Abstract
Fission yeast phosphate acquisition genes pho1, pho84, and tgp1 are repressed in phosphate-rich medium by transcription of upstream lncRNAs. Here, we show that phosphate homeostasis is subject to metabolite control by inositol pyrophosphates (IPPs), exerted through the 3′-processing/termination machinery and the Pol2 CTD code. Increasing IP8 (via Asp1 IPP pyrophosphatase mutation) de-represses the PHO regulon and leads to precocious termination of prt lncRNA synthesis. pho1 de-repression by IP8 depends on cleavage-polyadenylation factor (CPF) subunits, termination factor Rhn1, and the Thr4 letter of the CTD code. pho1 de-repression by mutation of the Ser7 CTD letter depends on IP8. Simultaneous inactivation of the Asp1 and Aps1 IPP pyrophosphatases is lethal, but this lethality is suppressed by mutations of CPF subunits Ppn1, Swd22, Ssu72, and Ctf1 and CTD mutation T4A. Failure to synthesize IP8 (via Asp1 IPP kinase mutation) results in pho1 hyper-repression. Synthetic lethality of asp1Δ with Ppn1, Swd22, and Ssu72 mutations argues that IP8 plays an important role in essential 3′-processing/termination events, albeit in a manner genetically redundant to CPF. Transcriptional profiling delineates an IPP-responsive regulon composed of genes overexpressed when IP8 levels are increased. Our results establish a novel role for IPPs in cell physiology.
INTRODUCTION
Cells respond to phosphate starvation by inducing the transcription of phosphate acquisition genes (1,2). The phosphate regulon in the fission yeast Schizosaccharomyces pombe comprises three genes that specify, respectively, a cell surface acid phosphatase Pho1, an inorganic phosphate transporter Pho84, and a glycerophosphate transporter Tgp1 (3). Expression of pho1, pho84 and tgp1 is actively repressed during growth in phosphate-rich medium by the transcription in cis of a long noncoding (lnc) RNA from the respective 5′ flanking genes prt, prt2 and nc-tgp1 (4–10). It is proposed that transcription of the upstream lncRNA interferes with expression of the downstream mRNA genes by displacing the activating transcription factor Pho7 from its binding site(s) in the mRNA promoters that overlap the lncRNA transcription units (8,10,11) (Figure 1A).
Figure 1.
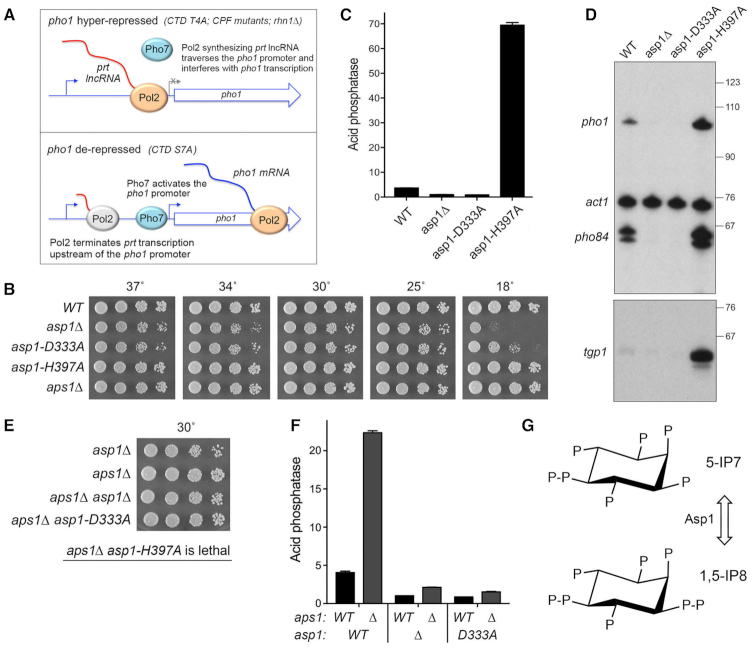
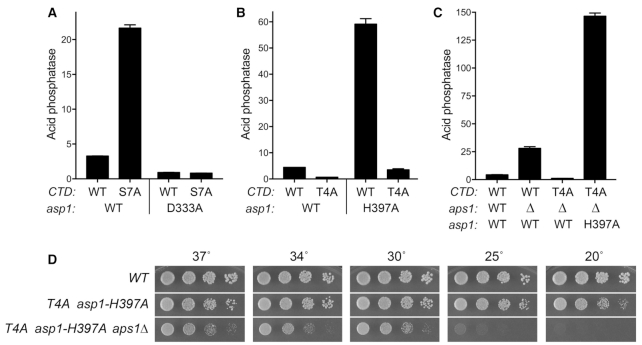
Opposite effects of IPP kinase and IPP pyrophosphatase mutations on pho1 expression. (A) Models for the pho1 hyper-repressed (top panel) and de-repressed (bottom panel) states of the prt–pho1 locus under phosphate-replete conditions. (B) Growth of S. pombe strains with the indicated asp1 and aps1 alleles. Cells were inoculated in YES broth and grown at 30°C. Exponentially growing cultures were adjusted to A600 of 0.1 and aliquots (3 μl) of serial 5-fold dilutions were spotted on YES agar and then incubated at the temperatures specified. (C) The indicated fission yeast strains were grown to A600 of 0.5 to 0.8 in liquid culture in YES medium at 30°C. Cells were then harvested, washed with water, and assayed for Pho1 acid phosphatase activity by conversion of p-nitrophenylphosphate to p-nitrophenol. Activity is expressed as the ratio of A410 (p-nitrophenol production) to A600 (input cells). Each datum in the bar graph is the average of assays using cells from at least three independent cultures ± SEM. (D) Total RNA from fission yeast cells with the indicated asp1 genotypes was analyzed by reverse transcription primer extension using a mixture of radiolabeled primers complementary to the pho84, act1, and pho1 mRNAs (Top panel) or the tgp1 mRNA (Bottom panel). The reaction products were resolved by denaturing PAGE and visualized by autoradiography. The positions and sizes (nt) of DNA markers are indicated on the right. (E) The indicated single and double mutant haploid progeny of genetic crosses were spot-tested for growth on YES agar at 30°C. The IPP pyrophosphatase-dead aps1Δ and asp1-H397A alleles were synthetically lethal. (F) The indicated strains were assayed for Pho1 acid phosphatase activity. The results show that the de-repression of Pho1 expression in aps1Δ cells (lacking Aps1 IPP pyrophosphatase) is erased by asp1Δ and asp1-D333A mutations that ablate Asp1 IPP kinase activity. (G) Structures of the predominant forms of IP7 and IP8 are shown. Asp1 kinase converts IP7 to IP8 and the Asp1 pyrophosphatase reverses this process (reviewed in (35)).
Recent studies highlight 3′ processing and transcription termination as a control point in the lncRNA-mediated repression of 3′-flanking gene expression. In particular, the prt-pho1 locus is established as a sensitive read-out of cellular influences on termination via their effect on the degree of interference with Pho1 expression by prt lncRNA transcription (12). Those influences include: the phosphorylation status of the RNA polymerase II (Pol2) carboxyl-terminal domain (CTD); the activity of protein kinases Csk1 and Cdk9; the Ctf1, Ssu72, Dis2, Ppn1 and Swd22 subunits of the CPF (cleavage polyadenylation factor) complex; and the transcription termination factor Rhn1 (3,7,8,12,13).
The S. pombe Pol2 CTD consists of tandem repeats of a heptapeptide Y1S2P3T4S5P6S7. CTD mutations S7A and S5A that prevent installation of the Ser7-PO4 or Ser5-PO4 marks de-repress pho1 and pho84 in phosphate-replete cells (7,8,10,13). By contrast, prevention of the Thr4-PO4 mark by T4A hyper-represses pho1 and pho84 under phosphate-rich conditions (7,13). Because such CTD mutations do not affect the activity of the lncRNA or mRNA gene promoters per se (8,10), it is proposed that CTD status affects Pol2 termination during lncRNA synthesis (Figure 1A). Specifically, it is hypothesized that loss of the Ser7-PO4 or Ser5-PO4 marks leads to precocious termination of prt lncRNA transcription prior to the pho1 promoter and loss of the Thr4-PO4 mark reduces termination and hence increases transcription across the pho1 promoter (8) (Figure 1A).
This termination-centric scenario is supported by recent findings that: (i) mutations of CPF subunits and Rhn1—proteins that normally promote 3′ processing/termination—result in hyper-repression of pho1 under phosphate-replete conditions; (ii) the de-repression of pho1 elicited by the CTD-S7A allele is erased by CPF and Rhn1 mutations; (iii) the pho1 hyper-repressive CTD-T4A allele is synthetically lethal with the pho1 hyper-repressive CPF alleles ppn1Δ and swd22Δ and (iv) the pho1 de-repressive CTD-S7A allele is synthetically lethal with the pho1 de-repressive cdk9 alleles T212A and T212E (12). We envision that there are additional influences on Pol2 transcription termination that are amenable to discovery via their impact on fission yeast phosphate homeostasis.
Inositol pyrophosphates (IPPs) are signaling molecules implicated in a broad range of physiological processes in eukaryal cells. IPPs participate in phosphate homeostasis in Saccharomyces cerevisiae and Arabidopsis thaliana (14). A role for IPPs in S. pombe phosphate gene regulation was suggested by the findings that a null allele of aps1 (which encodes a Nudix-family IPP pyrophosphatase) de-repressed Pho1 under phosphate-replete conditions, whereas a null allele of asp1 (which encodes a kinase that synthesizes IPP) hyper-repressed Pho1 (15,16). It was proposed that IP7 was the relevant metabolite that elicited de-repression of Pho1, based on the assumption that Asp1 kinase is responsible for IP7 synthesis from IP6 (16). This view is challenged by recent studies of the properties of fission yeast Asp1 and its effects on IPP levels in vivo (17–20). To wit: (i) Asp1 is actually a bifunctional enzyme composed of an N-terminal IPP-generating kinase domain and a C-terminal IPP-degrading pyrophosphatase domain; (ii) the in vivo effect of an asp1Δ null allele or a kinase-dead asp1-D333A allele was to eliminate intracellular IP8 and to increase the level of IP7; (iii) overexpression of the isolated Asp1 pyrophosphatase domain severely depressed intracellular IP8 and concomitantly increased IP7 and (iv) the in vivo effect of a pyrophosphatase-dead asp1-H397A allele was to increase the level of IP8 without affecting the level of IP7 (19). These results signify that the function of the Asp1 kinase is to generate IP8 via phosphorylation of its substrate IP7 and the function of the Asp1 pyrophosphatase is to convert its substrate IP8 back to IP7 (Figure 1G).
IPP dynamics reflect a balance between a kinase that converts IP7 to IP8 and pyrophosphatases that hydrolyze IP8 to IP7. Here, we focus on how genetic perturbations of IP8/IP7 metabolism impact the expression of phosphate-responsive genes during phosphate-replete growth. Our findings implicate IP8 in promoting precocious termination of PHO-regulatory lncRNA transcription. Shared phenotypes, epistasis, and synthetic lethalities establish a novel interactome embracing IPPs, RNA 3′ processing and termination factors, and the Pol2 CTD.
MATERIALS AND METHODS
Mutational effects on fission yeast growth
Cultures of S. pombe strains (Supplementary Figure S7) were grown in liquid medium until A600 reached 0.6–0.8. The cultures were adjusted to a final A600 of 0.1, and 3 μl aliquots of serial 5-fold dilutions were spotted on YES agar. The plates were photographed after incubation for 2 days at 34°C, 2.5 days at 30°C and 37°C, 4 days at 25°C, 6 days at 20°C, and 8 days at 18°C.
