Abstract
The aim of this study was to develop and validate a new non-invasive artificial intelligence (AI) model based on preoperative computed tomography (CT) data to predict the presence of liver metastasis (LM) in colon cancer (CC). A total of forty-eight eligible CC patients were enrolled, including twenty-four patients with LM and twenty-four patients without LM. Six clinical factors and one hundred and fifty-two tumor image features extracted from CT data were utilized to develop three models: clinical, radiomics, and hybrid (a combination of clinical and radiomics features) using support vector machines with 5-fold cross-validation. The performance of each model was evaluated in terms of accuracy, specificity, sensitivity, and area under the curve (AUC). For the radiomics model, a total of four image features utilized to construct the model resulting in an accuracy of 83.87% for training and 79.50% for validation. The clinical model that employed two selected clinical variables had an accuracy of 69.82% and 69.50% for training and validation, respectively. The hybrid model that combined relevant image features and clinical variables improved accuracy of both training (90.63%) and validation (85.50%) sets. In terms of AUC, hybrid (0.96; 0.87) and radiomics models (0.91; 0.85) demonstrated a significant improvement compared with the clinical model (0.71; 0.69), and the hybrid model had the best prediction performance. In conclusion, the AI model developed using preoperative conventional CT data can accurately predict LM in CC patients without additional procedures. Furthermore, combining image features with clinical characteristics greatly improved the model’s prediction performance. We have thus generated a promising tool that allows guidance and individualized surveillance of CC patients with high risks of LM.
Keywords: Colon cancer, liver metastasis, computed tomography, radiomics analysis, artificial intelligence, prediction
Introduction
Colon cancer (CC) is one of the leading cause of cancer-related deaths worldwide [1]. Even though the prognosis is generally favorable for early-stage CC patients with radical resection, the treatment performance of metastatic CC is disappointing, with a 5-year survival rate of only 13% [2]. Liver metastasis (LM) is the most common form of distant metastases in CC patients. Approximately 15-20% of the patients have synchronous LM at the time of the diagnosis, and the 5-year cumulative metachronous LM rate was 3.7% for TNM stage I tumors, 13.3% for stage II, and 30.4% for stage III [3]. Besides, LM is a well-known indicator of poor prognosis for cancer patients and is also one of the most challenging issues encountered during the treatment of CC [4]. Therefore, early identification of high-risk LM patients is crucial for the development of effective treatment plans and improving the clinical outcomes.
Non-invasive approaches, including computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET), are widely used for preoperative clinical evaluation of LM; however, the diagnostic accuracies of these imaging techniques are not satisfactory [5-7]. Therefore, the utilization of clinicopathological features to predict LM is currently very popular [8,9]. Chuang et al. previously showed that depth of tumor invasion, lymph node metastasis, and vascular invasion were independent factors for LM on univariate analysis [10]. However, these indicators are only available after a radical resection procedure. Therefore, new non-invasive, low-cost, and accurate approaches for the preoperative prediction of LM in CC patients are still being investigated.
Radiomics is a new field that produces high-throughput data from quantitative images and clinical data to improve the precision of disease diagnosis and prognosis using multi-modality imaging techniques [11-13]. Leveraging analysis of non-invasive data to support the process of clinical decision, radiomics was widely used in clinical studies especially in the evaluation of cancer stage and prediction of prognosis [14-16]. Our previous study demonstrated that CT-based image features can be helpful for predicting the metastatic status of the nodes and the stage of the CC [17]. Furthermore, some studies have used the image analysis approach to identify the association between tumor tissue and distant metastases in different malignancies [18-20]. Specifically, Chen et al. showed that a CT-based radiomics model demonstrated good performance in the prediction of brain metastasis (BM) in lung cancer patients [21]. Other studies also reported that radiomics signature has the potential for predicting LM in rectal and esophagogastric cancer [22-24].
Although the prediction of LM has been briefly studied for several diseases, the role of image features in predicting the presence of LM in CC patients still remains unclear. Therefore, the aim of this study is to investigate different CT algorithms and imaging sequences for prediction of LM in CC patients. Furthermore, we will evaluate the predictive performance of artificial intelligence (AI) models for diagnosis of LM in CC patients.
