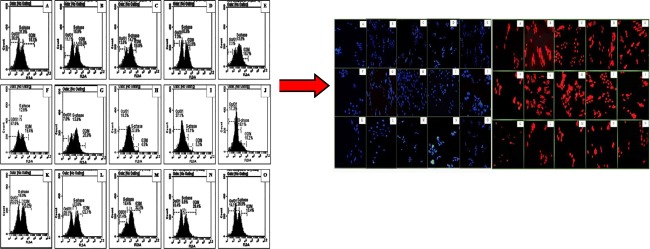
Abstract
Background
Among the different types of cancers, breast cancer, bone cancer and cervical cancer are the most common gender specific cancer types that are affecting the women worldwide. Currently, many enzymatic and cellular pathways are known as drug targets for the treatment of cancer. Even though many improvements have been made in the therapy of various types of cancer, but the major disadvantage of available anti-cancer drugs is their non-selective behavior towards cancer cells as well as normal cells.
Objectives
In the light of this fact, the searching of new compounds with selective behavior only towards cancer cells is critically important. Previously, we have identified several series of compounds as the potential inhibitors of these families.
Methods
Herein, we investigate quinolones and quinolines for their anti-cancer activity against breast cancer cells (MCF–7), bone marrow cancer cells (K–562) and cervical cancer cells (HeLa) by MTT assay. The most effective derivatives were further subjected to flow cytometry analysis followed by fluorescence microscopic analysis by using 4´,6-diamidine-2´-phenylindole (DAPI) and propidium staining (PI) staining.
Results
All the tested compounds were found selective only towards cancer cells. The identified compounds also induced either G2 or S-phase cell cycle arrest within the respective cancer cell line, chromatin condensation and the nuclear fragmentation, as well as maximum interaction with DNA.
Conclusions
These results provide evidence that the characteristic chemical features of attached groups are the key factors for their anticancer effects and play a useful role in revealing the mechanisms of action in relation to the known compounds in future research programs.
Graphical abstract.
Flow cytometric analysis of cell cycle using propidium iodide staining. Cell apoptosis observed under fluorescence microscope using DAPI and PI staining.
Keywords: Quinolones, Quinolines, Breast cancer cells (MCF–7), Bone marrow cells (K–562), Cervical cancer cells (HeLa), Cell-cycle
Introduction
Now a days, cancer is one of the most leading cause of death worldwide. It is characterized by disturbances and alterations in molecular events including uncontrolled cell proliferation [1], cellular transformation, improper regulation of cell cycle [2], angiogenesis and increased invasion ensuing metastases [3]. Moreover, a variety of factors are known to contribute towards cancer; many cellular/enzymatic pathways have been discovered as an important drug targets for the treatment of cancer [4]. Although many anticancer agents are known, but most of them are cytotoxic not only to the cancerous cells but also to the normal cells [5]. As a result, they cause severe side effects including nausea, hair loss, weight loss, fatigue, skin rashes and loss of appetite [6]. In the light of this fact, the search for novel chemotherapeutic agents is a motivating and continually sprouting field in cancer research. Still there is a need of developing more effective and less toxic agents that may lead as best anticancer effects with defined molecular targets.
In the course of identifying various chemical substances which may serve as leads for designing novel antitumor agents, many classes of organic compounds have been tested [7], with special attention being paid to quinolones and quinolines. These are the most common condensed heterocyclic aromatic compounds, containing nitrogen in their structure and are known for their diverse therapeutic potential [8]. Moreover, the quinolone and quinoline nucleus occurs in several natural compounds and pharmacologically active substances where they are responsible for broad spectrum of biological activity [9]. Previously we have identified several quinolones and quinolines derivatives; isoquinoline derivatives (4a-p), quinoline–4–carboxylic acid derivatives (3a-j) and 4–quinolone derivatives (2a-7d) as potent and selective inhibitors of alkaline phosphatases (specifically tissue non-specific alkaline phosphatase; TNAP) and NPPs (NPP1; ecto-nucleotide pyrophosphatases/phosphodiesterases1) with lower micromolar potency [10–12]. These members of ecto-nucleotidases possesses different enzymatic and cellular expression properties [13]. Moreover, they are involved in regulation of various signalling pathways especially purinergic signalling pathway though which they are responsible for the maintenance of nucleotide and nucleoside level [14]. The abberrant level of these ecto-nucleotidases is associated with various types of cancers, especially breast cancer as its associated malagnancies. Therefore, the inhibitors of these ecto-nucleotidases could play a promising role in the treatment of cancer [15].
Herein we performed extensive studies to investigate anticancer potential of quinolones and quinolines derivatives; isoquinoline derivatives (4a-p), quinoline–4–carboxylic acid derivatives (3a-j) and 4–quinolone derivatives (2a-7d) against different cancer cell lines i.e., breast cancer cells (MCF–7), bone-marrow cancer cells (K–562) and cervical cancer cells (HeLa) by using the flow cytometry, fluorescence microscopy and DNA-binding studies. The screening of compounds against healthy baby hamster kidney cells (BHK-21) was also performed. The results were found significant as they showed selective cytotoxicity against cancerous cells only. Moreover, the data obtained from both studies suggested that quinolone and quinoline derivatives, synthesized from different strategies could be beneficial for the treatment of various types of cancer. Further investigations may help researchers to synthesize such types of compounds with minimum toxicity.
Material and methods
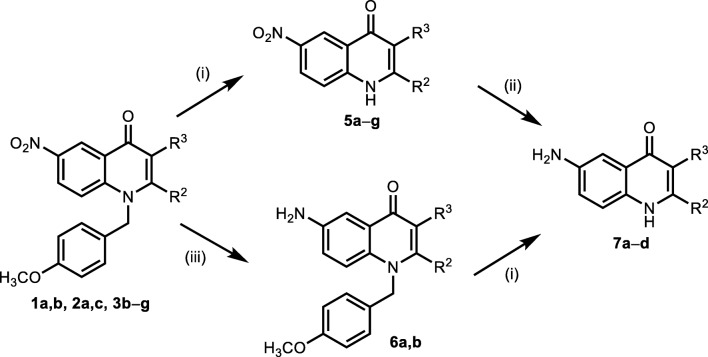
Synthesis of Isoquinoline, Quinoline–4–carboxylic acid and 4–quinolone compounds
The general procedure for the synthesis of isoquinoline derivatives (4a-p), quinoline–4–carboxylic acid derivatives (3a-j) and 4–quinolone derivatives (2a-7d) have been reported in our previous work [10–12], respectively.
Cell lines and cell cultures
Two cancer cell lines breast adenocarcinoma (MCF–7) cell line (ATCC® HTB–22™) human cervical cancer (HeLa) cell line (ATCC–USA) were kept in DMEM media [having heat–inactivated fetal bovine serum (10%), 100 U/mL penicillin and 100 μg/mL streptomycin] and bone marrow lymphoblast (K–562) cell line (ATCC® CCL–243™) was kept in RPMI–1640 [having heat–inactivated fetal bovine serum (15%), 100 U/mL penicillin and 100 μg/mL streptomycin] in T–75 cm2 sterile tissue culture flasks in a 5% CO2 incubator at 37 °C. For cell viability assay, these cell lines were grown in different 96–well plates by inoculating 2.5 × 103 cells/100 μL/well at 37 °C for 24 h in a CO2 (5%) incubator and then treated with selected compounds (10 μL). For cell cycle analysis and microscopic experiments, these cell lines (2.5 × 105 cells/mL) were incubated at 37 °C overnight in 6–well plate and then were treated with the most potent cytotoxic compounds (according to GI50 values) and positive control, i.e., carboplatin [16].
