Abstract
Cell type identification is essential for single-cell RNA sequencing (scRNA-seq) studies, currently transforming the life sciences. CHETAH (CHaracterization of cEll Types Aided by Hierarchical classification) is an accurate cell type identification algorithm that is rapid and selective, including the possibility of intermediate or unassigned categories. Evidence for assignment is based on a classification tree of previously available scRNA-seq reference data and includes a confidence score based on the variance in gene expression per cell type. For cell types represented in the reference data, CHETAH’s accuracy is as good as existing methods. Its specificity is superior when cells of an unknown type are encountered, such as malignant cells in tumor samples which it pinpoints as intermediate or unassigned. Although designed for tumor samples in particular, the use of unassigned and intermediate types is also valuable in other exploratory studies. This is exemplified in pancreas datasets where CHETAH highlights cell populations not well represented in the reference dataset, including cells with profiles that lie on a continuum between that of acinar and ductal cell types. Having the possibility of unassigned and intermediate cell types is pivotal for preventing misclassification and can yield important biological information for previously unexplored tissues.
INTRODUCTION
Single-cell RNA-sequencing (scRNA-seq) is transforming our ability to study heterogeneous cell populations (1–6). While tools to help interpret scRNA-seq data are developing rapidly (7–14), challenges in data analysis remain (15), with cell type identification a prominent example. Accurate cell type identification is a prerequisite for any study of heterogeneous cell populations, both when the focus is on subsets of a particular cell type of interest or when investigating the population structure as a whole (16–20). The introduction of single cell RNA sequencing has paved the way for rapidly discovering previously uncharacterized cell types (21–23) and this application too would greatly benefit from efficient identification of known cell types prior to focusing on new types.
Research into tumor composition presents an even more challenging setting, as the RNA expression profile of malignant cells is often different from any known cell type, as well as unique to the patient or even to the biopsy (24,25). Malignant cells can sometimes be identified in scRNA-seq data (26) but this is not always feasible or even possible, for instance with tumors that do not harbor easily identified copy number variations. In both cases, a first sign of the malignancy of cells in the sample is their imperviousness to classification, simply because their expression profiles do not resemble that of any known, healthy cell type.
Cell type identification in scRNA-seq studies is currently often done manually, starting by identifying transcriptionally similar cells using clustering. This is frequently followed by differential expression analysis of the resulting cell clusters combined with visual marker gene inspection (4,24,25,27–29). Such manual cell type identification is time-consuming and often subjective due to the choice of clustering method and parameters for example, or to the lack of consensus regarding which marker gene to use for each cell type. Such analyses are becoming more complex given the fast-expanding catalogue of defined cell types (15). Canonical cell surface markers are also not always suitable in scRNA-seq studies because the transcripts of these genes may not be measurable in the corresponding cell type owing to low expression or to degradation of the mRNA. This is aggravated by technical difficulties (drop-out) and, more generally, by the poor correlation between protein expression and mRNA abundances (22).
Recently, a number of cell type identification algorithms have emerged to address these problems. Automated methods such as scmap (30) and SingleR (31) base their cell type call on comparisons with annotated reference data using automatically chosen genes that optimally discriminate between cell types. A good cell type identification method should be both sensitive and selective. That is, it should correctly identify as many cells as possible, while not classifying cells when based on insufficient evidence. If the cell being identified is of a type that is not represented in the reference, such misclassification can easily occur. This is a concern when studying malignant cells which are often too heterogeneous to include in the reference data. To avoid overclassification, methods such as scmap (30) therefore leave cells unclassified if they are too dissimilar to any reference data.
Both the complete lack of classification as well as overclassification is unsatisfactory. For example, if the evidence for a very specific cell type assignment such as effector CD8 T-cell is not strong enough, a more general, less specific assignment such as T cell may still legitimately be made and might still be useful. The reason for such an intermediate cell type assignment could be that the correct T-cell subtype is not part of the reference dataset, or even that there is insufficient read-depth for the more specific call to be made. An even more interesting case is that of cells that are biologically of an intermediate type such as differentiating cells or cells undergoing transdifferentiation.
Here we present CHETAH (CHaracterization of cEll Types Aided by Hierarchical classification), an algorithm that explicitly allows the assignment of cells to an intermediate or unassigned type. The unassigned and intermediate types are inferred using a tree that is constructed from the reference data and which guides the classification. CHETAH’s classification is a stepwise process that traverses the tree and, depending on the available evidence, ends at one of the reference cell types or halts at the unassigned or one of the intermediate types. CHETAH is able to correctly classify published datasets and, in comparison to other methods, performs equally or better when considering cells whose type is represented in the reference data. For cells of an unknown type, CHETAH is more selective, yielding a classification that is as fine-grained as is justified by the available data. The benefit of calling unassigned and intermediate types is highlighted in several tumor datasets, showing CHETAH is consistently selective. This makes CHETAH a powerful tool for identifying cells that are not in the reference, such as malignant tumor cells, novel or intermediate cell types. The latter is shown in an analysis of published pancreas datasets, where a manifest expression gradient of cells with types varying between acinar and duct cell is described. CHETAH is implemented in R (32), is available at github.com/jdekanter/CHETAH, and has been incorporated in Bioconductor (33) release 3.9. It comes with an extensive Shiny application that makes exploration of the cell type identification process and the gene expression differences that support the classification very intuitive. CHETAH has been created bearing tumor analyses in mind, but as is demonstrated, it also complements existing methods for exploring previously uncharacterized non-cancerous tissues and cell types.
MATERIALS AND METHODS
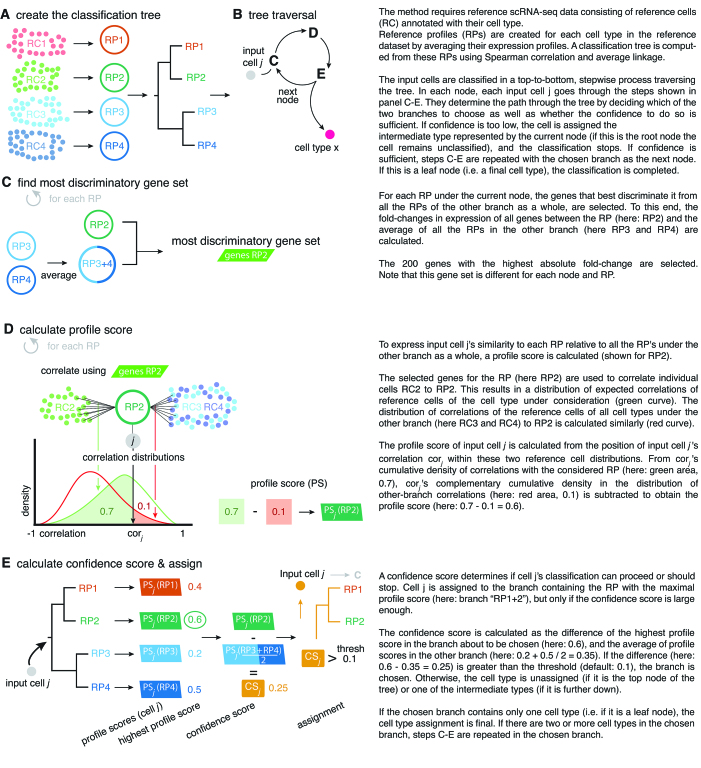
An outline of the CHETAH algorithm is depicted in Figure 1. The method requires a reference scRNA-seq dataset, annotated for cell type. Throughout this study, the reference dataset is completely independent from the dataset that is being classified. First a hierarchical classification tree is constructed from the reference scRNA-seq data (Figure 1A). Each input cell is classified individually by traversing the tree (Figure 1B). At each step of the process the cell to be classified is correlated to the expression profiles of the reference data cell types. This is done by first selecting the set of genes that best discriminates each reference cell type from all the cell types, collectively, in the opposite branch of the tree (Figure 1C). The input cell correlation to a reference cell type is compared to the distribution of correlations of the reference cells to assess whether there is enough evidence to allow this cell to take the next step (Figure 1D and E). If the confidence threshold is not passed, further classification of the cell stops and it is marked as unassigned if the evidence runs out at the top of the tree, or as intermediate if this happens within the classification tree. Classification also stops when a cell reaches one of the leaves of the tree, yielding assignment to a specific cell type.
