Summary
Age-related neurodegenerative disorders are characterized by slow, persistent accumulation of aggregated proteins. Although cells can elicit physiological responses to enhance cellular clearance and counteract accumulation, it is unclear how pathogenic proteins evade this process in disease. We find that Parkinson’s disease a-synuclein perturbs the physiological response to lysosomal stress by impeding SNARE protein ykt6. Cytosolic ykt6 is normally autoinhibited by a unique farnesyl-mediated regulatory mechanism however during lysosomal stress, it activates and redistributes into membranes to preferentially promote hydrolase trafficking and enhance cellular clearance. a-Synuclein aberrantly binds and deactivates ykt6 in patient-derived neurons, thereby disabling the lysosomal stress response and facilitating protein accumulation. Activating ykt6 by small-molecule farnesyltransferase inhibitors restores lysosomal activity and reduces a-synuclein in patient-derived neurons and mice. Our findings indicate that a-synuclein creates a permissive environment for aggregate persistence by inhibiting regulated cellular clearance, and provide a therapeutic strategy to restore protein homeostasis by harnessing SNARE activity.
Keywords: Parkinson’s disease, synucleinopathy, lysosomal storage disease, protein aggregation, induced pluripotent stem cells, lysosomal stress, proteomic stress
eTOC blurb
Cuddy et al. found that SNARE protein ykt6 plays a crucial role in proteostasis and lysosomal function by enhancing hydrolase trafficking under stressful conditions. Parkinson’s disease a-synuclein impedes ykt6, causing imbalanced proteostasis and self-propagating protein accumulation. Ykt6 can be therapeutically targeted by farnesyltransferase inhibitors that restore trafficking and lysosomal function.
Introduction
Age-related neurodegenerative disorders are characterized by the progressive accumulation of misfolded and aggregated proteins. In Parkinson’s disease (PD) and Dementia with Lewy bodies (DLB), the pre-synaptic protein alpha-synuclein (a-syn) forms insoluble inclusions and accumulates within Lewy bodies that histopathologically define the disease (Spillantini et al., 1997). Pathological a-syn aggregates persist for decades in PD patients and emerge in a predictable manner within circumscribed regions of the nervous system, including midbrain dopamine neurons of the substantia nigra that mediate movement (Braak et al., 2003). Genetic studies have provided clues into the etiology of PD and further implicate a-syn accumulation in disease pathogenesis. For example rare, early-onset familial PD-dementia is caused by increasing a-syn synthesis through SNCA triplication (Singleton et al., 2003), or point mutations such as A53T that enhance stability and aggregation (Conway et al., 1998; Li et al., 2004; Polymeropoulos et al., 1997; Stojkovska et al., 2017). GWAS has identified SNCA variants as risk factors in common sporadic forms of PD, some of which have been shown to elevate a-syn expression (Nalls et al., 2014; Soldner et al., 2016). While there is strong evidence that imbalanced protein homeostasis and a-syn aggregation are involved in familial and sporadic PD pathogenesis, the downstream toxic mechanisms induced by a-syn are not completely understood.
Protein homeostasis (proteostasis) relies on a careful balance of synthesis and degradation machinery. Cells that experience aggregation-induced proteomic stress can normally elicit a homeostatic response that activates adaptive cellular clearance pathways involving the lysosomal system, in an attempt to degrade accumulated material and restore proteostasis (Settembre et al., 2013). These pathways are controlled by transcription factor EB (TFEB), which responds to lysosomal stress through activating the transcription of autophagic and lysosomal proteins (Sardiello et al., 2009; Settembre et al., 2011). While the mechanisms that control the synthesis of lysosomal components under stressful conditions are known, the regulated trafficking pathways that respond to and synergize with this pathway are not completely understood. Such pathways must be capable of rapidly responding to changing cellular environments, in order to enhance the delivery of lysosomal machinery through the early secretory pathway between the endoplasmic reticulum (ER) and Golgi, and then finally into lysosomal compartments. Vesicular transport of newly synthesized lysosomal proteins require the pairing of cognate soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) proteins, which promote membrane fusion and delivery of cargo from the ER into the Golgi (Hay et al., 1997; Sollner et al., 1993). The mechanisms involved in constitutive transport at the early secretory pathway are well described, however regulated SNARE-mediated transport that occurs in response to proteomic stress is not completely understood. Furthermore, it is unclear how disease-linked proteins such as a-syn can persist and evade regulated clearance pathways in synucleinopathies.
Here, we sought to determine the mechanisms of aggregate persistence using patient-derived midbrain cultures that harbor endogenous SNCA mutations, and can accumulate a-syn for hundreds of days (Mazzulli et al., 2016a). We identify synaptobrevin-2 homolog ykt6 as a critical SNARE involved in the lysosomal stress response. Ykt6 is an unusual SNARE, since it lacks a transmembrane domain that is required for membrane association. Instead, post-translational modifications, including farnesylation and palmitoylation, control its membrane association and fusogenic activity (Daste et al., 2015). Ykt6 occurs within a cytosolic reserve pool in an autoinhibited, closed conformation that is mediated by intramolecular farnesyl moiety (Fukasawa et al., 2004; Hasegawa et al., 2003; McNew et al., 1997; Wen et al., 2010). Upon activation by a heretofore unknown stimuli, ykt6 opens into an extended conformation to promote membrane association, SNARE binding, and vesicular transport (Daste et al., 2015; Tochio et al., 2001). The majority of studies show that ykt6 plays a key role in ER-Golgi trafficking (Fukasawa et al., 2004; Liu and Barlowe, 2002; McNew et al., 1997; Zhang and Hong, 2001), and can also rescue a-syn-induced toxicity in yeast and cell lines (Cooper et al., 2006; Thayanidhi et al., 2010). However, ykt6 can also facilitate other transport steps depending on the cell type examined, including intra-Golgi (Xu et al., 2002), endosome-Golgi (Tai et al., 2004), and autophagosome formation (Matsui et al., 2018; Nair et al., 2011; Takats et al., 2018). Here, we examined the role of ykt6 in PD patient cultures and evaluated its potential as a therapeutic target to enhance lysosomal activity and reduce protein accumulation.
Results
Lysosomal dysfunction occurs subsequent to a-syn aggregation in PD patient midbrain cultures
To gain mechanistic insights into the effects of pathological aggregates on protein homeostasis, we examined lysosomal function in PD patient-derived iPSC-midbrain neurons (iPSn). Differentiated iPSCs from patients expressing A53T a-syn and isogenic controls showed expression of midbrain markers FOXA2, tyrosine hydroxylase (TH), and beta-iii-tubulin (Figure S1A), and accumulated pathological a-syn in an age-dependent manner between day 60 and 110 (Figure S1B–E). Lysosomal function was measured in living cultures using an in situ beta-glucocerebrosidase (GCase) activity assay that is compartment-specific (Mazzulli et al., 2016a). This revealed an age-dependent decline in lysosomal activity between day 75 and 130 (Figure S2A). The lysosomal activity of other hydrolases, including hexosaminidase, sulfatase, and beta-galactosidase, was also decreased (Figure S2B). A similar age-dependent decline in GCase activity was observed in a distinct SNCA triplication (trp) line compared to a healthy control line (Figure S2C). The reduction in GCase activity was sufficient to induce glycosphingolipid substrate accumulation (Figure S2D, E), confirming lysosomal dysfunction. To probe the mechanism of this effect, we measured the levels of lysosomal proteins by western blot. This indicated that total levels of GCase and the lysosomal marker LAMP1 were not depleted as expected, but were elevated in A53T iPSn compared to isogenic controls (Figure S2F). The molecular weight of GCase and LAMP1 increases as it matures through the ER and Golgi by protein glycosylation (Bergmann and Grabowski, 1989; D’Souza and August, 1986), and maturation can be estimated by molecular weight analysis. Western blot analysis showed the accumulation of low molecular weight, immature ER forms (corresponding to <62 kDa for GCase, and <98kDa for LAMP1) suggesting perturbations in protein maturation (Figure S2F). Consistent with disrupted GCase trafficking, measurement of in situ GCase activity from non-lysosomal compartments of living iPSn indicated a two-fold elevation in A53T lines (Figure S2G, left). In vitro activity assays of whole cell lysates, containing a mix of lysosomal and non-lysosomal forms, revealed a similar elevation of activity (Figure S2G, right). We attributed the elevated activity of whole-cell lysates to the accumulation of ER-GCase that is liberated during cell lysis and artificially activated in the acidic buffer that is optimized for the assay (Figure S2G, right). Together, these data indicate that A53T a-syn aberrantly redistributes GCase activity outside of lysosomal compartments resulting in lysosomal dysfunction.
We confirmed disrupted protein trafficking in PD iPSn by analysis of nicastrin, a protein that matures similarly to GCase and LAMP1 (Bagshaw et al., 2003; Yang et al., 2002), which also showed accumulation of immature ER forms at day 60 (Figure S2H). Together, this suggests that lysosomal dysfunction in A53T iPSn does not result from a depletion in lysosomal machinery, but is associated with disrupted protein maturation consistent with previous studies (Cooper et al., 2006). We also found that A53T iPSn could stimulate the synthesis of lysosomal components at the transcriptional level, since TFEB translocated into the nucleus of inclusion-bearing cells, and GBA1 mRNA was elevated compared to isogenic controls (Figure S2I, J). To correlate lysosomal phenotypes with cell viability, we assessed neurite content by neurofilament staining, and cell body toxicity by counting TH / FOXA2 / b-iii-Tubulin + neurons. Neurotoxicity of PD iPSn was not evident until day 100 and beyond (Figure S2K–M), suggesting that lysosomal dysfunction does not occur as a result of non-specific toxicity. Collectively, this indicates that lysosomal dysfunction occurs subsequent to a-syn aggregation and is associated with perturbations in protein trafficking.
Ykt6 is disrupted by a-Synuclein and is deficient in PD brain
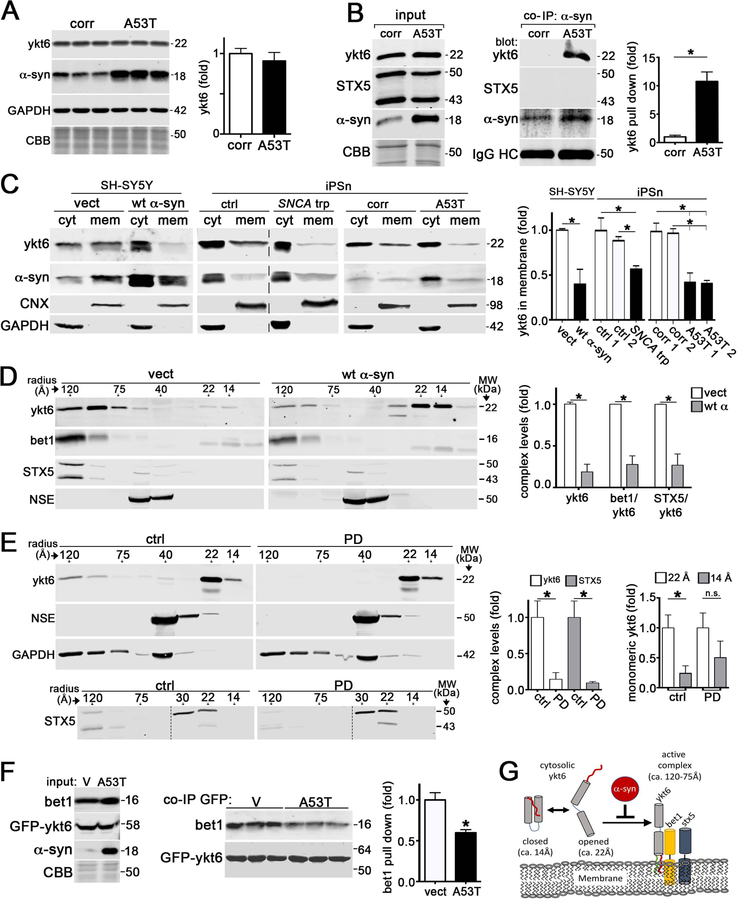
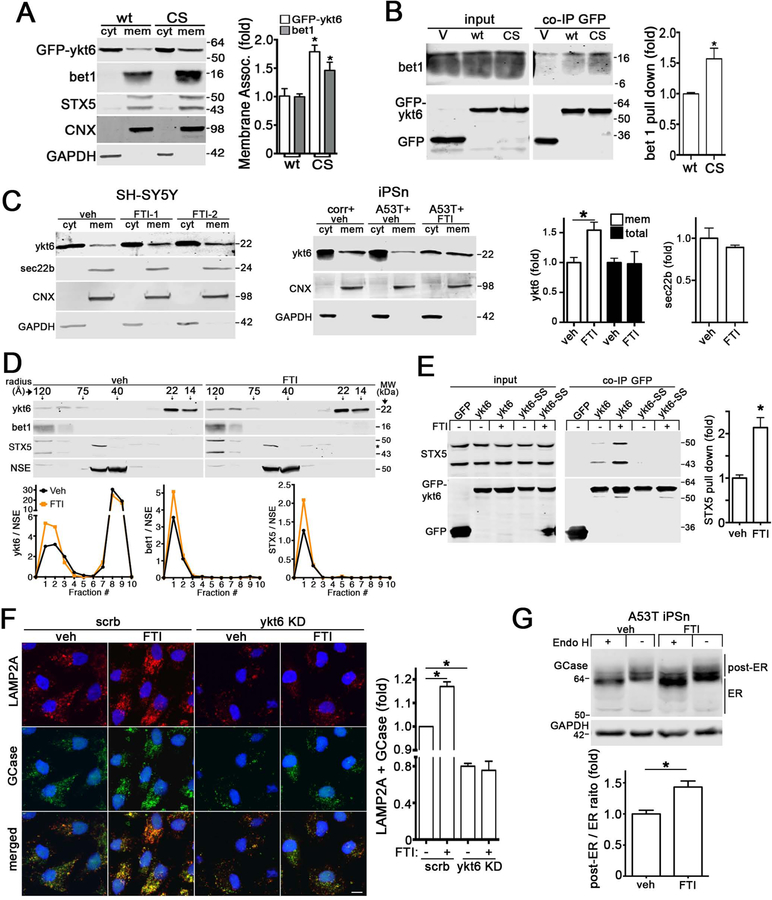
Since our data indicated disruptions in protein maturation, we examined the levels of SNAREs that facilitate trafficking between the ER and Golgi where sec22b, ykt6, syntaxin 5 (STX5), and bet1 are critical (Hay et al., 1997; Nichols and Pelham, 1998). Although ykt6 and sec22b are in distinct SNARE complexes, they can both bind STX5 and bet1 to form a functional complex (Xu et al., 2000; Zhang and Hong, 2001). Western blot analysis of A53T iPSn indicated a subtle elevation in sec22b but no change in other SNAREs, suggesting that perturbations in maturation are not associated with reduced SNARE levels (Figure 1A, Figure S3A, B). a-Syn can interact with synaptobrevin-2 (Burre et al., 2010; Sun et al., 2019), which shares sequence homology with ykt6 and sec22b. Therefore we assessed whether a-syn could form a complex with ykt6 or sec22b by co-immunoprecipitation (co-IP) in an established inducible H4 cell model of a-syn overexpression (Mazzulli et al., 2011). This showed that a-syn could co-IP with ykt6 but not sec22b (Figure S3C, D).
Figure 1. a-Synuclein interacts with ykt6 and disrupts SNARE complex assembly.
A) Western blot of ykt6 in A53T or isogenic corrected (corr) iPSn at day 75 (n=6). GAPDH or Coomassie brilliant blue (CBB) are loading controls. B) Co-immunoprecipitation (co-IP) of a-syn and ykt6 in d75 iPSn. Right, quantification (n=3). C) Analysis of cytosolic (cyt) or membrane bound (mem) ykt6 in wt a-syn SH-SY5Y cells, or d90 iPSn from A53T and SNCA triplication (trp) patients compared to multiple controls. Calnexin (CNX) and GAPDH are loading controls (n=3–4). The dashed line indicates cropped out replicates. D) SEC/western blot of ykt6 SNARE complexes in SH-SY5Y cells. Neuron specific enolase (NSE) is a loading control (n=3). E) SEC analysis of PD brain lysates (n=3). F) Co-IP/ western blot from transfected HEK cells (empty vector (v) or A53T a-syn, with GFP-ykt6) (n=3). G) Schematic showing the effects of a-syn on ykt6 function. Farnesyl, red; Palmitoyl, green. Molecular weights (MW) are in kilodaltons (kDa), molecular radius is in angstroms (A). Values are the mean +/− SEM, *p<0.05. See also Figures S1–3.
