Abstract
The psychiatric risk-associated transcription factor 4 (TCF4) is linked to schizophrenia. Rare TCF4 coding variants are found in individuals with Pitt-Hopkins syndrome—an intellectual disability and autism spectrum disorder. TCF4 contains a C-terminal basic-helix-loop-helix (bHLH) DNA binding domain which recognizes the enhancer-box (E-box) element 5′-CANNTG-3′ (where N = any nucleotide). A subset of the TCF4-occupancy sites have the expanded consensus binding specificity 5′-C(A/G)-CANNTG-3′, with an added outer Cp(A/G) dinucleotide; for example in the promoter for CNIH3, a gene involved in opioid dependence. In mammalian genomes, particularly brain, the CpG and CpA dinucleotides can be methylated at the 5-position of cytosine (5mC), and then may undergo successive oxidations to the 5-hydroxymethyl (5hmC), 5-formyl (5fC), and 5-carboxyl (5caC) forms. We find that, in the context of 5′-0CG-1CA-2CG-3TG-3′(where the numbers indicate successive dinucleotides), modification of the central E-box 2CG has very little effect on TCF4 binding, E-box 1CA modification has a negative influence on binding, while modification of the flanking 0CG, particularly carboxylation, has a strong positive impact on TCF4 binding to DNA. Crystallization of TCF4 in complex with unmodified or 5caC-modified oligonucleotides revealed that the basic region of bHLH domain adopts multiple conformations, including an extended loop going through the DNA minor groove, or the N-terminal portion of a long helix binding in the DNA major groove. The different protein conformations enable arginine 576 (R576) to interact, respectively, with a thymine in the minor groove, a phosphate group of DNA backbone, or 5caC in the major groove. The Pitt-Hopkins syndrome mutations affect five arginine residues in the basic region, two of them (R569 and R576) involved in 5caC recognition. Our analyses indicate, and suggest a structural basis for, the preferential recognition of 5caC by a transcription factor centrally important in brain development.
INTRODUCTION
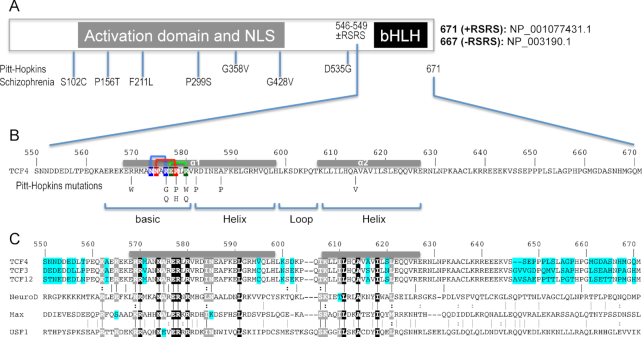
Human transcription factor 4 (TCF4) is essential for brain development, memory and cognition, and has been associated with schizophrenia, autism-spectrum intellectual disability, and Pitt-Hopkins syndrome (1). Together with TCF3 and TCF12, TCF4 is a member of the class I basic-helix-loop-helix (bHLH) family of transcription factors (2), homologous to the Drosophila sex determination factor daughterless (3), and also called an Enhancer-box binding protein (4,5). In fact, TCF4 is one of the few—if not only—E-proteins expressed throughout the adult mouse brain (6), where it affects morphology (7) and regulates synaptic plasticity, DNA methylation, and memory function (8). Common variants in human TCF4 were among the first genes to reach significance in genome-wide association studies of schizophrenia (9), and rare coding TCF4 variants outside of the bHLH domain were identified in individual schizophrenia patients by deep sequencing (10,11) (Figure 1A). In contrast, rare TCF4 mutations associated with Pitt-Hopkins syndrome (12)—a genetic disorder characterized by intellectual disability, distinct facial features, development delay and autonomic dysfunction—occur almost exclusively within the bHLH domain (13–18) (Figure 1B). The basis for association of common TCF4 variants with schizophrenia is currently unknown. Our study suggests structural effects of Pitt-Hopkins syndrome-associated mutations in the bHLH domain—the only functionally characterized domain of TCF4.
Figure 1.
Schematic of human TCF4 and sequence alignment of bHLH domains. (A) Human TCF4 transcripts potentially generate 18 isoforms with different N-termini (73), but all TCF4 isoforms contain the C-terminal bHLH DNA binding domain. In-frame alternative splicing increases the number of TCF4 isoforms. For example, alternative splicing at exon 18 of TCF4 leads to the presence or absence of two RS repeats––containing arginine (R) and serine (S)––immediately prior to the C-terminal bHLH domain. For the study described here, we use the residue numbering of +RSRS isoform (NP_001077431.1) for the bHLH domain. (B) Pitt-Hopkins mutations in bHLH that alter either the basic arginine residues at the protein–DNA interface or alanine residues that coordinate the dimerization. Three pairs of intra-molecular interactions exist in the major groove of DNA: N573•••R576 (blue), N574•••R578 (red) and E577•••R580 (green). (C) TCF3, TCF4, and TCF12 are Class I bHLH proteins, also called E-box binding proteins, and share high sequence identity within their bHLH domains, except for 7 positions (colored cyan). In contrast, other three representative proteins (NeuroD1, Max and USF1) used in the alignment shares only 9 invariant residues (white letters in black background) within the bHLH. White letters in grey background indicate conserved variation (R and K; I and L; T and S; L and M).
There is no structural information currently available for TCF4. The corresponding bHLH domain of TCF3 (also known as E47) has been structurally characterized as a homodimer (19), though the structural coordinates are not yet available in the Protein Data Bank (PDB), and as a heterodimer with NeuroD1 (20) (PDB 2QL2). The bHLH proteins can heterodimerize with other family members [>60 exist (21,22)], and TCF4 is no exception (1).
Recent chromatin immunoprecipitation sequencing of TCF4 in neuroblastoma cells (SH-SY5Y) and blastic plasmacytoid dendritic cell neoplasm cells (Cal-1 and Gen2.2) revealed, as expected, strong enrichment of the E-box motif CANNTG (N = any nucleotide) within the TCF4 binding sites (23–25). A subset of E-boxes has the palindromic sequence CACGTG, and it is significant that the central CpG and two outer CpA dinucleotides are common DNA methylation sites. Methylated cytosine, 5-methylcytosine (5mC or M), can be successively oxidized by Tet proteins to become 5-hydroxymethylated (5hmC or H), 5-formylated (5fC or F), and 5-carboxylated cytosine (5caC) (26,27) (Figure 2A). In addition to recognizing specific DNA sequences, a growing number of transcriptional regulators are being found to respond to different cytosine modification states (28–30). These transcription factors are potentially acting as direct epigenetic sensors. For example, like TCF4, the oncogenic MYC and its binding partner MAX are bHLH transcription factors; they preferentially recognize the E-box sequence CACGTG (31). MAX binds an unmodified E-box, and methylation of the central CpG greatly inhibits its binding (32), while oxidation to 5caC restores its binding to the level of unmodified C (33). For TCF4, a previous report suggested that 5hmC in two E-box sequences, ACACGTG and ACATGTG, when presented in hemi-modified fashion (i.e. modification on one strand only), increased DNA-binding of the TCF4 bHLH domain (34). Another report from the same group suggested that 5caC in a CpG dinucleotide immediately outside of E-box (CGCAGGTG) enhanced binding of TCF4, as well as that of other immediate family members (35). Here, to understand the effects of DNA sequence and modification on the binding of this important disease-associated transcription factor, we use the isolated TCF4 bHLH DNA binding domain to understand how the TCF4 protein recognizes E-box sequence, and how it ‘senses’ the oxidation status of cytosine.
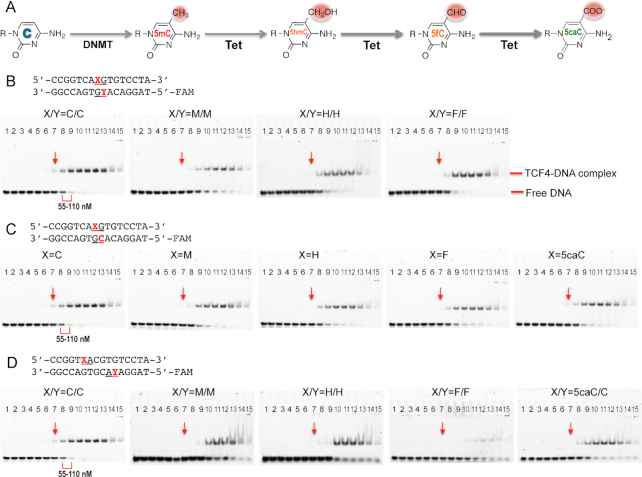
Figure 2.
