Abstract
Zinc finger E-box binding homeobox 1 (ZEB1), as a typical transcription inhibitory factor of E-cadherin, plays a major role in stimulating the invasion and metastasis of tumors via modulating the epithelial-mesenchymal transition (EMT) signal. However, its function and modulatory mechanisms in endometrial carcinoma (EC) remain unclear. In this study, silencing ZEB1 significantly reduced EC cell migration, invasion, and metastasis, as well as enhanced the sensitivity of EC cells to cisplatin (cDDP) in vitro and in vivo. Mechanism analysis indicated that ZEB1 interacts with hepatoma-derived growth factor (HDGF) and co-localizes in the nucleus. In addition, ZEB1 as a transcription factor binds to the promoter of HDGF to stimulate HDGF transcription. Furthermore, suppression of HDGF in ZEB1-overexpressed EC cells not only reduced the expression of β-catenin, TCF4, and ZEB1, but also repressed β-catenin translocation from the cytoplasm into the nucleus and further downregulated the combination with TCF4. Interestingly, the β-catenin/TCF4 signaling feedback stimulates ZEB1 transcription and therefore constitutes a positive feedback loop. In clinical samples, ZEB1 positively correlates with HDGF expression, and co-expression of ZEB1 and HDGF promotes the pathogenesis of EC. In summary, our study demonstrated that the positive feedback loop of ZEB1/HDGF/β-catenin/TCF4 plays an unfavorable role in the metastasis of endometrial carcinoma.
Keywords: ZEB1, HDGF, endometrial carcinoma, invasion, metastasis
Introduction
Endometrial carcinoma (EC) is one of the three major gynecological malignancies, and with the aging of the population and the increase in obesity, the current incidence and mortality of this disease is rapidly increasing [1,2]. Exposure to endogenous and exogenous estrogens is the main risk factor for the development of EC [3-6]. Other risk factors include obesity, diabetes, early age at menarche, nulliparity, late-onset menopause, older age (above 55 years), and use of tamoxifen [5,7]. The main treatment is total hysterectomy and bilateral salpingo-oophorectomy by minimally invasive approaches (endoscopic). The prognosis is good for those who are diagnosed with early stage EC, but for those patients with recurrent or metastatic disease, the median overall survival (OS) remains short, with 5-year overall survival rates as low as 16% [8,9]. Timeless efforts to unravel the pathogenesis of recurrent EC has been made, but the progress in targeted or personalized therapy is limited [8,10]. Therefore, elucidating the molecular mechanism of invasion and metastasis of endometrial carcinoma can reveal new targets for its treatment.
As a member of the zinc finger E-box-binding protein (ZEB) family, zinc finger E-box binding homeobox 1 (ZEB1) is an important nuclear transcription factor of E-cadherin that is associated with the cellular epithelial-mesenchymal transition (EMT) [11-13]. Mechanistically, the expression of E-cadherin is repressed by ZEB1 via binding two of the E-box sequences in its promoter region [14,15]. Studies have clarified that ZEB1 induces EMT and promotes tumor invasion in epithelial cells [16,17]. In addition, non-coding RNAs of the miR-200 family and miR-205 reduce the EMT of the epithelial phenotype in cancer cells by repressing the expression of ZEB1 and SIP1 [18,19]. In turn, as a transcriptional repressor, ZEB1 represses the transcription of miR-200 family members, therefore establishing an EMT regulatory loop [20]. All of the studies mentioned above indicate that ZEB1 promotes EMT in tumors.
Previous studies have proven that ZEB1 is aberrantly expressed in aggressive uterine cancers and plays a role in promoting EMT in epithelial cells [21,22]. Moreover, researchers found that ZEB1 was expressed at higher levels in type I endometrial cancers compared to hyperplastic or normal endometrium, and it may be a biomarker of aggressive endometrial cancers at high risk of recurrence and lymph node metastases [23,24]. The studies above show that ZEB1 is closely related to the metastasis of endometrial cancer, but the detailed mechanism of ZEB1 in endometrial carcinoma has not been thoroughly studied. Here, we found a novel molecular regulation mechanism by which ZEB1 promotes endometrial carcinoma metastasis via motivating hepatoma-derived growth factor (HDGF)/β-catenin/TCF4 signal and regulating sensitivity to cisplatin (DDP).
Materials and methods
Cell culture and sample collection
Three EC cell lines Ishikawa, RL-95-2 and HEC-1B were purchased from the Chinese Academy of Sciences (Shanghai, China) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone, Logan, UT) supplemented with 10% fetal calf serum (ExCell, Shanghai, China). All cell lines were maintained at 37°C in a humidified atmosphere of 5% CO2.
30 fresh NE tissues, 22 fresh primary EC tissues were obtained from the Third Affiliated Hospital of Southern Medical University. The tissues included 21 normal formalin-fixed and 98 paraffin-embedded samples of endometrial carcinoma (EC) were obtained in the Third Affiliated Hospital of Guangzhou Medical School, Guangzhou City, China from 2002 to 2008. Prior consent from the patients, and the approval of the Ethics Committee of this hospital was obtained for research purposes. All samples were pathological diagnosed and were staged according to 2009 FIGO.
Lentivirus infection
Lentiviral particles encoding the shRNA targeting HDGF, ZEB1 and the control sequences (shPLVctr) were constructed (Supplementary Table 1). Ov-LV-ZEB1 and negative control sequences (ov-NC) were constructed in the same manner, and all vectors were then amplified and transfected into EC cells using Polybrene reagent (Sigma-Aldrich, St. Louis, MO, USA).
Cell transfection
SiRNAs targeting the HDGF, ZEB1 and the control sequences (si-NC) were designed and synthesized by RiboBio Inc. (Guangzhou, China). All siRNA sequences are listed in Supplementary Table 2. HDGF, ZEB1, β-catenin, TCF4 plasmids were purchased in Vigene Biosciences. Before transfection, EC cells were plated onto specified well plate (Nest, Biotech, China) at 30-50% confluence. SiRNA and plasmid were then transfected using Lipofectamine TM 2000 (Invitrogen Biotechnology) according to the manufacturer’s protocol. Cells were harvested after 48-72 h for further experiments.
RNA isolation, reverse transcription, and QRT-PCR
RNA was extracted from the fresh NE tissues, and fresh primary EC tissues, and EC cells using Trizol (Takara Bio, Inc, Shiga, Japan), and the RNA transcribed into cDNA and amplified with specific sense primers. ARF genes were used as internal controls. The assays were performed in accordance with manufacturer’s instructions (Takara Bio, Inc., Shiga, Japan). PCR reactions for each gene were repeated three times. Specific sense primers for HDGF, ZEB1 and ARF are shown in Supplementary Table 3.
Western blotting
Whole cell or tissue extracts were prepared in lysis buffer on ice for 30 min and the protein concentration was measured using BCA protein assay. After determining the concentration, 30 μg of protein were separated on SDS-PAGE and transferred onto poly vinylidene fluoride membranes. Then the membranes were blocked with 5% BSA and incubated with primary antibodies overnight at 4°C. Next day, the membrane was incubated with secondary antibodies and visualized using an enhanced chemiluminescence reagent (Thermo Scientific, Waltham, MA, USA). Dilutions and sources of antibodies are shown in Supplementary Table 4. Original blotting images were shown in Supplementary Figure 3.
Cell migration and invasion assays
Transwell apparatus (Corning, NY, USA) were utilized to the migration and invasion assays. With 100 ml serum-free medium containing 1 × 105 exponentially growing cells were seeded into the top chamber, and 500 μl 10% FBS culture medium was added to the lower chamber. After the cells were incubated for 10~12 h for invasion assay, the migrated cells were stained with Giemsa solution, and photographed under the microscope. All assays were repeated at least three times independently.