To determine if endogenously expressed a-syn and ykt6 form a complex, we analyzed PD patient iPSn lysates from A53T, SNCA trp, PARK9, or GBA1 mutation carriers, as well as idiopathic PD neurons that accumulate pathological a-syn at the cell body (Mazzulli et al., 2016a; Mazzulli et al., 2016b). This showed that a-syn pulled down ykt6 in all PD iPSn lines, but not in controls (Figure 1B, S3E–F). a-Syn did not pull down the ykt6 cognate SNARE binding partner STX5 (Figure 1B, S3G), suggesting that a-syn may form a complex with ykt6 in the cytosol prior to SNARE assembly. Consistent with this, co-IP from isolated cytosol and membrane fractions revealed that a-syn-ykt6 complexes occurred only in the cytosol (Figure S3H).
We then determined if a-syn could influence the function of ykt6 by altering its cytosol-membrane distribution. Membrane-associated ykt6 was reduced in differentiated human neuroblastoma cells (SH-SY5Y) that overexpress wt a-syn compared to empty vector control lines, a finding that was validated in SNCA trp and A53T iPSn (Figure 1C). Immunostaining showed that a-syn overexpression reduced ykt6 colocalization with the Golgi marker GM130 (Figure S3I). To determine if a-syn disrupted ykt6 SNARE complexes, we used size exclusion chromatography (SEC) to separate monomers from high molecular weight assemblies. Western blot of the collected fractions indicated that ykt6 complexes (120–75 Angstrom (A) radius species) were reduced, while monomeric ykt6 at 22 and 14 A were elevated in SH-SY5Y a-syn cells, suggesting break down of SNARE assemblies (Figure 1D). The levels of STX5 and bet1, binding partners of ykt6 specifically at the ER-Golgi step, were also decreased (Figure 1D). This result was confirmed in PD iPSn (Figure S3J).
We next analyzed brain lysates from pathologically confirmed PD samples (Mazzulli et al., 2011) by SEC and found that ykt6 and STX5 complexes were depleted (Figure 1E). SEC analysis of open (22A) and closed (14A) monomeric forms of ykt6 (Pylypenko et al., 2008) also indicated a minor change in the proportion of 22A and 14A species, where the 14A compact monomer was slightly elevated in PD brain (Figure 1E). Changes in ykt6 were specific, since control proteins GAPDH and NSE showed the expected elution profiles (Figure 1E, Figure S3K). Finally, we confirmed that a-syn disrupted ykt6 SNARE complexes in a-syn overexpressing cells by co-IP, since ykt6 pulled down less bet1 compared to controls (Figure 1F). Together, this shows that a-syn reduces membrane-associated ykt6 and SNARE complexes (Figure 1G).
Ykt6 knock-down preferentially disrupts lysosomal protein trafficking and function
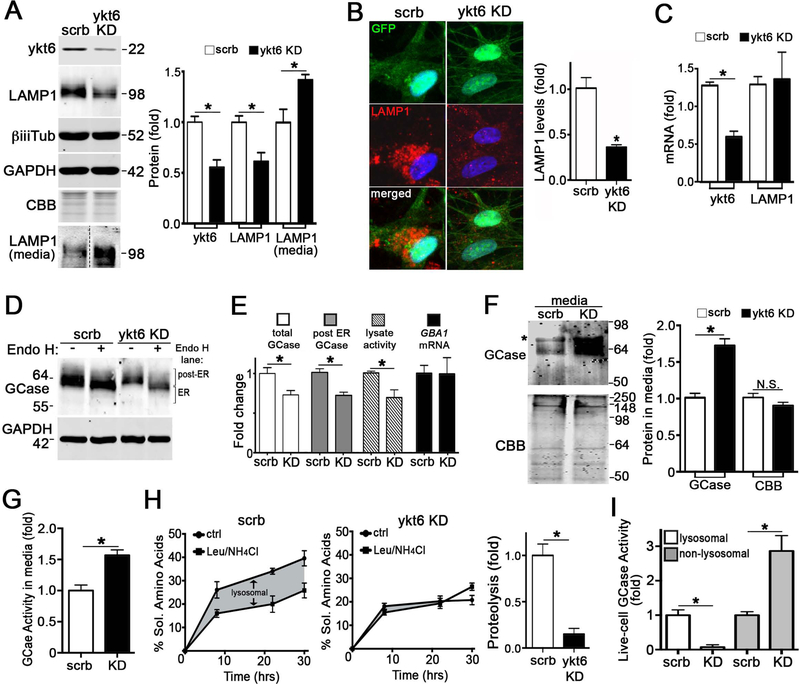
Since our data suggested an association between a-syn-induced lysosomal dysfunction and perturbations in ykt6, we next determined the effect of depleting ykt6 in healthy control iPSn on the lysosomal system. shRNA-mediated knock-down (KD) of ykt6 reduced intracellular LAMP1 protein by 50%, as shown by western blot and immunofluorescence analysis (Figure 2A, B). This effect was post-transcriptional, since LAMP1 mRNA did not change (Figure 2C). LAMP1 protein was elevated in the conditioned media of ykt6 KD iPSn, indicating that the decline of intracellular LAMP1 likely occurred by aberrant secretion into the media (Figure 2A). Ykt6 KD also reduced the levels of total intracellular GCase protein and activity (Figure 2D, E). The level of mature (post-ER) GCase was determined by resistance to endoglycosidase H (endo H), and enzyme that only cleaves high mannose residues of immature proteins, but not complex oligosaccharides of mature proteins (Tarentino et al., 1972). This showed that ykt6 KD reduced mature, endo H resistant GCase (Figure 2D, E). GCase depletion also occurred post-transcriptionally, since GBA1 mRNA was unchanged (Figure 2E), and elevated levels of GCase protein and activity were found in the conditioned media of ykt6 KD iPSn (Figure 2F, G). Lysosomal function was directly assessed by measuring the degradation rates of long-lived proteins, and revealed a decline in proteolysis upon ykt6 KD (Figure 2H). We also found reduced GCase activity within lysosomal compartments while non-lysosomal GCase activity was elevated (Figure 2I), further suggesting hydrolase mistrafficking. Consistent with lysosomal dysfunction, we found that prolonged ykt6 KD resulted in a-syn accumulation (Figure S4A). By sampling media from the same culture wells as used for proteolysis, we measured constitutive secretion of total proteins and found that ykt6 KD had no effect (Figure S4B, left). Similarly, we found no changes in protein secretion when ykt6 was reduced in iPSn that were stressed with lysosomal inhibitors (Figure S4B, right). Together, these data indicate that ykt6 is essential for lysosomal function. Partial ykt6 KD has dramatic effects on the lysosomal system, but no effect on constitutive protein secretion, indicating that ykt6 preferentially functions to traffic lysosomal machinery in iPSn.
Figure 2. Ykt6 is required for lysosomal function in human midbrain neurons.
A) Lentiviral mediated knock-down (KD) of ykt6 by shRNA (MOI3, dpi7) in control iPSn (scrb, scrambled) followed by western blot of lysates and media. b-iii-tubulin (biiiTub) and GAPDH are loading controls (n=8 lysate, n=3 media). The dashed line indicates cropped out replicates. B) Immunofluorescence of LAMP1 in control iPSn infected with lenti-ykt6 shRNA. GFP indicates infected neurons (n=3). C) mRNA quantification by Q-RT-PCT (n=4). D) Analysis of GCase maturity by endoglycosidase H (endo H) digestion of iPSn lysates. E) Quantification of total and endo H resistant GCase by western blot (from panel D), GCase activity from whole cell lysates, and GBA1 mRNA levels by Q-RT-PCR (n=4). F) Western blot of media from iPSn after ykt6 KD (n=3). *, band detected in media alone. G) GCase activity in media from scrb of ykt6 KD iPSn. H) Proteolysis quantification in living iPSn by measuring soluble (sol.) amino acids released into the media over time. The shaded area shows the response to lysosomal inhibitors, leupeptin (Leu) and NH4Cl (quantified on the right (n=4)). I) GCase activity was measured after ykt6 KD in differentiated SH-SY5Y a-syn cells (n=4). Values are the mean +/− SEM, *p<0.05. See also Figure S4.
Ykt6 is required for the physiological response to lysosomal stress in human midbrain neurons
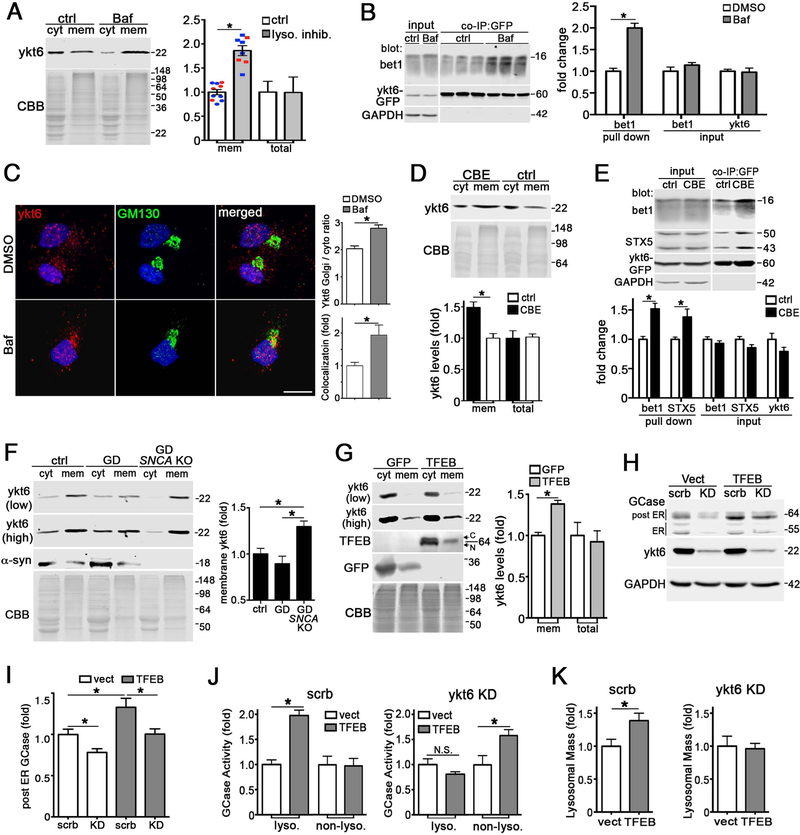
Our data indicates a preferential role for ykt6 in trafficking lysosomal proteins, and previous structural studies suggested tight regulatory features of ykt6 (Tochio et al., 2001) including an inactive reserve pool within the cytosol (Hasegawa et al., 2003; Thayanidhi et al., 2012; Tochio et al., 2001). While the cellular signals that activate ykt6 are unknown, we hypothesized that it may redistribute from the cytosol into membranes to promote trafficking of lysosomal machinery during periods of lysosomal stress. To test this, we treated healthy iPSn with lysosomal inhibitors bafilomycin A1 (baf) or NH4Cl, then measured ykt6 membrane distribution. Lysosomal inhibition caused a shift of ykt6 from the cytosol into membrane fractions, without changing total ykt6 (Figure 3A). Baf treatment promoted the assembly of ykt6 ER-Golgi SNARE complexes and increased its localization to the Golgi (Figure 3B, C). We confirmed that ykt6 responded to distinct initiators of lysosomal stress, including a chemically induced iPSn model of Gaucher disease (GD) using the GCase inhibitor CBE (Zunke et al., 2018), and sucrose-induced vacuolization (DeCourcy and Storrie, 1991). Both CBE and sucrose caused ykt6 to shift into membranes (Figure 3D, S4C), and increased ykt6 SNARE complexes (Figure 3E). Using the sucrose vacuolization iPSn model, we found that LAMP1 and GCase trafficking were elevated in a ykt6 dependent manner (Figure S4D, E). To test a more natural, chronic model of lysosomal storage, we assessed ykt6 membrane association in a GD iPSn. Compared to healthy controls, GD iPSn showed only subtle changes in cytosolic and membrane ykt6 (Figure 3F). This GD iPSn line accumulates pathogenic a-syn in a chronic manner (Mazzulli et al., 2016a), which may impede the response of ykt6. Therefore, we knocked-out (KO) SNCA in GD iPSn by CRISPR/Cas9 using a previously described editing strategy (Zunke et al., 2018). Analysis of off-target effects indicated a selective disruption of the SNCA gene (Figure S4F), and western blot validated that no a-syn protein was made (Figure 3F). GD-SNCA KO lines demonstrated a significant shift of ykt6 into membranes when compared to both its parental GD line and a healthy control (Figure 3F). Together, this indicates that lysosomal stress activates ykt6, which results in enhanced trafficking of lysosomal components.
Figure 3. Ykt6 is activated in response to lysosomal stress.
A) Ykt6 membrane shift analysis in bafA1 (200nM, 2 hrs) or NH4Cl (1M, 12 hrs) treated iPSn (d60). CBB, coomassie brilliant blue, (red=BafA1; blue= NH4Cl, n=7–10). B) Co-IP of GFP-ykt6 and bet1 in control iPSn (d60) treated with bafA1 (n=3). C) Ykt6 and GM130 immunofluorescence analysis of baf A1-treated H4 cells (n=9). Scale bars=10um. D) Ykt6 membrane shift analysis in CBE-treated SH-SY5Y cells (50mM, 2d, n=3). E) Co-IP of GFP-ykt6, bet1 and STX5 in CBE-treated SH-SY5Y cells (n=5). F) Ykt6 membrane shift analysis in control or Gaucher disease (GD) iPSn (d100). a-Syn was knocked-out (SNCA KO) in GD lines by CRISPR/Cas9. Two exposures are included to better visualize cyt and mem ykt6 (n=4). G) Ykt6 membrane shift analysis in lenti-infected SH-SY5Y cells (moi3, dpi 10) (n=3). C, cytosolic; N, nuclear. H) GCase maturation was assessed by endo H digest in lenti-infected SH-SY5Y cells. Empty vector (vect), scrambled (scrb). I) Control iPSn were infected as in H and analyzed for GCase maturation (n=3). J) Live cell GCase activity in lenti-infected iPSn (n=4). K) Lysosomal mass measurement in control iPSn (n=4). Values are the mean +/− SEM, *p<0.05. See also Figure S4.
Since TFEB responds to lysosomal stress by inducing lysosomal biogenesis (Sardiello et al., 2009), we next determined if ykt6 was involved in hydrolase trafficking upon TFEB overexpression. We found increased levels of membrane-associated ykt6 upon TFEB overexpression (Figure 3G), and enhanced GCase trafficking that was reduced by ykt6 KD (Figure 3H, I). TFEB elevated GCase activity within lysosomes, however ykt6 KD abolished this effect, and instead elevated activity in non-lysosomal compartments (Figure 3J). TFEB increased lysosomal mass as expected, and this effect was also diminished by ykt6 KD (Figure 3K). Together, this indicates that ykt6 responds to distinct stress stimuli that converge on lysosomal pathways, by synergizing with TFEB to enhance cellular clearance during stressful metabolic conditions.
