Summary
Hexanucleotide GGGGCC repeat expansion in C9ORF72 is the most prevalent genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). One pathogenic mechanism is the aberrant accumulation of dipeptide repeat (DPR) proteins produced by the unconventional translation of expanded RNA repeats. Here, we performed genome-wide CRISPR-Cas9 screens for modifiers of DPR protein production in human cells. We identified DDX3X, an RNA helicase, suppresses the repeat-associated non-AUG translation of GGGGCC repeats. DDX3X directly binds to (GGGGCC)n RNAs, but not antisense (CCCCGG)n RNAs. Its helicase activity is essential for the translation repression. Reduction of DDX3X increases DPR levels in C9ORF72-ALS/FTD patient cells and enhances (GGGGCC)n-mediated toxicity in Drosophila. Elevating DDX3X expression is sufficient to decrease DPR levels, rescue nucleocytoplasmic transport abnormalities, and improve survival of patient iPSC-differentiated neurons. This work identifies genetic modifiers of DPR protein production and provides potential therapeutic targets for C9ORF72-ALS/FTD.
Graphical Abstract
In Brief
DPR proteins produced by the unconventional translation of expanded RNA repeats contributes to neurodegeneration in C9ORF72-ALS/FTD. Cheng and Wang et al. identify DDX3X as a repressor of r(GGGGCC)n translation. Elevating DDX3X decreases DPRs, rescues pathological phenotypes and improves neuron survival.
The hexanucleotide GGGGCC (G4C2) repeat expansion in the C9ORF72 gene is the most common genetic cause of both Amyotrophic lateral sclerosis (ALS) and Frontotemporal degeneration (FTD) (DeJesus-Hernandez et al., 2011; Renton et al., 2011). One leading hypothesis of the disease mechanism is gain of toxicity from the repeat-containing RNA. On one hand, the expanded repeats form intranuclear RNA foci which may sequester RNA binding proteins (RBPs) and lead to their loss of function. On the other hand, the repeat-containing RNA can encode dipeptide repeat (DPR) proteins in multiple reading frames by repeat-associated non-AUG (RAN) translation. Multiple different DPR proteins translated from six reading frames of both sense GGGGCC (poly-GA, poly-GR, poly-GP) (Ash et al., 2013; Mackenzie et al., 2013; Mann et al., 2013; Mori et al., 2013b; Zu et al., 2013) and antisense CCCCGG (poly-PA, poly-PR, poly-GP) (Gendron et al., 2014; Gendron et al., 2013; Mori et al., 2013a; Zu et al., 2013) repeat RNAs are found in C9ORF72-ALS/FTD patients. Although it remains to be fully elucidated exactly how each DPR protein is synthesized in patient neurons (Cleary and Ranum, 2014; Gao et al., 2017), several toxic pathways have been proposed to be induced by the expanded repeat RNA and/or their translation products, including nucleocytoplasmic transport defects (Freibaum et al., 2015; Jovicic et al., 2015; Shi et al., 2017; Zhang et al., 2015; Zhang et al., 2016), nucleolar dysfunction (Kwon et al., 2014; Mizielinska et al., 2017), oxidative stress (Lopez-Gonzalez et al., 2016), stress granule dysfunction (Boeynaems et al., 2017; Lee et al., 2016; Zhang et al., 2018), altered ER homeostasis (Dafinca et al., 2016; Kramer et al., 2018) and mitochondrial defects (Choi et al., 2019).
Several lines of evidence indicate that different DPR proteins cause varying degrees of toxicity when overexpressed by AUG-driven translation using coding sequences without the repeats (Freibaum and Taylor, 2017). An approach to decrease the levels of these toxic DPRs by inhibiting their production holds potential therapeutic promise. Moreover, methods to reduce DPRs without changing the RNA repeats will help dissect the pathogenic pathways mediated by RNA versus DPRs. Recently, we and others showed that the translation of C9ORF72 GGGGCC repeats can initiate without the 5’-cap (Cheng et al., 2018; Sonobe et al., 2018) and is enhanced by stress stimuli through eIF2α phosphorylation (Cheng et al., 2018; Green et al., 2017; Sonobe et al., 2018; Westergard et al., 2019). This feature connects to the internal ribosome entry site (IRES)-mediated translation used widely by many viral RNAs and a handful of cellular RNAs (Komar and Hatzoglou, 2011; Stoneley and Willis, 2004). The level of IRES-translation can be regulated by IRES-transacting factors (ITAFs) that do not have the same function in canonical translation (Komar and Hatzoglou, 2011; Stoneley and Willis, 2004). RNA-binding proteins (RBPs) have been shown to modulate IRES-translation through altering RNA structures and the affinity between RNA structures and translation factors (Komar and Hatzoglou, 2011). This suggests the exciting possibility that translation of the expanded GGGGCC repeats might also be modulated by specific RBPs. Genetic approaches to define such regulators could be immensely informative.
We now performed comprehensive CRISPR-Cas9 genome-wide knockout (KO) screens (Koike-Yusa et al., 2014; Shalem et al., 2014; Shalem et al., 2015; Wang et al., 2014; Zhou et al., 2014) in human cells using fluorescence reporter cells to identify genetic modifiers of DPR production from the C9ORF72 GGGGCC repeats. Genes involved in RNA nuclear export, translation and proteasome pathways are highly enriched. In particular, we found a DEAD-box RNA helicase, DDX3X, binds the GGGGCC repeat RNA and represses the RAN translation from all three reading frames. The requirement of helicase activity suggests DDX3X unwinds or alters the repeat RNA structure, which is essential for RAN translation initiation. Decreasing DDX3X increased endogenous DPRs in C9ORF72-ALS/FTD patient lymphoblast and induced pluripotent stem cells (iPSCs). Conversely, exogenous expression of DDX3X reduces DPR levels, rescues nucleocytoplasmic transport abnormalities and glutamate-induced excitotoxicity of patient iPSC-differentiated neurons (iPSNs). These findings indicate that strategies to increase DDX3X expression or activity may have therapeutic potential for C9ORF72-ALS/FTD.
Results
CRISPR-Cas9 screens for modifiers of C9ORF72 DPR protein production
To identify potential genetic modifiers of DPR protein production, we established stable reporter cell lines suitable for unbiased genome-wide CRISPR-Cas9 screening. We fused (GGGGCC)70 repeats (including preceding intronic sequences) with EGFP in the GA-frame (Figure 1A) and engineered this reporter in retinal pigment epithelium (RPE-1) cells, a near-diploid human cell line (Figure 1B). Another fluorescent protein RFP670 was expressed by canonical AUG-translation as an internal control. We then established single cell clones that stably express sharp signals of both EGFP (representing DPR proteins by RAN translation) and RFP670 (representing canonical translation) revealed by flow cytometry (Figure 1C). Immunoprecipitation for the C-terminal EGFP or MYC tag followed by immunoblotting with either GA antibody or GFP antibody revealed a predominant product corresponding to GA70-EGFP (Figure S1A), confirming EGFP can represent the DPR protein level. This allows us to perform Fluorescence Activated Cell Sorting (FACS)-based forward genetic screening for candidate genes that can influence DPR protein production (Figure 1D).
Figure 1: Genome-wide CRISPR-Cas9 knockout screens in human cells identify modifiers of DPR protein production from C9ORF72 GGGGCC repeats.
(A) The diagram of the C9R-EGFP reporter construct.
(B) Flow cytometry of a single cell clone expressing C9R-EGFP in the GA frame.
(C) Flow cytometry of a single cell clone expressing C9R-EGFP for RAN translation and RFP670 for AUG-translation.
(D) Flow chart of the CRISPR-Cas9 screening procedure. RPE-1 reporter cells expressing Cas9 were infected with the lentiviral sgRNA KO library. The total infected cells before FACS, and the top GFP-high and GFP-low cell populations collected by FACS, were subjected to deep sequencing and statistical analysis for sgRNA distribution.
(E) The volcano plot visualizes gene knockout enrichment in cells with different DPR expression levels. Red: genes conferring up-regulation of GA-GFP levels when knocked out (10% FDR). Blue: genes conferring down-regulation of GA-GFP levels when knocked out (10% FDR).
(F) Functional enrichment of candidate modifiers identified from the genome-wide knockout screens.
(G) Validation of screen hits in the independent dual-luciferase reporter cells by siRNA knockdown. HeLa Flp-In monocistronic reporter cells were induced to express translation reporters by doxycycline after two days of siRNA transfection, and luciferase activities were measured after another 24 hours. The Nanoluc luciferase represents RAN translation of C9ORF72 GGGGCC repeats (C9R-NLuc), and the Firefly Luciferase is produced by canonical translation (AUG-FLuc). The relative expression was compared to non-targeting siRNA control. Data are mean ± s.e.m. from three biological replicates.
We stably expressed Cas9 nuclease in the reporter cells and conducted the genome-wide CRISPR-Cas9 knockout screening using a lentiviral single guide RNA (sgRNA) library comprising 10 sgRNAs per gene and targeting 20,567 protein-coding genes in the human genome, along with ~10,000 negative control sgRNAs (Morgens et al., 2017). The cells were maintained at least 1000-fold of the library complexity and the top 10% GFP-low and top 10% GFP-high cell populations with similar RFP670 levels were collected by FACS in duplicates (Figure 1D). We also used two sub-libraries enriched in genes involved in RNA metabolisms and performed the screening with more stringent condition (collecting the top 2% GFP-low and top 2% GFP-high cells). We then performed high-throughput sequencing to analyze and compare the distribution of each sgRNA among the GFP-low, GFP-high and the whole population of knockout cells (Figure 1D). sgRNAs that increase the GFP-GA level are expected to be enriched in the GFP-high cells and depleted in the GFP-low cells, and vice versa for sgRNAs that decrease the GFP-GA level (Figure 1D). We used the Cas9 high-throughput maximum-likelihood estimator (casTLE) algorithm (Morgens et al., 2016) to identify statistically significant suppressors and enhancers of DPR protein production (Figure 1E). With a 10% false discovery rate (FDR) and combining the two rounds of genomic and sub-library screens, we identified 221 modifiers of GFP-GA level, 76 enhancer genes (sgRNAs enriched in GFP-low) and 145 suppressor genes (sgRNAs enriched in GFP-high) (Table S1). Gene ontology analysis revealed enrichment of genes involved in translation and RNA transport (Figure 1F), two key RNA processing steps that can influence final protein expression levels. In addition, we also found significant enrichment of proteasome components, especially in the 145 suppressor genes, indicating GA-GFP might be turned over by proteasome degradation. As the readout of the screen is steady-state protein levels, enrichment of RNA processing and protein degradation-related genes indicates the potency of this screen approach.
We further used an independent approach to validate the candidate genes using the previously established inducible dual-luciferase reporter cells (Cheng et al., 2018). The Nanoluc Luciferase is generated through RAN translation of C9ORF72 GGGGCC repeats (C9R-NLuc), and the Firefly Luciferase is produced by canonical translation (AUG-FLuc) (Cheng et al., 2018). This system can better distinguish repeat-dependent activities from general effects on canonical pathways, and allows validation in multiple reading frames. As we are mostly interested in the regulation of DPR protein production, we primarily focused on genes involved in RNA metabolism and performed validation by knocking down each candidate gene using siRNA transfection. We tested 48 candidate genes, and validated 38 of them could affect the C9R-NLuc levels (Table S2). Among these, 19 candidate genes specifically or preferentially influenced the RAN translation reporter in both GA and GP reading frames (Figure 1G and Table S2). We focused on these genes as they are more likely to be directly involved in repeat RNA processing.
