Abstract
Viroids are naked RNAs that do not code for any known protein and yet are able to infect plants causing severe diseases. Because of their RNA nature, many studies have focused on the involvement of viroids in RNA-mediated gene silencing as being their pathogenesis mechanism. Here, the alterations caused by the Citrus exocortis viroid (CEVd) on the tomato translation machinery were studied as a new aspect of viroid pathogenesis. The presence of viroids in the ribosomal fractions of infected tomato plants was detected. More precisely, CEVd and its derived viroid small RNAs were found to co-sediment with tomato ribosomes in vivo, and to provoke changes in the global polysome profiles, particularly in the 40S ribosomal subunit accumulation. Additionally, the viroid caused alterations in ribosome biogenesis in the infected tomato plants, affecting the 18S rRNA maturation process. A higher expression level of the ribosomal stress mediator NAC082 was also detected in the CEVd-infected tomato leaves. Both the alterations in the rRNA processing and the induction of NAC082 correlate with the degree of viroid symptomatology. Taken together, these results suggest that CEVd is responsible for defective ribosome biogenesis in tomato, thereby interfering with the translation machinery and, therefore, causing ribosomal stress.
INTRODUCTION
Viroids are the smallest known plant pathogens, consisting of naked, single-stranded, closed circular RNAs with a high degree of secondary structure which do not code for any known proteins or peptides (1). These pathogens are capable of causing very severe diseases in numerous plants, diseases which result in a wide variety of symptoms that often resemble to those produced by viral infections (2–5). Viroids can be classified into two families: (i) the Pospiviroidae family, whose type member is the Potato spindle tuber viroid (PSTVd), and whose members have rod-type structures and replicate in the nucleus; and, (ii) the Avsunviroidae family, whose type member is the Avocado sunblotch viroid (ASBVd) and whose members possess showing hammerhead self-cleaving structures and replicate in the chloroplasts (6).
The mechanism by which viroids induce pathogenesis remains elusive. Due to the high degree of internal base-pairing and stable secondary structures, viroids are both inducers and targets of RNA silencing mechanisms since they are substrates for the Dicer-like endoribonucleases (7). The accumulation of viroid derived small RNAs (vd-sRNA) of 21- to 24-nucleotides (nt) in length has been detected in viroid infected plants, and the involvement of several silencing components in the viroid pathogenesis has been reported in different host–viroid combinations (8–20). Moreover, the RNAi-mediated down-regulation of the host's mRNAs by the direct interaction of such vd-sRNAs has been studied by several groups (21–25).
Despite their lack of protein-coding capacity, several lines of evidence have related viroid infections with changes in the translational machinery. Accordingly, alterations in the accumulation of the ribosomal proteins S3, S5 and L10, or in the eukaryotic translation factors such as eEF1A, eEF2 and eIF5A, have been detected in tomato plants infected with CEVd (26). Additionally, some viroids have been found to interact with either eEF1A or with the ribosomal protein L5 (26–28). Hop stunt viroid (HSVd) induces changes in the dynamic DNA methylation of the ribosomal RNA genes in the host plants, and that the accumulation levels of some rRNA-derived sRNAs are higher in infected as compared to control plants (29,30). In addition, PSTVd infection triggers the degradation of the ribosomal protein S3a-like mRNAs in tomato plants (24). The possible effect of these changes on the translational regulation has not yet been explored.
Ribosomopathies are human diseases associated with defects in ribosomal functioning, and are normally due to a ‘haploinsufficiency’ of ribosomal proteins or to defect in ribosome biogenesis (31). A number of studies of distinct ribosomopathies have revealed that ‘ribosomal stress’ signals converge on the p53 signalling pathway, leading to cell cycle arrest and apoptosis in the affected cells and tissues (31). This pathway seems to be absent in the plant kingdom. However, several studies have described anomalies in plant ribosome biogenesis that are associated with serious developmental alterations (32–35). For example, mutations in the plant-specific mitoribosome (also known as mitochondrial ribosome) genes have been described as resulting in both death and severe growth delay in Arabidopsis thaliana (36). On the other hand, the NAC transcription factor ANAC082 behaves as a ribosomal stress response mediator in A. thaliana, acting downstream of the perturbation of the biogenesis of the ribosome and leading to growth defects and developmental alterations (37).
Eukaryotic ribosomes are formed by two subunits that together constitute the functional 80S ribosome. The small subunit (40S) contains the 18S rRNA along with a total of 33 ribosomal proteins and functions as a decoder of mRNAs. The large subunit (60S) includes 25/28S, 5.8S and 5S rRNAs and approximately 47 ribosomal proteins, and is responsible for the peptidyl transferase activity. With the recent visualization of the assembly intermediaries through electronic cryomicroscopy (CryoEM), a detailed image of the assembled ribosome is available (38,39). In plants, the CryoEM structure of the large subunit of the spinach (Spinacia oleracea) chloroplast ribosome was recently reported (40).
The biogenesis of eukaryotic ribosomes is a fundamental process involving RNA polymerases and a large number of factors (41,42). The assembly process initiates in the nucleolus. While the 5S rRNA is transcribed independently by a type III RNA polymerase, the rest of the rRNAs are transcribed as a single primary transcript by an RNA polymerase I. In this primary transcript, the 18S, 5.8S and 25/28S rRNAs are separated by internal transcribed spacers (ITS1 and ITS2), and are flanked by external transcribed spacers (5′- and 3′-ETS). Both the ITS and ETS regions must be processed in a coordinated manner for the production of mature rRNAs. The processing of this pre-rRNA has been described in detail in both yeast and humans, but to date there is no clear outline in plants. However, possible models for A. thaliana (35) and Oryza sativa (43) have been proposed.
Several plant-specific ribosome biogenesis factors have been described as being essential in A. thaliana rRNA processing (44). In A. thaliana the resulting pre-rRNA contains the complete 5′ ETS region, including the A123B cluster and a 1083-nt insertion of unknown function. After a splicing-like process, XNR2 and a non-coding nucleoar RNA (U3snoRNA) participate in the processing at the P site, giving rise to the precursor 35S pre-rRNA. At this point, two alternative routes are proposed, both including the processing event in B2 of the 35S pre-rRNA. In one of the pathways (5′-ETS-first), the 5′ ETS region is eliminated by cutting at the P’ and P2 sites, producing the 33S and 32S pre-rRNAs, respectively. Then, the processing of the A2 site separates the 32S pre-rRNA into the 27SA2 and the 20S pre-rRNA which finally yield the mature 18S, 5.8S and 25S rRNA. The second pathway (ITS1-first) begins with the processing of the A3 site that produce the P-A3 and 27SA3 pre-rRNAs. After several processing events, the P-A3 generates the 18S rRNA while the 27SA3 yields the 5.8S and 25S rRNAs (35,45). In tomato plants, the precise 35S pre-rRNA processing pathway is not yet known, although some features such as the primary processing site, have been determined (46). The coexistence of both routes of rRNA processing in A. thaliana could serve to guarantee the biogenesis of a sufficient number of functional ribosomes, or, alternatively, both routes could be differentially regulated in different tissues or upon stress situations (35), for example in the case of biotic stresses caused by pathogens.
The regulation of translation finely modulates gene expression under biotic stress, such as that produced by viruses, which ensure the production of the virally encoded proteins by using the plant's machinery (47). Because of their non-coding nature, much less is known on how viroid interferes with the translation machinery of the host plant. In this study, alterations caused by viroids on the tomato translational machinery are reported, uncovering a novel mechanism of viroid–plant interaction.
MATERIALS AND METHODS
Plant material and viroid inoculation
Tomato plants (Solanum lycopersicum L. cv Rutgers, Rio Grande and UC82, epi mutants and their VFN8 parentals (48)) were cultivated in a growth chamber at 30°C for 16 h with fluorescent light, and at 25°C for 8 h in darkness. The inoculation of the plants with CEVd was carried out by puncturing the stems of the seedlings with a needle dipped in either buffer or the nucleic acid preparation as described by Bellés et al. (49). Leaf tissue was collected 3 weeks post-infection (wpi).
For PSTVd inoculation, Nicotiana benthamiana plants were cultivated in a growth chamber at a temperature of 25°C with 16 h of light and 8 h of darkness (50). A dimeric construct of PSTVd-RG1 (GenBank Accession No. U23058) was used to synthesize infectious dimeric transcripts as described previously (23), and the resulting PSTVd RNA transcripts were used to inoculate the plants.