Ykt6 rescues lysosomal function in PD iPSn and is enhanced by blocking its farnesylation
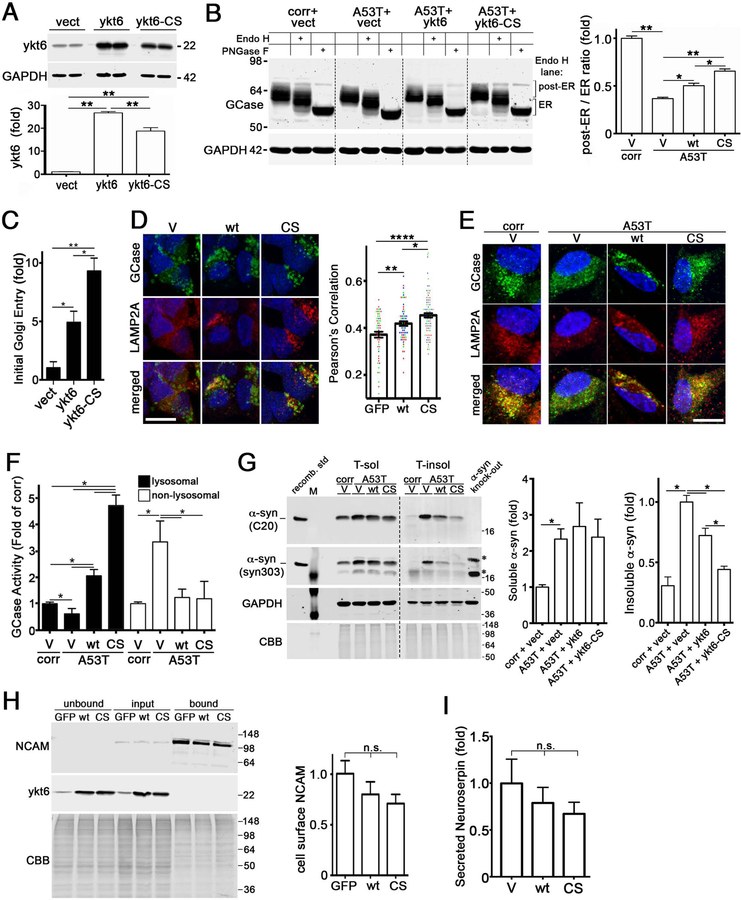
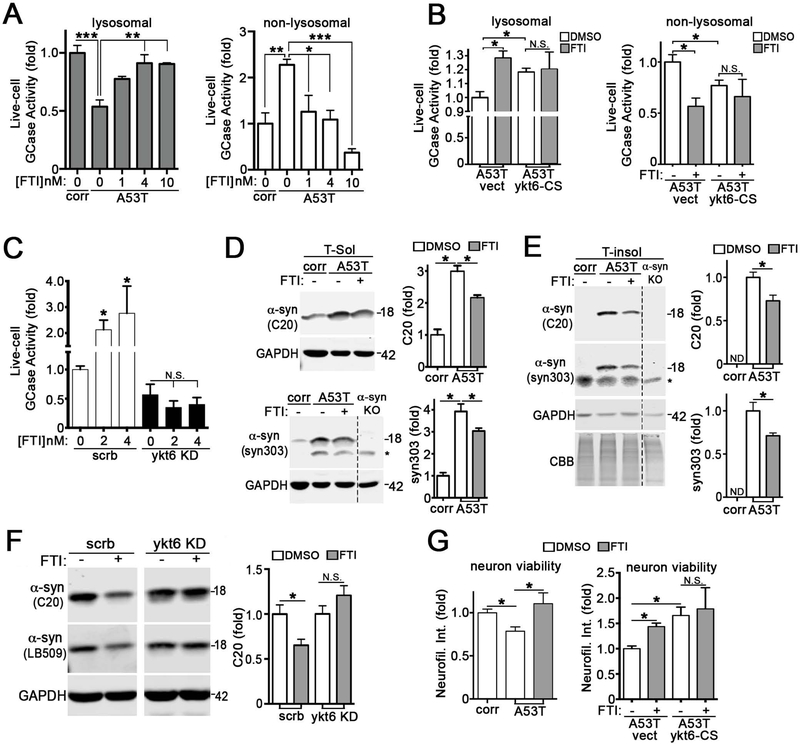
To determine if overexpressing ykt6 could restore lysosomal function in PD iPSn, we overexpressed either wild-type (wt) ykt6 or a mutant form that cannot be farnesylated (C195S, or CS) by lentivirus. We reasoned that blocking farnesylation of ykt6 may enhance its function by promoting an open, active conformation (Wen et al., 2010). Expression of wt ykt6 rescued GCase trafficking in PD iPSn and blocking its farnesylation enhanced this effect, despite the lower expression levels of titer-matched ykt6-CS (Figure 4A, B). This effect was reproducible in multiple synucleinopathy models (Figure S5A–D). Ykt6 also improved the maturation of hexosaminidase A and iduronate-2-sulfatase, indicating a general effect for lysosomal hydrolases (Figure S5E). ER-Golgi transport was directly assessed using a live-cell trafficking assay (Gordon et al., 2010). We found that wt ykt6 enhanced ER-Golgi transport, and this effect was nearly doubled by ykt6-CS (Figure 3C; S5F, G). Colocalization of GCase with LAMP2A was increased upon expression of ykt6 wt or CS in both SH-SY5Y cells and A53T iPSn (Figure 4D, E), consistent with increased targeting to lysosomes. Ykt6 enhanced GCase activity within lysosomes while decreasing non-lysosomal activity in PD iPSn and cell lines, an effect that was augmented by expression of ykt6-CS (Figure 4F; Figure S5H). Finally, we tested if ykt6 could reduce pathological a-syn in PD iPSn, since previous studies showed that enhancing GCase activity could clear a-syn (Aflaki et al., 2016; Mazzulli et al., 2016b; Rocha et al., 2015; Sardi et al., 2013). We found that insoluble a-syn was decreased by ~25% with wt ykt6 and ~60% with ykt6-CS, while no consistent change was found in the soluble a-syn fraction (Figure 4G).
Figure 4. Ykt6 rescues lysosomal function and is activated by blocking its farnesylation.
A) A53T iPSn and isogenic corrected lines (corr) were lenti-infected with empty vector (vect) or ykt6 constructs (wt or CS that cannot be farnesylated) and analyzed by western blot (moi 3, dpi 42). B) GCase maturation analysis in lenti-infected A53T iPSn by endo H digest / western blot (n=12). C) Analysis of ER-Golgi trafficking in living H4 cells transfected with ykt6 wt or CS (n=4). D) Immunostaining analysis of GCase and LAMP2A in SH-SY5Y a-syn cells (n=3 wells, values from individual cells are plotted). Scale bars=10um. E) Immunostaining analysis of GCase and LAMP2A in A53T iPSn at 42 dpi. Scale bars=5um. F) GCase activity was assessed in living A53T iPSn (n=6). G) Sequential extraction / western blot analysis of a-syn using C20 (total a-syn) or syn303 (oxidized a-syn) (n=7). Recombinant a-syn was used as a positive control, and a-syn knock-out iPSn were used as a negative control. *, non-specific bands; M, MW marker. N=4, normalized to total protein (CBB). H) Cell-surface biotinylation assay in SH-SY5Y a-syn cells, expressing wt or CS ykt6, followed by ykt6 and NCAM western blot. Bound lanes represent biotinylated proteins on the cell surface (n=6). I) Quantification of neuroserpin levels in culture media by ELISA of lenti-infected SH-SY5Y cells (n=3). Values are the mean +/= SEM, *p<0.05, **p0.01, ****p<0.0001. The dashed lines indicate cropped out replicates. See also Figure S5.
We next examined the specificity of ykt6 in enhancing lysosomal protein trafficking, by measuring the effect on cell surface or secreted proteins that also mature through the early secretory pathway. The cell surface levels of neural cell adhesion molecule (NCAM) was determined by surface biotinylation assays, and revealed a downward, non-significant trend when ykt6 wt or CS was expressed (Figure 4H). Measurement of total cell surface proteins also showed no change (Figure S5I). We then measured the levels of secreted neuroserpin in conditioned media, and found no change upon ykt6 wt or CS expression (Figure 4I). This indicates that ykt6 preferentially enhances the trafficking of lysosomal proteins in PD culture models.
Since we found that expression of the non-farnesyl-ykt6 (CS) augmented lysosomal activity compared to wt ykt6, we next examined the effect of blocking ykt6 farnesylation on membrane binding and SNARE complex formation. Ykt6-CS was elevated in membrane fractions compared to wt, along with ER-Golgi binding partners bet1 and STX5 (Figure 5A). Ykt6-CS also pulled down more bet1 compared to wt, consistent with increased membrane association (Figure 5B). This indicates that blocking ykt6 farnesylation enhances membrane association and cognate SNARE binding, which then augments the trafficking of lysosomal hydrolases.
Figure 5. Reducing farnesyl-ykt6 enhances SNARE assembly and improves the lysosomal targeting of GCase.
A) Ykt6 membrane shift analysis of SH-SY5Y cells expressing GFP-ykt6 wt or CS (n=3). B) Co-IP of GFP-ykt6 and bet1 in A53T iPSn (n=3). C) Ykt6 membrane shift analysis in FTI-treated SH-SY5Y wt a-syn cells or A53T iPSn (5nM farnesyltransferase inhibitors (FTI) LNK-754 (FTI-1) or LNK-3248 (FTI-2) for 5 days) (n=4). Sec22b was measured to assess specificity. D) SEC / western blot of FTI-1 treated SH-SY5Y cells. Quantification is shown below. E) Co-IP of GFP-ykt6 wt or mutant ykt6-SS, (cannot be palmitoylated or farnesylated) in FTI-treated SH-SY5Y cells (5nM LNK-754, 5d) (n=2). F) Immunofluorescence of GCase and LAMP2A in FTI-treated H4 a-syn cells transfected with scrambled (scrb) or ykt6 shRNA (n=3). Scale bar=10um. G) GCase maturation analysis in FTI-treated A53T iPSn by endo H (n=3). Values are the mean +/− SEM, *p<0.05. See also Figure S6.
Ykt6 can be therapeutically targeted by farnesyltransferase inhibitors
We next determined if ykt6 could be targeted by small molecule farnesyltransferase inhibitors (FTIs). We utilized a previously characterized FTI, LNK-754, that was developed for clinical use as potential cancer therapy, and has shown selective in vivo target engagement of farnesyltransferase (Downward, 2003; Moulder et al., 2004). Upon culturing cells with the FTI, we found that it reduced the farnesyl-ykt6 signal by 40% (Figure S6A). FTI treatment increased membrane association of ykt6 in cell lines and PD patient neurons, while sec22b levels did not change (Figure 5C). The FTI-mediated increase of membrane ykt6 occurred through cysteine 195, since ykt6-CS was not affected by FTI treatment (Figure S6B). SEC analysis showed that the FTI elevated the levels of ykt6 SNARE complexes in SH-SY5Y cells, a finding that was confirmed in A53T iPSn (Figure 5D, S6C). Co-IP showed increased ykt6 SNARE complexes with FTI treatment, while no effect was seen with a ykt6 mutant lacking lipid modification sites (ykt6-SS) (Figure 5E). Immunostaining for lysosomal markers indicated that FTIs elevated the levels of LAMP2A and GCase, which was abolished by ykt6 KD (Figure 5F; Figure S6D, E). Consistent with this, FTI treatment significantly elevated mature, endo H-resistant GCase in A53T iPSn and SH-SY5Y cells (Figure 5G, S6F). Together, this indicates that ykt6 activity can be modulated by FTIs, resulting in enhanced trafficking of lysosomal GCase.
We next determined if FTI treatment could improve lysosomal GCase activity in PD iPSn. GCase activity was partially rescued with concentrations as low as 1nM, and higher concentrations of 4 and 10nM resulted in a near-complete rescue (Figure 6A). This effect was likely due to reducing the farnesylation of ykt6, as opposed to other proteins, since FTIs could not further enhance lysosomal activity in ykt6-CS or ykt6 KD iPSn (Figure 6B, C). We also found that 5nM FTI treatment reduced soluble and insoluble levels of a-syn in A53T iPSn using the C20 antibody that detects total a-syn, as well as with syn303 that prominently reacts with pathological a-syn (Duda et al., 2002) (Figure 6D–E, S6G). Importantly, a-syn reduction by the FTI was dependent on ykt6 (Figure 6F). FTI treatment also improved neuron viability of A53T iPSn when analyzed at day 100, but had no effect on cells expressing ykt6-CS (Figure 6G). Together, these data indicate that ykt6 can be activated by FTI, and can rescue cellular pathologies in PD patient neurons.
Figure 6. FTIs enhance lysosomal GCase activity and reduce pathological a-syn by blocking ykt6 farnesylation.
A) GCase activity in FTI-treated A53T iPSn at d80 (LNK-754 5nM, 5d). B) GCase activity in FTI-treated A53T iPSn expressing empty vector (vect) or ykt6-CS (n=4). C) Lysosomal GCase activity was assessed in FTI-treated SH-SY5Y cells transfected with scrambled or ykt6 shRNA (KD) (n=3). D) Quantification of Triton X-100 soluble (T-sol) a-syn by western blot from FTI-treated A53T iPSn (5nM, 7d). *, non-specific band. (n=3). E) Quantification of insoluble a-syn from A53T iPSn as in D (n=3). F) Insoluble a-syn in FTI-treated H4 cells (5nM, 5 days) transfected with either scrambled or ykt6 shRNA (n=6). G) Neuron viability was assessed in FTI-treated A53T iPSn (5nM, 14d from d86-d100) by quantification of neurofilament protein. Right, neuron viability in FTI-treated A53T iPSn expressing empty vector or ykt6-CS (n=4). Values are the mean +/− SEM, *p<0.05, **p<0.01, ***p<0.001. The dashed line indicates cropped out replicates. See also Figure S6.
Farnesyltransferase inhibition activates ykt6 in vivo and reduces pathological aggregation in mice
We next sought to determine if FTIs could activate ykt6 in the brains of mice by first testing their effect in wild-type animals. Pharmacokinetic studies revealed that after a single oral dose, the FTI was cleared from plasma within 20 hours and could rapidly cross the blood-brain-barrier (Figure S7A). Daily intraperitoneal (i.p.) injection of wild-type mice showed that the FTI could increase the levels of membrane associated ykt6 and STX5, while no change was observed for sec22b (Figure S7B). SEC indicated that FTI treatment did not change the total levels of ykt6, but shifted its distribution from monomers into high molecular weight complexes that co-eluted with STX5 (Figure S7C, D). We also found elevated levels of mature, endo H resistant GCase and higher enzyme activity in the brain of FTI treated mice (Figure S7E, F). Consistent with lysosomal activation, FTI treatment also reduced a-syn levels (Figure S7G). Together, these data indicate that i.p. injection of FTIs can activate ykt6 in the brains of wild-type mice, resulting in lysosomal enhancement and a-syn reduction.
