Abstract
Colorectal cancer is one of the most commonly diagnosed malignancies among males and females worldwide. Although China is a country with a low incidence of colorectal cancer, with the improvement of China’s economy and lifestyle changes, the incidence rate in China has generally increased in recent years, and the morbidity and mortality of colorectal cancer rank fifth among those of all malignant tumours. Furthermore, despite recent improvements in screening strategies and treatments for colorectal cancer, the prognosis of advanced colorectal cancer is still poor, mainly due to the recurrence or distant metastasis of this disease. Thus, colorectal cancer still seriously threatens the health and life of people and is a major public health problem worthy of further study. Recently, accumulating evidence has revealed that colorectal carcinogenesis might be a multistep process driven by progressive genetic abnormalities, including changes in lncRNA expression. Moreover, a large number of studies have discovered and studied the abnormal expression of lncRNAs in colorectal cancer, providing a promising target for the diagnosis and treatment of colorectal cancer, which will promote human understanding of the pathogenesis of colorectal cancer and improve diagnosis and treatment. Therefore, in the present review, we mainly summarize the present status of colorectal cancer, the characteristics, functions and clinical perspectives of lncRNAs, and the current therapeutic methods used for colorectal cancer, especially the application of lncRNAs in the treatment of colorectal cancer. It is hoped that this review will give readers a new understanding of the roles of lncRNAs in colorectal cancer.
Keywords: Colorectal cancer, long non-coding RNAs (lncRNAs), radiation therapy, chemotherapy, small-molecule therapy
Introduction
Colorectal cancer
Cancer is a class of malignant disease that is characterized by uncontrolled cell proliferation, resistance to cell death and the ability to invade or migrate to other parts of the body by forming metastases, and it is the second cause of death affecting both men and women worldwide, following cardiovascular diseases [1]. Moreover, it has been estimated that the number of deaths due to cancers is expected to rise to 13.2 million in 2030 [2,3]. Therefore, cancer has caused a large global economic burden and has had a remarkable impact on public health [4]. Among different types of cancers, human colorectal cancer has been ranked the third most frequently diagnosed malignancy worldwide and one of the leading causes of cancer-related mortality detected in Western countries, exceeded only by lung, liver and stomach cancers [5]. Current management strategies for colorectal cancer treatment mainly include surgery, chemotherapy, radiotherapy, and immunotherapy [6,7]. Clinically, different treatment options are generally selected according to the state of the patient, the location, and stage of the colorectal cancer, patient age, and patient health [8]. Usually, patients with colorectal cancer have an excellent prognosis if the disease is diagnosed and surgically treated at an early stage (before metastasis), with surgery being the treatment of choice for patients with localized disease [9]. If colorectal cancer patients have entered the advanced stage, surgery is no longer the best treatment, and other treatments are given clinically [10]. However, with the major advances in these treatment methods, the overall survival rate of colorectal cancer patients has been notably improved, but many adverse effects of these treatments still exist [11]. For example, multi-drug resistance sometimes causes chemotherapy failure; adverse reactions induced by chemotherapy or radiotherapy, such as chronic diarrhoea or intestinal obstructions, conspicuously reduce the quality of life of patients; and individual differences are also an important challenge for immunotherapy [12]. Moreover, the most important point is that these treatments do not prevent the recurrence and metastasis of colorectal cancer [13,14]. In addition, the incidence of colorectal cancer has increased alarmingly in recent decades and will continue to increase in the future. Hence, significant efforts are urgently needed to understand colorectal carcinogenesis, but the underlying biological processes and pathogenesis mechanisms of colorectal cancer have still not been fully illuminated over the past several years [13].
On the basis of global epidemiological and scientific studies, several risk factors have been identified for colorectal cancer, such as sex, ethnicity, diet, obesity, older age, family history, genetic mutations, inflammatory processes, and the gut microbiota [15,16]. Recent findings indicate that among these factors, genetic factors, especially non-coding RNAs (ncRNAs), might largely contribute to the progression of cancer and tumour formation [17]. Furthermore, a large amount of evidence has clearly reported that tumorigenesis is a multistep process that results from a stepwise accumulation of genetic and epigenetic alterations, eventually triggering the normal colonic epithelial cells to transform into cancer cells [18]. Thus, many investigators have devoted great efforts to exploring the biological functions of ncRNAs in the occurrence, development and progression of colorectal cancer in the past few decades. These studies will open up new avenues for screening strategies, early diagnosis, prognosis and subsequent development of targeted drugs for colorectal cancer [19].
Long non-coding RNAs (lncRNAs)
The Encyclopedia of DNA Elements (ENCODE) project, which is the most comprehensive effort yet for surveying transcription in human eukaryotic cells, has precisely revealed that most of the eukaryotic genome can be transcribed, with approximately 20,000~25,000 genes encoding proteins according to high-throughput genome-sequencing technologies, whereas the vast majority of untranslated fractions of the transcriptome (accounting for about 98% of the total human genome) are transcribed as ncRNAs, which were formerly regarded as spurious “garbage” or scrambled transcriptional “noise” for decades [20]. However, these ncRNAs have recently been considered a novel class of RNA molecules and have challenged the traditional central dogma [21]. Additionally, it was also found that ncRNAs could be involved in multiple biological processes by directly or indirectly interfering with gene expression, such as cell proliferation, cell fate determination, apoptosis, signal transduction, organ development, and cell differentiation, and even the pathogenesis of many human diseases, especially cancers [17,21,22]. Generally, ncRNAs can be systematically classified into two groups according to their product size: small ncRNAs (sncRNAs) and lncRNAs. The sncRNAs are usually shorter than 200 nucleotides (nt), and this group contains microRNAs (miRNAs), small interfering RNAs (siRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), PIWI-interacting RNAs (piRNAs), ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and other sncRNAs. LncRNAs are longer than 200 nt and include intergenic lncRNAs, intronic lncRNAs, bidirectional lncRNAs, sense lncRNAs and antisense lncRNAs, which are classified based on their genomic localization and orientation according to the neighbouring protein-coding gene) (shown in Figure 1A) [21,23]. Among such a wide variety of ncRNAs, the potential roles of miRNAs and lncRNAs in the progression of cancers have received much attention from many investigators and have been extensively studied in recent years [24,25]. For example, miR-155 regulates the proliferation and apoptosis of pancreatic cancer cells by targeting suppressor of cytokine signalling 3 (SOCS3) [26]. High expression of miR-155 is closely associated with the recurrence or metastasis of non-small-cell lung cancer [27]. The cell survival, growth, and chemosensitivity of breast cancer were modulated by miR-155 via direct targeting of forkhead box O3a (FOXO3a) [28]. Therefore, these studies suggest that miR-155 might function as an oncomiR gene in various cancers. Additionally, the lncRNA H19/miR-29b-3p regulatory axis participates in epithelial-mesenchymal transition and the metastasis of bladder cancer [29]; the lncRNA H19 promotes the progression of lung adenocarcinoma by directly targeting methylation-dependent repression of the cadherin 1 (CDH1) promoter [30], and H19 affects epithelial-mesenchymal transition and metastasis by regulating the STAT3/EZH2 axis in oesophageal cancer [31]. Therefore, these data indicate that H19 might exert a crucial role in various cancers. In fact, in the study of carcinogenesis, lncRNA has been more favoured by researchers than miRNA in recent years [24]. The lncRNA maternally expressed gene 3 (MEG3), which is located at the DLK1-MEG3 site of chromosome 14q32.3, was the first lncRNA discovered to inhibit tumour growth, and its discovery promoted a surge of research regarding the relationship between lncRNAs and cancers [32]. Compared with sncRNAs, lncRNAs have a more complex spatial structure; therefore, lncRNAs can positively or negatively play a regulatory role in protein-RNA, RNA-RNA and RNA-DNA interactions through their special secondary and tertiary spatial structures [23]. In addition, compared with the protein-coding genes, although lncRNAs are expressed at a lower level, they have more tissue specificity and temporal specificity, and they are expressed in both the nucleus and the cytoplasm [33]. Moreover, lncRNAs are involved in every aspect of biological activity, from the growth and development of organisms to the proliferation and differentiation of cells, endocytosis, and neurotransmitter transmission [34]. Therefore, lncRNAs have potential future clinical implications in terms of diagnosis, prognosis and therapeutic strategies for cancer patients.
Figure 1.