Figure 1.
The CHETAH algorithm.
Reference data
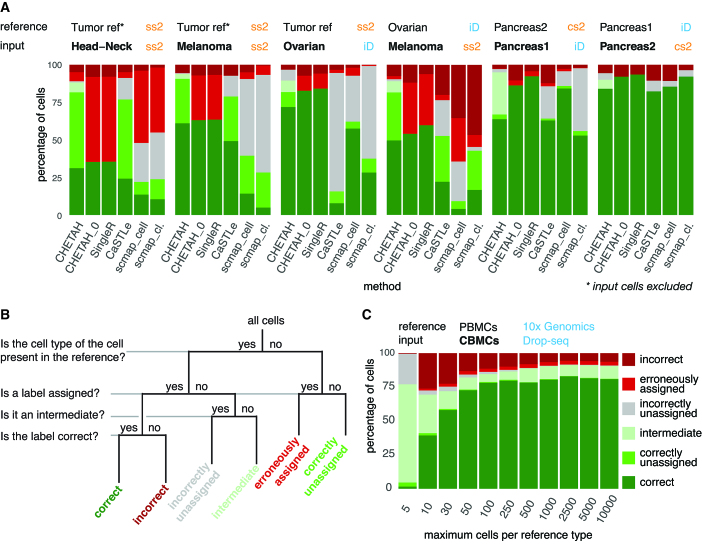
In order to classify input cells CHETAH requires scRNA-seq reference data along with cell type labels for each reference cell. Here, the reference dataset is always a completely independently generated dataset, from a different study and in several cases using a different scRNA-seq platform. The reference data needs to be normalized to an identical total number of transcripts per cell and should be expressed in log-scale. Malignant cells are best left out of the reference because they are too ill-defined and too patient-specific (34). In all the reference datasets used here, such cells cluster by patient whereas non-malignant cells largely cluster by cell type. The reference must contain at least 10 cells per cell type to adequately represent its transcriptional program as well as its variance (Figure 3C). More than 10 cells per reference cell type improves performance. More than 200 cells per cell type is superfluous. Since this also increases the computational burden it is useful to restrict the number of cells per reference cell type to a maximum of 500. This does not restrict the number of cells that can be analyzed in the query dataset. The selection of genes from the reference dataset for classification at each step (Figure 1B) is aimed at finding those with the highest discriminatory power. When using a reference dataset with a high drop-out rate, i.e. low transcript coverage per cell, it is advocated to remove highly expressed genes such as ribosomal protein genes from reference datasets beforehand, since their drop-out can reduce classification accuracy. For the sake of uniformity, ribosomal protein genes were removed here from all reference datasets although this only increased classification accuracy in one case (Figure 3C).
Figure 3.
CHETAH compared with other methods (bottom labels), across six combinations of input and reference datasets (top labels, including the corresponding scRNA-seq platform: ss2: Smart-seq2; iD: inDrops; cs2: CEL-seq2. Microfluidics methods in blue, well-plate methods in orange). For scmap, both the ‘cell’ mode (scmap_cell) and ‘cluster’ mode (scmap_cl.) where evaluated. CHETAH was run with default settings, but also with a zero confidence score threshold (CHETAH_0), thus forcing it to classify all cells to a final type. (A) Percentages of cells per classification result category as shown in (B). (B) Classification result categories used in A. (C) The influence of the number of cells per reference cell type on CHETAH’s classification performance was investigated as follows. The 7830 cells of the (Drop-seq protocol) CITE-seq study (39), were classified with reference cells from the PBMC dataset (27), generated with the 10× Genomics platform. This dataset contained a total of 68 579 cells. The numbers on the y-axis are the number of (randomly sampled) cells per reference cell type taken to classify the input dataset. Classification results were divided into the six categories depicted in (B). Besides investigating the influence of the number of cells in a reference type, this analysis also serves as an example of cross-platform performance, as well as an example using datasets with large numbers of cells. More details of the datasets used can be found in Table 1. Note that in the other analyses reported throughout, no limitation is placed on the number of cells per reference type.
Unless stated otherwise, the reference dataset used here, called ‘Tumor ref.’, consists of a combination of datasets of Colorectal (35), Breast (36), Melanoma (24) and Head-Neck tumors (25). The data for all these studies was generated using the Smart-seq2 platform. The cell types of the reference dataset were based on published manual classification of cell clusters using marker gene expression. The Melanoma and Head-Neck studies discuss the T cells in terms of their CD4+, CD8+ and T-reg subtypes but not all of these labels are available for all the cells in the online material of these publications. These reference cells were therefore classified manually using the same marker genes as used in the publications. Cells of a dataset being classified are of course excluded from the reference. In other words, all results reported are by classification using reference datasets completely independent from the query dataset. When comparing CHETAH and SingleR (31) results, the latter was run with averaged single-cell data because SingleR uses bulk, rather than single-cell expression profiles as its reference. Further details of the datasets used in this work, including pre-processing, are given in Table 1.
Table 1.
Datasets used in this study
| Name (publication) | Protocol | No. of healthy cells | No. of tumor cells | No. of cell types | Pre-processing after download | Biopsies | No. of donors |
|---|---|---|---|---|---|---|---|
| Melanoma (24) | Smart-seq2 | 3262 | 1251 | 9 | discarded cells without annotation | melanoma | 19 |
| Head-Neck (25) | Smart-seq2 | 3345 | 2215 | 12 | discarded cells without annotation | primary head and neck squamous cell carcinoma | 18 |
| Colorectal (35) | Smart-seq2 | 272 | 92 | 7 | no | colorectal cancer | 11 |
| Breast (36) | Smart-seq2 | 198 | 317 | 3 | no | breast cancer | 11 |
| Tumor ref. (24,25,35,36) | Smart-seq2 | 6122 | none | 12 | combined Melanoma, Head-Neck, Breast and Colorectal datasets, discarding malignant cells. | Detailed above | 19+18+11+11 |
| Ovarian (18) | InDrops | 2814 | 300 | 9 | no | ovarian cancer ascites | 4 |
| PBMC (27) | 10X Genomics | 68579 | none | 16 | no | healthy PBMC cells | 1 |
| CBMC (39) | Drop-seq | 7830 | none | 13 | as described (39) | cord blood CBMCs | unknown |
| Pancreas1 (16) | inDrops | 8569 | none | 14 | no | healthy pancreas | 4 |
| Pancreas2 (41) | CEL-seq2 | 2292 | none | 9 | no | healthy pancreas | 4 |
The PBMC and CBMC datasets were labeled identically to ensure comparability of annotated cell types.