Anticancer assays
Cell viability assays (MTT assay)
The cytotoxic potentials of the test compounds were evaluated in human breast adenocarcinoma cells (MCF-7), human myelogenous leukemia cells (K-562), human cervical adenocarcinoma cells (HeLa) by MTT (Dimethyl–2–thiazolyl–2,5–diphenyl–2H–tetrazolium bromide)–based cell viability assay as described earlier [17–19]. The effect of these derivatives was also examined against normal cells i.e., baby hamster kidney cells (BHK–21). Briefly, the cells (2.5 × 104/mL) were cultured in a volume of 90 μL in each well of a 96-well flat-bottom culture plate and kept in 5% CO2 incubator at 37 °C. After an overnight incubation, the cells were treated with the test compounds (100 μM) and incubated for 24 h. The media was poured out and 10 μL MTT reagent (5 mg/mL) was added to each well to crystallize the viable cells. The plate was incubated for 4 h at 37 °C and 5% CO2, followed by the addition of 100 μL of the reagent (1:1 solution of 50% isopropanol and 10% sodium dodecyl sulfate). The plate was further incubated for 30 min at room temperature and the optical density was observed at 570 nm wavelength by subtracting the absorbance at 630 nm, using microplate reader (Bio-Tek ELx 800™, Winooski, USA). The percent growth inhibition values of the each compound was calculated in reference to the negative control (blank) with the mean of three independent values (± SEM). The experiment was carried out in triplicate and derivatives exhibited equal to or more than 50% inhibition were further evaluated for their growth inhibitory (GI50) values.
Cell cycle analysis assay
The treated cells were subjected to cell cycle analysis by fow cytometry (BD Accuri™, United States), using the method as reported earlier [19, 20]. Initially, the cells (25 × 104 cells/mL) were treated with the most potent derivatives obtained from MTT assay from the selected series and incubated over night at 37 °C. Then the pallets of cells were attained by centrifugation at 4000 g (for 5 min) and after that the pallet was resuspended in 3% FBS solution (200 μL) that contained 5 μL of propidium iodide (20 μg/mL), 0.1% (v/v) Triton X-100 and ribonuclease A (10 μL, 10 mg/mL). The samples were analyzed by BD accuri flow cytometry as described earlier [21].
Microscopic analysis of apoptosis
The microscopic analysis of most potent derivatives was carried out to support the flow cytometry results as discussed previously [21, 22]. The confluent cells (2.5 × 105 cells/well) were treated with the compound and kept in incubator (5% CO2) at 37 °C. After 24 h, culture medium was removed and cells were washed thrice with cold PBS. The cells were further treated with 4% formalin and 0.1% Triton X–100. Finally, after 5 min incubation at room temperature, 10 μL (0.1 mg/mL) of 4′,6-diamidino-2-phenylindole (DAPI) or propidium iodide (PI) dye was mixed to stain the nuclear material and images were captured by using the fluorescence microscope ((Nikon ECLIPSE Ni–U), Japan) at excitation/emission wavelength of 350/460 nm and 493/632 for DAPI and PI, respectively.
DNA interaction and docking studies
To determine the mechanism of inhibition of the most potent compounds as well as to support the above results, the DNA interaction studies were performed according to the previously reported method [21, 23]. Briefly, the concentration of mammalian DNA was estimated at 260 nm wavelength using FLUOstar Omega (BMG Labtech, Germany). The test compound (100 μM) was treated with different concentrations of DNA from 0 μM to 340 μM, whereas the same concentrations of DNA were used in respective reference solutions. After an incubation for 10 min, the absorption spectra were recorded in 96 well plates with the path length of 5.5 mm.
The DNA docking studies were performed by Molecular Operating Environment (MOE, version 2014) software [24] as reported by our group, previously [21]. Firstly, the binding pockets in the DNA (minor and major groves) and the intercalating sites in the DNA were determined by the known compounds. Then, the 3D structures of compounds and 3D structure of DNA were further processed as discussed previously. A total 100 poses for each compound were generated to get accurate binding interactions. The best pose was then selected and visualized through Discovery Studio [25].
Results
Chemistry
Synthesis of Isoquinoline derivatives
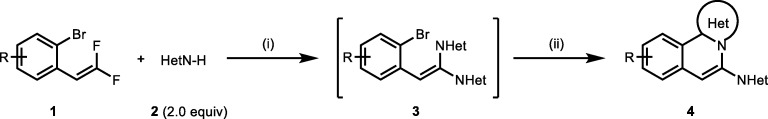
Isoquinoline ring system was synthesized in one–pot reaction by initially forming C–N bonds using catalyst–free defluorination of 1–bromo–2–(2,2–difluorovinyl)benzenes 1 in reaction with various N–H heterocycles 2. The α,α–dihetaryl substituted alkene intermediate 3 was then subjected to Pd(OAC)2 catalyzed intramolecular C2 arylation to give nitrogen–fused isoquinoline derivatives 4 as given in Scheme 1 [10].
Scheme 1.
One–pot two–step synthesis of N–fused isoquinolines. (Reaction conditions: 1) difluoroalkene1, heterocycle 2 (2 equiv.), K3PO4 (4 equiv.), in DMF (5 mL), 100 °C, 12 h; 2) Pd(OAc)2 (5 mol%), PPh3 (10 mol%), 140 °C) [10]
Synthesis of Quinoline–4–carboxylic Acid derivatives
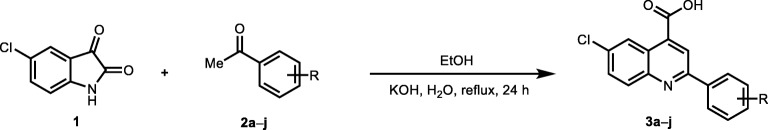
Quinoline–4–carboxylic acid derivatives (3a–j) were synthesized by the reaction of 5–chloro–isatin 1 with corresponding aryl substituted acetophenones (2a–j) in the presence of potassium hydroxide followed by acidification as given in Scheme 2 [11].
Scheme 2.
Synthesis of quinoline–4–carboxylic acids (3a–j) (Reaction conditions: 1) 5-chloro-isatin (1.0 mmol), corresponding aryl substituted acetophenones (1.1 mmol), KOH (3.0 mmol), in ethanol (5 mL), mixture was refluxed for 24 h, acidified with 2 M aqueous hydrochloric acid, recrystallized from ethanol to get compunds (3a-j) [11]
Synthesis of 4–Quinolone derivatives
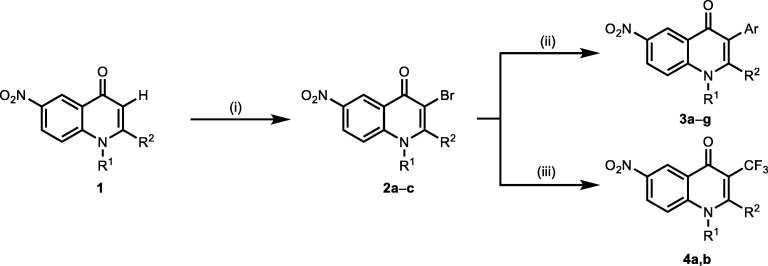
The synthesis pathway of 4-quinolones is depicted in Schemes 3 and 4. Nucleophilicity of the unsubstituted C–3 atom in a quinolone molecule 1 gave an option for an expanding of the molecule complexity. This could be demonstrated by a good reactivity with electrophilic agents. For example, utility of the N–bromsuccinimide resulted in products 2 brominated at their C–3 position. In this manner we obtained 3–bromoquinolones 2 as a platform for further functionalization (Schemes 3 & 4) [12].
