Abstract
Genomic identification of driver mutations and genes in cancer cells are critical for precision medicine. Due to difficulty in modelling distribution of background mutation counts, existing statistical methods are often underpowered to discriminate cancer-driver genes from passenger genes. Here we propose a novel statistical approach, weighted iterative zero-truncated negative-binomial regression (WITER, http://grass.cgs.hku.hk/limx/witer or KGGSeq,http://grass.cgs.hku.hk/limx/kggseq/), to detect cancer-driver genes showing an excess of somatic mutations. By fitting the distribution of background mutation counts properly, this approach works well even in small or moderate samples. Compared to alternative methods, it detected more significant and cancer-consensus genes in most tested cancers. Applying this approach, we estimated 229 driver genes in 26 different types of cancers. In silico validation confirmed 78% of predicted genes as likely known drivers and many other genes as very likely new drivers for corresponding cancers. The technical advances of WITER enable the detection of driver genes in TCGA datasets as small as 30 subjects and rescue of more genes missed by alternative tools in moderate or small samples.
INTRODUCTION
It is well known that genomic aberration in somatic cells makes important contribution to development of cancers (1). Mutations that confer selective growth advantage to cancer cells are called as cancer-driver mutations (2,3); a gene harboring driver-mutations is named as a cancer-driver gene. It has been established, for example, that mutations in the two driver genes TP53 and PIK3CA contribute to many types of cancer (4). However, cancers are also known to be highly heterogeneous (5) and many driver genes of most cancers-specific remain to be identified. Therefore, a full landscape of driver-genes remains unavailable for most cancers, although the genes are fundamental for early diagnosis, identification of effective drug targets and precise treatments (2).
There are generally two existing strategies to detect cancer driver genes with somatic mutations, background mutation rate (BMR) and ratiometric. The BMR-based methods evaluate whether a gene has more somatic mutations than expected; examples include MutSigCV (6) and MuSiC (7). The expected number of mutations is estimated by multiple predictors including base context, gene size and other variables of genes. The ratiometric-based methods detect cancer-driver genes according to the composition of mutation types normalized by the total number of mutations in a gene. For instance, the ratiometric 20/20 rule assessed the proportion of inactivating mutations (including synonymous mutations) and missense mutations (3). Oncodrive-fm (8) and OncodriveFML (9) integrated mutations’ functional impact into the ratiometric evaluation. OncodriveCLUST considered the positional clustering of mutation patterns (10). A method named 20/20 plus (11) extended the ratiometric idea in the 20/20 rule and integrated 18 additional features to predict cancer-driver genes by a machine learning approach. It also generated statistical P-values of the prediction scores by Monte Carlo simulations. Recently, negative binomial (NB) regression was used to estimate the synonymous mutations and then test the dN/dS ratio for detecting driver genes in multiple studies (12–14).
Although the general principles of both strategies are simple, three key technical issues remain unsolved. First, the statistical P-values produced by existing cancer-driver gene methods did not follow uniform distribution generally (11), implying the underfitting of background mutations. Although simulation or permutation can be used to produce valid P-values, a good fitting of background genes is critical for accurate discrimination of true driver genes from noise background genes. Second, existing statistical tests are generally underpowered to detect driver-genes with small or moderate effect size. This issue will become more severe when the sample is not large. It seems the NB regression is a promising framework for estimating cancer-driver genes (12–14). However, when sample size is not large, an inflation of zero-mutation genes may distort the NB distribution and their estimated dN/dS ratio may be unstable. The above-mentioned supervised machine learning approaches’ integration of common gene features beyond local samples may help enhance the power (11). However, given the high heterogeneity in cancers (6), adding more common features may not work for unique driver genes of a cancer. The trained model for known driver genes may also have limited power for detecting new driver genes. Finally, the predicted cancer-driver genes by different tools do not generally agree with each other (2). It is often laborious and subjectively biased to combine their results. Therefore, more powerful methods are pressingly needed for unraveling a full spectrum of cancer-driver genes.
Here, we describe an extended negative-binomial regression, weighted iterative zero-truncated negative-binomial regression (WITER), to detect cancer-driver genes by somatic mutations [including single nucleotide variants (SNVs) and short insertions and deletions (InDels)]. This approach belongs to the unsupervised method category and therefore does not suffer from training bias. The method has a unique three-tier structure for fitting the somatic mutations of background genes even in small samples. We then systematically compare its performance with alternative methods in 11 cancers. A comprehensive landscape of driver-genes is constructed by WITER and analyzed to investigate the common and unique insights across 26 cancers.
MATERIALS AND METHODS
The statistical framework for detecting cancer-driver genes by somatic mutations in cancers
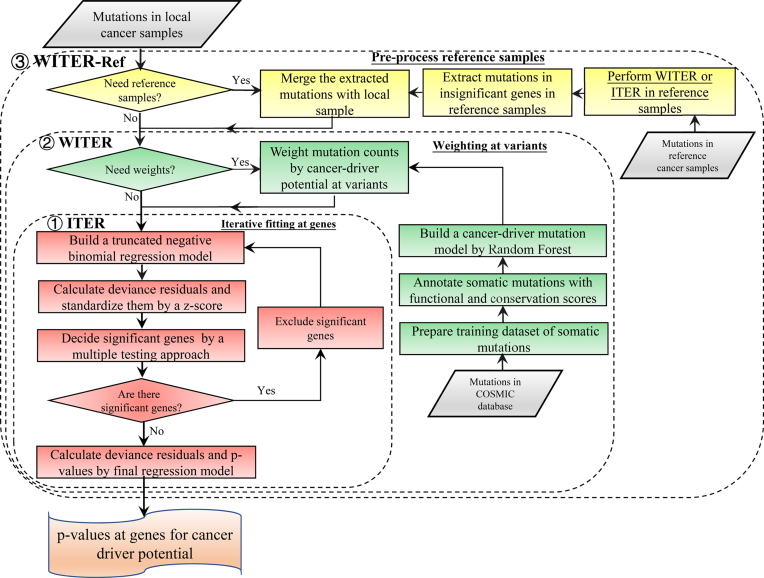
We propose a statistical framework, WITER, for detecting cancer-driver genes by somatic mutations in cancers. The main input is somatic mutations (including SNVs and InDels) in samples from cancer patients. The main output is a list of P-values for excess of somatic mutations at genes; a significant P-value suggests a gene that has excessive somatic mutations probably because these mutations confer selective growth advantages to cancer cells. The framework has a three-tier structure to improve the modelling of background mutations in passenger genes for a more powerful estimation of driver genes (See the diagram in Figure 1). In theory, the framework and the model are independent of types of somatic variants to be tested. However, the present paper focuses on non-synonymous and splicing variants because of abundant validation data and resource data in public domains. The approach and auxiliary functions have been implemented into a user-friendly software tool which is publicly available at http://grass.cgs.hku.hk/limx/witer.
Figure 1.
The diagram of the statistical framework for detecting cancer-driver genes. This framework includes three tiers denoted by the dashed rectangles. The first tier is an ITER. The second tier is a WITER. The third tier is the integration of reference samples. The unique components of each tier are marked by different colors. The major inputs are somatic mutations in different cancer patients. The outputs are P-values for excess of somatic mutations of individual gene in the cancer samples.
Tier I: an iterative zero-truncated negative-binomial regression to model background somatic mutations
The core method in the frame is an iterative zero-truncated negative-binomial regression (ITER), which estimates baseline somatic mutation counts of genes on the genome. The difference between the observed mutation counts and the estimated counts of a gene measures the excess of somatic mutations at a gene in a cancer. The assumption is that a gene with significant excess of somatic mutations may confer selective growth advantage in cancer as a driver gene (6). Denote the mutant allele counts at a variant j in a background gene i as 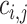 and the total alleles of
and the total alleles of 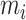 variants in this gene is, yi. We observed a large over dispersion in the distribution of yi (See examples in Supplementary Figure S1). Therefore, we assume yi follows an NB distribution:
variants in this gene is, yi. We observed a large over dispersion in the distribution of yi (See examples in Supplementary Figure S1). Therefore, we assume yi follows an NB distribution:
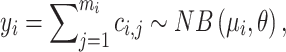 |
where 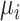 is the expected number of mutations and
is the expected number of mutations and 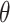 is a dispersion parameter. The probability mass function (PMF) is
is a dispersion parameter. The probability mass function (PMF) is 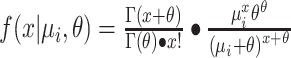 , where
, where 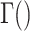 is the gamma function and x = 0,1,2, ….
is the gamma function and x = 0,1,2, ….
