outline of the manuscript. MQ, SL, FY-L, TA, and J-PM contributed to writing and revisions of the manuscript. SB and RB-G contributed to figure design and revisions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by funding from the Belgium “Fonds National de la Recherche Scientifique (FRS-FNRS)” (grant no. CDR J.0136.19).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Jennifer Mach for language improvement. RB-G is grateful to the FSR (Fonds special de recherché) for the award of a research fellowship. This work was published with the support of the University Foundation of Belgium.
References
- Adamski N. M., Anastasiou E., Eriksson S., O’Neill C. M., Lenhard M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. PNAS 106, 20115–20120. 10.1073/pnas.0907024106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adato A., Mandel T., Mintz-Oron S., Venger I., Levy D., Yativ M., et al. (2009). Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PloS Genet. 5, e1000777. 10.1371/journal.pgen.1000777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S., Rao A. V. (2000). Tomato lycopene and its role in human health and chronic diseases. CMAJ 163, 739–744. [PMC free article] [PubMed] [Google Scholar]
- Alba R., Payton P., Fei Z., McQuinn R., Debbie P., Martin G. B., et al. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17, 2954–2965. 10.1105/tpc.105.036053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert E., Gricourt J., Bertin N., Bonnefoi J., Pateyron S., Tamby J.-P., et al. (2016a). Genotype by watering regime interaction in cultivated tomato: lessons from linkage mapping and gene expression. Theor. Appl. Genet. 129, 395–418. 10.1007/s00122-015-2635-5 [DOI] [PubMed] [Google Scholar]
- Albert E., Segura V., Gricourt J., Bonnefoi J., Derivot L., Causse M. (2016b). Association mapping reveals the genetic architecture of tomato response to water deficit: focus on major fruit quality traits. J. Exp. Bot. 67, 6413–6430. 10.1093/jxb/erw411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander L., Grierson D. (2002). Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 53, 2039–2055. 10.1093/jxb/erf072 [DOI] [PubMed] [Google Scholar]
- Alseekh S., Tohge T., Wendenberg R., Scossa F., Omranian N., Li J., et al. (2015). Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 27, 485–512. 10.1105/tpc.114.132266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseekh S., Tong H., Scossa F., Brotman Y., Vigroux F., Tohge T., et al. (2017). Canalization of tomato fruit metabolism. Plant Cell 29, 2753–2765. 10.1105/tpc.17.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya T., Kanayama Y., Yamaki S., Yamada K., Shiratake K. (2006). Fruit-specific V-ATPase suppression in antisense-transgenic tomato reduces fruit growth and seed formation. Planta 223, 1272–1280. 10.1007/s00425-005-0176-x [DOI] [PubMed] [Google Scholar]
- Anastasiou E., Kenz S., Gerstung M., MacLean D., Timmer J., Fleck C., et al. (2007). Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13, 843–856. 10.1016/j.devcel.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Atkinson N. J., Dew T. P., Orfila C., Urwin P. E. (2011). Influence of combined biotic and abiotic stress on nutritional quality parameters in tomato (Solanum lycopersicum). J. Agric. Food Chem. 59, 9673–9682. 10.1021/jf202081t [DOI] [PubMed] [Google Scholar]
- Azzi L., Deluche C., Gévaudant F., Frangne N., Delmas F., Hernould M., et al. (2015). Fruit growth-related genes in tomato. J. Exp. Bot. 66, 1075–1086. 10.1093/jxb/eru527 [DOI] [PubMed] [Google Scholar]
- Baldwin E. A., Scott J. W., Shewmaker C. K., Schuch W. (2000). Flavor trivia and tomato aroma: biochemistry and possible mechanisms for control of important aroma components. HortScience 35, 1013–1022. 10.21273/hortsci.35.6.1013 [DOI] [Google Scholar]
- Ballester A.-R., Tikunov Y., Molthoff J., Grandillo S., Viquez-Zamora M., de Vos R., et al. (2016). Identification of loci affecting accumulation of secondary metabolites in tomato fruit of a Solanum lycopersicum × Solanum chmielewskii introgression line population. Front. Plant Sci. 7, 1428. 10.3389/fpls.2016.01428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero L. S., Cong B., Wu F., Tanksley S. D. (2006). Developmental characterization of the fasciated locus and mapping of Arabidopsis candidate genes involved in the control of floral meristem size and carpel number in tomato. Genome 49, 991–1006. 10.1139/g06-059 [DOI] [PubMed] [Google Scholar]
- Barry C. S., Giovannoni J. J. (2006). Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc. Natl. Acad. Sci. U.S.A. 103, 7923–7928. 10.1073/pnas.0602319103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastías A., López-Climent M., Valcárcel M., Rosello S., Gómez-Cadenas A., Casaretto J. A. (2011). Modulation of organic acids and sugar content in tomato fruits by an abscisic acid-regulated transcription factor. Physiol. Plant. 141, 215–226. 10.1111/j.1399-3054.2010.01435.x [DOI] [PubMed] [Google Scholar]
- Bauchet G., Grenier S., Samson N., Segura V., Kende A., Beekwilder J., et al. (2017). Identification of major loci and genomic regions controlling acid and volatile content in tomato fruit: implications for flavor improvement. New Phytol. 215, 624–641. 10.1111/nph.14615 [DOI] [PubMed] [Google Scholar]
- Beauvoit B. P., Colombié S., Monier A., Andrieu M.-H., Biais B., Bénard C., et al. (2014). Model-assisted analysis of sugar metabolism throughout tomato fruit development reveals enzyme and carrier properties in relation to vacuole expansion. Plant Cell 26, 3224–3242. 10.1105/tpc.114.127761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckles D. M., Hong N., Stamova L., Luengwilai K. (2012). Biochemical factors contributing to tomato fruit sugar content: a review. Fruits 67, 49–64. 10.1051/fruits/2011066 [DOI] [Google Scholar]
- Bemer M., Karlova R., Ballester A. R., Tikunov Y. M., Bovy A. G., Wolters-Arts M., et al. (2012). The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24, 4437–4451. 10.1105/tpc.112.103283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biais B., Bénard C., Beauvoit B., Colombié S., Prodhomme D., Ménard G., et al. (2014). Remarkable reproducibility of enzyme activity profiles in tomato fruits grown under contrasting environments provides a roadmap for studies of fruit metabolism. Plant Physiol. 164, 1204–1221. 10.1104/pp.113.231241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bino R. J., Ric de Vos C. H., Lieberman M., Hall R. D., Bovy A., Jonker H. H., et al. (2005). The light-hyperresponsive high pigment-2dg mutation of tomato: alterations in the fruit metabolome. New Phytol. 166, 427–438. 10.1111/j.1469-8137.2005.01362.x [DOI] [PubMed] [Google Scholar]
- Bollier N., Sicard A., Leblond J., Latrasse D., Gonzalez N., Gévaudant F., et al. (2018). At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. Plant Cell 30, 83–100. 10.1105/tpc.17.00653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesi E., Ferrante A., Gordillo B., Rodríguez-Pulido F. J., Cocetta G., Trivellini A., et al. (2016). Comparative physiology during ripening in tomato rich-anthocyanins fruits. Plant Growth Regul. 80, 207–214. 10.1007/s10725-016-0158-y [DOI] [Google Scholar]
- Bovy A., Schijlen E., Hall R. D. (2007). Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): the potential for metabolomics. Metabolomics 3, 399. 10.1007/s11306-007-0074-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy A. G., Gómez-Roldán V., Hall R. D. (2010). “Strategies to Optimize the Flavonoid Content of Tomato Fruit,” in Recent Advances in Polyphenol Research eds V. Lattanzio, C. Santos‐Buelga, M. T. Escribano‐Bailon and V. Lattanzio (Oxford, UK: Blackwell Publishing Ltd.), p. 138–162. 10.1002/9781444323375.ch5 [DOI] [Google Scholar]
- Buta J. G., Spaulding D. W. (1994). Changes in indole-3-acetic acid and abscisic acid levels during tomato (Lycopersicon esculentum Mill.) fruit development and ripening. J. Plant Growth Regul. 13, 163. 10.1007/BF00196382 [DOI] [Google Scholar]
- Buttery R. G., Teranishi R., Flath R. A., Ling L. C. (1989). “Fresh Tomato Volatiles,” in Flavor Chemistry ACS Symposium Series (Wasington, DC: American Chemical Society; ), p. 213–222. 10.1021/bk-1989-0388.ch017 [DOI] [Google Scholar]
- Cara B., Giovannoni J. J. (2008). Molecular biology of ethylene during tomato fruit development and maturation. Plant Sci. 175, 106–113. 10.1016/j.plantsci.2008.03.021 [DOI] [Google Scholar]
- Cardon G., Höhmann S., Klein J., Nettesheim K., Saedler H., Huijser P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237, 91–104. 10.1016/S0378-1119(99)00308-X [DOI] [PubMed] [Google Scholar]