NXF1-NXT1 RNA export pathway modulates DPR protein production
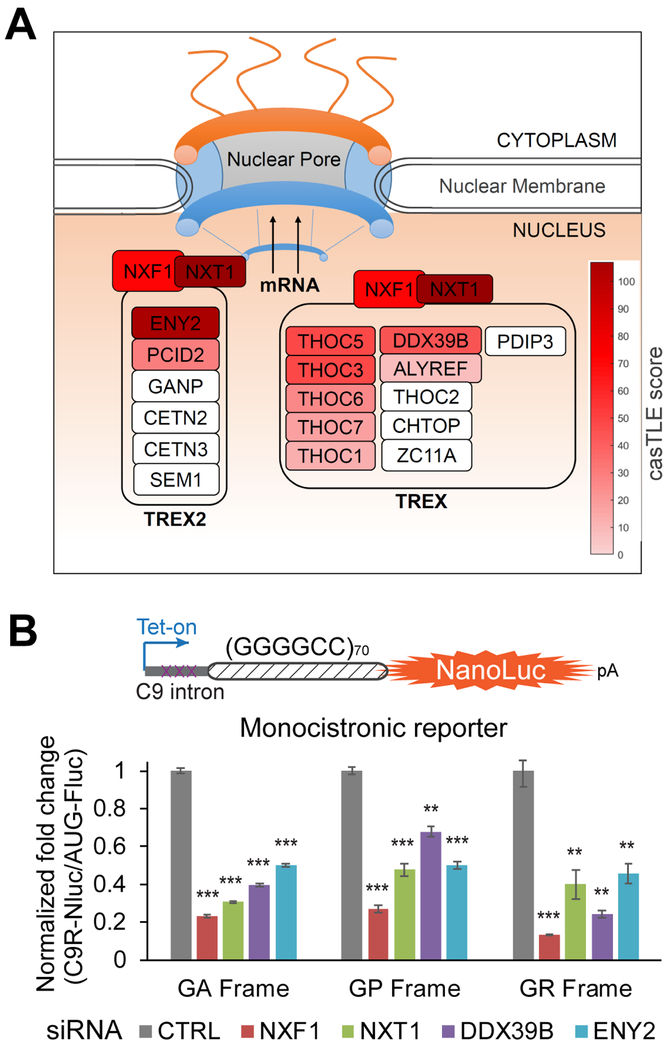
Mature mRNAs need to be exported from the nucleus to the cytoplasm to be translated into proteins. mRNA export is mainly mediated by nuclear RNA export factor 1 (NXF1) and its cofactor p15 (also known as NXT1), which are the key components of two conserved complexes: transcription-export complex (TREX) and TREX-2 (Wickramasinghe and Laskey, 2015). sgRNAs targeting several genes in the TREX and TREX-2 complexes were found to be enriched in the GFP-low cells from the screen (Figure 2A), strongly suggesting this pathway is essential for repeat RNA export, thereby affecting DPR protein production from all the reading frames. To confirm this finding, we generated new dual-luciferase reporter cell lines for the GR frame, using the same strategy as for GA and GP (Cheng et al., 2018). The RAN translation product of GR-frame is much lower than GA and GP (Figure S1B), which is consistent with previous reports (Green et al., 2017; Tabet et al., 2018). Using siRNAs to knock down each candidate gene in the NXF1-NXT1 pathway, we observed more substantial reduction of C9R-NLuc than AUG-FLuc in all three reading frames (Figure 2B). We further fractionated cells to separate the nucleus and the cytoplasm to test the distribution of GGGGCC repeat-containing RNAs. GAPDH pre-mRNA was highly enriched in nucleus and the mitochondria 12S RNA MT-RNR1 was predominantly in cytoplasm, confirming the successful fractionation (Figure S1C). The relative cytosolic amount of C9R-NLuc RNA was reduced when NXT1 was knocked down, but the FLuc RNA distribution was not altered (Figure S1C), consistent with the changes of the two luciferase activities (Figure 1G). This result confirms that the NXF1-NXT1 pathway mediates the nuclear export of GGGGCC repeat-containing RNA, thereby influencing the DPR protein production level.
Figure 2: NXF1-NXT1 RNA export pathway modulates DPR protein production.
(A) Schematic of the NXF1-NXT1 RNA export pathway. The components identified from the screens were colored with red, the gradient of which is proportional to the enrichment significance (casTLE score).
(B) HeLa Flp-In monocistronic reporter cells were induced to express translation reporters by doxycycline after two days of siRNA transfection, and luciferase activities were measured after another 24 hours. NLuc signals were normalized to FLuc in each sample and the relative expression was compared to non-targeting siRNA control. *P<0.05, **P<0.01, ***P<0.001, two-tailed t-test. Data are mean ± s.e.m. from three biological replicates.
See also Figure S1.
DDX3X is a modifier of DPR protein production
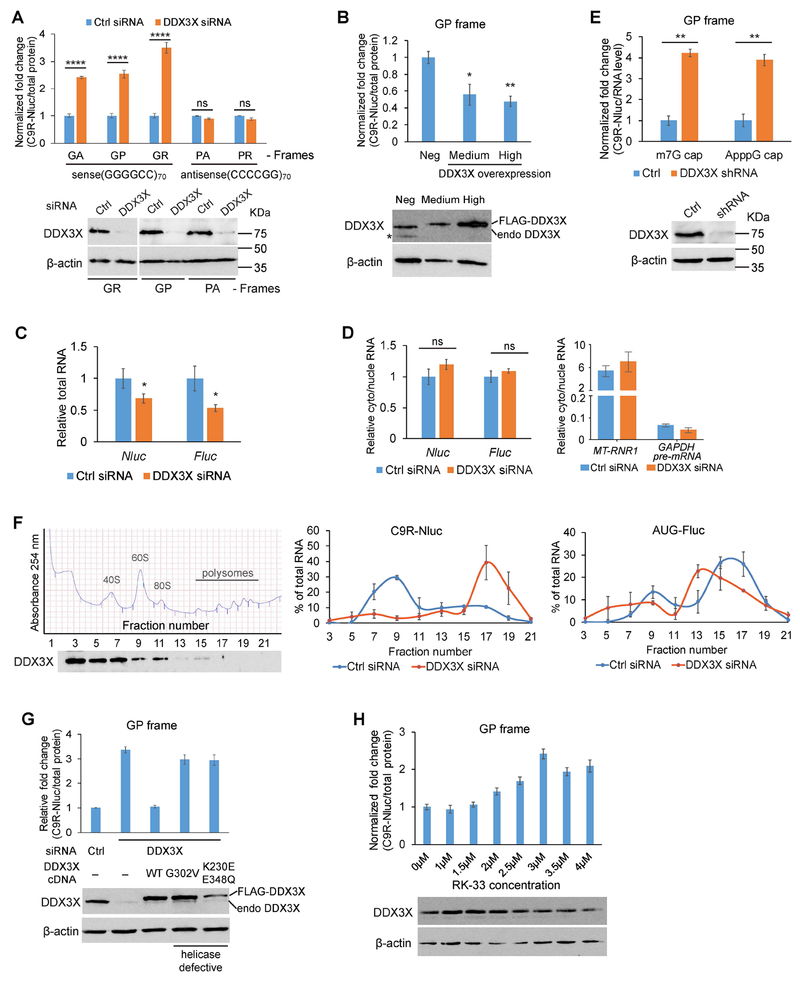
Genes with helicase activities are highly enriched in the CRISPR-Cas9 screen (Figure 1F). DDX3X, a DEAD-box RNA helicase, showed the strongest and most specific effect in the siRNA validation (Figure 1G and Table S2). DDX3X uses ATPase activity to remodel RNA structures and RNA-protein complexes, and has been implicated in multiple aspects of RNA metabolism, including pre-mRNA splicing, mRNA turnover, export, and translation (Linder and Jankowsky, 2011; Tarn and Chang, 2009). In order to evaluate the effect of DDX3X on the repeat RNA more precisely, we normalized NLuc to total protein amount. Reduction of DDX3X dramatically increased C9R-NLuc in all the three reading frames of sense repeat RNA (Figure 3A). Conversely, overexpression of DDX3X repressed the production of DPR proteins (Figure 3B). Interestingly, knocking down DDX3X did not change the production of poly-PA and poly-PR from antisense repeats (Figure 3A), suggesting its specific effect on the GGGGCC repeats.
Figure 3: DDX3X represses RAN translation of C9ORF72 GGGGCC repeats.
(A) After two days of siRNA transfection, HeLa Flp-In dual-luciferase reporter cells were induced to express translation reporters by doxycycline and luciferase activities were measured after another 24 hours. Top: NLuc signals were normalized to total protein in each sample and the relative expression was compared to non-targeting siRNA control. ****P<0.0001, two-tailed t-test. Data are mean ± s.e.m. from three biological replicates. Bottom: Immunoblotting of DDX3X showed the knockdown efficiency. β-actin was blotted as internal control.
(B) FLAG-DDX3X and GFP constructs were co-transfected into the GP reporter cells. After 24 h induction, the 20% highest GFP-expressing cells, 20% medium GFP-expressing cells, and GFP-negative cells were collected by FACS for subsequent assays. Top: NLuc signals were normalized to total protein in each sample and the relative expression was compared to negative control. *P<0.05, **P<0.01, two-tailed t-test. Data are mean ± s.e.m. from three biological replicates. Bottom: Immunoblotting of DDX3X showed the overexpression level. * non-specific product. β-actin was blotted as internal control.
(C) C9R-NLuc and FLuc total RNAs were measured by qRT-PCR in control or DDX3X siRNA transfected reporter cells. *P<0.05, two-tailed t-test. Error bars represent s.e.m. in three biological replicates.
(D) Cells were fractionated to separate nucleus and cytoplasm. The levels of nuclear marker GAPDH pre-mRNA and cytosolic marker mitochondria RNA MT-RNR1 (Right), as well as C9R-NLuc and FLuc RNA (Left), were measured by qRT-PCR and normalized to GAPDH mRNA in each fraction. The ratio of cytosolic/nuclear RNA showed the sub-cellular distribution of each RNA. Error bars represent s.e.m. in three biological replicates.
(E) In vitro transcribed C9R-Nluc RNA (frame-GP) with either 5’-m7G cap or ApppG cap analogue was transfected into either control or DDX3X knockdown HeLa cells. Top: The NLuc luciferase was normalized to RNA level in each condition. **P<0.01, two-tailed t-test. Data are mean ± s.e.m. from three biological replicates. Bottom: Immunoblotting of DDX3X showed the knockdown efficiency. β-actin was blotted as internal control.
(F) Reduction of DDX3X enhances the polysome association of C9R-NLuc RNA. Left: UV absorbance (254 nm) profile of sucrose gradient fractionations of cytoplasmic extracts from HeLa RAN translation reporter cells. The distribution of DDX3X across the ribosome profile was shown by immunoblotting. Middle and right panels: Quantification of C9R-NLuc RNA (middle) and AUG-FLuc RNA (right) distribution in polysome gradients. Each RNA was normalized to the spike-in CLuc internal control, and the relative levels in each fraction were calculated as percentage of the total levels from all the fractions. Data are mean ± s.e.m. from three biological replicates.
(G) siRNA and plasmid DNA encoding FLAG-tagged wild type or mutant DDX3X were co-transfected into HeLa Flp-In GP reporter cells. Immunoblotting of DDX3X showed the knockdown efficiency and exogenous expression levels. NLuc signals were normalized to total protein in each sample and the relative expression was compared to non-targeting siRNA control. Data are mean ± s.e.m. from three biological replicates.
(H) Reporter cells were treated with increasing dosage of DDX3X ATPase inhibitor RK-33 for 24 h. Immunoblotting showed the total DDX3X levels did not change significantly. NLuc signals were normalized to total protein in each sample and the relative expression was compared to DMSO treatment control. Data are mean ± s.e.m. from three biological replicates.
