Abstract
miR-29b has been identified as a rapamycin-induced microRNA (miRNA) in Tsc2-deficient, mTORC1-hyperactive cells. The biological significance of this induction of miR-29b is unknown. We have found that miR-29b acts as an oncogenic miRNA in Tsc2-deficient cells: inhibition of miR-29b suppressed cell proliferation, anchorage-independent cell growth, cell migration, invasion, and the growth of Tsc2-deficient tumors in vivo. Importantly, the combination of miR-29b inhibition with rapamycin treatment further inhibited these tumor-associated cellular processes. To gain insight into the molecular mechanisms by which miR-29b promotes tumorigenesis, we used RNA sequencing to identify the tumor suppressor retinoid receptor beta (RARβ) as a target gene of miR-29b. We found that miR-29b directly targeted the 3′UTR of RARβ. Forced expression of RARβ reversed the effects of miR-29b overexpression in proliferation, migration, and invasion, indicating that it is a critical target. miR-29b expression correlated with low RARβ expression in renal clear cell carcinomas and bladder urothelial carcinomas, tumors associated with TSC gene mutations. We further identified growth family member 4 (ING4) as a novel interacting partner of RARβ. Overexpression of ING4 inhibited the migration and invasion of Tsc2-deficient cells while silencing of ING4 reversed the RARβ-mediated suppression of cell migration and invasion. Taken together, our findings reveal a novel miR-29b/RARβ/ING4 pathway that regulates tumorigenic properties of Tsc2-deficient cells, and that may serve as a potential therapeutic target for TSC, lymphangioleiomyomatosis (LAM), and other mTORC1-hyperactive tumors.
Introduction
Tuberous sclerosis complex (TSC) is an autosomal dominant syndrome that affects multiple organ systems and manifests as hamartomatous tumors of the brain, heart, kidney, skin, and lungs [1]. TSC is caused by germline loss-of-function mutations in one of the two tumor suppressor genes, TSC1 or TSC2. The TSC1 and TSC2 proteins function within a complex that integrates signals from the cellular environment, including growth factors and nutrients, to regulate the activity of mammalian/mechanistic target of rapamycin complex 1 (mTORC1) [1]. mTORC1 controls numerous essential metabolic processes, including protein and lipid synthesis, nucleotide synthesis, glycolysis, and autophagy [2]. Mutational inactivation of TSC1 or TSC2 results in hyperactivation of mTORC1 [3–5].
Pivotal clinical trials have shown that mTORC1 inhibitors (sirolimus and everolimus) are effective agents for the treatment of several manifestations of TSC, including renal angiomyolipomas, subependymal giant cell astrocytomas, and pulmonary lymphangioleiomyomatosis (LAM). Partial responses are typically observed, with tumor regrowth upon treatment cessation; thus, continual lifelong therapy appears to be required, often beginning in early childhood [6–10].
MicroRNAs, also known as miRNAs or miRs, are short noncoding single-stranded RNA species that can negatively regulate gene expression. Through an RNA-induced silencing complex, miRNAs bind to the 3′-untranslated region of their target genes, either by perfect base pairing resulting in mRNA degradation or by imperfect base pairing to block translation. Because a single miRNA can bind to several different mRNA transcripts and one mRNA transcript is often targeted by multiple miRNA species, small changes in miRNA levels can have large downstream effects on phenotypes that can include proliferation, cell cycle progression, differentiation, migration, apoptosis, and metabolism [11].
miR-29b is one of the three members of the miR-29 family, which differ from each other by two or three bases. miR-29b-1 and miR-29b-2 are encoded by two separated genes on chromosome 7q32.3 and 1q32.2, respectively in human cells. Thus, two distinct precursor sequences (a pre-miR-29b-1 and pre-miR-29b-2) are produced, however the mature miR-29b sequence resulting from the precursors is identical [12, 13]. miR-29b has well-documented tumor suppressive activity, influencing cell proliferation, apoptosis, differentiation, metastasis, and chemotherapy sensitivity [14]. The expression of miR-29b is downregulated in multiple tumor types, including gastric cancer, prostate cancer, breast cancer, and lung cancer, consistent with a tumor suppressor mechanism [15]. However, miR-29b can have tumor-promoting activity in certain tissue and cell types [14].
Previously, miR-29b was found to be upregulated upon rapamycin treatment in TSC2-deficient patient-derived angiomyolipoma cells [16]. The goal of this study was to investigate the biological role of miR-29b in Tsc2-deficient cells. We have demonstrated that miR-29b functions as an oncomiR in Tsc2-deficient cells, promoting cell growth, migration, and invasion. We identified retinoic acid receptor beta (RARβ) as a novel direct target of miR-29b and found that RARβ is a positive regulator of the tumor suppressor inhibitor of growth family member 4 (ING4) via protein–protein interaction. Importantly, miR-29b inhibition suppressed the growth of Tsc2-deficent cells in a xenograft mouse model of TSC. Finally, we found a significant negative correlation between miR-29b and RARβ expression in renal clear cell carcinomas and bladder urothelial carcinomas (BLCA), two tumors that are associated with mutational inactivation of the TSC genes. Taken together, our findings contribute to a better understanding of the mechanisms through which miR-29b promotes tumorigenesis. Targeting miR-29b represents a novel therapeutic strategy for TSC and other tumors with mTORC1 hyperactivation.
Results
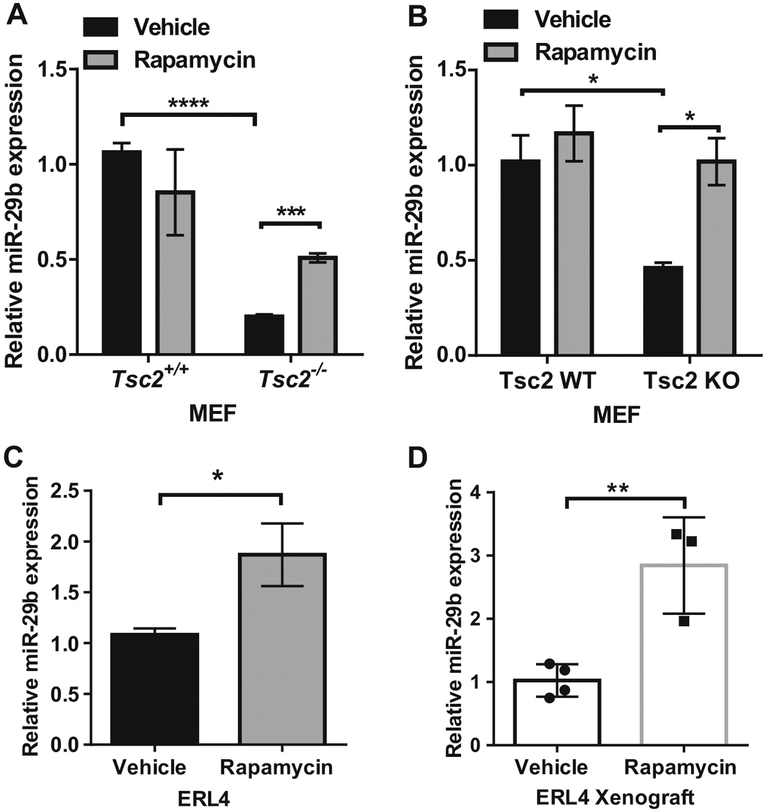
Rapamycin upregulates miR-29b expression in vitro and in vivo in Tsc2-deficient but not Tsc2-expressing cells
We previously found that miR-29b is upregulated by rapamycin in human TSC2-deficient angiomyolipoma-derived 621–101 cells [16]. To determine whether rapamycin-induced miR-29b expression is observed in other Tsc2-deficient models, we treated two pairs of Tsc2 wild-type and Tsc2-knockout mouse embryonic fibroblasts (MEFs) (referred to as Tsc2+/+ and Tsc2−/− MEFs; Tsc2 WT and Tsc2 KO MEFs) with rapamycin (20 nM) for 24 h. Using RT-qPCR, we found that miR-29b expression was upregulated by ~2.5-fold (P < 0.001) in Tsc2−/− MEFs (Fig. 1a) and by twofold (P < 0.05) in Tsc2-KO MEFs (Fig. 1b), upon rapamycin treatment. Unexpectedly, miR-29b expression was higher in wild-type MEFs compared with Tsc2-deficient MEFs. Rapamycin treatment of Tsc2-expressing wild-type cells, however, did not affect miR-29b levels (Fig. 1a, b). Rapamycin treatment of ERL4 cells (Tsc2-deficient ELT3 cells derived from an Eker rat uterine leiomyomas and stably expressing luciferase) resulted in 1.7-fold (P < 0.05) miR-29b upregulation (Fig. 1c).
Fig. 1.