- Carrara S., Pardosi A., Soldatini G. F., Tognoni F., Guidi L. (2001). Photosynthetic activity of ripening tomato fruit. Photosynthetica 39, 75–78. 10.1023/A:1012495903093 [DOI] [Google Scholar]
- Carrari F., Fernie A. R. (2006). Metabolic regulation underlying tomato fruit development. J. Exp. Bot. 57, 1883–1897. 10.1093/jxb/erj020 [DOI] [PubMed] [Google Scholar]
- Carrari F., Asis R., Fernie A. R. (2007). The metabolic shifts underlying tomato fruit development. Plant Biotechnol. 24, 45–55. 10.5511/plantbiotechnology.24.45 [DOI] [Google Scholar]
- Carrera E., Ruiz-Rivero O., Peres L. E., Atares A., Garcia-Martinez J. L. (2012). Characterization of the procera tomato mutant shows novel functions of the SlDELLA protein in the control of flower morphology, cell division and expansion, and the auxin-signaling pathway during fruit-set and development. Plant Physiol. 160, 1581–1596. 10.1104/pp.112.204552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno D. C., Osorio S., Nunes-Nesi A., Bertolo A. L. F., Carneiro R. T., Araújo W. L., et al. (2011). Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening. Plant Cell 23, 162–184. 10.1105/tpc.109.072231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabouni S., Jones B., Delalande C., Wang H., Li Z., Mila I., et al. (2009). Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J. Exp. Bot. 60, 1349–1362. 10.1093/jxb/erp009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti M., Zhang N., Sauvage C., Muños S., Blanca J., Cañizares J., et al. (2013). A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. U. S. A. 110, 17125–17130. 10.1073/pnas.1307313110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles M. T., Rolland D., Roussel D., Merisier M. J., Charlebois D., Arul J. (2015). Assessment of changes in organic acid and sugar profiles of tomato fruits induced by uv-c hormesis. Acta Hortic. 1079, 159–164. 10.17660/ActaHortic.2015.1079.16 [DOI] [Google Scholar]
- Chung M.-Y., Vrebalov J., Alba R., Lee J., McQuinn R., Chung J.-D., et al. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64, 936–947. 10.1111/j.1365-313X.2010.04384.x [DOI] [PubMed] [Google Scholar]
- Clevenger J. P., Van Houten J., Blackwood M., Rodríguez G. R., Jikumaru Y., Kamiya Y., et al. (2015). Network analyses reveal shifts in transcript profiles and metabolites that accompany the expression of sun and an elongated tomato fruit. Plant Physiol. 168, 1164–1178. 10.1104/pp.15.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong B., Liu J., Tanksley S. D. (2002). Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. PNAS 99, 13606–13611. 10.1073/pnas.172520999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz A. B., Bianchetti R. E., Alves F. R. R., Purgatto E., Peres L. E. P., Rossi M., et al. (2018). Light, ethylene and auxin signaling interaction regulates carotenoid biosynthesis during tomato fruit ripening. Front. Plant Sci. 9, 1370. 10.3389/fpls.2018.01370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Mendívil A., López-Valenzuela J. A., Calderón-Vázquez C. L., Vega-García M. O., Reyes-Moreno C., Valdez-Ortiz A. (2015). Transcriptional changes associated with chilling tolerance and susceptibility in ‘Micro-Tom’ tomato fruit using RNA-Seq. Postharvest Biol. Technol. 99, 141–151. 10.1016/j.postharvbio.2014.08.009 [DOI] [Google Scholar]
- D’Ambrosio C., Stigliani A. L., Giorio G. (2018). CRISPR/Cas9 editing of carotenoid genes in tomato. Transgenic Res. 27, 367–378. 10.1007/s11248-018-0079-9 [DOI] [PubMed] [Google Scholar]
- Davies J. N., Hobson G. E. (1981). The constituents of tomato fruit–the influence of environment, nutrition, and genotype. Crit. Rev. Food Sci. Nutr. 15, 205–280. 10.1080/10408398109527317 [DOI] [PubMed] [Google Scholar]
- de Jong M., Mariani C., Vriezen W. H. (2009). The role of auxin and gibberellin in tomato fruit set. J. Exp. Bot. 60, 1523–1532. 10.1093/jxb/erp094 [DOI] [PubMed] [Google Scholar]
- Ding J., Chen B., Xia X., Mao W., Shi K., Zhou Y., et al. (2013). Cytokinin-induced parthenocarpic fruit development in tomato is partly dependent on enhanced gibberellin and auxin biosynthesis. PloS One 8 (7), e70080. 10.1371/journal.pone.0070080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diouf I. A., Derivot L., Bitton F., Pascual L., Causse M. (2018). Water deficit and salinity stress reveal many specific qtl for plant growth and fruit quality traits in tomato. Front. Plant Sci. 9, 279. 10.3389/fpls.2018.00279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson E. M., Bovy A., Manning K., Harrison L., Andrews J., Silva J. D., et al. (2004). Effect of the colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 136, 4184–4197. 10.1104/pp.104.045765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Zamir D. (1994). Introgressions from Lycopersicon pennellii can improve the soluble-solids yield of tomato hybrids. Theoret. Appl. Genet. 88, 891–897. 10.1007/BF01254002 [DOI] [PubMed] [Google Scholar]
- Fahad S., Bajwa A. A., Nazir U., Anjum S. A., Farooq A., Zohaib A., et al. (2017). Crop production under drought and heat stress: plant responses and management options. Front. in Plant Sci. 8, 1147. 10.3389/fpls.2017.01147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT (2019). Available at: http://www.fao.org/faostat/en/#home [Accessed April 15, 2019].
- Fei Z., Tang X., Alba R., Giovannoni J. (2006). Tomato Expression Database (TED): a suite of data presentation and analysis tools. Nucleic Acids Res. 34, D766–D770. 10.1093/nar/gkj110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C. Y., Han J. X., Han X. X., Jiang J. (2015). Genome wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 573, 261–272. 10.1016/j.gene.2015.07.055 [DOI] [PubMed] [Google Scholar]
- Flores P., Hernández V., Hellín P., Fenoll J., Cava J., Mestre T., et al. (2016). Metabolite profile of the tomato dwarf cultivar Micro-Tom and comparative response to saline and nutritional stresses with regard to a commercial cultivar. J. Sci. Food Agric. 96, 1562–1570. 10.1002/jsfa.7256 [DOI] [PubMed] [Google Scholar]
- Frary A., Nesbitt T. C., Grandillo S., Knaap E., Cong B., Liu J., et al. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. 10.1126/science.289.5476.85 [DOI] [PubMed] [Google Scholar]
- Fraser P. D., Truesdale M. R., Bird C. R., Schuch W., Bramley P. M. (1994). Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol. 105, 405–413. 10.1104/pp.105.1.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman E., Carrari F., Liu Y.-S., Fernie A. R., Zamir D. (2004). Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305, 1786–1789. 10.1126/science.1101666 [DOI] [PubMed] [Google Scholar]
- Friedman M. (2002). Tomato glycoalkaloids: role in the plant and in the diet. J. Agric. Food Chem. 50, 5751–5780. 10.1021/jf020560c [DOI] [PubMed] [Google Scholar]
- Friedman M. (2015). Chemistry and anticarcinogenic mechanisms of glycoalkaloids produced by eggplants, potatoes, and tomatoes. J. Agric. Food Chem. 63, 3323–3337. 10.1021/acs.jafc.5b00818 [DOI] [PubMed] [Google Scholar]
- Fujisawa M., Shima Y., Higuchi N., Nakano T., Koyama Y., Kasumi T., et al. (2011). Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235, 1107–1122. 10.1007/s00425-011-1561-2 [DOI] [PubMed] [Google Scholar]
- García-Hurtado N., Carrera E., Ruiz-Rivero O., López-Gresa M. P., Hedden P., Gong F., et al. (2012). The characterization of transgenic tomato overexpressing gibberellin 20-oxidase reveals induction of parthenocarpic fruit growth, higher yield, and alteration of the gibberellin biosynthetic pathway. J. Exp. Bot. 63, 5803–5813. 10.1093/jxb/ers229 [DOI] [PubMed] [Google Scholar]
- Garceau D. C., Batson M. K., Pan I. L. (2017). Variations on a theme in fruit development: the PLE lineage of MADS-box genes in tomato (TAGL1) and other species. Planta 246, 313–321. 10.1007/s00425-017-2725-5 [DOI] [PubMed] [Google Scholar]
- Gerszberg A., Hnatuszko-Konka K. (2017). Tomato tolerance to abiotic stress: a review of most often engineered target sequences. Plant Growth Regul. 83, 175–198. 10.1007/s10725-017-0251-x [DOI] [Google Scholar]
- Gharbi E., Martínez J.-P., Benahmed H., Lepoint G., Vanpee B., Quinet M., et al. (2017). Inhibition of ethylene synthesis reduces salt-tolerance in tomato wild relative species Solanum chilense . J. Plant Physiol. 210, 24–37. 10.1016/j.jplph.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Gilbert L. (2009). Étude de la biosynthèse de l’ascorbate et des métabolismes associés chez la Tomate : rôle de la L-galactono-1,4-lactone déshydrogénase et de la GDP-D-mannose-3’,5’-épimérase. Available at: http://www.theses.fr/2009BOR21668 [Accessed May 29, 2019].