Materials and methods
Patients
This retrospective study was approved by the ethics committee of the Affiliated Hospital of Qingdao University. A total of 620 consecutive CC patients were received radical colectomy with lymph node dissection between October 2015 and July 2018. The clinicopathological and radiological databases were retrospectively reviewed. The patients with LM inclusion criteria were as follows: (a) patients with CC diagnosis on pathology, (b) patients with no history of previous or coexisting other malignancies, (c) patients who underwent preoperative enhanced CT for local CC staging and for LM diagnosis, and (d) patients with metastatic CC who underwent synchronous resection of primary tumor and LM. The exclusion criteria is as follows: (a) patients who underwent treatment (radiotherapy, chemotherapy or chemoradiotherapy) before the baseline CT examination, (b) poor image quality that is unqualified for image analysis, and (c) patients with LM who did not receive synchronous resection of the primary tumor and LM. The twenty-four patients with LM who received simultaneous resection of primary tumor and metastases were enrolled in the LM group; in order to match the study, another twenty-four patients without LM who underwent radical colectomy and lymph node dissection were selected as the non-LM group. Finally, a total of forty-eight consecutive patients were incorporated into our study. The clinical characteristics of the patients for each experiment were presented in Table 1. Our study framework steps are demonstrated in Figure 1.
Table 1.
Clinical characteristics of the forty-eight CC patients with and without LM
| Characteristics | LM (n = 24) | Non-LM (n = 24) | P |
|---|---|---|---|
| Age, years (Mean ± SD) | 63.33±11.21 | 59.71±13.86 | 0.197 |
| Gender, n (%) | 1 | ||
| Male | 15 (62.50) | 15 (62.50) | |
| Female | 9 (37.50) | 9 (37.50) | |
| Tumor site, n (%) | 0.057 | ||
| Right | 14 (58.33) | 20 (83.33) | |
| Left | 10 (41.67) | 4 (16.67) | |
| Histologic grade, n (%) | 0.383 | ||
| Moderately differentiated | 15 (62.50) | 12 (50.00) | |
| Poorly differentiated | 9 (37.50) | 12 (50.00) | |
| Tumor diameter, cm (Mean ± SD) | 7.97±3.12 | 7.31±1.55 | 0.385 |
Note: n, number; LM, liver metastasis; SD, standard deviation; tumor size was measured at the thickest part of the colon lesion vertical to the bowel wall on the cross-sectional image; P value was derived from the univariable association analyses between each characteristic, P < 0.05 was considered statistically significant.
Figure 1.
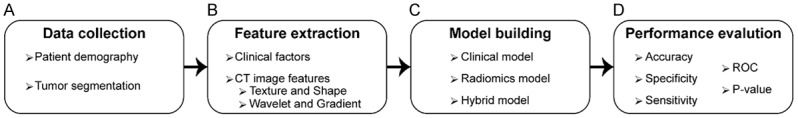
The workflow of the study. The collection of the clinical characteristics of the patients and segmentation of tumor regions on CT data (A). Obtaining clinical factors and extraction of CT image features (B). Building three AI models-clinical, radiomics, and hybrid (combination of clinical and radiomics features) - for prediction of liver metastasis (C). The evaluation of performances of the generated models in terms of accuracy, sensitivity, specificity, and area under the curve (D).
CT image acquisition
All patients underwent contrast-enhanced abdominal and pelvic CT using a two 64-detector row spiral CT system (Discovery 750 HD, GE Medical System, USA or Volume Ultra, GE Medical Systems, USA). CT scan was performed 65 s after intravenous injection of 100 ml iopromide (Uitravist-300; Bayer Schering Pharma, Berlin Germany) at 3 ml/s for enhancement. Scanning parameters were as follows: 120 kv; 160 mAs; 0.6 s rotation time and an imaging matrix of 512×512. Portal venous phase images at 1.0 mm thickness were retrieved for radiomics analysis.
Tumor segmentation and feature extraction
Two radiologists with more than 10 years of experience in abdominal radiology interpretation examined all the slices of CT data to find the slice with the maximum tumor diameter. Later, tumor regions were outlined by the first radiologist using open-source software (ITK-SNAP) [25], and the second radiologist validated the drawn region of interest (ROI) for the tumor tissue (Figure 2).
Figure 2.
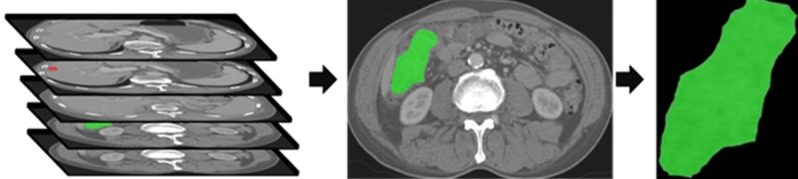
The CT images with identified tumor regions. An example of manual segmentation of primary tumor (green regions) and liver metastasis (red regions) on colon cancer CT images.