Classification tree
The first step is to create a reference profile (RP) for each cell type in the set of reference cells by averaging, for each cell type, the logged gene expression over all cells of that type (Figure 1A). The RPs are subsequently clustered hierarchically using Spearman correlation and average linkage to obtain the classification tree.
Hierarchical classification
The classification of input cells proceeds in a stepwise fashion, from the root to the leaves of the classification tree. At each step, the branch is selected that contains the reference cell type most likely to be the correct one, but the classification stops if the confidence in this decision becomes too low (see confidence score below). As described under profile score, the choice of the most likely cell type and therefore which branch to choose, is based on the cell's similarities to each of the individual RPs under each branch. The similarity of a cell to a RP under consideration (called the candidate RP), in the branch under consideration (called the candidate branch), is always in relation to all the RPs under the (so-called) other branch. During the classification process, only the leaf node data (i.e. from all cells of a particular reference cell type) are used. Any details of the tree topology under either branch are ignored, i.e. no hypothetical expression profiles are inferred for the intermediate tree nodes. After calculating the cell's similarities to all RPs under both branches, the cell is assigned to the branch that contains the cell type to which it is most similar, provided the evidence is strong enough based on the confidence score.
Feature selection
The similarity of a cell to a reference profile is based on their Spearman correlation. This choice is based on its identical performance to other correlation methods (Supplementary Figure S1A) and on the fact that there is no assumption about the underlying distribution. The correlation is calculated using the subset of genes that best discriminates between the candidate RP in the candidate branch, and the average expression profile of the other branch as a whole. (The latter is calculated as the mean of all RPs under that branch). The selection of the best subset of genes, a process known as feature selection, is not critical and good results are achieved when simply using the 200 genes that have the largest absolute fold-change between the candidate RP and the average expression profile of the other branch. This choice is based on a variety of parameter sweeps and shown in Supplementary Figure S1. It is important to note that the feature set, i.e. the subset of genes used to calculate similarities, is different for each RP and for each node of the classification tree. Many different feature selection methods work well (Supplementary Figure S1). The use of different discriminatory gene sets at each decision node and for each RP is an important, novel aspect of the method.
Similarities
The similarity of a cell to an RP in the candidate branch is of course reflected in their correlation, but the values of these correlations to the various RPs cannot be directly used for comparisons. The reason is that the subset of genes used for each correlation is generally different for each RP and for each node. The similarity of an input cell j to candidate RP x is therefore cast in relative terms by using the cumulative probabilities of this correlation within two different distributions of correlations. The first one is the distribution of self-correlations, that is, the distribution of the correlations of the individual cells constituting the candidate RP to that candidate RP itself. These self-correlations represent the typical correlation values for a cell that is really of that type. The second distribution is that of the non-self correlations. They are the correlations, again to the candidate RP, of all the individual reference cells under the other branch. They represent the correlation values that can be expected for cells that are not of any type under the candidate branch. By contrasting the two cumulative probabilities a profile score is obtained that robustly points the way through the classification tree.
Profile scores
The two cumulative probabilities just defined are used to define the profile score 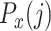 , representing cell j’s similarity to candidate RP x, as follows:
, representing cell j’s similarity to candidate RP x, as follows:
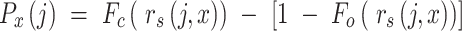 |
(1) |
with
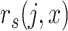 the Spearman correlation of input cell
the Spearman correlation of input cell 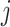 ’s expression with candidate reference profile
’s expression with candidate reference profile 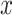
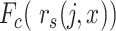 the cumulative probability of j’s correlation within the distribution of self-correlations
the cumulative probability of j’s correlation within the distribution of self-correlations 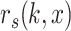 , that is, of all reference cells
, that is, of all reference cells 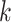 of type
of type 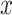 with their ‘own’ candidate reference profile
with their ‘own’ candidate reference profile 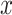
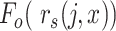 the cumulative probability of j’s correlation within the distribution of correlations
the cumulative probability of j’s correlation within the distribution of correlations 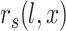 of all reference cells
of all reference cells 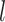 under the other branch, again with reference profile
under the other branch, again with reference profile 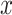
The profile score 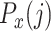 has a value between 1 and −1 and is, in a particular node, a measure for the likelihood that cell j is of type x. A value of 1 means that cell j is much more likely to be of type x (and therefore belong to its branch) rather than any of the types in the other branch and, conversely, −1 represents the lowest likelihood of this being so, and therefore cell j is much more likely to belong in the other branch. In each node, one set of genes is selected for each RP under that node. This gene set is used for all the correlations (of both input and reference cells) needed to calculate the profile scores. Note that due to the different gene subsets used in each step of the tree traversal, the most similar RP for a cell may change during the steps of the classification process. For example, during the first few steps a cell that in reality is of type CD4 T-cell could initially, and incorrectly, appear more similar to a CD8 T cell than to the expected CD4 T-cell type. This would however still lead to the correct branch choice, namely that of all T cells. In later steps the similarity to the actual CD4 T-cell type would become strongest, guiding the cell to a correct final CD4 T-cell label.
has a value between 1 and −1 and is, in a particular node, a measure for the likelihood that cell j is of type x. A value of 1 means that cell j is much more likely to be of type x (and therefore belong to its branch) rather than any of the types in the other branch and, conversely, −1 represents the lowest likelihood of this being so, and therefore cell j is much more likely to belong in the other branch. In each node, one set of genes is selected for each RP under that node. This gene set is used for all the correlations (of both input and reference cells) needed to calculate the profile scores. Note that due to the different gene subsets used in each step of the tree traversal, the most similar RP for a cell may change during the steps of the classification process. For example, during the first few steps a cell that in reality is of type CD4 T-cell could initially, and incorrectly, appear more similar to a CD8 T cell than to the expected CD4 T-cell type. This would however still lead to the correct branch choice, namely that of all T cells. In later steps the similarity to the actual CD4 T-cell type would become strongest, guiding the cell to a correct final CD4 T-cell label.