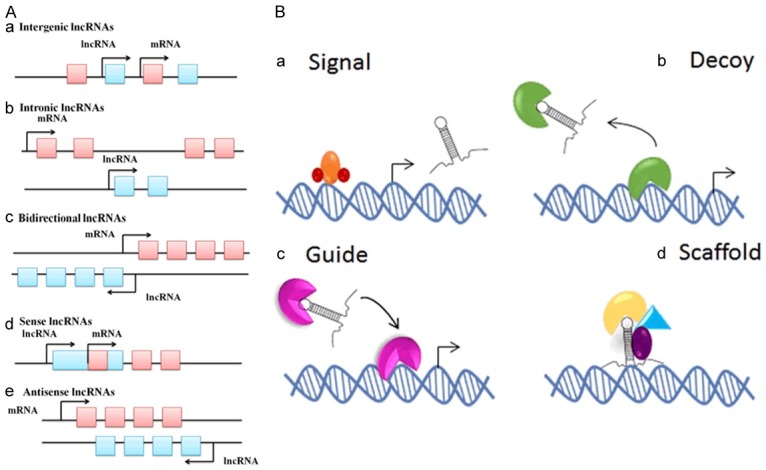
A. The classification of lncRNAs [37]. a. Intergenic lncRNAs: the entire sequence of the lncRNA falls between two protein-coding genes as a distinct unit; b. Intronic lncRNAs: the entire sequence of the lncRNA falls within the intron of a protein-coding gene; c. Bidirectional lncRNAs: the expression of the lncRNA and its neighbouring protein-coding genes on the opposite strand is initiated in close genomic proximity; d. Sense lncRNAs: lncRNAs are found inside or to the 5’ side of a protein-coding gene and are transcribed in the same direction of protein-coding genes; these ultimately overlap at least one protein-coding exon; e. Antisense lncRNAs: lncRNAs are found inside or to the 3’ side of a protein-coding gene and are transcribed in the opposite direction of protein-coding genes; these ultimately overlap at least one protein-coding exon. B. The biological roles of lncRNAs [38]. a. Signalling molecule: activate gene expression; b. Decoy molecule: suppress gene expression; c. Guide molecule: promote chromatin modification; d. Scaffold molecule: act on chromatin structure.
In the past decade, advances in genome sequencing and analysis have made the discovery of tens of thousands of lncRNAs easier, and the process of lncRNA biogenesis was also revealed [35]. First, most lncRNAs can be transcribed from intergenic, exonic or distal protein-coding regions of the genome by the enzyme RNA polymerase II, and only a small portion of them are synthesized by the RNA polymerase III (Pol III) complex or the single-polypeptide nuclear RNA polymerase IV (spRNAP IV) complex. Second, the pre-mature lncRNA undergoes 5’-capping with methyl-guanosine, takes on a multi-exonic structure and undergoes 3’-poly-adenylation. Third, lncRNAs continue to undergo alternative splicing modifications and RNA editing procedures to generate diversity. Finally, mature lncRNAs are released and transported to subcellular locations based on their functions [36]. Currently, it was concluded that lncRNAs can carry out their functions in four different ways, namely, as signals, decoys, guides, and scaffolds (Figure 1B) [24]. As a signal molecule, lncRNA is an indicator of transcriptional activity, which can activate or deactivate the natural functions of target proteins. As a decoy molecule, lncRNA can negatively regulate an effector by preventing the access of regulatory proteins to DNA. As a guide molecule, lncRNA is required for the proper localization of specific proteins, a process that is closely associated with cancer-related gene expression and can ultimately cause the formation of cancers. As a scaffold molecule, lncRNA is equal to an adaptor or an assembly platform to bind two or more protein partners and serves a structural role [23]. Hence, the multifunctional regulatory mechanisms of lncRNAs, their roles in human diseases, and their potential diagnostic and therapeutic applications will become the future research directions of researchers [33].
LncRNAs and colorectal cancer
According to preliminary statistics, approximately 90% of colorectal cancers are sporadic, and only a few cases (<10%) are hereditary [39]. Furthermore, there is growing evidence that colorectal cancer is a result of multiple factors and multiple gene interactions, and variations in molecular signalling pathways exert an essential role during the occurrence and development of colorectal cancer [40,41]. Although the current research on the molecular signalling pathways of colorectal cancer has made some progress, the underlying mechanisms of colorectal cancer during its occurrence, development and metastasis still cannot be explained [42]. However, in recent years, with the rise in lncRNA research, the mystery of lncRNA has also been gradually revealed [24]. Although lncRNAs cannot directly encode the synthesis of proteins, they are closely related to the regulation of gene expression at the epigenetic level, transcriptional level and post-transcriptional level, and they have been shown to participate in the occurrence and development of colorectal cancer [43].
LIT1/KCNQ1OT1 was detected for the first time in the genome of patients with colorectal cancer and was the first lncRNA discovered in colorectal cancer [44]. Since then, an increasing number of studies have focused on lncRNAs that are differentially expressed during the pathogenesis of colorectal cancer, and some specific lncRNAs associated with the biological processes and clinical aspects of colorectal cancer have been gradually identified [42]. For instance, the lncRNA CTA-941F9.9 is frequently found to be downregulated and may be regarded as a biomarker for carcinogenesis in colorectal cancer [45]; the lncRNA FYVE, RhoGEF and PH domain-containing 5 antisense RNA 1 (FGD5-AS1) accelerates cell proliferation, migration, and invasion via sponging miR-302e as an endogenous competing mechanism and further upregulating cell division cycle-associated 7 (CDCA7) in colorectal cancer [46]; the lncRNA small nucleolar RNA host gene 6 (SNHG6) inhibits cell proliferation and metastasis by targeting ETS proto-oncogene 1 (ETS1), which then regulates the PI3K/AKT/mTOR pathway to play a role in colorectal cancer [47]; the lncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via the miR-508-3p/partition-defective 3 (PARD3) axis [48]; and the lncRNA zinc finger antisense 1 (ZFAS1) participates in biological processes in colorectal cancer, such as cell proliferation, migration, invasion, and apoptosis, by mediating miR-7-5p [49]. In addition, many lncRNAs have been found to play a role in the initiation, progression and metastasis of colorectal cancer (as illustrated in Table 1) [24,36,43]. Thus, the pathophysiological significance of lncRNAs in colorectal cancer is undoubtedly an important future research direction [16]. At the same time, detecting and identifying the lncRNAs with potential functions in colorectal cancer are the only way to study lncRNAs, and these processes are a prerequisite for the use of lncRNAs in the diagnosis and treatment of tumours.
Table 1.
