Abstract
The ligands for the natural killer group 2 (NKG2D) protein render tumor cells susceptible to NKG2D-dependent immune cell attack. However, cancer cells escape from immune surveillance by downregulating NKG2D ligands. We previously discovered that engagement of activated CD8+ T cells and tumor cells induces NKG2D ligands on tumor cells, but the underlying mechanism remains to be defined. Both in vivo mouse tumor models and in vitro cell assays were performed to study the downstream signaling. Our results supported the notion that, upon engagement with the cognate receptors, CD137 ligand and CD40 initiates activation of nuclear factor-kappa B (NF-κB) signaling in tumor cells even in the absence of CD8+ T cells. Like tumor and CD8+ T cell contact-dependent NKG2D ligand induction, this CD137L/CD40-mediated signaling activation was associated with elevated levels of acetyltransferase P300/CBP-associated factor (PCAF), whereas inhibition of phosphorylated NF-κB abrogated PCAF induction. Although stimulation of CD137L/CD40-mediated signaling is vital, inflammatory cytokines, including interferon gamma (IFNγ) and TNFα, also facilitate NKG2D ligand–induced immune surveillance via both facilitating T cell chemotaxis and CD137L/CD40 induced NF-κB/PCAF activation. Collectively, our results unveil a novel mechanism of NKG2D ligand upregulation involving reverse signaling of CD40 and CD137L on tumor cells which, along with inflammatory cytokines IFNγ and TNFα, stimulate downstream NF-κB and PCAF activation. Understanding this mechanism may help in development of induced NKG2D ligand–dependent T cell therapy against cancers.
Keywords: NKG2D ligands, CD8+ T cells, TNFRSF, NF-κB, PCAF
Introduction
The natural killer group 2, member D (NKG2D) protein is expressed on all natural killer cells and activated CD8+ T cells, as well as some natural killer T cells, γδ T cells, and CD4+ T cells1. NKG2D-expressing immune cells recognize and eradicate tumor cells and infected cells by interacting with its cognate ligands. These ligands are expressed at undetectable levels in normal cells, but their expression is elevated by infection and oncogenic transformation2. In humans, the diverse NKG2D ligands belong to two families: the major histocompatibility complex (MHC) class I chain-related family (MICA and MICB) and the UL16-binding protein family (ULBP1–6). In mice, MICA and MICB have no cognate proteins, and the ULBP family is subdivided into the retinoic acid early inducible-1 (Rae-1), histocompatibility 60, and transmembrane protein murine UL16-binding protein-like transcript 1 subfamilies3, 4.
Because of its role in enhancing tumor immune surveillance, understanding how NKG2D ligand expression is regulated in tumor cells is critical. The established mechanisms of NKG2D ligand regulation include DNA damage pathway–mediated stabilization of NKG2D ligand mRNA5, 6, an E2 factor–dependent hyperproliferative mechanism7, the nuclear factor-kappa B (NF-κB) signaling–induced inflammation mechanism8, 9, and the recently identified histone acetyltransferase (HAT) CREB-binding protein (CBP)/p300–engaged epigenetic mechanism10. These mechanisms were verified in vitro, but the in vivo mechanisms have not yet been reported in the literature. Our group recently discovered CD8+ T cell–dependent upregulation of NKG2D ligands on tumor cells in vitro and in vivo11. We found that treating solid tumors with interleukin (IL)-12 plus doxorubicin may recruit CD8+ T cells to engage with tumor cells, thereby increasing NKG2D ligand expression on the tumor cell surface. However, the precise mechanism by which CD8+ T cells induce NKG2D ligands on tumor cells is yet to be identified.
Although the co-stimulatory molecules are not widely expressed on all types of cancer cells, they are present on certain tumor cells and have an impact on tumor immunity. For instance, CD40, CD70, and CD137 ligands, of the tumor necrosis factor receptor (TNFR) superfamily (TNFRSF), are expressed on a variety of solid tumor cells12–17. In addition to stimulating immune cells, engagement of TNFRSF receptors by T cells transports positive signals back to tumor cells. These signals regulate gene expression and cellular processes in tumor cells16 through intracellular interaction with TNFR-associated factors (TRAFs)17, 18 and further potentiate a number of signaling pathways. The best-characterized signaling activation upon TNFRSF ligation is that of the NF-κB pathway, which regulates gene expression involved in the inflammatory response and in cell survival19–22.
NF-κB was reported to regulate human NKG2D ligand MICA8. There also is evidence that the NF-κB heterodimer p50-p65 was associated with HAT (CBP/p300) and recruited the HAT complex member p300/CBP-associated factor (PCAF)23–25, which in turn facilitated the activation of p6524. HATs, therefore, are coactivators to bridge NF-κB and expression of its target genes23. Nevertheless, whether NF-κB signaling accounts for HAT-mediated NKG2D ligand upregulation has yet to be reported.
We hypothesized that engagement between CD8+ T cells and tumor cells stimulates NF-κB signaling, which recruits HAT to induce NKG2D ligand expression on tumor cells. This study shows that TNFRSF member CD137 ligand and CD40 on tumor cells interact with their cognate receptors on CD8+ T cells and transmit stimulatory signals in tumor cells to trigger the intracellular NF-κB pathway. The activated NF-κB subsequently stimulates the HAT PCAF, thereby upregulating NKG2D ligand expression on the tumor cell surface. Meanwhile, CD8+ T cells release inflammatory cytokines that augment the production of chemoattractants to recruit abundant effector immune cells to the tumor site, thereby amplifying the signals of NKG2D ligand induction and strengthening tumor immune surveillance.
Results
Engagement via TNFRSF is necessary for Rae-1 upregulation on tumor cells
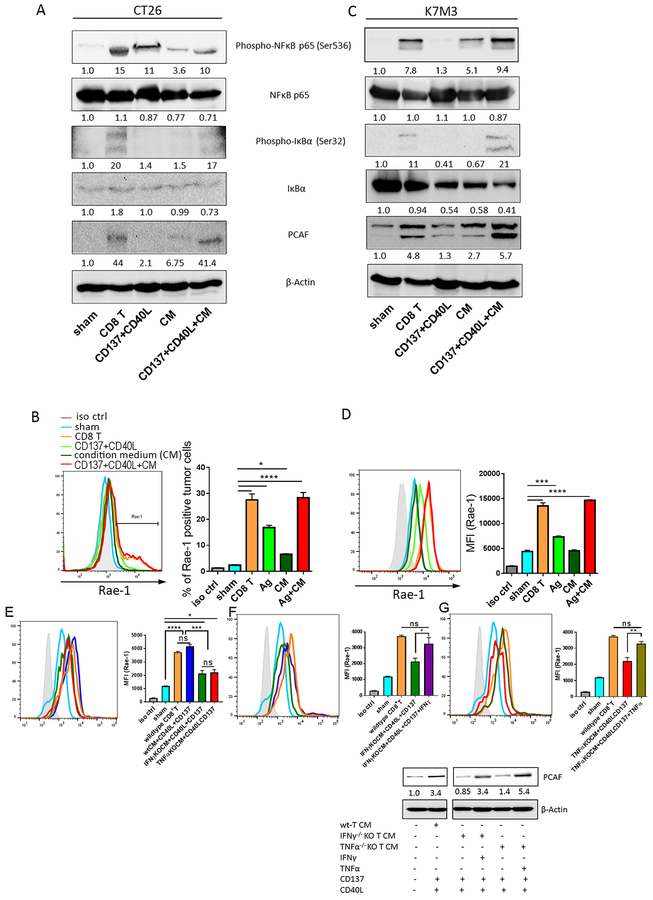
We previously discovered that NKG2D ligands can be upregulated on mouse tumor cell surfaces by CD8+ T cells and that this upregulation is associated with HAT induction11. This induction was independent of the known stress-induced NKG2D ligand induction and is long-lasting11. Since results from our earlier study showed that only depletion of CD8+ T cells abolished Rae-1 upregulation, and co-culturing only with CD8+ T cells, but not any other subtypes of immune cells, induced Rae-1 on tumor cells, we solely focused on CD8+ T cells in this study. We tested this model in vitro by co-culturing IL-12–stimulated CD8+ T cells and tumor cells and detected dramatic NKG2D ligand induction on a variety of mouse and human tumor cells (data not shown). A possible mechanism for this upregulation was that the inflammatory cytokines secreted by CD8+ T cells trigger the activation of HATs and induce NKG2D ligand expression. However, we observed in vitro that exposure to medium conditioned by CD8+ T cells was insufficient to induce expression of NKG2D ligands (Fig. 1A, 1B). Instead, direct contact with CD8+ T cells was required to upregulate expression of the NKG2D ligand Rae-1 on CT26 (mouse colon carcinoma) cells and K7M3 (mouse osteosarcoma) cells (Fig. 1A, 1B), suggesting that direct ligand/cognate receptor interaction between tumor cells and CD8+ T cells stimulates certain downstream signaling in tumor cells.