DDX3X represses (GGGGCC)n RAN translation
To determine how DDX3X modulates DPR levels, we systematically measured the levels of repeat-containing RNA and examined its subcellular distribution. Endogenous DDX3X is mainly localized in the cytoplasm (Figure S2A), and reduction of DDX3X did not change the number and size of nuclear RNA foci (Figure S2B,C). qRT-PCR using total RNA showed that there was a slight reduction of C9R-NLuc RNA in the DDX3X knockdown cells (Figure 3C), in contrast with the upregulation of luciferase activity (Figure 3A). The relative amount of C9R-NLuc and FLuc RNAs in the cytosolic versus nuclear fractions did not change dramatically after reducing DDX3X (Figure 3D), suggesting the effect on DPR production is not due to reduced RNA export.
We next used two independent approaches to further explore whether DDX3X affects RAN translation directly. First, we in vitro transcribed the C9R-NLuc RNA and transfected into cells with or without stable DDX3X knockdown by shRNA. We observed significant higher amount of DPR proteins produced in the DDX3X knockdown cells compared to control (Figure 3E). The upregulation is similar on the RNAs with and without the functional 5’-cap (Figure 3E), indicating the 5’-cap is not essential for the translation changes caused by DDX3X. Second, we used sucrose gradients to directly analyze the distribution of C9R-NLuc RNA on polyribosomes in reporter cells. We fractionated the cytoplasmic extracts on 10-50% sucrose gradients and measured RNA levels in each fraction by qRT-PCR. In contrast to FLuc RNA, which peaked in the heavy polyribosome fractions, the main peak of C9R-NLuc RNA in control cells was in the monoribosome fractions (Figure 3F). This distribution is consistent with the low efficiency of RAN translation compared to canonical AUG-translation. DDX3X knockdown shifted the C9R-NLuc RNA peak toward the polyribosome fractions, demonstrating reduction of DDX3X enhances the translation initiation efficiency of the repeat RNA (Figure 3F).
DDX3X alteration in the HeLa reporter cells did not lead to changes on cell proliferation rate (Figure S3A,B) or apoptosis (Figure S3C). There was also no induction of stress granules (Figure S3D) or elevation of the phospho-eIF2α level (Figure S3E) upon DDX3X knockdown, indicating the upregulation of RAN translation is not due to the integrated stress response (Cheng et al., 2018; Green et al., 2017). We also used puromycin incorporation to measure all the actively translating nascent polypeptides (Schmidt et al., 2009), and observed no major changes in control versus DDX3X knockdown or overexpressing cells (Figure S3F,G). This indicates the changes of DDX3X levels in our experiments did not affect global protein synthesis and general cell status. Altogether, these data suggest that the translational regulation of (GGGGCC)n repeat RNA is directly modulated by DDX3X.
DDX3X helicase activity is required for RAN translation repression
DDX3X was predominantly found in the monoribosome fractions (Figure 3F), implicating its role in the translation initiation step. We speculate that the secondary structure of GGGGCC repeats is essential for RAN translation, and the RNA unwinding activity of DDX3X might disrupt or alter this structure and repress this non-canonical translation initiation. As the RNA unwinding function requires ATP hydrolysis by DDX3X (Tarn and Chang, 2009), we examined whether the ATPase activity of DDX3X is required for the RAN translation repression. First, we expressed siRNA-resistant cDNAs of either wild type or ATPase defective mutants in DDX3X knockdown cells and examined their rescue activities. We used two DDX3X mutants, G302V and K230E/E348Q, which were reported to be defective in RNA-stimulated ATP hydrolysis activity (Russell, 2015; Shih et al., 2008; Yedavalli et al., 2004). They both have similar expression patterns and subcellular localization as the wild type protein (Figure S4A). The upregulation of RAN translation was only reduced by expression of the wild type DDX3X, but not by the two mutants (Figure 3G). Alternatively, we treated cells with a small molecule RK-33, which was reported to specifically bind the ATP-binding cleft of DDX3X and inhibits its helicase activity (Bol et al., 2015a). The RAN translation products showed dosage-dependent increase although the total DDX3X level was not significantly changed (Figure 3H, S4B). Taken together, these findings establish that DDX3X helicase activity is essential to repress RAN translation of the (GGGGCC)n repeat RNA.
DDX3X directly binds GGGGCC repeat RNA
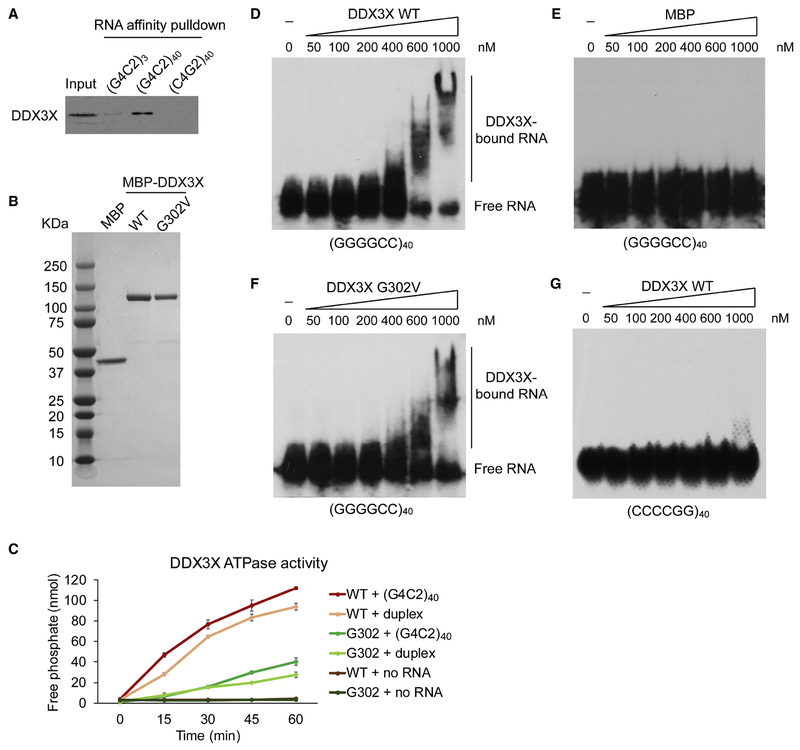
We next examined the interaction between DDX3X and the GGGGCC repeats. We synthesized (GGGGCC)40 or (CCCCGG)40 RNA repeats excluding the surrounding C9ORF72 intron sequences by in vitro transcription. RNA affinity pulldown using biotin-labeled RNA showed that only the GGGGCC repeats pulled down the DDX3X protein from cell extract, but the antisense CCCCGG repeats did not (Figure 4A), consistent with the specific effect of DDX3X on the sense repeat RNA translation (Figure 3A). We next purified the recombinant DDX3X protein (Figure 4B) and tested its RNA binding properties in vitro. The ATPase activity of DDX3X was reported to be stimulated by a hybrid double-stranded/single-stranded RNA duplex substrate, but not by single-stranded RNA (ssRNA) or blunt double-stranded RNA (dsRNA) (Epling et al., 2015). We observed that its ATPase activity was dramatically stimulated by the (GGGGCC)40 RNA, comparable to the effect induced by the positive control of partial duplex RNA (Figure 4C). As expected, the G302V mutant exhibited extremely low RNA-stimulated ATPase activities (Figure 4C). We also performed electrophoretic gel mobility shift assay (EMSA) to visualize the direct binding between RNA repeats and DDX3X. The sense (GGGGCC)40 RNA shifted to reduced mobility (RNA-DDX3X complexes) with increasing protein concentrations (Figure 4D), but not with the MBP tag alone (Figure 4E). The G302V mutant showed similar binding affinity on the (GGGGCC)40 repeats (Figure 4F), suggesting the ATPase and helicase activity is not essential for binding to the repeat RNA. Consistent with previous experiments (Figure 3A and 4A), the antisense (CCCCGG)40 RNA cannot be bound and shifted by DDX3X (Figure 4G). Altogether, these results indicate that DDX3X directly and selectively binds to the GGGGCC repeat RNA, which activates its ATPase activity for RNA structure unwinding.
Figure 4: DDX3X directly binds GGGGCC repeat RNA.
(A) Affinity pulldown of biotin-labeled repeat RNAs and their interacting proteins from HeLa cell extract, followed by immunoblotting with DDX3X antibody.
(B) MBP and MBP-tagged DDX3X wild type or G302V mutant were expressed and purified from bacteria, separated on SDS-PAGE and stained with Coomassie blue.
(C) Time-dependent RNA-stimulated ATPase activity of 1 pmol DDX3X WT or G302 mutant in the presence of 2 pmol RNA and 0.1 mmol ATP was measured by quantification of phosphate release. Data are mean ± s.e.m. from three biological replicates.
(D-G) EMSA of in vitro transcribed radiolabeled RNA repeats incubated with increasing doses of purified DDX3X proteins. (D) EMSA for MBP-DDX3X WT with (GGGGCC)40 RNA. (E) EMSA for MBP with (GGGGCC)40. (F) EMSA for MBP-DDX3X G302V with (GGGGCC)40.
(G) EMSA for MBP-DDX3X WT with antisense repeat (CCCCGG)40 RNA.
See also Figure S4.
Several reports suggest that (GGGGCC)n RNA forms G-quadruplex structures (Fratta et al., 2012; Haeusler et al., 2014; Reddy et al., 2013; Su et al., 2014), and also likely mixes with alternative structures including hairpins (Su et al., 2014). Interestingly, besides the well-known binding activity on partial dsRNA duplex structures (Epling et al., 2015), DDX3X is also recently revealed as a novel interactor of G-quadruplex-containing transcripts (Herdy et al., 2018). We therefore tested whether DDX3X binds the G-quadruplex structure of the GGGGCC repeats. We performed the gel shift assay in the presence of LiCl instead of KCl, as it is known that K+ stabilizes G-quadruplexes while Li+ destabilizes the structure (Hardin et al., 1992). Surprisingly, DDX3X binds the (GGGGCC)n repeat RNAs with similar affinity in the presence of Li+ compared to K+ (Figure S4C), suggesting the binding is not mediated through the G-quadruplex structure.
Loss of DDX3X enhances repeat-mediated toxicity in Drosophila
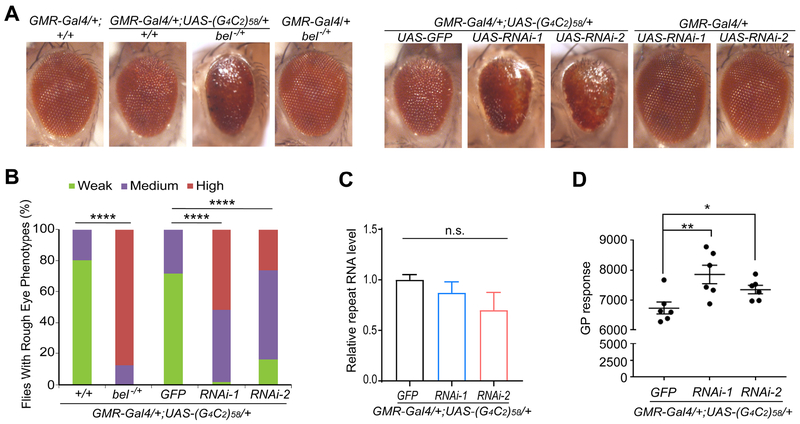
Next, to test the effect of modulating DDX3X levels in vivo, we used a transgenic fly line engineered to express 58 copies of the GGGGCC repeat under the control of the GMR-Gal4 driver., Expression of the expanded repeats caused a retinal degeneration phenotype (Figure 5A), as previously shown (Freibaum et al., 2015). Partial loss of bel, the Drosophila orthologue of DDX3X, either through the mutant allele (belEY08943) or RNAi knockdown (belJF02884 and belGL00205), exacerbated the retinal degeneration phenotype induced by expanded GGGGCC repeats (Figure 5A,B); heterozygous mutant flies or flies expressing RNAi alone did not show retinal degeneration (Figure 5A). The ectopic expression of Bel partially rescued the enhanced retinal degeneration induced by bel knockdown in flies expressing expanded repeats (Figure S5). Correlating with the modifier effect of bel, we found that Bel knockdown increased DPR protein production as indicated by elevated poly(GP) level without affecting the level of RNA containing GGGGCC repeats (Figure 5C,D). Therefore, bel is a genetic modifier of the GGGGCC repeats-mediated toxicity in vivo.