The CT image characteristics were obtained using six feature extraction methods (first-order statistic, FoS; co-occurrence matrix, CoM; run-length matrix, RLM; local binary patterns, LBP; fractal analysis, FA; and tumor structure, TS) and three transformations (gradients, GD; histogram of oriented gradients, HoG; and wavelet transform, WT) with in-house implemented scripts in Matlab® (MathWorks, MA) [17,26]. Six first-order statistical features were calculated from gray-level pixel intensity histogram to capture a summary of the intensity distribution of the lesions without considering spatial positioning. To measure joint probabilities of pixels with different intensities, we generated co-occurrence matrix in four directions (0°, 90°, 180°, 270°) and extracted six features by averaging each co-occurrence matrix after merging spatial direction features as averaging. The run-length matrix were computed to characterize coarseness of the textures using gray-level intensity runs and calculate seven features from the run-length matrix. LBP technique analyzes the image texture by comparing the association between neighboring pixels in a region and generates a histogram for this relationship. In our study, we computed 10 LBP features of tumor tissues. The complexity of the texture structure in CT image were reflected by measuring the fractal dimension. Besides, gradient image was generated to describe coarseness or fineness of the structures leveraging intensity variation in specific directions. A total of nineteen features were calculated from gradient images employing statistics (FoS) and texture (CoM and RLM) methods. In additon, six additional features were obtained from gradient histograms that were generated with the HoG approach by clustering gradients into nine directional groups. To extract spatial characteristics of the texture, we calculated wavelet detail images utilizing Daubechies function resulting in four sub-images. We calculated statistical (FoS) and textural features (GLCM and GLRM) in addition to power, variance, and average signal magnitudes of the wavelet images.
Following the extraction of 152 image features from preoperative conventional CT data, we standardized the features using the z-score normalization approach for stable convergence while optimizing the hyperparameters (kernel parameter and penalty) of the generated classifiers. In order to identify the relevancy and space dimensionality of image features, we employed the RELIEFF algorithm which determined the ranking of the features by analyzing the differences among nearest-neighbor pairs [27].
Classifier model building
In order to evaluate the prediction accuracy of radiomics features and clinical variables, we assessed the importance of the variables individually to build three separate AI models (radiomics, clinical, and hybrid) using a kernel-based support vector machines (SVM) method with 5-fold cross-validation. Using the SVM technique, the separating hyperplane was generated by optimizing the minimal distance between separating plane and closest samples. During SVM optimization, sequential minimal optimization method was utilized by mapping image features to high dimensional space with radial basis function kernels that lead to a non-linear classification of the samples. Prior to the development of the models, the data was randomly separated into five groups (four were used for training and the remaining for validation of the model). The process was repeated five times with different clusters used for training and validation to better evaluate the models. The distribution of the patients in training and validation sets after combining groups of five experiments are presented in Table 2.
Table 2.
The distribution of the patient characteristics in 5-fold cross-validation
| Characteristics | Training cohort | Validation cohort | ||
|---|---|---|---|---|
|
|
|
|||
| LM | Non-LM | LM | Non-LM | |
| Gender | ||||
| Male | 60/75 | 60/75 | 15/75 | 15/75 |
| Female | 36/45 | 36/45 | 9/45 | 9/45 |
| Tumor site | ||||
| Left | 40/50 | 17/21 | 10/50 | 4/21 |
| Right | 56/70 | 79/99 | 14/70 | 20/99 |
| Histologic grade | ||||
| Moderately differentiated | 56/70 | 52/65 | 14/70 | 13/65 |
| Poorly differentiated | 40/50 | 44/55 | 10/50 | 11/55 |
Note: LM, liver metastasis.
For the clinical model, we obtained the demographics (age and gender) and tumor-related (location and histological status) characteristics from patient records after the removal of personal identifiers. Besides, tumor size were measured in the longest and shortest diameter using CT data. To determine how clinical variables correlate with tumor characteristics, we examined all the features in an exhaustive search approach evaluating the average performance of the AI models for training and validation sets in terms of accuracy and area under the receiver operating curve (AUC). The clinical model was generated using the variable set that demonstrated the best performance.
For the radiomics model, we included six features with the highest rankings identified by the RELIEEF algorithm [27]. Using the same approach as the developing clinical model, the set of features that reflect the biological changes of metastasis were identified. Afterwards, the selected clinical and CT image features were further analyzed by generating a hybrid model that combined clinical and imaging variables with the best performance using the exhaustive search approach.