Expression of lncRNAs associated with human colorectal cancer
| LncRNAs | Location | Expression | Samples or models | Function in tumorigenesis | References |
|---|---|---|---|---|---|
| ADAMTS9-AS2 (ADAMTS9 antisense RNA 2) | Chromosome 3 | Decreased | Colorectal cancer patients | ADAMTS9-AS2 overexpression in colorectal cancer cells inhibited cell proliferation, migration, and invasion, while suppression of ADAMTS9-AS2 showed opposite effects. | [50] |
| CCAL (colorectal cancer-associated lncRNA) | NA | Increased | Colorectal cancer cell lines | CCAL transferred from fibroblasts via exosomes strengthens chemoresistance in colorectal cancer cells. | [51] |
| CCAT1 (colon cancer-associated transcript-1) | 8q24.21 | Decreased | Colorectal cancer patients | CCAT1 exerts an essential role in the genesis, development, invasion and metastasis of colorectal cancer, and it also mediates epithelial-mesenchymal transition (EMT) in colorectal cancer. | [52] |
| CCAT1-L (colon cancer-associated transcript-1 ligand) | 8q24 | Increased | Colorectal cancer patients | Lower CCAT1-L decreased the long-range interactions between the MYC promoter and its enhancers. Meanwhile, CCAT1-L also acted on CTCF and further changed the chromatin conformation at these loop regions. | [53] |
| CCAT2 (colon cancer-associated transcript-2) | 8q24.21 | Increased | Colorectal cancer patients and cell lines | CCAT2 played a crucial role in colorectal cancer pathogenesis through regulating MYC and WNT, which could provide an alternative explanation of the SNP-conferred cancer risk. | [54] |
| CRNDE (colorectal neoplasia differentially expressed) | 16q12.2 | Increased | Colorectal cancer patients and cell lines | The CRNDE/miR-181a-5p regulatory axis motivated cell proliferation and chemoresistance in colorectal cancer by modulating the Wnt/β-catenin signalling pathway. | [55] |
| CTA-941F9.9 | NA | Decreased | Colorectal cancer patients and cell lines | CTA-941F9.9 was discovered to be reduced in colorectal cancer compared with expression in non-tumour adjacent tissues and was speculated to be involved in colorectal cancer carcinogenesis. | [45] |
| DUXAP8 (double homeobox A pseudogene 8) | Chromosome 22 | Increased | Colorectal cancer patients and cell lines | The STAT3-induced upregulation of the lncRNA DUXAP8 functioned as a ceRNA for miR-577 to facilitate tumour metastasis in colorectal cancer through the regulation of RAB14. | [56] |
| FGD5-AS1 (FGD5 antisense RNA 1) | 3p25.1 | Increased | Colorectal cancer cell lines | FGD5-AS1 accelerated colorectal cancer progression through forming a miR-302e sponge, which further regulated its downstream target CDCA7. | [46] |
| FOXP4-AS1 (FOXP4 antisense RNA 1) | Chromosome 6 | Increased | Colorectal cancer patients and cell lines | FOXP4-AS1, which is considered an unfavourable prognostic factor, triggered cell proliferation and apoptosis in colorectal cancer. | [57] |
| GAS5 (growth arrest-specific 5) | 1q25.1 | Decreased | Colorectal cancer patients and cell lines | The lncRNA GAS5 promoted PTEN expression by functioning as a competing endogenous RNA (ceRNA) of miR-222-3p, thus inhibiting cell metastasis and promoting cell autophagy during the development of colorectal cancer. | [58] |
| GLCC1 | NA | Increased | Colorectal cancer patients | GLCC1 protected c-Myc transcriptional factors from ubiquitination by directly acting on the HSP90 chaperone, which further altered the transcriptional pattern of c-Myc target genes, such as LDHA, and consequently reprogrammed the glycolytic metabolism of colorectal cancer cell proliferation. | [59] |
| H19 (human homologue 19) | 11p15.5 | Increased | Colorectal cancer patients and cell lines | Abnormal H19 expression resulted in EMT of colorectal cancer cells by mediating the miR-29b-3p/PGRN axis and continuing to act on Wnt signalling. | [60] |
| HNF1A-AS1 (HIF1A antisense RNA 1) | Chromosome 14 | Increased | Colorectal cancer cell lines | HNF1A-AS1 has been reported to be involved in carcinogenesis via activation of the Wnt/β-catenin signalling pathway, which indicates a poor prognosis in colorectal cancer. | [61] |
| HOTAIR (HOX transcript antisense RNA) | 12q13.13 | Increased | Colorectal cancer patients and cell lines | HOTAIR participated in the progression and chemoresistance of colorectal cancer via increasing miR-203a-3p expression and activating the Wnt/β-catenin signalling pathway. | [62] |
| ITGB1 (integrin subunit beta 1) | Chromosome 10 | Increased | Colorectal cancer patients and cell lines | ITGB1 enhanced colorectal cancer cell migration and invasion via upregulating BDNF. | [63] |
| LINC02418 (long intergenic non-protein-coding RNA 2418) | 12q24.33 | Increased | Colorectal cancer patients | LINC02418 served as a ceRNA to further promote MELK expression through sponging miR-1273g-3p and might serve as a diagnostic marker for colorectal cancer. | [64] |
| MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) | 11q13.1 | Increased | Colorectal cancer patients and cell lines | MALAT1 triggered autophagy, facilitated cell proliferation, and hindered apoptosis by sponging miR-101 in colorectal cancer cells. | [65] |
| MEG3 (maternally expressed gene 3) | 14q32.2 | Increased | Colorectal cancer patients | MEG3 controlled cellular biological functions by directly regulating adenosine deaminase’s effect on RNA 1 in colorectal cancer. | [66] |
| MIR17HG (miR-17-92a-1 cluster host gene) | Chromosome 13 | Increased | Colorectal cancer patients and cell lines | MIR17HG promoted colorectal cancer progression via miR-17-5p and played an oncogenic role in colorectal cancer. | [67] |
| MIR503HG (MIR503 host gene) | Chromosome X | Decreased | Colorectal cancer cell lines | Forced MIR503HG expression suppressed colorectal cancer cell migration and invasion mediated by TGF-β2. | [68] |
| PRNCR1 (prostate cancer-associated non-coding RNA 1) | 8q24.21 | Increased | Colorectal cancer cell lines | PRNCR1, a potential oncogene, accelerated cell proliferation in colorectal cancer. | [69] |
| PVT-1 (Pvt1 oncogene) | 8q24 | Increased | Colorectal cancer cell lines | PVT-1 might be a prognostic indicator for colorectal cancer patients based on its antiapoptotic activity in colorectal cancer. | [70] |
| RHBDD1 (rhomboid domain-containing 1) | Chromosome 2 | Increased | Colorectal cancer patients | RHBDD1 caused tumour metastasis via activating the Wnt signalling pathway and its downstream target ZEB1 in colorectal cancer. | [71] |
| ROR1-AS1 (ROR1 antisense RNA 1) | 1p31.3 | Increased | Colorectal cancer patients and cell lines | Triggering of the Wnt/β-catenin signalling pathway by ROR1-AS1 enhanced cell metastasis and proliferation in colorectal cancer. | [72] |
| RUNX1-IT1 (RUNX1 intronic transcript 1) | Chromosome 21 | Decreased | Colorectal cancer cell lines | RUNX1-IT1, as a tumour-suppressive gene, participated in the progression of colorectal cancer by restraining cell proliferation and migration. | [73] |
| SLCO4A1-AS1 (SLCO4A1 antisense RNA 1) | 20q13.33 | Increased | Colorectal cancer cell lines | SLCO4A1-AS1 induced autophagy via mediating the miR-508-3p/PARD3 axis, which ultimately promoted colorectal cancer cell proliferation. | [48] |
| SNHG6 (small nucleolar RNA host gene 6) | Chromosome 8 | Increased | Colorectal cancer cell lines | SNHG6 directly enhanced the TGF-β/Smad signalling pathway via targeting UPF1 and promoted EMT via regulation of ZEB, which ultimately led to cell proliferation, invasion and migration in colorectal cancer. | [74] |
| SNHG14 (small nucleolar RNA host gene 14) | Chromosome 15 | Increased | Colorectal cancer patients | The SNHG14/hsa-miRNA-3940-5p/NAP1L2 axis had more advantages than CEA or CA19.9 in differentiating colorectal cancer from controls. | [75] |
| SH3PXD2A-AS1 (SH3PXD2A antisense RNA 1) | Chromosome 10 | Increased | Colorectal cancer patients and cell lines | SH3PXD2A-AS1 facilitated cancer progression partly by the targeted inhibition of P57 and KLF2 expression in colorectal cancer. | [76] |
| TP73-AS1 (TP73 antisense RNA 1) | Chromosome 1 | Increased | Colorectal cancer patients and cell lines | TP73-AS1 directly bound miR-194 and accelerated cell proliferation, migration and invasion via elevating TGF-α expression in colorectal cancer. | [77] |
| TUG1 (taurine upregulated 1) | Chromosome 22 | Increased | Colorectal cancer cell lines | TUG1 upregulated KIAA1199 expression by sponging miR-600, which further promoted cell metastasis and epithelial-mesenchymal transition in colorectal cancer. | [78] |
| XIRP2-AS1 (XIRP2 antisense RNA 1) | 2q24.3 | Decreased | Colorectal cancer patients and cell lines | XIRP2-AS1 impeded cell proliferation and invasion by targeting miR-182 in in vitro and in vivo models. Moreover, the clinical sample analysis showed that XIRP2-AS1 might be a favourable factor for evaluating the overall survival and progression-free survival of patients with colon cancer. | [79] |
Treatment strategies for colorectal cancer
Colorectal cancer, originating from the colon and the rectum, represents a serious health concern, with approximately one million new cases of colorectal cancer diagnosed worldwide, and half a million people dying from colorectal cancer every year [16]. Although there has been a dramatic improvement in the survival rates of colorectal cancer in the past 10 years due to the development of new therapeutic strategies, the incidence of colorectal cancer still remains high [6,15]. Thus, the treatment strategies for colorectal cancer have always been an area of interest for clinicians [10]. Recently, holistic treatment options, including surgery, radiation therapy, chemotherapy, immunotherapy, and targeted drugs, have been established [41,80]. In addition, small-molecule therapy programmes are also gradually emerging [18,19].