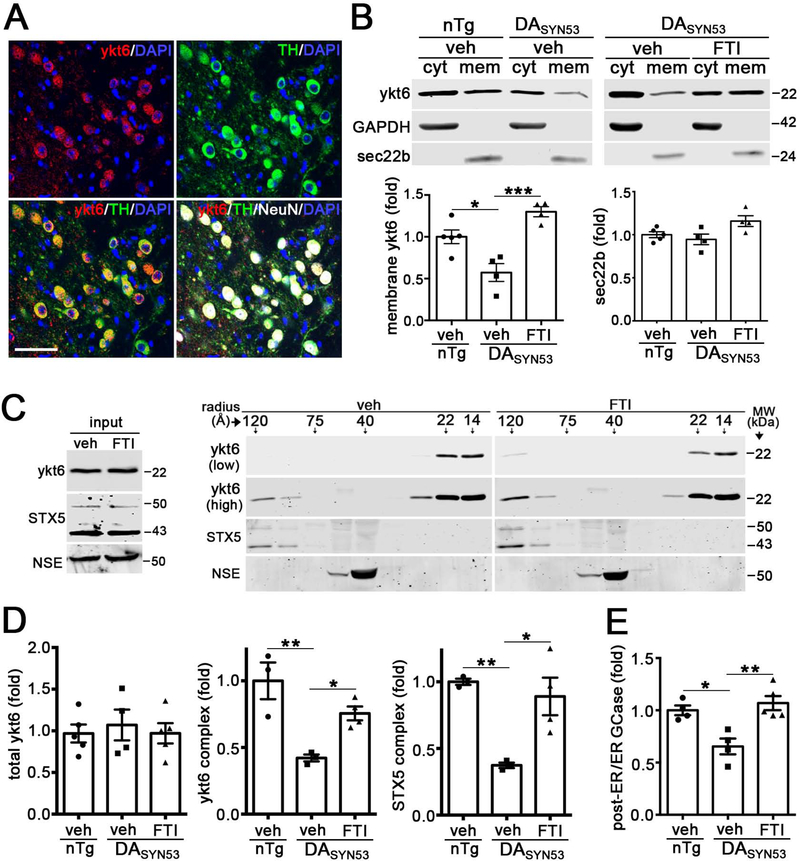
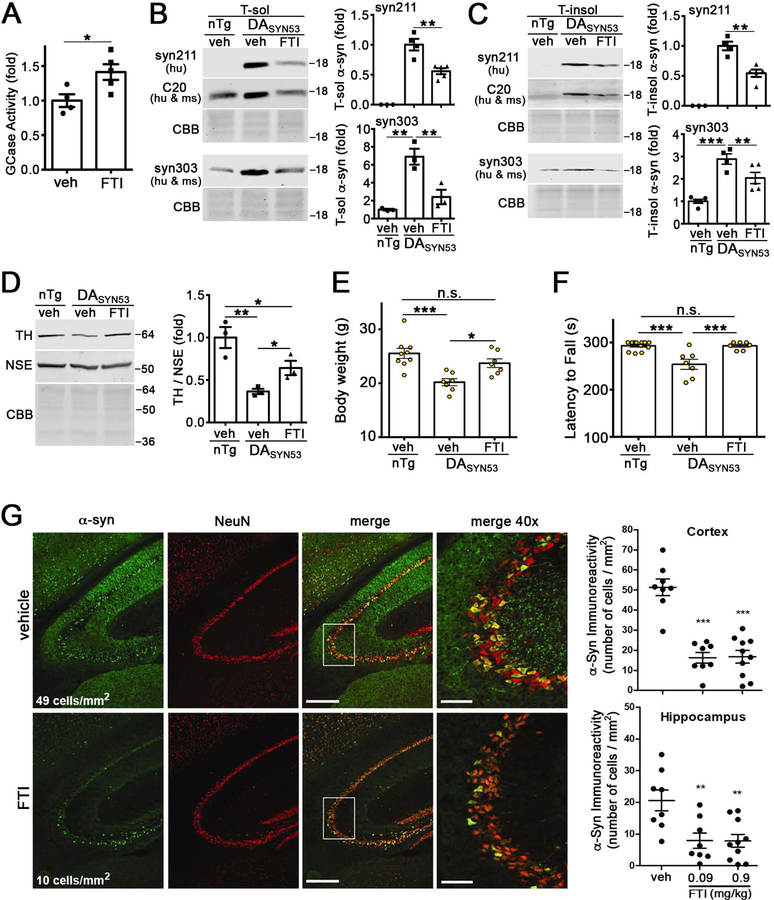
Having demonstrated that the FTI can activate ykt6 in vivo, we then tested its ability to rescue pathological phenotypes of an established mouse model that expresses human A53T a-syn within dopaminergic neurons (DASYN53) (Chen et al., 2015). We first examined the expression pattern of ykt6 within the midbrain and found that it was robustly expressed in TH+ neurons of the substantia nigra (Figure 7A). DASYN53 mice demonstrated reduced membrane-associated ykt6 and SNARE complexes compared to non-transgenic (nTg) littermates, and FTI treatment by i.p. injection reversed these effects (Figure 7B–D). FTI treatment enhanced GCase trafficking, as indicated by increased mature, endo H resistant forms of the protein (Figure 7E). To determine the functional effects of FTIs on the lysosomal system, we measured GCase activity in midbrain of DASYN53 mice and found that the FTI enhanced activity by ca. 40% (Figure 8A). Importantly, this level of activity enhancement was sufficient to reduce soluble and insoluble A53T a-syn in midbrain extracts (Figure 8B, C). We then assessed the level of TH protein in the midbrains of DASYN53 mice, since previous studies have shown that reduction of TH is tightly associated with neurodegeneration and occurs subsequent to A53T a-syn accumulation (Chen et al., 2015). We confirmed that TH protein was reduced in the midbrain of DASYN53 mice compared to nTg mice, and found that FTI treatment could partially restore TH levels (Figure 8D). We also documented a mild but significant reduction in body weight and motor performance of DASYN53 mice compared to nTg animals, which was rescued by FTI treatment (Figure 8E, F). Finally, we tested if FTIs could reduce pathology in a distinct synucleinopathy model that expresses human wt-syn under the PDGF-b promoter (Masliah et al., 2000). Since previous studies showed a progressive accumulation of a-syn within the hippocampus and cortex between 6 and 9 months of age (Amschl et al., 2013), FTI treatment was initiated at 6 months and continued for 3 months. Here, the FTI was administered to mice twice per day orally, to more closely resemble the projected route that could be employed in humans. Immunohistochemical analysis of human wt a-syn at 9 months revealed a significant decline in a-syn immunoreactivity in the cortex and hippocampus, while the neuronal marker, NeuN, was unchanged (Figure 8G). Together, these data indicate that the FTI activates ykt6 in the brains of mice to stimulate the lysosomal system and reduce pathological a-syn. FTI-mediated a-syn reduction in multiple iPSn and in vivo models indicates a strong therapeutic potential of this pathway for the treatment of synucleinopathies.
Figure 7. FTI treatment restores functional membrane-associated ykt6 complexes in DASYN53 mice.
A) Immunohistochemistry in the midbrain of mice showing ykt6 (red), tyrosine hydroxylase (TH, green), NeuN (white), and nuclei (DAPI, blue). Scale bar, 50mm. B) Ykt6 membrane shift analysis in olfactory bulbs of FTI (LNK-754)-injected DASYN53 mice (12–14 months, i.p. 26d, 0.9mg/kg) (n=5, non-transgenic (nTg) +veh; n=4, DASYN53 + veh; n=4 DASYN53 + FTI). GAPDH and sec22b are loading controls. C) SEC / western blot analysis of midbrains of FTI-treated DASYN53 mice. Inputs are shown on the left. NSE is a loading control. Low exposure of ykt6 shows changes in monomers (22–14A); high exposures shows changes in complexes (120A). (n=3, nTg + veh; n=4, DASYN53 + veh; n=4 DASYN53 + FTI). D) Quantification of complexes from SEC in C. E) GCase maturation was assessed in midbrain lysates by endo H resistance (n=4, nTg + veh; n=4, DASYN53 + veh; n=4 DASYN53 + FTI). For all quantifications, each point represents a measurement from an individual animal. Values are the mean +/− SEM, *p<0.05; **p<0.01, ***p<0.001, ANOVA with Tukey’s post-hoc test. See also Figure S7.
Figure 8. FTI treatment enhances GCase activity and reduces pathological a-syn in transgenic mice.
A) GCase activity in the midbrain of FTI (LNK-754)-treated DASYN53 mice (treated as in fig. 7) (n=4, veh; n=5, FTI). B & C) Quantification of soluble and insoluble a-syn from midbrains of FTI-treated DASYN53 mice normalized to total protein (CBB). Syn211 is selective for human (hu) a-syn; C20 and syn303 detect mouse (ms) and hu a-syn, (n=3, nTg +Veh; n=4, DASYN53 + veh; n=5 DASYN53 + FTI). D) Midbrain TH levels were quantified by western blot of FTI-treated DASYN53 mice (n=3). E) Body weight of FTI-treated DASYN53 mice (n=9, nTg +Veh; n=7, DASYN53 + veh; n=7 DASYN53 + FTI). F) Rotarod test of FTI-treated DASYN53 mice (n=12, nTg +Veh; n=7, DASYN53 + veh; n=7 DASYN53 + FTI). G) Immunohistochemistry of a-syn in the hippocampus of PDGF-b wt-a-syn mice treated for 3 months (between 6 and 9 months) via oral gavage at 0.09 mg/kg (n=8) and 0.9 mg/kg (n=10). (0.9 mg/kg). a-Syn, green; NeuN, red. Scales bars = 0.6mm and 0.15mm (40X). Each point represents a measurement from an individual animal. Values are the mean +/−SEM. *p<0.05, **p<0.01, and *** p<0.001. Student’s t-test, panel A; ANOVA with Tukey’s post-hoc test, panels B-F; ANOVA with Newman-Kuels post hoc test compared to vehicle, panel G. See also Figure S7.
Discussion
We have identified a novel regulatory trafficking pathway that senses and controls the physiological response to lysosomal stress. Previous studies have delineated regulatory gene networks that are required for the synthesis of lysosomal components upon metabolic stress (Sardiello et al., 2009; Settembre et al., 2011). Our studies indicate that ykt6 may be preferentially involved and synergistically interact with these networks by enhancing the transport of lysosomal machinery to promote cellular clearance. Interestingly, TFEB does not appear to regulate the transcription of ykt6 (Palmieri et al., 2011), and our data indicates that total ykt6 protein levels are unaffected by lysosomal stressors or TFEB overexpression (Figure 3). Instead, our studies indicate that ykt6 responds to lysosomal stress by mechanisms that are mediated by post-translational prenylation, which may enable its rapid response to changing cellular environments. The non-functional cytosolic reserve pool of ykt6 likely acts as a buffer that senses metabolic stress and moves into membranes to promote hydrolase trafficking and lysosomal activity.
Our studies in human midbrain models indicate that lysosomal function is highly sensitive to ykt6 levels, since even partial KD could dramatically affect the transport and activity of lysosomal machinery, while constitutive protein secretion was not changed (Figure 2, S4B). Previous studies have also noted a partial association of ykt6 with lysosomes (Hasegawa et al., 2003). Although this suggests that ykt6 may be preferentially involved in lysosomal clearance pathways, we were unable to achieve complete depletion of ykt6. It is possible that more efficient knock-down could affect the maturation of other proteins that are processed through the early secretory pathway. It is also possible that ykt6 may have a specialized role in trafficking lysosomal proteins within human iPSn, or its activity could be distributed at different cellular compartments depending on metabolic requirements. For example, other studies have shown that ykt6 could affect constitutive protein secretion in non-neuronal cells or in Drosophila (Gordon et al., 2010; Gross et al., 2012). Ykt6 may have a more prominent secretory function in cells that exhibit high levels of basal constitutive secretion, such as rapidly dividing neuroendocrine or HEK cell lines. It is also important to note that previous studies have established redundancy in SNARE mediated trafficking pathways (Liu and Barlowe, 2002). Human iPSC-midbrain neurons may not possess a redundant SNARE pathway that can stimulate the trafficking of lysosomal proteins in the absence of ykt6, whereas secretory or plasma membrane protein trafficking may be compensated for by other SNARE complexes when ykt6 is depleted. Together, our data underscore the complexity of protein transport mechanisms utilized under basal vs. stressful conditions, and suggests that distinct SNARE complexes mediate different cargoes depending on the metabolic needs of the cell.
Previous studies have shown that the autophagic-lysosomal system is activated during periods of a-syn accumulation (Ebrahimi-Fakhari et al., 2011; Mak et al., 2010), and elevated expression of lysosomal proteins occurs in response to pathogenic protein accumulation in other neurodegenerative diseases (Cataldo et al., 1995). Consistent with this, we find that TFEB can translocate into the nucleus and elevate the mRNA and protein of lysosomal components in PD patient neurons that exhibit cellular inclusions (Figure S2). This suggests that lysosomal dysfunction does not occur from reduced synthesis of lysosomal proteins, but instead occurs from impeded trafficking as suggested by previous studies (Chung et al., 2013; Cooper et al., 2006; Gitler et al., 2008; Mazzulli et al., 2016a). Our findings suggest that once a-syn accumulation is initiated, it can disable the physiological response to lysosomal stress by blocking vesicular trafficking pathways that are essential for lysosomal function (Figure S8). Since lysosomes are important for a-syn clearance (Cuervo et al., 2004), disabling this pathway could provide a permissive environment for aggregates to persist and continue to grow in the cell. This may also lead to favorable conditions for aggregate self-replication, by increasing opportunities for seed fibrils to interact and convert newly made, physiological a-syn into additional pathogenic species. Self-replication of amyloid fibrils by templated conformational conversion is a well established phenomenon (Jarrett and Lansbury, 1993), and early studies have shown that pathological a-syn fibrils can replicate by conversion of soluble monomers through a nucleation dependent mechanism (Wood et al., 1999). Similarly, more recent in vivo and culture studies have shown that a-syn fibrils can propagate within and between neurons (Luk et al., 2012; Volpicelli-Daley et al., 2011). Our studies are consistent with the notion that pathological a-syn possesses self-reproducing features by disabling clearance mechanisms that are meant to eliminate it from the cell (Figure S8).
Our data also have important implications for the development of novel therapies. We find that overexpression and activation of ykt6 alone is sufficient to restore lysosomal activity in PD patient neurons, leading to a reduction of pathological a-syn. Ykt6 overexpression preferentially rescues lysosomal function, and does not change the levels of plasma membrane proteins or constitutive secretion (Figure 4). Our data indicates that ykt6 rescues lysosomal function by enhancing ER-Golgi transport and that blocking farnesylation of ykt6 could further enhance membrane association and binding of ER-Golgi SNAREs (Figure 5A, B). Important for the development of ykt6 as a therapeutic target, we identify a druggable method to harness its fusogenic activity through the use of a validated FTI that exhibits drug-like properties. The FTI potently influences GCase activity by correcting its distribution into lysosomes at concentrations as low as 1nM, resulting in reduced a-syn in PD patient neurons (Figure 5C–G, Figure 6). FTI treatment rescued phenotypes in DASYN53 mice, but also enhanced lysosomal activity in healthy wild-type mice (Figure S7). This indicates the potential of this pathway for the clinical advancement of not only synucleinopathies, but possibly of other diseases characterized by protein aggregation such as Alzheimer’s disease. Strategies centered on enhancing protein trafficking and lysosomal activity may provide the most effective disease-altering therapies for synucleinopathies, since they target two pathways that synergize and are considered critical for pathogenesis (Abeliovich and Gitler, 2016).