Wound healing assay
Cells were seeded in a 6-well plate and grown to confluence. Cells were washed three times with the PBS and the bottom of well was scratched with a 10 μl pipette tip. Images were captured by microscopy, the gap distance was measured, and the average was calculated for each plate.
In vivo tumorigenesis and metastasis assays
Female BALB/c nude mice at 4-weeks old were purchased from Nanjing University, Jiangsu, China. A total of 4 × 106 cells/mouse were intravenously injected into the mice and they were sacrificed 21 days after the delivery of tumor cells. Hematoxylin-Eosin (HE) staining was used to detect metastatic lung by using a dissecting microscope and the number of lung metastasis in mice were calculated. All animal studies were conducted in accordance with the principles and procedures outlined in the Southern Medical University Guide for the Care and Use of Animals under assurance number SCXK (Guangdong) 2008-0002.
Immunohistochemistry
Paraffin sections prepared from in vivo experiments were used for immunohistochemistry assays to detect protein expression levels of HDGF, ZEB1, Ki67, PCNA proteins. Immunohistochemistry was carried out according as described [25] and the antibodies used were anti-ZEB1, HDGF, PCNA, Ki67 (Supplementary Table 4).
Immunofluorescence
EC cells were seeded on coverslips and cultured overnight rinsed with PBS and fixed with cold 4% paraformaldehyde for 5 min at RT. Subsequently, cells were permeabilized in PBS solution and 0.2% Triton X-100 at room temperature for 30 min and incubated with primary antibodies HDGF, TCF4, ZEB1, β-catenin, E-cadherin, Vimentin (Supplementary Table 4) in PBS for 1 hour at RT. Then washing with PBS, then the coverslips were incubated for 1 h in the dark room with Alexa Fluor 488 goat anti-rabbit and 594 goat anti-mouse (1:500, Bioworld Technology, Inc). Finally, the coverslips were then mounted onto slides with mounting solution containing 0.2 mg/ml DAPI and sealed with nail polish. Slides were stored at 4°C in a dark box and Confocal laser scanning microscopy images were captured under a Zeiss LSM 880 confocal microscope.
CoIP assay
EC cells cultured in six-well plates were lysed with 600 μl Pierce IP Lysis buffer (Thermo Scientific) containing protease and phosphatase inhibitor cocktails (Thermo Scientific). Co-immunoprecipitation (co-IP) experiments was performed by using the Thermo Scientific Pierce co-IP kit following the manufacturer’s protocol. The specific anti-HDGF, TCF4, ZEB1, β-catenin, Flag, His or normal rabbit IgG (Supplementary Table 4) antibodies were first immobilized for 2 h using AminoLink Plus coupling resin respectively. The resin was then washed and incubated with cells lysate overnight. The resin was washed again and protein eluted using elution buffer after incubation. Subsequently western blot analyses were performed as described previously.
ChIP assay
According to the manufacturer’s instructions, ChIP assay was performed to examine whether ZEB1 combined to HDGF promoter by a ChIP assay kit (Millipore, catalog: 17-371). Firstly, The EC cells transfected with ZEB1 were fixed with 1% formaldehyde to covalently crosslink proteins to DNA and then chromatin was harvested from the EC cells. Crosslinked DNA was sheared to 200~1,000 base pairs in length with sonication and then subjected to an immmmunoselection process, which required the use of anti-ZEB1. Finally, real time-PCR was done using specific primers of HDGF predicted by JASPAR database (Supplementary Table 3).
Luciferase reporter assay
EC cells (5 × 104) were cultured in 24-well plates for 24 h, and the cells were transfected with 200 ng TOP/FOP reporter luciferase plasmid (GeneChem Technologies, Shanghai, China), and 10 ng of pRL Renilla luciferase (as an internal control group) using Lipofectamine TM 2000. Cells were subjected to luciferase reporter assay 24 h after transfection, using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Statistical analysis
The SPSS 20.0 statistical software package (SPSS Inc. Chicago, IL, USA) were used for statistical analysis. The data are expressed as the mean ± sd. from at least three independent experiments. The statistical significance was compared by Student’s two-tailed t-test for two groups and one-way ANOVA for multiple groups. Spearman’s correlation coefficient was used for analyzing association between ZEB1 and HDGF. Survival analysis was performed using Kaplan-Meier survival curves. All statistical tests were two-sided and a P value of < 0.05 was considered statistically significant. *P < 0.05, **P < 0.01 and ***P < 0.001.
Results
Suppressing ZEB1 inhibits EC cell metastasis and chemoresistance
To assess its biological function, we infected EC cells with lentivirus expressing short hairpin RNA targeting ZEB1 and the negative control (shPLVctr) (Supplementary Figure 1A, 1B). With efficient knockdowns from shZEB1-2 and shZEB1-3 in the HEC-1B cell line, the same fragment in the Ishikawa cell line was separately detected by Western blotting or RT-PCR assays, compared to the shPLVctr group (Supplementary Figure 1C, 1D). Efficient cells were selected for subsequent studies.
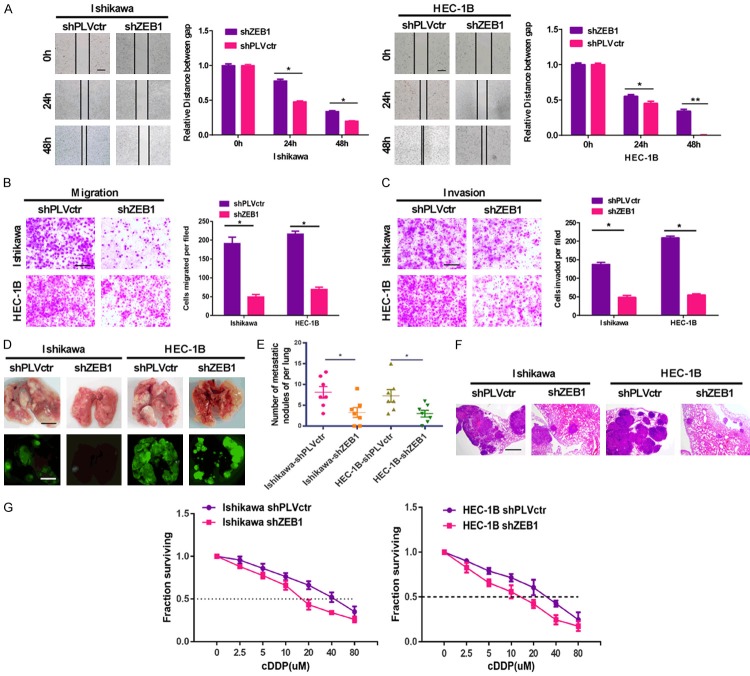
Scratch, transwell and Boyden assays were performed to separately test the ability of invasion and migration in shZEB1 and shPLVctr EC cells. In the scratch and transwell assay, ZEB1 knockdown reduced the migration ability of EC cells compared with the control group (Figure 1A, 1B). The invasiveness of shZEB1 EC cells was significantly decreased relative to negative control cells in the Boyden chamber assays (Figure 1C). Moreover, we injected stable transfected shZEB1 EC cells into nude mice via the tail vein and supervised the development of metastasis nodules in the lungs. There were less and smaller lung metastatic nodules in the shZEB1 group as compared to the control group, in which mice carried shPLVctr EC cells (Figure 1D). In addition, the number of tumor nodules in the shZEB1 group was lower than that of the control group (Figure 1E). Hematoxylin and eosin (HE) staining of dissected lung tissue confirmed the presence of nodules (Figure 1F).
Figure 1.
