Abstract
Many Burkholderia spp. produce in vitro secondary metabolites with relevant biological activities and potential practical applications. Burkholderia gladioli pv. agaricicola (Bga) possess promising biological activities regulated by N-Acyl homoserine lactones (N.AHLs) based quorum sensing (QS) mechanism. In the current study, N.AHLs-deficient (ICMP11096glad-I) and N.AHLs-complemented (ICMP11096glad-IR) mutants were constructed in which the gene coding for AHL synthase was inactivated by allelic exchange in glad I mutant strain. The aims of this research were to (i) assess the antagonistic activity of the wild type (WT) and the glad-I mutant of Bga against Bacillus megaterium (G+ve) and Escherichia coli (G−ve), (ii) screen their hydrolytic enzymes and hemolytic substances, (iii) monitor the pathogenic effect against Agaricus bisporus, and finally (iv) analyze the bioactive secondary metabolites produced by WT and mutant strain using high performance liquid chromatography (HPLC). Results showed that N.AHLs-deficient mutant exhibited high reduction of antagonistic activity against the tested microorganisms and notable reduction of chitinolytic, proteolytic and glucanolytic activities and complete absence of hemolytic activity, and the glad-IR complemented mutant was able to regain the major part of these activities. Furthermore, N.AHLs-deficient mutant strain was unable to degrade flesh cubes pseudo-tissues of A. bisporus. On the other hand, the virulence effect of complemented mutant was like to the parental WT strain. HPLC analysis revealed that some of the single components produced by WT strain were absent in N.AHLs-deficient mutant and others were highly reduced. The out-findings of the current research gave a spot into the regulatory role of N.AHLs and QS phenomenon in the biological activity of Bga bacterium.
Keywords: phytopathogenic bacteria, antagonistic activity, Agaricus bisporus, secondary metabolites, quorum sensing, HPLC
Introduction
Many bacteria regulate their biological activities and interaction with surrounding conditions through a particular mechanism called quorum sensing (QS) (Steindler and Venturi, 2007). This mechanism enables the bacterial cells to communicate to each other by responding to different signal molecules (Withers et al., 2001).
Quorum sensing mechanism in bacteria is classified into three main classes: LuxI/LuxR in Gram-negative (G−ve) bacteria; oligopeptide-two-component G+ve bacteria; and luxS-encoded autoinducer 2 (AI-2) in both G−ve and G+ve bacteria (Withers et al., 2001; Li and Tian, 2012). The signal molecules are also divided into two main groups: N-Acyl homoserine lactones (N.AHLs) in G−ve and small peptides in G+ve bacteria (Davey and O’Toole, 2000; Costerton et al., 2003). QS regulates the production of virulence factors, bioactive metabolites, symbiosis, biofilm formation and motility for colony escape (Kumari et al., 2006; Duerkrop et al., 2007; Zhang et al., 2012). N.AHLs mediated QS have already been reported as regulators for several primary and secondary metabolites in some G−ve bacteria belonging to Pseudomonas and Burkholderia species and to Agrobacterium genus (Zhou et al., 2003; Dong and Zhang, 2005).
Burkholderia gladioli Yabuuchi is a widespread G−ve bacteria in large variety of sources, including soil, water, and vegetation (Yabuuchi et al., 1992; Coenye and Vandamme, 2003; Stoyanova et al., 2007). Originally B. gladioli was considered a causal agent of cavity disease of mushroom (Chowdhury and Heinemann, 2006). It was also considered a plant growth promoting rhizobacterium (Karakurt and Aslantas, 2010; Elshafie et al., 2017b). B. gladioli is also considered an important human pathogen causing fibrocystic lung disease (Wilsher et al., 1997).
Burkholderia gladioli pv. agaricicola (Bga) is one of the most serious pathogens in the mushroom industry (Gill, 1995). It causes soft rot on a number of commercially important mushrooms such as Lentinula edodes, Pleurotus ostreatus, Flammulina velutipes, Pholiota nameko, Hypsizygus marmoreus, and Grifola frondosa in Japan and different cultivated Agaricus species in New Zealand and Europe (Chowdhury and Heinemann, 2006).