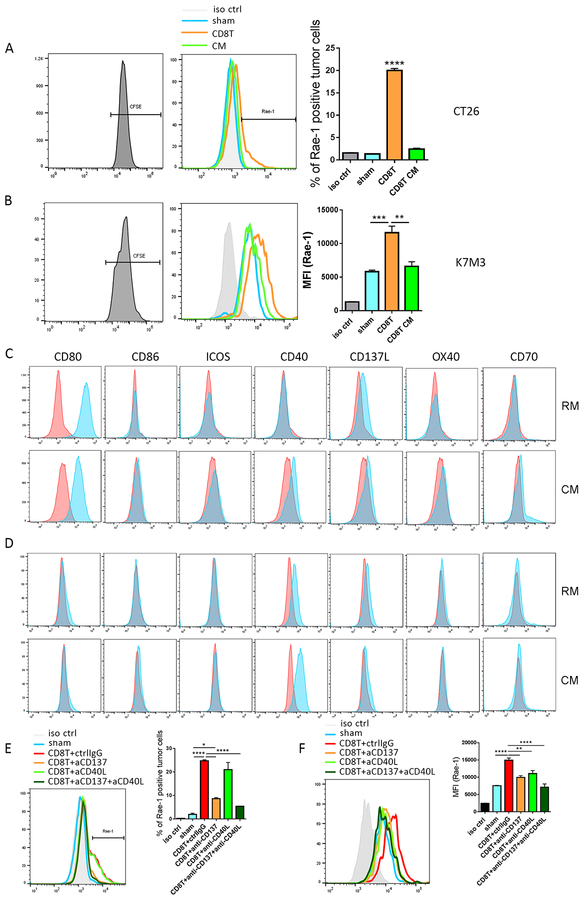
Fig. 1. CD8+ T cells engage with tumor cells through TNFRSF to boost NKG2D ligand expression.
(A, B) Murine CT26 colon carcinoma (A) and K7M3 osteosarcoma (B) cells were stained with CFSE and co-incubated with sham, activated CD8+ T cells (CD3/CD8 Dynabeads, IL-2 [50 U/mL], and IL-12 [10 ng/mL]) at a 1:1 tumor to T cell ratio, or CD8+ T cell–conditioned medium (CM) for 24 h. Rae-1 expression on tumor cells was determined by flow cytometry. Bar graphs show means ± standard error of the mean (SEM). MFI, mean fluorescence intensity. (C, D) CT26 (C) and K7M3 (D) cells were stained with CFSE and cultured in regular medium (RM) or CD8+ T cell CM. Expression of CD80, CD86, ICOSL, CD40, CD137L, OX40, and CD70 was determined by flow cytometry. (E, F) Activated CD8+ T cells were pretreated with control IgG, anti-CD137 (5 ng/mL), anti-CD40L (5 ng/mL), or anti-CD137 plus anti-CD40L for 3 h. CT26 (E) and K7M3 (F) cells were stained with CFSE and co-incubated with sham or pretreated CD8+ T cells for 24 h. Rae-1 expression on tumor cells was determined by flow cytometry. iso ctrl: isotype control. Bar graphs show means ± SEM. Results are representative of three repeated experiments. * P<0.05; ** P<0.01; *** P<0.005; **** P<0.001; ns, no statistical significance.
We next aimed to define the interacting molecules through which CD8+ T cells transduce the signals to tumor cells. It has been accepted that the T cell co-stimulatory receptors CD80 and CD86, CD137 ligand, ICOSL, and CD40, CD70, and CD252 are expressed on certain tumor cells12, 16, 26, 27 and interact with CD8+ T cells via cognate ligands. We therefore screened the expression of these receptors on CT26 and K7M3 cells in regular culture medium or in CD8+ T cell–conditioned medium, which contains secreted inflammatory signals from T cells (Fig. 1C, 1D). The B7 family receptor CD80 was highly expressed on CT26 cells, but expression levels of CD80 and CD86 on K7M3 cells were very low. These results suggested that neither receptor is associated with Rae-1 (one of the NKG2D ligands) regulation because Rae-1 expression was upregulated on both tumor cell lines; in fact, Rae-1 induction was higher on K7M3 cells than on CT26 cells (Fig. 1A, 1B). Notably, the TNFR family CD137 ligand was expressed on both tumor cell lines. Another TNFR family member, CD40, was highly expressed on K7M3 cells (Fig. 1D); although it was not expressed on CT26 cells in the regular medium, it was induced on CT26 cells by the conditioned medium (Fig. 1C). These results suggested that higher expression levels of CD40 and CD137L were associated with the stronger induction of Rae-1 on K7M3 cells than on CT26 cells.
To validate the roles played by CD137 ligand and CD40 in NKG2D ligand upregulation, we pretreated activated CD8+ T cells with control (IgG), anti-CD137, anti-CD40 ligand, or anti-CD137 plus anti-CD40 ligand neutralizing antibodies for 24 h before co-incubation with CFSE-labeled tumor cells (1:1 tumor cell-to-T cell ratio). Notably, the CD8+ T cells treated with control IgG induced Rae-1 expression on tumor cell surfaces. In contrast, disruption of CD137 ligand signaling partially abrogated the Rae-1 upregulation caused by CD8+ T-cell stimulation. Moreover, blocking both CD137 ligand and CD40 signaling synergistically abrogated Rae-1 upregulation on CT26 and K7M3 cells (Fig. 1E, 1F). These results established that CD8+ T cells interact with tumor cells through TNFRSF members to upregulate Rae-1 expression.
Inhibition of NF-κB pathway abolished CD8+ T cell–mediated NKG2D ligand upregulation in mouse and human tumor cells
Given that CD8+ T cells transmit their stimulation signals into tumor cells through the engagement between TNFRSF members and cognate ligands, we sought to identify the signaling pathway through which Rae-1 expression is upregulated on the tumor cell surface. Normally, TNFRSF receptors recruit TRAFs to trigger complex signaling networks in cellular processes19, including NF-κB, MAPK, and PI3K. TNFRSF receptors also recruit the TRAF-independent Janus family kinase 3 and signal transducer and activator of transcription 3 (STAT3) pathways. We therefore employed specific inhibitors to identify the most crucial signaling pathways for Rae-1 regulation. CT26 and K7M3 mouse tumor cells were pretreated with vehicle control or with one of the following potent signaling inhibitors: STAT3 inhibitor S3I-201 (50 μM for 24 h), p38 inhibitor PH797804 (1 μM for 4 h), MAPK kinase inhibitor PD98059 (2 μM for 4 h), PI3K inhibitor PKI-587 (5 nM for 4 h), or NF-κB inhibitor QNZ (100 nM for 4 h). The treated cells were then labeled with CFSE and co-incubated with IL-12–stimulated mouse CD8+ T cells (1:1 tumor cell-to-T cell ratio). Rae-1 expression on the tumor cell surface was determined via flow cytometry (Fig. 2). Our results show that the STAT3 inhibitor S3I-201 did not affect CD8+ T cell–mediated Rae-1 induction on either CT26 or K7M3 cells, suggesting that Rae-1 upregulation is independent of STAT3 signaling. In striking contrast, however, the NF-κB inhibitor QNZ dramatically abrogated Rae-1 induction in both tumor cell lines, indicating that NF-κB signaling plays an essential role in Rae-1 regulation. Treatment with PH797804 or PKI-587 partially abolished Rae-1 upregulation in CT26 cells, as did PD98059 or PKI-587 in K7M3 cells, suggesting that different cognate signaling pathways may contribute to boosting Rae-1 expression via crosstalk effects. Nevertheless, only NF-κB signaling blockade significantly impaired induction of Rae-1 expression in both tumor cell lines.