Scheme 3.
Modification strategies at the C–3 position in the 4–quinolinones. (Reagents and conditions: (i) 1.45 equiv. of NBS, CH3COOH, 20 °C, 1.5 h; (ii) 1.2 equiv. of aryl boronic acid, 0.1 equiv. of Pd(PPh3)4 10 equiv. K2CO3 in 5.5 mL of toluene with 1 mL of H2O and 1.5 mL of MeOH at 90 °C for 4 h; (iii) CF3COONa 4 equiv., CuI 8 equiv., DMA, 120 °C 6 h) [12]
Scheme 4.
Functionalization of 2, 3 and 4 derivatives. (Reagents and conditions: (i) CF3COOH, reflux 2–10 h; (ii) Methanol: AcOH 1:1, 0.1 equiv. Pd/C (10%), H2, 2–3 h; (iii) Methanol, 0.1 equiv. Pd/C (10%), H2, 5 h) [12]
Biological results
Cytotoxic potential of Compounds by MTT assay
Isoquinoline derivatives
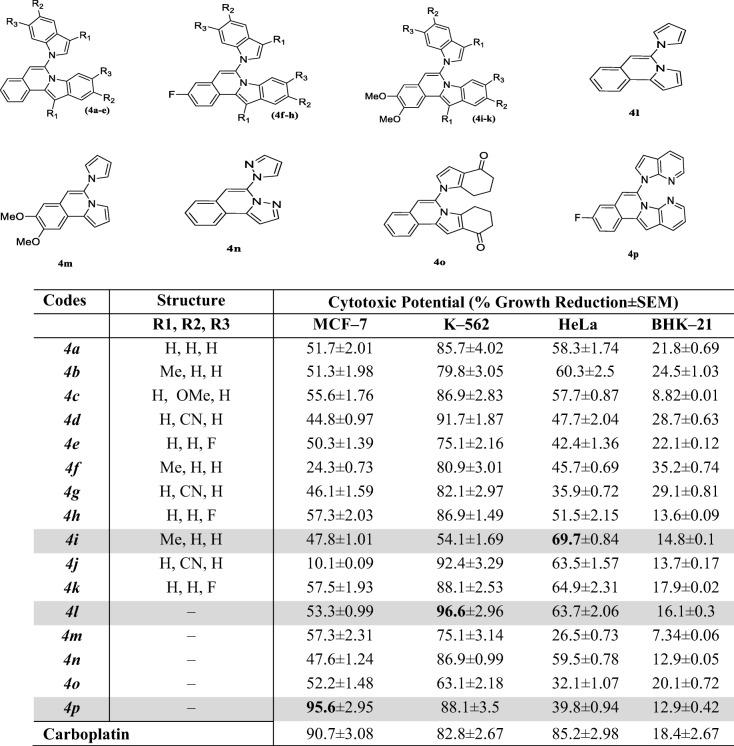
The cytotoxic potential of different isoquinoline derivatives (4a-p) is illustrated in Table 1 as a percent growth reduction. The study was carried out by using three cancer cell lines (HeLa, MCF–7 and K–562) and one normal cell line (BHK–21). The cell viability was evaluated by treating the different cells with 100 μM of each derivative for 24 h. In addition, it was found from the results that these derivatives remained inactive against normal cell line and possessed promising anticancer effects (Table 1).
Table 1.
Cytotoxic potential of isoquinoline derivatives (4a-p) against cancerous and normal cell lines
The cytotoxic potential of tested compounds was measured at the final concentration of 100 μM. Results represented here as the mean (S.E.M) of three independent determinations
The potent derivatives were further evaluated for the determination of growth inhibitory values (GI50) values towards MCF–7, K–562 and HeLa cells, respectively (Table 2).
Table 2.
Growth inhibitory values GI50 ± SEM (μM) of compounds 4i, 4 l and 4p against respective cell lines
| Code | MCF–7 | K–562 | HeLa |
|---|---|---|---|
| GI50 ± SEM (μM) | |||
| 4i | >100 | 17.1 ± 2.66 | 1.43 ± 0.26 |
| 4 l | 7.25 ± 0.86 | 10.9 ± 1.04 | 7.19 ± 0.78 |
| 4p | 0.49 ± 0.02 | 13.6 ± 1.78 | >100 |
| Carboplatin | 3.91 ± 0.32 | 4.11 ± 0.78 | 5.13 ± 0.45 |
GI50 denotes compound concentrations that result in a 50% decrease in the cell number compared to non–treated controls and were derived after 24 h treatment
The bold entries in the Table represent the GI50 ± SEM (μM) for the potent compounds among the series against each cell line
Quinoline–4–carboxylic derivatives
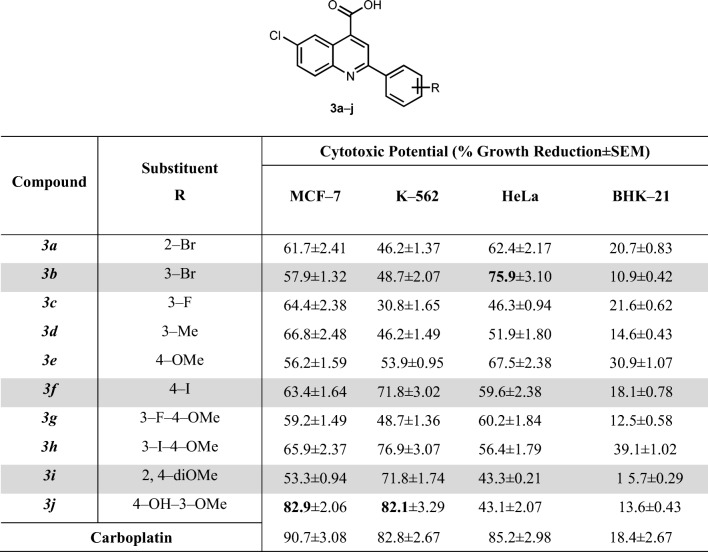
The cytotoxic potential of Quinoline–4–carboxylic acid derivatives (3a-j) are depicted in Table 3.
Table 3.
Cytotoxic potential of quinoline-4-carboxylic acid derivatives (3a-j) against cancerous and normal cell lines
The cytotoxic potential of tested compounds was measured at the final concentration of 100 μM. Results represented here as the mean (SEM) of three independent determinations
The growth inhibitory concentrations (GI50) of the most potent derivatives were further evaluated in the respective cell lines that are given in Table 4.
Table 4.