Recent investigations have shown that Bga produces N.AHLs (Prashanth et al., 2011) but their regulatory role in virulence and other phenotype traits and biological activity are not yet known (Andolfi et al., 2008; Cimmino et al., 2008; Elshafie et al., 2012). In particular, Elshafie et al. (2017a) have studied the in vitro antimicrobial effects of four wild type (WT) strains of Bga and their hemolytic, enzymatic and virulence activities, and results showed that all WT strains inhibited Bacillus megaterium and E. coli growth. Results showed also that Bga ICMP11096 showed the highest antimicrobial activity toward both target microorganisms, was able to haemolyze erythrocytes cell membrane, and produces the hydrolytic enzymes (chitinase, glucanase, and protease). Moreover, the virulence effect of the four studied strains have been verified against Agaricus bisporus tissues (Elshafie et al., 2017a).
The main objective of the current research was to construct N.AHLs-deficient (ICMP11096glad-I) and N.AHLs-complemented (ICMP11096glad-IR) mutants of Bga ICMP11096 and to study their antimicrobial, hydrolytic, hemolytic, and virulence activities compared to WT strain and to analyze also the produced bioactive fractions of the WT and ICMP11096glad-I mutant using high performance liquid chromatography (HPLC).
Materials and Methods
Bacterial Strains
The WT strain of Bga ICMP11096 has been described in detail in previous works as reported by Elshafie et al. (2017a), and was maintained as lyophils at 4°C, and later on subcultures were obtained on the King B medium (KB) for 48 h at 25°C. Modified N.AHLs-deficient mutant of the WT strain (ICMP11096glad-I) and the complemented mutant (ICMP11096glad-IR) were prepared at the International Centre for Genetic Engineering and Biotechnology (ICGEB), Trieste, Italy. Chromobacterium violaceum CV026 sensor was grown in Luria-Bertani medium (LB) at 30°C. The target microorganisms Bacillus megaterium ITM100 (B. megaterium) and Escherichia coli ITM103 (E. coli) were previously identified and stored as pure freeze-dried cultures at −20°C in the collection present at the School of Agricultural, Forestry, Food and Environmental Sciences (SAFE), University of Basilicata, Potenza, Italy. Conjugations in B. gladioli strains were performed by triparental mating using E. coli DH5 (pRK2013) as helper and incubated at 30°C for 12 h. Transconjugants were selected on KB medium containing the appropriate antibiotics. The plasmids in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description | Source or reference |
| B. gladioli pv agaricicola strains | ||
| ICMP 11096 | ||
| ICMP 11096glad-I | gladI:Km of B. gladioli 11096 | This study |
| ICMP 11096glad-IR | ICMP 11096gladI complemented with pBBgladIR | This study |
| CV026 | C. violaceum AHLs sensor strain; double transposon mutant of ATCC31532, violacein and AHL negative | McClean et al., 1997 |
| pGEM2T easy | Cloning vector AmpR | Promega |
| pLAFR3 | Broad-host-range cloning vector, IncP1, TcR | Staskawicz et al., 1987 |
| pRK2013 | KmR Tra + Mob +, ColE1 replicon | |
| pKNOCK-Km | Conjugative suicide vector KmR | Alexeyev, 1999 |
| pBBRMCS-5 | Cloning vector GmR | Kovach et al., 1995 |
| pKmgladI | glad I internal fragment cloned in pKNOCK-Km | This study |
| pLgladIR | pLAFR3 containing B. gladioli ICMP11096 | This study |
| Genomic DNA | ||
| pBBgladIR | pBBRMCS-5 carrying a 3544 bp fragment containing B. gladioli quorum sensing genes | This study |
Recombinant DNA Techniques
Recombinant DNA techniques, including digestion with restriction enzymes, agarose gel electrophoresis, Southern blot analysis of restriction digested DNA, ligation with T4 DNA ligase, and transformation of E. coli were performed as previously described (Sambrook et al., 1989). Plasmids were purified by using EuroGold plasmid miniprep kit, and agarose gel electrophoresis purification of DNA fragments was performed with EuroGold gel purification kit (EuroClone, Italy). Genomic DNA was isolated by sarkosyl-pronase lysis. PCR amplifications were made using GoTaq DNA polymerase (Promega). DNA sequencing was performed by Macrogen Europe.