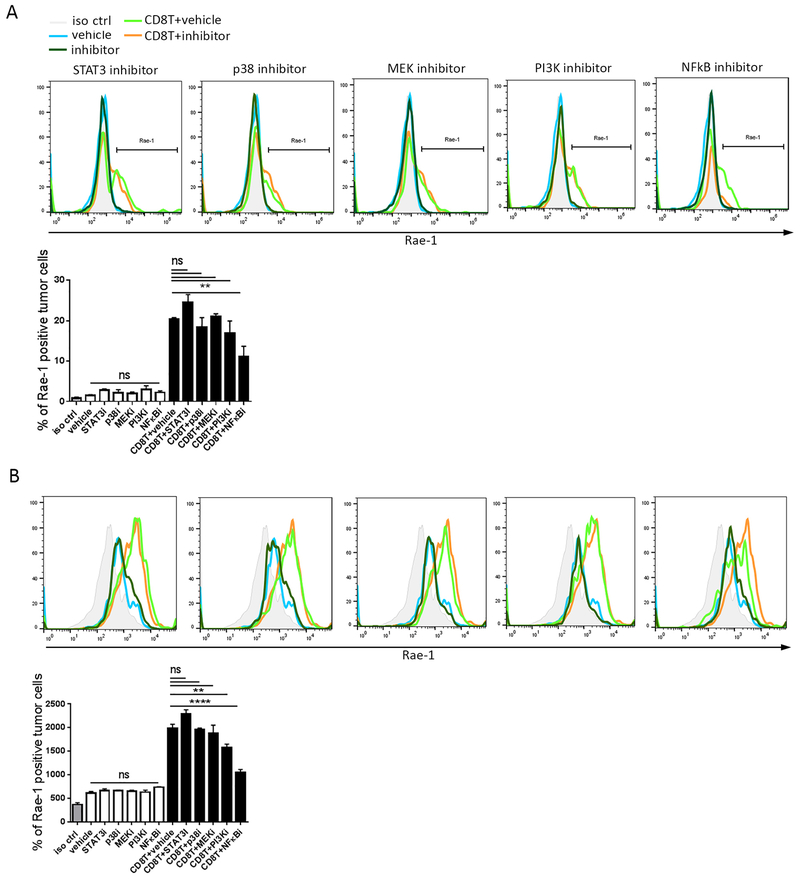
Fig. 2. Inhibition of the NF-κB pathway abolished CD8+ T cell–mediated NKG2D ligand upregulation.
CT26 (A) and K7M3 (B) cells were pretreated with an NF-κB pathway inhibitor: STAT inhibitor S3I-201 (50 μM for 24 h), p38 inhibitor PH797804 (1 μM for 4 h), MAPK kinase (MEK) inhibitor PD98059 (2 μM for 4 h), PI3K inhibitor PKI-587 (5 nM for 4 h), or NF-κB inhibitor QNZ (100 nM for 4 h). The pretreated cells were then labeled with CFSE and cultured in the presence or absence of stimulated mouse CD8+ T cells (1:1 tumor cell-to-T cell ratio). Rae-1 expression on the tumor cell surface was determined via flow cytometry. iso ctrl: isotype control. Bar graphs show means ± SEM. Results are representative of three repeated experiments. ** P<0.01; **** P<0.001; ns, no statistical significance
To determine whether the mechanism is human relevant, we characterized the critical role of NF-κB signaling in human tumor cell lines by pre-treating HCT116 human colon carcinoma cells and CCH.OS.O human osteosarcoma cells with vehicle control or 100 nM QNZ for 4 h and labeled them with CFSE before co-incubation with IL-12–stimulated human CD8+ T cells (1:1 tumor cell-to-T cell ratio). In parallel with the results from the mouse cell lines, inhibition of NF-κB signaling abolished CD8+ T cell–induced ULBP1 upregulation, suggesting a similar mechanism for NKG2D ligand regulation in human cancer cells through NF-κB signaling (Fig. S1A, S1B).
NF-κB signaling and PCAF are induced in tumor cells upon CD8+ T cell engagement in vitro and in vivo
Having established that Rae-1 upregulation in mouse tumor cells requires activation of NF-κB and HATs28, we hypothesized that the engagement of CD8+ T cells with tumor cells should simultaneously activate NF-κB and PCAF. We previously reported that treatment with IL-12–encoding DNA plus doxorubicin enhanced the accumulation of CD8+ T cells in tumors and led to Rae-1 upregulation on tumor cells, whereas disruption of PCAF in tumors abolished Rae-1 induction29. In the current study, we found that treatment with IL-12 and doxorubicin triggered NF-κB activation and induced PCAF expression in both LLC and K7M3 tumors in vivo (Fig. 3A, 3B). Likewise, immunoblotting showed that the in vitro encounter of CD8+ T cells and K7M3 tumor cells also resulted in phosphorylation of the NF-κB subunit p65 and upregulation of PCAF (Fig. 3C). To understand whether NF-κB activation induced PCAF expression, we applied QNZ, which inhibits NF-κB phosphorylation, to K7M3 cells before co-incubating them with CD8+ T cells. We found that inhibition of NF-κB activation abrogated the induction of PCAF expression, which subsequently impaired Rae-1 upregulation (Fig. 3C). In agreement with our previous observation in vivo11, knocking down PCAF from K7M3 cells totally abrogated CD8+ T cell-induced Rae-1 expression (Fig. 3D). Overall, these results suggested that NF-κB activation leads to PCAF induction, which in turn upregulates Rae-1 expression on tumor cells.
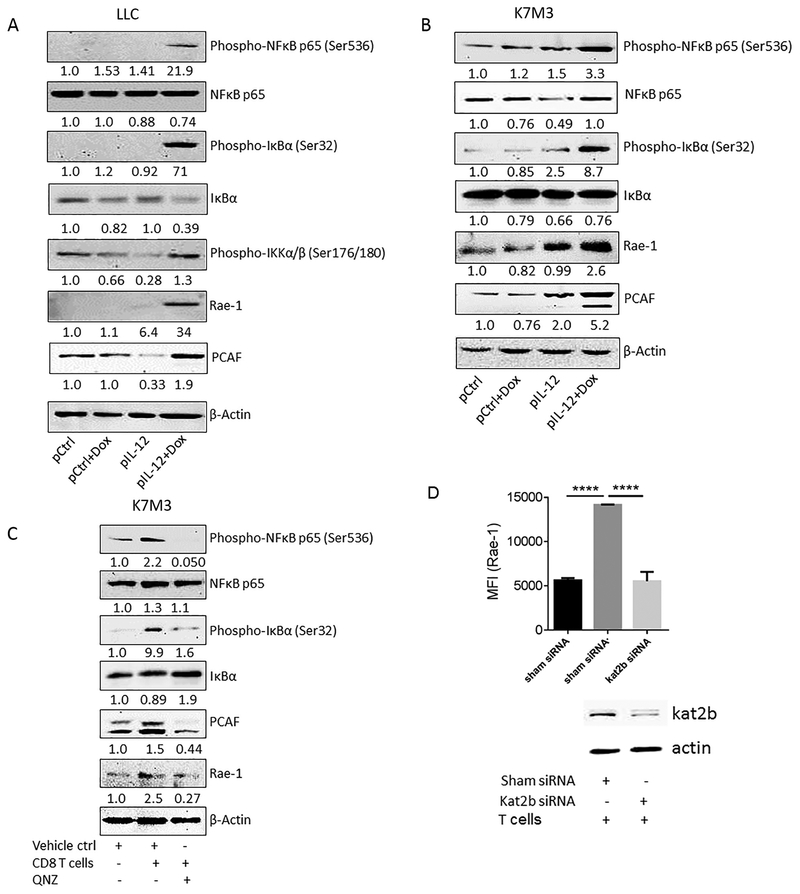
Fig. 3. NF-κB signaling and PCAF are induced in tumor cells upon CD8+ T cell engagement in vitro and in vivo.