- Gillaspy G., Ben-David H., Gruissem W. (1993). Fruits: a developmental perspective. Plant Cell 5, 1439–1451. 10.1105/tpc.5.10.1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez E., Pineda B., Capel J., Antón M. T., Atarés A., Pérez-Martín F., et al. (2010). Functional analysis of the arlequin mutant corroborates the essential role of the ARLEQUIN/TAGL1 gene during reproductive development of tomato. PloS One 5, e14427. 10.1371/journal.pone.0014427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez E., Castañeda L., Pineda B., Pan I. L., Moreno V., Angosto T., et al. (2016). TOMATO AGAMOUS1 and ARLEQUIN/TOMATO AGAMOUS-LIKE1 MADS-box genes have redundant and divergent functions required for tomato reproductive development. Plant Mol. Biol. 91, 513–531. 10.1007/s11103-016-0485-4 [DOI] [PubMed] [Google Scholar]
- Giovannoni J. J. (2004). Genetic regulation of fruit development and ripening. Plant Cell 16, S170–S180. 10.1105/tpc.019158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. in Plant Biol. 10, 283–289. 10.1016/j.pbi.2007.04.008 [DOI] [PubMed] [Google Scholar]
- Goldental-Cohen S., Israeli A., Ori N., Yasuor H. (2017). Auxin response dynamics during wild-type and entire flower development in tomato. Plant Cell Physiol. 58, 1661–1672. 10.1093/pcp/pcx102 [DOI] [PubMed] [Google Scholar]
- Gonzalo M. J., Brewer M. T., Anderson C., Sullivan D., Gray S., van der K. E. (2009). Tomato fruit shape analysis using morphometric and morphology attributes implemented in tomato analyzer software program. J. Am. Soc. Hortic. Sci. 134, 77–87. 10.21273/JASHS.134.1.77 [DOI] [Google Scholar]
- Guillon F., Philippe S., Bouchet B., Devaux M.-F., Frasse P., Jones B., et al. (2008). Down-regulation of an Auxin Response Factor in the tomato induces modification of fine pectin structure and tissue architecture. J. Exp. Bot. 59, 273–288. 10.1093/jxb/erm323 [DOI] [PubMed] [Google Scholar]
- Guo M., Simmons C. R. (2011). Cell number counts – The fw2.2 and CNR genes and implications for controlling plant fruit and organ size. Plant Sci. 181, 1–7. 10.1016/j.plantsci.2011.03.010 [DOI] [PubMed] [Google Scholar]
- Gur A., Zamir D. (2004). Unused natural variation can lift yield barriers in plant breeding. PloS Biol. 2, e245. 10.1371/journal.pbio.0020245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Hu G., Breitel D., Liu M., Mila I., Frasse P., et al. (2015). Auxin response factor SlARF2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PloS Genet. 11, e1005649. 10.1371/journal.pgen.1005649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández V., Hellín P., Fenoll J., Flores P. (2015). Increased temperature produces changes in the bioactive composition of tomato, depending on its developmental stage. J. Agric. Food Chem. 63, 2378–2382. 10.1021/jf505507h [DOI] [PubMed] [Google Scholar]
- Hetherington S. E., Smillie R. M., Davies W. J. (1998). Photosynthetic activities of vegetative and fruiting tissues of tomato. J. Exp. Bot. 49, 1173–1181. 10.1093/jxb/49.324.1173 [DOI] [Google Scholar]
- Hu J., Israeli A., Ori N., Sun T. P. (2018). The interaction between DELLA and ARF/IAA mediates crosstalk between gibberellin and auxin signaling to control fruit initiation in tomato. Plant Cell 30, 1710–1728. 10.1105/tpc.18.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., van der Knaap E. (2011). Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11. Theor. Appl. Genet. 123, 465–474. 10.1007/s00122-011-1599-3 [DOI] [PubMed] [Google Scholar]
- Huang Y.-X., Goto Y., Nonaka S., Fukuda N., Ezura H., Matsukura C. (2015). Overexpression of the phosphoenolpyruvate carboxykinase gene (SlPEPCK) promotes soluble sugar accumulation in fruit and post-germination growth of tomato (Solanum lycopersicum L.). Plant Biotechnol. 32, 281–289. 10.5511/plantbiotechnology.15.1019a [DOI] [Google Scholar]
- Ikeda H., Shibuya T., Imanishi S., Aso H., Nishiyama M., Kanayama Y. (2016). Dynamic metabolic regulation by a chromosome segment from a wild relative during fruit development in a tomato introgression line, IL8-3. Plant Cell Physiol. 57, 1257–1270. 10.1093/pcp/pcw075 [DOI] [PubMed] [Google Scholar]
- Itkin M., Seybold H., Breitel D., Rogachev I., Meir S., Aharoni A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60, 1081–1095. 10.1111/j.1365-313X.2009.04064.x [DOI] [PubMed] [Google Scholar]
- Ito Y., Kitagawa M., Ihashi N., Yabe K., Kimbara J., Yasuda J., et al. (2008). DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55, 212–223. 10.1111/j.1365-313X.2008.03491.x [DOI] [PubMed] [Google Scholar]
- Ito Y., Nishizawa-Yokoi A., Endo M., Mikami M., Shima Y., Nakamura N., et al. (2017). Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat. Plants 3, 866. 10.1038/s41477-017-0041-5 [DOI] [PubMed] [Google Scholar]
- Jiang F., Lopez A., Jeon S., de Freitas S. T., Yu Q., Wu Z., et al. (2019). Disassembly of the fruit cell wall by the ripening-associated polygalacturonase and expansin influences tomato cracking. Hortic. Res. 6, 17. 10.1038/s41438-018-0105-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B., Frasse P., Olmos E., Zegzouti H., Li Z. G., Latché A., et al. (2002). Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J. 32, 603–613. 10.1046/j.1365-313X.2002.01450.x [DOI] [PubMed] [Google Scholar]
- Jones M. G. (1987). Gibberellins and the procera mutant of tomato. Planta 172, 280–284. 10.1007/BF00394598 [DOI] [PubMed] [Google Scholar]
- Kahlau S., Bock E. (2008). Plastid transcriptomics of tomato fruit development and chloroplast-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20, 856–874. 10.1105/tpc.107.055202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama Y. (2017). Sugar metabolism and fruit development in the tomato. Horti. J. 86, 417–425. 10.2503/hortj.OKD-IR01 [DOI] [Google Scholar]
- Karlova R., Rosin F. M., Busscher-Lange J., Parapunova V., Do P. T., Fernie A. R., et al. (2011). Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23, 923–941. 10.1105/tpc.110.081273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R., Chapman N., David K., Angenent G. C., Seymour G. B., de Maagd R. A. (2014). Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 65, 4527–4541. 10.1093/jxb/eru316 [DOI] [PubMed] [Google Scholar]
- Kataoka K., Yashiro Y., Habu T., Sunamoto K., Kitajima A. (2009). The addition of gibberellic acid to auxin solutions increases sugar accumulation and sink strength in developing auxin-induced parthenocarpic tomato fruits. Sci. Hortic. 123, 228–233. 10.1016/j.scienta.2009.09.001 [DOI] [Google Scholar]
- Kevany B. M., Tieman D. M., Taylor M. G., Cin V. D., Klee H. J. (2007). Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 51, 458–467. 10.1111/j.1365-313X.2007.03170.x [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Kim S.-K., Jung J., Jeong M.-J., Ryu C.-M. (2018). Exploring the sound-modulated delay in tomato ripening through expression analysis of coding and non-coding RNAs. Ann. Bot. 122, 1231–1244. 10.1093/aob/mcy134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kläring H.-P., Klopotek Y., Krumbein A., Schwarz D. (2015). The effect of reducing the heating set point on the photosynthesis, growth, yield and fruit quality in greenhouse tomato production. Agric. For. Meteorol. 214–215, 178–188. 10.1016/j.agrformet.2015.08.250 [DOI] [Google Scholar]
- Klee H. J., Giovannoni J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59. 10.1146/annurev-genet-110410-132507 [DOI] [PubMed] [Google Scholar]
- Klee H., Tieman D. (2002). The tomato ethylene receptor gene family: Form and function. Physiol. Plant. 115, 336–341. 10.1034/j.1399-3054.2002.1150302.x [DOI] [PubMed] [Google Scholar]
- Klee H. J. (2010). Improving the flavor of fresh fruits: genomics, biochemistry, and biotechnology. New Phytol. 187, 44–56. 10.1111/j.1469-8137.2010.03281.x [DOI] [PubMed] [Google Scholar]