Identification and Inactivation of Quorum Sensing
Cosmid Library
A cosmid library was constructed to identify the QS genes of WT Bga strain. More specifically, partial EcoRI digested genomic DNA from Bga strain ICMP11096 was cloned into the cosmid vector pLAFR3 and packaged into phage particles by using the Gigapack Gold packaging extract (Stratagene). The recombinant phage particles were transduced to E. coli HB101. The cosmid library was conjugated en masse into the C. violaceum CV026 AHL biosensor (McClean et al., 1997). One transconjugant which showed restoration of QS regulated violacein production, presumably containing the Bga QS genes, was further isolated to purify the pLgladIR cosmid. The pLgladIR cosmid was cut with different restriction enzymes and analyzed by Southern blot using the tofI gene from Burkholderia glumae as a probe (Devescovi et al., 2007). The tofI gene is coding for the acyl homoserine lactone synthase in B. glumae and it was chosen because of the high phylogenetic closeness of the two species. The gladI gene was localized on a XhoI fragment of approximately 3500 bp. This fragment was further cloned in pBBRMCS-5, to generate pBBgladIR. When conjugated in CV026, pBBgladIR was able to restore the violacein production. The plasmid was partially sequenced and the glad-I and a luxR homolog (glad-IR) were identified.
Construction of Glad I QS Mutants
To generate the knockout mutant of gladI gene, an internal fragment of gladI gene (261 bp) was PCR amplified by using the following primers generated in the current study gladI_Fw 5′-TGCGCGCGACTATTGCCGAC-3′ and gladI_Rev 5′-GAACAGCCGCTCGATACTGC-3′. The primers were chosen from the partially sequenced pBBgladIR. The fragment was then cloned in pGEM vector (Promega) sequenced and subsequently cloned as an EcoRI fragment in the pKNOCK-Km vector (Alexeyev, 1999) generating pKmgladI. This plasmid was then used as a suicide delivery system and conjugated in the WT Bga strain in order to obtain the genomic mutant ICMP11096glad-I. The fidelity of the marker exchange event was confirmed by PCR analysis. To complement the glad-I mutation, the plasmid pBBgladIR was transferred to ICMP11096glad-I by triparental mating, generating ICMP11096glad-IR.
Antimicrobial Effect
The N.AHLs-deficient mutant (ICMP11096glad-I) and the complemented one (ICMP11096glad-IR) were evaluated for their ability to inhibit the growth of B. megaterium and E. coli in dual agar plate assay following the method of Lavermicocca et al. (1997) as reported by Camele et al. (2019). In particular, small masses of fresh bacterial cultures were transferred in the center of 90 mm Ø MMA Petri dishes. All plates were then sprayed with the single target bacterial suspension containing 108 Colony Form Unit (CFU)/ml. The antagonistic activity was determined by measuring the diameter of inhibition zones and was expressed as the antagonistic capacity percentage. The test was repeated twice with three replicates.
Hemolytic Effect
The hemolytic effect of the above mentioned two mutants was evaluated against the cell membrane of erythrocyte’s (RBCs) as explained by Sakr et al. (2018) following the method of Munsch and Alatossava (2002). The hemolysis ability was indicated by measuring the diameter of transparent area in comparison to WT strain as reported in the previous study by Elshafie et al. (2017a). The test was repeated twice with three replicates.
Extracellular Hydrolytic Enzymes
The possible hydrolytic enzymes that might be produced by the two studied mutants of Bga was screened as specified by Elshafie et al. (2017a). In particular, chitinase and protease activities were determined according to Tahtamouni et al. (2006). Cellulase activity was detected following the method of Essghaier et al. (2009) and glucanase activity was detected according to the method of Teather and Wood (1982). Amylase, pectinase and polygalacturanase activities were detected following the methods of Sung et al. (1993) and Bhardwaj and Garg (2010). The enzymatic activity was evidenced by the appearance of clear halos around the colonies and their diameters were measured in (mm) compared to WT stain.
Pathogenicity Assay
Pathogenicity test of WT and the two studied mutants of Bga was performed on A. bisporus flesh pseudo tissues as following: 20 μl drops of each bacterial suspensions at 106, 104, and 103 CFU/ml were deposited on carpophores of A. bisporus and then were incubated for 96 h at 22 ± 25°C (Elshafie et al., 2017c). Carpophores treated with 20 μl drops of sterile distilled water were used as control. The virulence effect was observed after 4 days of incubation through the evaluation of the discoloration intensity. The virulence effect of WT Bga strain, obtained in the previous study by Elshafie et al. (2017a), was used here only for a comparison purposes.