Performance evaluation and statistical analysis
For performance evaluation of the three models, we utilized accuracy, sensitivity, specificity, and receiver operating characteristic (ROC) curves for training and validation. In the 5-fold cross-validation approach, the performance metrics of generated models were calculated by averaging them over the five experiments. While selecting the potential model parameters and feature sets, the generated models were sorted according to the accuracy and AUC by validation sets as removing the models that generate a difference more than 7% accuracy between training and validation. To compare the superiority of the generated models (radiomics, clinical and hybrid), multiple Student’s t-test using AUC values obtained from five experiments for each model were performed. P < 0.05 was considered statistically significant.
For patient demographic information, Student’s t-test for continuous variables and Chi-square test were performed to evaluate relationships for categorical factors. We also presented continuous variables as mean ± standard deviation.
Results
Clinical characteristics of patients
A total of forty-eight CC patients were incorporated into our research including twenty-four patients with LM and twenty-four patients without LM. There were no statistically significant difference in clinical characteristics between patients with LM and without LM.
Feature selection
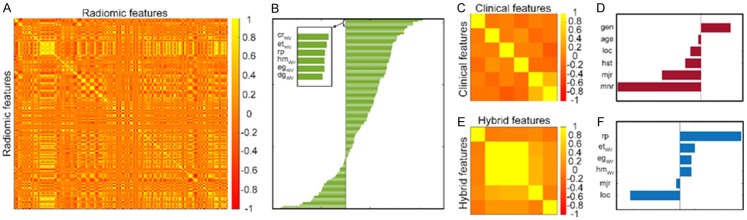
The radiomics model was constructed with four features (entropy, energy, and homogeneity of the vertical wavelet image and low gray level run emphasis of raw image) following detailed assessment of top-ranked features in terms of accuracy and AUC measures of the validation. In Figure 3A, we visualized the correlation between the features and showed the ranking of the features in Figure 3B. For the clinical model, six features were evaluated for accuracy and AUC in the validation sets, and the final model was generated using the two features (tumor site and diameter of tumor tissue) showing the best performance. The correlation of the clinical features are shown in Figure 3C, and the ranking of the features is presented in Figure 3D. For the hybrid model, a total of six features selected via clinical and radiomics models were evaluated. Afterward, the final model was generated with four selected features (heterogeneity, entropy, and energy of vertical wavelet image and diameter of tumor). Figure 3E shows the correlation values among the features for the hybrid model; the rankings of the selected features are presented in Figure 3F.
Figure 3.
The correlation and ranking of the selected features. A and B demonstrate the cross-correlation and ranking of the image features used for radiomics model; C and D show the cross-correlation and ranking of the clinical factors used for the clinical model; E and F present cross-correlation and ranking of the features used for the hybrid model.
Evaluation of the models for predicting liver metastasis
In the present study, we generated three AI models to predict the presence of LMs: clinical, radiomics and hybrid. For each model, all combinations of the features were evaluated in an exhaustive search procedure to identify significant variables that allowed prediction of LM and also evaluated the performance of the generated models in terms of accuracy and AUC measurements (Figure 4).
Figure 4.
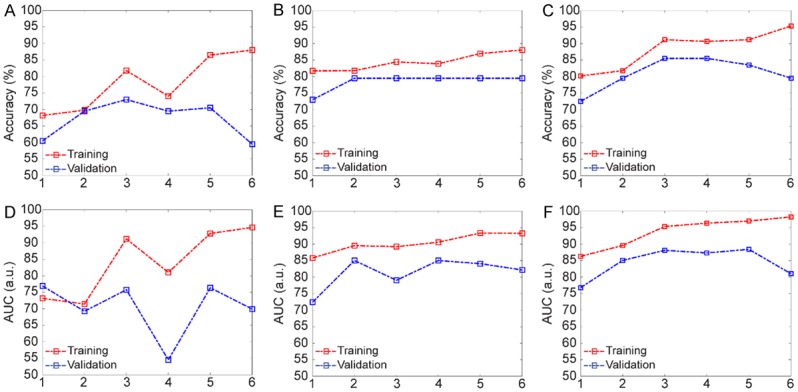
The performance evaluation of the generated models with respect to a number of features. A and D describe the accuracy and AUC in the clinical model; B and E show the performances of the radiomics models; C and F demonstrate the accuracy and the AUC in the hybrid model.