Rapamycin upregulates miR-29b expression in Tsc2-deficient but not Tsc2 wild-type cells in vitro and in vivo. miR-29b expression was assessed by RT-qPCR in Tsc2+/+ or Tsc2−/− MEFs (a), Wild-type or Tsc2 knockout MEFs (b), and in Tsc2-deficient rat ERL4 cells (c), treated with vehicle (DMSO) or rapamycin (20 nM) for 24 h. Data are presented as mean fold change in miR-29b expression ± SD relative to Tsc2+/+ MEFs, Tsc2 WT MEFs, or ERL4 treated with vehicle. Results are from n = 3 biological replicates. Data are presented as mean ± SD. Data for bar graphs were calculated using two-way ANOVA followed by Bonferroni’s posttest for multiple comparisons. d Mice bearing ERL4 xenograft tumors were treated intraperitoneally with rapamycin (3 mg/kg) or vehicle every other day for 6 days (vehicle: n = 3, rapamycin: n = 4). Tumors were harvested 4 h after the last treatment and miR-29b expression was examined by RT-qPCR. Data are presented as mean ± SD. Statistical significance was determined by Mann–Whitney’s U test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
To determine if rapamycin-dependent upregulation of miR-29b occurs in vivo, we treated Tsc2-deficient ERL4 xenograft tumors with rapamycin (3 mg/kg) every 2 days for 6 days (three treatments). miR-29b expression was increased by 2.8-fold (P < 0.01) compared with vehicle-treated tumors (Fig. 1d). These results demonstrate that miR-29b expression is upregulated by rapamycin treatment selectively in Tsc2-deficient cells, but not in Tsc2-expressing cells, both in vitro and in vivo.
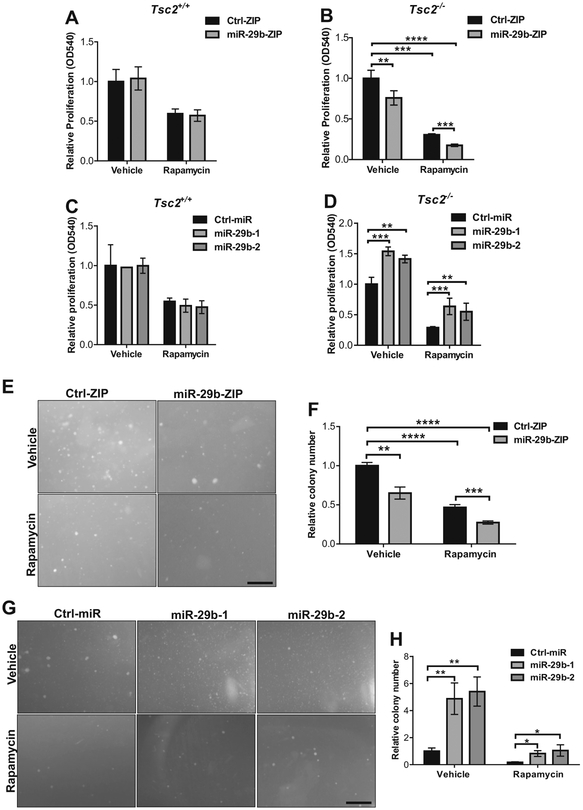
miR-29b promotes proliferation and anchorage-independent cell growth of Tsc2-deficient cells
To understand the functional significance of miR-29b in Tsc2-deficient cells, we examined the effects of miR-29b knockdown on cell proliferation in Tsc2+/+ and Tsc2−/− MEFs. Cells were transduced with miR-29b-ZIP lentivirus; a lentivirus expressing anti-miR-29b small RNAs (for miR-29b expression knockdown), or empty miR-Ctrl-ZIP (as a mock control). Four days after virus infection, the cultured cells expressed green fluorescent protein (GFP), a marker encoded by the miR-Zip vector. The GFP-positive cells expressing miR-29b-ZIP or miR-Ctrl-ZIP were sorted to enrich for miR-29b-ZIP or miR-Ctrl-ZIP-expressing population and confirmed by flow cytometry (Supplementary Fig. 1a). We verified that the levels of miR-29b are suppressed in the presence or absence of rapamycin in Tsc2-deficient MEFs-expressing miR-29b-ZIP or miR-Ctrl-ZIP by quantifying the absolute amounts of miR-29b (Supplementary Fig. 1b). Cells were treated with vehicle or rapamycin for 24 h and then stained with crystal violet. miR-29b inhibition did not affect the proliferation of Tsc2+/+ MEFs (Fig. 2a). In contrast, cell proliferation in Tsc2−/− MEFs stably expressing miR-29b-ZIP was reduced by 25% (P <0.01) (Fig. 2b). Rapamycin treatment further suppressed the proliferation of Tsc2-deficient cells expressing miR-29b-ZIP by 80% (P < 0.0001) compared with vehicle-treated control cells (Fig. 2b). Similarly, miR-29b knockdown with miR-29b-ZIP in Tsc2−/− MEFs further enhanced the inhibition of cell proliferation by the ATP-competitive inhibitors of mTOR, Torin 1 or PP242, compared with vehicle-treated cells (Supplementary Fig. 2b, d). These inhibitors had no effect on proliferation levels in Tsc2+/+ MEFs (Supplementary Fig. 2a, c).
Fig. 2.
miR-29b regulates cell growth in Tsc2-deficient, but not Tsc2 wild-type cells. Cell proliferation was assessed by crystal violet staining in Tsc2+/+ (a) or Tsc2−/− (b) MEFs stably expressing miR-29b-ZIP or Ctrl-ZIP following treatment with rapamycin (20 nM) or vehicle (DMSO) for 72 h. Data are presented as mean fold change in OD 540 ± SD relative to Ctrl-ZIP cells treated with vehicle of n = 3 biological replicates. Cell proliferation was assessed by crystal violet staining in Tsc2+/+ (c) or Tsc2−/− (d) MEFs stably overexpressing pre-miR-29b-1 (miR-29b-1), pre-miR-29b-2 (miR-29b-2) or Ctrl-miR following treatment with rapamycin (20 nM) or vehicle (DMSO) for 72 h. Data are presented as mean fold change in OD 540 ± SD relative to Ctrl-miR cells treated with vehicle of three independent experiments. e Representative images of Tsc2−/− MEFs stably expressing Ctrl-ZIP or miR-29b-ZIP grown in soft agar with addition of vehicle (DMSO) or rapamycin (5 nM) for 2 weeks. f Data are presented as mean fold change in colony number ± SD relative to Ctrl-ZIP treated with vehicle of n = 3 biological replicates. g Representative images of Tsc2−/− MEFs stably overexpressing pre-miR-29b-1 (miR-29b-1), pre-miR-29b-2 (miR-29b-2), or Ctrl-miR grown in soft agar with addition of vehicle (DMSO) or rapamycin (5 nM) for 2 weeks. h Data are presented as mean fold change in colony number ± SD relative to Ctrl-miR treated with vehicle of n = 3 biological replicates. Data for bar graphs were calculated using two-way ANOVA followed by Bonferroni’s posttest for multiple comparisons. Scale bar = 500 μm. *P < 0.05; **P < 0.01; ***P < 0.001
We next examined the effect of miR-29b overexpression on cell proliferation. Tsc2+/+ or Tsc2−/− MEFs stably expressing one of the two precursors of miR-29b (miR-29b-1 or miR-29b-2) were generated. The relative and absolute levels of mature miR-29b were measured by qPCR (Supplementary Fig. 3a, b). miR-29b overexpression levels in Tsc2−/− MEFs were 2–2.5-fold higher compared with Tsc2−/− MEFs expressing miR-Ctrl (Supplementary Fig. 3a, b). Overexpression of miR-29b did not impact the proliferation of Tsc2+/+ MEFs (Fig. 2c), but increased the proliferation of Tsc2−/− MEFs even in the context of rapamycin treatment (Fig. 2d).
Consistent with these results, anchorage-independent growth assays of Tsc2−/− MEFs revealed that miR-29b inhibition results in a 35% decrease in colony number (P <0.01) (Fig. 2e, f). The combination of rapamycin and miR-29b inhibition further reduced colony formation by 73% (P < 0.0001) compared with control (Fig. 2f). In parallel, we found that overexpression of miR-29b increased colony number by fivefold in vehicle-treated Tsc2−/− cells (P < 0.01) and by twofold in rapamycin-treated Tsc2−/− cells (P < 0.05) (Fig. 2g, h). These data demonstrate that miR-29b is a critical regulator of cell proliferation and anchorage-independent growth in Tsc2-deficient cells and suggest that inhibition of miR-29b may enhance the therapeutic effect of rapamycin.
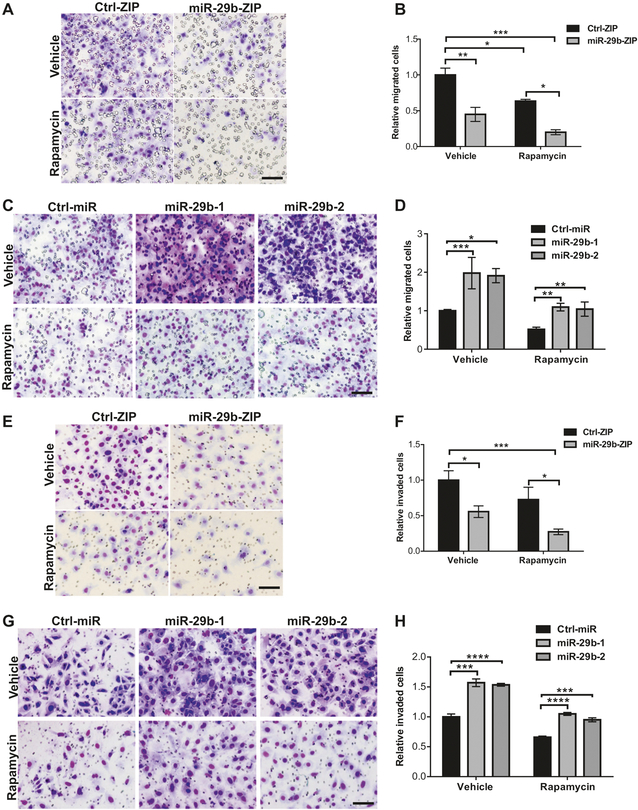
miR-29b promotes cell migration and invasion in Tsc2-deficient cells
To investigate the role of miR-29b in Tsc2-deficient cell migration, we used Tsc2−/− MEFs stably expressing miR-29b-ZIP or Ctrl-ZIP in a transwell migration assay. miR-29b knockdown decreased cell migration by 55% (P <0.05). The combination of miR-29b inhibition and rapamycin treatment reduced migration even further (81%, p <0.0001) (Fig. 3a, b). Consistent with these data, over-expression of miR-29b in Tsc2−/− MEFs enhanced cell migration by twofold (P < 0.05) compared with Ctrl-ZIP following vehicle or rapamycin treatment (Fig. 3c, d).