Confidence score
Each input cell is assigned to the branch containing the candidate reference cell type for which it has the highest profile score. This assignment represents the choice between the left and right branch, but a key design goal of the algorithm is its ability to stop classification at an intermediate node. The choice for each cell j, between stopping classification or continuing to the next round, is based on its confidence score 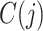 defined as
defined as
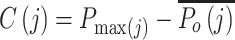 |
(2) |
with 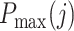 the highest profile score for cell j in the branch about to be chosen and
the highest profile score for cell j in the branch about to be chosen and 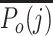 the mean of the profile scores in the other branch, i.e. the branch not containing the reference profile having the highest profile score (Figure 1C). Equation (2) is always positive because branches leading to a negative score are by definition never chosen by the algorithm. The confidence score is a measure for the evidence to assign a cell to a branch, with 2 representing maximal evidence and 0 representing no evidence. The confidence score has an easy explanation. If it is close to 0, the best candidate cell type in the branch about to be chosen is as good as the average of the cell types in the other branch. This implies that there is no basis to justify the choice between either branch, so none should be taken and classification of the cell should therefore stop in the current node. In contrast, a large score represents good support to continue the classification because there is a cell type in the candidate branch that has a much better profile score than the average profile score of the other branch. By default, cells are assigned to the branch if the confidence score is >0.1, but different values can also be specified in the algorithm's parameters. Cells that remain in a non-leaf node of the tree are called unassigned or of intermediate type whereas cells assigned to a leaf-node are of a final type. The labels for the intermediate types are generated automatically (Node1, Node2, etc.) but biologically meaningful names such as T cell can often readily be given. By choosing a cut-off >0.1, only the more confident calls will be made, hence more cells will be labeled as being unassigned or of intermediate type. Conversely, by lowering the confidence cut-off, the algorithm will classify more cells to a final type, however such calls are supported by less evidence. A cut-off of 0.0 forces the method to classify all cells to a final type, as is exemplified later. The above stepwise calculations of the profile scores and confidence scores yield an elegant and, importantly, transparent algorithm.
the mean of the profile scores in the other branch, i.e. the branch not containing the reference profile having the highest profile score (Figure 1C). Equation (2) is always positive because branches leading to a negative score are by definition never chosen by the algorithm. The confidence score is a measure for the evidence to assign a cell to a branch, with 2 representing maximal evidence and 0 representing no evidence. The confidence score has an easy explanation. If it is close to 0, the best candidate cell type in the branch about to be chosen is as good as the average of the cell types in the other branch. This implies that there is no basis to justify the choice between either branch, so none should be taken and classification of the cell should therefore stop in the current node. In contrast, a large score represents good support to continue the classification because there is a cell type in the candidate branch that has a much better profile score than the average profile score of the other branch. By default, cells are assigned to the branch if the confidence score is >0.1, but different values can also be specified in the algorithm's parameters. Cells that remain in a non-leaf node of the tree are called unassigned or of intermediate type whereas cells assigned to a leaf-node are of a final type. The labels for the intermediate types are generated automatically (Node1, Node2, etc.) but biologically meaningful names such as T cell can often readily be given. By choosing a cut-off >0.1, only the more confident calls will be made, hence more cells will be labeled as being unassigned or of intermediate type. Conversely, by lowering the confidence cut-off, the algorithm will classify more cells to a final type, however such calls are supported by less evidence. A cut-off of 0.0 forces the method to classify all cells to a final type, as is exemplified later. The above stepwise calculations of the profile scores and confidence scores yield an elegant and, importantly, transparent algorithm.
Parameters
CHETAH comes with an extensive Shiny application, is implemented in R (32), is available at github.com/jdekanter/CHETAH and has been incorporated in Bioconductor (33) release 3.9. Easily selectable parameters include the choice of correlation measure (default: Spearman), the discriminatory gene set selection method (default: the 200 genes with largest absolute expression difference between reference cell type under consideration and all reference cell types in the opposite branch), the hierarchical clustering method (default: Spearman, average linkage) and the confidence score threshold (default: 0.1). These default settings are uniformly applied throughout this study.
RESULTS
The CHETAH algorithm is summarized in the first paragraph of the ‘Materials and Methods’ section and in Figure 1. The method makes use of a reference dataset with cell type annotations. Throughout this study the reference dataset is always a completely independently generated dataset, from a different study and in several cases using a different scRNA-seq platform. Reference cell types are hierarchically clustered into a classification tree which guides the cell type identification process (Figure 1A). The classification tree aids cell type identification but is not intended as a recapitulation of cell taxonomy. The cells to be classified are shunted from the root of this tree to its leaves (Figure 1B), but only to the most specific tree node that is still supported by the available evidence, as quantified by a robust measure of confidence (Figure 1D and E). Confidence is based on passing a threshold. This is determined by the degree to which the input cell's correlation to a reference cell type fits with the distribution of correlations of reference cells of the same type and contrasted with the degree to which it fits with that of other reference cells to this type (Figure 1D and E). The genes on which the classification is based are selected to be those that are most discriminatory for each step in the classification (Figure 1C). This too is an important aspect of the method. Many parameters such as correlation measure, number of discriminatory genes, number of cells per reference type, etc., are selectable by the user (Materials and Methods) and the choice for the single set of default parameters used throughout this study is explained in the ‘Materials and Methods’ section. Cells for which classification confidence runs out are typically of a type that is not present in the reference dataset, and are said to be either unassigned or of an intermediate type. Intermediate entails that classification has halted at a node due to lack of confidence to proceed. Unassigned entails that this already occurred at the first step in the classification tree. Note that there are several intermediate types, each corresponding to one of the internal nodes of the classification tree.
CHETAH’s accuracy is investigated by comparing its classifications with published cell type labels. The aim is to reproduce these using only the reference data. The reference datasets used here are always from a source that is completely independent of the query dataset, ensuring that the reported accuracies do not reflect bias from over-fitting. Since the accuracy might be lower if the scRNA-seq technology of the input data and the reference differs, cross-platform results are also examined. CHETAH is subsequently compared to other cell type identification methods and the effectiveness of the intermediate cell type assignments is also demonstrated in an analysis of previously published pancreas datasets. For an overview of the datasets see Table 1.
Accuracy
The performance of CHETAH is first evaluated by applying it to Melanoma (24) and Head-Neck (25) cancer datasets. The classifications of the these datasets is shown in Figure 2 and Supplementary Figure S2, summarized in Table 2. Throughout this study all classification results are obtained by applying CHETAH on a new query dataset, with a completely different, independent dataset as reference. The reported results are therefore without bias toward the query dataset, as could be the case if reference and query datasets are the same. Since the reference datasets do not contain malignant tumor cells, such cells should not be classified to any final type, but as unassigned or any of the intermediate types instead. CHETAH correctly classifies practically all (mean > 99%) malignant cells as unassigned or intermediate types. Note that in the published data the classifications were manual while the identification of tumor cells was based on estimated copy number variations. In contrast, CHETAH’s type assignments are fully automatic and the aberrant nature of the malignant cells is indicated by their classification as unassigned or intermediate. This selectivity is an important quality of the method, essential for preventing the type of misclassification that readily occurs when methods forcefully assign every cell to a type regardless of the evidence. Selectivity is especially relevant when dealing with tumor samples, as well as with samples containing cell types not present in the reference dataset.
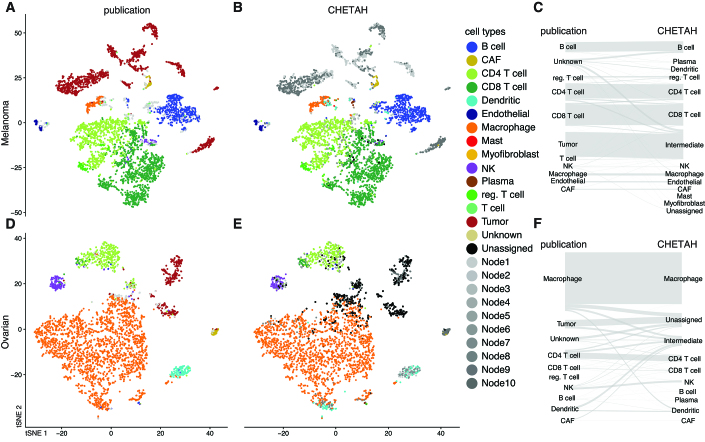
Figure 2.