Figure 5: Partial loss of bel function enhanced expanded GGGGCC repeat toxicity in Drosophila.
(A) Representative images of external eye phenotypes of adult files. (G4C2)58 expression driven by the GMR-Gal4 driver induces a rough eye phenotype. Partial loss of bel function either through the mutant allele (belEY08943) or RNAi knockdown (belJF02884 and belGL00205) enhances (G4C2)58 toxicity. UAS-GFP was used as control for UAS-RNAi lines. Partial loss of bel by itself has no phenotype. The genotypes of all flies examined here are described in detail in the Methods section.
(B) Quantification of the rough eye phenotypes of flies with various genotypes as presented in Panel (A). ****P < 0.0001, by chi-square analysis. The number of flies analyzed for each genotype ranges from 50 to 200.
(C) Repeat RNA level was measured by qRT-PCR and is shown to be not changed by RNAi knockdown (belJF02884 and belGL00205). Values are mean ± s.e.m. from four independent experiments, analyzed by one-way ANOVA.
(D) ELISA analysis of poly(GP) protein levels in the heads of 5-day-old flies. GP levels are increased by RNAi knockdown of bel expression. Values are mean ± s.e.m. of six independent experiments, analyzed by one-way ANOVA with Dunn’s post hoc test.
See also Figure S5.
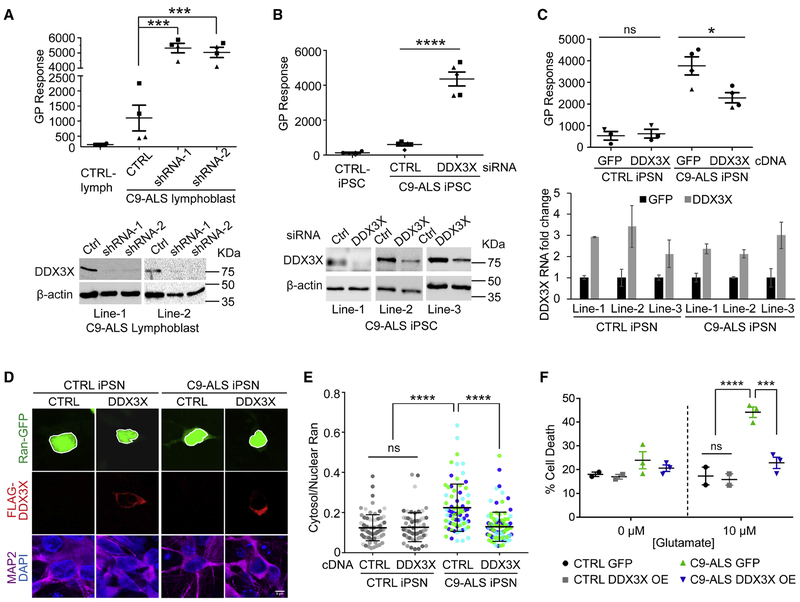
DDX3X modifies endogenous DPR level and toxicity in patient cells
We further validated the findings on the endogenous DPR protein production and toxicity in patient cells. We developed an enzyme-linked immunosorbent assay (ELISA) to measure poly-GP levels quantitatively (Figure S6A). DDX3X was reduced by two individual shRNAs in lymphoblast cells derived from C9ORF72-ALS patients. The ELISA assay showed that GP levels dramatically increased upon DDX3X knockdown in two independent patient cell lines (Figure 6A). We also tested the effect in three lines of iPSCs from ALS patients carrying C9ORF72 repeat expansion (Donnelly et al., 2013; Zhang et al., 2015). DDX3X knockdown by siRNA transfection induced dramatic increase of GP levels (Figure 6B), without influencing iPSC proliferation or apoptosis (Figure S6B,C). We also differentiated the iPSCs into a mixed population of neurons containing about 20-30% islet-1-positive motor neurons (Figure S6D), which yields much higher GP amount than iPSCs (Figure 6C). A 2-3-fold overexpression of exogenous DDX3X leads to significant reduction of GP levels (Figure 6C). In both knockdown and overexpression conditions, the RNA levels of the repeat-containing C9ORF72 intron were not or only modestly affected (Figure S6E,F), which cannot account for the changes in DPR protein levels. These findings strongly suggest that DDX3X represses RAN translation of C9ORF72 sense repeats in patient cells.
Figure 6: DDX3X modulates DPR protein levels and DPR-mediated toxicity in patient cells.
(A) DDX3X in two C9ORF72-ALS patient lymphoblast cell lines was knocked down by two individual shRNAs. (Top) Poly-GP was measured by ELISA. Different shapes of data points represent different lymphoblast lines. ***P<0.001, two-tailed t-test. Data are mean ± s.e.m. from two lines with two biological replicates each. (Bottom) Immunoblotting of DDX3X showed the knockdown efficiency. β-actin was blotted as internal control.
(B) C9ORF72-ALS iPSCs were transfected with either non-targeting control or DDX3X siRNA. (Top) Poly-GP was measured by ELISA. Different shapes of data points represent different iPSC lines. ****P<0.0001, two-tailed t-test. Data are mean ± s.e.m. from three C9ORF72-ALS cell lines and two with two biological replicates. (Bottom) Immunoblotting of DDX3X showed the knockdown efficiency. β-actin was blotted as internal control.
(C) iPSNs were infected with lentivirus expressing either DDX3X or GFP as negative control. (Top) Poly-GP was measured by ELISA. Different shapes of data points represent different lines. *P<0.05, two-tailed t-test. Data are mean ± s.e.m. from three control and three C9ORF72-ALS cell lines and one with two biological replicates. (Bottom) qRT-PCR of DDX3X RNA showed the fold of overexpression compared to the endogenous level in each line. Data are mean ± s.e.m. from two technical replicates.
(D) Immunofluorescence of exogenously expressed FLAG-tagged DDX3X, Ran-GFP reporter, and MAP2 as neuronal marker in iPSC-derived neurons. The scale bar represents 5 μm.
(E) Quantification of neuronal C/N Ran ratios in (D) showed increased cytoplasmic Ran levels in C9ORF72-ALS iPSN compared to control, and overexpression of DDX3X decreased cytoplasmic Ran. Neurons expressing both Ran-GFP and FLAG-DDX3X were measured and compared with the negative control. Data points of different colors represent individual cells from different cell lines. 25-40 neurons were quantified at each condition. ****P<0.0001, one-way ANOVA with Dunn’s post hoc test. Data are mean ± s.d. from three C9ORF72-ALS iPSN lines and three CTRL iPSN lines.
(F) Glutamate-induced excitotoxicity of C9ORF72 iPSNs was significantly higher than control iPSNs. Exogenous expression of DDX3X prevents glutamate-induced C9ORF72 iPSN cell death. Each data point represents one individual line. About 500 neurons were quantified at each condition. ***P<0.001, ****P<0.0001, two-way ANOVA with Tukey’s post hoc test. Data are mean ± s.d. from two CTRL iPSN lines and three C9ORF72-ALS iPSN lines.
See also Figure S6.
We next examined whether ectopic expression of DDX3X can have beneficial effects on C9ORF72 patient cells. Nucleocytoplasmic transport has been shown to be dysfunctional in C9ORF72-ALS patient cells (Freibaum et al., 2015; Jovicic et al., 2015; Zhang et al., 2015; Zhang et al., 2016). A pathological phenotype in the C9ORF72-ALS iPSNs is the alteration of cytoplasmic-nuclear (C/N) distribution of Ran protein (Zhang et al., 2015). We therefore examined whether DDX3X overexpression can rescue the impaired Ran gradient in patient iPSNs. We co-transfected cDNA constructs encoding DDX3X and Ran-GFP reporter (Zhang et al., 2015) into the differentiated neurons and quantified C/N ratio of Ran via immunofluorescence after 20-22 additional days of differentiation and maturation. Compared to non-ALS control, C9ORF72 iPSNs showed increased C/N Ran gradient as reported (Figure 6D,E) (Zhang et al., 2015). Exogenous DDX3X expression decreased the cytosolic fraction of Ran and rescued the Ran gradient abnormalities (Figure 6D,E). Similar rescue effect was also observed using the NLS-tdTomato-NES nucleus-cytoplasm transport reporter (Figure S6G,H), which has been used to monitor nuclear-cytoplasmic trafficking and was shown to be dysregulated in C9ORF72-ALS iPSNs (Zhang et al., 2015). Finally, we tested whether DDX3X overexpression can improve neuron survival. It has been reported that C9ORF72-ALS iPSNs are more sensitive to glutamate-induced excitotoxicity than controls (Donnelly et al., 2013). We expressed the exogenous DDX3X or GFP by high efficient lentivirus infection (Figure S6I), and measured the percentage of cell death after glutamate treatment. DDX3X overexpression reduced the glutamate-induced neuron death and improved survival of C9ORF72-ALS iPSNs (Figure 6F). Altogether, these results strongly indicate that DDX3X can modulate the repeat-mediated toxicity in patient cells by regulating the DPR production from RAN translation.
Discussion
We performed FACS-based CRISPR-Cas9 KO screens to discover genetic modifiers of C9ORF72 DPR production derived from sense GGGGCC repeats. Identification of genes enriched in RNA transport, translation and proteasome pathways strongly indicates the specificity and promise of using this comprehensive screening approach to uncover disease mechanisms. We identified several factors in the NXF1-NXT1 mRNA export machinery, and their reduction leads to lower DPR production. A recent report also showed the NXF1-dependent nuclear export of GGGGCC repeat-containing transcript using a candidate approach (Hautbergue et al., 2017). The convergent findings showed that the unbiased CRISPR screening is highly effective to identify novel genetic modifiers of DPR production. Discovery of specific modifiers of repeat RNA export and RAN translation pathways implicates that the repeat RNA processing can be regulated and potentially targeted for therapeutic utility, and variation of DPR levels in different cell types or ages might contribute to disease vulnerability.
RNA helicases are highly conserved enzymes that use ATP to remodel RNA structures or ribonucleoprotein complexes (RNPs) (Jankowsky, 2011). In eukaryotes, they participate in almost all aspects of RNA metabolism. The (GGGGCC)n RNA has been reported to form complex structures, including G-quadruplex and hairpins (Fratta et al., 2012; Haeusler et al., 2014; Reddy et al., 2013; Su et al., 2014). Therefore, it is exciting to identify helicases as potential modifiers of repeat RNA metabolism. DDX3X is a prime example of the DEAD box family of RNA helicases, and has been implicated in multiple RNA processes including transcription, mRNA export, translation and decay (Russell, 2015; Sharma and Jankowsky, 2014). The exact role of DDX3X on global canonical translation remains to be clarified as both stimulation and inhibition have been suggested, and several studies showed that only specific subsets of mRNAs are affected by loss of DDX3X (Guenther et al., 2018; Russell, 2015; Sharma and Jankowsky, 2014; Valentin-Vega et al., 2016). This is consistent with our observation that global translation was not altered by DDX3X knockdown. Studies of DDX3X RNA binding profiles in mammalian cells revealed ~4000 peaks on mRNAs, with enrichment at 5’-UTR (Herdy et al., 2018; Valentin-Vega et al., 2016). The translation of only selected RNA targets were shown to be reduced upon loss of DDX3X (Valentin-Vega et al., 2016). One proposed mechanism is that DDX3X promotes canonical translation by destabilizing complex RNA structures at 5’ UTR regions through interacting with the cap-dependent translation initiation complex eIF4F (Soto-Rifo et al., 2012). Our work identified that DDX3X directly binds on the GGGGCC repeat RNA, and functions as a repressor for RAN translation independent of 5’-cap, indicating a different mechanism. The requirement of helicase activity suggests that the repeat RNA structure is essential to promote initiation of RAN translation, and alteration or disruption of the structure by helicases can decrease the initiation efficiency. Interestingly, repressing Ded1p (DDX3 orthologue in yeast) was shown to lead to accumulation of RNA structure and increase near-cognate initiation codon usage (Guenther et al., 2018), an effect consistent with the RAN translation changes. The gel mobility shift assay indicates that DDX3X does not bind the G-quadruplex structure of GGGGCC repeats, but probably other structures, such as hairpins. Consistent with our finding, a recent work also reported that the hairpin form of (GGGGCC)n RNA is involved in RAN translation and is a target for small molecules (Wang et al., 2019). As the endogenous RNA structure modulator, the cellular helicases also have the potential to be therapeutically relevant targets.