Total ribosome isolation and purification by gel filtration
The total ribosomes and polysomes were prepared as previously described (51), with the following modifications. Actively growing leaf samples (25 g) were washed with DEPC-treated water, frozen in liquid nitrogen and macerated to a fine powder using a cold mortar and pestle. The resulting powder was then homogenized in 2 volumes of cold plant extraction buffer (50 mM Tris–HCl pH 9.0, 30 mM MgCl2, 400 mM KCl, 17% [w/w] sucrose) and clarified by filtering through DEPC-treated Miracloth cheesecloth. The resulting extracts were centrifuged at 1000 × g for 7 min at 4°C. One tenth of a volume of 20% Triton X-100 was added to the supernatants, and they were then centrifuged at 15 500 × g for 20 min. The clear supernatants were then carefully layered on 60% (w/v) sucrose cushions (20 mM Tris–HCl pH 7.6, 5 mM MgCl2, 510 mM NH4Cl, 60% (w/v) sucrose) and centrifuged at 140 000 × g for 19 h using a SW28 rotor in a Beckman Coulter ultracentrifuge. The resulting pellets were carefully rinsed with resuspension buffer (50 mM KCl, 20 mM Tris–HCl pH 7.6, 5 mM MgCl2), and were then resuspended in 200 μl of the same buffer.
For further ribosome purification by gel filtration (52), MicroSpin S-400 HR columns were used according to manufacturer's instructions (GE Healthcare UK).
Polysome profiling
Ribosomal fractions from both control and CEVd-infected tomato leaves were separated by ultracentrifugation on a sucrose density gradient according to a modification of the protocol described by Mustroph et al. (53). Briefly, the tissues (300 mg) were homogenized in 1 ml of extraction buffer (200 mM Tris–HCl pH 9.0, 200 mM KCl, 5 mM EGTA, 35 mM MgCl2, 4% (w/v) Triton X-100, 1% (w/v) sodium deoxycholate, 5 mM DTT and 0.5 mg/ml heparin). Sucrose gradients (10–60% (w/v)) were then prepared and 300 μl of each homogenate were placed on top of them. Following ultracentrifugation at 210 000 × g for 3 h and 40 min at 4°C, the absorbance at 254 nm was measured for each of the collected fractions.
RNA extraction
In order to detect either PSTVd or CEVd in total ribosomes, the RNA was extracted with 5.5 M guanidine–HCl, followed by acid phenol:chloroform extraction and lastly by re-extraction of the supernatant with an equal volume of chloroform. RNAs were ethanol precipitated, dissolved in RNAse-free water and then treated with DNase I according to the manufacturer's instructions (Promega).
In order to detect CEVd or vd-sRNA in the different sucrose gradient fractions, RNA extraction was performed by adding 100 μl of a solution containing 5% SDS and 0.2 M EDTA to 1 ml of each fraction. Subsequently, 1 volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added, the samples were then mixed and centrifuged at 12 000 × g for 2 min. The aqueous phase was collected and the RNA precipitated by the addition of absolute ethanol. The resulting RNA pellets were dissolved in 20 μl of RNase-free water.
RNA extraction from plant tissues was performed using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and following the manufacturer's instructions. RNAs used for the Bioanalyzer (Agilent 2100 Bioanalyser; Agilent, Palo Alto, CA, USA) and the quantitative RT-PCR (RT-qPCR) analyses were further precipitated by adding one volume of 6 M LiCl. After incubating for 4 h on ice bath, RNA pellets were recovered by centrifugation at 12 000 × g for 10 min, washed with 3 M LiCl and dissolved in RNase-free water to a concentration of 1 μg/μl.
Northern blot hybridization
In order to detect either PSTVd or CEVd in total ribosomes, 5.0 μg of the total RNA samples extracted from the 3 wpi (weeks post-inoculation) plants were denatured at 65°C for 10 min using 2.5 volumes of sample buffer (50% formamide, 6% formaldehyde in 200 mM MOPS, 50 mM sodium acetate, and 10 mM EDTA, pH 7), and were then separated on 1.2% (for PSTVd) or 1.5% (for CEVd) agarose-formaldehyde gels prepared in 200 mM MOPS, 50 mM sodium acetate and 10 mM EDTA pH 7 buffer. The Northern blot hybridizations were performed as described previously (23). For PSTVd detection, a dimeric (–) PSTVd riboprobe was used. The accumulation of the 5S rRNA was used as loading control (the full length 5S rRNA that had been amplified from tomato cv Rutgers and cloned into pBluescript KS+ vector was used (50)). In order to prepare the riboprobes, either the T3 or the T7 MAXIscript kit (Ambion) was used after linearizing the plasmid with the appropriate restriction endonuclease. In order to detect CEVd in total ribosomes, CEVdRT and 5S oligonucleotide probes, which were generated using a polynucleotide kinase and [γ-32P]-ATP, were used. The probes were mixed to simultaneously detect 5S and viroid RNA in the same membrane.
In order to perform the Northern hybridizations of the different gradient fractions, the RNAs were separated by denaturing PAGE electrophoresis on either 5% gels for CEVd detection, or 17% gels for vd-sRNA detection. The RNAs were visualised by ethidium bromide staining, and were then electrotransferred onto Nytran membranes and cross-linked by UV irradiation (54). Minus (–) strand specific riboprobes for CEVd generated by in vitro transcription using viroid cDNA clones in the presence of [α-32P] UTP (54) were used to detect the viroid in the different fractions. Prehybridization, hybridization and washes were carried out according to Martinez de Alba et al. (9) at either 70°C for CEVd detection or at 35°C for the sRNAs. Equal loading between paired fractions corresponding to control and infected leaves was confirmed by ethidium bromide staining and UV irradiation.
Northern blot hybridizations of total RNAs from plant tissues were performed according to Missbach et al. (55,56). The oligonucleotide probes used, which were also prepared using a polynucleotide kinase and [γ-32P]-ATP, are listed in Supplementary Table S1.
RT-PCR and RT-qPCR
For the RT-PCR, first strand cDNA was synthesized from 5 μg of total RNA obtained from the tomato tissues using Moloney murine leukemia virus reverse transcriptase (Promega) and either the 18Srev, CEVdRT, Vid-RE (57) or the 25Srev primer. Five microliters of a 1:10 dilution of the reverse transcriptase reactions were used for the PCR in a Perkin-Elmer thermocycler with the following program: 25 cycles at 95°C for 1 min, 55°C for 1 min and 72°C for 2 min followed by a final extension at 72°C for 5 min. The PCR primers used are listed in Supplementary Table S1.
For the quantitative RT-PCR (RT-qPCR) analysis, 2 U of TURBO DNase (Ambion, Austin, TX, USA) were added per microliter of total RNA. Then, total RNA (1 μg) was used as a template to obtain the corresponding cDNA using an oligo(dT)18 primer and the PrimeScript RT reagent kit (Perfect Real Time, Takara Bio Inc., Otsu, Shiga, Japan) following the manufacturer's directions. qPCR was carried out as previously described (58). The housekeeping gene transcript actin (Accession number: Solyc04g011500) was used as the endogenous reference. The PCR primers used are listed in Supplementary Table S1.
For the RT-qPCR analysis of total RNA extracted from Nicotiana benthamiana plants, total RNA was used for the cDNA synthesis as described previously (50), and the obtained cDNA was used in qPCR experiments together with the appropriate primer combinations in order to verify the accumulation of the NbNAC082 mRNA. Actin mRNA was used as the housekeeping gene. A negative control was maintained for every primer pair, and these were consistently negative. All of the RT-qPCR analyses were performed commercially at the Laboratoire de Génomique Fonctionnelle de l’Université de Sherbrooke, Sherbrooke, QC, Canada (https://palace.lgfus.ca). The PCR primers used are listed in Supplementary Table S1.
Statistics and image analysis
For statistical analysis, t-test analyses were performed using the SPSS v.19 package (IBM). A P value <0.05 was considered significant. Image analyses used to quantify the Northern blot bands were performed by using the ImageJ (https://imagej.nih.gov/ij/) image processing software.
RESULTS
CEVd and PSTVd are found in the total ribosome fraction and in the purified ribosomes of infected tomato plants
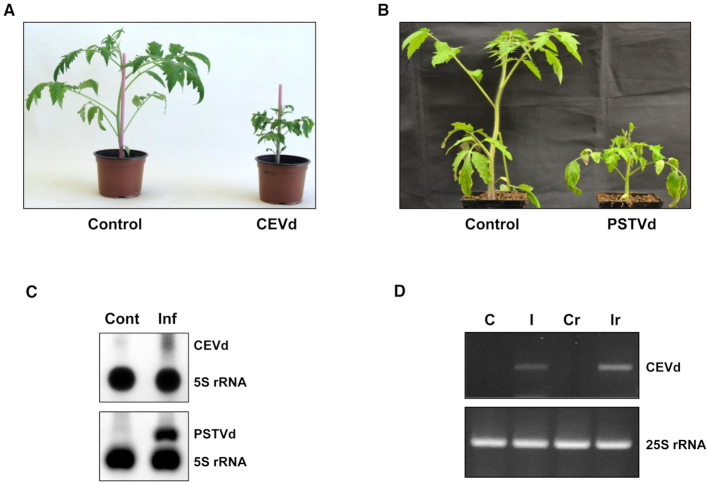
In order to elucidate the molecular details of the association of viroids with the host during infection, tomato plants cv. Rutgers were infected with either CEVd or PSTVd. As can be seen in Figure 1, tomato plants infected with either CEVd or PSTVd showed severe disease symptoms, including stunting and epinasty, at 3 wpi when compared to mock-inoculated plants (Figure 1A and B). In order to monitor viroid accumulation, total RNA extracted from the leaf samples collected at 3 wpi was analyzed by both Northern blot hybridization and RT-PCR amplification in order to confirm the presence of either CEVd or PSTVd in tomato, observing the accumulation of both viroids (Supplementary data Figure S1).