As somatic mutation may be a rare event, many genes have no somatic mutations in a sample of typical size. The proportions of genes without somatic mutations in real data are then much larger than the probability of the NB model at x = 0 (See examples in Supplementary Figure S2). Therefore, we proposed to use a zero-truncated negative binomial (ZTNB) distribution to model the mutant allele counts of background gene i. We showed the ZTNB model fitted the mutations counts better than the NB and other alternative models in the ‘Results’ section (Table 2). The PMF of ZTNB is:
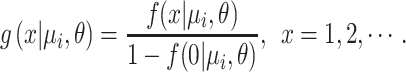 |
Table 2.
Akaike information criterion (AIC) of the various regression models
| Poisson | Zt- Poisson | Negative Binomial | Zt-Negative Binomial | |
|---|---|---|---|---|
| BLCA | 50439.28 | 49991.30 | 42766.17 | 40965.04 |
| BRCA | 93502.18 | 93343.70 | 60491.40 | 58701.48 |
| COAD | 55069.59 | 54675.87 | 40495.83 | 38459.15 |
| HNSC | 91759.62 | 91608.29 | 60737.90 | 59131.56 |
| KIRC | 49696.06 | 49152.82 | 38404.63 | 36484.36 |
| LUAD | 140194.62 | 140168.95 | 79081.09 | 78048.05 |
| LUSC | 79647.23 | 79409.43 | 55650.58 | 54129.60 |
| OV | 53288.73 | 52812.71 | 40753.37 | 38623.84 |
| SKCM | 126186.28 | 126133.21 | 70968.39 | 69622.20 |
| STAD | 63094.87 | 62754.31 | 48865.94 | 47114.14 |
| UCEC | 66024.20 | 65665.04 | 48584.07 | 46526.46 |
Note: The glm() function in R was used to fit the generalized linear model (GLM) of Poisson distribution. The glm.nb() function in the R package of MASS was used to fit the GLM of NB distribution. The other two models were fitted by the R package of countreg. The full names of cancers are in Supplementary Table S1.
Based on the ZTNB, we constructed a generalized linear regression model to estimate mutant allele of non-synonymous and splicing variants in a gene i by multiple covariables:
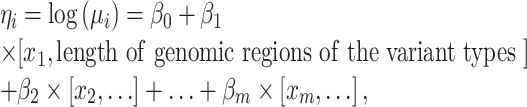 |
where 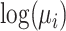 is the link function and the
is the link function and the 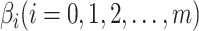 are the regression coefficients.
are the regression coefficients.
The ZTNB regression is proposed to model somatic mutations in background or passenger genes. Besides the length of genomic regions, predictors in the regression model are flexible and depend on mutation types. Previous studies showed somatic mutation rates tend to be higher in genes with low expression levels, repressed chromatin, DNA modification and late replication times in cancer cells (6,15–17). Therefore, the four types of predictors are adopted in the prediction models. Besides, copy number variations (CNVs) occur in cancer cells frequently (18). It may be necessary to recalibrate the background mutation rate by the CNVs of background genes although the CNVs of driver genes may also contribute to cancers (18). In addition, assuming the synonymous and non-synonymous mutations in the same passenger genes have similar mutation rate for passenger genes, we also consider the number of synonymous mutations in local cancer samples as a predictor. Besides, we add gene's constraint scores for non-synonymous mutations in natural populations as a predictor (19), which assumes a gene having higher mutation potential in germline cells tends to have higher mutation potential in somatic cells as well. There are in total 10 predictors in the present paper, half of which are cancer-specific (See details in Supplementary Table S2). All the 10 predictors are used to estimate 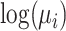 . This model is also open for other types covariables as long as they can improve the prediction accuracy. Statistically insignificant predictors will have little contribution to the estimation.
. This model is also open for other types covariables as long as they can improve the prediction accuracy. Statistically insignificant predictors will have little contribution to the estimation.
The coefficients of the predictors are estimated by maximum likelihood with a quasi-Newton method. With the estimated coefficients, the deviance residues are calculated and standardized as 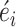 (See the detailed methods in the supplementary notes). A large
(See the detailed methods in the supplementary notes). A large 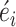 means the observed number of somatic mutations is larger than the expected mutation counts under null hypothesis. The corresponding P-value is then approximated by,
means the observed number of somatic mutations is larger than the expected mutation counts under null hypothesis. The corresponding P-value is then approximated by,
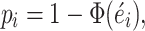 |
where 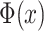 is the cumulative distribution function of the standard normal distribution. We demonstrate the P-values follow uniform distribution in real and simulated data approximately [Figures 2 and 3A; Supplementary Figures S3 and S4].
is the cumulative distribution function of the standard normal distribution. We demonstrate the P-values follow uniform distribution in real and simulated data approximately [Figures 2 and 3A; Supplementary Figures S3 and S4].
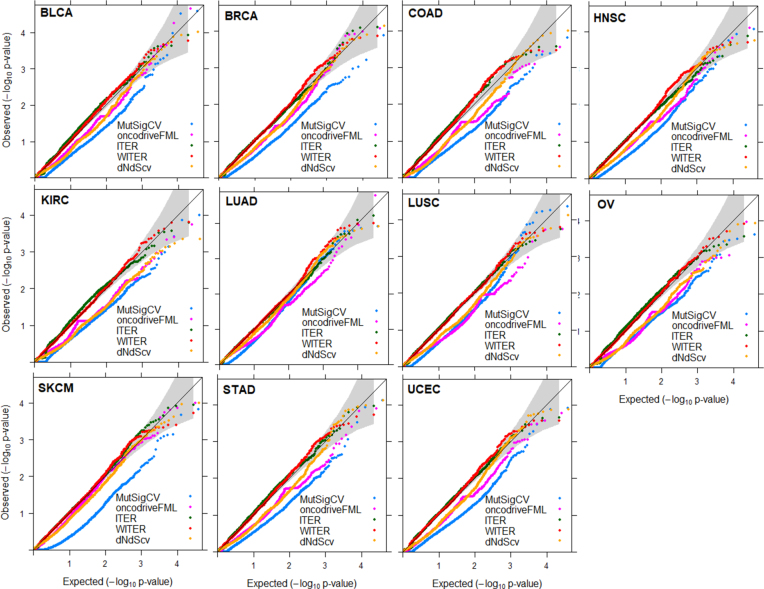
Figure 2.
QQ plot of background gene P-values produced by four methods in 11 cancers. The P-values less than a cutoff according to FDR q 0.05 were excluded. Among the 34 collected cancers, 11 cancers have 25 000 variants with somatic mutations in the datasets and are used for the comparison.
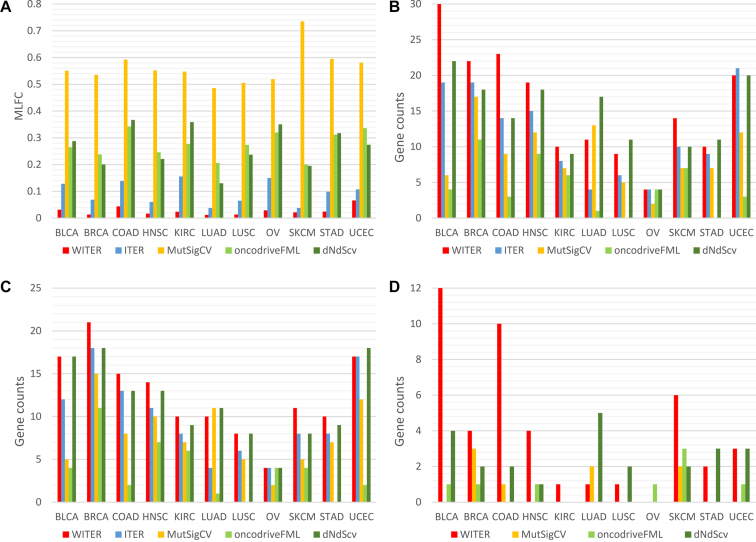
Figure 3.
Performance comparison of different methods for detecting cancer driver mutation in 11 cancers. (A) The MLFC of four methods; (B) the number of significant genes; (C) cancer consensus significant genes; (D) the number of unique significant genes. The P-values less than a cutoff according to FDR q 0.05 are excluded. The full names of cancers are in Supplementary Table S1.
Note the above ZTNB-based regression model estimates somatic non-synonymous mutations among passenger genes. The mutations in true driver genes, although they may be only a small fraction of the entire genome, might harm the estimation to some extent. In order to reduce distortion from driver genes, we further propose to perform the regression under an iterative procedure:
Step 1: fit the ZTNB regression model and calculate P-vales for all genes.
Step 2: exclude significant genes by a loose P-value cutoff corresponding to Benjamini–Hochberg (BH) false discovery rate (FDR) q value ≤ 0.1.
Step 3: fit ZTNB regression model with the retained genes and calculate P-values.
Step 4: repeat Step 2 and 3 until there is no extra significant genes according to the same P-value cutoff.