There have been extensive studies on the toxicities induced by DPR proteins. Most of the previous work used exogenous AUG-driven expression of DPR proteins with relatively short length, which might not accurately reflect the toxic contribution from the endogenous DPR proteins in ALS/FTD. Our observation that inhibition on RAN translation by DDX3X reduces the pathological phenotypes and neuron death of C9ORF72-ALS iPSNs supports the pathogenic contribution from DPRs in patient cells. This study raises the possibility of changing repeat RNA metabolisms by manipulating cellular helicases as a strategy to decrease toxicities caused by repeat expansion. It is also likely to be informative to explore whether DDX3X (and other candidate genes revealed from the genetic CRISPR screening) could be a risk factor that can contribute to neurodegeneration by modulating DPR levels in different cell types or brain regions. Strategies to increase DDX3X expression or activity in selectively vulnerable neurons have the therapeutic potential for C9ORF72-ALS/FTD. It is noted that mutations of DDX3X have been linked to cancer (Jones et al., 2012; Robinson et al., 2012), but with opposing view that DDX3 could act as both a putative oncogene and tumor suppressor depending on the context (Bol et al., 2015b). More recently, de novo loss of function mutations were found to be causative for intellectual disability (Snijders Blok et al., 2015), a developmental disease. It will be important to explore how DDX3X elevation influences post-mitotic neurons in adult stage when pursuing its therapeutic utility for C9ORF72-ALS/FTD.
STAR METHODS
Detailed methods are provided in the online version of this paper and include the following
LEAD CONTACT AND MATERIALS AVAILABILITY
Human lymphoblast cells
Further information and requests for resources and reagents should be directed to, and will be fulfilled by the Lead Contact, Shuying Sun (shuying.sun@jhmi.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human cell lines
RPE-1 cells were grown in DMEM/F12 supplemented with 10% (v/v) FBS, 1.2 mg/ml Sodium Biocarbonate, 2mM L-Glutamine, Non-essential amino acid (NEAA), 100 U/ml penicillin and 100 μg/ml streptomycin.
HeLa Flp-In dual luciferase reporter cells (Cheng et al., 2018) were grown in DMEM supplemented with 10% (v/v) FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. All cells are maintained at 37°C in a humidified incubator supplemented with 5% CO2.
Human lymphoblast cells
Patient lymphoblast cells were obtained from Coriell Institute (Table S3). The cells were grown in RPMI1640 supplemented with 10% (v/v) FBS, 2mM L-Glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin.
Human iPSCs and diMNs
iPSCs were grown in mTeSR medium (StemCell Technologies, 85850) on matrigel coated plates. Peripheral blood mononuclear cell (PBMC)-derived iPSC lines from C9ORF72-ALS patients and non-neurological disease controls (Table S3) were obtained from the Cedars-Sinai Answer ALS repository. Presence or absence of the C9ORF72 repeat expansion was verified by repeat-primed PCR (RP-PCR) (Renton et al., 2011).
iPSCs were maintained and differentiated into spinal motor neurons according to the publicly-available 18-day ‘diMNs’ (direct iMN) protocol, which generates a mixed population consisting of 20-30% islet-1 positive motor neurons (http://neurolincs.org/pdf/diMN-protocol.pdf and a manuscript under review) (Sances et al., 2016).
Fruit flies
Fruitflies (Drosophila melanogaster) were raised at 25°C on standard medium. For all experiments, we used both adult male and female flies, between 5 and 10 days old. The UAS/GAL4 system was used to express transgene using tissue-specific promoter drivers.
METHOD DETAILS
Plasmids
For the C9R-EGFP reporter, EGFP lacking ATG start codon was subcloned downstream of the (GGGGCC)70 repeats in frame with GA by replacing the NLuc reporter (Cheng et al., 2018), which was cloned in the pCAG vector for constitutive expression. RFP670 was subcloned into retroviral vector pBABE via BamHI and SalI sites. The DDX3X cDNA was subcloned into pcDNA 3.1+ vector with N-terminal FLAG tag via BamHI and XhoI sites, and into pcDNA3.1-vector with N-terminal HA tag via XhoI and BamHI sites. The G302V, K230E/E348Q, and siRNA-resistant silent mutations were introduced by QuickChange mutagenesis. For recombinant protein expression and purification from Escherichia coli, DDX3X was cloned into pMAL-c2X (Addgene #72586) at the XbaI site using In-Fusion cloning (Takara) to generate MBP-DDX3X-6HIS. DDX3X G302V was generated from MBP-DDX3X-6HIS using QuickChange II site directed mutagenesis (Agilent). Lenti-Cas9 was acquired from Addgene (#52962). Lentiviral vectors expressing DDX3X or GFP were engineered by replacing Cas9 from Lenti-Cas9 vector with DDX3X with FLAG tag in the C-terminal or GFP cDNA by AgeI and BamHI sites. For in vitro transcription template DNA, (GGGGCC)40 was cloned into the pBluescirpt SK+ vector by XhoI + SalI. The DDX3X shRNA-1 and shRNA-2 constructs were obtained from Dharmacon (TRCN0000000003, TRCN0000000004).
Cell culture and transfection
To generate stable cell lines, RPE-1 cells were transfected with the C9R-EGFP reporter plasmid by nucleofection (Lonza, VACA-1003), followed by 400 μg/ml G418 selection. The cells were infected with retrovirus expressing RFP670, packaged by 293 Phenix cells, and infected cells were enriched by fluorescence-activated cell sorting (FACS). Single cell clones were subcultured and screened to establish ones with stable and sharp peak of GFP signal. The correct clones were infected with lentivirus expressing Cas9 and selected by 15μg/ml Blasticidin. 293T cells were used to package lentiviruses expressing Cas9, DDX3X shRNAs, DDX3X cDNA or CRISPR sgRNA library. 293T cells were grown in DMEM supplemented with 10% (v/v) FBS, 100 U/ml penicillin and 100 μg/ml streptomycin.
TransIT-LTI (Mirus) was used to transfect plasmid DNAs; Lipofectamine RNAiMAX (Invitrogen) was used to transfect siRNAs; TransIT-X2 (Mirus) was used to co-transfect plasmids and siRNAs for the rescue experiment. The reporter gene expression in HeLa Flp-In cells was induced with 2μg/ml doxycycline at 48 h after transfection, and samples were collected after another 24 h. siGENOME pooled siRNAs and siGENOME Non-Targeting siRNA (GE Dharmacon) were transfected at 25nM. A single siRNA of DDX3X (GCAAATACTTGGTGTTAGA) was used for the rescue experiment. For DDX3X overexpression experiment, a GFP-expressing plasmid was co-transfected into luciferase reporter cells, and the highest 20% and medium 20% GFP-expressing cells were isolated by FACS for subsequent assays. For RNA transfection experiment, the RNA was in vitro transcribed as described previously (Cheng et al., 2018) and transfected into cells by TransIT-mRNA transfection kit (Mirus), and samples were collected after 24 h. RK-33(SelleckChem) were added to cells 24h before harvest at 1, 1.5, 2, 2.5, 3, 3.5, 4μM concentration. Three wells of biological replicates were prepared at each condition for all dual-luciferase reporter experiments. The NLuc and FLuc luciferase activities were measured blindly by Nano-Glo Dual Luciferase Assay (Promega, N1620) on Tecan Infinite 200 PRO. NLuc levels were normalized to FLuc, or both were normalized to total protein amounts for each sample. Protein lysates were quantified by BCA Assay (ThermoFisher Scientific).
Lymphoblast cells were infected by lentivirus expressing DDX3X shRNAs to knock down DDX3X. The infected cells were selected by 1 μg/ml puromycin and harvested for protein and RNA measurement after 10 days.
For siRNA transfection in iPSCs, the dissociated cells were resuspended with the RNAiMAX/siRNA (350 nM) mixture and incubate at room temperature for 10mins before seeding in full volume of mTeSR medium (18 nM final siRNA concentration).
At day 12 post-differentiation, iPSNs were dissociated and transfected with Ran-GFP (or NLS-tdTomato-NES) (Zhang et al., 2015) and DDX3X-FLAG (or DDX3X-HA) plasmids or vector control using P3 Primary Cell 4D X Kit L (Lonza, V4XP-3024) by Amaxa 4D-Nucleofector (Lonza, AAF-1002B). Subsequently, the transfected cells were seeded on top of a confluent monolayer of mouse astrocytes. After another 20-22 days of differentiation, cells were harvested for immunofluorescence analysis. To overexpress DDX3X in iPSNs, the neurons were infected with lentivirus expressing GFP or DDX3X at day 18 post-differentiation, and then harvested 7 days after infection for protein and RNA assays. To test the glutamate-induced excitotoxicity, control and C9ORF72-ALS iPSNs with either GFP-FLAG or DDX3X-FLAG overexpression (18 days after infection) were treated with 10μM L-glutamate for 4hrs. Then the cells were stained with Hoechst33342 (5μg/ml) and Propidium Iodide (1μg/ml) for 30 min to visualize total and dead cells, respectively. Afterwards, the cells were washed with PBS to remove Propidium Iodide and followed by immunofluorescence of FLAG tag to confirm the infection efficiency.
CRISPR-Cas9 screens
The genome-wide CRISPR/Cas9 deletion library with ten sgRNAs per gene(Morgens et al., 2017) was packaged into lentivirus and infected into Cas9-expressing C9R-EGFP reporter RPE-1 cells. About 150 million cells were infected with the library at an MOI < 0.4. Infected cells underwent puromycin selection at 5 μg/ml for 3 d. Cells were maintained at 1,000×coverage (~1,000 cells containing each sgRNA) at all the steps during screening. 14 d and 17 d post-infection, 250 million cells were analyzed by FACS and the top 10% GFP-high and 10% GFP-low cell populations were collected. Genomic DNA was extracted for all screen populations separately using the QIAamp Blood Maxi Kit and Blood Mini Kit (Qiagen). The fragments containing sgRNAs were amplified by PCR and subjected to deep sequencing on an Illumina Nextseq platform to monitor the library composition. Sufficient sgRNA library representation was confirmed before and after FACS. Guide compositions between GFP-low, GFP-high, and total populations was compared using casTLE (Morgens et al., 2016). Briefly, the enrichment of individual sgRNAs was calculated as the log ratios different conditions, and gene-level effects were calculated from ten guides targeting each gene. A confidence score was then derived as a log-likelihood ratio describing the significance of the gene-level effect, and P values were then calculated as previously described (Morgens et al., 2016). Gene ontology analysis was performed using DAVID (https://david.ncifcrf.gov/).