- Kou X., Liu C., Han L., Wang S., Xue Z. (2016). NAC transcription factors play an important role in ethylene biosynthesis, reception and signaling of tomato fruit ripening. Mol. Genet. Genomics 291, 1205–1217. 10.1007/s00438-016-1177-0 [DOI] [PubMed] [Google Scholar]
- Krauss S., Schnitzler W. H., Grassmann J., Woitke M. (2006). The influence of different electrical conductivity values in a simplified recirculating soilless system on inner and outer fruit quality characteristics of tomato. J. Agric. Food Chem. 54, 441–448. 10.1021/jf051930a [DOI] [PubMed] [Google Scholar]
- Ku H.-M., Doganlar S., Chen K.-Y., Tanksley S. D. (1999). The genetic basis of pear-shaped tomato fruit. Theor. Appl. Genet. 99, 844–850. 10.1007/s001220051304 [DOI] [Google Scholar]
- Kumar R., Tamboli V., Sharma R., Sreelakshmi Y. (2018). NAC-NOR mutations in tomato Penjar accessions attenuate multiple metabolic processes and prolong the fruit shelf life. Food Chem. 259, 234–244. 10.1016/j.foodchem.2018.03.135 [DOI] [PubMed] [Google Scholar]
- Lecourieux F., Lecourieux D., Vignault C., Delrot S. (2010). A sugar-inducible protein kinase, vvsk1, regulates hexose transport and sugar accumulation in grapevine cells. Plant Physiol. 152, 1096–1106. 10.1104/pp.109.149138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Chamley M., Petit J., Garcia V., Just D., Baldet P., Germain V., et al. (2005). Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiol. 139, 750–769. 10.1104/pp.105.063719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg C. H., Eissler C. L., Wang X., Johns M. A., Duvall M. R., Mao L. (2008). Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 59, 2253–2265. 10.1093/jxb/ern094 [DOI] [PubMed] [Google Scholar]
- Levin I., Gilboa N., Yeselson E., Shen S., Schaffer A. A. (2000). Fgr, a major locus that modulates the fructose to glucose ratio in mature tomato fruits. Theor. Appl. Genet. 100, 256–262. 10.1007/s001220050034 [DOI] [Google Scholar]
- Li Y., Zhu B., Xu W., Zhu H., Chen A., Xie Y., et al. (2007). LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep. 26, 1999–2008. 10.1007/s00299-007-0394-8 [DOI] [PubMed] [Google Scholar]
- Li Z., Palmer W. M., Martin A. P., Wang R., Rainsford F., Jin Y., et al. (2012). High invertase activity in tomato reproductive organs correlates with enhanced sucrose import into, and heat tolerance of, young fruit. J. Exp. Bot. 63, 1155–1166. 10.1093/jxb/err329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Tao X., Li L., Mao L., Luo Z., Khan Z. U., et al. (2016a). Comprehensive RNA-Seq Analysis on the Regulation of Tomato Ripening by Exogenous Auxin. PloS One 11, e0156453. 10.1371/journal.pone.0156453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Yin F., Song L., Zheng X. (2016b). Alleviation of chilling injury in tomato fruit by exogenous application of oxalic acid. Food Chem. 202, 125–132. 10.1016/j.foodchem.2016.01.142 [DOI] [PubMed] [Google Scholar]
- Li J., Tao X., Bu J., Ying T., Mao L., Luo Z. (2017). Global transcriptome profiling analysis of ethylene-auxin interaction during tomato fruit ripening. Postharvest Biol. Technol. 130, 28–38. 10.1016/j.postharvbio.2017.03.021 [DOI] [Google Scholar]
- Li S., Xu H., Ju Z., Cao D., Zhu H., Fu D., et al. (2018a). The RIN-MC fusion of mads-box transcription factors has transcriptional activity and modulates expression of many ripening genes. Plant Physiol. 176, 891–909. 10.1104/pp.17.01449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang H., Zhang Y., Martin C. (2018b). Can the world’s favorite fruit, tomato, provide an effective biosynthetic chassis for high-value metabolites?. Plant Cell Rep. 37, 1443–1450. 10.1007/s00299-018-2283-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Chen K., Grierson D. (2019a). A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 221, 1724–1741. 10.1111/nph.15545 [DOI] [PubMed] [Google Scholar]
- Li Y., Lu Y., Li L., Chu Z., Zhang H., Li H., et al. (2019b). Impairment of hormone pathways results in a general disturbance of fruit primary metabolism in tomato. Food Chem. 274, 170–179. 10.1016/j.foodchem.2018.08.026 [DOI] [PubMed] [Google Scholar]
- Lin Z., Hong Y., Yin M., Li C., Zhang K., Grierson D. (2008). A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55, 301–310. 10.1111/j.1365-313X.2008.03505.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Eck J. V., Cong B., Tanksley S. D. (2002). A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. PNAS 99, 13302–13306. 10.1073/pnas.162485999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Kim Y. J., Müller R., Yumul R. E., Liu C., Pan Y., et al. (2011). AGAMOUS terminates floral stem cell maintenance in arabidopsis by directly repressing WUSCHEL through recruitment of polycomb group proteins. Plant Cell 23, 3654–3670. 10.1105/tpc.111.091538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Pirrello J., Kesari R., Mila I., Roustan J.-P., Li Z., et al. (2013). A dominant repressor version of the tomato Sl-ERF.B3 gene confers ethylene hypersensitivity via feedback regulation of ethylene signaling and response components. Plant J. 76, 406–419. 10.1111/tpj.12305 [DOI] [PubMed] [Google Scholar]
- Liu M., Pirrello J., Chervin C., Roustan J.-P., Bouzayen M. (2015). Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol. 169, 2380–2390. 10.1104/pp.15.01361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Alseekh S., Brotman Y., Zheng Y., Fei Z., Tieman D. M., et al. (2016). Identification of a Solanum pennellii Chromosome 4 Fruit Flavor and Nutritional Quality-Associated Metabolite QTL. Front. Plant Sci. 7, 1671. 10.3389/fpls.2016.01671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Huang B., Chen L., Xian Z., Song S., Chen R., et al. (2018). Genome-wide identification, phylogenetic analysis, and expression profiling of polyamine synthesis gene family members in tomato. Gene 661, 1–10. 10.1016/j.gene.2018.03.084 [DOI] [PubMed] [Google Scholar]
- Luengwilai K., Saltveit M., Beckles D. M. (2012). Metabolite content of harvested Micro-Tom tomato (Solanum lycopersicum L.) fruit is altered by chilling and protective heat-shock treatments as shown by GC–MS metabolic profiling. Postharvest Biol. Technol. 63, 116–122. 10.1016/j.postharvbio.2011.05.014 [DOI] [Google Scholar]
- Luo J. (2015). Metabolite-based genome-wide association studies in plants. Curr. Opin. Plant Biol. 24, 31–38. 10.1016/j.pbi.2015.01.006 [DOI] [PubMed] [Google Scholar]
- Lupi A. C. D., Lia B. S., Gramegna G., Trench B., Alves G. R. R., Demarco D., et al. (2019). Solanum lycopersicum GOLDEN2-LIKE 2 transcription factor affects fruit quality in a light- and auxin-dependent manner. PloS One 14, e212224. 10.1371/journal.pone.0212224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytovchenko A., Eickmeier I., Pons C., Osorio S., Szecowka M., Lehmberg K., et al. (2011). Tomato fruit photosynthesis is seemingly unimportant in primary metabolism and ripening but plays a considerable role in seed development. Plant Physiol. 157, 1650–1663. 10.1104/pp.111.186874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Feng H., Meng X., Li D., Yang D., Wu C., et al. (2014). Overexpression of tomato SlNAC1transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 14, 351. 10.1186/s12870-014-0351-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamatha H., Srinivasa Rao N. K., Laxman R. H., Shivashankara K. S., Bhatt R. M., Pavithra K. C. (2014). Impact of elevated CO2 on growth, physiology, yield, and quality of tomato (Lycopersicon esculentum Mill) cv. Arka Ashish. Photosynthetica 52, 519–528. 10.1007/s11099-014-0059-0 [DOI] [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A. J., King G. J., et al. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38, 948–952. 10.1038/ng1841 [DOI] [PubMed] [Google Scholar]