Growth inhibitory values GI50 ± SEM (μM) for compounds 3b and 3j against respective cell lines
| Code | MCF–7 | K–562 | HeLa |
|---|---|---|---|
| GI50 ± SEM (μM) | |||
| 3b | 4.11 ± 0.99 | >100 | 1.15 ± 0.65 |
| 3j | 0.54 ± 0.05 | 25.8 ± 0.79 | >100 |
| Carboplatin | 3.91 ± 0.32 | 4.11 ± 0.78 | 5.13 ± 0.45 |
GI50 denotes compound concentrations that result in a 50% decrease in the cell number compared to non–treated controls and were derived after 24 h treatment
The bold entries in the Table represent the GI50 ± SEM (μM) for the potent compounds among the series against each cell line
4–quinolone derivatives
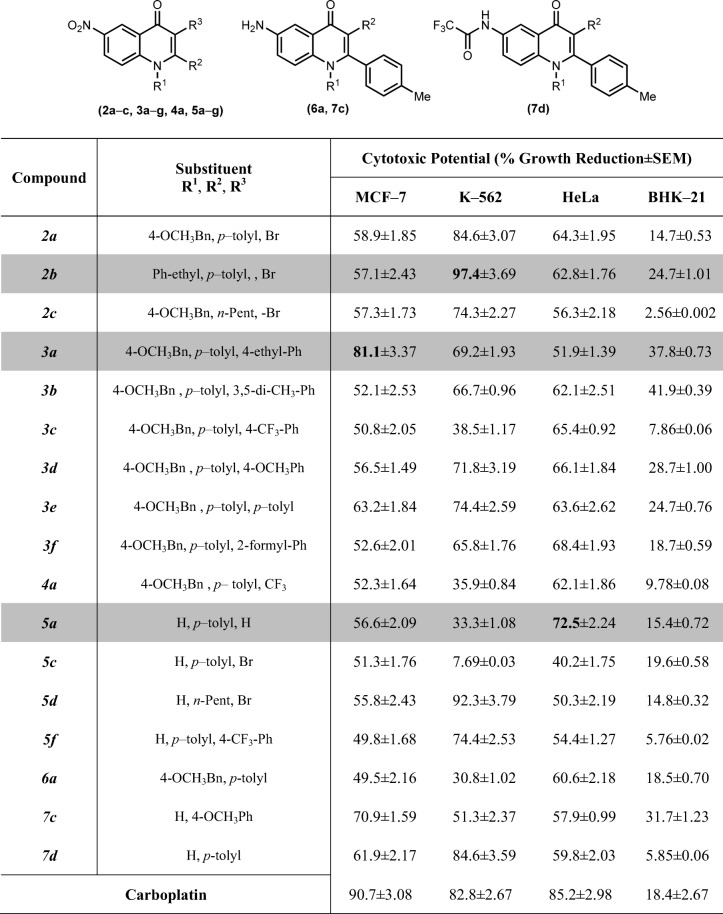
The 4–quinolone derivatives (2a-c, 3a-g, 4a, 5a-g, 6a, 7c-d) exhibited moderate to excellent cytotoxic behavior against different cell lines as compared to normal BHK–21 cells, as depicted in Table 5.
Table 5.
Cytotoxic potential of 4-quinolone derivatives (2a-c, 3a-g, 4a, 5a-g, 6a, 7c,d) against cancerous and normal cell lines
The cytotoxic potential of tested compounds was measured at the final concentration of 100 μM. Results represented here as the mean (SEM) of three independent determinations
The most potent compounds were further evaluated for the determination of GI50 values against MCF–7, K–562 and HeLa cells (Table 6).
Table 6.
Growth inhibitory values GI50 ± SEM (μM) for compounds 2b, 3a and 5a against respective cell lines
| Code | MCF–7 | K–562 | HeLa |
|---|---|---|---|
| GI50 ± SEM (μM) | |||
| 2b | 6.22 ± 0.77 | 7.91 ± 0.62 | 7.65 ± 0.97 |
| 3a | 0.61 ± 0.02 | 13.5 ± 1.67 | 9.88 ± 1.23 |
| 5a | 6.45 ± 0.87 | >100 | 2.02 ± 0.11 |
| Carboplatin | 3.91 ± 0.32 | 4.11 ± 0.78 | 5.13 ± 0.45 |
GI50 denotes compound concentrations that result in a 50% decrease in the cell number compared to non–treated controls and were derived after 24 h treatment
The bold entries in the Table represent the GI50 ± SEM (μM) for the potent compounds among the series against each cell line
Cell cycle analysis and detection of apoptosis by flow cytometry
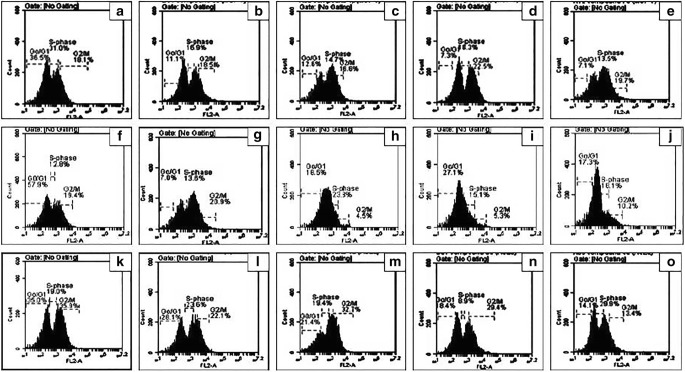
The most effective cytotoxic derivatives from quinolones and quinoline series were selected for flow cytometry analysis to further investigate their effect on cell cycle progression and apoptosis in the three mentioned cell lines i.e., HeLa, K-562 and MCF–7. Figs. 1 and 2 showed the DNA content within the dividing cells as estimated by propidium iodide staining, whereas the percent cell population within the cell cycle phases i.e., Go – G1, S, and G2/ M phases.
Fig. 1.
Flow cytometric analysis of cell cycle using propidium iodide (PI) staining. (a) untreated MCF–7 cells (b-e) MCF–7 cells treated with carboplatin, 4p, 3j and 3a, respectively. (f) untreated K–562 cells (g-j) K–562 cells treated with carboplatin,4 l, 3j and 2b, respectively. (k) untreated HeLa cells (l-o) HeLa cells treated with carboplatin,4i, 3b and 5a, respectively
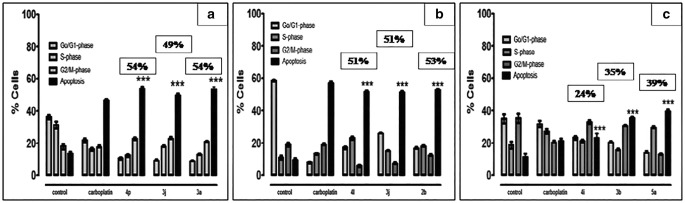
Fig. 2.