Immunoprecipitation
RPE-1 reporter cells were lysed in IP lysis buffer. The lysates were clarified by centrifugation at 13,000g for 20 min at 4 °C. Dynabeads Protein G were washed twice with PBST, and incubated with GFP antibody (MCKCC) or MYC antibody (Sigma) for 1 h at room temperature. The beads were washed twice in PBST, and once in IP lysis buffer (0.3% (v/v) NP-40, 200 mM NaCl, 50 mM Tris, pH 7.4, 1 mM DTT, 0.1 mM EDTA, and protease-inhibitor cocktail), and then incubated with the cleared lysates at 4 °C overnight. After washing three times in lysis buffer, the beads were resuspended in SDS-containing gel sample buffer and electrophoresed on an SDS-PAGE gel.
Immunofluorescence, immunoblotting and antibodies
Cells were fixed with 4% (v/v) para-formaldehyde in PBS for 20 min, permeabilized in 0.2% (v/v) Triton X-100 for 5 min, blocked in 1% bovine serum albumin and 2% goat serum for 30 min, incubated with primary antibodies for 1 h, washed with PBS, and finally incubated with Alexa Fluor 488/546/647 conjugated secondary antibodies (ThermoFisher Scientific). Nuclei were counterstained with DAPI. Cells were imaged with a fluorescence microscope (Zeiss Axiophot). For immunoblotting, goat anti-mouse or anti-rabbit IgG HRP-conjugated (GE healthcare) was used along with chemiluminescent detection reagents (Thermo Scientific). The primary antibodies included DDX3X (Proteintech, 1:1000 for immunoblotting), DDX3X (Santa Cruz, 1:300 for immunofluorescence), β-actin (Cell Signaling, 1:1000 for immunoblotting), FLAG (Sigma, 1:1000 for immunoblotting, 1:200 for immunofluorescence), poly-GA (Rb4334, 1:1000 for immunoblotting), Cleaved-Caspase-3 (Cell Signaling, 1:400 for immunofluorescence), phospho-eIF2α (Cell Signaling, 1:1000 for immunoblotting), G3BP (BD, 1:300 for immunofluorescence), β3-Tubulin (Cell Signaling, 1:200 for immunofluorescence), MAP2 (Cell Signaling, 1:200 for immunofluorescence). For puromycin incorporation assay, cells were treated with 1μM puromycin for 30mins before harvesting. Puromycin-labelled protein was analyzed by immunoblotting with anti-puromycin (Sigma, 1:500).
For Ran gradient and NLS-tdTomato-NES nucleocytoplasmic transport reporter analysis in iPSC-neurons, cells were stained with MAP2 and DAPI to distinguish cytoplasm and nucleus. Z-stack images were taken blindly on a Zeiss LSM700 laser scanning confocal microscope, with consistent exposure time are laser setting without saturation. Full Z-stacks were taken at 0.5μm intervals and the individual planes were then projected into maximum intensity images for further image quantification.
Cell Cycle analysis
A total of 1×106 cells from each group were collected as cell suspensions, washed with cold hank’s balanced salt solution (HBSS) containing 2% FBS, fixed in 70% ethanol overnight on ice. Cells were then washed twice with HBSS containing 2% FBS and incubated with DAPI (10μg/ml) at room temperature for 30min. After staining, cells were filter through 40μm mesh filter and analyzed using an BD LSR II flow cytometer with a UV laser.
Generation of poly-GP antibody
Two rabbits were immunized with the peptide antigen C-Ahx-(GP)8-amide (21st Century Biochemicals). Specificity of antiserum versus pre-immune serum was verified by peptide dot blot and Western blot of HEK cells overexpressing GFP-tagged dipeptide repeat proteins (Figure S6A) (plasmids kindly provided by L. Petrucelli, Mayo) (Gendron et al., 2013). Antibodies were affinity purified before use.
Meso Scale Discovery (MSD) ELISA
96-well small spot streptavidin coated plates were blocked overnight at 4°C in 1% blocker A in PBS (all reagents from MSD). Following three washes in PBS-0.05% Tween (PBST), wells were coated with 0.35 μg/mL biotinylated rabbit anti-GP antibody for 1 hour at room temperature. After three PBST washes, cell lysates were added in duplicate, along with a standard curve of GP8 peptide. Following a 2-hour incubation at room temperature, wells were washed three times, and sulfo-tagged detection antibody added at 0.25 μg/mL for 1 hour. Following the final set of PBST washes, 150 μL read buffer was added and samples immediately imaged using MESO QuickPlex SQ 120. Specificity was verified using lysates of HEK cells overexpressing GFP-tagged dipeptide repeat proteins and linearity across range of interest assessed by serial dilution (Figure S6A). The assay was performed by a technician blinded to the experimental groups.
RNA isolation and qRT-PCR
To isolate total RNA from cells, Trizol (Invitrogen) and treatment with RQ1 DNase I (Promega) was used. For first-strand cDNA synthesis, random hexamers were used with High-capacity cDNA reverse transcription kit (Applied Biosystems).
All qRT-PCR reactions were performed with three biological replicates for each group and two technical replicates using the iQ SYBR green supermix (Bio-Rad) on the CFX96 real-time PCR detection system (Bio-Rad). The data were analyzed using the CFX96 optical system software (Bio-Rad; version 1.1). Expression values were normalized to GAPDH mRNA. Inter-group differences were assessed by two-tailed Student’s t-test. The C9ORF72 repeat RNA level in patient cells was detected by TaqMan gene expression assay using TaqMan fast advanced master mix (Thermo Fisher Scientific), and normalized to β-actin mRNA. Custom primer sequences are listed in Table S4. Human ACTB primer set (Hs.PT.56a.40703009.g) was purchased from Integrated DNA technologies.
For cell fractionation, cells were lysed in gentle lysis buffer (20 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.3% (v/v) NP-40). Nuclei were pelleted at 2300 g for 5 min at 4 °C, and supernatant (cytosolic fraction) were transferred to a new tube. The nuclei were re-suspended in gentle lysis buffer and spun down again to collect the pellet (nuclear fraction). Trizol was directly added to the two fractions for subsequent RNA extraction.
Sucrose gradient assay
10% (w/v) and 50% sucrose was prepared in 20 mM Tris-HCl pH 8.0, 150 mM KCl, 5 mM MgCl2, 0.5mM DTT, 100 μg /ml cycloheximide. Cells were treated with 100 μg/ml cycloheximide at 37 °C for 30 min before harvesting. Cells were lysed in 500 μl of polysome-extraction buffer (20 mM Tris pH 8.0, 1.5 mM MgCl2, 140 mM KCl, 1% (v/v) Triton X-100, 100 μg /ml cycloheximide). The lysates were spun at 13,000 g for 10 min, after a 10-min incubation on ice, and laid onto 10-50% sucrose gradients and centrifuged at 4 °C in a Sorvall SW41 rotor at 40,000 rpm for 3 h. Fractions were collected from the top using a gradient fractionation system (Brandel) while measuring the OD at 254 nm. 1 μg control CLuc RNA was spiked into each fraction and used for internal normalization in the qRT-PCR assay. Two-volume of 100% Ethanol was added to each fractions, and incubated at −80°C overnight. The fractions were spun at 16000g for 20min, the pellet was air dry after discarding the supernatant. 1 ml Trizol or 30 μl 2xLaemmli sample buffer (Biorad) was add to the pellet to extract RNA or protein. The RNA distribution across the fractions was analyzed by qRT-PCR, and the protein distribution was analyzed by immunoblotting.
RNA affinity pulldown
The (GGGGCC)40 construct was linearized with HindIII digestion to synthesize sense (GGGGCC)40 RNA by T7 RNA polymerase (Promega), and with ACC65I digestion to generate antisense (CCCCGG)40 RNA by T3 RNA polymerase (Promega). Biotin-UTP (Sigma) was used in the in vitro transcription reaction to label the adjacent sequences of the repeat RNAs. Biotinylated RNA was folded in RNA structure buffer (10 mM HEPES pH 7.5, 100 mM KCl, 5 mM MgCl2) by heating at 90°C for 2 min and slowly cooled down to room temperature. Whole cell extracts were prepared from HeLa cell by resuspending the cell pellet in RNase-free lysis buffer containing 10 mM HEPES-KOH pH7.4, 100 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.25% (v/v) NP-40, protease inhibitor (Pierce). After incubated on ice for 5min, the lysates were passed through a 25G syringe 10-20 times, and centrifuged at 14000rpm for 10min at 4°C. The folded biotin-RNA probe was bound on the streptavidin beads (Invitrogen) in RNA structure buffer for 30 min at room temperature using gentle rotation. Then the beads were incubated with the cell extracts at 30°C for 1 h. After washing five times in buffer D (10 mM HEPES pH7.4, 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.5% PMSF, 10% glycerol), the proteins specifically bound on the RNA were released by incubating the beads with 1000U RNase I (Invitrogen) in Tris-HCl (pH7.4) buffer. The elution was subsequently analyzed by immunoblotting.
DDX3X protein expression and purification from Escherichia coli
MBP-HIS, MBP-DDX3X-6HIS and MBP-DDX3XG302V-6HIS plasmids were transformed to BL21 DE3 RIL cells and grown in LB media with 0.2% dextrose and 100 μg/ml ampicillin at 37 °C shaking at 250 rpm. to an optical density at 600 nm of ~0.5. Protein expression was induced with 500 μM IPTG at 15 °C with shaking at 250 rpm overnight. Bacteria were collected by centrifugation and lysed by sonication in 20 ml per 1 L culture volume of binding buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol, 0.5% NP-40, 2.5 mM imidazole, 2.5 mM β-mercaptoethanol) plus protease inhibitor cocktail (Roche) and PMSF. Cell lysates were centrifuged at 30,000 x g for 20 minutes. Clarified lysate was bound in batch to NiNTA resin (Qiagen) for 30 minutes at 4°C. Resin was washed with 10-20 column volumes (CV) of wash buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol, 0.5% NP-40, 10 mM imidazole, 2.5 mM β-mercaptoethanol). Protein was eluted from a column in 10 CV of elution buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol, 0.5% NP-40, 200 mM imidazole, 2.5 mM β-mercaptoethanol). Eluted protein was bound to amylose resin (New England Biolabs), washed in wash buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM DTT,) and eluted in wash buffer containing 10 mM maltose. Eluted proteins were concentrated and buffer exchanged in wash buffer to remove maltose using Amicon Ultra-15 centrifugal filters (Millipore). Protein concentration was determined by absorbance at 280nM and purity was assayed by resolving 1.5 μg of each protein on a 4-20% SDS-PAGE gel and visualizing with Coomassie blue stain.
Electrophoretic gel mobility shift assay (EMSA)
The RNA was in vitro transcribed as above in the presence of α-32P-CTP. The RNA was incubated with varying concentrations of recombinant DDX3X in binding buffer (50 mM HEPES-KOH pH 7.3, 100 mM KCl or LiCl, 5 mM MgCl2, 0.22 mg/ml tRNA, 0.1 μg/μl BSA) at 30°C for 1 h. Samples were then loaded onto 6% native polyacrylamide gel in 0.5×TBE and electrophoresed for 2.5 h at room temperature. The gel was dried by Gel dryer (Model 583, Bio-Rad) and bands were detected by exposure to the film.
ATPase Assay
ATPase experiments were performed at 30 °C in 50 μL reactions containing 20 mM HEPES pH 8.5, 13 mM KCl, 5 mM MgCl2, 5 mM ATP, 2 μM RNA and 1 μM recombinant protein. RNA duplex (sense: 5’-AGCACCGUAAAGACGC-3’; antisense: 5’-GCGUCUUUACGGUGCUUAAAACAAAACAAAACAAAACAAAA-3’) and RNA repeats were folded in RNA annealing buffer (10mM Tris pH7.4, 100mM KCl) by heating at 90°C for 2 min and slowly cooled down to room temperature. Time course experiments were performed in duplicates by taking out 5 μL aliquot from the reaction every 15 min until the 1 h end point. Released phosphate was quantified with the Biomol Green detection kit (Enzolifesciences). Standard curve reactions were prepared with NaH2PO4 as a phosphate standard in identical buffer conditions.