Electrophoretic mobility shift assay of TCF4 bHLH protein binding to oligos containing a single E-box. (A) Schematic of chemical reactions of DNA cytosine methylation by DNMT and 5mC oxidations by Tet enzymes. (B) The central CpG dinucleotides are unmodified (C/C) or fully modified (M/M, H/H, F/F; where M = 5mC, H = 5hmC, and F = 5fC). (C) The central CpG dinucleotides are hemi-modified (M/C, H/C, F/C or 5caC/C; where 5caC = 5-carboxyC). (D) The two outer CpA dinucleotides are unmodified (C/C), fully modified (M/M, H/H, F/F) or hemi-modified (5caC/C). The protein concentrations used were a maximum of 7 μM (the right most lane 15 of each panel) followed by serial 2-fold dilutions (from right to left). The arrows indicated a reference point where the shift was observed for the unmodified oligo. The same samples were quantified by fluorescence polarization (Supplementary Figure S1).
MATERIALS AND METHODS
Protein expression and purification
Human TCF4 C-terminal bHLH domain (residues 569–628; NP_001077431.1) was cloned into a modified pET28b vector as an N-terminal 6xHis-SUMO tag (pXC2002). After it was transformed into Escherichia coli BL21-Codon Plus(DE3)-RIL (Stratagene), bacterial cells were cultured in a shaking incubator in LB medium at 37°C until the culture density reached A600 nm = 0.4, at which time the temperature of the incubator was changed to 16°C, and at A600 nm ∼0.8, isopropyl β-d-1-thiogalactopyranoside (IPTG) at 0.4 mM was added for induction and cultures allowed to grow overnight. Harvested cells were lysed by sonication in 20 mM Tris (pH 7.5), 300 mM NaCl, 20 mM imidazole, 5% (v/v) glycerol, 0.5 mM tris (2-carboxyethyl) phosphine (TCEP) and 0.1 mM phenylmethylsulfonyl fluoride. The lysate was treated with polyethylenimine added drop by drop until a concentration of 0.15% (w/v) was attained (36) and then centrifuged at 47 850 x g for 30 min at 4°C. The supernatant containing the 6xHis-SUMO tagged protein was collected and subjected to a four-column chromatography protocol, conducted in a BIO-RAD NGC™ system. The sample was loaded onto a 5-ml HisTrap column (GE Healthcare), the column washed, and the protein eluted with a linear imidazole gradient from 20 to 500 mM. This eluted protein was incubated with Ulp1 protease (purified in-house) at 4°C overnight, to remove the 6xHis-SUMO tag, leaving two additional N-terminal residues (His-Met) fused to the recombinant protein. The cleaved protein was loaded onto HiTrap Q-SP tandem column (GE Healthcare) (36) and eluted from the SP column with a 50 ml linear gradient from 300 mM to 1 M NaCl. The peak fractions were concentrated to 5 ml and loaded onto a HiLoad Superdex 75 (16/60) column (GE healthcare) and eluted as a single peak in 20 mM Tris (pH 8.0), 250 mM NaCl, 5% (v/v) glycerol and 0.5 mM TCEP. The quality and size of the final purified protein were confirmed by 18% SDS PAGE. Due to the lack of aromatic residues, the protein concentration was estimated by absorbance at 205 nm [ε205 = 27+120*(A280/A205)] (37).
The mutant protein R569-to-Trp (R569W; pXC2095) was expressed and purified similarly in the absence of the last step of gel filtration chromatography. The mutant protein was soluble up to ∼2.7 mg/ml. We used A205 nm and A280 nm (ε280 = 0.770) to estimate the concentration and both gave similar results.
DNA binding assays
We used three classic DNA binding assays: (i) fluorescence polarization, (ii) electrophoretic mobility shift and (iii) isothermal titration calorimetry.
Fluorescence polarization (FP) assays were performed using a Synergy 4 microplate reader (BioTek) to measure DNA binding affinity. The 6-carboxy-fluorescein (FAM)-labeled double strand DNA probe (5 nM) were incubated with an increasing amount of protein (monomer concentration 0.4 nM to 7 μM) for 15 min in 20 mM Tris (pH 7.5), 5% (v/v) glycerol, 150 mM NaCl. GraphPad Prism software (version 7.0) was used to do curve fitting. KD values were calculated as [mP] = [maximum mP] x [C]/ (KD + [C]) + [baseline mP], where mP is millipolarization, [C] is protein concentration and ΔmP = ([mP] – [baseline mP]). The reported mean ± SEM of the interpolated KD values were calculated from two independent experiments each performed in duplicate.
Electrophoretic mobility shift assays (EMSA) were performed with the same set of samples used in the FP assays. An aliquot of 10 μl of reaction was loaded onto 8% native PAGE gels, and run at 150 V for 30 min in 0.5× TBE buffer. The images were scanned by Typhoon (GE-Dynamic phosphor/fluorescence imager).
Isothermal titration calorimetry (ITC) experiments were performed at 25°C using a MicroCal PEAQ-ITC automated system (Malvern instrument Ltd). Double-stranded oligonucleotides (3.3 μM) were maintained in the sample cell, and the protein (77.7 μM) was injected into the cell using a syringe. The amount of each injection was 2.5 μL with continuous stirring (750 rpm), and the reference power was set as 8 μcal/s. The duration of each injection was fixed at 4 s and the spacing time between the injections was 200 s in order to achieve equilibrium. For each oligo, a reference titration of buffer (20 mM Tris, pH 7.5 and 150 mM NaCl) without protein was subtracted from experimental data as a control for the heat of dilution and non-specific binding. Binding constants were calculated by fitting the data using the ITC data analysis module ‘one set of sites’ supplied by the manufacturer.
Crystallography
We crystallized TCF4 bHLH domain in complex with 11-to-13-mer self-annealed oligos (Supplementary Table S1) by the sitting drop vapor diffusion method, at room temperature (∼19°C). We incubated purified protein (dimer) and double-stranded oligonucleotide in a ratio up to 1.5:1 at 4°C for 30 min in 20 mM Tris (pH 7.5), 150 mM NaCl, 5% (v/v) glycerol and 0.5 mM TCEP before crystallization (monomer concentration at 0.67 mM and DNA at 0.19 mM). An Art Robbins Phoenix Crystallization Robot was used to set up screens. For reported datasets in Supplementary Table S1, X-ray diffraction data were collected from crystals that formed in solutions of 20% (w/v) polyethylene glycol (PEG) 4000, 10% (v/v) 2-propanol and 0.1 M sodium citrate tribasic dihydrate pH 5.6 (11-mer oligo), 25% PEG 3350, 0.1 M Tris pH 8.5 and 0.2 M sodium chloride (13-mer oligo), and 30% PEG 4000, 0.1 M Tris pH 8.5, and 0.2 M lithium sulfate monohydrate or 0.2 M sodium acetate trihydrate (5caC-containing 12-mer oligo). Crystals were cryo-protected by soaking in mother liquor supplemented with 20% (v/v) ethylene glycol before plunging into liquid nitrogen.
X-ray diffraction datasets were collected at the SERCAT beamline 22ID of Advanced Photon Source at Argonne National Laboratory. Crystals were rotated a total of 400° in 0.25° increments to achieve good redundancy. The diffraction intensities were reduced and scaled using HKL-2000 (38). During scaling, particularly attention was paid to the space group determination: for a higher symmetry space group, if a significantly large number of reflections were rejected and the R-merge values were abnormally higher in lower resolution shells, the data were reprocessed in a lower symmetry space group. For instance, crystals with 13-mer oligo could be processed in C2, but statistics for processing data in the P1 space group were significantly better (PDB: 6OD3). We note that while the twin operator (39) shows a significant twinning fraction in PDB 6OD4 and 6OD5, other criteria (L-test; (40)) do not indicate this is a potential problem. As a test, we performed a round of refinement with the option of a twin law and fraction in the PHENIX refinement module (41) but observed worse electron density maps. We concluded that this apparent twinning is probably the result of two nearly identical axes (b = 43.60 Å and c = 43.56 Å in PDB 6OD4 and a = 44.68 Å and b = 44.76 Å in PDB 6OD5), as well as a non-crystallographic symmetry rotational axis that is parallel to a crystallographic axis (a axis in PDB 6OD4 and c axis in PDB 6OD5).