CHETAH’s classification of two tumor sample datasets is nearly identical to the published manual classification. The t-SNE plots depict each cell as a dot, with the colors representing the inferred cell type shown in the legend. Gray colors indicate intermediate cell types which are labeled automatically as Node1, Node2, etc. For the corresponding classification trees see Supplementary Figure S2. Rows of panels: datasets classified (A-C: Melanoma, Tirosh et al. (24); D-F: Ovarian, Schelker et al. (18)); columns: classification method. For an overview of the datasets see Table 1.
Table 2.
Percentages of cell type labels as inferred by CHETAH, compared with the published cell types
| Reference dataset | Input dataset | % correctly unassigned | % identical final type | % intermediate type, same lineage | % intermediate type, different lineage | % Incorrectly unassigned | % different final type |
|---|---|---|---|---|---|---|---|
| Tumor ref. without Melanoma | Melanoma (24) | 99.5 | 86.9 | 4.1 | 0.8 | 0.5 | 7.7 |
| Tumor ref. without Head-Neck | Head-Neck (25) | 89.2 | 71.2 | 14.6 | 1.1 | 1.9 | 11.2 |
| Tumor ref. | Ovarian (18) | 99.3 | 79.9 | 9.0 | 0.7 | 6.8 | 3.6 |
The reference and input datasets are shown. For an overview of the datasets see Table 1. When classifying the Melanoma and Head-Neck datasets, these datasets are left out of the ‘Tumor ref.’ reference, as indicated. The column correctly unassigned shows the percentage of cells of a type that was absent from the reference that were classified as unassigned or any of the intermediate types. The other columns refer to sample cells of a type represented in the reference that should therefore be assigned and contain percentages of cells of final or intermediate type, summing to 100%. The term lineage refers to the classification tree determined by CHETAH.
CHETAH classifies the majority (mean 79%) of non-malignant cells the same as in the original publication. Of the cells classified differently, the majority (mean 61%) are classified as an intermediate type. In the inferred classification tree these intermediate assignments are overwhelmingly in the correct classification lineage (85, 91 and 95% for Melanoma, Head-Neck and Ovarian, respectively). Only a small number of cells are labeled differently by CHETAH. For many of them there is in fact strong evidence from established marker gene expression that the assignment from CHETAH is correct (Supplementary Figures S4 and 5). Taken together, these results show that the selective approach works well. Cells of an established type that are present in the reference dataset are classified correctly. Samples cells of a new or aberrant type, not represented in the reference dataset are either not assigned to a type or are classified as an intermediate type, an outcome that should indeed be regarded as a pointer for a more detailed inspection.
Cross-platform classification
The data from the Melanoma and Head-Neck studies were obtained using Smart-seq2 (37) and were also classified using reference data originating from the same platform. To evaluate CHETAH’s performance across platforms, an Ovarian dataset (18) produced on the inDrops platform (38) was analyzed with CHETAH using the ‘Tumor ref.’ reference (Smart-seq2-based) and conversely, the Melanoma dataset (Smart-seq2-based) was classified using the Ovarian dataset as a reference. The results, presented in Figures 2D–F and 3A, respectively, show a performance similar to that obtained within one platform. Taking the first of the two cross-platform classifications as an example (Figure 2F), it is clear that the majority of cells (79.9%) that are not tumor or unknown retain the published labels. Of all the cells getting a different cell type label most become unassigned or intermediate (87.3%) and this is especially true for the Unknown (80.0%) and tumor cells (99.3%), in line with expectation. The robustness of the cross-platform classification is probably due to the use of rank-based similarities, implying that other combinations of scRNA-seq technologies will likely yield similar good results. This is further exemplified by accurate classification of a Drop-seq dataset (39) using a Chromium 10× Genomics dataset (27) (Figure 3C).
Comparison with existing methods
The important challenge of cell type identification has recently also started to be addressed through the development of other automated approaches. CHETAH was therefore next compared to the state-of-the-art methods CaSTLe (40), scmap (30) (both versions, i.e. scmap_cell and scmap_cluster) and SingleR (31) by running these programs with standard settings on the Ovarian, Melanoma and Head-Neck datasets (Figure 3A). To evaluate the performance also on non-tumor tissues, two pancreas datasets, Pancreas1 (16) and Pancreas2 (41) were included and mutually classified using the other as the reference. The ground truth for the classifications are the cell type labels from the original publications, but without the malignant cells from the tumor datasets. They are not part of the reference data and should therefore be considered an unknown cell type and should remain unassigned or intermediate.
To compare methods, two classes of input cells can be distinguished, namely, (i) the cells that are of a type that is present in the reference and (ii) cells for which no reference is available (Figure 3B). For the first class it is meaningful to assess the correctness of the classification, because an optimal method should correctly identify all such cells. Those cell type inferences can therefore be correct or incorrect, corresponding to the true and false positives respectively (Figure 3B). In addition, the categories intermediate or unassigned are allowed, to accommodate methods such as scmap and CHETAH that produce intermediate and unassigned calls. The second class of input cells, those of a type absent from the reference, should not be classified by an optimal cell type identification. These are therefore divided into correctly unassigned cells which can be considered true negatives, and their false positive counterparts, here called erroneously assigned, i.e. cells that were, but should not have been, classified.
In the cancer datasets, CHETAH generally outperforms other methods (Figure 3A) in terms of combined true positives (correct assignments) and true negatives (cells correctly left unclassified). This is particularly important for studies of cancer since malignant cells are typically very patient specific and would almost always be misclassified by greedy methods. SingleR, having no classification cut-off, always classifies all cells to a final type, leading to a large number of erroneously assigned cells in cancer samples with many malignant cells (Figure 3). For example, both the cancer-associated fibroblasts (CAFs) and malignant cells are all classified as CAFs by SingleR. In datasets containing many unknown cells such as the malignant cells in the cancer samples, such approaches would therefore require very careful post hoc inspection of the classification on a per cell or per cluster basis, an approach that automated methods are meant to obviate. The selective nature of CHETAH makes the analysis much more efficient. As anticipated, forcing CHETAH to become greedy and classify all cells by applying a confidence score threshold of zero, yields a performance almost identical to SingleR’s (Figure 3).
In contrast to the cancer datasets, the pancreas data are less complex, containing cell types with strong differential gene expression and few unknown cells. Note that a perfect classifier should leave none of the cells in the Pancreas2 dataset unidentified, because all its cell types are represented in the Pancreas1 reference. The converse is not true because for some of the cell types no distinction is made in Pancreas1. This is one reason that all the methods perform better on Pancreas2 (Figure 3A). An additional reason is the low expression of standard Pancreas markers in one of the donor samples included in the Pancreas1 dataset (Supplementary Figure S8). In the comparison on non-cancer datasets, CHETAH’s forte of rarely classifying cells without sufficient resemblance to the reference cell types is diminished. This results in a performance similar to that of the other methods (Figure 3). However, as is exemplified below, the inclusion of an intermediate assignment can have benefits for such datasets too.