Figure 1.
Detection of viroids in the purified ribosomes of the total ribosome fraction of infected plants. (A) The symptoms of tomato cv Rutgers infected with CEVd at 3 wpi as compared to those of mock-inoculated (Control) tomato plants. (B) The symptoms of tomato cv Rutgers infected with PSTVd-RG at 3 wpi as compared to those in mock-inoculated (Control) tomato plants. (C) Northern blot hybridization analysis of the isolated total ribosomes of both CEVd (upper panel) and PSTVd-RG (lower panel) infected tomato plants (Inf) and their corresponding control plants (Cont). 5S rRNA was used as a control. (D) RT-PCR of the total RNAs from control (C) and CEVd-infected (I) tomato leaves, or of the RNAs from MicroSpin S-400 purified ribosomes from control (CR) and CEVd-infected (IR) tomato leaves. The RT reactions were performed with either the CEVdRT primer (upper panel) or the 25Srev primer (lower panel). The PCRs were performed with either the primers CEVddir and CEVdrev (upper panel) or 25Sdir and 25Srev (lower panel).
To evaluate the possible association of viroids with the host's ribosomes, young leaves collected from both mock- and viroid-inoculated plants were used for total ribosome isolation. Northern blot hybridizations were performed using both CEVd and PSTVd specific probes that detected the presence of viroid molecules in the total ribosome fraction of the infected plants (Figure 1C). In addition, the presence of CEVd in purified tomato ribosomes was confirmed by RT-PCR analysis of the RNAs extracted after purification of the ribosomes by gel filtration (52). For this purpose, MicroSpin S-400 columns were used to purify ribosomes from the total ribosome fractions of both control and CEVd-infected tomato leaves. A band corresponding to the full length CEVd (371 bp) was found to be amplified by RT-PCR only in the ribosomes obtained from CEVd infected tomato leaves (Figure 1D). Taken together, these results reveal that, despite its lack of protein-coding information, viroids are associated with the ribosomes of infected plants. As similar results were obtained with both CEVd and PSTVd infected plants, efforts were further focused on the molecular characterization of the association of CEVd with the tomato ribosomal machinery.
CEVd produces alterations in tomato polysome profiles
In order to study the translational profile of tomato plants infected with CEVd, ribosome fractions from both control and infected tomato leaves were separated by ultracentrifugation on a sucrose density gradient, and the fractions corresponding to the small subunits (40S), large subunits (60S), monosomes (80S) and polysomes were collected and analyzed. Figure 2 illustrates the polysome profiles for both control and CEVd-infected tomato leaves. Evident differences can be observed between both profiles, with a reproducible reduction in the abundance of polysomes in the infected leaves. Moreover, the fractions corresponding to both the 40S ribosomal subunit and the 80S monosomes were also found to be less abundant in the infected tomato leaves, while the 60S subunit appeared to be unaffected. These results indicate that the CEVd infection provokes alterations in the tomato polysome profiles, specifically affecting the 40S ribosomal subunit.
Figure 2.
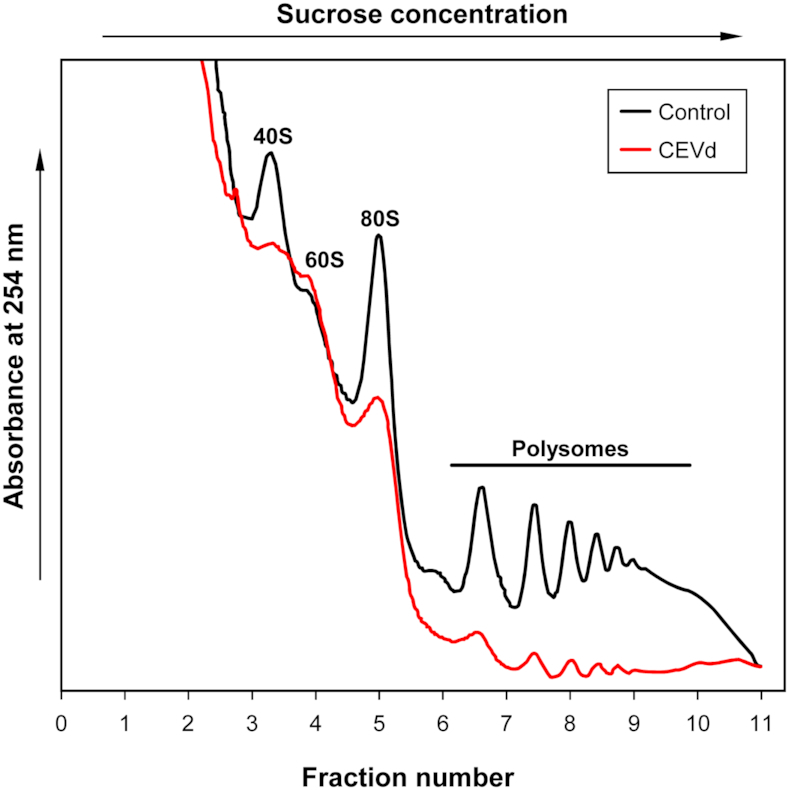
CEVd produces alterations in tomato polysome profiles. Overlay of sucrose density gradient analysis of polysomes from the leaves of control (black line) and CEVd-infected (red line) tomato plants. The positions of the 40S, 60S and 80S ribosomal subunits indicated in the panel were located based on the A260 peaks. The experiments were repeated at least three times and a representative result is shown.
CEVd and vd-sRNA associate with different tomato ribosome fractions
In order to study the presence of CEVd in the different tomato ribosome fractions, the RNA from both the control and the CEVd-infected fractions were isolated. A Northern blot hybridization analysis was performed by using 20 μl of combined gradient fractions (i.e. fractions 1–2, 3–4, 5–6, 7–8 and 9–11) which were separated by 5% polyacrylamide denaturing gel electrophoresis (PAGE). The presence of CEVd was detected in every gradient fraction derived from the infected leaves, with the signal being more intense in the lower density fractions corresponding to the small (40S) and the large (60S) free ribosomal subunits, as well as to the 80S monosomes (Figure 3A). These results confirm that CEVd associates in vivo with tomato ribosomes.
Figure 3.
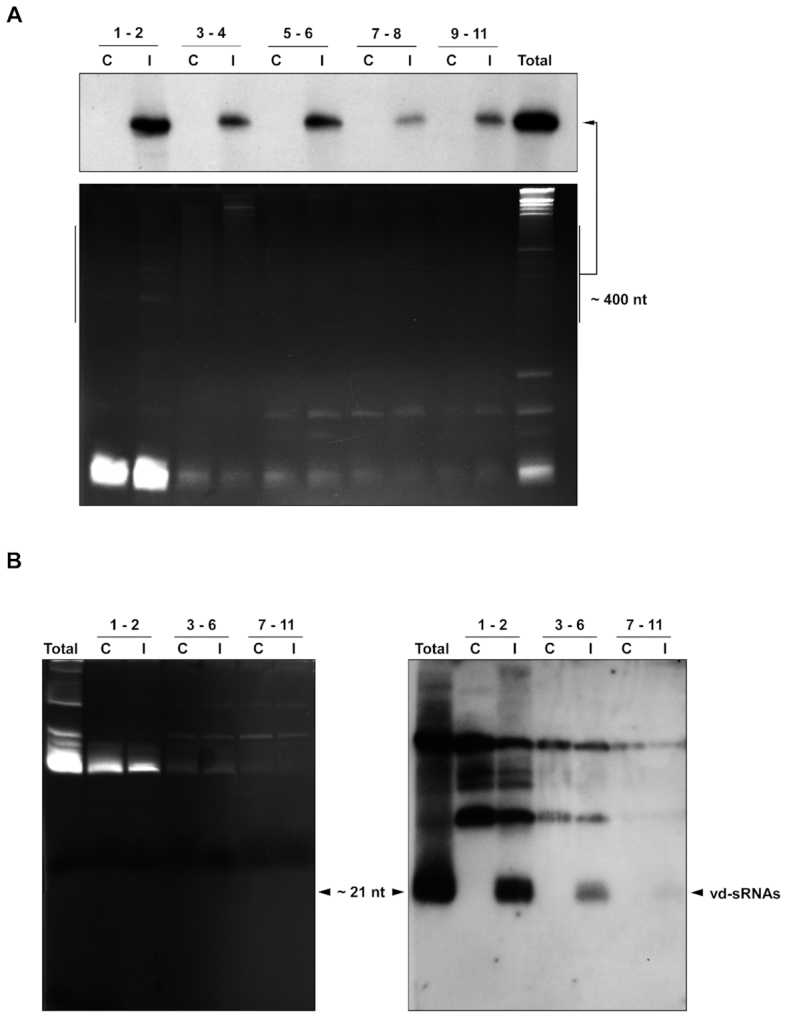
CEVd and vd-sRNAs associate with tomato ribosomes. (A) Detection of CEVd by Northern blot hybridization in the ribosomal fractions from the control (C) and infected (I) tomato plant leaves from Figure 2. RNAs prepared from the ribosomal fractions were separated on a 5% denaturing polyacrylamide gel, stained with ethidium bromide (lower panel), transferred to nylon membranes and hybridized with radioactive (–) CEVd riboprobes (upper panel). Total RNA from CEVd-infected tomato leaves was used as a positive control (last lane). (B) Detection of CEVd-specific small RNAs (vd-sRNA) in the tomato ribosomal fractions from Figure 2 by northern blot hybridization. RNA preparations from both control and CEVd-infected tomato ribosomal fractions corresponding to the different fractions were pooled into three sets (fractions 1–2, 3–6 and 7–11). The samples were then separated on a 17% denaturing polyacrylamide gel, stained with ethidium bromide (left panel), transferred to nylon membrane and hybridized with (–) CEVd riboprobes (right panel). The positive polarity 21–24 nt vd-sRNAs are indicated with an arrow. Total RNA from CEVd-infected tomato leaves was used as a positive control (first lane).