STAR METHODS
Materials Availability:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joseph R. Mazzulli, PhD, Email: jmazzulli@northwestern.edu
Unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement. Some of the materials utilized in this study are the property of other investigators and therefore requests for these materials will be referred to the relevant institutions.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to corresponding author Joseph R Mazzulli (jmazzulli@northwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Overview of models employed in this study
Inducible human H4 neuroglioma cells expressing a-syn under the control of a tetracycline-responsive promoter (tet-off) (Mazzulli et al., 2011) and iPSC-derived dopaminergic neurons (iPSn; from control, PD and GD patients) were utilized as cell models to study a-syn aggregation and structure. iPS cells models have been previously authenticated (Mazzulli et al., 2016a). The authentication procedure is based on genotyping for common disease-causing mutations (published in (Mazzulli et al., 2011)), pluripotency analysis, karyotype analysis, efficient differentiation into midbrain dopamine neurons, and absence of mycoplasma. H4 cells have been previously authenticated (Mazzulli et al., 2011) by analyzing expression of a-synuclein (a-syn) and absence of mycoplasma. The gender of cell lines is listed in the “Key Resources Table“ document. Randomization of samples or cell cultures during analysis of HPLC, cell toxicity assays, and biochemical assays was done to account for any technical variations as noted below. Blinding of the samples to the experimenter was done when possible and noted below. In some cases, obvious differences in culture behavior, morphology, solution turbidities, color, or other easily identifiable features made blinding difficult.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-alpha synuclein (LB509) | Abcam | Cat #ab27766 RRID:AB_727020 |
| Chicken polyclonal NeuN | EMD Millipore | Cat# ABN91 |
| Rabbit polyclonal anti-alpha synuclein (C-20) | Santa Cruz | Cat #SC-7011-R RRID:AB_2192953 |
| Rabbit polyclonal anti-GFP | Sigma Aldrich | Cat #G1544 RRID: AB_439690 |
| Mouse monoclonal anti-Bet1 (17) | Santa Cruz | Cat #SC-136390 RRID: AB_10838396 |
| Mouse monoclonal anti-syntaxin 5 (B-8) | Santa Cruz | Cat #SC-365124 RRID: AB_10709311 |
| Mouse monoclonal anti-sec22b | Santa Cruz | Cat #SC-101267 RRID: AB_2186088 |
| Mouse monoclonal anti-ykt6 (discontinued item) | Abcam | Cat #ab77150 RRID: AB_1566879 |
| Mouse monoclonal anti-ykt6 | Santa Cruz | Cat #SC-365732 RRID: AB_10859388 |
| Rabbit monoclonal anti-GM130 | Abcam | Cat #ab52649 RRID: AB_880266 |
| Rabbit polyclonal anti-LAMP2 | Thermofisher Scientific | Cat #51–2210 RRID: AB_2297357 |
| Mouse monoclonal anti-LAMP1 | Santa Cruz | Cat #SC-20011 RRID: AB_626853 |
| Mouse monoclonal anti-GCase | N/A (gift of J. Aerts) | N/A |
| Rabbit polyclonal anti-TFEB | Bethyl Laboratories | Cat #A303–673A RRID: AB_11204751 |
| Mouse monoclonal anti-Neuronal Class III Beta-tubulin (TUJ1) | Covance | Cat # MMS-P435-P100 RRID: |
| Rat monoclonal anti-alpha synuclein (15G7) | Enzo Life Sciences | Cat #ALX-804–258-LC05 RRID: AB_11180660 |
| Mouse monoclonal anti-alpha synuclein (303) | Biolegend | Cat #824301 RRID: AB_2564879 |
| Rabbit polyclonal anti-calnexin | Cell Signaling Technologies | Cat #2433S RRID: AB_2243887 |
| Mouse monoclonal anti-GAPDH (6C5) | Millipore | Cat #CB1001 RRID:AB_2107426 |
| Mouse monoclonal anti-neurofilament | Dev. Studies Hybrid Bank, U. Iowa | Cat #2H3 RRID:AB_531793 |
| Rabbit polyclonal anti-neuron specific enolase (NSE) | Polysciences Inc. | Cat #17437 |
| Mouse monoclonal anti-NCAM | Sigma Aldrich | Cat #C9672 RRID: AB_1079450 |
| Mouse monoclonal anti-tyrosine Hydroxylase (TH-2) | Sigma | Cat #T1299 RRID:AB_477560 |
| Rabbit polyclonal anti-nicastrin antibody | Cell Signaling | Cat # 3632 |
| Secondary antibody: IRdye 800-conjugated IgG antibodies | Licor Biosciences | Cat #P/N 925–32210 RRID:AB_2687825 |
| Secondary antibody: IRdye 680-conjugated IgG antibodies | Thermo Life Technologies | Cat #A32734 RRID:AB_2633283 |
| Secondary antibody: Alexa Fluor 680 Goat anti-mouse IgG (H+L) | Life Tech | Cat #A-21058 RRID:AB_2535724 |
| Secondary antibody: Alexa Fluor 568 Goat anti-mouse IgG (H+L) | Life Tech | Cat #A11031 RRID: AB_144696 |
| Secondary antibody: Alexa Fluor 488 Goat anti-rabbit IgG (H+L) | Life Tech | Cat #A11034 RRID: AB_2576217 |
| Secondary antibody: Alexa Fluor 350 Goat anti-rabbit IgG (H+L) | Life Tech | Cat #A21068 RRID: AB_141378 |
| CellLight Golgi-GFP BacMam 2.0 | Life Tech | Cat #C10592 |
| Bacterial and Virus Strains | ||
| lenti-pER (HIV, replication incompetent) | Mazzulli et al., 2011 | N/A |
| Wild-type ykt6 lentivirus | This paper | |
| Ykt6-CS lentivirus | This paper | |
| Biological Samples | ||
| Brain homogenates of transgenic A53T-a-syn mice | Chen et al. 2015 | N/A |
| Brain slices of wildtype a-syn Tg mice | Masliah et al. 2000 | N/A |
| Brain slices and homogenates of wildtype mice | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Doxycycline hydrochloride (DOX) | Sigma | Cat #D3447 |
| Geneticin (G418) | Thermo Fisher Scientific | Cat #10131027 |
| Hygromycin | Thermo Fisher Scientific | Cat #10687010 |
| Puromycin | Thermo Fisher Scientific | |
| Penicillin/streptomycin | Thermo Fisher Scientific | Cat #10378016 |
| Opti-MEM | Thermo Fisher Scientific | Cat # 51985091 |
| all-trans-Retinoic acid | Sigma | Cat# R2625 |
| Farnesyltransferase inhibitor LNK-754 | Link Medicine | N/A |
| Farnesyltransferase inhibitor LNK-3248 | Link Medicine | N/A |
| Human recombinant alpha-synuclein | Expressed in e.coli; Mazzulli et al., 2007 | N/A |
| EZ-Link™ NHS-Biotin | Thermo Fisher Scientific | Cat #20217 |
| Pierce™ High Capacity NeutrAvidin™ Agarose beads | Thermo Fisher Scientific | Cat #29204, |
| D/D solubilizer, iDimerize system | Takara | Cat# 635067 |
| Critical Commercial Assays | ||
| Genomic DNA isolation kit (DNeasy Blood and Tissue Kit) | Qiagen | Cat #69504 |
| Maxiprep Kit (endotoxin-free) | Qiagen | Cat #12362 |
| T7 Endonuclease I | New England Biolabs | Cat #M0302 |
| PureLink RNA Mini Kit | Thermo Fisher Scientific | Cat #12183018A |
| QuickChange II Kit | Agilent | Cat #200521 |
| CellTagTM 700 | Licor Biosciences | Cat # 926–41090 |
| Quantitative RT-PCR: LAMP1 (ID:Hs00174766_m1) | Thermo Fisher Scientific | N/A |
| Quantitative RT-PCR: GBA1 (ID:Hs00164683_m1) | Thermo Fisher Scientific | N/A |
| Quantitative RT-PCR: YKT6 (ID:Hs01127135_m1) | Thermo Fisher Scientific | N/A |
| Quantitative RT-PCR: GAPDH (ID:Hs02758991_g1) | Thermo Fisher Scientific | N/A |
| HIV1-P24 ELISA kit | Zeptometrix | Cat # 0801111 |
| Human neuroserpin ELISA development kit | LSBio | Cat #LS-F31370 |
| Deposited Data | ||
| N/A | ||
| Experimental Models: Cell Lines | ||
| H4 neuroglioma cells (sex origin unknown) | Mazzulli et al., 2011; From: Pamela McLean (Mayo Clinic, Jacksonville, Florida, USA) | N/A |
| iPS-derived midbrain neurons: Control WT, Male origin (C3) | C3; Mazzulli et al., 2011, 2016a; Somatic cells originally from: Hedrich et al, 2006, Archives of Neurology 63 (6):833–838 | N/A |
| iPS-derived midbrain neurons: PD SNCA trp Patient (ND34391), Female sex | This paper; Mazzulli et al., 2016a; Coriell cell repository | N/A |
| iPS-derived midbrain neurons: A53T a-syn and isogenic controls | Soldner et al. 2011 | N/A |
| SH-SY5Y cells, female origin | ATCC | Cat #CRL22–66 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| CRISPR/Cas9n; SNCA gene guide RNA 1: 5’-AGCAGCCACAACTCCCTCCTTGG-3’ | This paper, designed with: crispr.mit.edu | N/A |
| CRISPR/Cas9n; SNCA gene guide RNA 2: 5’- TGAGAAAACCAAACAGGGTGTGG −3’ | This paper, designed with: crispr.mit.edu | N/A |
| PCR primer to validate PGK-puro insertion (on genomic DNA): 5’ Fwd: CATAAAATCTGTCTGCCCGCTCTC | This paper, generation: Integrated DNA Technologies | N/A |
| PCR primer to validate PGK-puro insertion (on genomic DNA): 5’ Rev: GTGGGCTTGTACTCGGTC | This paper, generation: Integrated DNA Technologies | N/A |
| PCR primer to validate PGK-puro insertion (on genomic DNA): 3’ Fwd: CTTCTACGAGCGGCTCGGCTT | This paper, generation: Integrated DNA Technologies | N/A |
| PCR primer to validate PGK-puro insertion (on genomic DNA): 3’ Rev: TGTGGTCATCCTCCACCTGACT | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (ADAMTSL4): Fwd: GGTGGTGTCTGGCGTTCTGT | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (ADAMTSL4): Rev: TCCCTCCTCCTTCCAAGTGCAG | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (ARHGAP32): Fwd: GGCCTGGGTTCCAATTCTGACT | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (ARHGAP32): Rev: GAACGTGCCCAGAACAACCGAA | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (ATG9B): Fwd: TTGCAGCTGCGCCACTTCAA | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (ATG9B): Rev: GCGCTTCACATCCATAAGGGCA | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (SEMA4B): Fwd: GAGAGGACCAGGGTGCAGTTAG | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (SEMA4B): Rev: GATCACCGAGGGTACCAGTCCC | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (SLC26A1): Fwd: CCTTCTACGAGGATGCCACAGAGT | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (SLC26A1): Rev: GCCTTCTGGAAACACAGAGACCCT | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (SLC26A3): Fwd: GCATTCACATGTGCATGGTGCC | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (SLC26A3): Rev: AGGAAGGAGGCATGGACAGTGA | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (SNCB): Fwd: AGCTGGGGAAGGGGATGGAAA | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (SNCB): Rev: CTTTCATCACTGCACTGGTCCCTG | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (SNCG): Fwd: ATCGGCGTCAATAGGAGGCATC | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (SNCG): Rev: GGCCTCTTCCTGCTGTGTCT | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (TSC2): Fwd: TGCTCTGCTCTCTGCTCCATGGTA | This paper, generation: Integrated DNA Technologies | N/A |
| Sequencing primer of off-targets on genomic DNA (TSC2): Rev: TCGCAGGTGAAGGGACAGTTTC | This paper, generation: Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| Cas9-nickase plasmid PX335 | Ran et al., 2013, Addgene | Cat #42335 |
| HR plasmid: PITX3–2A-eGFP-PGK-Puro backbone | Addgene | Cat #31943 |
| pCDNA3.1 | Invitrogen | Discontinued |
| pCDNA3.1-wt-a-syn | This paper | |
| pEGFP-wt-ykt6 | This paper | |
| pEGFP-ykt6-CS | This paper | |
| pEGFP | Clontech | Discontinued |
| MISSION shRNA sequences in pLKO.1 (Clone # TRCN0000059765) | Sigma-Aldrich | |
| Software and Algorithms | ||
| Snapgene software | www.snapgene.com | N/A |
| GraphPad Prism software V6 | www.graphpad.com/scientific-software/prism | N/A |
| Odyssey software (Image Studio V3.1.4) | Licor Biosciences; https://www.licor.com/bio/products/software/image_studio/ | N/A |
| Adobe Photoshop CS | Adobe; https://www.adobe.com/products/photoshop.html | N/A |
| ImageJ | https://imagej.net | N/A |
| Other | ||
| Lipofectamine 3000 | Thermo Fisher Scientific | Cat #L3000008 |
| Taq DNA polymerase | New England Biolabs | Cat #E5000S |
| T7 Endonuclease | New England Biolabs | Cat #M0302S |
| Q5 High Fidelity DNA polymerase | New England Biolabs | Cat #M0491 |
| Matrigel (Corning) | Fisher Scientific | Cat #CB-40234 |
| mTeSR1 | Stemcell Technologies | Cat #85850 |
| Neurobasal media | Thermo Fisher Scientific | Cat #21103–049 |
| NeuroCult SM1 supplement | Stemcell Technologies | Cat #05711 |
| GFP-trap agarose beads | Chromotek | Cat #GTA-20 |
H4 cell culture
The culture of inducible human H4 neuroglioma cells express a-syn under the control of a tetracycline-responsive promoter (tet-off) were previously described (Mazzulli et al., 2011). Cells were grown in Optimem media containing 5% fetal bovine serum (FBS), 200 mg/ml Geneticin and Hygromycin, and 1% penicillin/streptomycin (from www.thermofisher.com). a-Syn expression was turned off by the addition of 1 mg/ml doxycycline (DOX) for 48 hours.
Generation of stable-transfected SH-SY5Y cell lines
Naïve SH-SY5Y cells (ATCC # CRL-2266, female origin) were cultured in a 10 cm dish in DMEM containing 10% FBS and 1% penicillin/streptomycin and transfected with pCDNA3.1, pCDNA3.1-wt-a-syn, pEGFP-wt-ykt6, pEGFP-ykt6-CS or pEGFP alone with lipofectamine 2000. After 48 hrs, cells were passaged into 10 ea 10cm dishes G418 was added to the media gradually in 50uM increments, starting at 50uM and gradually increasing to 800uM over 2 weeks. Remaining colonies (approx. 20 per line) were picked, expanded, and analyzed for a-syn expression, morphology, and the ability to differentiate with retinoic acid. The most stable clones were chosen for vector or wt-a-syn expressing cells and used for studies and cultured in the above media containing 200uM G418. For each experiment, SH-SY5Y cells were differentiated with all trans retinoic acid (10uM) for 5 days.
iPS cell culture and neuronal differentiation
iPS cell culture procedures and differentiation into midbrain dopaminergic neurons have been described in detail previously (Mazzulli et al., 2016a). Human iPSCs were maintained in mTeSR1 media (http://www.stemcell.com/en/Products/All-Products/mTeSR1.aspx) on matrigel (Thermofisher Scientific) coated dishes. Established iPSC lines from a healthy control was used (line “C3, 2135” from (Mazzulli et al., 2016a)). Parkinson’s disease patient derived lines expressing A53T a-syn and matching isogenic corrected lines were generously provided by Dr. R. Jaenisch (Whitehead Institute of MIT) and characterized previously (Soldner et al., 2011). iPSC lines harboring the SNCA gene triplication were previously described and extensively characterized (Mazzulli et al., 2016a). iPSCs were differentiated into midbrain DA neurons using an established protocol (Kriks et al., 2011). Neurons were differentiated for 40 days in a cocktail of growth factors (Kriks et al., 2011) followed by withdrawal of growth factors from day 40–60. They were maintained in neurobasal media (Thermofisher Scientific, #21103–049) containing NeuroCult SM1 supplement (Stemcell Technologies #05711) and 1 % penicillin/streptomycin until used for experiments. Maturation and quality control of iPSC-neurons was analyzed on each differentiation batch by the location of a-synuclein into synapse, by colocalization with synapsin, and the ratios of biii-Tubulin / GAPDH to assess the relative levels and efficiency of differentiation between batches. A full and extensive characterization of iPSCs and iPSC-derived midbrain neurons has previously been done in our recent study (Mazzulli et al., 2016a). iPSC lines were cultured for 60–130 days for each experiment, and each assay was done with at least 3 technical replicates as indicated in figure legends, and confirmed in 2 to 3 independent differentiation batches.
Control and transgenic alpha-synuclein mouse lines
Wild type C57Bl/6 mice were purchased at 3 months of age (equal male and female groups) from Charles River (Strain code 027) and housed in accordance with the US National Institutes of Health Guide to the Care and Use of Laboratory Animals and Society for Neuroscience guidelines. Mice were provided standard rodent chow and water ad libitum.
Transgenic mice expressing human A53T-a-syn under the control of the dopamine transporter promoter (DASYN53) were previously described (Chen et al., 2015). Single-transgenic mice between the ages of 12 and 14 months (equal male and female groups) were utilized for experiments. Mice were bred and housed according to the Institutional Animal Care and Use Committee at Northwestern University guides and handled in accordance with the US National Institutes of Health Guide to the Care and Use of Laboratory Animals and Society for Neuroscience guidelines. Mice were provided standard rodent chow and water ad libitum. Genotyping of animals was performed as a service provided by Transnetyx (https://www.transnetyx.com) by tail clipping and were numbered consecutively by ear tags. Non-transgenic control littermates were always compared to mice expressing A53T a-synuclein mice. These mice were employed for studies shown in Figures 7 and 8. The use of wild-type and transgenic mice were approved under Northwestern IACUC protocol number IS00011551.
Transgenic mice that overexpressed wild-type alpha-synuclein under the PDGF-b promotor were also used (Masliah et al., 2000). Mice were raised according to the animal welfare regulations of the Austrian guidelines for the care and use of laboratory animals at the QPS research animal facility under standardized conditions. Animals were housed at 24 °C with a re lative humidity between 40–70% with a constant light cycle (12 hours light/dark). Mice were provided standard rodent chow (Altromin, Germany) and normal tap water ad libitum. Genotyping of animals was performed by tail clipping and were numbered consecutively by classical ear markings. Mice were approximately six months old at the beginning of the study. These mice were employed for studies shown in Figures 8.
METHOD DETAILS
CRISPR/Cas9n of iPSCs
T7 Endonuclease I Assay for the analysis of off-target effects of alpha-syn knock-out iPSC
These procedures have been previously described (Zunke et al., 2018). Genomic regions of the top 9 homologous regions to SNCA were amplified using Q5 High Fidelity DNA polymerase (New England Biolabs). PCR amplicons were denatured and hybridized in a thermal cycler: 95°C for 10 minutes, 95–85°C (ramp rate −2C/sec), and 85–25°C (ramp rate −0.2°C/sec). The hybridized PCR product was digested using T7 Endonuclease I (New England Biolabs) for 1 hour at 37°C. Equal volumes of undigested and digested PCR products were resolved on 1.5% agarose gel along with positive control (Genecopoeia).
Biochemistry and cell biology
Generation of plasmids
pENTR223-ykt6 containing cDNA plasmids were obtained from the Harvard plasmid ID repository (https://plasmid.med.harvard.edu/PLASMID/) and subcloned into either pCDNA3.1 for transfections or pER4 for lentiviral generation. GFP fusions were added by subcloning ykt6 into pEGFP-C1 (Clontech). Mutations in ykt6 were generated by site directed mutagenesis using the QuikChange kit from Agilent (www.agilent.com).