Intermediate types
In data from tumor samples the classification to an intermediate type suggests, by exclusion, that a cell is aberrant and therefore potentially malignant. The position in the classification tree, of the node of an intermediate type may shed further light on the biology of these cells. For example, in the Melanoma and Head-Neck datasets, 54 and 74%, respectively, of the malignant cells, classify to the node directly above endothelial. This suggests that the expression pattern of these cells shares characteristics with endothelial and fibroblast types (see Supplementary Figure S3A and B for the classification trees). Conversely, these cells display no affinity with the hematopoietic lineage, which is consistent with these tumors not being of hematopoietic origin. Classification to an intermediate type in combination with the position in the classification tree is therefore useful for analysis of cancer datasets.
Assignment to an intermediate cell type can also be useful in non-cancer datasets. This is demonstrated by two examples. In the Pancreas1 dataset, two kinds of stellate cells were originally identified, both of which are of mesenchymal origin (42). PDGFRA and RGS5 were applied as marker genes for activated and quiescent stellate cells respectively. Pancreas2 only contains the more general label mesenchymal, and the corresponding cells only exhibit expression of PDGFRA but not RGS5 (Supplementary Figure S6), implying that these reference cells more closely resemble activated rather than quiescent stellate cells. When CHETAH classifies the Pancreas1 dataset using the more limited Pancreas2 reference data, it correctly identifies the Pancreas1 activated stellate cells as mesenchymal while leaving the quiescent stellate cells unassigned, or assigning them to the node directly above the endothelial and mesenchymal types (Supplementary Figure S8B), correctly determining that these cells are of a mesenchymal type not represented in the reference.
Acinar–duct cell gradient in pancreas data
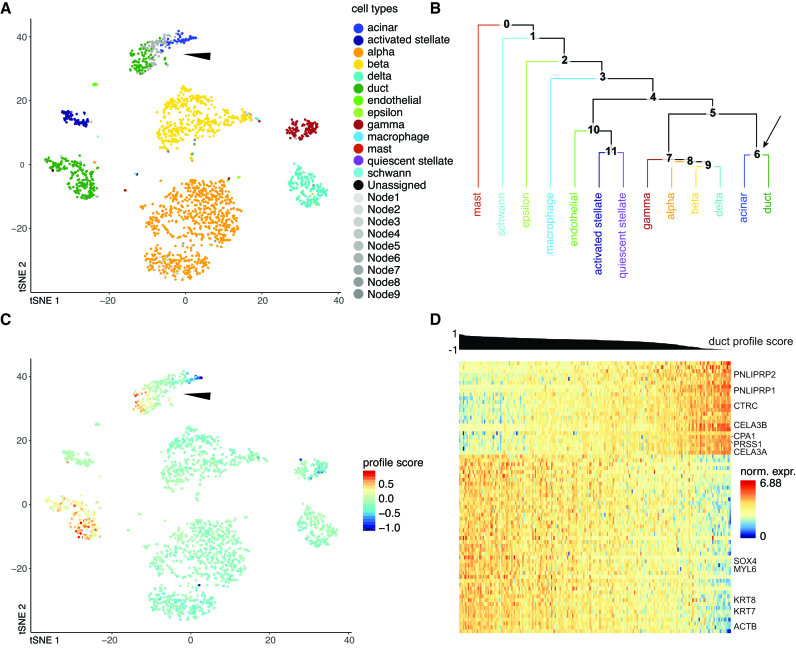
Another useful consequence of allowing an intermediate type is exemplified in Figure 4. Some cells in a cluster identified as acinar in the Pancreas2 publication are labeled ductal by CHETAH (Figure 4A), while conversely the cluster called ductal in the Pancreas1 study is partly classified as acinar (Supplementary Figure S8A). The presence of these mixed acinar–ductal groups in both datasets suggests a shared underlying phenomenon. Acinar and ductal cells arise from the same progenitors and are closely related (43). They are separated by only one node in CHETAH’s classification tree (Figure 4B and Supplementary Figure S8B), which is the intermediate type to which CHETAH assigns the remaining cells of these clusters. When visualizing the profile score for duct cell in this intermediate node (arrows in Figure 4B and Supplementary Figure S8B), a smooth gradient is clear in both clusters (Figure 4C and Supplementary Figure S8C).
Figure 4.
CHETAH identifies opposing gradients of duct and acinar cell marker genes in the Pancreas2 dataset (41). (A) t-SNE plot of the Pancreas2 dataset as classified by CHETAH, with colors representing the inferred cell types. The arrowhead indicates a population that was labeled as acinar cell in the publication, but is classified to a mixture of duct cell (blue), acinar cell (green) and intermediate Node 6 (gray) by CHETAH. (B) The classification tree used for (A), based on the Pancreas1 dataset. The arrow indicates the acinar/ductal intermediate node (Node 6) for which the profile score of duct cells is shown in (C). (C) As (B), but with all cells colored by the profile score for ductal cell in Node 6. The cells in the cluster of interest show a gradient of the profile score. (D) Heatmap showing the normalized expression of the genes (rows) used by CHETAH to calculate the profile score plotted in (C), for the cells (columns) in the cluster indicated by the arrowhead in panel (A). Only genes (rows) having an absolute correlation >0.5 with the profile score are shown. The cells are sorted by the duct cell profile score in Node 6 which is shown above the heatmap. Well-known acinar (top) and ductal marker genes (bottom) are labeled (see main text). For the heatmap with all genes annotated see Supplementary Figure S7.
A heatmap of the expression of the genes most strongly (anti)correlating with this profile score shows the well-known cell type markers for these cell types (Figure 4D and Supplementary Figure S8D). These cell type-specific markers again exhibit a gradient of decreasing ductal and increasing acinar expression. Among the negatively correlating genes are acinar markers like CPA1, PRSS1 and CTRC (44,45) and among the positively correlating genes are pancreas duct cell markers like KRT7 and KRT19 (46). A similar gradient in the expressions of genes having unusually large loadings in the first principal component of their ductal cell population has been reported previously (16). This is a different manifestation of the fact that, for these cells, there is no dichotomy between acinar and ductal. Instead, the type of these cells is best described as lying on a continuum between acinar and ductal. The intermediate type assignment and profile score provide a direct and intuitive visualization highlighting such cases and the utility of the approach taken by CHETAH.
DISCUSSION
Classification of cell types in scRNA-seq data is an essential step that was by necessity initially performed manually (18,24–26). Owing to the subjective and time-consuming nature of manual approaches, automated approaches have recently been developed (30,31,40). CHETAH has several features which work in its favor. Importantly, it compares input cell data with real, rather than imputed reference cell profiles. Moreover, besides using correlations, the classification decision is also based on a confidence score determined by the degree to which an input cell matches the expression variance embodied by the cumulative distribution function of the correlations to the reference cells. This facilitates the highly selective nature of CHETAH, underlying the ability to classify cells as specifically as the input and reference data allows, but without greedy over-classification, as controlled through the confidence score threshold. One consequence is the assignment to intermediate or unassigned cell types for input cells not present in the reference data. The assignment to an intermediate or unassigned type is essential to prevent overclassification and acts as an automated flag to more closely inspect such cells. The importance of this is evident both from the tumor datasets for which the method was initially devised, but also for non-cancer datasets as is also exemplified. In the tumor datasets analyzed here practically all malignant cells were classified to intermediate types. Although genetic lesions such as copy number variations can be used to identify malignant cells (24), this does represent an additional step. Moreover, such aberrations are not necessarily present (as in many pediatric tumors (47,48)) and/or may not be readily detectable. Automated highlighting of malignant tumor cells by CHETAH through classification as an intermediate or unassigned cell type is a significant improvement compared to blind misclassification. CHETAH’s confidence threshold can be adjusted to the needs of the dataset at hand, making it a flexible tool for research. The method is made available as the R (32) package CHETAH in Bioconductor (33) release 3.9, useful for application in conjunction with tools such as SCENIC (9), Scater (10), Census (11), Monocle (12), Seurat (13) and CellBIC (14). The CHETAH package additionally includes a Shiny app for intuitive visualization of the type labels, profile and confidence scores in a t-SNE (49) plot, as well as the inferred classification tree and expression heatmaps of discriminatory genes.