Surgery, radiation therapy, and chemotherapy
At present, the treatment of colorectal cancer is still based on surgery, and surgical treatment is also recognized as the only way to cure colorectal cancer for patients who are diagnosed early (who have an excellent prognosis) [9]. Although we have not seen breakthroughs in the field of surgery in the past few years, surgeons have thoroughly compared existing surgical methods [81]. We have evolved from traditional open surgery to a variety of surgical procedures, including endoscopic surgery, laparoscopic surgery, transanal total mesorectal excision, and robotic surgery [81,82]. After a large number of clinical studies have obtained similar oncology and survival outcomes, surgeons will focus on controlling surgical complications and improving postoperative quality of life [83]. In addition to surgical treatment, radiotherapy is the most effective and valuable treatment for local and non-metastatic colorectal cancer patients but is inefficient when the cancer has spread throughout the body [84,85]. If regional or distant metastases are discovered at the time of diagnosis, clinicians may use surgery combined with other treatments, such as chemotherapy [86]. Chemotherapy, as the major therapeutic strategy, mainly utilizes different drugs or drug combinations and delivers these drugs to the metastatic site to further inhibit the rapid proliferation of cancer cells or reduce cancer cell division [87]. However, due to the inhibition of cell growth, which is required for the maintenance of hair follicles, bone marrow and gastrointestinal tract cells, patients ultimately suffer from unwanted side effects, such as hand-foot syndrome, diarrhoea, gastrointestinal toxicity, mucositis, anaemia, neutropenia, vomiting, nausea, fatigue, haematologic disorders and liver toxicity [88,89]. Moreover, with the frequent use of chemotherapy drugs, drug resistance has also emerged, which leads to limited chemotherapy efficacy [90]. As such, to overcome the underlying side effects, better treatment options for colorectal cancer urgently need to be explored.
Immunotherapy
With the rapid development and cross-infiltration of related disciplines such as oncology, immunology and molecular biology, research on tumour immunotherapy has advanced by leaps and bounds [91]. Immunotherapy has become an important treatment for cancer after surgery, radiotherapy and chemotherapy [91,92]. Currently, immunotherapy methods for colorectal cancer mainly include cancer vaccines, cell therapy, cytokine treatment, and immunological checkpoint inhibitor treatment [93]. Cancer vaccines enhance and improve the killing effect of the immune system on tumour cells by regulating the interaction between antigen-presenting cells and T lymphocytes [94]. To date, cancer vaccines for colorectal cancer have consisted of whole tumour cell vaccines, peptide vaccines, DNA vaccines, dendritic cell (DC) vaccines, and viral vector vaccines [95]. Cellular therapy, also known as adoptive cellular immunotherapy (ACT), uses external T cells to influence the ability of patient T cells to specifically recognize tumour cells, thereby producing a killing effect on tumour cells. Widely studied immune effector cells include autologous lymphokine-activated killer (LAK) cells, autologous tumour-infiltrating lymphocytes (TILs), natural killer (NK) cells, cytotoxic T lymphocytes (CTLs), and genetically modified T cells, such as chimeric antigen receptor (CAR) T cells, and T cell receptor chimeric (TCR) T cells [96]. In regards to cytokine treatment, cytokines are pleiotropic proteins that effectively activate tumour immune cells and fight tumour immune suppression [97]. They play key roles in all aspects of innate and specific immune responses and can kill tumour cells by activating the body’s immune system and can also directly interfere with tumour cell proliferation. Studies have confirmed that cytokines that have inhibitory effects on gastrointestinal malignancies include interleukins (such as IL-2, IL-15, and IL-21), interferons (such as IFN-γ and IFN-α), and granulocyte-macrophage colony-stimulating factor (GM-CSF) [98]. An immune checkpoint refers to protein molecules expressed on tumour cells and/or immune cells that can regulate the initiation/activation processes of T cells [99]. Presently, inhibitory checkpoints have been found to include programmed cell death protein 1 (PD-1), PD-1/2 ligands (PD-L1/2), cytotoxic T lymphocyte antigen 4 (CTLA-4), lymphocyte activation gene 3, B/T lymphocyte-weakening factor, T cell immunoglobulin and mucin 3 [99,100]. These co-suppressor molecules can induce T cell apoptosis and inhibit the function of activated T cells, while tumour cells can use this inhibition of the immune system to escape the specific killing effect of immune cells in the tumour microenvironment [100]. Immunological checkpoint inhibitors that have been recently approved by the Food and Drug Administration (FDA) for the treatment of malignant tumours include anti-PD-1 antibodies (e.g., nivolumab and pembrolizumab), anti-PD-L1 antibodies (e.g., atezolizumab, avelumab and durvalumab), and anti-CTLA-4 antibodies (e.g., ipilimumab), but there are few immunological checkpoint inhibitors that can be used for colorectal cancer treatment or that have demonstrated efficacy in clinical trials. In fact, with the development of modern medicine, immunotherapy has gradually become an area of interest for cancer treatment; immunotherapy is also a new direction for the treatment of colorectal cancer and has improved the prognosis of some patients to some extent [101]. However, the existing immunotherapy regimen in colorectal cancer shows a good immune response rate in only some subtypes of colorectal cancer, and most patients still have a low immune response rate. Multi-centre, large-sample-size studies are needed to increase the effectiveness and safety of immunotherapies in colorectal cancer patients, which could make more patients obtain more benefits [102]. Hence, colorectal cancer immunotherapy is a very promising treatment option and deserves further research and exploration.
Targeted drugs
With the advancements in tumour biomedicine, especially the development of tumour-targeted therapies and individualized precision therapies based on genotyping, patients with advanced colorectal cancer are expected to receive further benefits [103]. Targeted treatments are aimed at blocking specific biologic transduction pathways or proteins that are involved in tumour growth and tumour metastasis and are altered (i.e., upregulated, downregulated or mutated) in cancers [104]. At present, the reason why targeted therapy is so favoured by researchers is mainly because targeted therapy has unique advantages compared to other treatments, such as minimizing the death of normal cells, avoiding undesirable side effects, and carrying low toxicity and high efficacy [105]. Epidermal growth factor receptor (EGFR) and vascular endothelial growth factor receptor (VEGFR) were the first protein targets developed by researchers [106]. Suppressing EGFR regulates tumour cell proliferation, invasion, metastasis, and tumour cell apoptosis via mediating the Ras/Raf/MEK/ERK-MAPK or PI3K/AKT/mTOR signalling pathways, while suppressing VEGFR promotes neovascularization, which can further provide nutrients for the proliferation of tumour cells [106].
A large number of studies have indicated that overexpression of EGFR is observed in colorectal cancer [107]. Cetuximab is a monoclonal antibody to EGFR that inhibits tumour cell proliferation, angiogenesis and metastasis, induces apoptosis, and hinders tumour growth. Therefore, cetuximab can be used to treat colorectal cancer at an advanced stage [108]. Studies have confirmed that cetuximab is more effective in RAS wild-type primary tumours located on the left side of the body than in metastatic colorectal cancer [109]. Additionally, panitumumab, which is a fully humanized, high-affinity IgG2 monoclonal antibody, can directly recognize EGFR [108]. Compared with cetuximab, panitumumab has a higher affinity for EGFR, a longer half-life and less allergic reactions [110]. Furthermore, it has been reported that panitumumab is not inferior to cetuximab in the treatment of patients with refractory KRAS wild-type metastatic colorectal cancer [110,111]. Activation of VEGFR can trigger vascular endothelial cell proliferation, differentiation, and tumour cell infiltration; thus, blocking the VEGFR-mediated signalling pathway might suppress tumour growth [112]. Bevacizumab is a recombinant humanized IgG1 monoclonal antibody that specifically blocks VEGFR, attenuates its binding to endogenous VEGFR, inhibits endothelial cell proliferation and tumour angiogenesis, and eventually inhibits tumorigenesis [113]. Compared with cetuximab, bevacizumab was not associated with RAS mutation status [114]. In addition, apatinib is a small-molecule anti-angiogenic drug that competes with for the ATP-binding site of VEGFR-2 in cells, blocks downstream signal transduction, and inhibits tumour neovascularization [115]. Accumulating evidence has demonstrated that apatinib is safe and effective in phase II and III clinical studies of various solid tumours, such as advanced gastric cancer, colorectal cancer, and lung cancer [116]. There have also been many therapeutic drugs developed to inhibit EGFR (i.e., trastuzumab, pertuzumab, trastuzumab-emtansine, and lapatinib) and VEGFR (i.e., bevacizumab, ramucirumab, sorafenib, and sunitinib) [107,112]. Moreover, nanocarriers, colloidal nano-scale systems capable of transporting anticancer agents, are emerging strategies in indirect cancer-targeted therapy [117]. With the in-depth study of the pathogenesis of colorectal cancer, more drugs may be developed for targeted therapy to achieve effective treatment of colorectal cancer in the future.
Small-molecule therapy
Ever since the discovery of lncRNAs, there is growing evidence that lncRNAs can be more exquisitely controlled and are restricted to specific cell types to a greater degree than mRNAs; therefore, lncRNAs have been regarded as potential markers for the diagnosis and treatment of colorectal cancer [6,18]. Moreover, the functions of some lncRNAs in colorectal cancer pathogenesis have been identified (shown in Figure 2) [19,42]. The Wnt/β-catenin signalling pathway, the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signalling pathway, the PI3K/AKT signalling pathway, the MAPK cascade, the p53 pathway, Notch signalling, nuclear factor κB (NF-κB) signalling, and other pathways participate in the pathogenesis of colorectal cancer via regulating cell proliferation, angiogenesis, metastasis, invasion and apoptosis, which are important events involved in the formation of colorectal cancer [5,118]. In fact, uncontrolled proliferation, migration, invasion, metastasis, and apoptosis are the main causes of failure in the treatment of colorectal cancer [119,120]. However, numerous lncRNAs participate in tumour proliferation, migration, invasion and metastasis in colorectal cancer [120,121]. Therefore, these dysregulated lncRNAs may be directly targeted by new drugs for colorectal cancer [121]. Small-molecule therapy, especially for the treatment of lncRNAs, will enable individualized treatment of colorectal cancer patients in the future [24,122].