Deletion of asp1 and aps1
PCR amplification and standard cloning methods were employed to construct plasmids in which an antibiotic-resistance cassette (kanMX or natMX) (21,22) is flanked by 560- to 680-bp gene-specific DNA segments corresponding to genomic sequences upstream and downstream of the asp1 or aps1 ORF. The disruption cassettes were excised and transfected into diploid S. pombe cells. Antibiotic-resistant transformants were selected and analyzed by Southern blotting to ensure correct integration of kanMX or natMX at the target locus, thereby deleting the entire asp1 or aps1 ORF. Heterozygous diploids were then sporulated and G418- or nourseothricin-resistant aps1Δ and asp1Δ haploids were isolated. A hygromycin-resistant aps1Δ strain was generated by marker switching (22).
Tests of mutational synergies
Standard genetic methods were employed to generate haploid strains harboring mutations/deletions in two (or three) differently marked genes. In brief, pairs of haploids with null or missense mutations were mixed on malt agar to allow mating and sporulation, and the mixture was then subjected to random spore analysis. Spores (∼1500) were plated on YES agar and also on media selective for marked mutant alleles; the plates were incubated at 30°C for up to 5 days to allow slow growing progeny to germinate and form colonies. At least 500 viable progeny were screened by replica-plating for the presence of the second (and then third) marker gene, or by sequentially replica-plating from YES to selective media. A finding that no haploids with two marker genes were recovered after 6–8 days of incubation at 30°C was taken to indicate synthetic lethality. [Note that by sequentially replica-plating and gauging the numbers of colonies at each step, we ensured that wild-type (unmarked) and the viable differentially marked single mutant alleles were recovered at the expected frequencies.] Growth phenotypes of viable double and triple mutant strains were assessed in parallel with the parental and wild-type cells at different temperatures (18–37°C) by spotting as described above. No synergy between two mutant alleles was indicated in cases where the double-mutant grew similar to one or both of the parent strains. Synergistic growth defects were scored as ts if the double-mutant cells failed to grow at high temperatures at which the single mutant strains formed colonies, but grew similarly to the single mutants at lower temperatures. Double-mutant cells that exhibit a growth defect at low (but not high) temperatures relative to the parent strains were scored as cs.
Acid phosphatase activity
Cells were grown at 30°C in YES medium. Aliquots of exponentially growing cultures were harvested, washed with water, and resuspended in water. To quantify acid phosphatase activity, reaction mixtures (200 μl) containing 100 mM sodium acetate (pH 4.2), 10 mM p-nitrophenylphosphate and cells (ranging from 0.01 to 0.1 A600 units) were incubated for 5 min at 30°C. The reactions were quenched by addition of 1 ml of 1 M sodium carbonate, the cells were removed by centrifugation, and the absorbance of the supernatant at 410 nm was measured. Acid phosphatase activity is expressed as the ratio of A410 (p-nitrophenol production) to A600 (cells). The data are averages (±SEM) of at least three assays using cells from three independent cultures.
prt-pho1 reporter plasmids and assays
The prt–pho1 reporter plasmid, marked with a kanamycin-resistance gene (kanMX), contains the tandem prt and pho1 genes spanning from 1831 nucleotides upstream of the pho1 ORF (comprising the prt promoter and prt lncRNA) to 647 nucleotide downstream of the pho1 ORF (8). [The prt transcription start site is located 1198 nucleotides upstream of the pho1 ORF, or 1147 nucleotides upstream of the pho1 transcription start site]. For reporter activity assays and RNA isolation, kanMX-marked plasmids were transfected into [prt2-pho84-prt-pho1]Δ cells (10) and transformants were selected on YES agar medium containing 150 μg/ml G418. Single colonies of individual transformants were pooled (≥20) and grown in liquid YES + G418 medium to A600 of 0.5–0.8. Aliquots were harvested by centrifugation for acid phosphatase activity measurements as described above or for RNA analysis as described below.
RNA analyses
Total RNA was extracted via the hot phenol method (23) from 10 to 20 A600 units of yeast cells that had been grown exponentially to A600 of 0.6–0.8 at 30°C. For analysis of specific transcripts by primer extension, aliquots (15 μg) of total RNA were used as templates for M-MuLV reverse transcriptase-catalyzed extension of 5′ 32P-labeled oligodeoxynucleotide primers complementary to the pho84, pho1, tgp1, or act1 mRNAs. The primer extension reactions were performed as described previously (24) and the products were analyzed by electrophoresis of the reaction mixtures through a 22-cm 8% polyacrylamide gel containing 7 M urea in 80 mM Tris-borate, 1.2 mM EDTA. The 32P-labeled primer extension products were visualized by autoradiography of the dried gel. The primer sequences were as follows: act1 5′-GATTTCTTCTTCCATGGTCTTGTC; pho1 5′-GTTGGCACAAACGACGGCC; pho84 5′-AATGAAGTCCGAATGCGGTTGC; tgp1 5′-GATTCATCCCAGCCACCAGAC. For Northern blotting, aliquots (10 μg) of total RNA were resolved by electrophoresis through a 1.2% agarose/formaldehyde gel. After photography under UV light to visualize ethidium bromide-stained rRNAs and tRNAs, the gel contents were transferred to a Hybond-XL membrane (GE Healthcare) and hybridization was performed as described previously (9) using a 32P-labeled single-strand DNA complementary to the segment of the prt RNA from nucleotides +159 to +198.
Transcriptome profiling by RNA-seq
RNA was isolated from S. pombe cells grown in liquid culture in YES medium at 30°C to an A600 of 0.5–0.6. Cells were harvested by centrifugation and total RNA was extracted using hot phenol. The integrity and quantity of total RNA was gauged with an Agilent Technologies 2100 Bioanalyzer. The Illumina TruSeq stranded mRNA sample preparation kit was used to purify poly(A)+ RNA from 500 ng of total RNA and to carry out the subsequent steps of poly(A)+ RNA fragmentation, strand-specific cDNA synthesis, indexing, and amplification. Indexed libraries were normalized and pooled for paired-end sequencing performed by using an Illumina HiSeq 4000 system. FASTQ files bearing paired-end reads of length 51 bases were mapped to the S. pombe genome (ASM294v2.28) using HISAT2–2.1.0 with default parameters (25). The resulting SAM files were converted to BAM files using Samtools (26). Count files for individual replicates were generated with HTSeq-0.10.0 (27) using exon annotations from Pombase (GFF annotations, genome-version ASM294v2; source ‘ensembl’). RPKM analysis and pairwise correlations (Supplementary Figures S4 and S6) were performed as described previously (13). Differential gene expression and fold change analysis was performed in DESeq2 (28). Cut-off for further evaluation was set for genes that were up or down by at least two-fold in mutant fission yeast strains versus wild-type, had a P-value (Benjamini-Hochberg adjusted) of ≤0.05, and had an average normalized count across all samples of ≥100. The list of genes that were dysregulated by these criteria in one or more of the mutant strains is compiled in Supplemental Table S1 (for WT, asp1-H397A, asp1-H397A aps1Δ ssu72-C13S, and ssu72-C13S) and Supplemental Table S2 (for WT, aps1Δ and asp1-D333A).
RESULTS
Opposite effects of IPP kinase and IPP pyrophosphatase mutations on pho1 expression
We surveyed fission yeast asp1Δ, asp1-D333A (IP7 kinase-dead), and asp1-H397A (IP8 pyrophosphatase-dead) alleles for their effects on growth on YES agar medium at 18 to 37°C (Figure 1B) and on pho1 expression during exponential growth at 30°C in liquid culture under phosphate-replete conditions (Figure 1C). The asp1-H397A strain grew as well as the wild-type control strain at 18–37°C, as gauged by colony size. By contrast, asp1Δ cells formed smaller colonies at 25°C to 37°C and failed to grow at 18°C. The asp1-D333A strain resembled asp1Δ, except that it was able to form small colonies at 18°C. Acid phosphatase activity (a gauge of Pho1 enzyme level that correlates with pho1 mRNA levels, as assayed by RT-qPCR, Northern blotting, primer extension, and RNA-seq [7,8,12,13,15]) was quantified by incubating suspensions of serial dilutions of the phosphate-replete cells for 5 min with p-nitrophenylphosphate and assaying colorimetrically the formation of p-nitrophenol. The basal Pho1 activity of wild-type cells was hyper-repressed by 4-fold in asp1Δ and asp1-D333A cells and de-repressed by 19-fold in asp1-H397A cells (Figure 1C). These results implicate: (i) increased IP8 levels (and/or increased IP8:IP7 ratio) in the asp1-H397A strain (19) as a trigger for de-repression of pho1 expression and (ii) IP8 absence and/or increased IP7 levels in the asp1Δ and asp1-D333A strains (19) as a driver of hyper-repression of pho1 expression under phosphate-replete conditions.
By performing primer extension analysis of pho1, pho84, and tgp1 mRNA levels in phosphate-replete wild-type, asp1Δ, asp1-D333A, asp1-H397A cells, we found that all three transcripts of the PHO regulon were increased in the IPP pyrophosphatase-defective asp1-H397A strain (Figure 1D). Conversely, the pho1, pho84, and tgp1 mRNA levels were reduced in the IPP kinase-defective asp1Δ and asp1-D333A strains (Figure 1D). Thus, perturbations of IPP status concordantly impact the entire PHO axis in fission yeast.
We also queried the effect of inactivation of a different fission yeast IPP pyrophosphatase enzyme, Aps1. Aps1 is a Nudix-family hydrolase that catalyzes hydrolysis of diadenosine polyphosphates (Ap6A and Ap5A) and IPPs (IP7 and IP8) in vitro (29). Intracellular levels of IP7 and IP8 in fission yeast (1–2 μM) greatly exceed that of Ap5A (4 nM) (30). Studies of the effects of aps1Δ and aps1 overexpression did not definitively assign a physiologic substrate, but did point to IPPs as the best candidates, rather than Ap5A (30). The aps1Δ strain grew as well as wild-type fission yeast on YES agar at 18–37°C (Figure 1B). We observed a 6-fold de-repression of Pho1 activity in aps1Δ cells (Figure 1F).
Epistasis relationships of the asp1 and aps1 alleles
Epistasis with respect to cell growth and pho1 expression was queried by attempting to construct all possible combinations of double-mutant strains via mating and sporulation. We thereby found (by random spore analysis; see Methods) that asp1Δ aps1Δ and asp1-D333A aps1Δ strains were viable and grew well on YES agar (Figure 1E). By contrast, the asp1-H397A and aps1Δ alleles were synthetically lethal; to wit, (i) we were unable to obtain viable double-mutants after screening a large population of haploid progeny of the genetic cross (Figure 1E) and (ii) wild-type progeny and the differentially marked asp1-H397A and aps1Δ single mutants were recovered at the expected frequencies. This result signifies that the IPP pyrophosphatases have essential but redundant functions in fission yeast and it suggests that accumulation of too much IP8 (or too high an IP8:IP7 ratio) is in some way toxic.
Analysis of Pho1 expression in the viable double-mutants grown under phosphate-replete conditions was highly instructive, insofar as the de-repression of Pho1 elicited by the aps1Δ allele was erased by its combination with the hyper-repressive IPP kinase-defective asp1Δ and asp1-D333A alleles (Figure 1F). We surmise that the de-repressive effect of loss of the Aps1 pyrophosphatase activity requires the presence of IP8 generated by the Asp1 kinase and we therefore infer that IP8 is a relevant substrate for the Aps1 pyrophosphatase with respect to phosphate homeostasis.