In the pancreas datasets, CHETAH uncovers a group of cells exhibiting a gradient of profiles between acinar and ductal, previously suggested to be centroacinar cells (16). An alternative explanation is that these are acinar cells undergoing acinar-to-ductal transdifferentiation or metaplasia (ADM) (45). This is commonly seen in acinar cells that, like those in both pancreas studies discussed here, are cultured for several days (50) or subjected to stresses or injury (51). Subtle phenomena such as the acinar-ductal gradient are easily overlooked by greedy methods and especially by (manual) methods that assign the same cell type to all cells of one apparent cluster. The accuracy of CHETAH is dependent on the availability of well-annotated reference datasets. It is firmly established that hierarchical trees derived from clustering gene expression data reflect many aspects of the underlying biology (52). Here such trees are applied as a guide for classification only, without surmising accurate cellular taxonomies. Detailed hierarchical trees that reflect all aspects of cell types and cell states will obviously perform better for classification. Although the concordance between cell type identification based on cell surface markers and gene expression appears to be good (39), it is important to point out that gene expression is only one way of characterizing cells. The definition of cell types and the difference with cellular state are receiving renewed interest and scrutiny with the advent of quantitative single-cell techniques such as scRNA-seq (see e.g. ref (53)). For the method presented here, the definition of cell type is pragmatic and can best be described as any group of cells annotated within a reference set as belonging together, and having sufficiently similar gene expression among themselves and sufficiently different gene expression with other types defined in the reference, so as to allow identification with high confidence. Classification of cells from diverse tissues, diseases and states will become easier with the increasing availability of well-annotated scRNA-seq datasets. Efforts like the Human Cell Atlas (HCA) (23) are aimed at generating scRNA-seq datasets for almost each (healthy) tissue and cell type. CHETAH’s accurate handling of unknown cell types should prove useful in discovering novel cell types in such data. Conversely, the annotated HCA data would be very suitable as a reference for CHETAH.
Approaches for analysis of scRNA-seq data are being developed at a rapid pace. A recent addition is SuperCT (54) which incorporates supervised classification into a framework for cell-type classification. Although complementary in application scope (the reference dataset is fixed), we nevertheless compared accuracies, with CHETAH performing at a similar level of 92% concordance as analyzed by the cross-validation method in the SuperCT study, albeit by necessity as tested on different datasets. CHETAH is not limited to the use of scRNA-seq and can likely be used with other quantitative single cell data such as those obtained using DNA accessibility (55,56), chromatin state (57), methylome (58), epitope (39) or RNA velocity (59) sequencing methods, provided sufficiently rich reference data is available. Although the full range of single cell genome-wide approaches can be expected to increase further in the near future, the need for methods such as CHETAH that improve the ease and precision of the analysis of the resulting data is evident.
SOFTWARE AVAILABILITY
CHETAH is available at github.com/jdekanter/CHETAH and through Bioconductor (33). All scripts that are needed to perform the analyses mentioned in this paper and to create the t-SNE plots using Seurat (13) are deposited at github.com/jdekanter/CHETAH_paper_figures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Holstege, Kemmeren and Molenaar groups at the Máxima Center for insightful discussions and comments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council (ERC) Advanced Grant [671174]; KIKA. Funding for open access charge: ERC; KIKA.
Conflict of interest statement. None declared.
REFERENCES
- 1. Islam S., Kjällquist U., Moliner A., Zajac P., Fan J.-B., Lönnerberg P., Linnarsson S.. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011; 21:1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saadatpour A., Lai S., Guo G., Yuan G.-C.. Single-cell analysis in cancer genomics. Trends Genet. 2015; 31:576–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grün D., van Oudenaarden A.. Design and analysis of single-cell sequencing experiments. Cell. 2015; 163:799–810. [DOI] [PubMed] [Google Scholar]
- 4. Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwé H., Pircher A., Eynde K.V. den et al.. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018; 24:1277–1289. [DOI] [PubMed] [Google Scholar]
- 5. Levitin H.M., Yuan J., Sims P.A.. Single-cell transcriptomic analysis of tumor heterogeneity. Trends Cancer. 2018; 4:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Svensson V., Vento-Tormo R., Teichmann S.A.. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018; 13:599–604. [DOI] [PubMed] [Google Scholar]
- 7. Moignard V., Woodhouse S., Haghverdi L., Lilly A.J., Tanaka Y., Wilkinson A.C., Buettner F., Macaulay I.C., Jawaid W., Diamanti E. et al.. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat. Biotechnol. 2015; 33:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lun A.T.L., McCarthy D.J., Marioni J.C.. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor [version 2; peer review: 3 approved, 2 approved with reservations]. F1000Research. 2016; 5:2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.-C., Geurts P., Aerts J. et al.. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017; 14:1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCarthy D.J., Campbell K.R., Lun A.T.L., Wills Q.F.. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017; 33:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qiu X., Hill A., Packer J., Lin D., Ma Y.-A., Trapnell C.. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 2017; 14:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C.. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017; 14:979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Butler A., Hoffman P., Smibert P., Papalexi E., Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018; 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim J., Stanescu D.E., Won K.J. CellBIC: bimodality-based top-down clustering of single-cell RNA sequencing data reveals hierarchical structure of the cell type. Nucleic Acids Res. 2018; 46:e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stegle O., Teichmann S.A., Marioni J.C.. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 2015; 16:133–145. [DOI] [PubMed] [Google Scholar]
- 16. Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A., Ryu J.H., Wagner B.K., Shen-Orr S.S., Klein A.M. et al.. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016; 3:346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng C., Zheng L., Yoo J.-K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q. et al.. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017; 169:1342–1356. [DOI] [PubMed] [Google Scholar]
- 18. Schelker M., Feau S., Du J., Ranu N., Klipp E., MacBeath G., Schoeberl B., Raue A.. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat. Commun. 2017; 8:2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lane K., Van Valen D., DeFelice M.M., Macklin D.N., Kudo T., Jaimovich A., Carr A., Meyer T., Pe’er D., Boutet S.C. et al.. Measuring signaling and RNA-Seq in the same cell links gene expression to dynamic patterns of NF-κB activation. Cell Syst. 2017; 4:458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vladoiu M.C., El-Hamamy I., Donovan L.K., Farooq H., Holgado B.L., Ramaswamy V., Mack S.C., Lee J.J., Kumar S., Przelicki D. et al.. Childhood cerebellar tumors mirror conserved fetal transcriptional programs. Nature. 2019; doi:10.1038/s41586-019-1158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grün D., Lyubimova A., Kester L., Wiebrands K., Basak O., Sasaki N., Clevers H., van Oudenaarden A.. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015; 525:251–255. [DOI] [PubMed] [Google Scholar]