(A, B) Mice (n=3/group) were treated twice (10 days apart) with one of the four indicated treatments: control, doxorubicin (Dox), IL-12–encoding DNA, or IL-12–encoding DNA plus doxorubicin. Tumors were collected on day 4 after the second treatment. Tumor samples were subjected to immunoblotting of phospho-NF-κB, NF-κB, phospho- IkBα, phospho- Ikkα/β, Rae-1, PCAF, and β-Actin. Values under each band indicate the density of protein expression. (C) K7M3 cells were pretreated with vehicle control or QNZ for 4 h, labeled with CFSE, and co-incubated with activated CD8+ T cells for 24 h. Tumor cells were isolated and subjected NF-κB and B and to immunoblotting of phospho-NF-κB, NF-κB, phospho-IkBα, IkBα, Rae-1, PCAF, and β-Actin. Values under each band indicate the density of protein expression. (D) K7M3 cells were transfected with control siRNA or PCAF (kat2b) siRNA, labeled with CFSE, and co-cultured with CD8+ T cells at the ratio of 1:1. The knockdown of PCAF was confirmed using immunoblotting, and Rae-1 levels on tumor cells were detected by using flow cytometry. MFI, mean fluorescence intensity. Bar graphs show means ± SEM. Results are representative of three repeated experiments. **** P<0.001.
Inflammatory cytokines are required for CD8+ T cell chemotaxis to upregulate Rae-1 in tumors
Next, we asked whether the inflammatory cytokines produced by CD8+ T cells are required for Rae-1 upregulation. In our in vivo and in vitro models for Rae-1 induction, CD8+ T cells are activated by IL-12, so the inflammatory cytokines comprise mainly IFNγ and TNFα. To test their roles in Rae-1 induction in tumors, IFNγ −/− and TNFα −/− and wildtype LLC tumor model mice were treated with IL-12 plus doxorubicin. Strikingly, the dramatic induction of Rae-1 in tumors observed in wildtype mice was abrogated in the absence of IFNγ or TNFα (Fig. 4A, 4B), revealing that inflammatory cytokines (IFNγ and TNFα) play a key role in CD8+ T cell–mediated NKG2D ligand upregulation. However, the question here is whether the inflammatory cytokines function as T cell recruiters to enhance the encounter between CD8+ T cells and tumors cells or directly trigger tumor cell signaling to induce Rae-1.
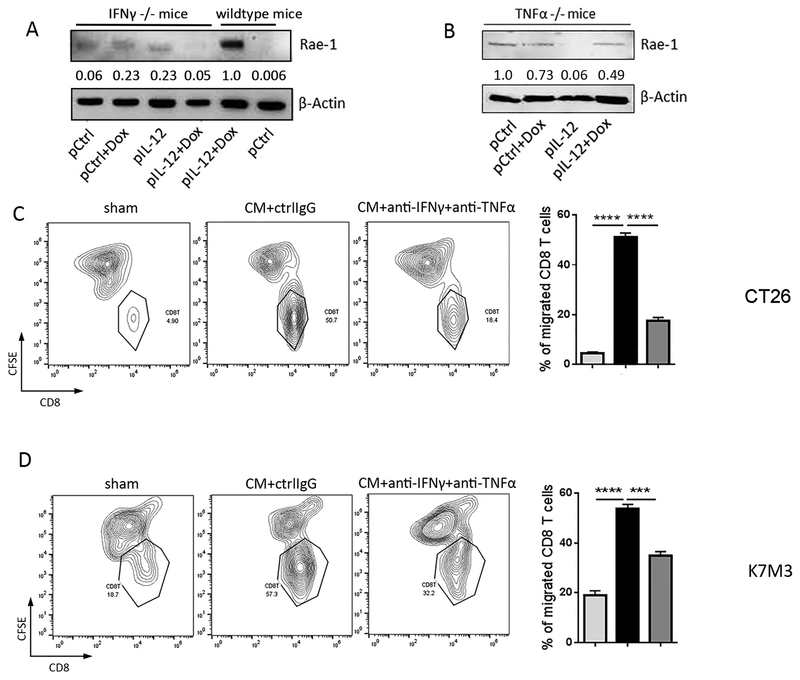
Fig. 4. Inflammatory cytokines IFNγ and TNFα facilitate CD8+ T cell migration to tumor cells.
(A, B) LLC tumor–bearing IFNγ−/− or TNFα−/− mice (n=3/group) were treated twice (10 days apart) with one of the four indicated treatments: control, doxorubicin (Dox), IL-12–encoding DNA, or IL-12–encoding DNA plus doxorubicin. Tumors were collected on day 4 after the second treatment. Tumors were subjected to immunoblotting of Rae-1 in comparison with wildtype littermates. Values under each band indicate the density of protein expression. (C, D) CT26 (C), K7M3 (D) tumor cells were cultured for 24 h in regular RPMI medium or with T cell–conditioned medium (CM) with or without anti-IFNγ (1 μg/mL) and anti-TNFα (0.5 μg/mL) neutralizing antibodies. CFSE-labeled tumor cells were plated in the lower chambers of Transwell plates, while activated CD8+ T cells (5 × 105/well) were plated in the upper wells of the insert. After 24 h of co-culture, CD8+ T cells that had migrated to the lower chambers were detected via flow cytometry. Results are representative of three repeated experiments. ** P<0.01; *** P<0.005; **** P<0.001.
In the tumor immune environment, inflammatory cytokines, in particular IFNγ and TNFα, may boost the production of T cell chemoattractants30, 31. We therefore hypothesized that the inflammatory cytokines in CD8+ T cell–conditioned medium enhance T cell chemotaxis and recruit a CD8+ T cell influx into tumors, where the CD8+ T cells interact with tumor cells to upregulate Rae-1 expression. To test this hypothesis, we assessed how efficiently CD8+ T cells migrated to tumor cells in response to exposure to inflammatory cytokines. CFSE-labeled CT26 or K7M3 mouse tumor cells and human colorectal adenocarcinoma cells HT29 were incubated with regular culture medium or CD8+ T cell–conditioned medium for 24 h in the presence or absence of IFNγ and TNFα neutralizing antibodies. Tumor cells and IL-12–stimulated CD8+ T cells were plated in Matrigel-coated Transwell chambers for cell invasion assays. After 24 h of incubation, the number of CD8+ T cells that had migrated to the conditioned medium–treated tumor cells was significantly higher than the number of CD8+ T cells that had migrated to the tumor cells treated with regular medium (Fig. 4C, 4D, S2), suggesting that the conditioned medium promoted T cell recruitment to tumors. Crystal violet staining of the bottom side of the Transwell inserts showed that dramatically higher numbers of CD8+ T cells had been attracted by conditioned medium–treated tumor cells and penetrated the extracellular matrix than had been attracted by tumor cells in regular medium (Fig. S3A–S3C). IFNγ and TNFα are known for promoting T cell chemotaxis32–34. We neutralized these cytokines with antibodies to determine the impact of these cytokines in condition medium on T cell recruitment to tumor cells. By striking contrast, neutralization of IFNγ and TNFα in the conditioned medium with the cognate antibodies significantly reduced chemotaxis (Fig. 4C, 4D), which demonstrated that IFNγ and TNFα in the conditioned medium played an essential role in enhancing T cell chemotaxis. In the immune context of the tumor environment, tumor-infiltrated CD8+ T cells secrete IFNγ and TNFα in response to IL-12 stimulation, augment effector T cell chemotaxis, and therefore promote CD8+ T cell and tumor cell contact-mediated Rae-1 induction. The essential role of IFNγ and TNFα in promoting chemotaxis also explains how our in vitro cell co-culture model can be duplicated in vivo.