HPLC Fractionation
Purification of Bioactive Substances
The purified filtrate of ICMP11096glad-I mutant was analyzed by HPLC as reported previously by Elshafie et al. (2017c). Aliquots (30 mg.ml–1) of lyophilized bioactive metabolites derived from 5 day old bacterial culture filtrates were diluted in sterile distilled water. An aliquot (10 ml) of the prepared mixture, containing 300 mg of lyophilized substances, was loaded on a cartridge syringe (Strata C18-T) prewashed with 2 ml of methanol and 2 ml of distilled water. The cartridge was subsequently washed with 1 ml distilled water and later the bioactive substances were recovered by adding 1 ml of methanol 98%.
HPLC Fractionation
The methanol fractions were analyzed in HPLC-Agilent 1200 series austere following the specific analytical procedures described below. The separation was obtained with an Agilent ECLIPSE XDB, C18 (4.6 × 150 mm, 5 μm). The injected volume was 20 μl; column and autosampler chamber temperatures were at 25 and 4°C, respectively. Flow rate was 1 ml.min–1 and mobile phase were A: 0.2% formic acid (FA) in H2O and B: 0.1% FA in Acetonitrile (CH3CN) with the following gradients (A%: B%) 80–20 from 0 to 5 min; 60–40 from 5 to 60 min and 80–20 from 60 to 65 min. The HPLC chromatogram was obtained at a wave length λ = 380 nm.
Statistical Analysis
The experimental outputs were statistically analyzed using statistical Package for the Social Sciences SPSS (version 13.0, Prentice Hall: Chicago, IL, United States, 2004). Experimental data was expressed as mean ± SD and comparisons were employed by a one way ANOVA followed by Tukey post hoc test for detecting any significance of the investigated data regarding the studied biological activities at P < 0.05.
Results and Discussion
Antimicrobial Activity
Results of ICMP11096glad-I mutant showed significant reduction of antimicrobial activity in MMA substrate in comparison with the WT strain (Table 2) and this reduction could be due to the absence of signal molecules N.AHLs which indicate their regulatory role in the QS process enhancing the production of bioactive secondary metabolites. On the other hand, ICMP11096glad-I complemented mutant explicated similar values of antimicrobial activity of the respective WT strain which indicates that it was able to regain its bioactivity (Table 2).
TABLE 2.
Antimicrobial, hemolytic, and enzymatic activities of wild type (WT), ICMP11096glad-I and ICMP11096glad-IR mutants of B. gladioli pv. agaricicola.
| Studied strains | Antimicrobial effecta | Erycrocyte cell lysisb | Extracellular hydrolytic enzymesc | |||||||
| B. meg | E. col | Protease | Cellulase | Chitinase | Amylase | Glucanase | Pectinase | Polygalacturonase | ||
| Bga WT | 35 ± 1.5 | 30 ± 2.7 | 15 ± 2.1 | 12 ± 1.3 | – | 25 ± 3.6 | – | 28 ± 3.5 | – | – |
| Bga gladI | 8 ± 0.7 | 5 ± 0.2 | – | 5 ± 0.4 | – | 5 ± 0.7 | – | 12 ± 2.1 | – | – |
| Bga gladIR complemented | 33 ± 2.4 | 28 ± 3.3 | 12 ± 1.5 | 9 ± 1.9 | – | 12 ± 1.7 | – | 22 ± 2.8 | – | – |
aAntimicrobial effect of wild type (WT) and mutants of Bga ICMP11096 strain against Bacillus megaterium (B. meg) and E. coli (E. col) expressed as antagonistic capacity percentage; bHemolytic effect against erythrocyte cell membrane of fresh bovine blood; cExtracellular hydrolytic enzymes (hydrolysis zones in mm) caused by WT, ICMP11096glad-I and ICMP11096glad-IR mutants.
Hemolytic Activity
Results of observations of ICMP11096glad-I mutant, expressed as average diameters of hemolysis zones (mm), did not show any hemolytic activity compared to WT strain, which has been investigated in the previous study, ranged between 31.0 ± 6.5 and 75.0 ± 11.5 mm (Elshafie et al., 2017a). The ICMP11096glad-IR complemented mutant was able to regain the majority of hemolytic effect level close to the respective WT Bga strain (Table 2). These outcomes indicated that N.AHLs plays the principal role in production or/and regulation of hemolysis factors in agreement with the hypothesis of Munsch and Alatossava (2002) who stated that several Pseudomonas and other related bacteria associated with the cultivated mushrooms A. bisporus, such as Burkholderia sp., are hemolytic.