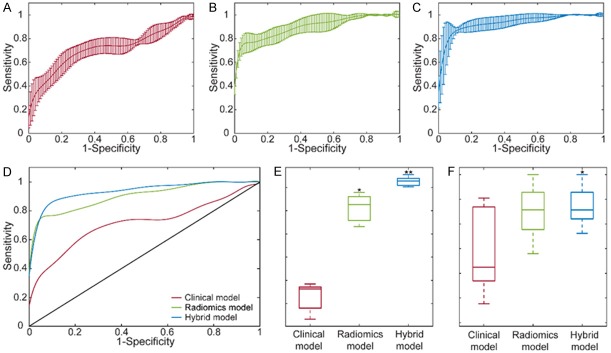
The clinical model was generated upon assessment of all the combinations of the variables while analyzing the performance of the candidate models. The behavior of the model was described in terms of accuracy and AUC metrics for the clinical model (Figure 4A and 4D). Upon evaluation of the results, the final clinical model generated with three features demonstrated an accuracy of 69.82% for training and 69.50% for validation. The model specificity was 83.31% for training and 83.14% for validation, while the sensitivity was 56.36% for training and 62.00% for the validation. Besides, the average AUC values were 0.71 for training and 0.69 for the validation. Figure 5A shows the ROC curve of the training model for each of five validation sets with a visualization of mean ± standard deviation.
Figure 5.
ROC curves of the final SVM classifiers for generated models with mean and standard deviation (A-C). As (D) reflect the comparison of the ROC curves for the generated models, (E, F) evaluate the difference between the models for training and validation, respectively. (* represents an improvement of the model was statistically significant than clinical model and ** demonstrates the statistical difference compared to clinical and radiomics models).
For the radiomics model, all the subsets of the six features were extensively evaluated to identify the relevant features that reflect the existence of LMs. In Figure 4B and 4E, the performances of the generated models with respect to a varying number of features were demonstrated. The results showed that the radiomics model with four features generated the highest performances for training and validation sets. Therefore, the model was constructed with four features presenting an accuracy of 83.87% for training and 79.50% for validation that increased the accuracy almost 10% compared to the clinical model. In addition, the specificity of the model was 96.59% for training and 86.48% for validation while the sensitivity of the model was 75.28% for training and 78.86% for the validation. Besides, the AUC values of the model were 0.91 for training and 0.85 for the validation (Figure 5B).
For the hybrid model, six features selected for clinical and radiomics models were analyzed with an assessment of all the classifiers with respect to increasing number of features. The accuracy and AUC of the classifiers are shown in Figure 4C and 4F. After evaluation of the performances of the training and validation sets, the hybrid model were constructed with four features which improved the prediction accuracy with an average of 6% (a total of 90.63% for training and 85.50% for validation) compared to radiomics model. The hybrid model demonstrated specificity of 95.41% for training and 94.29% for validation while the sensitivity of the model was 86.04% for training and 79.33% for the validation. Besides, the AUC value of this model was 0.96 for training and 0.87 for the validation. The ROC curve of the five training sets was shown in the form of mean ± standard deviation in Figure 5C.
After generating the final classifiers for the models, we evaluated the prediction performances in terms of average AUC measures of the models (Figure 5D). The radiomics model demonstrated a statistically significant improvement (P < 0.001) compared to the clinical model (Figure 5E). In addition, hybrid model presented a significant improvement compared to clinical (P < 0.001) and radiomics models (P = 0.002) for the training set. Although statistical significance was not observed in AUC measurements for the validation data (except among hybrid and clinical model, P = 0.041), prediction performance was improved by combining the key features from radiomics and clinical models (Figure 5F).
Discussion
In the present study, we developed three AI models to estimate the individual risks of the LM in CC patients using clinical characteristics and preoperative CT images features: clinical, radiomics, and hybrid models. We further evaluated the predictive accuracy of these models to detect high-risk patients with LM. Our results revealed that the hybrid model (85.50%) showed the best discriminative ability to detect high-risk patients with LM compared with the radiomics (79.50%) and clinical models (69.50%). Therefore, a non-invasive model can be developed as an individualized visual tool for detecting LM and helping clinicians select appropriate surveillance and intervention plans for CC patients with high risks of LM.
The liver is the most common site for distant metastases of CC, and up to 70% of all CC patients may develop LM eventually [28]. Furthermore, LM is the leading cause of death and also a key point in the treatment of cancer patients. Currently, hepatic resection is considered to be the only treatment that can provide long-term survival in CC [29]. The patients with resectable primary tumor and LM have a better 5-year survival rate compared to patients with palliative treatment (50% vs 5%) [30]. With the continuous improvement of hepatectomy technique and perioperative care, complication and mortality rate have dropped dramatically. Meanwhile, simultaneous resection of the primary tumor and LM is recommended in clinical practice of CC patients for optimal outcomes [31]. Unfortunately, only about 25% of CC patients can undergo simultaneous hepatic resection [32]. In this study, 13.2% (82/620) of patients have synchronous LM, and only 24 (29.3%) of these patients received simultaneous resection of the primary tumor and LM. These patients had no complications after surgery and were discharged smoothly. Therefore, screening for high-risk factors and early detection of LM will provide opportunities for surgical resection and improve the prognosis of CC patients.