To decipher the direct impact of inflammatory cytokines on tumor cells, CT26 and K7M3 cells were cocultured with stimulated CD8+ T cells or, alternatively, CD8+ T cells were substituted by cognate ligands of CD40 and CD137L (CD40L and CD137) recombinant proteins in the presence or absence of CD8+ T cell–conditioned medium. NF-κB signaling, PCAF expression, and Rae-1 expression were assessed via immunoblotting and flow cytometry, respectively. In theory, CD137 and CD40L play the same role as CD8+ T cells in terms of interacting with tumor cells to transduce stimulating signals. We found that stimulation with TNFRSF ligands CD40L or CD137 alone induced moderate activation of NF-κB signaling and minor upregulation of PCAF expression (Fig. 5A, 5C). In contrast, TNFRSF stimulation in the presence of the CD8+ T cell–conditioned medium activated NF-κB signaling to a similar or greater extent than did CD8+ T-cell co-culture (Fig. 5A, 5C). Accordingly, the induction of Rae-1 expression on tumor cells correlated with the levels of phospho-NF-κB and PCAF (Fig. 5B, 5D). Our results imply that the secreted components in CD8+ T cell–conditioned medium, especially inflammatory cytokines, may facilitate the activation of CD40 and CD137L downstream signaling and subsequently trigger PCAF and NKG2D ligand induction in tumor cells.
Fig. 5. Inflammatory cytokines are crucial for CD8+ T cell–mediated Rae-1 upregulation on tumor cells.
(A-D) CT26 (A-B) and K7M3 (C-D) cells were co-cultured with stimulated CD8+ T cells or, for comparison, treated with TNFSRF CD137 and CD40 ligand recombinant proteins (Ag) in the presence or absence of CD8+ T cell–conditioned medium (CM). NF-κB signaling and Rae-1 expression were assessed via immunoblotting (A, C) and flow cytometry (B, D), respectively. MFI, mean fluorescence intensity. Bar graphs show means ± SEM. Values under each band indicate the density of protein expression. Results are representative of three repeated experiments. (E) LLC tumor cells were labeled with CFSE and cultured in medium conditioned with wildtype CD8+ T cells, IFNγ-knockout (KO) CD8+ T cells, or TNFαKO CD8+ T cells in the presence of recombinant CD40L (0.5 μg/mL) and CD137 (10 μg/mL) proteins. LLC cells alone were used as a negative control, and LLCs co-incubated with wildtype CD8+ T cells were used as a positive control. Rae-1 expression was determined by using flow cytometry. (F, G) LLC cells were labeled with CFSE and cultured in IFNγKO CD8+ T cell CM (F) or TNFαKO CD8+ T cell CM (G) in the presence or absence of IFNγ (10 ng/mL) (F) or TNFα (10 ng/mL) (G), respectively. LLC cells alone were used as a negative control, and LLCs co-incubated with wildtype CD8+ T cells were used as a positive control. Rae-1 expression was determined by using flow cytometry. iso ctrl: isotype control. Immunoblots of PCAF after each treatment was at the bottom panel. Bar graphs show means ± SEM. Results are representative of three repeated experiments. * P<0.05; ** P<0.01; *** P<0.005; **** P<0.001; ns, no statistical significance.
Further, we validated that IFNγ and TNFα are critical for TNFRSF ligand–induced Rae-1 induction by CD8+ T cell–conditioned medium depleted of IFNγ or TNFα. In the presence of CD137 and CD40 ligand, the effect of IFNγ−/− or TNFα−/− CD8+ T cell–conditioned medium on NF-κB signaling activation was impaired, reducing the levels of Rae-1 upregulation compared to wildtype CD8+ T cell–conditioned medium, as determined by flow cytometry assay (Fig. 5E). Remarkably, adding IFNγ or TNFα to the IFNγ−/− or TNFα−/− CD8+ T cell–conditioned medium, respectively, along with CD137 and CD40 ligand, substantially elevated PCAF and Rae-1 expression on LLC cells, as did wildtype CD8+ T cell–conditioned medium (Fig. 5F, 5G). These results demonstrate that, in addition to increasing chemotaxis, IFNγ and TNFα in CD8+ T cell–conditioned medium facilitated PCAF and Rae-1 induction on tumor cells. Based on these results, we can simplify the CD8+ T cell and tumor cell co-culture model for NKG2D ligand induction to include these crucial components: CD40 and CD137L signaling as well as IFNγ and TNFα to upregulate NKG2D ligands via NF-κB signaling and PCAF activation (Fig. 6).
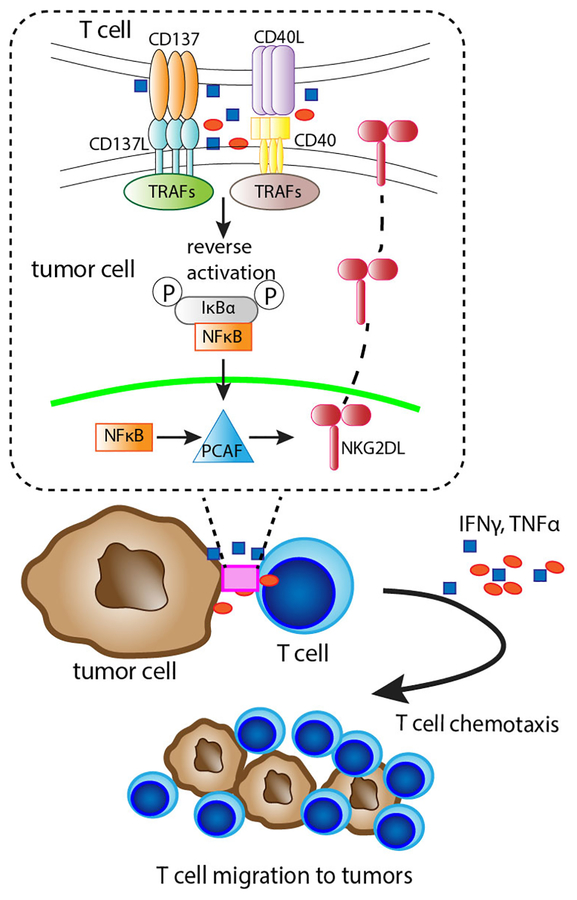
Fig 6. Illustration of the mechanism of CD8+ T cell engagement-induced Rae-1 expression on tumor cells.
Rae-1 upregulation was through CD137L/CD40 downstream NF-κB signaling and PCAF activation on tumor cells.
Discussion
Taken together, our results demonstrate that CD8+ T cell–mediated NKG2D ligand upregulation on tumor cells is initiated upon contact between TNFRSF members on tumor cells and their cognate ligands on CD8+ T cells. This contact transmits signals to activate the NF-κB signaling pathway, which in turn boosts PCAF expression, resulting in the further induction of NKG2D ligand expression on tumor cells. The inflammatory cytokines IFNγ and TNFα produced by CD8+ T cells promote T cell chemotaxis in the tumor microenvironment, reinforcing CD8+ T cell engagement in a positive feedback loop, and also augment the stimulus to facilitate PCAF and NKG2D ligand upregulation. According to this mechanism, CD8+ T cells in this model can, alternatively, be replaced by CD40 and CD137L reverse signaling, activating IFNγ and TNFα to substantially upregulate NKG2D ligands on tumor cells.