Do Asp1 perturbations affect the prt lncRNA or pho1 mRNA promoters?
In principle, manipulation of IPP metabolism by mutations in Asp1 could affect lncRNA-mediated repression of the downstream PHO genes by: (i) modulating the activity of the lncRNA promoter or (ii) affecting the intrinsic activity of the mRNA promoter (independent of lncRNA synthesis). To address these issues, we employed a plasmid-borne prt–pho1 reporter (Supplementary Figure S1A) that was introduced into fission yeast cells in which the chromosomal pho1 gene was deleted. This reporter faithfully reflects known homeostatic controls on the native pho1 locus (8). Here we found that the prt–pho1 reporter is responsive to Asp1 activity perturbations, whereby Pho1 reporter expression under phosphate-replete conditions is hyper-repressed by 4-fold in asp1-D333A cells and de-repressed by 8-fold in asp1-H397A cells compared to the wild-type asp1+ control (Supplementary Figure S1D). We proceeded to test a mutated version of the prt–pho1 reporter construct in which the prt promoter is inactivated by nucleotide changes in the HomolD and TATA box elements that drive prt lncRNA synthesis (Supplementary Figure S1B) (8). This mutant reporter provides a readout of the intrinsic activity of the pho1 promoter, freed from interference by transcription of the flanking prt lncRNA. The Pho1 activity of the mutant plasmid in wild-type cells is high (i.e. de-repressed) and is not different from the Pho1 activity in asp1-H397A cells (Supplementary Figure S1E). These results signify that the de-repressive effect of asp1-H397A on pho1 expression from the wild-type prt–pho1 locus is not caused by upregulation of the pho1 promoter per se. Note that the Pho1 activity of the mutant reporter in asp1-D333A cells was 50% higher than in wild-type cells (Supplementary Figure S1E), an effect opposite to the repressive impact of asp1-D333A on pho1 expression when the prt gene is transcribed (Supplementary Figure S1D).
The effects of Asp1 mutations on the prt promoter were assessed using a different plasmid reporter (Supplementary Figure S1C) in which the prt promoter directly drives expression of the pho1 ORF (8). Pho1 expression from this plasmid was virtually identical in wild-type and asp1-H397A cells (Supplementary Figure S1F), signifying that the de-repression of native pho1 by asp1-H397A is not caused by decreased activity of the prt promoter. The Pho1 activity from the prt promoter reporter in asp1-D333A cells was 56% higher than in wild-type cells (Supplementary Figure S1F); this effect is consonant with that seen for the pho1 promoter reporter (Supplementary Figure S1E).
De-repression of pho1 expression by asp1-H397A and aps1Δ depends on CPF subunits and Rhn1
Absent an effect of the asp1-D333A and asp1-H397A alleles on the isolated prt and pho1 promoters sufficient to explain their impact on phosphate homeostasis, we hypothesized that IPPs affect transcription termination during prt lncRNA synthesis and hence the degree of interference with the pho1 promoter. Specifically, we envision that increased IP8 levels in the asp1-H397A strain lead to precocious termination during prt lncRNA synthesis, by enhancing the responsiveness of elongating Pol2 to the action of cleavage/polyadenylation and termination factors. If this is the case, there are clear predictions concerning epistasis relationships between asp1-H397A and the 3′ processing/termination machinery. Loss-of-function mutations in fission yeast proteins that promote cotranscriptional 3′ processing and transcription termination hyper-repress Pho1 under phosphate-replete conditions (12). This hyper-repression is observed in knockout strains lacking the Dis2, Ctf1, Ppn1, or Swd22 subunits of the CPF complex, a strain with a catalytically dead (C13S) version of the Ssu72 protein phosphatase subunit of CPF, and a strain that lacks the transcription termination factor Rhn1 (12; and Figure 2A).
Figure 2.
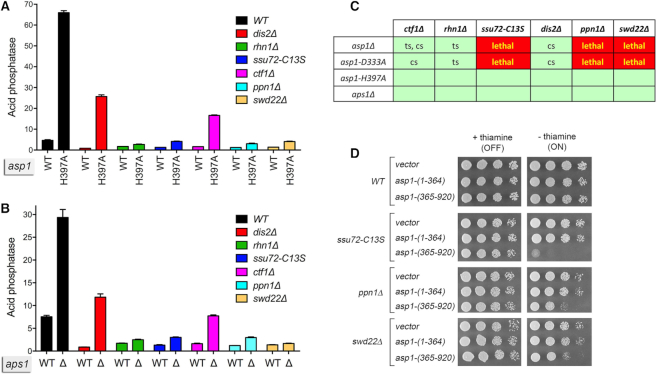
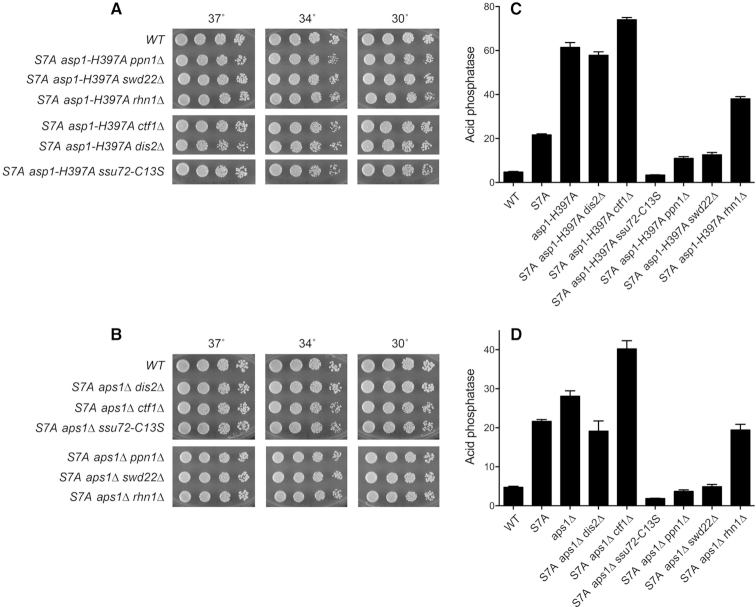
Genetic and functional interactions between Asp1, Aps1, CPF subunits, and Rhn1. (A and B) De-repression of Pho1 expression by asp1-H397A and aps1Δ depends on CPF subunits and Rhn1. S. pombe strains bearing the indicated asp1 alleles (wild-type or H397A; panel A) or aps1 alleles (wild-type or aps1Δ; panel B) in combination with CPF subunit or Rhn1 mutations as specified were grown in liquid culture at 30°C and assayed for acid phosphatase activity. (C) Synthetic lethalities and growth defects. Synthetically lethal pairs of alleles are highlighted in red boxes. Viable double-mutants without a synthetic defect are indicated by plain green boxes. Viable double-mutants that displayed temperature-sensitive (ts) or cold-sensitive (cs) defects are annotated as such in their respective green boxes. (D) Asp1 pyrophosphatase domain overexpression inhibits growth of CPF subunit mutants. Wild-type, ssu72-C13S, ppn1Δ, and swd22Δ strains were transformed with plasmids overexpressing the Asp1 pyrophosphatase domain (aa 365–920) or the Asp1 kinase domain (aa 1–364) under the control of the thiamine-regulated nmt1 promoter (19). Control cells were transformed with the empty plasmid vector. After growth in PMG–Leu medium containing thiamine, serial dilutions were spotted on PMG–Leu medium that either contained 15 μM thiamine (nmt1 promoter OFF) or lacked thiamine (nmt1 promoter ON) and incubated at 30°C.
To address epistasis, we introduced the asp1-H397A allele into the CPF subunit mutant strains; the asp1-H397A allele did not affect their growth (Figures 2C and 5A). It was noteworthy that introducing the asp1-H397A allele into the rhn1Δ strain suppressed the ts growth defect of rhn1Δ at 37°C (Supplementary Figure S2). We then assessed Pho1 expression under phosphate-replete conditions. The instructive findings were that the de-repression of Pho1 by asp1-H397A was effaced in rhn1Δ, ssu72-C13S, ppn1Δ, and swd22Δ cells and was partially blunted in dis2Δ and ctf1Δ cells (Figure 2A). Thus, the increase in Pho1 expression in asp1-H397A cells requires CPF subunits and Rhn1, consistent with the precocious termination model cited above.
Figure 5.
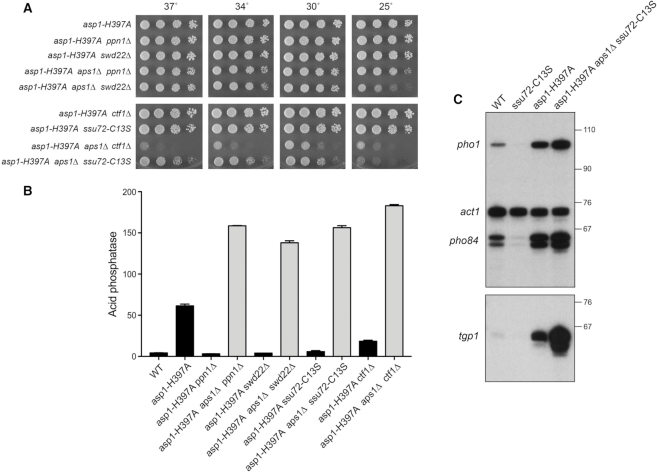
asp1-H397A aps1Δ lethality is rescued by ppn1Δ, swd22Δ, ssu72-C13S, and ctf1Δ. (A) Strains with the asp1-H397A allele in various combinations with aps1Δ and CPF subunit mutations were spot tested for growth at the temperatures specified. (B) The indicated strains were grown in liquid culture at 30°C and assayed for acid phosphatase activity. Each datum in the bar graph is the average of assays using cells from at least three independent cultures ± SEM. (C) Total RNA from fission yeast cells with the indicated genotypes was analyzed by reverse transcription primer extension using a mixture of radiolabeled primers complementary to the pho84, act1, and pho1 mRNAs (Top panel) or the tgp1 mRNA (Bottom panel). The reaction products were resolved by denaturing PAGE and visualized by autoradiography. The positions and sizes (nt) of DNA markers are indicated on the right. The results in B and C show that inactivation of the Asp1 and Aps1 pyrophosphatases additively de-represses pho1, pho84, and tgp1 expression in a manner that is not prevented by CPF mutations.
We extended this epistasis analysis by introducing aps1Δ into the various CPF subunit and Rhn1 mutant backgrounds. Note that aps1Δ also suppressed the ts growth defect of rhn1Δ (Supplementary Figure S2). The de-repression of Pho1 by aps1Δ was eliminated in rhn1Δ, ssu72-C13S, ppn1Δ, and swd22Δ cells and attenuated in dis2Δ and ctf1Δ cells (Figure 2B). These results echo the effects of CPF and Rhn1 mutations on phosphate homeostasis in asp1-H397A cells (Figure 2A) and fortify the precocious termination model.