Figure 2.
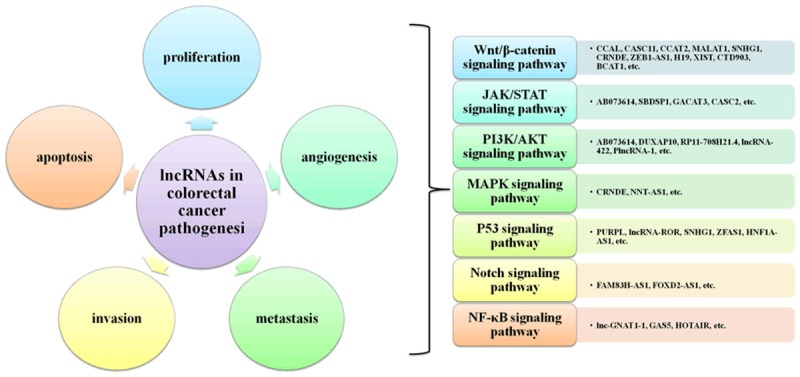
LncRNAs involved in the formation of colorectal cancer.
Conclusion
Colorectal cancer is one of the most common deadly cancers worldwide [16]. The incidence rate in China is generally on the rise, and the morbidity and mortality rate rank fifth among those of all malignant tumours [6,15]. Presently, the treatment of colorectal cancer is mainly surgery, and with the advancement of surgical techniques, especially the extensive application of total mesorectal excision, the recurrence rate and mortality of colorectal cancer have been greatly reduced [10,80]. However, local anatomy, lymphatic drainage, and tumour biological features of colorectal cancer limit the prospects of surgical techniques in improving patient outcomes and long-term survival [9,104]. Therefore, the treatment of colorectal cancer tends to emphasize the multi-disciplinary, comprehensive treatment of surgically resected tumours, including radiotherapy, chemotherapy, immunotherapy and traditional Chinese treatments, which are mainly for improving the rate of surgical resection and reducing the rates of recurrence and mortality [104]. In recent years, with the clinical application of antibody drugs and anti-angiogenic drugs, therapeutic effects in colorectal cancer have been significantly improved; but in the treatment of patients with advanced-stage disease, postoperative recurrence and chemotherapy resistance are still difficult points, and the prognosis of these patients remains very poor, which seriously affects the quality of life of patients and causes significant economic losses to patients’ families and national health insurance funds [102]. Despite continuous research on the risk factors, pathogenesis, diagnosis, treatment and prognosis of colorectal cancer in recent years, the molecular and genetic mechanisms of colorectal cancer are still not fully understood, and therapeutic effects have not been satisfactory [39]. These shortcomings are considered to be the result of a combination of environmental factors and genetic characteristics [2]. Multiple factors, such as activation of tumour susceptibility genes, inactivation of tumour suppressor gene, abnormal regulation of signalling pathways and the complexity of ncRNA regulation of protein-coding genes, are highly related to the development of colorectal cancer [17]. Among these factors, the deregulation of lncRNAs is regarded as an important change during the carcinogenesis of colorectal cancer [18,24]. At the same time, growing evidence has demonstrated that lncRNAs can promote or inhibit the generation of colorectal cancer at different levels and through different mechanisms of action [42]. Thus, in this review, we summarized the functions of identified lncRNAs in the pathogenesis of colorectal cancer. Moreover, based on the roles of lncRNAs, the therapeutic potential of lncRNAs has become a new area of interest for research, which will open new directions for the treatment of colorectal cancer and bring new hope to patients with colorectal cancer. Nevertheless, it is undeniable that although lncRNAs may bring accurate treatment to patients with colorectal cancer, they still have certain challenges in that lncRNAs that are successful in cell and animal models do not necessarily translate into the clinic. In conclusion, further comprehensive study of lncRNAs will probably provide the ultimate solution for colorectal cancer diagnostics and therapeutics.
Acknowledgements
This study is supported by Fund from Xisike-Hausen Oncology Research Foundation (No. Y-HS2017-059), Qingdao Annual Industrial Cultivation Plan for Science and Technology (No. 19-6-1-27-nsh) and Qingdao Expert Workstation.
Disclosure of conflict of interest
None.
References
- 1.Pérez-Herrero E, Fernández-Medarde A. Advanced targeted therapies in cancer: drug nanocarriers, the future of chemotherapy. Eur J Pharm Biopharm. 2015;93:52–79. doi: 10.1016/j.ejpb.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 5.Zoratto F, Rossi L, Verrico M, Papa A, Basso E, Zullo A, Tomao L, Romiti A, Lo Russo G, Tomao S. Focus on genetic and epigenetic events of colorectal cancer pathogenesis: implications for molecular diagnosis. Tumour Biol. 2014;35:6195–206. doi: 10.1007/s13277-014-1845-9. [DOI] [PubMed] [Google Scholar]
- 6.Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ihnát P, Vávra P, Zonča P. Treatment strategies for colorectal carcinoma with synchronous liver metastases: which way to go? World J Gastroenterol. 2015;21:7014–21. doi: 10.3748/wjg.v21.i22.7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Leersum NJ, Aalbers AG, Snijders HS, Henneman D, Wouters MW, Tollenaar RA, Eddes EH. Synchronous colorectal carcinoma: a risk factor in colorectal cancer surgery. Dis Colon Rectum. 2014;57:460–6. doi: 10.1097/DCR.0000000000000068. [DOI] [PubMed] [Google Scholar]
- 9.Akgül Ö, Çetinkaya E, Ersöz Ş, Tez M. Role of surgery in colorectal cancer liver metastases. World J Gastroenterol. 2014;20:6113–22. doi: 10.3748/wjg.v20.i20.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derakhshankhah H, Izadi Z, Alaei L, Lotfabadi A, Saboury AA, Dinarvand R, Divsalar A, Seyedarabi A, Barzegari E, Evini M. Colon cancer and specific ways to deliver drugs to the large intestine. Anticancer Agents Med Chem. 2017;17:1317–1327. doi: 10.2174/1871520617666170213142030. [DOI] [PubMed] [Google Scholar]
- 11.Krishnaiah YS, Khan MA. Strategies of targeting oral drug delivery systems to the colon and their potential use for the treatment of colorectal cancer. Pharm Dev Technol. 2012;17:521–40. doi: 10.3109/10837450.2012.696268. [DOI] [PubMed] [Google Scholar]
- 12.Chourasia MK, Jain SK. Pharmaceutical approaches to colon targeted drug delivery systems. J Pharm Pharm Sci. 2003;6:33–66. [PubMed] [Google Scholar]
- 13.Gulbake A, Jain A, Jain A, Jain A, Jain SK. Insight to drug delivery aspects for colorectal cancer. World J Gastroenterol. 2016;22:582–99. doi: 10.3748/wjg.v22.i2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C, Ng HL, Pan W, Chen H, Zhang G, Bian Z, Lu A, Yang Z. Exploring different strategies for efficient delivery of colorectal cancer therapy. Int J Mol Sci. 2015;16:26936–52. doi: 10.3390/ijms161125995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore JS, Aulet TH. Colorectal cancer screening. Surg Clin North Am. 2017;97:487–502. doi: 10.1016/j.suc.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 17.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149:1204–1225. e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zamani M, Hosseini SV, Mokarram P. Epigenetic biomarkers in colorectal cancer: premises and prospects. Biomarkers. 2018;23:105–114. doi: 10.1080/1354750X.2016.1252961. [DOI] [PubMed] [Google Scholar]
- 20.ENCODE Project Consortium. The ENCODE (ENCyclopedia of DNA elements) project. Science. 2004;306:636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 21.Daugaard I, Hansen TB. Biogenesis and function of ago-associated RNAs. Trends Genet. 2017;33:208–219. doi: 10.1016/j.tig.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Xu J, Shao T, Zhang Y, Chen H, Li X. RNA function prediction. Methods Mol Biol. 2017;1654:17–28. doi: 10.1007/978-1-4939-7231-9_2. [DOI] [PubMed] [Google Scholar]
- 23.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 24.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15:429. doi: 10.2174/138920101505140828161335. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Guo J, Fan H. MiR-155 regulates the proliferation and apoptosis of pancreatic cancer cells through targeting SOCS3. Eur Rev Med Pharmacol Sci. 2019;23:5168–5175. doi: 10.26355/eurrev_201906_18181. [DOI] [PubMed] [Google Scholar]