Co-immunoprecipitation analysis
To co-immunoprecipitate (co-IP) a-syn and ykt6, cultures were extracted in 0.3% CHAPS buffer (0.3% CHAPS, 40mM HEPES pH 7.4, 120 mM NaCl, 1mM EDTA, 10% v/v glycerol) by homogenization and incubation on ice / water slurry for 30 min. Lysates were cleared by centrifugation at 21,000 x g for 20 min and pre-cleared by incubation with normal rabbit IgG (Santa Cruz) + pre-blocked protein A/G beads (Santa Cruz). 500ug of lysate was incubated with 2ug of anti a-syn antibody (C-20, Santa Cruz), ykt6 antibody (ab77150, abcam, www.abcam.com), or 2ug of normal rabbit IgG to control for non-specific binding, plus 30ul of pre-blocked protein A/G beads overnight at 4°C with rotation. Beads were collected by centrifugation at 1000 x g for 5 min, washed three times in 0.3% CHAPS lysis buffer, and complexes were eluted by boiling in 2X Laemmeli sample buffer. Samples were loaded onto 18% SDS-PAGE gels and analyzed by western blot using anti-a-syn (C-20), anti-sec22b (29-F7, SC-101267, Santa Cruz), anti-ykt6 (abcam), anti-bet1 (Santa Cruz, SC-136–390), anti-syntaxin 5 (Santa Cruz, SC-365124). Corresponding immunodepleted lysates were also analyzed by western blot to confirm interactions.
To co-IP ykt6 SNARE assemblies, approximately 106–107 cells expressing GFP-tagged YKT6, were harvested in 1 ml of PBS, scraped off the plate and transferred to centrifuge tubes. The cells were subjected to centrifugation 200Xg for 5m at 4°C. The cell pellet was resuspended in ~ 200µl co-IP buffer (10mM Tris-HCl pH 7.5, 150mM NaCl, 0.5M EDTA and 0.5% NP-40 containing protease inhibitor cocktail, PMSF and NaF ) and subjected homogenization (iPSC-neurons) or extensive pipetting (cell lines). The cells were incubated on ice water-slurry for 30 min on a rocker. The cell lysates were cleared by centrifugation at 21,000Xg for 30 min, 4°C and supernatants were subjected to protein assay. A total of 1mg of protein was adjusted to 500–1000µl with dilution buffer (Co-IP buffer without 0.5% NP-40). 25µl of GFP-trap agarose beads (GTA-20, Chromotek, www.chromotek.com) were equilibrated by mixing beads with 500µl dilution buffer followed by centrifugation at 2500Xg for 2 min. The recovered beads were subjected to same procedure 2 more times. Equilibrated beads were mixed with the diluted lysate and incubated overnight at 4°C under constant shaking. GFP trap beads were recovered by centrifugation and subjected to washing with dilution buffer 3 times. Proteins bound to GFP were eluted in 2X Laemmeli sample buffer and boiling the samples at 95°C for 10 min. Eluates were analyzed by western blot as described below. The samples were not analyzed in a blinded manner.
Separation of lysates into cytosol and membrane fractions
Cell cultures from either a 10cm dish (SH-SY5Y) or 6 well (iPSn) were scraped in cold PBS and pelleted by centrifugation at 400xg, 5 min, 4°C. The supernatant was discarded and cell pellets were homogenized in 200–400ul of cold PBS containing protease inhibitor cocktail mix (1 mM phenylmethanesulfonyl fluoride (PMSF), 50 mM NaF, 2 mM Na orthovanadate, and a protease inhibitor cocktail (Roche diagnostics, http://www.roche.com, # 11–836-170–001)) using a Teflon homogenizer in a conical glass vessel for 20–40 times, using a motor driven spindle at 4000 rpm (GlasCol, https://www.glascol.com/homogenizers). The extract was pelleted at 2,500 x g, 4°C for 10 min, the supernatant was saved, and the pellet was re-extracted as before. The supernatants were then centrifuged at 100,000 x g, 4°C for 30 min and saved as the cytosolic fractions. The remaining pellet was extracted in 1% Triton X-100 buffer (1% Triton X-100, 20 mM HEPES pH 7.4, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1.5 mM MgCl2, 1 mM phenylmethanesulfonyl fluoride (PMSF), 50 mM NaF, 2 mM Na orthovanadate, and a protease inhibitor cocktail (Roche diagnostics, http://www.roche.com, # 11–836-170–001)) by pipetting and 3 freeze / thaw cycles, incubated on and ice-water slurry for 20 minutes and ultracentrifuged at 100,000 x g, 4°C for 30 minutes. The triton-soluble fraction was saved as the membrane containing fraction. Fractions were then analyzed by western blot analysis as described below. The samples were not analyzed in a blinded manner.
Size exclusion chromatography analysis
Cultures or PD brain (cingulate cortex, previously described (Mazzulli et al., 2011)) were homogenized as described above in 1% Triton buffer, centrifuged at 100,000 x g for 30 min at 4°C. 1mg of soluble lysate was injected on a Superdex 200 HR 10/300 gel filtration column (www.gelifesciences.com) using a mobile phase of phosphate buffered saline at pH 7.4 (PBS) (sample injection volume.: 800mg lysate into 250 ml, flow rate at 0.3 ml/min; 0.5 ml sized fractions, sample loop maximum volume = 400ul), using an Agilent HPLC 1200 series pumps, autoinjector, UV/vis detector, and fraction collector. Samples were concentrated using 10,000 MWCO filters from Millipore, mixed with sample buffer, boiled, and loaded onto SDS-PAGE gels for western blot analysis. The sample identity was blinded for human samples and cell cultures.
Knock-down of ykt6 shRNA expressing constructs
MISSION shRNA sequences in pLKO.1 vectors targeting ykt6 were obtained from Sigma-Aldrich and tested for efficiency in transfected HEK cells by ykt6 western blot analysis. Clone # TRCN0000059765 was found to achieve the most efficient knock-down in the absence of toxicity and was used for further experiments. This construct was packaged into lentiviral particles by transfecting HEK293T cells with FuGENE HD (Promega) and virused was used at MOI 3, dpi 7 in iPSC-neurons or SH-SY5Y cells. Constructs were also transfected into SH-SY5Y cells and stable cell lines were selected with puromycin at 1–2 ug/ml.
Measurement of farnesyl-ykt6 in SH-SY5Y cells
The method used was based off of a previously published protocol (Kho et al., 2004). SH-SY5Y stable lines expressing GFP alone or GFP-ykt6 were cultured on 10 cm dishes until ~75% confluent. Media was replaced on all cultures with media containing 20 uM lovastatin and 20 uM farnesyl-alcohol azide (Cayman Chemical Company). Either 10 nM FTI (LNK-754) or an equivalent volume of DMSO was then added. After 48 hours of incubation, protein was extracted in 400 uL RIPA lysis buffer supplemented with fresh protease inhibitor cocktail, 50 mM NaF, 0.2 mM sodium orthovanadate, and 1 mM PMSF. 1 mg total protein in 1 mL Dilution/Wash buffer (10 mM Tris/Cl; 150 mM NaCl; 0.5 mM EDTA) was incubated with 20 uL GFP-Trap _A beads (Chromotek) for 1 hour at 4°C rotating end over end. Beads were collected by centrifugation at 2500x g for 2 minutes at 4°C following the provided protocol. Beads were washed 3X in Dilution/Wash buffer before resuspension in 500 uL Dilution/Wash buffer containing 50 uM phosphine-biotin (Cayman Chemical Company). Reactions were shaken horizontally overnight at room temperature. Beads were collected, washed 3X, and boiled for 10 minutes in 50 uL 2X sample buffer. The mixture was finally centrifuged at 2500x g for 2 minutes, and the supernatant was collected for western blot analysis using streptavidin-IRDye 800 conjugated detection reagent to detect farnesyl-ykt6. Values were normalized to total amount of GFP-ykt6 precipitated using anti-GFP antibodies (Sigma) or anti-ykt6 antibodies. The sample identity was not blinded.
Measurement of Neuroserpin, LAMP1, GCase protein and activity from culture media
Media from cultures where ykt6 was depleted was conditioned for 3 days, collected and concentrated, and used for analysis. As a negative control, media not exposed to cultures was analyzed in parallel. For analysis of LAMP1, GCase, and total N-linked glycosylated proteins, 500µl of conditioned media was mixed with 20µg/ml biotinylated Concanavalin (CON-A) and the reaction mixture was incubated overnight at 4°C with gentle rotation. CON-A bound proteins were recovered using neutrAvidin agarose beads (29204, ThermoFisher, Scientific). NeutrAvidin (25µl) was added to the reaction mix and incubated at 4°C for 1h. Beads were collected by centrifugation at 2500Xg for 2 min. This was followed by washing with PBS 3 times. N-glycosylated proteins were eluted in 2X Laemmeli sample buffer and boiling the samples at 95°C for 10 min. Samples were analyzed by western blot for LAMP1, GCase, or total N-glycosylated proteins by Coomassie brilliant blue staining of the pulled-down samples.
Neuroserpin levels were measured in the conditioned media using LSBio Human neuroserpin ELISA development kit (LS-F31370). Conditioned media was collected and subjected to centrifugation at 200Xg for 5m at 4°C to remove any dead cells. 1 ml media was concentrated to 20ul using Amicon ultra centrifugal filter units (UFC50108K, Millipore). Concentrated media (10µl) were subjected to neuroserpin ELISA following the manufacturer’s protocol. The neuroserpin levels in the media were calculated using a standard curve and normalized to the total cellular protein levels.
Analysis of GCase activity was done in 100ul of conditioned media that was concentrated to 20ul using Amicon ultra centrifugal filter units (UFC50108K, Millipore) and buffer exchanged into GCase activity assay buffer ((0.25% (v/v) Triton X-100 (Sigma-Aldrich #T-8787), 0.25% (w/v) Taurocholic acid (Sigma-Aldrich, # T9034), 1 mM EDTA, in citrate/phosphate buffer, pH 5.4). 4-methylumbelliferyl β-D-glucopyranoside (4-MU-Gluc; Sigma-Aldrich) was added and incubated at 37°C for 2 hours, followed by addition of an equal amount of stopping solution (1 M glycine, pH 12.5). 4-MU-Gluc fluorescence (ex = 355 nm, em = 460) was detected in a Molecular Devices i3 microplate reader using fluoro plates (Nunc, #475515). The sample identities were not blinded.
Measurement of mRNA
Total RNA was extracted and isolated from ykt6 shRNA infected iPSC neurons at 7 dpi using the PureLink RNA Mini Kit (Thermofisher Scientific). cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermofisher Scientific). Real-time PCR was performed on a Applied Biosytems 7500 Fast system using pre-designed Taqman-primer probe sets: LAMP1 (Hs00174766_m1), GBA1 (Hs00164683_m1), YKT6 (Hs01127135_m1), and GAPDH (Hs02758991_g1). The quantification is represented as fold change of target mRNA expression normalized to GAPDH levels by delta-Ct method. The values are mean and s.e.m of three biological replicates (n=3) with three technical replicates for each. The sample identities were not blinded.
Analysis of post-ER GCase
40ug of lysates were digested with Endoglycosidase H (Endo H) according to the manufacturer’s protocol (New England Biolabs, www.neb.com) for 2 hours. In parallel, samples without enzyme were incubated under the same conditions as a control. Laemmeli sample buffer was added and digests were run on 10% SDS-PAGE gels for 3–4 hours at 120V. Gels were subjected to western blot analysis by incubation with GCase (8E4) antibodies (Gift of J. Aerts, Leiden University). Endo H resistant bands migrating at 62–64 kDa were measured as post-ER forms of GCase, and normalized to GAPDH levels. The sample identities were not blinded.
Sequential biochemical extraction for the analysis of pathological a-synuclein
Cells were harvested in phosphate buffered saline (PBS), pH 7.4, and pelleted by centrifugation at 400x g for 5 minutes. Cell pellets were extracted in 1% Triton X-100 buffer (1% Triton X-100, 20 mM HEPES pH 7.4, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1.5 mM MgCl2, 1 mM phenylmethanesulfonyl fluoride (PMSF), 50 mM NaF, 2 mM Na orthovanadate, and a protease inhibitor cocktail (Roche diagnostics, http://www.roche.com, # 11–836-170–001)) by homogenization, incubated on and ice-water slurry for 20 minutes and ultracentrifuged at 100,000 x g, 4°C for 30 minutes. Supernatant (Triton-soluble fraction) was used for Bradford or BCA (Pierce) protein assay. The Triton-insoluble pellets were dissolved in SDS-lysis buffer (2% SDS, 50 mM Tris, pH7.4 and a protease inhibitor cocktail (Roche diagnostics)). The samples were boiled for 10 min, sonicated three times for 3 seconds, boiled again for 10 min and centrifuged at 100,000xg for 30 min at 22°C. Pr otein concentration was measured by BCA assay (Pierce). The sample identities were not blinded.
Western blot analysis
20–40ug of total lysate or immunoprecipitated material was loaded into tris-glycine SDS-PAGE gels (10–15% acrylamide, depending on the assay) and run at 150V for 1.5 hrs for most assays, or for 3–4 hours for GCase post-ER / ER ratio analysis, using Thermofisher Scientific Mini Gel Tank module (#A25977). Gels were transferred onto Millipore immobilon-FL PVDF membranes with low-autofluorescence using Thermofisher Scientific Miniblot (# B1000) at 20V for 1 hour, post-fixed in 0.4% PFA (for a-syn) or 100% MeOH (all other applications), and blocked in Odyssey blocking buffer (Licor). Primary antibodies were incubated overnight at 4oC, washed 3 times with TBS-Tween (0.1%), and secondary antibodies (Alexa 680, Molecular Probes; or IRDye800, Licor) were added at 1:10,000 in blocking buffer for 1–2 hours. The blots were washed as before and scanned / analyzed using an Odyssey infrared imaging system with Image Studio Software. All blots were normalized to loading controls including GAPDH, biii-Tubulin, or total protein (Coomassie stain from the gel left over after the transfer). The sample identities were not blinded.
Measurement of neuron viability
iPSC-neurons were seeded on a PDL/laminin coated 96 well plate and analyzed and treated with either lentivirus expressing ykt6 CS vs empty vector, or DMSO vs 5nM FTI (LNK-754) for 14 days. Cells were fixed in 4% paraformaldehyde for 20 minutes and incubated with PBS containing 0.3% TritonX-100 for 20 minutes, then blocked with Odyssey blocking buffer (Li-cor) for 1 hour. Anti-neurofilament antibody (1:1000, mouse IgG 2H3, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) was incubated overnight in blocking buffer at 4°C, followed by washing in PBS with 0.1% Tween for 20 minutes. IRdye 800-conjugated anti-mouse IgG antibodies (1:1000 dilution, Li-cor) was incubated in blocking buffer for 1hour, and CellTag™ 700 (Li-cor) was added with the secondary antibody to use for cell normalization. Cells were washed four times in PBS- 0.1% Tween and scanned on an Odyssey infrared imaging system (Li-cor). Neurofilament intensity was determined using Image Studio software (version 2.1 Li-cor) and normalized to cell volume. Replicates represent values from four individual culture wells and were analyzed by Student’s T-test using GraphPad Prism software V 6.0. The sample identities were not blinded.
Cell surface biotinylation assay
For biotinylation assays, cells were cultured on 10cm dishes close to 90–100% confluency. Cells were then rinsed with cold PBS containing calcium (2.66mM) two times. Cell surface proteins were subjected to biotinylation using EZ-Link™ NHS-Biotin (20217, The rmo-fisher scientific), 500µg/ml in PBS containing calcium. The reaction was set up at 4°C for half an hour on a rocker at low speed. Excess biotin was removed by washing cells with PBS containing calcium 3 times, 5 min each. This was followed by quenching excess biotin by washing cells with glycine 100mM 20 min at 4°C on a rocker. The cells were washed in PBS and harvested by centrifugation at 200Xg for 5 min. The cell pellet was lysed in RIPA buffer (10mM Tris/Cl pH 7.5, 150mM NaCl, 5mM EDTA, 0.1% SDS, 1% Triton-100 and 1% deoxycholate) and subjected to centrifugation at 21,000Xg for 20 min at 4°C. The supernatants were subjected to a DCA protein assay (Biorad), and biotinylated proteins were precipitated with Pierce™ High Capaci ty NeutrAvidin™ Agarose beads (29204, ThermoFisher, Scientific). The desired amount of protein (500µg) in a volume of 500µl adjusted with lysis buffer was incubated with 50µl of equilibrated NeutrAvidin beads, at 4°C overnight rotating end over end. Beads were recovered by centrifugation at 2500xg, 5 min, 4°C, and subjected to washing with lysis buffer 3 times. Proteins bound to NeutrAvidin beads were eluted in 2X Laemmeli sample buffer and boiled 95°C for 10 min. and analyzed by western blot using fluorescent labeled streptavidin antibody or anti-NCAM antibodies (Sigma, C9672). The sample identities were not blinded.