RNA fluorescent in situ hybridization (FISH)
LNA-incorporated DNA probe with TYE-563 dye (Exiqon) (Lagier-Tourenne et al., 2013) was used against the (GGGGCC)n RNA repeats. Cells growing on coverslips were fixed with 4% PFA at room temperature for 15min, and permeabilized with 0.25% Triton X-100 in PBS for another 15 min at room temperature. After washing with PBS, they were incubated with pre-hybridization buffer (10% deionized formamide in 2x SSC) at room temperature for 30 min. Then FISH probes were diluted to 200 nM in hybridization buffer (10% Dextran Sulfate, 10% deionized formamide in 2x SSC supplemented with 0.1mg/mL E.coli tRNA, 10mM Ribonucleoside Vanadyl Complex (NEB), and SUPERaseIn RNase inhibitor (Ambion)) and incubated with the cells at 66°C for 3 hours. After washed with washing solution (10% formamide in 2x SSC) three times at 66°C (10 min each time), cells were stained with DAPI and mounted for imaging. FISH labeled cells were imaged blindly by a Zeiss LSM700 laser scanning confocal microscope and a total of 6μm Z-stacks with 0.3μm intervals were acquired.
Drosophila genetics and rough eye phenotype analysis
All the fly stocks and crosses were raised at 25°C on a standard fly medium. w1118, GMR-Gal4, UAS-GFP, bel mutant allele (BL19945), UAS-bel-RNAi-1 (BL28049), UAS-bel-RNAi-2 (BL35302) lines were obtained from the Bloomington Drosophila Stock Center. The UAS-(G4C2)58-GFP/TM6,Tb fly line was described previously(Freibaum et al., 2015). The UAS-bel-GFP line was kindly provided by Fr. Deng WM, Florida State University, Tallahassee, FL.
GMR-Gal4/CyO; UAS-(G4C2)58-GFP/TM6 flies were crossed to w1118, UAS-GFP, bel mutant allele line and bel RNAi lines. The effects of bel mutant allele and RNAi lines on the retinal degeneration phenotype of 5-day-old adult F1 flies were examined with a dissecting microscope (Nikon SMZ-1500).
For the bel overexpression experiment, fly line UAS-bel-GFP/CyO; UAS-bel-RNAi/TM6B was generated in this study. Then fly lines UAS-bel-RNAi or UAS-RFP or UAS-bel-GFP/CyO; UAS-bel-RNAi/TM6B were crossed with GMR-Gal4/CyO; UAS-(G4C2)58-GFP/TM6 flies to generate flies with the desired genotypes.
QUANTIFICATION AND STATISTICAL ANALYSIS
For all the data quantification, statistical analysis was performed with Prism 7.0 (GraphPad Software). P value inferior to 0.05 was taken as statistically significant. Different experimental groups were measured blindly and decoded after the assays. The details of the statistical analysis are listed below and can also be found in the figure legends.
Luciferase assay and qRT-PCR
For all the luciferase assays, three biological replicates were used for quantification. For qRT-PCR, three biological replicates with each containing two technical replicates were used for quantification. Inter-group differences were assessed by two-tailed Student’s t-test after F-test. Data are mean ± s.e.m.
Cell Cycle analysis
Three biological replicates were used for the reporter cells. Three CTRL lines and three C9ORF72-ALS cell lines and each line with three biological replicates were used for iPSCs. The area parameter histogram of DAPI was used to determine the percentage of cells in G1, S and G2 phases by Flowjo. Data are mean ± s.e.m.
Poly(GP) ELISA analysis
For quantification of poly(GP) protein level in flies, six independent replicates were used. For lymphoblasts, two individual lines each with two biological replicates were used. For iPSCs, three individual lines and two of them with two biological replicates were used. For iPSNs, three CTRL lines and three C9ORF72-ALS cell lines (one with two biological replicates) were used. Data are mean ± s.e.m.
RNA foci quantification
Cell number (DAPI-stained nucleus) was counted automatically by the ImageJ Analyze Particles plugin. RNA foci were identified and the size was quantified by ImageJ 3D Object Counter plugin. The intensity threshold was set as 2 times of the mean nuclear fluorescence intensity, and the size filter was set as larger than 100 voxels (voxel size 108nm×108nm×200nm). RNA foci number per cell was then calculated by using the total foci number divided by the total cell number. More than 200 cells were counted in each replicate. Data are mean ± s.e.m from three biological replicates.
Drosophila rough eye phenotype analysis
Flies were grouped by severity of the rough eye phenotype (weak, medium, and high), and the percentage of flies in each group was calculated. The results were analyzed by chi-square test. The number of flies analyzed for each genotype ranges from 50 to 200.
Ran gradient and NLS-tdTomato-NES reporter analysis
The nuclear region of iPSN was determined using DAPI and the cytoplasm boundary was determined using MAP2, which also showed the neuronal identity. Images were quantified using ImageJ and the mean pixel intensit/μm2 (Integrated density -(area ×mean fluorescence of background)) of GFP/tdTomato was quantified blindly to determine the nuclear/cytoplasmic ratio of Ran gradient (Zhang et al., 2015). Three C9ORF72-ALS iPSN lines and three CTRL iPSN lines were included for quantification. 25-40 neurons were quantified at each condition for each line. One-way ANOVA coupled with Brown Forsythe test were used followed by Dunnett’s post hoc test to determine statistical significance. Data are mean ± s.d.
Neurotoxicity assay
For quantification of glutamate-induced excitotoxicity in iPSNs, two CTRL iPSN lines and three C9ORF72-ALS iPSN lines were used. Images were taken at 10X and then analyzed blindly. 500 neurons were quantified per treatment. Two-way ANOVA with Tukey’s post hoc test was used. Data are mean ± s.d.
DATA AND CODE AVAILABILITY
No unpublished critical custom code/algorithm was generated in this study.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-DDX3X | Proteintech | Cat#11115-1-AP; RRID:AB_10896499 |
| Mouse anti-DDX3X | Santa Cruz | Cat# sc-365768; RRID:AB_10844621 |
| Mouse anti-β-actin | Cell Signaling | Cat#3700; RRID:AB_2242334 |
| Mouse anti-FLAG | Sigma | Cat#F1804; RRID:AB_262044 |
| Rabbit anti-poly-GA | Provided by Don Cleveland lab (Jiang et al., 2016) | N/A |
| Rabbit anti-poly-GP | This paper (Rb5278) | N/A |
| Mouse anti-GFP | Memorial Sloan Kettering Cancer Center Monoclonal Antibody Core Facility | Cat#Htz-GFP19C8; RRID:AB_2716737 |
| Mouse anti-MYC | Millipore | Cat#05-724; RRID:AB_11211891 |
| Rabbit anti-Cleaved-Caspase-3 | Cell Signaling | Cat#9664; RRID:AB_2070042 |
| Rabbit anti-phospho-elF2α | Cell Signaling | Cat#9721; RRID:AB_330951 |
| Mouse anti-G3BP | BD | Cat#611126; RRID:AB_398437 |
| Mouse anti-β3-Tubulin | Cell Signaling | Cat#4466; RRID:AB_1904176 |
| Rabbit anti-MAP2 | Cell Signaling | Cat#8707; RRID:AB_2722660 |
| Mouse anti-puromycin | Sigma | Cat#MABE343; RRID:AB_2566826 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TranslT-LT1 | Mirus | Cat#2305 |
| TranslT-X2 | Mirus | Cat#6004 |
| RNAiMAX | Invitrogen | Cat# 13778075 |
| TransIT-mRNA transfection kit | Mirus | Cat# 2225 |
| RK-33 | SelleckChem | Cat# S8246 |
| Ribonucleoside Vanadyl Complex | NEB | Cat#S1402S |
| SUPERaseln RNase inhibitor | Ambion | Cat#AM2694 |
| Critical Commercial Assays | ||
| Nano-Glo Dual Luciferase Assay | Promega | Cat# N1620 |
| Biomol Green detection kit | BML-AK111 | Cat#BML-AK111 |
| Experimental Models: Cell Lines | ||
| Human: RPE-1 stably expressing C9R-EGFP, RFP670 and Cas9 | This paper | N/A |
| Human: HeLa Flp-ln dual luciferase reporter cells | Cheng et al., 2018 | N/A |
| Human: C90RF72-ALS patient lymphoblast cell ND11411 | Coriell Institute | https://coriell.org/ |
| Human: C90RF72-ALS patient lymphoblast cell ND12455 | Coriell Institute | https://coriell.org/ |
| Human: iPSC CS7VCZiALS | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.org/research/areas/biomanufacturing/ipsc.html |
| Human: iPSC CS2YNUALS | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.org/research/areas/biomanufacturing/ipsc.html |
| Human: iPSC CS0NKCiALS | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.org/research/areas/biomanufacturing/ipsc.html |
| Human: iPSC CS0594iCTR | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.org/research/areas/biomanufacturing/ipsc.html |
| Human: iPSC CS8PAAiCTR | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.org/research/areas/biomanufacturing/ipsc.html |
| Human: iPSC CS0702iCTR | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.org/research/areas/biomanufacturing/ipsc.html |
| Human: iPSC CS0BUUiALS | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.org/research/areas/biomanufacturing/ipsc.html |
| Human: iPSC CS1ATZiCTR | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.org/research/areas/biomanufacturing/ipsc.html |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: w[1118] | Bloomington Drosophila Stock Center (BDSC) | BDSC: 3605 |
| D. melanogaster: w[1118]; P{GMR-GAL4.w[-]} 2/CyO | BDSC | BDSC: 9146 |
| D. melanogaster: UAS-GFP | BDSC | BDSC: 6874 |
| D. melanogaster: y[1] w[67c23]; P{w[+mC] y[+mDint2] = EPgy2}bel[EY08943] (bel mutant allele) | BDSC | BDSC: 19945 |
| D. melanogaster: y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.JF02884}attP2 (bel RNAi line) | BDSC | BDSC: 28049 |
| D. melanogaster: y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8] = TRiP.GL00205}attP2 (bel RNAi line) | BDSC | BDSC: 35302 |
| D. melanogaster: UAS-(G4C2)58-GFP | Freibaum et al., 2015 | N/A |
| D. melanogaster: GMR-Gal4/CyO; UAS-(G4C2)58-GFP/TM6B, Tb | This paper (after recombining UAS-(G4C2)58-GFP with line BDSC #9146) | N/A |
| D. melanogaster: UAS-bel-GFP | Gift from Dr. Wu-Min Deng (Lo et al., 2016) | N/A |
| D. melanogaster: w[*]; P{w[+mC] = UAS-mCD8.ChRFP}2 | BDSC | BDSC: 27391 |
| Oligonucleotides | ||
| Non-Targeting siRNA | GE Dharmacon | Cat#D-001810-10-05 |
| Pooled DDX3X siRNA | GE Dharmacon | Cat#L-006874-02-0005 |
| Single siRNA of DDX3X (GCAAATACTTGGTGTTAGA) | Sigma | N/A |
| LNA probe targeting GGGGCC repeats | Exiqon | Cat#607323 |
| ACTB TaqMan qPCR primers | Integrated DNA Technologies | Hs.PT.56a.40703009.g |
| Custom qPCR primers | Integrated DNA Technologies; Sigma | See Table S4 |
| Recombinant DNA | ||
| Plasmid: pCAG-(GGGGCC)70-EGFP (frame GA) | This paper | N/A |
| Plasmid: pBABE-RFP670 | This paper | N/A |
| Plasmid: pcDNA3.1-FLAG-DDX3X | This paper | N/A |
| Plasmid: pcDNA3.1-HA-DDX3X | This paper | N/A |
| Plasmid: pcDNA3.1-FLAG-DDX3X(G302V) | This paper | N/A |
| Plasmid: pcDNA3.1-FLAG-DDX3X (K230E/E348Q) | This paper | N/A |
| Plasmid: pMAL-c2X-MBP-DDX3X-6HIS | This paper | N/A |
| Plasmid: pMAL-c2X-MBP-DDX3X (G302V)-6HIS | This paper | N/A |
| Plasmid: Lenti-DDX3X-FLAG | This paper | N/A |
| Plasmid: Lenti-GFP | This paper | N/A |
| Plasmid: pBluescript SK(+)-(GGGGCC)40 | This paper | N/A |
| Plasmid: pBluescript SK(+)-(GGGGCC)8 | This paper | N/A |
| Plasmid: pLKO.1-DDX3X shRNA-1 | Dharmacon | TRCN0000000003 |
| Plasmid: pLKO.1-DDX3X shRNA-2 | Dharmacon | TRCN0000000004 |
| Plasmid: pLenti-RAN-GFP | Zhang et al., 2015 | N/A |
| Plasmid: pLenti-NLS-tdTomato-NES | Zhang et al., 2015 | N/A |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Illustrator | Adobe | RRID:SCR_010279 |
| FlowJo | FowJo, LLC | https://www.flowjo.com/ |
Highlights.