Cell cycle profile by the most potent derivatives against respective cell lines (a) MCF–7 cells treated with compound 4p, 3j and 3a(b) K–562 cells treated with compound 4 l, 3j and 2b(c) HeLa cells treated with compound 4i, 3b and 5a. Data were analyzed by one way analysis of variance (ANOVA) using Graphpad PRISM® 5 (GraphPad, San Diego, California, USA). The mean of three experiments ± S.D. is shown and statistical differences between the groups was found significant ***p < 0.0001
Microscopic analysis (using DAPI & PI)
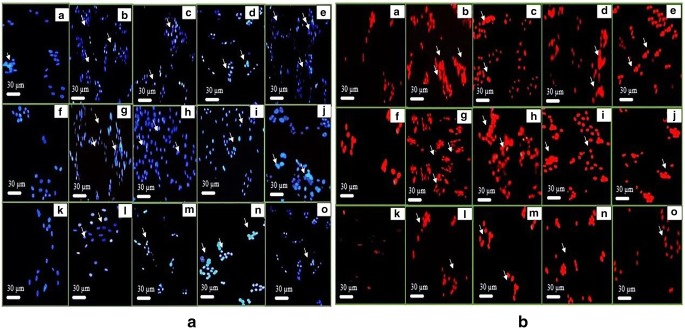
MCF-7, K-562 and HeLa cells were treated for with their respective potent derivatives from all selected series as mentioned above. The images were captured after 24 h, illustrating the cell death mechanism. DAPI and its counter stained PI were used for staining the nuclear material (Fig. 3).
Fig. 3.
Cell apoptosis observed under fluorescence microscope using DAPI (a) and PI (b) staining respectively. In both (a) and (b), (A) untreated MCF–7 cells (B-E) MCF–7 cells treated with carboplatin, 4p 3j and 3a, respectively. (F) untreated K–562 cells (G-J) K–562 cells treated with carboplatin,4 l, 3j and 2b, respectively. (K) untreated HeLa cells (L-O) HeLa cells treated with carboplatin,4i, 3b and 5a, respectively
DNA interaction studies
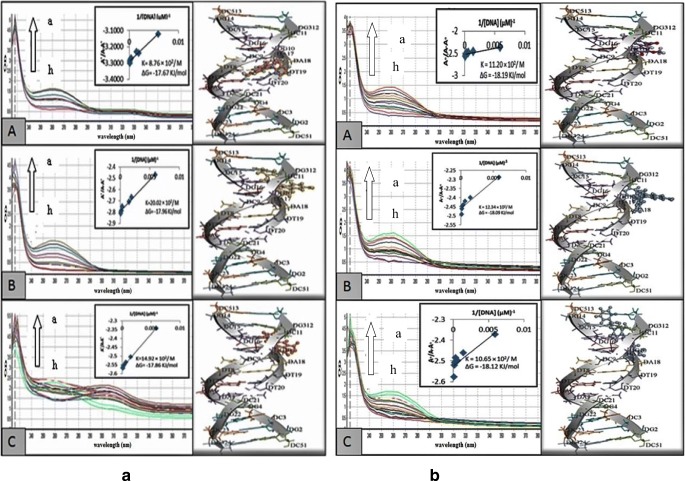
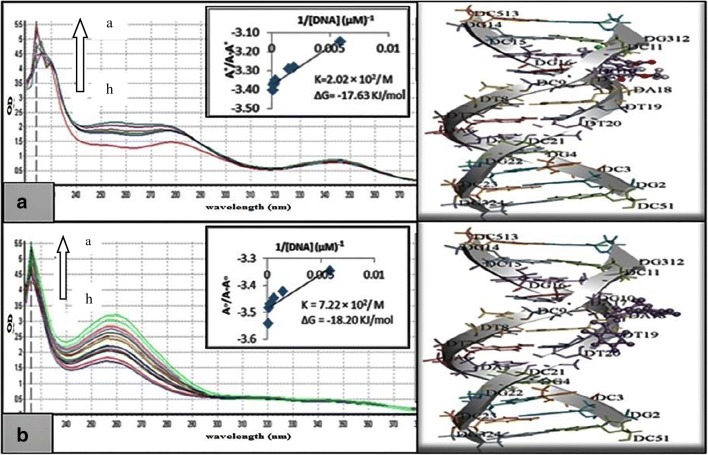
The most potent derivatives against MCF–7 cells (4p,3j and 3a), K–562 cells (4 l,3j and 2b) and HeLa cells (4i,3b and 5a) were further evaluated to determine their mechanism of action by interacting with HS–DNA in UV–visible range (Figs. 4 and 5).
Fig. 4.
Absorption spectra of 40 μM of (a) isoquinoline (4p(A),4 l(B) and 4i(C)) and (b) 4–quinolone derivatives (3a(A),2b(B) and5a(C)) in absence and presence of increasing concentration of DNA (indicated with arrow). The graph within the figure is the plot of A°/ (A–A°) vs 1/[DNA] plot for the determination of Gibb’s free energy as well as the binding constant for the tested derivatives
Fig. 5.
Absorption spectra of 40 μM of quinoline–4–carboxylic acid derivatives (3j (A) and 3b (B)) in absence and presence of in absence and presence of increasing concentration of DNA (indicated with arrow). The graph within the figure is the plot of A°/ (A–A°) vs 1/[DNA] plot for the determination of Gibb’s free energy as well as the binding constant for the tested derivatives
Discussion
The isoquinoline ring system is highly active pharmacophore and is responsible for various pharmaccological activities. The were synthesized in one-pot reaction by N-vinylation and Pd-catalysed C-H arylation reactions. Different N-H heterocycles were used and different functional groups were allowed, which resulted in wide range of products (4a-p) as previously reported earlier by us [10]. The quinoline–4–carboxylic derivatives (3a–3j) were synthesized by the reaction of 5-chloro-isatin 1 with corresponding aryl substituted acetophenones in the presence of potassium hydroxide followed by acidification as reported earlier by our group [11]. The 4-quinolone derivatives were synthesized via sequential derivatization methods in order to get C-1, C-3 and C-6 substituted 4-quinolone (2a-c, 3a-g, 4a, 5a-g, 6a, 7c-d) as reported earlier by us [12].
Biological results
Cytotoxic potential by MTT assay
The derivatives from three different series showed different behavior as discussed below:
Isoquinoline derivatives
The cytotoxic potential of different isoquinoline derivatives (4a-p) is illustrated in Table 1 as a percent growth reduction. Among all derivatives, derivatives 6-(4-Oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)-9,10-dihydroindolo[2,1-a]isoquinolin-11(8H)-one (4p), 9-Fluoro-6-(6-fluoro-1H-indol-1-yl)-[1,3]dioxolo[4,5-g]indolo[2,1- a]isoquinoline (4 l) and 2,3-Dimethoxy-12-methyl-6-(3-methyl-1H-indol-1-yl)indolo[2,1-a]isoquinoline (4i), induced an approximately 95%, 97% and 70% inhibition in the growth of breast cancer cells (MCF–7), bone–marrow cancer cell line (K–562) and human cervical cancer cell line (HeLa) cells, respectively. All the derivatives were found to inhibit the only cencerous cell lines selectively. None of the derivative exhibited cytotoxic effect towards the normal cell line. Different functional groups attached to the ring are responsible for such variable effect on cell lines. Moreover, it can be suggested from the detailed structure–activity relationship of derivative 9-Fluoro-6-(6-fluoro-1H-indol-1-yl)-[1,3]dioxolo[4,5-g]indolo[2,1-a]isoquinoline (4 l) and 6-(4-Oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)-9,10-dihydroindolo[2,1-a]isoquinolin-11(8H)-one (4p), that the presence of pyrol, pyrolo–pyridine rings as well as fluorine substitution in case of 6-(4-Oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)-9,10-dihydroindolo[2,1-a]isoquinolin-11(8H)-one (4p) might be responsible for this maximum anticancer potential towards MCF–7 and K–562 cell lines, respectively. Another quinolone derivative i.e., 2,3-Dimethoxy-12-methyl-6-(3-methyl-1H-indol-1-yl)indolo[2,1-a]isoquinoline (4i), showed maximum inhibitory response towards HeLa cell which is in accordance with its significant inhibitory effects on h–NPP1 [10]. The potent derivatives were further evaluated for the determination of growth inhibitory values (GI50) values towards MCF–7, K–562 and HeLa cells, respectively (Table 2).