Suppressing ZEB1 inhibits EC cell metastasis and chemoresistance. A. Scratch migration assay indicated that transfection of shZEB1 into EC cells for 48 h resulted in an impaired migration capacity, being compared to the negative control group. Scale bar: 200 µm. B. Down-regulating ZEB1 stably reduced the migration ability of EC cells in vitro by transwell assay. Scale bar: 250 µm. C. Stably suppressed ZEB1 reduced in vitro invasiveness of EC cells by boyden assay. Scale bar: 250 µm. D. External fluorescence images of lungs of mice were obtained 2 months after tail vein respectively. E. The number of lung metastatic nodules in each group. Scale bar: 5 mm. F. H&E staining of lung metastatic nodules from different experimental groups. Scale bar: 1 mm. G. Dose-response curves of Ishikawa and HEC-1B treated by shZEB1 and PLVctr respectively following 48 h treatment with DDP. The data are expressed as mean ± sd. of three independent experiments. *P < 0.05; **P < 0.01.
EC cells with stable silenced ZEB1 exhibited significantly increased sensitivity to cisplatin (DDP). EC cells were treated with different concentrations of DDP after 48 h, and the cell growth inhibition rates were calculated after ZEB1 silencing. The IC50 of DDP was 41.3 μM in the parental Ishikawa cells but decreased to 21.01 μM in ZEB1-silenced Ishikawa cells (P < 0.05), and a similar IC50 reduction from 27.69 μM to 13.06 μM occurred in HEC-1B cells (Figure 1G).
Suppressing ZEB1 blocks EMT
For further study how ZEB1 controls EC migration and invasion, we examined the expression of cell cycle and EMT markers after ZEB1 silencing in EC cells. A Western blot analysis showed that the epithelial marker E-cadherin was upregulated in stable knocked down ZEB1 EC cells. However, the mesenchyme markers N-cadherin, β-catenin, and vimentin were downregulated (Figure 2A). These results were confirmed by immunofluorescence (Figure 2B) analyses.
Figure 2.
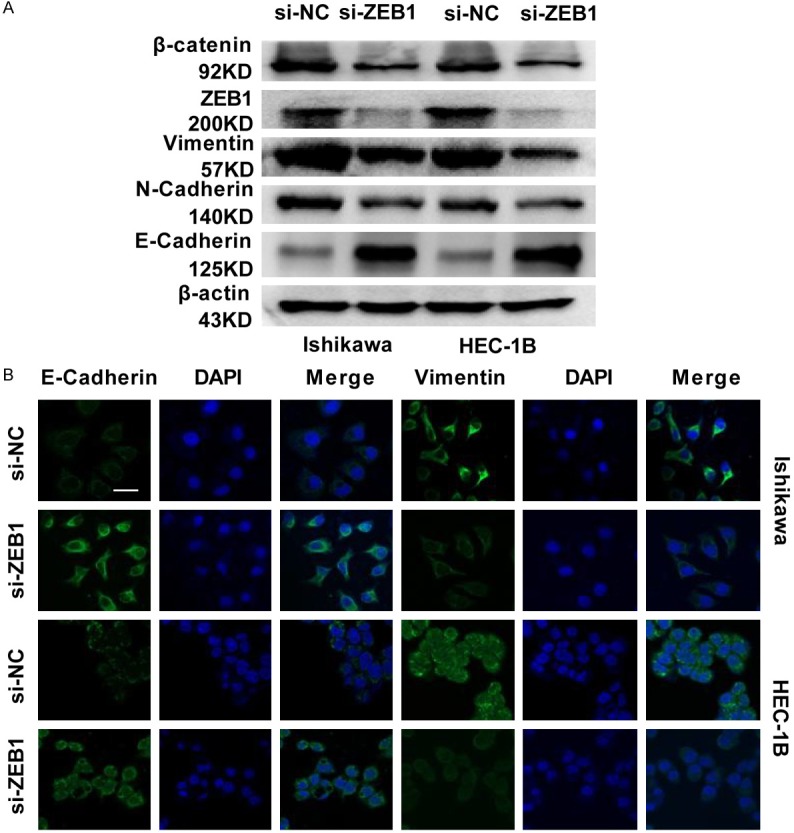
Suppressing ZEB1 reduces EMT signal. A. E-cadherin, N-cadherin, β-catenin and vimentin expression was evaluated by western blotting in ZEB1-silenced EC cells. B. Immunofluorescence analysis of E-cadherin and vimentin expression in ZEB1-silenced EC cells. Scale bar: 25 µm.
ZEB1 interacts with HDGF
To clarify how ZEB1 promotes EMT in EC cells, the DOMINE Database (domain-domain interactions) was used to search for proteins that interact with ZEB1, and it predicted a direct interaction between the ZEB1 and HDGF protein. Interestingly, the HDGF and ZEB1 interaction in EC cells was detected by exogenous and endogenous co-immunoprecipitation (co-IP) assay (Figure 3A, 3B). Immunofluorescence analyses showed that the ZEB1 and HDGF proteins co-localized in the nucleus (Figure 3C). In addition, overexpressed ZEB1 upregulated the mRNA and protein level of HDGF in EC cells. On the contrary, knocking down ZEB1 decreased the expression of HDGF on the mRNA and protein level (Figure 3D, 3E).
Figure 3.
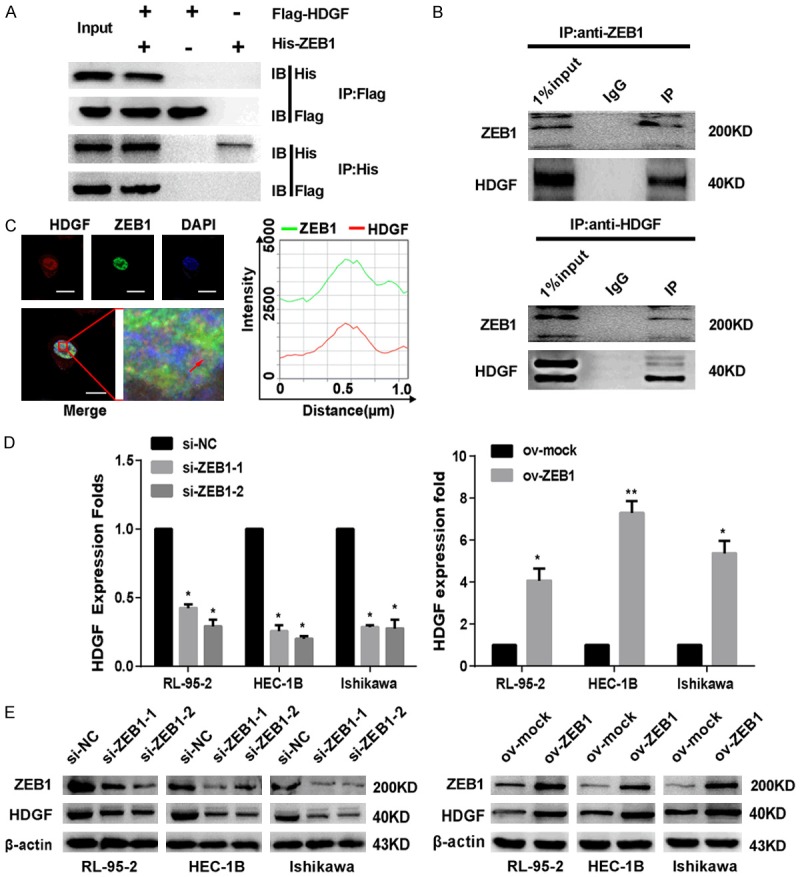
ZEB1 interacts with HDGF. A. Co-IP detected the interaction of exogenous ZEB1 and HDGF in EC cells. B. Reciprocal Co-IP of endogenous proteins showed a strong interaction between ZEB1 and HDGF. C. EC cells were immunostained with anti-HDGF (594; red), anti-ZEB1 (488; green) antibody and visualized. Scale bar: 20 µm. D. The transcriptional levels of the HDGF gene were shown after transfecting with ZEB1 plasmid or si-RNA (si-NC, si-ZEB1). ARF was used as a loading control. E. Western blotting showed HDGF protein expression levels after treated with ZEB1 plasmid or si-RNA (si-NC, si-ZEB1) in EC cells. The data are expressed as mean ± s.d. of three independent experiments. *P < 0.05; **P < 0.01.