The fitted ZTNB regression model in the last iteration is closest to the null hypothesis model and is then used to re-calculate deviance residuals and P-values of all genes (including the ones excluded during iteration).
Tier II: a weighting scheme to prioritize variants of high somatic mutation potential in cancer samples
As the mutations are not functionally equal, we further extend ITER to WITER which integrates prior weights of mutations to boost power. Assume a variant j of gene i has a score, 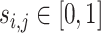 , indicating its cancer driver potential. We bin
, indicating its cancer driver potential. We bin 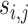 as an integer score, wi,i, by the ceiling function of
as an integer score, wi,i, by the ceiling function of 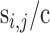 , i.e.,
, i.e., 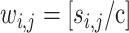 , where
, where 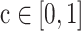 . The integer scores are then used as prior weights for the mutation. The weighted mutation allele count is:
. The integer scores are then used as prior weights for the mutation. The weighted mutation allele count is:
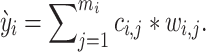 |
We also assume the weighted counts 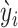 approximately follow a ZTNB distribution:
approximately follow a ZTNB distribution:
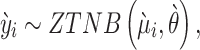 |
where 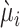 is the expected weighted count of mutations and
is the expected weighted count of mutations and 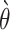 is a dispersion parameter of the ZTNB distribution. The ITER is a special case of WITER when
is a dispersion parameter of the ZTNB distribution. The ITER is a special case of WITER when 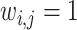 for all variants. After replacement of original counts (yi) with weighted counts (
for all variants. After replacement of original counts (yi) with weighted counts (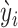 ), the above iterative ZTNB regression procedure is carried out to test whether a gene has excess of weighted mutation alleles. The bin width c is empirically determined by an automated exploration procedure in 100 values from 0.01 to 1.00 with an interval of 0.01. An optimal c is defined as the value minimizes the departure of P-values from uniform distribution measured by the mean absolute log2 fold change (MLFC) (11) and maximizes number of significant genes (FDR q ≤ 0.05), according to a balanced ranking.
), the above iterative ZTNB regression procedure is carried out to test whether a gene has excess of weighted mutation alleles. The bin width c is empirically determined by an automated exploration procedure in 100 values from 0.01 to 1.00 with an interval of 0.01. An optimal c is defined as the value minimizes the departure of P-values from uniform distribution measured by the mean absolute log2 fold change (MLFC) (11) and maximizes number of significant genes (FDR q ≤ 0.05), according to a balanced ranking.
In the present study, we built a model to predict cancer driver potential of variants, by a random forest model trained with a large cancer somatic mutation database, COSMIC (V83). (See details in the Supplementary Notes). One can also resort to other approaches to produce the prior weights.
Tier III: a strategy of integrating independent reference samples to stabilize the regression model for small samples
When the sample size is small, it is difficult to build a stable regression model. Note that the key idea of ITER and WITER is to build a prediction model for background passenger genes. When the mutation rates of passenger genes of two cancers are similar, it may be workable to integrate background genes of one cancer into another cancer. So we propose a strategy to integrate somatic mutations from reference sample for constructing a stable ITER or WITER model in small samples. This is carried out at two stages.
The above ITER or WITER is used to produce P-values for excess of somatic mutations at genes in a reference sample which have enough variants. Genes with P-values less than a very loose cutoff, say FDR q 0.3, are then excluded.
The somatic mutations of retained genes are integrated with the local small sample and input into ITER or WITER to build a new model. The excess of somatic mutations and corresponding P-values at genes are calculated based on the new model.
Curation of cancer-specific predictors of somatic mutations
We collected four types of cancer-specific predictors for somatic mutations, CNV, gene expression, DNA methylation and chromatin accessibility by ATAC-Seq. All data were produced from TCGA cohorts and the pre-processed data were downloaded from https://xenabrowser.net. The download links and processing methods of the data are described in the Supplementary Notes.
Performance comparison with alternative tools
There have been multiple tools for detecting cancer-driver genes (2). According to an evaluation study (11), three tools (MutSigCV (6), OncodriveFML (9) and 20/20plus (11)) having relatively better performance were chosen for comparisons in the present study. We compared their P-value distributions and number of significant genes to ITER and WITER. As MutSigCV and OncodriveFML were also developed under an unsupervised strategy, we chose them as the main comparison targets. The 20/20plus belongs to a supervised strategy which may be more suitable for known cancer-driver genes. To be fair, we only used it as supplementary comparison. In addition, a recent tool dNdScv (12) which used NB regression to model synonymous mutations for cancer-driver gene estimation was also considered for the comparison. The details of the usage of the alternative methods are described in Supplementary Notes.
Evaluation metrics in the performance comparison
We adopted four evaluation metrics for performance comparison, observed versus theoretical P-values, number of significant genes, the number and percentage of significant genes registered in Cancer Gene Census (CGC) (20). The former three were also major metrics in an evaluation framework of cancer driver gene prediction method (11). The CGC dataset contained 699 manually curated cancer genes. The departure of P-values from uniform distribution was measured by MLFC (11). When there is no departure, the MLFC is zero. An approximately valid statistical test should lead to a MLFC close to zero in background (or passage) genes. MLFC greater than 0.3 indicates serious deflation or inflation problem (11). We also used the distribution of Quantile-Quantile (QQ) plot to examine the distribution of P-values, particularly that of the small P-values. The Benjamini–Hochberg procedure was used to control false discovery rate (q≤0.05) for reporting significant genes.
Dataset of somatic mutations
We partitioned a curated full somatic mutation dataset by Tokheim et al., (11) into 34 sub-datasets according to the cancer types (See Supplementary Table S1). Eleven cancer types contained 2,800 or more variants (See the full list in Supplementary Table S1). These cancer samples were called relatively larger dataset throughout the paper and used for the method comparison. Their sample sizes ranged from 142 to 1093. The ratios of variant number to sample size in the 11 cancers ranged from 50 to 327. The remaining 23 cancers with a smaller number of variants were only used in the application analysis. The names, variant numbers and sample sizes of all the cancers can be seen in Supplementary Table S1.
In silico validation by PubMed search
We used PubMed search function to coarsely validate the implication of detected significant genes to the corresponding cancers. The underlying assumption is that the papers co-mentioning a gene and a cancer name in the title or abstract are likely to implicate the relatedness between the gene and the cancer. The more hit papers, the more likely the gene is related to the cancer. This is a quick in-silico validation although it may be rough. We employed the web application programming interfaces (APIs) of PubMed to execute the search. The search link was, http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=‘DiseaseNames (inlcuding homonymies)’[tiab]%29+AND+‘GeneSymbol (including RefSeq mRNA IDs)’ [tiab]. The search terms of each cancer types are in Supplementary Table S1. The query engine responded PubMed IDs and relevant data of the papers, if available.
RESULTS
Features correlated with somatic mutations of genes in cancer samples
We first investigated association of the 10 explanatory features (or predictors) with somatic mutation counts under the ZTNB regression in 11 cancers (See coefficients and P-values in Table 1). While confirming previous findings that somatic mutations tend to be more frequent in genes with low expression levels, repressed chromatin, and late replication times in most cancer (6,15–17), there were also four other interesting patterns. First, for gene expression and chromatin state, it seemed the cancer non-specific features were more significantly correlated with the somatic mutations than the cancer-specific ones in most cancers. For example, the averaged expression level in cancer cell lines had much more significant P-values in 10 out of the 11 cancers than the gene expression level in the matched cancer tissues. The chromatin state assayed by HiC from the K562 cell line was much more significant than that assayed by cancer-matched ATAC-Seq in 10 out of the 11 cancers. The absolute coefficient values of the former were also larger than that of the latter in the 10 cancers. Note all the feature values were standardized to make the coefficients comparable. Second, the significance level of most features varied from cancers to cancers. This was particularly true for three features, CNV, methylation and constraint score. For instance, CNV had a very significant P-value, 9.71E-20, in LUAD while its P-value was only 0.16 in BLCA. For methylation, its smallest P-value occurred in KIRC (P = 1.57E-9) while it had large P-values (>0.3) in four cancers. Third, besides the varied significance level, some features’ association directions were also different in different cancer types. The constraint score had positive significant coefficients in BRCA, KIRC and UCEC while it had negative significant coefficients in LUAD, LUSC and SKCM. While genes with higher CNV tended to have more somatic mutations in four cancers (BRCA, HNSC, LUAD and LUSC), the tendency got reversed in KIRC. The underlying mechanisms are unclear. Fourth, five cancer non-specific features (exon length, number of synonymous mutations, replication timing and cell line expression levels and HiC) had extremely significant P-values for all cancers. These features dominated the prediction performance in the ZTNB model. The dominant impact of these cancer non-specific features makes the proposed method independent of cancer-specific resources and thus more feasible in practice.