Subsequently, the presence or absence of vd-sRNA in the ribosomes was also analyzed. RNAs isolated from the different fractions were pooled into three sets (fractions 1–2, 3–6 and 7–11) and were then separated by 17% denaturing PAGE. The Northern blot hybridization was then performed in order to detect the presence of vd-sRNA (9). A strong hybridization signal indicating the presence of CEVd-infected material was observed in fractions 1 and 2 (Figure 3B). This signal decreased in fractions 3–6, and was almost negligible in fractions 7–11. Clearly, tomato ribosomes from infected leaves appear to contain vd-sRNA.
CEVd produces alterations in the rRNA processing of tomato plants
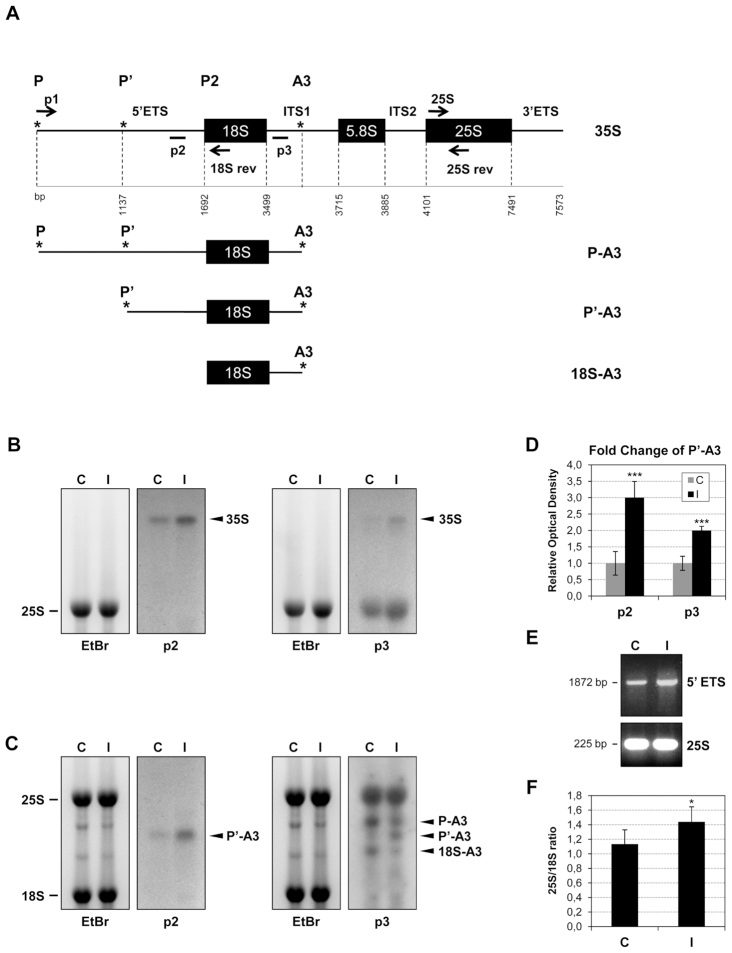
To study whether the observed alteration in the tomato ribosome profiles could be related with an impairment of the rRNA processing, different probes for Northern blot hybridization were located according to the schematic diagram of the tomato pre-rRNA sequence depicted in Figure 4A. The p2 and p3 probes were designed in the 5′ETS and ITS1 regions, respectively. RNA isolated from both control and CEVd-infected tomato plants was separated by agarose gel electrophoresis (Figures 4B and C). The ethidium bromide staining visualized the abundant mature cytoplasmic 25S and 18S rRNAs. Comparison of the signals in the Northern blots for the control and the CEVd-infected tomato pre-rRNAs revealed some significant alterations. Specifically, a clear over-accumulation of the 35S band in the CEVd-infected tomato plants was observed with both the p2 and p3 riboprobes (Figure 4B). Moreover, a prominent over-accumulation of a band which corresponded to P’-A3 was also detected with both probes in the CEVd-infected tomato leaves (Figure 4C). Concomitant to the increase of the P’-A3 band, a visible reduction in the final processed 18S-A3 band could be observed with riboprobe p3. The accumulation of the P’-A3 band was quantified from three independent experiments, and the differences were judged to be statistically significant (Figure 4D). These results suggest that CEVd produces alterations in the processing of the pre-rRNA of tomato plants, probably by impairing the P2 processing site located in the 5′-ETS region (Figure 4A).
Figure 4.
CEVd produces alterations in the rRNA processing of tomato plants. (A) Scheme of tomato pre-rRNA indicating the priming sites for the Northern probes (p2 and p3) and the primers used for the RT-PCR experiment (arrows; p1, 18Srev, 25S and 25Srev). The tomato GENBANK sequences used were the following ones: complete rRNA (X52215), 18S sequence (X51576), 5.8S sequence (X52265) and 25S sequence (X13557). Initiation and processing sites are marked with asterisks (*) according to Perry et al. (1990). The scheme was adapted from Missbach et al. (55). (B) RNAs from control (C) and CEVd-infected (I) tomato leaves were separated on an agarose-formadehyde gel, stained with ethidium bromide (left) for visualization of the mature 25S rRNA and then Northern hybridization was performed with the probes p2 or p3 (right) in order to detect the 35S pre-RNA. (C) RNAs from control (C) and CEVd-infected (I) tomato leaves were separated on an agarose gel, stained with ethidium bromide (left) in order to visualize the mature 25S and 18S rRNAs and then Northern hybridization was performed with the probes p2 or p3 (right) in order to detect the pre-rRNAs. (D) The quantification of P’-A3 in both control (C) and CEVd-infected (I) tomato RNAs was performed with at least 3 biological replicates. The values are represented as the mean ± SD. The triple asterisk (***) indicates statistically significant differences between the control and the treated plants with P < 0.001. (E) RT-PCR from control (C) and CEVd-infected (I) tomato leaves. The RT reactions were performed with the 18Srev primer. The PCRs were performed with either primers p1 and 18Srev (1872 pb, upper panel) or 25S and 25Srev (225 bp, lower panel). A representative gel of the RT-PCR fractionation is shown. (F) Total RNAs from control (C) and CEVd-infected (I) tomato leaves were resolved on a Bioanalyzer and the 25S/18S ratios were calculated. 25S/18S ratios are represented as the mean ± SD of at least three biological analyses. The asterisk (*) indicates statistically significant differences between the control and the treated plants with P < 0.05.
Since the accumulation of the P’-A3 suggests an impairment of the 5′-ETS processing in the CEVd infected plants, RT-PCR amplifications were performed with primers specific for that sequence. An over-accumulation of the 5′-ETS was detected in the infected plants (Figure 4E), thus indicating that the viroid could somehow be interfering with the processing of this region. Moreover, since the interference with the 5′-ETS processing could affect the final amount of properly processed 18S rRNA, the 25S/18S signal ratios were calculated in both control and CEVd-infected tomato plants. As shown in Figure 4F and Supplementary Figure S2, an increase in the 25S/18S ratio was observed in CEVd-infected tomato plants. These data suggest that the viroid-induced alteration of the pre-RNA processing in the 5′-ETS leads to an impairment in 18S rRNA accumulation.