Effect of IPP pyrophosphatase-dead mutations on prt lncRNA
The prt lncRNA derived from the chromosomal prt–pho1 locus in logarithmically growing vegetative cells is rapidly degraded by the nuclear exosome under the direction of DSR (determinant of selective removal) elements in the prt RNA (4,5,8). However, the increased gene dosage of the prt–pho1 cassette on the reporter plasmid in pho1Δ cells has permitted analysis of internally terminated prt transcripts by Northern blotting and the identification of two internal prt poly(A) sites, PAS and PAS2, by 3′-RACE (12). The prt locus gives rise to three classes of poly(A)+ RNA: (i) a ∼2.5 kb RNA corresponding to a prt–pho1 read-through transcript; (ii) a ∼0.4 kb species, prt PAS, that corresponds to prt RNA that was cleaved and polyadenylated at the +351 PAS site and (iii) a ∼0.6 kb species, prt PAS2, that corresponds to prt RNA that was cleaved and polyadenylated at the +589 PAS2 site (Figure 3A). These three classes of transcripts are seen here in a Northern blot of RNAs isolated from two independent cultures of asp1+ pho1Δ cells bearing the prt–pho1 reporter plasmid (Figure 3B, lanes WT). We find that the prt–pho1 read-through transcript is strongly suppressed in reporter-bearing asp1-H397A and aps1Δ cells, whereas the internally terminated transcripts are comparatively spared (Figure 3B). This result is consistent with the idea that increased IPP levels in IPP pyrophosphatase-dead mutants enhances the propensity of Pol2 to terminate prt transcription prior to traversal of the pho1 gene. Yet, it is not the case that the decrement in the long prt-pho1 read-through transcript in IPP pyrophosphatase-dead mutants is accompanied by an increase in the steady-state levels of the short prt PAS and prt PAS2 RNAs. It is conceivable that the asp1-H397A and aps1Δ mutations elicit termination/polyadenylation at diffuse sites within the prt gene (precluding detection as discrete species on a Northern blot) or that these alleles promote turnover of transcripts precociously terminated at PAS and PAS2.
Figure 3.
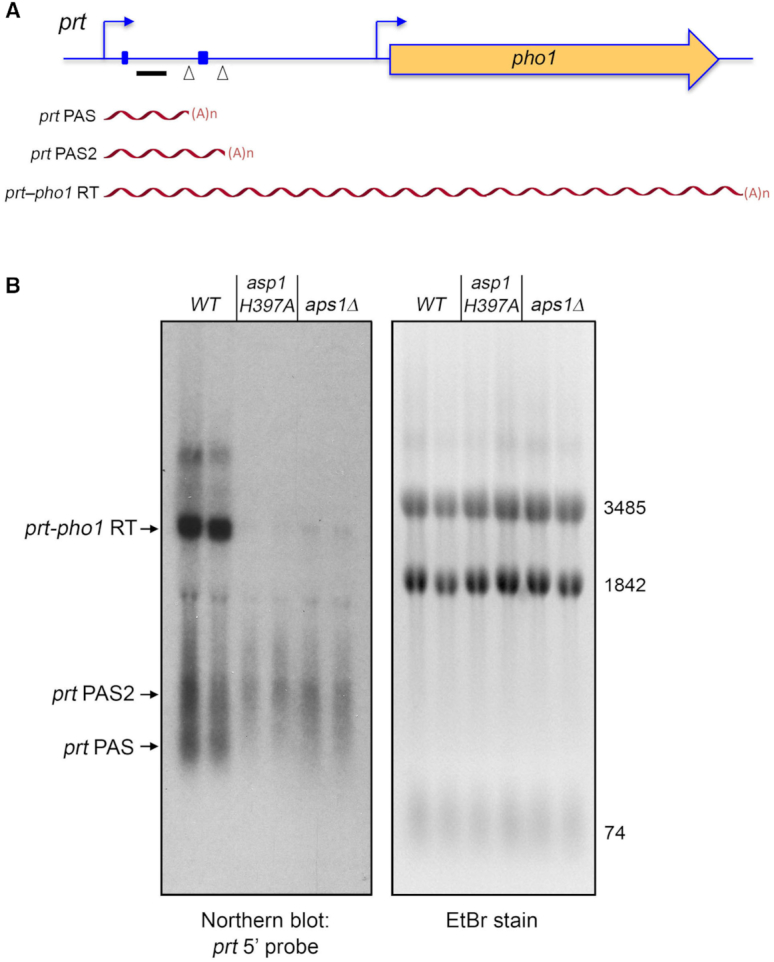
Northern analysis of prt RNA in IPP pyrophosphatase-dead mutants. (A) Schematic of the prt–pho1 locus in the reporter plasmid. Transcription start sites are indicated by bent blue arrows. Triangles denote internal poly(A) sites PAS and PAS2. DSR element clusters are indicated by small blue boxes. The gene-specific probe for prt (a 32P-labeled ssDNA complementary to the segment of the prt RNA from nucleotides +159 to +198) is denoted by a horizontal black bar. Three classes of poly(A)+prt transcripts described previously (12) are depicted as red wavy lines below the prt–pho1 locus. (B) RNA was isolated from two independent cultures of pho1Δ cells bearing the prt–pho1 reporter plasmid; the cells were either wild-type with respect to the Asp1 and Aps1 IPP pyrophosphatases or were pyrophosphatase-defective asp1-H397A or aps1Δ mutants as indicated. Right panel. The RNAs were resolved by formaldehyde-agarose gel electrophoresis and stained with ethidium bromide to visualize 28S and 18S ribosomal RNAs (3485 and 1842 nucleotides, respectively) and tRNAs (74 nucleotides). Left panel. The RNAs in the gel were transferred to membrane and hybridized to the prt probe. Annealed probe was visualized by autoradiography. The three classes of prt transcripts are indicated on the left. The PAS and PAS2 transcripts are assuredly derived from the prt–pho1 cassette, insofar as control experiments showed that they are undetectable by Northern analysis of RNA isolated from yeast strains bearing the empty plasmid vector.
De-repression of pho1 by asp1-H397A depends on DSR elements in the prt lncRNA
The DSR clusters in the prt transcript are implicated in prt termination (4). Therefore we tested a version of the prt–pho1 reporter (Figure 4A) in which the consensus DSR sequences in the two DSR clusters are altered by base substitutions (8). A prt-probed Northern blot of RNAs isolated from three independent cultures of asp1+ pho1Δ and asp1-H397A pho1Δ cells showed that formation of the prt–pho1 read-through transcript was restored in the asp1-H397A strain by virtue of the DSR point mutations (Figure 4B, compare to Figure 3B). Moreover, the de-repression of Pho1 expression elicited by the pyrophosphatase-dead asp1-H397A allele was erased by the DSR point mutations (Figure 4C). The evidence above collectively points to lncRNA termination as the target of the IPP pyrophosphatase-dead mutations on phosphate homeostasis.
Figure 4.
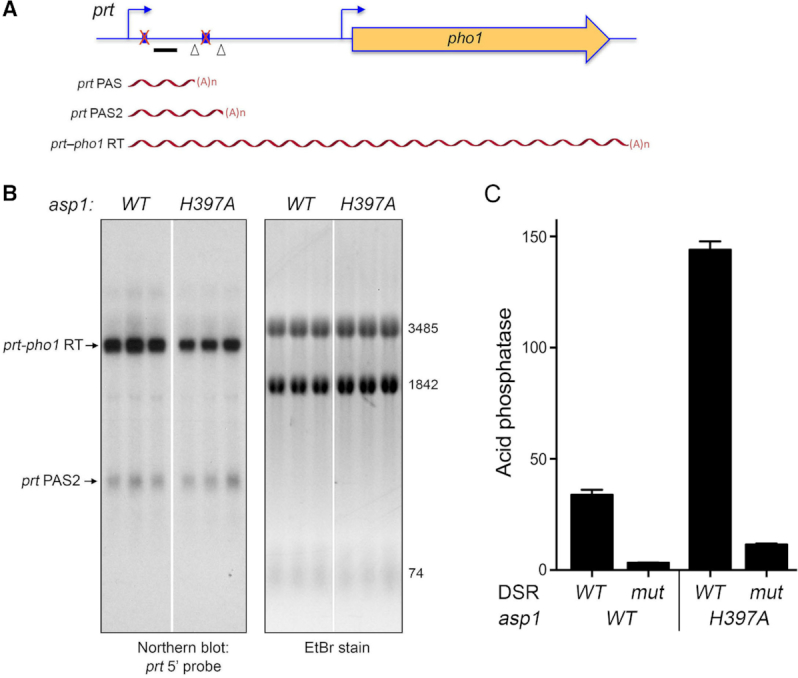
Northern analysis of prt RNA from a prt–pho1 reporter with mutant DSRs. (A) Schematic of the prt–pho1 locus in the reporter plasmid as in Figure 3, except that both DSR clusters are mutated (indicated by a red X over the blue boxes) as described (8). (B) RNA was isolated from three independent cultures of pho1Δ cells (either asp1 WT or H397A as specified) bearing the DSR mutant prt–pho1 reporter plasmid. Right panel. The RNAs were resolved by formaldehyde-agarose gel electrophoresis and stained with ethidium bromide to visualize 28S and 18S ribosomal RNAs (3485 and 1842 nucleotides, respectively) and tRNAs (74 nucleotides). Left panel. The RNAs in the gel were transferred to membrane and hybridized to the prt probe. Annealed probe was visualized by autoradiography. The WT and asp1-H397A RNAs were analyzed in parallel on the same agarose gel and blotted on the same membrane; the white space indicates that intervening lanes were cropped to bring the RNAs of interest close together for the purpose of composing the figure. (C) The reporter plasmids with wild-type or mutated DSRs were transfected into asp1+ (WT) or asp1-H397A strains in which the chromosomal pho1 locus was deleted. Acid phosphatase activity was determined as described in Supplementary Figure S1. Each datum in the bar graph is the average of assays using cells from three independent cultures ± SEM.
Synthetic genetic interactions of asp1 alleles with CPF and Rhn1 mutants
To further probe genetic connections between the IPP kinase and pyrophosphatase activities and the 3′-processing/termination machinery, haploid strains with the asp1Δ, asp1-D333A, asp1-H397A and aps1Δ alleles (marked with a 3′ flanking natMX or kanMX gene) were mated to haploid strains with null or missense mutations in cleavage/termination factors (marked with 3′ flanking antibiotic-resistance or ura4 genes). The resulting heterozygous diploids were sporulated and, for each allelic pair, a random collection of 500–1000 viable haploid progeny were screened by serial replica-plating for the presence of the flanking markers. A failure to recover any viable haploids with both markers was taken as evidence of synthetic lethality between the test alleles. [In such cases, the wild-type haploid progeny and the differentially marked single mutants were recovered at the expected frequencies.] The lethal allelic pairs are indicated by red boxes in the matrix shown in Figure 2C. The double-mutant haploids that passed selection were spotted on YES agar at 20–37°C in parallel with the component single mutants. Viable double-mutants without a synthetic defect are indicated by plain green boxes in Figure 2C. Viable double-mutants that displayed temperature-sensitive (ts) or cold-sensitive (cs) defects are annotated as such in their respective green boxes.
The results of the synthetic genetic array highlight allele-specific interactions of IPP metabolizing enzymes that provide new insights into the function of IPPs in fission yeast. To wit, we see that asp1-H397A and aps1Δ displayed no synthetic growth defects with the CPF subunit or Rhn1 mutants, consistent with our inferences from the phosphate homeostasis experiments that inactivation of IPP pyrophosphatases triggers precocious prt transcription termination and the CPF and Rhn1 mutants diminish prt termination. The finding that the two IPP pyrophosphatase-dead alleles reversed the ts growth defect of rhn1Δ (Supplementary Figure S2) suggests that increased IP8 ameliorates the impact of Rhn1 absence. By contrast, the IPP kinase-absent asp1Δ and asp1-D333A alleles—which we suggest are antagonists of lncRNA termination in the prt–pho1 system—are synthetically lethal with null alleles of CPF subunits Ppn1 or Swd22 and when the Ssu72 protein phosphatase is crippled (Figure 2C). Moreover, the asp1Δ and asp1-D333A mutations elicit conditional growth defects (ts or cs) in the ctf1Δ, dis2Δ, and rhn1Δ genetic backgrounds (Figure 2C). We surmise from the synthetic lethality observed here that IP8 (synthesized by the Asp1 kinase) and the Ppn1/Swd22/Ssu72 subunits of CPF play important but genetically redundant roles in promoting essential 3′ processing/transcription termination events in fission yeast.