Fig. 3.
miR-29b regulates cell migration and invasion in Tsc2-deficient cells. a Representative images of the transwell assay with decreased migration towards 10% FBS-containing growth medium of Tsc2−/− MEFs with miR-29b inhibition (miR-29b-ZIP) treated with vehicle (DMSO) or rapamycin (20 nM) for 6 h. b Mean fold change of the number of migrated cells ± SD relative to vehicle-treated Ctrl-ZIP of n = 3 biological replicates. c Representative images of the transwell assay with increased migration towards 10% FBS-containing growth medium of Tsc2−/− MEFs stably overexpressing miR-29b-1 or miR-29b-2. d Mean fold change of number of migrated cells ± SD relative to vehicle-treated Ctrl-miR of n = 3 biological replicates. e Representative images of the Boyden chamber invasion assay, showing decreased invasive capacity of Tsc2−/− MEFs with miR-29b inhibition (miR-29b-ZIP) treated with vehicle (DMSO) or rapamycin (20 nM) for 24 h. f Mean fold change of number of invaded cells ± SD relative to vehicle-treated Ctrl-ZIP of n = 3 biological replicates. g Representative images of the Boyden chamber invasion assay, showing increased invasive capacity of Tsc2−/− MEFs stably overexpressing miR-29b-1 or miR-29b-2. h Mean fold change of number of invaded cells ± SD relative to vehicle-treated Ctrl-miR of n = 3 biological replicates. Data for bar graphs were calculated using two-way ANOVA followed by Bonferroni’s posttest for multiple comparisons. Scale bar = 85 μm. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
Similarly, miR-29b inhibition markedly suppressed the invasive capacity of Tsc2−/− MEFs (45%, P < 0.01), as demonstrated using a Boyden chamber invasion assay, while the combination of miR-29b inhibition and rapamycin treatment further inhibited invasive capacity (73%, P <0.001) (Fig. 3e, f). Overexpression of miR-29b in Tsc2−/− MEFs enhanced cell invasion by 1.5-fold (P < 0.001); rapamycin-treated miR-29b overexpressing cells still showed a 30% increase in invasion (P < 0.001) compared with Ctrl-miR (Fig. 3g, h). These results demonstrate that miR-29b promotes the migratory and invasive behaviors of Tsc2-deficient cells, which may be enhanced with rapamycin treatment.
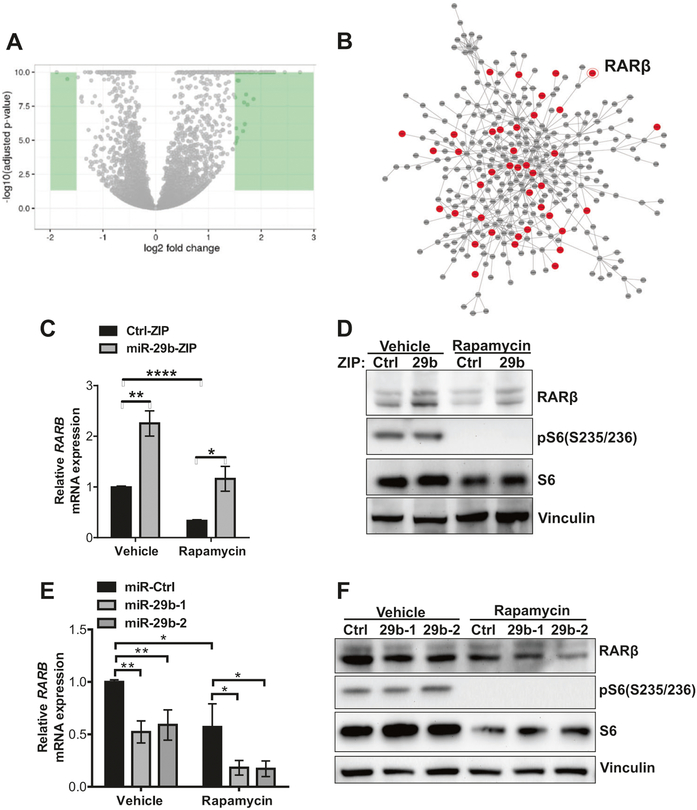
RNAseq analysis identifies RARβ as a candidate target of miR-29b
To investigate the molecular mechanism underlying these miR-29b-mediated cellular processes, we used RNA sequencing to analyze the transcriptional profiles of rapamycin-treated Tsc2−/− MEFs expressing either miR-29b-ZIP or Ctrl-ZIP (20 nM, 24 h). Transcripts that were differentially regulated by log2 fold > 1.5 are highlighted in green (Fig. 4a). Out of 15,613 genes that were detected, 730 genes were upregulated by at least 1.3-fold (P < 0.05) (Supplementary Fig. 4a and Supplementary Table 1) and 49 genes downregulated by at least 50% (P < 0.05) (Supplementary Fig. 4a and Supplementary Table 2). We next carried out a gene ontology analysis using DAVID bioinformatics resources. We focused on upregulated genes: since miRNAs downregulate their target mRNA expression, we expect miR-29b inhibition lead to upregulation of its direct targets. The upregulated genes were significantly enriched in multiple molecular functions, including heparin binding, sequence-specific DNA binding, metallopeptidase activity, growth factor activity and chemorepellent activity with FDR < 0.05 (Supplementary Fig. 4b). To visualize and catalog more comprehensively the links among miR-29b-regulated genes, we mapped the functional connections among differentially expressed genes and their first-degree interactors using data from a consolidated interactome and observed a number of proteins involved in sequence-specific DNA binding in the network, including RARβ (Fig. 4b, Supplementary Tables 1 and 2).
Fig. 4.
RNA sequencing identifies RARβ is a candidate target of miR-29b. a Volcano plot comparing changes in gene expression (as log2 fold change of fragments per kilobase of transcript per million mapped reads) of Tsc2−/− MEFs stably expressing miR-29-ZIP versus Ctrl-ZIP treated with rapamycin (20 nM) for 24 h. Green color highlights genes that were differentially regulated by log2 fold change > 1.5. b Interactome analysis of transcripts that are differentially regulated by miR-29b knockdown. Red indicates predicted direct targets and gray indicates indirect targets of miR-29b. c–f RARβ mRNA and protein levels were assessed in Tsc2−/− MEFs stably expressing miR-29b-ZIP or Ctrl-ZIP (c, d), or overexpressing miR-29b-1, miR-29b-2, or miR-Ctrl (e–f) treated with vehicle (DMSO) or rapamycin (20 nM) for 24 h. Data are presented as mean fold change ± SD relative to vehicle-treated controls of n = 3 biological replicates. Data for bar graphs were calculated using two-way ANOVA followed by Bonferroni’s posttest for multiple comparisons. *P <0.05; **P < 0.01; ****P <0.0001
To identify direct targets of miR-29b, we performed 3′ UTR-binding site prediction by TargetScan 6.2 and found 63 predicted targets among the 730 upregulated genes (red circles, Fig. 4b). We then focused on genes whose expression was upregulated by miR-29b inhibition and suppressed by rapamycin, and identified RARβ, whose expression is downregulated by rapamycin and is restored by miR-29b inhibition (Fig. 4c, d). Furthermore, miR-29b over-expression downregulated RARβ expression at both mRNA and protein levels, with partial restoration following rapamycin treatment (Fig. 4e, f). In summary, RNAseq complemented by RT-qPCR and western blot analyses revealed that RARβ expression is reciprocally regulated by miR-29b and rapamycin.
RARβ is a direct and functionally important target of miR-29b
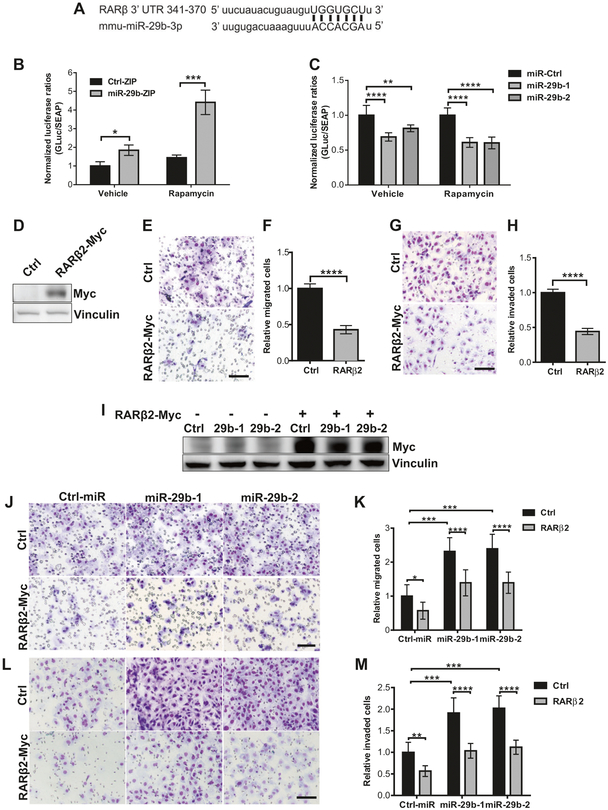
We next sought to determine whether the effects of miR-29b on RARβ are mediated via direct binding of miR-29b to the RARβ 3′UTR region, using a secreted dual-luciferase assay. The 3′UTR of RARβ contains a complementary site for the seed region of miR-29b (Fig. 5a). Knockdown of miR-29b in Tsc2−/− MEFs increased the relative secreted luciferase activity of RARβ 3′UTR by fourfold (P < 0.001) following rapamycin treatment (Fig. 5b). Similarly, miR-29b overexpression suppressed the relative-secreted luciferase activity by 50% (P < 0.0001) (Fig. 5c). These data indicate that RARβ is a direct target of miR-29b.