The molecular replacement method gave initial phasing of TCF4 with an 11-bp oligo using the PHASER module in PHENIX (42). The PHYRE2 server (43) was utilized for generating an initial search model based on TCF3 (PDB: 2QL2). A B-form DNA was generated by the make-na server (http://structure.usc.edu/make-na/server.html), which was used as a secondary search model. The molecular replacement solution was improved by a density-guided morphing procedure of PHENIX (44). COOT (45) was used for model building and corrections between refinement rounds. Structure quality was analyzed during refinements with PHENIX.refine (46) and later validated by the PDB validation server. Subsequent structure determinations were made with the 13-bp oligo and the 12-bp oligo containing 5caC, followed by the 11-bp oligo using molecular replacement. Molecular graphics were generated using PyMol (Schrödinger, LLC).
Analysis of ChIP profiles within TCF4-binding sites
To build TCF4-binding consensus sites from the published Chip-seq study in neuroblastoma cells (SH-SY5Y) and blastic plasmacytoid dendritic cell neoplasm cells (Cal-1 and Gen2.2) (24,25), GSM datasets were downloaded from the NCBI database with indicated accession numbers. The raw data were mapped to the hg19 version human reference genome using Bowtie2 (47). ChIP-seq peaks were identified using MACS2 (48). The DNA sequences within identified peak regions were extracted using BEDtools getfasta (49). An in-house python script was used to identify peaks with the 5′-C(A/G)-CANNTG-3′ or 5′-CANNTG-(T/C)G-3′ motif. Cistrome Data Browser (50) was used to visualize ChIP-seq and ATAC-seq data at the corresponding genomic regions.
RESULTS
TCF4 is insensitive to the central CpG modifications
We first measured the binding of human TCF4 bHLH domain to double-stranded oligonucleotides (oligos) containing the E-box consensus recognition sequence CACGTG, in which the status of the cytosine residues of the central CpG on both strands was either unmodified C, 5mC (M)-modified, or oxidized to 5hmC (H) and 5fC (F). [Due to synthetic limitations, the 5caC modification is incompatible with a FAM label on the same oligo, thus the 5caC-containing double-stranded oligo was only hemi-modified, see below]. We used fluorescence polarization (FP) to quantitatively measure the dissociation constants (KD). TCF4 DNA-binding domain bound the unmodified oligo with a KD of 110 nM (Table 1A and Supplementary Figure S1A). Under the same conditions, the modified oligos (containing M, H or F) have slightly reduced binding affinity (within a factor of 2) (Table 1A). Because the original observation made by Khund-Sayeed et al. (34) was based on microarrays containing DNA probes, with modifications on one strand and unmodified cytosine on the other strand, we repeated the binding experiments with hemi-modified oligos which allowed us to include the 5caC modification (Table 1B and Supplementary Figure S1B). Again, we observed marginally decreased binding with various cytosine modifications. The same samples used for the FP assays were used for electrophoretic mobility shift assay (EMSA) (Figure 2B–D), confirming that the binding affinity of TCF4 with the unmodified oligo is between 55 and 110 nM, and that the binding was decreased by only a little with the central CpG methylation and oxidations.
Table 1.
Summary of KD values measured by FP or ITC
| 5′-CCGGT-CAXGTG-TCCTA-3′ 3′-GGCCA-GTGYAC-AGGAT-5′-FAM | FP assay (*) (Fig. S1A, B) | |||||
| (A) | X = Y = C | X = Y = M | X = Y = H | X = Y = F | ||
| K D (μM) | 0.11±0.04 | 0.17±0.04 | 0.20±0.07 | 0.19±0.06 | ||
| (B) | X = Y = C | X = M, Y = C | X = H, Y = C | X = F, Y = C | X = 5caC, Y = C | |
| K D (μM) | 0.11±0.04 | 0.16±0.04 | 0.16±0.05 | 0.19±0.09 | 0.14±0.05 | |
| (C) | 5′-CCGGT-XACGTG-TCCTA-3′ 3′-GGCCA-GTGCAY-AGGAT-5′-FAM | FP assaya (Fig. S1C) | ||||
| X = Y = C | X = Y = M | X = Y = H | X = Y = F | X = 5caC, Y = C | ||
| K D (μM) | 0.11±0.04 | 0.6±0.2 | 0.6±0.2 | 0.4±0.1 | 0.3±0.1 | |
| (D) | 5′-ATA-XAXGTG-TAT-3′ 3′-TAT-GTGXAX-ATA-5′ | ITC assay (Fig. S1D) | ||||
| X = C | X = M | X = H | ||||
| K D (μM) | 0.35±0.10 | No binding | No binding | b | ||
| (E) | 5′-AXG-CACGTG-XGT-3′ 3′-TGX-GTGCAC-GXA-5′ | ITC assay (Fig. S2) | ||||
| X = C | X = M | X = H | X = F | X = 5caC | ||
| K D (μM) | 0.37±0.10 | 0.18±0.03 | 0.49±0.12 | 0.18±0.04 | 0.027±0.009 | |
b By adjusting slightly the buffer condition used in the ITC measurement, the binding affinities were about the same (0.11 μM by FP and 0.10 μM by ITC) for the two unmodified oligos (see Supplementary Figure S1E).
TCF4 binding to DNA is negatively influenced by CpA modifications within the E-box
We next examined the effect of CpA modifications on binding affinity. Full methylation (M), full hydroxymethylation (H) and full formylation (F) of both CpA sites (on opposite strands) resulted in approximately 4–6 fold weaker binding (Table 1C and Figure 2D). The 5caC-containing double-stranded oligo, which was only hemi-modified, resulted in ∼3-fold reduced binding, and we expect that a fully modified 5caC oligo might have even lower binding affinity to the TCF4 homodimer. Because our results of reduced binding affinity by cytosine modifications within the E-box sequence differ from those of Khund-Sayeed et al. (34), who suggested that 5mC decreased and 5hmC increased DNA binding of TCF4 respectively, we used a third traditional biophysical method, isothermal titration calorimetry (ITC), to quantitatively evaluate the DNA binding by TCF4. We again found that hydroxymethylation decreased binding. The differing results might reflect the assays: Khund-Sayeed et al. used 60 mer microarray probes of which 35 bp were hemi-modified, and a GST fusion to the bHLH domain; whereas we used 16-bp duplexes and untagged bHLH protein. Nevertheless, we find that the cumulative effect of modifications (5mC or 5hmC) at all cytosines within the E-box is to decrease DNA binding (Table 1D and Supplementary Figure S1D). [We note that the absolute KD values, measured by FP (0.11 μM) or ITC (0.35 μM), were different for the unmodified oligos. This difference was not due to the variation in oligo length and/or sequence outside of the E-box (see below); instead it reflects sensitivity to the buffer conditions (NaCl and glycerol) used in the ITC experiments (Supplementary Figure S1E).]