Table 1.
The significance level of covariates in 11 cancer datasets
| Predictors | BLCA | BRCA | COAD | HNSC | KIRC | LUAD | LUSC | OV | SKCM | STAD | UCEC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RegionLength | 0.23 (0) | 0.27 (0) | 0.23 (0) | 0.27 (0) | 0.21 (0) | 0.29 (0) | 0.26 (0) | 0.21 (0) | 0.29 (0) | 0.24 (0) | 0.24 (0) |
| SynonymousVariant | 0.31 (1.37E-247) | 0.43 (0) | 0.35 (3.65E-225) | 0.47 (0) | 0.31 (1.13E-264) | 0.65 (0) | 0.53 (0) | 0.33 (8.95E-230) | 0.73 (0) | 0.43 (0) | 0.34 (7.49E-265) |
| TissueExpression | 0.04 (7.09E-05) | 0.06 (1.92E-11) | -0.08 (6.78E-09) | -0.05 (8.97E-07) | 0.07 (4.02E-11) | -0.21 (1.16E-99) | -0.17 (1.42E-52) | -0.02 (0.10) | -0.36 (6.06E-166) | -0.09 (2.35E-14) | 0.07 (4.16E-13) |
| CellLineExpression | -0.07 (2.52E-14) | -0.12 (4.12E-40) | -0.20 (2.94E-79) | -0.21 (2.49E-137) | -0.04 (1.15E-05) | -0.32 (0) | -0.28 (3.95E-235) | -0.08 (9.26E-17) | -0.28 (2.97E-206) | -0.23 (1.43E-154) | -0.11 (4.79E-33) |
| ReplicationTiming | 0.10 (9.00E-19) | 0.16 (5.05E-56) | 0.25 (1.90E-90) | 0.28 (1.89E-172) | 0.04 (1.37E-04) | 0.43 (0) | 0.37 (2.63E-295) | 0.13 (1.99E-29) | 0.39 (2.92E-283) | 0.30 (1.51E-177) | 0.12 (6.69E-26) |
| CNV | 0.01 (0.16) | 0.02 (1.95E-04) | -5.25E-03 (0.30) | 0.04 (8.51E-10) | -0.04 (3.44E-14) | 0.07 (9.71E-20) | 0.07 (7.16E-18) | -8.85E-03 (0.48) | NA | 0.02 (1.35E-03) | 9.71E-03 (0.04) |
| HiC | -0.08 (7.05E-18) | -0.13 (5.99E-43) | -0.19 (4.53E-59) | -0.23 (1.16E-127) | -0.02 (0.03) | -0.34 (0) | -0.30 (3.16E-213) | -0.10 (1.47E-20) | -0.30 (2.05E-233) | -0.24 (3.87E-129) | -0.07 (1.42E-14) |
| ConstraintScore | 0.02 (0.02) | 0.05 (5.10E-11) | 0.02 (0.02) | -8.64E-03 (0.26) | 0.04 (2.63E-06) | -0.05 (1.23E-12) | -0.07 (8.70E-17) | 9.34E-03 (0.31) | -0.09 (6.37E-31) | -0.01 (0.07) | 0.11 (5.25E-39) |
| ATACSeq | -9.23E-03 (0.02) | -0.02 (2.66E-11) | -0.02 (1.56E-06) | -0.03 (1.27E-13) | 1.65E-03 (0.67) | -0.06 (7.49E-47) | -0.04 (1.59E-21) | NA | -0.05 (2.13E-24) | -0.03 (1.67E-11) | -5.32E-03 (0.13) |
| Methylation | -2.27E-04 (8.63E-03) | -3.35E-04 (1.08E-06) | 4.04E-05 (0.67) | 2.74E-05 (0.73) | -4.26E-04 (1.57E-09) | 2.55E-04 (0.02) | -6.81E-05 (0.34) | -3.05E-04 (7.06E-04) | -1.83E-04 (2.49E-03) | 9.46E-05 (0.41) | -2.14E-04 (0.01) |
Note: The coefficients are derived under the ZTNB model. The predictors are centered at median and scaled by median absolute deviation for standardization. The values in the brackets are P-values for the significance of the coefficients calculated by Wald test. The full names of cancers are in Supplementary Table S1.
Distributions of P-values and fitness of the ZTNB model for background mutation genes
We then investigated the type 1 errors of WITER and ITER according to the distribution of P-values. When the overall deviation from uniform distribution was measured as the MLFC for insignificant P-values (FDR q>0.05) (11), the WITER had very small MLFC (<0.066) in all the 11 tested cancers (Figure 3A). The smaller MLFC, the closer to uniform distribution. We also chose two alternative approaches [MusigCV (6) and OncodriveFML (9)] which achieved the best performance among seven widely used unsupervised tools (11) and one recent popular method [dNdScv (12)] for the comparison. Among the five compared methods, ITER had the second smallest MLFC. MusigCV (6) had the largest MLFC values, in which most values were over than 0.5. In terms of the MLFC, the OncodriveFML was better than MusigCV but worse than ITER. dNdScv had similar MLFC with OncodriveFML. As shown in the QQ plot (Figure 2), the main problem in MusigCV, OncodriveFML and dNdScv is their deflated P-values although the deflation at small P-values genes is mitigated. Therefore, the proposed ZTNB model produces relatively more valid uniform distribution of P-values, which is a challenge in most existing approaches (11).
We also compared the goodness-of-fit of the ZTNB model with three widely-used alternative models for mutation counts of genes (Table 2). It turned out the ZTNB model had the smallest Akaike information criterion (AIC) values in all the 11 cancers, suggesting the best goodness of fit among the four models (Table 2). The NB distribution was the second-best model although its AIC values were over 1000 larger than that of the ZTNB model in all cancers. This comparison suggested the zero-truncation effectively enhanced the fitting. The Poisson, either in the original version or the zero-truncated version, fitted data poorly. Their AIC values are much larger than that of the ZTNB. This comparison suggested the usage of NB distribution modeled the overdispersion better than the Poisson distribution.
WITER detected more significant genes in 11 cancers
We then compared the number of significant genes detected by the five unsupervised approaches (WITER, ITER, MutSigCV, OncodriveFML and dNdScv) in the 11 cancer datasets. Instead of following the conventional ‘pancancer’ (all cancers) evaluation strategy (11), we made the comparison for individual cancers, a more challenging scenario because of smaller sample sizes. The significant genes are determined according to FDR q-value 0.05. Among four approaches (WITER, ITER, MutSigCV and OncodriveFML), WITER and ITER estimated the largest and the second largest number of significant genes in 10 cancers (Figure 3B), respectively. WITER also detected more significant genes than dNdScv and equal number of significant genes in six and two cancers, respectively. dNdScv was ranked in the second place according to the number of significant genes and was followed by ITER. The OncodriveFML detects the minimal number of significant genes in 10 out of the 11 cancers although it also integrates function prediction score CADD as priors (21). Given the correct type 1 errors, the more significant genes by WITER than ITER suggests the integration of prior weights has a potential to improve statistical power. Note that all the subjects in the testing cancer datasets were excluded from the COSMIC database to reduce circular reasoning when building the prior weights for WITER.
A critical next question is that whether the increased number of significant genes by WITER or ITER are true driver genes. As there are almost few true answers in the real data, we adopted the Cancer Gene Census (CGC) list (20) to partly answer this question. Among the four approaches (WITER, ITER, MutSigCV and OncodriveFML), WITER always led to the largest number of CGC significant genes in 10 cancers (Figure 3C). It detected on average 4.5 and 8.7 more CGC significant genes in the 11 cancers than MutSigCV and OncodriveFML, respectively. It also detected on average 0.8 more CGC significant genes in the 11 cancers than dNdScv. dNdScv was still the second-best method according to the number of CGC genes. It detected the largest number of CGC genes in two cancers. Again, ITER detected more CGC significant genes than MutSigCV and OncodriveFML. Moreover, besides the absolute number of CGC significant genes, we also checked their percentage, which might be proportional to the false positive rates to some extent. The percentage varied from cancers to cancers generally (See details in Table 3). Compared to MutSigCV and dNdScv, WITER had higher or equal percentage in 6 out of the 11 cancers. However, the averaged percentages of the five tools in the cancers are comparable generally while MutSigCV and ITER had slightly higher percentage, 90.9% and 88.5%, respectively.
Table 3.