A prediction of the latter inference is that depletion of Asp1-synthesized IPPs via another means (not involving crippling of the Asp1 kinase) would elicit a synthetic phenotype in one or more of the CPF mutant backgrounds. Previous studies showed that this state could be achieved in asp1+ cells by overexpressing the Asp1 pyrophosphatase domain (aa 365–920) under the control of the thiamine-regulated nmt1 promoter, which depletes intracellular IP8 and concomitantly increases IP7 (19). We found that whereas overexpression of Asp1-(365–920) under thiamine deprivation had no effect on wild-type fission yeast growth on agar medium, it prevented growth of ssu72-C13S cells, and slowed growth of ppn1Δ and swd22Δ cells (Figure 2D). Controls showed that growth inhibition was specific for Asp1-(365–920), insofar as: (i) there was no effect of the nmt1 vector or of nmt1-driven expression of the Asp1 kinase domain, Asp1-(1–364); and (ii) growth inhibition by the Asp1 pyrophosphatase domain was seen in the absence of thiamine when the nmt1 promoter is on, but not in the presence of thiamine when the promoter is off (Figure 2D).
Lethality of asp1-H397Aaps1Δ is rescued by ppn1Δ, swd22Δ, ssu72-C13S, and ctf1Δ
As noted above, the synthetic lethality of asp1-H397A and aps1Δ implies that accumulation of too much IP8 is in some way toxic to fission yeast. Given the potentially broad impact of IPP signaling on cellular physiology, the key question in the present context is whether the lethality of the asp1-H397A aps1Δ strain arises from unconstrained precocious transcription termination. If this were the case, then it might be expected that the deleterious effects of too much IP8 in the asp1-H397A aps1Δ context would be ameliorated by null alleles of CPF subunits Ppn1 or Swd22 or the catalytically dead CPF subunit Ssu72-C13S (each of which is synthetically lethal in the context of a kinase-dead Asp1). To test this idea, we crossed asp1-H397A ppn1Δ with aps1Δ ppn1+, asp1-H397A swd22Δ with aps1Δ swd22+ and asp1-H397A ssu72-C13S with aps1Δ ssu72+, then sporulated the resulting diploids, and screened random spores for each of the differentially marked loci of interest. In this way, we recovered viable asp1-H397A aps1Δppn1Δ, asp1-H397A aps1Δswd22Δ, and asp1-H397A aps1Δssu72-C13S haploid strains. (By contrast, we recovered no viable asp1-H397A aps1Δppn1+, asp1-H397A aps1Δswd22+, or asp1-H397A aps1Δssu72+ haploids.) The asp1-H397A aps1Δppn1Δ and asp1-H397A aps1Δswd22Δ cells grew well on YES agar at 30°C to 37°C, but were slow growing at 25°C (Figure 5A). The asp1-H397A aps1Δssu72-C13S strain grew poorly at 30°C to 37°C and was grossly defective at 25°C (Figure 5A). We extended this approach to the null allele of CPF core subunit Ctf1 and recovered a viable asp1-H397Aaps1Δctf1Δ haploid strain that grew slowly at 30°C and was very sick at higher and lower temperatures (Figure 5A). These results suggest that the synthetic lethality of asp1-H397A aps1Δ is a consequence of IP8-driven precocious termination that depends on the Ppn1, Swd22, and Ctf1 subunits of CPF and the Ssu72 phosphatase activity. Whereas viability of asp1-H397A aps1Δ was restored when the aforementioned CPF subunits are deleted or inactive, additional genetic crosses showed that null alleles of dis2 or rhn1 were unable to suppress the synthetic lethality of asp1-H397A aps1Δ.
Altered phosphate homeostasis after rescue of asp1-H397A aps1Δ by CPF mutants
The rescue of asp1-H397A aps1Δ synthetic lethality by ppn1Δ, swd22Δ, ssu72-C13S, and ctf1Δ affords an opportunity to gauge the effect of inactivating both IPP pyrophosphatases on pho1 expression in phosphate-replete cells. The instructive findings were that: (i) Pho1 levels were significantly higher (2.2–3-fold) in every one of the four viable asp1-H397A aps1Δ strains than in the asp1-H397A single-mutant (Figure 5B); and (ii) adding the aps1Δ allele to the asp1-H397A double-mutants with ppn1Δ, swd22Δ, ssu72-C13S, and ctf1Δ overrode the antagonistic effects of the CPF mutants on pho1 de-repression by asp1-H397A (Figure 5B). We conclude that inactivation of the Asp1 and Aps1 pyrophosphatases additively de-represses pho1 expression in a manner that is not prevented by CPF mutations. The implication is that precocious termination of prt lncRNA synthesis is especially sensitive to increased IP8 in asp1-H397A aps1Δ cells.
Primer extension analysis of pho1 mRNA levels in wild-type, ssu72-C13S, asp1-H397A, and asp1-H397A aps1Δ ssu72-C13S triple mutant cells affirmed the acid phosphatase results, i.e. that pho1 was hyper-repressed by ssu72-C13S, de-repressed by asp1-H397A, and additively de-repressed in the rescued asp1-H397A aps1Δ ssu72-C13S background (Figure 5C). The same was true of the pho84 and tgp1 genes of the phosphate regulon (Figure 5C).
Lethality of dis2Δ ssu72-C13S is rescued by asp1-H397A
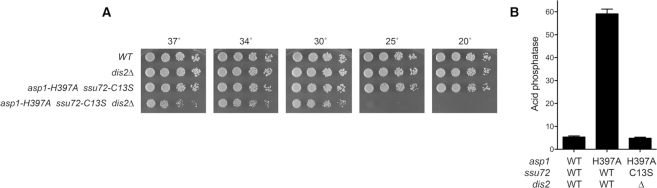
The synthetic lethality accompanying simultaneous inactivation of any of seven pairwise combinations of CPF subunits and/or Rhn1 (these being ctf1Δ ppn1Δ, ctf1Δ swd22Δ, ppn1Δ rhn1Δ, swd22Δ rhn1Δ, ppn1Δ ssu72-C13S, swd22Δ ssu72-C13S, and dis2Δ ssu72-C13S; (12)) is posited to be the consequence of a severe termination defect impacting the expression of essential S. pombe genes. We asked whether such a lethal termination defect in a double-mutant might be ameliorated by a third mutation that exerts an opposite effect, e.g. the asp1-H397A allele that we hypothesize promotes precocious termination of the lncRNAs that control phosphate homeostasis. To test this idea, we crossed viable double-mutants of asp1-H397A plus ctf1Δ, rhn1Δ, and ssu72-C13S (Figure 2A, C) with differentially marked single mutants ppn1Δ, swd22Δ, and dis2Δ and then screened by random spore analysis for viable triple mutants. Only in the cross of asp1-H397A ssu72-C13S with dis2Δ did we recover a viable asp1-H397A dis2Δ ssu72-C13S triple mutant. The asp1-H397A dis2Δ ssu72-C13S strain grew slower than the asp1-H397A ssu72-C13S double mutant on YES agar at 30°C, 34°C and 37°C (as gauged by colony size) and displayed a tight cs defect at 25°C and 20°C (Figure 6A). With respect to Pho1 expression, we saw that the asp1-H397A dis2Δ ssu72-C13S triple-mutant phenocopied the asp1-H397A ssu72-C13S double mutant in erasing the de-repression of Pho1 caused by asp1-H397A (Figure 6B).
Figure 6.
Lethality of dis2Δ ssu72-C13S is rescued by asp1-H397A. (A) Strains with the indicated genotypes were spot tested for growth at the temperatures specified. (B) The indicated strains were grown in liquid culture at 30°C and assayed for acid phosphatase activity.
Genetic interactions of Asp1 and Aps1 with the Pol2 CTD impact phosphate homeostasis
The de-repressive effect of asp1-H397A on pho1 expression and its genetic reliance on CPF subunits and Rhn1 reported here is similar to the CPF/Rhn1-dependent de-repression of pho1 observed in rpb1-CTD-S7A cells (12). To address whether the de-repression elicited by S7A depends on IPP status, we constructed a rpb1-CTD-S7A asp1-D333A double-mutant and found that the de-repression of Pho1 by S7A was erased when the IPP kinase activity was crippled (Figure 7A). In the opposite vein, the hyper-repression of pho1 in asp1Δ and asp1-D333A cells mimics that seen in rpb1-CTD-T4A cells (12,13). A doubly mutated rpb1-CTD-(T4A-S7A) strain maintains a hyper-repressed pho1 status, implying that the negative effects of T4A on prt termination win out over the precocious prt termination elicited by S7A (13). A pertinent question is whether T4A exerts a similar effect on the pho1 de-repression seen in asp1-H397A cells.
Figure 7.
Genetic interactions of Asp1 and Aps1 with the Pol2 CTD impact phosphate homeostasis. (A–C) Strains with the indicated genotypes were grown in liquid culture at 30°C and assayed for acid phosphatase activity. (A) The results show that de-repression of Pho1 by CTD-S7A requires the IPP kinase activity of Asp1. (B) The results show that CTD Thr4 is necessary for de-repression of Pho1 by the asp1 pyrophosphatase-dead H397A allele. (C) The results show that: (i) CTD Thr4 is necessary for de-repression of Pho1 by aps1Δ; and (ii) simultaneous inactivation of Asp1 and Aps1 pyrophosphatases overrides the hyper-repression of Pho1 by CTD-T4A. (D) Strains with the indicated genotypes were spot tested for growth on YES agar at the temperatures specified.
We addressed this issue by constructing an asp1-H397A CTD-T4A strain; this double mutant thrived at 25°C, 30°C, 34°C, and 37°C, but was slow growing at 20°C (Figure 7D). The CTD-T4A allele eliminated the large increase in Pho1 activity caused by asp1-H397A (Figure 7B). Similarly, CTD-T4A negated the de-repression of Pho1 by aps1Δ (Figure 7C). These results indicate that the precocious prt termination prompted by loss of the Asp1 or Aps1 pyrophosphatase activities per se, or loss of the Ser7 ‘letter’ of the CTD code, depends stringently on the Thr4 letter (either via the Thr4-PO4 mark or the Thr4 hydroxyl group).
It was instructive that the synthetic lethality of the asp1-H397A aps1Δ double-mutant was rescued by CTD-T4A (Figure 7D) and that adding aps1Δ to the asp1-H397A CTD-T4A double-mutant overrode the repressive effect of T4A and resulted in 2.5-fold greater Pho1 activity in the asp1-H397A asp1ΔCTD-T4A triple-mutant (Figure 7C) vis-à-vis the asp1-H397A single mutant (Figure 7B).
Synthetic lethalities and their suppression connect IPP status to the Pol2 CTD
Whereas neither asp1-H397A nor CTD-S7A affects growth of S. pombe at 30°C, we were unable to recover an asp1-H397A CTD-S7A double-mutant via mating and sporulation, nor were we able to recover an aps1ΔCTD-S7A double mutant. These findings signify that IPP accumulation (via inactivation of IPP pyrophosphatases) and loss of the CTD Ser7 mark are synthetically lethal. To query if the synthetic lethality of asp1-H397A or aps1Δ plus S7A mutations reflects a toxic level of precocious termination, we attempted to circumvent the lethality by introduction of mutations in CPF subunits and Rhn1. The striking findings were that: (i) the synthetic lethalities were reversed across the board by ctf1Δ, ssu72-C13S, swd22Δ, ppn1Δ, dis2Δ, and rhn1Δ and (ii) the rescued triple-mutants grew well at 30°C, 34°C and 37°C (Figure 8A, B).