Extracellular Hydrolytic Enzymes
The ICMP11096glad-I mutant showed a significant reduction of enzymatic activity especially with protease, chitinase and glucanase enzymes (Table 2). There is no production of cellulase, amylase, pectinase, and polygalacturonase. Furthermore, the reduction of proteolytic, chitinolytic, and glucanolytic activities could contribute to lowering the virulence effect of the mutant strain against A. bisporus tissues in agreement with Ordentlich et al. (1988). Furthermore, ICMP11096glad-IR complemented mutant showed closed values of hydrolysis effect to the WT strain of Bga (Table 2).
Pathogenicity Assay
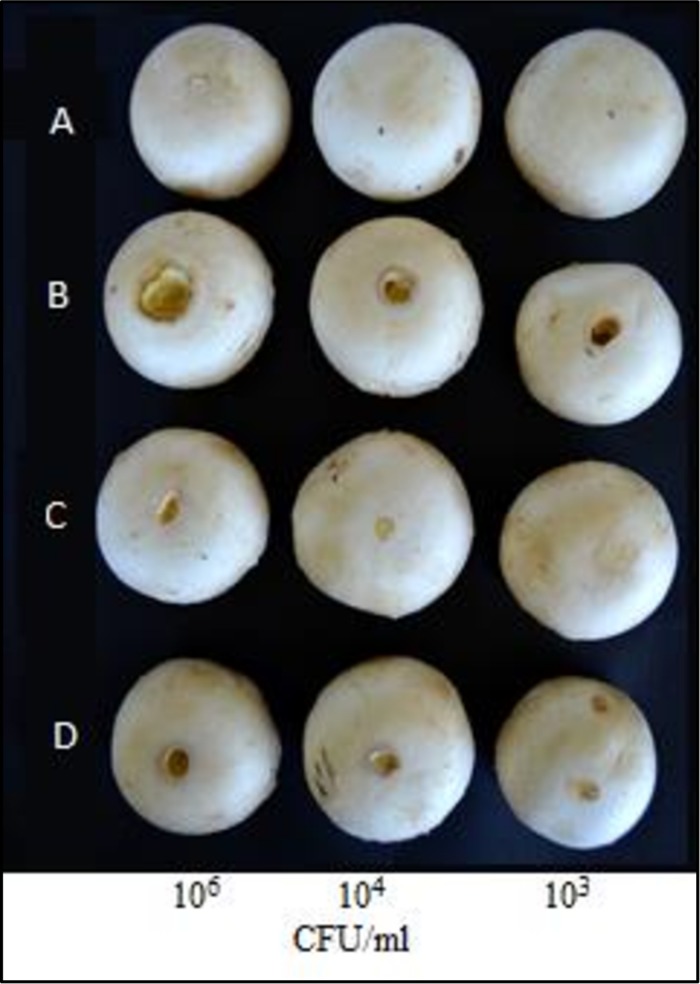
The ICMP11096glad-I mutant strain did not show any color change of the artificially infected A. bisporus tissues. The ICMP11096glad-IR complemented mutant was able to regain the virulence effect similar to WT strain (Figure 1). This result indicates that N.AHLs signal molecule is involved in the regulation of virulence factors which might be related also to the hydrolytic enzymes in accordance with Chowdhury and Heinemann (2006).
FIGURE 1.
Pathogenicity assay on carpophores of Agaricus bisporus. A = control (sterile distilled water); B = wild type (WT) strain ICMP 11096; C = N.AHLs-deficient mutant (ICMP11096glad-I); D = complemented plasmid mutant (ICMP11096glad-IR); 106, 104, and 103 CFU/ml are the tested concentration of bacterial suspensions.