- Mariotti L., Picciarelli P., Lombardi L., Ceccarelli N. (2011). Fruit-set and early fruit growth in tomato are associated with increases in indoleacetic acid, cytokinin, and bioactive gibberellin contents. J. Plant Growth Regul. 30, 405. 10.1007/s00344-011-9204-1 [DOI] [Google Scholar]
- Marsic N. K., Vodnik D., Mikulic-Petkovsek M., Veberic R., Sircelj H. (2018). Photosynthetic traits of plants and the biochemical profile of tomato fruits are influenced by grafting, salinity stress, and growing season. J. Agric. Food Chem. 66, 5439–5450. 10.1021/acs.jafc.8b00169 [DOI] [PubMed] [Google Scholar]
- Martí C., Orzáez D., Ellul P., Moreno V., Carbonell J., Granell A. (2007). Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52, 865–876. 10.1111/j.1365-313X.2007.03282.x [DOI] [PubMed] [Google Scholar]
- Martí R., Roselló S., Cebolla-Cornejo J. (2016). Tomato as a source of carotenoids and polyphenols targeted to cancer prevention. Cancers (Basel) 8, E58. 10.3390/cancers8060058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Pizarro C., Posé D. (2018). Genome editing as a tool for fruit ripening manipulation. Front. Plant Sci. 9, 1415. 10.3389/fpls.2018.01415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C., Vrebalov J., Tafelmeyer P., Giovannoni J. J. (2011). The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157, 1568–1579. 10.1104/pp.111.181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata C. I., Fabre B., Parsons H. T., Hertog M. L. A. T. M., Van Raemdonck G., Baggerman G., et al. (2018). Ethylene receptors, ctrs and ein2 target protein identification and quantification through parallel reaction monitoring during tomato fruit ripening. Front. Plant Sci. 9, 1626. 10.3389/fpls.2018.01626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matas A. J., Gapper N. E., Chung M.-Y., Giovannoni J. J., Rose J. K. (2009). Biology and genetic engineering of fruit maturation for enhanced quality and shelf-life. Curr. Opin. in Biotechnol. 20, 197–203. 10.1016/j.copbio.2009.02.015 [DOI] [PubMed] [Google Scholar]
- Matas A. J., Yeats T. H., Buda G. J., Zheng Y., Chatterjee S., Tohge T., et al. (2011). Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23, 3893–3910. 10.1105/tpc.111.091173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu S., Cin V. D., Fei Z., Li H., Bliss P., Taylor M. G., et al. (2009). Flavour compounds in tomato fruits: identification of loci and potential pathways affecting volatile composition. J. Exp. Bot. 60, 325–337. 10.1093/jxb/ern294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo S., Kikuchi K., Fukuda M., Honda I., Imanishi S. (2012). Roles and regulation of cytokinins in tomato fruit development. J. Exp. Bot. 63, 5569–5579. 10.1093/jxb/ers207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattos L. M., Moretti C. L., Jan S., Sargent S. A., Lima C. E. P., Fontenelle M. R. (2014). “Chapter 19 - Climate changes and potential impacts on quality of fruit and vegetable crops,” in Emerging Technologies and Management of Crop Stress Tolerance. Eds.Ahmad P., Rasool S. (San Diego:Academic Press; ), p. 467–486. 10.1016/B978-0-12-800876-8.00019-9 [DOI] [Google Scholar]
- McAtee P., Karim S., Schaffer R. J., David K. (2013). A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front. Plant Sci. 4, 79. 10.3389/fpls.2013.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurchie E. J., McGlasson W. B., Eaks I. L. (1972). Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 237, 235–236. 10.1038/237235a0 [DOI] [PubMed] [Google Scholar]
- Meng C., Yang D., Ma X., Zhao W., Liang X., Ma N., et al. (2016). Suppression of tomato SlNAC1 transcription factor delays fruit ripening. J. Plant Physiol. 193, 88–96. 10.1016/j.jplph.2016.01.014 [DOI] [PubMed] [Google Scholar]
- Minoia S., Petrozza A., D’Onofrio O., Piron F., Mosca G., Sozio G., et al. (2010). A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 3, 69. 10.1186/1756-0500-3-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S., Mandel T., Rogachev I., Feldberg L., Lotan O., Yativ M., et al. (2008). Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol. 147, 823–851. 10.1104/pp.108.116004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron D., Schaffer A. A. (1991). Sucrose phosphate synthase, sucrose synthase, and invertase activities in developing fruit of Lycopersicon esculentum Mill. and the sucrose accumulating Lycopersicon hirsutum Humb. and Bonpl. Plant Physiol. 95, 623–627. 10.1104/pp.95.2.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti C. L., Mattos L. M., Calbo A. G., Sargent S. A. (2010). Climate changes and potential impacts on postharvest quality of fruit and vegetable crops: a review. Food Res. Int. 43, 1824–1832. 10.1016/j.foodres.2009.10.013 [DOI] [Google Scholar]
- Mounet F., Moing A., Garcia V., Petit J., Maucourt M., Deborde C., et al. (2009). Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiol. 149, 1505–1528. 10.1104/pp.108.133967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounet F., Moing A., Kowalczyk M., Rohrmann J., Petit J., Garcia V., et al. (2012). Down-regulation of a single auxin efflux transport protein in tomato induces precocious fruit development. J. Exp. Bot. 63, 4901–4917. 10.1093/jxb/ers167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Q., Huang Z., Chakrabarti M., Illa-Berenguer E., Liu X., Wang Y., et al. (2017). Fruit weight is controlled by cell size regulator encoding a novel protein that is expressed in maturing tomato fruits. PloS Genet. 13, e1006930. 10.1371/journal.pgen.1006930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muños S., Ranc N., Botton E., Bérard A., Rolland S., Duffé P., et al. (2011). Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 156, 2244–2254. 10.1104/pp.111.173997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshed R., Lopez-Lauri F., Sallanon H. (2013). Effect of water stress on antioxidant systems and oxidative parameters in fruits of tomato (Solanum lycopersicon L, cv. Micro-tom). Physiol. Mol. Biol. Plants 19, 363–378. 10.1007/s12298-013-0173-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakuduti S. S., Holdsworth W. L., Klein C. L., Barry C. S. (2014). KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 78, 1022–1033. 10.1111/tpj.12529 [DOI] [PubMed] [Google Scholar]
- Nashilevitz S., Melamed-Bessudo C., Izkovich Y., Rogachev I., Osorio S., Itkin M., et al. (2010). An orange ripening mutant links plastid NAD(P)H dehydrogenase complex activity to central and specialized metabolism during tomato fruit maturation. Plant Cell 22, 1977–1997. 10.1105/tpc.110.074716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C. V., Vrebalov J., Gapper N. E., Zheng Y., Zhong S., Fei Z., et al. (2014). Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell 26, 585–601. 10.1105/tpc.113.118794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Quoc B., Foyer C. H. (2001). A role for “futile cycles” involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J. Exp. Bot. 52, 881–889. 10.1093/jexbot/52.358.881 [DOI] [PubMed] [Google Scholar]
- Nitsch L., Kohlen W., Oplaat C., Charnikhova T., Cristescu S., Michieli P., et al. (2012). ABA-deficiency results in reduced plant and fruit size in tomato. J. Plant Physiol. 169, 878–883. 10.1016/j.jplph.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Ntagkas N., Woltering E., Nicole C., Labrie C., Marcelis L. F. M. (2019). Light regulation of vitamin C in tomato fruit is mediated through photosynthesis. Environ. Exp. Bot. 158, 180–188. 10.1016/j.envexpbot.2018.12.002 [DOI] [Google Scholar]
- Obiadalla-Ali H., Fernie A. R., Lytovchenko A., Kossmann J., Lloyd J. R. (2004). Inhibition of chloroplastic fructose 1,6-bisphosphatase in tomato fruits leads to decreased fruit size, but only small changes in carbohydrate metabolism. Planta 219, 533–540. 10.1007/s00425-004-1257-y [DOI] [PubMed] [Google Scholar]