The percentage of the cancer consensus gene in the significant genes by different methods
| WITER | ITER | MutSigCV | oncodriveFML | dNdScv | |
|---|---|---|---|---|---|
| BLCA | 56.7 | 63.2 | 83.3 | 100 | 77.3 |
| BRCA | 95.5 | 94.7 | 88.2 | 100 | 100 |
| COAD | 65.2 | 92.9 | 88.9 | 66.7 | 92.9 |
| HNSC | 73.7 | 73.3 | 83.3 | 77.8 | 72.2 |
| KIRC | 100 | 100 | 100 | 100 | 100 |
| LUAD | 90.9 | 100 | 84.6 | 100 | 64.7 |
| LUSC | 88.9 | 100 | 100 | NA | 72.7 |
| OV | 100 | 100 | 100 | 100 | 100 |
| SKCM | 78.6 | 80 | 71.4 | 57.1 | 80 |
| STAD | 100 | 88.9 | 100 | NA | 81.8 |
| UCEC | 85 | 81 | 100 | 66.7 | 90 |
| Average | 85 | 88.5 | 90.9 | 87.7 | 84.7 |
Note: The full names of cancers are in Supplementary Table S1.
Note that the genes beyond CGC list are not necessarily spurious driver genes although the high percentage and number of CGC genes are a strong sign of higher power. Take two non-CGC genes for examples. The AJUBA gene (P = 7.0E-9 in HNSC) is involved in the regulation of NOTCH/CTNNB1 signaling and is an important driver gene of HNSC (22,23). TLR4 (P = 5.0E-5 in STAD) is an important member of Toll-like receptor (TLR) pathway and mutations in the gene may disrupt innate immune signaling and promote a microenvironment that favors tumorigenesis (24) and it was associated with gastric cancer (25).
Uniquely significant genes by individual approaches
We also investigated genes only significant in one of the compared tools. WITER detected the largest number of uniquely significant genes (FDR q≤0.05) in 7 out of the 11 cancer types. These genes were insignificant by MutSigCV, OncodriveFML and dNdScv and would be ignored (Figure 3D, See details in Supplementary Table S8). It also detected the largest total number of uniquely significant genes, 44, in all the 11 cancers, among which 21 (48%) genes were in the CGC list, (enrichment P < 1E-16, by hypergeometric distribution test in 19 198 protein coding genes). The BLCA had the largest number of uniquely significant genes, 12, by WITER among which four genes were CGC genes. Some genes were well-known driver genes for BLCA, e.g. ERBB2 (26,27). ERBB2 had 10 non-synonymous somatic mutant alleles in the BLCA samples. WITER calculated a P-value 7.5E-5 at this gene. The P-values by MutSigCV, OncodriveFML and dNdScv were 0.08, 0.96 and 0.003, respectively. dNdScv detected the second-largest number of uniquely significant genes, 21, in all cancers among which 16 ( = 76%) genes were in the CGC list. For example, FAT1 was a dNdScv-unique significant and CGC gene in 3 cancers (BLCA, LUSC and UCEC). However, it had a long coding region (13.9 kb) and repressed chromatin state (HiC score −13). After correction by the covariables, WITER produced insignificant P-values 0.30,0.76 and 0.37 in BLCA, LUSC and UCEC, respectively. Therefore, it may be questionable whether FAT1 is a true driver for the three cancers. MutSigCV did not detect unique significant genes in seven cancers. It detected in total eight uniquely significant genes in four cancers among which two ( = 25%) genes were in the CGC list. For example, the gene, FBN2, had 76 non-synonymous or splicing mutant alleles in the LUAD samples and MutSigCV produced a P-value 4.28E-07. However, it had a 9.1 kb coding region, 10 synonymous mutant alleles and late replication timing and low expression in LUAD, WITER produced an insignificant P-value 0.17 for its excess of the adjusted non-synonymous or splicing mutant alleles by the regression model. OncodriveFML also detected in total eight uniquely significant genes in the cancers, among which four (50%) genes were in the CGC gene list. In the comparison, we ignored ITER because all significant genes by ITER were also significant by WITER.
Rescued significant genes in smaller samples
As above CGC genes and literature survey might be biased to known cancer genes, we further designed a sample-size rescue experiment to compare their performance. We investigated how many significant genes detected in a large sample were missed in smaller samples by a tool can be rescued by another tool in the same small samples. We randomly drew six sub-samples of half size from the largest dataset, BRCA and estimated cancer-driver genes by four tools, MutSigCV, OncodriveFML, dNdScv and WITER. As shown in Supplementary Table S6, using half samples, WITER missed only ∼22% genes on average which were significant in the full sample by the same method. The usage of three other tools in the sub-sample rescued <1 of the genes missed by WITER on average. dNdScv missed ∼33% genes on average which were significant in the full sample by the same method. When the same half-size samples were analyzed by WITER, it rescued ∼44% of the missed genes by dNdScv. MutSigCV and OncodriveFML almost rescued no genes missed by dNdScv on average. MutSigCV missed ∼46% genes on average which were significant in the full sample by the same method. When the same half-size samples were analyzed by WITER, it rescued ∼47% of the missed genes by MutSigCV. The usage of dNdScv and OncodriveFML rescued 30% and 4% of genes missed by MutSigCV. OncodriveFML missed ∼39% genes on average which were significant in the full sample by the same method. When the same half-size samples were analyzed by WITER, it rescued ∼65% of the genes missed by OncodriveFML. The usage of MutSigCV and dNdScv rescued ∼19% genes missed by OncodriveFML. The higher proportions of significant genes and rescued genes by WITER in the sub-samples again showed that WITER had enhanced power to detect driver genes that would be missed by alternative methods.
Performance in 23 cancer datasets with relatively small samples
Another important advantage of WITER is its ability to detect cancer-driver genes in small samples with a usage of reference samples. We applied it to other 23 cancers of small samples and variants. We deliberately used two different reference cancers samples (BRCA and SKCM) with low and high-background mutation rates to investigate how WITER was sensitive to the reference samples. The results showed three major patterns. First, the usage of the reference datasets substantially improved the distribution of P-values. According to the QQ plots (Supplementary Figure S4), the P-value distributions of the background genes (FDR q>0.05) with reference samples were very close to the uniform distributions. In contrast, the P-values of the background genes without reference sample were weird and did not follow the uniform distribution. Second, WITER with reference samples detected significant genes even in cancers with very small sample size (See the results in Supplementary Table S7). Among the 18 cancers with one or more significant genes (FDR q≤0.05), two cancers had <60 subjects, i.e. LUSE (n = 30) and DLBCL (n = 56). Third, it seemed the difference in reference samples had small and simple influence on the number of significant genes. A low background mutation rate reference sample (e.g. the BRCA) led to slightly more significant genes than the one (SKCM) of high-background mutation rate for the tested sample. Moreover, we noted that almost all significant genes according to the high background mutation rate reference sample were also significant according to the low background-mutation rate reference sample. Therefore, false positive findings can be easily controlled by using a high background mutation rate reference sample in practice although this may result in false negatives. Anyhow, the overlapping of the significant genes according to the two extreme references was high.
It should be also noted that the additional significant genes according to the low background mutation rate reference sample are not necessarily spurious driver genes. For instance, KIT is a significant driver gene of LAML based on the BRCA reference (P = 5.43E-5) but insignificant based on SKCM reference (P = 8.9E-4). KIT is a well-known driver gene of acute lymphoblastic leukemia (28,29). To reduce possible false positive results rigorously, we still used the results from the conservative reference sample, SKCM, for the subsequent analysis.
Factors influencing power of WITER
We also investigated factors influencing the number of significant genes by WITER, which implies factors affecting its power. In a linear regression model, the sample size was correlated with the number of significant genes, with a coefficient of determination R2, 0.19 (Supplementary Figure S5). According to the fitted model, an estimation of sample size, ∼410, was required to detect 10 significant genes by WITER. Meanwhile, the number of significant genes was also related with the exome-wide background mutation rate, R2 = 0.12 (Supplementary Figure S6). For example, the LUSC had a high exome-wide background mutation rate, 305 mutations per exome. A relatively smaller sample, 175, had led to detection of 9 significant genes in real data. Therefore, when the sample size and number of mutations per sample enter as explanatory variables in a linear regression model, both have significant and positive correlation with the number of significant genes (P = 1.8E-4 at the sample size and P = 1.8E-4 at number of mutations per exome). The coefficient of determination R2 increased to 0.63.
The landscape of driver-genes at 26 cancer types
WITER’s effectiveness in small samples enabled the production of a comprehensive landscape of driver-genes in multiple cancer types. It detected one or more significant driver genes in 26 cancer types, out of which 13 cancer types had more than 5 genes (FDR q<0.05, See details in Figure 4 and the Supplementary Excel File). Three cancers had over 20 significant genes. The cancer with high-background mutation rate, BLCA, had the largest number of significant genes, 30. The cancer with the largest sample, BRCA, had 22 significant genes. COAD also had 23 significant genes. Thirty-five genes were significant simultaneously in two or more cancers. However, the number of overlapped significant genes (Supplementary Table S3) in the cancers were too small to produce sensible cancer clusters. As expected, the famous tumor suppressor gene, TP53, was the commonest significant genes (in 24 cancer types), followed by PIK3CA, PTEN, KRAS, RB1 and NRAS, each of which was estimated as driver genes for six or more cancer types.