Figure 8.
Synthetic lethalities and their suppression connect IPP status to the Pol2 CTD. (A and B) Strains with the indicated genotypes were spot tested for growth at the temperatures specified. The results show that the synthetic lethality of asp1-H397A or aps1Δ plus S7A mutation was reversed by ctf1Δ, ssu72-C13S, swd22Δ, ppn1Δ, dis2Δ, and rhn1Δ. (C and D) The indicated strains were grown in liquid culture at 30°C and assayed for acid phosphatase activity.
We proceeded to gauge Pho1 expression in the rescued CTD-S7A, IPP pyrophosphatase-defective, CPF/Rhn1 triple-mutants, the key question being whether the de-repressive effects of S7A and the asp1-H397A or aps1Δ alleles together ‘win out’ over the hyper-repressive effects of mutations in CPF subunits or Rhn1. The results were remarkable insofar as: (i) CPF/Rhn1 alleles affected Pho1 similarly in the S7A asp1-H397A and S7A aps1Δ backgrounds (compare Figure 8C and D) and (ii) there was a clear distinction between the dis2Δ, ctf1Δ, and rhn1Δ alleles that did not reverse Pho1 de-repression in S7A asp1-H397A and S7A aps1Δ cells and the ssu72-C13S, ppn1Δ, and swd22Δ alleles that eliminated or severely attenuated Pho1 de-repression (Figure 8C,D). The abilities of ssu72-C13S, ppn1Δ, and swd22Δ to dominate over S7A asp1-H397A and S7A aps1Δ (Figure 8C,D) contrast with the ‘loser’ status of these same alleles with respect to pho1 expression in asp1-H397A aps1Δ cells (Figure 5B), suggesting that the loss of the two IPP pyrophosphatases exerts a stronger effect on precocious termination than does the loss of one IPP pyrophosphatase in combination with CTD-S7A.
RNA-seq analysis defines an IPP-responsive gene set
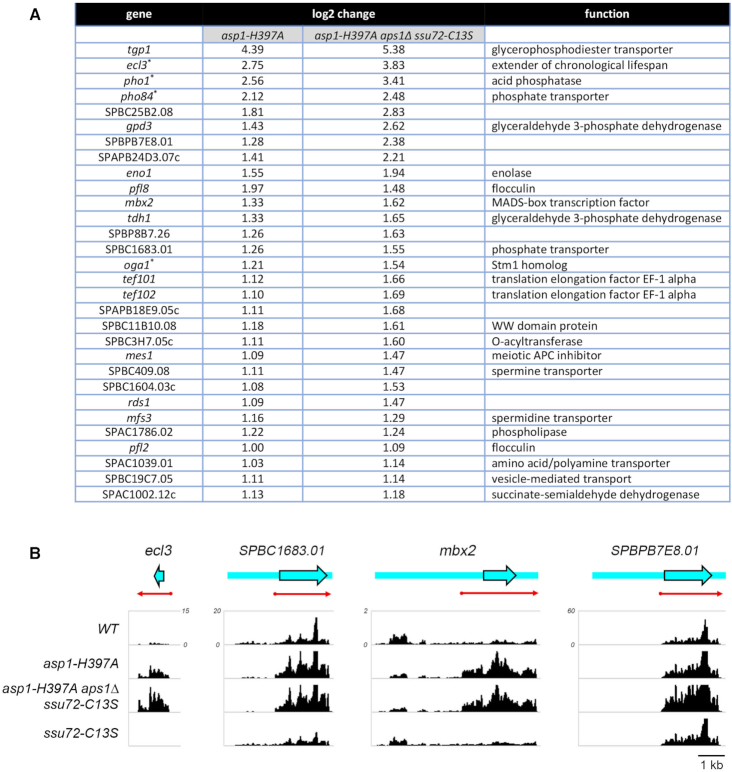
The concordant response of the three fission yeast PHO regulon genes to IPP pyrophosphatase-inactivating mutations raised the prospect that other genes might be up-regulated when IPP levels are increased in the asp1-H397A and asp1-H397A aps1Δ ssu72-C13S genetic backgrounds. To explore this idea, we performed RNA-seq on poly(A)+ RNA isolated from wild-type, asp1-H397A, asp1-H397A aps1Δssu72-C13S and ssu72-C13S cells. cDNAs obtained from three biological replicates (using RNA from cells grown to mid-log phase in YES medium at 30°C) were sequenced for each strain. In 11/12 of the datasets, 96–97% of the reads were mapped to unique genomic loci; in one dataset, 90% of the reads mapped to unique loci (Supplementary Figure S3). Read densities (RPKM) for individual genes were highly reproducible between biological replicates (Pearson coefficients of 0.98–0.99; Supplementary Figure S4). As internal controls, we affirmed that: (i) all of the reads for the asp1 transcript in the asp1-H397A strains had the intended His-to-Ala codon mutation; (ii) all the reads for the ssu72 transcript in the ssu72-C13S strains had the Cys-to-Ser codon mutation; and (iii) there were no reads for the deleted aps1 coding sequence in the asp1-H397A aps1Δ ssu72-C13S strain. A cutoff of ±2-fold change in normalized transcript read level and a corrected P-value of ≤0.05 were the criteria applied to derive initial lists of differentially expressed annotated loci in the mutants versus wild-type. We then focused on differentially expressed coding genes with average normalized read counts across all samples of ≥100 (DESeq2 baseMean parameter), in order to eliminate the many, mostly non-coding, transcripts that were expressed at very low levels in vegetative cells. The list of genes that were dysregulated by these criteria in one or more of the mutant strains is compiled in Supplemental Table S1.
Figure 9A shows the list of 30 annotated protein-coding genes that were up-regulated in asp1-H397A and asp1-H397A aps1Δssu72-C13S cells, but not in ssu72-C13S cells. This gene set comprises a putative IP8-responsive regulon. The ‘top hits’ with respect to fold upregulation included the three known phosphate-regulated genes: tgp1 (up 21-fold in asp1-H397A and 42-fold in asp1-H397A aps1Δssu72-C13S); pho1 (up 7-fold in asp1-H397A and 14-fold in asp1-H397A aps1Δssu72-C13S); and pho84 (up 4-fold in asp1-H397A and 6-fold in asp1-H397A aps1Δssu72-C13S). Thus, the RNA-seq data independently affirms the conclusions from assays of Pho1 enzyme activity, and of pho1, pho84, and tgp1 mRNA levels assayed by primer extension, that the PHO regulon is coordinately de-repressed when Asp1 pyrophosphatase is defective. Note: it is not the case that the PHO genes were overexpressed as a secondary effect of altered expression of other genes involved in PHO transcription/homeostasis or of genes encoding the 3′ processing/termination machinery. RNA-seq of asp1-H397A and asp1-H397A aps1Δssu72-C13S cells revealed insignificant effects on the transcripts encoding Pho7 (the transcription factor that drives pho1, pho84, and tgp1 mRNA synthesis), the Rpb1 subunit of Pol2, protein kinases Csk1 and Cdk9 (mutations of which de-repress pho1), cleavage/polyadenylation factors Ctf1, Cft1, Cft2, Dis2, Iss1, Pfs2, Pla1, Pta1, Ppn1, Rna14, Ssu72, Swd22, Ysh1, Yth1, Pcf11 and termination factors Rhn1, Seb1 and Dhp1.
Figure 9.
RNA-seq defines an IPP-responsive gene set. (A) List of annotated protein coding genes that were up-regulated at least two-fold in asp1-H397A and asp1-H397A aps1Δ ssu72-C13S cells. Any genes that were also upregulated two-fold in ssu72-C13S cells were excluded. The four genes denoted by asterisks (pho1, pho84, ecl3, and oga1) were down-regulated by at least two-fold in the ssu72-C13S strain. (B) Strand-specific RNA-seq read densities (counts/base/million, averaged in a 25 nucleotide window) of the indicated S. pombe strains are plotted on the y-axis as a function of position across the gene loci (x-axis). The read densities were determined from cumulative counts of all three RNA-seq replicates for each S. pombe strain. The y-axis scale for all tracks of each individual gene is shown next to the WT track. The common x-axis scale is shown on the bottom right. ORFs are indicated by blue arrowheads with black outline and UTRs by blue bars extending from the ORF, shown as they are annotated in Pombase. Red arrows indicate the predicted mRNA as judged from the RNA-seq data.
Another top upregulated hit in the IPP-responsive gene set (Figure 9A) was ecl3 (up 7-fold in asp1-H397A and 14-fold in asp1-H397A aps1Δssu72-C13S), which encodes an 89-amino acid protein that extends fission yeast chronological lifespan when overexpressed (31). It is noteworthy that: (i) the ecl3 gene is located on chromosome II, adjacent to and in opposite orientation to the prt2 lncRNA gene of the phosphate-regulated prt2–pho84–prt–pho1 gene cluster (10) and (ii) expression of the ecl3 paralogs ecl1 and ecl2, which are located at distant sites in the genome (31), is not affected in asp1-H397A and asp1-H397A aps1Δssu72-C13S cells. We speculate that ecl3 up-regulation by inactivation of IPP pyrophosphatase might reflect its proximity to prt2, e.g. that the prt2 promoter might be bidirectional (à la the nc-tgp1 promoter; ref. 6), driving transcription of the prt2 lncRNA that regulates pho84 and an oppositely transcribed lncRNA that interferes with ecl3 transcription. Consistent with the idea that de-repression of ecl3 by IPP pyrophosphatase inactivation might be connected to lncRNA transcription termination, the RNA-seq analysis showed that the ecl3 transcript was downregulated in ssu72-C13S cells (by 3-fold), concordant with the downregulation seen in ssu72-C13S cells for pho1 (by 5-fold) and pho84 (by 5-fold). Annotation of the ecl3 gene in PomBase (www.pombase.org) has no information other than the dimensions of the open reading frame (ORF, denoted by the blue arrow in Figure 9B, left panel). Plots of the strand-specific RNA-seq read density across this region in the WT, asp1-H397A, asp-H397A aps1Δ ssu72-C13S, and ssu72-C13S strains affirmed that ecl3 expression is upregulated by IPP pyrophosphatase inactivation and downregulated by Ssu72 inactivation (Figure 9B). Moreover, the read density in the upregulated strains clearly delineates the approximate margins of the ecl3 mRNA upstream and downstream of the ORF (red arrow in Figure 9B).
The IPP-responsive gene set also includes SPBC1683.01, which encodes a 573-amino acid putative inorganic phosphate transporter that is 84% identical in primary structure to the 572-amino acid Pho84 protein. The SPBC1683.01 mRNA is annotated in PomBase as a 3923-nucleotide transcript that includes a 1950-nucleotide 5′-UTR (Figure 9B). Because this long 5′-UTR contains 36 AUG codons upstream of the bona fide translation start site of the transporter ORF, we are skeptical of the transcript annotation. Rather, as we showed for prt2–pho84 (10), we suspect that the ∼2-kb region upstream of the transporter ORF specifies an independently transcribed lncRNA that regulates in cis the expression of the downstream SPBC1683.01 phosphate transporter. This idea is supported by the strand-specific RNA-seq read density across this gene in the WT, asp1-H397A, asp-H397A aps1Δ ssu72-C13S and ssu72-C13S strains (Figure 9B), insofar as there is a selective increase in RNA reads over the ORF upon IPP pyrophosphatase inactivation that is accompanied by a decrease in reads over the so-called 5′-UTR. By contrast, Ssu72 inactivation reduced reads over the ORF and tended to even out the read density across the locus (Figure 9B). The read density in the upregulated strains demarcates the approximate margins of the SPBC1683.01 mRNA (red arrow in Figure 9B). The results suggest that the putative lncRNA is a read-through transcript.