The mechanisms for NKG2D ligand regulation are often context-dependent and are still being discovered. We reported that engagement of CD8+ T cells upregulates the expression of NKG2D ligands on the tumor cell surface29. To better understand this mechanism, we sought to identify the stimulus that alters tumor cells to upregulate Rae-1 expression. In the context of the tumor microenvironment, direct contact between tumor cells and CD8+ T cells and/or inflammatory cytokines produced by infiltrated CD8+ T cells could provide such a stimulus.
Since contact between tumor and CD8+ T cells is crucial for the upregulation of Rae-1, we identified the interacting receptors on these cells. In general, CD8+ T cells engage in contact with antigen-presenting cells through the T cell receptor–MHC and co-stimulatory or inhibitory receptors to form an “immune synapse”35. Although tumor cells express low levels of MHC I, most types of tumor cells express the co-inhibitory receptors programmed death ligand 1, programmed death ligand 2, B7-H3, and B7-H4. Nevertheless, the expression of co-stimulatory receptors on tumor cells depends on the type of tumor. Previous studies indicated that CD80 and CD86 are found on colon cancer cells26, CD40L is expressed on bladder and cervical cancer cells12, 13, and CD70 plays different roles in lymphoma, pancreatic cancer, renal cancer, melanoma, and ovarian cancer15, 17. In the process of screening, we used three criteria to select the receptors for testing. First, the receptors had to be expressed on both CT26 and K7M3 cells, which we have been using throughout this study. Second, the receptors had to transmit activating signals to CD8+ T cells. Third, the receptors also had to transport signals back to tumor cells. Using these criteria, we excluded the B7 family of inhibitory receptors, which do not meet the second criterion. We also eliminated CD80, which is found only on CT26 cells and whose expression was dramatically reduced by CD8+ T cell–conditioned medium. Therefore, we focused on the TNFRSF co-stimulatory receptors, which are often expressed on antigen-presenting cells and are also present on various types of cancer cells. In tumor cells, TNFRSF members CD137 ligand and CD40 trigger reverse signaling through TRAFs, adaptor proteins that initiate the network of downstream signaling pathways, including p3836, ERK37, c-Jun N-terminal kinase38, PI3K-AKT39, and NF-κB40, some of which ultimately convert to NF-κB–dependent activating mechanisms41. Our finding that inhibition of NF-κB activation obstructed Rae-1 upregulation in the presence of CD8+ T cells but not in their absence revealed that NF-κB activation plays an essential role in CD8+ T cell–mediated Rae-1 upregulation.
The role of inflammatory cytokines in NKG2D ligand regulation is controversial. Bui et al. showed that mouse NKG2D ligand H60 can be diminished by interferons on sarcoma cells42. In contrast, Lin et al. demonstrated that human NKG2D ligand MICA on endothelial cells may be induced by TNFα9. We discovered that loss of IFNγ or TNFα abrogated the CD8+ T cell–mediated upregulation of Rae-1 on tumor cells both in vitro29 and in vivo, suggesting that inflammatory cytokines play critical roles in regulating expression of CD8+ T cell–induced NKG2D ligands. In fact, our data showed two ways for IFNγ and TNFα to contribute to NKG2D ligand induction. On one hand, they are crucial cytokines that stimulate NFκB signaling, which in turn induces the NKG2D ligand. On the other hand, as shown in Fig. 4C and 4D, IFNγ and TNFα have additional roles in facilitating T cell chemotaxis to tumor cells, which is also required for NKG2D ligand induction. In IFNγ−/− or TNFα−/− mice, as shown in Fig. 4A and 4B, not only is IFNγ- or TNFα-mediated T cell chemotaxis mostly impaired, but more importantly, IFNγ- and TNFα- induced NFκB signaling and PCAF activation are abrogated, resulting in complete failure of NKG2D ligand induction. Therefore, our study shows that both direct cell-to-cell contact with CD8+ T cells and the presence of inflammatory cytokines resulted in remarkably more robust Rae-1 upregulation in tumor cells.
Given that CD8+ T cell–dependent Rae-1 upregulation requires the activity of the HATs29, we sought in this study to decipher the connection between CD8+ T cell engagement and HAT activation. Abundant evidence shows that co-activators such as CBP/p300 and PCAF are crucial for activating NF-κB–mediated gene expression23, 24, 43, 44. In particular, phosphorylated p65 enhanced the engagement of NF-κB with the co-activator complex containing multiple HATs45. Sheppard et al. emphasized that, among the 20 co-activator proteins, NF-κB requires PCAF HAT activity to regulate gene expression24. In the present study, we demonstrated not only that the status of NF-κB activation was associated with the expression level of PCAF but also that both TNFRSF stimulation and inflammatory cytokines were required to induce PCAF. These results suggest that, besides NF-κB stimulation, inflammatory cytokines may play additional roles in PCAF activation, which needs to be further validated.
Conclusions
In conclusion, we discovered a novel mechanism of NKG2D ligand regulation on tumor cells. This mechanism is initiated by reverse signaling activation in tumor cells through TNFRSF members and depends on phosphorylated NF-κB and the HAT activity of PCAF. This unique interaction could be used to “label” tumor cells so that they can be efficiently recognized and eliminated by CD8+ T cells. Furthermore, stimulation by CD137 may promote the long-term survival of tumor-infiltrating CD8+ T cells.
Materials and Methods
Animal studies
Wild-type C57BL/6, BALB/C, IFNgKO (B6.129S7-Ifngtm1Ts/J), and TNFα− (B6.129S-Tnftm1Gkl/J) mice, 6 to 8 weeks old, were purchased from The Jackson Laboratory (Bar Harbor, ME). The mouse care and handling procedures were approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center. To create the mouse tumor models, Lewis lung carcinoma (LLC) tumor cells (1.5 × 105 cells in 30μL PBS) or CT26 tumor cells (2.0 × 105 in 30μL PBS) were inoculated subcutaneously into the C57BL/6 or BALB/C mice, respectively. K7M3 tumor cells (1.0 × 105 in 10μL PBS) were inoculated intraosseously into BALB/C mice. All animal studies were single blinded during group allocation and data analysis. Tumor-bearing mice were randomly allocated to treatment groups and subjected to treatment on day 7 after inoculation with control DNA (10 μg/mouse), control DNA plus doxorubicin (1 mg/kg), IL-12–encoding DNA (10 μg/mouse), or IL-12–encoding DNA plus doxorubicin via electroporation; a second treatment was administered 10 days later as described previously46.
Immune cells
Buffy coats from de-identified healthy human blood donors were purchased from the Gulf Coast Regional Blood Center (Houston, TX); their acquisition or this purpose was approved by the MD Anderson Institutional Review Board. Peripheral blood mononuclear cells were isolated from buffy coat samples via centrifugation using the Ficoll-Paque method. Human CD8+ T cells were enriched from peripheral blood mononuclear cells using an EasySep human CD8+ T cell isolation kit (Stemcell Technologies, Vancouver, BC, Canada). These cells were cultured in a mixture comprising 45% RPMI-1640 medium, 45% Click medium (Sigma-Aldrich, St. Louis, MO), and 10% fetal bovine serum, referred to in some experiments as regular medium. Mouse CD8+ T cells were enriched from splenocytes using an EasySep mouse CD8+ T cell isolation kit (Stemcell Technologies). Mouse T cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, and were activated with CD3/CD8 Dynabeads (Thermo Fisher Scientific, Waltham, MA), IL-12 (10 ng/mL), and IL-2 (50 U/mL) recombinant proteins. Supernatants of mouse T cell cultures after 24 h were used as conditioned medium.
Cell lines
CT26 (mouse colon cancer), K7M3 (mouse osteosarcoma) and LLC (mouse Lewis lung carcinoma), HCT116 (human colon cancer), HT29 (human colon cancer), and CCH.OS.D (human osteosarcoma) were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. All the cell lines were characterized by DNA fingerprinting within 6 months before initiating the experiments at MD Anderson Cancer Center’s Characterized Cell Line Core Facility.