TCF4 is highly selective for 5caC immediately adjacent to the E-box
We then analyzed the effect of the two base pairs immediately outside of the E-box. First, we compared the sequences TA versus CG (TA-CACGTG and CG-CACGTG). As expected, given that ChIP-seq motifs showed variable sequence immediately outside of E-box (23–25), TCF4 protein bound the two unmodified oligos with the same affinity under the same laboratory conditions (KD = 0.35 and 0.37 μM, respectively) in ITC measurements (Table 1D and E). Second, we reanalyzed previously-published ChIP-seq datasets of TCF4 (24,25), asking how many binding sites were bearing CpG or CpA at positions immediately next to the E-box. We found ∼15% in SH-SY5Y cells and 22–25% in Cal-1/Gen2.2 cells (Supplementary Figure S3). The frequency of Cp(A/G) at the positions is higher than expected (6.25%), which suggests a binding preference of TCF4 for these sites. This added Cp(A/G) dinucleotide potentially allows the cytosine methylation/oxidation status to play a regulatory role. We thus generated a set of oligos with modifications at the outer CpG site (0CG-1CA-2CG-3TG, with the numbering to indicate specific dinucleotides) and repeated the DNA binding assays. Compared to the unmodified oligo, methylation (M) and formylation (F) of the °CpG increased DNA binding by approximately 2-fold, while 5hmC (H) did not affect binding significantly (∼1.3×) (Table 1E and Supplementary Figure S2). In contrast, TCF4 affinity for 5caC increased by ∼14 fold (KD value decreased from 370 to 27 nM) (Table 1E). This result is in agreement with a previous study by Golla et al. who used conventional EMSA (35). In sum, modification of the central E-box 2CpG has very little effect on TCF4 binding, E-box 1,3CpA/TpG modification has a negative influence on binding, while modification of a flanking 0CpG, particularly carboxylation, has a strong positive impact on TCF4 binding to DNA.
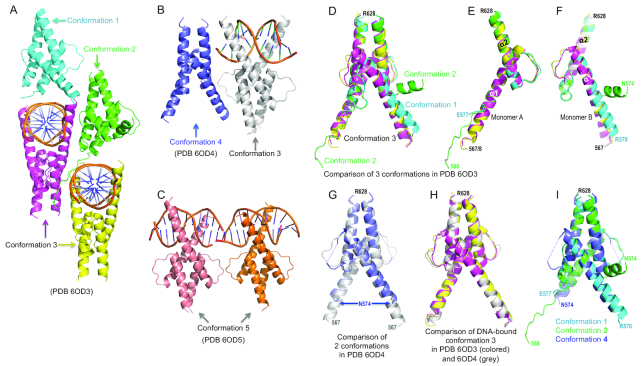
Five structural conformations of TCF4 bHLH domain
To understand how TCF4 binds DNA, and why it recognizes 5caC preferentially, we next co-crystallized the TCF4 bHLH domain with duplex oligos (11-to-13 bp) containing either TpA or 5caCpG immediately outside of E-box. The complexes crystallized in space group P1, resulting in three structures determined to resolutions of 1.5, 1.7 and 2.0 Å, respectively (Supplementary Table S1). We initially made the protein–DNA complexes with excess of protein (∼1.5:1), resulting in only half of the dimers bound with DNA (Figure 3A and B). When we decreased the molar ratio to approximately 1 dimer per duplex, all dimers had bound DNA molecules (Figure 3C). We will first describe the highest-resolution structure, with unmodified DNA (PDB ID 6OD3) at 1.5 Å resolution, and discuss the differences with the second structure containing unmodified DNA (PDB ID: 6OD4) and the 5caC-containing DNA (PDB ID: 6OD5).
Figure 3.
Five structural conformations of TCF4 bHLH domain. (A) In PDB 6OD3, the crystallographic unit contains four dimers exhibited in three conformations (1 to 3). (B) In PDB 6OD4, the crystallographic unit contains two dimers, with and without bound DNA. (C) In PDB 6OD5, the crystallographic unit includes two dimers bound to DNA containing the 5caC modification. (D) Superimposition of the four dimers in PDB 6OD3. (E, F) The two monomers (A and B) forming each dimer adopted dissimilar conformations. (G) Superimposition of two dimers in PDB 6OD4. (H) Superimposition of three DNA-bound conformations. (I) Superimposition of three conformations in the absence of cognate DNA.
As expected from the bHLH domains, the structure of each monomer when complexed with DNA comprises two long helices connected by a loop. The basic region in the N-terminal portion of the first helix (α1) creates a clamp-like basic environment, ideal for electrostatic interaction with the negatively charged DNA sugar-phosphate backbone. The second helix (α2), together with the C-terminal portion of helix α1, forms a four-helix bundle and mediates dimer interaction via hydrophobic residues. The three crystal complexes included five distinct conformations. In conformations 1 and 2, TCF4 approaches the DNA from the minor groove (Figure 3A); in conformation 3 TCF4 binds in the major groove of the DNA (Figure 3A and B); conformation 4 is TCF4 alone in the absence of DNA (Figure 3B); and in conformation 5 TCF4 recognizes a 5caC modification in the DNA (Figure 3C). In all cases, the dimer interactions mediated by the second helix (α2) and the C-terminal portion of helix α1 are largely unchanged (root-mean-square-deviation = 0.3 Å). In contrast, the N-terminal portion of the first helix undergoes dramatic conformational changes in the absence and presence of DNA (Figure 3D–F).
In the structure PDB 6OD3, the crystallographic unit contains four dimers (Figure 3A). Two of these dimers are in conformation 3 (magenta and yellow) and are bound to cognate DNA, while two additional dimers (cyan in conformation 1 and green in conformation 2) approach the DNA from the opposite direction (Figure 3A). Superimposition of the four dimers revealed that the two monomers forming each dimer (A and B) adopted distinct conformations (Figure 3E and F). The basic region of monomer A presents in three different ways: disordered in conformation 1, an extended loop in conformation 2, or as part of the long helix α1 in conformation 3 (Figure 3E). In contrast, in monomer B the corresponding helix α1 is broken into two shorter helices in conformation 2, while the carboxyl end of the helix is one turn shorter in both conformations 1 and 2, resulting in a different configuration of the loop connecting the two helices α1 and α2 (Figure 3F).
In the structure PDB 6OD4, the crystallographic unit contains just two dimers, one with bound DNA (conformation 3) and the other without DNA (conformation 4) (Figure 3B). Superimposition of the two dimers revealed no obvious differences between them. In the absence of DNA (conformation 4), the basic region is disordered, with helix α1 having been shortened by two turns, and a disordered loop connects the two helices (Figure 3G). The DNA-bound conformation 3 is highly similar in both structures (6OD3 and 6OD4), with root-mean-square-deviations of 0.5 Å comparing 109 pairs of Cα atoms (Figure 3H). In contrast, to summarize, the three conformations seen in the absence of DNA or approaching DNA from the minor groove (1, 2 and 4) have varied presentations of the basic region and the loop between the two helices (Figure 3I).
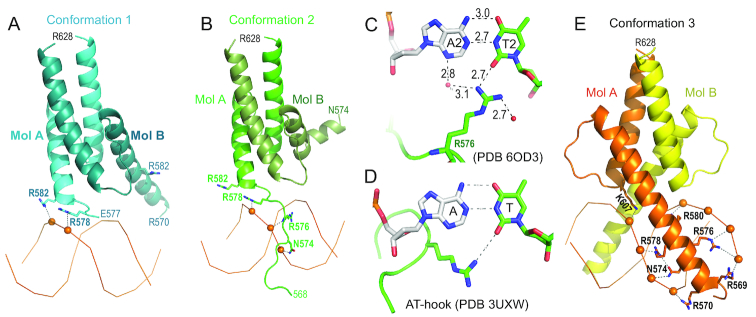
Interactions with DNA
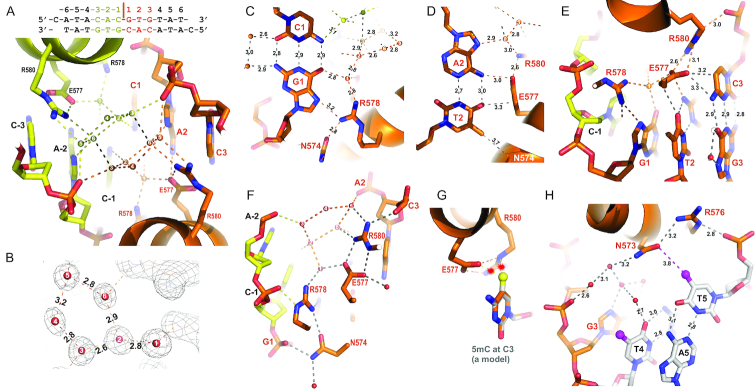
In the structure of PDB 6OD3, the two dimers in conformations 1 and 2 have asymmetric interactions with DNA backbone phosphate groups from the minor groove side (Figure 4A and B). In conformation 1, R578 and R582 of monomer A interacts with two neighboring phosphate groups, while the corresponding arginine residues of monomer B are far away from the DNA (Figure 4A). In conformation 2, the basic region of monomer A forms an extended loop going through the minor groove and interacts with three consecutive phosphate groups respectively by N574, R576 (via the main-chain amide nitrogen atom), and R578 (Figure 4B). Interestingly, the side chain of R576 forms a direct hydrogen bond with the A:T base pair in the minor groove (Figure 4C). This interaction is reminiscent of an AT-hook in the DNA minor groove, such as that of HMGA1 bound to an AATT sequence (Figure 4D) (51).