Fig. 5.
RARβ is a direct and functionally important target of miR-29b. a Sequence of mouse RARβ 3′UTR (nucleotide 341–370) showing the putative miR-29b-3p-binding site. Matching regions are indicated by lines. Dual-luciferase assay of Tsc2−/− MEFs stably expressing miR-29b-ZIP or Ctrl-ZIP (b), or overexpressing miR-29b-1, miR-29b-2 or miR-Ctrl (c) transfected with secreted Gaussia luciferase constructs containing the 3′UTR of RARβ for 24 h. Cells were then treated with vehicle (DMSO) or rapamycin (20 nM) for another 24 h. Secreted Gaussia luciferase activity was normalized to secreted alkaline phosphatase activity. Data are presented as relative luciferase activity ± SD relative to Ctrl-ZIP or Ctrl-miR treated with vehicle or rapamycin of n = 3 biological replicates. Data for bar graphs were calculated using one-way ANOVA followed by Bonferroni’s posttest for multiple comparisons. d Western blot analysis of Tsc2−/− MEFs transiently transfected with control vector or RARβ2-Myc-DDK. Vinculin was used as a loading control. e Representative images of migrated Tsc2−/− MEFs transiently transfected with control vector or RARβ2-Myc-DDK towards 10% FBS-containing growth medium. f Mean fold change of number of migrated cells ± SD relative to vector controls of n = 3 biological replicates and statistical significance was determined by two-tailed student’s t-test. g Representative images Boyden chamber invasion assays of Tsc2−/− MEFs transiently overexpressing RARβ2-Myc-DDK showing decreased cell invasion. h Mean fold change of number of invaded cells ± SD relative to vector controls of n = 3 biological replicates and statistical significance was determined by two-tailed student’s t-test. i Western blot analysis showing successful restoration of RARβ2 in Tsc2−/− MEFs overexpressing miR-29b-1 or miR-29b-2 transfected with RARβ2-Myc-DDK. Vinculin was used as a loading control. j Representative images of the transwell migration assay of Tsc2−/− MEFs overexpressing miR-29b-1 or miR-29b-2 with and without restoration of RARβ2. k Mean fold change of number of migrated cells ± SD relative to vector controls/control siRNA of n = 3 biological replicates. l Representative images of Boyden chamber invasion assays demonstrating RARβ restoration decreased the invasion of Tsc2−/− MEFs overexpressing miR-29b-1 or miR-29b-2. m Mean fold change of number of migrated cells ± SD relative to vector controls/control siRNA of n = 3 biological replicates. Scale bar = 85 μm. Data for bar graphs were calculated using two-way ANOVA followed by Bonferroni’s posttest for multiple comparisons. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
We next investigated whether RARβ has functional importance in Tsc2-deficient cells. In the mouse, the RARB gene produces four isoforms (RARβ1, RARβ2, RARβ3, and RARβ4) [17], only two of which (RARβ1 and RARβ2) are detected by our anti-RARβ antibodies as shown in Fig. 4c. We focused on RARβ2 since it is expressed in normal human adult tissue in contrast to RARβ1, which is primarily expressed during embryogenesis [18]. Overexpression of RARβ2 in Tsc2−/− MEFs (Fig. 5d) suppressed cell migration by 55% (P < 0.001) compared with cells transfected with vector controls (Fig. 5e, f), and inhibited cell invasion by 50% (P < 0.001) (Fig. 5g, h). Next, we transiently transfected Myc-tagged RARβ2 in Tsc2−/− MEFs overexpressing miR-29b (Fig. 5i). As expected, and opposite to miR-29b, forced expression of RARβ2 in Tsc2−/− MEFs overexpressing miR-29b inhibited cell migration by 40% (P < 0.0001) (Fig. 5j, k) and invasion by 47% (P < 0.0001) (Fig. 5l, m) compared with miR-29b overexpressing cells transfected with control vectors. Therefore, RARβ2 is critical target of miR-29b and regulates miR-29b-mediated migration and invasion of Tsc2-deficient cells.
ING4 physically interacts with RARβ
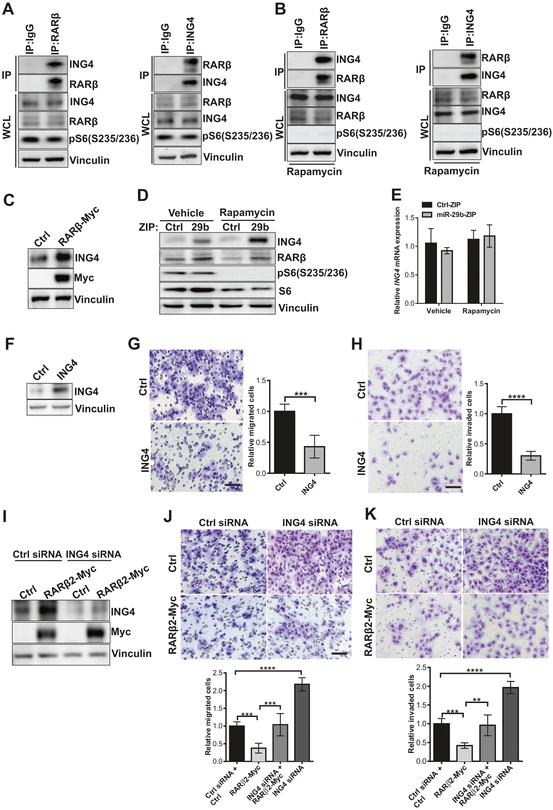
To identify targets of RARβ that may play a role in TSC, we used the publicly available BioGrid database of Protein, Chemical, and Genetic interactions (BioGrid) and found that ING4 is an unconfirmed candidate for RARβ-interacting partner, identified by a yeast two-hybrid screen [19]. We performed coimmunoprecipitation assays under native conditions to preserve protein–protein interactions, which revealed that ING4 is present in RARβ immunoprecipitates (Fig. 6a, left panel), and RARβ is present in ING4 immunoprecipitates (Fig. 6a, right panel). Next, we investigated whether the physical interaction between RARβ and ING4 is stable in the presence of rapamycin. Given that rapamycin can suppress the protein expression of RARβ, we performed the endogenous coimmunoprecipitation experiments using Tsc2-deficient cells expressing miR-29b-ZIP, to ensure an adequate amount of RARβ protein. Coimmunoprecipitation demonstrated that RARβ remains associated with ING4 following rapamycin treatment (Fig. 6b).
Fig. 6.
ING4 physically interacts with RARβ. a Western blots of Tsc2 −/− MEFs lysed and immunoprecipitated with RARβ, ING4, or rabbit IgG overnight, then incubated with Protein G Sepharose for 2 h. Immunoprecipitated proteins (IP) or whole cell lysates (WCL) were immunoblotted with RARβ, ING4, pS6, or vinculin. Results are representative of three independent experiments. b Tsc2−/− MEFs stably expressing miR-29b-ZIP were treated with rapamycin (20 nM) for 24 h. Cells were lysed, immunoprecipitated, and immunoblotted as described in a. c Tsc2−/− MEFs transiently transfected with control vector or RARβ2-Myc-DDK were analyzed by immunoblotting using Myc, ING4 or vinculin antibodies. d Immunoblot analysis of ING4 protein expression in Tsc2−/− MEFs stably expressing miR-29b-ZIP or Ctrl-ZIP treated with vehicle or rapamycin (20 nM) for 24 h. e ING4 mRNA expression was assessed by RT-qPCR in Tsc2−/− MEFs stably expressing miR-29b-ZIP or Ctrl-ZIP treated with vehicle or rapamycin (20 nM) for 24 h. Data are presented as mean ± SD relative to vehicle-treated Ctrl-ZIP of n = 3 biological replicates. Data for bar graphs were calculated using two-way ANOVA followed by Bonferroni’s posttest for multiple comparisons. f Tsc2−/− MEFs transiently transfected with control vector or ING4 were analyzed by immunoblotting using ING4 or vinculin antibodies. g Migration of Tsc2−/− MEFs transiently transfected with control vector or ING4 towards 10% FBS-containing growth medium. Data are presented as mean ± SD relative to vehicle-treated Ctrl of n = 3 biological replicates. Statistical significance was determined by two-tailed student’s t-test. h Invasion of Tsc2−/− MEFs overexpressing ING4 or control towards 10% FBS-containing growth medium. Data are presented as mean ± SD relative to vehicle-treated Ctrl of n = 3 biological replicates. Statistical significance was determined by two-tailed student’s t-test. i Tsc2−/− MEFs transiently transfected with the indicated constructs and/or siRNA were analyzed by immunoblotting using ING4, myc or vinculin antibodies. j Migration of Tsc2−/− MEFs transiently transfected with the indicated constructs and/or siRNA towards 10% FBS-containing growth medium. Data are presented as mean ± SD relative to vehicle-treated Ctrl siRNA + Ctrl of n = 3 biological replicates. Statistical significance was determined by one-way ANOVA followed by Bonferroni’s posttest for multiple comparisons. k Invasion of Tsc2−/− MEFs transiently transfected with the indicated constructs and/or siRNA towards 10% FBS-containing growth medium. Data are presented as mean ± SD relative to vehicle-treated Ctrl siRNA + Ctrl of n = 3 biological replicates. Statistical significance was determined by one-way ANOVA followed by Bonferroni’s posttest for multiple comparisons. Scale bar = 85 μm. **P < 0.01; ***P < 0.001; ****P <0.0001
To study the effects of RARβ on ING4, we over-expressed RARβ2 in Tsc2−/− MEFs and examined ING4 protein expression. As shown in Fig. 6c, RARβ2 over-expression increased ING4 protein levels. We next analyzed expression of ING4 in Tsc2−/− MEFs with miR-29b inhibition in the presence and absence of rapamycin treatment. miR-29b inhibition increased ING4 protein level in both vehicle-treated and rapamycin-treated cells (Fig. 6d). However, miR-29b inhibition and/or rapamycin treatment did not alter ING4 mRNA levels (Fig. 6e), suggesting that miR-29b indirectly regulates ING4 via RARβ.