Taken together, the results of this section indicate that CEVd impairs the maturation of the 18S rRNA, more specifically by interfering with the processing of 5′ETS. Since the 18S rRNA is a main constituent of the 40S subunit, this impairment could be related to the observed reduced levels of this subunit in the polysome profiles.
Alterations in the rRNA processing of tomato plants correlate with the symptomatology
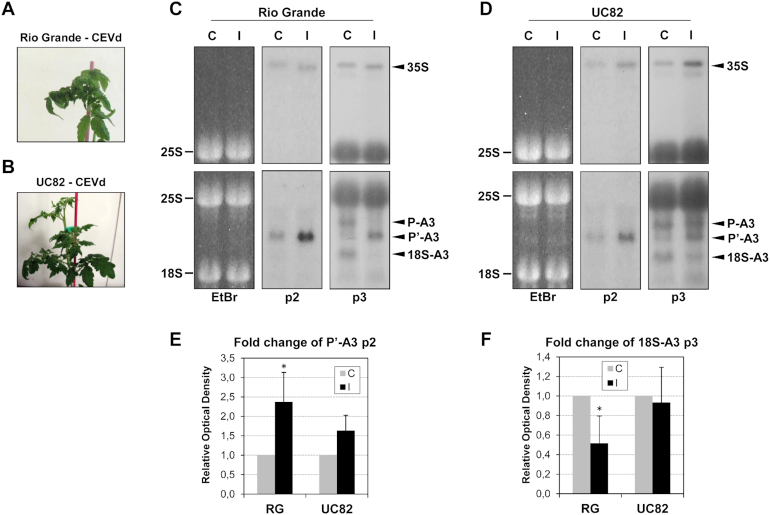
To address whether or not the impairment in the rRNA processing is associated with the viroid symptomatology, a Northern blot hybridization was performed using RNAs derived from the different infected tomato varieties, including Rio Grande and UC82, which display strong and mild symptoms, respectively (Figure 5A and B). Northern blots using the p2 and p3 riboprobes showed a correlation between the CEVd symptomatology and the impairment in the rRNA maturation, as the differences in the accumulation level of the P’-A3 band were less evident in the UC82 tomato variety which displays milder symptomatology, as compared to the more sensitive Rio Grande variety (Figure 5C and D). The quantification of the blots corresponding to several independent experiments showed that, while the accumulation of P’-A3 in Rio Grande infected plants was more than twice that observed in control plants, this over-accumulation was not statistically significant in UC82 plants (Figure 5E). In order to better demonstrate this correlation, the reduction of the 18S-A3 band was also quantified in both Rio Grande and UC82 tomato plants, and was found to be more remarkable in the more sensitive variety (Rio Grande; see Figure 5F).
Figure 5.
Alterations in tomato rRNA processing correlate with CEVd symptomatology. (A) Representative severe symptomatology displayed by CEVd-infected Rio Grande tomato plants. (B) Representative mild symptomatology displayed by CEVd-infected UC82 tomato plants. (C) RNAs from Rio Grande (RG), control (C) and CEVd infected (I) tomato leaves were separated on an agarose-formadehyde gel, stained with ethidium bromide (left) for visualization of the mature 25S and then were Northern blot analyzed with probes p2 or p3. (D) The RNAs from UC82 control (C) and CEVd infected (I) tomato leaves were separated on an agarose-formadehyde gel, stained with ethidium bromide (left) for visualization of the mature 25S and then were Northern blot analyzed with probes p2 or p3. (E) The quantification of P’-A3 was performed with at least 3 biological replicates in control (C), CEVd infected (I), Rio Grande (RG) and UC82 (UC82) tomato plants. The relative values are represented as the mean ± SD. The asterisk (*) indicates statistically significant differences between the control and the treated plants with P < 0.05. (F) The quantification of 18S-A3 was performed with at least three biological replicates in control (C), CEVd infected (I), Rio Grande (RG) the UC82 (UC82) tomato plants. The relative values are represented as the mean ± SD. The asterisk (*) indicates statistically significant differences between the control and the treated plants with P < 0.05.
CEVd induces SlNAC082 expression in a symptom-depending manner
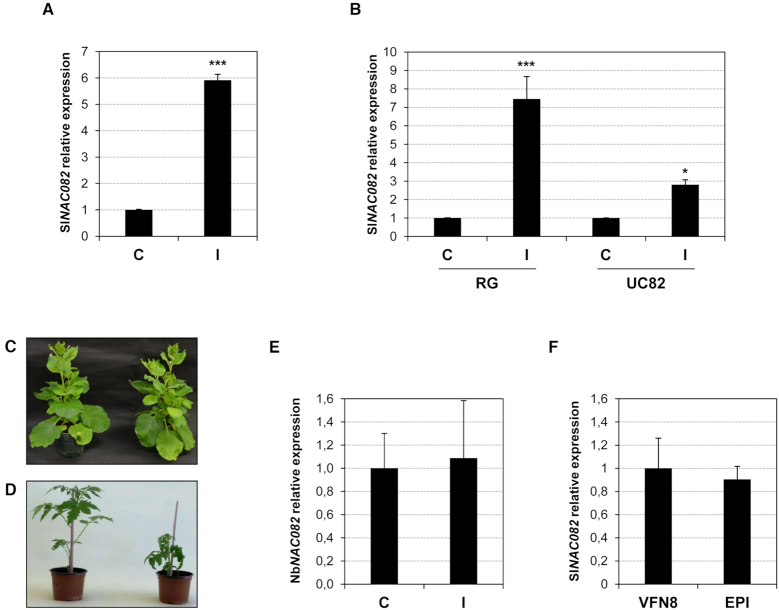
In order to confirm the observed ribosomal stress caused by CEVd in tomato plants, the induction of the ribosomal stress response mediator NAC082 (37) was studied in infected tomato plants. For this purpose, the closest tomato homologue to the A. thaliana transcription factor was identified (Supplementary Figure S3) and specific oligonucleotides were designed (Supplementary Table S1). Expression analysis by RT-qPCR amplification indicated a significant induction of the SlNAC082 mRNA in CEVd-infected tomato plants (Figure 6A), while the next closest tomato homologue (Supplementary Figure S3) that was used as a control remained unaltered (Supplementary Figure S4).
Figure 6.
NAC082 induction in tomato and N. benthamiana plants. (A) Total RNAs were isolated from control (C) and CEVd-infected (I) Rutgers tomato plants and were subjected to RT-qPCR analysis with primers specific for SlNAC082. The values were normalized to the tomato actin gene. The expression levels are represented as the mean ± SD of at least three biological replicates. The triple asterisk (***) indicates statistically significant differences between the control and the treated plants with P < 0.001. (B) Total RNAs were isolated from control (C) and CEVd-infected (I) Rio Grande (RG) and UC82 (UC82) tomato plants and were subjected to RT-qPCR analysis with primers specific for SlNAC082. The values were normalized to the tomato actin gene (Solyc04g011500). The expression levels are represented as the means ± SD of at least three biological replicates. Single (*) or triple asterisks (***) indicate statistically significant differences between the control and the treated plants with P < 0.05 or P < 0.001, respectively. (C) Representative symptomless N. benthamiana plant infected with PSTVd-RG (right), and the corresponding non-infected control plant (left). (D) Phenotype of tomato epi mutants (right), and the corresponding parental plants VFN8 (left). (E) Total RNAs were isolated from control (C) and PSTVd-RG1-infected (I) N. benthamiana plants and were subjected to RT-qPCR analysis with primers specific for NbNAC082. Data obtained for actin mRNA was used for normalization. The expression levels are represented as the means ± SD of at least three biological replicates. (F) Total RNAs were isolated from control (C) and CEVd-infected (I) epi and VFN8 tomato plants and were subjected to RT-qPCR analysis with primers specific for SlNAC082. The values were normalized to the tomato actin gene (Solyc04g011500). The expression levels are represented as mean ± SD of at least three biological replicates.