Figure 4.
Protein-phosphate interactions. (A) The dimer in conformation 1 binds DNA from the minor groove side. (B) The dimer in conformation 2 binds DNA with the extended N-terminal basic loop going through the minor groove. (C) R576 forms a H-bond with thymine in the minor groove. (D) An AT-hook of HMGA1 forms an H-bond with thymine in the minor groove (PDB: 3UXW). (E) The dimer in conformation 3 binds DNA in the major groove.
In the DNA-bound conformation 3, two monomers have symmetric interactions with DNA. From the N-terminal portion of helix α1 of each monomer of the dimer, six basic residues (R569, R570, R576, R578, R580 and K607) and one polar residue (N574) bind in the major groove of the DNA and contact seven phosphate groups, four and three respectively on each strand (Figure 4E). Among the different conformations, N574 and R578 have phosphate interactions along one DNA strand either from major or minor grooves, even though these residues reside in very different secondary structures (helix or extended loop) (comparing Figure 4B–E).
Besides phosphate contacts, the two monomers in conformation 3 have perfectly symmetrical interactions, many of which are mediated by water molecules, with the palindromic DNA sequence used for co-crystallization (Figure 5A). In the central CpG dinucleotide of the E-box lays a network of (at least) 12 water molecules (6 for each half) that connect protein side chains, DNA bases and phosphate groups (Figure 5A and B). In other words, there are few direct protein-base hydrogen bonds (H-bonds). At the central G1:C1 base pair, the only direct interaction is a weak H-bond (3.2 Å) formed between the N7 atom of guanine G1 and R578, and there is no direct interaction with the paired cytosine (Figure 5C). The single H-bond between G1 and R578 does not determine the guanine specificity, and is in agreement with the fact that TCF4 recognizes E-box sequences having variable nucleotides in the center and is insensitive to cytosine modification at C1. The single H-bond between G1 and R578-equivalent in TCF3 did exist in one monomer when TCF3 homodimer was bound with an asymmetric central sequence (5′-CACCTG-3′) (19), but was absent in the TCF3-NeuroD1 heterodimer bound with an asymmetric sequence (5′-CATCTG-3′) (20) (Supplementary Figure S4).
Figure 5.
Protein-DNA base interactions. (A) An ordered network of water molecules (numbered 1–6 for each set) occupies the major groove of central CpG dinucleotides. (B) Electron density (2Fo-Fc) shows the pentagonal interactions formed by the five water molecules numbered 2–6. (C) R576 interacts with G1. (D) E577 interacts with A2:T2. (E) E577 interacts with C3. (F) N574•••R578 and E577•••R580 are part of the water-mediated network that interact with two phosphate groups of each strand. (G) Methylation of C3 by modeling a methyl group (in yellow ball) onto unmodified C3 potentially results in repulsion (indicated by stars) with the E577 and R580 in the C-specific conformation. (H) N573•••R576 interacts with the phosphate group of T5.
The next base pair, T2:A2, is engaged in the most direct contacts (Figure 5D). First, the adenine forms one H-bond with E577 (via the exocyclic N6-amino group). Second, the paired thymine at T2 forms a weak O••• H-C type H-bond (52), and its 5-position methyl group is within van der Waals contact distance of Cα atom of N574 (Figure 5D). The next base pair, G3:C3 (Figure 5E), has two direct contacts with E577, but the interactions are focused on cytosine C3, the N4 and C5 atoms are within weak H-bond distance (3.2–3.3 Å) from one of the carboxylate oxygen atoms of E577. Among the interactions described above, the negatively-charged residue E577 bridges between two neighboring base pairs, A2:T2 and C3:G3; and at the same time its carboxylate group forms two H-bonds with the guanidino group of R580, which in turn interacts with the phosphate group of nucleotide C3 (Figure 5F). Thus the protein-DNA interface includes a network of interactions involving inter- and intra-molecular contacts (some via water molecules) that saturate the H-bonding and electrostatic potential of the E577•••R580 pair (Figure 5F). There are two roughly pentagonal interactions formed by, first, the five water molecules numbered 2–6, and second by the three waters numbered 1, 2, and 6 together with the side chains of E577 and R580 (Figure 5F). The water molecules within this network are also each saturated with four H-bonds. Under the circumstance of saturated interactions, the side chains of E577 and R580 are interlocked into a rigid configuration. Significantly, modification of C3 (5mC or 5hmC) in the CpA dinucleotide would introduce an additional methyl group or hydroxymethyl group that would clash with the rigid E577•••R580 pair, resulting in repulsion between protein and DNA (Figure 5G) and reduced binding affinity. The pentagonally-shaped water network was partially preserved in the structure at 1.7-Å resolution (PDB 6OD4), but was not visible in the structure of lower, 2.0-Å resolution (PDB 6OD5).
Besides the aforementioned E577•••R580 stabilizing force, for which the corresponding interactions are also preserved in the structures of TCF3 homodimer and heterodimer (19,20), there are two Asn-Arg pairs that might perform similar stabilizing interactions at the protein-DNA interface. The N574•••R578 interaction bridges two phosphate groups neighboring the central cytosine (Figure 5F). N573•••R576 bridges between the phosphate group of nucleotide T5 and, through a network of water molecules, nucleotides of G3 and T4 (Figure 5H). There is no direct contact with base pairs immediately outside of the E-box at positions 4 and 5. The closest distance is 3.8 Å between N573 and the 5-position methyl group of T5 (Figure 5H).
Recognition of 5caC modification
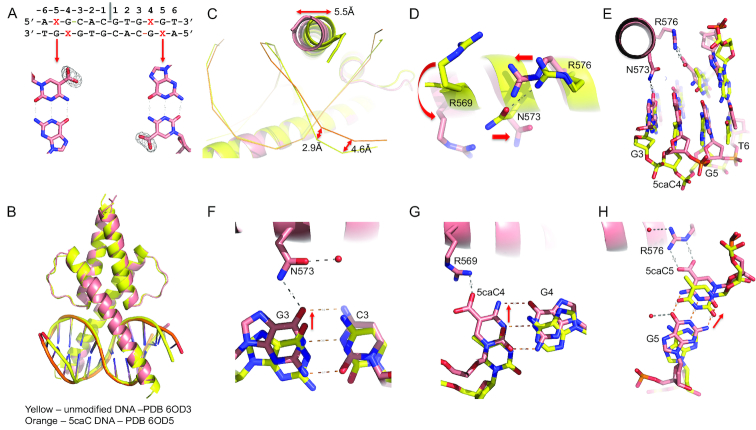
To understand the enhanced affinity of TCF4 for 5caC in E-box-flanking DNA, we next co-crystallized the TCF4 bHLH domain with a 12-base pair palindromic sequence containing a fully modified 5caCpG immediate outside the E-box element (5caCGCACGTG; Figure 6A). The overall structure is essentially identical to that of the TCF4-unmodified DNA complex (Figure 6B). The largest differences are localized to the N-terminal tip of the basic region, and the two base pairs where the modification occurs (Figure 6C). Three residues undergo large conformational changes in response to 5caC modification: R569 moves towards the DNA vs. pointing away, and the N573•••R576 bridge breaks as the two residues move in opposite directions (Figure 6D). Concurrently, the central base pairs of DNA (bp 1–2) have the least conformational difference, while the remaining base pairs (bp 3–6) have an increased shift towards the protein, due to gained interactions (Figure 6E). The acquired additional interactions include N573 interacting with guanine G3, and positively charged R569 and R576 interacting with the negatively charged carboxylate group of 5caC at C4 and C5 respectively (Figure 6F-H). The gained H-bonds and electrostatic interactions give rise to favorable binding of 5caC DNA as indicated by the change of binding entropy factor (-TΔS in Supplementary Figure S2).