To explore the functional significance of ING4 in Tsc2-deficient cells, we next overexpressed ING4 in Tsc2−/− MEFs (Fig. 6f) and found that cell migration was strikingly decreased, by 60% (P < 0.001) (Fig. 6g), and cell invasion was suppressed by 75% (P < 0.0001) (Fig. 6h). Since we had found earlier that overexpression of RARβ2 suppressed cell migration and invasion (Fig. 5d–h), we hypothesized that knockdown of ING4 in RARβ2 overexpressing cells could elevate migratory/invasive phenotypes. Inhibition of ING4 expression with four individual siRNA duplexes from SMARTpools (Fig. 6i) increased both the migration and the invasion of RARβ2 overexpressing Tsc2-deficient MEFs (Fig. 6j, k). Taken together, these results indicate that the interaction between ING4 and RARβ is important for inhibiting cell migration and invasion in Tsc2-deficient cells.
miR-29b inhibition suppresses tumor growth in vivo and miR-29b expression negatively correlates with RARβ in human kidney and bladder cancer
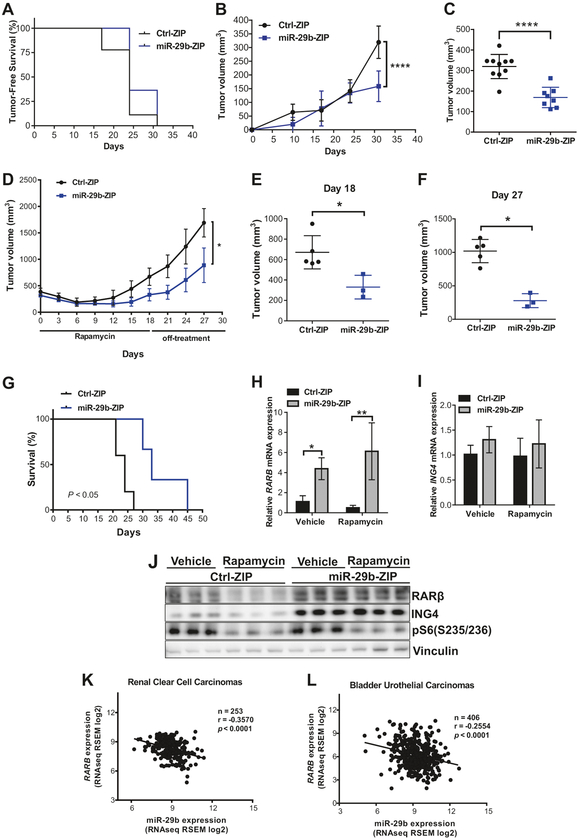
To determine how miR-29b impacts Tsc2-deficient cell growth in vivo, we generated Tsc2-deficient ERL4 cells expressing either Ctrl-ZIP or miR-29b-ZIP (miR-29b knockdown) and inoculated the cells subcutaneously in immunodeficient mice. miR-29b inhibition did not affect tumor-free survival (Fig. 7a). However, inhibition of miR-29b significantly decreased tumor growth (Fig. 7b): at 30 days post injection, miR-29b-knockdown cells formed significantly smaller tumors (P < 0.0001) (Fig. 7c). To examine the effect of combined rapamycin treatment with miR-29b inhibition, we treated mice carrying Ctrl-ZIP or miR-29b-ZIP tumors of 300–400 mm3 with rapamycin (3 mg/kg) every 2 days for 3 weeks and assessed tumor growth on and off treatment. We found that combination therapy of rapamycin plus miR-29b inhibition suppressed tumor regrowth by 56% (P < 0.01) compared with mice carrying Ctrl-ZIP tumors receiving rapamycin alone (Fig. 7d, e). The tumor growth rate was remarkably slower in miR-29b-ZIP xenografts compared with Ctrl-ZIP xenografts following rapamycin cessation, with a net decrease of 75% in tumor volume at 14 days following the last treatment (P < 0.05) (Fig. 7f). The combination of rapamycin and miR-29b inhibition also extended the median survival of the mice from 25 to 34 days (Fig. 7g).
Fig. 7.
miR-29b functions as an oncomiR in vivo and miR-29b levels are inversely correlated with RARβ expression in human tumors. a–c Mice were subcutaneously injected with 2 × 106 Tsc2-deficient ERL4 cells expressing miR-29b-ZIP or Ctrl-ZIP. miR-29b-ZIP (n = 19) expressing tumors showed no difference in tumor-free survival compared with Ctrl-ZIP (n = 20). Statistical significance was determined by Log-rank test (a). Tumor growth was analyzed by caliper measurement over 30 days (b). Mean tumor volume ± SD 30 days post injection indicated miR-29b-ZIP-expressing cells formed smaller tumors. Statistical significance was determined by Mann–Whitney’s U test (c). d–g miR-29b-ZIP (n = 3) or Ctrl-ZIP (n = 5) tumors 300–400 mm3 in size were subjected to rapamycin treatment (3 mg/kg, every 2 days for 3 weeks and 9 treatments in total). Tumor growth was analyzed by caliper measurement during rapamycin treatment and 9 days following rapamycin cessation and data are presented as mean ± SD. Statistical significance was determined by Mann–Whitney’s U test (d). Mean individual tumor volume ± SD 18 days after rapamycin treatment began. Statistical significance was determined by Mann–Whitney’s U test (e). The average tumor volume difference of miR-29b-ZIP or Ctrl-ZIP after the last rapamycin treatment (day 18) and the final measurement (day 27) is shown and data are presented as mean ± SD. Statistical significance was determined by Mann–Whitney’s U test (f). Survival analysis of mice-bearing miR-29b-ZIP or Ctrl-ZIP tumors treated by rapamycin and statistical significance was determined by Log-rank test (g). h–i miR-29b-ZIP or Ctrl-ZIP tumors 300–400 mm3 in size were subjected to short-term vehicle or rapamycin treatment (3 mg/kg, every 2 days for 3 treatments). Tumor RNA was extracted and analyzed for mRNA expression of RARβ (h) or ING4 (i). Data are presented as mean ± SD relative to vehicle-treated Ctrl-ZIP of n = 3. Data for bar graphs were calculated using two-way ANOVA followed by Bonferroni’s posttest for multiple comparisons. Tumor tissue lysates were prepared and analyzed by immunoblotting with RARβ, ING4, pS6, or vinculin antibodies. *P < 0.05; **P < 0.01; ****P < 0.0001. Comparison of miR-29b and RARβ expression levels in renal clear cell carcinomas (n = 253) (k) and bladder urothelial carcinomas (n = 406) (l) from TCGA. Pearson correlation coefficient (r) and values are indicated
To determine whether miR-29b inhibition and rapamycin treatment affect RARβ and ING4 protein expression, we treated mice-bearing tumors of 300–400 mm3 in size with rapamycin (3 mg/kg) every 2 days for 3 treatments in total. RT-qPCR analysis of tumor RNA demonstrated that miR-29b knockdown increased RARβ mRNA levels by fourfold (P < 0.05) in vehicle-treated tumors and by sixfold (P <0.01) in rapamycin-treated tumors (Fig. 7h). As expected, ING4 mRNA levels were unchanged in both vehicle-treated and rapamycin-treated tumors (Fig. 7i). Western blotting of tumor lysates confirmed that rapamycin downregulated RARβ protein expression while miR-29b inhibition restored its expression (Fig. 7j). These data support the model suggested by our in vitro data, that miR-29b inhibition impedes Tsc2-deficient cell growth and that RARβ and ING4 are downstream targets of miR-29b in vivo.