Plasmids, reagents, and drugs
The IL-12 DNA construct was purchased from Valentis, Inc. (Vilnius, Lithuania) and was confirmed in-house by sequence analyses. DNA was prepared by using an endotoxin-free Mega preparation kit from Qiagen, Inc. (Valencia, CA) according to the manufacturer’s instructions. Doxorubicin (Bedford Laboratories, Bedford, OH) was purchased from the pharmacy at MD Anderson Cancer Center. NF-κB pathway inhibitors S3I-201, PH797804, PD98059, PKI-587, and QNZ were purchased from Selleckchem (Houston, TX). IL-12 and IL-2 recombinant proteins were purchased from R&D Systems (Minneapolis, MN). Carboxyfluorescein succinimidyl ester (CFSE) was purchased from Life Technologies (Grand Island, NY).
Antibodies
Phycoerythrin-conjugated anti-mouse CD80 and CD86; CD137 ligand; CD40, CD70, ICOS ligand and OX40 antibodies; phycoerythrin-CY7–conjugated anti-mouse CD8 antibody; and isotype control antibodies were purchased from BioLegend (San Diego, CA). Anti-mouse phospho-NF-κB p65 (Ser536), NF-κB p65, phospho-IkBα (Ser32), IkBα (Ser32), phospho-Ikkα/β (Ser176/180), Ikkα, PCAF, and β-actin antibodies were purchased from Cell Signaling Technologies (Danvers, MA). The anti-mouse Rae-1 antibody was developed by the Monoclonal Antibody Core Facility at MD Anderson Cancer Center and validated in our previous study47. Goat anti-mouse Alexa Fluor 405 antibodies were purchased from Life Technologies. Horseradish peroxidase–conjugated anti-mouse immunoglobulin G (IgG) and anti-rabbit IgG were purchased from Cell Signaling Technologies. Anti-mouse CD137 and CD40 ligand-blocking antibodies were purchased from R&D Systems. A recombinant anti-mouse CD137 antibody was purchased from BioLegend, and a recombinant anti-mouse CD40 ligand was purchased from eBioscience (San Diego, CA). Anti-mouse IFNγ- and TNFα-neutralizing antibodies were purchased from R&D Systems.
Flow cytometry analysis
Tumor cells were sequentially incubated with primary and secondary antibodies for 30 min each at 4°C. Stained cells were analyzed using an Attune acoustic focusing cytometer (Applied Biosystems, Foster City, CA). Flow cytometry data were analyzed using the FlowJo software program (BD Biosciences, San Jose, CA).
Immunoblotting assay
Four days after the second treatment, the mice were killed; their tumors were collected and snap frozen in liquid nitrogen. Frozen tissue samples were smashed before being homogenized using a minibead beater with 5 to 8 silicone beads (BioSpec Products, Bartlesville, OK) in 0.4 mL of ice-cold lysis buffer. The tumor cells were subjected to lysis with RIPA buffer. The protein extracts were separated from the tissue residues by centrifugation at the maximum speed for 20 min at 4°C. Forty micrograms of total protein from each sample was separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes using the iBlot gel transfer device (Invitrogen, Grand Island, NY). The membranes were blotted with primary and secondary antibodies to detect the proteins of interest.
T cell migration assay
The capacity of T cells to penetrate the extracellular matrix and migrate to tumor cells in vitro was assessed using BioCoat Matrigel invasion assay (BD Biosciences) according to the manufacturer’s instructions. CT26, K7M3 and HT29 tumor cells were cultured for 24 h in regular RPMI medium, in RPMI and CD8+ T cell–conditioned medium at a ratio of 1:1 plus control IgG, or in RPMI and conditioned medium at a ratio of 1:1 plus anti-IFNγ (1 μg/mL) and anti-TNFα (0.5 μg/mL) neutralizing antibodies. The tumor cells (1 × 105 per well) were then stained with CFSE and plated in the lower chambers of a 24-well Transwell plate. IL-12–stimulated CD8+ T cells (5 × 105 per well) were plated in the wells of the upper Transwell insert. After 24 h of co-culture, the insert was removed from the Transwell plate. The cells in the lower chambers were collected and stained with an anti-CD8 antibody for quantification of CD8+ T cells via flow cytometry. The remaining cells and medium were removed from the upper wells of the insert using cotton swabs. The cells on the lower surfaces of the insert wells were fixed with 4% formaldehyde for 15 min and stained with 0.2% crystal violet for 20 min. The inserts were then dipped in distilled water to remove excess crystal violet and allowed to air dry. The crystal violet stain on the membrane was dissolved in 250 μL methanol and read with a microplate reader at 540 nm. The total number of penetrated CD8+ T cells was the sum of the cells in the lower chamber and the cells attached to the lower surfaces of the insert.
Statistical analysis
The directly measured outcomes were analyzed using a two-sided Student t-test to compare two treatment groups or one-way analysis of variance to compare more than two treatment groups. The statistical analyses were conducted using the GraphPad Prism software program (GraphPad Software, La Jolla, CA). P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
The following Cancer Center Support Grant core resources were used: Genetically Engineered Mouse Facility and Monoclonal Antibody Core Facility with Cancer Center Support Grant P30 CA016672. The authors would like to thank Ms. Kathryn L Hale from the Department of Scientific Publications for editing the manuscript.
Funding: This study was supported by the National Institutes of Health through grant R01 CA200574.
Footnotes
Competing interests: The authors have no potential conflicts of interest to declare.
References
- 1.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 1999; 285: 727–729. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. NKG2D Receptor and Its Ligands in Host Defense. Cancer Immunol Res 2015; 3: 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carayannopoulos LN, Naidenko OV, Fremont DH, Yokoyama WM. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J Immunol 2002; 169: 4079–4083. [DOI] [PubMed] [Google Scholar]
- 4.Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH et al. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity 2000; 12: 721–727. [DOI] [PubMed] [Google Scholar]
- 5.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 2005; 436: 1186–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasser S, Raulet D. The DNA damage response, immunity and cancer. Semin Cancer Biol 2006; 16: 344–347. [DOI] [PubMed] [Google Scholar]
- 7.Jung H, Hsiung B, Pestal K, Procyk E, Raulet DH. RAE-1 ligands for the NKG2D receptor are regulated by E2F transcription factors, which control cell cycle entry. J Exp Med 2012; 209: 2409–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molinero LL, Fuertes MB, Girart MV, Fainboim L, Rabinovich GA, Costas MA et al. NF-kappa B regulates expression of the MHC class I-related chain A gene in activated T lymphocytes. J Immunol 2004; 173: 5583–5590. [DOI] [PubMed] [Google Scholar]
- 9.Lin D, Lavender H, Soilleux EJ, O’Callaghan CA. NF-kappaB regulates MICA gene transcription in endothelial cell through a genetically inhibitable control site. J Biol Chem 2012; 287: 4299–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauer M, Schuldner M, Hoffmann N, Cetintas A, Reiners KS, Shatnyeva O et al. CBP/p300 acetyltransferases regulate the expression of NKG2D ligands on tumor cells. Oncogene 2017; 36: 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J, Bernatchez C, Zhang L, Xia X, Kleinerman ES, Hung MC et al. Induction of NKG2D Ligands on Solid Tumors Requires Tumor-Specific CD8(+) T Cells and Histone Acetyltransferases. Cancer Immunol Res 2017; 5: 300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke PW, James ND, Ganesan R, Wallace M, Burton A, Young LS. CD40 expression in bladder cancer. J Pathol 1999; 188: 38–43. [DOI] [PubMed] [Google Scholar]