- 27.Xu S, Shi L. High expression of miR-155 and miR-21 in the recurrence or metastasis of non-small cell lung cancer. Oncol Lett. 2019;18:758–763. doi: 10.3892/ol.2019.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869–79. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Lv M, Zhong Z, Huang M, Tian Q, Jiang R, Chen J. lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta Mol Cell Res. 2017;1864:1887–1899. doi: 10.1016/j.bbamcr.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Gao LM, Xu SF, Zheng Y, Wang P, Zhang L, Shi SS, Wu T, Li Y, Zhao J, Tian Q, Yin XB, Zheng L. Long non-coding RNA H19 is responsible for the progression of lung adenocarcinoma by mediating methylation-dependent repression of CDH1 promoter. J Cell Mol Med. 2019;23:6411–6428. doi: 10.1111/jcmm.14533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen MJ, Deng J, Chen C, Hu W, Yuan YC, Xia ZK. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int J Biochem Cell Biol. 2019;113:27–36. doi: 10.1016/j.biocel.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Zhou C, Huang C, Wang J, Huang H, Li J, Xie Q, Liu Y, Zhu J, Li Y, Zhang D, Zhu Q, Huang C. LncRNA MEG3 downregulation mediated by DNMT3b contributes to nickel malignant transformation of human bronchial epithelial cells via modulating PHLPP1 transcription and HIF-1alpha translation. Oncogene. 2017;36:3878–3889. doi: 10.1038/onc.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Fan S, Song E. Noncoding RNAs: new players in cancers. Adv Exp Med Biol. 2016;927:1–47. doi: 10.1007/978-981-10-1498-7_1. [DOI] [PubMed] [Google Scholar]
- 35.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 36.Dhanoa JK, Sethi RS, Verma R, Arora JS, Mukhopadhyay CS. Long non-coding RNA: its evolutionary relics and biological implications in mammals: a review. J Anim Sci Technol. 2018;60:25. doi: 10.1186/s40781-018-0183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–51. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.García Torrecillas JM, Ferrer Márquez M, Reina Duarte Á, Rubio-Gil F. Epidemiological investigation of colorectal cancer: perspective, prospective and challenges in a big data context. Semergen. 2016;42:509–513. doi: 10.1016/j.semerg.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Song M, Garrett WS, Chan AT. Nutrients, foods, and colorectal cancer prevention. Gastroenterology. 2015;148:1244–60. e16. doi: 10.1053/j.gastro.2014.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akin H, Tözün N. Diet, microbiota, and colorectal cancer. J Clin Gastroenterol. 2014;48(Suppl 1):S67–9. doi: 10.1097/MCG.0000000000000252. [DOI] [PubMed] [Google Scholar]
- 42.Tang X, Qiao X, Chen C, Liu Y, Zhu J, Liu J. Regulation mechanism of long noncoding RNAs in colon cancer development and progression. Yonsei Med J. 2019;60:319–325. doi: 10.3349/ymj.2019.60.4.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim T, Croce CM. Long noncoding RNAs: undeciphered cellular codes encrypting keys of colorectal cancer pathogenesis. Cancer Lett. 2018;417:89–95. doi: 10.1016/j.canlet.2017.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakano S, Murakami K, Meguro M, Soejima H, Higashimoto K, Urano T, Kugoh H, Mukai T, Ikeguchi M, Oshimura M. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci. 2006;97:1147–54. doi: 10.1111/j.1349-7006.2006.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Z, Zhou C, Zhong X, Shi J, Wu Z, Tang K, Wang Z, Song Y. The long noncoding RNA CTA-941F9.9 is frequently downregulated and may serve as a biomarker for carcinogenesis in colorectal cancer. J Clin Lab Anal. 2019;25:e22986. doi: 10.1002/jcla.22986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li D, Jiang X, Zhang X, Cao G, Wang D, Chen Z. Long noncoding RNA FGD5-AS1 promotes colorectal cancer cell proliferation, migration, and invasion through upregulating CDCA7 via sponging miR-302e. In Vitro Cell Dev Biol Anim. 2019;55:577–585. doi: 10.1007/s11626-019-00376-x. [DOI] [PubMed] [Google Scholar]
- 47.Meng S, Jian Z, Yan X, Li J, Zhang R. LncRNA SNHG6 inhibits cell proliferation and metastasis by targeting ETS1 via the PI3K/AKT/mTOR pathway in colorectal cancer. Mol Med Rep. 2019;20:2541–2548. doi: 10.3892/mmr.2019.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Jin J. LncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508-3p/PARD3 axis. Aging (Albany NY) 2019;11:4876–4889. doi: 10.18632/aging.102081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mo D, Liu W, Li Y, Cui W. Long non-coding RNA zinc finger antisense 1 (ZFAS1) regulates proliferation, migration, invasion, and apoptosis by targeting MiR-7-5p in colorectal cancer. Med Sci Monit. 2019;25:5150–5158. doi: 10.12659/MSM.916619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bu X, Qin A, Luo Z, Hu Y. Overexpression of the long non-coding RNA ADAMTS9-AS2 suppresses colorectal cancer proliferation and metastasis. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2019;44:741–748. doi: 10.11817/j.issn.1672-7347.2019.190142. [DOI] [PubMed] [Google Scholar]
- 51.Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng H, Zhang X, Kong F, Guan M. Long non-coding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer. 2019 doi: 10.1002/ijc.32608. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 52.Ye Z, Zhou M, Tian B, Wu B, Li J. Expression of lncRNA-CCAT1, E-cadherin and N-cadherin in colorectal cancer and its clinical significance. Int J Clin Exp Med. 2015;8:3707–15. [PMC free article] [PubMed] [Google Scholar]
- 53.Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, Yang L, Chen LL. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–31. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–61. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, Yu ZW, Jia YH, Bai XF, Li L, Liu YL, Cui BB. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du C, Wang HX, Chen P, Chen CH. STAT3-induced upregulation of lncRNA DUXAP8 functions as ceRNA for miR-577 to promote the migration and invasion in colorectal cancer through the regulation of RAB14. Eur Rev Med Pharmacol Sci. 2019;23:6105–6118. doi: 10.26355/eurrev_201907_18424. [DOI] [PubMed] [Google Scholar]
- 57.Li J, Lian Y, Yan C, Cai Z, Ding J, Ma Z, Peng P, Wang K. Long non-coding RNA FOXP4-AS1 is an unfavourable prognostic factor and regulates proliferation and apoptosis in colorectal cancer. Cell Prolif. 2017;50 doi: 10.1111/cpr.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu L, Wang HJ, Meng T, Lei C, Yang XH, Wang QS, Jin B, Zhu JF. lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via the GAS5/PTEN-signaling pathway in CRC. Mol Ther Nucleic Acids. 2019;17:644–656. doi: 10.1016/j.omtn.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Tang J, Yan T, Bao Y, Shen C, Yu C, Zhu X, Tian X, Guo F, Liang Q, Liu Q, Zhong M, Chen J, Ge Z, Li X, Chen X, Cui Y, Chen Y, Zou W, Chen H, Hong J, Fang JY. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat Commun. 2019;10:3499. doi: 10.1038/s41467-019-11447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding D, Li C, Zhao T, Li D, Yang L, Zhang B. LncRNA H19/miR-29b-3p/PGRN axis promoted epithelial-mesenchymal transition of colorectal cancer cells by acting on Wnt signaling. Mol Cells. 2018;41:423–435. doi: 10.14348/molcells.2018.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang X, Xiong Y, Tang F, Bian Y, Chen Y, Zhang F. Long noncoding RNA HNF1A-AS1 indicates a poor prognosis of colorectal cancer and promotes carcinogenesis via activation of the Wnt/beta-catenin signaling pathway. Biomed Pharmacother. 2017;96:877–883. doi: 10.1016/j.biopha.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 62.Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, Guo X, Zhang Y. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via MiR-203a-3p-mediated Wnt/ß-catenin signaling pathway. Cell Physiol Biochem. 2018;46:1275–1285. doi: 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- 63.Wan WB, Kong QL. Knockdown of long noncoding RNA linc-ITGB1 inhibits tumor metastasis in colorectal cancer through suppressing BDNF. Eur Rev Med Pharmacol Sci. 2019;23:6453–6458. doi: 10.26355/eurrev_201908_18528. [DOI] [PubMed] [Google Scholar]