Finally, we examined whether miR-29b expression correlates with RARβ in human cancer specimens using TCGA RNA-sequencing data from the renal clear cell carcinomas and bladder urothelial carcinomas. We focused on these tumors since they can be both associated with TSC gene mutations [20, 21]. In agreement with our experimental data, this database also showed a significant negative correlation of miR-29b expression with RARβ in renal clear cell carcinomas (r = −0.3537, P < 0.0001) (Fig. 7k) and bladder urothelial carcinomas (r = −0.2554, P < 0.0001) (Fig. 7l).
Discussion
miRNAs are involved in many aspects of tumor biology, and while the TSC pathway is directly involved in microRNA biogenesis via Drosha [22, 23], relatively little is known about the role of miRNA in the pathogenesis and therapy of TSC and LAM. We have previously discovered that miR-29b is a rapamycin-regulated miRNA, but the functional significance and therapeutic impact of miR-29b induction by rapamycin has not been previously explored in TSC. Here, we report that miR-29b functions as an oncogenic miRNA in Tsc2-deficient cells by promoting cell proliferation, migration, invasion and in vivo tumor growth. We demonstrate a novel mechanism of miR-29b-promoted Tsc2-deficient tumor growth by targeting the tumor suppressors RARβ and ING4, and underscore miR-29b as a new promising therapeutic target for TSC/LAM. Given that mTORC1 hyperactivation and miR-29b dysregulation are relevant to many human tumors [15], the mechanistic insights gained from this study have broad potential implications.
Our previous screen in which miR-29b was identified as a rapamycin-regulated microRNA (RapamiR) included only a single cell type (angiomyolipoma-derived cells). In this study, we found that rapamycin increased miR-29b expression in multiple Tsc2-deficient cells lines. We also found that miR-29b is expressed at lower levels in Tsc2-deficient cells compared with Tsc2-expressing wild-type cells, consistent with TSC-and mTORC1-dependence and with a prior report that mTORC1 is a negative regulator of miRNA biosynthesis [23]. In contrast, another RapamiR, miR-21, which is also upregulated by rapamycin, is not Tsc2 dependent [16, 24] highlighting the complexity of miRNA regulation by the TSC/mTORC1 pathway.
Our results indicate that miR-29b is a positive regulator of cell growth, migration, invasion, and in vivo tumorigenesis in Tsc2-deficient cells. The existing literature suggests that miR-29b plays a dual role in tumor progression, with both tumor suppressive and tumor-promoting activities. In many studies, miR-29b is described as a tumor suppressive miRNA by inhibiting cell proliferation [25], migration [26], invasion [27, 28], and DNA methylation [29, 30] as well as promoting apoptosis [31, 32] and differentiation [33]. In contrast, there is growing evidence that miR-29b has oncogenic effects. In acute myeloid leukemia miR-29b targets an important tumor suppressor, TET2 for degradation [34]. Inhibition of miR-29b in bladder cancer cells decreased growth [35], and overexpression of miR-29b in breast cancer enhanced cell migration, invasion, and resistance to apoptosis as a results of targeting PTEN, C1q and tumor necrosis factor-related protein 6 (C1QTNF6), SPARC, and COL4A2 [36, 37]. In Tsc2-deficient cells, miR-29b clearly functions as an oncogenic miRNA, suggesting for the first time that the genetic context influences miR-29’s actions.
Rapamycin has a cytostatic impact on Tsc2-deficient cells. In TSC clinical trials and in preclinical studies, there is an incomplete tumor regression, with tumor regrowth upon treatment discontinuation. We hypothesize that upregulation of miR-29b, which is protumorigenic, is one of the key mechanisms driving this cytostatic, partial response to rapamycin. This hypothesis is supported by our finding that suppression of miR-29b combined with rapamycin treatment further decreased Tsc2-deficient cell growth, migration, and invasion. Therefore, the combination of rapamycin and miR-29b inhibition could have therapeutic benefit in TSC and other mTORC1-hyperactive tumors. Combination therapeutic strategies such as this, that potentiate the actions of rapamycin and thereby decrease the need for continuous, lifelong therapy, could lead to significant clinical benefit for both children and adults living with TSC.
The mechanisms through which miR-29b executes its tumor-promoting function in TSC involve inhibition of the RARβ, a previously unidentified direct target of miR-29b. RARβ expression is upregulated in Tsc2-deficient cells with knockdown of miR-29b and upregulated in cells over-expressing miR-29b; this is the result of targeting of the 3′ UTR of the RARβ transcript by miR-29b. RARβ is a member of the steroid hormone receptor superfamily of nuclear transcriptional regulators. Like other members of this family, RARβ binds retinoic acid, the biologically active form of vitamin A [38]. Loss of RARβ2 expression has been associated with carcinogenesis in humans, and resistance to retinoid acid treatment [39]. In Tsc2-deficient cells, forced-expression of RARβ2 in the context of miR-29b overexpression inhibited cell migration and invasion, consistent with a model in which down regulation of RARβ2 is necessary for the oncogenic actions of miR-29b.
The mechanisms through which RARβ2 acts as a tumor suppressor include its ability to inhibit the expression of EGFR, activating protein-1 (AP-1), COX-2, and ERK1/2 [40]. Many of these studies focused on genes regulated via RARβ2’s transcriptional activity, either directly or indirectly. We have identified a novel mechanism through which RARβ2 can mediate tumor suppression, via forming a protein complex with the ING4, which is also a tumor suppressor. We found that RARβ binds to ING4 in endogenous coimmunoprecipitation analyses. Furthermore, this interaction has functional relevance, as RARβ2-mediated inhibition of cell migration and invasion was elevated by siRNA knockdown of ING4. These studies suggest that ING4 is a previously unrecognized critical downstream effector of RARβ.
ING4, a member of the ING tumor suppressor family, is involved in many cancer-related cellular processes including proliferation, migration, apoptosis, angiogenesis, hypoxia, DNA damage response, and contact inhibition [41]. Interestingly, ING4 has been demonstrated to mediate chromatin remodeling by interacting with tri-methylated histone H3 at lysine 4 [42] and histone acetyltransferase binding to ORC histone acetyltransferase complex (HBO1 HAT) [43]. Furthermore, ING4 is a negative regulator of nuclear factor-κB (NF-κB) activity [44] by acting as a E3 ubiquitin ligase to induce NF-κB/p65 degradation [45]. Of particular relevance to our findings, ING4 suppresses melanoma cell migration and invasion by regulating Kang-Ai 1, a well-known tumor metastasis suppressor in prostate cancer via the regulation of NF-κB/p65 [46, 47]. In addition, ING4 interacts with liprin α1 and inhibits liprin α1–induced cell motility [48]. We speculate that the interaction between RARβ and ING4 suppresses Tsc2-deficient cell migration and invasion via KAI, NF-κB/p65, or liprin α1, but further studies will be needed to clarify these mechanisms. This miR-29b/RARβ/ING4 axis could be relevant to pulmonary LAM, which is believed to result from the metastasis of benign smooth muscle-like cells to the lung. These cells can lead to cystic lung destruction, oxygen dependency, and death [49–51]. Thus, restoration of RARβ2 and ING4 by inhibiting miR-29b could be beneficial in reducing the migratory and invasive potential of LAM cells. RARβ2 can be activated by all-trans retinoic acid, which is already approved by US Food and Drug Administration for the treatment of acute promyelocytic leukemia.
In conclusion, miR-29b is upregulated by rapamycin in Tsc2-deficient cells. Inhibition of miR-29b suppresses Tsc2-deficient tumor progression by downregulating cell growth, migration, and invasion via RARβ, a direct and novel target of miR-29b, and ING4, a tumor suppressor that directly interacts with RARβ. These findings may have clinical relevance for the adults and children with TSC and for the women with LAM, and for sporadic tumors with mTORC1 hyperactivation (estimated to represent 50% of all human malignancies).
Materials and methods
Lentiviral gene delivery and establishment of stable cell lines
To make lentivirus, plasmids encoding Ctrl-ZIP, miR-29b-ZIP, miR-Ctrl, miR-29b-1, or miR-29b-2 (System Biosciences, USA) were cotransfected with the package (pCMV-dR8.91) and envelop (pCMV-VSVG) plasmids into 293T cells. The virus-containing supernatant was harvested, filtered with a 0.45 μm filter, and used to infect cells with 10 μg/mL polybrene.
Heterogeneous pools of miR-29b-knockdown cells were generated by transducing Ctrl-ZIP or miR-29b-ZIP lentivirus into MEFs or ERL4 cells. Clones were selected in complete growth medium containing 10 μg/ml of puromycin for 4 days, FACS sorted for GFP expression by the ZIP construct then maintained in 5 μg/ml of puromycin. Heterogeneous pools of miR-29b overexpressing cells were generated, selected, and maintained as described above by introducing miR-Ctrl, miR-29b-1, or miR-29b-2 lentivirus into MEFs.
Transfection
For overexpression studies, 3 μg of mouse pCMV6-RARβ2-Myc-DDK, pCMV6-ING4, or pCMV6 control vector was transiently transfected into Tsc2−/− MEFs using lipofectamine 3000 transfection kit (Invitrogen, USA). After 24 h, cells were lysed to analyze protein expression or plated for cell migration or invasion assays. To overexpress RARβ2 and to knockdown ING4, Tsc2−/− MEFs were cotransfected with 3 μg of pCMV6-RARβ2-Myc-DDK or 25 nM SMARTpool ING4 siRNA (Dharmacon, USA), respectively. Forty-eight hours following transfection, cells were lysed to analyze protein expression or plated for cell migration and invasion assays.