Whether the tomato ribosomal stress indicator correlated with the CEVd symptomatology was also examined. More specifically, the induction of SlNAC082 was analyzed in CEVd-infected Rio Grande and UC82 tomato plants, as well as in PSTVd-infected Nicotiana benthamiana plants. The variant of PSTVd-RG (359-nt in length) used is known to induce severe symptoms in tomato (Figure 1B) and to be symptomless in N. benthamiana (Figure 6C) (24,59). For this purpose, the SlNAC082 protein sequence was used to query in the Solanaceae databases in order to identify the closest homologue in N. benthamiana. Specific oligonucleotides for this gave then were designed (Supplementary Table S1). As Figure 6B shows, an evident correlation between viroid symptoms and ribosomal stress was observed, since the induction of SlNAC082 was stronger in Rio Grande infected tomato plants, than in the mild-symptom variety UC82. In agreement with this, no induction of NbNAC082 was detected in the asymptomatic PSTVd-RG1-infected N. benthamiana plants (Figure 6E). Finally, in order to test whether or not the presence of the viroid is required for the observed ribosomal stress, epinastic (epi) tomato mutants that overproduce ethylene, and their parental VFN8 tomato plants, were used (48,60). epi plants display a phenotype that mimics viroid infection, showing a strong epinasty and stunting (Figure 6E). Interestingly, no difference in SlNAC082 expression was observed in the epi mutant when it was compared to its parental VFN8 plant (Figure 6F), thus confirming that the induction of this ribosomal stress mediator requires the presence of the viroid. All of these results support the idea that CEVd causes ribosomal stress in tomato plants, and that this disorder directly correlates with the presence of the viroid and its symptomatology.
DISCUSSION
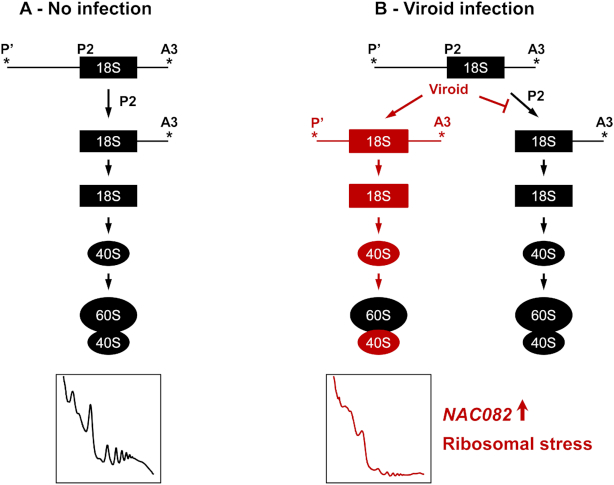
Viroids are non-coding circular, RNA pathogens that were discovered in the early 1970s (61). Despite their lack of protein coding capacity (62), studies have shown that viroid infection could affect either the host's translational machinery, or it could modulate translation directly (24,26–30). In the present study, CEVd and PSTVd, both members of the Pospiviroidae family, were shown to be associated with tomato ribosomes. Based on the data presented here, a model explaining how CEVd causes ribosomal stress in tomato plants is presented (Figure 7). The results indicate that CEVd produces alterations in both the processing of the 35S pre-rRNA and in the translational profiles of tomato plants. Specifically, an alteration in the processing of the 5′ETS rRNA was observed. Most likely this was caused by an impairment in the cleavage of the P2 site as both an over-accumulation of the P’-A3 band and a decrease in the 18S-A3 band were observed in CEVd-infected tomato plants (see Figure 4C). This impairment could affect the maturation of the 18S rRNA, and, therefore, the biogenesis of the 40S subunit which has been observed to be particularly affected in the polysome profiles of the CEVd-infected plants. In addition, the observed induction of the ribosomal stress mediator SlNAC082 in the infected tomato leaves supports the idea that this non-coding pathogen provokes alterations in the biogenesis of ribosomes. Finally, it was demonstrated that the observed ribosomal stress depends on the viroid's presence, and that it correlates with the viroid symptomatology.
Figure 7.
Proposed model for the tomato ribosomal stress produced by viroids. (A) Normal biogenesis of ribosomes in non-infected tomato plants. (B) Alterations in ribosome biogenesis produced by viroid infection (in red). Viroids produce impairment in the cleavage of the P2 site which in turn affects both the maturation of the 18S subunit and the biogenesis of the 40S subunit, and, therefore, alters the polysome profiles. The ribosomal stress marker NAC082 is induced in viroid infected plants.
The results presented here are in agreement with previous studies based on sequence homologies which suggested that viroids could exert their pathogenesis by interfering with the processing of the pre-rRNAs in nucleoli (63), and with others that compared their unique secondary structures to those displayed by rRNAs (64) and with an U3B snRNA (65). The U3 small nucleolar RNA (snoRNA), and their ribonucleoprotein complexes, are required for nucleolar processing of pre-18S rRNA (66–69). The (+) strand of PSTVd has been described as causing the redistribution of an U3 snoRNA in N. benthamiana cells (70). In a similar manner, CEVd could be producing a redistribution of an U3 snoRNA, thus causing an impairment of the 18S rRNA processing in tomato plants. The observed differences of the viroid effect on the tomato cultivars UC82 and Rio Grande suggest a RNA sequence-based mechanism. Future sequence analyses will hopefully provide a mechanistic explanation of the reported observations.
Similar to the results presented here, defects in both 18S rRNA synthesis and in small ribosomal subunit production have been described in human cells depleted in RPS19, a protein associated with the ribosomopathy known as Diamond Blackfan Anemia (71). Alterations in the translational profiles were observed in Pescadillo deficient N. benthamiana plants, which exhibit a delayed maturation of the 25S rRNA (72). In addition, alterations in plant ribosome biogenesis associated with developmental defects, such as those produced by CEVd, have been observed in several A. thaliana mutants (55,73–75). Here, it was observed that the detected ribosomal stress in CEVd-infected tomato plants depends on the presence of the viroid since epi mutants, which mimic the viroid infection phenotype, did not display changes in SlNAC082 induction. Finally, the results presented here are also in agreement with the previously described accumulation of CEVd in the nucleolus (76) where the ribosome biogenesis occurs (77).
Additionally, viroids have common genetic features with miRNAs (21,78). Translation inhibition is a major, but poorly understood mode of action of miRNA (79). In bacteria, sRNAs repress translation by pairing to the ribosome binding site and competing with the initiating ribosomes, an event that is often followed by rapid mRNA decay (80). In Drosophila melanogaster, miRNAs act to block the assembly of the eIF4F complex during translation initiation (81). Similarly to what was observed here for vd-sRNA, miRNAs have been described as associating with polysomes in A. thaliana (82). The authors proposed as the mechanism for the translation inhibition the binding of the AGO1-RISC complex to the target mRNA which sterically blocks the recruitment or movement of ribosomes (83). In particular, the double-stranded RNA binding cofactor DRB2 has been described as controlling the miRNA-guided translational inhibition, thereby allowing for the active selection of miRNA regulatory action (84). Interestingly, we have observed that vd-sRNA associate with the ribosome. Thus, in a way similar to that described for miRNA, vd-sRNA could also be interfering with the assembly of the translation machinery.
In order to avoid or mitigate the losses resulting from viroid infections, several strategies have been developed by researchers. These include: (i) searching for resistant cultivars; (ii) inducing cross-protection by infection with mild strains; (iii) the use of ribonucleases that act on double-stranded RNAs, or of catalytic antibodies endowed with intrinsic ribonuclease activity; and, (iv) the employment of antisense and sense RNAs, hammerhead self-cleaving RNA molecules and RNA interference (85). Here, new aspects of viroidal pathogenesis that could allow for the development of new biotechnological alternatives are explored. Further studies will help understand the underlying molecular mechanism of viroid action beyond the observations reported here.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Pr. Ricardo Flores (IBMCP-UPV-CSIC, Valencia, Spain) for kindly providing the pSTP19-CEVd plasmid used to prepare the viroid transcription probe, and for helpful discussions. We also thank Dr. Susana Tárraga, (Proteomics Service, IBMCP, UPV-CSIC), Dr Lorena Latorre (Genomics Service, IBMCP, UPV-CSIC), Deniz Streit, Jelena Kovacevic (Goethe University; Frankfurt, Germany) and Mustafa Malik-Ghulam (Université de Sherbrooke, Canada) for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Science, Innovation and Universities [BIO2009-11818, BIO2015-70483-R to A.F.]; Spanish Ministry of Science, Innovation and Universities [BFU2009-11958]; Generalitat Valenciana (Valencia, Spain) [AICO/2017/048]; Natural Sciences and Engineering Research Council of Canada [155219-17 to J.-P.P.]; The RNA group is supported by a grant from the Université de Sherbrooke; J.-P.P. holds the Research Chair of the Université de Sherbrooke in RNA Structure and Genomics, and is a member of the Centre de Recherche du CHUS; B.B.-P. was a recipient of a VALi+d postdoctoral contract of the Generalitat Valenciana [APOSTD/2017/039]; Schleiff group is funded through the Deutsche Forschungsgemeinschaft [SFB 902]. Funding for open access charge: Spanish Ministry of Science, Innovation and Universities.