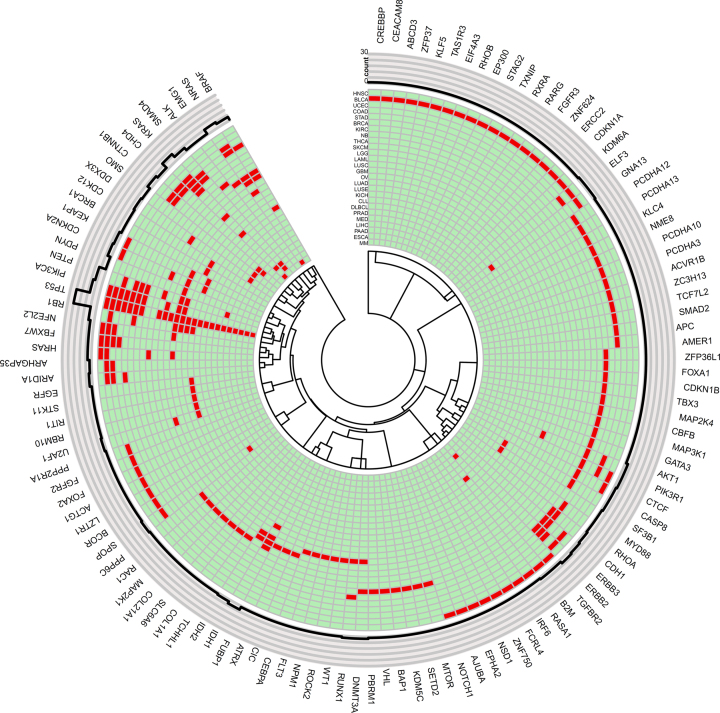
Figure 4.
Circos plot displays 178 significant genes in 26 cancers. Notes: The innermost ring denotes dendrogram of genes. The next ring contains significant genes (marked in red) of corresponding cancers. It is followed by a ring of cancers in which the genes are significant. The outmost ring contains gene symbols. The full names of cancers are in Supplementary Table S1.
Eighty-six genes were significant in only one of the 26 cancers (See details in Supplementary Table S4 and S5). Cancers with more significant genes tended to have more such specifically significant genes, implying high-genetic heterogeneity. In the in silico validation analysis, 61 ( = 71%) out of the 86 genes had one or more hit papers for the corresponding cancer types in the NCBI PubMed database (Supplementary Table S4 and S5). Forty-seven ( = 55%) genes had more than three hit papers. There were 14 genes having even 100 or more hit papers. The strong literature supporting rate suggested the high accuracy of WITER for estimating driver-genes at individual cancers even in small samples. It should be noted that the specifically significant genes of a cancer in the present analysis did not necessarily mean specific driver genes of the cancer. As sample sizes increase, some genes may become significant in other cancer types. However, the specifically significant genes of a cancer may imply genes' relatively higher contribution to the corresponding cancer. For instance, the BRCA had nine uniquely significant genes. Six out of the eight genes had at least 20 hit papers in the NCBI PubMed database, an indication of the strong relationship between the genes and BRCA. Interestingly, although BRCA1 is an important tumor suppressor gene for familiar BRCA, it was uniquely significant in OV. While BRCA1 had an insignificant P-value (P = 2.17E-4) in the BRCA dataset, it achieved a P-value 3.36E-10 in the OV dataset. BRCA1 also had 100 hit papers for OV in the NCBI PubMed database. Probably, for sporadic cases, BRCA1 was a stronger driver for OV than BRCA.
There were 39 specifically significant genes having only three or fewer hit papers in NCBI PubMed database. The BLCA had 12 such genes. RARG, for example, encodes a retinoic acid receptor that belongs to the nuclear hormone receptor family. This gene was linked to leukemia repeatedly by multiple studies (e.g. (30,31)). This is the first time that it is suggested as a driver gene of BLCA. ZNF624 is another interesting example gene, which encodes a zinc finger protein 624. It had 19 non-synonymous mutations and a high functional gene score 42 (P = 8.64E-10 by WITER). This gene has not been well-studied yet. There were few papers mentioning this gene in the PubMed database. The COAD had eight such genes. The ZC3H13, a canonical CCCH zinc finger protein, is a promising colorectal cancer specific candidate driver gene. A very recently study suggested it may suppress colorectal cancer proliferation and invasion via inactivating Ras-ERK signaling (32). The HNSC had four such genes. Among the four genes, ZNF750 is also a gene encoding a zinc finger protein. The hit paper reported a downregulation of ZNF750 by gene fusions in human papillomavirus (HPV) positive HNSC samples (33). ZNF750 has been suggested to be associated with esophageal squamous cell carcinoma (34), which indirectly supports its cancer driver potential to HNSC. Both SKCM and UCEC had 3 such genes. The specifically significant gene of SKCM, SLC6A6, encodes protein transports taurine and beta-alanine. A recent study found elevated SLC6A6 expression drove tumorigenesis and affected clinical outcomes in gastric cancer (35). However, no studies linked this gene to SKCM. The specifically significant gene of UCEC, LZTR1, was reported in multiple papers in Science journal very recently (36–38). These studies suggested LZTR1 acted as a conserved regulator of RAS ubiquitination and MAPK pathway activation; and mutations in this gene may dysregulate RAS ubiquitination and lead to impaired protein degradation of oncoproteins (e.g., Ras GTPase RIT1). The significant mutation burden in UCEC added a direct evidence on its contribution to cancers. Both KIRC and LGG had 2 specific genes with few hit papers. A specifically significant gene, KDM5C, of KIRC has been implicated to other cancers. For instance, Xu L et al. suggested that overexpression of KDM5C can enhance the proliferation and invasion of gastric cancer cells by decreasing p54 expression (39). A specifically significant gene of LGG, CIC, encodes capicua transcriptional repressor. A recent study suggested that CIC may work with ATXN1-ATXN1L as a potent regulator of the cell cycle related with development of cancers (40). Four cancers had only 1 specific gene with few hit papers. Although no publication directly linked the genes to corresponding cancers, a search in PubMed database showed all the four genes have been implicated to other cancers. Given the literature evidences, it is very unlikely that these genes had significant mutation burden for the corresponding cancers just by chance and they were potential driver genes for these cancers.
Among the above 39 uniquely significant genes, 24 also had mutation burden P-values by MutSigCV, OncodriveFML and dNdScv. Most of the genes had insignificant P-values by MutSigCV and OncodriveFML in the corresponding cancers(See details in Supplementary Tables S8). The significance level by dNdScv was higher than MutSigCV and OncodriveFML at most of the genes but was lower than WITER and ITER. For instance, WITER led to a significant P-value 2.95E-06 at the ZNF750 for HNSC, which was down regulated by gene fusions in HPV positive HNSC samples (33). The P-values by MutSigCV, OncodriveFML and dNdScv were only suggestively significant, 0.0044, 0.0011 and 4.69E-04. For the remaining 15 genes, the driver gene analysis in the corresponding cancers was not performed by the three alternative tools because the sizes of cancer samples were too small to generate a reliable estimation. WITER had the unique advantage of integrating reference samples to produce stable estimation in small local samples.
DISCUSSION
Accurately modelling counts of somatic mutations at background genes has long been a fundamental technique challenge in genomic characterization of cancer-driver genes (2,11). The proposed approach, WITER, has four technical advances to address this issue. The first one is the advanced model, ZTNB regression, that fits the number of somatic mutations in background genes better. In samples of a typic size, one often sees an inflation of zero mutation genes (Supplementary Figure S2), which makes it difficult to fit the distribution of genomic counts. The ZTNB distribution properly addresses the zero-inflation issue. This is demonstrated by the results that ZTNB model always achieved the minimal AIC among four alternative models (Table 2) and statistically valid P-values distribution (Figure 2 and Supplementary Figure S4). The valid P-values distribution solves the common drawback in alternative methods that resort to time-consuming simulation or permutation for statistical inference. The other three technical advances include iterative regression, integrating reference samples and imposing prior weights at variants. The iterative regression relieves distortion of driver genes to the background baseline estimation so that the residues of mutation counts at driver genes are not shrunk. Its allowance of integrating reference samples ensures a stable resulting model and thus a valid estimation in small sample. We find that the number of significant genes detected by WITER is generally not sensitive to the reference samples in most cancers (Supplementary Table S7). We showed that imposing prior weights at variants enables detection of more cancer consensus driver genes in all the tested datasets (Figure 3). These four technical advances together determine the enhanced power of WITER for detecting more cancer-driver genes than alternative methods while effectively controlling statistical type 1 error.