Live-cell assays
Live cell proteolysis assay and analysis of constitutive protein secretion
This assay was performed as described previously(Mazzulli et al., 2016a). iPSn were infected with either scrambled or shRNA sequences against ykt6, incubated for 7 days, then pulse-labeled with 3H-leucine for 48 hours. Half of the cultures were treated with leupeptin at 200uM for 2 days added at the same time as 3H-leucine, and 1M NH4Cl was added for 16 hours (diluted from 20mM stock made fresh in media) the night before the chase. After pulse-labeling for 48 hours, cells were washed and incubated with chasing medium (Neuralbasal media with SM1 neurocult, containing 2.8 mM cold leucine). Medium was changed one more time to exclude proteolysis that may occur through short-lived pathways (proteasome). Media aliquots of 70ul were removed at each time point (0, 8, 24, and 30 hours) and added directly to tubes containing 2mg/ml final BSA / 20% final trichloroacetic acid, vortexed, and incubated overnight at 4°C. Samples were centrifuged at 21,000 x g, 4°C for 20 minutes to separate insoluble protein from soluble amino acids. Soluble fractions were analyze in a Perkin Elmer Scintillation counter for the presence 3H-Leucine, which represents degraded proteins. Labeled protein pellets were solubilized in 0.1N NaOH/0.1% Na Deoxycholate and quantified by scintillation as secreted proteins. The adherent cultures were extracted in 0.1N NaOH/0.1% Na Deoxycholate and quantified by scintillation as the total labeled protein pool. 3H leucine quantified in the media was normalized to total protein detected from adherent cultures, and the data were expressed as % proteolysis or secretion over time. Sample identities were blinded.
Live cell GCase activity assay
This assay was performed using the artificial GCase substrate 5-(pentafluoro-benzoylamino)fluorescein di-ß-D-glucopyranoside (PFB-FDGluc) from Thermofish er. Fluorescent signal was quantified over time and normalized to total cell volume using CellTag 700 (Licor) or lysosomal mass by using cascade dextran blue (#D-1976, Thermofisher). The procedure and analysis method has been described previously in detail, within the supplemental information of ref (Mazzulli et al., 2016a). The sample identities were not blinded.
Live cell ER-Golgi trafficking assay
H4 cells were transfected with an reporter protein that matures through the secretory pathway in an inducible manner. The reporter is retained in the ER by reversible dimerization of FM domains and release from the ER can be temporally controlled by addition of a D/D solubilizer (iDimerize system, Takara, www.takarabio.com) that binds to FM domains and solubilizes the reporter as previously described (Rivera et al., 2000). Cells were infected with CellLight Golgi marker (Thermofisher), and colocalization was analyzed using live cell time lapse imaging on a confocal microscope (approx. one image every 10–20 seconds). Images were analyzed my Image J software to obtain a Pearson’s value at each time point and plotted against time to obtain kinetic values of transport. Initial Golgi entry was obtained by quantifying the area under kinetic curves between 0 and 200 seconds. The assay was repeated in 3 distinct culture wells per condition. The sample identities were not blinded.
Image analyses
Immunofluorescence analysis of cell cultures
For LAMP1, GM130, and ykt6 staining, cultures were plated on PDL/ laminin coated coverglasses and were fixed in 4% paraformaldehyde (PFA) in PBS for 20 minutes, washed 3 times in cold PBS, and permeabilized / blocked in blocking buffer (0.3% Triton X-100 buffer made in PBS with 2% BSA (fatty acid free) and 5% normal goat serum (NGS, Jackson immunofluorescence)) for 30 min. Primary antibodies were added at 1:100 dilutions in blocking buffer for LAMP1 (Santa Cruz, mouse monoclonal, #sc-20011), ykt6 (mouse monoclonal, Abcam #ab77150), and GM130 (rabbit polyclonal, Abcam, #ab52649) overnight at 4°C. Cells were washed 3 times for 20 min each in 0.3% Triton / PBS and secondary antibodies were added at 1:300 dilutions in blocking buffer for 2 hours, washed as before, mounted and examined by confocal microscopy on a Leica confocal microscope (Leica TCS SPE laser at 25–50% power; CTR4000 / DMI4000B microscope) through a 10um section (z-series at 1um per section). Pearson’s correlation values were used for colocalization and obtained with Image J software using the coloc 2 plugin. Individual cells were outlined as defined ROIs and calculated individually.
For ykt6 / GM130 colocalization analysis after stress, it was found that brief detergent treatment was required to better reveal changes in ykt6 localization. Cells were treated with 200 nM bafilomycin A1 for 2 hours, washed one time with cold PBS, then permeabilized for 2 minutes with 50ug/ml digitonin as described (Thayanidhi et al., 2012). Cells were then fixed in 4% PFA as described above, blocked for 30 minutes in PBS with 2% BSA and 5% NGS, and primary antibodies were incubated at 1:100 dilutions (ykt6, mouse monoclonal, Santa Cruz # sc-365732; GM130 rabbit polyclonal, Abcam, #ab52649) in PBS with 2% BSA and 5% NGS for 48 hours at 4°C in a humidified chamber. Cells were then washed 3 X 20 minutes in PBS, and secondary antibodies were added in the same blocking buffer for 2 hours. Cells were mounted and analyzed by confocal microscopy as described above.
For GCase / LAMP2 colocalization, cultures were fixed in 4% PFA in PBS for 20 minutes, then permeabilized in 0.2% saponin in 0.2% glycine buffer as described previously(Mazzulli et al., 2016b). Primary antibodies (anti-GCase 8E4, gift of Johannes Aerts, University of Leiden, NL; anti-LAMP2, Invitrogen (# 51–2210)) were diluted 1:100 in blocking buffer, incubated overnight at 4°C, washed and incubated with secondary antibodies (Alexa 568 conjugated anti-mouse, and Alexa 488-conjugated anti-rabbit, 1:400 dilution). Cells were mounted in DAPI-Fluoromount G (Southern Biotech) and analyzed by confocal microscopy as above. Colocalization was analyzed in individual cells as above using Pearson’s correlation value. Images were analyzed by independent blinded observers.
Immunohistochemistry of DASYN53 mice
Mice were perfused and right hemibrains were collected and fixed in 10% formalin followed by preservation in 30% sucrose, 1X PBS solution. 30 µM sections were s erially harvested into a 12-well plate from coronal or sagittal hemi-brain tissue using a freezing-sliding microtome and sections were stored in cryoprotective solution (1XPBS, 30% sucrose, 30% ethylene glycol) at −20°C until use. Free-floating sections were then washed three times in tris-buffered saline (TBS) followed by incubation in 16mM glycine in TBS for 1 hour at room temperature. Sections were then washed three additional times in TBS and blocked in 5% goat serum in 0.25% triton X-100 in TBS for 2 hours at room temperature. Sections were incubated overnight in primary antibodies (Anti-NeuN (Chicken EMD Millipore), 1:1000, Anti-ykt6 (Santa Cruz rabbit polyclonal (discontinued), 1:100, anti-tyrosine hydroxylase 1:1000 (Sigma mAb)) in 1% BSA, 0.25% triton X-100 and 1 X TBS at 4°C followe d by immunostaining using donkey Alexafluor-labeled secondary antibodies (Thermo Fisher Scientific). Sections were mounted using ProLong Gold (#P36934, Thermo Fisher Scientific) and imaged on a Nikon A1 laser scanning confocal microscope at Northwestern University Center for Advanced Microscopy and Nikon Imaging Centre.
Quantitative immunohistochemistry of PDGF-a-synuclein Tg mice.
The right hemisphere was immersion fixed in freshly prepared 4% paraformaldehyde/PBS (pH 7.4) for one hour at room temperature. Brains were then transferred to a 15% sucrose/PBS solution for 24 hours to prepare for cryoprotection. On the following day brains were flash frozen in liquid isopentane before storing at −80 °C. 10 µm thick sagittal sections of the cortex and hippoc ampus were obtained by cryostat sectioning, and quantitation of alpha-synuclein immunoreactive cells was done on a section at ca. +900mm medio-lateral distance as described in detail previously (Amschl et al., 2013). Sections were stained with a NeuN primary antibody (1:800; Chemicon, MAB377) and a Cy3 secondary (1:500; Jackson ImmunoResearch, 111–165-003) and with a human alpha-synuclein specific antibody (Clone 15G7, 1:5; Enzo Life Sciences, ALX-804–258-L001) and a Cy2 secondary antibody (Jackson ImmunoResearch, cat # 705–165-147). Fluorescence measurements used an exposure time of 400 ms at 100-fold magnification. Quantitation of human alpha-synuclein immunoreactive cells was performed with Image Pro Plus (version 4.5.1.29). Up to 100 single images at 100-fold magnification were assembled into a single image (real size about 3 × 1 m). Cells are identified by a macro-based procedure. In a detection step, all assembled images are contrasted manually and cells above an intensity threshold were selected within an outlined region of the brain section. A minimum size of 30 µm2 was applied to exclude transversely cut neuritic processes and peripheral cell cuts from cells in consecutive slices. The outlines of the object counts were then saved. Selected objects were then extracted from the original, contrast-free, image using the saved outlines and assembled according to object size in a sorted object image. Objects were then evaluated based on a roundness restriction (Lower limit: 1 Upper limit: 1.5) to partition alpha-synuclein positive cells from bias objects. A manual step using visual inspection of the objects was then used to manually correct the count for cells not round enough or separable from the background. In order to determine the immunoreactive cells per unit area, the area measured in each slice was calculated. Data was analyzed by one-way ANOVA followed by Newman-Kuels post hoc using GraphPad Prism.
Lentivirus treatment of cultures
Lentiviral transduction of cell lines and iPSC-neurons.
Lentivirus containing wild-type ykt6, ykt6-CS or GFP fusions were subcloned into the pER4 vector backbone driven by the PGK promoter(Deglon et al., 2000), packaged as described previously(Mazzulli et al., 2016a), and titered with a Zeptometrix HIV1-P24 ELISA kit. iPSC-neurons were infected at day 60 at a multiplicity of infection (MOI) of 2 to 3, depending on the batch, and harvested at either 6 or 8 weeks post infection. SH-SY5Y cells were similarly infected and analyzed at 7 days post infection (dpi).
FTI treatment of cultures and mice
Treatment of cultures with farnesyltransferase inhibitors
Farnesyltransferase inhibitors LNK-754 and LNK-3248 were dissolved in DMSO and added to cultures between 1 and 10 nM, with equivalent volumes of DMSO used as vehicle controls. For cell lines, the compound was added every day for 5–7 days. For iPSC-neurons, the compound was replaced every 48 hrs. Cultures were harvested and analyzed as described in the above sections.
Treatment of C57Bl/6 and DASYN53 mice with farnesyltransferase inhibitors
3 mo old C57BL6 mice were i.p. injected daily with 0.9mg/kg LNK-754 that was formulated in a vehicle of 0.5% sodium carboxymethylcellulose and filtered before use for 14 days. DASYN53 mice were i.p. injected daily with 0.9mg/kg LNK-754 as above. Mice were perfused with PBS and the brains (cortex) were rapidly dissected and frozen until analysis. DASYN53 mice were euthanized after 26 days of treatment, perfused with PBS, and the brain was rapidly dissected. The midbrain and olfactory bulbs were dissected and frozen for biochemical analysis.
Experimental design of FTI treatment in wild-type a-syn transgenic mice driven by the PDGF-b promoter
Transgenic mice divided into the following groups: vehicle (2.5% beta-cyclodextrin, n =8). 0.9 milligrams per kilogram (mpk) LNK-754 (n=10), and 0.09 mpk LNK-754 (n=8). Dosing was performed by oral gavage, twice per day, for 90 days between 6 and 9 months of age. Some mice (n=4) were sacrificed before dosing, at 6 months of age, to provide a baseline level of a-syn at the start of treatment (a-syn immunoreactivity was found to increase in untreated mice by ca. 50% between 6 and 9 months, consistent with previous studies (Amschl et al., 2013). Mice were euthanized by isoflurane overdose and transcardially perfused with 0.9% saline. Brains were then extracted, hemisected, and processed for immunohistochemistry as indicated under ‘ imaging analysis.’
Pharmacokinetic analysis
Pharmacokinetic analysis was performed as a service provided by Medicilon, Inc. Shanghai 585 Chuanda Road Pudong, Shanghai 201200, P.R.CHINA in male C57 BL/6 mice after a single dose of 9 mg/kg LNK-754 by oral gavage. LNK-754 was dissolved in 5% beta-cyclodextrin and stored at 50–60 °C until use. After collection of blood, animals were euthanized with carbon dioxide and perfused with saline.
Blood samples (250 µL) were collected via the retro-orbital vein and placed into tubes containing sodium heparin and centrifuged under refrigerated conditions at 8000 rpm for 6 minutes to separate plasma from the samples. Following centrifugation, the resulting plasma were transferred to clean tubes and stored frozen at −20 °C until bioanalysis. After collecti on of blood, animals were euthanized with carbon dioxide and perfused with saline. Whole brain was collected and rinsed with saline. Brain samples were then homogenized in saline (5 ml/g tissue) and stored at −20 °C unti l bioanalysis.
Plasma samples (50 µl) extracted with 500 µl methyl tert-butyl ether in the presence of internal standard. Samples were vortexed for 1 min, then centrifuged (3’ x 1,500 g). Supernatant was evaporated to dryness, then resuspended in 75 µl methanol and transferred to a glass autosampler vial for injection.
Homogenate was extracted with 500 µl methyl tert-bu tyl ether in the presence of internal standard. Samples were vortexed for 1 min, then centrifuged (3’ x 1,500 g). Supernatant was evaporated to dryness, then resuspended in 100 µl methanol and transferred to a glass autosampler vial for injection.
LC-MS was performed on an Agilent 1100 series liquid chromatography and an AB Inc API4000 (triple-quadrupole) with an ESI interface. Chromatography was performed on a Venusil HILIC column (5 m, 3.0 × 100 mm) using a mobile phase of 25:75 0.1% Formic Acid:Methanol at a flow rate of 350 µl/min at 25 °C. Injection volume was 5 µl.
Behavioral Analysis
Behavioral analysis of DASYN53 mice
These assays were done as previously described (Tsika et al., 2010). Body weight was taken daily before each injection of the FTI until the end of the injection protocol (nTg age-matched littermates + Vehicle, n=9; A53T + Vehicle, n=7; A53T + FTI, n=7). Reported body weights were from the end of the injection protocol (day 23–24). Each group had approximately equal male and female mice and no gender differences were noted. Balance and motor behavior testing was done at the end of the injection protocol (between days 24–26) and determined using a Rotarod apparatus (Ugo Basile) with acceleration from 4–40 rpm over a 300 second period. Latency to fall was recorded over 4 trials per mouse, and the data were presented as the average of 3 trials (excluding the first trial as a training session). For the rotorod study, mice were tested at in two separate groups from 4 different litters (nTg age-matched littermates + Vehicle, n=12; A53T + Vehicle, n=7; A53T + FTI, n=7). Each group had approximately equal male and female mice and no gender differences were noted. Animal numbers were obtained from pilot studies of rotorod performance by using the following equation : n number =2(Zα + Z1-β)2 (standard dev.)2 / (Effect size)2, where Zα is 1.96 (two tailed T-test with alpha=0.05) and Z1-β =1.2816 (power > 80%), and incorporating a 10% drop out frequency. Animal genotype and treatments were not blinded.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical tests were performed using GraphPad Prism software V6 (http://www.graphpad.com/scientific-software/prism/). ANOVA with Tukey’s post-hoc test was used when comparing more than two samples, while Student’s t-test (two-sided) was when comparing two samples.