Figure 6.
Recognition of 5caC modification. (A) The 5caC modification (X in the sequence) used for co-crystallization. Omit electron density (Fo-Fc) contoured at 5σ above the mean is shown for omitting the carboxylate groups of 5caC base. (B) Superimposition of two dimers in complex with unmodified DNA (yellow) and 5caC DNA (orange). Note the changes in the ends of DNA where modifications occur. (C) The movements of the tip of the basic region in helix α1 and DNA phosphate groups. (D) Movements of three residues (R569, N573 and R576) upon binding 5caC modified DNA. (E) The ends of DNA move towards protein. (F) N573 interacts with G3. (G) R569 interacts with the carboxylate group at C4. (H) R576 interacts with the carboxylate group at C5.
TCF4 mutations in Pitt-Hopkins syndrome
The TCF4 missense mutants in the HLH domain associated with Pitt-Hopkins syndrome include five arginine residues in the basic region (R569, R576, R578, R580 and R582), and two alanine residues involved in dimerization (A587 and A614) (Figure 1B and Supplementary Figure S5A). Of these seven, six are highly conserved in all classes of Vertebrata, and even in related transcription factors such as TCF12 and Atonal (Supplementary Figure S6). Among these seven residues, three have two substitutions each associated with Pitt-Hopkins syndrome, resulting in total of 10 mutant proteins. Previously, Sepp et al. examined the effect of five mutants on DNA binding using in vitro translated proteins (R576Q, R578H, R580W, R582P and A614V) (17). Among them, the four mutations of arginine abrogated DNA binding completely, and binding by A614V was severely reduced (17). When the Pitt-Hopkins syndrome-associated mutations of TCF4 were introduced to Drosophila daughterless, R580W and R582P mutants act dominant negatively, whereas R578H and A614V mutants were hypomorphic and less potent in induction of diseased phenotype (53).
The substitutions of arginine with large aromatic tryptophan (R569W and R580W) and smaller glycine (R576G) or rigid proline (R578P and R582P) would bring hydrophobicity to the basic region or altered structure in the middle of long helix α1, suggesting that it is loss of function alterations in TCF4 that are important in disease. This is consistent with the high conservation of these residues (Supplementary Figure S6). Indeed, the R569W mutant exhibited concentration-dependent aggregation, and diminished in DNA binding (Supplementary Figure S5B-S5C). This is reminiscent of MAX R36W (a related bHLH protein), of which is associated with myeloma (33). However, while the DNA binding is decreased, R569W retains its preference, albeit much reduced, for fully-carboxylated sequences (Supplementary Figure S5C), suggesting that R576 (the second 5caC-interacting arginine) is still partially functional for carboxylate group recognition in the context of R569W mutation.
Among the arginine residues mutated in the Pitt-Hopkins syndrome, R582 is the only arginine not involved in binding of either base or phosphate backbone in the cognate complex (Supplementary Figure S5D). However, R582 contacts a phosphate group from the minor groove side in the non-cognate complex (Figure 4A), which might indicate that R582 plays a role in guiding TCF4′s transition from nonspecific to specific DNA interactions along the substrate-recognition pathway.
Substitution of the positively-charged arginine with a polar glutamine or histidine occurs at TCF4 residues 576 (R576Q), 578 (R578H) and 580 (R580Q). Although the mutant residues retain the ability to make H-bonds, lack of a positive charge could affect interactions with the negatively-charged DNA phosphate group, and/or the shortened side chain could disrupt R•••E/N interactions (Figure 5). In the case of MAX R60Q, a mutation found in a wide variety of cancers including endometrial, glioma, acute myeloid leukemia, colorectal and stomach, the mutant protein had significantly reduced DNA binding (33). Two additional missense mutations that were observed in the helical regions (A587P and A614V) (Supplementary Figure S5E) might break helix α1 and/or affect dimerization.
DISCUSSION
Hydroxymethylcytosine (5hmC) is highly abundant in the brain (54), accumulating during lineage specification of neural stem cells (55) as well as in brain tumor cells (56). Tet dioxygenases are responsible for successive 5mC oxidations to 5hmC, 5fC and 5caC (26,27). Indeed, the levels of 5mC undergo dynamic change in brain development from fetus to young adult (57). Moreover, genome-wide mapping of Tet3 binding sites in mouse neural progenitor cells are selectively enriched for sequences containing an E-box motif (5′-tCACGTGa-3′) (58). Besides the dioxygenase activity that generates 5caC, Tet3 contains a DNA-binding CXXC domain, which binds 5caC (58,59). 5caC, the final product of Tet-mediated oxidation, together with 5fC, is found at much lower levels at steady state (1–10% that of 5hmC), preferentially accumulates at enhancers and other distal regulatory regions (such as promoter), and correlates with chromatin accessibility (60–66), implying that the dynamic turnover of 5mC may be particularly important in enhancer function. A recent study in human preimplantation embryos suggested that both 5fC and 5caC were present in the chromosomes during the whole period of preimplantation development, and each modification had its own pattern (67). ChIP-seq of TCF4 in neural-derived cells indicates TCF4 binding sites are enriched at active enhancers (23,24).
Here, we provide in vitro evidence that TCF4 binding is enhanced by the presence of 5caC immediately adjacent to the E-box. To our knowledge, there are no currently available data showing specific binding of 5caC-marked DNA, for TCF4 or any other transcription factor, in a cellular context. Genes potentially regulated directly by the binding of TCF4 include CNIH3, a gene involved in opioid dependence (68), LMX1A, a LIM homeobox transcription factor that drives Cux2 expression in the forebrain (69), TRIM8, an E3 ubiquitin ligase that regulates glioma-driving genes and functions related to the central nervous system (70), IDH2, a key metabolic enzyme that generates α-ketoglutarate, and TET1, one of the three 5mC dioxygenases that uses α-ketoglutarate as enzymic cofactor (Supplementary Figure S3C). It remains to demonstrate, in a cellular context, that the regulation of gene expression at these loci involves TCF4 binding to DNA marked by 5caC. We note that a previous example illustrated the consequence of protein-5caC interaction that the presence of 5caC on a template DNA strand reduced rate and substrate specificity of RNA polymerase II transcription (71).
We showed previously that another bHLH transcription factor, MAX, exhibits the greatest affinity for a 5caCpG in the central E-box (5′-CA-CG-TG-3′) (33). MAX arginine R36 recognizes 5caC; the corresponding residue in TCF4 is a hydrophobic valine (V581; see Figure 1C). In contrast, TCF4 demonstrated the highest affinity for a 5caCpG immediately adjacent to an E-box (5′-CG-CACGTG-3′) and R569 and R576 recognize, respectively, two 5caC residues of a fully-modified CpG site (Figure 6G and H). The corresponding residues in MAX are a lysine and a leucine (Figure 1C). Thus, two closely-related bHLH proteins use unique arginine residues for recognition of 5caC located at different positions of E-box elements. Furthermore, our analyses show the position-dependent effect of differential DNA modification at two cytosine residues on the binding affinity of TCF4. Specifically, TCF4 binding to DNA is affected differently by the modification at the outer CpG and its immediate neighbor CpA. In sum, these data raise the possibility that the role of DNA modification (methylation and/or oxidation) in regulating transcription factor activity is likely to be more refined than a simple on-off switch. Our study suggests that gene expression could plausibly be controlled by a combination of DNA sequence variations in the recognition sequence, patterns of DNA modification, and variable structural architectures of DNA-binding proteins, such as the basic region of bHLH when encountering DNA from either minor or major grooves. These observations imply existence of multiple layers of subtlety and versatility in epigenetic regulatory processes.
Although the principle of proteins recognizing DNA sequences by contacts in the major groove has been known for decades (72), our study on a relatively simple bHLH domain revealed five unique conformations along the protein-recognition pathway from nonspecific to specific complexes. Nonspecific interactions (conformations 1 and 2) can occur in the DNA minor groove, whereas direct interactions with bases occur only in the DNA major groove (conformation 3). The basic region of the recognition helix is intrinsic flexible, adopts multiple conformations ranging from a disordered, extended loop to part of a rigid helix (conformation 4). Two arginine residues (R569 and R576) can switch roles from a purely electrostatic interaction with a DNA phosphate to a highly specific binding mode with base modification (5caC) (conformation 5). This switch effectively increased binding affinity as well as extending the recognition sequence from 6-bp to an 8-bp extended E-box. Our data suggest a temporal order for the formation of specific contacts during the search along the DNA.