HPLC Analysis
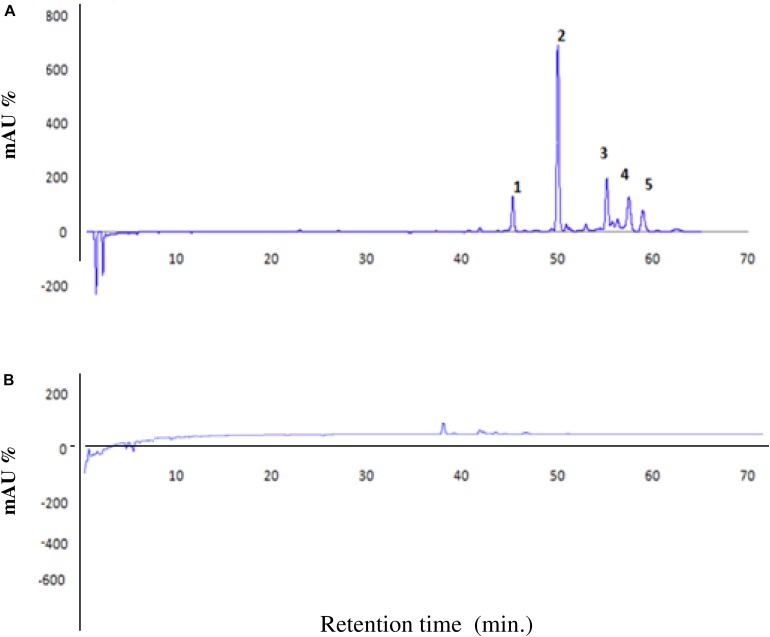
The HPLC chromatographic analysis of N.AHLs-deficient mutant glad I demonstrated a highly reduction of the five principal peaks typically present in WT strain from quantitative point of view (Elshafie et al., 2017c; Figure 2). The absence or trace level of the fractions is also in agreement with the above results of the pathogenicity assay which assumed that the lack of bioactive compounds in mutant strain might be linked to the absence of virulence factors.
FIGURE 2.
High performance liquid chromatography (HPLC) chromatogram of WT strain Bga ICMP11096 (A) and N.AHLs-deficient mutant (ICMP11096glad-I) at 380 nm (B).
Conclusion
As previously described by Zhou et al. (2003) in Burkholderia ambifaria BC-F, a biocontrol strain reported previously to exhibit broad-spectrum antifungal activity, the inactivation of the QS genes N.AHL synthase (bafI) and N.AHL-binding transcriptional activator (bafR) leads to a decreased antifungal activity. Similarly, the inactivation of the AHL synthase (gladI) in Bga caused the loss of the antibacterial activity and other phenotypic traits. The N.AHLs-deficient mutant showed a high reduction of the antimicrobial activity, complete absence of hemolytic effect and a significant reduction of hydrolytic enzymes (chitinase, protease, and glucanase). It is also concluded that there are various factors for regulating the genes implicated in the production of antimicrobial substances different from those responsible for the hemolytic substances. The above mentioned hydrolytic enzymes in WT strain could be strongly implicated in soft rot disease of A. bisporus. The outcomes from the current study are promising in controlling mushroom soft rot disease and clarifying the regulatory role of N.AHLs-mediated QS in biological traits of Bga are mostly likely important in the pathogen virulence.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
HE and SB: conceptualization. GD and VV: DNA techniques. SB and IC: data curation and review the manuscript. HE, IC, and GD: writing the original draft and editing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Alexeyev M. F. (1999). The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26 824–828. 10.2144/99265bm05 [DOI] [PubMed] [Google Scholar]
- Andolfi A., Cimmino A., Lo Cantore P., Iacobellis N. S., Evidente A. (2008). Bioactive and structural metabolites of Pseudomonas and Burkholderia species causal agents of cultivated mushrooms diseases. Pers. Med. Chem. 2 81–112. [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V., Garg N. (2010). “Exploitation of micro-organisms for isolation and screening of pectinase from environment,” in Proceedings of 8th International Conference, Making Innovation Work for Society: Linking, Leveraging and Learning, (Kuala Lumpur: University of Malaya; ), 1–3. [Google Scholar]
- Camele I., Elshafie H. S., Caputo L., Sakr S. H., De Feo V. (2019). Bacillus mojavensis: biofilm formation and biochemical investigation of its bioactive metabolites. J. Biol. Res. 92 39–45. 10.4081/jbr.2019.8296 [DOI] [Google Scholar]
- Chowdhury P. R., Heinemann J. A. (2006). The general secretory pathway of Burkholderia gladioli pv. agaricicola BG164R is necessary for cavity disease in white button mushrooms. Appl. Environ. Microbiol. 72 3558–3565. 10.1128/AEM.72.5.3558-3565.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A., Lo Cantore P., Karapetyan G., Kaczynski Z., Iacobellis N. S., Holst O., et al. (2008). “Characterization of antimicrobial and structural metabolites from Burkholderia gladioli pv. agaricicola,” in Pseudomonas syringae Pathovars and Related Pathogens – Identification, Epidemiology and Genomics, eds Fatmi M., Collmer A., Iacobellis N. S., Mansfield J. W., Murillo J., Schaad N. W. (Dordrecht: Springer; ), 347–355. 10.1007/978-1-4020-6901-7_36 [DOI] [Google Scholar]