- 64.Zhao Y, Du T, Du L, Li P, Li J, Duan W, Wang Y, Wang C. Long noncoding RNA LINC02418 regulates MELK expression by acting as a ceRNA and may serve as a diagnostic marker for colorectal cancer. Cell Death Dis. 2019;10:568. doi: 10.1038/s41419-019-1804-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Si Y, Yang Z, Ge Q, Yu L, Yao M, Sun X, Ren Z, Ding C. Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell Mol Biol Lett. 2019;24:50. doi: 10.1186/s11658-019-0175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang W, Xie Y, Chen F, Liu X, Zhong LL, Wang HQ, Li QC. LncRNA MEG3 acts a biomarker and regulates cell functions by targeting ADAR1 in colorectal cancer. World J Gastroenterol. 2019;25:3972–3984. doi: 10.3748/wjg.v25.i29.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu J, Meng Q, Li X, Yang H, Xu J, Gao N, Sun H, Wu S, Familiari G, Relucenti M, Zhu H, Wu J, Chen R. Long non-coding RNA MIR17HG promotes colorectal cancer progression via miR-17-5p. Cancer Res. 2019;79:4882–4895. doi: 10.1158/0008-5472.CAN-18-3880. [DOI] [PubMed] [Google Scholar]
- 68.Chuo D, Liu F, Chen Y, Yin M. LncRNA MIR503HG is downregulated in Han Chinese with colorectal cancer and inhibits cell migration and invasion mediated by TGF-beta2. Gene. 2019;713:143960. doi: 10.1016/j.gene.2019.143960. [DOI] [PubMed] [Google Scholar]
- 69.Yang L, Qiu M, Xu Y, Wang J, Zheng Y, Li M, Xu L, Yin R. Upregulation of long non-coding RNA PRNCR1 in colorectal cancer promotes cell proliferation and cell cycle progression. Oncol Rep. 2016;35:318–24. doi: 10.3892/or.2015.4364. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, Yamamoto H, Doki Y, Mori M, Mimori K. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164–71. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang M, Miao F, Huang R, Liu W, Zhao Y, Jiao T, Lu Y, Wu F, Wang X, Wang H, Zhao H, Ju H, Miao S, Wang L, Song W. RHBDD1 promotes colorectal cancer metastasis through the Wnt signaling pathway and its downstream target ZEB1. J Exp Clin Cancer Res. 2018;37:22. doi: 10.1186/s13046-018-0687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao T, Maierdan SL, Lv C. ROR1-AS1 promotes tumorigenesis of colorectal cancer via targeting Wnt/beta-catenin. Eur Rev Med Pharmacol Sci. 2019;23(Suppl):217–223. doi: 10.26355/eurrev_201908_18650. [DOI] [PubMed] [Google Scholar]
- 73.Shi J, Zhong X, Song Y, Wu Z, Gao P, Zhao J, Sun J, Wang J, Liu J, Wang Z. Long non-coding RNA RUNX1-IT1 plays a tumour-suppressive role in colorectal cancer by inhibiting cell proliferation and migration. Cell Biochem Funct. 2019;37:11–20. doi: 10.1002/cbf.3368. [DOI] [PubMed] [Google Scholar]
- 74.Wang X, Lai Q, He J, Li Q, Ding J, Lan Z, Gu C, Yan Q, Fang Y, Zhao X, Liu S. LncRNA SNHG6 promotes proliferation, invasion and migration in colorectal cancer cells by activating TGF-beta/Smad signaling pathway via targeting UPF1 and inducing EMT via regulation of ZEB1. Int J Med Sci. 2019;16:51–59. doi: 10.7150/ijms.27359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matboli M, Shafei AE, Ali MA, El-Din Ahmed TS, Naser M, Abdel-Rahman T, Anber N, Ali M. Role of extracellular LncRNA-SNHG14/miRNA-3940-5p/NAP12 mRNA in colorectal cancer. Arch Physiol Biochem. 2019;9:1–7. doi: 10.1080/13813455.2019.1650070. [DOI] [PubMed] [Google Scholar]
- 76.Ma Z, Peng P, Zhou J, Hui B, Ji H, Wang J, Wang K. Long non-coding RNA SH3PXD2A-AS1 promotes cell progression partly through epigenetic silencing P57 and KLF2 in colorectal cancer. Cell Physiol Biochem. 2018;46:2197–2214. doi: 10.1159/000489589. [DOI] [PubMed] [Google Scholar]
- 77.Cai Y, Yan P, Zhang G, Yang W, Wang H, Cheng X. Long non-coding RNA TP73-AS1 sponges miR-194 to promote colorectal cancer cell proliferation, migration and invasion via up-regulating TGFalpha. Cancer Biomark. 2018;23:145–156. doi: 10.3233/CBM-181503. [DOI] [PubMed] [Google Scholar]
- 78.Sun J, Hu J, Wang G, Yang Z, Zhao C, Zhang X, Wang J. LncRNA TUG1 promoted KIAA1199 expression via miR-600 to accelerate cell metastasis and epithelial-mesenchymal transition in colorectal cancer. J Exp Clin Cancer Res. 2018;37:106. doi: 10.1186/s13046-018-0771-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Zhou F, Shen F, Zheng Z, Ruan J. The LncRNA XIRP2-AS1 predicts favorable prognosis in colon cancer. Onco Targets Ther. 2019;12:5767–5778. doi: 10.2147/OTT.S215419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kozovska Z, Gabrisova V, Kucerova L. Colon cancer: cancer stem cells markers, drug resistance and treatment. Biomed Pharmacother. 2014;68:911–6. doi: 10.1016/j.biopha.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 81.Kim NK, Kim YW, Han YD, Cho MS, Hur H, Min BS, Lee KY. Complete mesocolic excision and central vascular ligation for colon cancer: Principle, anatomy, surgical technique, and outcomes. Surg Oncol. 2016;25:252–62. doi: 10.1016/j.suronc.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 82.Klaver CEL, Kappen TM, Borstlap WAA, Bemelman WA, Tanis PJ. Laparoscopic surgery for T4 colon cancer: a systematic review and meta-analysis. Surg Endosc. 2017;31:4902–4912. doi: 10.1007/s00464-017-5544-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Justiniano CF, Becerra AZ, Xu Z, Aquina CT, Boodry CI, Schymura MJ, Boscoe FP, Noyes K, Temple LK, Fleming FJ. A population-based study of 90-day hospital cost and utilization associated with robotic surgery in colon and rectal cancer. J Surg Res. 2019;245:136–144. doi: 10.1016/j.jss.2019.07.052. [DOI] [PubMed] [Google Scholar]
- 84.Sekiya S, Imamura K, Takeuchi S, Teramura K, Watanabe Y, Tamoto E, Takada M, Kinoshita Y, Anbo Y, Nakamura F, Kashimura N, Noguchi H, Miura K, Hirano S. Pathological complete response of locally advanced colon cancer after preoperative radiotherapy: a case report and narrative review of the literature. Surg Case Rep. 2018;4:58. doi: 10.1186/s40792-018-0466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou J, Guo Z, Yu W, Li S, Qiao W. Clinical evaluation of preoperative radiotherapy combined with FOLFOX chemotherapy on patients with locally advanced colon cancer. Am Surg. 2019;85:313–320. [PubMed] [Google Scholar]
- 86.Kannarkatt J, Joseph J, Kurniali PC, Al-Janadi A, Hrinczenko B. Adjuvant chemotherapy for stage II colon cancer: a clinical dilemma. J Oncol Pract. 2017;13:233–241. doi: 10.1200/JOP.2016.017210. [DOI] [PubMed] [Google Scholar]
- 87.Meyers BM, Cosby R, Quereshy F, Jonker D. Adjuvant chemotherapy for stage II and III colon cancer following complete resection: a cancer care ontario systematic review. Clin Oncol (R Coll Radiol) 2017;29:459–465. doi: 10.1016/j.clon.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 88.Cappell MS. Pathophysiology, clinical presentation, and management of colon cancer. Gastroenterol Clin North Am. 2008;37:1–24. doi: 10.1016/j.gtc.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 89.Karim S, Booth CM, Brennan K, Peng Y, Siemens DR, Krzyzanowska MK, Mackillop WJ. Estimating the optimal rate of adjuvant chemotherapy utilization for stage III colon cancer. Cancer Med. 2019;8:5590–5599. doi: 10.1002/cam4.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karthika C, Sureshkumar R. Can curcumin along with chemotherapeutic drug and lipid provide an effective treatment of metastatic colon cancer and alter multidrug resistance? Med Hypotheses. 2019;132:109325. doi: 10.1016/j.mehy.2019.109325. [DOI] [PubMed] [Google Scholar]
- 91.Tomášek J, Kiss I. Immunotherapy of colorectal and anal carcinoma. Klin Onkol. 2017;30:62–65. doi: 10.14735/amko20173S62. [DOI] [PubMed] [Google Scholar]