ZEB1 promotes HDGF expression by binding to the promoter region of HDGF
Notably, ZEB1 is a classic transcription inhibitory factor. Given that ZEB1 and HDGF co-localized in the nucleus, we speculated that ZEB1 may be a transcription factor of HDGF. To verify this assumption, we used the JASPAR open access database as bioinformatics software to analyze a 2-kb region upstream of the transcription start site of HDGF. There were putative ZEB1-binding elements in three regions (-1233~-1225, -490~482 and -54~-46) of the human HDGF promoter region by sequence alignment analysis using the JASPAR database of transcription factor binding profiles (Figure 4A). Chromatin immunoprecipitation (ChIP) assay was used to confirm whether antibody-specific ZEB1 protein could bind to these three regions of the HDGF promoter in EC cells. Being compared to the negative control group, ZEB1 protein was recruited to three binding sites (ZEB1-A, ZEB1-B, and ZEB1-C) in the putative HDGF promoter in two EC cells lines, which was confirmed by ChIP, real-time PCR, and agarose gel electrophoresis (Figure 4B, 4C).
Figure 4.
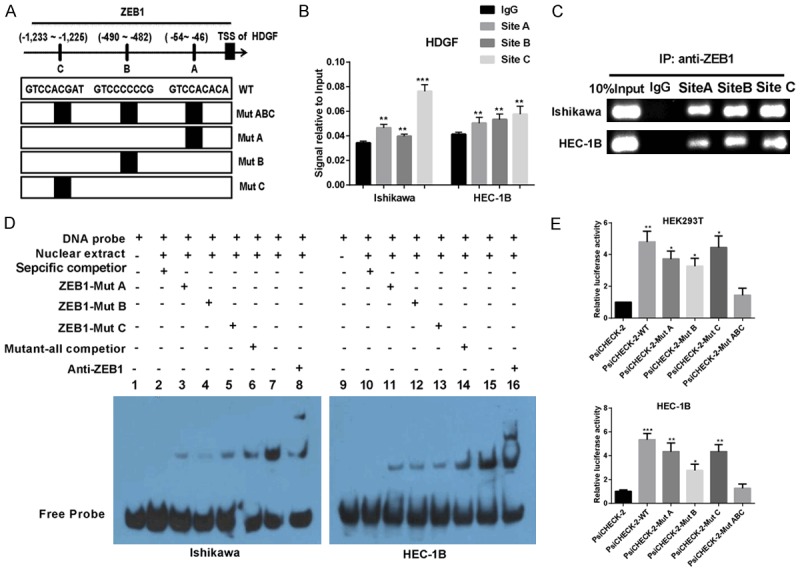
ZEB1 promotes HDGF expression by binding to the promoter region of HDGF. (A) Schematic diagram of the promoter regions of HDGF with the putative ZEB1 TFBSs (A-C), and the structure of the wild-type (WT) and binding sites mutant (MutA, MutB, MutC and MutABC). (B) PCR gel showed amplification of ZEB1-binding sites A and B after ChIP using antibody against ZEB1. (C) The gel figures were accompanied by the locations of molecular weight markers. (D) EMSA result was shown from nuclear proteins extracted from Ishikawa and HEC-1B cells after incubation with individual DIG-ddUTP-labeled oligonucleotide probes (lanes 2-8, 10-16). The free probe of labeled ZEB1 was run in lanes 1 and 9 as a control. A 100-fold excess of unlabelled ZEB1-WT was used to compete with ZEB1 binding (lanes 6 and 14, compared with lanes 2 and 10). A 100-fold excess of unlabelled mutated ZEB1-A, ZEB1-B and ZEB1-C was used to compete with binding of respective labeled probes (lanes 3-5 and lanes 11-13 compared with lanes 2 and 10). (E) Relative luciferase activity of the indicated promoter vectors in HEK293T, and HEC-1B cells transfected with ZEB1 plasmids. Mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
Moreover, an electrophoresis mobility shift assay (EMSA) experiment was used to detect if ZEB1 was bound to HDGF, as shown in Figure 4D. When EC cell nuclear extracts were incubated with biotin-labeled probe, a migrating complex appeared in lines 7 and 15, whereas there was no migration formation when the unlabelled wild-type probe of ZEB1 was added as binding competition (lanes 2 and lanes 10). When mutated A, B, or C was added to compete with biotin-labelled A, B, or C, respectively, in Ishikawa cells and HEC-1B cells (lanes 3-5, lanes 11-13), bands were not affected, and the bands unaffected in all the three sites were mutated groups (lanes 6 and lanes 14). The EMSA assay results above indicated that the three predicted ZEB1-binding sites in the promoter region of HDGF were functional.
For further examination of the result above, a dual-luciferase reporter assay was utilized to test the HDGF transcriptional activity. Compared to the wild-type of HDGF promoter, HDGF transcriptional activity significantly decreased in HEK293T and EC cells transfected with the mutant HDGF promoter luciferase reporter (Figure 4E).
HDGF mediates ZEB1 to promote EC migration and invasion by modulating β-catenin-induced EMT signal
To explore the precise molecular mechanisms and relevant interactions between HDGF and ZEB1, we examined the mRNA and protein levels of ZEB1 in si-HDGF or ov-HDGF EC cells, and the results showed that there was no significant difference in the mRNA levels of ZEB1 (Supplementary Figure 1E). Interestingly, HDGF overexpression elevated the protein level of ZEB1. On the contrary, HDGF knockdown decreased the expression of ZEB1 protein (Supplementary Figure 1F).
Given that ZEB1 is a well-known downstream target of the Wnt/β-catenin signaling pathway, which regulates EMT, we predicted that HDGF interacts with β-catenin to regulate the expression of ZEB1. In accordance with our predictions, HDGF interacted with TCF4 and β-catenin, as revealed by an endogenous co-immunoprecipitation (CoIP) assay and immunofluorescence analysis, respectively (Figure 5A-C). Subsequently, CoIP assay indicated that downregulated HDGF induced less β-catenin protein binding to TCF4. Additionally, less TCF4 interacted with β-catenin after silencing HDGF with siRNA (Figure 5D). Further study showed that the expression of nuclear and cytoplasmic β-catenin was dramatically decreased after silencing HDGF in EC cells, while overexpressed HDGF caused the opposite results (Figure 5E, 5F). Finally, a luciferase reporter assay was used to examine whether the transcriptional activity of β-catenin and TCF4 would be affected by interacting with HDGF. Silencing HDGF had the opposite effect (Figure 5G).
Figure 5.