- Obroucheva N. V. (2014). Hormonal regulation during plant fruit development. Russian J. Dev. Biol. 45, 11–21. 10.1134/S1062360414010068 [DOI] [PubMed] [Google Scholar]
- Odanaka S., Bennett A. B., Kanayama Y. (2002). Distinct physiological roles of fructokinase isozymes revealed by gene-specific suppression of Frk1 and Frk2 expression in Tomato. Plant Physiol. 129, 1119–1126. 10.1104/pp.000703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olimpieri I., Caccia R., Picarella M. E., Pucci A., Santangelo E., Soressi G. P., et al. (2011). Constitutive co-suppression of the GA 20-oxidase1 gene in tomato leads to severe defects in vegetative and reproductive development. Plant Sci. 180, 496–503. 10.1016/j.plantsci.2010.11.004 [DOI] [PubMed] [Google Scholar]
- Osorio S., Alba R., Damasceno C. M. B., Lopez-Casado G., Lohse M., Zanor M. I., et al. (2011). Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and Ethylene Receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157, 405–425. 10.1104/pp.111.175463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S., Scossa F., Fernie A. (2013). Molecular regulation of fruit ripening. Front. Plant Sci. 4, 198. 10.3389/fpls.2013.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan I. L., McQuinn R., Giovannoni J. J., Irish V. F. (2010). Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 61, 1795–1806. 10.1093/jxb/erq046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patanè C., Tringali S., Sortino O. (2011). Effects of deficit irrigation on biomass, yield, water productivity and fruit quality of processing tomato under semi-arid Mediterranean climate conditions. Sci. Hortic. 129, 590–596. 10.1016/j.scienta.2011.04.030 [DOI] [Google Scholar]
- Pattison R. J., Catalá C. (2012). Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 70, 585–598. 10.1111/j.1365-313X.2011.04895.x [DOI] [PubMed] [Google Scholar]
- Pem D., Jeewon R. (2015). Fruit and vegetable intake: benefits and progress of nutritioneducation interventions- narrative review article. Iran J. Public Health 44, 1309–1321. [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín F., Yuste-Lisbona F. J., Pineda B., Angarita-Díaz M. P., García-Sogo B., Antón T., et al. (2017). A collection of enhancer trap insertional mutants for functional genomics in tomato. Plant Biotechnol. J. 15, 1439–1452. 10.1111/pbi.12728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P., Mizzotti C., Colombo M., Masiero S. (2014). Genetic regulation and structural changes during tomato fruit development and ripening. Front. Plant Sci. 5, 124. 10.3389/fpls.2014.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L., Hareven D., Rounsley S. D., Yanofsky M. F., Lifschitz E. (1994). Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6, 163–173. 10.1105/tpc.6.2.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A. L. T., Nguyen C. V., Hill T., Chen K. L., Figueroa-Balderas R., Aktas H., et al. (2012). Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336, 1711–1715. 10.1126/science.1222218 [DOI] [PubMed] [Google Scholar]
- Qin G., Wang Y., Cao B., Wang W., Tian S. (2012). Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 70, 243–255. 10.1111/j.1365-313X.2011.04861.x [DOI] [PubMed] [Google Scholar]
- Raffo A., Baiamonte I., Nardo N., Nicoli S., Moneta E., Peparaio M., et al. (2018). Impact of early harvesting and two cold storage technologies on eating quality of red ripe tomatoes. Eur. Food Res. Technol. 244, 805–818. 10.1007/s00217-017-2996-x [DOI] [Google Scholar]
- Raiola A., Rigano M. M., Calafiore R., Frusciante L., Barone A. (2014). Enhancing the health-promoting effects of tomato fruit for biofortified food. Mediators Inflammation 2014. 10.1155/2014/139873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambla J. L., Tikunov Y. M., Monforte A. J., Bovy A. G., Granell A. (2014). The expanded tomato fruit volatile landscape. J. Exp. Bot. 65, 4613–4623. 10.1093/jxb/eru128 [DOI] [PubMed] [Google Scholar]
- Ren Z., Wang X. (2016). SlTIR1 is involved in crosstalk of phytohormones, regulates auxin-induced root growth and stimulates stenospermocarpic fruit formation in tomato. Plant Sci. 253, 13–20. 10.1016/j.plantsci.2016.09.005 [DOI] [PubMed] [Google Scholar]
- Ren Z., Li Z., Miao Q., Yang Y., Deng W., Hao Y. (2011). The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. J. Exp. Bot. 62, 2815–2826. 10.1093/jxb/erq455 [DOI] [PubMed] [Google Scholar]
- Rodríguez G. R., Muños S., Anderson C., Sim S.-C., Michel A., Causse M., et al. (2011). Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 156, 275–285. 10.1104/pp.110.167577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Leal D., Lemmon Z. H., Man J., Bartlett M. E., Lippman Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480.e8. 10.1016/j.cell.2017.08.030 [DOI] [PubMed] [Google Scholar]
- Rothan C., Diouf I., Causse M. (2019). Trait discovery and editing in tomato. Plant J. 97, 73–90. 10.1111/tpj.14152 [DOI] [PubMed] [Google Scholar]
- Rounis V., Skarmoutsos K., Tsaniklidis G., Nikoloudakis N., Delis C., Karapanos I., et al. (2015). Seeded and parthenocarpic cherry tomato fruits exhibit similar sucrose, glucose, and fructose levels, despite dissimilarities in UGPase and SPS gene expression and enzyme activity. J. Plant Growth Regul. 34, 47–56. 10.1007/s00344-014-9441-1 [DOI] [Google Scholar]
- Ruan Y.-L., Patrick J. W. (1995). The cellular pathway of postphloem sugar transport in developing tomato fruit. Planta 196, 434–444. 10.1007/BF00203641 [DOI] [Google Scholar]
- Sánchez-Rodríguez E., Leyva R., Constán-Aguilar C., Romero L., Ruiz J. M. (2012a). Grafting under water stress in tomato cherry: improving the fruit yield and quality. Ann. Appl. Biol. 161, 302–312. 10.1111/j.1744-7348.2012.00574.x [DOI] [Google Scholar]
- Sánchez-Rodríguez E., Ruiz J. M., Ferreres F., Moreno D. A. (2012b). Phenolic profiles of cherry tomatoes as influenced by hydric stress and rootstock technique. Food Chem. 134, 775–782. 10.1016/j.foodchem.2012.02.180 [DOI] [PubMed] [Google Scholar]
- Sagar M., Chervin C., Mila I., Hao Y., Roustan J.-P., Benichou M., et al. (2013). SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 161, 1362–1374. 10.1104/pp.113.213843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagor G. H. M., Berberich T., Tanaka S., Nishiyama M., Kanayama Y., Kojima S., et al. (2016). A novel strategy to produce sweeter tomato fruits with high sugar contents by fruit-specific expression of a single bZIP transcription factor gene. Plant Biotechnol. J. 14, 1116–1126. 10.1111/pbi.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Fukuda N., Matsukura C., Nishimura S. (2009). Effects of salinity on distribution of photosynthates and carbohydrate metabolism in tomato grown using nutrient film technique. J. Jpn. Soc Hortic. Sci. 78, 90–96. 10.2503/jjshs1.78.90 [DOI] [Google Scholar]
- Saliba-Colombani V., Causse M., Langlois D., Philouze J., Buret M. (2001). Genetic analysis of organoleptic quality in fresh market tomato. 1. Mapping QTLs for physical and chemical traits. Theor. Appl. Genet. 102, 259–272. 10.1007/s001220051643 [DOI] [Google Scholar]
- Sangwanangkul P., Bae Y.-S., Lee J.-S., Choi H.-J., Choi J.-W., Park M.-H. (2017). Short-term pretreatment with high CO2 alters organic acids and improves cherry tomato quality during storage. Hortic. Environ. Biotechnol. 58, 127–135. 10.1007/s13580-017-0198-x [DOI] [Google Scholar]
- Schaffer A. A., Petreikov M., Miron D., Fogelman M., Spiegelman M., Bnei-Moshe Z., et al. (1998). Modification of carbohydrate content in developing tomato fruit. HortScience 34, 1024–1027. [Google Scholar]
- Schauer N., Zamir D., Fernie A. R. (2005). Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicum complex. J. Exp. Bot. 56, 297–307. 10.1093/jxb/eri057 [DOI] [PubMed] [Google Scholar]