The depth of tumor invasion and lymph node metastasis are two components of the TNM system which are considered to be high-risk factors for the development of LM [33]. Carcinoembryonic antigen (CEA) level was also recognized as an independent factor for LM of CC [34]. Chuang et al. demonstrated that preoperative serum CEA level, depth of tumor invasion, lymph node metastasis, and vascular invasion were independent factors that correlate with LM by investigating 1,099 CC cases over a 7-year follow up [10]. Although these clinicopathologic factors have a good predictive performance of LM in CC, these factors are only available after invasive procedures. In our study, we attempted to establish a clinical model using non-invasive preoperative factors, including age, gender, tumor site, and histologic grade to predict LM in CC patients. However, the accuracy and sensitivity were only 69.82% and 56.36% respectively, and the prediction performance of the clinical model was not satisfactory.
CT, MRI, and PET-CT are common imaging examinations which can be helpful in the diagnosis of LM in CC preoperatively. However, the sensitivity and accuracy of their diagnosis are not as perfect as we expected. According to the meta-analysis with 20-year study, the detection sensitivity of LM in CT, MRI, and PET were 74.4%, 80.3%, and 81.4%, respectively [35]. With the rapid development of radiomics in recent years, many studies have used radiomics signature extracted from images to predict distant metastases in different cancers [18,20,36]. Shu et al. reported that MRI-based image features can provide non-invasive information to predict the risk of synchronous and metachronous LM in rectal cancer patients with satisfactory results [22,23]. CT scans are routinely performed preoperatively in CC patients for assessment and have higher potential to extract more significant and useful image features [12,37]. Therefore, in this study, we generated a radiomics model using quantitative image features extracted from CT of CC patients with a prediction accuracy of 79.50%, which corresponded with an average increase of 10% compared to the clinical model. Furthermore, we combined clinical and image features following the analyses and obtained a much better predictive outcome (85.50%). The results proved that the generated hybrid model may serve as an accurate and reliable detection tool for LM in CC patients.
The present study also has some limitations. First, this investigation is based on a single institutional retrospective analysis with small-sample study. Further investigations including multi-institutional prospective studies will expand patient cohorts and improve generalization of the outcome. Second, image features were extracted from the venous phase of multiphase enhancement sequences; arterial phase and delayed phase image features should be further investigated. Finally, our study lacked postoperative follow-up data, hence we could not investigate the relationship between models and survival outcomes of the patients. Despite these limitations, we hope that our result will contribute to accurate prediction of the LM in CC patients.
In conclusion, we established a new prediction model based on CT image that can successfully and accurately predict the presence of LM in CC patients. Furthermore, combining image features with clinical characteristics greatly improved the model’s prediction performance. This AI model may serve as a promising tool for individualized surveillance of CC patients with high risks of LM and also provide the basis for clinicians to improve treatment decisions for these patients.
Acknowledgements
This study was supported by the National Cancer Institute (grants R01CA209886, R01CA196967), by 2019 Harold E. Eisenberg Foundation Scholar Award, by the Fishel Fellowship Award at the Robert H. Lurie Comprehensive Cancer Center and by the key research and development program of Tianjin Municipal Science and Technology Commission (17YFZCSY00870).