DATA AVAILABILITY
The X-ray structures (coordinates and structure factor files) of TCF4 bHLH domain with bound DNA have been submitted to PDB under accession number 6OD3 (13-mer), 6OD4 (11-mer) and 6OD5 (12-mer with 5caC).
Supplementary Material
ACKNOWLEDGEMENTS
We thank B. Baker of New England Biolabs for synthesizing the oligonucleotides and Dongxue Wang of Emory University for initial cloning and protein expression.
Authors contributions: J.Y. performed protein expression and purification, DNA binding experiments, mutagenesis and crystallization. J.R.H. performed X-ray data collection and structure determination. J.L. and Y.H. analyzed published TCF4 ChIP-seq data. X.Z. and X.C. organized and designed the scope of the study; R.M.B. performed data analysis and assisted in preparing the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health (NIH) [GM049245-24 to X.C. and HL134780 to Y.H.]; Cancer Prevention and Research Institute of Texas [RR160029 to X.C.]; American Cancer Society [RSG-18-043-01-LIB to Y.H.]. The open access publication charge for this paper has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal
Conflict of interest statement. None declared.
This paper is linked to: https://doi.org/10.1093/nar/gkz557.
REFERENCES
- 1. Forrest M.P., Hill M.J., Quantock A.J., Martin-Rendon E., Blake D.J.. The emerging roles of TCF4 in disease and development. Trends Mol. Med. 2014; 20:322–331. [DOI] [PubMed] [Google Scholar]
- 2. Massari M.E., Murre C.. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell Biol. 2000; 20:429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caudy M., Vassin H., Brand M., Tuma R., Jan L.Y., Jan Y.N.. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell. 1988; 55:1061–1067. [DOI] [PubMed] [Google Scholar]
- 4. Murre C., McCaw P.S., Baltimore D.. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989; 56:777–783. [DOI] [PubMed] [Google Scholar]
- 5. Henthorn P., Kiledjian M., Kadesch T.. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990; 247:467–470. [DOI] [PubMed] [Google Scholar]
- 6. Brzozka M.M., Radyushkin K., Wichert S.P., Ehrenreich H., Rossner M.J.. Cognitive and sensorimotor gating impairments in transgenic mice overexpressing the schizophrenia susceptibility gene Tcf4 in the brain. Biol. Psychiatry. 2010; 68:33–40. [DOI] [PubMed] [Google Scholar]
- 7. Crux S., Herms J., Dorostkar M.M.. Tcf4 regulates dendritic spine density and morphology in the adult brain. PLoS One. 2018; 13:e0199359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kennedy A.J., Rahn E.J., Paulukaitis B.S., Savell K.E., Kordasiewicz H.B., Wang J., Lewis J.W., Posey J., Strange S.K., Guzman-Karlsson M.C. et al.. Tcf4 Regulates Synaptic Plasticity, DNA Methylation, and Memory Function. Cell Rep. 2016; 16:2666–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stefansson H., Ophoff R.A., Steinberg S., Andreassen O.A., Cichon S., Rujescu D., Werge T., Pietilainen O.P., Mors O., Mortensen P.B. et al.. Common variants conferring risk of schizophrenia. Nature. 2009; 460:744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu X., Zhang B., Liu W., Paciga S., He W., Lanz T.A., Kleiman R., Dougherty B., Hall S.K., McIntosh A.M. et al.. A survey of rare coding variants in candidate genes in schizophrenia by deep sequencing. Mol. Psychiatry. 2014; 19:857–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Basmanav F.B., Forstner A.J., Fier H., Herms S., Meier S., Degenhardt F., Hoffmann P., Barth S., Fricker N., Strohmaier J. et al.. Investigation of the role of TCF4 rare sequence variants in schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2015; 168B:354–362. [DOI] [PubMed] [Google Scholar]
- 12. Pitt D., Hopkins I.. A syndrome of mental retardation, wide mouth and intermittent overbreathing. Aust. Paediatr. J. 1978; 14:182–184. [DOI] [PubMed] [Google Scholar]
- 13. Amiel J., Rio M., de Pontual L., Redon R., Malan V., Boddaert N., Plouin P., Carter N.P., Lyonnet S., Munnich A. et al.. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am. J. Hum. Genet. 2007; 80:988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zweier C., Peippo M.M., Hoyer J., Sousa S., Bottani A., Clayton-Smith J., Reardon W., Saraiva J., Cabral A., Gohring I. et al.. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome). Am. J. Hum. Genet. 2007; 80:994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brockschmidt A., Todt U., Ryu S., Hoischen A., Landwehr C., Birnbaum S., Frenck W., Radlwimmer B., Lichter P., Engels H. et al.. Severe mental retardation with breathing abnormalities (Pitt-Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum. Mol. Genet. 2007; 16:1488–1494. [DOI] [PubMed] [Google Scholar]
- 16. de Pontual L., Mathieu Y., Golzio C., Rio M., Malan V., Boddaert N., Soufflet C., Picard C., Durandy A., Dobbie A. et al.. Mutational, functional, and expression studies of the TCF4 gene in Pitt-Hopkins syndrome. Hum. Mutat. 2009; 30:669–676. [DOI] [PubMed] [Google Scholar]
- 17. Sepp M., Pruunsild P., Timmusk T.. Pitt-Hopkins syndrome-associated mutations in TCF4 lead to variable impairment of the transcription factor function ranging from hypomorphic to dominant-negative effects. Hum. Mol. Genet. 2012; 21:2873–2888. [DOI] [PubMed] [Google Scholar]
- 18. Forrest M., Chapman R.M., Doyle A.M., Tinsley C.L., Waite A., Blake D.J.. Functional analysis of TCF4 missense mutations that cause Pitt-Hopkins syndrome. Hum. Mutat. 2012; 33:1676–1686. [DOI] [PubMed] [Google Scholar]
- 19. Ellenberger T., Fass D., Arnaud M., Harrison S.C.. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994; 8:970–980. [DOI] [PubMed] [Google Scholar]
- 20. Longo A., Guanga G.P., Rose R.B.. Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition. Biochemistry. 2008; 47:218–229. [DOI] [PubMed] [Google Scholar]
- 21. Murre C., Bain G., van Dijk M.A., Engel I., Furnari B.A., Massari M.E., Matthews J.R., Quong M.W., Rivera R.R., Stuiver M.H.. Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta. 1994; 1218:129–135. [DOI] [PubMed] [Google Scholar]
- 22. De Masi F., Grove C.A., Vedenko A., Alibes A., Gisselbrecht S.S., Serrano L., Bulyk M.L., Walhout A.J.. Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res. 2011; 39:4553–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forrest M.P., Hill M.J., Kavanagh D.H., Tansey K.E., Waite A.J., Blake D.J.. The psychiatric risk gene transcription factor 4 (TCF4) regulates neurodevelopmental pathways associated With Schizophrenia, autism, and intellectual disability. Schizophr. Bull. 2018; 44:1100–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xia H., Jahr F.M., Kim N.K., Xie L., Shabalin A.A., Bryois J., Sweet D.H., Kronfol M.M., Palasuberniam P., McRae M. et al.. Building a schizophrenia genetic network: transcription factor 4 regulates genes involved in neuronal development and schizophrenia risk. Hum. Mol. Genet. 2018; 27:3246–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ceribelli M., Hou Z.E., Kelly P.N., Huang D.W., Wright G., Ganapathi K., Evbuomwan M.O., Pittaluga S., Shaffer A.L., Marcucci G. et al.. A Druggable TCF4- and BRD4-dependent transcriptional network sustains malignancy in blastic plasmacytoid dendritic cell neoplasm. Cancer Cell. 2016; 30:764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L. et al.. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009; 324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y.. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011; 333:1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ren R., Horton J.R., Zhang X., Blumenthal R.M., Cheng X.. Detecting and interpreting DNA methylation marks. Curr. Opin. Struct. Biol. 2018; 53:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., Das P.K., Kivioja T., Dave K., Zhong F. et al.. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017; 356:eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kribelbauer J.F., Laptenko O., Chen S., Martini G.D., Freed-Pastor W.A., Prives C., Mann R.S., Bussemaker H.J.. Quantitative Analysis of the DNA Methylation Sensitivity of Transcription Factor Complexes. Cell Rep. 2017; 19:2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blackwood E.M., Eisenman R.N.. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991; 251:1211–1217. [DOI] [PubMed] [Google Scholar]