- Schijlen E. G. W. M., Beekwilder J., Hall R. D., van der Meer I. M. (2008). Boosting beneficial phytochemicals in vegetable crop plants. CAB Rev.: Perspect. In Agric. Vet. Sci. Nutr. Natural Resour. 3, 21. 10.1079/PAVSNNR20083025 [DOI] [Google Scholar]
- Schnitzler W. H., Krauss S. (2010). Quality and health promoting compounds of tomato fruit (Lycopersicum esculentum Mill) under salinity. Acta Hortic. 856, 21–30. 10.17660/ActaHortic.2010.856.2 [DOI] [Google Scholar]
- Schouten R. E., Woltering E. J., Tijskens L. M. M. (2016). Sugar and acid interconversion in tomato fruits based on biopsy sampling of locule gel and pericarp tissue. Postharvest Biol. Technol. 111, 83–92. 10.1016/j.postharvbio.2015.07.032 [DOI] [Google Scholar]
- Schroeder J. I., Delhaize E., Frommer W. B., et al. (2013). Using memvbrane transporters to improve crops for sustainable food production. Nature 497, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevillano L., Sanchez-Ballesta M. T., Romojaro F., Flores F. B. (2009). Physiological, hormonal and molecular mechanisms regulating chilling injury in horticultural species. Postharvest technologies applied to reduce its impact. J. Sci. Food Agric. 89, 555–573. 10.1002/jsfa.3468 [DOI] [Google Scholar]
- Seymour G. B., Chapman N. H., Chew B. L., Rose J. K. C. (2013). Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol. J. 11, 269–278. 10.1111/j.1467-7652.2012.00738.x [DOI] [PubMed] [Google Scholar]
- Shammai A., Petreikov M., Yeselson Y., Faigenboim A., Moy-Komemi M., et al. (2018). Natural genetic variation for expression of a SWEET transporter among wild species of Solanum lycopersicum (tomato) determines the hexose composition of ripening tomato fruit. Plant J. 96, 343–357. 10.1111/tpj.14035 [DOI] [PubMed] [Google Scholar]
- Shao G., Wang M., Liu N., Yuan M., Kumar P., She D.-L. (2014). Growth and comprehensive quality index of tomato under rain shelters in eesponse to different irrigation and drainage treatments. Sci. World J. 10.1155/2014/457937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y., Kitagawa M., Fujisawa M., Nakano T., Kato H., Kimbara J., et al. (2013). Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Mol. Biol. 82, 427–438. 10.1007/s11103-013-0071-y [DOI] [PubMed] [Google Scholar]
- Shin J.-H., Mila I., Liu M., Rodrigues M. A., Vernoux T., Pirrello J., et al. (2019). The RIN-regulated Small Auxin-Up RNA SAUR69 is involved in the unripe-to-ripe phase transition of tomato fruit via enhancement of the sensitivity to ethylene. New Phytol. 222, 820–836. 10.1111/nph.15618 [DOI] [PubMed] [Google Scholar]
- Shinozaki Y., Hao S., Kojima M., Sakakibara H., Ozeki-Iida Y., Zheng Y., et al. (2015). Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J. 83, 237–251. 10.1111/tpj.12882 [DOI] [PubMed] [Google Scholar]
- Shinozaki Y., Ezura H., Ariizumi T. (2018a). The role of ethylene in the regulation of ovary senescence and fruit set in tomato (Solanum lycopersicum). Plant Signaling Behav. 13, e1146844. 10.1080/15592324.2016.1146844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki Y., Nicolas P., Fernandez-Pozo N., Ma Q., Evanich D. J., Shi Y., et al. (2018b). High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat. Commun. 9, 364. 10.1038/s41467-017-02782-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki Y., Ezura K., Hu J., Okabe Y., Bénard C., Prodhomme D., et al. (2018c). Identification and functional study of a mild allele of SlDELLA gene conferring the potential for improved yield in tomato. Sci. Rep. 8, 12043. 10.1038/s41598-018-30502-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden C. J., Thomas B., Baxter C. J., Smith J. A. C., Sweetlove L. J. (2015). A tonoplast Glu/Asp/GABA exchanger that affects tomato fruit amino acid composition. Plant J. 81, 651–660. 10.1111/tpj.12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich M., Je B. I., Simon R., Jackson D. (2016). CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143, 3238–3248. 10.1242/dev.133645 [DOI] [PubMed] [Google Scholar]
- Sorrequieta A., Ferraro G., Boggio S. B., Valle E. M. (2010). Free amino acid production during tomato fruit ripening: a focus on L-glutamate. Amino Acids 38, 1523–1532. 10.1007/s00726-009-0373-1 [DOI] [PubMed] [Google Scholar]
- Soyk S., Lemmon Z. H., Oved M., Fisher J., Liberatore K. L., Park S. J., et al. (2017). Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 169, 1142–1155.e12. 10.1016/j.cell.2017.04.032 [DOI] [PubMed] [Google Scholar]
- Srivastava A., Handa A. K. (2005). Hormonal regulation of tomato fruit development: A Molecular perspective. J. Plant Growth Regul. 24, 67–82. 10.1007/s00344-005-0015-0 [DOI] [Google Scholar]
- Su L., Bassa C., Audran C., Mila I., Cheniclet C., Chevalier C., et al. (2014). The auxin Sl-IAA17 transcriptional repressor controls fruit size via the regulation of endoreduplication-related cell expansion. Plant Cell Physiol. 55, 1969–1976. 10.1093/pcp/pcu124 [DOI] [PubMed] [Google Scholar]
- Sun B., Xu Y., Ng K.-H., Ito T. (2009). A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 23, 1791–1804. 10.1101/gad.1800409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Sun Y., Zhang M., Wang L., Ren J., Cui M., et al. (2012). Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, zlters fruit texture in transgenic tomato. Plant Physiol. 158, 283–298. 10.1104/pp.111.186866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Looi L.-S., Guo S., He Z., Gan E.-S., Huang J., et al. (2014). Timing mechanism dependent on cell division is invoked by polycomb eviction in plant stem cells. Science 343, 1248559. 10.1126/science.1248559 [DOI] [PubMed] [Google Scholar]
- Suresh B. V., Roy R., Sahu K., Misra G., Chattopadhyay D. (2014). Tomato genomic resources database: an integrated repository of useful tomato genomic information for basic and applied research. PloS One 9, e86387. 10.1371/journal.pone.0086387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadmor Y., Fridman E., Gur A., Larkov O., Lastochkin E., Ravid U., et al. (2002). Identification of malodorous, a wild species allele affecting tomato aroma that was aelected against during domestication. J. Agric. Food Chem. 50, 2005–2009. 10.1021/jf011237x [DOI] [PubMed] [Google Scholar]
- Tamasi G., Pardini A., Bonechi C., Donati A., Pessina F., Marcolongo P., et al. (2019). Characterization of nutraceutical components in tomato pulp, skin and locular gel. Eur. Food Res. Technol. 245, 907–918. 10.1007/s00217-019-03235-x [DOI] [Google Scholar]
- Tanksley S. D. (2004). The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16, S181–S189. 10.1105/tpc.018119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D. M., Zeigler M., Schmelz E. A., Taylor M. G., Bliss P., Kirst M., et al. (2006). Identification of loci affecting flavour volatile emissions in tomato fruits. J. Exp. Bot. 57, 887–896. 10.1093/jxb/erj074 [DOI] [PubMed] [Google Scholar]
- Tieman D., Zhu G., Resende M. F. R., Lin T., Nguyen C., Bies D., et al. (2017). A chemical genetic roadmap to improved tomato flavor. Science 355, 391–394. 10.1126/science.aal1556 [DOI] [PubMed] [Google Scholar]
- Tigchelaar E. C., Tomes M., Kerr M., Barman R. (1973). A new fruit ripening mutant, non-ripening (nor) . Rep. Tomato Genet. Coop 23, 33. [Google Scholar]
- Tohge T., Fernie A. R. (2015). Metabolomics-inspired insight into developmental, environmental and genetic aspects of tomato fruit chemical composition and quality. Plant Cell Physiol. 56, 1681–1696. 10.1093/pcp/pcv093 [DOI] [PubMed] [Google Scholar]
- Tohge T., de Souza L. P., Fernie A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68, 4013–4028. 10.1093/jxb/erx177 [DOI] [PubMed] [Google Scholar]
- Tomato Genetics Resource Center (2019). Available at: https://tgrc.ucdavis.edu/[Accessed April 16, 2019].