The mbx2 gene that is upregulated in asp1-H397A and asp1-H397A aps1Δssu72-C13S cells encodes a 372-amino acid MADS-box transcription factor that positively regulates invasive growth and flocculation by driving the expression of cell surface flocculin proteins (32,33). The IPP-responsive upregulated gene set includes the Mbx2-regulated flocculin genes pfl8 and pfl2 (Figure 9A). Moreover, the flocculin genes gsf2 and pfl3 are upregulated in asp1-H397A and asp1-H397A aps1Δssu72-C13S cells (though they are also upregulated in the ssu72-C13S strain). The increased expression of the Mbx2 transcription factor and several flocculin proteins neatly accounts for the findings that asp1-H397A cells display a strong flocculation phenotype and hyper-invasive growth vis-à-vis asp1+ cells (17). It is noteworthy that mbx2 mRNA is annotated in PomBase as a 6079-nucleotide transcript with a 4059-nucleotide 5′-UTR (Figure 9B) that contains 69 AUG codons upstream of the translation start site of the Mbx2 ORF. Thus, we speculate that the ∼4-kb region upstream of the Mbx2 ORF specifies a lncRNA that regulates the expression of the Mbx2 mRNA. The RNA-seq read density in the upregulated IPP pyrophosphatase-inactive strains indicates the dimensions of the true mbx2 mRNA (red arrow in Figure 9B).
Yet another example of such potential regulation in the IPP-responsive gene set is SPBPB7E8.01, which encodes an uncharacterized 569-amino acid protein. The SPBPB7E8.01 mRNA is annotated as a 4959-nucleotide transcript with a 2698-nucleotide 5′-UTR (Figure 9B) that contains 41 AUG codons upstream of the translation start site of the SPBPB7E8.01 ORF. Again, we envision that the ∼2.7-kb region upstream of the SPBPB7E8.01 ORF is transcribed independently as a lncRNA that regulates the expression of the SPBPB7E8.01 mRNA. The RNA-seq read densities clearly delineate the margins of the SPBPB7E8.01 mRNA (red arrow in Figure 9B) and show that it does not include the annotated long 5′-UTR.
In sum, the above genome-wide RNA-seq analysis affirms the conclusions from the preceding gene-specific experiments (at RNA and protein level) that the fission yeast PHO regulon is de-repressed by IPP pyrophosphatase inactivation and it delineates a novel IPP-responsive gene regulon, many members of which are known to be (e.g. tgp1, pho1, pho84), or are likely to be, under repressive control by an upstream flanking lncRNA.
Transcriptome analysis of aps1Δ accords with that of asp1-H397A
To gauge the effect of loss of the Aps1 pyrophosphatase activity per se on gene expression, we performed RNA-seq of poly(A)+ RNA isolated from wild-type and aps1Δ cells, sequencing cDNAs obtained from three biological replicates (see Supplementary Figures S5 and S6 for compilation of read counts and reproducibility of read densities between biological replicates). Figure 10 shows the list of 19 annotated protein-coding genes that were up-regulated in aps1Δ cells. Nine of these 19 genes (denoted by asterisks in Figure 10) were upregulated in asp1-H397A cells (Figure 9A). The co-regulated gene set includes the three members of the PHO regulon and the prt2-adjacent ecl3 gene. The tgp1, ecl3, pho1, and pho84 transcripts were increased by 7-fold, 4-fold, 3-fold and 3-fold, respectively, in aps1Δ cells. We also observed upregulation of mbx2 and of several of the flocculin genes under its transcriptional control. The concordant overlapping set of upregulated genes in two different genetic backgrounds in which distinct IPP pyrophosphatase activities are missing fortifies the case for an IPP-responsive transcriptional program in fission yeast.
Figure 10.
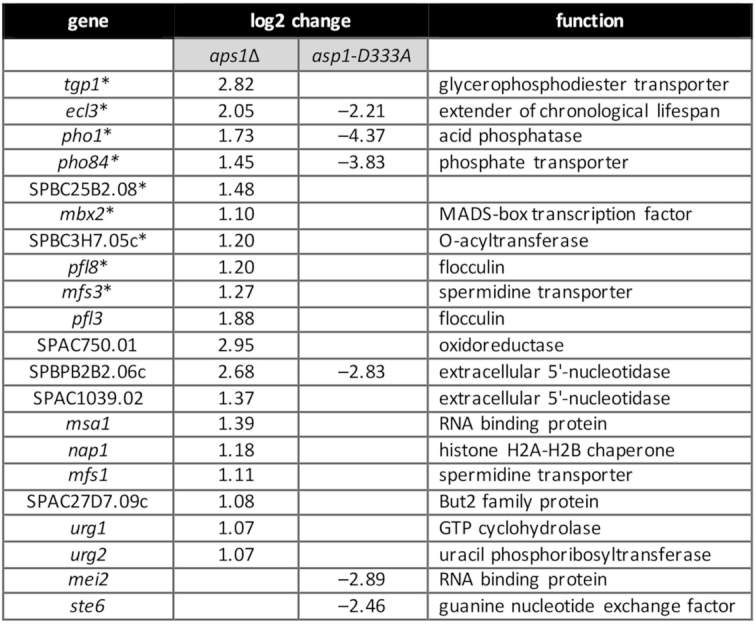
Transcriptome profile of aps1Δ and asp1-D333A cells. List of 19 annotated protein-coding genes that were up-regulated at least 2-fold in aps1Δ cells and six annotated protein-coding genes that were downregulated at least four-fold in asp1-D333A cells. Nine genes that were coordinately upregulated in aps1Δ and asp1-H397A IPP pyrophosphatase-deficient cells are denoted by asterisks.
Transcriptome analysis of asp1-D333A
If, as we hypothesize, increased levels of IP8 in the IPP pyrophosphatase-defective strains is responsible for the observed increases in a limited set of mRNAs, then we might expect to see opposite effects on some of these IPP-responsive transcripts when IP8 synthesis is precluded by the kinase-defective asp1-D333A allele. RNA-seq analysis of poly(A)+ RNA from asp1-D333A cells identified six protein-coding genes that were down-regulated by at least 4-fold compared to the wild-type control (Figure 10), four of which correspond to transcripts that were upregulated in aps1Δ and/or asp1-H397A cells. [As an internal control, we affirmed that all of the reads for the asp1 transcript in the asp1-D333A strains had the intended Asp-to-Ala codon mutation.] The two top hits in the down-regulated asp1-D333A gene set were pho1 and pho84, with decrements of 20-fold and 14-fold, respectively. The ecl3 transcript was reduced 5-fold in asp1-D333A cells. SPBPB2B2.06c encoding a putative extracellular 5′ nucleotidase was 7-fold down in asp1-D333A cells. Of the other aps1Δ or asp1-H397A up-regulated genes, only mfs3 was appreciably down-regulated in asp1-D333A cells (by 3-fold). Thus, there appear to be two classes of IPP-responsive genes: those that require IP8 for normal expression and are overexpressed when IP8 levels are elevated above normal (e.g. the PHO genes); and those that do not require IP8 for normal expression but are overexpressed when IP8 levels are elevated.
DISCUSSION
Fission yeast phosphate acquisition genes pho1, pho84, and tgp1 are repressed during growth in phosphate-rich medium by the transcription in cis of upstream flanking lncRNAs. This transcription interference phenomenon is tuned by the phosphorylation status of the RNA polymerase II CTD and the RNA 3′ processing/termination factors CPF and Rhn1 in a manner whereby genetic changes that elicit precocious termination of lncRNA synthesis lead to de-repression of downstream mRNA expression (e.g. rpb1-CTD-S7A) and changes that diminish lncRNA termination hyper-repress mRNA expression (e.g. rpb1-CTD-T4A, CPF subunit mutations, rhn1Δ). In the present study, we show that fission yeast phosphate homeostasis is subject to metabolite control by inositol pyrophosphates (IPPs), exerted via the 3′ processing/termination machinery and the Pol2 CTD.
The genetic evidence implicating IPPs, particularly IP8, as a new player in the interactome of the CTD code with 3′ processing and termination, stems from the coherent set of biochemical phenotypes and mutational synergies elicited by manipulations of the kinase and pyrophosphatase enzymes that convert IP7 to IP8 and vice versa. It is already established that: (i) inactivation of the Asp1 kinase component eliminates IP8 and increases intracellular IP7 and (ii) inactivation of the Asp1 pyrophosphatase module increases IP8 without much effect on IP7 (19). Here we find that increasing IP8 via IPP pyrophosphatase mutation de-represses the PHO regulon. Focusing on pho1 de-repression, we show that increased IP8 (or increased IP8:IP7 ratio) leads to precocious termination during prt lncRNA synthesis, and that pho1 de-repression depends on CPF subunits, Rhn1, and the Thr4 letter of the CTD code. Conversely, a failure to synthesize IP8 (via IPP kinase mutation) results in pho1 hyper-repression, thereby phenocopying mutations of CPF subunits, Rhn1, and CTD-T4A. These findings suggest that IP8 enhances the responsiveness of elongating Pol2 to the action of cleavage/polyadenylation and termination factors. Indeed, the de-repression of pho1 caused by the mutating CTD Ser7 to alanine depends on the Asp1 kinase that generates IP8. Moreover, the synthetic lethality of asp1Δ (no IP8) with mutations of CPF subunits Ppn1, Swd22 and Ssu72 argues that IP8 plays an important role in promoting essential 3′ processing/transcription termination events in fission yeast, albeit in a manner that is genetically redundant to CPF.
Too much IP8 is apparently toxic to fission yeast, insofar as simultaneous inactivation of the asp1 and aps1 IPP pyrophosphatase enzymes is lethal. That this lethality is tied to RNA 3′ processing/termination and the CTD code is established by the finding that asp1-H397A aps1Δ inviability is suppressed by mutations of CPF subunits Ppn1, Swd22, Ssu72, and Ctf1 and by CTD-T4A. In the same vein, increasing IP8 by inactivating the Asp1 pyrophosphatase is lethal in combination with CTD-S7A, implying that these changes additively promote precocious termination to the point that it becomes toxic. Here, too, the lethality of asp1-H397A CTD-S7A is suppressed by mutations of CPF subunits and termination factor Rhn1. Lastly, the yin-yang relationship of IPP pyrophosphatase mutations (that elicit precocious termination) and CPF mutations (that diminish termination) is cemented by the finding that the synthetic lethality of pairwise mutations of CPF subunits Dis2 and Ssu72 is suppressed by asp1-H397A allele that increases IP8.
Whereas IPPs have been implicated in a broad range of physiological processes (34,35), the present study is, to our knowledge, unique in forging a link between perturbations of IPP dynamics and RNA 3′ processing/transcription termination. The immediate target(s) of IP8 in eliciting the effects we describe here are as yet unknown. Two distinct themes have been invoked previously to account for IPP effects, whereby IPPs either: (i) bind to target proteins and allosterically enhance or inhibit their functions or (ii) serve as phosphate donors for transfer of an IPP β-phosphate to a previously mono-phosphorylated protein target leading to a covalent pyrophosphate modification of the target (34–37). Potential mechanisms for IP8 to promote 3′-processing/termination include: (i) allosteric binding or covalent modification of the processing/termination machinery so as to enhance its action on elongating Pol2; (ii) allosteric interaction with, or covalent modification of Pol2 to make it more responsive to the processing/termination machinery. Given the connections established here between IP8 and the Pol2 CTD phospho-site code, an enticing speculation is that increased IP8 levels elicit pyrophosphorylation of the phospho-CTD. There is precedent for allosteric interactions of IPPs with protein kinases that result in inhibition of kinase activity (36). The fission yeast Csk1 kinase is critical for repression of pho1 under phosphate-replete conditions, i.e., pho1 is de-repressed in a csk1Δ strain (15) in a manner that depends on Ssu72 and Rhn1 (12). Because the csk1Δ and IPP pyrophosphatase-dead mutations have overlapping phosphate homeostasis phenotypes, we reasoned that if the effect of increasing IP8 was transduced via inhibition of Csk1 kinase, then there would be no mutational synergy in a csk1Δ asp1-H397A double mutant. However, this model was vitiated by our finding that csk1Δ and asp1-H397A were synthetically lethal (not shown).