The proposed approach is different from existing NB models for estimating cancer-driver genes (12–14). First, WITER is characterized with the four technique innovations as mentioned above. The NB model is merely the basic framework. It was these innovations that led to an improved driver-gene estimation in moderate and small samples. Second, the modelling targets of WITER and the existing methods are also different. WITER used NB distribution to model the distribution of mutation counts at non-synonymous variants of background mutations. It then directly uses non-synonymous mutation burden to estimate driver potential of genes. What these existing methods modeled with the NB distribution was the synonymous variants of genes. With the estimated synonymous mutation counts, the dN/dS ratio at genes was then estimated. The dN/dS = 1 was then tested by likelihood ratio approach for cancer-driver genes (12). So, these existing methods had more parameters in the analysis, which may be less effective in moderate or small samples. Therefore, WITER produced more valid P-value distribution and detected more significant cancer-driver genes.
The ZTNB model and three-tier framework are generic and can be extended to other types of mutations. In the present paper, we focus on the non-synonymous and splicing variants (including SNVs and InDels). This is because the availability of abundant data (e.g. exome sequencing data) in the public domains greatly facilitates the methodological validation. It is true that non-coding variants (usually discovered by whole genome sequencing) also contribute to development of cancers (41). However, the public resources of whole-genome sequencing are much fewer than that of the exome sequencing. Theoretically, one can replace the non-synonymous variants with non-coding variants like upstream or downstream variants while the predictors are replaced correspondingly. Nik-Zainal et al. suggested the feasibility of NB regression to estimate cancer driver genes with non-coding mutations (14). Similar to coding variants, the four technical innovations on NB regression may also have the potential to enhance power for non-coding variants. As the cost of high-coverage whole genome sequencing is decreasing, more data will be available for an evaluation of the method in non-coding variants in the future.
A slightly unexpecting finding is that some genomic features (e.g. gene expression and chromatin accessibility) from cancer non-specific cell lines are generally more relevant to somatic mutations than that from cancer-matched primary tumors. Lawrence et al. stated that matched normal tissues led to similar results as the cancer non-specific cell lines for the estimation of correlation between cancer somatic mutation frequency and gene expression level in independent normal tissues (6).The present study uses matched cancer tissues instead of matched normal tissues, which may be the cause of the difference. Polak et al. suggested cell-of-origin chromatin features (including chromatin accessibility) were stronger determinants of cancer mutation profiles of the entire genome than chromatin features of matched cancer cell lines (15). In the present study, our finding is in chromatin accessibility in coding regions, which is not equivalent to Polak et al. (15). Although the underlying causes of the differences are subject to more and deeper study in the future, the strong correlation between somatic mutations and the cancer non-specific predictors makes WITER flexible in practice.
We compared the proposed method with three widely used and well-performed approaches (11), all of which belong to the unsupervised category. Another category of methods is the supervised approaches for detecting cancer driver genes. According to Tokheim et al. (11), the supervised method 20/20plus outperformed the best unsupervised methods at that time (including MutSigCV and OncodriveFML) in terms of P-value distributions and the number of significant genes. However, a supervised strategy has learning bias toward the training samples in nature (42). If the training sample is not representative of all diseased sample, the trained model may have low power in new samples. This would be particularly true for cancers because of their high-genetic heterogeneity (5). Second, the 20/20plus also used many common genomic features of a gene (e.g. evolutionary conservation, predicted functional impact of variants, and gene interaction network connectivity) in the prediction (11). Although the usage of common genomic features will add information to prioritize common cancer-driver genes, it also runs the risk of diluting the information in local sample for identifying unique cancer driver genes, which would be important for a precision diagnosis and treatment of a specific cancer. At last, the 20/20plus resorts to time-consuming permutation procedure to generate P-values for statistical test. In contrast, the WITER and ITER are much faster than 20/20plus because it calculates P-values analytically. Nevertheless, we also made additional comparisons between WITER and 20/20plus approach in the 11 cancers. The P-value distributions of background genes produced by both methods are similar and approximately follow uniform distribution. (See QQ plots in Supplementary Figure S7). Although WITER detected more significant and cancer-consensus genes than 20/20plus in only 2 cancers (See details in Supplementary Figure S8), it rescued more missed genes in the half sample size experiment (See details in Supplementary Table S6).
WITER used a new way of integrating prior weights into the NB distributions. The rationale is to amplify the mutations with high driver potential so that the true driver gene can achieve higher statistical significance. This is similar (though not equal) to a scenario in which genes with higher driver potential mutations are stratified and are obtained a less stringent multiple testing threshold to identify more driver genes. The effectiveness of the prior-weight usage was demonstrated by the controlled type 1 errors (see non-inflated P-value in Figures 2, 3A and Supplementary Figure S4) and more significant genes (Figure 3B–D). However, our proposed integration strategy does not exclude alternative ways of using prior weights. Alternatively, the Bayesian approaches (43), stratified FDR or independent hypothesis weighting approach (44) may be also potentially workable strategies. However, stratified FDR has the disadvantage that the weights must take a small number of discrete values (to ensure large strata), whereas our proposed method uses continuously distributed weights. The independent hypothesis weighting approach (44) adjusts for multiple testing at the same time as introducing weights. This means that P-values are differentially adjusted toward 1, so that simple diagnostics such as Q-Q plot would no longer be meaningful. In contrast, for our method, the adjusted P-values can be checked for consistency with a uniform distribution by QQ-plot. Moreover, a tricky point is that the available weights are often at mutation level while the hypothesis are carried out at gene level. How the mutation level weights can be optimally used for gene level hypothesis tests under the conventical integration framework would be an interesting future work.
A limitation of the present study is that many true cancer-driver genes are generally unraveled for most cancers. Most results suggesting the enhanced power of WITER in the paper are indirect. We did not simulate datasets to quantify the increased power of WITER relative to the alternative methods by artificially setting ‘true’ driver genes because there are many unknown factors shaping the landscape of somatic mutations. An artificial model in simulation is often too subjective to represent the reality. The usage of real data and in silico validation is widely adopted and effective in methodological studies (11). In the present study we showed WITER detected more significant genes than alternative methods in most tested cancers. High coverages of these genes in CGC list and co-mentioned with the corresponding cancers in titles or abstracts of more than three papers in PubMed database were shown. The enhanced power of WITER was further confirmed with more rescued genes compared to alternative methods in half-size samples. These results strongly suggested that there was a high true discovery rate in the significant genes by WITER and it detected more genuine cancer-driver genes than alternative methods.
ITER and WITER are designed for individual cancer types, which is usually a more challenging scenario than for Pancancer due to much smaller sample sizes. ITER’s and WITER’s basic assumption is that a small fraction of genes are driver genes and their regression models are built on background genes. When data of all cancers are merged into a large Pancancer dataset, the proportion of cancer-driver genes will increase substantially and the trained model will shift from background genes toward driver genes. The shifting may diminish the difference between observed mutation counts and the estimated background baseline mutations counts at cancer-driver genes, which may reduce the power subsequently. To avoid miss-leading, we did not estimate driver genes in a Pancancer dataset. For the large Pancancer dataset, many existing tools have already had a good statistical performance (11).