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre L, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Robinson JR, Newcomb PA, Hardikar S, Cohen SA, Phipps AI. Stage IV colorectal cancer primary site and patterns of distant metastasis. Cancer Epidemiol. 2017;48:92–95. doi: 10.1016/j.canep.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244:254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, Fan J, Qin X, Cai J, Gu J, Wang S, Wang X, Zhang S, Zhang Z China CRLM Guideline Group. Chinese guidelines for the diagnosis and comprehensive treatment of colorectal liver metastases (version 2018) J Cancer Res Clin Oncol. 2019;145:725–736. doi: 10.1007/s00432-018-2795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SJ, Zea R, Kim DH, Lubner MG, Deming DA, Pickhardt PJ. CT texture features of liver parenchyma for predicting development of metastatic disease and overall survival in patients with colorectal cancer. Eur Radiol. 2018;28:1520–1528. doi: 10.1007/s00330-017-5111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee KH, Lee JM, Park JH, Kim JH, Park HS, Yu MH, Yoon JH, Han JK, Choi BI. MR imaging in patients with suspected liver metastases: value of liver-specific contrast agent gadoxetic acid. Korean J Radiol. 2013;14:894–904. doi: 10.3348/kjr.2013.14.6.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kijima S, Sasaki T, Nagata K, Utano K, Lefor AT, Sugimoto H. Preoperative evaluation of colorectal cancer using CT colonography, MRI, and PET/CT. World J Gastroenterol. 2014;20:16964–16975. doi: 10.3748/wjg.v20.i45.16964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mantke R, Schmidt U, Wolff S, Kube R, Lippert H. Incidence of synchronous liver metastases in patients with colorectal cancer in relationship to clinico-pathologic characteristics. Results of a German prospective multicentre observational study. Eur J Surg Oncol. 2012;38:259–265. doi: 10.1016/j.ejso.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Wu JB, Sarmiento AL, Fiset PO, Lazaris A, Metrakos P, Petrillo S, Gao ZH. Histologic features and genomic alterations of primary colorectal adenocarcinoma predict growth patterns of liver metastasis. World J Gastroenterol. 2019;25:3408–3425. doi: 10.3748/wjg.v25.i26.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang SC, Su YC, Lu CY, Hsu HT, Sun LC, Shih YL, Ker CG, Hsieh JS, Lee KT, Wang JY. Risk factors for the development of metachronous liver metastasis in colorectal cancer patients after curative resection. World J Surg. 2011;35:424–429. doi: 10.1007/s00268-010-0881-x. [DOI] [PubMed] [Google Scholar]
- 11.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, Bellomi M. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2:36. doi: 10.1186/s41747-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 14.Wu S, Zheng J, Li Y, Yu H, Shi S, Xie W, Liu H, Su Y, Huang J, Lin T. A Radiomics nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. Clin Cancer Res. 2017;23:6904–6911. doi: 10.1158/1078-0432.CCR-17-1510. [DOI] [PubMed] [Google Scholar]
- 15.Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X, Ma ZL, Liu ZY. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J. Clin. Oncol. 2016;34:2157–64. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 16.Rahmim A, Bak-Fredslund KP, Ashrafinia S, Lu L, Schmidtlein CR, Subramaniam RM, Morsing A, Keiding S, Horsager J, Munk OL. Prognostic modeling for patients with colorectal liver metastases incorporating FDG PET radiomic features. Eur J Radiol. 2019;113:101–109. doi: 10.1016/j.ejrad.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Eresen A, Lu Y, Yang J, Shangguan J, Velichko Y, Yaghmai V, Zhang Z. Radiomics signature for the preoperative assessment of stage in advanced colon cancer. Am J Cancer Res. 2019;9:1429–1438. [PMC free article] [PubMed] [Google Scholar]
- 18.Fan L, Fang M, Tu W, Zhang D, Wang Y, Zhou X, Xia Y, Li Z, Liu S. Radiomics signature: a biomarker for the preoperative distant metastatic prediction of stage I nonsmall cell lung cancer. Acad Radiol. 2019;26:1253–1261. doi: 10.1016/j.acra.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Zhang C, Wang L, Luo R, Li J, Zheng H, Yin Q, Zhang Z, Duan S, Li X, Wang D. MRI radiomics analysis for predicting preoperative synchronous distant metastasis in patients with rectal cancer. Eur Radiol. 2019;29:4418–4426. doi: 10.1007/s00330-018-5802-7. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Dong D, Li H, Tian J, Ouyang F, Mo X, Zhang B, Luo X, Lian Z, Pei S, Dong Y, Huang W, Liang C, Liu J, Zhang S. Development and validation of a magnetic resonance imaging-based model for the prediction of distant metastasis before initial treatment of nasopharyngeal carcinoma: a retrospective cohort study. EBioMedicine. 2019;40:327–335. doi: 10.1016/j.ebiom.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen A, Lu L, Pu X, Yu T, Yang H, Schwartz LH, Zhao B. CT-based radiomics model for predicting brain metastasis in category T1 lung adenocarcinoma. AJR Am J Roentgenol. 2019:1–6. doi: 10.2214/AJR.18.20591. [DOI] [PubMed] [Google Scholar]