Genome-wide CRISPR-Cas9 screens to identify regulators of DPR protein production
The RNA helicase DDX3X suppresses RAN translation of C9ORF72 (GGGGCC)n repeats
Elevating DDX3X expression decreases DPR levels in C9ORF72-ALS patient cells
Elevating DDX3X rescues pathological features and improves survival of patient iPSNs
Acknowledgements
We thank Dr. Rachael Green’s lab for the help with sucrose gradient assay. We thank Dr. Lyle Ostrow, Dr. Leonard Petrucelli, Dr. Mercedes Prudencio and Dr. Tania Gendron for suggestions on patient samples. We thank the Sun lab members for helpful discussion. S.S. receives funding from the NIH (R01NS107347), Target ALS, the Packard Center for ALS Research and ALS Association. F.-B.G. receives funding from NIH (R01NS101986), the Packard Center for ALS Research, the Target ALS Foundation and the Muscular Dystrophy Association. M.C.B receives funding from Target ALS. L.R.H. is a recipient of AAN/ALSA Clinician Scientist Development Award in ALS Research. J.S. receives funding from the Target ALS Foundation, ALS Association and the Packard Center for ALS Research. B.P. is a recipient of American Heart Association post-doctoral fellowship (19POST34430035) and BrightFocus Foundation post-doctoral fellowship (A2019612F). Z.Q. receives funding from NIH (R35GM124824).
Footnotes
ADDITIONAL RESOURCES
Not applicable.
Supplementary Materials:
Table S1: Candidate genes as modifiers of GA-GFP levels by CRISPR-Cas9 KO screens. Related to Figure 1.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing financial interests.
References
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R, et al. (2013). Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH, et al. (2017). Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Molecular cell 65, 1044–1055 e1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol GM, Vesuna F, Xie M, Zeng J, Aziz K, Gandhi N, Levine A, Irving A, Korz D, Tantravedi S, et al. (2015a). Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO molecular medicine 7, 648–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol GM, Xie M, and Raman V (2015b). DDX3, a potential target for cancer treatment. Mol Cancer 14, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Wang S, Mestre AA, Fu C, Makarem A, Xian F, Hayes LR, Lopez-Gonzalez R, Drenner K, Jiang J, et al. (2018). C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2alpha phosphorylation. Nature communications 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Lopez-Gonzalez R, Krishnan G, Phillips HL, Li AN, Seeley WW, Yao WD, Almeida S, and Gao FB (2019). C9ORF72-ALS/FTD-associated poly(GR) binds Atp5a1 and compromises mitochondrial function in vivo. Nat Neurosci 22, 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, and Ranum LP (2014). Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders. Current opinion in genetics & development 26, 6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafinca R, Scaber J, Ababneh N, Lalic T, Weir G, Christian H, Vowles J, Douglas AG, Fletcher-Jones A, Browne C, et al. (2016). C9orf72 Hexanucleotide Expansions Are Associated with Altered Endoplasmic Reticulum Calcium Homeostasis and Stress Granule Formation in Induced Pluripotent Stem Cell-Derived Neurons from Patients with Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Stem Cells 34, 2063–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. (2013). RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epling LB, Grace CR, Lowe BR, Partridge JF, and Enemark EJ (2015). Cancer-associated mutants of RNA helicase DDX3X are defective in RNA-stimulated ATP hydrolysis. J Mol Biol 427, 1779–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, and Isaacs AM (2012). C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Scientific reports 2, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. (2015). GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, and Taylor JP (2017). The Role of Dipeptide Repeats in C9ORF72-Related ALS-FTD. Front Mol Neurosci 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Richter JD, and Cleveland DW (2017). Rethinking Unconventional Translation in Neurodegeneration. Cell 171, 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Belzil VV, Zhang YJ, and Petrucelli L (2014). Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol 127, 359–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. (2013). Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 126, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, Goldstrohm AC, Barmada SJ, and Todd PK (2017). RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nature communications 8, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther UP, Weinberg DE, Zubradt MM, Tedeschi FA, Stawicki BN, Zagore LL, Brar GA, Licatalosi DD, Bartel DP, Weissman JS, et al. (2018). The helicase Ded1p controls use of near-cognate translation initiation codons in 5’ UTRs. Nature 559, 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. (2014). C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin CC, Watson T, Corregan M, and Bailey C (1992). Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG). Biochemistry 31, 833–841. [DOI] [PubMed] [Google Scholar]
- Hautbergue GM, Castelli LM, Ferraiuolo L, Sanchez-Martinez A, Cooper-Knock J, Higginbottom A, Lin YH, Bauer CS, Dodd JE, Myszczynska MA, et al. (2017). SRSF1-dependent nuclear export inhibition of C9ORF72 repeat transcripts prevents neurodegeneration and associated motor deficits. Nature communications 8, 16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdy B, Mayer C, Varshney D, Marsico G, Murat P, Taylor C, D’Santos C, Tannahill D, and Balasubramanian S (2018). Analysis of NRAS RNA G-quadruplex binding proteins reveals DDX3X as a novel interactor of cellular G-quadruplex containing transcripts. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E (2011). RNA helicases at work: binding and rearranging. Trends Biochem Sci 36, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, et al. (2012). Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW 3rd, Sun S, Herdy JR, Bieri G, et al. (2015). Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18, 1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, and Yusa K (2014). Genomewide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature biotechnology 32, 267–273. [DOI] [PubMed] [Google Scholar]
- Komar AA, and Hatzoglou M (2011). Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer NJ, Haney MS, Morgens DW, Jovicic A, Couthouis J, Li A, Ousey J, Ma R, Bieri G, Tsui CK, et al. (2018). CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, and McKnight SL (2014). Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M, et al. (2013). Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A 110, E4530–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. (2016). C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell 167, 774–788 e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, and Jankowsky E (2011). From unwinding to clamping - the DEAD box RNA helicase family. Nature reviews Molecular cell biology 12, 505–516. [DOI] [PubMed] [Google Scholar]
- Lo PK, Huang YC, Poulton JS, Leake N, Palmer WH, Vera D, Xie G, Klusza S, and Deng WM (2016). RNA helicase Belle/DDX3 regulates transgene expression in Drosophila. Dev Biol 412, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, Petrucelli L, Miller BL, Almeida S, and Gao FB (2016). Poly(GR) in C9ORF72-Related ALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in iPSC-Derived Motor Neurons. Neuron 92, 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D, et al. (2013). Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126, 859–879. [DOI] [PubMed] [Google Scholar]
- Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M, et al. (2013). Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta neuropathologica communications 1, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Ridler CE, Balendra R, Thoeng A, Woodling NS, Grasser FA, Plagnol V, Lashley T, Partridge L, and Isaacs AM (2017). Bidirectional nucleolar dysfunction in C9orf72 frontotemporal lobar degeneration. Acta neuropathologica communications 5, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Deans RM, Li A, and Bassik MC (2016). Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nature biotechnology 34, 634–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Wainberg M, Boyle EA, Ursu O, Araya CL, Tsui CK, Haney MS, Hess GT, Han K, Jeng EE, et al. (2017). Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nature communications 8, 15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, et al. (2013a). Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol 126, 881–893. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. (2013b). The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SY, Macgregor RB Jr., and Pearson CE (2013). The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. The Journal of biological chemistry 288, 9860–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. (2012). Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R (2015). Unwinding the mechanisms of a DEAD-box RNA helicase in cancer. J Mol Biol 427, 1797–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sances S, Bruijn LI, Chandran S, Eggan K, Ho R, Klim JR, Livesey MR, Lowry E, Macklis JD, Rushton D, et al. (2016). Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat Neurosci 19, 542–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, and Pierre P (2009). SUnSET, a nonradioactive method to monitor protein synthesis. Nature methods 6, 275–277. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, and Zhang F (2015). High-throughput functional genomics using CRISPR-Cas9. Nature reviews Genetics 16, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, and Jankowsky E (2014). The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit Rev Biochem Mol Biol 49, 343–360. [DOI] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, et al. (2017). Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci U S A 114, E1111–E1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JW, Tsai TY, Chao CH, and Wu Lee YH (2008). Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27, 700–714. [DOI] [PubMed] [Google Scholar]
- Snijders Blok L, Madsen E, Juusola J, Gilissen C, Baralle D, Reijnders MR, Venselaar H, Helsmoortel C, Cho MT, Hoischen A, et al. (2015). Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. American journal of human genetics 97, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonobe Y, Ghadge G, Masaki K, Sendoel A, Fuchs E, and Roos RP (2018). Translation of dipeptide repeat proteins from the C9ORF72 expanded repeat is associated with cellular stress. Neurobiol Dis 116, 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D, and Ohlmann T (2012). DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. The EMBO journal 31, 3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, and Willis AE (2004). Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 23, 3200–3207. [DOI] [PubMed] [Google Scholar]
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, et al. (2014). Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 83, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee CZ, Lin CC, Jiang J, Jansen-West K, Abou-Hamdan H, et al. (2018). CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nature communications 9, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, and Chang TH (2009). The current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol 6, 17–20. [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Wang YD, Parker M, Patmore DM, Kanagaraj A, Moore J, Rusch M, Finkelstein D, Ellison DW, Gilbertson RJ, et al. (2016). Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Scientific reports 6, 25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, and Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZF, Ursu A, Childs-Disney JL, Guertler R, Yang WY, Bernat V, Rzuczek SG, Fuerst R, Zhang YJ, Gendron TF, et al. (2019). The Hairpin Form of r(G4C2)(exp) in c9ALS/FTD Is Repeat-Associated Non-ATG Translated and a Target for Bioactive Small Molecules. Cell Chem Biol 26, 179–190 e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergard T, McAvoy K, Russell K, Wen X, Pang Y, Morris B, Pasinelli P, Trotti D, and Haeusler A (2019). Repeat-associated non-AUG translation in C9orf72-ALS/FTD is driven by neuronal excitation and stress. EMBO molecular medicine 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe VO, and Laskey RA (2015). Control of mammalian gene expression by selective mRNA export. Nature reviews Molecular cell biology 16, 431–442. [DOI] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, and Jeang KT (2004). Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119, 381–392. [DOI] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. (2015). The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen-West K, Zhang X, Prudencio M, Chew J, Cook CN, et al. (2018). Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nature medicine 24, 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. (2016). C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci 19, 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, and Wei W (2014). High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509, 487–491. [DOI] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. (2013). RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A 110, E4968–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No unpublished critical custom code/algorithm was generated in this study.