- 13.Altenburg A, Baldus SE, Smola H, Pfister H, Hess S. CD40 ligand-CD40 interaction induces chemokines in cervical carcinoma cells in synergism with IFN-gamma. J Immunol 1999; 162: 4140–4147. [PubMed] [Google Scholar]
- 14.Gallagher NJ, Eliopoulos AG, Agathangelo A, Oates J, Crocker J, Young LS. CD40 activation in epithelial ovarian carcinoma cells modulates growth, apoptosis, and cytokine secretion. Mol Pathol 2002; 55: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly JM, Darcy PK, Markby JL, Godfrey DI, Takeda K, Yagita H et al. Induction of tumor-specific T cell memory by NK cell-mediated tumor rejection. Nat Immunol 2002; 3: 83–90. [DOI] [PubMed] [Google Scholar]
- 16.Salih HR, Kosowski SG, Haluska VF, Starling GC, Loo DT, Lee F et al. Constitutive expression of functional 4–1BB (CD137) ligand on carcinoma cells. J Immunol 2000; 165: 2903–2910. [DOI] [PubMed] [Google Scholar]
- 17.Trebing J, El-Mesery M, Schafer V, Weisenberger D, Siegmund D, Silence K et al. CD70-restricted specific activation of TRAILR1 or TRAILR2 using scFv-targeted TRAIL mutants. Cell Death Dis 2014; 5: e1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauer J, Puschner S, Ramakrishnan P, Simon U, Bongers M, Federle C et al. TNF receptor (TNFR)-associated factor (TRAF) 3 serves as an inhibitor of TRAF2/5-mediated activation of the noncanonical NF-kappaB pathway by TRAF-binding TNFRs. Proc Natl Acad Sci U S A 2005; 102: 2874–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev 2009; 229: 152–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol 2004; 25: 280–288. [DOI] [PubMed] [Google Scholar]
- 21.Saoulli K, Lee SY, Cannons JL, Yeh WC, Santana A, Goldstein MD et al. CD28-independent, TRAF2-dependent costimulation of resting T cells by 4–1BB ligand. J Exp Med 1998; 187: 1849–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Forero I, Azpilikueta A, Bolanos-Mateo E, Nistal-Villan E, Palazon A, Teijeira A et al. T cell costimulation with anti-CD137 monoclonal antibodies is mediated by K63-polyubiquitin-dependent signals from endosomes. J Immunol 2013; 190: 6694–6706. [DOI] [PubMed] [Google Scholar]
- 23.Na SY, Lee SK, Han SJ, Choi HS, Im SY, Lee JW. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor kappaB-mediated transactivations. J Biol Chem 1998; 273: 10831–10834. [DOI] [PubMed] [Google Scholar]
- 24.Sheppard KA, Rose DW, Haque ZK, Kurokawa R, McInerney E, Westin S et al. Transcriptional activation by NF-kappaB requires multiple coactivators. Mol Cell Biol 1999; 19: 6367–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werbajh S, Nojek I, Lanz R, Costas MA. RAC-3 is a NF-kappa B coactivator. FEBS Lett 2000; 485: 195–199. [DOI] [PubMed] [Google Scholar]
- 26.Tirapu I, Huarte E, Guiducci C, Arina A, Zaratiegui M, Murillo O et al. Low surface expression of B7–1 (CD80) is an immunoescape mechanism of colon carcinoma. Cancer Res 2006; 66: 2442–2450. [DOI] [PubMed] [Google Scholar]
- 27.Held-Feindt J, Mentlein R. CD70/CD27 ligand, a member of the TNF family, is expressed in human brain tumors. Int J Cancer 2002; 98: 352–356. [DOI] [PubMed] [Google Scholar]
- 28.Hu JB, Song GL, Liu D, Li SJ, Wu JH, Kang XQ et al. Sialic acid-modified solid lipid nanoparticles as vascular endothelium-targeting carriers for ischemia-reperfusion-induced acute renal injury. Drug Deliv 2017; 24: 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Hu J, Liu C, Guo Q, Yang J, Okoli CP, Lang Y et al. Characteristics, source, and potential ecological risk assessment of polycyclic aromatic hydrocarbons (PAHs) in the Songhua River Basin, Northeast China. Environ Sci Pollut Res Int 2017; 24: 17090–17102. [DOI] [PubMed] [Google Scholar]
- 30.Antonelli A, Ferrari SM, Fallahi P, Frascerra S, Santini E, Franceschini SS et al. Monokine induced by interferon gamma (IFNgamma) (CXCL9) and IFNgamma inducible T-cell alpha-chemoattractant (CXCL11) involvement in Graves’ disease and ophthalmopathy: modulation by peroxisome proliferator-activated receptor-gamma agonists. J Clin Endocrinol Metab 2009; 94: 1803–1809. [DOI] [PubMed] [Google Scholar]
- 31.Mohan K, Ding Z, Hanly J, Issekutz TB. IFN-gamma-inducible T cell alpha chemoattractant is a potent stimulator of normal human blood T lymphocyte transendothelial migration: differential regulation by IFN-gamma and TNF-alpha. J Immunol 2002; 168: 6420–6428. [DOI] [PubMed] [Google Scholar]
- 32.Murdoch C, Finn A. Chemokine receptors and their role in inflammation and infectious diseases. Blood 2000; 95: 3032–3043. [PubMed] [Google Scholar]
- 33.Baggiolini M. Chemokines and leukocyte traffic. Nature 1998; 392: 565–568. [DOI] [PubMed] [Google Scholar]
- 34.Moser B, Loetscher M, Piali L, Loetscher P. Lymphocyte responses to chemokines. Int Rev Immunol 1998; 16: 323–344. [DOI] [PubMed] [Google Scholar]
- 35.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM et al. The immunological synapse: a molecular machine controlling T cell activation. Science 1999; 285: 221–227. [PubMed] [Google Scholar]
- 36.Cannons JL, Choi Y, Watts TH. Role of TNF receptor-associated factor 2 and p38 mitogen-activated protein kinase activation during 4–1BB-dependent immune response. J Immunol 2000; 165: 6193–6204. [DOI] [PubMed] [Google Scholar]
- 37.Sabbagh L, Pulle G, Liu Y, Tsitsikov EN, Watts TH. ERK-dependent Bim modulation downstream of the 4–1BB-TRAF1 signaling axis is a critical mediator of CD8 T cell survival in vivo. J Immunol 2008; 180: 8093–8101. [DOI] [PubMed] [Google Scholar]
- 38.Kim HH, Kwack K, Lee ZH. Activation of c-jun N-terminal kinase by 4–1BB (CD137), a T cell co-stimulatory molecule. Mol Cells 2000; 10: 247–252. [PubMed] [Google Scholar]
- 39.Lee DY, Choi BK, Lee DG, Kim YH, Kim CH, Lee SJ et al. 4–1BB signaling activates the t cell factor 1 effector/beta-catenin pathway with delayed kinetics via ERK signaling and delayed PI3K/AKT activation to promote the proliferation of CD8+ T Cells. PLoS One 2013; 8: e69677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arch RH, Thompson CB. 4–1BB and Ox40 are members of a tumor necrosis factor (TNF)-nerve growth factor receptor subfamily that bind TNF receptor-associated factors and activate nuclear factor kappaB. Mol Cell Biol 1998; 18: 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartkowiak T, Curran MA. 4–1BB Agonists: Multi-Potent Potentiators of Tumor Immunity. Front Oncol 2015; 5: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bui JD, Carayannopoulos LN, Lanier LL, Yokoyama WM, Schreiber RD. IFN-dependent down-regulation of the NKG2D ligand H60 on tumors. J Immunol 2006; 176: 905–913. [DOI] [PubMed] [Google Scholar]
- 43.Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science 1997; 275: 523–527. [DOI] [PubMed] [Google Scholar]
- 44.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 2004; 23: 2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell 1998; 1: 661–671. [DOI] [PubMed] [Google Scholar]
- 46.Zhu S, Waguespack M, Barker SA, Li S. Doxorubicin directs the accumulation of interleukin-12 induced IFN gamma into tumors for enhancing STAT1 dependent antitumor effect. Clin Cancer Res 2007; 13: 4252–4260. [DOI] [PubMed] [Google Scholar]
- 47.Han S, Zhang C, Li Q, Dong J, Liu Y, Huang Y et al. Tumour-infiltrating CD4(+) and CD8(+) lymphocytes as predictors of clinical outcome in glioma. Br J Cancer 2014; 110: 2560–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.