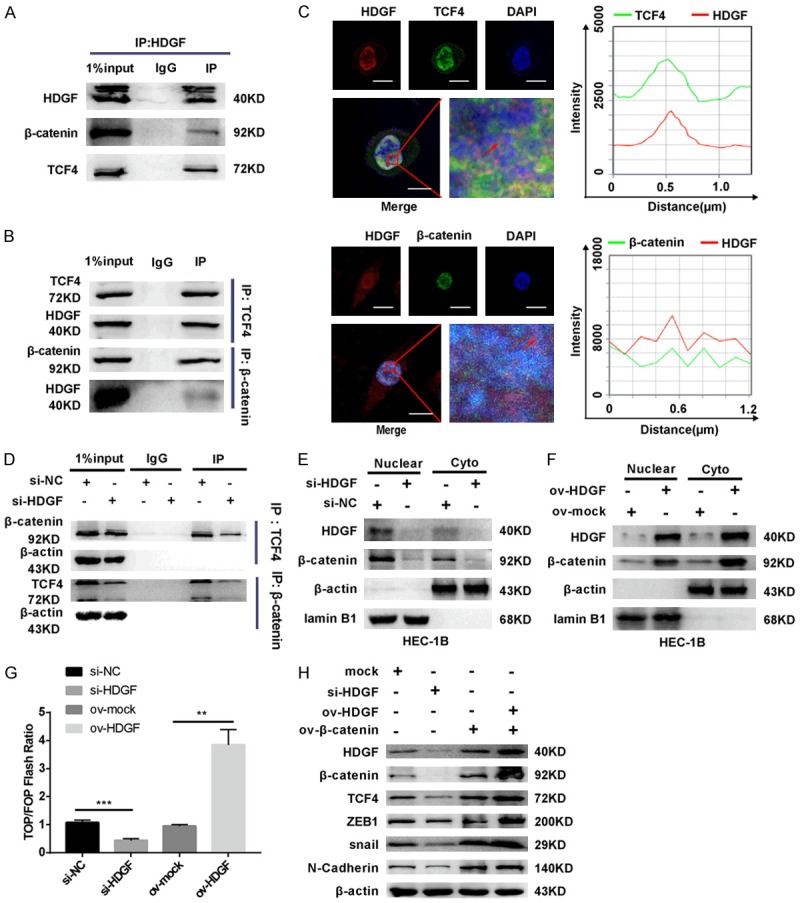
HDGF mediates ZEB1 to promote EC migration and invasion by modulating β-catenin-induced EMT signal. A and B. Strong interaction between HDGF and β-catenin, TCF4, showing by reciprocal CoIP through using antibody (anti-TCF4, anti-β-catenin) respectively. C. EC cells were immunostained with anti-HDGF, TCF4, β-catenin and visualized under fluorescence microscope. Scale bar: 20 µm. D. CoIP assay showed that less TCF4 bound to β-catenin after suppression of HDGF compared to control group. Similarly, decreased HDGF protein induced less β-catenin binding to TCF4 by CoIP assay. β-actin served as internal control. E and F. Suppress HDGF decreased nuclear translocation of β-catenin, while HDGF over-expression had the opposite effect. β-actin served as the cyto internal control. Lamin B1 was considered as nuclear loading control. G. HDGF over expression enhanced whereas HDGF silencing suppressed TOP flash reporter activity, respectively. H. Western blotting analysis for the expression levels of HDGF, β-catenin, TCF4, ZEB1, and EMT-associated proteins in EC cells with different treatments. Data were shown as the mean ± SD, **P < 0.01; ***P < 0.001.
To determine the regulating relationship between HDGF, ZEB1, and β-catenin, we used siRNA (si-ZEB1) and plasmids (ov-HDGF or ov-β-catenin) with Western blot analysis to detect the change in HDGF- and EMT-associated factors (Figure 5H). Loss of HDGF resulted in decreased levels of β-catenin, ZEB1, TCF4, Snail, and N-cadherin. Overexpressed β-catenin resulted in increased expression of the proteins mentioned above in EC cells. Furthermore, overexpressed β-catenin in HDGF-overexpressed EC cells elevated the expression of EMT-associated factors, of which there were many more than those in overexpressed β-catenin EC cells.
Previous studies had revealed that HDGF promoted cell growth, migration, and invasion in tumors [26-28]. To further illuminate the involvement of HDGF in ZEB1-mediated cell invasion and metastasis, si-RNA (si-HDGF and si-NC) were transfected into ZEB1-overexpressed EC cells. Functional analysis by scratch, transwell, and Boyden assays revealed that si-HDGF weakened migration and invasion of ZEB1-overexpressed EC cells (Supplementary Figure 2A-C).
β-catenin/TCF4 induces ZEB1 and HDGF expression in EC cells
To determine the regulating relationship between ZEB1, HDGF, β-catenin, and TCF4, we upregulated β-catenin and TCF4 in EC cells to detect the expression of HDGF and ZEB1 by Western blot analysis. The results showed that the expression of HDGF and ZEB1 was increased compared to the control group (Figure 6A).
Figure 6.

β-catenin/TCF4 induces ZEB1 and HDGF expression in EC cells. A. The protein levels of the ZEB1 and HDGF gene were examined after transfecting with β-catenin or TCF4 plasmids by Western blotting. B. The over-expression of β-catenin was effective by Immunofluorescent staining assay. Scale bar: 5 µm. C. The over-expression of TCF4 were effective by Immunofluorescent staining assay. Scale bar: 7.5 µm. D and E. Immunofluorescent staining assay show that ZEB1 and HDGF were elevated after up-regulated β-catenin or TCF4. Scale bar: 20 µm.
Correspondingly, immunofluorescence staining showed that the expression of ZEB1 and HDGF was elevated in EC cells after transfection with β-catenin or TCF4 plasmids compared with the control groups (Figure 6B-E). These results demonstrated that β-catenin and TCF4 induces the expressions of HDGF and ZEB1.
The expression of HDGF and ZEB1 and their relationship with the clinical pathological factors of endometrial carcinoma
Immunohistochemistry assay was used to examine ZEB1 and HDGF expression in 98 endometrial cancer tissues, compared with 21 normal endometrial tissues. Using serial sections of paraffin-embedded EC tissues we observed that tumor tissues with high expression level of ZEB1 have higher expressions level of HDGF compare with low expressions level ZEB1 tumor tissues. In addition, both of HDGF and ZEB1 are mainly expressed in nucleus (Figure 7A). Furthermore, patients with tumors exhibiting high levels of HDGF and distinguished levels of ZEB1 had shorter survival times (Figure 7B, 7C). More interestingly, patients with high HDGF and ZEB1 expression had the shortest survival time (Figure 7D). QPCR analysis indicated that the ZEB1 expression level was significantly decreased in 30 endometrial cancer tissues in comparison to 22 normal endometrial tissues (P < 0.05) (Figure 7E). Western blot analysis of fresh endometrial carcinoma samples and fresh normal endometrial samples showed that both HDGF and ZEB1 were highly expressed in endometrial cancer in comparison with normal endometrial tissue (Figure 7F).
Figure 7.