- Tsaniklidis G., Kotsiras A., Tsafouros A., Roussos P. A., Aivalakis G., Katinakis P., et al. (2016). Spatial and temporal distribution of genes involved in polyamine metabolism during tomato fruit development. Plant Physiol. Biochem. 100, 27–36. 10.1016/j.plaphy.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Van de Poel B., Bulens I., Markoula A., Hertog M. L. A. T. M., Dreesen R., Wirtz M., et al. (2012). Targeted systems biology profiling of tomato fruit reveals coordination of the yang cycle and a distinct regulation of ethylene biosynthesis during postclimacteric ripening. Plant Physiol. 160, 1498–1514. 10.1104/pp.112.206086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E., Chakrabarti M., Chu Y. H., Clevenger J. P., Illa-Berenguer E., Huang Z., et al. (2014). What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5, 227. 10.3389/fpls.2014.00227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meulebroek L., Bussche J. V., De Clercq N., Steppe K., Vanhaecke L. (2015). A metabolomics approach to unravel the regulating role of phytohormones towards carotenoid metabolism in tomato fruit. Metabolomics 11, 667–683. 10.1007/s11306-014-0728-9 [DOI] [Google Scholar]
- Viuda-Martos M., Sanchez-Zapata E., Sayas-Barberá E., Sendra E., Pérez-Álvarez J. A., Fernández-López J. (2014). Tomato and tomato byproducts. Human health benefits of lycopene and its application to meat products: a review. Crit. Rev. In Food Sci. Nutr. 54, 1032–1049. 10.1080/10408398.2011.623799 [DOI] [PubMed] [Google Scholar]
- Vogel J. T., Tieman D. M., Sims C. A., Odabasi A. Z., Clark D. G., Klee H. J. (2010). Carotenoid content impacts flavor acceptability in tomato (Solanum lycopersicum). J. Sci. Food Agric. 90, 2233–2240. 10.1002/jsfa.4076 [DOI] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. 10.1126/science.1068181 [DOI] [PubMed] [Google Scholar]
- Vrebalov J., Pan I. L., Arroyo A. J. M., McQuinn R., Chung M., Poole M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF Gene TAGL1 . Plant Cell 21, 3041–3062. 10.1105/tpc.109.066936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Schauer N., Usadel B., Frasse P., Zouine M., Hernould M., et al. (2009). Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21, 1428–1452. 10.1105/tpc.108.060830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Lu G., Hou Z., Luo Z., Wang T., Li H., et al. (2014). Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. J. Exp. Bot. 65, 3005–3014. 10.1093/jxb/eru137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Baldwin E. A., Zhao W., Plotto A., Sun X., Wang Z., et al. (2015). Suppression of volatile production in tomato fruit exposed to chilling temperature and alleviation of chilling injury by a pre-chilling heat treatment. LWT - Food Sci. Technol. 62, 115–121. 10.1016/j.lwt.2014.12.062 [DOI] [Google Scholar]
- Wang Y., Luo Z., Lu C., Zhou R., Zhang H., Zhao L., et al. (2019). Transcriptome profiles reveal new regulatory factors of anthocyanin accumulation in a novel purple-colored cherry tomato cultivar Jinling Moyu. Plant Growth Regul. 87, 9–18. 10.1007/s10725-018-0444-y [DOI] [Google Scholar]
- Wei Z., Du T., Li X., Fang L., Liu F. (2018). Interactive effects of elevated CO2 and N fertilization on yield and quality of tomato grown under reduced irrigation regimes. Front. Plant Sci. 9, 328. 10.3389/fpls.2018.00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Xiao H., Cabrera A., Meulia T., van der Knaap E. (2011). SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 157, 1175–1186. 10.1104/pp.111.181065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Li H., Zhang J., Chen R., Zhang Y., Ouyang B., et al. (2006). Dissection of GA 20-oxidase members affecting tomato morphology by RNAi-mediated silencing. Plant Growth Regul. 50, 179–189. 10.1007/s10725-006-9117-3 [DOI] [Google Scholar]
- Xiao H., Radovich C., Welty N., Hsu J., Li D., Meulia T., et al. (2009). Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biol. 9, 49. 10.1186/1471-2229-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. L., Gauthier L., Desjardin Y., Gosselin A. (1997). Photosynthesis in leaves, fruits, stem and petioles of greenhouse-grown tomato plants. Photosynthetica 33, 113–123. 10.1023/A:1022135507700 [DOI] [Google Scholar]
- Xu C., Liberatore K. L., MacAlister C. A., Huang Z., Chu Y.-H., Jiang K., et al. (2015). A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47, 784–792. 10.1038/ng.3309 [DOI] [PubMed] [Google Scholar]
- Yáñez M., Cáceres S., Orellana S., Bastías A., Verdugo I., Ruiz-Lara S., et al. (2009). An abiotic stress-responsive bZIP transcription factor from wild and cultivated tomatoes regulates stress-related genes. Plant Cell Rep. 28, 1497–1507. 10.1007/s00299-009-0749-4 [DOI] [PubMed] [Google Scholar]
- Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldmann K. A., Meyerowitz E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. 10.1038/346035a0 [DOI] [PubMed] [Google Scholar]
- Yelle S., Chetelat R. T., Dorais M., DeVerna J. W., Bennett A. B. (1991). Sink metabolism in tomato fruit: IV. Genetic and biochemical analysis of sucrose accumulation. Plant Physiol. 95, 1026–1035. 10.1104/pp.95.4.1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H. C., Lee S., Tanksley S. D., Lanahan M. B., Klee H. J., Giovannoni J. J. (1995). The tomato Never-ripe locus regulates ethylene-inducible gene expression and is linked to a homolog of the Arabidopsis ETR1 gene. Plant Physiol. 107, 1343–1353. 10.1104/pp.107.4.1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H. C., Shelton B. A., Howard L. R., Lee S., Vrebalov J., Giovannoni J. J. (1997). The tomato high-pigment (hp) locus maps to chromosome 2 and influences plastome copy number and fruit quality. Theor. Appl. Genet. 95, 1069–1079. 10.1007/s001220050664 [DOI] [Google Scholar]
- Yuan X.-Y., Wang R.-H., Zhao X.-D., Luo Y.-B., Fu D.-Q. (2016). Role of the tomato Non-Ripening mutation in regulating fruit quality elucidated using iTRAQ protein profile analysis. PloS One 11, e0164335. 10.1371/journal.pone.0164335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanor M. I., Rambla J.-L., Chaïb J., Steppa A., Medina A., Granell A., et al. (2009). Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. J. Exp. Bot. 60, 2139–2154. 10.1093/jxb/erp086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Yuan B., Leng P. (2009). The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J. Exp. Bot. 60, 1579–1588. 10.1093/jxb/erp026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Butelli E., Alseekh S., Tohge T., Rallapalli G., Luo J., et al. (2015). Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 6, 8635. 10.1038/ncomms9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.-F., Gong Z.-H., Wu M.-B., Chan H., Yuan Y.-J., Tang N., et al. (2019). Integrative comparative analyses of metabolite and transcript profiles uncovers complex regulatory network in tomato (Solanum lycopersicum L.) fruit undergoing chilling injury. Sci. Rep. 9, 4470. 10.1038/s41598-019-41065-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yuan X., Chen S., Meng L., Fu D. (2018). Role of the tomato TAGL1 gene in regulating fruit metabolites elucidated using RNA sequence and metabolomics analyses. PloS One 13, e0199083. 10.1371/journal.pone.0199083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Sauvage C., Zhao J., Bitton F., Bauchet G., Liu D., et al. (2019). Meta-analysis of genome-wide association studies provides insights into genetic control of tomato flavor. Nat. Commun. 10, 1534. 10.1038/s41467-019-09462-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Huang G., Jia D., Wang J., Mota M., Pereira L. S., et al. (2013). Responses of drip irrigated tomato (Solanum lycopersicum L.) yield, quality and water productivity to various soil matric potential thresholds in an arid region of Northwest China. Agric. Water Manage. 129, 181–193. 10.1016/j.agwat.2013.08.001 [DOI] [Google Scholar]
- Zhong S., Fei Z., Chen Y.-R., Zheng Y., Huang M., Vrebalov J., et al. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31, 154–159. 10.1038/nbt.2462 [DOI] [PubMed] [Google Scholar]
- Zhou R., Kong L., Wu Z., Rosenqvist E., Wang Y., Zhao L., et al. (2019). Physiological response of tomatoes at drought, heat and their combination followed by recovery. Physiol. Plant 165, 144–154. 10.1111/ppl.12764 [DOI] [PubMed] [Google Scholar]
- Zhu M., Chen G., Zhou S., Tu Y., Wang Y., Dong T., et al. (2014). A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 55, 119–135. 10.1093/pcp/pct162 [DOI] [PubMed] [Google Scholar]
- Zhu G., Wang S., Huang Z., Zhang S., Liao Q., Zhang C., et al. (2018). Rewiring of the fruit metabolome in tomato breeding. Cell 172, 249–261.e12. 10.1016/j.cell.2017.12.019 [DOI] [PubMed] [Google Scholar]
- Zouine M., Maza E., Djari A., Lauvernier M., Frasse P., Smouni A., et al. (2017). TomExpress, a unified tomato RNA-Seq platform for visualization of expression data, clustering and correlation networks. Plant J. 92, 727–735. 10.1111/tpj.13711 [DOI] [PubMed] [Google Scholar]
- Zushi K., Matsuzoe N. (2015). Metabolic profile of organoleptic and health-promoting qualities in two tomato cultivars subjected to salt stress and their interactions using correlation network analysis. Sci. Hortic. 184, 8–17. 10.1016/j.scienta.2014.12.030 [DOI] [Google Scholar]