Supplementary Material
Highlights.
Ykt6 responds to lysosomal stress by enhancing hydrolase trafficking
a-Synuclein impedes the lysosomal stress response by blocking ykt6 in patient neurons
Reducing the farnesylation of ykt6 enhances hydrolase trafficking and lysosomal function
Farnesyltransferase inhibitors activate ykt6 and lysosomes in patient neurons and mice
Acknowledgments:
Work was supported by NINDS grants R01NS092823 (JRM), R21NS107770 (JRM), WCAS Undergraduate Research Grant Program (KT). We thank Taiji Tsunemi (Juntendo Univ., Japan) for providing TFEB expression plasmids, Xiaoxi Zhuang (U. Chicago) for providing DASYN53 mice, Puneet Opal (Northwestern U.) for use of the rotarod apparatus, and Birgit Hutter-Paier. Manuela Prokesch, Daniel Havas (QPS Research, Graz, Austria) for support on the wild-type a-synuclein mouse studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: JRM is a scientific co-founder of Lysosomal Therapeutics, INC
DATA AND SOFTWARE AVAILABILITY
Data supporting the conclusions of this study can be requested from lead contact at jmazzulli@northwestern.edu.
REFERENCES
- Abeliovich A, and Gitler AD. (2016). Defects in trafficking bridge Parkinson’s disease pathology and genetics. Nature 539, 207–216. [DOI] [PubMed] [Google Scholar]
- Aflaki E, Borger DK, Moaven N, Stubblefield BK, Rogers SA, Patnaik S, Schoenen FJ, Westbroek W, Zheng W, Sullivan P, et al. (2016). A New Glucocerebrosidase Chaperone Reduces alpha-Synuclein and Glycolipid Levels in iPSC-Derived Dopaminergic Neurons from Patients with Gaucher Disease and Parkinsonism. J Neurosci 36, 7441–7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amschl D, Neddens J, Havas D, Flunkert S, Rabl R, Romer H, Rockenstein E, Masliah E, Windisch M, and Hutter-Paier B. (2013). Time course and progression of wild type alpha-synuclein accumulation in a transgenic mouse model. BMC Neurosci 14, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw RD, Pasternak SH, Mahuran DJ, and Callahan JW. (2003). Nicastrin is a resident lysosomal membrane protein. Biochemical and biophysical research communications 300, 615–618. [DOI] [PubMed] [Google Scholar]
- Bergmann JE, and Grabowski GA. (1989). Posttranslational processing of human lysosomal acid beta-glucosidase: a continuum of defects in Gaucher disease type 1 and type 2 fibroblasts. Am J Hum Genet 44, 741–750. [PMC free article] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, and Braak E. (2003). Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging 24, 197–211. [DOI] [PubMed] [Google Scholar]
- Burre J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, and Sudhof TC. (2010). Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 329, 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Barnett JL, Berman SA, Li J, Quarless S, Bursztajn S, Lippa C, and Nixon RA. (1995). Gene expression and cellular content of cathepsin D in Alzheimer’s disease brain: evidence for early up-regulation of the endosomal-lysosomal system. Neuron 14, 671–680. [DOI] [PubMed] [Google Scholar]
- Chen L, Xie Z, Turkson S, and Zhuang X. (2015). A53T human alpha-synuclein overexpression in transgenic mice induces pervasive mitochondria macroautophagy defects preceding dopamine neuron degeneration. J Neurosci 35, 890–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, Baru V, Lou Y, Freyzon Y, Cho S, et al. (2013). Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science 342, 983–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway KA, Harper JD, and Lansbury PT. (1998). Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med 4, 1318–1320. [DOI] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, et al. (2006). Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science (New York, NY) 313, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, and Sulzer D. (2004). Impaired Degradation of Mutant -Synuclein by Chaperone-Mediated Autophagy. Science 305, 1292–1295. [DOI] [PubMed] [Google Scholar]
- D’Souza MP, and August JT. (1986). A kinetic analysis of biosynthesis and localization of a lysosome-associated membrane glycoprotein. Archives of biochemistry and biophysics 249, 522–532. [DOI] [PubMed] [Google Scholar]
- Daste F, Galli T, and Tareste D. (2015). Structure and function of longin SNAREs. J Cell Sci 128, 4263–4272. [DOI] [PubMed] [Google Scholar]
- DeCourcy K, and Storrie B. (1991). Osmotic swelling of endocytic compartments induced by internalized sucrose is restricted to mature lysosomes in cultured mammalian cells. Exp Cell Res 192, 52–60. [DOI] [PubMed] [Google Scholar]
- Deglon N, Tseng JL, Bensadoun JC, Zurn AD, Arsenijevic Y, Pereira de Almeida L, Zufferey R, Trono D, and Aebischer P. (2000). Self-inactivating lentiviral vectors with enhanced transgene expression as potential gene transfer system in Parkinson’s disease. Hum Gene Ther 11, 179–190. [DOI] [PubMed] [Google Scholar]
- Downward J. (2003). Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3, 11–22. [DOI] [PubMed] [Google Scholar]
- Duda JE, Giasson BI, Mabon ME, Lee VM, and Trojanowski JQ. (2002). Novel antibodies to synuclein show abundant striatal pathology in Lewy body diseases. Ann Neurol 52, 205–210. [DOI] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Cantuti-Castelvetri I, Fan Z, Rockenstein E, Masliah E, Hyman BT, McLean PJ, and Unni VK. (2011). Distinct roles in vivo for the ubiquitin-proteasome system and the autophagy-lysosomal pathway in the degradation of alpha-synuclein. J Neurosci 31, 14508–14520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa M, Varlamov O, Eng WS, Söllner TH, and Rothman JE. (2004). Localization and activity of the SNARE Ykt6 determined by its regulatory domain and palmitoylation. Proceedings of the National Academy of Sciences of the United States of America 101, 4815–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, Caldwell KA, Caldwell GA, Rochet JC, McCaffery JM, et al. (2008). The Parkinson’s disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci U S A 105, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DE, Bond LM, Sahlender DA, and Peden AA. (2010). A targeted siRNA screen to identify SNAREs required for constitutive secretion in mammalian cells. Traffic (Copenhagen, Denmark) 11, 1191–1204. [DOI] [PubMed] [Google Scholar]
- Gross JC, Chaudhary V, Bartscherer K, and Boutros M. (2012). Active Wnt proteins are secreted on exosomes. Nature cell biology 14, 1036–1045. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Zinsser S, Rhee Y, Vik-Mo EO, Davanger S, and Hay JC. (2003). Mammalian ykt6 is a neuronal SNARE targeted to a specialized compartment by its profilin-like amino terminal domain. Molecular biology of the cell 14, 698–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Chao DS, Kuo CS, and Scheller RH. (1997). Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell 89, 149–158. [DOI] [PubMed] [Google Scholar]
- Jarrett JT, and Lansbury PT Jr. (1993). Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell 73, 1055–1058. [DOI] [PubMed] [Google Scholar]
- Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, et al. (2004). A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci U S A 101, 12479–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, et al. (2011). Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 480, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lesuisse C, Xu Y, Troncoso JC, Price DL, and Lee MK. (2004). Stabilization of alpha-synuclein protein with aging and familial parkinson’s disease-linked A53T mutation. J Neurosci 24, 7400–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, and Barlowe C. (2002). Analysis of Sec22p in endoplasmic reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol Biol Cell 13, 3314–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Kehm V, Carroll J, Zhang B, O’Brien P, Trojanowski JQ, and Lee VM. (2012). Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak SK, McCormack AL, Manning-Bog AB, Cuervo AM, and Di Monte DA. (2010). Lysosomal degradation of alpha-synuclein in vivo. J Biol Chem 285, 13621–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E, Rockenstein E, Veinbergs I, Mallory M, Hashimoto M, Takeda A, Sagara Y, Sisk A, and Mucke L. (2000). Dopaminergic loss and inclusion body formation in alpha-synuclein mice: implications for neurodegenerative disorders. Science 287, 1265–1269. [DOI] [PubMed] [Google Scholar]
- Matsui T, Jiang P, Nakano S, Sakamaki Y, Yamamoto H, and Mizushima N. (2018). Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J Cell Biol 217, 2633–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, Sidransky E, Grabowski GA, and Krainc D. (2011). Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell 146, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzulli JR, Zunke F, Isacson O, Studer L, and Krainc D. (2016a). alpha-Synuclein-induced lysosomal dysfunction occurs through disruptions in protein trafficking in human midbrain synucleinopathy models. Proc Natl Acad Sci U S A 113, 1931–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzulli JR, Zunke F, Tsunemi T, Toker NJ, Jeon S, Burbulla LF, Patnaik S, Sidransky E, Marugan JJ, Sue CM, et al. (2016b). Activation of beta-Glucocerebrosidase Reduces Pathological alpha-Synuclein and Restores Lysosomal Function in Parkinson’s Patient Midbrain Neurons. J Neurosci 36, 7693–7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Sogaard M, Lampen NM, Machida S, Ye RR, Lacomis L, Tempst P, Rothman JE, and Sollner TH. (1997). Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J Biol Chem 272, 17776–17783. [DOI] [PubMed] [Google Scholar]
- Moulder SL, Mahany JJ, Lush R, Rocha-Lima C, Langevin M, Ferrante KJ, Bartkowski LM, Kajiji SM, Noe DA, Paillet S, et al. (2004). A phase I open label study of the farnesyltransferase inhibitor CP-609,754 in patients with advanced malignant tumors. Clin Cancer Res 10, 7127–7135. [DOI] [PubMed] [Google Scholar]
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen W-L, Griffith J, Nag S, Wang K, Moss T, et al. (2011). SNARE Proteins Are Required for Macroautophagy. Cell 146, 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, et al. (2014). Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nature Genetics 46, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BJ, and Pelham HR. (1998). SNAREs and membrane fusion in the Golgi apparatus. Biochim Biophys Acta 1404, 9–31. [DOI] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, and Ballabio A. (2011). Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20, 3852–3866. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. (1997). Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047. [DOI] [PubMed] [Google Scholar]
- Pylypenko O, Schonichen A, Ludwig D, Ungermann C, Goody RS, Rak A, and Geyer M. (2008). Farnesylation of the SNARE protein Ykt6 increases its stability and helical folding. J Mol Biol 377, 1334–1345. [DOI] [PubMed] [Google Scholar]
- Rivera VM, Wang X, Wardwell S, Courage NL, Volchuk A, Keenan T, Holt DA, Gilman M, Orci L, Cerasoli F Jr., et al. (2000). Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science 287, 826–830. [DOI] [PubMed] [Google Scholar]
- Rocha EM, Smith GA, Park E, Cao H, Brown E, Hayes MA, Beagan J, McLean JR, Izen SC, Perez-Torres E, et al. (2015). Glucocerebrosidase gene therapy prevents alpha-synucleinopathy of midbrain dopamine neurons. Neurobiol Dis 82, 495–503. [DOI] [PubMed] [Google Scholar]
- Sardi SP, Clarke J, Viel C, Chan M, Tamsett TJ, Treleaven CM, Bu J, Sweet L, Passini MA, Dodge JC, et al. (2013). Augmenting CNS glucocerebrosidase activity as a therapeutic strategy for parkinsonism and other Gaucher-related synucleinopathies. Proceedings of the National Academy of Sciences 110, 3537–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. (2011). TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, and Ballabio A. (2013). Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. (2003). alpha-Synuclein locus triplication causes Parkinson’s disease. Science 302, 841. [DOI] [PubMed] [Google Scholar]
- Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. (2011). Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 146, 318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, Goldmann J, Myers RH, Young RA, and Jaenisch R. (2016). Parkinson-associated risk variant in distal enhancer of alpha-synuclein modulates target gene expression. Nature 533, 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, and Rothman JE. (1993). SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, and Goedert M. (1997). Alpha-synuclein in Lewy bodies. Nature 388, 839–840. [DOI] [PubMed] [Google Scholar]
- Stojkovska I, Krainc D, and Mazzulli JR. (2017). Molecular mechanisms of alpha-synuclein and GBA1 in Parkinson’s disease. Cell Tissue Res. [DOI] [PMC free article] [PubMed]
- Sun J, Wang L, Bao H, Premi S, Das U, Chapman ER, and Roy S. (2019). Functional cooperation of alpha-synuclein and VAMP2 in synaptic vesicle recycling. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed]
- Tai G, Lu L, Wang TL, Tang BL, Goud B, Johannes L, and Hong W. (2004). Participation of the syntaxin 5/Ykt6/GS28/GS15 SNARE complex in transport from the early/recycling endosome to the trans-Golgi network. Mol Biol Cell 15, 4011–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S, Glatz G, Szenci G, Boda A, Horvath GV, Hegedus K, Kovacs AL, and Juhasz G. (2018). Non-canonical role of the SNARE protein Ykt6 in autophagosome-lysosome fusion. PLoS Genet 14, e1007359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarentino AL, Plummer TH Jr., and Maley F. (1972). A re-evaluation of the oligosaccharide sequence associated with ovalbumin. J Biol Chem 247, 2629–2631. [PubMed] [Google Scholar]
- Thayanidhi N, Helm JR, Nycz DC, Bentley M, Liang Y, and Hay JC. (2010). Alpha-synuclein delays endoplasmic reticulum (ER)-to-Golgi transport in mammalian cells by antagonizing ER/Golgi SNAREs. Molecular biology of the cell 21, 1850–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayanidhi N, Liang Y, Hasegawa H, Nycz DC, Oorschot V, Klumperman J, and Hay JC. (2012). R-SNARE ykt6 resides in membrane-associated protease-resistant protein particles and modulates cell cycle progression when over-expressed. Biology of the Cell 104, 397–417. [DOI] [PubMed] [Google Scholar]
- Tochio H, Tsui MM, Banfield DK, and Zhang M. (2001). An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science 293, 698–702. [DOI] [PubMed] [Google Scholar]
- Tsika E, Moysidou M, Guo J, Cushman M, Gannon P, Sandaltzopoulos R, Giasson BI, Krainc D, Ischiropoulos H, and Mazzulli JR. (2010). Distinct region-specific alpha-synuclein oligomers in A53T transgenic mice: implications for neurodegeneration. J Neurosci 30, 3409–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley LA, Luk KC, Patel TP, Tanik SA, Riddle DM, Stieber A, Meaney DF, Trojanowski JQ, and Lee VM. (2011). Exogenous alpha-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron 72, 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Yu J, Pan L, Wei Z, Weng J, Wang W, Ong YS, Tran TH, Hong W, and Zhang M. (2010). Lipid-Induced conformational switch controls fusion activity of longin domain SNARE Ykt6. Molecular cell 37, 383–395. [DOI] [PubMed] [Google Scholar]
- Wood SJ, Wypych J, Steavenson S, Louis JC, Citron M, and Biere AL. (1999). alpha-synuclein fibrillogenesis is nucleation-dependent. Implications for the pathogenesis of Parkinson’s disease. J Biol Chem 274, 19509–19512. [DOI] [PubMed] [Google Scholar]
- Xu D, Joglekar AP, Williams AL, and Hay JC. (2000). Subunit structure of a mammalian ER/Golgi SNARE complex. J Biol Chem 275, 39631–39639. [DOI] [PubMed] [Google Scholar]
- Xu Y, Martin S, James DE, and Hong W. (2002). GS15 forms a SNARE complex with syntaxin 5, GS28, and Ykt6 and is implicated in traffic in the early cisternae of the Golgi apparatus. Mol Biol Cell 13, 3493–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DS, Tandon A, Chen F, Yu G, Yu H, Arawaka S, Hasegawa H, Duthie M, Schmidt SD, Ramabhadran TV, et al. (2002). Mature glycosylation and trafficking of nicastrin modulate its binding to presenilins. J Biol Chem 277, 28135–28142. [DOI] [PubMed] [Google Scholar]
- Zhang T, and Hong W. (2001). Ykt6 forms a SNARE complex with syntaxin 5, GS28, and Bet1 and participates in a late stage in endoplasmic reticulum-Golgi transport. J Biol Chem 276, 27480–27487. [DOI] [PubMed] [Google Scholar]
- Zunke F, Moise AC, Belur NR, Gelyana E, Stojkovska I, Dzaferbegovic H, Toker NJ, Jeon S, Fredriksen K, and Mazzulli JR. (2018). Reversible Conformational Conversion of alpha-Synuclein into Toxic Assemblies by Glucosylceramide. Neuron 97, 92–107 e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.