Quinoline–4–carboxylic acid derivatives
The results showed (Table 3) that these derivatives possessed variable degree of inhibition against the different cell lines (HeLa, MCF–7, K–562) as compared to normal cells. These derivatives were also found selective only towards cancer cell lines. Considering the breast cancer cells (MCF–7), the compound 6-Chloro-2-(4-hydroxy-3-methoxyphenyl)quinoline-4-carboxylic acid (3j) displayed 82.9% reduction in cellular growth under favorable conditions. These results here support the enzymatic inhibitory potential (alkaline phosphatase) of same compound i.e., 6-Chloro-2-(4-hydroxy-3-methoxyphenyl)quinoline-4-carboxylic acid (3j) which has been discussed in our previous data [11]. The potent inhibitor of h–TNAP, displayed the maximum growth reduction of breast cancer cell line (MCF–7). The detailed study of this derivative structure suggested that the maximum activity might be due to the presence of electron donating methyl group at position 3 (Tables 3 and 4). Against MCF–7 cells, the other derivatives also exhibited significant growth inhibition, suggesting the effectiveness of these derivatives against breast cancer.
These derivatives showed moderate sensitivity towards the bone marrow lymphoblast K–562 cells and HeLa cells by exhibiting % growth reduction values in range of 53% to 76% and 51% to 68%, respectively. Five derivatives (3a, 3b, 3c, 3d, 3 g) remained inactive against K–562 while two derivatives 2-(3-Bromophenyl)-6-chloroquinoline-4-carboxylic acid (3b) and 6-Chloro-2-(2,4-dimethoxyphenyl)quinoline-4-carboxylic acid (3i) showed minimum response against HeLa cells.
4–quinolone derivatives
The 4–quinolone derivatives (2a-7d) exhibited moderate to excellent cytotoxic behavior against different cell lines, as depicted in Table 5. Overall, many derivatives were found selective inhibitor of K–562 as compared to MCF–7 cells. The compound 3-(4-Ethylphenyl)-1-(4-methoxybenzyl)-6-nitro-2-p-tolylquinolin-4(1H)-one (3a) exhibited maximum inhibitory activity against MCF–7 cells while other compounds 3-Bromo-6-nitro-1-(1-phenylethyl)-2-p-tolylquinolin-4(1H)-one (2b) and 6-Nitro-2-p-tolylquinolin-4(1H)-one (5a) were more active against K–562 and HeLa cells, respectively. Additionally, the growth inhibitory activity of these compounds against cancer cells also showed that these compounds are more sensitive towards cancer cells and are less sensitive towards normal cells. From Table 5, it was found that the cytotoxicity effect of these derivatives against MCF–7 was moderate except 3-(4-Ethylphenyl)-1-(4-methoxybenzyl)-6-nitro-2-p-tolylquinolin-4(1H)-one (3a). The structure activity relationship (SAR) of these derivatives clearly demonstrated the effect of different functional groups at R1, R2 and R3 position affects the cytotoxic potential.
Flow cytometric analysis of cell cycle
The most potent derivatives from quinolones and quinoline series displayed promising cytotoxic behavior on the MCF–7 cells, suggesting that these derivatives have much potential to inhibit growth of breast cancer cells. Briefly, compound 3-(4-ethylphenyl)-1-(4-methoxybenzyl)-6-nitro-2-p-tolylquinolin-4(1H)-one (3a) from 4–quinolone showed maximum apoptosis (54.4%) in comparison to 6-(4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)-9,10-dihydroindolo[2,1-a]isoquinolin-11(8H)-one (4p) from isoquinoline and 6-chloro-2-(4-hydroxy-3-methoxyphenyl)quinoline-4-carboxylic acid (3j) from quinoline–4–carboxylic acid. Both derivatives induced maximum apoptotic potential such as 51.4% and 51.9%, respectively, within the respective series and resulted in equipotent apoptosis as that of positive control i.e., carboplatin (50.1%). The cell cycle analysis suggested the possible interference and inhibition of mitotic spindling that finally resulted in DNA disruption and cell arrest in G2 phase. Therefore, these derivatives resulted in amassing of cells at G2 phase. Among these potent derivatives (3a, 4p, 3j), the compound 3-(4-ethylphenyl)-1-(4-methoxybenzyl)-6-nitro-2-p-tolylquinolin-4(1H)-one (3a) from 4–quinolone and 6-chloro-2-(4-hydroxy-3-methoxyphenyl)quinoline-4-carboxylic acid (3j) from quinoline–4–carboxylic acid depicted the higher apoptotic cell death i.e., 19.7% and 22.5%, respectively, within G2 phase. Fig. 1 showed the percentage apoptosis within different cell cycle phases i.e., Go– G1, S, and G2/ M, of these potent derivatives (4p, 3jand3a). When the effect was observed against K–562 cells, the potent derivatives from each series (4 l, 3j, 2b) exhibited promising and comparable apoptosis as that of positive control, carboplatin.
Against K–562 cells, the compound 9-fluoro-6-(6-fluoro-1H-indol-1-yl)-[1,3]dioxolo[4,5-g]indolo[2,1-a]isoquinoline (4 l) from isoquinolines, 6-chloro-2-(4-hydroxy-3-methoxyphenyl)quinoline-4-carboxylic acid (3j) from quinoline–4–carboxylic acid and 3-bromo-6-nitro-1-(1-phenylethyl)-2-p-tolylquinolin-4(1H)-one (2b) from 4–quinolone exhibited percentage apoptotic value i.e., 54.3%, 51.1% and 57.9%, respectively. The positive control, carboplatin induced 58.1% apoptosis against K–562 cells. Compound compound 9-fluoro-6-(6-fluoro-1H-indol-1-yl)-[1,3]dioxolo[4,5-g]indolo[2,1-a]isoquinoline (4 l) from isoquinolines exhibited higher S phase apoptotic cell death (approx. 24%) among all derivatives but its overall apoptosis (54.3%) is slightly less than other derivative 3-bromo-6-nitro-1-(1-phenylethyl)-2-p-tolylquinolin-4(1H)-one (2b) from 4–quinolone. In S phase, these derivatives intercalated with DNA molecule and undergone programmed cell death (apoptosis). The cell cycle analysis suggested that these derivatives are every effective with respect to S phase and ultimately more cell death occurred in this phase as compared to other phases (Figs. 1 and 2).