The SPX protein domain has been identified as an inositol polyphosphate/inositol pyrophosphate sensor found in plant and budding yeast proteins involved in phosphate homeostasis (38,39). Fission yeast have six annotated SPX domain-containing proteins (www.pombase.org). However, an SPX domain is not associated with any fission yeast proteins implicated in 3′ processing or transcription termination.
The ENTH protein domain involved in membrane traffic binds to inositol polyphosphates (40). The α-helical fold of the ENTH domain is very similar to that of the VHS domain (41) and to that of the Pol2 CTD-interaction domain (CID) found in several components of the mRNA transcription/processing apparatus (42). Fission yeast CID-containing proteins include CPF subunit Pcf11 and termination factors Rhn1 and Seb1. CID proteins have not (to our knowledge) been reported to bind IPPs. Our inspection of the structure of S. pombe Seb1 (43) suggests that it may have a counterpart of the lysine-rich surface motif responsible for inositol polyphosphate binding in the ENTH domain (40). Seb1 interacts physically with the fission yeast CPF complex (43). It is conceivable that IPP interaction with Seb1 (or one of the other fission yeast CID proteins) enhances 3′-processing/termination.
Finally, transcriptional profiling of IPP pyrophosphatase-defective S. pombe strains delineated what we construe to be an IP8-responsive regulon composed of genes that are overexpressed when IP8 levels are increased. The IP8-responsive gene set includes the phosphate acquisition regulon, which is repressed by upstream lncRNA synthesis, as well as other genes that are plausible candidates for similar lncRNA control.
DATA AVAILABILITY
RNA-seq data in this publication have been deposited in NCBI’s Gene Expression Omnibus. GSE127550, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127550, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131237.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Ursula Fleig (Heinrich Heine Universität) for providing the fission yeast asp1-D333A and asp1-H397A strains and the expression plasmids for the Asp1 kinase domain and pyrophosphatase domain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH grants [R01-GM52470 and R35-GM126945]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Tomar P., Sinha H.. Conservation of PHO pathway in ascomycetes and the role of Pho84. J. Biosci. 2014; 39:525–536. [DOI] [PubMed] [Google Scholar]
- 2. Jain A., Nagarajan V.K., Raghothama K.G.. Transcriptional regulation of phosphate acquisition by higher plants. Cell Mol. Life Sci. 2012; 69:3207–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carter-O’Connell I., Peel M.T., Wykoff D.D., O’Shea E.K.. Genome-wide characterization of the phosphate starvation response in Schizosaccharomyces pombe. BMC Genomics. 2012; 13:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shah S., Wittmann S., Kilchert C., Vasiljeva L.. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 2014; 28:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee N.N., Chalamcharia V.R., Reyes-Turce F., Mehta S., Zofall M., Balachandran V., Dhakshnamoorthy J., Taneja N., Yamanaka S., Zhou M. et al.. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013; 155:1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ard R., Tong P., Allshire R.C.. Long non-coding RNA-mediate transcriptional interference of a permease gene confers drug tolerance in fission yeast. Nat. Commun. 2014; 5:5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwer B., Sanchez A.M., Shuman S.. RNA polymerase II CTD phospho-sites Ser5 and Ser7 govern phosphate homeostasis in fission yeast. RNA. 2015; 21:1770–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chatterjee D., Sanchez A.M., Goldgur Y., Shuman S., Schwer B.. Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho-sites govern the repression of pho1 gene expression under phosphate-replete conditions in fission yeast. RNA. 2016; 22:1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sanchez A.M., Shuman S., Schwer B.. Poly(A) site choice and Pol2 CTD Serine-5 status govern lncRNA control of phosphate-responsive tgp1 gene expression in fission yeast. RNA. 2018; 24:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garg A., Sanchez A.M., Shuman S., Schwer B.. A long noncoding (lnc) RNA governs expression of the phosphate transporter Pho84 in fission yeast and has cascading effects on the flanking prt lncRNA and pho1 genes. J. Biol. Chem. 2018; 293:4465–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwer B., Sanchez A.M., Garg A., Chatterjee D., Shuman S.. Defining the DNA binding site recognized by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis. mBio. 2017; 8:e01218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanchez A.M., Shuman S., Schwer B.. An RNA polymerase II CTD genetic interaction network with 3′-processing and termination factors in fission yeast and its impact on phosphate homeostasis. Proc. Natl Acad. Sci. U.S.A. 2018; 115:E10652–E10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwer B., Bitton D.A., Sanchez A.M., Bähler J., Shuman S.. Individual letters of the RNA polymerase II CTD code govern distinct gene expression programs in fission yeast. Proc. Natl Acad. Sci. U.S.A. 2014; 111:4185–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Azevedo C., Saiardi A.. Eukaryotic phosphate homeostasis: the inositol pyrophosphate perspective. Trends Biochem. Sci. 2017; 42:219–231. [DOI] [PubMed] [Google Scholar]
- 15. Henry T.C., Power J.E., Kerwin C.L., Mohammed A., Weissman J.S., Cameron D.M., Wykoff D.D.. Systematic screen of Schizosaccharomyces pombe deletion collection uncovers parallel evolution of the phosphate signal pathways in yeasts. Euk. Cell. 2011; 10:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Estill M., Kerwin-Iosue C.L., Wykoff D.D.. Dissection of the PHO pathway in Schizosaccharomyces pombe using epistasis and the alternative repressor adenine. Curr. Genet. 2015; 61:175–183. [DOI] [PubMed] [Google Scholar]
- 17. Pöhlmann J., Fleig U.. Asp1, a conserved 1/3 inositol polyphosphate kinase, regulates the dimorphic switch in Schizosaccharomyces pombe. Mol. Cell Biol. 2010; 30:4535–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Topolski B., Jakopec J., Künzel N.A., Fleig U.. Inositol pyrophoshate kinase Asp1 modulate chromosome segregation fidelity and spindle function in Schizosaccharomyces pombe. Mol. Cell Biol. 2016; 36:3128–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pascula-Ortiz M., Saiardi A., Walla E., Jakopec V., Künzel N.A., Span I., Vangala A., Fleig U.. Asp1 bifunctional activity modulates spindle function via controlling cellular inositol pyrophosphate levels in Schizosaccharomyces pombe. Mol. Cell Biol. 2018; 38:e00047-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H., Nair V.S., Holland A.A., Capolicchio S., Jessen H.J., Johnson M.K., Shears S.B.. Asp1 from Schizosaccharomyces pombe binds a [2Fe-2S]2+ cluster which inhibits inositol pyrophosphate 1-phosphatase activity. Biochemistry. 2015; 54:6462–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A. 3rd, Steever A.B., Wach A., Philippsen P., Pringle J.R.. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998; 14:943–951. [DOI] [PubMed] [Google Scholar]
- 22. Hentges P., Van Driessche B., Tafforeau L., Vandenhaute J., Carr A.M.. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005; 22:1013–1019. [DOI] [PubMed] [Google Scholar]
- 23. Herrick D., Parker R., Jacobson A.. Identification and comparison of stable and unstable RNAs in Saccharomyces cerevisiae. Mol. Cell Biol. 1990; 10:2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwer B., Mao X., Shuman S.. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 1998; 26:2050–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pyl P.T., Anders S., Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Safrany S.T., Ingram S.W., Cartwright J.L., Falck J.R., McLennan A.G., Barnes L.D., Shears S.B.. The diadenosine hexaphosphate hydrolase from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human disphoshoinositol polyphosphate phosphohydrolase: overlapping substrate specificities in a MutT-type protein. J. Biol. Chem. 1999; 274:21735–21740. [DOI] [PubMed] [Google Scholar]
- 30. Ingraham S.W., Safrany S.T., Barnes L.D.. Disruption and overexpression of the Schizosaccharomyces pombe aps1 gene, and effect on growth rate, morphology and intracellular diadenosine 5′,5‴-P1,P5-pentaphosphate and diphosphoinositol polyphosphate concentrations. Biochem. J. 2003; 369:519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohtsuka H., Ogawa Y., Mizuno H., Mita S., Aiba H.. Identification of Ecl family genes that extend chronological lifespan in fission yeast. Biosci. Biotechnol. Biochem. 2009; 73:885–889. [DOI] [PubMed] [Google Scholar]
- 32. Matsuzawa K.T., Yoritsune K., Takegawa K.. MADS box transcription factor Mbx2/Pvg4 regulates invasive growth and flocculation by inducing gsf2+ expression in fission yeast. Eukaryot. Cell. 2012; 11:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kwon E.J., Laderoute A., Chatfield-Reed K., Vachon L., Karagiannis J., Chua G.. Deciphering the transcriptional-regulatory network of flocculation in Schizosaccharomyces pombe. PLos Genet. 2012; 8:e1003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saiardi A. Cell signalling by inositol pyrophosphates. Subcell. Biochem. 2012; 59:413–443. [DOI] [PubMed] [Google Scholar]
- 35. Shears S.B. Intimate connections: inositol pyrophosphates at the interface of metabolic regulation and cell signaling. J. Cell Physiol. 2018; 233:1897–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee Y.S., Huang K., Quiocho F.A., O’Shea E.K.. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 2008; 4:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu M., Chong L.S., Perlman D.H., Resnick A.C., Fiedler D.. Inositol polyphosphates intersect with signaling and metabolic networks via two distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6757–E6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wild R., Gerasimaite R., Kung J.L., Truffault V., Pavlovic I., Schmidt A., Saiardi A., Hessen H.J., Poirer Y., Hothorn M. et al.. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science. 2016; 352:986–990. [DOI] [PubMed] [Google Scholar]
- 39. Jung J.Y., Ried M.K., Hothorn M., Poirer Y.. Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain. Curr. Opin. Biotechnol. 2018; 49:156–162. [DOI] [PubMed] [Google Scholar]
- 40. Ford M.G.J., Pearse B.M.F., Higgins M.K., Vallis Y., Owen D.J., Gibson A., Hopkins C.R., Evans P.R., McMahon H.T.. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001; 291:1051–1055. [DOI] [PubMed] [Google Scholar]
- 41. De Camilli P., Chen H., Hyman J., Panepucci E., Bateman A., Brunger A.T.. The ENTH domain. FEBS Lett. 2002; 513:11–18. [DOI] [PubMed] [Google Scholar]
- 42. Corden J.L. RNA polymerase II C-terminal domain: tethering transcription to transcript and template. Chem. Rev. 2013; 113:8423–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wittmann S., Renner M., Watts B.R., Adams O., Huyesin M., Baejen C., El Omari K., Kilchert C., Heo D.H., Kecman T. et al.. The conserved protein Seb1 drives transcription termination by binding RNA polymerase II and nascent RNA. Nat. Commun. 2017; 8:14861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data in this publication have been deposited in NCBI’s Gene Expression Omnibus. GSE127550, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127550, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131237.