- Coenye T., Vandamme P. (2003). Diversity and significance of Burkholderia species occuping diverse ecological niches. Environ. Microbiol. 5 719–729. 10.1046/j.1462-2920.2003.00471.x [DOI] [PubMed] [Google Scholar]
- Costerton W., Veeh R., Shirtliff M., Pasmore M., Post C., Ehrlich G. (2003). The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Invest. 112 1466–1477. 10.1172/JCI20365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. E., O’Toole G. A. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64 847–867. 10.1128/mmbr.64.4.847-867.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devescovi G., Bigirimana J., Degrassi G., Cabrio L., LiPuma J. J., Kim J., et al. (2007). Involvement of a quorum-sensing-regulated lipase secreted by a clinical isolate of Burkholderia glumae in severe disease symptoms in rice. Appl. Environ. Microbiol. 73 4950–4958. 10.1128/AEM.00105-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y. H., Zhang L. H. (2005). Quorum sensing and quorum-quenching enzymes. J. Microbiol. 43 101–109. [PubMed] [Google Scholar]
- Duerkrop B. A., Ulrich R. L., Greenberg E. P. (2007). Octanoyl-homoserine lactone is the cognate signal for Burkholderia mallei BmaR1-BmaI1 quorum sensing. J. Bacteriol. 189 5034–5040. 10.1128/JB.00317-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshafie H. S., Camele I., Racioppi R., Scrano L., Bufo S. A. (2012). In vitro antifungal activity of Burkholderia gladioli pv. agaricicola against some phytopathogenic fungi. Int. J. Mol. Sci. 13 16291–16302. 10.3390/ijms131216291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshafie H. S., Racioppi R., Bufo S. A., Camele I. (2017a). In vitro study of biological activity of four strains of Burkholderia gladioli pv. agaricicola and identification of their bioactive metabolites using GC-MS. Saudia J. Biol Sci. 24 295–301. 10.1016/j.sjbs.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshafie H. S., Sakr S., Bufo S. A., Camele I. (2017b). An attempt of biocontrol the tomato-wilt disease caused by Verticillium dahliae using Burkholderia gladioli pv. agaricicola and its bioactive secondary metabolites. Int. J. Plant Biol. 8 57–60. 10.4081/pb.2017.7263 [DOI] [Google Scholar]
- Elshafie H. S., Viggiani L., Mostafa M. S., El-Hashash M. A., Bufo S. A., Camele I. (2017c). Biological activity and chemical identification of ornithine lipid produced by Burkholderia gladioli pv. agaricicola ICMP 11096 using LC-MS and NMR analyses. J. Biol. Res. 90 96–103. 10.4081/jbr.2017.6534 [DOI] [Google Scholar]
- Essghaier B., Fardeau M. L., Cayol J. L., Haijaoui M. R., Boudabous A., Jijakli H., et al. (2009). Biological control of grey mould in strawberry fruits by halophilic bacteria. J. Appl. Microbiol. 106 833–846. 10.1111/j.1365-2672.2008.04053.x [DOI] [PubMed] [Google Scholar]
- Gill W. M. (1995). Bacterial diseases of Agaricus Mushrooms. Rep. Tottori. Mycol. Inst. 33 34–55. [Google Scholar]
- Karakurt H., Aslantas R. (2010). Effects of some plant growth promoting Rhizobacteria (PGPR) strains on plant growth and leaf nutrient content of apple. J. Fruit Ornamen. Plant Res. 18 101–110. [Google Scholar]
- Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166 175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- Kumari A., Pasini P., Deo S. K., Flomenhoft D., Shashidhar S., Daunert S. (2006). Biosensing systems for the detection of bacterial quorum signaling molecules. Anal. Chem. 78 7603–7609. 10.1021/ac061421n [DOI] [PubMed] [Google Scholar]
- Lavermicocca P., Iacobellis N. S., Simmaco M., Graniti A. (1997). Biological properties and spectrum of activity of Pseudomonas syringae pv. syringae toxins. Physiol. and Mol. Plant Pathol. 50 129–140. 10.1006/pmpp.1996.0078 15688938 [DOI] [Google Scholar]