Conflict of interest statement. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- 1. Palukaitis P. What has been happening with viroids. Virus Genes. 2014; 49:175–184. [DOI] [PubMed] [Google Scholar]
- 2. Di Serio F., Flores R.. Viroids Molecular implements for dissecting RNA trafficking in plants. RNA Biol. 2008; 5:128–131. [DOI] [PubMed] [Google Scholar]
- 3. Ding B. Annual Review of Phytopathology. 2009; 47:105–131. [DOI] [PubMed] [Google Scholar]
- 4. Flores R., Owens R.A., Taylor J.. Pathogenesis by subviral agents: viroids and hepatitis delta virus. Curr. Opin. Virol. 2016; 17:87–94. [DOI] [PubMed] [Google Scholar]
- 5. Steger G., Riesner D.. Viroid research and its significance for RNA technology and basic biochemistry. Nucleic Acids Res. 2018; 46:10563–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Di Serio F., Flores R., Verhoeven J.T., Li S.F., Pallas V., Randles J.W., Sano T., Vidalakis G., Owens R.A.. Current status of viroid taxonomy. Arch. Virol. 2014; 159:3467–3478. [DOI] [PubMed] [Google Scholar]
- 7. Vogt U., Pelissier T., Putz A., Razvi F., Fischer R., Wassenegger M.. Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J. Cell Mol. Biol. 2004; 38:107–118. [DOI] [PubMed] [Google Scholar]
- 8. Itaya A., Folimonov A., Matsuda Y., Nelson R.S., Ding B.. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant Microbe Interact. 2001; 14:1332–1334. [DOI] [PubMed] [Google Scholar]
- 9. Martinez de Alba A.E., Flores R., Hernandez C.. Two chloroplastic viroids induce the accumulation of small RNAs associated with posttranscriptional gene silencing. J. Virol. 2002; 76:13094–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Markarian N., Li H.W., Ding S.W., Semancik J.S.. RNA silencing as related to viroid induced symptom expression. Arch. Virol. 2004; 149:397–406. [DOI] [PubMed] [Google Scholar]
- 11. Vogt U., Pelissier T., Putz A., Razvi F., Fischer R., Wassenegger M.. Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J. 2004; 38:107–118. [DOI] [PubMed] [Google Scholar]
- 12. Gomez G., Pallas V.. Mature monomeric forms of Hop stunt viroid resist RNA silencing in transgenic plants. Plant J. 2007; 51:1041–1049. [DOI] [PubMed] [Google Scholar]
- 13. Carbonell A., Martinez de Alba A.-E., Flores R., Gago S.. Double-stranded RNA interferes in a sequence-specific manner with the infection of representative members of the two viroid families. Virology. 2008; 371:44–53. [DOI] [PubMed] [Google Scholar]
- 14. St-Pierre P., Hassen F., Thompson D., Perreault J.P.. Characterization of the siRNAs associated with peach latent mosaic viroid infection. Virology. 2009; 383:178–182. [DOI] [PubMed] [Google Scholar]
- 15. Martinez G., Donaire L., Llave C., Pallas V., Gomez G.. High-throughput sequencing of Hop stunt viroid-derived small RNAs from cucumber leaves and phloem. Mol. Plant Pathol. 2010; 11:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ivanova D., Milev I., Vachev T., Baev V., Yahubyan G., Minkov G., Gozmanova M.. Small RNA analysis of Potato Spindle Tuber Viroid infected Phelipanche ramosa. Plant Physiol. Biochem. 2014; 74:276–282. [DOI] [PubMed] [Google Scholar]
- 17. Islam W., Noman A., Qasim M., Wang L.. Plant responses to pathogen attack: small RNAs in focus. Int. J. Mol. Sci. 2018; 19:E515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Minoia S., Carbonell A., Di Serio F., Gisel A., Carrington J.C., Navarro B., Flores R.. Specific argonautes selectively bind small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo. J. Virol. 2014; 88:11933–11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katsarou K., Mavrothalassiti E., Dermauw W., Van Leeuwen T., Kalantidis K.. Combined activity of DCL2 and DCL3 is crucial in the defense against potato spindle tuber viroid. PLoS Pathog. 2016; 12:e1005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dadami E., Boutla A., Vrettos N., Tzortzakaki S., Karakasilioti I., Kalantidis K.. DICER-LIKE 4 but not DICER-LIKE 2 may have a positive effect on potato spindle tuber viroid accumulation in Nicotiana benthamiana. Mol. Plant. 2013; 6:232–234. [DOI] [PubMed] [Google Scholar]
- 21. Navarro B., Gisel A., Rodio M.E., Delgado S., Flores R., Di Serio F.. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012; 70:991–1003. [DOI] [PubMed] [Google Scholar]
- 22. Eamens A.L., Smith N.A., Dennis E.S., Wassenegger M., Wang M.B.. In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of Potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology. 2014; 450–451:266–277. [DOI] [PubMed] [Google Scholar]
- 23. Adkar-Purushothama C.R., Brosseau C., Giguere T., Sano T., Moffett P., Perreault J.P.. Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell. 2015; 27:2178–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adkar-Purushothama C.R., Iyer P.S., Perreault J.-P.. Potato spindle tuber viroid infection triggers degradation of chloride channel protein CLC-b-like and Ribosomal protein S3a-like mRNAs in tomato plants. Sci. Rep. 2017; 7:8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carbonell A., Daros J.A.. Artificial microRNAs and synthetic trans-acting small interfering RNAs interfere with viroid infection. Mol. Plant Pathol. 2017; 18:746–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lisón P., Tarraga S., Lopez-Gresa P., Sauri A., Torres C., Campos L., Belles J.M., Conejero V., Rodrigo I.. A noncoding plant pathogen provokes both transcriptional and posttranscriptional alterations in tomato. Proteomics. 2013; 13:833–844. [DOI] [PubMed] [Google Scholar]
- 27. Dube A., Bisaillon M., Perreault J.P.. Identification of proteins from prunus persica that interact with peach latent mosaic viroid. J. Virol. 2009; 83:12057–12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eiras M., Nohales M.A., Kitajima E.W., Flores R., Daros J.A.. Ribosomal protein L5 and transcription factor IIIA from Arabidopsis thaliana bind in vitro specifically Potato spindle tuber viroid RNA. Arch. Virol. 2011; 156:529–533. [DOI] [PubMed] [Google Scholar]
- 29. Martinez G., Castellano M., Tortosa M., Pallas V., Gomez G.. A pathogenic non-coding RNA induces changes in dynamic DNA methylation of ribosomal RNA genes in host plants. Nucleic Acids Res. 2014; 42:1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Castellano M., Martinez G., Marques M.C., Moreno-Romero J., Kohler C., Pallas V., Gomez G.. Changes in the DNA methylation pattern of the host male gametophyte of viroid-infected cucumber plants. J. Exp. Bot. 2016; 67:5857–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mills E.W., Green R.. Ribosomopathies: There's strength in numbers. Science. 2017; 358:eaan2755. [DOI] [PubMed] [Google Scholar]
- 32. Mayer C., Grummt I.. Cellular stress and nucleolar function. Cell Cycle. 2005; 4:1036–1038. [DOI] [PubMed] [Google Scholar]
- 33. Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I.. The nucleolus under stress. Mol. Cell. 2010; 40:216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohbayashi I., Sugiyama M.. Plant nucleolar stress response, a new face in the NAC-dependent cellular stress responses. Front. Plant Sci. 2018; 8:2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weis B.L., Kovacevic J., Missbach S., Schleiff E.. Plant-specific features of ribosome biogenesis. Trends Plant Sci. 2015; 20:729–740. [DOI] [PubMed] [Google Scholar]
- 36. Waltz F., Nguyen T.-T., Arrivé M., Bochler A., Chicher J., Hammann P., Kuhn L., Quadrado M., Mireau H., Hashem Y. et al.. Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants. 2019; 5:106–117. [DOI] [PubMed] [Google Scholar]
- 37. Ohbayashi I., Lin C.Y., Shinohara N., Matsumura Y., Machida Y., Horiguchi G., Tsukaya H., Sugiyama M.. Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in arabidopsis. Plant Cell. 2017; 29:2644–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kressler D., Hurt E., Bassler J.. A puzzle of life: crafting ribosomal subunits. Trends Biochem. Sci. 2017; 42:640–654. [DOI] [PubMed] [Google Scholar]
- 39. Rorbach J., Aibara S., Amunts A.. Ribosome origami. Nat. Struct. Mol. Biol. 2017; 24:879–881. [DOI] [PubMed] [Google Scholar]
- 40. Ahmed T., Yin Z., Bhushan S.. Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci. Rep. 2016; 6:35793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Henras A.K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y.. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci.: CMLS. 2008; 65:2334–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bassler J., Hurt E.. Eukaryotic ribosome assembly. Annu. Rev. Biochem. 2018; 88:281–306. [DOI] [PubMed] [Google Scholar]