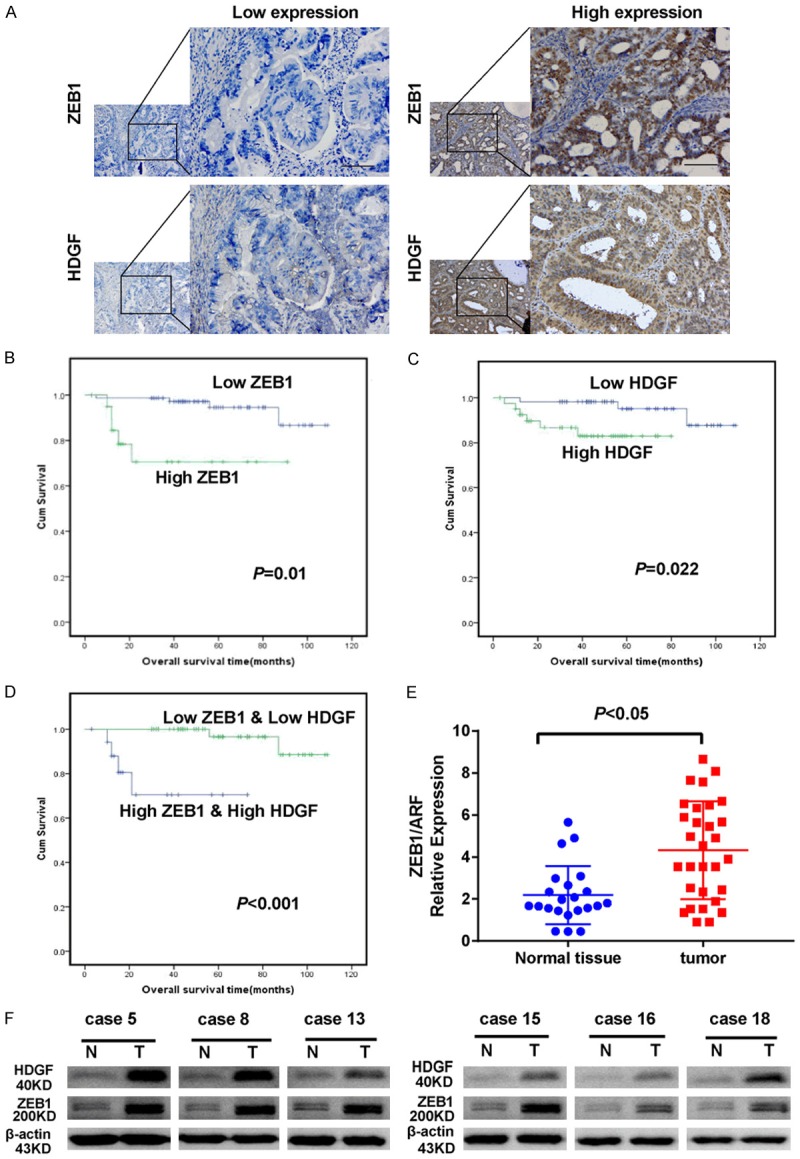
Expression levels of HDGF and ZEB1 genes in endometrial carcinoma and normal endometrial tissue. A. Expression of ZEB1 and HDGF in serial EC sample sections; Scale bar: 25 µm. B. Higher levels of ZEB1 reduced the survival time of EC patients. C. Higher levels of HDGF reduced the survival time of EC patients. D. Kapla-Meier analysis of overall survival in patients with variable expression of HDGF and ZEB1. E. Compared with normal endometrial tissues, ZEB1 expression was markedly increased in endometrial carcinoma (P < 0.05). F. Western blot analysis of ZEB1 and HDGF expression in 6 cases of endometrial carcinoma tissue samples compared with 6 normal endometrial tissue. β-actin was used as a loading control.
To investigate the relationship between the expression of ZEB1 or HDGF and their clinicopathological features, an immunostaining score was used to evaluate the expression of HDGF and ZEB1. As shown in Table 1, ZEB1 protein expression in EC was significantly correlated with FIGO stage (P < 0.001), depth of myometrial invasion (P = 0.015), and lymph node metastasis (P < 0.001). HDGF immunostaining was significantly associated with FIGO stage (P < 0.001), histological grading (P < 0.001), and lymph node metastasis (P < 0.001). However, it was not associated with patient’s age, menopausal status, histological grading, or depth of myometrial invasion. Furthermore, univariate analysis indicated that FIGO stage, histological grading, lymph node metastasis, depth of myometrial invasion, and expression of ZEB1 and HDGF were also significantly correlated with patients’ survival (P < 0.001, P < 0.001, P < 0.001, P = 0.009, P = 0.003, and P = 0.041, respectively). Multivariate analysis showed that ZEB1 expression was not an independent prognostic factor for EC patients (Table 2). Correlation analysis showed that there was a positive correlation between HDGF and ZEB1 staining in endometrial carcinoma tissues (r = 0.465, P < 0.001) (Table 3).
Table 1.
Correlation between the ZEB1 and HDGF protein expression with the clinicopathologic Characteristics in EC
| Characteristics | ZEB1 (%) | HDGF (%) | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| N | Negative | Positive | P | N | Negative | Positive | P | |
| Age | ||||||||
| < 50 | 29 | 22 (75.86) | 7 (24.14) | 0.672 | 29 | 16 (55.17) | 13 (44.83) | 0.697 |
| ≥ 50 | 69 | 55 (79.71) | 14 (20.29) | 69 | 41 (59.42) | 28 (40.58) | ||
| Menopausal status | ||||||||
| Premenopausal | 49 | 39 (79.59) | 10 (20.41) | 0.806 | 49 | 31 (63.27) | 18 (36.73) | 0.306 |
| Postmenopausal | 49 | 38 (77.55) | 11 (22.45) | 49 | 26 (53.06) | 23 (46.94) | ||
| FIGO stage | ||||||||
| I-II | 79 | 73 (92.40) | 6 (7.60) | < 0.001 | 79 | 56 (70.89) | 23 (29.11) | < 0.001 |
| III | 19 | 4 (21.05) | 15 (78.95) | 19 | 1 (5.26) | 18 (94.74) | ||
| Histological grading | ||||||||
| G1+G2 | 85 | 69 (81.18) | 16 (18.82) | 0.108 | 85 | 55 (64.71) | 30 (35.29) | < 0.001 |
| G3 | 13 | 8 (61.54) | 5 (38.46) | 13 | 2 (15.38) | 11 (84.62) | ||
| Depth of myometrial invasion | ||||||||
| < 50% | 64 | 55 (85.94) | 9 (14.06) | 0.015 | 64 | 41 (64.06) | 23 (35.94) | 0.104 |
| ≥ 50% | 34 | 22 (64.71) | 12 (35.29) | 34 | 16 (47.06) | 18 (52.94) | ||
| Lymph node status | ||||||||
| Negative | 82 | 75 (91.46) | 7 (8.54) | < 0.001 | 82 | 56 (68.29) | 26 (31.71) | < 0.001 |
| Positive | 16 | 2 (12.5) | 14 (87.5) | 16 | 1 (6.25) | 15 (93.75) | ||
Table 2.
Summary of univariate and multivariate Cox regression analysis of overall survival duration
| Parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| P | HR | 95% CI | P | HR | 95% CI | |
| Age | ||||||
| < 50 versus ≥ 50 | 0.709 | 1.341 | 0.277-6.4 99 | |||
| Family history of tumor | ||||||
| Negative versus positive | 0.384 | 0.037 | 0.001-62.56 | |||
| Education | ||||||
| < Graduation versus ≥ graduation | 0.460 | 24.96 | 0.005-9059.13 | |||
| Health Insurance | ||||||
| No versus yes | 0.094 | 0.168 | 0.021-1.358 | |||
| Career | ||||||
| ≤ Worker versus > worker | 0.362 | 21.409 | 0.021-3710.32 | |||
| Menopausal status | ||||||
| Premenopausal versus postmenopausal | 0.686 | 0.762 | 0.204-2.850 | |||
| Complications | ||||||
| With versus without | 0.397 | 0.506 | 0.105-2.445 | |||
| FIGO stags | ||||||
| I-II versus III | < 0.001 | 27.139 | 5.125-143.712 | 0.721 | 0.006 | 0.001-37.56 |
| Histological grading | ||||||
| G1-G2 versus G3 | < 0.001 | 25.884 | 5.109-131.139 | 0.003 | 37.455 | 3.328-421.54 |
| Lymph node status | ||||||
| Negative versus positive | < 0.001 | 88.99 | 9.074-872.781 | 0.452 | 1.72 | 0.012-8.357 |
| Depth of myometrial invasion | ||||||
| < 50% versus ≥ 50% | 0.009 | 8.143 | 1.682-39.421 | 0.122 | 4.754 | 0.659-34.299 |
| Postoperative irradiation | ||||||
| Yes versus no | 0.468 | 2.175 | 0.266-17.779 | |||
| Postoperative chemotherapy | ||||||
| Yes versus no | 0.096 | 3.426 | 0.805-14.589 | |||
| ZEB1 expression | ||||||
| Negative versus positive | 0.003 | 7.458 | 1.951-28.509 | 0.231 | 0.200 | 0.014-2.78 |
| HDGF expression | ||||||
| Negative versus positive | 0.041 | 5.35 | 1.071-2.723 | 0.384 | 0.219 | 0.007-6.703 |
Table 3.