While using the HeLa cells, among 4–quinolones derivatives, the compound 6-nitro-2-p-tolylquinolin-4(1H)-one (5a) showed maximum apoptotic percentage (41.8%). While, 2-(3-bromophenyl)-6-chloroquinoline-4-carboxylic acid;3b (35.1%) and 2,3-dimethoxy-12-methyl-6-(3-methyl-1H-indol-1-yl)indolo[2,1-a]isoquinoline;4i (22.8%) of quinoline–4–carboxylic acid and isoquinoline derivatives, respectively, caused maximum apoptotic potential within their series. These potent derivatives intercalated with DNA molecule during S phase and resulted in programmed cell death (apoptosis). The compound 5a resulted in significant S phase apoptotic cells arrest as well as their accumulation as compared to other derivatives 2-(3-bromophenyl)-6-chloroquinoline-4-carboxylic acid (3b) from quinoline–4–carboxylic acid and 2,3-dimethoxy-12-methyl-6-(3-methyl-1H-indol-1-yl)indolo[2,1-a]isoquinoline (4i) from isoquinoline. Figure 2 represented the percentage cell apoptosis of potent compounds 2,3-dimethoxy-12-methyl-6-(3-methyl-1H-indol-1-yl)indolo[2,1-a]isoquinoline (4i) from isoquinoline, 2-(3-bromophenyl)-6-chloroquinoline-4-carboxylic acid (3b) and compound 6-nitro-2-p-tolylquinolin-4(1H)-one (5a) during different cell cycle phase (Go– G1, S, and G2/ M). Hence, the apoptotic cell death with respect to the total cell count indicated the compound 6-nitro-2-p-tolylquinolin-4(1H)-one (5a) from 4–quinolones as most potent derivative possessing higher pro–apoptotic activity than 2,3-dimethoxy-12-methyl-6-(3-methyl-1H-indol-1-yl)indolo[2,1-a]isoquinoline (4i) from isoquinoline and 2-(3-bromophenyl)-6-chloroquinoline-4-carboxylic acid (3b) from quinoline–4–carboxylic acid, which are in relation with the results obtained from MTT assay, discussed earlier.
The scattering of cell cycle phases as Go – G1, S, and G2 - M in the MCF-7 cells by carboplatin (11.1%, 16.9% and 18.5%) and compounds 4p, 3j and 3a indicated prominent increase in DNA content in G2 - M phase and resultant decrease in G0 - G1 phase as compared to the untreated cells. However, in K-562 cells, most prominent changes in Go – G1, S and G2 - M phases were shown by carboplatin (7.0%, 13.6% and 20.9%). Furthermore, the cell cycle progression of the HeLa cells (when treated with carboplatin (28.1%, 23.6% and 22.1%) and compounds 4i,3b and 5a) was particularly promoted the transition of G1/S phase and entry to the S phase (Fig. 1) and the distributions of DNA content was observed.
Microscopic analysis (using DAPI & PI)
The findings from the morphological analysis were in close agreement with the anti-proliferative assay as well as flow cytometry results. After 24 h of incubation, the images under fluorescent microscope exhibited the apoptotic changes within the cells like fragmented and condensed nuclei. In apoptotic cells, there is shrinkage and fragmentations of nuclei occur whereas in viable cells, the nuclear material is regular, oval shape with homogeneous chromatin. The images containing cells treated with DAPI staining (Fig. 3a) revealed bluish intact nuclei in the control whereas bright fragmented nuclei in treated cells with different derivatives (4p, 3j, 3a, 4 l, 2b, 3b and 5a). Similarly, PI treated viable cells revealed nuclei without staining because PI is impermeable to nuclear membrane. However, in case of dead cells, the compromised nuclear membrane allows the permeability of PI with resultant image of red nuclei in front of black background (Fig. 3b). The potent derivatives (3a,2b and 5a) from 4–quinolone series resulted in higher apoptosis in their respective cell lines, as represented by DAPI and PI stained images, with relation to positive control, carboplatin.
DNA interaction studies
It was found from Figs. 4 and 5 that with the increase in HS–DNA concentration there is increase in absorbance with resultant no shift of band positions. Hence this hyperchromic effect exhibited the non–covalent interaction of various derivatives with HS–DNA. Against MCF–7 cells, the derivative 3a showed maximum inhibitory activity along with higher DNA interactions having Gibbs free energy Δ–18.19 KJ/mol. Moreover, compound 3-bromo-6-nitro-1-(1-phenylethyl)-2-p-tolylquinolin-4(1H)-one (2b) from 4–quinolone exhibited higher inhibitory potential as compared to other derivatives (9-fluoro-6-(6-fluoro-1H-indol-1-yl)-[1,3]dioxolo[4,5-g]indolo[2,1-a]isoquinoline (4 l) from isoquinolines, 6-chloro-2-(4-hydroxy-3-methoxyphenyl)quinoline-4-carboxylic acid (3j)) against K–562 with Gibbs free energy Δ–18.09 KJ/mol. Among the potent derivatives of HeLa cells (4i, 3b and 5a), 2-(3-bromophenyl)-6-chloroquinoline-4-carboxylic acid (3b) resulted in maximum interactions with HS-DNA with Gibbs free energy Δ–18.20 KJ/mol. Except the potent derivatives, other derivatives were also studied for DNA binding interaction and concluded that few of them resulted in higher interaction. From the docking studies, the results were in agreement with the experimental data. Furthermore, it was evident that the potent compounds from all three series indicated strong hydrogen bond interactions with the minor groove of DNA (Figs. 4 and 5).
It was revealed from the above analysis that few compounds from isoquinolines, quinoline–4–carboxylic acid and 4–quinolone, which were found potent and selective inhibitors of APs and NPPs [10–12], are found here with strong anticancer potential. The cytotoxic results obtained from MTT assay confirmed that these compounds are selective towards cancerous cells only. Anitcancer mechanisms; cell cycle arrest and apoptosis induced by the identified compounds were confirmed by flow cytometry using PI staining and by fluorescence microscopy using two nucleus staining dyes, i.e., DAPI and PI. Furthermore, the mechanism of cytotoxic compound was further determined by DNA interaction studies and it was found that the most potent inhibitors exhibited the non–covalent mode of interaction with the herring sperm –DNA.
Conclusion
In continuation of more biological investigations we have evaluated the anticancer activities of previously reported ecto-nucleotidase (alkaline phosphatase, nucleotide pyrophosphatase/phosphodiesterase) inhibitors against different cancer cell lines. Herein, the reported results have shown that most of the derivatives from both series have promising anticancer potential against used cell lines i.e., MCF–7, K–562 and HeLa. Moreover, the identified derivatives induced promising cell apoptosis in the S phase of cell cycle through DNA intercalation. Therefore, it can be suggested that these compounds may have considerable importance in future research suggesting further testing of these compounds for their anti-cancer effect in advanced animal models.
Acknowledgements
J.I. is thankful to the Higher Education Commission of Pakistan for the financial support through Project No.Ph-V-MG-3/Peridot/R&D/HEC/2019 and 6927/NRPU/R&D/17.
Authors’ contribution
Syeda Abida Ejaz performed the biochemical assays and drafted the manuscript under the supervision of Jamshed Iqbal. Imtiaz Khan, Elina Ausekle, Mariia Miliutina and Peter Langer provided the compounds for the study. All the authors checked and finally approved the draft before submission.