Western blot analysis
Cells were lysed using 1% Triton in TBS containing protease and phosphatase inhibitors. Tumors were lysed using 1X RIPA buffer containing protease and phosphatase inhibitors. Equal amounts of total protein were separated by electrophoresis on a 4–12% Bis-Tris gel and transferred to a PVDF membrane. Blots were blocked in 5% BSA, then incubated with primary antibodies (See Supplementary Table 3) overnight at 4 °C. The membranes were washed and then incubated with horseradish peroxidase-conjugated secondary antibodies at room temperature for 1 h. Bands were detected using SuperSignal West Pico Plus chemiluminescence substrate (Thermo Fisher Scientific, USA) and photographed with G:BOX blot imaging system (Syngene, UK).
RNA isolation, miRNA isolation, and RT-qPCR
Total RNA from cells or tumors was extracted using RNeasy RNA isolation kit (Qiagen, Germany). MiRNA was extracted using miRCURY RNA isolation kit (Exiqon, USA). Extracted total RNA was reverse transcribed into cDNA using High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, USA) and miRNA was reverse transcribed using TaqMan Advanced miRNA cDNA synthesis kit (Thermo Fisher Scientific, USA). For gene expression, quantitative real-time PCR (qPCR) was performed using Taqman Universal Master Mix II, with UNG and for miRNA expression qPCR was performed using Taqman Fast Advanced Master mix (Thermo Fisher Scientific, USA). Real-time PCR was performed using an Applied Biosystems StepOnePlus Real-time PCR system (Thermo Fisher Scientific, USA) with miRNA-specific primers or gene-specific primers (see Supplementary Table 4). Values represent the average of three independent experiments, normalized to β-actin for gene expression or miR-320a for miR-29b expression.
To quantify the absolute amounts of miR-29b, a synthesized Caenorhabditis elegans microRNA mimic (Thermo Fisher Scientific, USA), cel-miR-39 was used as the spike-in control. miRNA was isolated from cells using the PureLink miRNA isolation kit (Thermo Fisher Scientific, USA) following the manufacturer’s protocol and cel-miR-39 (250 pmol) was added to each sample when cells were lysed. Extracted miRNA was reverse transcribed as described above. In addition, synthetic miRVana mmu-miR-29b-3p (ID: MC10103) and cel-miR-39–3p (ID: MC10956) mimics (Thermo Fisher Scientific, USA) were serially diluted to final concentrations of 2000 nM, 200 nM, 20 nM, 2 nM, 0.2 nM, 0.02 nM, 2 pM, 0.2 pM. Serial dilutions of hsa/mmu-miR-29b and cel-miR-39 mimics were reverse transcribed and assayed concurrently with miRNA extracted from cells. For each qPCR plate, standard curves for hsa/mmu-miR-29b and cel-miR-39 were included to convert the cycle threshold (Ct) values of each sample into the corresponding number of miRNA copies. Finally, the exact copies of miR-29b are calculated by the yield ratios of added cel-miR-39 to obtain the absolute amounts of miR-29b in cells.
Crystal violet proliferation and anchorage-independent growth assay
Cells were seeded (0.5 × 103 per well) in 96-well plates in 75 μL growth media (without selection antibiotics) on day 0. Rapamycin (LC laboratories, USA) (20 nM) or vehicle (DMSO) was added when cells were attached. Cells were fixed in 10% formalin for 10 min then stained with 0.5% crystal violet for 20 min. Excess crystal violet was removed, and cells were washed with H2O. Then, 100% of methanol was added to dissolve crystal violet and absorbance was read at 540 nm using a Synergy HT Multi-Mode Microplate Reader (BioTek, USA). Experiments were performed in triplicate.
Anchorage-independent growth assays were done in sixwell 35 mm tissue culture dishes containing a bottom layer of 0.7% (w/v) agarose in complete growth medium and a top layer of 0.3% agarose solution mixed with 1.5 × 104 Tsc2−/− MEFs in presence of rapamycin (5 nM) or vehicle (DMSO) in triplicate and incubated at 37 °C for 2 weeks. Colonies were manually scored using ImageJ analysis software from images taken using an Olympus SZH10 Stereo microscope (Olympus, Japan) and a Nikon D3000 camera (Nikon, Japan).
Migration and invasion assays
For migration assays, cells were seeded at a density of 5 × 104 cells in the upper chamber of a 6.5-mm Transwell with 8.0-μm pore polycarbonate membrane inserts (Corning, USA) in 100 μl of serum-free DMEM containing rapamycin (20 nM) or vehicle (DMSO) for 6 h at 37 °C. Complete growth medium was placed in the bottom compartment as a chemoattractant. After incubation, nonmigrated cells were wiped off from the upper surface using cotton swabs. The migrated cells on the lower surface were fixed and stained using the Differential stain kit (Newcomer Supply, USA). Five random fields of migrated cells were imaged using a ×15 objective on an Olympus FSX100 microscope; migrated cells were quantified using ImageJ analysis software. The experiments were performed in duplicate.
For invasion assays, cells were seeded at a density of 2.5 × 104 cells in the upper chamber of a 6.5-mm Transwell with 8.0-μm PET membrane precoated with Matrigel (Corning, USA) in 100 μl of serum-free DMEM-containing rapamycin (20 nM) or vehicle (DMSO) for 24 h at 37 °C. Invaded cells were fixed, stained, imaged, and counted as described above.
Co-immunoprecipitation
Cells were harvested, washed with PBS and lysed in 1X RIPA buffer (Cell Signaling, USA) containing protease and phosphatase inhibitor cocktails with constant rotation at 4 °C for 30 min. Cell lysates (2000–2200 μg) were incubated with RARβ, ING4, or rabbit IgG control antibodies together with protein G Sepharose beads (GE healthcare, USA) at 4 °C overnight. The immune complexes were isolated via centrifugation and washed four times with 1X RIPA buffer. The proteins were then separated with an SDS/polyacrylamide gel.
Luciferase reporter assay
Ctrl-ZIP, miR-29b-ZIP, Ctrl-miR, miR-29b-1, or miR-29b-2 stable expressing Tsc2−/− MEFs were seeded in sixwell dishes, then transfected using lipofectamine 3000 (Thermo Fisher Scientific, USA) with 2 μg of the 3′UTR dual reporter plasmid (pEZX-MT05–3′UTR RARβ), purchased from GeneCopoeia, USA. Cells were treated with rapamycin (20 nM) or vehicle (DMSO) 24 h following transfection. The luciferase activity of the cultured supernatant was measured 48 h after transfection using Secrete-Pair™ Dual Luminescent Kit (GeneCopoeia, USA) according to the manufacturer’s instructions.
In vivo tumorigenicity assay
All studies involving mice were undertaken after receiving ethical approval from Boston Children’s Hospital Animal Care and Use Committee. ERL4 cells stably expressing Ctrl-ZIP or miR-29b-ZIP (2.0 × 106) were mixed with phenol-red free Matrigel (BD Biosciences, USA) 1:1 (v/v) to obtain a final volume of 150 μl. The cell/Matrigel mix was injected unilaterally into 7–8-week-old female athymic Balbc nu/nu mice (Charles River, USA) using a 21G needle. Once the tumors become palpable, the width and length of palpable tumors were measured with digital calipers. Tumor volume was calculated using the equation (length) × (width)2/2. When tumors reached 300–400 mm3 in volume, mice were treated with rapamycin (3 mg/kg), every 2 days for a total of 3 treatments or every 2 days for a total of 9 treatments over 3 weeks. Four hours following the last treatment, tumors were harvested, snap frozen then stored at −80 °C for RNA and protein analysis. For all the animal studies, we have chosen to use at least three mice per group to ensure adequate power to detect difference between groups. All animal studies were blinded and conducted under the guidelines of Boston Children’s Hospital Animal Care and Use Committee.
Data source
To analyze the expression levels of RARβ and miR-29b, mRNA-seq and miRNA-seq data of kidney renal clear cell carcinomas and bladder urothelial carcinomas cohorts were obtained from the Broad Institute GDAC (TCGA data version 20150601).
Statistical analysis
Data are presented as the mean ± SEM. All statistical analyses and P values were determined by an unpaired Student’s t-test, Mann–Whitney’s U test, one-way ANOVA or two-way ANOVA followed by Bonferroni’s posttest using GraphPad Prism 6.0 (GraphPad software). The mRNA and miRNA expression data obtained from TCGA were analyzed for Pearson’s correlation coefficients. A P value <0.05 was regarded as significant.
Supplementary Material
Acknowledgements
We thank Lorena Pantano of the Harvard Chan Bioinformatics Core for assistance with RNAseq analysis. This work was supported by the Engles Program in TSC and LAM research, Department of Defense grant W81XWH-15–1–0263 TS14003 (EPH) and a postdoctoral Fellowship from Tuberous Sclerosis Alliance (HJL).
Footnotes
Ethical approval All studies involving mice were undertaken after receiving ethical approval from Boston Children’s Hospital Animal Care and Use Committee.
Conflict of interest The authors declare that they have no conflict of interest.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information The online version of this article (https://doi.org/10.1038/s41388-019-0957-5) contains supplementary material, which is available to authorized users.