- 92.Toh JWT, de Souza P, Lim SH, Singh P, Chua W, Ng W, Spring KJ. The potential value of immunotherapy in colorectal cancers: review of the evidence for programmed death-1 inhibitor therapy. Clin Colorectal Cancer. 2016;15:285–291. doi: 10.1016/j.clcc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 93.Bever KM, Le DT. An expanding role for immunotherapy in colorectal cancer. J Natl Compr Canc Netw. 2017;15:401–410. doi: 10.6004/jnccn.2017.0037. [DOI] [PubMed] [Google Scholar]
- 94.Jiang S, Good D, Wei MQ. Vaccinations for colorectal cancer: progress, strategies, and novel adjuvants. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parizadeh SM, Jafarzadeh-Esfehani R, Ghandehari M, Rezaei-Kalat A, Parizadeh SMR, Javanbakht A, Hassanian SM, Ferns GA, Khazaei M, Avan A. Personalized peptide-based vaccination for treatment of colorectal cancer: rational and progress. Curr Drug Targets. 2019;20:1486–1495. doi: 10.2174/1389450120666190619121658. [DOI] [PubMed] [Google Scholar]
- 96.Takimoto R, Kamigaki T, Okada S, Matsuda E, Ibe H, Oguma E, Naitoh K, Makita K, Goto S. Prognostic factors for colorectal cancer patients treated with combination of immune-cell therapy and first-line chemotherapy: a retrospective study. Anticancer Res. 2019;39:4525–4532. doi: 10.21873/anticanres.13629. [DOI] [PubMed] [Google Scholar]
- 97.Yun JW, Lee S, Kim HM, Chun S, Engleman EG, Kim HC, Kang ES. A novel type of blood biomarker: distinct changes of cytokine-induced STAT phosphorylation in blood T cells between colorectal cancer patients and healthy individuals. Cancers (Basel) 2019;11 doi: 10.3390/cancers11081157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xie Y, Huang L, Chen L, Lin X, Chen L, Zheng Q. Effect of dendritic cell-cytokine-induced killer cells in patients with advanced colorectal cancer combined with first-line treatment. World J Surg Oncol. 2017;15:209. doi: 10.1186/s12957-017-1278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Passardi A, Canale M, Valgiusti M, Ulivi P. Immune checkpoints as a target for colorectal cancer treatment. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18061324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koi M, Carethers JM. The colorectal cancer immune microenvironment and approach to immunotherapies. Future Oncol. 2017;13:1633–1647. doi: 10.2217/fon-2017-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yaghoubi N, Soltani A, Ghazvini K, Hassanian SM, Hashemy SI. PD-1/ PD-L1 blockade as a novel treatment for colorectal cancer. Biomed Pharmacother. 2019;110:312–318. doi: 10.1016/j.biopha.2018.11.105. [DOI] [PubMed] [Google Scholar]
- 102.Ciardiello D, Vitiello PP, Cardone C, Martini G, Troiani T, Martinelli E, Ciardiello F. Immunotherapy of colorectal cancer: challenges for therapeutic efficacy. Cancer Treat Rev. 2019;76:22–32. doi: 10.1016/j.ctrv.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 103.Bignucolo A, De Mattia E, Cecchin E, Roncato R, Toffoli G. Pharmacogenomics of targeted agents for personalization of colorectal cancer treatment. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18071522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Geng F, Wang Z, Yin H, Yu J, Cao B. Molecular targeted drugs and treatment of colorectal cancer: recent progress and future perspectives. Cancer Biother Radiopharm. 2017;32:149–160. doi: 10.1089/cbr.2017.2210. [DOI] [PubMed] [Google Scholar]
- 105.Banerjee A, Pathak S, Subramanium VD, G D, Murugesan R, Verma RS. Strategies for targeted drug delivery in treatment of colon cancer: current trends and future perspectives. Drug Discov Today. 2017;22:1224–1232. doi: 10.1016/j.drudis.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 106.Hodgkinson N, Kruger CA, Abrahamse H. Targeted photodynamic therapy as potential treatment modality for the eradication of colon cancer and colon cancer stem cells. Tumour Biol. 2017;39:1010428317734691. doi: 10.1177/1010428317734691. [DOI] [PubMed] [Google Scholar]
- 107.Miyamoto Y, Suyama K, Baba H. Recent advances in targeting the EGFR signaling pathway for the treatment of metastatic colorectal cancer. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sehnalová I, Říhová B, Němeček R, Kintrová K, Demlová R. The pharmacoeconomic analysis of cetuximab and panitumumab in the 1st line treatment of mCRC in real clinical practice in the Czech Republic. Klin Onkol. 2019;32:288–293. doi: 10.14735/amko2019288. [DOI] [PubMed] [Google Scholar]
- 109.Kim S, Kim N, Kang K, Kim W, Won J, Cho J. Whole transcriptome analysis identifies TNS4 as a key effector of cetuximab and a regulator of the oncogenic activity of KRAS mutant colorectal cancer cell lines. Cells. 2019;8 doi: 10.3390/cells8080878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, Šmakal M, Canon JL, Rother M, Williams R, Rong A, Wiezorek J, Sidhu R, Patterson SD. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 111.Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, Cabiddu M, Iacovelli R, Bossi I, Lonati V, Ghilardi M, de Braud F, Barni S. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer. 2015;51:587–94. doi: 10.1016/j.ejca.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 112.Terme M, Pernot S, Marcheteau E, Sandoval F, Benhamouda N, Colussi O, Dubreuil O, Carpentier AF, Tartour E, Taieb J. VEGFA-VEGFR pathway blockade inhibits tumor-induced regulatory T-cell proliferation in colorectal cancer. Cancer Res. 2013;73:539–49. doi: 10.1158/0008-5472.CAN-12-2325. [DOI] [PubMed] [Google Scholar]
- 113.Rosen LS, Jacobs IA, Burkes RL. Bevacizumab in colorectal cancer: current role in treatment and the potential of biosimilars. Target Oncol. 2017;12:599–610. doi: 10.1007/s11523-017-0518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qu CY, Zheng Y, Zhou M, Zhang Y, Shen F, Cao J, Xu LM. Value of bevacizumab in treatment of colorectal cancer: a meta-analysis. World J Gastroenterol. 2015;21:5072–80. doi: 10.3748/wjg.v21.i16.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liao X, Li H, Liu Z, Liao S, Li Q, Liang C, Huang Y, Xie M, Wei J, Li Y. Clinical efficacy and safety of apatinib in patients with advanced colorectal cancer as the late-line treatment. Medicine (Baltimore) 2018;97:e13635. doi: 10.1097/MD.0000000000013635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hu Y, Zhou P, Lin Y, Yang D, Wang B. Anti-colorectal cancer effect via application of polyethylene glycol modified liposomal Apatinib. J Biomed Nanotechnol. 2019;15:1256–1266. doi: 10.1166/jbn.2019.2770. [DOI] [PubMed] [Google Scholar]
- 117.Prados J, Melguizo C, Ortiz R, Perazzoli G, Cabeza L, Alvarez PJ, Rodriguez-Serrano F, Aranega A. Colon cancer therapy: recent developments in nanomedicine to improve the efficacy of conventional chemotherapeutic drugs. Anticancer Agents Med Chem. 2013;13:1204–16. doi: 10.2174/18715206113139990325. [DOI] [PubMed] [Google Scholar]
- 118.Yang Y, Du Y, Liu X, Cho WC. Involvement of non-coding RNAs in the signaling pathways of colorectal cancer. Adv Exp Med Biol. 2016;937:19–51. doi: 10.1007/978-3-319-42059-2_2. [DOI] [PubMed] [Google Scholar]
- 119.Liao C, Huang X, Gong Y, Lin Q. Discovery of core genes in colorectal cancer by weighted gene co-expression network analysis. Oncol Lett. 2019;18:3137–3149. doi: 10.3892/ol.2019.10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li H, Ma SQ, Huang J, Chen XP, Zhou HH. Roles of long noncoding RNAs in colorectal cancer metastasis. Oncotarget. 2017;8:39859–39876. doi: 10.18632/oncotarget.16339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong H, Zhou JY, Yang SM. Long non-coding RNAs in colorectal cancer. Oncotarget. 2016;7:5226–39. doi: 10.18632/oncotarget.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Luo J, Qu J, Wu DK, Lu ZL, Sun YS, Qu Q. Long non-coding RNAs: a rising biotarget in colorectal cancer. Oncotarget. 2017;8:22187–22202. doi: 10.18632/oncotarget.14728. [DOI] [PMC free article] [PubMed] [Google Scholar]