Compliance with ethical standards
Conflict of interest
The authors confirm that this article content has no conflict of interest. The authors also declare no competing financial interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nepali K, Sharma S, Sharma M, Bedi PMS, Dhar KL. 2014. Rational approaches, design strategies, structure activity relationship and mechanistic insights for anticancer hybrids. Eur J Med Chem. 2014;77(4):422–487. doi: 10.1016/j.ejmech.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Fortin S, Bérubé G. Advances in the development of hybrid anticancer drugs. Expert Opinion Drug Discov. 2013;8(2):1029–1047. doi: 10.1517/17460441.2013.798296. [DOI] [PubMed] [Google Scholar]
- 3.Raj T, Bhatia RK, Sharma M, Saxena AK, Ishar MPS. Cytotoxic activity of 3-(5-phenyl-3H-[1, 2, 4] dithiazol-3-yl) chromen-4-ones and 4-oxo-4H-chromene-3-carbothioic acid N-phenylamides. Eur J Med Chem. 2010;45(4):790–794. doi: 10.1016/j.ejmech.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Danaei G, Vander Hoorn S, Lopez AD, Murray CJ, Ezzati M. Causes of cancer in the world: comparative risk assessment of nine behavioural and environmental risk factors. Lancet. 2005;366(2):1784–1793. doi: 10.1016/S0140-6736(05)67725-2. [DOI] [PubMed] [Google Scholar]
- 5.Kateb B, Chiu K, Black KL, Yamamoto V, Khalsa B, Ljubimova JY, Ding H, Patil R, Portilla-Arias JA, Modo M, Moore DF. Nanoplatforms for constructing new approaches to cancer treatment, imaging, and drug delivery: what should be the policy? Neuroimage. 2011;54(2):S106–S124. doi: 10.1016/j.neuroimage.2010.01.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nepali K, Sharma S, Kumar D, Budhiraja A, Dhar KL. Anticancer hybrids-a patent survey. Recent Pat Anti-cancer Drug Discov. 2014;9(3):303–339. doi: 10.2174/1574892809666140520150459. [DOI] [PubMed] [Google Scholar]
- 7.Zimmermann H. Extracellular metabolism of ATP and other nucleotides. Naunyn Schmiedeberg's Arch Pharmacol. 2000;362(4–5):299–309. doi: 10.1007/s002100000309. [DOI] [PubMed] [Google Scholar]
- 8.Vitaku E, Smith DT, Njardarson JT. Analysis of the structural diversity, substitution patterns, and frequency of nitrogen heterocycles among US FDA approved pharmaceuticals: miniperspective. J Med Chem. 2014;57(24):10257–10274. doi: 10.1021/jm501100b. [DOI] [PubMed] [Google Scholar]
- 9.Sondhi SM, Johar M, Rajvanshi S, Dastidar SG, Shukla R, Raghubir R, Lown JW. Anticancer, anti-inflammatory and analgesic activity evaluation of heterocyclic compounds synthesized by the reaction of 4-isothiocyanato-4-methylpentan-2-one with substituted o-phenylenediamines, o-diaminopyridine and (un) substituted o. Aust J Chem. 2001;54(1):69–74. doi: 10.1071/CH00141. [DOI] [Google Scholar]
- 10.Zimmermann H, Zebisch M, Sträter N. Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signal. 2012;8(3):437–502. doi: 10.1007/s11302-012-9309-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yegutkin GG. Enzymes involved in metabolism of extracellular nucleotides and nucleosides: functional implications and measurement of activities. Crit Rev Biochem Mol Biol. 2014;49(6):473–497. doi: 10.3109/10409238.2014.953627. [DOI] [PubMed] [Google Scholar]
- 12.Ausekle E, Ejaz SA, Khan SU, Ehlers P, Villinger A, Lecka J, Sévigny J, Iqbal J, Langer P. New one-pot synthesis of N-fused isoquinoline derivatives by palladium-catalyzed C–H arylation: potent inhibitors of nucleotide pyrophosphatase-1 and-3. Org Biomol Chem. 2016;14(48):11402–11414. doi: 10.1039/C6OB02236G. [DOI] [PubMed] [Google Scholar]
- 13.Khan I, Shah SJ, Ejaz SA, Ibrar A, Hameed S, Lecka J, Millán JL, Sévigny J, Iqbal J. Investigation of quinoline-4-carboxylic acid as a highly potent scaffold for the development of alkaline phosphatase inhibitors: synthesis, SAR analysis and molecular modelling studies. RSC Adv. 2015;5(79):64404–64413. doi: 10.1039/C5RA12455G. [DOI] [Google Scholar]
- 14.Miliutina M, Ejaz SA, Khan SU, Iaroshenko VO, Villinger A, Iqbal J, Langer P. Synthesis, alkaline phosphatase inhibition studies and molecular docking of novel derivatives of 4-quinolones. Eur J Med Chem. 2017;126:408–420. doi: 10.1016/j.ejmech.2016.11.036. [DOI] [PubMed] [Google Scholar]
- 15.Orimo H. The mechanism of mineralization and the role of alkaline phosphatase in health and disease. J Nippon Med Sch. 2010;77(1):4–12. doi: 10.1272/jnms.77.4. [DOI] [PubMed] [Google Scholar]
- 16.Wanichpakorn S, Kedjarune-Laggat U. Primary cell culture from human oral tissue: gingival keratinocytes, gingival fibroblasts and periodontal ligament fibroblasts. SJST. 2010;32(4):327–331. [Google Scholar]
- 17.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 18.Niks M. Towards an optimized MTT assay. J Immunol Methods. 1990;130(2):149–151. doi: 10.1016/0022-1759(90)90309-J. [DOI] [PubMed] [Google Scholar]
- 19.Hassan S, Ejaz SA, Saeed A, Shehzad M, Khan SU, Lecka J, Sévigny J, Shabir G, Iqbal J. 4-Aminopyridine based amide derivatives as dual inhibitors of tissue non-specific alkaline phosphatase and ecto-5′-nucleotidase with potential anticancer activity. Bioorg Chem. 2018;76(5):237–248. doi: 10.1016/j.bioorg.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Saito Y, Uchida N, Tanaka S, Suzuki N, Tomizawa-Murasawa M, Sone A, Najima Y, Takagi S, Aoki Y, Wake A, Taniguchi S. Induction of cell cycle entry eliminates human leukemia stem cells in a mouse model of AML. Nature biotechnol. 2010;28(3):275–280. doi: 10.1038/nbt.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iqbal J, Ejaz SA, Saeed A, al Rashida M. Detailed investigation of anticancer activity of sulfamoyl benz (sulfon) amides and 1H–pyrazol–4–yl benzamides: An experimental and computational study. Eur J Pharmacol. 2018;832:11–24. doi: 10.1016/j.ejphar.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Lin GJ, Jiang GB, Xie YY, Huang HL, Liang ZH, Liu YJ. Cytotoxicity, apoptosis, cell cycle arrest, reactive oxygen species, mitochondrial membrane potential, and Western blotting analysis of ruthenium (II) complexes. J Biol Inorg Chem. 2013;18(8):873–882. doi: 10.1007/s00775-013-1032-2. [DOI] [PubMed] [Google Scholar]
- 23.Sirajuddin M, Ali S, McKee V, Zaib S, Iqbal J. Organotin (IV) carboxylate derivatives as a new addition to anticancer and antileishmanial agents: design, physicochemical characterization and interaction with Salmon sperm DNA. RSC Adv. 2014;4(2):57505–57521. doi: 10.1039/C4RA10487K. [DOI] [Google Scholar]
- 24.MOE, version 2014.0901, Chemical Computing Group (CCG), Montreal, Canada, http://www.chemcomp.com/MOEMolecular_Operating_Environment.html. Accessed August 2016.
- 25.Visualizer, D.S. 2005. Accelrys Software Inc, Discovery Studio Visualizer, 2.