- 22. Yang J., Tanaka Y., Seay M., Li Z., Jin J., Garmire L.X., Zhu X., Taylor A., Li W., Euskirchen G. et al.. Single cell transcriptomics reveals unanticipated features of early hematopoietic precursors. Nucleic Acids Res. 2017; 45:1281–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M. et al.. The human cell atlas. Elife. 2017; 6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G. et al.. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016; 352:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S. et al.. Single-Cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017; 171:1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L. et al.. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014; 344:1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. et al.. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017; 8:ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lyons Y.A., Wu S.Y., Overwijk W.W., Baggerly K.A., Sood A.K.. Immune cell profiling in cancer: molecular approaches to cell-specific identification. NPJ Precis. Oncol. 2017; 1:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F. et al.. Mapping the mouse cell atlas by Microwell-Seq. Cell. 2018; 172:1091–1107. [DOI] [PubMed] [Google Scholar]
- 30. Kiselev V.Y., Yiu A., Hemberg M.. scmap: projection of single-cell RNA-seq data across data sets. Nat. Methods. 2018; 15:359–362. [DOI] [PubMed] [Google Scholar]
- 31. Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R. et al.. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019; 20:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. R Core Team R: A Language and Environment for Statistical Computing. 2018; Vienna: R Foundation for Statistical Computing. [Google Scholar]
- 33. Huber W., Carey J.V., Gentleman R., Anders S., Carlson M. et al.. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015; 12:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yuan H., Yan M., Zhang G., Liu W., Deng C., Liao G., Xu L., Luo T., Yan H., Long Z. et al.. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019; 47:D900–D908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H., Courtois E.T., Sengupta D., Tan Y., Chen K.H., Goh J.J.L., Kong S.L., Chua C., Hon L.K., Tan W.S. et al.. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 2017; 49:708–718. [DOI] [PubMed] [Google Scholar]
- 36. Chung W., Eum H.H., Lee H.-O., Lee K.-M., Lee H.-B., Kim K.-T., Ryu H.S., Kim S., Lee J.E., Park Y.H. et al.. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017; 8:15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Picelli S., Faridani O.R., Björklund Å.K., Winberg G., Sagasser S., Sandberg R.. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014; 9:171–181. [DOI] [PubMed] [Google Scholar]
- 38. Zilionis R., Nainys J., Veres A., Savova V., Zemmour D., Klein A.M., Mazutis L.. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 2017; 12:44–73. [DOI] [PubMed] [Google Scholar]
- 39. Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P.. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017; 14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lieberman Y., Rokach L., Shay T.. CaSTLe—classification of single cells by transfer learning: Harnessing the power of publicly available single cell RNA sequencing experiments to annotate new experiments. PLoS One. 2018; 13:e0205499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Muraro M.J., Dharmadhikari G., Grün D., Groen N., Dielen T., Jansen E., van Gurp L., Engelse M.A., Carlotti F., de Koning E.J.P. et al.. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016; 3:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Erkan M., Adler G., Apte M.V., Bachem M.G., Buchholz M., Detlefsen S., Esposito I., Friess H., Gress T.M., Habisch H.-J. et al.. StellaTUM: current consensus and discussion on pancreatic stellate cell research. Gut. 2012; 61:172–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reichert M., Rustgi A.K.. Pancreatic ductal cells in development, regeneration, and neoplasia. J. Clin. Invest. 2011; 121:4572–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Athwal T., Huang W., Mukherjee R., Latawiec D., Chvanov M., Clarke R., Smith K., Campbell F., Merriman C., Criddle D. et al.. Expression of human cationic trypsinogen (PRSS1) in murine acinar cells promotes pancreatitis and apoptotic cell death. Cell Death Dis. 2014; 5:e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Askan G., Deshpande V., Klimstra D.S., Adsay V., Sigel C., Shia J., Basturk O.. Expression of markers of hepatocellular differentiation in pancreatic acinar cell neoplasms A potential diagnostic pitfall. Am. J. Clin. Pathol. 2016; 146:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bouwens L. Cytokeratins and cell differentiation in the pancreas. J. Pathol. 1998; 184:234–239. [DOI] [PubMed] [Google Scholar]
- 47. Gröbner S.N., Worst B.C., Weischenfeldt J., Buchhalter I., Kleinheinz K., Rudneva V.A., Johann P.D., Balasubramanian G.P., Segura-Wang M., Brabetz S. et al.. The landscape of genomic alterations across childhood cancers. Nature. 2018; 555:321–327. [DOI] [PubMed] [Google Scholar]
- 48. Ma X., Liu Y., Liu Y., Alexandrov L.B., Edmonson M.N., Gawad C., Zhou X., Li Y., Rusch M.C., Easton J. et al.. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018; 555:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maaten L. van der, Hinton G.. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008; 9:2579–2605. [Google Scholar]
- 50. Houbracken I., de Waele E., Lardon J., Ling Z., Heimberg H., Rooman I., Bouwens L.. Lineage tracing evidence for transdifferentiation of acinar to duct cells and plasticity of human pancreas. Gastroenterology. 2011; 141:731–741. [DOI] [PubMed] [Google Scholar]
- 51. Strobel O., Dor Y., Alsina J., Stirman A., Lauwers G., Trainor A., Castillo C.F., Warshaw A.L., Thayer S.P.. In vivo lineage tracing defines the role of Acinar-to-Ductal transdifferentiation in inflammatory ductal metaplasia. Gastroenterology. 2007; 133:1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eisen M.B., Spellman P.T., Brown P.O., Botstein D.. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Clevers H., Rafelski S., Elowitz M., Klein A., Shendure J., Trapnell C., Lein E., Lundberg E., Uhlen M., Martinez-Arias A. et al.. What is your conceptual definition of “cell type” in the context of a mature organism. Cell Syst. 2017; 4:255–259. [DOI] [PubMed] [Google Scholar]
- 54. Xie P., Gao M., Wang C., Zhang J., Noel P., Yang C., Von Hoff D., Han H., Zhang M.Q., Lin W. SuperCT: a supervised-learning framework for enhanced characterization of single-cell transcriptomic profiles. Nucleic Acids Res. 2019; 47:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buenrostro J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P., Chang H.Y., Greenleaf W.J.. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015; 523:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lake B.B., Chen S., Sos B.C., Fan J., Kaeser G.E., Yung Y.C., Duong T.E., Gao D., Chun J., Kharchenko P.V. et al.. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 2018; 36:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rotem A., Ram O., Shoresh N., Sperling R.A., Goren A., Weitz D.A., Bernstein B.E.. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015; 33:1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hou Y., Guo H., Cao C., Li X., Hu B., Zhu P., Wu X., Wen L., Tang F., Huang Y. et al.. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016; 26:304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Manno G.L., Soldatov R., Hochgerner H., Zeisel A., Petukhov V., Kastriti M., Lonnerberg P., Furlan A., Fan J., Liu Z. et al.. RNA velocity in single cells. Nature. 2018; 560:494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.