- 43. Hang R., Wang Z., Deng X., Liu C.Y., Yan B., Yang C., Song X., Mo B., Cao X.. Ribosomal RNA biogenesis and its response to chilling stress in oryza sativa L. Plant Physiol. 2018; 177:381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Palm D., Streit D., Shanmugam T., Weis B.L., Ruprecht M., Simm S., Schleiff E.. Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing. Nucleic Acids Res. 2018; 47:1880–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tomecki R., Sikorski P.J., Zakrzewska-Placzek M.. Comparison of preribosomal RNA processing pathways in yeast, plant and human cells - focus on coordinated action of endo- and exoribonucleases. FEBS Lett. 2017; 591:1801–1850. [DOI] [PubMed] [Google Scholar]
- 46. Perry K.L., Palukaitis P.. Transcription of tomato ribosomal DNA and the organization of the intergenic spacer. Mol. Gen. Genet.: MGG. 1990; 221:103–112. [DOI] [PubMed] [Google Scholar]
- 47. Echevarria-Zomeno S., Yanguez E., Fernandez-Bautista N., Castro-Sanz A.B., Ferrando A., Castellano M.M.. Regulation of translation initiation under biotic and abiotic stresses. Int. J. Mol. Sci. 2013; 14:4670–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z.F., Ying T.J., Bao B.L., Huang X.D.. Characteristics of fruit ripening in tomato mutant epi. J. Zhejiang Univ. Sci. B. 2005; 6:502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Belles J.M., Carbonell J., Conejero V.. Polyamines in plants infected by citrus exocortis viroid or treated with silver ions and ethephon. Plant Physiol. 1991; 96:1053–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adkar-Purushothama C.R., Perreault J.P.. Alterations of the viroid regions that interact with the host defense genes attenuate viroid infection in host plant. RNA Biol. 2018; 15:955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rivera M.C., Maguire B., Lake J.A.. Isolation of ribosomes and polysomes. Cold Spring Harbor Protoc. 2015; 2015:293–299. [DOI] [PubMed] [Google Scholar]
- 52. Hsu P.Y., Calviello L., Wu H.L., Li F.W., Rothfels C.J., Ohler U., Benfey P.N.. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. PNAS. 2016; 113:E7126–E7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mustroph A., Juntawong P., Bailey-Serres J.. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol. Biol. 2009; 553:109–126. [DOI] [PubMed] [Google Scholar]
- 54. Lopez-Gresa M.P., Lison P., Yenush L., Conejero V., Rodrigo I., Belles J.M.. Salicylic acid is involved in the basal resistance of tomato plants to citrus exocortis viroid and tomato spotted wilt virus. PLoS One. 2016; 11:e0166938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Missbach S., Weis B.L., Martin R., Simm S., Bohnsack M.T., Schleiff E.. 40S ribosome biogenesis co-factors are essential for gametophyte and embryo development. PLoS One. 2013; 8:e54084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sambrook J, R.R. Molecular Cloning: A Laboratory Manual. 2001; 3rd ednNY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 57. Verhoeven J.T.T., Jansen C.C.C., Willemen T.M., Kox L.F.F., Owens R.A., Roenhorst J.W.. Natural infections of tomato by Citrus exocortis viroid, Columnea latent viroid, Potato spindle tuber viroid and Tomato chlorotic dwarf viroid. Eur. J. Plant Pathol. 2004; 110:823–831. [Google Scholar]
- 58. Campos L., Granell P., Tarraga S., Lopez-Gresa P., Conejero V., Belles J.M., Rodrigo I., Lison P.. Salicylic acid and gentisic acid induce RNA silencing-related genes and plant resistance to RNA pathogens. Plant Physiol. Biochem.: PPB. 2014; 77:35–43. [DOI] [PubMed] [Google Scholar]
- 59. Kalantidis K., Denti M.A., Tzortzakaki S., Marinou E., Tabler M., Tsagris M.. Virp1 is a host protein with a major role in Potato spindle tuber viroid infection in Nicotiana plants. J. Virol. 2007; 81:12872–12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Barry C.S., Fox E.A., Yen H., Lee S., Ying T., Grierson D., Giovannoni J.J.. Analysis of the ethylene response in the epinastic mutant of tomato. Plant Physiol. 2001; 127:58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Diener T.O. Discovering viroids—a personal perspective. Nat. Rev. Microbiol. 2003; 1:75–80. [DOI] [PubMed] [Google Scholar]
- 62. Ding B., Itaya A.. Viroid: a useful model for studying the basic principles of infection and RNA biology. Mol. Plant-Microbe Interact.: MPMI. 2007; 20:7–20. [DOI] [PubMed] [Google Scholar]
- 63. Jakab G., Kiss T., Solymosy F.. Viroid pathogenicity and pre-rRNA processing: A model amenable to experimental testing. Biochim. Biophys. Acta (BBA) - Gene Struct. Expression. 1986; 868:190–197. [Google Scholar]
- 64. Meduski C.J., Velten J.. PSTV sequence similarity to large rRNA. Plant Mol. Biol. 1990; 14:625–627. [DOI] [PubMed] [Google Scholar]
- 65. Kiss T., Posfai J., Solymosy F.. Sequence homology between potato spindle tuber viroid and U3B snRNA. FEBS Lett. 1983; 163:217–220. [DOI] [PubMed] [Google Scholar]
- 66. Hughes J.M., Ares M. Jr.. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991; 10:4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sharma K., Tollervey D.. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell Biol. 1999; 19:6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y. et al.. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002; 417:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dutca L.M., Gallagher J.E., Baserga S.J.. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011; 39:5164–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Qi Y., Ding B.. Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell. 2003; 15:2566–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Idol R.A., Robledo S., Du H.Y., Crimmins D.L., Wilson D.B., Ladenson J.H., Bessler M., Mason P.J.. Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol. Dis. 2007; 39:35–43. [DOI] [PubMed] [Google Scholar]
- 72. Cho H.K., Ahn C.S., Lee H.S., Kim J.K., Pai H.S.. Pescadillo plays an essential role in plant cell growth and survival by modulating ribosome biogenesis. Plant J. 2013; 76:393–405. [DOI] [PubMed] [Google Scholar]
- 73. Weis B.L., Missbach S., Marzi J., Bohnsack M.T., Schleiff E.. The 60S associated ribosome biogenesis factor LSG1-2 is required for 40S maturation in Arabidopsis thaliana. Plant J.: Cell Mol. Biol. 2014; 80:1043–1056. [DOI] [PubMed] [Google Scholar]
- 74. Kojima K., Tamura J., Chiba H., Fukada K., Tsukaya H., Horiguchi G.. Two nucleolar proteins, GDP1 and OLI2, function as ribosome biogenesis factors and are preferentially involved in promotion of leaf cell proliferation without strongly affecting leaf adaxial-abaxial patterning in arabidopsis thaliana. Front. Plant Sci. 2017; 8:2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Maekawa S., Ishida T., Yanagisawa S.. Reduced expression of APUM24, encoding a novel rRNA processing factor, induces sugar-dependent nucleolar stress and altered sugar responses in arabidopsis thaliana. Plant Cell. 2018; 30:209–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bonfiglioli R.G., McFadden G.I., Symons R.H.. In-situ hybridization localizes avocado sunblotch viroid on chloroplast thylakoid membranes and coconut cadang cadang viroid in the nucleus. Plant J. 1994; 6:99–103. [Google Scholar]
- 77. Taliansky M.E., Brown J.W.S., Rajamaki M.L., Valkonen J.P.T., Kalinina N.O.. Maramorosch K, Shatkin AJ, Murphy FA. Chapter 5 - Involvement of the Plant Nucleolus in Virus and Viroid Infections: Parallels with Animal Pathosystems. Advances in Virus Research. 2010; 77:119–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hill J.M., Zhao Y., Bhattacharjee S., Lukiw W.J.. miRNAs and viroids utilize common strategies in genetic signal transfer. Front. Mol. Neurosci. 2014; 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li S., Liu L., Zhuang X., Yu Y., Liu X., Cui X., Ji L., Pan Z., Cao X., Mo B. et al.. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013; 153:562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Desnoyers G., Bouchard M.P., Masse E.. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet. 2013; 29:92–98. [DOI] [PubMed] [Google Scholar]
- 81. Fukaya T., Iwakawa H.O., Tomari Y.. MicroRNAs block assembly of eIF4F translation initiation complex in drosophila. Mol. Cell. 2014; 56:67–78. [DOI] [PubMed] [Google Scholar]
- 82. Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crete P., Voinnet O., Robaglia C.. Biochemical evidence for translational repression by arabidopsis MicroRNAs. Plant Cell. 2009; 21:1762–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Iwakawa H., Tomari Y.. Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell. 2013; 52:591–601. [DOI] [PubMed] [Google Scholar]
- 84. Reis R.S., Hart-Smith G., Eamens A.L., Wilkins M.R., Waterhouse P.M.. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat. Plants. 2015; 1:14027. [DOI] [PubMed] [Google Scholar]
- 85. Flores R., Navarro B., Kovalskaya N., Hammond R.W., Di Serio F.. Engineering resistance against viroids. Curr. Opin. Virol. 2017; 26:1–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.