Applying the powerful approach, WITER, we generated a landscape of driver genes in 26 cancers. Its unique advantage of integrating reference sample enables detection of driver genes in samples of size as small as 30 although more driver genes will be detected in larger samples. The analysis revealed many genes which are common driver genes for multiple cancers. Most majority of the genes have many literature supports. The common driver genes may be effective drug targets for treatment of cancers. Meanwhile, there are also a lot of significant genes which are unique for a single cancer. Some of these genes may be specific major driver genes of the corresponding cancers although increased sample sizes of other cancers may change the results. The cancer-specific driver genes are potentially effective for a precision diagnosis and treatment of corresponding tumors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tokheim and colleagues for sharing the high-quality curated somatic mutations in 32 cancers from multiple resources. We gratefully acknowledge TCGA consortium for the multi-omics data of cancers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2018YFC0910500]; National Natural Science Foundation of China [31771401]; Science and Technology Program of Guangzhou [201803010116]; Hong Kong Health and Medical Research Fund [02132236]; Hong Kong General Research Fund [17124017, 17121414, TRS T12C-714/14-R]. Funding for open access charge: National Key R&D Program of China [2018YFC0910500]; National Natural Science Foundation of China [31771401].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bunting S.F., Nussenzweig A.. End-joining, translocations and cancer. Nat. Rev. Cancer. 2013; 13:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bailey M.H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M.C., Kim J., Reardon B. et al.. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018; 173:371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A. Jr, Kinzler K.W.. Cancer genome landscapes. Science. 2013; 339:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim S. New and emerging factors in tumorigenesis: an overview. Cancer Manag. Res. 2015; 7:225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cyll K., Ersvaer E., Vlatkovic L., Pradhan M., Kildal W., Avranden Kjaer M., Kleppe A., Hveem T.S., Carlsen B., Gill S. et al.. Tumour heterogeneity poses a significant challenge to cancer biomarker research. Br. J. Cancer. 2017; 117:367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A. et al.. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013; 499:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dees N.D., Zhang Q., Kandoth C., Wendl M.C., Schierding W., Koboldt D.C., Mooney T.B., Callaway M.B., Dooling D., Mardis E.R. et al.. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012; 22:1589–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gonzalez-Perez A., Lopez-Bigas N.. Functional impact bias reveals cancer drivers. Nucleic Acids Res. 2012; 40:e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mularoni L., Sabarinathan R., Deu-Pons J., Gonzalez-Perez A., Lopez-Bigas N.. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016; 17:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tamborero D., Gonzalez-Perez A., Lopez-Bigas N.. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013; 29:2238–2244. [DOI] [PubMed] [Google Scholar]
- 11. Tokheim C.J., Papadopoulos N., Kinzler K.W., Vogelstein B., Karchin R.. Evaluating the evaluation of cancer driver genes. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:14330–14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martincorena I., Raine K.M., Gerstung M., Dawson K.J., Haase K., Van Loo P., Davies H., Stratton M.R., Campbell P.J.. Universal patterns of selection in cancer and somatic tissues. Cell. 2017; 171:1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Imielinski M., Guo G., Meyerson M.. Insertions and deletions target Lineage-Defining genes in human cancers. Cell. 2017; 168:460–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C. et al.. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016; 534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Polak P., Karlic R., Koren A., Thurman R., Sandstrom R., Lawrence M., Reynolds A., Rynes E., Vlahovicek K., Stamatoyannopoulos J.A. et al.. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015; 518:360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woo Y.H., Li W.H.. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat. Commun. 2012; 3:1004. [DOI] [PubMed] [Google Scholar]
- 17. Schuster-Bockler B., Lehner B.. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012; 488:504–507. [DOI] [PubMed] [Google Scholar]
- 18. Zack T.I., Schumacher S.E., Carter S.L., Cherniack A.D., Saksena G., Tabak B., Lawrence M.S., Zhsng C.Z., Wala J., Mermel C.H. et al.. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013; 45:1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnstrom K., Mallick S., Kirby A. et al.. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014; 46:944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R.. A census of human cancer genes. Nat. Rev. Cancer. 2004; 4:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beck T.N., Golemis E.A.. Genomic insights into head and neck cancer. Cancers Head Neck. 2016; 1:doi:10.1186/s41199-016-0003-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pickering C.R., Zhou J.H., Lee J.J., Drummond J.A., Peng S.A., Saade R.E., Tsai K.Y., Curry J.L., Tetzlaff M.T., Lai S.Y. et al.. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin. Cancer Res. 2014; 20:6582–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fels Elliott D.R., Perner J., Li X., Symmons M.F., Verstak B., Eldridge M., Bower L., O’Donovan M., Gay N.J., Consortium O. et al.. Impact of mutations in Toll-like receptor pathway genes on esophageal carcinogenesis. PLoS Genet. 2017; 13:e1006808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garza-Gonzalez E., Bosques-Padilla F.J., Mendoza-Ibarra S.I., Flores-Gutierrez J.P., Maldonado-Garza H.J., Perez-Perez G.I.. Assessment of the toll-like receptor 4 Asp299Gly, Thr399Ile and interleukin-8 -251 polymorphisms in the risk for the development of distal gastric cancer. BMC Cancer. 2007; 7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Glaser A.P., Fantini D., Shilatifard A., Schaeffer E.M., Meeks J.J.. The evolving genomic landscape of urothelial carcinoma. Nat. Rev. Urol. 2017; 14:215–229. [DOI] [PubMed] [Google Scholar]
- 27. Tsai Y.S., Tzai T.S., Chow N.H., Wu C.L.. Frequency and clinicopathologic correlates of ErbB1, ErbB2, and ErbB3 immunoreactivity in urothelial tumors of upper urinary tract. Urology. 2005; 66:1197–1202. [DOI] [PubMed] [Google Scholar]
- 28. Ayatollahi H., Shajiei A., Sadeghian M.H., Sheikhi M., Yazdandoust E., Ghazanfarpour M., Shams S.F., Shakeri S.. Prognostic importance of c-KIT mutations in core binding factor acute myeloid Leukemia: a systematic review. Hematol. Oncol. Stem Cell Ther. 2017; 10:1–7. [DOI] [PubMed] [Google Scholar]
- 29. Bordin F., Piovan E., Masiero E., Ambesi-Impiombato A., Minuzzo S., Bertorelle R., Sacchetto V., Pilotto G., Basso G., Zanovello P. et al.. WT1 loss attenuates the TP53-induced DNA damage response in T-cell acute lymphoblastic leukemia. Haematologica. 2018; 103:266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ha J.S., Do Y.R., Ki C.S., Lee C., Kim D.H., Lee W., Ryoo N.H., Jeon D.S.. Identification of a novel PML-RARG fusion in acute promyelocytic leukemia. Leukemia. 2017; 31:1992–1995. [DOI] [PubMed] [Google Scholar]
- 31. Qiu J.J., Zeisig B.B., Li S., Liu W., Chu H., Song Y., Giordano A., Schwaller J., Gronemeyer H., Dong S. et al.. Critical role of retinoid/rexinoid signaling in mediating transformation and therapeutic response of NUP98-RARG leukemia. Leukemia. 2015; 29:1153–1162. [DOI] [PubMed] [Google Scholar]
- 32. Zhu D., Zhou J., Zhao J., Jiang G., Zhang X., Zhang Y., Dong M.. ZC3H13 suppresses colorectal cancer proliferation and invasion via inactivating Ras-ERK signaling. J. Cell Physiol. 2019; 234:8899–8907. [DOI] [PubMed] [Google Scholar]
- 33. Guo T., Gaykalova D.A., Considine M., Wheelan S., Pallavajjala A., Bishop J.A., Westra W.H., Ideker T., Koch W.M., Khan Z. et al.. Characterization of functionally active gene fusions in human papillomavirus related oropharyngeal squamous cell carcinoma. Int. J. Cancer. 2016; 139:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin D.C., Hao J.J., Nagata Y., Xu L., Shang L., Meng X., Sato Y., Okuno Y., Varela A.M., Ding L.W. et al.. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 2014; 46:467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dazhi W., Jing D., Chunling R., Mi Z., Zhixuan X.. Elevated SLC6A6 expression drives tumorigenesis and affects clinical outcomes in gastric cancer. Biomark. Med. 2019; 13:95–104. [DOI] [PubMed] [Google Scholar]
- 36. Castel P., Cheng A., Cuevas-Navarro A., Everman D.B., Papageorge A.G., Simanshu D.K., Tankka A., Galeas J., Urisman A., McCormick F.. RIT1 oncoproteins escape LZTR1-mediated proteolysis. Science. 2019; 363:1226–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bigenzahn J.W., Collu G.M., Kartnig F., Pieraks M., Vladimer G.I., Heinz L.X., Sedlyarov V., Schischlik F., Fauster A., Rebsamen M. et al.. LZTR1 is a regulator of RAS ubiquitination and signaling. Science. 2018; 362:1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Steklov M., Pandolfi S., Baietti M.F., Batiuk A., Carai P., Najm P., Zhang M., Jang H., Renzi F., Cai Y. et al.. Mutations in LZTR1 drive human disease by dysregulating RAS ubiquitination. Science. 2018; 362:1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu L., Wu W., Cheng G., Qian M., Hu K., Yin G., Wang S.. Enhancement of proliferation and invasion of gastric cancer cell by KDM5C via decrease in p53 expression. Technol. Cancer Res. Treat. 2017; 16:141–149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40. Wong D., Lounsbury K., Lum A., Song J., Chan S., LeBlanc V., Chittaranjan S., Marra M., Yip S.. Transcriptomic analysis of CIC and ATXN1L reveal a functional relationship exploited by cancer. Oncogene. 2018; 38:273–290. [DOI] [PubMed] [Google Scholar]
- 41. Khurana E., Fu Y., Chakravarty D., Demichelis F., Rubin M.A., Gerstein M.. Role of non-coding sequence variants in cancer. Nat. Rev. Genet. 2016; 17:93–108. [DOI] [PubMed] [Google Scholar]
- 42. DesJardins M., Gordon D.F.. Evaluation and selection of biases in machine learning. Machine Learning. 1995; 20:5–22. [Google Scholar]
- 43. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014; 46:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ignatiadis N., Klaus B., Zaugg J.B., Huber W.. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods. 2016; 13:577–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.