- Li Y. H., Tian X. (2012). Quorum sensing and bacterial social interactions in biofilms. Sensors 12 2519–2538. 10.3390/s120302519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean K. H., Winson M. K., Fish L. (1997). Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acyl homoserine lactones. Microbiology 143 3703–3711. 10.1099/00221287-143-12-3703 [DOI] [PubMed] [Google Scholar]
- Munsch P., Alatossava T. (2002). Several pseudomonads, associated with the cultivated mushrooms Agaricus bisporus or Pleurotus sp., are haemolytic. Microbiol. Res. 157 311–315. 10.1078/0944-5013-00159 [DOI] [PubMed] [Google Scholar]
- Ordentlich A., Elad Y., Chet I. (1988). The role of chitinase of Serratia marcescens in biocontrol of Sclerotium rolfsii. Phytopathology 78 84–92. 10.1094/Phyto-78-84 [DOI] [Google Scholar]
- Prashanth S. N., Bianco G., Cataldi T. R., Iacobellis N. S. (2011). Acyl homoserine lactone production by bacteria associated with cultivated Mushrooms. J. Agri. Food Chem. 59 11461–11472. 10.1021/jf202313j [DOI] [PubMed] [Google Scholar]
- Sakr S. H., Elshafie H. S., Camele I., Sadeek S. A. (2018). Synthesis, spectroscopic, and biological studies of mixed ligand complexes of gemifloxacin and glycine with Zn(II), Sn(II), and Ce(III). Molecules 23 1–17. 10.3390/molecules23051182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook I., Fritsch F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. (1987). Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169 5789–5794. 10.1128/jb.169.12.5789-5794.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler L., Venturi V. (2007). Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 266 1–9. 10.1111/j.1574-6968.2006.00501.x [DOI] [PubMed] [Google Scholar]
- Stoyanova M., Pavlina I., Moncheva P., Bogatzevska N. (2007). Biodiversity and incidence of Burkholderia species. Biotechnol. Biotechnol. 47 306–310. 10.1080/13102818.2007.10817465 [DOI] [Google Scholar]
- Sung N., Keun K., Sung H. (1993). Isolation of soil bacteria secreting. Raw-starch-digesting enzyme and the enzyme production. J. Microbiol. Biotechnol. 3 99–107. [Google Scholar]
- Tahtamouni M. E. W., Hameed K. M., Saadoun I. M. (2006). Biological control of Sclerotinia sclerotiorum using indigenous chitolytic actinomycetes in Jordan. J. Plant Pathol. 22 107–114. 10.5423/PPJ.2006.22.2.107 [DOI] [Google Scholar]
- Teather R. M., Wood P. J. (1982). Use of Congo red-polysacchatide interaction in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43 777–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsher M. L., Kolbe J., Morris A. J., Welch D. F. (1997). Nosocomial acquisition of Burkholderia gladioli in patients with cystic fibrosis. Am. J. Resp. Crit. Care Med. 156 1436–1440. 10.1164/ajrccm.155.4.9105090 [DOI] [PubMed] [Google Scholar]
- Withers H., Swift S., William P. (2001). Quorum sensing as an integral component of gene regulatory. Curr. Opin. Microbiol. 4 186–193. 10.1016/S1369-5274(00)00187-9 [DOI] [PubMed] [Google Scholar]
- Yabuuchi E., Kosako Y., Oyaizu H., Yano I., Hotta H., Hashimoto Y., et al. (1992). Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36 1251–1275. 10.1111/j.1348-0421.1992.tb02129.x [DOI] [PubMed] [Google Scholar]
- Zhang G., Zhang F., Ding G., Li J., Guo X., Zhu J., et al. (2012). Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 6 1336–1344. 10.1038/ismej.2011.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Yao F., Roberts D. P., Lessie T. G. (2003). AHL-deficient mutants of Burkholderia ambifaria BC-F have decreased antifungal activity. Curr. Microbiol. 47 174–179. 10.1007/s00284-002-3926-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.