- 22.Shu Z, Fang S, Ding Z, Mao D, Cai R, Chen Y, Pang P, Gong X. MRI-based Radiomics nomogram to detect primary rectal cancer with synchronous liver metastases. Sci Rep. 2019;9:3374. doi: 10.1038/s41598-019-39651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang M, Cai Z, Zhang H, Huang C, Meng Y, Zhao L, Li D, Ma X, Zhao X. Machine learning-based analysis of rectal cancer MRI radiomics for prediction of metachronous liver metastasis. Acad Radiol. 2019;26:1495–1504. doi: 10.1016/j.acra.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 24.Klaassen R, Larue RTHM, Mearadji B, van der Woude SO, Stoker J, Lambin P, van Laarhoven HWM. Feasibility of CT radiomics to predict treatment response of individual liver metastases in esophagogastric cancer patients. PLoS One. 2018;13:e0207362. doi: 10.1371/journal.pone.0207362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Eresen A, Alic L, Birch SM, Friedeck W, Griffin JF 4th, Kornegay JN, Ji JX. Texture as an imaging biomarker for disease severity in golden retriever muscular dystrophy. Muscle Nerve. 2019;59:380–386. doi: 10.1002/mus.26386. [DOI] [PubMed] [Google Scholar]
- 27.Kononenko I, Šimec E, Robnik-Šikonja M. Overcoming the myopia of inductive learning algorithms with RELIEFF. Applied Intelligence. 1997;7:39–55. [Google Scholar]
- 28.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 29.Misiakos EP, Karidis NP, Kouraklis G. Current treatment for colorectal liver metastases. World J Gastroenterol. 2011;17:4067–4075. doi: 10.3748/wjg.v17.i36.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.House MG, Ito H, Gonen M, Fong Y, Allen PJ, DeMatteo RP, Brennan MF, Blumgart LH, Jarnagin WR, D’Angelica MI. Survival after hepatic resection for metastatic colorectal cancer: trends in outcomes for 1,600 patients during two decades at a single institution. J Am Coll Surg. 2010;210:744–752. 752–755. doi: 10.1016/j.jamcollsurg.2009.12.040. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe T, Muro K, Ajioka Y, Hashiguchi Y, Ito Y, Saito Y, Hamaguchi T, Ishida H, Ishiguro M, Ishihara S, Kanemitsu Y, Kawano H, Kinugasa Y, Kokudo N, Murofushi K, Nakajima T, Oka S, Sakai Y, Tsuji A, Uehara K, Ueno H, Yamazaki K, Yoshida M, Yoshino T, Boku N, Fujimori T, Itabashi M, Koinuma N, Morita T, Nishimura G, Sakata Y, Shimada Y, Takahashi K, Tanaka S, Tsuruta O, Yamaguchi T, Yamaguchi N, Tanaka T, Kotake K, Sugihara K Japanese Society for Cancer of the Colon and Rectum. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2016 for the treatment of colorectal cancer. Int J Clin Oncol. 2018;23:1–34. doi: 10.1007/s10147-017-1101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simmonds PC, Primrose JN, Colquitt JL, Garden OJ, Poston GJ, Rees M. Surgical resection of hepatic metastases from colorectal cancer: a systematic review of published studies. Br J Cancer. 2006;94:982–999. doi: 10.1038/sj.bjc.6603033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enquist IB, Good Z, Jubb AM, Fuh G, Wang X, Junttila MR, Jackson EL, Leong KG. Lymph node-independent liver metastasis in a model of metastatic colorectal cancer. Nat Commun. 2014;5:3530. doi: 10.1038/ncomms4530. [DOI] [PubMed] [Google Scholar]
- 34.Iizasa T, Suzuki M, Yoshida S, Motohashi S, Yasufuku K, Iyoda A, Shibuya K, Hiroshima K, Nakatani Y, Fujisawa T. Prediction of prognosis and surgical indications for pulmonary metastasectomy from colorectal cancer. Ann Thorac Surg. 2006;82:254–260. doi: 10.1016/j.athoracsur.2006.02.027. [DOI] [PubMed] [Google Scholar]
- 35.Niekel MC, Bipat S, Stoker J. Diagnostic imaging of colorectal liver metastases with CT, MR imaging, FDG PET, and/or FDG PET/CT: a meta-analysis of prospective studies including patients who have not previously undergone treatment. Radiology. 2010;257:674–684. doi: 10.1148/radiol.10100729. [DOI] [PubMed] [Google Scholar]
- 36.Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RT, Hermann G, Lambin P, Haibe-Kains B, Mak RH, Aerts HJ. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114:345–350. doi: 10.1016/j.radonc.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dighe S, Swift I, Brown G. CT staging of colon cancer. Clin Radiol. 2008;63:1372–1379. doi: 10.1016/j.crad.2008.04.021. [DOI] [PubMed] [Google Scholar]