- 32. Prendergast G.C., Lawe D., Ziff E.B.. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991; 65:395–407. [DOI] [PubMed] [Google Scholar]
- 33. Wang D., Hashimoto H., Zhang X., Barwick B.G., Lonial S., Boise L.H., Vertino P.M., Cheng X.. MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma. Nucleic Acids Res. 2017; 45:2396–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Khund-Sayeed S., He X., Holzberg T., Wang J., Rajagopal D., Upadhyay S., Durell S.R., Mukherjee S., Weirauch M.T., Rose R. et al.. 5-Hydroxymethylcytosine in E-box motifs ACAT|GTG and ACAC|GTG increases DNA-binding of the B-HLH transcription factor TCF4. Integr. Biol. (Camb.). 2016; 8:936–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Golla J.P., Zhao J., Mann I.K., Sayeed S.K., Mandal A., Rose R.B., Vinson C.. Carboxylation of cytosine (5caC) in the CG dinucleotide in the E-box motif (CGCAG|GTG) increases binding of the Tcf3|Ascl1 helix-loop-helix heterodimer 10-fold. Biochem. Biophys. Res. Commun. 2014; 449:248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patel A., Hashimoto H., Zhang X., Cheng X.. Characterization of how DNA modifications affect DNA binding by C2H2 zinc finger proteins. Methods Enzymol. 2016; 573:387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scopes R.K. Measurement of protein by spectrophotometry at 205 nm. Anal. Biochem. 1974; 59:277–282. [DOI] [PubMed] [Google Scholar]
- 38. Otwinowski Z., Borek D., Majewski W., Minor W.. Multiparametric scaling of diffraction intensities. Acta Crystallogr. A. 2003; 59:228–234. [DOI] [PubMed] [Google Scholar]
- 39. Yeates T.O. Detecting and overcoming crystal twinning. Methods Enzymol. 1997; 276:344–358. [PubMed] [Google Scholar]
- 40. Padilla J.E., Yeates T.O.. A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr. D Biol. Crystallogr. 2003; 59:1124–1130. [DOI] [PubMed] [Google Scholar]
- 41. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Terwilliger T.C., Read R.J., Adams P.D., Brunger A.T., Afonine P.V., Grosse-Kunstleve R.W., Hung L.W.. Improved crystallographic models through iterated local density-guided model deformation and reciprocal-space refinement. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 46. Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zheng R., Wan C., Mei S., Qin Q., Wu Q., Sun H., Chen C.H., Brown M., Zhang X., Meyer C.A. et al.. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019; 47:D729–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fonfria-Subiros E., Acosta-Reyes F., Saperas N., Pous J., Subirana J.A., Campos J.L.. Crystal structure of a complex of DNA with one AT-hook of HMGA1. PLoS One. 2012; 7:e37120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Horowitz S., Trievel R.C.. Carbon-oxygen hydrogen bonding in biological structure and function. J. Biol. Chem. 2012; 287:41576–41582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tamberg L., Sepp M., Timmusk T., Palgi M.. Introducing Pitt-Hopkins syndrome-associated mutations of TCF4 to Drosophila daughterless. Biol .Open. 2015; 4:1762–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kriaucionis S., Heintz N.. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009; 324:929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wheldon L.M., Abakir A., Ferjentsik Z., Dudnakova T., Strohbuecker S., Christie D., Dai N., Guan S., Foster J.M., Correa I.R. Jr et al.. Transient accumulation of 5-carboxylcytosine indicates involvement of active demethylation in lineage specification of neural stem cells. Cell Rep. 2014; 7:1353–1361. [DOI] [PubMed] [Google Scholar]
- 56. Ramsawhook A., Lewis L., Coyle B., Ruzov A.. Medulloblastoma and ependymoma cells display increased levels of 5-carboxylcytosine and elevated TET1 expression. Clin. Epigenet. 2017; 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D. et al.. Global epigenomic reconfiguration during mammalian brain development. Science. 2013; 341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jin S.G., Zhang Z.M., Dunwell T.L., Harter M.R., Wu X., Johnson J., Li Z., Liu J., Szabo P.E., Lu Q. et al.. Tet3 Reads 5-Carboxylcytosine through Its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep. 2016; 14:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu Y., Xu C., Kato A., Tempel W., Abreu J.G., Bian C., Hu Y., Hu D., Zhao B., Cerovina T. et al.. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012; 151:1200–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shen L., Wu H., Diep D., Yamaguchi S., D’Alessio A.C., Fung H.L., Zhang K., Zhang Y.. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013; 153:692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song C.X., Szulwach K.E., Dai Q., Fu Y., Mao S.Q., Lin L., Street C., Li Y., Poidevin M., Wu H. et al.. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013; 153:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu H., Wu X., Shen L., Zhang Y.. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol. 2014; 32:1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lu X., Han D., Zhao B.S., Song C.X., Zhang L.S., Dore L.C., He C.. Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res. 2015; 25:386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun Z., Dai N., Borgaro J.G., Quimby A., Sun D., Correa I.R. Jr, Zheng Y., Zhu Z., Guan S.. A Sensitive Approach to Map Genome-wide 5-Hydroxymethylcytosine and 5-Formylcytosine at Single-Base Resolution. Mol. Cell. 2015; 57:750–761. [DOI] [PubMed] [Google Scholar]
- 65. Neri F., Incarnato D., Krepelova A., Rapelli S., Anselmi F., Parlato C., Medana C., Dal Bello F., Oliviero S.. Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep. 2015; 10:674–683. [DOI] [PubMed] [Google Scholar]
- 66. Wu X., Inoue A., Suzuki T., Zhang Y.. Simultaneous mapping of active DNA demethylation and sister chromatid exchange in single cells. Genes Dev. 2017; 31:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pendina A.A., Efimova O.A., Krapivin M.I., Mekina I.D., Tikhonov A.V., Koltsova A.S., Petrovskaia-Kaminskaia A.V., Chiryaeva O.G., Kogan I.Y., Gzgzyan A.M. et al.. Genomic distribution of 5-formylcytosine and 5-carboxylcytosine in human preimplantation embryos. Mol. Reprod. Dev. 2018; 85:893–895. [DOI] [PubMed] [Google Scholar]
- 68. Nelson E.C., Agrawal A., Heath A.C., Bogdan R., Sherva R., Zhang B., Al-Hasani R., Bruchas M.R., Chou Y.L., Demers C.H. et al.. Evidence of CNIH3 involvement in opioid dependence. Mol. Psychiatry. 2016; 21:608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fregoso S.P., Dwyer B.E., Franco S.J.. Lmx1a drives Cux2 expression in the cortical hem through activation of a conserved intronic enhancer. Development. 2019; 146:dev170068. [DOI] [PubMed] [Google Scholar]
- 70. Venuto S., Castellana S., Monti M., Appolloni I., Fusilli C., Fusco C., Pucci P., Malatesta P., Mazza T., Merla G. et al.. TRIM8-driven transcriptomic profile of neural stem cells identified glioma-related nodal genes and pathways. Biochim. Biophys. Acta Gen. Subj. 2019; 1863:491–501. [DOI] [PubMed] [Google Scholar]
- 71. Kellinger M.W., Song C.X., Chong J., Lu X.Y., He C., Wang D.. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2012; 19:831–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Seeman N.C., Rosenberg J.M., Rich A.. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sepp M., Kannike K., Eesmaa A., Urb M., Timmusk T.. Functional diversity of human basic helix-loop-helix transcription factor TCF4 isoforms generated by alternative 5′ exon usage and splicing. PLoS One. 2011; 6:e22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The X-ray structures (coordinates and structure factor files) of TCF4 bHLH domain with bound DNA have been submitted to PDB under accession number 6OD3 (13-mer), 6OD4 (11-mer) and 6OD5 (12-mer with 5caC).