References
- 1.Henske EP, Jozwiak S, Kingswood JC, Sampson JR, Thiele EA. Tuberous sclerosis complex. Nat Rev Dis Prim. 2016;2:16035. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Sahra I, Manning BD. mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol. 2017;45:72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheadle JP, Reeve MP, Sampson JR, Kwiatkowski DJ. Molecular genetic advances in tuberous sclerosis. Hum Genet. 2000;107:97–114. [DOI] [PubMed] [Google Scholar]
- 4.Carsillo T, Astrinidis A, Henske EP. Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. Proc Natl Acad Sci USA. 2000;97:6085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosset C, Netto CBO, Ashton-Prolla P. TSC1 and TSC2 gene mutations and their implications for treatment in tuberous sclerosis complex: a review. Genet Mol Biol. 2017;40:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissler JJ, McCormack FX, Young LR, Elwing JM, Chuck G, Leonard JM, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358:140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bissler JJ, Kingswood JC, Radzikowska E, Zonnenberg BA, Frost M, Belousova E, et al. Everolimus for angiomyolipoma associated with tuberous sclerosis complex or sporadic lymphangioleiomyomatosis (EXIST-2): a multicentre, randomised, double-blind, placebo-controlled trial. Lancet. 2013;381:817–24. [DOI] [PubMed] [Google Scholar]
- 8.Franz DN, Belousova E, Sparagana S, Bebin EM, Frost M, Kuperman R, et al. Efficacy and safety of everolimus for sub-ependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet. 2013;381:125–32. [DOI] [PubMed] [Google Scholar]
- 9.Krueger DA, Care MM, Holland K, Agricola K, Tudor C, Mangeshkar P, et al. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N Engl J Med. 2010;363:1801–11. [DOI] [PubMed] [Google Scholar]
- 10.Young L, Lee HS, Inoue Y, Moss J, Singer LG, Strange C, et al. Serum VEGF-D a concentration as a biomarker of lymphangioleiomyomatosis severity and treatment response: a prospective analysis of the Multicenter International Lymphangioleiomyomatosis Efficacy of Sirolimus (MILES) trial. Lancet Respir Med. 2013;1:445–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Disco. 2017;16:203–22. [DOI] [PubMed] [Google Scholar]
- 12.Yan B, Guo Q, Fu FJ, Wang Z, Yin Z, Wei YB, et al. The role of miR-29b in cancer: regulation, function, and signaling. Onco Targets Ther. 2015;8:539–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mott JL, Kurita S, Cazanave SC, Bronk SF, Werneburg NW, Fernandez-Zapico ME. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J Cell Biochem. 2010;110:1155–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–71. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Zhang G, Wu JH, Jiang CP. Diverse roles of miR-29 in cancer (review). Oncol Rep. 2014;31:1509–16. [DOI] [PubMed] [Google Scholar]
- 16.Trindade AJ, Medvetz DA, Neuman NA, Myachina F, Yu J, Priolo C, et al. MicroRNA-21 is induced by rapamycin in a model of tuberous sclerosis (TSC) and lymphangioleiomyomatosis (LAM). PLoS ONE. 2013;8:e60014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmer A, Zimmer A. Induction of a RAR beta 2-lacZ transgene by retinoic acid reflects the neuromeric organization of the central nervous system. Development. 1992;116:977–83. [DOI] [PubMed] [Google Scholar]
- 18.de The H, Marchio A, Tiollais P, Dejean A. Differential expression and ligand regulation of the retinoic acid receptor alpha and beta genes. EMBO J. 1989;8:429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogorek B, Lam HC, Khabibullin D, Liu HJ, Nijmeh J, Triboulet R, et al. TSC2 regulates microRNA biogenesis via mTORC1 and GSK3beta. Hum Mol Genet. 2018;27:1654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye P, Liu Y, Chen C, Tang F, Wu Q, Wang X, et al. An mTORC1-Mdm2-Drosha axis for miRNA biogenesis in response to glucose- and amino acid-deprivation. Mol Cell. 2015;57:708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam HC, Liu HJ, Baglini CV, Filippakis H, Alesi N, Nijmeh J, et al. Rapamycin-induced miR-21 promotes mitochondrial homeostasis and adaptation in mTORC1 activated cells. Oncotarget. 2017;8:64714–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong JN, Yu J, Lin HS, Zhang XH, Yin XL, Xiao Z, et al. The role, mechanism and potentially therapeutic application of microRNA-29 family in acute myeloid leukemia. Cell Death Differ. 2014;21:100–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita T, Nohata N, Hanazawa T, Kikkawa N, Yamamoto N, Yoshino H, et al. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br J Cancer. 2013;109:2636–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poudyal D, Cui X, Le PM, Hofseth AB, Windust A, Nagarkatti M, et al. A key role of microRNAfigure-29b for the suppression of colon cancer cell migration by American ginseng. PLoS One. 2013;8:e75034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ru P, Steele R, Newhall P, Phillips NJ, Toth K, Ray RB. miRNA-29b suppresses prostate cancer metastasis by regulating epithelialmesenchymal transition signaling. Mol Cancer Ther. 2012;11:1166–73. [DOI] [PubMed] [Google Scholar]
- 29.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. [DOI] [PubMed] [Google Scholar]
- 33.Rossi M, Pitari MR, Amodio N, Di Martino MT, Conforti F, Leone E, et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol. 2013;228:1506–15. [DOI] [PubMed] [Google Scholar]
- 34.Cheng J, Guo S, Chen S, Mastriano SJ, Liu C, D’Alessio AC, et al. An extensive network of TET2-targeting MicroRNAs regulates malignant hematopoiesis. Cell Rep. 2013;5:471–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu F, Zhang Q, Cheng W, Zhang Z, Wang J, Ge J. Effect of miR-29b-1* and miR-29c knockdown on cell growth of the bladder cancer cell line T24. J Int Med Res. 2013;41:1803–10. [DOI] [PubMed] [Google Scholar]
- 36.Wang C, Bian Z, Wei D, Zhang JG. miR-29b regulates migration of human breast cancer cells. Mol Cell Biochem. 2011;352:197–207. [DOI] [PubMed] [Google Scholar]
- 37.Wang C, Gao C, Zhuang JL, Ding C, Wang Y. A combined approach identifies three mRNAs that are down-regulated by microRNA-29b and promote invasion ability in the breast cancer cell line MCF-7. J Cancer Res Clin Oncol. 2012;138:2127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun SY, Wan H, Yue P, Hong WK, Lotan R. Evidence that retinoic acid receptor beta induction by retinoids is important for tumor cell growth inhibition. J Biol Chem. 2000;275:17149–53. [DOI] [PubMed] [Google Scholar]
- 39.Ren M, Pozzi S, Bistulfi G, Somenzi G, Rossetti S, Sacchi N. Impaired retinoic acid (RA) signal leads to RARbeta2 epigenetic silencing and RA resistance. Mol Cell Biol. 2005;25:10591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song S, Lippman SM, Zou Y, Ye X, Ajani JA, Xu XC. Induction of cyclooxygenase-2 by benzo[a]pyrene diol epoxide through inhibition of retinoic acid receptor-beta 2 expression. Oncogene. 2005;24:8268–76. [DOI] [PubMed] [Google Scholar]
- 41.Cui S, Gao Y, Zhang K, Chen J, Wang R, Chen L. The emerging role of inhibitor of growth 4 as a tumor suppressor in multiple human cancers. Cell Physiol Biochem. 2015;36:409–22. [DOI] [PubMed] [Google Scholar]
- 42.Palacios A, Munoz IG, Pantoja-Uceda D, Marcaida MJ, Torres D, Martin-Garcia JM, et al. Molecular basis of histone H3K4me3 recognition by ING4. J Biol Chem. 2008;283:15956–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. [DOI] [PubMed] [Google Scholar]
- 44.Byron SA, Min E, Thal TS, Hostetter G, Watanabe AT, Azorsa DO, et al. Negative regulation of NF-kappaB by the ING4 tumor suppressor in breast cancer. PLoS ONE. 2012;7:e46823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou Y, Zhang Z, Xu Q, Wang H, Xu Y, Chen K. Inhibitor of growth 4 induces NFkappaB/p65 ubiquitin-dependent degradation. Oncogene. 2014;33:1997–2003. [DOI] [PubMed] [Google Scholar]
- 46.Tang Y, Cheng Y, Martinka M, Ong CJ, Li G. Prognostic significance of KAI1/CD82 in human melanoma and its role in cell migration and invasion through the regulation of ING4. Carcinogenesis. 2014;35:86–95. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Martinka M, Li G. Role of ING4 in human melanoma cell migration, invasion and patient survival. Carcinogenesis. 2008;29:1373–9. [DOI] [PubMed] [Google Scholar]
- 48.Shen JC, Unoki M, Ythier D, Duperray A, Varticovski L, Kumamoto K, et al. Inhibitor of growth 4 suppresses cell spreading and cell migration by interacting with a novel binding partner, liprin alpha1. Cancer Res. 2007;67:2552–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henske EP. Metastasis of benign tumor cells in tuberous sclerosis complex. Genes Chromosomes Cancer. 2003;38:376–81. [DOI] [PubMed] [Google Scholar]
- 50.McCormack FX, Travis WD, Colby TV, Henske EP, Moss J. Lymphangioleiomyomatosis: calling it what it is: a low-grade, destructive, metastasizing neoplasm. Am J Respir Crit Care Med. 2012;186:1210–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertolini F, Casarotti G, Righi L, Bollito E, Albera C, Racca SA, et al. Human renal angiomyolipoma cells of male and female origin can migrate and are influenced by microenvironmental factors. PLoS ONE. 2018;13:e0199371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.