Correlation between ZEB1 and HDGF expressions in EC
| HDGF | r | p | |||
|---|---|---|---|---|---|
|
| |||||
| Negative | Positive | ||||
| ZEB1 | Negative | 54 | 23 | 0.465 | < 0.001 |
| Positive | 3 | 18 | |||
Discussion
In this study, we found that downregulated ZEB1 decreased the ability of EC cells to invade and metastasize in vivo and in vitro. Furthermore, reducing the expression of ZEB1 enhanced the chemotherapy sensitivity of EC cells to cDDP. It is well known that the EMT signaling is a key event for inducing tumor invasion, metastasis, and chemotherapy resistance [29-31]. We observed that N-cadherin, β-catenin, and vimentin were decreased and E-cadherin was elevated in shZEB1 EC cells, which supports the results of previous studies [32].
Previous research found that ZEB1 cooperates with CtBP to repress the expression of E-cadherin, which regulates the development of tumors [33]. However, other unknown proteins that interact with ZEB1 and promote tumor invasion and metastasis remain to be discovered. To better understand its molecular mechanisms, we searched the DOMINE database (a website that predicts protein domain interaction) and found that HDGF is a potential interactive protein of ZEB1. Furthermore, we used CoIP and confocal microscopy to confirm that ZEB1 combines to HDGF and they colocalize in the nucleus, which demonstrated that ZEB1 interacts with HDGF in EC.
In prior studies, ZEB1 as a transcription factor has been reported to bind to the promoter region of E-cadherin and miR-200 family members and repress their transcription [14,20,30,31]. In the current study, we predicted three ZEB1-binding regions in the HDGF promoter by using the JASPAR database (http://jaspar.genereg.net), which hinted that ZEB1 induced HDGF expression at the transcription level. We firstly observed that silencing ZEB1 decreased the expression of HDGF at the mRNA and protein levels in EC cells. Furthermore, ChIP, luciferase, and EMSA assays confirmed that ZEB1 binds the HDGF promoter and induces its promoter activity. These results demonstrated that ZEB1 as the transcription factor of HDGF promotes its expression in EC.
HDGF is an important oncogene that is related to proliferation, invasion, and metastasis in liver cancer, stomach cancer, prostate cancer, and non-small cell lung cancer [27,34-36]. Our previous study indicated that patients with EC had a poor prognosis when expression of HDGF was abnormal [37]. These data demonstrated the significance of HDGF in EC pathogenesis. Nonetheless, the details explaining how HDGF induces the invasion and metastasis of EC are still unclear.
In a previous study, HDGF had been shown to interact with β-catenin [38]. Interestingly, β-catenin interacts with TCF4 [39,40] and the latter further transcribed ZEB1 to stimulate EMT signal [39] in tumors. Thus, we assumed that HDGF might mediate ZEB1 to induce the invasion and metastasis of EC by stimulating the β-catenin/TCF4/ZEB1 pathway and its downstream EMT signal. In accordance with the speculation, we observed that silencing HDGF in overexpressed ZEB1 EC cells reversed the ZEB1-induced promotion of migration and invasion in EC cells. Mechanism analysis indicated that knocking down HDGF inactivated the expression of the β-catenin and its downstream EMT signal in ZEB1-overexpressed EC cells. Furthermore, knocking down β-catenin and TCF4 reduced ZEB1 and HDGF expression, respectively. These data demonstrate that HDGF not only mediates ZEB1, but it also promotes EC migration and invasion by modulating β-catenin/TCF4/ZEB1 and its downstream EMT signal. In a word, ZEB1, HDGF, β-catenin, and TCF4 form a positive feedback loop that participates in EC metastasis.
Consistent with their known roles in EC cells, we performed Western blotting and then observed elevated expression of ZEB1 and HDGF in EC tissues compared with normal endometrium tissues. High expression of ZEB1 was positively correlated with FIGO stage, depth of myometrial invasion, and lymph node status, revealing that ZEB1 promoted the progression of EC. Similarly, high expression of HDGF was positively correlated with FIGO stage, histological grading, and lymph node metastasis, suggesting that HDGF was involved in the progression of EC. Patients with high ZEB1 or HDGF expression had poorer overall survival by Kaplan-Meier analysis. Moreover, ZEB1 expression was positively correlated with HDGF expression in EC tissues. EC patients with high expression of both ZEB1 and HDGF showed the worst prognoses for survival compared with other groups. These data further illustrate that increased ZEB1 and HDGF expression are important factors that synergistically promote the development and spread of EC.
Here, a regulatory model is shown in Figure 8 as a summary. ZEB1 induces its expression by interacting with HDGF, which in turn regulates the expression of β-catenin and TCF4, and ultimately induces its own expression. The worst prognosis for endometrial cancer patients resulted from highly expressed ZEB1 as well as high expression of HDGF. Together, these results confirm that ZEB1, HDGF, TCF4, and β-catenin form a loop that promotes the occurrence and progression of endometrial cancer via promoting its invasion, metastasis, and chemotherapy resistance. In summary, these findings indicate that ZEB1 combined with HDGF targeting may be a useful therapeutic option for the treatment of EC.
Figure 8.
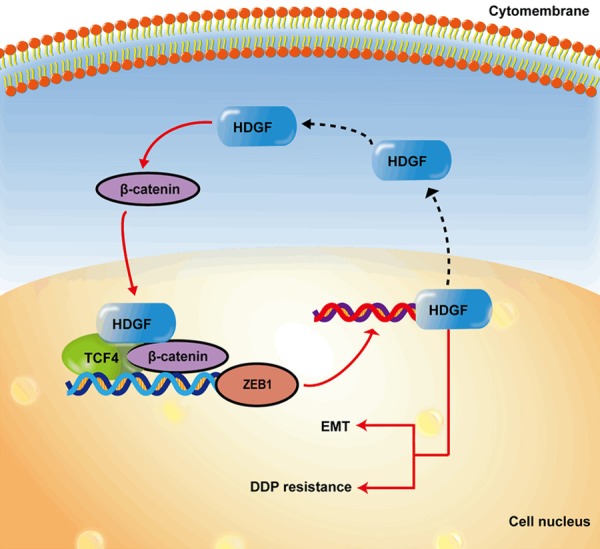
A schematic diagram for an atypical HDGF-β-catenin/TCF4-ZEB1 feedback loop.
Acknowledgements
This research was supported by the National Natural Science Foundation of China Medical Project in 2018-Clinical Medicine Research Special Fund (No: L2017084), Natural Science Fund of Guangdong Province (No: 2015A030313240), Medical research fund of Guangdong Province (No. A2015467) and Science and Technology Projects from Guangdong Province (2017ZC0100).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Felix AS, Brasky TM, Cohn DE, Mutch DG, Creasman WT, Thaker PH, Walker JL, Moore RG, Lele SB, Guntupalli SR, Downs LS, Nagel C, Boggess JF, Pearl ML, Ioffe OB, Deng W, Miller DS, Brinton LA. Endometrial carcinoma recurrence according to race and ethnicity: an NRG oncology/gynecologic oncology group 210 study. Int J Cancer. 2018;142:1102–1115. doi: 10.1002/ijc.31127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Key TJ, Pike MC. The dose-effect relationship between ‘unopposed’ oestrogens and endometrial mitotic rate: its central role in explaining and predicting endometrial cancer risk. Br J Cancer. 1988;57:205–212. doi: 10.1038/bjc.1988.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pike MC, Peters RK, Cozen W, Probst-Hensch NM, Felix JC, Wan PC, Mack TM. Estrogen-progestin replacement therapy and endometrial cancer. J Natl Cancer Inst. 1997;89:1110–1116. doi: 10.1093/jnci/89.15.1110. [DOI] [PubMed] [Google Scholar]
- 5.Kaaks R, Lukanova A, Kurzer MS. Obesity, endogenous hormones, and endometrial cancer risk: a synthetic review. Cancer Epidemiol Biomarkers Prev. 2002;11:1531–1543. [PubMed] [Google Scholar]
- 6.Grady D, Gebretsadik T, Kerlikowske K, Ernster V, Petitti D. Hormone replacement therapy and endometrial cancer risk: a meta-analysis. Obstet Gynecol. 1995;85:304–313. doi: 10.1016/0029-7844(94)00383-O. [DOI] [PubMed] [Google Scholar]
- 7.Purdie DM, Green AC. Epidemiology of endometrial cancer. Best Pract Res Clin Obstet Gynaecol. 2001;15:341–354. doi: 10.1053/beog.2000.0180. [DOI] [PubMed] [Google Scholar]
- 8.Connor EV, Rose PG. Management strategies for recurrent endometrial cancer. Expert Rev Anticancer Ther. 2018;18:873–885. doi: 10.1080/14737140.2018.1491311. [DOI] [PubMed] [Google Scholar]
- 9.McAlpine JN, Temkin SM, Mackay HJ. Endometrial cancer: not your grandmother’s cancer. Cancer. 2016;122:2787–2798. doi: 10.1002/cncr.30094. [DOI] [PubMed] [Google Scholar]
- 10.Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387:1094–1108. doi: 10.1016/S0140-6736(15)00130-0. [DOI] [PubMed] [Google Scholar]
- 11.Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. Delta-crystallin enhancer binding protein delta EF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 12.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14:481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X, Wang D, Ding Y, Zhou J, Liu G, Ji Z. lncRNA ZEB1-AS1 promotes migration and metastasis of bladder cancer cells by post-transcriptional activation of ZEB1. Int J Mol Med. 2019;44:196–206. doi: 10.3892/ijmm.2019.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 15.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell. 2007;18:3533–3544. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, Sommergruber W, Schweifer N, Wernitznig A, Beug H, Foisner R, Eger A. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, Kirchner T, Behrens J, Brabletz T. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 18.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 19.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spoelstra NS, Manning NG, Higashi Y, Darling D, Singh M, Shroyer KR, Broaddus RR, Horwitz KB, Richer JK. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 2006;66:3893–3902. doi: 10.1158/0008-5472.CAN-05-2881. [DOI] [PubMed] [Google Scholar]
- 22.Caramel J, Ligier M, Puisieux A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476. [DOI] [PubMed] [Google Scholar]
- 23.Singh M, Spoelstra NS, Jean A, Howe E, Torkko KC, Clark HR, Darling DS, Shroyer KR, Horwitz KB, Broaddus RR, Richer JK. ZEB1 expression in type I vs type II endometrial cancers: a marker of aggressive disease. Mod Pathol. 2008;21:912–923. doi: 10.1038/modpathol.2008.82. [DOI] [PubMed] [Google Scholar]
- 24.Feng G, Wang X, Cao X, Shen L, Zhu J. ZEB1 expression in endometrial biopsy predicts lymph node metastases in patient with endometrial cancer. Dis Markers. 2014;2014:680361. doi: 10.1155/2014/680361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo S, Xiao Y, Li D, Jiang Q, Zhu L, Lin D, Jiang H, Chen W, Wang L, Liu C, Fang W, Lin L. PGK1 and GRP78 overexpression correlates with clinical significance and poor prognosis in Chinese endometrial cancer patients. Oncotarget. 2018;9:680–690. doi: 10.18632/oncotarget.23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Q, Song X, Liu Z, Deng X, Luo R, Ge C, Li R, Li Z, Zhao M, Chen Y, Lin X, Zhang Q, Fang W. miRomics and proteomics reveal a miR-296-3p/PRKCA/FAK/Ras/c-Myc feedback loop modulated by HDGF/DDX5/beta-catenin complex in lung adenocarcinoma. Clin Cancer Res. 2017;23:6336–6350. doi: 10.1158/1078-0432.CCR-16-2813. [DOI] [PubMed] [Google Scholar]
- 27.Yang GY, Zhang AQ, Wang J, Li CH, Wang XQ, Pan K, Zhou C, Dong JH. Hepatoma-derived growth factor promotes growth and metastasis of hepatocellular carcinoma cells. Cell Biochem Funct. 2016;34:274–285. doi: 10.1002/cbf.3189. [DOI] [PubMed] [Google Scholar]
- 28.Chen SC, Kung ML, Hu TH, Chen HY, Wu JC, Kuo HM, Tsai HE, Lin YW, Wen ZH, Liu JK, Yeh MH, Tai MH. Hepatoma-derived growth factor regulates breast cancer cell invasion by modulating epithelial--mesenchymal transition. J Pathol. 2012;228:158–169. doi: 10.1002/path.3988. [DOI] [PubMed] [Google Scholar]
- 29.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han B, Zhang YY, Xu K, Bai Y, Wan LH, Miao SK, Zhang KX, Zhang HW, Liu Y, Zhou LM. NUDCD1 promotes metastasis through inducing EMT and inhibiting apoptosis in colorectal cancer. Am J Cancer Res. 2018;8:810–823. [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang J, Wang K, Chen Y, Chen H, Nice EC, Huang C. Redox regulation in tumor cell epithelial-mesenchymal transition: molecular basis and therapeutic strategy. Signal Transduct Target Ther. 2017;2:17036. doi: 10.1038/sigtrans.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi T, Gust KM, Wyatt AW, Goriki A, Jager W, Awrey S, Li N, Oo HZ, Altamirano-Dimas M, Buttyan R, Fazli L, Matsubara A, Black PC. Not all NOTCH is created equal: the oncogenic role of NOTCH2 in bladder cancer and its implications for targeted therapy. Clin Cancer Res. 2016;22:2981–2992. doi: 10.1158/1078-0432.CCR-15-2360. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Tillo E, Liu Y, de Barrios O, Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A, Postigo A. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci. 2012;69:3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto S, Tomita Y, Hoshida Y, Takiguchi S, Fujiwara Y, Yasuda T, Doki Y, Yoshida K, Aozasa K, Nakamura H, Monden M. Expression of hepatoma-derived growth factor is correlated with lymph node metastasis and prognosis of gastric carcinoma. Clin Cancer Res. 2006;12:117–122. doi: 10.1158/1078-0432.CCR-05-1347. [DOI] [PubMed] [Google Scholar]
- 35.Sun B, Gu X, Chen Z, Xiang J. MiR-610 inhibits cell proliferation and invasion in colorectal cancer by repressing hepatoma-derived growth factor. Am J Cancer Res. 2015;5:3635–3644. [PMC free article] [PubMed] [Google Scholar]
- 36.Ren H, Tang X, Lee JJ, Feng L, Everett AD, Hong WK, Khuri FR, Mao L. Expression of hepatoma-derived growth factor is a strong prognostic predictor for patients with early-stage non-small-cell lung cancer. J. Clin. Oncol. 2004;22:3230–3237. doi: 10.1200/JCO.2004.02.080. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Jiang Q, Hua S, Zhao M, Wu Q, Fu Q, Fang W, Guo S. High nuclear expression of HDGF correlates with disease progression and poor prognosis in human endometrial carcinoma. Dis Markers. 2014;2014:298795. doi: 10.1155/2014/298795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang J, Shi H, Li H, Zhen T, Dong Y, Zhang F, Yang Y, Han A. The interaction of hepatoma-derived growth factor and beta-catenin promotes tumorigenesis of synovial sarcoma. Tumour Biol. 2016;37:10287–10301. doi: 10.1007/s13277-016-4905-5. [DOI] [PubMed] [Google Scholar]
- 39.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 40.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.